Summary
Carotenoids contribute to fruit coloration and are valuable sources of provitamin A in the human diet. Abscisic acid (ABA) plays an essential role in fruit coloration during citrus fruit ripening, but little is known about the underlying mechanisms. Here, we identified a novel bZIP transcription activator called CsbZIP44, which serves as a central regulator of ABA‐mediated citrus carotenoid biosynthesis. CsbZIP44 directly binds to the promoters of four carotenoid metabolism‐related genes (CsDXR, CsGGPPs, CsBCH1 and CsNCED2) and activates their expression. Furthermore, our research indicates that CsHB5, a positive regulator of ABA and carotenoid‐driven processes, activates the expression of CsbZIP44 by binding to its promoter. Additionally, CsHB5 interacts with CsbZIP44 to form a transcriptional regulatory module CsHB5‐CsbZIP44, which is responsive to ABA induction and promotes carotenoid accumulation in citrus. Interestingly, we also discover a positive feedback regulation loop between the ABA signal and carotenoid biosynthesis mediated by the CsHB5‐CsbZIP44 transcriptional regulatory module. Our findings show that CsHB5‐CsbZIP44 precisely modulates ABA signal‐mediated carotenoid metabolism, providing an effective strategy for quality improvement of citrus fruit and other crops.
Keywords: citrus, abscisic acid (ABA), carotenoid, CsbZIP44, CsHB5, transcriptional regulatory module
Introduction
For fleshy fruits, the ripening process plays important roles in determining their quality, economic benefits and nutrition in human diets (Adams‐Phillips et al., 2004). Therefore, it is crucial to elucidate the regulatory mechanisms of fruit ripening. Fruit ripening involves several notable changes, such as fruit coloration, fruit softening, accumulation of soluble sugars and volatile compounds, as well as changes in phytohormone levels (Forlani et al., 2019; Karlova et al., 2014; Li et al., 2017).
Carotenoid metabolism can directly determine citrus fruit coloration, with significant impacts on the product quality and consumer acceptance (Carmona et al., 2012; Sun et al., 2021). Citrus (Citrus spp.) is one of the world's most important fleshy fruit crops with approximately 115 different types of carotenoids, showing the highest diversity of carotenoids among fruits (Fanciullino et al., 2006; Ikoma et al., 2016). The carotenoids accumulated in citrus fruits enhance the fruit nutritional value and promote human health as essential diet components (Sun et al., 2018). The carotenoid metabolism pathway has been well characterized in higher plants including citrus, particularly the key enzymes and their functions (Ikoma et al., 2014; Kato et al., 2004; Rodrigo et al., 2013).
Within the carotenoid biosynthesis pathway, 1‐Deoxy‐D‐xylulose 5‐phosphate reductoisomerase (DXR), as a rate‐limiting enzyme of the methylerythritol 4‐phosphate pathway, determines the abundance of precursors required for carotenoid biosynthesis. Geranylgeranyl diphosphate synthase (GGPPs) catalyses the synthesis of geranylgeranyl diphosphate (GGPP), which serves as the foundational step in carotenoid metabolism. Through subsequent desaturation, isomerization and cyclization reactions, the carotenoid pathway differentiates into the α‐ and β‐branch. Key enzymes in citrus, such as β‐carotene hydroxylases (BCHs) and 9‐cis‐epoxycarotenoid dioxygenases (NCEDs), play crucial roles in the biosynthesis of xanthophylls, contributing to the coloration of citrus fruits.
Carotenoid metabolism in citrus is influenced by multiple factors, including light, temperature, biotic and abiotic stresses and phytohormones (Gong et al., 2021; Luan et al., 2020; Sun et al., 2021). Citrus fruit is typically categorized as a non‐climacteric fruit, and extensive research has demonstrated the crucial role of abscisic acid (ABA) in regulating the formation of several ripening‐related traits, particularly carotenoid biosynthesis (Rodrigo et al., 2003; Romero et al., 2019; Wu et al., 2014). These studies suggest the existence of an integrated regulatory network controlling ABA‐mediated carotenoid metabolism in citrus. However, the molecular mechanisms underlying this process remain relatively poorly understood.
An increasing body of evidence supports the involvement of transcription factors (TFs) in regulating carotenoid and ABA metabolisms. For instance, the peach ERF TF, PpERF3, has been shown to positively regulate ABA biosynthesis by activating the expression of PpNCED2/3, leading to fruit coloration during ripening (Wang et al., 2019). In tomato fruit, the expression of SlERF6 and SlDET1 is negatively correlated with carotenoid levels (Davuluri et al., 2005; Lee et al., 2012). In Citrus reticulate, the TF CrMYB68 acts as a negative regulator of ABA and carotenoid metabolisms by downregulating the expression of CrBCH2 and CrNCED5 (Zhu et al., 2017). Another TF, CsHB5, which belongs to the homeodomain leucine zipper I (HD‐ZIP I) family, activates the expression of CsBCH1 and CsNCED2 by directly binding to their promoters, thereby regulating ABA‐induced ripening in citrus fruit (Zhang et al., 2021). Despite extensive research on the transcriptional regulation of carotenoid and ABA metabolisms in various plants, the carotenoid biosynthesis mediated by ABA signalling remains poorly understood.
Among various TFs, the basic region/leucine zipper (bZIP) TF stands out as one of the largest gene families in plants, playing pivotal roles in response to ABA signals and regulating pigment metabolisms. For instance, in apple, the bZIP TF MdbZIP4‐like directly binds to the promoter of MdMYB114, enhancing its transcript level and promoting anthocyanin accumulation (Jiang et al., 2021). Another ABA‐induced bZIP TF, MdbZIP44, acts as a partner with MdMYB1 to regulate anthocyanin biosynthesis in apple fruit (An et al., 2018). Additionally, the bZIP TF MdABI5 positively regulates the ABA‐induced regulatory module MdMYB1‐MdbHLH3, enhancing anthocyanin accumulation (An et al., 2021). These studies highlight the vital regulatory roles of bZIP TFs in modulating ABA‐induced anthocyanin metabolism. However, the involvement of bZIP TFs in ABA‐mediated carotenoid biosynthesis remains largely unknown.
In this study, we discovered the crucial role of a bZIP TF, CsbZIP44, in determining ABA‐mediated carotenoid biosynthesis by activating the expression of carotenogenic genes. Additionally, we observed that CsHB5 interacts with and activates CsbZIP44, which serves as a positive regulator of ABA biosynthesis and fruit ripening. The interaction and transcriptional activation between CsHB5 and CsbZIP44 further enhance their regulatory functions in promoting ABA‐induced carotenoid accumulation. Furthermore, we found a positive feedback regulation loop between the ABA signal and functions of the CsHB5‐CsbZIP44 module. Our findings prove that the transcriptional regulator module CsHB5‐CsbZIP44 plays a positive role in regulating carotenoid biosynthesis under ABA signalling.
Results
ABA‐mediated carotenoid biosynthesis is essential for citrus fruit coloration
Citrus fruits are non‐climacteric fruits, and ABA plays a crucial role in their coloration during development and maturation (Rodrigo et al., 2006; Romero et al., 2019; Wu et al., 2014). Here, we investigated the connection between ABA and carotenoid metabolisms by measuring endogenous ABA and carotenoid levels in citrus peel throughout fruit ripening. The total carotenoid and ABA content exhibited continuous increases from 170 DAFB (days after full blossom) to 230 DAFB, coinciding with peel coloration (Figure 1a–c). Additionally, a strong positive correlation (R 2 = 0.9127) was observed between the ABA content and total carotenoid content during citrus fruit ripening (Figure 1d). These findings suggest a close relationship between ABA and fruit coloration.
Figure 1.
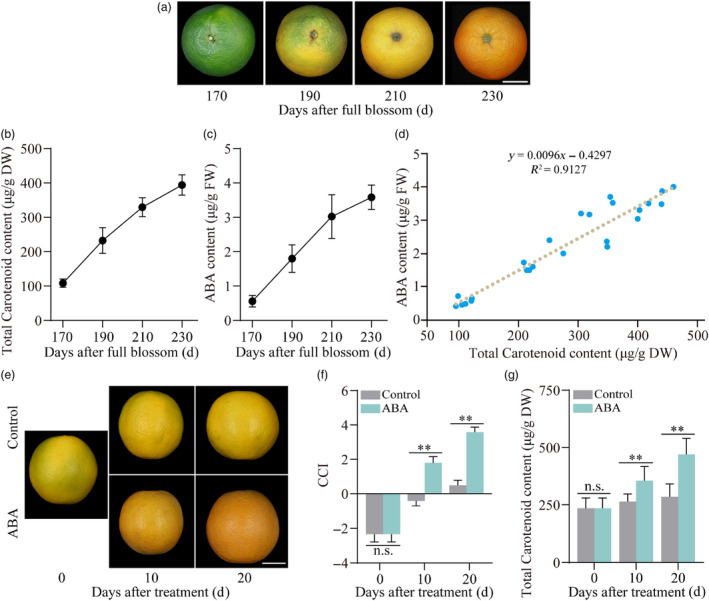
Abscisic acid (ABA) is closely related to peel coloration and carotenoid biosynthesis. (a–c) Changes in fruit coloration, carotenoid and ABA contents during ‘Valencia’ orange fruit (Citrus sinensis Osbeck.) ripening. (a) Fruit coloration. Bars = 3 cm. Contents of total carotenoid (μg/g DW) (b) and ABA (μg/g FW) (c). (d) Correlation analysis of ABA and carotenoid content during ‘Valencia’ orange fruit ripening. (e–g) ABA treatment promotes peel coloration and carotenoid accumulation. (e) Phenotype of ‘Valencia’ orange fruit under various treatments. Bars = 3 cm. The same set of citrus fruits used in the control and ABA‐treated groups on day 0 of treatment. (f) Effect of ABA treatment on citrus colour index (CCI). Positive values for red‐yellow, negative values for blue‐green and 0 for an intermediate mixture of red, yellow and blue‐green. (g) Total carotenoid content (μg/g DW). Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
To further investigate the impact of ABA on fruit coloration, different treatments were administered to citrus fruits on the tree at 190 DAFB. ABA treatment significantly enhanced peel coloration when compared to the control group (sprayed with sterile water). Conversely, no similar changes were observed in the NDGA (nordihydroguaiaretic acid, an ABA antagonist) treatment at 10 days after treatment (DAT; Figure S1A). The citrus colour index (CCI) value and total carotenoid content consistently demonstrated identical outcomes (Figure S1B,C). These findings further highlight the essential role of ABA in fruit coloration during citrus fruit maturation.
