Abstract
The Polycomb group (Pc-G) constitutes an important, functionally conserved group of proteins, required to stably maintain inactive homeobox genes repressed during development. Drosophila extra sex combs (esc) and its mammalian homolog embryonic ectoderm development (eed) are special Pc-G members, in that they are required early during development when Pc-G repression is initiated, a process that is still poorly understood. To get insight in the molecular function of Eed, we searched for Eed-interacting proteins, using the yeast two-hybrid method. Here we describe the specific in vivo binding of Eed to Enx1 and Enx2, two mammalian homologs of the essential Drosophila Pc-G gene Enhancer-of-zeste [E(z)]. No direct biochemical interactions were found between Eed/Enx and a previously characterized mouse Pc-G protein complex, containing several mouse Pc-G proteins including mouse polyhomeotic (Mph1). This suggests that different Pc-G complexes with distinct functions may exist. However, partial colocalization of Enx1 and Mph1 to subnuclear domains may point to more transient interactions between these complexes, in support of a bridging role for Enx1.
Two groups of genes, the Polycomb group (Pc-G) and trithorax group (trx-G), were first identified in Drosophila as providing a transcriptional memory mechanism for key developmental regulators such as the homeotic genes. The initial parasegment-specific expression pattern of homeotic genes is initiated by the gap and pair-rule proteins at 2 h of embryogenesis (20, 34). At about 4 h of development, the gap and pair-rule proteins decay. It is at this time that the Pc-G and trx-G proteins first become essential to the stable maintenance of gene expression patterns throughout subsequent cell divisions. Their gene products are thought to act in multiprotein complexes at the level of chromatin structure, where Pc-G proteins maintain inactive homeotic genes in a repressed state whereas trx-G proteins ensure maintenance of the active state (reviewed in references 18, 22, and 23). Since the Pc-G and trx-G proteins are ubiquitously expressed, even in domains where homeotic genes are active or repressed, respectively, the Pc-G and trx-G complexes cannot themselves convey positional information (22). A central but largely unanswered question is therefore how Pc-G and trx-G complexes are able to recognize and discriminate between the specific gene expression patterns initiated by the gap and pair-rule gene products. Careful analysis of Pc-G and trx-G mutant phenotypes in both the fly and the mouse provided important insights in that not all the Pc-G or trx-G genes have identical functions and different subgroups can be assigned on the basis of the presence or absence of genetic interactions between specific mutants (5, 15, 17, 26, 32). Of special interest in this regard is the extra sex combs (esc) gene (29). Elegant studies pioneered by Struhl and coworkers with esc temperature-sensitive alleles have shown that esc function is required during the first 3 to 6 h of embryogenesis (30). This contrasts with the requirement for other Pc-G products such as Polycomb (Pc), that have to be present not only during early development but also during later development to maintain the proper homeotic expression patterns (27, 30). Thus, a critical role for esc lies at the transition stage, when the gap and pair-rule gene products decay and Pc-G and trx-G have to take over. Together, these results led to the proposal of bridging models, suggesting that esc may on the one hand interact either directly or indirectly with early gap gene-encoded repressors such as Hunchback (Hb) and Krüppel (Kr) while on the other hand providing a recruitment site for other Pc-G proteins (12, 20, 27).
Recently, we and others have demonstrated a remarkable degree of functional conservation of the Pc-G and trx-G genes in mice (2, 8, 26, 31, 35). This is underscored by the partial rescue of Drosophila Pc mutant flies by introduction of a mouse Pc homolog, M33 (21). A further telling example is provided by the positional cloning of a classical mouse gastrulation mutant, eed (embryonic ectoderm development) (26). Sequence analysis indicated that eed is the mouse homolog of Drosophila esc, revealing an overall 55% identity and 74% similarity comprising 83% of the protein sequence with no gaps or insertions. This high degree of conservation includes all five WD40 repeats, which are thought to be an integral part of protein-protein interactions (see below) (26). eed-null mutant mice die around day 8.5 of gestation and display disrupted anteroposterior patterning of the primitive streak. This is before initiation of expression of most of the homeobox (hox) genes, which first occurs during middle to late gastrulation (26). This phenotype is much more severe than what is observed in other single- or double-Pc-G-null mutant mice described to date (reviewed in references 25 and 32), indicating that there is also a special, early requirement for eed in the mouse. To increase our understanding of initiation of mouse Pc-G repression and the special role of eed therein, we screened for Eed-interacting proteins by using the yeast two-hybrid system (7). If the bridging models are valid in mammals, such a screen could in principle detect both early repressors required for initiating hox gene repression and other Pc-G proteins necessary for propagation and maintenance of repression. Here we report on the results of such screens.
MATERIALS AND METHODS
Yeast two-hybrid screens and plasmids.
Yeast strains Y190 and MAV103, which contain two chromosomally located Gal4-inducible reporter genes, HIS3 and lacZ, were transformed with Eed bait plasmids as specified below in pPC97 or a derivative in which the LEU2 marker was changed in TRP1 (7). Production of the GAL4 DNA binding domain (DBD) fusion protein was confirmed by Western blot analysis. The bait-containing strains were subsequently transformed by the lithium acetate method with a 14.5-day CD1 mouse embryo cDNA library fused to the GAL4 transactivation (TA) domain (7) or a day 7.5 mouse embryo cDNA library in pGAD10 (Clonetech). One million transformants were selected for growth on plates lacking histidine and supplemented with 25 mM 3-aminotriazole. HIS+ colonies were subsequently analyzed for β-galactosidase (β-gal) activity by a colony lift assay. In the first screen (strain Y190, Eed5′GAL4DBD bait, day 14.5 library), 3.5 × 106 transformants gave rise to 150 HIS+ colonies, of which 18 were β-gal+. Of the 18, 4 represented clone Enx1/1.1 (see Fig. 1A). In the second screen (strain MAV103, EedΔN6 bait, day 14.5 embryo library), 2.6 × 106 transformants yielded 114 HIS+ colonies, of which 3 were β-gal+. Two of the three represented laminin, and the third was identical to Enx1/1.1. In the third screen (strain Y190, Eed3′GAL4DBD bait, day 7.5 mouse embryo library) 25 HIS+ colonies of 3 × 105 transformants were obtained, of which 1 was β-gal+. This clone contained Enx2/30.1. To map the interaction domains on Eed and Enx, fragments generated by restriction enzyme digests or PCR were subcloned in the GAL4-DBD and GAL4-TA vector and cotransformed to Y190. The resulting yeast colonies were then assayed for β-gal activity and growth on plates lacking histidine, as described above. The Eed-null mutant vector was generated by replacing an N-terminal eed fragment of Eed5′GAL4DBD with a fragment harboring the ENU-induced T1040-C transition, cloned from the original mutant mouse cDNA. This results in a single amino acid change, L196-P, in the second WD40 repeat (26). Details of the subcloning strategies are available on request. The HA-Eed expression contruct was generated by ligating the full-length Eed in frame with the HA epitope peptide in the pMT2SM-HA expression vector, driving expression from the adenovirus major-late promoter and harboring the simian virus 40 (SV40) small t-intron, a poly(A) signal, and the SV40 replication origin. p(Myc)3-EZH2 and p(HA)3-EZH2, containing the full-length human EZH2/ENX1 cDNA N-terminally tagged with a triple Myc tag or a triple-HA tag, respectively, in SV40 T-promoter/SV40-ori-containing expression plasmids, were obtained from T. Jenuwein, IMP, Vienna, Austria.
