Abstract
Vacuolar-type H+-ATPase (V-ATPase) is a multimeric complex present in a variety of cellular membranes that acts as an ATP-dependent proton pump and plays a key role in pH homeostasis and intracellular signalling pathways. In humans, 22 autosomal genes encode for a redundant set of subunits allowing the composition of diverse V-ATPase complexes with specific properties and expression. Sixteen subunits have been linked to human disease.
Here we describe 26 patients harbouring 20 distinct pathogenic de novo missense ATP6V1A variants, mainly clustering within the ATP synthase α/β family-nucleotide-binding domain. At a mean age of 7 years (extremes: 6 weeks, youngest deceased patient to 22 years, oldest patient) clinical pictures included early lethal encephalopathies with rapidly progressive massive brain atrophy, severe developmental epileptic encephalopathies and static intellectual disability with epilepsy. The first clinical manifestation was early hypotonia, in 70%; 81% developed epilepsy, manifested as developmental epileptic encephalopathies in 58% of the cohort and with infantile spasms in 62%; 63% of developmental epileptic encephalopathies failed to achieve any developmental, communicative or motor skills. Less severe outcomes were observed in 23% of patients who, at a mean age of 10 years and 6 months, exhibited moderate intellectual disability, with independent walking and variable epilepsy. None of the patients developed communicative language. Microcephaly (38%) and amelogenesis imperfecta/enamel dysplasia (42%) were additional clinical features. Brain MRI demonstrated hypomyelination and generalized atrophy in 68%. Atrophy was progressive in all eight individuals undergoing repeated MRIs.
Fibroblasts of two patients with developmental epileptic encephalopathies showed decreased LAMP1 expression, Lysotracker staining and increased organelle pH, consistent with lysosomal impairment and loss of V-ATPase function. Fibroblasts of two patients with milder disease, exhibited a different phenotype with increased Lysotracker staining, decreased organelle pH and no significant modification in LAMP1 expression. Quantification of substrates for lysosomal enzymes in cellular extracts from four patients revealed discrete accumulation. Transmission electron microscopy of fibroblasts of four patients with variable severity and of induced pluripotent stem cell-derived neurons from two patients with developmental epileptic encephalopathies showed electron-dense inclusions, lipid droplets, osmiophilic material and lamellated membrane structures resembling phospholipids. Quantitative assessment in induced pluripotent stem cell-derived neurons identified significantly smaller lysosomes.
ATP6V1A-related encephalopathy represents a new paradigm among lysosomal disorders. It results from a dysfunctional endo-lysosomal membrane protein causing altered pH homeostasis. Its pathophysiology implies intracellular accumulation of substrates whose composition remains unclear, and a combination of developmental brain abnormalities and neurodegenerative changes established during prenatal and early postanal development, whose severity is variably determined by specific pathogenic variants.
Keywords: epileptic encephalopathy, ATP6V1A, lysosomal disorder, progressive brain atrophy, developmental delay
Guerrini et al. describe the phenotypic spectrum of 26 patients harbouring 20 distinct pathogenic de novo missense variants in ATP6V1A, and show that the underlying pathology includes abnormal endo-lysosomal acidification and ultrastructural hallmarks of lysosomal storage disease.
See Groszer (https://doi.org/10.1093/brain/awac255) for a scientific commentary on this article.
See Groszer (https://doi.org/10.1093/brain/awac255) for a scientific commentary on this article.
Introduction
Vacuolar-type H+-ATPase (V-ATPase) is a multimeric complex present in a variety of cellular membranes that acts as an ATP-dependent proton pump and is responsible for acidification of intracellular organelles, membrane trafficking processes, such as receptor-mediated endocytosis, intracellular trafficking of lysosomal enzymes and protein degradation, in all eukaryotic cells.1,2 The role of V-ATPase in pH homeostasis and intracellular signalling pathways is ubiquitous, but it is highly significant in neurons where it plays additional role in neurotransmitter loading into synaptic vesicles and synaptic transmission.3,4
V-ATPase is composed by V1 cytosolic and V0 membrane complexes that allow ATP hydrolysis (V1) and hydrogen transport (V0). In humans, 22 autosomal genes encode for a redundant set of subunits allowing the composition of diverse V1 and V0 complexes with specific properties and tissue expression. Among these, 16 have been associated with human disease (Supplementary Table 1). ATP6V1A codes for the ubiquitously expressed A subunit belonging to the V1 complex.
A biallelic form of ATP6V1A-related multisystemic disorder has been observed in three children from two consanguineous families, exhibiting cutis laxa, cardiac abnormalities, dysmorphic facial features, severe hypotonia and early seizures.5 Subsequently, Vogt et al. identified ATP6V1A recessive variants in three affected individuals from two families exhibiting autosomal recessive cutis laxa type 2D and an overall progeroid appearance, a typical triangular facial gestalt with cataracts, limb contractures and in some lowered levels of plasma proline, arginine or ornithine.6 Two patients were compound heterozygous for a missense and a stopgain variant, the third was homozygous for a novel missense variant.6
In a previous study, we linked de novo heterozygous ATP6V1A pathogenic variants to a developmental and epileptic encephalopathy (DEE) whose pathomechanisms involve the effects of V-ATPase on pH homeostasis and neuronal connectivity.7
Here, we describe the clinical spectrum of ATP6V1A encephalopathy in 26 patients, harbouring 20 different de novo missense pathogenic variants clustering within the ATP synthase α/β family, nucleotide-binding domain (ATP-synt_ab). The phenotypic spectrum ranges from rapidly progressive early lethal encephalopathies to moderate intellectual disability and epilepsy with no obvious signs of progression. The underlying pathology, as demonstrable in patients’ fibroblasts and induced pluripotent stem cell (iPSC)-derived neurons (iNs), includes abnormal endo-lysosomal acidification and ultrastructural hallmarks of lysosomal storage diseases.
Materials and methods
Patients
We established a cohort of patients with pathogenic or likely pathogenic ATP6V1A variants from multiple diagnostic and research series from across Europe, the USA and Japan. After our initial report associating de novo missense ATP6V1A pathogenic variants with a DEE in four patients,7 the pathogenic role of ATP6V1A variants was increasingly recognized and a group of families was established (www.facebook.com/groups/ATP6V1A), which helped in assembling the cohort presented here. This study includes the four patients described in our initial report together with 22 newly identified patients, of whom 20 are sporadic and two are monozygotic twins.
We obtained written informed consent from all participants or their legal guardians according to local requirements. The study was approved by the Paediatric Ethics Committees of the Tuscany Region, Italy, in the context of the DESIRE FP7 EU project and its extension by the DECODE-EE project.
Detailed materials and methods for MRI investigations, genetic analysis, homology modelling and structural analysis, western blotting, immunocytochemistry, fluorescence and transmission electron microscopy, glycosylation, enzyme activity and biomarker analysis, and human iPSC differentiation into induced neurons are reported in the Supplementary material.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. Further derived data not provided in the article due to space limitations are available from the corresponding author on request.
Results
Clinical, imaging, neurophysiological and genetic information on the 26 patients exhibiting de novo ATP6V1A pathogenic variants are summarized in Fig. 1 and Supplementary Table 2, and are presented in detail in the following sections.
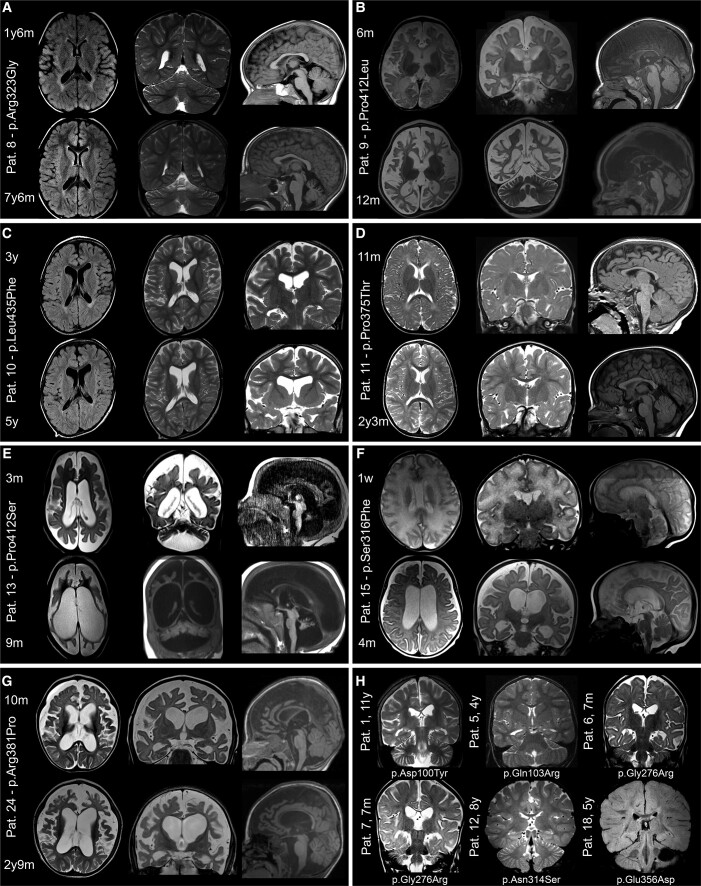
Figure 1.
Brain MRI in patients with ATP6V1A pathogenic variants. MRIs of patients who were imaged at least twice are shown in A–G. Images were taken from the initial and last follow-up investigations at 1.5 to 3 T and include T1- or T2-weighted and FLAIR sequences. Structural abnormalities include a combination of cerebellar and brainstem atrophy, dilated ventricles and subarachnoid spaces, thinning of the corpus callosum and hypomyelination. Comparison of initial and follow-up images demonstrates various rates of progression and anatomic involvement. In Patient (Pat.) 8 (A), at age 1 year 6 months, hypomyelination was the only feature, while cerebellar atrophy was present at 7 years 6 months. In Patients 9 (B) and 13 (E), MRI scan at age 6 months and 3 months, respectively, showed severe signs of atrophy that involved the cerebral cortex and white matter in Patient 9 and all brain, brainstem and cerebellar structures in Patient 13. Six months later, in Patient 9, at 12 months of age, atrophy had remarkably progressed, yet with minimal cerebellar involvement; in Patient 13, at 9 months, dramatic generalized shrinking of all brain structures had occurred. In Patient 15 (F), severe hypomyelination was present 1 week after birth followed, 4 months later, by severe brain atrophy. In Patient 24 (G) moderate brain atrophy with hypomyelination was present at 10 months, which became more severe at 2 years 9 months. In Patients 10 (C) and 11 (D), mild atrophy became apparent from age 3 to 5 years (Patient 10) and 11 months to 2 years 3 months (Patient 11). Coronal sections of the brain of six patients who had only one MRI scan are presented in H. These images, taken at different ages, show hypomyelination in all patients and minor atrophic changes in two (Patients 6 and 7).
