Abstract
The frequency of locally transmitted dengue virus (DENV) infections has increased in Europe in recent years, facilitated by the invasive mosquito species Aedes albopictus, which is well established in a large area of Europe. In Italy, the first indigenous dengue outbreak was reported in August 2020 with 11 locally acquired cases in the Veneto region (northeast Italy), caused by a DENV-1 viral strain closely related to a previously described strain circulating in Singapore and China. In this study, we evaluated the vector competence of two Italian populations of Ae. albopictus compared to an Ae. aegypti lab colony. We performed experimental infections using a DENV-1 strain that is phylogenetically close to the strain responsible for the 2020 Italian autochthonous outbreak. Our results showed that local Ae. albopictus is susceptible to infection and is able to transmit the virus, confirming the relevant risk of possible outbreaks starting from an imported case.
Keywords: dengue virus, mosquito, arboviruses, Italy, Aedes albopictus, experimental infection
1. Introduction
Dengue fever is a mosquito-borne tropical disease caused by four distinct but closely related serotypes of Dengue virus (DENV; family Flaviviridae, genus Flavivirus) that are transmitted by Aedes mosquitoes [1]. The species Aedes aegypti, widely spread in endemic areas, is considered the main vector of DENV. Although considered a secondary vector of DENV, Ae. albopictus is however associated with virus transmission in several areas of the world, including Europe [2]. The first Italian indigenous DENV outbreak was reported in Montecchio Maggiore (Vicenza province, Veneto region) in August 2020 with 11 cases secondary to an imported case from Indonesia [3]. To evaluate the role of Ae. albopictus in DENV transmission, we analyzed the vector competence, through experimental infections, of two Italian populations of Ae. albopictus. Potential vertical (transovarial) transmission of DENV was also evaluated. Since it was not possible to isolate the DENV-1 strain responsible for the 2020 Italian outbreak, either from mosquito pools or from human sera, for the experimental infections, we used a phylogenetically highly related DENV-1 strain circulating in Singapore.
2. Materials and Methods
2.1. Virus and Mosquito Populations
DENV-1 isolate SG (EHI)D1/30889Y14 (accession number MG097876) was selected based on the high genetic homology of 98.27% with the DENV-1 strain of the 2020 Italian autochthonous outbreak (namely VI/Italy/2020, accession number MZ291446; Supplementary File). The dengue strain was kindly provided by the Environmental Health Institute, National Environment Agency, Singapore for the purposes of the study. The virus was grown in VERO cells and titrated via plaque assay [4]. Two geographically different Ae. albopictus populations were collected from Rome (Lazio region, Central Italy) and Montecchio Maggiore (Veneto region, northeast Italy), the latter being the town where the 2020 DENV-1 outbreak occurred. A long-established Ae. aegypti laboratory colony (collected in Reynosa, Mexico, in 1998) was used as the reference. The eggs of two Ae. albopictus populations were collected by using ovitraps and were reared in the insectarium of the Istituto Superiore di Sanità in Rome. Adults were maintained before the test for a few generations (F3–F5) in climatic chambers under the following conditions: 26 ± 1 °C temperature; 70% relative humidity (RH); and a 14:10 h light/dark photoperiod. To check that the two Ae. albopictus populations were virus free, 5 pools of 20 specimens (males and females) per pool for each F0 offspring generation were analyzed for DENV via real-time PCR (qRT-PCR) [5].
2.2. Genetic Similarity Analysis
The sequence of the DENV-1 strain of the 2020 Italian autochthonous outbreak (VI/Italy/2020; accession number MZ291446) [3] was compared to DENV-1 sequences available in a sequence repository by using the BLASTN tool in order to identify the most related viral strains. We found more than 200 isolates showing high homology with MZ291446 (<98% homology). Few of these were cultured. MG097876, circulating in Singapore where Ae. albopictus was described, was selected because it was highly related (98.27%) and isolated in cells.
