Abstract
Objectives
Management of colorectal cancer warrants mutational analysis of KRAS/NRAS when considering anti–epidermal growth factor receptor therapy and BRAF testing for prognostic stratification. In this multicenter study, we compared a fully integrated, cartridge-based system to standard-of-care assays used by participating laboratories.
Methods
Twenty laboratories enrolled 874 colorectal cancer cases between November 2017 and December 2018. Testing was performed on the Idylla automated system (Biocartis) using the KRAS and NRAS-BRAF cartridges (research use only) and results compared with in-house standard-of-care testing methods.
Results
There were sufficient data on 780 cases to measure turnaround time compared with standard assays. In-house polymerase chain reaction (PCR) had an average testing turnaround time of 5.6 days, send-out PCR of 22.5 days, in-house Sanger sequencing of 14.7 days, send-out Sanger of 17.8 days, in-house next-generation sequencing (NGS) of 12.5 days, and send-out NGS of 20.0 days. Standard testing had an average turnaround time of 11 days. Idylla average time to results was 4.9 days with a range of 0.4 to 13.5 days.
Conclusions
The described cartridge-based system offers rapid and reliable testing of clinically actionable mutation in colorectal cancer specimens directly from formalin-fixed, paraffin-embedded tissue sections. Its simplicity and ease of use compared with other molecular techniques make it suitable for routine clinical laboratory testing.
Keywords: Colorectal cancer, Molecular pathology, Precision medicine, KRAS, NRAS, BRAF
Key Points.
The Idylla has a rapid and accurate turnaround time.
Actionable mutations in KRAS, NRAS, and BRAF can be detected by this cartridge-based system.
Ease of use makes this very suitable for laboratories that may not have much molecular expertise.
In 2009, KRAS exon 2 testing was established in both European and US clinical practice guidelines as a predictive marker of response to anti–epidermal growth factor receptor (EGFR) therapy in patients with metastatic colorectal cancer. Evidence from the PRIME, CRYSTAL, OPUS, and other studies of EGFR monoclonal antibody therapies has shown that mutations in KRAS exons 3 and 4 and NRAS exons 2, 3, or 4 are also predictive of a lack of response to the anti-EGFR antibody therapies cetuximab (Erbitux; Lilly) and panitumumab (Vectibix; Amgen).1-4 In 2015, the American Society of Clinical Oncology (ASCO) provided a provisional clinical update to include recommendations for expanded RAS (KRAS and NRAS) mutation testing in all patients with mCRC at diagnosis of stage IV disease when considering use of EGFR-targeted therapies, and a guideline statement was provided by the American Society for Clinical Pathology (ASCP), College of American Pathologists (CAP), Association for Molecular Pathology (AMP), and ASCO in 2017.5,6
Further analyses of the CRYSTAL study data and the COIN trial data showed that BRAF mutation at codon 600 is an indicator of poor prognosis for patients with mCRC as it is associated with shorter progression-free survival and overall survival, regardless of treatment.7,8 With the evidence produced by these study data, the 2017 ASCP/CAP/AMP/ASCO guidelines for colorectal cancer were updated to also include recommendations for BRAF V600 mutational analysis in all patients with mCRC at the time of diagnosis of stage IV disease for prognostic assessment.6
Determination of biomarker mutational status in patients with mCRC has been facilitated by the implementation of commercial and laboratory-developed assays employing polymerase chain reaction (PCR), Sanger sequencing, or next-generation sequencing (NGS) technologies.9-11 While beneficial in being able to provide a patient’s biomarker status, these assays and technologies have limitations, including physical separation of laboratory space for sample preparation and analyses, the cost and infrastructure needed to implement these technologies, and/or the requirement of highly experienced molecular laboratory personnel to run these assays.
Several manufacturers attempted to develop NGS platforms more suitable for small sample batch size runs and reagent kits that target small numbers of cancer-related genes, some of which are clinically actionable and others that have a potential to become clinically actionable. In addition, vendors have developed informatics pipelines to annotate and curate NGS data. While some claims for a 5-day turnaround time have been made, most laboratories return results within 7 to 14 days. This window of time is clinically relevant as it also depicted the time interval from diagnosis to first visits with oncologists to determine treatment strategies. As the field progresses, increasing numbers of tumors are being sent for genomic profiling, new therapies continue to be introduced, and claims for these and older therapies to be used in other tumor types and at earlier intervals are being approved. These advances have challenged the workflow and turnaround times set by most laboratories and have prompted laboratories to look for mechanisms to streamline operations and to meet growing clinical needs.
In 2017, Biocartis introduced the Idylla system to the US market. This platform is a simple, cartridge-based system capable of qualitative detection of a select number of somatic variants in clinically actionable cancer-related genes.12 The test system uses a single formalin-fixed, paraffin-embedded (FFPE) tissue section placed directly into a test cartridge, which is then inserted into the instrument with biomarker results returned within 2 hours.
Here we describe the RAS/BRAF multicenter biomarker study that was initiated by Biocartis and sponsored by Amgen in June 2017. The primary objectives of the study were to assess the time to result between the participating institutions’ standard-of-care (SOC) testing and the Biocartis Idylla system for KRAS, NRAS, and BRAF variant testing in colorectal cancer (CRC) and to measure the gene variant concordance rates between the SOC technologies and the Idylla system.
Materials and Methods
Study Design
This multicenter study included participation of 20 sites throughout the United States and Puerto Rico with an enrollment of 874 CRC samples. This was an observational study, and none of the Idylla results were reported for diagnostic or treatment decisions. A Clinical Research Organization partner (Vital Systems) was enlisted to facilitate Western Institutional review board/institutional review board (IRB) approvals at the sites and to manage the data collected during the study. Idylla systems were installed at each site, and laboratory personnel assigned to the study were trained on the platform and completion of the case report forms.
