Abstract
Forkhead box protein 3 expressing (FOXP3+) regulatory T cells (Treg cells) suppress conventional T cells and are essential for immunological tolerance. FOXP3, the master transcription factor (TF) of Treg cells, controls the expression of multiples genes to guide Treg cell differentiation and function. However, only a small fraction (<10%) of Treg cell-associated genes are directly bound by FOXP3 and FOXP3 alone is insufficient to fully specify the Treg cell programme, indicating a role for other accessory TFs operating upstream, downstream and/or concurrently with FOXP3 to direct Treg cell specification and specialized functions. Indeed, the heterogeneity of Treg cells can be at least partially attributed to differential expression of TFs that fine-tune their trafficking, survival and functional properties, some of which are niche specific. In this Review, we discuss the emerging roles of accessory TFs in controlling Treg cell identity. We specifically focus on members of the basic helix-loop-helix family (AHR), basic leucine zipper family (BACH2, NFIL3 and BATF), CUT homeobox family (SATB1), zinc finger domain family (BLIMP1, Ikaros and BCL-11B) and interferon regulatory factor family (IRF4), as well as lineage-defining TFs (T-bet, GATA3, RORγt and BCL-6). Understanding the imprinting of Treg cell identity and specialized function will be key to unravelling basic mechanisms of autoimmunity and identifying novel targets for drug development.
Introduction
CD4+ regulatory T cells (Treg cells), characterized by expression of CD25 (IL-2 receptor α-chain) and forkhead box p3 (FOXP3), represent one of the key cellular mechanisms for peripheral tolerance induction in mammals1-3. The transcription factor (TF) FOXP3, a member of the forkhead-winged-helix family, is constitutively expressed in Treg cells and is essential both for their specification and function2-4. The critical roles of FOXP3 and Treg cells themselves are illustrated by mammalian Treg cell-deficiency diseases that manifest as fatal multiorgan autoimmune inflammation. These include the human syndrome X-linked immunodysregulation polyendocrinopathy and enteropathy (IPEX) and the Scurfy mouse model, in which mutations in the forkhead domain of FOXP3, which is responsible for nuclear import and DNA binding, result in Treg cell deficiency and failure to restrict self-reactive conventional T cells5-7. Despite its importance, FOXP3 is insufficient by itself to specify the complete Treg cell transcriptome8,9, exemplified by the lack of suppressive capability of FOXP3-expressing activated human conventional T cells. Indeed, chromatin immunoprecipitation with high-throughput sequencing (CHIP-seq) studies that pinpoint genome-wide FOXP3 binding in Treg cells indicate that only a small proportion of genes that are dependent on intact FOXP3 expression are bound by FOXP310,11. This suggests that a substantial part of the Treg cell transcriptional programme is regulated by other TFs, either alone or in combination with FOXP3.
Recent advances in single cell transcriptomics and emerging concepts of mammalian T helper cell polarization suggest that it is common for “master” lineage-specifying TFs to be co-expressed. Thus, T cell behaviour and stability might best be understood when considering transcriptional outputs produced by interactions between gradients of competing TFs, non-coding RNAs and cellular epigenetics12-15. Conceptually, a modular model offers the most accurate framework for describing the Treg cell lifecycle, with specific functions “added” to the essential FOXP3 programme by accessory TFs. In this Review, we focus on the emerging roles of some of the accessory TFs that control Treg cell specification and/or maturation (Figure 1 and Tables 1-2). Although there are many such TFs, we restrict the discussion to members of the basic helix-loop-helix family (AHR), basic leucine zipper (bZIP) family (BACH2, NFIL3 and BATF), CUT homeobox family (SATB1), zinc finger domain family (BLIMP1, Ikaros and BCL-11B), interferon regulatory factor family (IRF4), as well as conventional T cell lineage-defining TFs (T-bet, GATA3, RORγt and BCL-6).
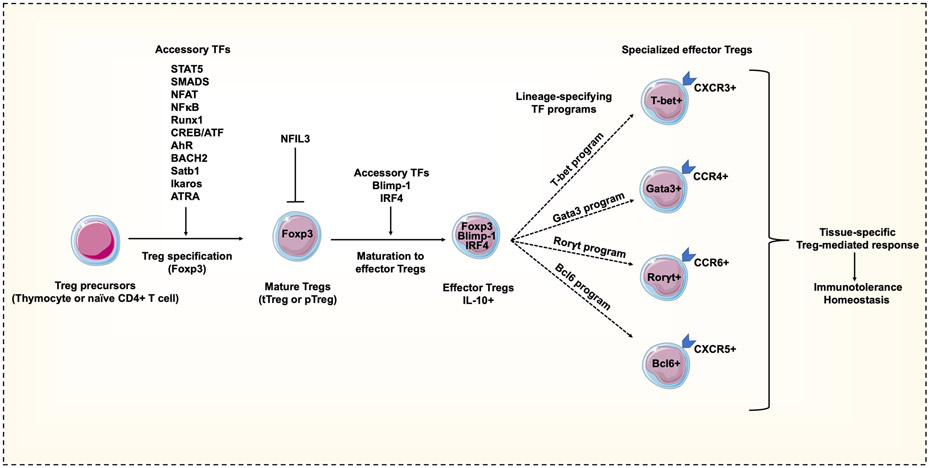
Figure 1. Accessory transcription factors in regulatory T cell specification and maturation.
Thymocytes or naive CD4+ T cells differentiate into regulatory T (Treg) cells following T cell receptor (TCR) engagement within microenvironments rich in Treg cell-inducing soluble factors, such as interleukin-2 (IL-2) and transforming growth factor-β (TGFβ). The coordinated integration of multiple accessory transcription factors (TFs) drive Treg cell specification and epigenetic changes (such as at the Treg cell-specific demethylated region (TSDR)) that are indispensable for stable expression of forkhead box protein 3 (FOXP3). Treg cell maturation to effector Treg cells is driven by FOXP3-dependent and FOXP3-independent accessory TFs that induce FOXP3 expression and enhance production of effector (suppressive) cytokines. Some TFs, such as NFIL3, have the ability to repress FOXP3 expression. Mature effector Treg cells can also be induced to express additional accessory programmes driven by TFs usually associated with lineage-specification in conventional T cells. These shape the unique features of specialized subpopulations of Treg cells, such as tissue homing.
Table 1 ∣.
Accessory and lineage-specifying transcription factors that shape regulatory T cell phenotype
| Transcription factor |
Transcription factor family |
Function in Treg cells | Location |
|---|---|---|---|
| FOXP3 | FOX protein family | Development and function | Expressed in CD4+ Treg cells |
| AHR | Class I basic helix-loop-helix transcriptional regulator | FOXP3 agonist in development and enhanced suppressive function (IL-10, GZMB and homing receptors) | Organ specific (central nervous system, gut-associated lymphoid tissue) |
| BACH2 | Basic leucine zipper transcriptional regulator | FOXP3 agonist in development, function, maintenance of steady state and suppressor of pro-inflammatory genes | Expressed in CD4+ Treg cells |
| SATB1 | CUT homeobox factor | FOXP3 expression in early developmental stages but antagonist in mature Treg cells | Expressed mostly in Treg cell precursors |
| BCL-11B * | Zinc finger domain protein | FOXP3 agonist in Treg cells; inter-dependent function with FOXP3 in Treg cells | Expressed in CD4+ Treg cells |
| Ikaros | Zinc finger domain protein | Development and differentiation of in vitro-induced Treg cells | Expressed in CD4+ Treg cells |
| IRF4 | Interferon regulatory factor | Generation of effector Treg cells; synergism with FOXP3 and BLIMP1 to transactivate IL10 | Mucosa and visceral adipose tissue |
| BLIMP1 | Zinc finger domain protein | Required for optimal function of effector Treg cells; synergism with FOXP3 and IRF4 to transactivate IL10, GZMB and suppress IL-17 production | Organs (kidney, pancreas, lung, central nervous system) and gut-associated lymphoid tissue |
| BATF | Basic leucine zipper transcriptional regulator | Development of non-lymphoid Treg cell precursors; growth and sustainability of tissue Treg cells | Non-lymphoid Treg cell precursors; tissue Treg cells |
| NFIL3 * | Basic leucine zipper transcriptional regulator | FOXP3 antagonist; directly binds the FOXP3 locus as well as FOXP3 protein | Inducible during chronic infections |
| T-bet | Nuclear receptor family | Enhances suppressive capacity and expression of tissue-specific homing receptors for sites of TH1-type inflammation | Organs (central nervous system, pancreas) and gut-associated lymphoid tissue |
| GATA3 | Nuclear receptor family | Enhanced suppressive capacity with expression of tissue-specific homing receptors | Organs (skin, kidney) and gut-associated lymphoid tissue |
| RORγt | Nuclear receptor family | Enhanced suppressive capacity and expression of tissue-specific homing receptors | Gut-associated lymphoid tissue |
| BCL-6 | Zinc finger domain protein | Expression of homing receptors for germinal centres; regulation of germinal centre reactions | Germinal centre Treg cells |
Transcription factors for which comparable human data are limited or not available. For all other transcription factors, there is functional evidence in both human and mouse Treg cells.
Table 2 ∣.