2.3. Experimental Infection
The experimental infections were performed in a biosafety level 3 laboratory with 5–10 days old female mosquitoes that were allowed to feed for 60 min using a membrane feeding apparatus containing a blood–DENV mixture. The virus was diluted in rabbit blood to a final virus concentration of 5 log10 plaque-forming units (PFU)/mL. The infectious blood was maintained at 37 °C through a warm water circulation system. Unfed and partially fed mosquitoes were excluded from the study; only completely engorged females were transferred to a climate chamber (set to the same environmental conditions as previously described) and were provided with a 10% sucrose solution. They were monitored for 28 days. At 0, 7, 14, 21, and 28 days post-infectious blood meal, 10–23 mosquitoes of each species and population were individually processed. To determine the vector competence, the whole body (head, thorax, and abdomen), legs plus wings, and saliva of the mosquitoes were screened for DENV RNA to estimate the infection, dissemination, and transmission rates (IR, DR, and TR), respectively [5]. Mosquito saliva was collected by inserting the entire proboscis into a single quartz capillary filled with 1 μL of Vaseline oil. Vaseline enables the clear identification of saliva droplets, helping to rule out the possibility that a negative result for the virus in a saliva sample is due to a lack of saliva production. One microliter of 1% pilocarpine, a saliva stimulant that is an analogue of acetylcholine [6], prepared in phosphate-buffered saline (PBS) at 0.1% Tween 80, was applied on the mosquito thorax. After 30 min, the medium containing the saliva was expelled under pressure from the capillary into a 1.5 mL tube containing 500 μL of mosquito diluent consisting of PBS, 20% heat-inactivated FBS, and a 1% penicillin/streptomycin/amphotericin B mix (Invitrogen Corp., Carlsbad, CA, USA; GIBCO Brl, Rockville, MD, USA). Virus titers were quantified by using crossing point values obtained from a qRT-PCR [7] and comparing them with a standard curve obtained from 10-fold serial dilutions of virus stock of known concentration [8,9]. Potential vertical transmission of DENV was also analyzed. For this, mosquitoes were allowed to lay eggs (first gonotrophic cycle—FGC) after the infectious blood meal. Larvae from the FGC were reared up to adulthood in the climatic chamber, and adults were tested for DENV RNA via qRT-PCR analysis on pools (5 pools for Ae. albopictus from Rome, 3 pools for Ae. albopictus from Montecchio Maggiore, and 4 pools for Ae. aegypti), consisting of 5 mosquitoes/pool.
2.4. Data Analysis and Statistics
Statistical significance tests were performed using a parametric Student’s t test. All statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). For all analyses, a p-value ≤ 0.05 was considered significant.
3. Results
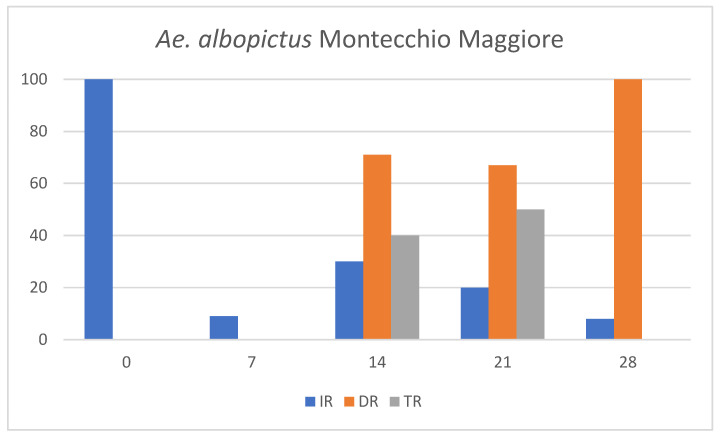
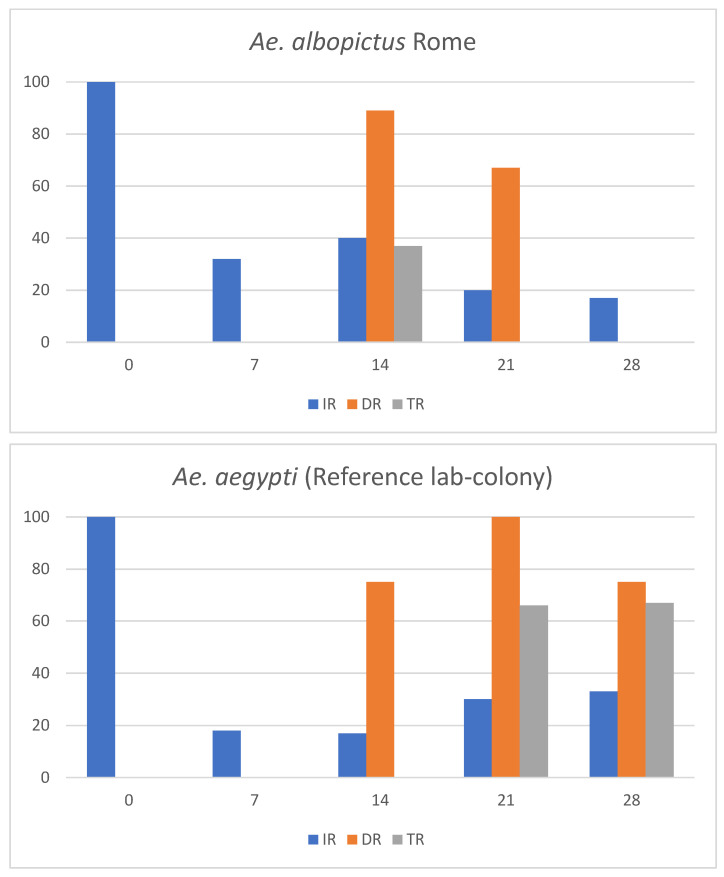
Both Ae. albopictus field populations collected in Montecchio Maggiore and Rome were DENV free as all tested pools were negative. All tested Aedes populations showed susceptibility to DENV-1 infection, allowing the virus to replicate and spread to the salivary glands. The values of IR, DR, TR, and the mean viral titers are shown in Figure 1 and Table 1, respectively.
Figure 1.
Infection (IR), dissemination (DR) and transmission rates (TR) in Aedes albopictus and Ae. aegypti colonies at different collection times postinfection with DENV-1.
Table 1.
Infection rate (IR): number of positive bodies/number of tested fed females; dissemination rate (DR): number of positive legs plus wings/number of positive bodies; transmission rate (TR): number of positive saliva/number of positive bodies.
