Abstract
The HOX11 homeobox gene was first identified through studies of the t(7;10) and t(10;14) chromosomal translocations of acute T-cell leukemia. In addition, analysis of Hox11−/− mice has demonstrated a critical role for this gene in murine spleen development. A possible mode of in vivo function for the HOX11 protein in these two situations is regulation of target genes following DNA binding via the homeodomain, but little is known about how HOX11 regulates transcription in vivo. By performing transcriptional studies in yeast and mammalian one-hybrid systems, a modular transcriptional transactivation region at the NH2 terminus of HOX11 has been functionally dissected from other parts of the protein. This NH2-terminal region includes the previously identified short conserved Hep motif, which itself activates transcription in one-hybrid assays. The importance of the NH2-terminal region for the function of HOX11 in vivo was assayed by activating a HOX11-dependent gene in NIH 3T3 cells. Activation of this gene was found to be dependent upon an intact homeodomain in HOX11, but maximal activation was obtained only when the NH2-terminal 50 amino acids of HOX11 was present, showing that this region of HOX11 is important for in vivo transcriptional control of a chromosomal target gene.
Chromosomal translocations in acute leukemias frequently involve activation of genes encoding proteins involved in transcription (2, 18, 19). The HOX11 homeobox gene was first identified by cloning the breakpoints of t(7;10)(q35;q24) and t(10;14)(q24;q11) chromosomal translocations, found in 5 to 10% of patients with acute T-cell leukemia (5, 9, 13, 15). These translocations result in the HOX11 gene, which is normally found on chromosome 10, band q24, being placed within the transcriptional control region of the T-cell receptor δ gene on chromosome 14 or the T-cell receptor δ gene on chromosome 7q35 (13, 29). The abnormal expression of HOX11 in T cells as a result of these chromosomal translocations in humans is thought to be a key step in the progression toward malignancy (5, 9, 13, 15), a view reinforced by the demonstration of oncogenic activity of HOX11 in transplant recipients of MSCV-HOX11-transduced bone marrow cells (10, 11).
The HOX11 gene belongs to a family of homeodomain-encoding genes which includes the related Hox11L1 and Hox11L2 genes first characterized in the mouse (4). The homeodomain is conserved within this family, and, like other homeodomain proteins, the HOX11 family has been implicated in the regulation of cellular growth and differentiation. In mice, Hox11 is essential for generation of the spleen (3, 21), promoting survival of the splenic precursor cells (3), and null mutation of the Hox11L1 gene results in myenteric neuronal hyperplasia and megacolon (25).
Since the homeodomain is a sequence-specific DNA-binding element, most homeodomain-containing proteins are believed to function through trans regulation (direct or indirect) of specific target genes (16). Consistent with this, HOX11 is a nuclear protein and can bind to DNA in vitro (4, 26) and a fusion protein consisting of HOX11 fused to the GAL4 DNA-binding domain (GAL4-DBD) was found to transactivate various promoters in both yeast and mammalian cells (28). Three distinct domains of HOX11 were found to be necessary for transactivation by this fusion protein (28), namely, the glycine-proline-rich region at the NH2 terminus, the glutamine-rich region at the COOH terminus, and the homeodomain itself. However, the only data which pertain to functional characteristics in non-artificial reporter assays are that the NH2- and COOH-terminal regions of HOX11 appear dispensable for transforming function (11) and that introduction of HOX11 into NIH 3T3 cells results in transcription of a HOX11-dependent gene (denoted Hdg-1 [8]). Therefore, we have assessed regions of HOX11 required for optimal in vivo transactivation of the endogenous Hdg-1 gene. In this stringent assay, we show that deletions in the HOX11 homeodomain (in either the NH2-terminal arm or the third helix) prevent induction of Hdg-1 by HOX11. We also show that the NH2-terminal 50- amino-acid stretch is crucial for optimal function of the HOX11 protein in vivo.
MATERIALS AND METHODS
Plasmids.
The yeast expression vector for the Y1 fusion protein was constructed by PCR amplification of the HOX11 sequence corresponding to amino acids 252 to 330 and cloning into NcoI-BamHI-digested pAS-CYH2 (a modified version of pAS [6]). This intermediate construct contained a unique KpnI site preceding the HOX11 sequence, and following digestion with NcoI and KpnI, a second HOX11 PCR product corresponding to amino acids 1 to 241 was inserted to create the Y1 expression vector. Y1 protein lacks amino acids 242 to 251 of HOX11, which are replaced by glycine and threonine residues from the introduced KpnI site. Mutants Y2 to Y11 were made by PCR amplification with primers designed to amplify the indicated regions of HOX11 and the Y1 expression vector as the template. Y12 to Y17 were constructed by cloning of annealed oligonucleotides.