Phenotypic spectrum
Mean age of the 26 patients at last follow-up was 7 years (extremes: 6 weeks for the youngest deceased patient to 22 years for the oldest living patient).
The most severe end of the phenotypic spectrum was manifested in four patients (Patients 9, 13, 15 and 26) who exhibited a profound, rapidly progressive encephalopathy, manifested since birth with severe hypotonia and absence of any developmental skill, leading to early death in three of them (Patient age at death: 9-2 years; 15-5 months, 26-6 weeks) (Supplementary Table 2). The most common first clinical manifestation in the cohort was early hypotonia, reported in 18 of the 24 patients (75%) for whom precise information on initial symptoms was available. In 10 of the 18 infants with early hypotonia, seizures were also considered an initial symptom, as they were noticed at the same time.
Twenty out of 25 patients with available clinical information exhibited epilepsy (80%). Convulsive seizures during fever were an initial manifestation in seven of those with epilepsy (Patients 1–4, 12, 21 and 22) and were not followed by epilepsy in one patient (Patient 11). On the basis of epilepsy outcome and associated intellectual disability, we classified 16 patients who survived after early infancy as having a DEE (Patients 1, 4–8, 10 and 17–25) (16/25, 64%; 16/21 with epilepsy, 76%; Supplementary Table 2), whose onset was in infancy in 13 and in childhood in three (Patients 8, 21 and 23). In the infantile onset DEE group, infantile spasms were the most prominent seizure type (11/13, 85% with DEE; 11/21, 52% of all patients with epilepsy) and appeared at a mean age of 7 months (range 2 to 13 months). Additional myoclonic, tonic, tonic-clonic, atonic, absence and focal seizures were reported later in these children. In one of the three patients with childhood onset DEE (Patient 8), intractable generalized tonic-clonic seizures appeared at age 7, followed by loss of acquired motor skills. All patients with DEE had been unsuccessfully treated with multiple antiepileptic medication trials.
Ten of the 16 patients with DEE (Patients 1, 4, 6, 7, 10, 17, 19, 20, 24 and 25) exhibited early profound developmental delay with hypotonic quadriplegia and failed to achieve any developmental, communicative or motor skill. The six remaining patients in this group (Patients 5, 8, 18 and 21–23) acquired motor skills, which were subsequently lost in two of them, after worsening of epilepsy (at 4 years, Patient 5) or after seizure onset (at 7 years, Patient 8). In this group, the only patients who acquired language skills, albeit poor, were the two whose epilepsy had started in childhood (Patients 8 and 21). Mean age of the 16 patients with DEE at last follow-up was 6 years and 5 months (extremes: 10 months to 14 years).
The six remaining patients exhibited an association of intellectual disability and epilepsy (Supplementary Table 2; Patients 2, 3, 11, 12, 14, and 16, 6/26, 23%), which represented the mild end of the phenotypic spectrum. Intellectual disability was moderate in five patients and mild–moderate in one (Patient 16); epilepsy was severe in two of these patients who, however, did not exhibit seizure-related progression of symptoms typical of DEE. Different seizure types were diagnosed in this group, comprising generalized tonic-clonic (3/6), focal (2/6), myoclonic (1/6) and absence (1/6). One patient had only experienced rare seizures during fever (Patient 11). Mean age at seizure onset was 27 months (range 15 months to 3 years). All six patients achieved independent walking, and four acquired simplified language skills. Three exhibited mild autistic traits (Patients 3, 12 and 16) and one an attention deficit hyperactivity disorder trait (Patient 16). Mean age of these six patients at the last follow-up was 10 years and 6 months (4–22 years).
Overall, language impairment was a highly prominent feature of this entire cohort since, of the 19 patients who were older than two years at last follow-up, 13 had remained nonverbal and six had poor language skills.
EEG recordings were available for 24 patients (Patients 1–13, 15–22 and 24–26), 19 of whom with epilepsy (Patients 1–8, 10, 12, 16–22, 24 and 25). A hypsarrhythmic EEG was reported in two patients (2/24, 8%; 2/19 with epilepsy, 10%), multifocal epileptiform discharges in 11 (11/24, 46%; 11/19 with epilepsy, 58%) and focal abnormalities in seven (7/24, 29%; 7/19 with epilepsy, 37%). Background EEG activity was described as abnormally slow in 13 patients (13/24, 54%), but we could not precisely correlate this finding with the clinical status of patients at the time of recordings.
Head circumference measurements, at different ages, were available for 24/26 patients. Microcephaly (head circumference < −2 SD) was present in nine patients (Patients 1, 4, 9, 13, 14, 17, 19, 24 and 25; 9/24, 38%), and could be demonstrated to be progressive in six (Patients 1, 4, 13, 19, 24 and 25). Head size measurements at birth, available in 12 patients, revealed negative SD scores in 10 (10/12, 83%) while values <−1SD were observed in 14 patients (Patients 1, 2, 4, 5, 7, 9, 11, 13–15, 17–19 and 25) and were already present in the first year of life in eight.
Amelogenesis imperfecta or enamel dysplasia were described in 11/26 patients (Patients 1, 2, 5, 8, 10–12, 14, 17, 19 and 22, 42%) who manifested combinations of eruption defects, defective enamel mineralization, excessive caries and crumbling teeth (Supplementary Fig. 1). Although information on dentition was not available for all patients, these findings were consistent and noteworthy. Additional clinical manifestations included lack of visual contact (17/26, 65%), dyskinetic movements (4/26, 15%), optic atrophy (4/26, 15%), cerebellar signs (3/26, 12%), congenital arthrogryposis of joints (3/26, 12%), coloboma and dysmorphic features in one patient each. Five patients were G-tube dependent (Patients 6, 7, 13, 17 and 25, 19%). No signs of osteopetrosis or renal acidosis, which have been associated with disease causing mutations of multiple V-ATPase subunits (Supplementary Table 1), emerged from the study of this series.
Neuroimaging
Brain MRI was available in 25/26 patients, examined between age 1 week and 11 years, and showed structural abnormalities in 20 (20/25, 80%) (Fig. 1).
Abnormal MRI showed atrophic changes variably affecting the cerebral cortex (Patients 1, 4, 6, 7, 9–11, 13–15, 17 and 24), brainstem, cerebellum or both (Patients 1, 5–8, 10–13, 15, 17 and 24), associated with dilated ventricles (Patients 1, 4, 6, 7, 9, 10, 13, 15, 17 and 24), thinning of the corpus callosum (Patients 6, 7, 9, 12, 13, 15 and 24) and hypomyelination (Patients 1, 4–13, 15, 18, 21 and 24).
In all eight individuals for whom repeated MRIs were available for comparison (Patients 5, 8–11, 13, 15 and 24; Fig. 1), there was worsening of atrophy, whose rate of progression and severity varied considerably. In Patients 9, 13 and 15, atrophy worsened dramatically within the first months of life, paralleling a clinical deterioration that eventually led to premature death in the first and second year of life in two of them (Patients 9 and 15). The remaining five patients with follow-up imaging showed worsening of structural changes over longer periods, differently affecting brain and cerebellum (Fig. 1). For example, in Patient 8 from the age of 1 year and 6 months to 7 years and 6 months, only mild cortical and cerebellar atrophy became apparent. In Patient 11, instead, after a normal MRI at 11 months, mild cortical atrophy became apparent at repeat MRI scan at age 2 years and 3 months, while the cerebellum remained normal and hypomyelination persisted unchanged. Abnormal high signal intensity of the white matter, attributed to hypomyelination, was observed at any age and showed no signs of progression.
ATP6V1A whole gene-level constraint and predicted transmission model
The ATP6V1A whole gene-level constraint showed a positive missense Z-score of 3.37 [Genome Aggregation Database (gnomAD) v.2.1.1] and a residual variation intolerance score (based on Exome Aggregation Consortium v.2) of −1.041 (corresponding to the 11th percentile) suggesting an increased constraint and therefore intolerance to variations. The DOMINO tool assessed a high likelihood (0.911) for ATP6V1A gene to harbour dominant changes.
Genetic variants in ATP6V1A
The 26 patients harboured 20 unique heterozygous ATP6V1A variants (Fig. 2 and Supplementary Table 2). Six variants had already been reported (p.Asp100Tyr; p.Asp349Asn; p.Asp371Gly; p.Pro27Arg; p.Asn314Asp and p.Pro249Arg), of which four, now included as Patients 1–4 (p.Asp100Tyr; p.Asp349Asn; p.Asp371Gly and p.Pro27Arg) in our initial description of ATP6V1A encephalopathy,7 and two in patients listed in large cohorts with developmental disabilities, with limited clinical information,8,9 now included as Patients 14 (p.Asn314Asp) and 17 (p.Pro249Arg), with more clinical detail for Patient 17. The remaining 14 variants are novel. All 20 variants were de novo and missense; five substitutions were recurrent in unrelated patients (p.Pro375Thr, Patients 11 and 22; p.Ser316Phe, Patients 15 and 26; p.Pro249Arg, Patients 17 and 23; p.Arg381Pro, Patients 19 and 24; p.Arg359Gly, Patients 20 and 21), whereas one substitution (p.Gly276Arg) occurred in a monozygotic twin pair (Patients 6 and 7). In four unrelated patients, two distinct substitutions involved each of two residues (p.Asn314Ser/Asp and p.Pro412Ser/Leu), resulting in 18 different residues being involved in the overall series. None of the 20 missense substitutions was observed in the Exome Aggregation Consortium, gnomAD, Human Genetic Variation Database and BRAVO population databases (Supplementary Table 3). Predictions obtained from the dbNSFP database (v.4.1a) were almost entirely concordant in defining a harmful effect on the ATP6V1A protein (Supplementary Table 4). The PolyPhen-2, Sorting Intolerant From Tolerant, PROVEAN, Combined Annotation Dependent Depletion, Mendelian Clinically Applicable Pathogenicity, MetaLR and MetaSVM in silico tools scores for ATP6V1A missense substitutions observed in patients and in gnomAD individuals, highlighted that all de novo variants identified in affected individuals cluster in the deleterious range for each score (Supplementary Fig. 2). Both missense badness, PolyPhen-2 and constraint and missense variant pathogenicity prediction scores obtained from patients’ variants were significantly different from those observed in gnomAD controls (Mann–Whitney’s U-test; missense badness, PolyPhen-2 and constraint P-value = 9.4 × 10−11 and missense variant pathogenicity prediction P = 4.1 × 10−8), as illustrated in the box plot graphs (Supplementary Fig. 3). Overall, these findings consistently supported a deleterious effect of the 20 variants.