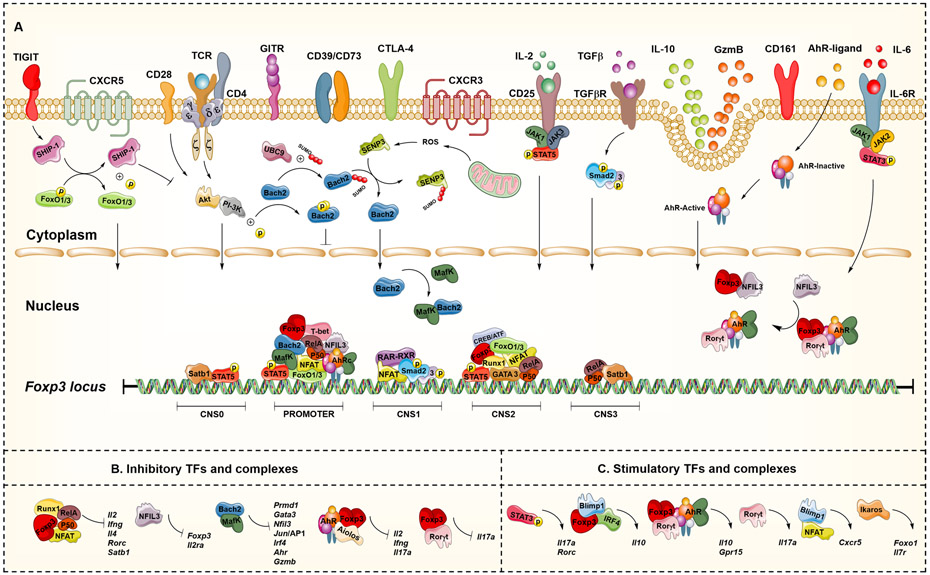
Inhibitory and stimulatory transcription factors and complexes in regulatory T cells
| TFs and TF complexes | Genes regulated |
|---|---|
| Inhibitory TFs and complexes | |
| AHR–FOXP3–Aiolos | IL2, IFNG, IL17A |
| BACH2–MAFK | PRMD1, GATA3, NFIL3, JUN–AP-1, IRF4, AHR, GZMB |
| FOXP3–RORγt | IL17A |
| FOXP3–RUNX1–RELA–p50–NFAT | IL2, IFNG, IL4, RORC, SATB1 |
| NFIL3 | FOXP3, IL2RA |
| Stimulatory TFs and complexes | |
| BATF | IL10, CTLA4, TIGIT, TNFRSF4, TNFRSF9, IL1RL1 |
| BCL11B | IL10, FOXP3 |
| BLIMP1–BCL-6–NFAT | CXCR5 |
| BLIMP1–FOXP3–IRF4 | IL10 |
| Ikaros | IL7R, FOXO1 |
| FOXP3 | BATF |
| FOXP3–RORγt–AHR | IL10, GPR15 |
| RORγt | IL17A |
| STAT3 | IL17A, RORC |
During regulatory T cell differentiation, transcriptional interactions result in the transient assembly of both inhibitory and stimulatory transcription factors (TFs) and complexes, which independently or in conjunction with FOXP3, repress or induce the expression of genes involved in the maintenance of hallmark regulatory T cell genes and specialized function within tissues.
Treg cell specification and maturation
The development of thymus-derived Treg cells, referred to as ‘tTreg cells’, takes place neonatally in the thymus. This is triggered by thymocytes that are activated through T cell receptors (TCRs) with an above-average affinity for self-antigens and by CD25 signalling, which lead to the expression of FOXP316-18. In addition, Treg cells also form postnatally in the periphery, known as ‘pTreg cells’. Peripheral specification is driven by the sensing of environmental signals, including TCR and CD25 signalling. In both tTreg cells and pTreg cells, TFs downstream of TCR engagement bind to the promoter and conserved non-coding sequence (CNS) regions of the FOXP3 gene locus, including a Treg cell-specific demethylated region (TSDR)16,17 (Figure 2). CpG dinucleotides at the TSDR are mostly demethylated in tTreg cells, but partially or completely methylated in in vitro-induced Treg cells (‘iTreg cells’) and conventional T cells19-23. While there are many mechanisms by which Treg cells repress immune cell activation24, most are attributed to stable FOXP3 expression24,25, which in turn is determined by specific epigenetic marks, such as those at the TSDR, and by the recruitment of multiple TFs driving FOXP3 expression26-28 (Figure 2). To imprint Treg cell specification and function, cells require a network of accessory TFs, including nuclear factor of activated T cells (NFAT)29, nuclear factor-κB (NF-κB)30, signal transducer and activator of transcription 5 (STAT5)27, runt-related transcription factor 1 (RUNX1)31, cAMP response element binding protein (CREB), activating transcription factor (ATF)32 and SMAD proteins33. These TFs operate in concert to specify the mature Treg cell programme (Table 2), which is characterized by expression of specific cell-surface molecules (such as CD25) and soluble factors and repression of genes associated with effector T cell function (such as IL2, IFNG and IL4)4,17,24,29,30 (Table 2). The intricate nature of these networks could be exemplified by RUNX1, which activates transcription of IL2 and IFNG when FOXP3 is absent31. Conversely, when FOXP3 is present, it interacts with RUNX1 and prevents induction of interleukin-2 (IL-2) and interferon-γ (IFNγ) and imprints Treg cell-associated molecules and suppressive function31. Thus, deletion of Runx1 in mouse naive CD4+ T cells permits unhindered cellular activation and cytokine production, resulting in spontaneous, catastrophic autoimmunity34. As expected, single nucleotide variants in the RUNX1 binding site are associated with susceptibility to autoimmunity (psoriasis) in humans35.
Figure 2. The coordinated network of accessory and lineage-specifying transcription factors regulating FOXP3 expression.
Engagement of T cell receptors (TCRs) by antigen-presenting cells (APCs) through the MHC class II-antigen complex, signalling of interleukin-2 (IL-2) via the CD25-STAT5 module, and activation of canonical transforming growth factor-β (TGFβ)-dependent SMAD pathways all work together to promote the differentiation of regulatory T cells (Treg cells) and expression of forkhead box protein 3 (FOXP3). Other signals, such as cytokines and endogenous chemical compounds present in the environment, are detected by specific cell-surface receptors and transcription factors, such as AHR and RORγt. These signals are integrated along with additional transcriptional regulators (such as BACH2, MAF, Ikaros, Aiolos, RUNX1, FOXO1, SATB1 and NFIL3) at conserved noncoding sequence (CNS) regions of FOXP3. Together, these regulate expression of FOXP3 and the intracellular mechanisms required to activate Treg cell suppressive functions through cell contact or soluble factors. These transcriptional regulators also form stimulatory and inhibitory complexes that regulate genes involved in the maintenance of hallmark and specialized genes expressed by Treg cells, as summarized in Table 2.
Treg cell maturation in peripheral tissues is shaped by activating signals in the environment, which induce an effector phenotype (characteristically CD62LlowCD44hiCCR7low) and suppressive markers such as IL-10 and programmed cell death 1 (PD1)22,36,37. Recruitment of accessory TFs superimpose supplementary gene modules or ‘programmes’ that are hallmarks of effector Treg cells and impart specialized function and specify trafficking to different tissues (Figure 1). Consistently, transcriptional profiling of FOXP3hiCD4+ subpopulations shows a small but reproducible (core) set of Treg cell-specific genes (such as Foxp3, Il2ra and Tnfrsf18 (which encodes GITR)) onto which additional programmes, sometimes with striking similarity to those observed in conventional T cells, are added38,39. Thus, there is substantial heterogeneity in Treg cell programmes involving interplay and interactions between a number of transcriptional regulators40.
Accessory transcription factors in Treg cell specification
AHR
Aryl hydrocarbon receptor (AHR) is a member of the class I bHLH proteins, serving as sensors of diverse environmental factors, such as xenobiotics, oxygen tension and endogenous ligands generated from host cells, diet and the microbiota41-43. Initially recognized as a mediator of the toxic effects of dioxins, AHR is widely expressed in immune and non-immune cells42,43. AHR demonstrates promiscuity for endogenous and exogenous ligands with different structures and physiochemical characteristics, thus can produce opposing effects in different cells44. In immune cells, AHR is an immunoregulatory TF, influencing T cell differentiation and cytokine production45. For example, in parent-into-F1 acute graft versus host disease models, AHR signalling suppresses cytotoxic T cells46 and generates CD4+CD25+ T cells with characteristics of suppressive Treg cells expressing cytotoxic T lymphocyte antigen 4 (CTLA4) and glucocorticoid-induced TNFR-related protein (GITR)47. Consistently, in vivo injection of mice with the high affinity AHR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induces Treg cells, suppressing experimental autoimmune encephalomyelitis (EAE)48 and experimental autoimmune uveoretinitis49. Approximately 40-50% of Ahr−/− mice die shortly after birth due to inflammatory infiltration in multiple organs50. These mice demonstrate ~30% reduction in Treg cells, with the residual Treg cells displaying decreased FOXP3 levels51. Interestingly, 6-formylindolo[3,2-b]carbazole (FICZ), a tryptophan-derived endogenous high affinity AHR ligand52, does not induce FOXP3 expression48. Instead, FICZ synergizes with transforming growth factor-β (TGFβ), IL-6 and IL-23 to induce expression of retinoic acid receptor-related orphan receptor-γt (RORγt), expansion of T helper 17 (TH17) cells48,53 and increased severity of EAE48.
The exact molecular mechanism or mechanisms of AHR in Treg cell development and function is incompletely understood. Inactive AHR resides in a cytoplasmic complex containing heat shock protein 90, AHR-interacting protein, p23 chaperone and c-SRC protein kinase. These components prevent AHR from ubiquitylation and degradation, thus maintaining steady-state protein expression. AHR agonists induce conformational changes promoting its nuclear translocation to target genes containing the AHR binding DNA motif (the dioxin response element (DRE))54,55 (Figure 2). The mouse Foxp3 promoter contains an evolutionarily conserved AHR-binding site and three non-evolutionarily conserved AHR-binding sites48. AHR binds both sites in TCDD-treated naive CD4+ T cells and transactivates Foxp3 transcription48. The Gpr15 locus also contains important AHR-binding sites56 and encodes an orphan G protein-coupled chemoattractant receptor, the key factor for T cell homing to the large intestine57. AHR binds two open chromatin regions in the Gpr15 locus to enhance its expression. FOXP3 also binds the same AHR-binding regions in Treg cells and physically interacts with AHR56. By contrast, RORγt overexpression competitively antagonizes AHR binding at the Gpr15 locus56. Interactions between AHR, FOXP3 and RORγt (identified by co-immunoprecipitation) indicate a Treg cell-specific network that precisely regulates Gpr15 expression56 (Table 2).