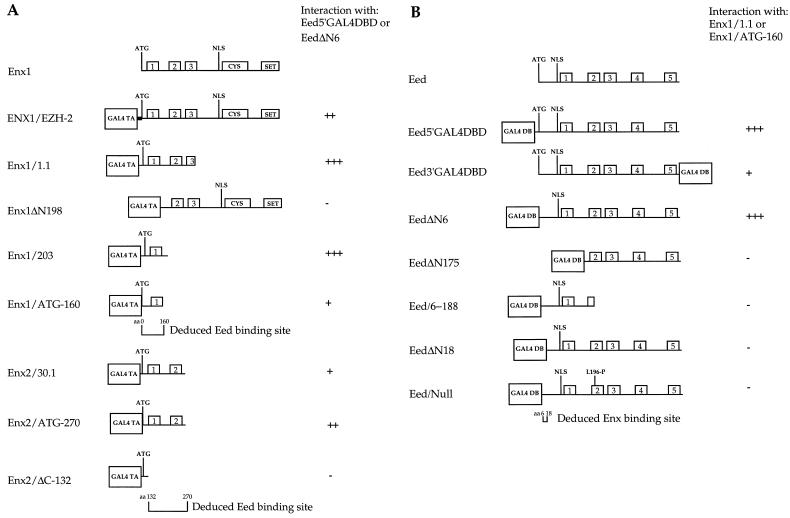
FIG. 1.
Mapping of binding domains on Enx1, Enx2, and Eed by using the yeast two-hybrid system. (A) Eed-binding regions on Enx1 and Enx2. The complete Enx1 coding region (746 aa) with conserved domains is schematically shown at the top for reference. The positions of boxes 1, 2, and 3 are indicated, together with cysteine-rich (CYS) and SET domains (14). NLS is the nuclear localization signal (note that the GAL4 TA domain also contains an independent nuclear localization signal), and ATG denotes the start codon. Clone ENX1/EZH-2 denotes the full-length N-terminally Myc-tagged EZH2 coding sequence N-terminally fused to the GAL4 TA domain (the Myc tag is indicated by the black box). Appropriate deletion constructs were generated in the GAL4 TA two-hybrid vectors pPC86 and pGAD10 and tested for interaction with Eed baits Eed5′GAL4DBD and EedΔN6; both gave identical results. The relative strength of interactions was determined by measuring β-gal activity. Enx1/1.1 (aa −29 [prior to ATG] to 287) was obtained in the two-hybrid screens; Enx1ΔN198 (lacks the N-terminal 198 aa) was obtained as described in reference 14; Enx1/203 is aa −29 to 203, and Enx1/ATG-160 is aa 1 to 160. Enx2/30.1 denotes the original identified two-hybrid clone (aa −26 to 270). Enx2/ATG-270 is aa 1 to 270; Enx2/ΔC-132 is aa −10 to 132. The deduced interaction domains are depicted below. (B). Enx-binding regions on Eed. The Eed coding region (441 aa) with conserved WD40 repeat domains (domains 1 to 5) is shown at the top. Appropriate deletion constructs were generated in the GAL4DBD pPC97 two-hybrid vectors or derivatives thereof and tested with the Enx1/1.1 or Enx1/ATG-160 GAL4TA plasmids. Both gave identical results. Eed5′GAL4DBD; aa −12 to 441; Eed3′GAL4DBD, aa 1 to 441; EedΔN6, aa 6 to 441; EedΔN175, aa 175 to 441; Eed/6–188, aa 6 to 188; EedΔN18, aa 18 to 441; Eed/Null, aa 1 to 441 (L196-P). Apart from requirement for intact WD40 repeats (see the text), the binding domain on Eed maps to aa 6 to 18, as indicated at the bottom.
Liquid β-gal assay.
Y190 yeast cells harboring the appropriate bait and prey constructs were grown to the log phase in selective minimal medium. The optical density at 600 nm (OD600) was measured to control for the number of cells. Cells from 1 ml of logarithmic culture were pelleted, washed once with Z-buffer, and resuspended in 150 μl of prewarmed (30°C) Z-buffer. To this was added 50 μl of chloroform and 20 μl of 0.1% sodium dodecyl sulfate (SDS), and the mixture was subjected to hard vortexing for 15 s. The reactions were started by adding 700 μl of prewarmed Z-buffer containing 1 mg of o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate, and they were timed. When visible yellowing of the tubes occurred, the reactions were terminated (with the time of the reaction recorded) by placing the tubes on ice and addition of 0.5 ml of Na2CO3. Cell debris was removed by centrifuging the tubes for 10 min at 21,000 × g, and the OD420 was recorded for each sample. Data are expressed as Miller units: [(OD420 × 1,000)/(OD600 × time in minutes × volume in milliliters)]. Each reaction was performed in triplicate and repeated in at least two independent experiments.
