Abstract
HEF1, p130Cas, and Efs/Sin constitute a family of multidomain docking proteins that have been implicated in coordinating the regulation of cell adhesion. Each of these proteins contains an SH3 domain, conferring association with focal adhesion kinase; a domain rich in SH2-binding sites, phosphorylated by or associating with a number of oncoproteins, including Abl, Crk, Fyn, and others; and a highly conserved carboxy-terminal domain. In this report, we show that the HEF1 protein is processed in a complex manner, with transfection of a single cDNA resulting in the generation of at least four protein species, p115HEF1, p105HEF1, p65HEF1, and p55HEF1. We show that p115HEF1 and p105HEF1 are different phosphorylation states of the full-length HEF1. p55HEF1, however, encompasses only the amino-terminal end of the HEF1 coding sequence and arises via cleavage of full-length HEF1 at a caspase consensus site. We find that HEF1 proteins are abundantly expressed in epithelial cells derived from breast and lung tissue in addition to the lymphoid cells in which they have been predominantly studied to date. In MCF-7 cells, we find that expression of the endogenous HEF1 proteins is cell cycle regulated, with p105HEF1 and p115HEF1 being rapidly upregulated upon induction of cell growth, whereas p55HEF1 is produced specifically at mitosis. While p105HEF1 and p115HEF1 are predominantly cytoplasmic and localize to focal adhesions, p55HEF1 unexpectedly is shown to associate with the mitotic spindle. In support of a role at the spindle, two-hybrid library screening with HEF1 identifies the human homolog of the G2/M spindle-regulatory protein Dim1p as a specific interactor with a region of HEF1 encompassed in p55HEF1. In sum, these data suggest that HEF1 may directly connect morphological control-related signals with cell cycle regulation and thus play a role in pathways leading to the progression of cancer.
Research over the last 3 decades suggests that environmental cues related to cell shape and cell-cell contacts play an important role in regulating the nuclear processes of cell cycle, gene expression, and cell viability (17, 20). Efforts to decipher routes for transmission of this environmental information have identified a number of mechanisms, generally involving the sequential activation of kinase cascades that are coordinated initially by assembly of cascade constituents at a cytoskeletal membrane focal site. For communication of signals across the nuclear membrane, in some cases particular activated kinases undergo translocation from the cytoplasm into the nucleus in response to proliferative signals (12). In other cases, signaling substrates such as transcription factors are initially cell membrane associated or cytoplasmic and become nuclear following activation (32). Based on the results of this study, we propose that the HEF1 protein might function in a novel pathway transmitting growth control signals between focal adhesions at the cell periphery and the mitotic spindle.
We recently described HEF1 (34), a novel member of a newly defined family of docking adapter proteins that also includes p130Cas (for Crk-associated substrate) (59) and Efs (33) (also designated as Sin [1]). The members of this family have a common domain structure, with an amino-terminal SH3 domain, a large central domain encompassing multiple SH2-binding sites, and a carboxy-terminal domain conserved among all members of the HEF1-p130Cas-Efs protein family but not similar to any other protein in GenBank. Because of the multiplicity of interactive domains encompassed in each of these proteins, they appear likely to play a central role in the coordination of growth signal processing.
As p130Cas was the first characterized member of the p130Cas-HEF1-Efs group, its study is most advanced and has established a framework for evaluation of the other family members. p130Cas was initially described as a protein phosphorylated in response to Crk transformation via Abl and Src kinases (34, 49, 53). p130Cas has been shown by immunolocalization to reside primarily at focal adhesions and along adhesion-proximal regions of stress fibers (52), where it associates with focal adhesion kinase (FAK) (53). During integrin-mediated adhesion to extracellular matrix, p130Cas is phosphorylated along with other cytoskeletal proteins including FAK, paxillin, tensin, and cortactin (45, 52, 69). A central p130Cas function in transformation of some cell types has been suggested by findings that expression of antisense constructs of p130Cas in Ras- or Src-transformed NIH3T3 cells is sufficient to revert cells morphologically to a flat cell phenotype (4). Cumulatively, these data support a model in which p130Cas acts together with paxillin, tensin, and talin to coordinate stable connections between integrin receptors and the actin cytoskeleton: as such, altered regulation of p130Cas function by oncogenic phosphorylation might play a central role in establishing the characteristically altered cell morphologies of transformed cells.
In our prior characterization of HEF1, we determined that the protein shares not only a domain structure but also a number of functional properties with p130Cas. The HEF1 SH3 domain confers association with FAK, and immunofluorescence detects a substantial quantity of HEF1 at focal adhesions (34). HEF1 associates with and is phosphorylated by Abl (34). HEF1 phosphorylation occurs in response to integrin ligation, resulting in association with the CrkL adapter protein (38) and signaling to C3G (3). However, HEF1 differs from p130Cas in several potentially important ways. While p130Cas is abundant in many cell types, HEF1 mRNA levels vary extensively between tissues. Based on a limited initial analysis, we had previously noted that HEF1 expression might be particularly abundant in epithelial cell populations (34), while we and others have determined that HEF1 is also highly expressed and is an important signaling intermediate in differentiating B (38) and T (41) cells. Further, in contrast to p130Cas, which localizes exclusively to focal adhesions and the cytoplasm, immunofluorescence localizes a pool of the HEF1 population in the nucleus. The previous studies seeking to compare HEF1 and p130Cas expression were, however, limited by the lack of reagents capable of distinguishing the two proteins, given their high degree of similarity.
In this study, we have sought to answer key questions about HEF1 expression and function. Using a panel of antibodies specific for HEF1, we have found that the full-length transfected HEF1 cDNA is processed into at least four specific protein products, p115HEF1, p105HEF1, p65HEF1, and p55HEF1. In contrast to earlier reports stating that HEF1 expression may be peculiar to hematopoietic cells, we demonstrate that the HEF1 protein is also abundant in cell lines and primary tissues derived from breast and lung epithelium. Expression and phosphorylation of the different HEF1 isoforms are cell growth and cell cycle regulated, with p105HEF1 and p115HEF1 appearing early following serum induction or release from the G1/S boundary, while p55HEF1 expression peaks at mitosis. In contrast, p130Cas levels remain essentially constant throughout the cell cycle. Using phosphatase treatment, we demonstrate that p115HEF1 arises from p105HEF1 via phosphorylation; using targeted mutagenesis, we demonstrate that p55HEF1 arises from full-length HEF1 as the result of cleavage at a DLVD candidate caspase motif. By cell fractionation and immunofluorescence, we show that, while p105HEF1 and p115HEF1 are predominantly cytoplasmic, p55HEF1 associates with the mitotic spindle. Supporting this localization, a two-hybrid library screen with HEF1 resulted in the identification of the mitotic spindle regulatory protein Dim1p as a specific interactor with a region encompassed by p55HEF1. These results suggest a novel model for HEF1 function, in which this protein integrates signals related to cell adhesion with those controlling progression of the cell cycle through mitosis.
MATERIALS AND METHODS
Cell lines.
MCF-7 and BT474 are human breast carcinoma cell lines. A549 and SKLU are human lung carcinoma cell lines. H9 is a human T-cell lymphoma cell line. Jurkat is a human acute T-cell leukemia cell line. Nalm-6 is a human pre-B-cell line. All cells were cultured under standard medium conditions prescribed by the American Type Culture Collection.
Plasmids.
For overexpression of HEF1 in mammalian cells, the plasmid pcDNA3-HEF1 was used. To construct this plasmid, an assembled full-length HEF1 cDNA encoding the 834-amino-acid (aa) HEF1 protein (34) was inserted into the EcoRI-XhoI-cut vector pcDNA3 (Invitrogen), which expresses the protein from the cytomegalovirus promoter. pcDNA3-HEF1DLVA was created by using oligonucleotide-directed PCR mutagenesis to create a D→A change at aa 363 of full-length HEF1; this construct is otherwise identical to pCMV-HEF1, with the coding region of HEF1 completely sequenced.
Antibodies.
The rabbit polyclonal antibody α-HEF1-R1 (previously designated α-HEF1-SB) has been described elsewhere (34). The antibody α-HEF1-R2 was similarly raised with the same multiple-antigen peptide (MAP) (8) in a different rabbit. The affinity purification of the α-HEF1 antibodies was performed as previously described (34). The rabbit polyclonal antibodies α-p130Cas-B (raised against p130Cas aa 318 to 486) and α-p130Cas-F (raised against p130Cas aa 670 to 896) were a generous gift of Amy Bouton and Thomas Parsons and have been described elsewhere (30). The mouse monoclonal antibody to p130Cas (raised against aa 644 to 819) was purchased from Transduction Laboratories. aa 318 to 486 of p130Cas (based on the original numbering of the 968-aa cDNA described in reference 59) maximally align with aa 163 to 318 of HEF1 (based on the original numbering of the 834-aa cDNA described in reference 34), aa 644 to 819 of p130Cas maximally align with aa 489 to 686 of HEF1, and aa 670 to 896 of p130Cas maximally align with aa 526 to 762 of HEF1. See Fig. 1C for a diagram of the above antibody epitopes.