Study Sites
Site enrollment was initiated in November 2017 and ended in December 2018. Twenty sites, including 11 (55%) medium- and large-sized hospitals, three (15%) specialty cancer centers, two (10%) small hospitals, and four (20%) pathology or oncology practices and reference laboratories were recruited and agreed to participate in this study. The sites used a diversity of technologies for SOC somatic variant testing—six (30%) of the sites used in-house NGS, nine (45%) sent out their NGS testing, and five (25%) used both in-house and send-out testing with a combination of PCR, NGS, and Sanger sequencing technologies. In this study, 75.1% of the SOC samples were analyzed by NGS, 15.4% by PCR, 3.8% by Sanger sequencing, and 5.6% by a combination of these or other technologies.
Seventy percent (14/20) of the sites routinely tested for KRAS, NRAS, and BRAF in their CRC cases while six of the sites tested for one or more of these genes separately depending on the ordering clinician. For this study, all sites tested for variants in all three genes using the Idylla system with the KRAS cartridge and the combined NRAS/BRAF gene cartridge. The Idylla system is a cartridge-based assay where the user inserts a single section of FFPE tissue sandwiched between two small pieces of filter paper section into the cartridge. The cartridge is then inserted into the Idylla instrument with results being available within 2 hours. All steps in these real-time PCR-based assays are performed within the single-use, disposable cartridge. One cartridge is used specifically for KRAS mutation detection, and a second cartridge is specific for BRAF/NRAS mutations.
Samples
All patients with CRC being tested for KRAS, NRAS, and/or BRAF somatic variants as per institutional SOC practices were eligible for this study. For the study, samples were selected with a minimum neoplastic tumor content of 10% to maintain parity between SOC technologies and Idylla. Sample collection began in January 2018 and ended in March 2019. A data collection sheet accompanied each sample collected and included demographic information as well as diagnosis and time to test results for SOC testing and Idylla testing. Workflow diagrams were created for each site to document the sample journey for both SOC and Idylla testing and to document the average and range of days required for receipt of results by the oncologist.
Results
Sample Tracking
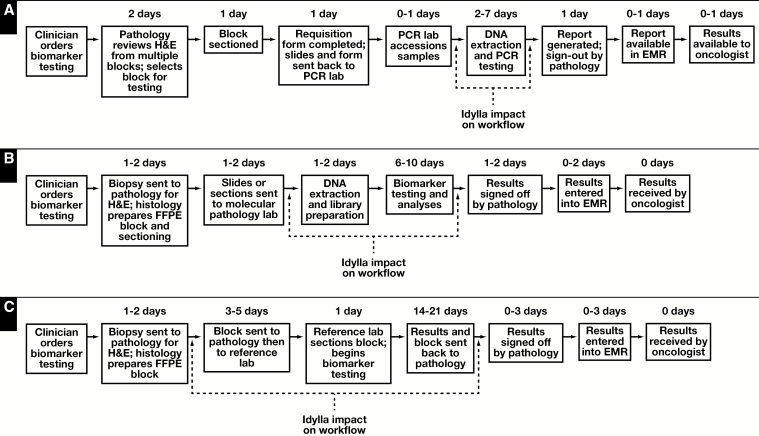
Representative study site workflow diagrams for in-house biomarker testing by PCR and NGS as well as send-out biomarker testing by NGS were developed Figure 1. SOC results from in-house biomarker testing were typically available within 7 to 14 days after physician ordering while send-out NGS test results were available within 20 to 30 days after initial ordering.
Figure 1.
A, Example of an in-house polymerase chain reaction–based workflow for RAS biomarker testing. Times for the various assay steps are averages or ranges. The Idylla impact on workflow is shown in brackets. B, Example of an in-house next-generation sequencing (NGS)–based workflow for RAS biomarker testing. Times for the various assay steps are averages or ranges. The Idylla impact on workflow is shown in brackets. C, Example of a send-out NGS-based workflow for RAS biomarker testing. Times for the various assay steps are averages or ranges. The Idylla impact on workflow is shown in brackets. EMR, electronic medical record; FFPE, formalin fixed, paraffin embedded.
Samples
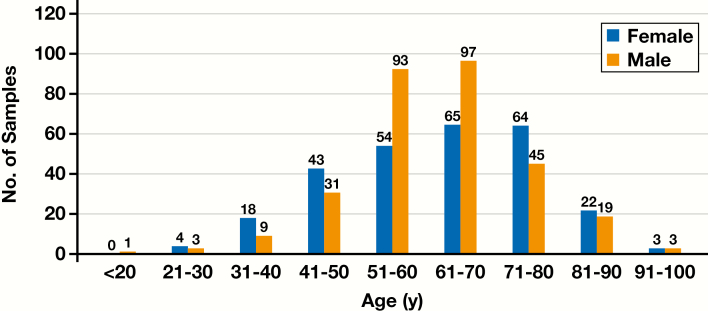
A total of 874 CRC samples were enrolled into the study. For the 574 samples in which biomarker test order date, age, and sex were reported, 273 (47.6%) were from women and 301 (52.4%) from men. Ages for 22 (8.1%) of the women and 13 (4.3%) of the men were younger than 40 years, 226 (82.8%) of the women and 266 (88.4%) of the men were between 41 and 80 years, and 25 (9.1%) of the women and 22 (7.3%) of the men were older than 81 years Figure 2. Ethnicity and race were not uniformly reported due to differences in the individual institutions’ IRB requirements.
Figure 2.
Age vs sex demographics for the 574 samples in which biomarker test order date, age, and sex were reported.