In addition, fruits harvested at 190 DAFB were subjected to various treatments to further elucidate the mechanism by which ABA mediates fruit coloration. All treated fruits were stored in a dark phytotron at room temperature for 20 days, and sampling was conducted at 10‐day intervals. Throughout the entire storage period, ABA treatment notably enhanced fruit coloration, particularly at 10 DAT (Figure 1e). To more comprehensively assess the impact of ABA treatment on peel colour, we measured the CCI values under treatment. After 10 days of ABA treatment, the CCI value was significantly higher than that of the control (Figure 1f). Since carotenoid content directly contributes to citrus peel colour, we further analysed the carotenoid levels in the peel of ABA‐treated citrus fruits using high‐performance liquid chromatography (HPLC). The results indicated a clear accumulation of total carotenoids after 10 days of ABA treatment (Figure 1g). ABA treatment continued to elevate the CCI value and carotenoid content in citrus peel at 20 DAT (Figure 1f,g). To further validate the effects of the above ABA treatment, citrus calli were treated with a medium supplemented with ABA. As a result, ABA‐treated calli exhibited a deep yellow colour and higher levels of carotenoids than the control (Figure S1D,E). These results demonstrate that ABA treatment significantly promotes carotenoid biosynthesis in citrus.
Identification of a candidate carotenoid regulator, CsbZIP44 , involved in ABA‐induced carotenoid biosynthesis in citrus
Our previous study has reported the crucial involvement of BCHs and NCEDs in ABA‐mediated fruit coloration in citrus (Zhang et al., 2021; Zhu et al., 2017, 2020). Furthermore, we observed a significant induction of CsBCH1 and CsNCED2 expression following ABA treatment (Figure S2). These findings prompted us to perform a yeast one‐hybrid (Y1H) screen using the promoters of CsBCH1 and CsNCED2 as baits to identify key TFs involved in regulating ABA‐induced carotenoid biosynthesis in citrus.
Following the Y1H screening, three TFs were identified. Only Cs3g25230 exhibited a significant up‐regulation in ABA‐treated citrus fruit and calli (Figure 2a,b; Figure S3), indicating its crucial involvement in ABA‐induced carotenoid biosynthesis. Furthermore, we investigated the spatial and temporal expression patterns of Cs3g25230. Quantitative reverse‐transcription PCR (RT‐qPCR) analysis revealed higher expression levels of Cs3g25230 in fruits compared to other tissues. Cs3g25230 expression peaked at 190 DAFB, followed by a gradual decline by 230 DAFB (Figure 2c), which closely correlates with fruit coloration, carotenoid accumulation and ABA production during fruit ripening (Figure 1a–c). Based on these findings, we identified Cs3g25230 as the key TF responsible for regulating ABA‐mediated carotenoid biosynthesis, which warrants further investigation.
Figure 2.
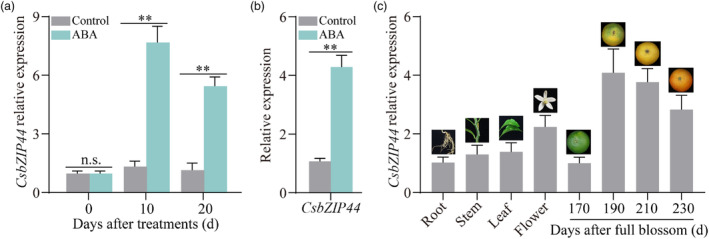
ABA‐activated CsbZIP44 is highly associated with citrus fruit coloration. Expression of CsbZIP44 under ABA treatment in citrus fruit (a) and calli (b). (c) Spatial and temporal expression analyses of CsbZIP44. Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
Sequence analysis of Cs3g25230 revealed a full‐length coding sequence (CDS) of 495 bp, encoding a protein comprising 164 amino acids. Gene conserved domain analysis conducted at the National Center for Biotechnology Information (NCBI) identified a basic/leucine zipper (bZIP) superfamily conserved domain within Cs3g25230 (Figure S4A). Phylogenetic analysis comparing Cs3g25230 with other bZIP family proteins from Arabidopsis thaliana revealed that Cs3g25230 clustered closely with AtbZIP44 (Figure S4B). Therefore, the designation of Cs3g25230 was changed to CsbZIP44. Furthermore, multiple sequence alignment demonstrated the presence of a bZIP superfamily conserved domain, comprising a ‘basic’ region and a ‘zipper’ region, in both the protein sequences of CsbZIP44 and AtbZIP44 (Figure S4C). These findings indicate that CsbZIP44 belongs to the bZIP TF family.
To determine the subcellular localization of CsbZIP44, a GFP‐CsbZIP44 fusion vector driven by the 35S promoter was constructed and transiently co‐expressed with NF‐YA‐mCherry, a nuclear marker in Nicotiana benthamiana (N. benthamiana) leaf epidermal cells. Confocal laser‐scanning microscopy revealed that the CsbZIP44‐GFP fusion protein was localized to the nucleus (Figure S5A). To investigate the influence of specific regions on CsbZIP44 transcriptional activation activity, the protein was divided into three parts based on the conserved domain: NTD (N‐terminal domain; amino acids 1 to 33), bZIP (bZIP conserved domain; amino acids 33 to 83), and CTD (C‐terminal domain; amino acids 83 to 165; Figure S5B). Analysis of individual, combined domain and full‐length fragments in a yeast assay demonstrated that the CTD is essential for the activation activity of CsbZIP44 (Figure S5C). To examine the functional mechanism of CsbZIP44, we tested its transcriptional activation activity in N. benthamiana. The pBD‐VP16 construct was included as a positive control, while the empty pBD vector was used as a negative control (Figure S5D). As shown in Figure S5E, the effector fused with CsbZIP44 exhibited significantly higher relative luciferase activity than the control, similar to pBD‐VP16. These findings collectively indicate that ABA‐activated CsbZIP44 functions as a nucleus‐localized transcriptional activator.
CsbZIP44 is essential for ABA‐mediated carotenoid biosynthesis in citrus
To assess the function of CsbZIP44, we generated a CsbZIP44‐overexpressing plasmid (PK7‐CsbZIP44) and transiently transformed it into ‘Valencia’ orange fruits (Citrus sinensis Osbeck.) considering the extended juvenile period of transgenic citrus plants (Figure 3a). The CsbZIP44‐overexpressing fruits exhibited enhanced red coloration and a greater accumulation of carotenoids around the infiltration site than control fruits injected with the empty vector PK7 (Figure 3a,b). RT‐qPCR analysis revealed a significant up‐regulation of carotenoid catabolism‐related genes, including CsDXR, CsGGPPs, CsBCH1 and CsNCED2, upon overexpression of CsbZIP44 (Figure 3c–g). These findings provide evidence that overexpression of CsbZIP44 remarkably enhances fruit coloration in citrus.
Figure 3.
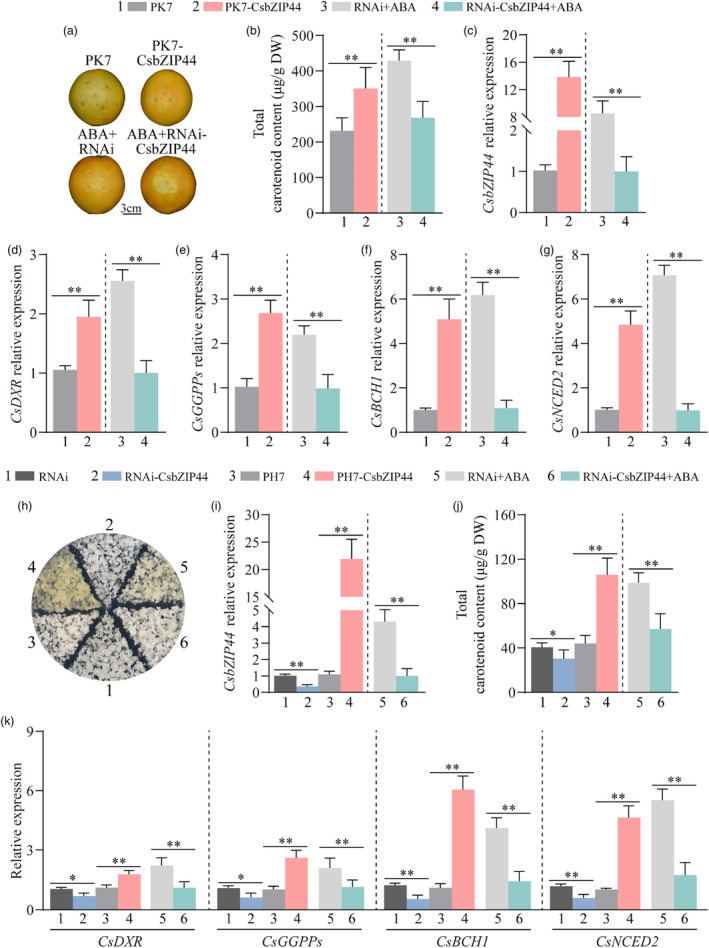
CsbZIP44 is essential for ABA‐induced carotenoid biosynthesis in citrus. (a–g) Transient expression of CsbZIP44 in ‘Valencia’ orange fruit. (a) Phenotypes. Empty vector PK7 and RNAi as control. PK7‐CsbZIP44 and RNAi‐CsbZIP44 indicate overexpressing and interfering CsbZIP44 respectively. Bars = 3 cm. Transcript levels of CsbZIP44 (c), CsDXR (d), CsGGPPs (e), CsBCH1 (f) and CsNCED2 (g). (b) Total carotenoid content (μg/g DW). (h–k) Stable transformation of CsbZIP44 in citrus calli. (h) Phenotypes. PH7‐CsbZIP44 and RNAi‐CsbZIP44 indicate overexpressing and interfering CsbZIP44 respectively. Empty vector PH7 and RNAi as control. Expression levels of CsbZIP44 (i) and CsDXR, CsGGPPs, CsBCH1 and CsNCED2 (k). (j) Total carotenoid content (μg/g DW). Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
To elucidate the role of CsbZIP44 in ABA‐induced carotenoid biosynthesis in citrus, we subjected CsbZIP44‐interfering fruits (RNAi‐CsbZIP44) and control fruits (RNAi) to ABA treatment (Figure 3a). Compared to the control, the CsbZIP44‐interfering fruits exhibited noticeably lighter coloration in the peel surrounding the injection sites (Figure 3a). Consistently, the carotenoid content and the expression levels of CsbZIP44 and carotenogenic genes (including CsDXR, CsGGPPs, CsBCH1 and CsNCED2) were significantly lower in the CsbZIP44‐interfering fruits than in the control (Figure 3b–g). These findings strongly suggest that CsbZIP44 plays a crucial role in ABA‐induced fruit coloration in citrus fruits.