TGFβ, a key cytokine influencing both in vitro and in vivo Treg cell differentiation, also has synergic effects with AHR58 (Figure 2). SMAD proteins, canonical signalling molecules of TGFβ, bind to the FOXP3 CNS1 locus and to IL2 promoter to induce and repress gene expression, respectively33,59,60. Thus, naive human CD4+ T cells activated in the presence of TGFβ express FOXP3 without necessarily demonstrating suppressive activity61. However, if AHR is concurrently active, they do gain both the phenotypic characteristics of FOXP3+ Treg cells (such as high FOXP3 expression, but low IFNA1, IL2 and IL17 expression) and CD39-dependent suppressive function58. TGFβ-induced AHR+ Treg cells have high expression of SMAD1 and Aiolos (Ikaros family zinc finger 3), which are involved in repression of IL2 transcription in Treg cells58. Conversely, naive CD4+ T cells stimulated through AHR alone develop a FOXP3− type 1 regulatory T cell phenotype, expressing the AHR-bound target IL1058,62,63. IL10 is also bound by MAF at a MAF-recognition element, also known as MARE. AHR and MAF, individually and synergistically, transactivate IL10 to control this key immunoregulatory cytokine58,64. Collectively, these data indicate that AHR is a FOXP3 agonist and that the AHR-regulated module aids development and enhances suppressive function of Treg cells. Some of this is mediated directly and some through synergistic effects with TGFβ and recruitment of additional transcriptional regulators.
BACH2
BTB domain and CNC homology 2 (BACH2) is a member of the BACH subfamily of bZIP TFs that function as transcriptional activators and repressors65. BACH proteins contain a cap'n'collar-type bZIP domain as well as an amino-terminal broad complex, tramtrack, bric-a-brac/poxvirus and zinc finger domain, which is typically a protein interaction motif66. BACH2 maintains the balance between networks of TFs that are critical to maturation and function of both T and B cells65,67,68. It modulates the differentiation and function of multiple immune cells, including Treg cells, TH1, TH2 and TH17 cells, CD8+ T cells and natural killer (NK) cells, prevents terminal exhaustion, supports quiescence and long-term maintenance of T cell subsets and enforces stem-like transcriptional programmes69-73. Consistent with this, CHIP-seq shows BACH2-binding sites at key genes driving conventional T cell differentiation in mice, including Jun, Prmd1, Gata3, Irf4, Nfil3 (see below), Ahr (see above) and Gzmb, which are targeted for repression69,74 (Table 2). This repressive function is aided by competition for genome occupancy between BACH2 and other bZIP TFs, such as the AP-1 family member JUND70,71,75.
Treg cells are high expressors of BACH2 both in the thymus and, more heterogeneously, in the periphery69,76. BACH2 is critical for Treg cell differentiation as demonstrated by BACH2 deficient mice that develop spontaneous lethal multi-organ (especially gut and lung) lymphocytic and macrophagic inflammation, together with antinuclear and anti-double-stranded DNA autoantibodies69. These mice are deficient in Treg cells, their residual Treg cells are low in FOXP3 and don’t prevent transfer colitis69. Bach2−/− conventional T cells display spontaneous activation and produce elevated TH1 and TH2 cytokines69, indicating de-repression of conventional T cell gene programmes. Moreover, Bach2−/− naive T cells differentiate poorly to Treg cells in response to TGFβ in vitro69. All these predicates indicate that BACH2 is required for efficient generation of Treg cells77. Indeed, nuclear BACH2 is an obligate homodimer66, which in turn can form heterodimers through the bZIP domain with small MAF proteins, including MAFF, MAFG and MAFK, allowing binding to MAREs66. One such locus of BACH2 binding is in the Foxp3 promoter in TGFβ-induced Treg cells78 (Figure 2).
BACH2 also has a functional role in fully differentiated mature FOXP3+ Treg cells, in which it is highly expressed76. Treg cells from mice with genetic ablation of Bach2 selectively in Treg cells (Bach2fl/flFoxp3Cre) demonstrate reduced capacity to regulate allergic inflammation in the lungs79. Consistent with the repressive function of BACH2, the actual level of BACH2 expression in mature Treg cells may also be important. For example, BACH2 expression is downregulated in activated or effector Treg cells, which explains why BACH2−FOXP3+ Treg cells express chemokine receptors, co-stimulatory molecules, inhibitory molecules and proteins involved in Treg cell function more highly than BACH2+FOXP3+ Treg cells76. This is also seen in subpopulations of (human) Treg cells with wound healing properties80. CD161+ Treg cells are highly suppressive retinoic acid-induced FOXP3+ human Treg cells that produce effector cytokines80,81 and a wound healing gene programme80. These are recruited to the gastrointestinal tract and are particularly enriched in inflammatory bowel diseases80,82. In these cells, BACH2 downregulation works in concert with at least three other TFs, RORγt, FOSL2 and AP-1, to permit the expression of genes involved in wound healing80. Another example where BACH2 functions within networks of multiple immunoregulatory TFs is the contraction programme of human TH1 cells. In this instance, BACH2 is recruited by signals initiated by vitamin D receptor (VDR) and cooperates with VDR, STAT3 and JUN to repress effector (TH1) programmes and induce IL-10 production83.
On a population level, BACH2 is associated with both polygenic and Mendelian diseases, indicating its critical immunoregulatory role. Genetic variations in the human BACH2 locus are associated with susceptibility to several autoimmune diseases, including rheumatoid arthritis84, Crohn disease85, multiple sclerosis86, type 1 diabetes87 and asthma88. The BACH2 gene is tightly regulated by an extensive regulatory region composed of multiple enhancers, collectively termed a super enhancer. Super enhancers ensure appropriate and finely tuned expression of critically important genes and polymorphisms within these loci associate with multiple autoimmune diseases65. BACH2 itself is recruited to other super enhancer loci within T cells to act as a ‘guardian’ TF preventing autoimmunity65. In turn, single nucleotide variants in BACH2 identified in genome-wide association studies commonly occur within its associated super enhancer region, some of which (such as rs72928038) functionally impair BACH2 expression89. Homozygous deficiency of BACH2 in human populations have not been described because this gene has low tolerance to loss of function. Nevertheless, rare patients with BACH2 haploinsufficiency do exist and present with monogenic BACH2-related immunodeficiency and autoimmunity syndrome, characterized by decreased FOXP3 expression in Treg cells, increased expression of T-bet and gut homing receptors (CCR9 and integrin-β7) on CD4+ T cells, and clinical colitis together with B cell immunodeficiency68.
BACH2 shuttles between the nucleus and cytoplasm guided by a carboxy-terminal nuclear localization signal90. Serine residues in BACH2 can also be phosphorylated by phosphoinositide 3-kinase–AKT signalling and regulate nuclear trafficking91, as can sumoylation or desumoylation events at lysine residues92. Sumoylation, the addition of small ubiquitin-like modifiers (SUMO) to proteins is a reversible post-translational modification regulating trafficking, stability and biology of TFs93. Desumoylation of BACH2 catalysed by SUMO-specific protease 3 (SENP3) prevents nuclear export, resulting in nuclear accumulation and stabilization of Treg cell-associated gene loci92. SENP3 deficiency, therefore, causes spontaneous autoimmunity and enhanced antitumour immunity92 from Treg cell dysregulation. Reactive oxygen species (ROS) induce SENP3 expression94, thus ROS-rich environments, such as in cancer, drive Treg cell-mediated tumour immunosuppression, whereas ROS-low states impair Treg cell function95 (Figure 2). Indeed, this mechanism represents a plausible explanation for the link between low ROS levels and increased susceptibility to autoimmunity96. Collectively, the BACH2 module enhances FOXP3 expression and suppresses pro-inflammatory genes, aiding the development, function and steady-state maintenance of Treg cells.
BCL-11B
BCL-11B, a C2H2 zinc finger TF, has essential roles in T cell specification. It is expressed in thymocytes at the double negative 2 stage, promotes T cell lineage commitment and represses alternative lineage specification, particularly NK cells97,98. BCL-11B suppresses TH2 cell programmes in mature lymphocytes to restrict TH17 cell plasticity99. Likewise, by enhancing group 2 innate lymphoid cell (ILC2) programmes (and repressing group 3 innate lymphoid cell programmes) BCL-11B maintains peripheral ILC2 populations100. BCL-11B has both transcriptional repressor and activator functions in association with the nucleosome remodeling and deacetylase (NuRD) complex (a key transcriptional corepressor) and histone acetyltransferases (HATs, such as p300), respectively101,102. Deletion of Bcl11b in T cells (Bcl11bfl/flCd4Cre) causes colitis and wasting disease, which is preventable by transferring wild-type Treg cells103. Treg cell-specific Bcl11b deletion (Bcl11bfl/flFoxp3Cre) results in lethal multi-organ autoimmunity similar to mice lacking Treg cells104,105. Treg cells of Bcl11bfl/flCd4Cre mice and Bcl11bfl/flFoxp3Cre mice demonstrate impaired103 and almost complete loss, of suppressive function104,105, respectively. Treg cells lacking BCL-11B have lower fitness than wild-type counterparts and significant loss of characteristic Treg cell-associated genes104,105, including Il10 and Foxp3 expression103-105. There is, in fact, considerable inter-dependence between the gene regulatory programmes of BCL-11B and FOXP3. Since BCL-11B directly binds the Il10 promoter, Foxp3 promoter and Foxp3 CNS0–CNS2 regions and is required for their efficient transcription103,105 (Figure 2 and Table 2), there is significant overlap between BCL-11B-bound and FOXP3-bound genes104,105. Mechanistically, FOXP3 is required for optimal recruitment of BCL-11B to its targets and BCL-11B is, in turn, required for optimal recruitment of FOXP3 to its target loci104,105. Thus, FOXP3 is misdirected to alternative loci when BCL-11B is absent and expression of characteristic Treg cell genes is lost. Collectively, BCL-11B is an essential TF supporting the Treg cell programme and restraining alternative lineages. The BCL-11B-regulated module enhances FOXP3 expression and Treg cell-associated genes and works cooperatively with FOXP3 protein.