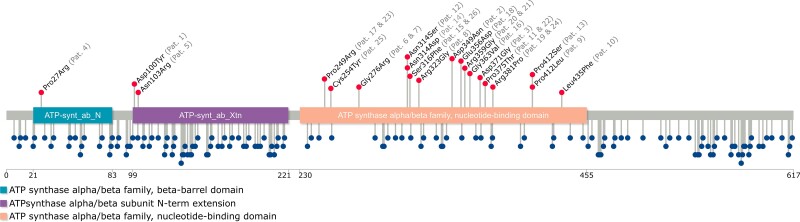
Figure 2.
Schematic representation of the ATP6V1A protein. The structure of ATP6V1A includes the ATP-synt_ab_N (light blue), ATP_synt_ab_Xtn (purple) and ATP-synt_ab (light orange) domains. The red lollipops show the location of the pathogenic variants identified in the patients in this series (patients’ identifiers are reported in grey between brackets), whereas the blue lollipops represent the missense variants reported in the gnomAD database.
We compared the presence and location of the 20 missense variants occurring in the 26 patients to those reported in the gnomAD population database using a lollipop diagram depicting the full ATP6V1A protein and its Pfam-identified domains (Fig. 2).
Analysis of the impact of these 20 variants, on the basis of regional depletion in the general population using the missense tolerance ratio (MTR)-Viewer tool, indicated that MTR scores were above the neutrality threshold for one ATP6V1A residue (Patient 4, Pro27 residue), above the 50th percentile (median), but below the neutrality threshold for five residues (Patient 1, Asp100; Patient 5, Gln103; Patient 8, Arg323; Patients 11/22, Pro375; Patients 12/14, Asn314); and below the median but above the 25th percentile for 10 residues (Patient 2, Asp349; Patient 3, Asp371; Patients 6/7, Gly276; Patient 10, Leu345; Patient 15/26, Ser316; Patient 16, Gly363; Patients 17/23, Pro249; Patient 18, Glu356; Patient 19, Arg381; Patients 20/21, Arg359; Patient 25, Cys254); one remaining residue (Patients 9/13, Pro412) fell below the 5th percentile (Supplementary Fig. 4).
Furthermore, we observed the regions with the lowest MTR scores, and therefore the highest intolerance to missense substitutions, to fall within the amino acid boundaries of the ATP-synt_ab domain (230–455 aa). Seventeen/20 substitutions were in the ATP-synt_ab domain, suggesting its enrichment for pathogenic ATP6V1A variants (Fig. 2). This domain is listed among the genome-wide significantly enriched domains for constraint (constrained coding regions: P-value of 0.0359; Bonferroni corrected across the 6060 Pfam known domains)10 (Supplementary Fig. 5). The two remaining ATP6V1A Pfam domains, namely ATP-synt_ab_N and ATP-synt_ab_Xtn, were not significantly enriched for constrained coding regions (Supplementary Fig. 5). The p.Pro412 residue involved in the missense substitutions (Patient 9: p.Pro412Leu and Patient 13: p.Pro412Ser) was located in one of the highly constrained ATP6V1A constrained coding regions not covered by missense depletion (Hg19:chr3:113514042-113514777; 98.53%ile10). Consistent with this observation, Patients 9 and 13 exhibited a strikingly similar phenotype with early-onset rapidly progressive encephalopathy and profound brain atrophy (Fig. 1).
Taken together, these findings strongly support pathogenicity of the ATP6V1A missense variants in this cohort and point to pathogenic variants clustering within the ATP-synt_ab domain.
Structural consideration of the V-ATPase substitutions
All substitutions but one (Pro27Arg, which maps in the N-terminal β-barrel domain) affect amino acids mapping in the N-terminal α/β, central domain surrounding the catalytic site (Fig. 3 and Supplementary Video 1). The effect of p.Pro27Arg (perturbation of the ATP6V1A/B interaction), p.Asp100Tyr (destabilization of the protein folding), p.Asp349Asn (impairment of the catalytic function) and p.Asp371Gly (impairment of the rotation process) had been discussed previously.7 For p.Asp349Asn, the predicted effect was based on structural observations only, but is in contrast with the previously reported5 and here confirmed cellular phenotype. To further investigate the effect of Asp349Asn [Asp329Asn in Enterococcus hirae (HE)], we performed molecular dynamics simulations of wild-type and mutated homologous EH A subunit with Mg2+-ATP bound, revealing that the mutation has no significant impact on the Mg2+-ATP structure and environment (Supplementary Fig. 5). In the mutated protein the conformation of ATP is preserved, as well as its interactions with surrounding amino acids, binding to Mg2+ and coordination of the latter by water molecules. The simulation thus suggests that Asp349Asn does not necessarily impair ATP binding and catalytic activity of ATP6V1A, albeit affecting it in a yet uncharacterized manner.
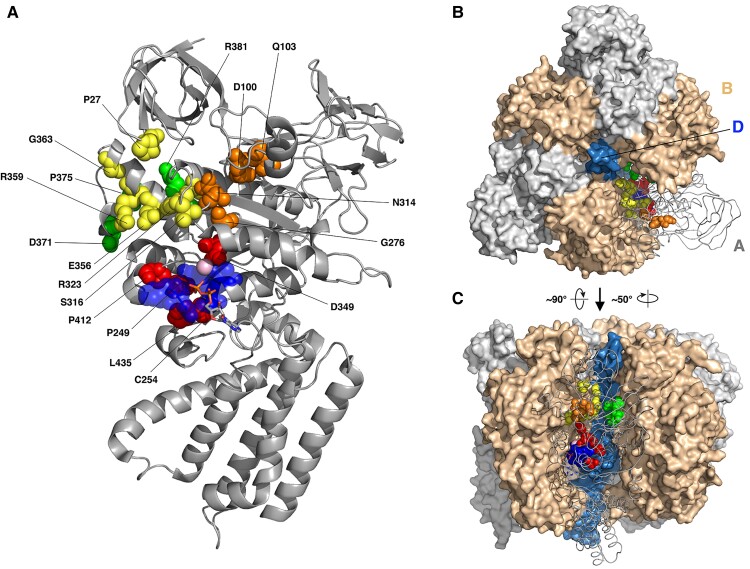
Figure 3.
Cryo-EM structure of the Atp6v1a. (A) The cryo-EM structure of the Atp6v1a subunit from the mammalian V-ATPase (PDB code 6VQ9). Pathogenic variant sites are drawn as red, yellow, green and orange spheres if they are near the phosphate binding loop (p-loop, blue spheres), A/B interface, A/D or non-catalytic A/B interface and uncharacterized locations, respectively. The pink sphere is the magnesium ion, and the bound ADP molecule is drawn as sticks. (B and C) Structure of the V1 domain from upper (B) and lateral (C) views. One A subunit is shown as grey tube and the other two as grey surface, and B and D subunits are shown as ochre and blue surfaces, respectively. Pathogenic variant sites are drawn as in A.
Among the newly identified substitutions, p.Pro249Arg, p.Cys254Thr, p.Pro412Leu/Ser and p.Leu435Phe (Fig. 3, red spheres) are located either on or near the P-loop (residues 250–257), which includes the ATP binding site and may affect ATP/V-ATPase binding; p.Ser316Phe, p.Arg323Gly, p.Glu356Asp, p.Arg359Gly and p.Gly363Val are at the interface between A and B subunits on the catalytic site side (Fig. 3, yellow spheres), while p.Pro375Thr and p.Arg381Pro are at the interface between A and B subunits on the opposite side (the non-catalytic interface) (Fig. 3, green spheres). All these pathogenic variants may perturb ATP6V1A/B interaction. The remaining substitutions (p.Gln103Arg, p.Asn314Asp/Ser, p.Gly276Arg) occur at sites whose structural characterization is less immediate (Fig. 3, orange spheres). Gln103 is only three positions away from Asp100, whose effect on the protein folding and degradation had been demonstrated previously7 and confirmed here (Fig. 4). Being surrounded by a cluster of hydrophobic residues, replacement of Gln103 with a bulkier, positively charged arginine is expected to strongly affect local amino acidic interactions. Asn314Asp substitution disrupts the H bond between the residue 314 and Cys277 (Supplementary Fig. 6). In addition, this substitution determines that the free carboxyl group of Asp314 maps very close (∼2.7 Å) to the carboxyl group of Asp100 (Supplementary Fig. 7). The Asn314Ser substitution is predicted to introduce an additional H bond between residue 314 and Cys277 (Supplementary Fig. 7). Thus, although Asn314 does not map in a specific functional region of the protein, the Asn314Asp/Ser substitutions might affect local amino acidic interactions. Gly276 is ∼5 Å away from the side chain of Asp349, which is in turn involved in the coordination of the Mg2+ ion at the ATP biding site.7 Hence, Gly276Arg is likely to perturb the catalytic site as well.
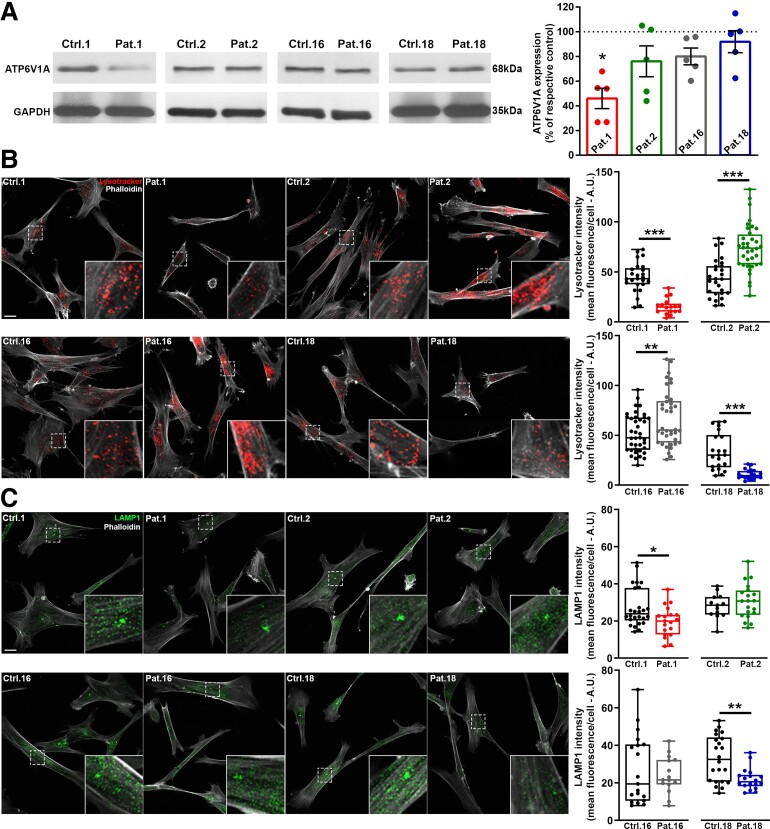
Figure 4.