Information collected on tumor stage by the sites was optional and available for 356 of 874 samples. Tumor stage included 32 (9.0%) cases classified as stage I, 79 (22.2%) cases classified as stage II, 82 (23.0%) cases classified as stage III, and 163 (45.8%) classified as stage IV. Sample enrollment was open to any CRC sample that was eligible for biomarker testing as determined by the ordering physician (including stages I and II). Although 31.2% of the samples had biomarker testing performed at stages I and II, this may not be indicative of reflex testing, as tumor staging data were available on less than half of the samples. However, the data are consistent with genetic testing mostly being performed in later stage advanced disease.
Furthermore, 745 (85.2%) of the 874 SOC samples were tested for all three biomarkers while 129 (14.8%) of the SOC samples only had partial biomarker testing performed. Interestingly, despite National Comprehensive Cancer Network (NCCN) guideline recommendations for testing CRC cases for variants in KRAS, BRAF, and NRAS, the practice is not uniform among institutions with respect to which cases and genes are tested.
Turnaround Times
For 18 of the 20 participating sites, on the case report forms, there were sufficient data on 780 samples to measure the time to result on the SOC and matching Idylla samples tested (the two sites without data were oncology clinics where it was difficult to obtain accurate information on sample shipped/testing start times).
Workflow turnaround times were calculated for the data set using the following time points: when the gene variant testing was ordered by the clinician, when the sample was shipped or if there was no shipment when the biomarker testing started, the results received by the pathologist, and the results received by the oncologist. For the Idylla turnaround times, it was assumed that preanalytic ordering, block preparation, sectioning, and sample review times remained the same as SOC workflow. Since no Idylla results were reported to the oncologist, postanalytic reporting timelines were also assumed to be the same as the SOC workflow.
Turnaround times for the different SOC technologies were also calculated for the 780-sample data set Table 1. In-house PCR-based assays were reported to have an average testing turnaround time of 5.6 days, send-out PCR of 22.5 days, in-house Sanger sequencing of 14.7 days, send-out Sanger sequencing of 17.8 days, in-house NGS of 12.5 days, and send-out NGS of 20.0 days. A combination of in-house and send-out testing or other methodologies had an average turnaround time of 11 days.
Table 1.
Average Turnaround Time for In-House and Send-Out SOC RAS Biomarker Testing by Technologya
| Technology | Average TAT Days | No. (%) of Samples |
|---|---|---|
| In-house PCR | 5.6 | 76 (9.7) |
| Send-out PCR | 22.5 | 42 (5.4) |
| In-house Sanger sequencing | 14.7 | 3 (0.4) |
| Send-out Sanger sequencing | 17.8 | 28 (3.6) |
| In-house NGS | 12.5 | 350 (44.9) |
| Send-out NGS | 20.0 | 268 (34.4) |
| Other (combinations: technologies, in-house/send-out) | 11.0 | 13 (1.7) |
| Total | 780 (100) |
PCR, polymerase chain reaction; NGS, next-generation sequencing; SOC, standard of care; TAT, turnaround time.
aThe 780-sample set from the testing timeline data was used for the analysis.
It should be noted that there were 14 invalid/quantity not sufficient (QNS) samples by SOC and 10 QNS samples for Idylla that were included in this data set. Timeline measurements for biomarker testing began when a sample was shipped or testing initiated regardless of technology and ended when the results were received by the clinicians. The QNS sample timelines were therefore both measurable and applicable to reporting a nonresult to the clinician.
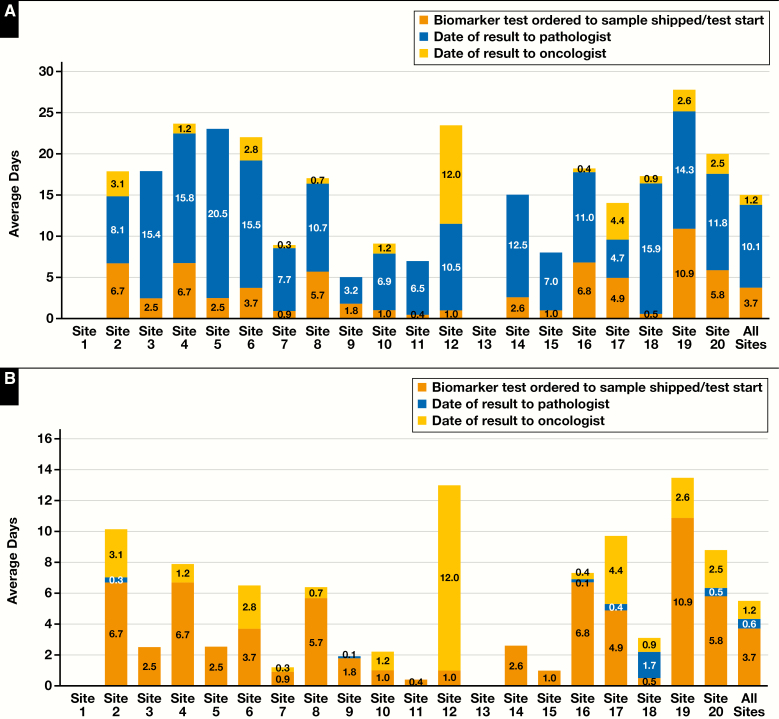
The expected benefit of the Idylla system is the ability to generate targeted yet clinically actionable somatic variant results very quickly compared with most other molecular-based methods that require DNA extraction prior to any type of downstream testing being performed. On average across all sites, the SOC time to result was 15.0 days—3.7 days from testing ordered to sample shipped/testing started, 10.1 days from sample shipped/testing started to result to pathologist, and 1.2 days from result to pathologist to result to oncologist Figure 3A. This is in stark contrast to the average Idylla time to result of 4.9 days, with a range of 0.4 to 13.5 days. Idylla results were, on average, available 10.1 days sooner with a range of 3.3 to 20.5 days of time saved depending on the SOC technology Figure 3B.
Figure 3.
