Abstract
Poly(ADP-ribose) polymerase (PARP; EC 2.4.2.30) is a zinc-finger DNA-binding protein that detects and signals DNA strand breaks generated directly or indirectly by genotoxic agents. In response to these breaks, the immediate poly(ADP-ribosyl)ation of nuclear proteins involved in chromatin architecture and DNA metabolism converts DNA damage into intracellular signals that can activate DNA repair programs or cell death options. To have greater insight into the physiological function of this enzyme, we have used the two-hybrid system to find genes encoding proteins putatively interacting with PARP. We have identified a physical association between PARP and the base excision repair (BER) protein XRCC1 (X-ray repair cross-complementing 1) in the Saccharomyces cerevisiae system, which was further confirmed to exist in mammalian cells. XRCC1 interacts with PARP by its central region (amino acids 301 to 402), which contains a BRCT (BRCA1 C terminus) module, a widespread motif in DNA repair and DNA damage-responsive cell cycle checkpoint proteins. Overexpression of XRCC1 in Cos-7 or HeLa cells dramatically decreases PARP activity in vivo, reinforcing the potential protective function of PARP at DNA breaks. Given that XRCC1 is also associated with DNA ligase III via a second BRCT module and with DNA polymerase β, our results provide strong evidence that PARP is a member of a BER multiprotein complex involved in the detection of DNA interruptions and possibly in the recruitment of XRCC1 and its partners for efficient processing of these breaks in a coordinated manner. The modular organizations of these interactors, associated with small conserved domains, may contribute to increasing the efficiency of the overall pathway.
The genomic integrity of cells is controlled by a network of protein factors that assess the status of the genome and either cause progression of proliferation or induce a halt in the cell cycle. In eukaryotes, DNA strand breaks, introduced either directly by ionizing radiation or indirectly following enzymatic incision of a DNA lesion, trigger the synthesis of poly(ADP-ribose) by the enzyme poly(ADP-ribose) polymerase (PARP) (1, 13, 39). At the site of breakage, PARP catalyzes the transfer of the ADP-ribose moiety from its substrate, NAD+, to a limited number of protein acceptors involved in chromatin architecture and DNA metabolism, including the enzyme itself. These modified proteins, which carry long chains of negatively charged ADP-ribose polymers, lose their affinity for DNA and are thus inactivated. The short half-life of the polymer is attributed to the high activity of poly(ADP-ribose) glycohydrolase, which cleaves the ribose-ribose bond (28, 30). Therefore, poly(ADP-ribosylation) is an immediate but transient postranslational modification of nuclear proteins, induced by DNA-damaging agents.
The physiological role of PARP has been much debated in the last decade, but recent molecular and genetic approaches, including expression of either a dominant-negative mutant (26, 36, 44) or antisense oligonucleotides (14), have clearly implicated PARP in the base excision repair (BER) pathway. A more definitive assessment of PARP function was recently provided by the generation of PARP-deficient mice by homologous recombination (35, 53). We found that PARP−/− mice are hypersensitive to monofunctional alkylating agents and γ-irradiation and display a marked genomic instability (sister chromatid exchanges and chromatid and chromosome breaks) following DNA damage (35). Interestingly, γ-irradiation of these mice causes acute toxicity of the epithelia of their small intestines (35), as has been observed with other DNA damage and signalling and repair enzyme deficiencies (2, 3), thus emphasizing the crucial function of DNA surveillance programs of rapidly dividing cells. Similar results indicating that PARP is important for the maintenance of genomic stability following environmental or experimental stress were recently obtained (54).
In this work, we have used the two-hybrid system to identify genes encoding proteins that putatively interact with PARP and are involved in its biological function. The human PARP cDNA fused to the LexA-encoding DNA-binding domain (DBD) was used as bait to screen a HeLa cDNA library fused with the activation domain of Gal4. This screening resulted in the identification of the BER pathway protein XRCC1 (X-ray repair cross-complementing 1) as a factor that associates with PARP. This interaction was further confirmed by in vivo experiments with glutathione S-transferase (GST)-tagged fusion proteins expressed in Cos-7 and HeLa cells. XRCC1 and PARP were found to interact via their respective BRCT (BRCA1 C terminus) modules (4, 9) and via an additional site located in the N-terminal zinc-finger domain of PARP. This association dramatically decreased the catalytic activity of PARP without modifying its nick sensor function. Therefore, the association of PARP with XRCC1, a partner of DNA ligase III (7, 8) and DNA polymerase β (25), is suggestive of a role in the detection and protection of a DNA strand break and the subsequent targeting of a BER complex to the damaged site.
MATERIALS AND METHODS
Bacterial and Saccharomyces cerevisiae strains and eukaryotic cells.
Escherichia coli DH5α F′ and HB 101 cells were used for subcloning of cDNA and for rescue of plasmids from yeast cells, respectively. For two-hybrid screening, the yeast reporter strain used (L40) has the following phenotype: MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ. Cos-7 cells and HeLa S3 cells were grown in 10% fetal calf serum-supplemented Dulbecco modified Eagle medium (Sigma Chemical Co., St. Louis, Mo.).
Plasmids.