| Species | Days Post Infection | IR | Main Value (PFU/mL) | DR | Main Value (PFU/mL) | TR | Main Value (PFU/mL) |
|---|---|---|---|---|---|---|---|
| Ae. albopictus Rome | 0 | 11/11 | 1.6 × 101 | - | - | ||
| 7 | 7/21 | 6.3 × 101 | 0/7 | 0/7 | |||
| 14 | 9/22 | 7.6 × 102 | 8/9 | 1.3 × 101 | 3/9 | 0.4 × 101 | |
| 21 | 3/15 | 3.0 × 102 | 2/3 | 0.7 × 101 | 0/3 | ||
| 28 | 2/12 | 4.1 × 102 | 0/2 | 0/2 | |||
| Ae. albopictus Montecchio M. | 0 | 12/12 | 1.9 × 101 | - | - | ||
| 7 | 2/22 | 2.4 × 101 | 0/2 | 0/2 | |||
| 14 | 7/23 | 3.8 × 102 | 5/7 | 0.2 × 101 | 2/7 | 0.1 × 101 | |
| 21 | 3/15 | 6.4 × 102 | 2/3 | 0.5 × 101 | 1/3 | 0.5 × 101 | |
| 28 | 1/12 | 7.4 × 102 | 1/1 | 4.4 × 101 | 0/1 | ||
| Ae. aegypti Reynosa | 0 | 12/12 | 3.5 × 101 | - | - | ||
| 7 | 4/22 | 1.9 × 101 | 0/4 | 0/4 | |||
| 14 | 4/23 | 4.2 × 101 | 3/4 | 0.5 × 101 | 0/4 | ||
| 21 | 3/10 | 1.4 × 103 | 3/3 | 1 × 101 | 2/3 | 0.2 × 101 | |
| 28 | 4/12 | 1.9 × 102 | 3/4 | 3.3 × 101 | 2/4 | 0.2 × 101 |
To confirm the ingestion of infectious viral particles, the engorged mosquitoes were analyzed immediately after the infectious blood meal. The results showed a viral titer of approximately 1 log10 PFU/mL in the tested specimens. The viral titers detected in the bodies of Ae. albopictus increased gradually in all mosquito specimens, reaching the highest mean values of 7.6 × 102 PFU/mL 14 days post-infection (dpi) and 7.4 × 102 PFU/mL 28 dpi in Rome and Montecchio Maggiore, respectively. In Ae. aegypti the highest mean titer was 1.4 × 103 PFU/mL achieved 21 dpi (Table 1). Viral titers were higher in Ae. aegypti compared to Ae. albopictus populations, in particular at 21 dpi. The analysis indicated a progressive increase in IR over time for both Ae. albopictus populations, reaching the maximum value 14 dpi (40% Rome, 30% Montecchio Maggiore); subsequently, IR values decreased at 21 and 28 dpi. Conversely, Ae. aegypti showed a steady increase in IR throughout the observation period. DENV-1 was detected in legs plus wings from 14 dpi in all tested populations. All three mosquito groups showed high DR values (range 67–100%). DENV-1 was detected in saliva 14 dpi in both Ae. albopictus populations, highlighting a shorter extrinsic incubation period (EIP) compared to Ae. aegypti, in which DENV-1 was first detected in saliva 21 dpi. However, it was not possible to detect DENV-1 in the saliva after 21 dpi in Ae. albopictus from Rome and 28 dpi in Ae. albopictus from Montecchio Maggiore. In contrast, DENV-1 was detected in the saliva of Ae. aegypti until 28 dpi. Nevertheless, DENV-1 titers were relatively low in saliva (0.1 × 101–0.5 × 101 PFU/mL) in all analyzed populations (Table 1). Cumulative values of IR, DR and TR calculated from 7 to 28 dpi are reported in Table 2.
Table 2.
Infection rate (IR): number of positive bodies/number of tested fed females; dissemination rate (DR): number of positive legs plus wings/number of positive bodies; transmission rate (TR): number of positive saliva/number of positive bodies. Vector competence index (VCI): maximum value 1.0.
|
Ae. albopictus Rome |
Ae. albopictus Montecchio M. |
Ae. aegypti Reference Lab-Colony |
|
|---|---|---|---|
| IR | 30% | 18% | 22% |
| DR | 48% | 61% | 60% |
| TR | 14% | 23% | 27% |
| VCI | 0.04 | 0.04 | 0.06 |
The Aedes albopictus population from Rome exhibited a higher IR (30%) compared to the other populations (18% for Ae. albopictus of Montecchio Maggiore and 22% for Ae. aegypti). However, the results highlighted a higher mean value of TR for Ae. aegypti (27%) compared to both Ae. albopictus populations (14% for Rome and 23% for Montecchio Maggiore). There was no evidence of vertical transmission in the offspring of the three Aedes populations.