A, Standard-of-care (SOC) RAS biomarker turnaround times for each study site and average timelines for sites that submitted data (n = 780). Timeline data were available for 18 of the 20 sites. Turnaround times were measured from biomarker testing ordered to sample shipped or testing started (BTO to SSTS), sample shipped or testing started to date of result to pathologist (SSTS to RTP), and date of result to pathologist to date of result to oncologist (RTP to RTO). B, Idylla RAS biomarker turnaround times for each study site and average timelines for sites that submitted data (n = 780). Timeline data were available for 18 of the 20 sites. Turnaround times for Idylla were created using a combination of SOC preanalytical and postanalytical testing timelines in conjunction with Idylla sample testing timelines. Preanalytical timelines were assumed to be the same as SOC for biomarker testing ordered to sample shipped or testing started (BTO to SSTS), and postanalytical timelines were assumed to be the same for SOC date of result to pathologist to date of result to oncologist (RTP to RTO). The sample shipped or testing started date to date of result to pathologist (SSTS to RTP) substituted the SOC actual testing times with the Idylla testing time on the corresponding sample.
Concordance
Of the 874 CRC samples analyzed for concordance, there were 14 invalid/QNS samples by SOC (1.6%) and 10 for Idylla (1.1%). These samples were removed from concordance analyses, resulting in an overall study sample size of 850. Biomarker concordance rates between the different SOC technologies and the Idylla system were analyzed on an individual mutation basis and on a clinical treatment basis. On an individual mutation basis, each sample was analyzed by SOC and Idylla to determine whether the sample was wild type or contained mutations for KRAS, NRAS, and/or BRAF. If a mutation or mutations were present in the sample, concordance was examined and recorded on a mutation variant level. Concordance on a mutation basis included variants that were not stated as being clinically relevant in the current NCCN CRC guidelines (eg, BRAF G469 or BRAF D594 mutations). On a clinical treatment basis, each sample was analyzed by SOC and Idylla to determine whether the sample biomarker status determined whether the patient would be eligible for targeted therapy. If the sample was RAS wild type or RAS wild type and BRAF positive, it was assumed that the patient would be eligible for targeted therapy such as Vectibix or Erbitux. If the sample contained a KRAS, NRAS, KRAS/BRAF, or NRAS/BRAF mutation, it was assumed that the patient would be eligible for nontargeted therapy.
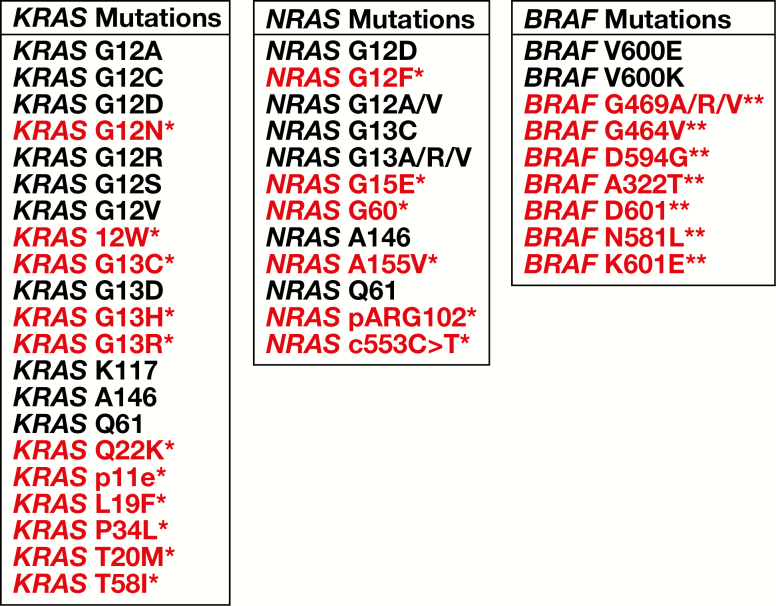
Accounting for all mutations including those not in the Idylla cartridge by design, on an individual mutation basis, the concordance across all sites was 88.6%, and on a clinical mutation treatment basis, the concordance across all sites was 94.3% Table 2. When the samples with mutations that were not in the Idylla cartridge or those samples discordant due to not being tested for one of the three genes as part of the SOC were removed from analysis, the individual mutation concordance increased to 93%, and the clinical concordance increased to 95.9% Table 3. All detected mutations in the study as well as mutations detected by other SOC technologies and not included in the Idylla cartridge by design are listed in Figure 4.
Table 2.
Average Concordance on an Individual Mutation Basis and the Average Clinical Concordance After Removing the Quantity Not Sufficient/Invalid Samplesa
| Characteristic | % |
|---|---|
| Average individual mutation agreement | 88.8 |
| Range overall agreement | 75-100 |
| Average clinical concordance | 94.3 |
| Range clinical concordance | 80-100 |
aRanges across all of the individual sites are also included.
Table 3.
Average Concordance on an Individual Mutation Basis and the Average Clinical Concordance After Removing the Quantity Not Sufficient/Invalid Samples, the Mutations Not in the Idylla Cartridge by Design, and Discrepant Samples Not Tested as Standard of Carea
| Characteristic | % |
|---|---|
| Average individual mutation agreement | 93 |
| Range overall agreement | 79-100 |
| Average clinical concordance | 95.9 |
| Range clinical concordance | 79-100 |
aRanges across all of the individual sites are also included.
Figure 4.
Mutations detected in the study and frequency of mutations expected in colorectal cancer. Mutations in red are not identified by Idylla. The highlighted RAS mutations (*) are rare variants with prevalence frequencies less than 1.0%, most of which are not identified in the COSMIC database. These RAS variants are not in the Idylla cartridge by design and only represent 1.4% of the concordance discrepancies. The highlighted BRAF mutations (**) are not in the current National Comprehensive Cancer Network guidelines as prognostic or predictive indicators of disease progression.