The two-hybrid vector pBTM116 containing the DBD of the transcription factor LexA (residues 1 to 211) is described elsewhere (52). The human cDNA of PARP and a catalytically inactive mutant (D993A) were cloned in frame with the DBD of LexA. The vector pGAD-GH contains the yeast transcription factor GAL4 activation domain II (amino acids 768 to 881). The pBC vector is a eukaryotic GST fusion vector (11). pBC-NLS is a derivative of pBC containing the simian virus 40 nuclear location signal (NLS) (22) in the NdeI restriction site. In-frame deletion mutations in pBC or pBC-NLS producing full-length and truncated forms of PARP and XRCC1 were carried out by standard procedures.
Two-hybrid assays.
Two-hybrid screening of a cDNA expression library from HeLa cells in the pGAD-GH vector, β-galactosidase filter assay, and rescue of plasmids from yeast clones into bacterial hosts were performed as described previously (12, 52).
Transfection and protein-protein binding assay.
Cos-7 cells were electroporated (500 μF, 240 V) with 5 μg of recombinant DNA. The binding affinity assay to probe overexpressed GST-tagged proteins was performed as described in reference 11.
PARP activity.
Cos-7 cells were transfected with either pBC-NLS or pBC XRCC1-(170–428). After 36 h, cells were harvested, washed in phosphate-buffered saline (PBS), pelleted, and resuspended in 20 mM Tris-HCl (pH 7.5)–0.4 M KCl–20% (vol/vol) glycerol–5 mM dithiothreitol supplemented with leupeptin, pepstatin, and aprotinin, each at 5 mg/ml. PARP activity in 25 μg of total proteins was quantified essentially as described in reference 38.
XRCC1 expression and purification.
The cDNA encoding the full-length XRCC1 factor (kindly donated by K. Caldecott), cloned in the pET 16b expression vector, was expressed in BL21 bacteria. His-tagged recombinant XRCC1 was affinity purified on a nickel column (immobilized metal ion adsorption chromatography; Pharmacia, Uppsala, Sweden) as previously described (8).
Rabbit antibody against His-tagged recombinant XRCC1.
Two rabbits were immunized by intramuscular injections (100 μg of purified recombinant protein/injection) in the presence of complete Freund’s adjuvant for the first inoculation (day 0) and incomplete Freund’s adjuvant for the following inoculations (days 15, 30, 45, and 60). From a week after the second injection, the rabbits were bled on a fortnightly basis until week 14. The animals were prebled, and these samples were tested as controls in the different assays.
Double indirect immunofluorescence.
Cells grown on coverslips were exposed to 1 mM H2O2 for 10 min at 37°C, washed three times with PBS, and fixed with methanol-acetone (1:1, vol/vol) for 10 min at 4°C. After being washed three times with PBS supplemented with 0.1% (vol/vol) Tween 20, cells were incubated overnight at 4°C with a mixture of monoclonal antibodies against GST (immunoglobulin G1 [IgG1], 1:1,000 dilution) and poly(ADP-ribose) (10H; IgG3, 1:200 dilution). After being washed, the coverslips were incubated in a mixture of secondary antibodies: Texas red-conjugated anti-mouse IgG1 and fluorescein isothiocyanate-conjugated anti-mouse IgG3 (1:200 dilution) for 4 h at room temperature. Immunofluorescence was evaluated with a Zeiss Axioplan microscope equipped with a model C5985 chilled charge-coupled device camera (Hamamatsu).
RESULTS
XRCC1 interacts with PARP by its domain homologous to rad4/cut5.
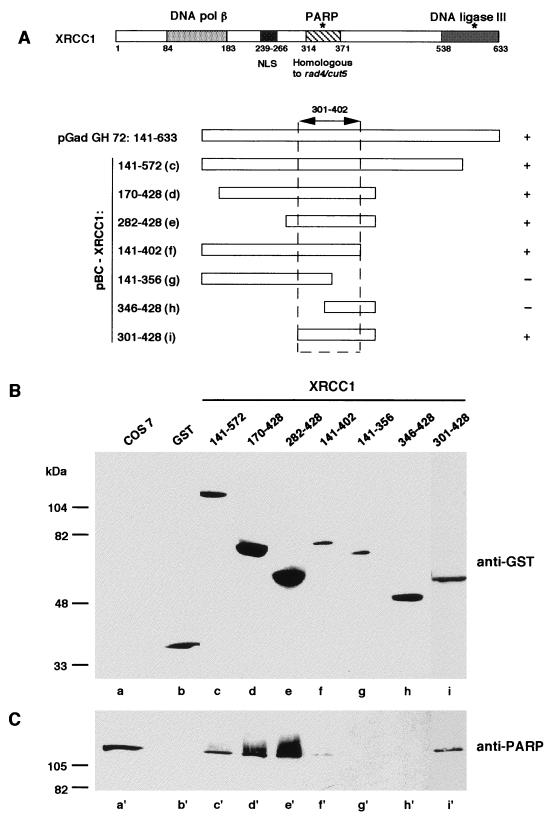
We have used the two-hybrid system to screen over 1 million yeast transformants for proteins that putatively interact with human PARP. An inactive mutant (PARP D993A) bearing a point mutation in the catalytic domain (46) was chosen as bait to avoid toxic effects due to the expression of an active PARP in yeast (21). Eighty-nine clones were isolated from a HeLa cDNA library that were both His3+ and LacZ+. Partial sequencing of cDNA revealed that among 15 independent clones, three were in computer databases and had identifiable sequences. One of them (pGAD-GH 72) had complete identity with a portion of the cDNA of the DNA repair factor XRCC1 (residues 141 to 633) (51). The rescued plasmid tested in the presence of the pBTM116 plasmid fused to the wild-type PARP activated HIS3 and lacZ reporter genes, demonstrating the nontoxicity of PARP in this system. To ascertain the specificity of this interaction, pGAD-GH 72 was cotransfected in yeast with pBTM116 fused to the genes for lamin C and Ras. The resulting phenotype was His3− LacZ− (data not shown), thus indicating that PARP specifically binds to XRCC1 in the two-hybrid system.