Mammalian expression vectors for GAL4-DBD-HOX11 fusions were also made by PCR amplification of the indicated regions of HOX11 and subsequent cloning in frame into BamHI-XbaI-digested pM1 vector (23). Mammalian expression vectors for HOX11, HOXΔN50, and HOX263C were constructed by PCR amplification of the indicated regions of HOX11 and cloning into the XbaI site of the pEFBOS vector (17). HOXΔN50 also contained a methionine residue as the first amino acid preceding the HOX11 sequence. HOXMHEP and HOXΔHEP were also made by PCR amplification of HOX11 sequence with primers designed to introduce mutations into the Hep motif (indicated in Fig. 3). pEFBOS-HOXΔH3 was made by PCR with the yeast expression vector for Y1 as the template. pEFBOS-HOXMDPA was made as follows. A HOX11 sequence corresponding to amino acids 174 to 330 containing the mutated FPWM motif at amino acids 174 to 177 (changed to MDPA and introducing a unique BamHI site) was PCR amplified and cloned into the XbaI site of pEFBOS (clone pEFBOS-ICMDPA). pEFBOS-ICMDPA contains two XbaI sites; however, the XbaI site C terminal to the HOX11 sequence can be blocked by dam methylation, allowing a second HOX11 PCR product corresponding to amino acids 1 to 173 to be cloned into XbaI-BamHI-digested pEFBOS-ICMDPA to create pEFBOS-HOXMDPA. The HOXΔKN mutation was made by using HOX11 sequences inserted at the BamHI-XhoI sites of pBluescript KS. Digestion with XbaI and BglII removed the sequence corresponding to the first 213 codons of HOX11. This was replaced by a PCR-generated mutant sequence containing an internal deletion of amino acids 198 to 204. pEFBOS-HOXΔKN was made by PCR with this pBluescript KS-HOXΔKN construct as template. The sequences of all the constructs were verified.
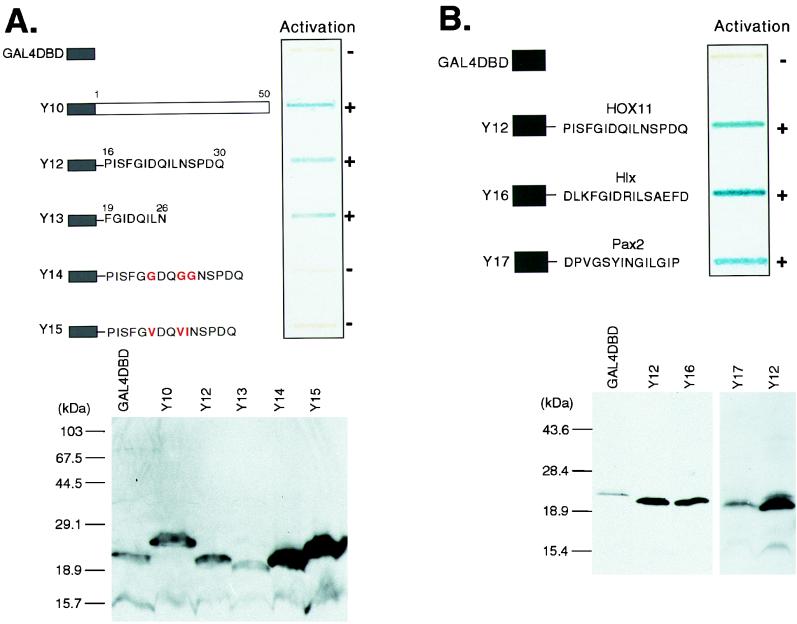
FIG. 3.
The Hep sequence can function as a sequence-specific activation domain. (A) The upper panel shows a yeast β-galactosidase slot blot assay of GAL4-DBD-Hep fusions. Fusions Y10 and Y12 to Y15 are represented schematically. The GAL4-DBD is fused to amino acids 1 to 50 of HOX11 (Y10), amino acids 16 to 30 of HOX11 (Y12), and amino acids 19 to 26 of HOX11 (Y13). Y14 and Y15 are based on Y12 but contain the amino acid substitutions shown. After incubation of the β-galactosidase assay mixture for 2 h, the ability of each fusion to activate the lacZ reporter gene was assessed by the blue color as indicated (when exposed for 12 h, Y14 and Y15 yielded blue coloration comparable to the 2-h exposure of Y12 and Y13). The lower panel shows Western blot detection of GAL4-DBD-HOX11 fusions in yeast extracts with a monoclonal GAL4-DBD antiserum. (B) The ability to activate in yeast is a conserved function of the Hep/octapeptide motif. The upper panel shows the results of a yeast β-galactosidase assay of GAL4-DBD-Hep/octapeptide fusions. The fusions Y12, Y16, and Y17 are represented schematically. GAL4-DBD is fused to a 15-amino-acid peptide encompassing the HOX11 Hep motif (Y12), the Hlx Hep motif (Y16), or the octapeptide sequence from Pax-2 (Y17). The lower panel shows Western blot detection of GAL4-DBD-Hep/octapeptide fusions in yeast extracts with a monoclonal GAL4-DBD antiserum.
Yeast transformation and β-galactosidase assay.
Yeast strain Hf7c (Clontech) was grown on yeast extract-peptone-dextrose (YEPD) plates or in supplemented Sabouraud dextrose (SD) medium and transformed by the lithium acetate method (22). Yeast cells were assayed for β-galactosidase activity by a modified filter assay (24).
Transient transfections and CAT assays.
Cos-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum. Lipofectin reagent (Gibco-BRL) was used for transient transfections. For each 10-cm tissue culture dish, 5 μg of the reporter plasmid pG5EC and 10 μg of each pM1-based expression vector were used. A 1-μg portion of pVP65, expressing a fusion of GAL4-DBD to the VP65 activation domain, was cotransfected with pG5EC as a positive control. Chloramphenicol acetyltransferase (CAT) assays were performed 36 h after transfection.
Extract preparation and Western blotting.