To further validate the function of CsbZIP44, we performed stable transformation of citrus calli using CsbZIP44‐overexpressing (PH7‐CsbZIP44) and CsbZIP44‐interfering (RNAi‐CsbZIP44) constructs, along with empty PH7 and RNAi vectors as controls (Figure 3h). The expression of CsbZIP44 was confirmed by RT‐qPCR analysis in the transgenic calli (Figure S6; Figure 3i). Compared to the control, the CsbZIP44‐overexpressing calli exhibited a distinct yellow coloration, while the CsbZIP44‐interfering calli appeared paler (Figure 3h). HPLC analysis revealed a higher accumulation of carotenoids in the CsbZIP44‐overexpressing calli while interfering with CsbZIP44 had the opposite effect (Figure 3j). RT‐qPCR analysis demonstrated that the expression patterns of CsDXR, CsGGPPs, CsBCH1 and CsNCED2 corresponded with the changes in calli colour and carotenoid content in the transgenic calli (Figure 3k). Additionally, to verify the essential role of CsbZIP44 in ABA‐induced carotenoid biosynthesis, we treated CsbZIP44‐interfering and control calli with an ABA solution. Following ABA treatment, the CsbZIP44‐interfering calli displayed a paler white colour than the control (Figure 3h). Moreover, the carotenoid content and the expression of CsbZIP44 and carotenogenic genes (including CsDXR, CsGGPPs, CsBCH1 and CsNCED2) in the CsbZIP44‐interfering calli were significantly lower than those in the control (Figure 3i,k). These results are consistent with the transient transformation performed in ‘Valencia’ orange fruits.
In summary, the results obtained thus far collectively demonstrate that CsbZIP44 plays a crucial role in promoting carotenoid accumulation by upregulating the expression of CsDXR, CsGGPPs, CsBCH1 and CsNCED2. These findings provide compelling evidence for the essential involvement of CsbZIP44 in ABA‐mediated citrus carotenoid biosynthesis.
CsbZIP44 directly binds the promoters of carotenogenic genes and significantly activates their transcription
The transcription levels of CsDXR, CsGGPPs, CsBCH1 and CsNCED2 were upregulated in CsbZIP44‐overexpressing fruit and calli. We speculated that CsbZIP44 directly regulates the transcription of the above carotenoid metabolism‐related genes. To verify this hypothesis, we first performed a yeast one‐hybrid (Y1H) assay to test whether CsbZIP44 binds to the promoters of these carotenogenic genes. As shown in Figure 4a, the Y1H assay confirmed that CsbZIP44 can bind to the promoters of CsDXR, CsGGPPs, CsBCH1 and CsNCED2.
Figure 4.
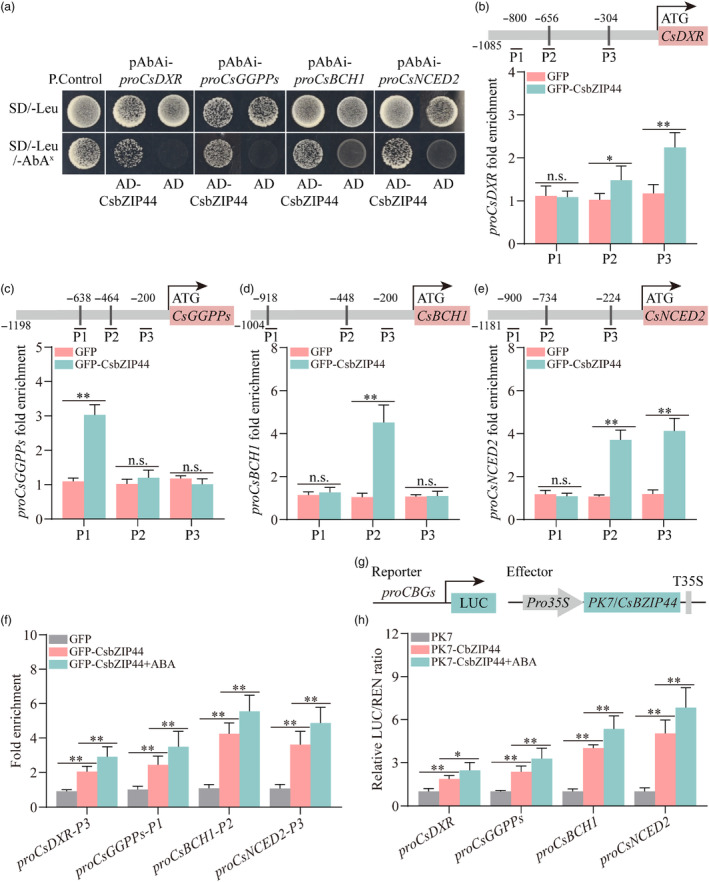
CsbZIP44 directly activates the transcription of carotenogenic genes. (a) Yeast one‐hybrid (Y1H) assay identified interactions of CsbZIP44 with target genes promoters. Empty PGADT7 + pAbAi‐proCBGs and PGADT7‐Rec‐p53 + p53‐AbAi as the negative (N. Control) and positive controls (P. Control) respectively. Aureobasidin A (AbA) is a yeast cell growth inhibitor. CBGs, carotenoid biosynthesis genes. SD/−Leu/AbAx, SD/−Leu medium supplemented with 200 ng ml−1 as the basal concentration of proCsDXR and proCsGGPPs. SD/−Leu/AbAx, SD/−Leu medium supplemented with 150 ng ml−1 as the basal concentration of proCsBCH1 and proCsNCED2. (b–e) Chromatin Immunoprecipitation (ChIP)‐PCR assay showed the interaction of CsbZIP44 with several regions in the promoters of CsDXR, CsGGPPs, CsBCH1 and CsNCED2 respectively. The grey lines represent the putative binding motif of bZIP family proteins in these promoters. Cross‐linked chromatin samples were extracted from GFP‐CsbZIP44 fruit calli and precipitated with an anti‐GFP antibody. The eluted DNA fragment was used to amplify by quantitative (q)‐PCR. (f) ChIP‐PCR assay showed that ABA treatment increases the binding of CsbZIP44 to the promoters of CsDXR, CsGGPPs, CsBCH1, CsNCED2. proCsDXR‐P3, proCsGGPPs‐P1, proCsBCH1‐P2 and proCsNCED2‐P3 refers to the promoter region of CsDXR, CsGGPPs, CsBCH1 and CsNCED2 in (b) to (e) respectively. Cross‐linked chromatin samples were extracted from GFP‐CsbZIP44 fruit calli treated with or without ABA (250 μm) and precipitated with an anti‐GFP antibody. Eluted DNA was used to amplify the sequences by q‐PCR. (g) Schematic representation of reporter and effector constructs used in dual‐luciferase assay. (h) Dual‐luciferase assay indicated that ABA treatment enhances the activation by CsbZIP44 of the CBG promoters. CBG, carotenoid biosynthesis gene. ABA treatment (100 μm) in the dual‐luciferase assay was conducted 3 h before determination. Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
To confirm the binding of CsbZIP44 to these promoters in vivo, we conducted chromatin immunoprecipitation (ChIP)‐PCR assays using 35S:CsbZIP44‐GFP transgenic citrus calli and empty vector GFP transgenic calli as controls. In CsbZIP44‐overexpressing calli, one or two fragments containing the potential binding elements of CsbZIP44 showed strong signals (Figure 4b–e), suggesting that CsbZIP44 directly binds to these promoters in vivo. Furthermore, ABA treatment further strengthened the binding signals of CsbZIP44 to the promoters of the target genes in vivo (Figure 4f).
Moreover, we performed electrophoretic mobility shift (EMSA) assays using the purified MBP‐CsbZIP44 protein and the purified empty MBP protein as a control. Probes were synthesized based on the fragment with the strongest binding signal in the ChIP‐PCR assay. The EMSA results further confirmed the binding of CsbZIP44 to these promoters in vitro (Figure S7).
To further test whether CsbZIP44 regulates the expression of these genes, we conducted a transient expression assay in N. benthamiana leaves using a luciferase system. The system consisted of LUC reporters driven by the endogenous promoters of CsDXR, CsGGPPs, CsBCH1 and CsNCED2, and the effector contained the full‐length coding sequence (CDS) of CsbZIP44 (Figure 4g). We observed that the CsbZIP44 effector significantly enhanced the luciferase intensities of the reporters driven by the endogenous promoters of these target genes compared to the empty effector (PK7). This finding indicates that CsbZIP44 positively regulates the promoter activity of the target genes (Figure 4h). Moreover, this activation was further enhanced by ABA treatment (Figure 4h). These results collectively demonstrate that ABA‐induced CsbZIP44 promotes the expression of CsDXR, CsGGPPs, CsBCH1 and CsNCED2 by directly binding to their promoters.
CsHB5 interacts with CsbZIP44 and activates CsbZIP44 expression
bZIP TFs have been widely reported to interact with other TFs to form regulatory complexes that respond to ABA and participate in fruit coloration and pigment metabolism (An et al., 2018, 2021; Bhagat et al., 2021; Chenge‐Espinosa et al., 2018). To further analyse the molecular mechanism of ABA‐mediated carotenoid metabolism regulated by CsbZIP44, we conducted a yeast two‐hybrid (Y2H) screen using CsbZIP44 as the bait. Before the Y2H screen, we confirmed that BD‐CsbZIP44 does not exhibit transcriptional self‐activation in yeast cells (Figure S8). The results of the Y2H screen identified an ABA‐activated homeodomain leucine zipper I (HD‐ZIP I) TF called CsHB5 (Figure S9).
We initially conducted a Y2H assay to verify the interaction between CsbZIP44 and CsHB5. As depicted in Figure 5a, yeast cells co‐transformed with BD‐CsbZIP44 and AD‐CsHB5, along with the positive control, grew successfully and displayed a blue colour on SD‐Leu/Trp/His/Ade medium. However, other combinations and the negative control did not grow or exhibit a blue colour on this medium. These results indicate that CsbZIP44 can interact with CsHB5. Next, we performed a pull‐down assay using fusion proteins of CsbZIP44‐MBP and CsHB5‐GST to further confirm this interaction. It was observed that CsHB5‐GST was pulled down by CsbZIP44‐MBP, confirming that CsbZIP44 directly interacts with CsHB5 in vitro (Figure 5b). To validate that this interaction also occurs in vivo, we constructed fusion constructs of CsbZIP44‐GFP and CsHB5‐Flag and conducted a co‐immunoprecipitation (Co‐IP) assay. Co‐expression of these constructs in N. benthamiana leaves demonstrated that CsHB5‐Flag was immunoprecipitated by CsbZIP44‐GFP but not by GFP, using an anti‐GFP antibody. This finding verifies the in vivo interaction between CsbZIP44 and CsHB5 (Figure 5c).