Individual Mutation Rates
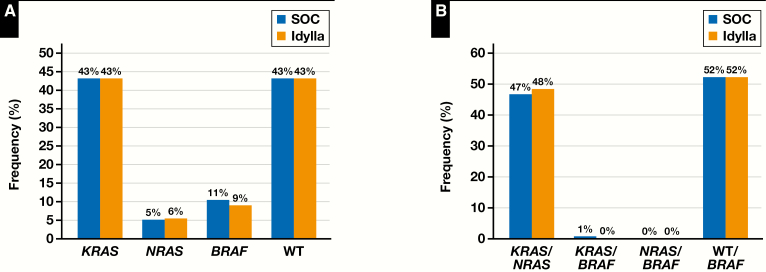
Data from COSMIC and My Cancer Genome websites suggest that the variant frequencies of the KRAS, NRAS, and BRAF genes in CRC are 36% to 40%, 1% to 6%, and 8% to 15%, respectively. Individual gene variant frequencies were very similar between routine SOC procedures and the Idylla cartridge-based assays, indicating that the targeted variants tested for in the cartridge were readily identified in CRC without the need for additional testing to detect other actionable mutations outside of those in the cartridge that may have been detected by SOCs. In the concordance data set (n = 850), both SOC and Idylla testing detected a 43% KRAS mutation rate; for NRAS, SOC and Idylla mutation detection rates were 5% and 6%, respectively; and for BRAF, SOC and Idylla detection rates were 11% and 9%, respectively Figure 5A. CRC cases that had no variants detected (classified as wild type) included 43% of samples tested in both SOC methods and the Idylla assays. On a clinical basis for this sample set, the KRAS/NRAS mutation frequency was 47% by SOC vs 48% by Idylla. The KRAS/BRAF mutation frequency by SOC was 1% vs 0% by Idylla, and RAS wild type/BRAF mutation frequency by both SOC and Idylla was 52% Figure 5B.
Figure 5.
A, Comparison of the frequency of detection of individual KRAS, NRAS, and BRAF mutations or wild-type (WT) results by standard of care (SOC) and Idylla (n = 850). B, Comparison of the frequency of detection of KRAS or NRAS mutations, KRAS or NRAS and BRAF mutations, and wild-type results or BRAF mutations by standard of care and Idylla (n = 850).
Invalids and Discordants
Analyzing all clinically relevant and nonclinically relevant mutations in the 850 concordance sample data set, there were 86 (10.1%) discordant samples compared with the validated SOC procedure Table 4. A discordant rate of 10.1% should not be considered excessive when comparing results across different detection technologies since there are variations in sensitivity (eg, NGS, PCR, and Sanger sequencing) and site determination of their SOC biomarker panels. Discordant samples were categorized as mutations not included in the Idylla cartridge by design, false positives, false negatives, sensitivity related where there were borderline results that were either close to assay cutoffs or not detectable due to technology limitations, variant cross-reactivity where genetic sequences were close together in the genome and identified different mutation variants but provided the same clinical result (eg, KRAS G12V and G12D), and unknown or undetermined reasons. For classification of false positives and false negatives, SOC NGS was considered the gold standard. However, for other SOC technologies to be considered as a gold standard in discrepancy adjudication, vendor information was reviewed to determine if the system could detect the biomarker in question. For example, other PCR systems could not detect the KRAS Q61H mutation as it is not in the vendor’s SOC biomarker panel by design and was therefore classified as a technology- or assay-related discordant. All discrepant results and data were reviewed by the sites, Idylla PCR curves were analyzed by Biocartis, and, whenever possible, the assay was rerun on Idylla and/or SOC if sufficient sample remained.
Table 4.
Number and Percentages of Discrepant Samples by Categorya
| Reasons for Discrepancy (All Mutations) | No. of Discrepant Individual Mutations (n = 91) | No. (%) of Discrepant Samples (n = 86) | % of Discrepant Samples in the Concordance Data Set (n = 850) |
|---|---|---|---|
| Mutations not in Idylla cartridge | 32 | 29 (33.7) | 3.4 |
| Biomarker not run as SOC or not in SOC panel | 14 | 14 (16.3) | 1.6 |
| Biomarkers run as SOC but discrepant; unknown | 9 | 9 (10.5) | 1.1 |
| Discrepant (false) positives | 15 | 15 (17.4) | 1.8 |
| Discrepant (false) negatives | 6 | 6 (7.0) | 0.7 |
| Sensitivity (SOC or Idylla technology) | 7 | 7 (8.1) | 0.8 |
| Possible variant cross-reactivity or identified in Idylla Explore software | 8 | 6 (7.0) | 0.7 |
| Total discrepant results | 10.1 |
SOC, standard of care.
aDiscordant samples were categorized as mutations not included in the Idylla cartridge by design, false positives, false negatives, sensitivity related where there were borderline results that were either close to assay cutoffs or not detectable due to technology limitations, variant cross-reactivity where genetic sequences were close together in the genome and identified different mutation variants but provided the same clinical result, and unknown or undetermined reasons. In the 850-sample data set, there were 91 instances of discordance within 86 samples. The additional five instances of discordance were due to double mutations detected in samples. For example, a double mutation could refer to both a KRAS Q22K mutation and a BRAF D594G mutation present in a sample and detected by SOC next-generation sequencing but not by Idylla.
Of the 86 discordant samples, 29 (33.7%) were due to mutations not included in the Idylla cartridge by design and did not have a significant impact on the clinical concordance. Fifteen (17.4%) were discrepant (false) positives, 14 (16.3%) were due to mutations identified by Idylla but not performed by the SOC method, nine (10.5%) instances of discordance were due to unknown reasons, six (9.3%) were due to possible variant cross-reactivity, seven (8.1%) were technology or assay sensitivity related, and six (7.0%) were discrepant (false) negatives. Also, within the 86 discordant sample set, there were five additional instances of discordance due to double mutations identified by NGS (eg, both KRAS Q22K and BRAF D594G mutations present in a sample and detected by NGS but not by Idylla).