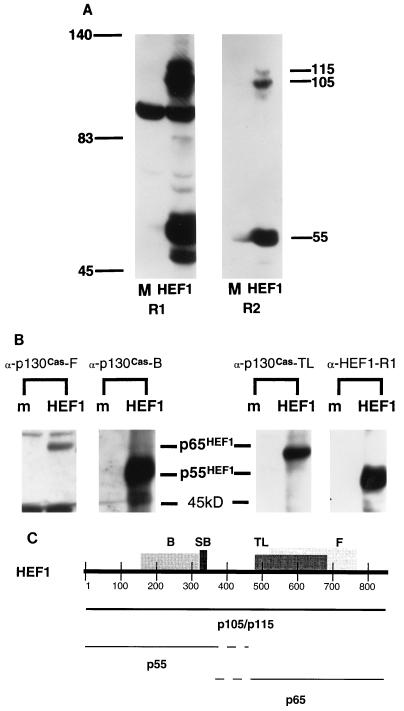
FIG. 1.
Transfection of full-length HEF1 cDNA into HeLa cells produces p105HEF1, p115HEF1, p65HEF1, and p55HEF1. (A) HEF1 isoforms detected by α-HEF1-SB antisera. HeLa cells were either mock transfected (M) or transfected with pCMV-HEF1 (HEF1), and crude lysates were analyzed by SDS-PAGE and Western blot analysis, with either α-HEF1-R1 (R1) or α-HEF1-R2 (R2) antiserum for visualization of HEF1 isoforms. Numbers outside the lanes indicate molecular mass in kilodaltons. (B) Epitope mapping detects p55HEF1 and p65HEF1. HeLa cells were either mock transfected (m) or transfected with pCMV-HEF1 (HEF1). Cell lysates were resolved by SDS-PAGE and probed with four antibodies reacting with HEF1: α-p130Cas-B, α-p130Cas-F, α-p130Cas-TL, and α-HEF1-R1 (described in Materials and Methods). (C) Locations of epitopes for HEF1-reactive antisera and predicted p55HEF1-p65HEF1 boundaries. Shown are the locations of epitopes for the antibodies α-p130Cas-B (B), α-p130Cas-F (F), α-p130Cas-TL (TL), and α-HEF1 (SB) on the full-length 834-aa HEF1 coding sequence; details are in Materials and Methods. Assignment of endpoints for p55HEF1 and p65HEF1 is approximate, based on patterns of reactivity demonstrated in panel B.
The antibody RC20 (Transduction Laboratories) was used to visualize phosphotyrosine. A mouse monoclonal antibody (Sigma) directed against tubulin was used in the immunofluorescent confocal microscopy. As a control for the cell synchronization studies, α-cyclin B1 (PharMingen) was used.
Manipulation of cells.
Transfection of cells with pcDNA3-HEF1 or pcDNA3-HEF1DLVA was done with Lipofectamine (Gibco BRL), under conditions as described by the manufacturer.
For serum starvation and induction, MCF-7 cells at ∼60 to 80% confluence were maintained for 48 h in Dulbecco’s modified Eagle’s medium (DMEM) with no serum and then induced by refeeding with DMEM plus 10% calf serum, with cell lysates made at times noted in the figure legends.
For cell synchronization by thymidine block and release, a previously described procedure for MCF-7 cells was followed almost in its entirety (14), with the exception that cell numbers plated were based on recommendations in reference 63. Both single- and double-thymidine blocked cultures were tested for cell synchronization and showed comparable results. Collection of a mitotic shakeoff was performed by gently striking tissue culture plates against a hard surface and subsequently harvesting medium containing nonadherent cells to be concentrated by centrifugation. Effective cell synchronization was confirmed by probing blots matching those used for HEF1 visualization with antibody to cyclin B1.
For cell synchronization in M phase by nocodazole block and release, the protocol described in reference 7 was generally followed. To select appropriate conditions for block of MCF-7 cells, a titration of 500 nM to 5 μM nocodazole was tested, and the lowest concentration allowing effective arrest was used for the experiments described below. Thus, cells were incubated for 14 h in medium containing 1 μM nocodazole and released into DMEM–10% fetal bovine serum.
Immunofluorescence and microscopy.
Cells were plated on coverslips 24 h before being processed for immunofluorescence. All steps were carried out at room temperature. Coverslips were washed twice in 1× phosphate-buffered saline (PBS) and then fixed in 3.5% paraformaldehyde for 7 min. Cells were permeabilized for 5 min in buffer B (0.1 M Tris [pH 7.5], 1.5 M NaCl, 1% bovine serum albumin) with 0.2% Triton X-100. Coverslips were then washed for 5 min in buffer B. The primary antibody incubations were done for 1 h at room temperature. The antibody dilutions were made in buffer B and were identical to those used for immunoblotting. Coverslips were washed for 5 min in buffer B plus Triton X-100 followed by a 5-min wash in buffer B alone. Secondary antibody (either fluorescein isothiocyanate-conjugated anti-mouse [Jackson Laboratories] or biotinylated anti-rabbit [Vector Laboratories]) was diluted in buffer B and incubated with slides for 1 h. Cells were washed twice for 5 min in buffer B, and those that had biotin-conjugated secondary antibodies were incubated with Texas red streptavidin (Vector Laboratories) for 15 min. Following this, the cells were washed twice with buffer B and then mounted with antifade mounting medium from Vector Laboratories.
Immunoprecipitations.
Cell lysates were prepared as follows with all steps done at 4°C. The cells were washed twice in 1× PBS and then scraped in buffer A-PTY buffer (50 mM HEPES [pH 7.5], 50 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7 plus 1 mM Na3VO4, 1 mM phenylmethylsulfonylfluoride, 0.01 mg of aprotinin per ml, 0.01 mg of leupeptin per ml). The lysates were then spun in a microcentrifuge for 10 min at ∼12,000 × g. Following this, the supernatant was removed and used in the immunoprecipitation experiments. Ten microliters of anti-HEF1 antiserum was incubated with 0.2 mg of cell lysate and 10 μl of 50% protein A–Sepharose beads (Sigma) overnight at 4°C. Beads were washed four times in 500 μl of buffer A and analyzed by immunoblotting. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels were used to resolve the products of the immunoprecipitation protocol.
Phosphatase treatment.
HeLa cells were transfected with pcDNA3-HEF1 under conditions previously noted. The transfectants were harvested in buffer A (lacking NaF and Na3VO4), and the soluble supernatant was used for the experiments. Fifteen micrograms of lysate was treated with 2,000 U of lambda phosphatase (New England Biolabs) in the provided buffer for 30 min at 30°C.
Cell fractionation.
Cell fractionation was performed essentially as described previously for HeLa cells (21). Protein concentration was assayed by a bicinchoninic acid assay (Pierce).
Immunohistochemistry.
Selected adult and fetal human tissues fixed in 10% phosphate-buffered formaldehyde embedded in paraffin were used to evaluate the immunohistochemical localization of HEF1. Antigen retrieval was accomplished by boiling deparaffinized 5-μm-thick paraffin sections for 10 min in distilled water with a 750-W microwave oven at a low setting. After preincubation in goat serum and peroxidase blocking, the sections were incubated overnight at 4°C with a polyclonal rabbit anti-HEF1 antiserum (α-HEF1-R1 or α-HEF1-R2) diluted 1:100. Negative controls were incubated overnight in PBS. After the sections were washed with PBS for 10 min, the immunohistochemical reaction was accomplished with a commercial avidin-biotin-peroxidase kit (Vectastain Elite; Vector) with diaminobenzidine as chromogen. Note that while α-HEF1-R1 recognized HEF1 more strongly by Western blot analysis, it also cross-reacted with at least one additional cellular protein, p95: α-HEF1-R2 was specific for HEF1 in Western blot analysis but generally of lower affinity. Therefore, only production of identical patterns of recognition by both antibodies was described as a positive result.
Two-hybrid library screening and analysis of specificity of Dim1p-HEF1 interaction.
A two-hybrid library screening was performed by standard protocols (26) with LexA-HEF1102–229 expressed from the plasmid pEG202 (26) as bait in the strain EGY48 containing lexAop-LacZ reporter construct pJK103 to screen a pJG4-5 HeLa cDNA library (28). Approximately 5 × 105 yeast transformants were screened; positives were confirmed by retransformation into naive EGY48/pJK103/LexA-HEF1102–229 yeast. To map the interaction of the positive hsDim1p with HEF1 and to analyze the specificity of interaction, the pJG4-5–hsDim1p activation domain fusion was independently tested against LexA-HEF11–154, LexA-HEF1151–229, LexA-HEF11–105; the series of GUS334 (LexA-p85ΔSH2), GUS370 (LexA-SHC), GUS365 (LexA-IRS1), and GUS307 (IR), which contain identical fragments of p85, SHC, IRS1, and IR as described for pJG4-5 fusions in references 27 and 48 (gifts of T. Gustafson); and pRFHMI (LexA-bicoid) (18), a gift of Russ Finley. Expression of all fusion proteins was assayed by Western blot analysis, and β-galactosidase values were calculated by a standard assay. Data points shown in Table 1 reflect the averages of six independent transformants, in a typical experiment out of two to three repetitions.
TABLE 1.