To confirm the physical interaction between PARP and XRCC1 in mammalian cells, XRCC1 (encoding the amino acids 141 to 572) was cloned in the eukaryotic expression vector pBC (11) in frame with GST, giving rise to the pBC XRCC1-(141–572) plasmid. The fusion protein and wild-type GST (as a control) were overproduced in Cos-7 cells and subsequently affinity purified on glutathione-agarose beads. Following washing, bound and associated proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotted (Fig. 1). PARP was associated with overproduced GST–XRCC1-(141–572) (Fig. 1B, lane c and 1C, lane c′) but not with GST alone (lanes b and b′). Furthermore, the PARP-XRCC1 complex was resistant to 1 M NaCl, indicating a high-affinity interaction between the two interactors. As a further confirmation, HeLa whole-cell extracts were immunoprecipitated either with a monoclonal anti-PARP antibody (27) or with PBS as a negative control. The presence of XRCC1 was revealed only in the immune complex (data not shown). Taken together, these results confirm that full-length PARP interacts with full-length XRCC1; this interaction occurs not only in the yeast two-hybrid system but also in a mammalian cellular context (see also Fig. 3). This interaction has been suggested by Caldecott et al., as PARP was coimmunoprecipitated by anti-XRCC1 antibodies along with DNA ligase III and appeared to interact with XRCC1 in the two-hybrid assay (6).
FIG. 1.
Interaction of PARP with GST-tagged XRCC1 domains. (A) Modular organization of XRCC1. Amino acids 84 to 183 make up the region that interacts with DNA polymerase (pol) β (25); amino acids 538 to 633 make up the region that interacts with DNA ligase III (37); amino acids 301 to 402 make up the region that interacts with PARP (cf. panels B and C); and amino acids 239 to 266 make up the NLS (cf. Fig. 2). The asterisks indicate the two BRCT modules (4, 9). The portion of XRCC1 encoded by the cDNA clone isolated in the two-hybrid procedure and the GST-tagged XRCC1 deletion mutants expressed in Cos-7 cells are also diagrammed. Expressed GST-fusion proteins and interacting endogenous proteins (PARP) were selectively extracted and analyzed by Western blotting as described in Materials and Methods with successively anti-GST (B) and anti-PARP (C) antibodies. Lanes a and a′ contain the total extract of control untransfected Cos-7 cells.
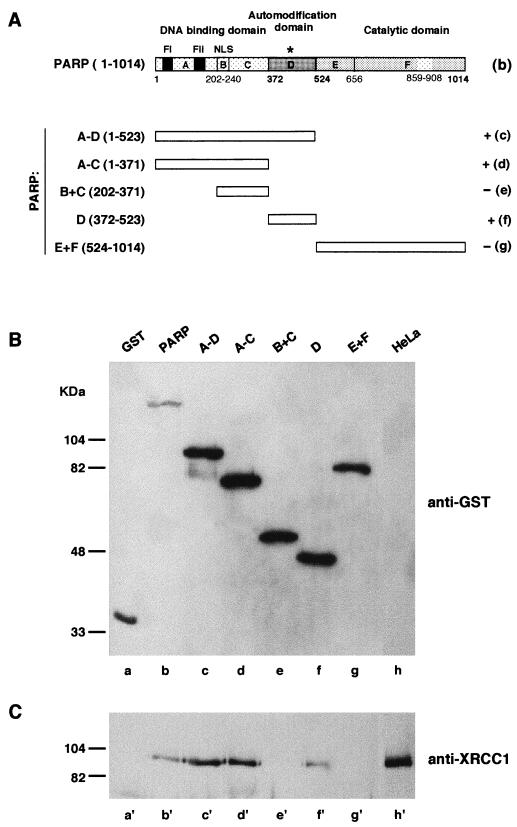
FIG. 3.
Interaction of XRCC1 with GST-PARP and PARP functional domains. (A) Modular organization of PARP (diagram b) and truncated forms of PARP (diagrams c to g) expressed as GST-fusion proteins in HeLa cells. The asterisk indicates the BRCT motif present in domain D of PARP; FI and FII correspond to the PARP zinc fingers (4, 9). Fusion proteins and interacting endogenous proteins (XRCC1) were selectively extracted and analyzed by Western blotting as described in Materials and Methods with successively anti-GST (B) and anti-XRCC1 (C) antibodies. Lanes h and h′ contain total extract of control untransfected HeLa cells.