ATP6V1A/LAMP1 expression and LysoTracker labelling in patient-derived fibroblasts. (A) Left: Representative western blots from fibroblasts lysates of patients (Patients 1, 2, 16 and 18) and the respective controls (Ctrl 1, 2 16 and 18). Right: Quantification of ATP6V1A expression levels normalized on GAPDH signal and expressed, for each patient, as percentage of the respective control. Data are means ± SEM from five independent experiments. *P < 0.05; Kruskal–Wallis/Dunn’s tests. (B) Left: Representative images of fibroblasts incubated with Lysotracker (red) and stained with phalloidin (white). Higher magnifications of the fields labelled by dotted squares are shown in the insets. Scale bar = 20 µm. Right: Quantification of Lysotracker intensity. The fluorescence signal was measured in the cell body identified by phalloidin staining. Each dot represents the mean fluorescence intensity of a single cell. Data are from 26/17 cells for Ctrl 1/Patient 1, 27/36 cells for Ctrl 2/Patient 2, 39/38 cells for Ctrl 16/Patient 16, 22/20 cells for Ctrl 18/Patient 18. (C) Left: Representative images of fibroblasts double stained with LAMP1 (green) and phalloidin (white). Higher magnifications of the fields labelled by dotted squares are shown in the insets. Scale bar = 20 µm. Right: Quantification of LAMP1 intensity. Immunoreactivity was measured in the cell body identified by phalloidin labelling. Each dot represents the mean fluorescence intensity of a single cell. Data are from 26/19 cells for Ctrl 1/Patient 1, 13/20 cells for Ctrl 2/Patient 2, 19/15 cells for Ctrl 16/Patient 16, 22/18 cells for Ctrl 18/Patient 18. *P < 0.05, **P < 0.01, ***P < 0.01; unpaired Student’s t-test/Mann–Whitney U-test.
Glycosylation analysis
Previous reports of altered glycosylation profiles in patients with biallelic ATP6V1A variants, resulting in cutis laxa with severe neurological impairment, prompted us to investigate glycosylation status in two severely affected patients in our cohort. Transferrin N-glycan MS profiles showed mild abnormalities, consisting in a mild increase of the monosialo-biantennary glycoforms at m/z 2431.3 (A2G2S1) and 2605.5 (FA2G2S1) in both patients, and of the disialo-triantennary species at m/z 3241.7 (A3G3S2) and 3415.9 (FA3G3S2) for Patients 18 and 1, respectively. A general fucosylation increase, due to the enhancement of the monofucosylated structures normally present in human serum transferrin, was also observed in Patient 1 (data not shown).
Total serum N-glycan analysis reflected the same glycosylation defects (Supplementary Fig. 8), with slightly increased hyposialylated and/or fucosylated structures in Patient 1’s (Supplementary Fig. 8B) and increased amount of the biantennary monosialo-glycoforms A2G2S1 and FA2G2S1 in Patient 18’s (Supplementary Fig. 8C) glycosylation profile.
Transferrin N-glycan fucosylation was investigated in both patients by Matrix-assisted laser desorption ionization–time of flight–tandem mass spectrometry, with the purpose of discriminating core- from antennary fucosylated isoforms and establishing whether changes in the relative amount of the respective structures occurred. The fragmentation spectra of the most abundant fucosylated structure of patients’ serum transferrin (namely FA2G2S2 at m/z 2966.5), revealed a ∼3-fold increase in the relative intensity of the antennary fucosylated isomer with respect to the reference range for controls (Supplementary Fig. 9). Matrix-assisted laser desorption ionization–mass spectrometry analysis of serum apolipoprotein CIII (ApoCIII) was performed in Patient 1 and showed no obvious alterations of the mucin type O-glycosylation profile (data not shown). In summary, we found N-glycosylation alterations due to small sialylation defects, in line with observations in patients with biallelic ATP6V1A variants5 and growth of antennary fucosylation.
Enzymatic activity and biomarker assay in fibroblasts
Patients had 56 ± 15–240 ± 42% mean normal control activity for alpha-l-iduronidase (IDUA), alpha-galactosidase and acid β-glucocerebrosidase (ABG) when measured in vitro at optimal enzyme-specific pH. We quantified enzyme-specific natural substrates heparan sulphate and dermatan sulphate for IDUA, the glucosylsphingosine (lyso-Gb1) for ABG and the globotriaosylsphingosine (lyso-Gb3) for alpha-galactosidase. Dermatan sulphate was slightly increased in Patient 18 and decreased in Patients 1, 2, 11 and 16. The increment of heparan sulphate was more evident (+69–240%) for all patients except Patient 16. The lyso-Gb1 (+21–223%) and the lyso-Gb3 (+30–80%) were higher in all patients with respect to controls (Supplementary Table 5).
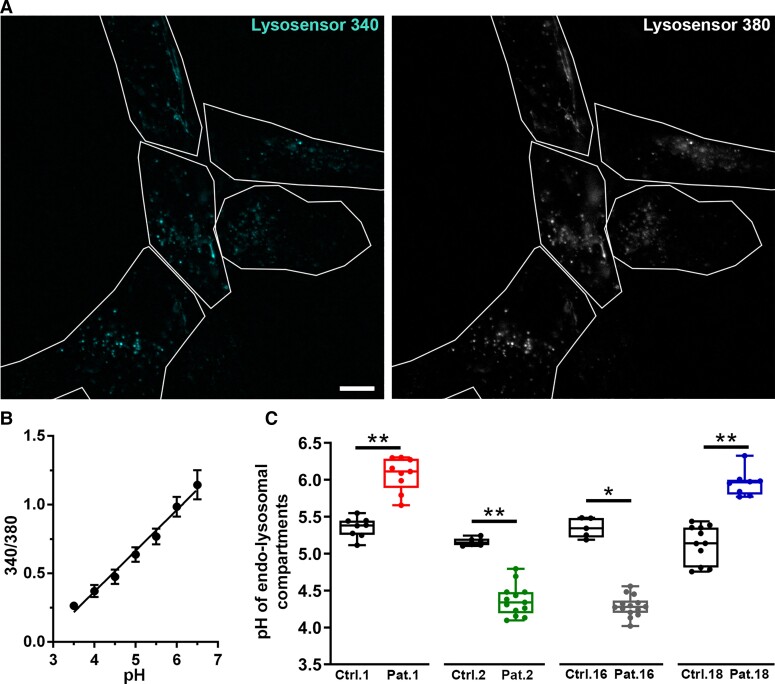
ATP6V1A expression, lysosomal labelling and pH measurements in patient-derived fibroblasts
We used patients’ fibroblasts to evaluate the effect of four of the mutations described here (i.e. Patients 1, 2, 16 and 18) on ATP6V1A and LAMP1 expression and on lysosomal pH. As controls, we used fibroblasts matched for passage in culture, age of biopsy and sex of healthy donors and gave them the same number as the respective patients. We first analysed ATP6V1A protein expression and observed a significant decrease in fibroblast lysates from Patient 1, confirming previous findings in different cellular systems.7 We observed no significant changes in ATP6V1A levels for Patients 2, 16 and 18 (Fig. 4A). These findings indicate that p.Asp100Tyr results in increased degradation of the ATP6V1A subunit, whereas p.Asp349Asn, p.Gly363Val and p.Glu356Asp are compatible with its physiological expression. We next assessed the expression of the lysosomal marker LAMP1 and the organelle acidification process using Lysotracker and Lysosensor dyes. In fibroblasts of Patients 1 and 18, with respect to controls we identified decreased LAMP1 expression, accompanied by a lower Lysotracker staining. Differently, fibroblasts of Patients 2 and 16 exhibited an increased Lysotracker staining, with no significant modification in LAMP1 expression when compared to their respective controls (Fig. 4B and C). A change in the non-ratiometric Lysotracker dye can reflect either a decrease in the cell density of acidic organelles or a change in their internal pH. To obtain an absolute measure of the intraorganellar pH, we used the ratiometric LysoSensor yellow/blue dextran. These experiments revealed a pH increase in fibroblasts of Patients 1 and 18 and an opposite pH decrease in those of Patients 2 and 16 (Fig. 5). For both Lysotracker/LAMP1 labelling and pH measurements, closely similar results were obtained when all controls were pooled together (Supplementary Fig. 10).
Figure 5.
Evaluation of lysosomal pH in patient-derived fibroblasts. (A) Representative images of fibroblasts incubated with LysoSensor yellow/blue dextran and visualized in live at 340 and 380 nm excitation. White lines represent cells detected in bright field and used as regions of interest for intensity measurement. Scale bar = 10 µm. (B) The calibration curve, obtained by plotting the fluorescence intensity 380/340 ratios as a function of pH, was fitted with linear regression. Data are means ± SEM from the five cells shown in A. (C) pH value derived from 340/380 ratio and relative calibration curve. Each dot represents the mean pH from a single coverslip. 5–14 coverslips have been analysed for each experimental group with an average of 14 cells analysed for each coverslip. *P ≤ 0.001; **P ≤ 0.0001 unpaired Student’s t-test/Mann–Whitney U-test.
These findings indicate that the p.Asp100Tyr and p.Glu356Asp substitutions cause impaired proton pumping, which results in a more basic lysosomal pH. This phenotype derives from either a loss of expression of the ATP6V1A subunit (p.Asp100Tyr) or loss of the catalytic function of the subunit. On the other hand, the p.Asp349Asn and p.Gly363Val substitutions resulted in a different cellular phenotype with no change in lysosomal marker expression and decreased intra-organellar pH, suggestive of increased proton pumping activity or lower ratios of endocytic versus lysosomal acidic organelles.