Immunoprecipitations.
U2-OS cell extracts were made by resuspending the cell pellets in 1 ml of ELB buffer (3), consisting of 5 mM EDTA, 0.5 mM dithiothreitol, 1 μg each of chymostatin, aprotinin, antipain, and leupeptin per ml, and 1 mM phenylmethylsulfonyl fluoride, and subjected to Dounce homogenization 10 times with a tight Eppendorf potter, sonicated for 20 s (50% duty cycle in a Branson sonifier), and centrifuged for 15 min at 21,000 × g. The supernatant of the lysate was used for immunoprecipitations. The protein concentration of the extracts was measured by the Bradford method. The lysate was precleared for 1 h with normal mouse or rabbit serum coupled to protein A-Sepharose beads, and 250 μl of the supernatant (protein concentration, 10 mg/ml) was used in immunoprecipitations with 3 μl of polyclonal anti-Enx1 antiserum (22) or 100 μl of tissue culture supernatant from hybridoma cells. After 1 h at 4°C, immune complexes were collected with 30 μl of protein A-Sepharose, washed four times (5 min each) in 1 ml of ELB buffer, and transferred to a fresh tube for a final wash. After addition of 30 μl of 2× SDS sample buffer, the immunoprecipitated proteins were separated on an SDS–9% polyacrylamide gel and transferred to nitrocellulose.
Western blot analysis.
Nitrocellulose membrane was blocked for 3 h at room temperature in PBST (phosphate-buffered saline [PBS], 0.05% Tween 20) containing 5% dried milk. The membrane was subsequently incubated for 1 h at room temperature with primary antibody (mouse monoclonal antibodies 12CA5 [1:500] and 9E10 [1:500], anti-GAL4DBD rabbit polyclonal antibody [1:200] [Santa Cruz]) diluted in PBST containing 1% dried milk. After being washed, the membrane was incubated for 1 h at room temperature with horseradish peroxidase-linked goat anti-rabbit immunoglobulin G IgG (Biosource) or goat anti-mouse IgG (Bio-Rad) as the second antibody diluted 1:10,000 in PBST containing 1% dried milk. Antibodies were detected by enhanced chemiluminescence (Amersham).
Transfection experiments.
COS7 cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. For transfections, cells were seeded at a density of 2 × 105 per well in six-well culture dishes. The next day, the cells were incubated with Lipofectamine (GIBCO-BRL) containing 2 μg of plasmid DNA/well as specified by the manufacturer. After 48 h, total protein extracts were prepared in ELB, as described above.
U2-OS cells were transfected (8 μg of plasmid DNA and 0.5 μg of puromycin- or G418-selectable marker DNA) by calcium phosphate precipitation by using standard procedures. One day after transfection, the cells were subjected to puromycin selection (8 μg/ml) or G418 selection (400 μg/ml). Colonies were isolated after 1 to 2 weeks of selection. Transfected cells were seeded on multiwell coverslips and processed for immunofluorescence as described below.
Immunofluorescence.
The osteosarcoma cell line U2-OS was grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum on multiwell coverslips to approximately 50% confluence, washed three times in PBS, and fixed in freshly prepared 2% formaldehyde–PBS for 15 min at room temperature. The cells were then incubated twice for 5 min with PBS, once for 5 min with PBS containing 0.5% Triton X-100, twice for 5 min with PBS; once for 5 min with 0.1 M glycine in PBS, and twice for 5 min with PBS. They were preblocked in blocking solution (PBS containing 5% fetal calf serum, 5% normal goat serum, and 0.02% Triton X-100) for 1 h at room temperature; 2 h at room temperature with 1:20-diluted affinity purified rabbit polyclonal Mph1/Rae-28 antibody or 1:40-diluted 12CA5 hybridoma supernatant in blocking solution; four times for 5 min with PBS plus 0.02% Triton X-100; and 1 h at room temperature with 1:100-diluted fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratories) or 1:1,000-diluted tetramethylrhodamine-5-isothiocyanate (TRITC)-conjugated goat anti-rabbit antibody in blocking solution. After four 5-min incubations with PBS plus 0.02% Triton X-100, the specimens were embedded in vectastain (with or without propidium iodide stain). Images of labelled cells were produced on a Bio-Rad confocal laser-scanning microscope with a 63x/1.35 oil immersion lens. FITC and TRITC images were recorded separately and then superimposed. When the primary antisera were omitted or replaced with normal rabbit or mouse serum (NRS or NMS) as specificity controls, no significant background signal was detected.
RT-PCR expression analysis.
A 5-μg sample of total RNA was used in a standard reverse transcriptase (RT) reaction with Superscript RT (GIBCO- BRL) in a total reaction volume of 20 μl, as specified by the manufacturer. A 0.5-μl volume of template was used in 50 μl of PCR mixture, containing 0.2 mM deoxynucleoside triphosphates, 0.5 μM each of the primers listed below, 1.5 mM MgCl2, and 1 U of Taq polymerase. The cycling program was optimized for the linear amplification range, and was 3 min at 94°C, followed by 35 cycles of 1 min at 94°C, 45 s at 55°C, and 2 min at 72°C, followed by a final extension of 10 min at 72°C. A 15-μl sample of each reaction mixture was loaded on a 2% NuSieve agarose gel and visualized with ethidium bromide (EtBr). The expected fragments are 176 nucleotides (nt) for HPRT, 248 nt for Enx1, and 480 nt for Enx2. The primers were 5′ HPRT (CCAGCAAGCTTGCAACCTTAACCA), 3′ HPRT (GTAATGATCAGTCAACGGGGGAC), 5′ Enx1 (AATGGAAATCCCTTGACATC), 3′ Enx1 (TTGAAAAATGTTACCATACTGC), 5′ Enx2 (ACGGACGTCTTCTAGCCCTC), and 3′ Enx2 (GGCTCTATGTTCACAGGATG).
RESULTS
Eed interacts with Enx1 and Enx2.