To identify the region in XRCC1 that mediates the interaction with PARP, deletion mutants of XRCC1 were overproduced in Cos-7 cells as fusions with the GST reporter protein and cellular extracts purified in batch by affinity on glutathione beads were analyzed by Western blotting as described above. The absence of an NLS in some constructs was offset by the addition of the simian virus 40 NLS (22) cloned in frame with GST into the pBC expression vector. As shown in Fig. 1B and C, XRCC1-(301–428) was sufficient for interacting with PARP whereas fragment XRCC1-(346–428) was not. These results indicate that the central portion of XRCC1 is involved in binding to PARP and suggest that amino acids within the region of residues 301 to 402 are required for the interaction. Interestingly, this domain is homologous to a duplicated sequence in the Schizosaccharomyces pombe rad4/cut5 gene (29), which is required for maintaining the dependency of mitosis on correct progression through the cell cycle (40). As is clearly visible in Fig. 1C (lanes c′, d′, and e′), oligo(ADP-ribosyl)ated PARP corresponding to a retarded species on SDS-polyacrylamide gels and immunostained by an anti-PARP antibody (46) was preferentially associated with XRCC1, suggesting that a conformational change induced by limited auto-poly(ADP-ribosyl)ation, increased the accessibility and/or the affinity of PARP for XRCC1. A similar conclusion has already been reached for the heteroassociation between PARP and HeLa nuclear proteins, including histones (18).
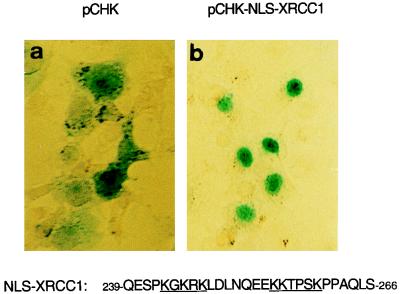
In the course of this study, we identified a functional nuclear localization signal in XRCC1. Computerized sequence analysis predicted a bipartite NLS at positions 255 to 274 (51); however, a larger region, positions 239 to 274, contained several clusters of basic residues, including the motif 244-GKRK-X7-KKTPSK-259, which resembles the bipartite NLSs of PARP (45) and of mKIN17 (34). Therefore, the peptide consisting of residues 239 to 266 was fused to β-galactosidase in the vector pCHK, which was subsequently transfected in Cos-7 cells as described previously (43). Localization of the β-galactosidase–XRCC1-(239–266) fusion protein was clearly nuclear (Fig. 2b), unlike the cytoplasmic localization of β-galactosidase alone (Fig. 2a), demonstrating that this sequence (239 to 266) encompasses a functional NLS.
FIG. 2.
XRCC1 NLS. Cos-7 cells were transfected with the empty vector pCHK (a) or with the construct pCHK-NLS-XRCC1-(239–266), which encodes the XRCC1 bipartite NLS fused to β-galactosidase (b). The subcellular localization of the recombinant proteins was assessed by histochemical staining with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Micrographs of Cos-7 cells transfected with the XRCC1 putative NLS exclusively displayed blue nuclei (b).
PARP interacts with XRCC1 by its zinc-finger domain and its central region.
To identify domains of PARP interacting with XRCC1, deletion mutants of PARP were overproduced as fusions with the GST reporter protein in HeLa cells containing a high amount of XRCC1. The cellular extracts purified in batch by affinity on gluthatione-agarose beads were analyzed by Western blotting with a polyclonal antibody raised against XRCC1 (Fig. 3C) and a monoclonal anti-GST antibody (Fig. 3B) to monitor the amounts of expressed GST fusion proteins. Full-length XRCC1 was revealed to be associated with overproduced GST–full-length PARP but not with GST alone (Fig. 3B, lanes a and b, and 3C, lanes a′ and b′). Furthermore, XRCC1 was also found associated with PARP domains A to D (lanes c and c′), domains A to C (lanes d and d′), and domain D (lanes f and f′) but not with domains B and C (lanes e and e′) or with domains E and F (lanes g and g′). Altogether, these results demonstrate that PARP contacts XRCC1 at least by two interfaces: one located in the 29-kDa N-terminal domain (most probably in zinc-finger domain A) and the second (domain D) located in the central region; both were recently found to contain protein-binding sites (18, 33).
Similar results (data not shown) were obtained with a glutathione-agarose bead extract of Cos-7 cells expressing GST–XRCC1-(282–428), which was incubated with cleared lysates of E. coli overproducing PARP deletion mutants (references 17 and 47 and unpublished results).
XRCC1 negatively regulates PARP activity in vivo and in vitro.