4. Discussion
Dengue fever is a mosquito-borne tropical disease caused by DENV that has become a major public health problem in recent years causing approximately 390 million infections globally every year [10]. Several DENV strains have been implicated in autochthonous transmission in Europe since 2010. In recent years, DENV-1 and -2 have been the most prevalent serotypes in infections among European travelers [11]. Although Ae. albopictus is considered a secondary vector of DENV, its widespread distribution and high density in temperate areas represent a real risk for local outbreaks originating from imported cases. Until November 2023, 126 autochthonous/non-travel associated dengue cases have been reported in Europe in Italy (82), France (41), and Spain (3) [12,13]. In the summer of 2022, 65 autochthonous cases of DENV transmitted by Ae. albopictus were reported in France [14]. The unexpectedly high number of indigenous cases was however associated with nine distinct virus introduction and transmission events. This demonstrates the high risk of autochthonous transmission from imported cases but also the relatively small number of secondary cases associated with each individual index case. The first dengue outbreak in Italy occurred in August–September 2020. The outbreak was geographically limited to a small town in the Veneto region and caused very few indigenous human cases. Aedes albopictus was the mosquito vector incriminated as it is present and abundant in the area. This event could have been affected by various concomitant biotic and abiotic factors such as the lower vectorial competence of the Ae. albopictus population compared to the global primary vector, Ae. Aegypti, or the sudden change in atmospheric conditions with heavy rains and a sudden drop in temperatures which, together with vector control operations, led to a drastic decrease in the mosquito density. Another important determinant was the concomitant SARS-Cov-2 pandemic, during which the health authorities imposed a lockdown, effectively limiting people’s movements. This is confirmed by the fact that the people infected with DENV-1 during the outbreak were all relatives and neighbors [3]. It is known that the pathogenetic variation in virus strains, geographic distribution, vector abundance, and climatic factors influence vector susceptibility to DENV [15]. Studies on the role of possible mutations responsible for the better adaptation of arboviruses in Ae. albopictus have recently been conducted. Bellone et al. demonstrated that consecutive in vivo passages in Ae. albopictus resulted in the emergence of specific DENV-1 strains exhibiting increased infectivity for this vector both in vivo and in cultured mosquito cells. These alterations were facilitated by numerous adaptive mutations in the virus genome [2]. A comparative examination of the CHIKVs responsible for the Italian epidemics in 2007 and 2017 revealed that only the 2007 strain possessed the adaptive mutation E1 A226V for Ae. albopictus. These results highlight the significance of genomic studies in elucidating the potential role of the mutations in determining the adaptive capacity of a virus to different vectors [16]. Moreover, arboviruses adapt to secondary vectors as exemplified by the adaptation of Chikungunya virus to Ae. albopictus [17]. Therefore, a similar scenario cannot be ruled out for DENV. Our findings demonstrated that the Asiatic DENV-1 strain that was closely related to the 2020 Italian outbreak