To identify Eed-interacting proteins, a day 14.5 mouse embryo cDNA library fused to the GAL4 TA domain in the cen,ars pPC67 vector (7) was screened twice with two independent Eed baits. In the first screen, a full-length eed cDNA was N-terminally fused to the GAL4-DBD in pPC97. Due to the way the fusion was made, this construct directs the expression of a linker peptide consisting of 10 amino acids derived from eed sequences prior to the original ATG as reported previously 26) and 12 amino acids derived from polylinker sequences (Fig. 1B, Eed5′GAL4DBD). Of 3.5 × 106 clones screened, 4 specific interacting clones were each identified multiple times. Because of potentially irrelevant interactions through the N-terminally fused linker peptide and because of potential interference with the binding of proteins to the N-terminal region due to proximity of the GAL4 DBD, a second Eed bait construct was generated in which eed starts at the ATG and is C-terminally fused to the GAL4-DBD (Fig. 1b, Eed3′GAL4DBD). Use of this bait in a retransformation experiment with the four originally obtained clones indicated that only one clone was capable of interacting. Sequence analysis revealed that this clone contained an N-terminal fragment of the Pc-G gene enx1, a mouse homolog of Drosophila Enhancer of Zeste [E(z)] (Fig. 1a, Enx1/1.1). Human ENX1/EZH2 was first identified in a two-hybrid screen as a specific interactor with the signal transduction protein and oncoprotein Vav (14). Subsequently, several groups have identified ENX1/EZH2 and ENX2/EZH1 as two E(z)-homologous genes in both mice and humans (1, 13, 14, 19) (see Discussion).
The C-terminal bait was also used to rescreen the same library. Of 2.6 × 106 clones, 2 positive clones were identified. One was identical to the previously isolated enx1 clone, and the second encoded the basement membrane-associated protein laminin α4, probably representing an irrelevant interaction. If eed, like its Drosophila counterpart esc, is required in a narrow window during early development when Hox gene expression patterns are determined, which in the mouse occurs around gastrulation, potential relevant interacting proteins might be underrepresented in a day 14.5 library. Therefore, we used the C-terminal Eed bait to screen a day 7.5 embryonic library fused to the GAL4 TA domain in a 2μm vector. Of 3 × 105 clones screened, 1 positive clone was obtained. This clone represented an N-terminal fragment of the second mouse E(z) homolog, enx2 (Fig. 1A, Enx2/30.1) (14, 19).
The Enx two-hybrid experiments were repeated with an Eed bait carrying the 17Rn51989SB allele, which represents a hypomorphic mutation in vivo (26). The only difference from the wild-type Eed bait is a T-A transversion at nt 1031, resulting in an N-to-I substitution at amino acid (aa) 193 in the second WD40 repeat. Interestingly, we observed a slightly weaker lacZ staining on filters. To quantify this difference, a liquid β-gal activity assay was used (see Materials and Methods). As indicated in Table 1, in two independent experiments a reproducible weaker interaction was indeed observed for the Eed-hypomorphic bait.
TABLE 1.
Quantified two-hybrid interactions between Eed and Enx1a
| Construct | β-gal activity (MU)b in:
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| Eed5′GAL4DBD + Enx1/1.1 | 34.2 (0.98) | 37.5 (0.15) |
| Eedhypomorph5′GAL 4DBD + Enx1/1.1 | 21 (0.23) | 20.0 (0.33) |
| Eedhypomorph5′GAL 4DBD + pPC86 | 0 | 0.1 (0.04) |
Quantitative liquid β-galactosidase assays were performed in triplicate, on log-phase yeast cells containing the indicated two-hybrid plasmids.
Data are presented in Miller units (MU) as explained in Materials and Methods, with standard deviations in parentheses.
Interacting domains on Enx1 and Enx2.
The mouse and human E(z) relatives enx1 and enx2 are highly conserved Pc-G members; there is an overall amino acid identity of 61% between mouse and Drosophila sequences. The regions of highest homology are found in five domains throughout both E(z) and the Enx proteins, suggesting conservation of distinct functional domains (13, 14, 16, 19). These comprise the N-terminally located boxes 1, 2, and 3, of which box 1 is the best conserved [95.4% between Enx1 and E(z) (14)]. These domains are followed by a centrally located cysteine-rich domain and a C-terminal SET domain. The latter represents a well-conserved domain which has also been found in other proteins implicated in transcriptional regulation at the level of chromatin, such as Suppressor-of-position-effect variegation 3-9 [Su(var)3-9] and trithorax (trx); this may suggest that interactions occur via the SET domain with a common partner (16).
The partial enx clones obtained in the two-hybrid screens implicated the N-terminal region as potential interaction domain, whereas the cysteine-rich region and SET domain are dispensable (Fig. 1A). Both the enx1 and enx2 clones represent fusions of leader sequences upstream of the ATG to the GAL4 TA. enx1 clone 1.1 (nt 46 to 996 of the published sequence [13]) is preceded by short in-frame CT and GA repeats that are not represented in the published sequence. These may represent a product of alternative splicing, since complex splicing patterns have been reported for the enx genes (1, 13, 19). Subsequent deletion analysis showed that this region is not implicated in Eed binding (Fig. 1A, Enx1/ATG-160). enx1 clone 1.1 encompasses the well-conserved box 1 and part of box 2 (Fig. 1A). enx2 clone 30.1 (nt 29 to 1051 of the published sequence) also retains box 1 and box 2 (Fig. 1A). An ENX1/EZH2 clone originally identified in the Vav two-hybrid screen was found to lack 198 N-terminal amino acids (14, 19). This clone does not interact with Eed (Fig. 1A, Enx1ΔN198). Subsequent deletion constructs localized the interaction domain on Enx1 to a region from the ATG to aa 160 (Fig. 1A, Enx1/ATG-160). The interaction domain on enx2 was similarly localized to an N-terminal region containing box 1 and box 2 (Fig. 1A). In addition, the less well-conserved sequences from the ATG to box 1 were insufficient in Eed binding (Fig. 1A, Enx2/ΔC-132). Taken together, the Enx1 and Enx2 mapping data strongly suggest that the region between aa 132 and 160, almost precisely containing box 1, is required for Eed binding. This domain is different from the Vav interaction domain, which was mapped to a region containing box 2 and box 3 on ENX1 (14).