Yeast extracts were prepared by sodium dodecyl sulfate extraction (22). Cos-7 and NIH 3T3 cell extracts were prepared with 300 μl of lysis buffer (10 mM HEPES [pH 7.6], 0.25 M NaCl, 0.5% Nonidet P-40, 5 mM EDTA) per 107 cells. Extracts and molecular weight standards (prestained protein molecular weight standards, 14,300- to 200,000-molecular-weight range; Gibco-BRL) were electrophoresed through sodium dodecyl sulfate–15% polyacrylamide gels and transferred to a nitrocellulose membrane (Schleicher & Schuell). The membranes were blocked for 1 h in 5% Marvel (Premier Beverages) solution and incubated with either a monoclonal GAL4-DBD antiserum (Santa Cruz Biotechnology) or a polyclonal HOX11 antiserum (see below). The membranes were incubated with horseradish peroxidase-conjugated secondary antibodies prior to visualization of proteins with enhanced chemiluminescence (ECL) detection reagents (Amersham Life Science).
The HOX11 antiserum was made from bacterially synthesized HOX11 protein. The HOX11 coding sequence was cloned into the pET-15b bacterial expression vector (Novagen). Purified His-tagged HOX11 protein was prepared and used for rabbit immunization.
Construction of NIH 3T3 clones.
NIH 3T3 cells were maintained in DMEM containing 10% fetal calf serum. The cells were cotransfected with linearized pEFBOS-based expression plasmid (10 μg) and pMC1neopolyA selection plasmid (0.5 μg) with Lipofectin reagent. At 24 h after transfection, selective medium (DMEM containing 10% fetal calf serum and 0.5 mg of G418 per ml) was added to the cells. After approximately 12 days in selective medium, G418-resistant clones were picked and screened by Western blotting for expression of HOX11 protein or by genomic PCR for the presence of integrated pEFBOS vector (data not shown).
Northern analysis.
Northern analysis was carried out as described previously (20) with 10 μg of total RNA per lane. Probes were labelled by random priming (7). The Hdg-1 cDNA probe was a 517-bp DpnII fragment of the Hdg-1 cDNA, and the mouse ATP synthase (subunit c) cDNA was a 246-bp DpnII fragment (8). Hybridization levels were quantitated with a model 300A computing densitometer (Molecular Dynamics).
RESULTS
The NH2-terminal region of HOX11 activates transcription in yeast.
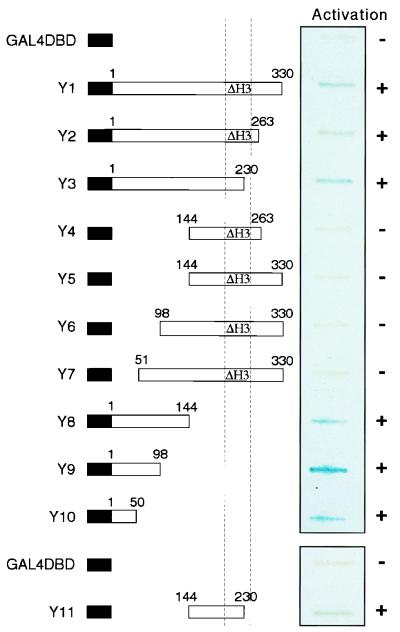
The ability of GAL4-DBD-HOX11 fusion proteins to activate the transcription of a reporter gene was tested in yeast cells. Various HOX11 sequences were fused to GAL4-DBD in the yeast expression vector pAS-CYH2 (a modified pAS [6]) and transformed into the Hf7c yeast strain (containing a lacZ reporter gene with upstream GAL4 DNA-binding sites). Although a fusion of the GAL4-DBD with the complete HOX11 sequence activated the lacZ reporter, the yeast exhibited severe growth problems (data not shown). Thereafter, we used a basic construct which contains a deletion of the third helix in the HOX11 homeodomain. This clone resulted in a detectable fusion protein with no obvious effect on yeast growth and which was still capable of transcriptional activation (Fig. 1, Y1).
FIG. 1.
Yeast one-hybrid assay of HOX11 transcriptional activation domains. GAL4-DBD-HOX11 fusions are represented schematically on the left. The position of the homeodomain is indicated by the dashed vertical lines, and mutants lacking the third helix of the homeodomain (amino acids 242 to 251) are marked by the symbol ΔH3. The ability of each fusion to activate the lacZ reporter gene, as assessed by the presence of a blue color in the slot blot β-galactosidase assay, is indicated on the right. Apparent differences in the intensity of blue coloration are ignored because although expression of the GAL4-DBD alone and fusions Y1 to Y11 was confirmed by Western blot analysis with a monoclonal GAL4-DBD antiserum, the levels of protein expression varied (data not shown).
A series of mutant HOX11 constructs were made, based on this helix three-deletion mutant as a starting point, for delineation of the activation functions of HOX11 (Fig. 1). Activation of the reporter gene was observed for constructs Y2, Y3, Y8, Y9, and Y10. Within these, the minimum region of overlap which retains the transactivation element is the first 50 amino acids of HOX11. Furthermore, deletion of this 50-residue stretch (clones Y4 to Y7) destroys the activation activity despite the retention of a segment which can also activate transcription, albeit weakly, when present alone (Y11). The data therefore indicate that at least two independent regions of the HOX11 protein can transactivate transcription in this yeast assay. A previous study identified three distinct regions of HOX11 required for optimal transactivation by a GAL4-DBD-HOX11 fusion (28). These were the glycine-proline-rich region at the NH2 terminus, the homeodomain, and the glutamine-rich region at the COOH terminus. The differing results may reflect our deletion of the third helix of the homeodomain.
The first 50 amino acids of HOX11 can activate transcription in mammalian cells.