strain equally infected both Ae. aegypti and two local populations of Ae. albopictus. Interestingly, the virus reached the salivary glands of Ae. albopictus earlier than in Ae. aegypti. However, the virus survived longer in Ae. aegypti than in both populations of Ae. albopictus, suggesting a longer transmission potential of DENV-1 in Ae. aegypti. In our study, strains of Ae. albopictus collected in different areas of the country were infected with the same titer of DENV-1 in the same environment and experimental conditions to evaluate their vector competence. Of note, in agreement with our results, the DENV-1 strain used in the present study belonged to an established lineage during the DENV-1 outbreak in Singapore in 2013–14 and demonstrated efficient infection of an Ae. Albopictus-derived cell line [18]. The IR, DR, and TR rates in our study were not significantly different between Ae. albopictus and Ae. aegypti populations. In addition, the viral titers detected in the Ae. albopictus and Ae. aegypti populations were comparable. This result highlights the potentially important role of Ae. albopictus in the transmission of DENV-1 in non-endemic areas. In fact, the shorter EIP observed in Ae. albopictus compared to Ae. aegypti is noteworthy with regard to its potential to transmit DENV-1 effectively. Dengue outbreaks in France and Italy in 2023 confirm the increasing risk of DENV transmission in Europe potentially by Ae. albopictus, originating from imported cases. Data on the presence, abundance, seasonal fluctuations, and evaluation of vectorial competence of invasive mosquito species circulating in a non-endemic country are pivotal to assess the risk of arboviral transmission chains and, together with strengthened surveillance systems, for implementation of prevention and control measures to mitigate the adverse impacts on human health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16020176/s1.
Author Contributions
Methodology, C.F. (Claudia Fortuna), M.D.L., F.S., L.T., G.M. and G.V.; experimental steps, C.F. (Claudia Fortuna), M.D.L., F.S., L.T., G.M., G.V., A.A., F.C. and I.B.; phylogenetic analysis, C.A.; data analysis, C.F. (Claudia Fortuna) and G.M.; writing—original draft preparation, C.F. (Claudia Fortuna) and M.D.L.; writing—review and editing, H.C.H., F.M. and D.B.; technical support, C.F. (Cristiano Fiorentini). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was supported by Bando Ricerca Indipendente ISS 2020-2022: Evaluation of the vector competence of Italian Aedes albopictus populations for dengue virus type 1 and 2 in order to contribute to the risk assessment of dengue in Italy (Project No: Iss20-80089926288b) and partially by EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Gubler D.J., Ooi E.E., Vasudevan S., Farrar J. Dengue and Dengue Hemorrhagic Fever. 2nd ed. CABI; Wallingford, UK: 2014. [Google Scholar]
- 2.Bellone R., Lequime S., Jupille H., Göertz G.P., Aubry F., Mousson L., Piorkowski G., Yen P.S., Gabiane G., Vazeille M., et al. Experimental adaptation of dengue virus 1 to Aedes albopictus mosquitoes by in vivo selection. Sci. Rep. 2020;10:18404. doi: 10.1038/s41598-020-75042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzarini L., Barzon L., Foglia F., Manfrin V., Pacenti M., Pavan G., Rassu M., Capelli G., Montarsi F., Martini S., et al. First autochthonous dengue outbreak in Italy, August 2020. Eurosurveillance. 2020;25:2001606. doi: 10.2807/1560-7917.ES.2020.25.36.2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAuley A.J., Beasley D.W.C. Propagation and Titration of West Nile Virus on Vero Cells. Methods Mol. Biol. 2016;1435:19–27. doi: 10.1007/978-1-4939-3670-0. [DOI] [PubMed] [Google Scholar]