HEF1 associates specifically with the mitotic regulatory protein hsDim1pa
| LexA fusion | Value for construct
|
|
|---|---|---|
| hsDim1p-activation domain | Activation domain vector | |
| Specificity of interaction | ||
| HEF1102–229 | 572 ± 324 | 21 ± 9 |
| p85ΔSH2 | 5 ± 2 | 4 ± 1 |
| SHC | 12 ± 3 | 9 ± 3 |
| IRS1 | 3 ± 1 | 4 ± 4 |
| Insulin receptor | 3 ± 1 | 4 ± 1 |
| Bicoid | 7 ± 4 | 6 ± 3 |
| Mapping of site on HEF1 | ||
| HEF11–105 | 5 ± 1 | 6 ± 1 |
| HEF11–124 | 153 ± 32 | 8 ± 1 |
| HEF11–154 | 109 ± 22 | 13 ± 2 |
| HEF1102–175 | 20 ± 4 | 5 ± 2 |
| HEF1102–229 | 640 ± 45 | 29 ± 2 |
| HEF1125–229 | 50 ± 5 | 18 ± 2 |
| HEF1151–229 | 45 ± 8 | 22 ± 4 |
| Bicoid | 4 ± 1 | 4 ± 0 |
| Alone | 4 ± 1 | 6 ± 0 |
EGY48 yeast cells were transformed with the pJK103 LexA operator-LacZ reporter and indicated combinations of LexA-fused and activation-domain-fused (pJG4-5) proteins. β-Galactosidase activities were calculated as the average values for six separate colonies for each data point. Numbers shown are the β-galactosidase values (± standard deviations) obtained for different constructs.
Sequence analysis.
Sequence alignments were done with the GCG package of programs (15). PEST motifs were identified with the program PESTfind (55), available at http://www.at.embnet.org/embnet/tools/bio/PESTfind/.
RESULTS
A single HEF1 cDNA is processed to p115, p105, p55, and p65.
HEF1 and p130Cas have similar electrophoretic mobilities, with each protein detected endogenously as multiple species migrating between ∼105 and ∼130 kDa. As a first step to characterizing HEF1 expression, we transfected HeLa cells with a plasmid in which the cytomegalovirus promoter expressed a full-length HEF1 cDNA. We then used two HEF1-directed affinity-purified polyclonal antisera (details in Materials and Methods) to determine the sizes of the resulting HEF1 protein products (Fig. 1A, R1 and R2).
The predicted molecular mass of the 834-aa HEF1 protein is 93 kDa. Transfection of the cDNA encoding this protein into HeLa cells resulted in the production of three detectable protein species, migrating at 115, 105, and 55 kDa, which were visualized on Western blots by both HEF1-specific antisera (Fig. 1A). All three species could also be immunoprecipitated by either of the HEF1-specific antisera. These proteins will be designated p115HEF1, p105HEF1, and p55HEF1 throughout this report. We note of the two HEF1-specific antibodies, that the first (α-HEF1-R1 [previously described in reference 34]) additionally recognizes an abundant ∼95-kDa cellular protein in crude cell lysates whose expression is not increased in cells transfected with the HEF1 cDNA and which is most likely unrelated to HEF1 (Fig. 1A, lane R1). In contrast, the second antibody (α-HEF1-R2, new in this study) recognized only the 115, 105, and 55-kDa species specific to the HEF1 cDNA. We have used both antibodies throughout this study, as α-HEF1-R1 has proven more efficient at detection of HEF1 species in Western blot analysis of lysates and immunoprecipitation experiments, while the exclusive specificity of α-HEF1-R2 has made it the antibody of choice for immunofluorescence and immunohistochemistry, although it produces a significantly weaker signal on a Western blot (see Materials and Methods for details of use).
The 115- and 105-kDa species detected following transfection of the HEF1 cDNA correspond to the doublet of HEF1 protein formerly detected endogenously (34) and are presumed to contain full-length coding sequences. The p55HEF1 species was unexpected and has not been described previously. As it appeared following transfection of a processed cDNA, it could not result from an alternative splice of the HEF1 gene. Thus, it was initially taken either to represent a product of specific protease cleavage or, alternatively, to derive from translational initiation at an internal ATG contained in the HEF1 cDNA.
The appearance of p55HEF1 implied that the HEF1 cDNA was being processed in a complex manner. Given that the HEF1 antibodies we used were directed against a short peptide sequence, we considered the possibility that processing of the HEF1 cDNA might result in the generation of some protein species not recognized by the HEF1-specific antibodies. We had previously shown that a number of antibodies generated against several domains of p130Cas cross-react with the p105 and p115 HEF1 species (34). We used three of these antibodies to examine products produced by the HEF1 cDNA (Fig. 1B and C). We found that, while α-HEF1 (R1 and R2) and α-p130Cas-B recognized a p55 species in HEF1-transfected cells, α-p130Cas-F and α-p130Cas-TL instead recognized an additional p65 (p65HEF1) species. Based on the localization of the epitopes against which the antibodies are directed, the p55 species contains amino-terminal HEF1 sequences (SH3 domain and SH2-binding site sequences), while the p65 species contains carboxy-terminal HEF1 sequences (Fig. 1C).
Abundant HEF1 expression in epithelial cells in vitro and in vivo.
We had previously performed a limited analysis showing that the HEF1 mRNA was particularly abundant in tissues including human lung and placenta (34). However, one research group has claimed that expression of HEF1 protein is specific to lymphocytes (e.g., description of Cas-L [41]), and we and others have shown that HEF1 protein function is particularly required for signaling in B and T cells (2, 38, 60). To resolve the issue of the HEF1 expression pattern prior to analysis of the endogenous protein’s regulation, we used α-HEF1 antibody to directly examine cell lines and primary tissues for HEF1 protein (Fig. 2).
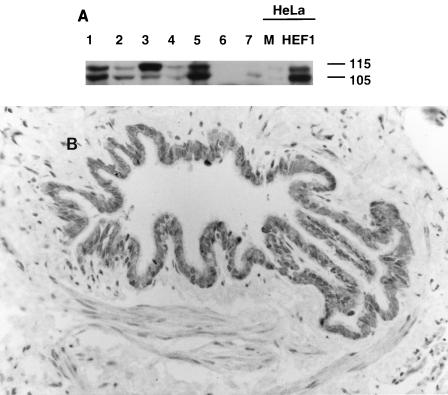
FIG. 2.
HEF1 protein isoforms are abundant in breast, lung, and lymphoid cell lines and in primary bronchial tissue. (A) Antibody α-HEF1 was used to visualize HEF1 protein in cell lysates prepared from multiple cell lines: MCF-7 (lane 1), BT474 (lane 2), A549 (lane 3), SKLU (lane 4), H9 (lane 5), Jurkat (lane 6), Nalm-6 (lane 7), mock-transfected HeLa (lane M), and HeLa transfected with pcDNA3-HEF1 (lane HEF1). For the M and HEF1 lanes, significantly less lysate was added. Numbers at right indicate molecular mass in kilodaltons. (B) Immunohistochemical detection of HEF1 in the human bronchiolar epithelium with antibody α-HEF1. Note that the immunostain is localized in the cytoplasm of the epithelial cells lining the lumen of the bronchiole. The stained nuclei seen in the epithelium and wall of this pulmonary structure are stained with hematoxylin and are HEF1 negative. The panel shows immunoperoxidase and hematoxylin stain of a paraffin section. Magnification, ca. × 52.
Based on the results of prior HEF1 mRNA Northern blot analysis (34, 35), we focused particularly on cell lines derived from mammary epithelial tissue (MCF-7 and BT474), lung epithelial tissue (A549 and SKLU), and B and T lymphocytes (H9, Nalm-6, and Jurkat) (Fig. 2A), while including other representative cell types (data not shown). Using the α-HEF1-R1 antibody for Western blot visualization, we determined that the previously described HEF1 protein species, migrating as a doublet at p105 and p115, are in fact present in most of the cell lines assayed and appear to be particularly abundant in breast and lung epithelial lines, as predicted by RNA analysis. To confirm that the HEF1 protein expression observed in cell lines reflected HEF1 abundance in particular tissues, rather than being specific to immortalized tumor cells maintained in vitro, we then used the HEF1-specific antibodies α-HEF1-R1 and α-HEF1-R2 to examine by immunohistochemistry a series of human paraffin-embedded tissue sections. For both HEF1-directed antibodies, the strongest staining was detected in the respiratory epithelium of airways from the main bronchi to the bronchioles in adult and fetal lung tissues. The positive stain was generally localized in the cell cytoplasm, with preference for the apical portion of cells (Fig. 2B and data not shown). These results were taken to establish that the HEF1 protein is strongly expressed in some epithelial lineages as well as in hematopoietic cells, guiding our choice of cell lines for further analysis.
Serum stimulation and cell cycle progression regulate expression of p115HEF1 and p105HEF1 in G1/S.
We had hypothesized that HEF1 proteins might coordinate signaling between multiple cellular compartments (34). Other proteins with similar properties have been shown to be regulated in response to cell growth stimulation and during the cell cycle (5, 51). We therefore characterized HEF1 protein expression during serum stimulation and throughout cell cycle progression in synchronized cells. MCF-7 breast carcinoma cells were chosen for this analysis, based on our identification of abundant HEF1 expression in this cell line and the tractability of this line for cell cycle and morphological studies.