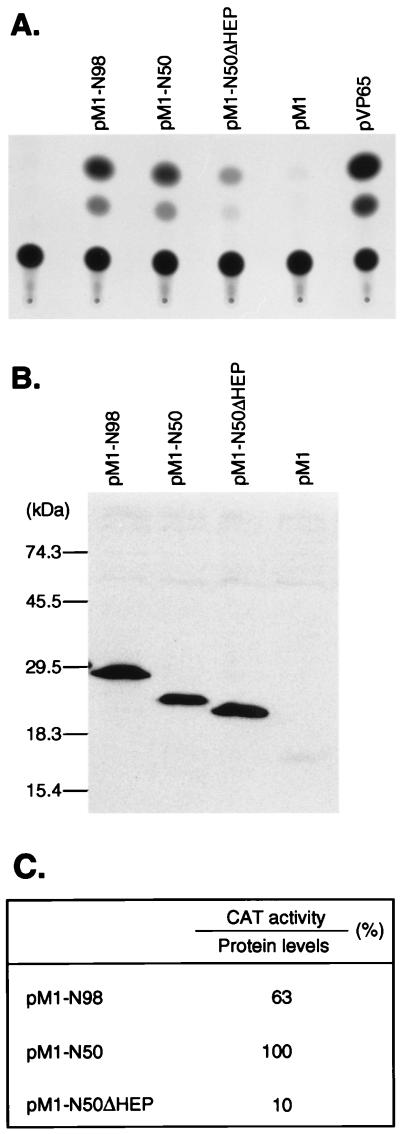
The ability of the first 50 amino acids of HOX11 to function independently as an activation domain was confirmed in mammalian cells. Sequences corresponding to amino acids 1 to 98 of HOX11 or amino acids 1 to 50 were fused to the GAL4-DBD in the mammalian expression vector pM1 (23) (to create pM1-N98 and pM1-N50, respectively [Fig. 2]). These plasmids were transfected into Cos-7 cells together with the pG5EC reporter construct (23). While the parent vector pM1 did not activate the reporter gene, transfection of either pM1-N98 or pM1-N50 resulted in similar levels of activation of the reporter (Fig. 2A). Therefore, the first 50 amino acids of HOX11 can function independently as a transcriptional transactivation domain in mammalian cells.
FIG. 2.
The NH2-terminal region of HOX11 can activate transcription in mammalian cells. (A) CAT assay of Cos-7 cells after transfection of GAL4-DBD-HOX11 fusion plasmids. Cos-7 cells were transfected with the pG5EC reporter construct alone or in combination with various GAL4-DBD expression vectors: pM1 (expressing the GAL4-DBD alone), pM1-N98 (expressing amino acids 1 to 98 of HOX11 fused to the GAL4-DBD), pM1-N50 (expressing amino acids 1 to 50 of HOX11 fused to the GAL4-DBD), pM1-N50ΔHEP (based on pM1-N50 but containing an internal deletion of the Hep motif at amino acids 19 to 26), or pVP65 (expressing the VP65 activation domain fused to the GAL4-DBD). (B) Western blot detection of GAL4-DBD-HOX11 fusions in Cos-7 cell extracts. Extracts were prepared 36 h after transfection and analyzed with a monoclonal GAL4-DBD antiserum. GAL4-DBD fusion proteins were detected more readily than was GAL4-DBD, for unknown reasons. (C) Chart showing the relative CAT activities of pM1-N98, pM1-N50, and pM1-N50ΔHEP after normalization for protein expression levels. CAT activity and protein levels were quantitated by densitometry. The values are expressed as percentages, with pM1-N50 activity assigned the value of 100%.
The HOX11 Hep sequence can facilitate transcriptional activation in both yeast and mammalian cells.
Examination of the amino acid sequence of the HOX11 NH2-terminal region for motifs which might be important in transcription did not reveal anything other than the previously noted Hep motif (9). The Hep sequence is found in several homeodomain proteins and is related to the octapeptide sequence found in many Pax proteins (1). An assessment of its role in transactivation by HOX11 was made by preparing an internal deletion of the Hep sequence in the expression clone pM1-N50 for transfection of Cos-7 cells. This analysis (Fig. 2A) showed that the deletion mutant (pM1-N50ΔHEP) was significantly reduced (10-fold [Fig. 2C]) in its ability to activate transcription of the cotransfected reporter plasmid. This diminution of function apparently occurs as a direct result of the Hep deletion and is not an effect of protein expression levels (as judged by CAT activities normalized to detectable protein levels [Fig. 2B; a quantitation is given in Fig. 2C]).
The isolated Hep sequence was then tested for the ability to mediate activation. A construct was made which encoded 15 amino acids encompassing the Hep motif, fused to the GAL4-DBD. This construct was tested in the yeast one-hybrid assay system and found to be sufficient for activation (Y12) (Fig. 3, upper panels), as was the core 8-amino-acid Hep sequence fused to the GAL4-DBD (Y13) (Fig. 3A, upper panel). The ability to activate reporter gene transcription was impaired when the three-core hydrophobic residues were mutated to glycine (Y14) (Fig. 3A, upper panel) or when two isoleucines were changed to valines and the leucine was changed to isoleucine (Y15) (Fig. 3A, upper panel). These varied activation effects were not due to reduced protein expression (Fig. 3A, lower panel).
The Hep sequence has been detected in several homeodomain proteins, and the so-called octapeptide sequences of Pax proteins are related to the Hep sequence (1). Although no specific function has been attributed to the Hep segments, our data on the ability of the HOX11 Hep motif to assist transactivation suggests that protein interactions with this segment may constitute one of the functions. As a means of testing a role of related Hep/octapeptide motifs in transcriptional activation, the Hep motif from the homeodomain protein Hlx and the octapeptide sequence from Pax-2 were fused to GAL4-DBD (Y16 and Y17). Figure 3B (upper panel) shows that both the Hlx and Pax2 sequences behave similarly to the HOX11 Hep motif in activating transcription in yeast at efficiencies comparable to that of the HOX11 Hep motif when protein levels are compared (Fig. 3B, lower panel).