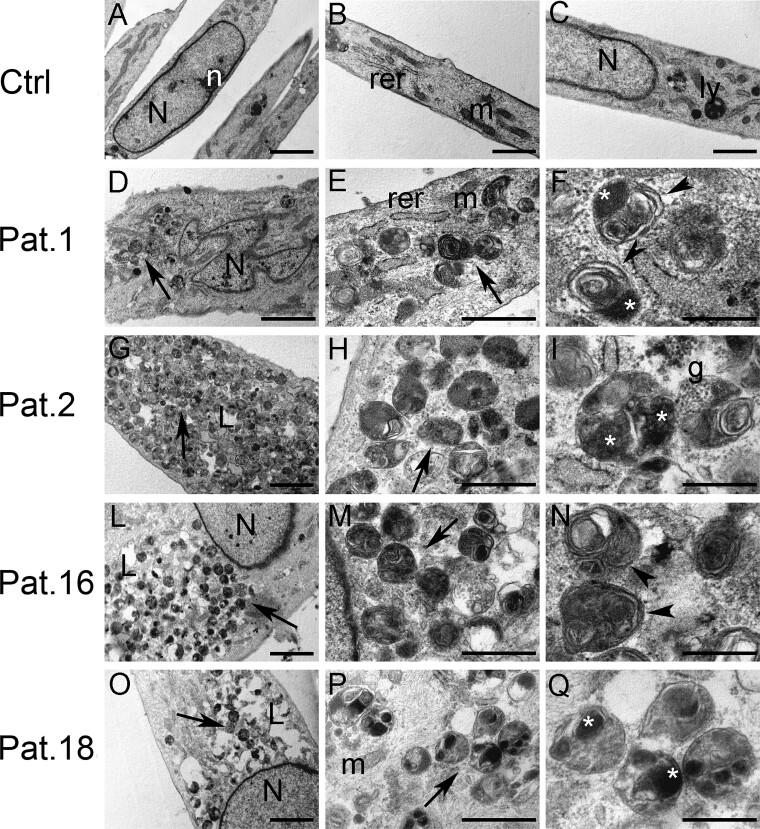
Ultrastructural analysis of patients-derived fibroblasts
Control fibroblasts showed a thin and elongated appearance with centrally located nuclei and well-organized nucleoli (Fig. 6A–C). Condensed mitochondria, slightly dilated endoplasmic reticulum and small lysosomes were observed in the cytoplasm. Conversely, fibroblasts from patients bearing ATP6V1A pathogenic variants (Patients 1, 2, 16 and 18) showed several cytoplasmic single membrane-bounded vacuoles filled with heterogeneous substances, resembling lysosomal structures (Fig. 6D–Q). Most vacuoles in fibroblasts from Patient 1 were packed with osmiophilic material and lamellated membrane structures resembling phospholipids (Fig. 6D and E). In Patient 2’s fibroblasts, the vacuolar structures were more heterogeneous and contained small electron-dense granular materials in addition to lamellated membrane structures and osmiophilic material. Furthermore, few lipid droplets were visible in the cytoplasm (Fig. 6G and H). In Patient 16’s fibroblasts, vacuoles were packed with lamellated membrane structures and abundant lipid droplets were present (Fig. 6L–N). In Patient 18’s fibroblasts, vacuolar structures were very heterogeneous and filled with osmiophilic material and substances with different electron densities. Lipid droplets were also present (Fig. 6O–Q).
Figure 6.
Ultrastructural analysis of patient-derived fibroblasts. (A–C) Control fibroblasts. (D–Q) Fibroblasts from Patient 1 (D and E), Patient 2 (G–I), Patient 16 (L–N) and Patient 18 (O–Q). Fibroblasts from patients bearing ATP6V1A pathogenic variants showed several cytoplasmic single membrane-bounded vacuoles filled with heterogeneous substances, resembling autolysosomes (arrows in D, E, G, H, L, M, O and P). Vacuoles from patients’ fibroblasts were filled with various substances, such as lamellated membrane structures (arrowheads in F and N), osmiophilic material (asterisks in F, I and Q), electron-dense granular material (g in I) and substances with different electron-density (Q). N = nucleus; n = nucleolus; rer = rough endoplasmic reticulum; m: mitochondria; ly = lysosome; L = lipid droplets; arrows = cytoplasmic single membrane-bounded vacuoles; arrowheads = lamellated membrane structures; asterisks = osmiophilic material; g = electron-dense granular material. Scale bars = 2 µm (A, D, G, L and O), 1 µm (B and C), 500 nm (E, H, M and P), 200 nm (F, I, N and Q).
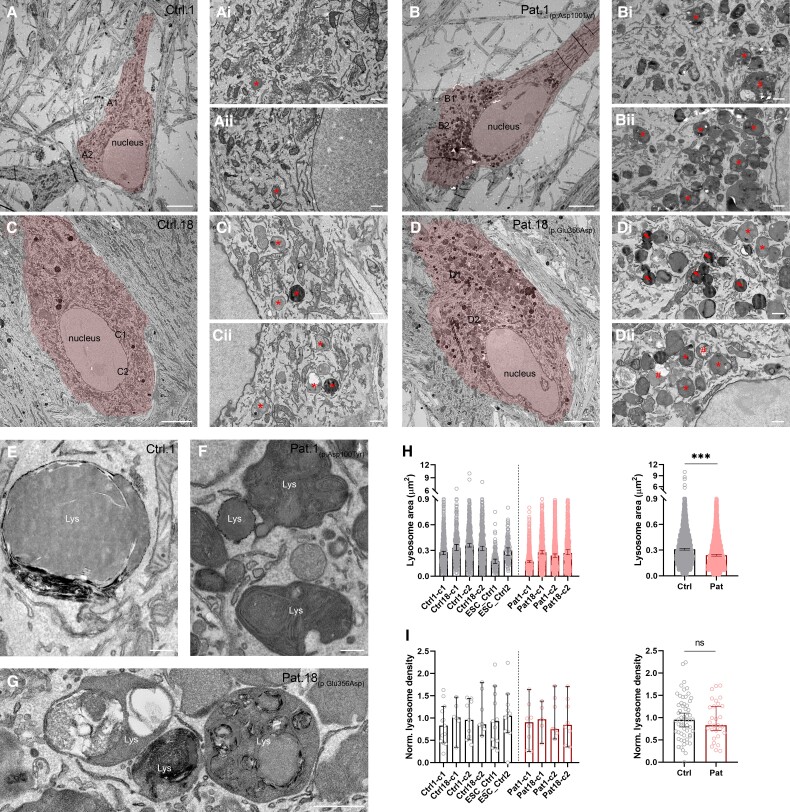
Ultrastructural analysis of human iPSC-derived neurons with ATP6V1A pathogenic variants
To study potential morphological consequences of ATP6V1A mutations in neuronal cells, we reprogrammed human fibroblasts to iPSCs and differentiated them to glutamatergic neurons [i.e. induced neurons (iNs)]. Quality of iPSC clones was confirmed by immunofluorescence analysis (Supplementary Fig. 12) and karyotyping that ruled out genomic rearrangements (Supplementary Fig. 13). We compared iNs obtained from Patients 1 and 18 with iNs obtained from two control subjects (i.e. Ctrls 1 and 18) and two independent clones of human embryonic stem cell-derived iNs (i.e. ESC_Ctrl 1 and 2). As expected, iNs from controls and patients showed Microtubule-Associated Protein 2 staining and immunoreactivity for pre- and postsynaptic markers (Supplementary Fig. 11). Next, we used transmission electron microscopy analysis to study the ultrastructure of lysosomes in the iN somata. Electron-dense lysosomes of various size and shape were often accumulated around the nucleus (Fig. 7B and D) of iNs from both patients. We observed three types of lysosome. Most of the lysosomes appeared as dark lamellar bodies, characterized by dense concentric lamellae inside their lumen (Fig. 7B1–2, D1–2, F and G; lysosomes marked by *). Other lysosomes appeared as irregular structures, filled with dark and light material resembling lipid droplets (Fig. 7B1, D2; lysosomes marked by #). A subset of lysosomes was small, round-shaped and filled with electron-dense homogenous material (Fig. 7D1, lysosomes marked by arrows). In contrast, neuronal somata from controls contained small to large and mostly round lysosomes filled with homogenous material (Fig. 7A1–2, C1–2 and E).
Figure 7.
Ultrastructural analysis of patient-derived iNs. (A–D) Representative electron micrographs showing somata and proximal dendrites of two iPSC-derived controls (Ctrls 1 and 18; A and C), and Patient 1 (B) and Patient 18 (D) iNs (highlighted in red). [A(i and ii) and C(i and ii)] High magnification images of the boxed regions in A and C, respectively. Asterisks indicate lysosomes in control iNs. [B(i and ii) and D(i and ii)] High magnification images of the boxed regions in B and D, respectively. Note the abundance of lysomes in Patient 1 (B) and Patient 18 (D) iN somata compared to control iNs. Asterisks indicate lysosomes with dense concentric lamellae, a number sign (#) points to irregular shaped lysosomes filled with heterogeneous material and lipid droplets, and arrows point to small round-shaped lysosomes filled with electron-dense material. (E) Large lysosome in a control iN. Note the homogenous and round structure of the organelle. (F) Lysosomes (Lys) in the iN soma of Patient 1. (G) Lysosomes with heterogeneous material and lipid droplets in the iN soma of Patient 18. Note the irregular shape of lysosomes filled with concentric lamellae. (H, left) Lysosomal areas in iNs from four control subjects [n = 2 independent iPSC-derived iN subclones for Ctrl 1 (-c1/c2) and Ctrl 18 (-c1/c2) and one subclone from two independent hESC-derived iNs (ESC_Ctrl 1 and 2)] and two patients [n = 2 independent iPSC-derived iN subclones per each subject (Patient 1−c1/c2 and Patient 18-c1/c2)]. Data are presented as medians with 95% confidence intervals (CIs). (H, right) Pooled area of lysosomes for control and patient samples (ncontrol = 2429 lysosomes from six samples; npatient = 2918 lysosomes from four samples; P < 0.001, Mann–Whitney’s U-test). Data are presented as medians with 95% CIs. (I, left) Density of lysosomes in iNs from four control subjects [n = 2 independent iPSC-derived iN subclones for Ctrl 1 (-c1/c2) and Ctrl 18 (-c1/c2) and one subclone from two independent hESC-derived iNs (ESC_Ctrl 1-c1 and ESC_Ctrl 2-c1) and two patients (n = 2 independent iPSC-derived iN subclones per each subject (Patient 1-c1/c2 and Patient 18-c1/c2)]. Data are presented as medians with 95% CIs. (I, right) Pooled densities of lysosomes in control and patient samples (ncontrol = 58 cells from six samples; npatient = 29 cells from four samples; P = 0.702, Mann–Whitney’s U-test). Data are presented as medians with 95% CIs. Lys = lysosome, Ctrl = control; Pat = patient; ns = not significant, *** P < 0.001. Scale bars = 5 µm (A–D), 500 nm (Ai–Di, Aii–Dii, G), 200 nm (E and F).
To quantitatively assess the observed ultrastructural differences between patients’ and controls’ iNs, we quantified the number and area of lysosomes on electron micrographs of iN somata and found the area of lysosomes in patient-derived iNs to be significantly smaller compared to controls (P < 0.001, Mann–Whitney U-test, Fig. 7H). To measure the density of lysosomes in iN somata, we normalized the number of lysosomes and soma area of each cell to the total measured lysosome number and soma area, respectively, and found that the densities were comparable between iNs from patients and controls (P = 0.702, Mann–Whitney U-test, Fig. 7I).