MCF-7 cells were brought to quiescence by maintenance for 2 days in serum-free DMEM and then induced by the replacement of culture medium with 10% calf serum (Fig. 3). p105HEF1 levels began to increase at 30 to 60 min following addition of serum and reached maximal levels at approximately 2 to 4 h following induction. p115HEF1 levels also increased, but more slowly: the first increase occurred at approximately 4 h following stimulation. By 24 h following stimulation, both p105HEF1 and p115HEF1 levels had returned to near baseline, although cells were still subconfluent and actively growing. In contrast, addition of fresh serum to cells that were exponentially growing in DMEM–10% calf serum resulted in no change in HEF1 protein expression (results not shown). These results implied a specific association of HEF1 induction with initiation of the cell cycle.
FIG. 3.
Induction of p105HEF1 and p115HEF1 following serum stimulation in MCF-7 cells. MCF-7 cells were brought to quiescence by starvation for serum (St.), then the medium was changed to DMEM–10% calf serum, and lysates were made at the times indicated after medium addition (15 or 30 min and 1, 2, 4, 6, or 24 h). Crude cell lysates were resolved by SDS-PAGE, and HEF1 species were visualized by α-HEF1; cross-reactive p95 confirms the equal loads of lanes. Numbers at right and left indicate molecular mass in kilodaltons.
To more closely examine HEF1 expression during the cell cycle and to compare HEF1 with p130Cas expression, MCF-7 cells were synchronized by thymidine block, followed by release and return to active growth (Fig. 4). Cell lysates were used for immunoprecipitation with α-HEF1 (specific for HEF1), α-p130Cas (cross-reactive with p130Cas and HEF1), or control, and immunoprecipitates were visualized with antibody to HEF1 or p130Cas. The time course of p105HEF1 and p115HEF1 upon release from thymidine block was similar to that after serum stimulation (Fig. 4A). Following initial low-level expression, p105HEF1 levels increased, with maximal expression at 3 to 6 h following release, while peak levels of p115HEF1 occurred at 6 to 9 h following release. In contrast, p130Cas was readily detectable in blocked cells, and p130Cas levels did not change for 12 h following release, although a slight reduction was observed at 24 h postrelease (Fig. 4B). This differential regulation of expression following growth stimulation suggested that HEF1 and p130Cas might possess alternative functions in the progression of the cell cycle.
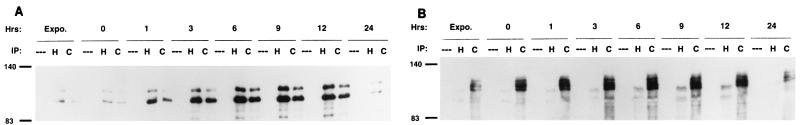
FIG. 4.
Induction of HEF1 but not p130Cas during reentry into cell cycle. Crude lysates were made from either exponentially growing MCF-7 cells (Expo.) or MCF-7 cells synchronized by thymidine block and released for the number of hours noted (0, 1, 3, 6, 9, 12, or 24). Lysates were immunoprecipitated by either control (---), α-HEF1 (H), or the α-p130Cas-TL antibodies (C). Lysates were resolved by SDS-PAGE and probed in Western blot analysis with α-HEF1 antibodies (A); the blot was then stripped and reprobed with α-p130Cas-TL antibody to p130Cas (B). Note that, although antibody to p130Cas is additionally cross-reactive with HEF1, because of the different electrophoretic mobilities of the two proteins, HEF1- or p130Cas-derived species can be readily discriminated by superimposing enhanced chemiluminescence-visualized Western blots sequentially probed with the two antibodies. IP, immunoprecipitation. Numbers at left of each panel indicate molecular mass in kilodaltons.
p115HEF1 is a phosphorylated modification of p105HEF1.
Both HEF1 and p130Cas are known to be phosphorylated on tyrosines during integrin engagement and following oncogenic transformation (reviewed in reference 29). To determine whether p115HEF1 might represent a tyrosine-phosphorylated form of p105HEF1 and to determine whether phosphorylation of HEF1 was regulated during the cell cycle, we assessed HEF1 phosphotyrosine content in MCF-7 cells synchronized by thymidine block. HEF1 was immunoprecipitated at 0, 1, 3, 6, 9, 12, or 24 h following release from block, and the phosphotyrosine (Fig. 5A) and HEF1 (Fig. 5B) content of immunoprecipitates was determined by visualization with the antiphosphotyrosine antibody RC20 or α-HEF1-R1, respectively. By this means, the p105HEF1 and p115HEF1 species appeared to be tyrosine phosphorylated to comparable levels and with similar time courses as the proteins accumulated.
FIG. 5.
(A and B) p105HEF1 and p115HEF1 are tyrosine phosphorylated to comparable extents. Crude lysates were made from MCF-7 cells synchronized by thymidine block and released for the number of hours noted (0, 1, 3, 6, 9, 12, or 24). Lysates were immunoprecipitated either by control (---) or α-HEF1 (H) antibody. These lysates were resolved by SDS-PAGE and probed in Western blot analysis with the RC20 antibody to phosphotyrosine (A); following stripping, the blot was reprobed with α-HEF1 antibody (B). (C) p115HEF1 levels are reduced by treatment with lambda phosphatase. Lysates from HeLa cells transfected with pCMV-HEF1 were treated either with (+) or without (--) lambda phosphatase, resolved by SDS-PAGE, and visualized with α-HEF1. Numbers in the margins of each panel represent molecular mass in kilodaltons.
To further pursue this issue, we then transfected HeLa cells with the pcDNA3-HEF1 construct; treated whole-cell lysates with lambda phosphatase, a broad-specificity phosphatase that targets tyrosines, serines, and threonines; and examined the abundance of the p105HEF1 and p115HEF1 species (Fig. 5C). In this assay, the p115HEF1 species was almost completely eliminated, while the p105HEF1 species was enriched following phosphatase treatment. Thus, the p115HEF1 species represents a phosphorylated form of p105HEF1; based on the phosphatase and antiphosphotyrosine results, this phosphorylation difference appears more likely to represent a serine-threonine phosphorylation event, although it may correspond to a tyrosine phosphorylation not detectable with the RC20 antibody.
Endogenous p55HEF1 specifically appears at mitosis and arises through cleavage of the full-length HEF1 protein at a candidate caspase site.
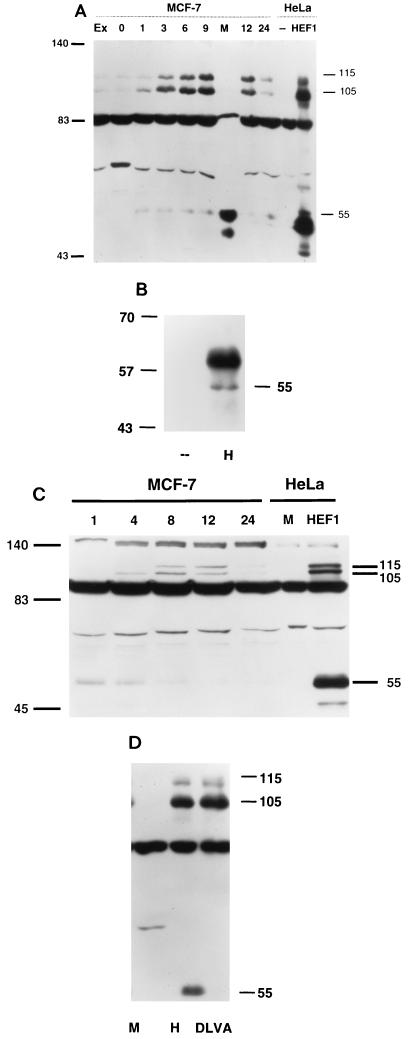
During assay of cell-synchronized MCF-7 cultures, we had noted the endogenous p55HEF1 as a relatively minor species that appeared to increase in abundance at later time points in the cell cycle than the p105HEF1 and p115HEF1 species. We considered that the relative paucity of this species in contrast to our initial results with transfected HEF1 cDNA (Fig. 1) might reflect an actual difference in steady-state synthesis of p55HEF1 from endogenous versus transfected full-length HEF1. Alternatively, if p55HEF1 were specifically abundant in mitotic cells, this might also explain the underrepresentation, as the initial wash steps in the lysis procedure clearly removed many tenuously attached mitotic cells, and as mitosis is of such short duration (∼30 min) that only a subpopulation of cells are in this phase of the cell cycle at any single time point. To test this second hypothesis, we directly analyzed lysates prepared from MCF-7 cells synchronized by thymidine block, released, and allowed to grow between 1 and 24 h, and in addition from cells prepared as mitotic shakeoffs from plates at 9 h after release from thymidine block (Fig. 6A), a time at which maximal numbers of cells should be entering mitosis (14). Results of this analysis were dramatic. Specifically, in cells prepared from mitotic shakeoff, p55HEF1 was strongly induced, while p105HEF1 and p115HEF1 were diminished, supporting the idea that the endogenous p55HEF1 protein might have a function specific to mitosis.
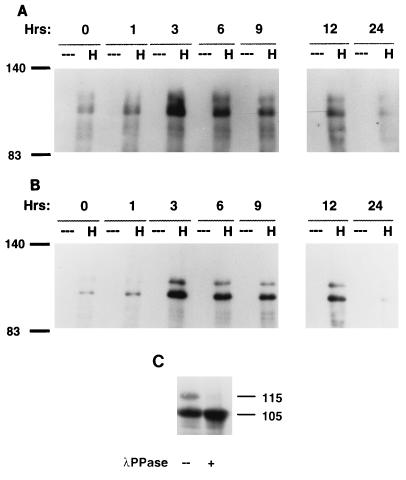
FIG. 6.