Ikaros
Members of the Ikaros zinc finger TF family, especially Helios and possibly EOS, have been associated with Treg cell biology and extensively discussed elsewhere106-112. More recently, Ikaros, a repressor of pro-inflammatory genes encoded by IKZF1, has been suggested as an essential TF in the induction of iTreg cells. Ikaros-deficient CD4+ T cells (Ikarosfl/flLckCre) can differentiate to TH1, TH2 and TH17 cell lineages in vitro but fail to generate iTreg cells113,114. In fact, Ikaros-deficient CD4+ T cells cultured under iTreg cell-polarizing conditions demonstrate aberrant production of IL-17 and IL-22 upon TCR activation113 and their fully differentiated TH17 cells take on the phenotype of pathogenic cells (RORγt+T-bet+IL-17+)114. In the absence of Ikaros, expression of another TF, FOXO1115, and IL-7RA decreases in CD4+ T cells116. FOXO1 promotes iTreg cell differentiation by binding to the promoter and CNS regions of Foxp3117 (Figure 2 and Table 2). T cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT), a receptor on Treg cells118, further regulates FOXO1 activity by increasing its availability in the nucleus via inhibition of AKT119. Ikaros is, in fact, required for development of all lymphoid lineages, thus mice that are deficient for all Ikaros isoforms are lymphopenic and fail to thrive from 1-3 weeks of life due to opportunistic infections120. Thus, mutations of IKZF1 strongly correlate with poor outcomes in high-risk acute lymphoblastic leukaemia121, single nucleotide variants in IKZF1 are associated with autoimmunity (such as systemic lupus erythematosis and Sjogren syndrome)122,123, and immune dysregulation characterized by abnormal T and B cell differentiation is a feature of Mendelian diseases mediated by haploinsufficient, dominant negative or gain-of-function mutations in IKZF1124,125. Although some of these patients exhibit reduced Treg cells, the broad immune dysregulation makes it currently unclear to what extent these clinical phenotypes are related to Treg cells, as opposed to other immune cells126. Collectively, Ikaros represses pro-inflammatory genes and provides an essential module for the induction of iTreg cells.
SATB1
DNA binding protein SATB1 controls transcriptional and epigenetic changes by forming long-range chromatin loops, bringing distal regions together and recruiting epigenetic modifying enzymes and transcriptional machineries to target gene loci127,128. SATB1 is highly expressed by thymocytes and is essential for controlling genes participating in T cell development and activation129,130. It is upregulated at the single-positive and double-positive stages of thymocytes development, where it binds to CNS0 and, with less affinity, to CNS3 of FOXP3, before being downregulated again in mature Treg cells131 (Figure 2). Mature (especially human) Treg cells express low levels of SATB1 compared with conventional T cells even after TCR and IL-2 stimulations132, because FOXP3 negatively regulates SATB1 expression. FOXP3 binds SATB1 in Treg cells and represses its expression whereas FOXP3 knockdown increases SATB1 in Treg cells132.
Satb1−/− mice are viable, but small, with small immune organs, and die at ~3 weeks of age, possibly from apoptosis of non-immune cells129. Mice with haematopoietic cell-specific Satb1 knockout (Stat1bfl/flVavCre) produce autoantibodies and develop spontaneous autoimmunity affecting multiple organs133. They have activated conventional T cells and reduced Treg cell number and function133. Selective deletion of Satb1 in CD4+ cells (Satb1fl/flCd4Cre) results in a significant reduction of CD4 single-positive thymocytes, but an almost complete absence of tTreg cells131. This is because a Treg cell-specific super enhancer is activated in Treg cell precursors in a SATB1-dependent manner and required for expression of signature genes attributed to Treg cells131. Thus, SATB1 is required for tTreg cell lineage specification and its deletion impairs tTreg cell development before FOXP3 expression. In mature Treg cells, FOXP3 represses Satb1 expression to prevent conventional T cell polarization and maintain stable Treg cell identity. Thus, overexpressing SATB1 in mature Treg cells results in loss of suppressive function and gain of conventional T cell programmes producing IFNγ, IL-4 and IL-17A132. SATB1 binds the promoter of Bhlhe40, which encodes a TF that drives granulocyte-macrophage colony-stimulating factor (GM-CSF) production, which is an essential TH17 cell pathogenic chemokine134. Thus, SATB1 in conventional T cells regulates pathogenic TH17 cells and its ablation in TH17 cells protects mice from EAE, due to marked reduction of GM-CSF134. Notably, SATB1 single nucleotide variants (rs11719975) associate with multiple sclerosis135. Likewise, high SATB1 expression and lineage instability is a feature of tumour-infiltrating Treg cells136. Conversely, Treg cells in chronic hepatitis B virus infection characteristically have low levels of SATB1137, which may contribute to impaired viral clearance138. Overall, SATB1 is critical for tTreg cell development prior to FOXP3 expression by controlling transcriptional and epigenetic changes. The SATB1 module in mature Treg cells is antagonistic to the Treg cell phenotype and fosters conventional T cell programmes that associate with autoimmunity.
Accessory transcription factor programmes in Treg cell maturation
IRF4
Interferon regulatory factor 4 (IRF4) is a TF involved in the expression of interferon-induced genes139 and found in a variety of cells, including B and T cells, macrophages and dendritic cells140-147. The protein is rapidly induced in T cells following TCR stimulation and subsequently regulates the commitment of T cells to TH2 and TH17 fates142,143,148. IRF4 expression in thymic epithelial cells in response to RANK signalling is crucial for inducing tTreg cells149 and IRF4-deficient thymic epithelial cells (Irf4fl/flFoxN1Cre) poorly stimulate tTreg cell differentiation, without affecting the pTreg cell pool149. Both thymic and peripheral mouse FOXP3+ Treg cells have high IRF4 levels because FOXP3 directly induces Irf4 expression after binding the Irf4 promoter, hence Foxp3 deletion markedly reduces Irf4 mRNA levels in Treg cells150. Deleting Irf4 in differentiated Treg cells (Irf4fl/flFoxp3Cre) causes spontaneous and lethal autoimmunity from 6-8 weeks of age150. In fact, Irf4 deficiency in Treg cells compromises maturation into effector Treg cells, illustrated by low expression of characteristic markers, such as inducible T cell costimulator (ICOS), CTLA4 and IL-10, as well as suppressive function36. Because the TF Pparg is directly induced by IRF4 and is essential for development of adipose-tissue Treg cells37, which are involved in preserving insulin sensitivity and glucose tolerance151, complete IRF4-deficient mice have a near total absence of this Treg cell subset37. These mice do not, however, develop overwhelming autoimmunity similar to Irf4fl/flFoxp3Cre strains, confirming that IRF4 also has key roles in conventional effector T cell programmes. Human IRF4 deficiency also leads to reduced frequency of Treg cells152. A subset of intratumoral human effector Treg cells that have high expression of IRF4 and potent suppressive function are associated with poor cancer prognosis136,153. This observation is corroborated by mice in which inducible deletion of Irf4 in Treg cells (Irf4fl/flFoxp3EGFPCre-ERT2 mice fed tamoxifen) accelerates tumour clearance153.
IRF4 interacts with FOXP3 in Treg cells (they co-immunoprecipitate from nuclear lysates150) and has a role in gene transcription, but its biology in Treg cells is not fully understood. IRF4 is in the nucleoplasm and makes homomeric and heteromeric interactions with other TFs, especially other members of the IRF and AP-1 families, through its C-terminal region139. These interactions are important to enhance its weak DNA binding and transactivation. The N-terminal DNA binding domain of IRF4 binds DNA consensus motifs akin to classical interferon-stimulated response elements139,154. One such locus is IL10, where it works cooperatively with B lymphocyte-induced maturation protein 1 (BLIMP1), which is encoded by PRDM1 (see below), or PU.1 to remodel active chromatin and transactivate gene transcription36,155 (Tables 1-2). It worth noting that IRF4 also binds the PRDM1 locus36 and induces BLIMP1 expression and that BLIMP1 is absent in IRF4-deficient Treg cells36. In summary, the IRF4-regulated module is essential for Treg cell maturation in the periphery into effector Treg cells and for synergism with FOXP3 and BLIMP1 to transactivate IL10.
BLIMP1
BLIMP1 is a transcriptional repressor interacting with other TFs, such as IRF4, through its proline-rich N-terminal domain156. It is well characterized as a master regulator orchestrating plasma cell development and immunoglobulin secretion157,158. It also has a role in T cells159,160, as seen in the association between PRDM1 polymorphisms and multiple autoimmune diseases, including inflammatory bowel diseases161-164 and development of colitis in mice with T cell ablation of Prdm1165. However, this TF is not essential for Treg cell differentiation as Treg cell numbers are generally unaltered in BLIMP1 deficiency. Rather, BLIMP1 is required for optimal function of activated and effector Treg cells. In fact, only 10-20% of mature Treg cells express BLIMP1 in lymphoid organs, whereas most Treg cells in tissues (such as the gut, lungs and central nervous system) do so, suggesting that its expression is likely to be key in specialized tissue-resident or effector Treg cells36,166. This is intuitive because BLIMP1 expression is induced by inflammatory signals, which are more likely to be present in tissues. These signals include factors such as IFNγ-induced STAT1 and IRF4, which directly bind to the promoter of Prdm136,166. In these Treg cells, BLIMP1 helps to maintain regulatory function by stabilizing Treg cell-associated genes and repressing conventional T cell-associated genes. For example, BLIMP1 in FOXP3+RORγt+ Treg cells is crucial for suppressing the production of TH17 cytokines and maintaining regulatory function (see below) by binding to CNS regions at the Il17a gene167. Consistently, EAE is exacerbated by Treg cell-selective deficiency of BLIMP1 (Prdm1fl/flFoxp3Cre) due to increased inflammatory features, such as IL-17 and IFNγ production, and loss of expression of classical Treg cell genes including Foxp3, Gzmb and Il10166.
As discussed previously, BLIMP1 works with IRF4 to activate IL10 transcription36 (Tables 1-2). IL-10 production is downregulated in Treg cells from BLIMP1-deficient mice and BLIMP1 overexpression rescues their suppressive function36,159. As expected, BLIMP1 deficiency leads to similar consequences as IL-10 insufficiency. For example, IL-10Rα signalling in adipocytes causes insulin resistance and glucose intolerance in mice by altering chromatin accessibility and repressing transcription of thermogenic genes168, but Prdm1fl/flFoxp3Cre mice are protected169. In contrast, MOG35–55-induced EAE and autoimmune diabetes in non-obese diabetic mice are more severe due to lack of IL-10 and expansion of TH1 and TH17 cells, respectively170,171. As anticipated, BLIMP1+ Treg cells in tissues have enhanced suppressive function, which can be both advantageous (as an important component of graft-infiltrating FOXP3+ Treg cells that maintain spontaneously induced kidney allograft tolerance172) or detrimental (as a mechanism for tumour immune evasion173). Furthermore, expression of BLIMP1 is a cardinal feature of a subset of Treg cells found in germinal centres, known as follicular regulatory T (TFR) cells, which are discussed below. In summary, the BLIMP1-programme is required for optimal function of activated and effector Treg cells and synergism with FOXP3 and IRF4 to transactivate Il10 and GzmB and suppress IL-17 production.