Figure 5.
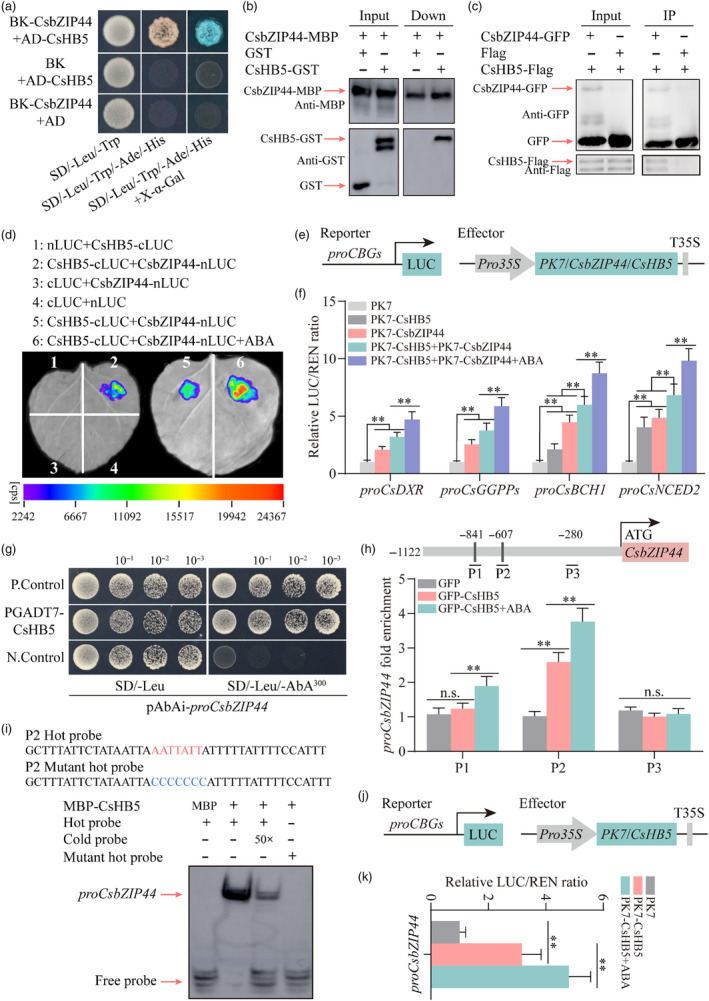
CsHB5 interacts with CsbZIP44 and transcriptionally activates CsbZIP44. (a) Yeast two‐hybrid (Y2H) assay revealing an interaction between CsbZIP44 and CsHB5. Yeast grown in SD/−Trp/−Leu medium and SD/−Trp/−Leu/−His/−Ade medium is shown. The interaction is indicated by yeast growth and X‐α‐Gal staining. (b) The interactions between CsbZIP44 and CsHB5 were analysed using a pull‐down assay. Fusion proteins GST‐CsHB5 and MBP‐CsbZIP44 were used in the pull‐down analysis. GST‐ and MBP‐antibodies were used for immunoblot analyses. The band detected by the GST antibody in the pull‐down protein sample indicates the interaction between CsbZIP44 and CsHB5. (c) The interaction between CsbZIP44 and CsHB5 was confirmed with a co‐immunoprecipitation (Co‐IP) assay. The fused constructs GFP‐tagged CsbZIP44, and flag‐tagged CsHB5 were co‐overexpressed in Nicotiana benthamiana leaves. GFP antibody beads were used for immunoprecipitation. GFP‐ and flag‐antibodies were used for immunoblot analyses. The band detected by the GFP antibody in the IP samples indicates an interaction between CsbZIP44 and CsHB5. (d) A luciferase complementation imaging assay shows that CsbZIP44 interacts with CsHB5, and this interaction is enhanced by ABA treatment. Agrobacterium tumefaciens strain GV3101 harbouring different constructs was infiltrated into different wild tobacco leaf regions. Luciferase activities were imaged in these regions 3 days after infiltration. cps, signal counts per second. (e) Schematic representation of reporter and effector constructs used in dual‐luciferase assay. (f) Dual‐luciferase assay showing that the interaction between CsbZIP44 and CsHB5 significantly increases the activation effect on the promoter activity of target genes, which is strengthened by ABA treatment. (g) Y1H assay showing interactions of CsHB5 with CsbZIP44 promoter. Empty PGADT7 + pAbAi‐proCsbZIP44 and PGADT7‐Rec‐p53 + p53‐AbAi as the negative (N. Control) and positive controls (P. Control) respectively. Aureobasidin A (AbA) is a yeast cell growth inhibitor. SD/−Leu/AbA250, SD/−Leu medium supplemented with 250 ng ml−1 as the basal concentration of proCsbZIP44. (h) ChIP‐PCR assay indicating the interaction of CsHB5 with several regions in the CsbZIP44 promoter in vivo. The grey lines represent putative binding motif of HD‐ZIP family proteins in the CsbZIP44 promoter. Cross‐linked chromatin samples were extracted from GFP‐CsHB5 fruit calli treated with or without ABA (250 μm) and precipitated with an anti‐GFP antibody. Eluted DNA was used to amplify the sequences by q‐PCR. (i) Electrophoretic mobility shift assay (EMSA) assay confirming that CsHB5 directly binds the binding element of HD‐ZIP TFs in the CsbZIP44 promoter in vitro. Purified MBP‐tagged CsHB5 protein was used in EMSA assay, and purified MBP protein was used as a negative control. Black arrows indicate the position of biotin‐labelled promoter fragment (hot probe) containing the putative binding motifs of HD‐ZIP family proteins. Red arrows indicate the positions of protein‐DNA complexes or free probes. Red letters represent the binding motifs, and blue letters indicate their corresponding mutant motifs. ‘+’ and ‘–’ the presence and absence of the indicated probe or protein respectively. Increasing amounts (50 folds) of the unlabelled DNA fragments (cold probe) were added as competitors. (j) Schematic representation of reporter and effector constructs used in dual‐luciferase assay. (k) Dual‐luciferase assay showing that CsHB5 enhances the activation of the CsbZIP44 promoter, and this activation is induced by ABA treatment. ABA treatment (100 μm) in the luciferase complementation imaging assay and the dual‐luciferase assay was conducted for 3 h before determination. Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
Additionally, we performed a LUC complementation experiment by co‐infiltrating N. benthamiana leaves with CsbZIP44‐nLUC and CsHB5‐cLUC constructs. Imaging results revealed a strong luminescence signal in the co‐expression region of CsbZIP44‐nLUC and CsHB5‐cLUC (Figure 5d, region 2), while no signal was detected in the negative control regions (Figure 5d, regions 1, 3 and 4). Furthermore, a more intense luminescence signal was observed in the ABA‐treated region (Figure 5d, region 6) compared to the untreated region (Figure 5d, region 5), indicating that ABA strengthens the interaction between CsbZIP44 and CsHB5. These results provide further evidence that CsbZIP44 interacts with CsHB5 in vivo, and this interaction is enhanced by ABA signal.
Our previous study reported that CsHB5 plays a positive role in fruit ripening and ABA metabolism by directly activating the transcription of CsBCH1 and CsNCED2 in citrus fruit (Zhang et al., 2021). To further determine whether the interaction complex between CsbZIP44 and CsHB5 affects the transactivation activity of CsbZIP44 and CsHB5 on target genes, we performed a transient expression assay using the dual‐luciferase system in N. benthamiana leaves (Figure 5e). We observed that the co‐expression of CsbZIP44 and CsHB5 significantly enhanced the luciferase intensities of the target gene promoters compared to the overexpression of CsbZIP44 or CsHB5 alone (Figure 5f). This effect was further amplified by ABA treatment (Figure 5f). These results indicate that the ABA‐induced interaction complex between CsbZIP44 and CsHB5 enhances the transactivation activity of both CsbZIP44 and CsHB5 on their target genes.
A previous study has demonstrated that the transcript level of CsHB5 increases during fruit ripening and ABA accumulation in citrus (Zhang et al., 2021), which is consistent with the observed pattern of CsbZIP44 during citrus fruit ripening (Figure 2c). Furthermore, CsBCH1 and CsNCED2 are target genes jointly regulated by both CsbZIP44 and CsHB5 (Figure 4a,d–h and Figure S7C,D; Zhang et al., 2021). These findings suggest the presence of protein interaction and regulation between CsbZIP44 and CsHB5.
Interestingly, we observed no significant difference in the expression of CsHB5 between the control and CsbZIP44‐overexpressing citrus calli (Figure S10A). However, the transcription level of CsbZIP44 was clearly increased in CsHB5‐overexpressing citrus calli (Figure S10B). Furthermore, we identified two potential cis‐elements (AATNATT) of CsHB5 in the CsbZIP44 promoter (Figure S11). Based on these findings, we speculated that CsbZIP44, as a downstream target gene of CsHB5, is upregulated by CsHB5. We initially performed a Y1H assay to verify this hypothesis to demonstrate that CsHB5 directly binds to the CsbZIP44 promoter (Figure 5g). ChIP‐PCR assays revealed that CsHB5 directly binds to these fragments containing the two potential binding elements in vivo, with stronger binding signals induced by ABA treatment (Figure 5h). EMSA experiments further confirmed the binding in vitro (Figure 5i). Moreover, a LUC activity assay indicated that CsHB5 significantly activates CsbZIP44 expression (Figure 5j,k), and this activation is significantly enhanced by ABA treatment (Figure 5k). These findings provide evidence that CsHB5 serves as the regulator of CsbZIP44 and activates its expression in an ABA‐induced manner.