(A) Specific appearance of p55HEF1 in mitotic shakeoff. Crude lysates were made from either exponentially growing cells (Ex) or cells synchronized by thymidine block and released for the number of hours notes (0, 1, 3, 6, 9, 12, or 24). At 9 h following release, a mitotic shakeoff was prepared (M). As a control to indicate the position of the p105HEF1, p115HEF1 and p55HEF1 species, HeLa cell lysates (known to express relatively low levels of endogenous HEF1) either mock transfected (--) or transfected with pcDNA3-HEF1 (HEF1) were also analyzed. The lysates were resolved by SDS-PAGE and probed in Western blot analysis with α-HEF1 antibodies. (B) Endogenous p55HEF1 can be immunoprecipitated by antibody to HEF1. Five hundred micrograms of whole-cell lysate prepared from mitotic shakeoffs of MCF-7 cells 9 h after release from thymidine block was used for immunoprecipitation with either control (--) or α-HEF1 antibodies (H), followed by visualization with α-HEF1. Note that the prominent diffuse band migrating at ∼59 to 64 kDa represents the immunoglobulin blob generally detected in immunoprecipitations. (C) p55HEF1 is abundant in nocodazole-blocked cells and is replaced by p105HEF1 and p115HEF1 following release. MCF-7 cells were blocked in mitosis by incubation in 1 μM nocodazole for 14 h and released. Cell lysates were prepared from cells at 1, 4, 8, 12, and 24 h after release. As before, as a control for sizes, an aliquot of mock-transfected (M) or HEF1-transfected (HEF1) HeLa cells was included. Lysates were resolved by SDS-PAGE and probed in Western blot analysis with α-HEF1. (D) Production of p55HEF1 results from a cleavage of the full-length HEF1 protein at a DLVD motif located at aa 360 to 363. PCR-based mutagenesis was used to alter DLVD360–363 to DLVA in the context of the full-length 834-aa HEF1 coding sequence, and the mutant was cloned into the pCMV expression vector. Whole-cell lysates from HeLa cells mock transfected (M), transfected with pcDNA3-HEF1 (H), or transfected with pcDNA3-HEF1DLVA (DLVA) were visualized with α-HEF1 antibodies. Numbers in the margins of each panel indicate molecular mass in kilodaltons.
To further test the idea that p55HEF1 was specifically produced in mitosis, we performed a number of additional experiments. First, to strengthen confidence that this p55 species detected endogenously by Western blot analysis corresponded to HEF1, as opposed to a spurious cross-reactive species detected only following protein denaturation, we additionally used antibody to HEF1 to perform immunoprecipitations from mitotic shakeoff populations (Fig. 6B). By this means, p55HEF1 was specifically immunoprecipitated, while p105HEF1 and p115HEF1 were not, confirming that the p55HEF1 species was detectable in native as well as denatured conformations by the α-HEF1 antibodies and additionally supporting the finding that p105HEF1 and p115HEF1 are absent in mitotic cells. Second, given the multiple connections among HEF1, p130Cas, and cell adhesion, we wished to exclude the possibility that p55HEF1 might be appearing in response to cell rounding and loss of contact with the culture plate occurring at mitosis. We therefore prepared lysates from trypsinized cells held in suspension and screened these for induction of the p55HEF1 species. No induction of p55HEF1 was observed (results not shown). Third, it was possible that p55HEF1 was being induced in dying cells that had lost contact with the culture plate and were floating in the culture medium. Therefore, we allowed some cells derived from mitotic shakeoff to replate for 60 min, after which the cells which had attached were lysed and analyzed. p55HEF1 was still abundant in attached cells, indicating that it was not specific to dying nonadherent cells (results not shown).
Finally, to confirm expression of p55HEF1 at mitosis by a second assay, cells were arrested with nocodazole at mitosis and then released and allowed to resume growth (Fig. 6C). Strikingly, nocodazole-treated extracts contained extremely high levels of p55HEF1 but only low levels of p105HEF1 and p115HEF1. Following release, p105HEF1 levels begin to increase at ∼2 to 4 h and p115HEF1 begins to appear after 4 to 8 h, a lag consistent with the results obtained with thymidine-blocked cells. In contrast, p55HEF1 levels begin to diminish at ∼4 h following release and are significantly reduced at 8 to 12 h. We take these results as strongly favoring the interpretation that expression of p55HEF1 is specifically induced at mitosis and is lost as cells proceed through the next round of cell division.
Based on its detection with antibodies directed against the amino-terminal end of the HEF1 coding sequence, the p55HEF1 species appeared more likely to derive from posttranslational processing of a larger form of HEF1, rather than from alternative translational initiation from an internal methionine. Based on epitope specificity of the antibody panel (Fig. 1C), cleavage producing p55HEF1 would have to occur between aa 340 and 525. Proteolytic cleavage systems implicated in the control of such processing include the proteasome (e.g., reference 49) and caspases, which although primarily functioning in apoptosis have also been shown to process some proteins in normally growing cells (e.g., reference 25). We therefore inspected HEF1 for candidate cleavage motifs and identified a DLVD sequence at aa 360 to 363 that was closely related to the DEVD sequence preferred by the Cpp32–Yama–Ced-3 caspase-3 family (66). To test the utilization of this site, we inserted a single amino acid mutation converting DLVD to DLVA in the full-length HEF1 coding sequence, eliminating the P-1 position aspartic residue required for cleavage. In parallel transfections of plasmids expressing full-length versus DLVA-mutated HEF1 into HeLa cells, while the two constructs synthesized comparable levels of p105HEF1 and p115HEF1, production of p55HEF1 was completely eliminated by the DLVA mutation (Fig. 6D), both confirming that this species arises as the result of a cleavage and implicating caspases as the cleaving enzymes.
p105HEF1 and p115HEF1 are predominantly cytoplasmic and associated with focal adhesions, whereas p55HEF1 associates with the mitotic spindle.
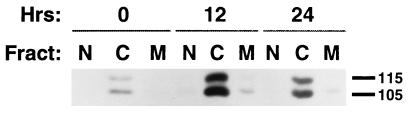
p130Cas localizes to focal adhesions and the cytoplasm (52), whereas we had previously used immunofluorescence to detect α-HEF1-reactive species at both the cell periphery and the nucleus (34). Based on our identification of multiple forms of HEF1 protein, with differential timing of expression, one possibility was that p55HEF1, p105HEF1, and p115HEF1 might localize to different intracellular compartments. Accordingly, we performed cell fractionation followed by Western blot analysis with α-HEF1-R1 to assay distribution of endogenous HEF1 proteins in MCF-7 cells (Fig. 7). By this means, p105HEF1 and p115HEF1 were found to localize predominantly to cytoplasmic fractions (which generally contain focal adhesion-associated proteins), with a minor population being detected in nuclear and membrane (combined Golgi and endoplasmic reticulum) fractions (visible on a longer exposure [data not shown]). This basic distribution did not change following growth induction.
FIG. 7.
The p105HEF1 and p115HEF1 species are predominantly cytoplasmic. MCF-7 cells were brought to quiescence by serum deprivation and then refed with DMEM-10% serum, and cell lysates were made at the times indicated (0, 12, and 24 h). Lysates were separated into nuclear (N), cytoplasmic (C), and combined membrane (M) fractions as described in Materials and Methods. Fractions were resolved by SDS-PAGE and probed in Western blot analysis with α-HEF1. Numbers at right indicate molecular mass in kilodaltons.
Analysis of the intracellular distribution of p55HEF1 was intrinsically more complex, because at the time of maximal induction of p55HEF1 at mitosis, nuclear envelope breakdown has occurred, confounding cell fractionation. Fractions prepared from mitotic shakeoffs indicated that p55HEF1 associated almost exclusively with the insoluble fraction that includes chromosomes and the spindle apparatus (results not shown) but provided poor resolution. Thus, to provide an accurate localization for this protein and directly assess p55HEF1 localization in mitotic cells, we used α-HEF1-R2 to perform immunofluorescence analysis on MCF-7 cells (Fig. 8).
FIG. 8.
p55HEF1 associates with the mitotic spindle. Cells in prophase (A to C), metaphase (D to F), late anaphase (G to I), and cytokinesis (J to L) were stained with α-HEF1-R2 antiserum to HEF1 and visualized with rhodamine (A, D, G, and J) or stained with α-tubulin and visualized with fluorescein isothiocyanate (B, E, H, and K); a merged image is shown in panels C, F, I, and L, with HEF1-tubulin colocalization shown in yellow. Note punctate staining of α-HEF1-R2, which does not colocalize with microtubules in nonmitotic cells. Focal adhesion staining is not visible in this optical section, although it is clearly present in nonmitotic cells. Identical results were obtained with α-HEF1-R2 in a single stain, excluding bleedover from tubulin as a source of HEF1 staining (data not shown).