BATF
Basic leucine zipper ATF-like transcriptional factor (BATF), an AP-1 subfamily bZIP TF, is a regulator of T cells, most notably in the differentiation of T follicular helper (TFH), TH2 and TH17 cells174-176. BATF is particularly prominent in precursors of Treg cells that share transcriptional programmes with TH2 cells (see below) and are found within tissues177. Some of these tissue-resident Treg cells express CC-chemokine receptor 8 (CCR8) and have tissue repair capacity178. Batf−/− mice have significantly reduced tissue-infiltrating Treg cells but no spontaneous autoimmunity37,177,179. Treg cells of these mice do not express ST2, the IL-33 receptor, required for development and maintenance of adipose-tissue Treg cells37. BATF and IRF4 (see above) both bind Il1rl1 (the gene encoding ST2) and induce ST2 expression37. Batf−/− mice lack autoimmunity most likely due to the important role it has in specifying inflammatory conventional T cell lineages, because Treg cell-specific Batf ablation (Batffl/flFoxp3Cre) causes spontaneous TH2 cell-dominant multi-organ inflammatory disorder180. Treg cells from these mice selectively fail to suppress TH2 cell inflammation, but function normally with respect to suppression of TH1 cells and conventional T cell proliferation180. This may in part be explained by excess production of TH2 cytokines by Batffl/flFoxp3Cre Treg cells themselves180. In humans with IPEX syndrome, an interesting FOXP3 mutation, FOXP3A384T, causes tissue-restricted autoimmunity, attributable to abnormally low BATF expression. This mutation has a gain-of-function effect, broadening DNA-binding specificity of FOXP3 and enhancing interaction with the BATF promoter181. The importance of BATF to tissue-infiltrating Treg cells is highlighted by comprehensive mapping through CHIP-seq and assay for transposase-accessible chromatin with sequencing (ATAC-seq) of human Treg cells in tumour microenvironments. These corroborate overlapping functions of BATF and IRF4 and indicate that BATF binds at key genetic loci, such as IL10, CTLA4, TIGIT and TNFRSF4, to enhance Treg cell fitness in the tumour microenvironment182 (Table 2). As anticipated, high and low BATF expression in these Treg cells correlate with poor and favourable prognoses, respectively182. To summarize, the BATF-driven module is necessary for the development of non-lymphoid tissue Treg cell precursors, as well as for the development and sustainability of Treg cells that reside in tissues.
NFIL3
Nuclear factor interleukin-3 (NFIL3) is a member of the bZIP family183 that was initially identified by its ability to bind and repress an E4 promoter sequence containing an ATF consensus site184 and later characterized as binding and activating transcription of IL3185. NFIL3 regulates transcription in different immune cells. In B cells it regulates IgE class switching186, in NK cells and dendritic cells it promotes development and function187-189 and in T cells it regulates cytokine production190,191. NFIL3 expression is induced in Treg cells during chronic infections192 and impairs Treg cell function by downregulating FOXP3 expression193. It has also been proposed as an early marker gene for non-lymphoid tissue Treg cells177. Expressions of NFIL3 and FOXP3 are reciprocally linked, as TGFβ, which drives iTreg cell differentiation33,59,60, represses NFIL3, whereas overexpression of NFIL3 in Treg cells represses FOXP3 expression and impairs suppressive function in vitro and in vivo193. NFIL3 directly binds the gene promoter and CNS elements of FOXP3 (including the TSDR), and physically interacts with FOXP3 protein itself193 (Figure 2 and Table 2). Moreover, NFIL3 binds the IL10 gene; thus, multiple immune cell subsets, including Treg cells, have defective IL-10 production in the NFIL3-deficient state194. In other immune lineages, including ILCs and dendritic cells, NFIL3 functions upstream of a transcriptional circuit involving other TFs, including DNA-binding protein inhibitor ID2 and zinc finger E-box-binding homeobox 2 (ZEB2), that imprint specification in precursors195. These TFs are also active in Treg cells. Notably, ID2 is required for differentiation of GATA3+ adipose tissue-associated Treg cells (see below) 196 and, together with ID3, for maintenance of Treg cells in general197. There are suggestions that ID2 expression may play a role in Treg cell plasticity (see below) and also negatively regulate IL10 transcription indirectly198,199. ZEB2 is a negative regulator of Treg cell function. Thus, iTreg cells differentiated from Zeb2fl/flERCre precursors treated with tamoxifen or mature Treg cells with short hairpin RNA-mediated Zeb2 knockdown exhibit enhanced suppressive function200. However, it remains unclear whether NFIL3 regulates either of these TFs in Treg cells. Humans with homozygous mutations in NFIL3 develop autoimmunity, notably juvenile idiopathic arthritis and autoimmune thyroiditis. Nfil3−/− mice phenocopy the arthritis susceptibility, and the disease mechanism in both humans and mice is attributable to myeloid cell dysregulation and IL-1β hyperproduction201. Numbers and function of Treg cells have not been reported in such patients. Although the mutation in NFIL3-deficient kindreds (c.G510A, p.M170I) reduces NFIL3 protein levels (~50%), these data suggest that immunomodulatory NFIL3 biology is more complex and intertwined with FOXP3. In summary, the NFIL3-driven module is a feature of chronic inflammatory states and impairs FOXP3 expression and antagonizes Treg cell function.
Lineage-specifying transcription factors
As discussed, Treg cells can take on effector phenotypes and co-opt additional transcriptional programmes that are typically restricted to other cell lineages, licensing them to enter specific tissues or endowing specialized function. Depending on the environment and tissue, Treg cells can express lineage-specifying TFs from alternative lineages, such as TH1, TH2, TH17 and TFH cells (see below) and produce cytokines traditionally considered to be restricted to conventional T cells, such as IFNγ and IL-17A. This raises the question of Treg cell “stability” and its role in autoimmune diseases and cell therapies202. As discussed above, stable FOXP3 expression is maintained by key TFs and epigenetic modifications including those at the FOXP3 CNS regions19-22. As expected, mouse iTreg cells, with higher TSDR methylation, lose Foxp3 expression more readily than tTreg cells20,203. Naive and memory human Treg cells cultured with IL-1β together with IL-2 or IL-6 downregulate FOXP3 expression and suppressive function and express characteristic TH17 cell-associated genes including IL-17 and CCR6204,205. Likewise, repeated stimulation of human Treg cells through the TCR alone leads to loss of FOXP3 expression206. Mouse Treg cells exposed to IL-6, with or without IL-1β, also produce IL-1760,207 as do Treg cells adoptively transferred into lymphopenic recipient mice208. In fact, fate mapping mice can identify ex-FOXP3+ Treg cells in vivo and some of these can be diabetogenic209. Similarly, excess IL-4 signalling can render mouse Treg cells allergenic210. There remains a debate on the degree of Treg cell plasticity and its contribution to inflammation in humans, particularly as other fate-mapping studies show Treg cells to be remarkably stable in vivo211. The arguments for and against the plasticity model have been rehearsed elsewhere202,212-214 but the full extent of Treg cell instability and its impact on human diseases is yet to be understood.
T-bet
T-bet is a well-established lineage-defining TF for TH1 cells215. It drives TH1 cell differentiation while repressing other lineages, such as TH2 cells215,216 and TH17 cells217. T-bet in TH1-polarized cells binds and transactivates the IFNG locus218 and binds FOXP3 promoter and represses its activation219 (Figure 2). However, many (~30-70%) effector Treg cells exhibit TH1 cell-like characteristics, defined by co-expression of T-bet and CXC-chemokine receptor 3 (CXCR3), which is regulated by T-bet, in lymphoid and non-lymphoid tissues. Similarly, intestinal Treg cells exhibit prevalent co-expression of T-bet and RORγt220. Such TH1-like Treg cells are readily identifiable in patients with multiple sclerosis221. Surprisingly, IFNγ induces FOXP3 expression in conventional T cells222 and chemically-induced colitis in mice is characterized by increased proportions of IFNγ-expressing TH1-like Treg cells in the lamina propria223. T-bet expression in Treg cells is likely induced in a similar manner to conventional T cells, where TH1 cell specification involves IFNγ signalling through STAT1 and IL-12 signalling through STAT4224. Specifically, CXCR3+T-bet+ Treg cells are dramatically reduced in mice lacking either STAT1 or IFNγ receptor225. Full TH1 cell specification in Treg cells is usually not completed because repressive histone 3 lysine 27 trimethylation (H3K27me3) epigenetic marks at Il12rb2 delay IL-12Rβ2 expression and prevent timely STAT4 signalling203.
T-bet regulates Treg cell function during TH1 cell responses, although the mechanism is not clear. T-bet-deficient mice lack CXCR3+FOXP3+ Treg cells225 and T-bet ablation (such as Tbx21−/− and Tbx21fl/flFoxp3Cre strains) causes spontaneous autoimmune disease220,225,226 and increased severity of Toxoplasma gondii infection227, similar to the exacerbated EAE seen in IFNγ-deficient mice222. T-bet-expressing Treg cells have high levels of CXCR3, GITR, CTLA4 and CD103 expression and abundant IL10 and TGFB mRNA225. Indeed, TGFβ expression increases T-bet+FOXP3+ Treg cells under TH1 cell-polarizing conditions228. CXCR3-expressing Treg cells suppress TH1 cell and CD8+ T cell proliferation219,225,229; CXCR3 expression is likely to be key, as it licenses Treg cells for access to sites of TH1 cell-mediated disease where they suppress inflammation225. However, T-bet in some models, such as in experimental colitis, has also been proposed to act as a pathological mediator in Treg cells223. In summary, the T-bet programme licenses Treg cells to enter sites of TH1 cell-associated inflammation where they are required for repressing TH1 cells (and CD8+ T cells). However, the role of T-bet as a potential pathogenic factor in Treg cells needs further investigation.