Maximal activation of the endogenous Hdg-1 gene by HOX11 in NIH 3T3 cells requires the NH2-terminal 50 amino acids.
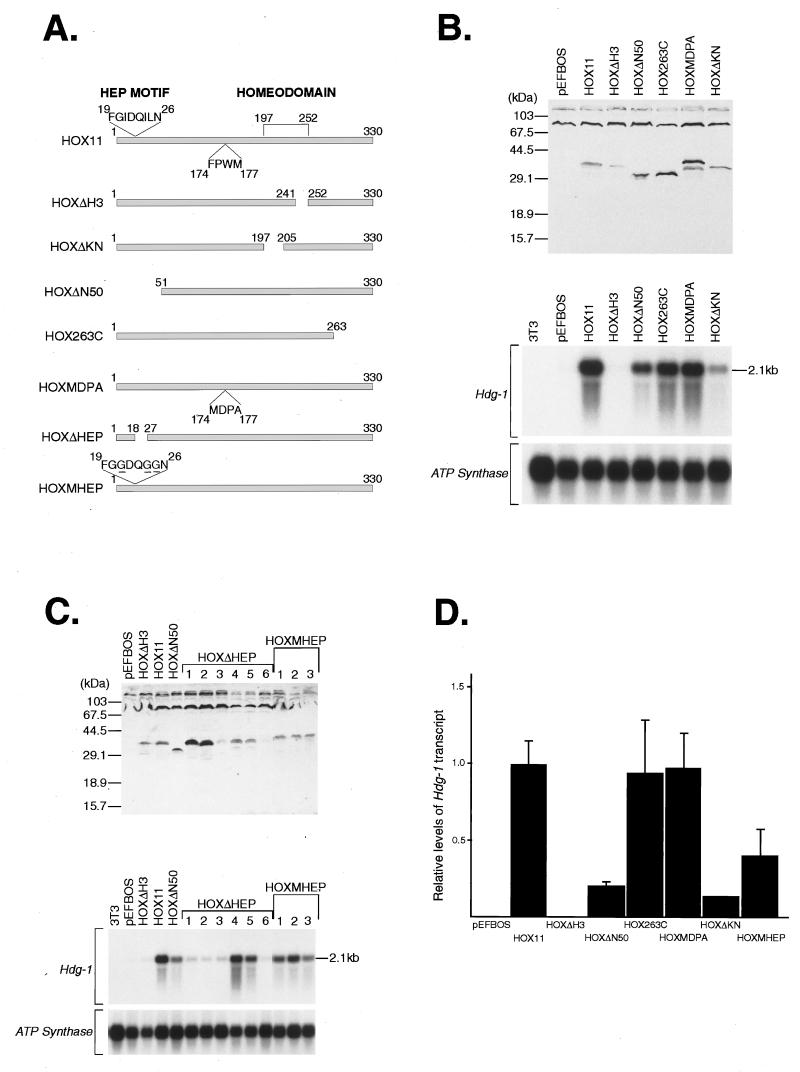
The one-hybrid, fusion protein approach used above is artificial in both the context of the mutants used (i.e., fusion with the GAL4-DBD) and the transcription-responsive assay used (i.e., a transient-reporter assay). A more physiologically relevant system would be the test of mutagenesis on HOX11 functions in vivo, via activation of a gene known to be regulated by HOX11. Such a gene has recently been identified (denoted Hdg-1) by cDNA representational difference analysis (8). Hdg-1 is upregulated in NIH 3T3 cells stably expressing HOX11 protein, and this provides an in vivo assay for HOX11 functional domains. NIH 3T3 cells were stably transfected with expression vectors coding for normal HOX11 proteins or mutant forms. NIH 3T3 clones which expressed mutant HOX11 proteins were isolated (Fig. 4A), and protein expression was confirmed by Western blotting with a polyclonal anti-HOX11 antiserum (Fig. 4B and C, upper panels).
FIG. 4.
The NH2-terminal 50 amino acids of HOX11 is required for optimal induction of Hdg-1 expression in NIH 3T3 cells. (A) Schematic representation of HOX11 wild-type and mutant proteins expressed from the indicated HOX11 constructs made in the pEFBOS vector. The positions of the Hep sequence, the conserved FPWM motif, and the homeodomain are indicated in the diagram of HOX11, as are the MDPA mutation in HOXMDPA and the mutation of the Hep sequence in HOXMHEP. The various constructs were cotransfected into NIH 3T3 cells with a neomycin resistance vector, and individual clones were picked. (B) The upper panel shows Western blot detection of HOX11 wild-type and mutant proteins in extracts from a representative set of NIH 3T3 clones with a polyclonal HOX11 antiserum. The lower panel shows Northern blot detection of the Hdg-1 transcript in RNA from a representative set of NIH 3T3 clones expressing wild-type and mutant HOX11 proteins. The 2.1-kb Hdg-1 transcript is detected in a clone expressing wild-type HOX11 but not in a clone selected to contain the empty expression vector (pEFBOS) or in normal NIH 3T3 cells. The effect of various HOX11 mutations (as represented in panel A) on the ability to induce Hdg-1 expression is shown. Although expressed at a level comparable to that of HOX11, HOXΔN50 exhibits a fourfold reduction in its ability to induce Hdg-1 (based on densitometric analysis, as shown in panel D). Hybridization with an ATP synthase probe was used as a control for loading of the Northern filter. (C) The upper panel shows Western blot detection of HOX11 Hep mutants in extracts from NIH 3T3 clones with a polyclonal HOX11 antiserum. Six independent clones expressing HOXΔHEP were obtained and analyzed. Three clones expressing HOXMHEP are shown. The lower panel shows Northern blot detection of the Hdg-1 transcript in RNA from NIH 3T3 clones expressing HOX11 Hep mutants. The 2.1-kb Hdg-1 transcript is detected in all three HOXMHEP clones but at levels approximately 2.5-fold lower than in HOX11 wild-type clones (based on densitometric analysis, as shown in panel D). The Hdg-1 transcript is detected in two of the six HOXΔHEP clones. Hybridization with an ATP synthase probe was used as a loading control for the Northern filter. (D) Histogram showing relative levels of Hdg-1 transcript in NIH 3T3 clones expressing wild-type and mutant HOX11 proteins. In each case, three independent clones were analyzed (except for the HOXΔKN mutant, which is based on data obtained with two independent clones). The relative signal values were obtained by densitometric scanning of autoradiographs and normalized for levels of HOX11 transcript.