- 5.Drosten C., Göttig S., Schilling S., Asper M., Panning M., Schmitz H., Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubrulle M., Mousson L., Moutailler S., Vazeille M., Failloux A.B. Chikungunya virus and Aedes mosquitoes: Saliva is infectious as soon as two days after oral infection. PLoS ONE. 2009;4:e5895. doi: 10.1371/journal.pone.0005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortuna C., Remoli M.E., Di Luca M., Severini F., Toma L., Benedetti E., Bucci P., Montarsi F., Minelli G., Boccolini D., et al. Experimental studies on comparison of the vector competence of four Italian Culex pipiens populations for West Nile virus. Parasites Vectors. 2015;8:463. doi: 10.1186/s13071-015-1067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J., Stanfield S.M., Duffy M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S.L., Mores C.N., Lord C.C., Tabachnick W.J. Impact of extrinsic incubation temperature and virus exposure on vector competence of Culex pipiens quinquefasciatus Say (Diptera: Culicidae) for West Nile virus. Vector Borne Zoonotic Dis. 2007;7:629–636. doi: 10.1089/vbz.2007.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda C., Martínez M.J., Montalvo T., Eritja R., Navero-Castillejos J., Herreros E., Marqués E., Escosa R., Corbella I., Bigas E., et al. Arbovirus surveillance: First dengue virus detection in local Aedes albopictus mosquitoes in Europe, Catalonia, Spain, 2015. Eurosurveillance. 2018;23:1700837. doi: 10.2807/1560-7917.ES.2018.23.47.1700837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Centre for Disease Prevention and Control Dengue Worldwide Overview. 2023. [(accessed on 30 December 2023)]. Available online: https://www.ecdc.europa.eu/en/dengue-monthly.
- 13.Epicentro Istituto Superiore di Sanità Arbovirosi in Italia. 2023. [(accessed on 30 December 2023)]. Available online: https://www.epicentro.iss.it/arbovirosi/dashboard.
- 14.Cochet A., Calba C., Jourdain F., Grard G., Durand G.A., Guinard A., Noël H., Paty M.C., Franke F. Autochthonous dengue in mainland France, 2022: Geographical extension and incidence increase. Eurosurveillance. 2022;27:2200818. doi: 10.2807/1560-7917.ES.2022.27.44.2200818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soni M., Khan S.A., Bhattacharjee C.K., Dutta P.J. Experimental study of dengue virus infection in Aedes aegypti and Aedes albopictus: A comparative analysis on susceptibility, virus transmission and reproductive success. Invertebr. Pathol. 2020;175:107445. doi: 10.1016/j.jip.2020.107445. [DOI] [PubMed] [Google Scholar]
- 16.Lindh E., Argentini C., Remoli M.E., Fortuna C., Faggioni G., Benedetti E., Amendola A., Marsili G., Lista F., Rezza G., et al. Open Forum Infectious Diseases. Oxford University Press; New York, NY, USA: 2018. The Italian 2017 Outbreak Chikungunya Virus Belongs to an Emerging Aedes albopictus-Adapted Virus Cluster Introduced from the Indian Subcontinent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khongwichit S., Chansaenroj J., Chirathaworn C., Poovorawan Y.J. Chikungunya virus infection: Molecular biology, clinical characteristics, and epidemiology in Asian countries. Biomed. Sci. 2021;28:84. doi: 10.1186/s12929-021-00778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo C., Ping Tien W., Xu H., Ong J., Rajarethinam J., Ling Lai Y., Ng L.C., Hapuarachchi H.C. Highly Selective Transmission Success of Dengue Virus Type 1 Lineages in a Dynamic Virus Population: An Evolutionary and Fitness Perspective. iScience. 2018;6:38–51. doi: 10.1016/j.isci.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the article.