Interacting domains on Eed: implication of WD40 repeats and an N-terminal domain.
Deletion of the first WD40 repeat of Eed abolishes binding to Enx1 (Fig. 1B, EedΔN175). In addition, an ENU-induced Eed-null mutant harboring a single amino acid substitution (L196-P) in the second WD40 repeat was defective in Enx binding, suggesting the importance of WD40 repeats for this interaction (Fig. 1B, Eed/Null). This probably reflects the unique conformation adopted by multiple WD40 repeat-containing proteins, involving the mutual interaction of WD40 β-strands to form a “β-propeller” structure, as was first demonstrated for the Gβ subunit of heterotrimeric G proteins (28, 33). Interestingly, apart from the necessity for intact WD40 repeats, the extreme N terminus of Eed is also important for binding to Enx, since deletion of 18 N-terminal amino acids abrogates binding (Fig. 1B, EedΔN18). By analogy, the Gβ N-terminal coiled-coil domain has also been shown to extend from the core β-propeller region and is implicated in interactions with the Gγ chain and effector proteins (28). As a control, protein extracts were prepared from all the bait-containing yeast clones, and proper expression of the GAL4-DBD fusion proteins was confirmed by Western blotting with an anti-GAL4DBD antiserum (data not shown).
In vivo interaction of Enx1 and Eed.
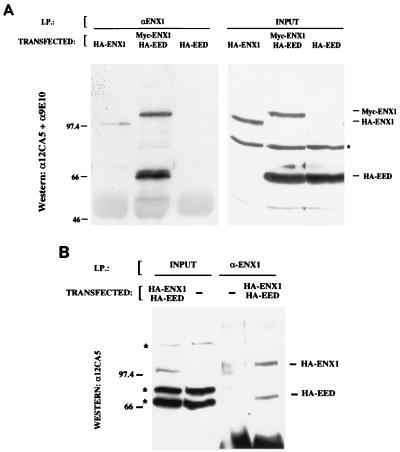
To verify the in vivo relevance of the interactions observed in yeast, an N-terminal HA epitope-tagged full-length eed expression construct was generated and cotransfected in COS7 cells with full-length N-terminally Myc- or HA-tagged EZH2, the human homolog of mouse enx1 (courtesy of T. Jenuwein). After 48 h, total protein extracts were prepared under mild conditions in ELB buffer and subjected to Western blotting to verify expression (Fig. 2A, right panel). Both the epitope-tagged Eed and ENX1 proteins run significantly slower in SDS-polyacrylamide gel electrophoresis than would be predicted from their calculated molecular masses: 50 kDa for Eed and 80 kDa for ENX1, respectively. This is caused in part by the addition of the epitope tags (note the difference in size between the triple Myc- or HA-tagged ENX1), but in Eed (single HA tag) it also probably reflects an aberrant mobility in SDS-polyacrylamide gel electrophoresis as has been observed for other Pc-G proteins (3, 4). Subsequently, the same lysates were used in immunoprecipitation experiments with a polyclonal rabbit serum directed against human ENX1/EZH2 (courtesy of O. Hobert [14]), or with NRS as a negative control. As depicted in Fig. 2a, left panel, the ENX1 serum specifically immunoprecipitated epitope-tagged ENX1 and is capable of coimmunoprecipitating the coexpressed HA-Eed protein (middle lane). NRS control immunoprecipitates yielded no signal (data not shown). Since the transiently transfected COS7 cells express relatively large amounts of cotransfected proteins, we verified the Enx1-Eed interactions in stably transfected human U2-OS osteosarcoma cell lines. U2-OS cells were transfected with the HA-eed construct, and clones were isolated and assayed for stable, low-level expression in all cells by indirect immunofluorescence detection of 12CA5 (anti-HA monoclonal antibody)-specific staining. (Detection on Western blots of HA-Eed was hampered by comigration of an anti-12CA5 cross-reactive band in this cell line [Fig. 2B, left panel].) Subsequently, the HA-eed-expressing cell line was transfected with the HA-ENX1 expression construct, and the resulting clones were verified by Western blotting for HA-ENX1 expression (Fig. 2B, left panel). Total protein extracts from mock-transfected cells or HA-Eed/HA-ENX1-transfected cells in ELB buffer were subjected to immunoprecipitation with the anti-ENX1 serum, and specific coimmunoprecipitation of HA-Eed was also observed in the stable low-level-expressing U2-OS cell line (Fig. 2B, right panel), indicating that Eed and ENX1 are capable of interacting in vivo.
FIG. 2.
In vivo interaction of Eed and Enx1. (A) Coimmunoprecipitation (I.P.) of Eed with Enx1 from transiently transfected COS cell extracts. (Right) Total COS cell extracts transfected with (HA)3-ENX1, (Myc)3-Enx1+HA-Eed, or HA-Eed were subjected to Western blotting with 12CA5 (anti-HA) and 9E10 (anti-Myc) monoclonal antibodies. The position and relative expression of epitope-tagged proteins are indicated on the right. (Left) The same extracts were subjected to immunoprecipitation with a polyclonal rabbit serum against Enx1 (14). Precipitated proteins were detected by Western blotting with 12CA5 and 9E10. The Enx1 serum specifically coimmunoprecipitates Eed (middle lane) and does not cross-react with Eed (left lane, specificity control). The 12CA5 monoclonal antibody cross-reacts on Western blots of input total lysates with an abundant protein of 74 kDa in most cell lines; this band is marked with an asterisk. (B) Coimmunoprecipitation (I.P.) of Eed with Enx1 from stably transfected U2-OS cell extracts. (Left) Extracts from a stable cell line expressing both HA-Eed and (HA)3-ENX1 or a mock-transfected cell line were subjected to Western blot analysis with the 12CA5 monoclonal antiserum. The position of HA-ENX1 is indicated; HA-Eed comigrates with an cross-detected endogenously expressed protein band; correct expression was verified by immunofluorescence (see the text and Fig. 3). Cross-detected bands in total lysates by the 12CA5 monoclonal antibody are marked by asterisks on the left. (Right) The same extracts were subjected to immunoprecipitation with the anti-Enx1 antiserum. HA-Eed is efficiently coprecipitated from ENX1-expressing cells (right lane).