Discussion
This series of 26 patients with pathogenic de novo ATP6V1A variants defines the phenotypic spectrum of a dominant encephalopathy with variable severity. While rapidly progressive and early lethal phenotypes are seen at the most severe end of the spectrum, DEE with progressive worsening over several years form a continuum with milder static encephalopathies with intellectual disability and epilepsy that represent the milder end. Progression of symptoms, when present, had a variable speed. DEE exhibited a relatively static course for years, or worsened at different ages, always resulting in a severe condition with no acquisition of communicative or motor skills. In the less severely affected group, including six patients only, epilepsy and cognitive impairment were moderately severe, poor language skills had emerged in some, but none developed communicative language. Although no clear signs of progression were obvious in this group at the last follow-up (mean age 10 years and 6 months; range 4–22 years), a worsening course remains possible in the longer term.
Epilepsy was a main feature in the whole series. Of the four patients with rapidly progressive course (Patients 9, 13, 15 and 26), two died between 6 weeks and 5 months having had no seizures, another died at 2 years soon after seizure onset and the fourth has had no seizures at 25 months. On the basis of MRI investigations in these patients, atrophic changes became apparent and rapidly evolved since the third month of life. Severe brain atrophy may cause fading of synchronized electrical brain activity and, paradoxically, reduce epileptogenesis. In the 20 remaining patients with epilepsy, multiple seizure types were observed with a clear prominence of infantile spasms (10/20) and early fever related seizures (8/20). Seizures were an initial symptom in 9/25, but were never considered as an isolated feature as they appeared in a context of early hypotonia in most and developmental delay in all.
A small head size (<−1 SD) could be demonstrated within the third year of life in 14/23 patients, reaching values consistent with microcephaly in at least 9/23. Although the randomness of head size measurements did not allow curves of head size growth to be precisely delineated, brain MRI showed signs of atrophy of brain and brainstem/cerebellar structures in 14/24 patients, accompanied by hypomyelination in most, or only hypomyelination in 2/24. All patients who had a repeat MRI scan, exhibited progressive atrophic changes, whose severity paralleled progression of symptoms. Clinical and imaging findings are consistent with a progressive neurodegenerative clinical course and indicate, in most patients, prenatal central nervous system impairment leading to symptoms onset early after birth or within the first year of life.
Abnormal dentition with enamel dysplasia, reported in 11 patients, is consistent with similar observations in patients with autosomal recessive osteopetrosis (OMIM #259700), caused by biallelic pathogenic variants of the TCIRG1 gene, encoding the A3 subunit of the vacuolar H+ ATPase. V-ATPase is significantly upregulated in enamel organ cells during maturation-stage11 and the R740S/R740S V-ATPase a3 osteopetrotic mouse exhibits hypoplastic and hypomineralized enamel.12 In these animals, ameloblasts contain numerous enlarged vacuoles, and osteoclasts have high lysosomal pH, which may interfere with proper enamel formation.12
How do the 20 pathogenic variants described in the 26 patients possibly alter ATP6V1A function? The ATP6V1A gene is constrained to genetic variations as suggested by the gnomAD missense Z-score and the residual variation intolerance score and is predicted by DOMINO to be compatible with a dominant inheritance model, as we demonstrated in a previous study.7 Here, we strengthen the association of the ATP6V1A gene with a dominant encephalopathy by describing 26 patients 20 of whom, harbouring 14 novel pathogenic variants, are newly reported.7–9 Overall, these 26 patients harbour 20 unique, and five recurrent, de novo missense substitutions not reported in the Exome Aggregation Consortium, gnomAD, Human Genetic Variation Database and BRAVO databases and predicted to be deleterious by in silico tools [PolyPhen-2, Sorting Intolerant From Tolerant (SIFT), PROVEAN, Combined Annotation Dependent Depletion (CADD), Mendelian Clinically Applicable Pathogenicity (M-CAP), MetaLR and MetaSVM]. Missense badness, PolyPhen-2 and constraint and missense variant pathogenicity prediction scores for these 20 pathogenic variants are significantly different from those observed for naturally occurring variants reported in gnomAD database, which further supports pathogenicity. In addition, MTR scores based on regional depletion in the general population, highlighted a below-neutrality threshold for 19/20 residues (all but p.Pro27Arg) affected by the de novo substitutions. The MTR score does not include information about purifying selection within protein tertiary structures and this could explain why the p.Pro27Arg variant is assigned a neutral score. Indeed, the structural modelling showed that p.Pro27Arg perturbates the ATP6V1A/B interaction.7 The regions with the lowest MTR scores were observed in the ATP-synt_ab domain, known to be genome-wide significantly constrained.10 Seventeen/20 substitutions (85%) were located in this domain, suggesting an enrichment of pathogenic ATP6V1A variants in it.
We previously mapped the four substitutions p.Pro27Arg, p.Asp100Tyr, p.Asp349Asn and p.Asp371Gly on the E. hirae crystal structure (PDB code 3VR6),13 the Saccharomyces cerevisiae cryo-EM structure (PDB code 3J9T),14 and a homology model of the human A subunit generated by us, showing that the variant sites are in the same locations in the three proteins.7 Recently, the cryo-EM structure of a V-ATPase from Rattus norvegicus has become available.15 The subunit A of this enzyme shows 98% sequence identity with the human ATP6V1A, and its structure differs from our homology model by a root mean squared deviation of 2.8Å (1.9Å without considering the helical C-terminal domain). Therefore, the four substitutions analysed in our initial study7 are in the same locations also in the R. norvegicus Atp6v1a subunit, and we resolved to map the entire set of substitutions on this structure.
When mapped on the rat structure, most of the pathogenic variants (13/20) resulted either close to the P-loop involved in ATP binding or in the A/B interface at the side of the catalytic site, thus possibly altering V-ATPase activity. Three other pathogenic variants mapped at the A/D and the non-catalytic A/B interface and could hence alter V-ATPase rotation or the interaction between subunits. The remaining four substitutions mapped in less specific regions of the protein but still appear to affect local amino acidic interactions in ways that could possibly result in decreased protein stability and expression. For Patient 1, bearing one of these pathogenic variants (Asp100Tyr), loss of expression of the ATP6V1A subunit was confirmed by western blot analysis, suggesting that in this patient (and possibly in all others harbouring pathogenic variants affecting protein stability), an impaired expression forms the basis of the pathogenic process.
The number of patients carrying the various pathogenic variants is not sufficient to validate genotype-phenotype correlation with statistics. However, by correlating the position of the pathogenic variants in the rat Atp6v1a 3D structure with phenotype severity, we found that variants affecting protein folding, perturbing the catalytic site or mapping near the P-loop containing the ATP binding site are more frequently associated with severe phenotypes (Supplementary Table 6). In addition, a correlation between the specific amino acid substitution and phenotype severity can be assumed. Indeed, Patient 12, carrying the p.Asn314Ser substitution, exhibited a milder phenotype than Patient 14, carrying the more perturbing p.Asn314Asp substitution. A strikingly severe phenotype requiring neonatal intensive care support at birth and resulting in early death was observed in both, unrelated, patients carrying the p.Ser316Phe substitution (Patients 15 and 26) The p.Pro412 residue, located in a highly constrained region of ATP-synt_ab domain and involved in the two different missense substitutions p.Pro412Leu and p.Pro412Ser (Patients 9 and 13), resulted in profound phenotypes with early progressive severe brain atrophy.
V-ATPase is known to be expressed at endo-lysosomal membranes to guarantee proper acidification of the degradative organelles for efficient enzymatic activity. Thus, acidification defects are usually linked with a progressive accumulation of non-degraded material inside lysosomes, a hallmark of lysosomal storage diseases. Accordingly, ultrastructural analysis of fibroblasts from Patients 1, 2, 16 and 18 showed enlarged single membrane intracellular structures, reminiscent of autolysosomes, containing heterogeneous materials, with accumulated lipids among other degradative substrates. We further confirmed these findings in iPSC-derived iNs. Particularly, in the somata of iNs from Patients 1 and 18, we found an accumulation of lysosomes with lamellar structures, lysosomes filled with dark and light material, resembling lipid droplets and many small lysosomes filled with electron-dense material. In addition, we found the area of lysosomes to be significantly decreased in patients’ iNs. Together with the ultrastructural diversity of iN lysosomes, this suggests that a functionally impaired ATP6V1A protein alters lysosomal maturation and function.
The overall clustering of the residues associated to pathogenic variants indicates that altered V-ATPase catalytic function or stability forms the basis of the abnormal phenotype and, together with ultrastructural data, clearly suggests the primarily lysosomal nature of pathology. Fibroblasts of two severely affected patients (Patients 1 and 18) exhibited reduced Lysotracker and LAMP1 labelling with increased endo-lysosomal pH, suggesting that the filled structures observed by EM experience increased pH and reduced expression of the glycoprotein LAMP1. Reduced proton pump activity may result from both loss of function of the ATP6V1A subunit as a consequence of impaired catalytic activity (in line with the Glu356Asp pathogenic variant of Patient 18 mapping at the A/B interface at the catalytic side), or from loss of expression due to protein degradation (as demonstrated for Asp100Tyr in Patient 1). Impaired lysosomal acidification occurs in several progressive lysosomal disorders16 and has been described in a patient with fulminant neurodegeneration carrying a de novo splice site pathogenic variant in the mammalian V-ATPase accessory factor ATP6AP2.17 Impaired lysosomal acidification with decreased Lysotracker signal was also recently shown in cells expressing ATP6V0A1 missense pathogenic variants, resulting in DEE.18 Fibroblasts of two less severely affected patients in our series (Patients 2 and 16), exhibiting mild–moderate intellectual disability and epilepsy, revealed instead increased Lysotracker signal with no alteration of LAMP1 labelling and decreased endo-lysosomal pH. Although the molecular mechanism leading to the observed cellular phenotype needs further investigation, these findings suggest an increased V-ATPase function with altered acidic composition of the endo-lysosomal compartment. More acidic pH at intracellular organelles has been reported to similarly result in cytoplasmic inclusion with non-degraded materials due to reduced lysosomal hydrolase activity at the lowered pH, accumulation of aberrant lysosomal structures or both.19 Hyperacidification of intracellular organelles has been associated with other neurodevelopmental disorders characterized by intellectual disability and seizures and caused by loss of function pathogenic variants in vesicular Na+/H+ exchanger genes.20
Our data indicate that dominant pathogenic variants in ATP6V1A lead to defective lysosomal function, due to altered acidification. It is expected that in an altered environment, outside the correct pH range, multiple enzymes operate sub-optimally leading to storage of the electron-dense macromolecules that appear as lamellar and lipidic material in lysosomes of fibroblasts and iNs. These findings are supported by the measurement of IDUA, ABG and alpha-galactosidase enzymatic activity. When enzyme activity was measured in vitro after fibroblasts lysis and at optimal pH guaranteed by the specific buffer used for quantification (3.4 for IDUA, 4.6 for alpha-galactosidase and 5.1 for ABG),21,22 no differences were observed between patients and controls, indicating that ATP6V1A mutations do not affect enzyme activity directly. Conversely, concentrations of the biomarkers we tested differed in patients’ versus controls’ cells, suggesting that, in vivo, altered acidification induced by ATP6V1A mutations creates a sub-optimal catalytic environment that affects enzyme activity. This is certainly but a key element in the chain of downstream effects leading to the cumulative differences observed in patients’ versus controls’ cells.