The transcriptional regulatory module CsHB5‐CsbZIP44 positively regulates carotenoid biosynthesis
The results presented above demonstrate that CsHB5 directly activates the expression of CsbZIP44 and interacts with CsbZIP44 to participate in ABA‐mediated carotenoid metabolism. To further investigate the role of the transcriptional regulatory module CsHB5‐CsbZIP44 in citrus, we conducted transient infiltration assays in ‘Valencia’ orange fruit (Figure 6a). The levels of CsbZIP44 and CsHB5 expression in the citrus peel surrounding the injection sites were determined using RT‐qPCR (Figure 6b,c). In comparison to the control fruit (empty vector PK7), the fruit co‐expressing CsbZIP44 and CsHB5 (PK7‐CsbZIP44 + PK7‐CsHB5) exhibited a greater degree of red coloration around the injection sites (Figure 6a). Similarly, the carotenoid content (Figure 6d) and the expression levels of CsbZIP44 (Figure 6b), CsDXR (Figure 6e), CsGGPPs (Figure 6f), CsBCH1 (Figure 6g) and CsNCED2 (Figure 6h) were higher in the areas of fruit co‐expressing CsbZIP44 and CsHB5 compared to those overexpressing only CsbZIP44. These findings suggest that CsHB5 enhances the function of CsbZIP44, promoting carotenoid biosynthesis and fruit coloration.
Figure 6.
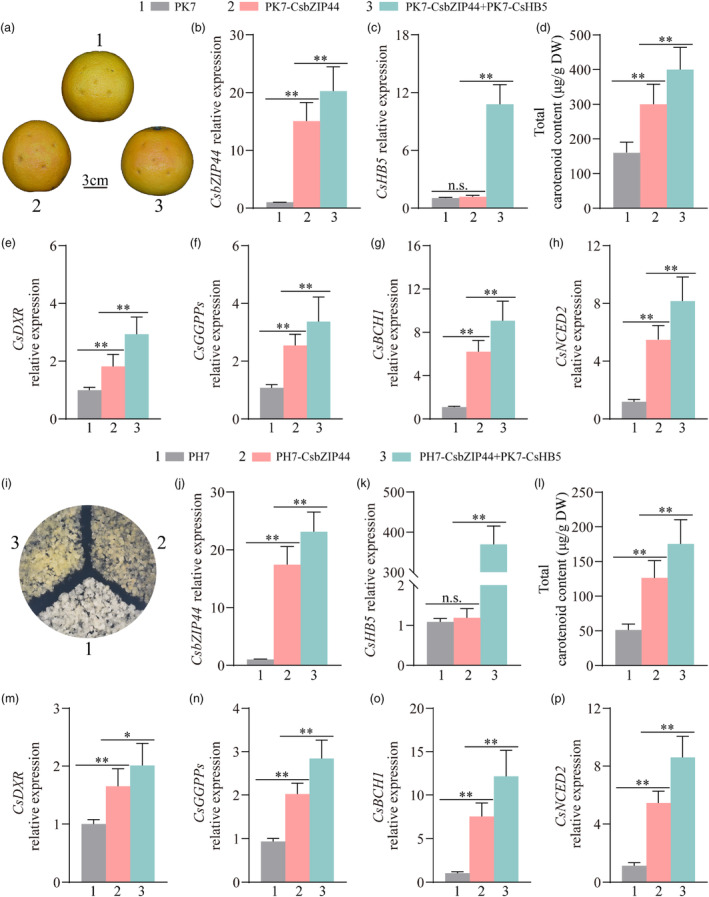
The transcriptional regulatory module CsbZIP44‐CsHB5 positively regulates carotenoid biosynthesis in citrus. (a–h) Transient expression of CsbZIP44 and CsHB5 in ‘Valencia’ orange fruit. (a) Phenotypes. Empty vector PK7 as control. PK7‐CsbZIP44 and RNAi‐CsbZIP44 indicates overexpressing CsbZIP44 and CsHB5 respectively. Bars = 3 cm. Transcript levels of CsbZIP44 (b), CsHB5 (c), CsDXR (e), CsGGPPs (f), CsBCH1 (g) and CsNCED2 (h). (d) Total carotenoid content (μg/g DW). (i–p) Stable transformation of CsbZIP44 and CsHB5 in citrus calli. (i) Phenotypes. PH7 indicates the transformation of PH7 empty vector in citrus calli as control. PH7‐CsbZIP44 indicates overexpressing CsbZIP44. PH7‐CsbZIP44 + PK7‐CsHB5 indicates co‐overexpressing CsbZIP44 and CsHB5. The expression levels of CsbZIP44 (j) CsHB5 (k), CsDXR (m), CsGGPPs (n), CsBCH1 (o) and CsNCED2 (p). (l) Total carotenoid content (μg/g DW). Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
We further confirmed the above conclusions by conducting a stable transformation of citrus calli. Calli co‐overexpressing CsbZIP44 and CsHB5 (PH7‐CsbZIP44 + PK7‐CsHB5) exhibited a distinct yellow coloration compared to calli with only CsbZIP44 overexpression (PH7‐CsbZIP44) and control calli (empty vector PH7; Figure 6i). This colour change was accompanied by increased expression of CsbZIP44 and CsHB5 in the transgenic calli (Figure 6j,k). Furthermore, the carotenoid content (Figure 6l) and the expression levels of CsbZIP44 (Figure 6j) and its target genes (Figure 6m–p) were significantly higher in the co‐overexpressing calli compared to the control and CsbZIP44‐only overexpressing calli. These results are consistent with those of the transient transformation experiments conducted in the ‘Valencia’ orange fruit. Overall, these findings demonstrate that the transcriptional regulatory module CsHB5‐CsbZIP44 plays a positive role in regulating citrus carotenoid biosynthesis.
Discussion
Carotenoids are natural terpenoid pigments that play essential roles in plant development and maturation and are important for human nutrition (Nisar et al., 2015). In addition to determining fruit coloration, carotenoids also enhance the nutritional and health benefits of citrus fruit for humans by providing antioxidants and precursors for vitamin A biosynthesis, thereby reducing the risk of various chronic diseases (Fraser and Bramley, 2004; Kato et al., 2004).
Citrus fruits are not only among the most important fleshy fruits in the world but also have the most diverse carotenoid composition, with a larger number of carotenoid species compared to other fruits (Fanciullino et al., 2006; Yuan et al., 2015). Extensive studies have indicated that ABA (abscisic acid) significantly affects carotenoid biosynthesis during citrus fruit ripening (Rodrigo et al., 2003; Romero et al., 2019; Wang et al., 2016). However, the underlying molecular mechanisms governing ABA‐mediated carotenoid biosynthesis remain poorly understood.
In this study, we identified a novel bZIP (basic leucine zipper) transcription factor, CsbZIP44, which is induced by ABA and positively regulates ABA‐mediated carotenoid biosynthesis. CsbZIP44 physically interacts with CsHB5 and functions as the target gene of CsHB5 in citrus. These findings define a novel role of CsbZIP44 in ABA signalling and carotenoid biosynthesis by associating with CsHB5, suggesting a possible molecular pathway for improving fruit coloration in citrus.
CsbZIP44 acts as the central regulator to determine the ABA‐mediated carotenoid biosynthesis
ABA is a well‐known inducer of fruit coloration during citrus fruit ripening by regulating carotenoid metabolism (Rodrigo et al., 2003; Romero et al., 2019). Previous studies have shown that the application of exogenous ABA or the use of inhibitors of ABA biosynthesis can significantly enhance or inhibit citrus fruit coloration (Wang et al., 2016; Zhu et al., 2020). In the present study, treatment with ABA or NDEG (an ABA biosynthesis inhibitor) resulted in a significant promotion or inhibition of fruit coloration and carotenoid accumulation in ‘Valencia’ orange fruit respectively (Figure S1A–C; Figure 1e–g). Additionally, we observed a higher level of carotenoids in ABA‐treated citrus calli, which was not previously reported (Figure S1D,E). These findings further support the essential role of ABA in inducing carotenoid biosynthesis in citrus fruit.
In recent years, several TFs have been reported to influence carotenoid biosynthesis in citrus fruit by regulating ABA metabolism. For instance, an R2R3‐MYB TF (CrMYB68) was found to delay ABA biosynthesis by reducing the expression of CrBCH2 and CrNCED5 in a stay‐green mutant of Citrus reticulata cv Suavissima (Zhu et al., 2017). Additionally, we previously identified a citrus HD‐ZIP I TF, CsHB5, which functions as an activator of ABA‐triggered fruit ripening by upregulating the expression of ABA and carotenoid metabolism genes (Zhang et al., 2021). However, the key TFs regulating ABA‐mediated carotenoid biosynthesis in citrus remain largely unknown.
In contrast to previous studies, our current research identifies and demonstrates that the ABA‐induced CsbZIP44 positively regulates citrus carotenoid accumulation by directly binding to the promoters of carotenoid metabolism‐related genes (including CsDXR, CsGGPPs, CsBCH1 and CsNCED2) and significantly activating their expression through a series of biochemical experiments (Figures 2 and 4; Figure S7). Transgenic assays further confirm that CsbZIP44, as an essential regulator, directly influences ABA‐mediated carotenoid biosynthesis in citrus fruit (Figure 3). These findings systematically and specifically reveal that CsbZIP44 acts as a necessary bridge between ABA signalling and carotenoid metabolism in citrus.
A phylogenetic analysis comparing CsbZIP44 with other bZIP family proteins from A. thaliana revealed that CsbZIP44 closely clustered with AtbZIP44 (Figure S4B). In A. thaliana, AtbZIP44 is primarily associated with abiotic stresses, such as cold and salinity stresses (Kilian et al., 2007; Weltmeier et al., 2009), seed germination (Iglesias‐Fernández et al., 2013) and auxin‐driven primary root growth (Weiste et al., 2017). In contrast to AtbZIP44, CsbZIP44 performs a novel role in citrus by regulating carotenoid metabolism in response to ABA signals. Another highly homologous TF, MdbZIP44, which shares similarity with AtbZIP44, is involved in ABA‐modulated anthocyanin accumulation in apples (An et al., 2018). We observed that both MdbZIP44 and CsbZIP44 respond conservatively to ABA signals and participate in regulating pigment metabolism in horticultural crops. These findings suggest that CsbZIP44 and its homologues have evolved novel and diverse regulatory functions across different species, thereby enriching the functional diversity of bZIP family TFs in plants.
The CsHB5‐CsbZIP44 regulatory module positively regulates ABA‐mediated carotenoid biosynthesis
Carotenoid metabolism plays a crucial role in plant growth and development, and as a result, plants have developed intricate regulatory mechanisms to effectively coordinate this process. Several studies have indicated that TFs act as important regulators in carotenoid metabolism by forming complexes with other TFs to synergistically regulate the expression of their target genes. For instance, in Medicago truncatula, MtWP1 associates with MtTT8 and MtWD40‐1 to regulate carotenoid metabolism (Meng et al., 2019). In papaya, two ethylene‐induced transcriptional regulatory modules, CpMADS4‐CpNAC3 (Fu et al., 2021) and CpEIN3a‐CpNAC2 (Fu et al., 2017) positively regulate carotenoid biosynthesis during fruit ripening. Additionally, in citrus, CrNAC036 interacts with CrMYB68 to jointly inhibit ABA biosynthesis and fruit coloration (Zhu et al., 2020). However, the specific transcriptional regulatory module underlying ABA‐mediated carotenoid biosynthesis remains poorly understood.