Since in earlier studies we demonstrated the presence of a human multiprotein complex containing several distinct Pc-G proteins in U2-OS cells (3, 4, 11), in this study we determined whether HA-Eed or HA-ENX1 immunopurifies with the previously detected complex. No evidence for coimmunoprecipitation was obtained when using antisera against the mouse posterior sex combs homologs Bmi1 and Mel18, the mouse polyhomeotic homolog Mph1/RAE28, and the mouse polycomb homologs M33 and MPc2, all of which reside in the previously detected complex (all mouse sera cross-detect their respective highly conserved human homologs [3, 4, 11]). Likewise, no evidence for interaction was obtained in direct yeast two-hybrid tests with the above-mentioned Pc-G genes and eed or ENX1 (data not shown).
Immunolocalization of HA-Eed and HA-ENX1 in U2-OS cells.
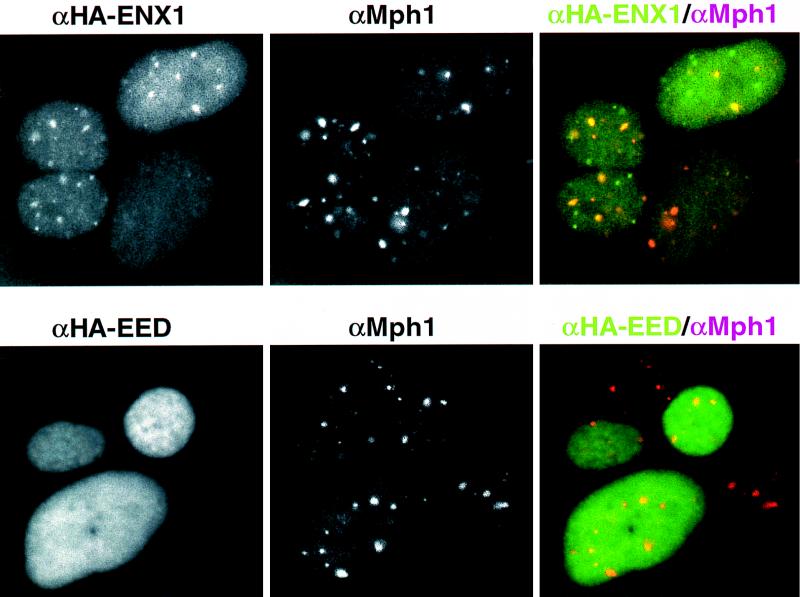
We previously reported the nonuniform distribution of Pc-G proteins in specific subnuclear domains in U2-OS interphase cells (3, 4). To determine the localization of Eed and ENX1 and to compare this pattern to the subnuclear staining of previously detected Pc-G complexes, stably transfected HA-Eed or HA-ENX1 U2-OS cell lines were fixed and costained with an affinity-purified rabbit anti-Mph1 antiserum and the 12CA5 monoclonal serum against the HA tag and observed by confocal laser-scanning microscopy. The results show that the HA-Eed protein is localized exclusively to the nucleus, in a uniform pattern, unlike the subnuclear pattern observed for MPH1 (Fig. 3, bottom). The specificity of the 12CA5 monoclonal antibody for the HA epitope is indicated by the absence of detectable signal in untransfected cells (Fig. 3, bottom; the untransfected cell in the middle and right panels displays only the red Mph1 signal). Interestingly, apart from a diffuse uniform staining like Eed, the HA-ENX1 protein also localizes in part to subnuclear domains (Fig. 3, top left panel). The superimposed image indicates that part of the ENX1 and MPH1 domains colocalize (Fig. 3, top right panel, overlap in yellow/orange). The subnuclear ENX1 staining is observed in approximately 70% of cells in an asynchronous population, whereas all cells also display a weak diffuse nuclear staining. These results indicate that Eed and ENX1 are nuclear proteins, and the ENX1 pattern suggests that perhaps a more transient interaction may occur between the previously detected Pc-G complexes and ENX1 in a subset of cells, which may be below the detection limit of our coimmunoprecipitation experiments.
FIG. 3.
Immunolocalization of HA-Eed, HA-ENX1, and the Pc-G complex protein Mph1 in U2-OS cells. (Top) U2-OS cells transfected with the (HA)3-ENX1 construct were fixed and incubated with both the 12CA5 monoclonal antibody and an affinity-purified rabbit anti-Mph1 serum. Detection was carried out with FITC-conjugated anti-mouse IgG and TRITC-conjugated anti-rabbit IgG, using confocal laser-scanning microscopy. The FITC signal (left) indicates a diffuse nuclear staining together with concentrations in subnuclear domains for ENX1. The TRITC signal (middle) detects endogenous expression of human MPH1 in subnuclear domains, characteristic of a previously described human Pc-G multiprotein complex (3). The merged image (right) shows a partial colocalization of ENX1 and MPH1 domains (ENX1 in green, MPH1 in red, overlap in yellow/orange). (Bottom) U2-OS cells transfected with the HA-Eed expression construct were fixed and stained with 12CA5 and anti-Mph1 sera as above. Eed is localized diffusely throughout the nucleus. Note the specificity of the 12CA5 serum for transfected cells: in two untransfected cells (merged images), only the TRITC signal is detected. Bar, 10 μm.
Developmental expression patterns of Enx1 and Enx2: overlap with Eed.