GATA3
GATA3 is the master lineage-specifying TF of TH2 cells230,231. Treg cell and TH2 cell biology shows overlap232, including high expression of GATA3 in dermal and intestinal Treg cells and its induction after TCR and IL-2 stimulation233. Some Treg cells even express higher GATA3 levels than conventional T cells234. GATA3-expressing Treg cells are marked by surface expression of ST2235, a cytokine receptor transcriptionally controlled by GATA3236. GATA3 expression in Treg cells is independent of TH2-polarizing cytokines but can be opposed by TH1-specifying or TH17-specifying cytokines, notably IL-12 and IL-6233, and is repressed by BCL-6237. This may explain why GATA3 binds and represses genes for lineage-specifying TFs of opposing conventional T cell lineages, notably TBX21 and RORC233. GATA3 binds to the TSDR in Treg cells (but not in conventional T cells) to transactivate FOXP3 transcription and also cooperates with FOXP3 protein to maintain FOXP3 expression (Figure 2); thus, FOXP3 mRNA is reduced in GATA3-deficient Treg cells233,234. Treg cell-specific GATA3 deficient (Gata3fl/flFoxp3EGFP-Cre) mice develop lymphadenopathy, splenomegaly and lymphocytic infiltration of multiple organs and show enhanced production of IFNγ, IL-4 and IL-17A by 16 weeks234. GATA3-deficient Treg cells can suppress conventional T cells but don’t accumulate in tissues, thus can’t prevent transfer colitis, and some even produce IL-17233,234. GATA3 expression is also important in Treg cells that resolve inflammation in acute kidney injury238. Depletion of Treg cells in the skin or Treg cell-specific Gata3 deletion (Gata3fl/flFoxp3CreERT2 mice fed tamoxifen), leads to enhanced TH2 cytokine expression, TH2 cell infiltration, fibroblast activation and production of pro-fibrotic genes239. In summary, the GATA3 programme stabilizes FOXP3 expression, enhances suppressive capacity and permits expression of tissue-specific homing receptors.
RORγt
RORγt is an isoform of RORγ which is encoded by RORC and found in immune cells during thymopoiesis and lymphopoiesis240,241. It is commonly expressed by (predominantly group 3) ILCs242,243 and CD4+ T cells including TH17 cells244 and Treg cells245-247. TH17 cell differentiation is orchestrated by RORγt, which induces expression of IL-17244,248. However, pathogenicity of TH17 cells depends on microenvironmental availability of additional STAT3-activating cytokines, such as IL-23249 or IL-1250. Mouse iTreg cells and tTreg cells have repressive H3K27me3 epigenetic modifications at Il17a, but permissive and bivalent histone 3 lysine 4 trimethylation (H3K4me3) modifications at Rorc, respectively251, theoretically permitting FOXP3 and RORγt co-expression following appropriate stimulation60,207. Unsurprisingly, a significant proportion of FOXP3+ Treg cells, especially those in the gastrointestinal tract, express RORγt, along with markers such as IL-10 and ICOS245,252. There is, in fact, large overlaps in gene expression profiles between FOXP3+RORγt− Treg cells, FOXP3+RORγt+ Treg cells and RORγt+ T cells, indicating high similarity between these populations253.
Treg and TH17 cell differentiation is induced from naive precursors. TGFβ induces FOXP3 expression via activation of SMAD proteins33,59, whereas IL-6-mediated activation of STAT3 is required for RORγt expression254. Interestingly, IL-6-deficient mice have fewer RORγt+ Treg cells252. The RORγt programme is peripherally induced, for example by the gut microbiota through short-chain fatty acids and retinoic acid, or via antigen presentation by RORγt-expressing antigen-presenting cells; thus, germ-free mice have fewer RORγt+ Treg cells246,252,255-257. Epigenetic studies of RORγt+ Treg cells indicate demethylation at Treg cell-specific signature genes253. This suggests a stable regulatory, rather than inflammatory, phenotype and that they may occupy an important immunoregulatory niche in the gut, for example to prevent inflammation in response to the microbiota. Indeed, colonic conventional T cells in unchallenged mice harbouring specific deletion of Rorc in Treg cells (Rorcfl/flFoxp3Cre) have dysregulated TH1 and TH17 cells and develop significantly more severe colitis when challenged with trinitrobenzenesulfonic acid246. Likewise, selective deletion of Stat3 in Treg cells (Stat3fl/flFoxp3Cre) causes spontaneous gastrointestinal inflammation through failure to regulate local TH17 cell-mediated inflammation258. In humans, RORγt+ Treg cells are induced by retinoic acid, are enriched in the gastrointestinal tract, produce IL-17 in a STAT3-dependent manner80,259 and can be identified by expression of CCR6205 and the C-type lectin-like receptor CD16180. These cells retain suppressive function and express a wound healing programme that is regulated by a TF network that includes BACH280,259 (see above).
The intracellular biology of RORγt in Treg cells is not fully understood. FOXP3 interacts with RORγt in co-expression studies conducted in HEK293T cells245 and, in fact, does so through the region encoded by FOXP3 exon 2, which is necessary to suppress RORγt-mediated IL17A promoter activation260 (Figure 2 and Table 2).
The RORγt programme in Treg cells overlaps with the MAF programme246,253,261,262, which induces IL-10 in multiple T helper cell subsets64,263,264. Helicobacter hepaticus is a pathobiont that induces multiple gut T cell lineages, including pTreg cells, TFH cells and pathogenic TH17 cells265 265 and drives enterocolitis in mice lacking immunoregulatory cytokines (such as Il10−/− strains)266. In H. hepaticus-challenged mice, Treg cell-specific deletion of Maf (Maffl/flFoxp3Cre) leads to colitis due to lack of IL-10 in pTreg cells, low levels of RORγt+ Treg cells and expansion of pathogenic TH17 cells265. However, Rorcfl/flFoxp3Cre mice are not susceptible to colitis, showing Treg cell-expressed MAF is non-redundant for immune tolerance to gut pathobionts265. In fact, MAF represses Il17a transcription in RORγt+ Treg cells, which become the main source of Treg cell-derived IL-17 in MAF-deficient Treg cells261. Collectively, the RORγt programme enhances the suppressive capacity of Treg cells and permits expression of tissue-specific homing receptors, for example to the gut, where they occupy an immunoregulatory niche to prevent or ameliorate inflammation in response to the microbiota or initiate wound healing.
BCL-6
BCL-6, a zinc finger TF, is essential for germinal centre formation and is critical both for germinal centre B cells and as the lineage-specifying TF of TFH cells267-270. It is also expressed in a subset of Treg cells found within germinal centres, known as TFR cells, which share characteristics with TFH cells, including expression of the chemokine receptor CXCR5. The entry of TFR cells into B cell follicles is facilitated by CXCR5. As discussed above, expression of BLIMP1 is a cardinal feature of TFR cells. Despite being normally repressed by BLIMP1, the presence of BCL-6, together with BLIMP1 and NFATc1–NFATαA complex allows for CXCR5 expression to occur in these cells271 (Table 2). TFR cells are absent in the thymus but induced in the periphery mostly from pre-existing Treg cells (rather than naive conventional T cells)272,273. Under some circumstances, TFR cells can also be induced from FOXP3− precursors274. Their function is to regulate germinal centre reactions to ensure dominance of antigen-specific B cell clones over self-reactive clones that may cause autoimmunity272,273,275,276. Indeed, mice with genetic ablation of Bcl6 in Treg cells (Bcl6fl/flFoxp3Cre) clear viral infections more effectively but develop spontaneous antibody-mediated autoimmunity276. Likewise, abnormalities in TFR cell proportions have been linked to antibody-mediated diseases in humans277. TFR cells are induced through some of the same developmental cues as TFH cells, including SLAM-associated protein-mediated signals272, but not through the same cytokines that induce BCL-6 in TFH− cells (IL-6 and IL-21) 273. In humans, TFR cells in the circulation are less mature and distinct from those in tissues, possibly representing spill-over from previous immunizations275,278. Thus, BCL-6 is a cardinal feature of TFR cells, which enter germinal centres via BCL-6-encoded CXCR5 and ensure dominance of antigen-specific B cell clones over self-reactive clones and prevent autoimmunity.
Concluding remarks
FOXP3+ Treg cells play a crucial role in maintaining peripheral tolerance by suppressing other immune cells. FOXP3 is clearly the non-redundant and key regulator of Treg cell development and function. However, only a proportion of the Treg cell-specific transcriptome can be directly attributed to areas that are bound by FOXP3 itself. This aligns with a modular framework for describing the Treg cell lifecycle in which specific functions are added to the essential FOXP3 programme by multiple accessory TFs to imprint and maintain the Treg cell phenotype and tissue-specific functions. Many of these are non-redundant, thus result in Treg cell deficiency when genetically ablated in mice. Some of these may represent normal responses to maintain tolerance to the microbiota and may potentially be abnormally appropriated to participate in the pathogenesis of pathogenic organisms or immunological evasion by cancers. It is anticipated that knowledge of Treg cell biology will expand with the help of high-throughput methods and screening technologies that can assess multiple TFs simultaneously and ranging from the single cell to the tissue level. Likewise, future studies and robust markers categorically distinguishing tTreg cells from pTreg cells may enable closer examination of the role of transcriptional regulators in Treg cells induced from precursors at different sites. A deeper understanding of Treg cells will bring insights into the basic mechanisms of disease and pave the way for next-generation therapies for autoimmunity and transplant rejection.
Acknowledgements
This work was supported by extramural research programmes of the US National Institutes of Health (R35GM138283 to M.K.). This research was supported (in part) by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases (project number ZIA/DK075149 to B.A.). The authors gratefully acknowledge support from the Purdue University Center for Cancer Research, P30CA023168. The authors also thank Dr Vanja Lazaveric (NIH) for constructive feedback on the first draft of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Sakaguchi S, Fukuma K, Kuribayashi K & Masuda T Organ-specific autoimmune diseases induced in mice by elimination of T cell subset. I. Evidence for the active participation of T cells in natural self-tolerance; deficit of a T cell subset as a possible cause of autoimmune disease. J Exp Med 161, 72–87 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Sakaguchi N, Asano M, Itoh M & Toda M Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol 155, 1151–1164 (1995). [PubMed] [Google Scholar]
- 3.Hori S, Nomura T & Sakaguchi S Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA & Rudensky AY Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Bennett CL et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet 27, 20–21 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Brunkow ME et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Wildin RS et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet 27, 18–20 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Hill JA et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity 27, 786–800 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto N. et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol 18, 1197–1209 (2006). [DOI] [PubMed] [Google Scholar]
- 10. Birzele F. et al. Next-generation insights into regulatory T cells: expression profiling and FoxP3 occupancy in Human. Nucleic Acids Res 39, 7946–7960 (2011). References 10 and 11 show that FOXP3-bound genes represent only a small fraction of the hallmark Treg cell genes directly regulated by FOXP3.