Expression of Hdg-1 was detected as a 2.1-kb transcript in Northern analysis of RNA from NIH 3T3 clones expressing wild-type HOX11 but not in NIH 3T3 cells or NIH 3T3 cells transfected with only the expression vector (Fig. 4B and C, lower panels). It was found that mutations of the homeodomain affected Hdg-1 gene activation. Deletion of the third helix of the homeodomain prevented Hdg-1 activation (HOXΔH3) (Fig. 4B and C, lower panels, and Fig. 4D), while deletion of the NH2-terminal arm of the homeodomain (also required for contacting DNA [16]) resulted in low levels of Hdg-1 expression (HOXΔKN) (Fig. 4B and C, lower panels, and Fig. 4D). The effects seen with these mutants indicate that an intact homeodomain is required by HOX11 to induce Hdg-1 expression, most probably therefore acting through DNA binding (although not necessarily binding to the Hdg-1 gene itself).
In relation to the NH2-terminal region of HOX11, which we found to be a modular activation domain, a deletion of the NH2-terminal 50 amino acids of HOX11 (HOXΔN50) (Fig. 4A) was found to impair Hdg-1 induction. Although HOXΔN50 and HOX11 protein levels were comparable (Fig. 4B and C, upper panels), HOXΔN50 induced lower levels of Hdg-1 RNA than did wild-type HOX11 (Fig. 4B and C, lower panels). Densitometric quantitation of induced RNA levels show an approximately fourfold reduction of Hdg-1 with HOXΔN50 (Fig. 4D). The NH2-terminal 50 amino acids of HOX11 therefore appears to be important for the optimal induction of Hdg-1 in NIH 3T3 cells, and residual levels of transcription observed after HOXΔN50 expression are presumably mediated by other regions of the HOX11 protein.
The role of the Hep sequence in induction of Hdg-1 expression was also assessed in NIH 3T3 clones expressing two different Hep mutants (HOXMHEP and HOXΔHEP [Fig. 4A]). HOXMHEP contains three amino acid substitutions in the HOX11 Hep sequence (a mutation which impairs the ability of the isolated Hep sequence to activate in yeast [Y14] [Fig. 3A]), and this protein induces Hdg-1 expression at levels 2.5-fold lower than HOX11 (Fig. 4C, lower panel, and Fig. 4D). However, the second Hep mutant, HOXΔHEP (containing an internal deletion of the Hep motif) produced clonal differences. Although HOXΔHEP was present (as judged by Western analysis [Fig. 4C, upper panel]), some NIH 3T3 clones of HOXΔHEP expressed levels of Hdg-1 RNA comparable to levels induced by wild-type HOX11 (HOXΔHEP clones 4 and 5 [Fig. 4C]), while others did not appear to express Hdg-1 at all (HOXΔHEP clones 1, 2, 3, and 6 [Fig. 4C]). No clonal variation was seen for any other HOX11 protein studied. It is possible that for HOXΔHEP, conditions of growth or specific cellular environment will explain this variation, and this is worthy of a separate investigation. However, the present data indicate that the Hep sequence plays a role in the transcriptional activation of Hdg-1 mediated by the NH2-terminal region of HOX11 in vivo.
Cooperative interactions between homeodomain proteins and Pbx/exd proteins appear to be mediated, at least in part, by a motif, FPWM, just upstream of the homeodomain (16). However, mutation of this motif at amino acids 174 to 177 (HOXMDPA [Fig. 4A]) had no effect on the ability to induce Hdg-1 expression (Fig. 4B, lower panel, and Fig. 4D). Therefore, if HOX11 does require interaction with a cofactor for Hdg-1 induction, other regions of the protein must be involved. In addition, the COOH-terminal region of HOX11, which was previously thought to contribute to the HOX11 activation potential (28), does not appear to be required for Hdg-1 induction. A deletion of 67 amino acids from the COOH terminus of HOX11 (HOX263C [Fig. 4A]) had no effect on Hdg-1 induction (Fig. 4B, lower panel, and Fig. 4D).
DISCUSSION
NH2-terminal activation domain and the Hep sequence.
HOX11 is assumed to be an important transcriptional regulator, both in the context of genes required for spleen development in the embryo and following deregulation of the HOX11 gene by chromosomal translocations, for genes involved in the development of T-lineage tumors. It is therefore important to identify regions of HOX11 required for activation of HOX11-dependent genes in vivo. The results presented here show that the HOX11 protein can activate endogenous gene expression (directly or indirectly) in a homeodomain-dependent manner and that an activation domain at the NH2 terminus of the protein is important for this.