A weakly α-HEF1-reactive species was detected at peripheral structures, presumably due to the presence of p105HEF1 and p115HEF1 (results not shown). Notably, the predominant signal detected with this antibody corresponded to the mitotic spindle. HEF1 protein first associates with the forming mitotic spindle in prophase as the centrioles and microtubule asters become organized and then remains associated with the mitotic spindle through late anaphase. The staining pattern of HEF1 (Fig. 8A, D, G, and J) is coincident with that of tubulin (Fig. 8B, E, H, and K) in the various mitotic figures, whereas in surrounding interphase cells in the culture, HEF1 staining is not colocalized to the microtubule networks (Fig. 8C and F). During cytokinesis (Fig. 8J to L), HEF1 and tubulin are coincident in the midbody region, where many proteins involved in regulating the cell cycle are degraded at the end of mitosis. In addition to the midbody staining, HEF1 appears in a punctate pattern that is diffuse throughout the cytoplasm, suggesting that it is beginning to separate from the spindle prior to degradation (Fig. 8B).
As controls for the specificity of the staining pattern observed, we performed several additional experiments. The mitotic spindle staining pattern was completely blocked by inclusion of the SB peptide to which α-HEF1-R2 was raised in staining reactions (results not shown). α-HEF1-R2 produced identical staining patterns on the spindle in multiple cell lines assayed, including HeLa, PtK, and others (data not shown). Finally, the α-HEF1-R1 antibody also stained the spindle of mitotic cells; however, this was against a background of diffuse staining, presumably attributable to cross-reactivity with p95, which we have shown to be present in all cellular compartments (data not shown). Further, use of a series of alternative fixation and permeabilization regimens did not alter the spindle staining pattern, supporting the idea that this localization does not reflect artifactual precipitation of antibody onto the mitotic spindle (data not shown). As noted above (Fig. 1 and 6B), we have shown that α-HEF1-R2 reacts only with HEF1 in Western blot analysis and solely immunoprecipitates a p55 protein species from mitotic cell lysates. While lack of availability of additional specific antibodies directed against the amino-terminal region of HEF1 prohibits independent confirmation of this staining pattern, these cumulative studies support the idea that it is specific for p55HEF1.
Association of HEF1 with Dim1p supports a p55HEF1 function at the mitotic spindle.
If the HEF1 protein possessed a function relevant to the mitotic spindle, this might reasonably be reflected in physical interaction between the region of HEF1 encompassed by p55HEF1 and other proteins with spindle-associated functions. To investigate this possibility, we performed a two-hybrid library screen with LexA-fused HEF1102–229, which excluded the HEF1 amino-terminal SH3 domain previously used for screening (34) but incorporated part of the previously defined SH2-binding site-rich substrate domain present in p55HEF1. As analysis with the α-phosphotyrosine antibody RC20 indicates that the p55HEF1 species possesses little if any tyrosine phosphorylation (35) while the p105HEF1 and p115HEF1 species are tyrosine phosphorylated (Fig. 5), it seemed reasonable that, in the absence of tyrosine phosphorylation expected in yeast, p55HEF1-interacting proteins might be enriched by this approach.
From a screen of ∼500,000 transformants of a HeLa cDNA library with LexA-HEF1102–229 as a bait, two classes of cDNAs were isolated. One corresponded to a novel protein currently under study; the second corresponded to the human homolog of the Dim1p protein (hsDim1p), recently described in Schizosaccharomyces pombe as a protein required for progression between the G2 and M phases of the cell cycle (6). Dim1p is notable for the very high level of conservation observed across species; the protein is identical in humans and in mice and maintains 65% identity between humans and Saccharomyces cerevisiae, while overexpression of a mammalian form of the protein was shown to complement a null allele of Dim1+ in S. pombe (6). Of particular interest, the morphological defect of cells containing weak alleles of Dim1p was specifically destabilization of the mitotic spindle (6), suggesting that Dim1p may either be a spindle component or regulate spindle assembly, making it a strong candidate for association with p55HEF1.
To demonstrate that the interaction of hsDim1p was specific for HEF1 and to delineate the region of HEF1 with which hsDim1p interacted, we screened activation-domain-fused hsDim1p against a set of other signaling proteins, including some incorporating one or more SH2 binding sites (Table 1), as well as against a series of truncations of HEF1 spanning the original bait protein (Table 1). From this experiment, we determined that, in contrast to the strong interaction observed with HEF1, no interaction above background was observed in a series of LexA fusions to p85, SHC, IRS1, and the insulin receptor, supporting the idea that the interaction is specific to HEF1 (Table 1). In analyzing the point of contact between HEF1 and hsDim1p (Table 1), hsDim1p interacted strongly with HEF1102–229; moderately with HEF11–124 and HEF11–154; and weakly if at all with HEF11–105, HEF1102–175, HEF1125–229, or HEF1151–229. This sum of results indicated that a major determinant of the interaction resides between aa 102 and 124 on HEF1 but that this region in itself is not sufficient to mediate association in the absence of additional amino-terminal or carboxy-terminal flanking sequences. As all constructs are comparably expressed, the flanking sequences are likely either to provide additional interactive contacts (particularly for sequences 151 to 229) or to contribute to correct protein folding.
DISCUSSION
In this study, we have identified a number of properties of the HEF1 gene that are likely to be important for understanding the activity of its protein products. First, we have shown that a single cDNA encoding an 834-aa protein species can be processed in vivo to produce at least four distinct protein species, p115HEF1, p105HEF1, p65HEF1, and p55HEF1. Second, we have demonstrated that at least three of these species (p115HEF1, p105HEF1, and p55HEF1) are abundant endogenously and that their expression is differentially regulated during cell cycle progression. Third, we have established the mechanism of production of these species, with p115HEF1 being a phosphorylated modification of p105HEF1 and p55HEF1 being cleaved at a DLVD caspase consensus from full-length HEF1. Fourth, we have shown that the different HEF1 species localize to different compartments of the cell, with p115HEF1 and p105HEF1 being predominantly cytoplasmic and associated with focal adhesions, while p55HEF1 is associated with the mitotic spindle. Fifth, we have identified the human homolog of S. pombe Dim1p as a HEF1-interacting protein, providing a second link to spindle function. These results indicate that HEF1 may connect control of cell attachment to substrate with regulation of mitotic spindle in G2/M, thus acting as a regulatory protein in the cellular decision to divide or initiate apoptosis.
p105HEF1 and p115HEF1 have recently become the topic of much study because of their implication as potentially key transducers of signaling related to integrin engagement during differentiation of B and T cells (2, 3, 38, 41, 60, 65). This study for the first time demonstrates that the abundance of these forms of HEF1 is cell cycle regulated, with expression being particularly high following initiation of cell division (Fig. 3 to 5). Our data indicate that p105 appears earlier in the cell cycle than does p115HEF1, raising the possibility that a stage-specific kinase may phosphorylate p105HEF1. However, this cannot be a complete explanation, as inspection of a panel of cell lines (Fig. 2) indicates that the relative abundance of the two forms varies between different lines of asynchronously growing cells. At this time, functional differences between the p105HEF1 and p115HEF1 forms of HEF1 remain unclear. Studies of the comparable p130Cas doublet indicate that the slower-migrating, more phosphorylated form of this protein is more tightly associated with the cytoskeleton (54), suggesting that p115HEF1 may function primarily at focal adhesions while the p105HEF1 form may additionally function in other cellular compartments. Elsewhere, we present evidence that the HEF1 protein encompasses a carboxy-terminal helix-loop-helix motif and that the p105HEF1, but not p115HEF1, species of HEF1 forms endogenous complexes with helix-loop-helix proteins (36), raising the intriguing possibility that this form of the protein may bridge cell adhesion and transcriptional control related to cell differentiation in a manner similar to that demonstrated for β-catenin (5).
Several points relative to the production of p55HEF1 and p65HEF1 are worthy of particular note. First, our data indicate that these species contain little if any tyrosine phosphorylation, despite the fact that p55HEF1 encompasses the SH2-binding site-rich substrate domain and is derived from the p105HEF1-p115HEF1 species. This difference suggests that processing by cellular phosphatases (e.g., reference 24) may be enhanced either accompanying or subsequent to cleavage from p105HEF1-115HEF1, perhaps as a result of removal from the concentration of tyrosine kinases residing at focal complexes (43). Alternatively, part of the p105HEF1-p115HEF1 population may exist in a non-tyrosine-phosphorylated form, and this may serve as the precursor to p55HEF1-p65HEF1. Second, a particularly intriguing aspect of the production of p55HEF1 is the implied involvement of caspases in the proteolytic cleavage of p105HEF1 and/or p115HEF1. The number of signaling molecules known to be targeted by caspase family members in apoptosis is rapidly increasing; at least some such targets include proteins either associated with HEF1, such as FAK (70), or in related signaling pathways, such as MEKK-1 (10). A much more limited set of molecules has been shown to be regulated by caspases in normally growing cells (e.g., reference 25), and the generality of this process has yet to be defined. The DLVD motif shown to be required for HEF1 cleavage is not conserved with p130Cas, emphasizing that this processing and release of p55HEF1 is likely to be a HEF1-specific phenomenon. Finally, it appears likely that HEF1 protein expression may be subject to additional forms of regulation; inspection for motifs (55) identifies a strongly predicted PEST sequence in the carboxy-terminal half of the protein (perhaps accounting for the very limited detection of endogenous p65HEF1 [35]), while in vivo labelling experiments with 35S have suggested that the protein possesses a short half-life (35), as has been observed for many proteins associated with cell cycle regulation processes. One as yet unresolved issue is that of why p55HEF1 is abundant following transfection of cDNA, whereas this species is restricted to mitotic cells endogenously; it may be that the transient overexpression obtained following transfection overwhelms intracellular mechanisms that negatively regulate posttranslational cleavage.