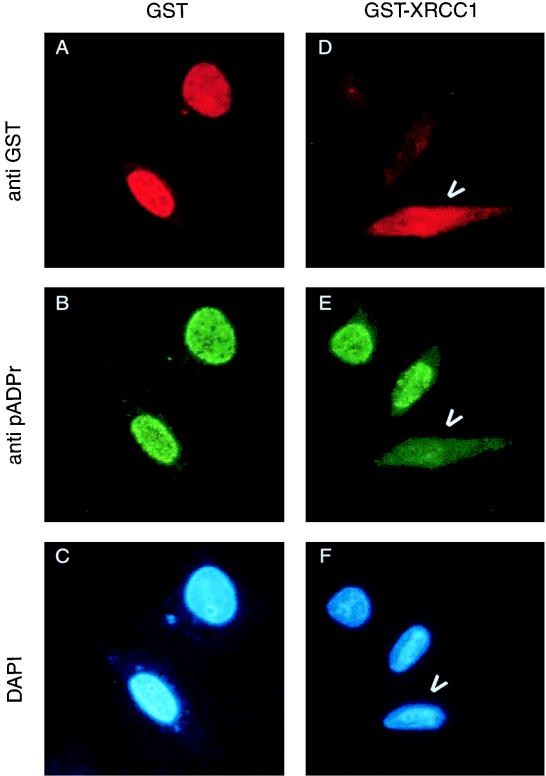
The functional consequences of the interaction between PARP and XRCC1 were investigated by two different methods. First, HeLa cells grown on coverslips and overproducing the fusion protein GST-XRCC1 or GST-NLS alone were exposed to oxidative stress (1 mM H2O2 for 10 min), which led to an immediate synthesis of poly(ADP-ribose). In situ polymer synthesis was visualized by double indirect immunofluorescence analysis with the monoclonal antibody 10H (green fluorescence; Fig. 4B and E), whereas the transfected cells were identified with an anti-GST antibody (red fluorescence; Fig. 4A and D). As shown in Fig. 4D to F, cells overproducing GST-XRCC1 were impaired in poly(ADP-ribose) synthesis, in response to hydrogen peroxide-generated DNA breaks, while the nontransfected cells visible in the same field exhibited normal PARP enzymatic activity. A similar result was obtained with Cos-7 cells expressing the fusion protein GST–XRCC1-(141–572) (data not shown). As a control, the GST-NLS fusion protein did not interfere with poly(ADP-ribose) synthesis induced by the same DNA damaging agent (Fig. 4A to C). This result strongly suggests that overexpression of full-length XRCC1 leads to inhibition of PARP activity stimulated by DNA breaks in vivo.
FIG. 4.
In vivo negative regulation of PARP activity by XRCC1. HeLa cells expressing either GST alone (A to C) or GST-XRCC1 (D to F) were treated with H2O2 for 10 min. After methanol-acetone fixation, the cells were incubated with a mixture of monoclonal antibodies against GST and against poly(ADP-ribose) (pADPr). The primary antibodies were detected with Texas red or fluorescein isothiocyanate-conjugated secondary antibodies. Arrowheads point out cells expressing GST-XRCC1 and lacking PARP activity (D to F). DAPI, 4′,6-diamidino-2-phenylindole.
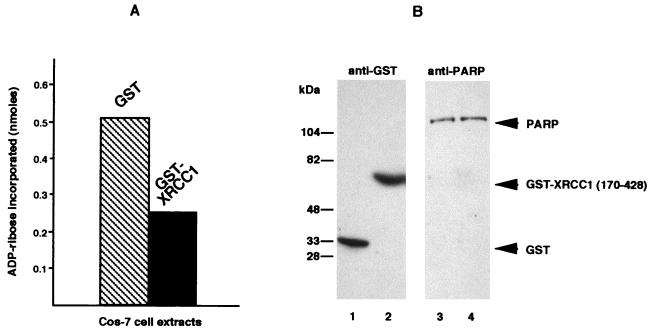
In a second set of experiments, whose results are displayed in Fig. 5, we measured under standard conditions (38) the global poly(ADP-ribosylation) activity of cellular PARP in whole-Cos-7-cell extracts overproducing either GST-NLS or GST–XRCC1-(170–428). The same amount of PARP was present in each extract, as shown in panel B. Although only 5 to 10% of cells were transfected and therefore overproduced the recombinant protein, PARP activity was dramatically reduced in XRCC1-(170–428)-containing crude extracts, thus confirming that XRCC1 may negatively regulate PARP activity.
FIG. 5.
In vitro negative regulation of PARP activity by XRCC1. (A) PARP activities were determined in total extracts of Cos-7 cells expressing either GST or GST–XRCC1-(170–428), as described in Materials and Methods. (B) Immunoblot detection of GST (lanes 1 and 2) and PARP (lanes 3 and 4) in extracts from Cos-7 cells transfected either with pBC-NLS (lanes 1 and 3) or with pBC XRCC1-(170–428) (lanes 2 and 4).
Affinity-purified XRCC1 can be oligo(ADP-ribosylated) and regulates negatively PARP automodification in vitro.
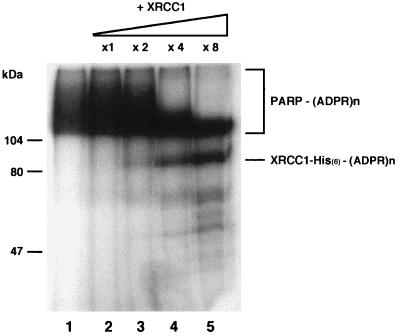
As a further confirmation of the down regulation of PARP activity mediated by XRCC1 interaction, PARP automodification was estimated by incorporation of [32P]NAD+ and visualized by SDS-polyacrylamide gel electrophoresis in the absence or in the presence of increasing amounts of affinity-purified XRCC1. As shown in Fig. 6, increasing amounts of XRCC1 strongly limit PARP automodification, in agreement with the results discussed above. At the same time, XRCC1 became oligo(ADP-ribosyl)ated, thus confirming a functional connection between the two interactors.
FIG. 6.