The NH2-terminal activation domain was initially delineated by the yeast one-hybrid assay. All fusion proteins containing the NH2-terminal 50 amino acids of HOX11 activated, including a fusion of amino acids 1 to 211 of HOX11 (data not shown), analogous to the fusion clone used in a two-hybrid assay (12). This mapping of an activation domain to the NH2-terminal 50 amino acids of HOX11 differs from the results of a previous study which identified multiple regions of HOX11 involved in transactivation, including an NH2-terminal glycine-proline-rich region of about 190 amino acids (28). In our yeast one-hybrid analysis, the glycine-proline-rich region does not appear to be important, with activation function being localized discretely to the NH2-terminal 50 amino acids.
Within the NH2-terminal 50 amino acids of HOX11 is a conserved Hep sequence for which no function has yet been assigned. Our data support a possible role for the Hep sequence in transcriptional activation. First, it appears to be a necessary component of the NH2-terminal activation region, and second, the Hep sequence itself is sufficient to support activation when used in isolation in one-hybrid assays, with contributions from conserved hydrophobic residues in the core sequence. In addition, the Hep motif of Hlx and the related octapeptide motif of Pax-2 are both capable of activation when fused to the GAL4-DBD. Previously, the Pax-2 octapeptide had been shown to act in cis as a transcriptional repressor domain (14). The Pax-2 octapeptide therefore appears to have the capacity for modular transcriptional function, being capable of repression when studied in the context of the Pax-2 protein (14) and activation when studied in isolation. The Hep sequence may also act in both positive and negative transcriptional regulation.
Regions of HOX11 required for Hdg-1 induction.
It has become apparent that protein activation domains mapped in reporter assays may not necessarily be important for all in vivo function; e.g., studies of the GATA-1 transcription factor have shown that an obligatory activation domain mapped by standard reporter assays is dispensable for GATA-1 function in terminal erythroid cell differentiation (27). The relevance of transcriptional assays which identify protein segments with activation potential in isolation from the intact native protein is therefore at issue. Although the NH2-terminal 50 amino acids of HOX11 could function as an activation domain in one-hybrid assays, it was therefore important to evaluate the role of this region in a functional assay that provides a stringent analysis of HOX11 protein function. In particular, such an assay could provide insight into the differing results obtained from one-hybrid analyses of HOX11 (reference 28 and data presented above). The gene Hdg-1 is activated after HOX11 expression in NIH 3T3 cells (8), and this chromosomal activation provides a powerful physiological model for analysis of HOX11 functional domains. Whether the induction of Hdg-1 expression is a direct effect of HOX11 on the Hdg-1 promoter is unknown. Mapping of the Hdg-1 promoter to identify the “HOX11-responsive element” is in progress to clarify this (8a).
The induction of Hdg-1 expression in transfected NIH 3T3 cells was found to be dependent on the intact homeodomain. When the NH2-terminal 50 amino acids of HOX11 was deleted, Hdg-1 induction was significantly impaired (fourfold), in agreement with mapping of this region as an activation domain in one-hybrid reporter assays. The NH2-terminal 50 amino acids of HOX11 is therefore important for in vivo transcriptional control of a chromosomal gene. The Hep sequence, located within the NH2-terminal 50 amino acids of HOX11, may play a role in this activation of Hdg-1 transcription. However, since regions of HOX11 other than the NH2 terminus contribute to Hdg-1 induction, the in vivo role of the Hep sequence is difficult to assess. If the Hep sequence is an important component of the NH2-terminal region, one would expect Hep mutants to exhibit a reduced level of Hdg-1 induction comparable to that of the HOXΔN50 mutant. Consistent with this, the HOXMHEP mutant (containing three amino acid substitutions within the conserved Hep sequence) activates Hdg-1, with the induced level close to that of HOXΔN50 (Fig. 4D).
Although the NH2-terminal 50 amino acids is important for Hdg-1 induction, other regions of the HOX11 protein are clearly involved. The homeodomain is essential, since deletions within this domain (either of the third helix or of the NH2-terminal arm) result in the loss of Hdg-1 induction. Both these regions of the homeodomain are predicted to contact DNA, and so deletions made presumably result in a loss of HOX11 DNA-binding activity. The HOX11 homeodomain has also been suggested to contain activation function (28) and may therefore be playing a dual role in Hdg-1 induction. In our yeast one-hybrid analysis, we were unable to study the intact HOX11 homeodomain due to growth retardation in the presence of the functional homeodomain, and it is therefore impossible for us to address this question unless specific mutations which ablate the activation function of the homeodomain while retaining DNA-binding activity can be identified. However, the COOH-terminal glutamine-rich region of HOX11, which previously had been thought to contain activation function (28), did not activate in our yeast one-hybrid analysis and also does not appear to be required for Hdg-1 induction.
Mechanism of Hdg-1 induction.
The importance of the NH2-terminal 50 amino acids of HOX11 for activation of a chromosomal gene in vivo may provide insights into the mechanisms of HOX11-mediated transcriptional regulation by helping to identify interacting factors. Apart from the conserved Hep sequence, the NH2-terminal 50 amino acids does not resemble any known transcriptional regulatory domain. However, since this region is involved in Hdg-1 induction (but not necessarily by direct binding to the Hdg-1 promoter), it presumably interacts, either directly or indirectly, with a component(s) of the basal transcriptional machinery. The identification of such factors should further delineate the role of the conserved Hep sequence and help to elucidate the functional role of HOX11 in both splenogenesis and tumorigenesis.