Through a yeast two‐hybrid (Y2H) screen, we identified a HD‐ZIP I TF, CsHB5, as an interacting partner of CsbZIP44. We further demonstrated that the interaction between CsbZIP44 and CsHB5 enhances their ability to activate target genes (Figure 5a–f). Moreover, in addition to the protein–protein interaction, CsHB5 directly binds to the promoter of CsbZIP44 and activates its expression (Figure 5g–k). Importantly, transgenic assays confirmed that the CsHB5‐CsbZIP44 transcriptional regulatory module plays a positive role in regulating carotenoid biosynthesis in citrus.
Previous studies have reported the involvement of transcriptional regulatory modules composed of bZIP TFs in the regulation of pigment metabolism (An et al., 2018, 2021; Bhagat et al., 2021; Chenge‐Espinosa et al., 2018). However, the CsHB5‐CsbZIP44 transcriptional regulatory module, which includes CsbZIP44, exhibits a higher level of complexity. It not only involves protein–protein interactions but also encompasses both upstream and downstream transcriptional regulation, distinguishing it from previously reported regulatory modules composed of bZIP TFs.
In our previous studies, we reported that CsHB5 regulates ABA and carotenoid metabolism by activating the transcription of CsBCH1 and CsNCED2 (Zhang et al., 2021). In the current study, we conducted a comparative analysis of CsHB5 and CsbZIP44. CsHB5 functions as a regulator of ABA‐triggered senescence and is involved in various senescence‐related processes, such as chlorophyll degradation, ABA metabolism, and reactive oxygen species (ROS) signal transduction (Zhang et al., 2021). Although CsHB5 is implicated in the regulation of carotenoid metabolism, we believe that this association is a secondary result related to CsHB5‐mediated citrus senescence. We propose that there are other more direct and essential transcription factors that act between CsHB5 and carotenoid metabolism, specifically regulating ABA‐mediated carotenoid biosynthesis.
In contrast to CsHB5, the ABA‐induced CsbZIP44 demonstrates a more crucial and efficient role in regulating citrus carotenoid biosynthesis. CsbZIP44 directly activates the expression of multiple carotenoid metabolism‐related genes, including CsDXR, CsGGPPs, CsBCH1 and CsNCED2 (Figures 3 and 4; Figure S7). Unlike CsHB5, CsbZIP44's role in regulating carotenoid metabolism is more direct. Biochemical experiments and transgenic assays have confirmed the positive regulation of carotenoid biosynthesis by the ABA‐induced CsHB5‐CsbZIP44 transcriptional regulatory module (Figures 5 and 6). This is the first report of ABA signal‐regulated carotenoid metabolism in plants, highlighting the significance of this regulatory module in the field.
There is a positive feedback regulation loop between CsHB5‐CsbZIP44 regulatory module‐regulated carotenoid biosynthesis and ABA signal
In addition to the increase in carotenoid content, we observed higher levels of ABA in citrus fruit and calli overexpressing CsbZIP44 compared to the control, while interfering with CsbZIP44 expression resulted in the opposite effect (Figure S12A,B). Furthermore, co‐overexpression of CsbZIP44 and CsHB5 significantly enhanced the ABA levels compared to the sole overexpression of CsbZIP44 in transgenic citrus fruit and calli (Figure S12C,D). These findings demonstrate that the CsHB5‐CsbZIP44 transcriptional regulatory module positively regulates not only carotenoid biosynthesis but also ABA metabolism in citrus.
ABA, as an oxygenated derivative of carotenoid, is influenced by carotenoid metabolism, as demonstrated in previous studies (Fang et al., 2008; Galpaz et al., 2008; Lindgren et al., 2003). Carotenoid biosynthesis directly impacts ABA content because carotenoids serve as the sole precursors for ABA biosynthesis (Du et al., 2010; Hirai et al., 2000). Among the genes involved in carotenoid metabolism, BCHs (Rodrigo et al., 2006; Wang et al., 2016; Zhang et al., 2021) and NCEDs (Zhu et al., 2017, 2020) play a critical role in ABA biosynthesis in citrus. Several TFs have been identified to affect ABA content by regulating the expression of NCEDs and BCHs. For example, CsERF061 promotes carotenoid biosynthesis and ABA accumulation by activating the expression of nine carotenogenic genes, including CsBCH and CsNCED3 (Zhu et al., 2021). In Citrus reticulate, CrNAC036 and CrMYB68 act as negative regulators of ABA biosynthesis by inhibiting the expression of CrNCED5 and CrBCH2 (Zhu et al., 2017, 2020). Our previous study demonstrated that overexpression of CsHB5 significantly enhances the transcript levels of CsBCH1 and CsNCED2, as well as ABA content in citrus (Zhang et al., 2021).
Similarly, this study revealed that the CsHB5‐CsbZIP4 transcriptional regulatory module directly binds and activates the promoters of CsBCH1 and CsNCED2 (Figures 4 and 5F; Figure S7C,D). Furthermore, the expression levels of CsBCH1 and CsNCED2 were significantly increased in citrus calli and fruit with either individual or combined overexpression of CsHB5 and CsbZIP4 (Figures 3f,g,k and 6g,h,o,p). Based on these findings, we propose two possible mechanisms for ABA accumulation. First, the CsHB5‐CsbZIP44 regulatory module promotes carotenoid accumulation, providing sufficient precursors for ABA biosynthesis, indirectly leading to an increase in ABA content. Second, the CsHB5‐CsbZIP44 regulatory module activates the expression of ABA metabolism genes, such as CsBCHs and CsNCEDs, directly promoting ABA biosynthesis. However, further studies are required to gain a deeper understanding of the precise mechanisms involved in this process.
In addition to being moderated by carotenoid metabolism, multiple studies have inferred that the ABA signal has a feedback regulation effect on carotenoid metabolism (Galpaz et al., 2008; Lu et al., 2018; Zhu et al., 2021). However, there is a lack of full and specific evidence to support these inferences. In this study, we utilized exogenous ABA treatment to investigate the feedback regulation of increased endogenous ABA induced by the CsHB5‐CsbZIP44 regulatory module in carotenoid metabolism. ABA treatment significantly induced carotenoid accumulation, as well as the expression of CsbZIP44, CsHB5, and their target genes involved in carotenoid metabolism in citrus fruit and calli (Figures 1e–g and 2a,b; Figures S1, S2 and S9). Furthermore, the interaction between CsbZIP44 and CsHB5, as well as their regulation on their target genes, was also strengthened under ABA treatment (Figures 4f–h and 5d–f,k). Our study specifically indicates that the CsHB5‐CsbZIP44 transcriptional regulatory module plays a central role in the positive feedback regulation loop between ABA‐mediated carotenoid biosynthesis and carotenoid accumulation‐promoted ABA metabolism.
Based on our studies, we have proposed a specific regulation model to analyse the mechanism of ABA‐mediated carotenoid biosynthesis (Figure 7). ABA stimulates CsbZIP44 and CsHB5 to directly bind the promoters of carotenogenic genes, activating their expression and promoting carotenoid accumulation. Additionally, CsHB5 interacts with CsbZIP44 and enhances CsbZIP44 expression by binding its promoter, forming the transcriptional regulatory module CsHB5‐CsbZIP44, which further strengthens their functions. Furthermore, the accumulation of ABA creates a positive feedback effect on the CsHB5‐CsbZIP44 regulatory module, regulating carotenoid biosynthesis. This regulatory mechanism significantly contributes to the enhancement of citrus fruit quality. For instance, by inducing the biosynthesis of endogenous ABA during citrus fruit development and maturation through adjustment of the cultivation environment or techniques, we can activate the CsHB5‐CsbZIP44 regulatory module and promote fruit coloration. Moreover, exogenous ABA treatment can be employed to improve citrus fruit colour during postharvest storage by activating the CsHB5‐CsbZIP44 regulatory module. Collectively, our work provides novel insight into the complex regulatory networks governing ABA‐mediated carotenoid biosynthesis for future research on the interplay between ABA signalling and carotenoid metabolism and offers valuable strategies for enhancing the visual quality of citrus fruits during fruit ripening and postharvest storage.
Figure 7.
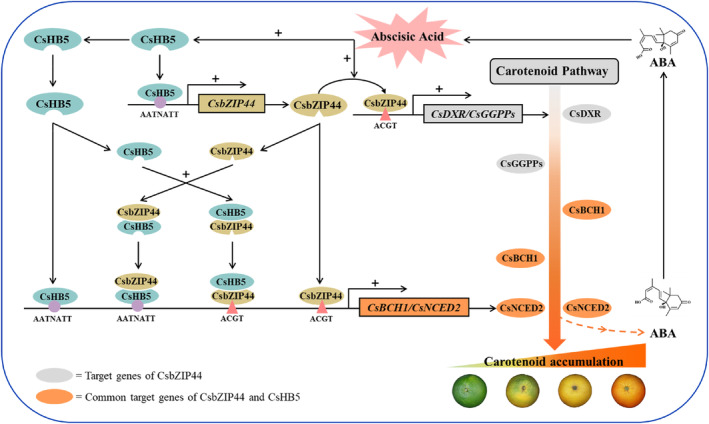
Model for the positive feedback regulatory loop between ABA and carotenoid metabolisms mediated by the transcriptional regulatory module CsbZIP44‐CsHB5 in citrus. ABA induces CsHB5 and activates CsbZIP44 expression through binding the AATNATT cis‐elements in the CsbZIP44 promoter, and then CsbZIP44 promotes carotenoid accumulation by directly enhancing the transcriptional levels of carotenogenic genes (including CsDXR, CsGGPPs, CsBCH1 and CsNCED2) in citrus fruit. Moreover, CsbZIP44 interacts with CsHB5 to further positively regulate carotenoid biosynthesis by directly activating the expressions of CsBCH1 and CsNCED2. In turn, ABA as ripening signal significantly induces the regulation and interaction of CsbZIP44 and CsHB5 in feedback regulation, thereby promoting carotenoid biosynthesis during citrus fruit ripening. ‘+’ represents promotion. CsDXR, 1‐deoxy‐D‐xylulose 5‐phosphate; CsGGPPs, geranylgeranyl diphosphates; CsBCH1, β‐carotene hydroxylase 1; CsNCED2, 9‐cisepoxycarotenoid dioxygenase 2.