One of the most surprising findings of this study is the colocalization of p55HEF1 with the mitotic spindle and the subsequent establishment of HEF1 interaction with Dim1p. This spindle association does not reflect a generic association of p55HEF1 with tubulin, as analysis of cells transfected with the full-length HEF1 cDNA and thus overexpressing p55HEF1 during interphase shows no indication of association with microtubule networks (35). Therefore, localization of p55HEF1 to the mitotic spindle seems most likely to indicate an affinity of the p55HEF1 species for either a bundled form of tubulin specific to mitosis or a spindle-associated protein(s). We believe the latter possibility is more likely, because scrutiny at high magnification of HEF1 immunofluorescence in mitotic cells (35) indicates that the protein is arrayed in a striated or punctate pattern along the spindle, similar to that previously reported for MAPs such as mitotic-specific motors and spindle assembly factors (13). Thus, HEF1 may dock with the spindle via association with MAPs or potentially hsDim1p (whose localization has not yet been determined); alternatively, a number of signaling and cell cycle control-related proteins have been reported to associate with the mitotic spindle, including among others p34cdc2 (57), MAP kinase (42), and the CAS protein (involved in regulation of apoptosis and cyclin B degradation) (62).
Following the early observations that morphological and adhesive properties of cells govern their viability and proliferative capacity (17, 20), multiple groups have focused on elucidating specific signaling pathways involved in such regulation. Cell adhesion, and in particular stimulation of the integrin receptor, is required in some cell lines for DNA synthesis (47); for appropriate differentiation of a cell type, as reflected by the induction of cell-type-specific gene expression (16, 64); and more recently, for prevention of anoikis, defined as programmed cell death induced by loss of attachment (22, 40, 58). Perhaps significantly, proteins known to associate with HEF1 or p130Cas have been implicated in some of these pathways. For example, disruption of contacts between the HEF1-p130Cas-Efs partner protein FAK and the integrin receptor results in induction of apoptosis (31), while expression of constitutively active FAK is protective against anoikis (23). HEF1 may function in activation of the MAP kinase pathway required for cell proliferation and gene expression, either through interaction with FAK (11, 37, 61) or through further association of the HEF1 partner Crk (3, 38, 41) with C3G (67), promoting activation of Ras, or by some other means. The data in this study allow us to propose a model in which p105HEF1 and p115HEF1 coordinate signaling complexes at focal adhesions in response to adhesion or growth factor signals initiating cell proliferation: cleavage of HEF1 at G2/M removes p105HEF1-p115HEF1 from focal adhesions, potentially disrupting these complexes; as cell attachment to substrate diminishes and cells round up for mitosis, addition of p55HEF1 to the mitotic spindle contributes to signals favoring cell cycle progression, potentially via interaction with the recently defined mitotic regulator hsDim1p. Based on the implication of caspases as HEF1-cleaving enzymes and the known association of HEF1 with FAK, aberrant HEF1 cleavage may additionally play a role in the progression of anoikis. Finally, the identification of HEF1 as a helix-loop-helix-containing protein with the capacity to associate with other helix-loop-helix factors, described elsewhere, indicates that HEF1 may couple integrin-related cell proliferation signals with regulation of cellular differentiation status. These connections to cell division, apoptosis, and differentiation position HEF1 to be an important central regulator of cell growth controls.
In a final summary, since the initial description of p130Cas, a number of studies have established mechanisms by which this protein complexes with signaling partners at focal adhesions to presumably regulate cell shape and motility during normal cell adhesion and oncogenesis (4, 9, 19, 24, 30, 39, 44–46, 50, 52–54, 56, 59, 68, 69). This current study departs from the preexisting literature on the p130Cas-HEF1-Efs family to suggest that, via regulated expression of synthesis, HEF1 in particular may function in part as a cell cycle sensor, whereas p130Cas does not. We further demonstrate that cleaved HEF1 is associated with the mitotic spindle, whereas our studies to date indicate that p130Cas is unlikely to be cleaved and is exclusively cytoplasmic. While p130Cas is abundant in many cell types, including fibroblasts, HEF1 is the predominant species in an alternative group of cell types, including some epithelial and hematopoietic lineages. Cumulatively, these results suggest that, for the HEF1-p130Cas-Efs family, sequence homology and conserved domain structure may nevertheless be adapted to diverse cellular functions.
ACKNOWLEDGMENTS
This research was supported by National Cancer Institute/NIH grant R29-CA63366 (to E.A.G.) and core funds CA-06927 (to Fox Chase Cancer Center) and by American Cancer Society grant CB-74749 to E.A.G. Over the course of this study, S.F.L. was supported by NIH postdoctoral training grant T32 CA09035, American Cancer Society fellowship PF-4383, and NIH fellowship F32 GM18223 and Y.-Z.Z. was supported by NIH training grant T32 CA09035.
Ying Tong Wang and Joanne Estojak provided outstanding technical help on this project. We are grateful to Serge Manie and Arnie Freedman for cell lines and much helpful discussion; to Maggie Kasten, Chuck Clevenger, and Mary Ann Sells for cell lines; and to T. Gustafson for LexA fusion constructs used in specificity tests. We are very grateful to Jonathan Boyd for help with confocal microscopy. We thank Jonathan Chernoff, David Wiest, Sarah Fashena, and Tim Yen for incisive comments on the manuscript.
REFERENCES
- 1.Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel, p130Cas-related protein, Sin. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 2.Astier A, Manie S, Avraham H, Hirai H, Law S F, Zhang Y, Golemis E A, Fu Y, Druker B J, Haghayeghi N, Freedman A S, Avraham S. The related adhesion focal tyrosine kinase differentially phosphorylates p130Cas and the Cas-like protein, p105HEF1. J Biol Chem. 1997;272:19719–19730. doi: 10.1074/jbc.272.32.19719. [DOI] [PubMed] [Google Scholar]
- 3.Astier A, Manie S N, Law S F, Canty T, Hagheyeghi N, Druker B J, Salgia R, Golemis E A, Freedman A S. Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in human B cells. Possible relevance to neoplastic lymphohematopoietic cells. Leuk Lymphoma. 1998;28:65–72. doi: 10.3109/10428199709058332. [DOI] [PubMed] [Google Scholar]
- 4.Auvinen M, Paasinen-Sohns A, Hirai H, Andersson L C, Holtta E. Ornithine decarboxylase- and ras-induced cell transformations: reversal by protein tyrosine kinase inhibitors and role of pp130CAS. Mol Cell Biol. 1995;15:6513–6525. doi: 10.1128/mcb.15.12.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interactions of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Berry L D, Gould K L. Fission yeast dim1+ encodes a functionally conserved polypeptide essential for mitosis. J Cell Biol. 1997;137:1337–1354. doi: 10.1083/jcb.137.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandeis M, Hunt T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 8.Briand J-P, Barin C, van Regenmortel M H V, Muller S. Application and limitations of the multiple antigen peptide (MAP) system in the production and evaluation of anti-peptide and anti-protein antibodies. J Immunol Methods. 1992;156:255–265. doi: 10.1016/0022-1759(92)90033-p. [DOI] [PubMed] [Google Scholar]
- 9.Burnham M R, Harte M T, Richardson A, Parsons J T, Bouton A H. The identification of p130Cas-binding proteins and their role in cellular transformation. Oncogene. 1996;12:2467–2472. [PubMed] [Google Scholar]
- 10.Cardone M H, Salvesen G S, Widmann C, Johnson G, Frisch S M. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Kinch M S, Lin T H, Burridge K, Juliano R L. Integrin-mediated cell adhesion activates mitogen-activated protein kinases. J Biol Chem. 1994;269:26602–26605. [PubMed] [Google Scholar]
- 12.Chen R H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleveland D W. Tubulin and associated proteins. In: Kreis T, Vale R, editors. Guidebook to the cytoskeletal and motor proteins. New York, N.Y: Oxford University Press; 1993. pp. 101–105. [Google Scholar]
- 14.Cos S, Fernandez F, Sanchez-Barcelo E J. Melatonin inhibits DNA synthesis in MCF-7 human breast cancer cells in vitro. Life Sci. 1996;58:2447–2453. doi: 10.1016/0024-3205(96)00249-4. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–397. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiPersio C M, Jackson D A, Zaret K S. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol Cell Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dulbecco R. Topoinhibition and serum requirement of transformed and untransformed cells. Nature. 1970;227:802–806. doi: 10.1038/227802a0. [DOI] [PubMed] [Google Scholar]
- 18.Finley R, Brent R. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flinn H M, Ridley A J. Rho stimulates tyrosine phosphorylation of focal adhesion kinase, p130 and paxillin. J Cell Sci. 1996;109:1133–1141. doi: 10.1242/jcs.109.5.1133. [DOI] [PubMed] [Google Scholar]
- 20.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 21.Frangioni J V, Beahm P H, Shifrin V, Jost C A, Neel B G. The non-transmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 22.Frisch S M, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frisch S M, Vuori K, Ruoslahti E, Chan-Hui P-Y. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garton A J, Flint A J, Tonks N K. Identification of p130cas as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, Kamen R, Tracey D, Allen H. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 26.Golemis E A, Serebriiskii I, Gyuris J, Brent R. Interaction trap/two-hybrid system to identify interacting proteins. In: Ausubel F M, Brent R, Kingston R, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1997. pp. 20.1.1–20.1.35. [Google Scholar]
- 27.Gustafson T A, He W, Schaub C D, Craparo A, O’Neill T J. Phosphotyrosine-dependent interaction of SHC and insulin receptor substrate 1 with the NPEY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyuris J, Golemis E A, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 29.Hanks S K, Polte T R. Signaling through focal adhesion kinase. Bioessays. 1997;19:137–145. doi: 10.1002/bies.950190208. [DOI] [PubMed] [Google Scholar]
- 30.Harte M T, Hildebrand J D, Burnham M R, Bouton A H, Parsons J T. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J Biol Chem. 1996;271:13649–13655. doi: 10.1074/jbc.271.23.13649. [DOI] [PubMed] [Google Scholar]
- 31.Hungerford J E, Compton M T, Matter M L, Hoffstrom B G, Otey C A. Inhibition of pp125FAK in cultured fibroblasts results in apoptosis. J Cell Biol. 1996;135:1383–1390. doi: 10.1083/jcb.135.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 33.Ishino M, Ohba T, Sasaki H, Sasaki T. Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which contains a Src homology 3 domain and associates with Fyn. Oncogene. 1995;11:2331–2338. [PubMed] [Google Scholar]
- 34.Law S F, Estojak J, Wang B, Mysliwiec T, Kruh G D, Golemis E A. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law, S. F., and E. A. Golemis. Unpublished results.