ACKNOWLEDGMENTS
N.M. was supported by an LRF fellowship, and W.G. was supported by a C. J. Martin fellowship.
We thank A. Forster for important technical help throughout this project.
REFERENCES
- 1.Allen J D, Lints T, Jenkins N A, Copeland N G, Strasser A, Harvey R P, Adams J M. Novel murine homeo box gene on chromosome 1 expressed in specific hematopoietic lineages and during embryogenesis. Genes Dev. 1991;5:509–520. doi: 10.1101/gad.5.4.509. [DOI] [PubMed] [Google Scholar]
- 2.Cleary M L. Oncogenic conversion of transcription factors by chromosomal translocations. Cell. 1991;66:619–622. doi: 10.1016/0092-8674(91)90105-8. [DOI] [PubMed] [Google Scholar]
- 3.Dear T N, Colledge W H, Carlton M B L, Lavenir I, Larson T, Smith A J H, Warren A J, Evans M J, Sofroniew M V, Rabbitts T H. The Hox11 gene is essential for cell survival during spleen development. Development. 1995;121:2909–2915. doi: 10.1242/dev.121.9.2909. [DOI] [PubMed] [Google Scholar]
- 4.Dear T N, Sanchez-Garcia I, Rabbitts T H. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc Natl Acad Sci USA. 1993;90:4431–4435. doi: 10.1073/pnas.90.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dube I D, Kamel-Reid S, Yuan C C, Lu M, Wu X, Corpus G, Raimondi S C, Crist W M, Carroll A J, Minowada J, Baker J B. A novel human homeobox gene lies at the chromosome 10 breakpoint in lymphoid neoplasias with chromosomal translocation t(10;14) Blood. 1991;78:2996–3003. [PubMed] [Google Scholar]
- 6.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A P, Vogelstein B A. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 8.Greene, W. K., S. Bahn, N. Masson, and T. H. Rabbitts. Unpublished data.
- 8a.Greene, W. K., and T. H. Rabbitts. Unpublished data.
- 9.Hatano M, Roberts C W M, Minden M, Crist W M, Korsmeyer S J. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukaemia. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 10.Hawley R G, Fong A Z C, Lu M, Hawley T S. The HOX11 homeobox-containing gene of human leukemia immortalizes murine hematopoietic precursors. Oncogene. 1994;9:1–12. [PubMed] [Google Scholar]
- 11.Hawley R G, Fong A Z, Reis M D, Zhang N, Lu M, Hawley T S. Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res. 1997;57:337–345. [PubMed] [Google Scholar]
- 12.Kawabe T, Muslin A J, Korsmeyer S J. HOX11 interacts with protein phosphatases PP2A and PP1 and disrupts a G2/M cell-cycle checkpoint. Nature. 1997;385:454–458. doi: 10.1038/385454a0. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy M A, Gonzalez-Sarmiento R, Kees U R, Lampert F, Dear N, Boehm T, Rabbitts T H. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc Natl Acad Sci USA. 1991;88:8900–8904. doi: 10.1073/pnas.88.20.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner M S, Dressler G R. Mapping of Pax-2 transcription activation domains. J Biol Chem. 1996;271:21088–21093. doi: 10.1074/jbc.271.35.21088. [DOI] [PubMed] [Google Scholar]
- 15.Lu M, Gong Z, Shen W, Ho A D. The tcl-3 proto-oncogene altered by chromosomal translocation in T-cell leukaemia codes for a homeobox protein. EMBO J. 1991;10:2905–2910. doi: 10.1002/j.1460-2075.1991.tb07840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann R S. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabbitts T H. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell. 1991;67:641–644. doi: 10.1016/0092-8674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- 19.Rabbitts T H. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 20.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 21.Roberts C W M, Shutter J R, Korsmeyer S J. Hox11 controls the genesis of the spleen. Nature. 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 22.Rose M D, Winston F, Hieter P, editors. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 23.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Garcia I, Axelson H, Rabbitts T H. Functional diversity of LIM proteins: amino-terminal activation domains in the oncogenic proteins RBTN1 and RBTN2. Oncogene. 1995;10:1301–1306. [PubMed] [Google Scholar]
- 25.Shirasawa S, Yunker A M, Roth K A, Brown G A, Horning S, Korsmeyer S J. Enx (Hox11L1)-deficient mice develop myenteric neuronal hyperplasia and megacolon. Nat Med. 1997;3:646–650. doi: 10.1038/nm0697-646. [DOI] [PubMed] [Google Scholar]
- 26.Tang S, Breitman M L. The optimal binding sequence of the Hox11 protein contains a predicted recognition core motif. Nucleic Acids Res. 1995;23:1928–1935. doi: 10.1093/nar/23.11.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss M J, Yu C, Orkin S H. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N, Shen W, Ho A D, Lu M. Three distinct domains in the HOX-11 homeobox oncoprotein are required for optimal transactivation. Oncogene. 1996;13:1781–1787. [PubMed] [Google Scholar]
- 29.Zutter M, Hockett R D, Roberts C W M, McGuire E A, Bloomstone J, Morton C C, Deaven L L, Crist W M, Carroll A J, Korsmeyer S J. The t(10;14)(q24;q11) of T-cell acute lymphoblastic leukemia juxtaposes the δ T-cell receptor with TCL3, a conserved and activated locus at 10q24. Proc Natl Acad Sci USA. 1990;87:3161–3165. doi: 10.1073/pnas.87.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]