Materials and methods
Plant materials
Citrus materials including ‘Valencia’ orange fruit (Citrus. sinensis Osbeck.), roots, stems, leaves and flower were randomly collected from different orientations on more than six mature trees growing in the National Center of Citrus Breeding, Huazhong Agricultural University, Wuhan, China. ‘Valencia’ orange fruit was sampled every 20 d from 170 DAFB (days after full blossom) to 230 DAFB. All samples without any damages and immediately transferred to the laboratory.
Citrus calli were subcultured at 20‐day intervals on solid Murashige Tucker (MT) medium in darkness at 25 °C. Wild tobacco (Nicotiana benthamiana) was planted in a standard greenhouse conditions at 23–25 °C with relative humidity of 85%–90% under 16 h light/8 h dark cycles.
Treatments of citrus fruit and calli
For details on treatments, see Supporting Information.
Determinations of citrus colour index and carotenoid content
The citrus colour index (CCI) was quantified according to the formula: CCI = 1000 × a/(L × b). The magnitude of the CCI value is proportional to the degree of coloration. Peel colour parameters of L (brightness index), a (red saturation index) and b (yellow saturation index) were measured as described previously using the KONICA MINOLTA CR‐400 (Japan) (Sun et al., 2021). Peel colour parameters of L, a and b were determined at six evenly distributed equatorial sites of the citrus fruit using the KONICA MINOLTA CR‐400 (Japan). Nine citrus fruits were used as one biological replicate, and a total of three biological replicates were performed. Carotenoid extraction and analysis of citrus fruit and calli were performed as described previously (Zhu et al., 2022). Carotenoid identification was conducted by comparing the characteristic spectral properties and typical retention times, as well as the carotenoid levels were quantified by comparing the calibration curves with authentic standards from CaroteNature (Lupsingen, Switzerland). At least three biological replicates from independent extractions were performed.
Endogenous ABA determination
Endogenous ABA extraction and measurement were performed as previously described (He et al., 2018; Zhang et al., 2021; Zhu et al., 2017). An Agilent 1100 HPLC system coupled to an Agilent API3000 (Agilent) was used for ABA analysis and 2H6‐ABA (Olomouc, Czech Republic) was used as internal standards for ABA. Particular procedures were followed according to above studies. At least three biological replicates from independent extractions were performed.
RNA extraction, cDNA synthesis and real‐time quantitative PCR (RT‐qPCR)
Total RNA extraction and cDNA synthesis were conducted according to the manufacturer's instructions of the RN38‐EASY RNA extraction kit (Aidlab Biotechnology, Beijing, China) and the HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme) respectively. RT‐qPCR was performed as previously reported (Lu et al., 2018; Zhang et al., 2021). For citrus fruit, the peel from three independent groups as three replications. For citrus calli, each line of transgenic calli as one biological replicate. Three replicates were used in each experiment. Each replicate was conducted in triplicates. The RT‐qPCR data were normalized to the expression of ACTIN and analysed using the E−ΔΔCt method. All gene‐specific primers used for RT‐qPCR are listed in Table S1.
Generations of transgenic citrus calli
See Supporting Information for detail. All primers are listed in Table S1.
Citrus fruit injection assays
See Supporting Information for detail. All primers are listed in Table S1.
Gene cloning and sequence analysis
The full‐length CDS and promoter sequence of CsbZIP44 from ‘Valencia’ orange fruit were amplified according to the reference genome of sweet orange (http://citrus.hzau.edu.cn/orange/) and CPBD (http://citrus.hzau.edu.cn/). Multiple sequence alignments were performed by CLUSTAL W and GENEDOC software programs. Phylogenetic tree was performed using MEGA 7.0. All primers are listed in Table S1.
Subcellular localization assay
The CsbZIP44 full‐length CDS was fused to the CsbZIP44‐GFP vector. The construct (35S::CsbZIP44‐GFP) and the control vector (35S::GFP) were co‐transformed with the nuclear marker NF‐YA4‐mCherry into N. benthamiana leaves by A. tumefaciens‐mediated transformation respectively. After incubation for 3 days, the florescence images were captured with the confocal microscope (TCS SP8, Leica, Germany). All primers are listed in Table S1.
Transcriptional activation assay
See Supporting Information for detail. All primers are listed in Table S1.
Dual luciferase reporter assay
See Supporting Information for detail. All primers are listed in Table S1.
Yeast one‐hybrid (Y1H) assay
See Supporting Information for detail. All primers are listed in Table S1.
Yeast two‐hybrid (Y2H) assay
See Supporting Information for detail. All primers are listed in Table S1.
Electrophoretic mobility shift (EMSA) assay
See Supporting Information for detail. All primers are listed in Table S1.
Chromatin immunoprecipitation (ChIP)‐PCR (ChIP‐PCR) assay
See Supporting Information for detail. Specific primers used in ChIP‐PCR assays were listed in Table S1.
Co‐immunoprecipitation (CoIP) assay
See Supporting Information for detail. All primers are listed in Table S1.
Pull‐Down assay
See Supporting Information for detail. All primers are listed in Table S1.
Luciferase complementation assay
See Supporting Information for detail. All primers are listed in Table S1.
Statistical analyses
The statistical analysis of data was performed by Microsoft Office 2010 and GraphPad 8.0 software. Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
Accession numbers
Sequence data from this study can be found in the reference genome of sweet orange (http://citrus.hzau.edu.cn/orange/) and CPBD (http://citrus.hzau.edu.cn/). All accession numbers for this study are listed in Table S1.
Conflict of interest
All authors have no conflict of interest to declare.
Author contributions
X.X.D. supervised the research; Q.S. and X.X.D. designed the experiments; Q.S. performed the experiments with contributions from Z.C.H. and R.R.W.; Y. Z. and Z.Z.X. provided the plant materials. Q.S. wrote the manuscript; X.X.D. revised the manuscript; J.L.Y., L.J.C., J.X., W.W.G., Y.J.C., Q.X. and D.G.H. provided critical comments on manuscript editing.
Supporting information
Figure S1 Abscisic acid (ABA) treatment promotes carotenoid biosynthesis in citrus.
Figure S2 ABA treatment significantly activates the expressions of CsBCH1 and CsNCED2 in citrus.
Figure S3 The expressions of alternative transcription factors (TFs) under ABA treatment.
Figure S4 Sequence alignment and phylogenetic analysis of CsbZIP44.
Figure S5 CsbZIP44 acts as a nucleus‐localized transcriptional activator.
Figure S6 Expression of CsbZIP44 in transgenic citrus calli.
Figure S7 CsbZIP44 directly binds the promoters of target genes that contribute to carotenoid accumulation.
Figure S8 CsbZIP44 has no transcriptional self‐activation in yeast cells.
Figure S9 ABA treatment activates the expression of CsHB5.
Figure S10 CsbZIP44 as a potential downstream target gene of CsHB5.
Figure S11 Schematic representation of potential binding elements (AATNATT) of CsHB5 in the CsbZIP44 promoter.
Figure S12 CsbZIP44 and CsHB5 positively regulate ABA biosynthesis in citrus.
Data S1 Detailed description of methods.
Table S1 Genes ID and the primers used for this study
Acknowledgements
We thank the Prof. Aide Wang (Shenyang Agricultural University, Shenyang, China) for providing the pRI101, pRI101‐GFP and NY‐YA4‐mCherry plasmids. We thank the Prof. Dagang Hu (Shandong Agricultural University, Taian, China) for providing critical suggestions on the ideas of this manuscript. This research was supported by the National Natural Science Foundation of China (Nos. 31930095, 32172527) and Modern Agro‐industry Technology Research System (CARS‐26).