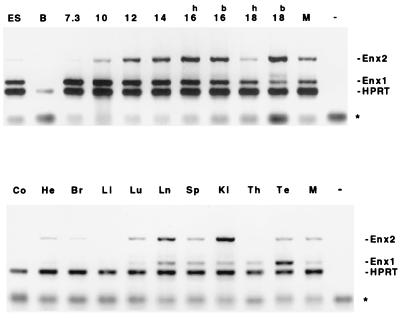
If the observed interactions between Enx1, Enx2, and Eed are relevant for their in vivo function, one would expect the expression patterns to coincide during development. Since the onset of Eed expression possibly occurs earlier than for most other Pc-G genes, we used a sensitive, semiquantitative RT-PCR assay to monitor the expression patterns of Enx1 and Enx2 during development and in adult mice. The same technique was previously used to determine the detailed expression pattern of Eed, allowing for a direct comparison (26). As shown in Fig. 4, whereas no expression was detected in blastocysts, Enx1 was already highly expressed in day 7.3 mouse embryos and became gradually more restricted to specific organs during development, with the highest expression being in the testis. In contrast, Enx2 expression was first detected on day 9 and continued to be expressed at moderate levels in most tissues (Fig. 4). These results are in good agreement with those of previous Northern blot and RNase protection experiments (13, 19), and they indicate that there is indeed extensive overlap in onset and tissue distribution between Enx1, Enx2, and Eed: the onset of Enx1 expression parallels that of Eed, whereas in adult mice Enx2 and Eed are expressed in most tissues (26) (Fig. 4).
FIG. 4.
Developmental expression patterns of Enx1 and Enx2. The contrast of the EtBr pictures was reversed, to facilitate visualization. The asterisk indicates migration of primers; the expected Enx1, Enx2, and HPRT fragments are indicated on the right. (Top) Semiquantitative RT-PCR detection of Enx1 and Enx2 expression during early mouse development. ES, embryonic stem cells; B, blastocysts; M, primary mouse embryo fibroblasts. Embryo RNAs from day 7.3 to day 18 postcoitum are indicated by numbers above the lanes. RNA from 16- and 18-day embryos was separately isolated from the head (h) and trunk (b). (Bottom) semiquantitative RT-PCR detection of Enx1 and Enx2 expression in organs of an adult mouse. Co, colon; He, heart; Br, brain; Li, liver; Lu, lung; Ln, lymph nodes; Sp, spleen; Ki, kidney; Th, thymus; Te, testis; M, mouse embryo fibroblasts. − denotes specificity control (no template). Similarly, no products were observed if RT was omitted from the cDNA reaction (data not shown).
DISCUSSION
Herein we describe the specific biochemical interaction of the essential Pc-G protein Eed, the mouse equivalent of Drosophila esc, with the E(z) homologs Enx1 and Enx2. These interactions are of special interest, since esc exerts a unique function in stably implementing Pc-G repression of homeotic genes that are initially repressed by gap gene products such as hunchback. Likewise, eed null-mutant mice reveal an essential, early requirement for eed during gastrulation, around which time mouse hox gene expression is initiated, a process of which little is known to date (26, 30). Both enx genes and eed encode the most highly conserved Pc-G members known to date. This conservation extends to the interaction domains that we identified, with the N-terminal Enx box 1 domain having 95% identity to its counterpart sequence on Drosophila E(z). This strongly suggests that Drosophila esc and E(z) will also interact through the same domains. A direct interaction between esc and E(z) correlates well with genetic interactions in Drosophila observed between Pc-G mutants: combinations of heterozygous mutations in the Pc-G genes Pc, Ph, Psc, Sce, Pcl, and Asx generally show synergistic enhancement of mutant phenotypes, suggesting the combined requirement of these genes for a common function (5, 17). Indeed, several of these gene products (and their respective homologs in mice) were subsequently found to interact in large Pc-G multiprotein complexes (3, 10). However, such strong genetic interactions were generally not observed between esc mutants and the above-mentioned Pc-G members, suggesting the presence of a separate function. Interestingly, one of the exceptions is the strong interaction between esc and E(z) mutants (5). These data raise the question whether separate Pc-G complexes with separate functions exist. Our coimmunoprecipitation experiments and direct two-hybrid screens did not reveal interactions between Eed or Enx1 and the above-mentioned Pc-G complex, consistent with the existence of separate complexes. However, a caveat in this interpretation is that some combinations of Pc-G proteins may not be compatible with two-hybrid detection, probably because of interference with the transactivation function of GAL4 (3; our unpublished observations). It is also possible, despite the mild conditions used, that direct biochemical interactions will be missed because of the limited sensitivity of coimmunoprecipitations or because of instability of larger protein complexes. Of interest in this regard are our observations that ENX1 partially colocalizes in distinct subnuclear domains with the previously described Pc-G complex, which is ubiquitously expressed in human U2-OS cells (Fig. 3). This could point to a more transient interaction, which may have escaped detection by coimmunoprecipitation experiments.
Whereas esc is specifically required during the transition between early repression and Pc-G-mediated repression, E(z) also appears to be required during subsequent development, much like other Pc-G proteins such as Polycomb. Interestingly, the E(z) protein colocalizes on polytene chromosomes to at least a subset of the Pc binding sites (6, 24), consistent with a possible bridging role for Enx/E(z). A central role for E(z) in Pc-G complex formation or maintenance is indicated by phenotypes observed in E(z) temperature-sensitive mutants: at the nonpermissive temperature, partial chromosome decondensation is observed, accompanied by loss of binding of Pc-G proteins to their specific polytene binding sites (6, 24). Ongoing screens for Enx-interacting proteins should help to further clarify this issue and may reveal as yet unknown interacting components of Pc-G repression complexes.
While this work was in progress, Denisenko and Bomsztyk described the isolation of Eed in a two-hybrid screen with the ribonucleoprotein K, which they suggest is a scaffold protein (9). Since the K protein has been found to bind to a wide range of different proteins, the relevance for Pc-G function is not immediately obvious, and this interaction awaits in vivo verification. Their data suggest that instead of the reported ATG (25), an upstream GTG is used to initiate Eed translation. The K binding domain was mapped to this upstream sequence (9). If K binding is relevant for Pc-G function, this is likely to be unique for mammalian Pc-G, since the leader sequence is not conserved in Drosophila esc (9, 12, 27). Perhaps most compellingly, a specific repression domain is localized to this N-terminal extension (9). Of note in this regard is that a slight extension of the Eed bait with 10 aa before the ATG revealed three extra interacting proteins, besides Enx1, in our first two-hybrid screen (see Results). The significance of these interacting proteins is currently under investigation. Notwithstanding these limitations, our results clearly identified an important interaction domain on the well-conserved mouse Pc-G proteins, Enx1, Enx2, and Eed. The necessity for intact WD40 repeats on Eed may not be surprising, given the importance of these domains for tertiary protein structure in forming a β-propeller core (28, 33). Besides this, the N-terminal 18 aa is also implicated in Enx interaction, stressing the importance of the Eed N terminus for protein-protein interactions (Fig. 1B). On the Enx proteins, the Eed interaction domain also maps to the N-terminal region, encompassing the highly conserved box 1 sequence (Fig. 1A). This domain is distinct from and adjacent to the previously reported Vav binding domain. Interestingly, a previously characterized Eed hypomorphic mutation results in a protein that binds significantly less avidly to Enx1 in yeast, as measured by a quantifiable liquid β-gal assay (Table 1). This suggests that the strength of the observed interaction between Eed and Enx1 parallels the in vivo functioning of Eed in regulation of development (26) (Table 1).