Analyzing the data to determine the clinical impact of the discordant samples—that is, whether the biomarker results would render the appropriate information to assist with therapy selection—the number of discordant samples with clinical impact was reduced from 86 to 50 Table 5. With 50 clinically discordant samples in the 850-sample data set, the discordant rate was 5.7%. Of the 50 discordant samples, 12 (24.0%) were due to mutations not included in the Idylla cartridge by design, 13 (26.0%) were due to mutations identified by Idylla but not performed by the SOC method, eight (16.0%) were due to unknown reasons, seven (14.0%) were technology or assay sensitivity related, six (12.2%) were discrepant (false) negatives, and four (8.0%) were discrepant (false) positives.
Table 5.
Analysis of the Discrepant Samples With Clinical Impacta
| Reasons for Discrepancy (Treatment/Prognosis Impacted) | No. of Discrepant Individual Mutations (n = 54) | No. (%) of Discrepant Samples (n = 50) | % of Discrepant Samples in the Concordance Data Set (n = 850) |
|---|---|---|---|
| Mutations not in Idylla cartridge | 50 | 12 (24.0) | 1.4 |
| Biomarker not run as SOC or not in SOC panel | 13 | 13 (26.0) | 1.5 |
| Biomarkers run as SOC but discrepant; unknown | 8 | 8 (16.0) | 0.9 |
| Discrepant (false) positives | 4 | 4 (8.0) | 0.47 |
| Discrepant (false) negatives | 6 | 6 (12.0) | 0.7 |
| Sensitivity (SOC or Idylla technology) | 7 | 7 (14.0) | 0.82 |
| Possible variant cross-reactivity or identified in Idylla Explore software | 1 | 0 (0) | 0.0 |
| Total discrepant results | 5.79 |
SOC, standard of care.
aDiscordant samples were categorized as mutations not included in the Idylla cartridge by design, false positives, false negatives, sensitivity related where there were borderline results that were either close to assay cutoffs or not detectable due to technology limitations, variant cross-reactivity where genetic sequences were close together in the genome and identified different mutation variants but provided the same clinical result, and unknown or undetermined reasons. In determining whether the discordant biomarker results would render the appropriate information to assist with therapy selection, the number of discordant samples with clinical impact was reduced from 86 to 50.
Discussion
Genomic profiling of tumors has become increasingly important in precision medicine. However, the process of identifying the genetic variants can require significant technical expertise, as well as be costly and labor-intensive. Turnaround time remains a major issue. Complex workflows, especially for NGS assays, can take several days, and samples have to be batched to be cost-effective. Because of its complexity, smaller centers generally outsource genotyping of tumors to external molecular pathology laboratories, which causes additional delays.
In this multicenter study, we investigated the utility of the fully automated Biocartis Idylla platform for biomarker testing in patients with CRC in a variety of clinical laboratory settings and its impact on the time to result compared with SOC methods. The study achieved the primary objectives of assessing the time to result between an institution’s SOC and the Idylla system for genetic biomarker testing in patients with CRC and concordance of molecular results produced. Turnaround time, regardless of how it was measured, was shortened significantly when KRAS, BRAF, and NRAS testing was performed with the Idylla system. On average, Idylla was able to provide biomarker results 10 days earlier than the current SOC technologies. The ability of the Idylla system to perform testing directly on a single FFPE tissue section provided as a curl or unstained slide eliminates the need for separate DNA extraction for traditional PCR or sequencing-based assays. In addition to the fully automated nature of the analysis, the reduced number of preanalytical steps enables significant time savings with minimal labor or hands-on time. The shortened turnaround time not only provides results faster but also improves workflow in the laboratory by completing case testing in a more timely fashion.
By measuring the process time from the placement of the order for biomarker test to results received, the study also identified areas for potential improvements in time savings, such as the time interval from biomarker test order to testing or shipment to an external reference laboratory (range, 0.4-11.1 days) and the time of result from the pathologist to reach to the oncologist (range, 0-12 days).
A second objective of the study was to demonstrate that the Idylla platform in comparison with different SOC technologies provided accurate, comprehensive analysis of actionable mutations in CRC samples. The data show that the Idylla system is highly comparable with current SOC testing methods achieving an average clinical concordance of 95.9%.
One may argue that the Idylla cartridge-based assays have a technical limitation, as only select variants can be detected with the different cartridges. Clinically, however, testing for only those variants deemed truly clinically actionable in accordance with the current NCCN colon cancer guidelines is a major advantage for test result interpretation.13 Thus, it is rare to receive a result that is questionable or of unknown significance with respect to response to a particular therapeutic. As laboratories continue to adopt NGS, rare and potentially actionable variants will continue to be detected more frequently and a system such as Idylla will need to modify the variant library within the cartridges.
One other important advantage of the Idylla system is that it was able to successfully analyze the RAS and BRAF mutational status using only a minimal sample input of one 5-μmol/L section of FFPE tissue per cartridge, resulting in an invalid rate of only 1.1%. As experienced throughout the study, samples for NGS SOC biomarker testing typically required about 10 sections of FFPE tissue for analysis. The Idylla platform offers significant tissue stewardship as an added advantage when specimen quantity is limited. One of the study sites also included several 1-mm CRC punch biopsy samples, indicating that even smaller tissue-based specimens can be successfully analyzed by Idylla.14 This is in line with findings of other studies showing successful mutational analysis for RAS, BRAF, and EGFR with Idylla using very low tissue input, including samples that were deemed unsuitable for NGS evaluation.15-21
For laboratories not performing NGS and for those not performing any molecular testing or are sending out molecular testing to external laboratories, rapid testing exemplified by the Idylla system helps to democratize testing capabilities and significantly improves turnaround times. As the majority of care given to patients with cancer in the United States is rendered in smaller hospital settings, this platform allows those institutions to perform the most clinically actionable testing at their facility with this user-friendly system and minimal hands-on time for testing and rapid result reporting. Decentralized genetic biomarker testing can significantly shorten the time from initial clinical diagnosis to the administration of an appropriate precision medicine therapy.