References
- Adams‐Phillips, L. , Barry, C. and Giovannoni, J. (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9, 331–338. [DOI] [PubMed] [Google Scholar]
- An, J.‐P. , Yao, J.‐F. , Xu, R.‐R. , You, C.‐X. , Wang, X.‐F. and Hao, Y.‐J. (2018) Apple bZIP transcription factor MdbZIP44 regulates abscisic acid‐promoted anthocyanin accumulation. Plant Cell Environ. 41, 2678–2692. [DOI] [PubMed] [Google Scholar]
- An, J.‐P. , Zhang, X.‐W. , Liu, Y.‐J. , Wang, X.‐F. , You, C.‐X. and Hao, Y.‐J. (2021) ABI5 regulates ABA‐induced anthocyanin biosynthesis by modulating the MYB1‐bHLH3 complex in apple. J. Exp. Bot. 72, 1460–1472. [DOI] [PubMed] [Google Scholar]
- Bhagat, P.K. , Verma, D. , Sharma, D. and Sinha, A.K. (2021) HY5 and ABI5 transcription factors physically interact to fine tune light and ABA signaling in Arabidopsis. Plant Mol. Biol. 107, 117–127. [DOI] [PubMed] [Google Scholar]
- Carmona, L. , Zacarías, L. and Rodrigo, M.J. (2012) Stimulation of coloration and carotenoid biosynthesis during postharvest storage of ‘Navelina’ orange fruit at 12°C. Postharvest Biol. Technol. 74, 108–117. [Google Scholar]
- Chenge‐Espinosa, M. , Cordoba, E. , Romero‐Guido, C. , Toledo‐Ortiz, G. and León, P. (2018) Shedding light on the methylerythritol phosphate (MEP)‐pathway: long hypocotyl 5 (HY5)/phytochrome‐interacting factors (PIFs) transcription factors modulating key limiting steps. Plant J. 96, 828–841. [DOI] [PubMed] [Google Scholar]
- Davuluri, G.R. , van Tuinen, A. , Fraser, P.D. , Manfredonia, A. , Newman, R. , Burgess, D. , Brummell, D.A. et al. (2005) Fruit‐specific RNAi‐mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23, 890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H. , Wang, N. , Cui, F. , Li, X. , Xiao, J. and Xiong, L. (2010) Characterization of the β‐carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol. 154, 1304–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciullino, A.‐L. , Dhuique‐Mayer, C. , Luro, F. , Casanova, J. , Morillon, R. and Ollitrault, P. (2006) Carotenoid diversity in cultivated citrus is highly influenced by genetic factors. J. Agric. Food Chem. 54, 4397–4406. [DOI] [PubMed] [Google Scholar]
- Fang, J. , Chai, C. , Qian, Q. , Li, C. , Tang, J. , Sun, L. , Huang, Z. et al. (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre‐harvest sprouting and photo‐oxidation in rice. Plant J. 54, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani, S. , Masiero, S. and Mizzotti, C. (2019) Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 70, 2993–3006. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D. and Bramley, P.M. (2004) The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43, 228–265. [DOI] [PubMed] [Google Scholar]
- Fu, C.‐C. , Han, Y.‐C. , Kuang, J.‐F. , Chen, J.‐Y. and Lu, W.‐J. (2017) Papaya CpEIN3a and CpNAC2 co‐operatively regulate carotenoid biosynthesis‐related genes CpPDS2/4, CpLCY‐e and CpCHY‐b during fruit ripening. Plant Cell Physiol. 58, 2155–2165. [DOI] [PubMed] [Google Scholar]
- Fu, C.‐C. , Chen, H.‐J. , Gao, H.‐Y. , Wang, S.‐L. , Wang, N. , Jin, J.‐C. , Lu, Y. et al. (2021) Papaya CpMADS4 and CpNAC3 co‐operatively regulate ethylene signal genes CpERF9 and CpEIL5 during fruit ripening. Postharvest Biol. Technol. 175, 111485. [Google Scholar]
- Galpaz, N. , Wang, Q. , Menda, N. , Zamir, D. and Hirschberg, J. (2008) Abscisic acid deficiency in the tomato mutant high‐pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730. [DOI] [PubMed] [Google Scholar]
- Gong, J. , Zeng, Y. , Meng, Q. , Guan, Y. , Li, C. , Yang, H. , Zhang, Y. et al. (2021) Red light‐induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J. Exp. Bot. 72, 6274–6290. [DOI] [PubMed] [Google Scholar]
- He, Y. , Han, J. , Liu, R. , Ding, Y. , Wang, J. , Sun, L. , Yang, X. et al. (2018) Integrated transcriptomic and metabolomic analyses of a wax deficient citrus mutant exhibiting jasmonic acid‐mediated defense against fungal pathogens. Hortic. Res. 5, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, N. , Yoshida, R. , Todoroki, Y. and Ohigashi, H. (2000) Biosynthesis of abscisic acid by the non‐mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Biochem. 64, 1448–1458. [DOI] [PubMed] [Google Scholar]
- Iglesias‐Fernández, R. , Barrero‐Sicilia, C. , Carrillo‐Barral, N. , Oñate‐Sánchez, L. and Carbonero, P. (2013) Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase‐encoding gene AtMAN7. Plant J. 74, 767–780. [DOI] [PubMed] [Google Scholar]
- Ikoma, Y. , Matsumoto, H. and Kato, M. (2014) The characteristics of carotenoid biosynthesis in citrus fruit. Jpn Agric. Res. Q. 48, 9–16. [Google Scholar]
- Ikoma, Y. , Matsumoto, H. and Kato, M. (2016) Diversity in the carotenoid profiles and the expression of genes related to carotenoid accumulation among citrus genotypes. Breed. Sci. 66, 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Sun, Q. , Zhang, T. , Liu, W. , Wang, N. and Chen, X. (2021) MdMYB114 regulates anthocyanin biosynthesis and functions downstream of MdbZIP4‐like in apple fruit. J. Plant Physiol. 257, 153353. [DOI] [PubMed] [Google Scholar]
- Karlova, R. , Chapman, N. , David, K. , Angenent, G.C. , Seymour, G.B. and de Maagd, R.A. (2014) Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541. [DOI] [PubMed] [Google Scholar]
- Kato, M. , Ikoma, Y. , Matsumoto, H. , Sugiura, M. , Hyodo, H. and Yano, M. (2004) Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiol. 134, 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, J. , Whitehead, D. , Horak, J. , Wanke, D. , Weinl, S. , Batistic, O. , D'Angelo, C. et al. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV‐B light, drought and cold stress responses. Plant J. 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Lee, J.M. , Joung, J.‐G. , McQuinn, R. , Chung, M.‐Y. , Fei, Z. , Tieman, D. , Klee, H. et al. (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 70, 191–204. [DOI] [PubMed] [Google Scholar]
- Li, T. , Xu, Y. , Zhang, L. , Ji, Y. , Tan, D. , Yuan, H. and Wang, A. (2017) The Jasmonate‐activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell, 29, 1316–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren, L.O. , Stålberg, K.G. and Höglund, A.‐S. (2003) Seed‐specific overexpression of an endogenous arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol. 132, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Zhang, Y. , Zhu, K. , Yang, W. , Ye, J. , Chai, L. , Xu, Q. et al. (2018) The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 176, 2657–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, Y. , Wang, S. , Wang, R. and Xu, C. (2020) Accumulation of red apocarotenoid β‐citraurin in peel of a spontaneous mutant of huyou (Citrus changshanensis) and the effects of storage temperature and ethylene application. Food Chem. 309, 125705. [DOI] [PubMed] [Google Scholar]
- Meng, Y. , Wang, Z. , Wang, Y. , Wang, C. , Zhu, B. , Liu, H. , Ji, W. et al. (2019) The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40‐1 to regulate carotenoid‐derived flower pigmentation in Medicago truncatula . Plant Cell, 31, 2751–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar, N. , Li, L. , Lu, S. , Khin, N.C. and Pogson, B.J. (2015) Carotenoid metabolism in plants. Mol. Plant, 8, 68–82. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.J. , Marcos, J.F. , Alférez, F. , Mallent, M.D. and Zacarías, L. (2003) Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit‐specific alteration that results in yellow pigmentation and decreased ABA content. J. Exp. Bot. 54, 727–738. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.‐J. , Alquezar, B. and Zacarías, L. (2006) Cloning and characterization of two 9‐cis‐epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 57, 633–643. [DOI] [PubMed] [Google Scholar]
- Rodrigo, M.J. , Alquézar, B. , Alós, E. , Lado, J. and Zacarías, L. (2013) Biochemical bases and molecular regulation of pigmentation in the peel of Citrus fruit. Sci. Hortic. 163, 46–62. [Google Scholar]
- Romero, P. , Lafuente, M.T. and Rodrigo, M.J. (2019) A sweet orange mutant impaired in carotenoid biosynthesis and reduced ABA levels results in altered molecular responses along peel ripening. Sci. Rep. 9, 9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. , Yuan, H. , Cao, H. , Yazdani, M. , Tadmor, Y. and Li, L. (2018) Carotenoid metabolism in plants: the role of plastids. Mol. Plant, 11, 58–74. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , He, Y. , Ye, J. , Zheng, X. , Zhou, C. , Fu, A. , Wei, R. et al. (2021) Storage with apple fruit to improve peel color and maintain freshness of Newhall navel orange. Sci. Hortic. 287, 110246. [Google Scholar]
- Wang, X. , Yin, W. , Wu, J. , Chai, L. and Yi, H. (2016) Effects of exogenous abscisic acid on the expression of citrus fruit ripening‐related genes and fruit ripening. Sci. Hortic. 201, 175–183. [Google Scholar]
- Wang, X. , Zeng, W. , Ding, Y. , Wang, Y. , Niu, L. , Yao, J.‐L. , Pan, L. et al. (2019) PpERF3 positively regulates ABA biosynthesis by activating PpNCED2/3 transcription during fruit ripening in peach. Hortic. Res. 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiste, C. , Pedrotti, L. , Selvanayagam, J. , Muralidhara, P. , Fröschel, C. , Novák, O. , Ljung, K. et al. (2017) The Arabidopsis bZIP11 transcription factor links low‐energy signalling to auxin‐mediated control of primary root growth. PLoS Genet. 13, e1006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier, F. , Rahmani, F. , Ehlert, A. , Dietrich, K. , Schütze, K. , Wang, X. , Chaban, C. et al. (2009) Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Xu, Z. , Zhang, Y. , Chai, L. , Yi, H. and Deng, X. (2014) An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late‐ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 65, 1651–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H. , Zhang, J. , Nageswaran, D. and Li, L. (2015) Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2, 15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, Y. , Sun, Q. , Lu, S. , Chai, L. , Ye, J. and Deng, X. (2021) Citrus transcription factor CsHB5 regulates abscisic acid biosynthetic genes and promotes senescence. Plant J. 108, 151–168. [DOI] [PubMed] [Google Scholar]
- Zhu, F. , Luo, T. , Liu, C. , Wang, Y. , Yang, H. , Yang, W. , Zheng, L. et al. (2017) An R2R3‐MYB transcription factor represses the transformation of α‐ and β‐branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate . New Phytol. 216, 178–192. [DOI] [PubMed] [Google Scholar]
- Zhu, F. , Luo, T. , Liu, C. , Wang, Y. , Zheng, L. , Xiao, X. , Zhang, M. et al. (2020) A NAC transcription factor and its interaction protein hinder abscisic acid biosynthesis by synergistically repressing NCED5 in Citrus reticulate . J. Exp. Bot. 71, 3613–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, K. , Sun, Q. , Chen, H. , Mei, X. , Lu, S. , Ye, J. , Chai, L. et al. (2021) Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 72, 3137–3154. [DOI] [PubMed] [Google Scholar]
- Zhu, K. , Chen, H. , Zhang, Y. , Liu, Y. , Zheng, X. , Xu, J. , Ye, J. et al. (2022) Chapter six—Carotenoid extraction, detection, and analysis in citrus. In Methods in Enzymology ( Wurtzel, E.T. , ed), p. 179, 212. Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Abscisic acid (ABA) treatment promotes carotenoid biosynthesis in citrus.
Figure S2 ABA treatment significantly activates the expressions of CsBCH1 and CsNCED2 in citrus.
Figure S3 The expressions of alternative transcription factors (TFs) under ABA treatment.
Figure S4 Sequence alignment and phylogenetic analysis of CsbZIP44.
Figure S5 CsbZIP44 acts as a nucleus‐localized transcriptional activator.
Figure S6 Expression of CsbZIP44 in transgenic citrus calli.
Figure S7 CsbZIP44 directly binds the promoters of target genes that contribute to carotenoid accumulation.
Figure S8 CsbZIP44 has no transcriptional self‐activation in yeast cells.
Figure S9 ABA treatment activates the expression of CsHB5.
Figure S10 CsbZIP44 as a potential downstream target gene of CsHB5.
Figure S11 Schematic representation of potential binding elements (AATNATT) of CsHB5 in the CsbZIP44 promoter.
Figure S12 CsbZIP44 and CsHB5 positively regulate ABA biosynthesis in citrus.
Data S1 Detailed description of methods.
Table S1 Genes ID and the primers used for this study