- 11.Zheng Y. et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007). [DOI] [PubMed] [Google Scholar]
- 12.O'Shea JJ & Paul WE Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirahara K. et al. Helper T-cell differentiation and plasticity: insights from epigenetics. Immunology 134, 235–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayamada S, Takahashi H, Kanno Y & O'Shea JJ Helper T cell diversity and plasticity. Curr Opin Immunol 24, 297–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar D, Sahoo SS, Chauss D, Kazemian M & Afzali B Non-coding RNAs in immunoregulation and autoimmunity: Technological advances and critical limitations. J Autoimmun 134, 102982 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharabi A. et al. Regulatory T cells in the treatment of disease. Nat Rev Drug Discov 17, 823–844 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Sakaguchi S, Yamaguchi T, Nomura T & Ono M Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Lio CW & Hsieh CS A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron U. et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol 37, 2378–2389 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Floess S. et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 5, e38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y. et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toker A. et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol 190, 3180–3188 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Vignali DA, Collison LW & Workman CJ How regulatory T cells work. Nat Rev Immunol 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akkaya B. et al. Regulatory T cells mediate specific suppression by depleting peptide-MHC class II from dendritic cells. Nat Immunol 20, 218–231(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkura N et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Burchill MA, Yang J, Vogtenhuber C, Blazar BR & Farrar MA IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol 178, 280–290 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Yao Z. et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y. et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126, 375–387 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Camperio C. et al. Forkhead transcription factor FOXP3 upregulates CD25 expression through cooperation with RelA/NF-kappaB. PLoS One 7, e48303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono M. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kim HP & Leonard WJ CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med 204, 1543–1551 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takimoto T. et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol 185, 842–855 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Wong WF et al. Runx1 deficiency in CD4+ T cells causes fatal autoimmune inflammatory lung disease due to spontaneous hyperactivation of cells. J Immunol 188, 5408–5420 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Helms C. et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet 35, 349–356 (2003). [DOI] [PubMed] [Google Scholar]
- 36. Cretney E. et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol 12, 304–311 (2011). This publication shows that Blimp1 and IRF4 co-operate to define a subset of specialized IL-10− producing Treg cells within the mucosal tissues.
- 37. Vasanthakumar A. et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat Immunol 16, 276–285 (2015). This study provides evidence that IL-33 drives the expression of IRF4 and BATF, which are essential for functional maintenance of visceral adipose tissue Treg cells.
- 38.Bhairavabhotla R. et al. Transcriptome profiling of human FoxP3+ regulatory T cells. Hum Immunol 77, 201–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zemmour D. et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu L, Barbi J & Pan F The regulation of immune tolerance by FOXP3. Nat Rev Immunol 17, 703–717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu YZ, Hogenesch JB & Bradfield CA The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol 40, 519–561 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Lamas B, Natividad JM & Sokol H Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol 11, 1024–1038 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Zhou L. AHR Function in Lymphocytes: Emerging Concepts. Trends Immunol 37, 17–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denison MS & Nagy SR Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43, 309–334 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Wang X. et al. AHR promoter variant modulates its transcription and downstream effectors by allele-specific AHR-SP1 interaction functioning as a genetic marker for vitiligo. Sci Rep 5, 13542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kerkvliet NI, Shepherd DM & Baecher-Steppan L T lymphocytes are direct, aryl hydrocarbon receptor (AhR)-dependent targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): AhR expression in both CD4+ and CD8+ T cells is necessary for full suppression of a cytotoxic T lymphocyte response by TCDD. Toxicol Appl Pharmacol 185, 146–152 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Funatake CJ, Marshall NB, Steppan LB, Mourich DV & Kerkvliet NI Cutting edge: activation of the aryl hydrocarbon receptor by 2,3,7,8-tetrachlorodibenzo-p-dioxin generates a population of CD4+ CD25+ cells with characteristics of regulatory T cells. J Immunol 175, 4184–4188 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Quintana FJ et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Zhang L. et al. Suppression of experimental autoimmune uveoretinitis by inducing differentiation of regulatory T cells via activation of aryl hydrocarbon receptor. Invest Ophthalmol Vis Sci 51, 2109–2117 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Salguero P et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 268, 722–726 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Elizondo G, Rodriguez-Sosa M, Estrada-Muniz E, Gonzalez FJ & Vega L Deletion of the aryl hydrocarbon receptor enhances the inflammatory response to Leishmania major infection. Int J Biol Sci 7, 1220–1229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wincent E. et al. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem 284, 2690–2696 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Veldhoen M. et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Nukaya M & Bradfield CA Conserved genomic structure of the Cyp1a1 and Cyp1a2 loci and their dioxin responsive elements cluster. Biochem Pharmacol 77, 654–659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez-Vazquez C & Quintana FJ Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 48, 19–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiong L. et al. Ahr-Foxp3-RORgammat axis controls gut homing of CD4(+) T cells by regulating GPR15. Sci Immunol 5, eaaz7277 (2020). This is a key publication showing that interactions between AHR, FOXP3 and RORγt finely regulate the expression of GPR15 at the epigenetic level to orchestrate Treg cell gut homing
- 57.Kim SV et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 340, 1456–1459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gandhi R. et al. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol 11, 846–853 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maruyama T, Konkel JE, Zamarron BF & Chen W The molecular mechanisms of Foxp3 gene regulation. Semin Immunol 23, 418–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang XO et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tran DQ, Ramsey H & Shevach EM Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood 110, 2983–2990 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okamura T et al. CD4+CD25−LAG3+ regulatory T cells controlled by the transcription factor Egr-2. Proc Natl Acad Sci U S A 106, 13974–13979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Groux H. et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 389, 737–742 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Apetoh L. et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11, 854–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vahedi G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oyake T. et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol 16, 6083–6095 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richer MJ, Lang ML & Butler NS T Cell Fates Zipped Up: How the Bach2 Basic Leucine Zipper Transcriptional Repressor Directs T Cell Differentiation and Function. J Immunol 197, 1009–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afzali B. et al. BACH2 immunodeficiency illustrates an association between super-enhancers and haploinsufficiency. Nat Immunol 18, 813–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Roychoudhuri R. et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498, 506–510 (2013). This paper shows a key role for BACH2 expression within Treg cells and its role in the prevention of autoimmunity.
- 70.Kuwahara M. et al. Bach2-Batf interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat Commun 7, 12596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roychoudhuri R. et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17, 851–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imianowski CJ et al. BACH2 restricts NK cell maturation and function, limiting immunity to cancer metastasis. J Exp Med 219, e20211476 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao C. et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8(+) T cells. Nat Immunol 22, 370–380 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sidwell T. et al. Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis. Nat Commun 11, 252 (2020). The authors provide evidence that BACH2 prevents premature differentiation of effector Treg cells.
- 75.Rincon M & Flavell RA AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J 13, 4370–4381 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grant FM et al. BACH2 drives quiescence and maintenance of resting Treg cells to promote homeostasis and cancer immunosuppression. J Exp Med 217, e20190711 (2020). This paper provides evidence that BACH2 prevents premature differentiation of effector Treg cells.
- 77.Kim EH et al. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol 192, 985–995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Do JS et al. Foxp3 expression in induced T regulatory cells derived from human umbilical cord blood vs. adult peripheral blood. Bone Marrow Transplant 53, 1568–1577 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Contreras A. et al. BACH2 in TRegs Limits the Number of Adipose Tissue Regulatory T Cells and Restrains Type 2 Immunity to Fungal Allergens. J Immunol Res 2022, 6789055 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Povoleri GAM et al. Human retinoic acid-regulated CD161(+) regulatory T cells support wound repair in intestinal mucosa. Nat Immunol 19, 1403–1414 (2018). This paper identifies CD161+ Treg cells as a highly suppressive subset of Treg cells that produce IL-17 and possess wound-healing properties in the gut.
- 81.Pesenacker AM et al. CD161 defines the subset of FoxP3+ T cells capable of producing proinflammatory cytokines. Blood 121, 2647–2658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rutgeerts P, Vermeire S & Van Assche G Mucosal healing in inflammatory bowel disease: impossible ideal or therapeutic target? Gut 56, 453–455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chauss D. et al. Autocrine vitamin D signaling switches off pro-inflammatory programs of T(H)1 cells. Nat Immunol 23, 62–74 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McAllister K. et al. Identification of BACH2 and RAD51B as rheumatoid arthritis susceptibility loci in a meta-analysis of genome-wide data. Arthritis Rheum 65, 3058–3062 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franke A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 42, 1118–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.International Multiple Sclerosis Genetics, C. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cooper JD et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 40, 1399–1401 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferreira MA et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 378, 1006–1014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mouri K. et al. Prioritization of autoimmune disease-associated genetic variants that perturb regulatory element activity in T cells. Nat Genet 54, 603–612 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoshino H. et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem 275, 15370–15376 (2000). [DOI] [PubMed] [Google Scholar]
- 91.Ando R. et al. The Transcription Factor Bach2 Is Phosphorylated at Multiple Sites in Murine B Cells but a Single Site Prevents Its Nuclear Localization. J Biol Chem 291, 1826–1840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yu X. et al. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat Commun 9, 3157 (2018). Here, the authors identify SENP3 as a key molecule regulating the function and stability of Treg cells by controlling the nuclear localization of BACH2.