- 36.Law, S. F., Y.-Z. Zhang, S. Fashena, G. Toby, J. Estojak, and E. A. Golemis. Dimerization of the docking/adaptor protein HEF1 via a carboxy-terminal helix-loop-helix domain. Submitted for publication. [DOI] [PubMed]
- 37.Lin T H, Aplin A E, Shen Y, Chen Q, Schaller M, Romer L, Aukhil I, Juliano R L. Integrin-mediated activation of MAP kinase is independent of FAK: evidence for dual integrin signaling pathways in fibroblasts. J Cell Biol. 1997;136:1385–1395. doi: 10.1083/jcb.136.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manie S N, Beck A R P, Astier A, Law S F, Canty T, Hirai H, Druker B J, Avraham H, Haghayegi N, Sattler M, Salgia R, Griffin J D, Golemis E A, Freedman A S. Involvement of p130Cas and p105HEF1, a novel Cas-like docking protein, in a cytoskeleton-dependent signaling pathway initiated by ligation of integrin or antigen receptor on human B cells. J Biol Chem. 1997;272:4230–4236. doi: 10.1074/jbc.272.7.4230. [DOI] [PubMed] [Google Scholar]
- 39.Mayer B J, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 40.Meredith J E, Jr, Fazeli B, Schwartz M A. The extracellular matrix as a cell survival factor. Mol Biol Cell. 1993;4:953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta-1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996;184:1365–1375. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minshull J, Sun H, Tonks N K, Murray A W. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto S, Teramoto H, Coso O A, Gutkind J S, Burbelo P D, Akiyama S K, Yamada K M. Integrin function: molecular hierarchies of cytoskeletal and signaling molecules. J Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamoto T, Sakai R, Ozawa K, Yazaki Y, Hirai H. Direct binding of the C-terminal region of p130-Cas to SH2 and SH3 domains of Src kinase. J Biol Chem. 1996;271:8959–8965. doi: 10.1074/jbc.271.15.8959. [DOI] [PubMed] [Google Scholar]
- 45.Nojima Y, Morino N, Mimura T, Hamasaki K, Furuya H, Sakai R, Sato T, Tachibana K, Morimoto C, Yazaki Y, Hirai H. Integrin-mediated cell adhesion promotes tyrosine phosphorylation of p130Cas, a Src homology 3-containing molecule having multiple Src homology 2-binding motifs. J Biol Chem. 1995;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa S, Toyoshima H, Kozutsumi H, Hagiwara K, Sakai R, Tanaka T, Hirano N, Mano H, Yazaki Y, Hirai H. The C-terminal SH3 domain of the mouse c-Crk protein negatively regulates tyrosine-phosphorylation of Crk associated p130 in rat 3Y1 cells. Oncogene. 1994;9:1669–1678. [PubMed] [Google Scholar]
- 47.O’Neill C, Jordan P, Ireland G. Evidence for two distinct mechanisms of anchorage stimulation in freshly explanted and 3T3 Swiss mouse fibroblasts. Cell. 1986;44:489–496. doi: 10.1016/0092-8674(86)90470-8. [DOI] [PubMed] [Google Scholar]
- 48.O’Neill T J, Craparo A, Gustafson T A. Characterization of an interaction between insulin receptor substrate 1 and the insulin receptor by using the two-hybrid system. Mol Cell Biol. 1994;14:6433–6442. doi: 10.1128/mcb.14.10.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 50.Parsons J T. Integrin-mediated signalling: regulation by protein tyrosine kinases and small GTP-binding proteins. Curr Opin Cell Biol. 1996;8:146–152. doi: 10.1016/s0955-0674(96)80059-7. [DOI] [PubMed] [Google Scholar]
- 51.Pendergast A M. Nuclear tyrosine kinases: from Abl to WEE1. Curr Opin Cell Biol. 1996;8:174–181. doi: 10.1016/s0955-0674(96)80063-9. [DOI] [PubMed] [Google Scholar]
- 52.Petch L A, Bockholt S M, Bouton A, Parsons J T, Burridge K. Adhesion-induced tyrosine phosphorylation of the p130src substrate. J Cell Sci. 1995;108:1371–1379. doi: 10.1242/jcs.108.4.1371. [DOI] [PubMed] [Google Scholar]
- 53.Polte T R, Hanks S K. Interaction between focal adhesion kinase and Crk-associated tyrosine kinase substrate p130Cas. Proc Natl Acad Sci USA. 1995;92:10678–10682. doi: 10.1073/pnas.92.23.10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polte T R, Hanks S K. Complexes of focal adhesion kinase (FAK) and Crk-associated substrate (p130Cas) are elevated in cytoskeleton-associated fractions following adhesion and Src transformation. J Biol Chem. 1997;272:5501–5509. doi: 10.1074/jbc.272.9.5501. [DOI] [PubMed] [Google Scholar]
- 55.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 56.Reynolds A B, Kanner S B, Wang H-C R, Parsons J T. Stable association of activated pp60src with two tyrosine-phosphorylated cellular proteins. Mol Cell Biol. 1989;9:3951–3958. doi: 10.1128/mcb.9.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riabowol K, Draetta G, Brizuela L, Vandre D, Beach D. The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell. 1989;57:393–401. doi: 10.1016/0092-8674(89)90914-8. [DOI] [PubMed] [Google Scholar]
- 58.Ruoslahti E, Reed J C. Anchorage independence, integrins, and apoptosis. Cell. 1994;77:477–478. doi: 10.1016/0092-8674(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 59.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sattler M, Salgia R, Shrikhande G, Verma S, Uemura N, Law S F, Golemis E A, Griffin J D. Differential signaling after beta1 integrin ligation is mediated through binding of CRKL to p120CBL and p110HEF1. J Biol Chem. 1997;272:14320–14326. doi: 10.1074/jbc.272.22.14320. [DOI] [PubMed] [Google Scholar]
- 61.Schlaepfer D D, Hanks S K, Hunter T, van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 62.Schwerf U, Pastan I, Willingham M C, Brinkmann U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc Natl Acad Sci USA. 1996;93:2670–2674. doi: 10.1073/pnas.93.7.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein G S, Stein J L, Lian J B, Last T J, Owen T, McCabe L. Synchronization of normal diploid and transformed mammalian cells. In: Celis J E, editor. Cell biology: a laboratory handbook. San Diego, Calif: Academic Press, Inc.; 1994. pp. 282–287. [Google Scholar]
- 64.Streuli C H, Bailey N, Bissell M J. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tachibana K, Urano T, Fujita H, Ohashi Y, Kamiguchi K, Iwata S, Hirai H, Morimoto C. Tyrosine phosphorylation of crk-associated substrates by focal adhesion kinase. A putative mechanism for the integrin-mediated tyrosine phosphorylation of crk-associated substrates. J Biol Chem. 1997;272:29083–29090. doi: 10.1074/jbc.272.46.29083. [DOI] [PubMed] [Google Scholar]
- 66.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nashima K, Michiyuki M. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vuori K, Hirai H, Aizawa S, Ruoslahti E. Induction of p130cas signaling complex formation upon integrin-mediated cell adhesion: a role for Src family kinases. Mol Cell Biol. 1996;16:2606–2613. doi: 10.1128/mcb.16.6.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vuori K, Ruoslahti E. Tyrosine phosphorylation of p130Cas and cortactin accompanies integrin-mediated cell adhesion to extracellular matrix. J Biol Chem. 1995;270:22259–22262. doi: 10.1074/jbc.270.38.22259. [DOI] [PubMed] [Google Scholar]
- 70.Wen L P, Fahrni J A, Troie S, Guan J L, Orth K, Rosen G D. Cleavage of focal adhesion kinase by caspases during apoptosis. J Biol Chem. 1997;272:26056–26061. doi: 10.1074/jbc.272.41.26056. [DOI] [PubMed] [Google Scholar]