In laboratories that routinely perform more comprehensive molecular testing for these genes as part of larger NGS panels, implementation of the Idylla system for KRAS, BRAF, and NRAS testing in colorectal cancers may still be advantageous. Several studies have shown the need and value of implementing parallel or sequential testing using a rapid biomarker assay next to NGS for several cancer types.16,17,21-24 Given the cost, labor, and state of reimbursement for NGS panels, screening for more common mutations using the Idylla platform may reduce the amount of NGS testing needed for certain tumor types and may help improve biomarker reimbursement rates while greatly decreasing the turnaround time for reporting actionable results.
This multicenter study demonstrated that the Idylla system significantly improves genetic biomarker testing turnaround times. The Idylla system requires minimal sample input and is robust with respect to the accurate identification of clinically actionable variants in the KRAS, BRAF, and NRAS genes. Its performance is reproducible in a variety of testing institutions with various levels of experience in genomic testing. While these assays are less expensive to perform than standard methods due to the labor savings and extent of multiplexing, we do not currently have good reimbursement data, although there has been some suggestion that single-gene Current Procedural Terminology codes for these genes are more reimbursable than other forms of testing.
Contributor Information
Gregory J Tsongalis, Clinical Genomics and Advanced Technology (CGAT) Laboratory, Department of Pathology and Laboratory Medicine, Dartmouth Hitchcock Health System, Lebanon, NH; Geisel School of Medicine at Dartmouth, Hanover, NH.
M Rabie Al Turkmani, Clinical Genomics and Advanced Technology (CGAT) Laboratory, Department of Pathology and Laboratory Medicine, Dartmouth Hitchcock Health System, Lebanon, NH; Geisel School of Medicine at Dartmouth, Hanover, NH.
Michael Suriawinata, Clinical Genomics and Advanced Technology (CGAT) Laboratory, Department of Pathology and Laboratory Medicine, Dartmouth Hitchcock Health System, Lebanon, NH; Geisel School of Medicine at Dartmouth, Hanover, NH.
Michael J Babcock, Bioinformatics & Molecular Pathology, Dahl-Chase Diagnostic Services & Pathology Associates, Bangor, ME.
Kristi Mitchell, Bioinformatics & Molecular Pathology, Dahl-Chase Diagnostic Services & Pathology Associates, Bangor, ME.
Yi Ding, Diagnostic Medicine Institute, Geisinger Medical Center, Danville, PA.
Lisa Scicchitano, Diagnostic Medicine Institute, Geisinger Medical Center, Danville, PA.
Adrian Tira, Department of Pathology, Rush University Medical Center, Chicago, IL.
Lela Buckingham, Department of Pathology, Rush University Medical Center, Chicago, IL.
Sara Atkinson, Department of Cytology, Cone Health Moses Cone Hospital, Greensboro, NC.
Amy Lax, Department of Cytology, Cone Health Moses Cone Hospital, Greensboro, NC.
Dara L Aisner, Colorado Molecular Correlates Laboratory (CMOCO), Department of Pathology, University of Colorado Anschutz Medical Campus, Aurora.
Kurtis D Davies, Colorado Molecular Correlates Laboratory (CMOCO), Department of Pathology, University of Colorado Anschutz Medical Campus, Aurora.
Holly N Wood, Colorado Molecular Correlates Laboratory (CMOCO), Department of Pathology, University of Colorado Anschutz Medical Campus, Aurora.
Stacey S O’Neill, Department of Pathology, Wake Forest School of Medicine, Winston-Salem, NC.
Edward A Levine, Division of Surgical Oncology, Department of Surgery, Wake Forest School of Medicine, Winston-Salem, NC.
Judy Sequeira, Department of Pathology and Laboratory Medicine, Comprehensive Care and Research Center, Cancer Treatment Centers of America Atlanta, Newnan, GA.
Shuko Harada, Molecular Diagnostics Laboratory, Department of Pathology, University of Alabama Birmingham School of Medicine, Birmingham.
Gina DeFrank, Molecular Diagnostics Laboratory, Department of Pathology, University of Alabama Birmingham School of Medicine, Birmingham.
Ravikumar Paluri, Department of Medicine, Division of Hematology/Oncology, University of Alabama Birmingham School of Medicine, Birmingham.
Bradford A Tan, Department of Pathology and Laboratory Medicine, Comprehensive Care and Research Center, Cancer Treatment Centers of America Chicago, Zion, IL.
Heather Colabella, Department of Pathology and Laboratory Medicine, Comprehensive Care and Research Center, Cancer Treatment Centers of America Chicago, Zion, IL.
Christopher Snead, CHRISTUS Cancer Treatment Center, Shreveport, LA.
Marcia Cruz-Correa, Pan American Center for Oncology Trials, Oncologic Hospital, Puerto Rico Medical Center, Rio Piedras, Puerto Rico.
Virginia Ramirez, Pan American Center for Oncology Trials, Oncologic Hospital, Puerto Rico Medical Center, Rio Piedras, Puerto Rico.
Arnaldo Rojas, Pan American Center for Oncology Trials, Oncologic Hospital, Puerto Rico Medical Center, Rio Piedras, Puerto Rico.
Huiya Huang, Department of Pathology, Medical College of Wisconsin, Milwaukee.
Alexander C Mackinnon, Department of Pathology, Medical College of Wisconsin, Milwaukee.
Fernando U Garcia, Department of Pathology and Laboratory Medicine, Comprehensive Care and Research Center, Cancer Treatment Centers of America Philadelphia, Philadelphia, PA.