ADP-ribosylation of XRCC1 and its effect on PARP auto-poly(ADP-ribosylation). Incubations of XRCC1 with 300 ng of purified PARP were performed with 1 μM [32P]NAD for 2 min at 25°C under standard conditions (38). Acid-insoluble products were separated by gel electrophoresis and revealed by autoradiography of the stained and dried gel. Lane 1, PARP alone; lanes 2 to 5, purified His-tagged XRCC1 added at the indicated molar ratios with respect to PARP. (ADPR)n, ADP-ribosylation.
DISCUSSION
In order to delineate the biochemical mechanisms that govern the processing of DNA strand breaks in eukaryotic cells, the two-hybrid system was used to identify genes encoding proteins putatively interacting with PARP. By this technique, we previously identified a human homolog of the yeast ubiquitin-conjugating enzyme Ubc9 (33) as a partner of PARP. In this work, we have demonstrated that XRCC1 is physically associated with PARP in vivo and in vitro. The DNA repair factor XRCC1 complements the CHO cell lines EM9 and EM-C11, which are hypersensitive to both monofunctional alkylating agents and γ rays (50, 51, 58). In response to genotoxic agents that induce DNA base damage, EM9 and EM-C11 exhibit 10-fold increases in the occurrence of sister chromatid exchanges and severe defects in the rejoining of DNA breaks (19, 31, 58). Consistent with its proposed role in mammalian BER, XRCC1, which has no demonstrated catalytic activity, is supposed to serve as a scaffold protein during the BER reaction through its interaction with DNA ligase III (7, 25, 37) and DNA polymerase β (25). In this study, PARP has been identified as a third partner of XRCC1, thus most likely linking poly(ADP-ribosylation) reactions to the BER pathway (26, 35, 36, 44).
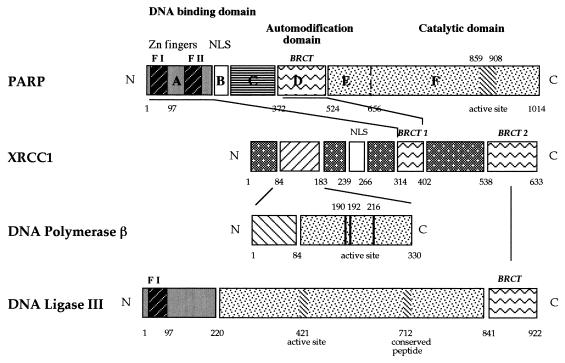
Interestingly, XRCC1 interacts with DNA ligase III and PARP through two BRCT modules (Fig. 1A and 7). The BRCT module is an autonomous folding unit of about 90 to 100 amino acids, first described for BRCA1 (breast cancer protein 1 carboxyl terminus) (24), and is widespread in a superfamily of DNA damage-responsive and cell cycle checkpoint proteins (4, 9). This motif, predicted to consist of four β-strands forming a core sheet structure and two α-helices, appears to be typical of domains involved in specific protein-protein interactions. Indeed, it has recently been demonstrated that XRCC1 and DNA ligase III interact through their respective C termini (37), containing BRCT modules located at positions 538 to 633 and 841 to 922, respectively (Fig. 7). Moreover, the alternative shorter form of DNA ligase III of 96 kDa, which is abundant in testes and which lacks the BRCT module (32, 55), failed to interact with XRCC1 (37), confirming the importance of this region for protein-protein interaction during male meiosis (5, 55).
FIG. 7.
Network of interactions between enzymes and factors participating in the BER pathway. BRCT modules involved in PARP-XRCC1 and in DNA ligase III-XRCC1 (37) interactions are indicated. XRCC1 contacts DNA polymerase β (25) by its N-terminal region. PARP and DNA ligase III have the same nick detection motif (zinc finger F1 [F I], amino acids 1 to 97).
Deletion analysis indicates that PARP contacts XRCC1 through its N-terminal zinc-finger domain or its central portion (Fig. 3). Both regions were recently found to be involved in histone and nuclear protein binding (18, 33). Quite interestingly, the central part in contact with XRCC1 contains the automodification domain (domain D) (Fig. 3A and 7), which comprises a BRCT motif at positions 384 to 476 (4, 9). Our results suggest that auto-poly(ADP-ribosylation), which takes place in domain D, modulates the interaction between PARP and XRCC1, as indicated by the preferential binding of XRCC1 to oligo(ADP-ribosylated) PARP (Fig. 1). In line with this, negative regulation of PARP activity was detected both in vitro and in vivo, suggesting that XRCC1 may limit PARP auto-(ADP-ribosylation) and that it therefore participates, indirectly, in the protection of DNA ends produced during DNA damage and repair. Conversely, in EM9 cells the absence of or a defect in XRCC1 may have contributed to increased PARP activity, compared to that of the parental cell line AA8, in time course experiments (20). Finally, the recruiting properties of XRCC1 may be essential to attract DNA polymerase β and ultimately DNA ligase III to the repair site, which occurs concomitantly with the release of PARP from the DNA lesion, itself implying a necessary high level of PARP auto-modification.