Since lysosomes are also key players in cellular signalling and nutrient sensing, multiple cell compartments may indeed suffer the downstream consequences of impaired endo-lysosomal acidification. Mouse embryonic fibroblasts lines obtained after V-ATPase inhibition or Atp6v1h silencing showed cellular iron deficiency, due to compromised iron release from lysosomes, resulting in impaired mitochondrial function and non-apoptotic cell death. These effects, accompanied by an inflammatory response, were also observed in a mouse model of impaired lysosomal acidification and were reversed by supplementation of the mouse diet with iron, which bypasses the endo-lysosomal pathway.23 Therefore, impaired endo-lysosomal acidification may also result in an impaired iron homeostatic cycle and cause dysfunctional lysosomes to result in dysfunctional mitochondria.
Lysosomal disorders are caused by pathogenic variants in genes encoding lysosomal proteins, such as lysosomal glycosidases, proteases, integral membrane proteins, transporters, enzyme modifiers or activators.24 Most are inherited as autosomal recessive traits, and only three are X-linked. Therefore, biallelic or hemizygous pathogenic variants are necessary to cause disease via a critical reduction of the resulting enzymatic activity. In patients with heterozygous ATP6V1A pathogenic variants, however, the consequences of the activity of multiple enzymes being simultaneously impaired are sufficient to cause disease.
Mutations affecting stability and assembly of V-ATPase subunits have been associated to altered glycosylation pathways, as supported by serum glycome investigations in patients with biallelic ATP6V0A2, ATP6V1E1 and ATP6V1A variants and complex multisystem phenotypes including cutis laxa, suggesting a defective glycan processing due to impaired Golgi trafficking. The reported glycosylation changes were less pronounced in ATP6V1A- than in ATP6V0A2- and ATP6V1E1-related disorders, probably depending on differential tissue-specific expression of the component subunits.5,25 These earlier results prompted us to verify the glycosylation status of two of our severely affected patients (Patients 1: p.Asp100Tyr and 18: p.Glu365Asp) (Supplementary Table 6). We found MS profiles of serum transferrin and total serum N-glycans to reveal minor N-glycome changes due to a small increase of hyposialylated glycoforms, mostly the biantennary monosialylated ones. This sialylation defect (more evident in Patient 18) was in line with previous findings in biallelic ATP6V1A patients (PIII:1 with homozygous Arg338Cys mutation and PIV:1 with Gly72Asp mutation), showing only minor transferrin hyposialylation caused by lack of a single sialic acid.5 In addition, we observed in both MS glycoprofiles slight increased fucosylation (more evident in Patient 1) that contributed to the general mild N-glycosylation disarrangement. A more in-depth serum transferrin MS/MS glycosylation analysis revealed a specific increase of the antennary fucosylation in our patients. As ATP6V1A is widely expressed, the observed deviation from physiological glycosylation could derive from either reduced activity of pH-dependent enzymes, such as sialyltransferases and fucosyltransferase, and/or from their mis-localization, due to non-optimal acidification of Golgi cisternae, or both.26 An additional downstream impact in Golgi homeostasis by the altered lysosomal acidification is possible, as also hypothesized for ATP6AP2 deficiency.27
Altered antennary fucosylation of plasma/serum glycoprotein has been documented in different diseases,28–30 including developmental disorders.31 Identification and quantitation of antennary fucosylation is gaining primary interest as it may reflect the total incidence of Lewis x/sialyl-Lewis x structural epitopes influencing glycoproteins functionality.32 Although the contribution of the mild and variable overall serum glycosylation changes to ATP6V1A phenotypes is difficult to ascertain, our findings further point to a link between lysosomal dysfunction and impaired glycosylation.
ATP6V1A is a endolysosomal membrane protein, and ATP6V1A encephalopathy features seizures, intellectual disability and motor disorders in line with other diseases resulting from pathogenic variants affecting this protein category.24 ATP13A2 is also a lysosomal membrane protein, but disorders caused by pathogenic variants in its coding gene cause a form of recessive parkinsonism with dementia of juvenile onset that is considered to be a neuronal ceroid lipofuscinosis due to electron microscopy findings.24,33 ATP13A2 has been established as a lysosomal polyamine exporter across lysosomes and early/late endosomes in neurons.34–36 Knockdown of the Atp13a2 gene in cultured neurons causes accumulation and enlargement of lysosomes, decreased lysosomal degradation and accumulation of autophagosomes and alpha-synuclein (OMIM #163890), resulting in neuronal toxicity.37 A role for specific heterozygous ATP13A2 variants in patients with early-onset Parkinson disease has been supported by rare reports and cellular studies.36 Pathogenic variants in other members/regulators of the V-ATPase complex have been recently associated with neurodevelopmental disorders and result in severely impaired brain development when modelled in mice.17,18 In addition, in a multiomics profiling from cortical areas of patients with Alzheimer’s disease, ATP6V1A deficit has been associated with neuronal impairment and neurodegeneration. In ATP6V1A silenced neurons, network activity was significantly reduced with increased immature spikes and alteration of synaptic proteins.38 This observation further suggests a role of ATP6V1A in neuronal maturation and activity, as we proposed for neurons expressing ATP6V1A pathogenic variants.7
In conclusion, ATP6V1A encephalopathy provides a new paradigm in the group of lysosomal diseases as it derives from a endo-lysosomal membrane protein dysfunction resulting in altered lysosomal homeostasis. Its pathophysiology is complex, as it implies intracellular accumulation of substrates whose composition remains unclear, as well as the combination of structural brain abnormalities that are established early during prenatal brain development and progressive neurodegenerative changes whose severity is variably dictated by specific pathogenic variants.
Supplementary Material
Acknowledgements
We thank all patients and family members for their participation in this study. We thank Melanie Pieraks and Eva Reinthaler (Neurolentech, Austria) for generating the human iPSC lines and for performing quality checks. We thank Vanessa Zheden and Daniel Gütl for their excellent technical support in the specimen preparation for transmission electron microscopy and Flavia Leite for preparing the lentiviruses. The support from Electron Microscopy Facility and Molecular Biology Services at IST Austria is greatly acknowledged. We would like to thank Doctors Jane Hurst and Richard Scott for their help in retrieving the detailed clinical information of Patient 17. The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. See Supplementary Material for Undiagnosed Disease Network consortium details. Genetic information on Patient 23 was made available through access to the data and findings generated by the 100 000 Genomes Project; www.genomicsengland.co.uk (to K.L.).
Abbreviations
- ATP-synt_ab
ATP synthase α/β family, nucleotide-binding domain
- DEE
developmental and epileptic encephalopathy
- iN
iPSC-derived neuron
- iPSC
induced pluripotent stem cell
- MTR
missense tolerance ratio
- V-ATPase
vacuolar-type (H+) ATPase
Appendix 1
ATP6V1A collaborators. Full details are provided in the Supplementary material.
Martino Montomoli, Claudia Bianchini, Damien Sternberg, Laurent Frobert and Elena Parrini.
Contributor Information
Renzo Guerrini, Neuroscience Department, Children's Hospital Meyer, University of Florence, Florence, Italy.
Davide Mei, Neuroscience Department, Children's Hospital Meyer, University of Florence, Florence, Italy.
Katalin Kerti-Szigeti, Institute of Science and Technology Austria (ISTA), Klosterneuburg, Austria.
Sara Pepe, Department of Experimental Medicine, University of Genoa, Italy; Center for Synaptic Neuroscience and Technology, Istituto Italiano di Tecnologia, Genova, Italy.
Mary Kay Koenig, Department of Pediatrics, Division of Child and Adolescent Neurology, The University of Texas McGovern Medical School, Houston, TX, USA.
Gretchen Von Allmen, Department of Pediatrics, Division of Child and Adolescent Neurology, The University of Texas McGovern Medical School, Houston, TX, USA.
Megan T Cho, GeneDx, Gaithersburg, MD 20877, USA.
Kimberly McDonald, Pediatric Neurology, University of Mississippi Medical Center, Jackson, MS, USA.
Janice Baker, Genetics and Genomics, Children's Minnesota, Minneapolis, MN, USA.
Vikas Bhambhani, Genetics and Genomics, Children's Minnesota, Minneapolis, MN, USA.
Zöe Powis, Ambry Genetics, Aliso Viejo, CA, USA.
Lance Rodan, Division of Genetics and Genomics and Department of Neurology, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA.
Rima Nabbout, Reference Centre for Rare Epilepsies, Department of Genetics, Necker Enfants Malades Hospital, APHP, member of ERN EpiCARE, Université de Paris, Paris, France.
Giulia Barcia, Reference Centre for Rare Epilepsies, Department of Genetics, Necker Enfants Malades Hospital, APHP, member of ERN EpiCARE, Université de Paris, Paris, France.
Jill A Rosenfeld, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
Carlos A Bacino, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
Cyril Mignot, APHP, Sorbonne Université, Départément de Génétique, Centre de Référence Déficiences Intellectuelles de Causes Rares, Paris, France; Institut du Cerveau (ICM), UMR S 1127, Inserm U1127, CNRS UMR 7225, Sorbonne Université, 75013 Paris, France.
Lillian H Power, Pediatric Neurology, Stead Family Department of Pediatrics, University of Iowa Stead Family Children’s Hospital, Iowa City, IA, USA.
Catharine J Harris, Department of Pediatric Genetics, University of Missouri Medical Center, Columbia, MO 65212, USA.
Dragan Marjanovic, Danish Epilepsy Centre Filadelfia, Adult Neurology, Dianalund, Denmark.
Rikke S Møller, Department of Epilepsy Genetics and Personalized Treatment, Danish Epilepsy Center Filadelfia, Dianalund, Denmark; Department of Regional Health Services, University of Southern Denmark, Odense, Denmark.
Trine B Hammer, Department of Epilepsy Genetics and Personalized Treatment, Danish Epilepsy Center Filadelfia, Dianalund, Denmark.
The DDD Study, Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK.
Riikka Keski Filppula, Department of Clinical Genetics, Oulu University Hospital, Medical Research Center Oulu and PEDEGO Research Unit, University of Oulu, Oulu, Finland.
Päivi Vieira, Clinic for Children and Adolescents, Oulu University Hospital, Medical Research Center Oulu and PEDEGO Research Unit, University of Oulu, Oulu, Finland.