Sharon M Cavone, Department of Pathology and Laboratory Medicine, Comprehensive Care and Research Center, Cancer Treatment Centers of America Philadelphia, Philadelphia, PA.
Mutasim Elfahal, Department of Pathology and Laboratory Medicine, Lahey Hospital and Medical Center, Beth Israel Lahey Health, Burlington, MA.
Gyorgy Abel, Department of Pathology and Laboratory Medicine, Lahey Hospital and Medical Center, Beth Israel Lahey Health, Burlington, MA.
Mohammad A Vasef, Department of Pathology, University of New Mexico Health Sciences Center, Albuquerque.
Andrew Judd, Department of Pathology, University of New Mexico Health Sciences Center, Albuquerque.
Mark W Linder, Department of Pathology and Laboratory Medicine, University of Louisville Hospital, Louisville, KY.
Khaled Alkhateeb, Department of Pathology and Laboratory Medicine, University of Louisville Hospital, Louisville, KY.
William L Skinner, Mercy Health–Paducah Medical Oncology and Hematology, Paducah, KY.
Ralph Boccia, The Center for Cancer and Blood Disorders, Bethesda, MD.
Kashyap Patel, Carolina Blood and Cancer Care Associates, PA, Rock Hill, SC.
References
- 1. Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697-4705. [DOI] [PubMed] [Google Scholar]
- 2. Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28:1713-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [DOI] [PubMed] [Google Scholar]
- 4. Bokemeyer C, Bondarenko I, Hartmann JT, et al. Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol. 2011;22:1535-1546. [DOI] [PubMed] [Google Scholar]
- 5. Allegra CJ, Rumble RB, Hamilton SR, et al. Extended RAS gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: American Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol. 2016;34:179-185. [DOI] [PubMed] [Google Scholar]
- 6. Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J Mol Diagn. 2017;19:187-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maughan TS, Adams RA, Smith CG, et al. ; MRC COIN Trial Investigators . Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wasan H, Meade AM, Adams R, et al. ; COIN-B Investigators . Intermittent chemotherapy plus either intermittent or continuous cetuximab for first-line treatment of patients with KRAS wild-type advanced colorectal cancer (COIN-B): a randomised phase 2 trial. Lancet Oncol. 2014;15:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fleming M, Ravula S, Tatishchev SF, et al. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3:153-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu B, O’Toole SA, Trent RJ. Somatic DNA mutation analysis in targeted therapy of solid tumours. Transl Pediatr. 2015;4:125-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125-136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Solassol J, Vendrell J, Märkl B, et al. Multi-center evaluation of the fully automated PCR-based Idylla™ KRAS mutation assay for rapid KRAS mutation status determination on formalin-fixed paraffin-embedded tissue of human colorectal cancer. PLoS One. 2016;11:e0163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. National Comprehensive Cancer Network. Colon Cancer (Version 4.2018). https://nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed June 2018.
- 14. Morlote D, Yemelyanova A, Guo R, et al. Validation of FFPE tissue punches for detection of KRAS and BRAF mutations with the Idylla PCR-based molecular diagnostics assay. J Molec Diagn. 2018;20:967 (ST004). [Google Scholar]
- 15. Pepe F, De Luca C, Smeraglio R, et al. Performance analysis of SiRe next-generation sequencing panel in diagnostic setting: focus on NSCLC routine samples. J Clin Pathol. 2019;72:38-45. [DOI] [PubMed] [Google Scholar]
- 16. Al-Turkmani MR, Schutz SN, Tsongalis GJ. Potential of STAT somatic mutation testing at resection. Clin Chem. 2018;64:865-866. [DOI] [PubMed] [Google Scholar]
- 17. Al-Turkmani MR, Godwin KN, Peterson JD, et al. Rapid somatic mutation testing in colorectal cancer by use of a fully automated system and single-use cartridge: a comparison with next generation sequencing. J Appl Lab Med. 2018;3:178-184. [DOI] [PubMed] [Google Scholar]
- 18. Colling R, Wang LM, Soilleux E. Validating a fully automated real-time PCR-based system for use in the molecular diagnostic analysis of colorectal carcinoma: a comparison with NGS and IHC. J Clin Pathol. 2017;70:610-614. [DOI] [PubMed] [Google Scholar]
- 19. De Luca C, Vigliar E, d’Anna M, et al. KRAS detection on archival cytological smears by the novel fully automated polymerase chain reaction-based Idylla mutation test. Cytojournal. 2017;14:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weyn C, Van Raemdonck S, Dendooven R, et al. Clinical performance evaluation of a sensitive, rapid low-throughput test for KRAS mutation analysis using formalin-fixed, paraffin-embedded tissue samples. BMC Cancer. 2017;17:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang H, Springborn S, Haug K, et al. Evaluation, validation, and implementation of the Idylla system as rapid molecular testing for precision medicine. J Mol Diagn. 2019;21:862-872. [DOI] [PubMed] [Google Scholar]
- 22. Franczak C, Dubouis L, Gilson P, et al. Integrated routine workflow using next-generation sequencing and a fully-automated platform for the detection of KRAS, NRAS and BRAF mutations in formalin-fixed paraffin embedded samples with poor DNA quality in patients with colorectal carcinoma. PLoS One. 2019;14:e0212801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilson P, Franczak C, Dubouis L, et al. Evaluation of KRAS, NRAS and BRAF hotspot mutations detection for patients with metastatic colorectal cancer using direct DNA pipetting in a fully-automated platform and next-generation sequencing for laboratory workflow optimisation. PLoS One. 2019;14:e0219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uguen A, Troncone G. A review on the Idylla platform: towards the assessment of actionable genomic alterations in one day. J Clin Pathol. 2018;71:757-762. [DOI] [PubMed] [Google Scholar]