For mammalian cells, two distinct branches of the BER pathway that differ in the type of DNA polymerase required have been reported (10, 15, 16). The short patch pathway involves DNA polymerase β, XRCC1, and DNA ligase III (23), while the long patch pathway requires DNA polymerase β or δ, PCNA, DNase IV (FEN1), and DNA ligase I (25). Although the entire process for uracil residues has been reconstituted in vitro with cell extracts (15, 16) or purified proteins (25), the recent disruption of genes involved in BER has contributed to establishing the importance of XRCC1 (56), which apparently is not absolutely required in in vitro BER assays but has turned out to be a factor as crucial (49) as DNA polymerase β (48) or human AP endonuclease (HAP1) (57) for animal viability. Similarly, PARP, which was found to negatively regulate BER in cell extracts (41, 42), recently appeared to be essential for the survival of mice under genotoxic stress (35).
In summary, enzymes and factors involved in BER appear to behave as multimodular polypeptides capable of various combinations in protein-protein contacts mediated by small specific domains, thus ensuring rapid recruitment and coordination of the different partners. The interaction network data combining PARP, XRCC1, DNA ligase III, and DNA polymerase β are summarized in Fig. 7. The absence of one of the constituents of the BER complex may drastically reduce the efficiency of the overall pathway, leading to genomic instability and a decrease in survival, as is the case for XRCC1 mutant cell lines (e.g., EM9 and EMC-11) and mouse embryonic fibroblasts derived from PARP knockout mice, which, interestingly, display similar phenotypes (51a).
ACKNOWLEDGMENTS
This work was supported by the Association pour la Recherche Contre le Cancer, Electricité de France, CNRS (grant ACC-SV radiations ionisantes), by the Foundation pour la Recherche Médicale, and by the Commissariat à l’Energie Atomique.
We are grateful to E. Flatter for excellent technical assistance and K. Caldecott, B. Chatton, Y. Lutz, M. Miwa, and T. Sugimura for generous gifts of constructs and reagents.
REFERENCES
- 1.Althaus F R, Richter C. ADP-ribosylation of proteins. Enzymology and biological significance. Mol Biol Biochem Biophys. 1987;37:1–237. [PubMed] [Google Scholar]
- 2.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shilo Y, Crawley J N, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 3.Biedermann K A, Sun J, Giaccia A J, Tosto L M, Brown J M. Scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 5.Brookman K W, Tebbs R S, Allen S A, Tucker J D, Swiger R R, Lamerdin J E, Carrano A V, Thompson L H. Isolation and characterization of mouse Xrcc-1, a DNA repair gene affecting ligation. Genomics. 1994;22:180–188. doi: 10.1006/geno.1994.1359. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott K, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase β and possibly poly(ADP-ribose)polymerase, and DNA ligase III is a novel molecular “nick sensor” in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldecott K W, McKeown C K, Tucker J D, Ljungquist S, Thompson L H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldecott K W, Tucker J D, Stanker L H, Thompson L H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callebaut I, Mornon J P. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 10.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 11.Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- 12.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Murcia G, Ménissier-de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 14.Ding R, Smulson M. Depletion of nuclear poly(ADP-ribose) polymerase by antisense RNA expression: influences on genomic stability, chromatin organization, and carcinogen cytotoxicity. Cancer Res. 1994;54:4627–4634. [PubMed] [Google Scholar]
- 15.Frosina G, Fortini P, Rossi O, Carrozzino F, Abbondandolo A, Dogliotti E. Repair of abasic sites by mammalian cell extracts. Biochem J. 1994;304:699–705. doi: 10.1042/bj3040699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox L S, Lane D P, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 17.Gradwohl G, Ménissier-de Murcia J, Molinete M, Simonin F, Koken M, Hoeijmakers J H, de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griesenbeck J, Oei S L, Mayer-Kuckuk P, Ziegler M, Buchlow G, Schweiger M. Protein-protein interaction of the human poly(ADP-ribose)polymerase depends on the functional state of the enzyme. Biochemistry. 1997;36:7297–7304. doi: 10.1021/bi962710g. [DOI] [PubMed] [Google Scholar]
- 19.Hoy C A, Fuscoe J C, Thompson L H. Recombination and ligation of transfected DNA in CHO mutant EM9, which has high levels of sister chromatid exchange. Mol Cell Biol. 1987;7:2007–2011. doi: 10.1128/mcb.7.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikejima M, Bohannon D, Gill D M, Thompson L H. Poly(ADP-ribose) metabolism appears normal in EM9, a mutagen-sensitive mutant of CHO cells. Mutat Res. 1984;128:213–220. doi: 10.1016/0027-5107(84)90109-x. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser P, Auer B, Schweiger M. Inhibition of cell proliferation in Saccharomyces cerevisiae by expression of human NAD+ ADP-ribosyltransferase requires the DNA binding domain (“zinc fingers”) Mol Gen Genet. 1992;232:231–239. doi: 10.1007/BF00280001. [DOI] [PubMed] [Google Scholar]