Clara Hildebrandt, Division of Genetics and Genomics, Metabolism Program, Boston Children's Hospital, Boston, MA, USA.
Stephanie Sacharow, Boston Children’s Hospital, Harvard Medical School, Boston, MA, USA.
Undiagnosed Diseases Network, Common Fund, National Institutes of Health, Bethesda, MD 20892, USA.
Luca Maragliano, Center for Synaptic Neuroscience and Technology, Istituto Italiano di Tecnologia, Genova, Italy; Department of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy.
Fabio Benfenati, Center for Synaptic Neuroscience and Technology, Istituto Italiano di Tecnologia, Genova, Italy; IRCCS Ospedale Policlinico San Martino, Genova, Italy.
Katherine Lachlan, Wessex Clinical Genetics Service, University Hospital Southampton NHS Foundation Trust, Southampton, UK; Human Development and Health, Faculty of Medicine University of Southampton, Southampton, UK.
Andreas Benneche, Department of Medical Genetics, Haukeland University Hospital, Bergen, Norway.
Florence Petit, CHU Lille, Clinique de Génétique, F-59000 Lille, France.
Jean Madeleine de Sainte Agathe, Laboratoire de Biologie Médicale Multi Sites SeqOIA, Laboratoire de Médecine Génomique, APHP. Sorbonne Université, Paris, France.
Barbara Hallinan, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA; Division of Child Neurology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Yue Si, GeneDx, Gaithersburg, MD 20877, USA.
Ingrid M Wentzensen, GeneDx, Gaithersburg, MD 20877, USA.
Fanggeng Zou, GeneDx, Gaithersburg, MD 20877, USA.
Vinodh Narayanan, Neurogenomics Division, Center for Rare Childhood Disorders, Translational Genomics Research Institute (TGen), Phoenix, AZ 85012, USA.
Naomichi Matsumoto, Department of Human Genetics, Yokohama City University Graduate School of Medicine, Yokohama, Japan.
Alessandra Boncristiano, Neuroscience Department, Children's Hospital Meyer, University of Florence, Florence, Italy.
Giancarlo la Marca, Newborn Screening, Clinical Chemistry and Pharmacology Laboratory, Meyer Children’s University Hospital, Florence, Italy; Department of Experimental and Clinical Biomedical Sciences, University of Florence, Florence, Italy.
Mitsuhiro Kato, Department of Pediatrics, Showa University School of Medicine and Epilepsy Medical Center, Showa University Hospital, Tokyo, Japan.
Kristin Anderson, Founder and Research Liaison, ‘ATP6V1A Families' Facebook group.
Carmen Barba, Neuroscience Department, Children's Hospital Meyer, University of Florence, Florence, Italy.
Luisa Sturiale, CNR, Institute for Polymers, Composites and Biomaterials, IPCB, 95126 Catania, Italy.
Domenico Garozzo, CNR, Institute for Polymers, Composites and Biomaterials, IPCB, 95126 Catania, Italy.
Roberto Bei, Department of Clinical Sciences and Translational Medicine, University of Rome ‘Tor Vergata', Rome, Italy.
Laura Masuelli, Department of Experimental Medicine, University of Rome ‘Sapienza', Rome, Italy.
Valerio Conti, Neuroscience Department, Children's Hospital Meyer, University of Florence, Florence, Italy.
Gaia Novarino, Institute of Science and Technology Austria (ISTA), Klosterneuburg, Austria.
Anna Fassio, Department of Experimental Medicine, University of Genoa, Italy; IRCCS Ospedale Policlinico San Martino, Genova, Italy.
Funding
This work was supported by the EU 7th Framework Programme (FP7) under the project DESIRE grant N602531 (to R.G.); the Regione Toscana under the Call for Health 2018 (grant DECODE-EE) (to R.G.); the ‘Brain Project’ by Fondazione Cassa di Risparmio di Firenze (to R.G.); IRCCS Ospedale Policlinico San Martino 5×1000 and Ricerca Corrente (to A.F. and F.B.). The European Reference Network (ERN) for rare and complex epilepsies (EpiCARE) provided financial support for meetings organization. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of Wellcome or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). This study makes use of DECIPHER (https://www.deciphergenomics.org), which is funded by Wellcome. K.K.-S. was supported by the ISTplus fellowship.
Competing interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories (J.A.R., C.A.B.). Y.S., I.M.W. and F.Z. are employees of GeneDx, Inc. Z.P. is a past employee of Ambry Genetics. All other authors declare they have no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Forgac M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. [DOI] [PubMed] [Google Scholar]
- 2. Cotter K, Capecci J, Sennoune S, et al. Activity of plasma membrane V-ATPases is critical for the invasion of MDA-MB231 breast cancer cells. J Biol Chem. 2015;290:3680–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morel N, Poëa-Guyon S. The membrane domain of vacuolar H(+)ATPase: a crucial player in neurotransmitter exocytotic release. Cell Mol Life Sci. 2015;72:2561–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodzęta A, Kahms M, Klingauf J. The presynaptic v-ATPase reversibly disassembles and thereby modulates exocytosis but is not part of the fusion machinery. Cell Rep. 2017;20:1348–1359. [DOI] [PubMed] [Google Scholar]
- 5. Van Damme T, Gardeitchik T, Mohamed M, et al. Mutations in ATP6V1E1 or ATP6V1A cause autosomal-recessive cutis laxa. Am J Hum Genet. 2017;100:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogt G, El Choubassi N, Herczegfalvi Á, et al. Expanding the clinical and molecular spectrum of ATP6V1A related metabolic cutis laxa. J Inherit Metab Dis. 2021;44:972–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fassio A, Esposito A, Kato M, et al. De novo mutations of the ATP6V1A gene cause developmental encephalopathy with epilepsy. Brain. 2018;141:1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosch DGM, Boonstra FN, de Leeuw N, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet. 2016;24:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deciphering Developmental Disorders Study . Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Havrilla JM, Pedersen BS, Layer RM, Quinlan AR. A map of constrained coding regions in the human genome. Nat Genet. 2019;51:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarkar J, Wen X, Simanian EJ, Paine ML. V-type ATPase proton pump expression during enamel formation. Matrix Biol. 2016:52-54:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson L, Ganss B, Wang A, et al. V-ATPases containing a3 subunit play a direct role in enamel development in mice. J Cell Biochem. 2017;118:3328–3340. [DOI] [PubMed] [Google Scholar]
- 13. Arai S, Yamato I, Shiokawa A, et al. Reconstitution in vitro of the catalytic portion (NtpA3-B3-D-G complex) of Enterococcus hirae V-type Na+-ATPase. Biochem Biophys Res Commun. 2009;390:698–702. [DOI] [PubMed] [Google Scholar]
- 14. Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. [DOI] [PubMed] [Google Scholar]
- 15. Abbas YM, Wu D, Bueler SA, Robinson CV, Rubinstein JL. Structure of V-ATPase from the mammalian brain. Science. 2020;367:1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification–the emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev. 2016;32:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirose T, Cabrera-Socorro A, Chitayat D, et al. ATP6AP2 variant impairs CNS development and neuronal survival to cause fulminant neurodegeneration. J Clin Invest. 2019;129:2145–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aoto K, Kato M, Akita T, et al. ATP6V0A1 encoding the a1-subunit of the V0 domain of vacuolar H+-ATPases is essential for brain development in humans and mice. Nat Commun. 2021;12:2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicoli E-R, Weston MR, Hackbarth M, et al. Lysosomal storage and albinism due to effects of a de novo CLCN7 variant on lysosomal acidification. Am J Hum Genet. 2019;104:1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kondapalli KC, Prasad H, Rao R. An inside job: how endosomal Na(+)/H(+) exchangers link to autism and neurological disease. Front Cell Neurosci. 2014;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Scott CR, Chamoles NA, et al. Direct multiplex assay of lysosomal enzymes in dried blood spots for newborn screening. Clin Chem. 2004;50:1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blanchard S, Sadilek M, Scott CR, Turecek F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: application to screening newborns for mucopolysaccharidosis I. Clin Chem. 2008;54:2067–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yambire KF, Rostosky C, Watanabe T, et al. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. eLife. 2019;8:e51031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Platt FM, D’Azzo A, Davidson BL, Neufeld EF, Tifft CJ. Lysosomal storage diseases. Nat Rev Dis Primers. 2018;4:27. [DOI] [PubMed] [Google Scholar]
- 25. Kornak U, Reynders E, Dimopoulou A, et al. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat Genet. 2008;40:32–34. [DOI] [PubMed] [Google Scholar]
- 26. Linders PTA, Peters E, Ter Beest M, Lefeber DJ, van den Bogaart G. Sugary logistics gone wrong: membrane trafficking and congenital disorders of glycosylation. Int J Mol Sci. 2020;21:4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rujano MA, Serio MC, Panasyuk G, et al. Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J Exp Med. 2017;214:3707–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Vroome SW, Holst S, Girondo MR, et al. Serum N-glycome alterations in colorectal cancer associate with survival. Oncotarget. 2018;9:30610–30623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doherty M, Theodoratou E, Walsh I, et al. Plasma N-glycans in colorectal cancer risk. Sci Rep. 2018;8:8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thanabalasingham G, Huffman JE, Kattla JJ, et al. Mutations in HNF1A result in marked alterations of plasma glycan profile. Diabetes. 2013;62:1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pivac N, Knezević A, Gornik O, et al. Human plasma glycome in attention-deficit hyperactivity disorder and autism spectrum disorders. Mol Cell Proteomics. 2011;10:M110.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider M, Al-Shareffi E, Haltiwanger RS. Biological functions of fucose in mammals. Glycobiology. 2017;27:601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bras J, Verloes A, Schneider SA, Mole SE, Guerreiro RJ. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipofuscinosis. Hum Mol Genet. 2012;21:2646–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Veen S, Martin S, Van den Haute C, et al. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature. 2020;578:419–424. [DOI] [PubMed] [Google Scholar]
- 35. Schultheis PJ, Fleming SM, Clippinger AK, et al. Atp13a2-deficient mice exhibit neuronal ceroid lipofuscinosis, limited α-synuclein accumulation and age-dependent sensorimotor deficits. Hum Mol Genet. 2013;22:2067–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Podhajska A, Musso A, Trancikova A, et al. Common pathogenic effects of missense mutations in the P-type ATPase ATP13A2 (PARK9) associated with early-onset parkinsonism. PLoS ONE. 2012;7:e39942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Usenovic M, Knight AL, Ray A, et al. Identification of novel ATP13A2 interactors and their role in α-synuclein misfolding and toxicity. Hum Mol Genet. 2012;21:3785–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang M, Li A, Sekiya M, et al. Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer’s disease. Neuron. 2021;109:257–272.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. Further derived data not provided in the article due to space limitations are available from the corresponding author on request.