Here, we provide the first evidence for direct biochemical interactions of the Pc-G proteins Enx1 and Enx2 with Eed, a special Pc-G member required during early mouse development. These results, together with the partial colocalization of Enx1 with other Pc-G proteins such as MPH1, are consistent with a central role for Eed/Enx in Pc-G protein complex assembly and, as such, fit at least half of the proposed bridging model. The two-hybrid screen performed with the day 7.5 cDNA embryo library was not saturating, which could be one explanation for the lack of detection of other Eed-interacting proteins, which may represent factors required at the initiation of hox gene expression. Another possibility is that the above-mentioned N-terminal extension initiating at the GTG codon is required for such interactions, which will be addressed in future experiments.
ACKNOWLEDGMENTS
We thank O. Hobert and A. Ullrich for generously sharing the Enx1 antiserum and the ΔENX1 two-hybrid construct, T. Jenuwein for providing the epitope-tagged ENX1/EZH2 cDNA constructs, and J. Jacobs for developing the RT-PCR protocol.
This work was supported in part by the Life Sciences Foundation (SLW) grant, which is subsidized by the Netherlands Organization for Scientific Research (NWO), to J.W.V., and by an NIH grant (HD24462) to T.M.
REFERENCES
- 1.Abel K J, Brody L C, Valdes J M, Erdos M R, McKinley D R, Castilla L H, Merajaver S D, Cough F J, Friedman L S, Ostermeyer E A, Lynch E D, King M C, Welch P L, Osborne-Lawrence S, Spillman M, Bowcock A M, Collins F S, Weber B L. Characterization of EZH1, a human homolog of Drosophila Enhancer of Zeste near BRCA1. Genomics. 1996;37:161–171. doi: 10.1006/geno.1996.0537. [DOI] [PubMed] [Google Scholar]
- 2.Akasaka T, Kanno M, Balling R, Miesa M A, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 3.Alkema M J, Bronk M, Verhoeven E, Otte A P, van’t Veer L J, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 4.Alkema M J, Jacobs J, Voncken J W, Jenkins N A, Copeland N G, Satijn D P E, Otte A P, Berns A, van Lohuizen M. MPc2, a new murine homolog of the Drosophila Polycomb protein is a member of the mouse Polycomb transcriptional repressor complex. J Mol Biol. 1997;273:993–1003. doi: 10.1006/jmbi.1997.1372. [DOI] [PubMed] [Google Scholar]
- 5.Campbell R B, Sinclair D A, Couling M, Brock H W. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol Gen Genet. 1995;246:291–300. doi: 10.1007/BF00288601. [DOI] [PubMed] [Google Scholar]
- 6.Carrington E A, Jones R S. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–4083. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- 7.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coré N, Bel S, Gaunt S J, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 9.Denisenko O N, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol Cell Biol. 1997;17:4707–4717. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke A, DeCamillis M, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunster M J, Satijn D P E, Hamer K M, den Blaauwen J L, de Bruijn D, Alkema M J, van Lohuizen M, van Driel R, Otte A P. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and the human homologs of Polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutjahr T, Frei E, Spicer C, Baumgartner S, White R A, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 1995;4:4296–4306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobert O, Sures I, Ciossek T, Fuchs M, Ullrich A. Isolation and developmental expression analysis of Enx-1, a novel mouse Polycomb group gene. Mech Dev. 1996;55:171–184. doi: 10.1016/0925-4773(96)00499-6. [DOI] [PubMed] [Google Scholar]
- 14.Hobert O, Jallal B, Ullrich A. Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol. 1996;16:3066–3073. doi: 10.1128/mcb.16.6.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones R S, Gelbart W M. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones R S, Gelbart W M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- 18.Kennison J A. Transcriptional activation of Drosophila homeotic genes from distant regulator elements. Trends Genet. 1993;9:75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- 19.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the polycomb-group gene Enhancer of Zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller J, Gaunt S, Lawrence P A. Function of the Polycomb protein is conserved in mice and flies. Development. 1995;121:2847–2852. doi: 10.1242/dev.121.9.2847. [DOI] [PubMed] [Google Scholar]
- 22.Paro R. Mechanisms of heritable gene repression during development of Drosophila. Curr Opin Cell Biol. 1993;5:999–1005. doi: 10.1016/0955-0674(93)90084-4. [DOI] [PubMed] [Google Scholar]
- 23.Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- 24.Rastelli L, Chan C S, Pirotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 26.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 27.Simon J, Bornemann D, Lunde K, Schwartz C. The extra sex combs product contains WD40 repeats and its time of action implies a role distinct from other Polycomb group products. Mech Dev. 1995;53:197–208. doi: 10.1016/0925-4773(95)00434-3. [DOI] [PubMed] [Google Scholar]
- 28.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Crystal structure of a Ga protein βγ dimer at 2.1Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 29.Struhl G. A gene required for the correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- 30.Struhl G, Brower D. Early role of the esc+ gene product in the determination of segments in Drosophila. Cell. 1982;31:285–292. doi: 10.1016/0092-8674(82)90428-7. [DOI] [PubMed] [Google Scholar]
- 31.van der Lugt N M T, Domen J, Linders K, van Roon M, Robanus Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, Berns A. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the Bmi-1 proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 32.van Lohuizen M. Functional analysis of mouse Polycomb-group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 34.White R A H, Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986;47:311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- 35.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]