- 22.Kalderon D, Richardson W D, Markham A F, Smith A E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 23.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin E V, Altschul S F, Bork P. Functional motifs. Nat Genet. 1996;13:266–267. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 25.Kubota Y, Nash R A, Klungland A, Barnes D, Lindahl T. Reconstitution of DNA base excision repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;23:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 26.Küpper J-H, Müller M, Jacobson M K, Tatsumi-Miyajima J, Coyle D L, Jacobson E L, Bürkle A. trans-Dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against γ-irradiation and N-methyl-N′-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol Cell Biol. 1995;15:3154–3163. doi: 10.1128/mcb.15.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamarre D, Talbot B, de Murcia G, Laplante C, Leduc Y, Mazen A, Poirier G G. Structural and functional analysis of poly(ADP-ribose) polymerase: an immunological study. Biochim Biophys Acta. 1988;950:147–160. doi: 10.1016/0167-4781(88)90007-3. [DOI] [PubMed] [Google Scholar]
- 28.Lautier D, Lagueux J, Thibodeau J, Menard L, Poirier G G. Molecular and biochemical features of poly (ADP-ribose) metabolism. Mol Cell Biochem. 1993;122:171–193. doi: 10.1007/BF01076101. [DOI] [PubMed] [Google Scholar]
- 29.Lehmann A R. Duplicated region of sequence similarity to the human XRCC1 DNA repair gene in the Schizosaccharomyces pombe rad4/cut5 gene. Nucleic Acids Res. 1993;21:5274. doi: 10.1093/nar/21.22.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin W, Amé J C, Aboul-Ela N, Jacobson E L, Jacobson M K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 31.Ljungquist S, Kenne K, Olsson L, Sandstrom M. Altered DNA ligase III activity in the CHO EM9 mutant. Mutat Res. 1994;314:177–186. doi: 10.1016/0921-8777(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 32.Mackey Z B, Ramos W, Levin D S, Walter C A, McCarrey J R, Tomkinson A E. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol Cell Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson M, Ménissier-de Murcia J, Mattei M J, de Murcia G, Niedergang C P. Poly(ADP-ribose)polymerase interacts with a novel human ubiquitin conjugating enzyme: hUbc9. Gene. 1997;190:287–296. doi: 10.1016/s0378-1119(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 34.Mazin A, Timchenko T, Menissier-de Murcia J, Schreiber V, Angulo J F, de Murcia G, Devoret R. Kin17, a mouse nuclear zinc finger protein that binds preferentially to curved DNA. Nucleic Acids Res. 1994;22:4335–4341. doi: 10.1093/nar/22.20.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ménissier de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Olivier F J, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molinete M, Vermeulen W, Burkle A, Ménissier de Murcia J, Kupper J H, Hoeijmakers J H, de Murcia G. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J. 1993;12:2109–2117. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nash R A, Caldecott K W, Barnes D E, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 38.Niedergang C, Okazaki H, Mandel P. Properties of purified calf thymus poly(adenosine diphosphate ribose) polymerase. Eur J Biochem. 1979;102:43–57. doi: 10.1111/j.1432-1033.1979.tb06261.x. [DOI] [PubMed] [Google Scholar]
- 39.Oei S L, Griesenbeck J, Schweiger M. The role of poly ADP-ribosylation. Rev Physiol Biochem Pharmacol. 1997;131:4135–4137. doi: 10.1007/3-540-61992-5_7. [DOI] [PubMed] [Google Scholar]
- 40.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 41.Satoh M S, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 42.Satoh M S, Poirier G G, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber V, de Murcia G, Ménissier-de Murcia J. A eukaryotic expression vector for the study of nuclear localization signals. Gene. 1994;150:411–412. doi: 10.1016/0378-1119(94)90466-9. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald D, de Murcia G, Ménissier de Murcia J. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc Natl Acad Sci USA. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber V, Molinete M, Boeuf H, de Murcia G, Ménissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992;11:3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simonin F, Briand J P, Muller S, de Murcia G. Detection of poly(ADP-ribose) polymerase in crude extracts by activity-blot. Anal Biochem. 1991;195:226–231. doi: 10.1016/0003-2697(91)90321-j. [DOI] [PubMed] [Google Scholar]
- 47.Simonin F, Poch O, Delarue M, de Murcia G. Identification of potential active-site residues in the human poly(ADP-ribose) polymerase. J Biol Chem. 1993;268:8529–8535. [PubMed] [Google Scholar]
- 48.Sobol R W, Horton J K, Kuhn R, Gu H, Singhal R K, Prasad R, Rajewsky K, Wilson S H. Requirement of mammalian DNA polymerase beta in base excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 49.Tebbs R S, Meneses J J, Pedersen R A, Thompson L H, Cleaver J E. XRCC1 knockout mice are nonviable. Environ Mol Mutagen. 1996;27:68. [Google Scholar]
- 50.Thompson L H, Brookman K V, Dillehay L E, Carrano A V, Mazrimas C L, Minkler J L. A CHO-cell line strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister chromatid exchange. Mutat Res. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 51.Thompson L H, Brookman K W, Jones N J, Allen S A, Carrano A V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Trucco, C., F. J. Javier Oliver, G. de Murcia, and J. Ménissier-de Murcia. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell ines. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 52.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E W. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei Y F, Robins P, Carter K, Caldecott K, Pappin D J C, Yu G L, Wang R P, Shell B K, Nash R A, Schär P, Barnes D E, Haseltine W A, Lindahl T. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and genetic recombination. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson D M, III, Thompson L H. Life without DNA repair. Proc Natl Acad Sci USA. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xanthoudakis S, Smeyne R J, Wallace J D, Curran T. The redox/DNA repair patch protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zdzienicka M Z, van der Schans G P, Natarajan A T, Thompson L H, Neuteboom I, Simons J W I M. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 1992;7:265–269. doi: 10.1093/mutage/7.4.265. [DOI] [PubMed] [Google Scholar]