- 93.Hay RT SUMO: a history of modification. Mol Cell 18, 1–12 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Huang C. et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J 28, 2748–2762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim HR et al. Reactive oxygen species prevent imiquimod-induced psoriatic dermatitis through enhancing regulatory T cell function. PLoS One 9, e91146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gelderman KA, Hultqvist M, Holmberg J, Olofsson P & Holmdahl R T cell surface redox levels determine T cell reactivity and arthritis susceptibility. Proc Natl Acad Sci U S A 103, 12831–12836 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Santo JP Immunology. A guardian of T cell fate. Science 329, 44–45(2010). [DOI] [PubMed] [Google Scholar]
- 98.Albu DI et al. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med 204, 3003–3015 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Califano D. et al. Diverting T helper cell trafficking through increased plasticity attenuates autoimmune encephalomyelitis. J Clin Invest 124, 174–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Califano D. et al. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity 43, 354–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cismasiu VB et al. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24, 6753–6764 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Cismasiu VB et al. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood 108, 2695–2702 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vanvalkenburgh J et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med 208, 2069–208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hasan SN et al. Bcl11b prevents catastrophic autoimmunity by controlling multiple aspects of a regulatory T cell gene expression program. Sci Adv 5, eaaw0706 (2019). Together with ref 105 (Drashansky et al.), this paper identifies a critical role for BCL-11B in enhancing FOXP3 expression and Treg cell-associated genes.
- 105.Drashansky TT et al. Bcl11b prevents fatal autoimmunity by promoting T(reg) cell program and constraining innate lineages in T(reg) cells. Sci Adv 5, eaaw0480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thornton AM et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim HJ et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 350, 334–339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chougnet C & Hildeman D Helios-controller of Treg stability and function. Transl Cancer Res 5, S338–S341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rieder SA et al. Eos Is Redundant for Regulatory T Cell Function but Plays an Important Role in IL-2 and Th17 Production by CD4+ Conventional T Cells. J Immunol 195, 553–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pan F et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raffin C. et al. Human memory Helios− FOXP3+ regulatory T cells (Tregs) encompass induced Tregs that express Aiolos and respond to IL-1beta by downregulating their suppressor functions. J Immunol 191, 4619–4627 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Gokhale AS, Gangaplara A, Lopez-Occasio M, Thornton AM & Shevach EM Selective deletion of Eos (Ikzf4) in T-regulatory cells leads to loss of suppressive function and development of systemic autoimmunity. J Autoimmun 105, 102300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heller JJ et al. Restriction of IL-22-producing T cell responses and differential regulation of regulatory T cell compartments by zinc finger transcription factor Ikaros. J Immunol 193, 3934–3946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lyon de Ana C, Arakcheeva K, Agnihotri P, Derosia N & Winandy S Lack of Ikaros Deregulates Inflammatory Gene Programs in T Cells. J Immunol 202, 1112–1123 (2019). This study shows that conditional deletion of Ikaros in CD4+ T cells impairs Treg cell differentiation and promotes TH17 cell-mediated autoimmunity.
- 115.Graves DT & Milovanova TN Mucosal Immunity and the FOXO1 Transcription Factors. Front Immunol 10, 2530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Agnihotri P, Robertson NM, Umetsu SE, Arakcheeva K & Winandy S Lack of Ikaros cripples expression of Foxo1 and its targets in naive T cells. Immunology 152, 494–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ouyang W. et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol 11, 618–627 (2010). [DOI] [PubMed] [Google Scholar]
- 118.Yu X. et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 10, 48–57 (2009). [DOI] [PubMed] [Google Scholar]
- 119.Lucca LE et al. TIGIT signaling restores suppressor function of Th1 Tregs. JCI Insight 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Georgopoulos K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994). [DOI] [PubMed] [Google Scholar]
- 121.Mullighan CG et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453, 110–114 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Yap WH, Yeoh E, Tay A, Brenner S & Venkatesh B STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. FEBS Lett 579, 4470–4478 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Qu S et al. Common variants near IKZF1 are associated with primary Sjogren's syndrome in Han Chinese. PLoS One 12, e0177320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoshino A. et al. Abnormal hematopoiesis and autoimmunity in human subjects with germline IKZF1 mutations. J Allergy Clin Immunol 140, 223–231 (2017). [DOI] [PubMed] [Google Scholar]
- 125.Hoshino A. et al. Gain-of-function IKZF1 variants in humans cause immune dysregulation associated with abnormal T/B cell late differentiation. Sci Immunol 7, eabi7160 (2022). [DOI] [PubMed] [Google Scholar]
- 126.Toubai T. et al. Ikaros deficiency in host hematopoietic cells separates GVL from GVHD after experimental allogeneic hematopoietic cell transplantation. Oncoimmunology 4, e1016699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yasui D, Miyano M, Cai S, Varga-Weisz P & Kohwi-Shigematsu T SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419, 641–645 (2002). [DOI] [PubMed] [Google Scholar]
- 128.Cai S, Lee CC & Kohwi-Shigematsu T SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38, 1278–1288 (2006). [DOI] [PubMed] [Google Scholar]
- 129.Alvarez JD et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev 14, 521–535 (2000). [PMC free article] [PubMed] [Google Scholar]
- 130.Dickinson LA, Joh T, Kohwi Y & Kohwi-Shigematsu T A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 (1992). [DOI] [PubMed] [Google Scholar]
- 131. Kitagawa Y. et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol 18, 173–183 (2017). This study indicates that SATB1 is a Treg cell-specific super enhancer crucial for Treg cell fate decision in thymocytes before FOXP3 expression.
- 132. Beyer M. et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol 12, 898–907 (2011). This paper shows that SATB1-mediated modulation of global chromatin remodeling is repressed by FOXP3-dependent mechanisms in mature Treg cells.
- 133.Kondo M. et al. SATB1 Plays a Critical Role in Establishment of Immune Tolerance. J Immunol 196, 563–572 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Yasuda K. et al. Satb1 regulates the effector program of encephalitogenic tissue Th17 cells in chronic inflammation. Nat Commun 10, 549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.International Multiple Sclerosis Genetics, C. et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 45, 1353–1360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Akimova T. et al. Human lung tumor FOXP3+ Tregs upregulate four "Treg-locking" transcription factors. JCI Insight 2, e94075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li XF et al. Inhibition of SATB1 expression in regulatory T cells contributes to hepatitis B virus-related chronic liver inflammation. Mol Med Rep 11, 231–236 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Wang Y. et al. Overexpression of SATB1 Gene Inhibits the Immunosuppressive Function of Regulatory T Cells in Chronic Hepatitis B. Ann Clin Lab Sci 47, 403–408 (2017). [PubMed] [Google Scholar]
- 139.Zhao GN, Jiang DS & Li H Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta 1852, 365–378 (2015). [DOI] [PubMed] [Google Scholar]
- 140.Klein U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 141.Sciammas R et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25, 225–236 (2006). [DOI] [PubMed] [Google Scholar]
- 142.Brustle A. et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol 8, 958–966 (2007). [DOI] [PubMed] [Google Scholar]
- 143.Man K. et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 144.Staudt V. et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202 (2010). [DOI] [PubMed] [Google Scholar]
- 145.Honma K. et al. Interferon regulatory factor 4 negatively regulates the production of proinflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci U S A 102, 16001–16006 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marecki S, Atchison ML & Fenton MJ Differential expression and distinct functions of IFN regulatory factor 4 and IFN consensus sequence binding protein in macrophages. J Immunol 163, 2713–2722 (1999). [PubMed] [Google Scholar]
- 147.Williams JW et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun 4, 2990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rengarajan J. et al. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med 195, 1003–1012 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Haljasorg U. et al. Irf4 Expression in Thymic Epithelium Is Critical for Thymic Regulatory T Cell Homeostasis. J Immunol 198, 1952–1960 (2017). [DOI] [PubMed] [Google Scholar]
- 150. Zheng Y. et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 458, 351–356 (2009). This paper shows that FOXP3 induces the expression of IRF4 in Treg cells and confers the ability to suppress TH2 cell responses.
- 151.Cipolletta D. et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bravo Garcia-Morato M et al. New human combined immunodeficiency caused by interferon regulatory factor 4 (IRF4) deficiency inherited by uniparental isodisomy. J Allergy Clin Immunol 141, 1924–1927 e1918 (2018). [DOI] [PubMed] [Google Scholar]
- 153.Alvisi G. et al. IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J Clin Invest 130, 3137–3150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Biswas PS, Bhagat G & Pernis AB IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis? Immunol Rev 233, 79–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ahyi AN, Chang HC, Dent AL, Nutt SL & Kaplan MH IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol 183, 1598–1606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Keller AD & Maniatis T Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev 5, 868–879 (1991). [DOI] [PubMed] [Google Scholar]
- 157.Shapiro-Shelef M. et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19, 607–620 (2003). [DOI] [PubMed] [Google Scholar]
- 158.Turner CA Jr., Mack DH & Davis MM Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77, 297–306 (1994). [DOI] [PubMed] [Google Scholar]
- 159.Bankoti R. et al. Differential regulation of Effector and Regulatory T cell function by Blimp1. Sci Rep 7, 12078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kallies A. et al. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol 7, 466–474 (2006). [DOI] [PubMed] [Google Scholar]
- 161.Imielinski M et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet 41, 1335–1340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gateva V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 41, 1228–1233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Han JW et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41, 1234–1237 (2009). [DOI] [PubMed] [Google Scholar]
- 164.Anderson CA et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43, 246–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Martins GA et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol 7, 457–465 (2006). [DOI] [PubMed] [Google Scholar]
- 166. Garg G. et al. Blimp1 Prevents Methylation of Foxp3 and Loss of Regulatory T Cell Identity at Sites of Inflammation. Cell Rep 26, 1854–1868 e1855 (2019). This study shows that BLIMP1 restrains methylation of CNS2 in the Foxp3 locus to preserve Treg cell stability in the brain.
- 167. Ogawa C. et al. Blimp-1 Functions as a Molecular Switch to Prevent Inflammatory Activity in Foxp3(+)RORgammat(+) Regulatory T Cells. Cell Rep 25, 19–28 e15 (2018). This paper shows that BLIMP1 binds to the IL17 locus and represses production of IL-17 in RORγt+FOXP3+ Treg cells.
- 168.Rajbhandari P. et al. IL-10 Signaling Remodels Adipose Chromatin Architecture to Limit Thermogenesis and Energy Expenditure. Cell 172, 218–233 e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Beppu LY et al. Tregs facilitate obesity and insulin resistance via a Blimp-1/IL-10 axis. JCI Insight 6, e140644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lin MH et al. T cell-specific BLIMP-1 deficiency exacerbates experimental autoimmune encephalomyelitis in nonobese diabetic mice by increasing Th1 and Th17 cells. Clin Immunol 151, 101–113 (2014). [DOI] [PubMed] [Google Scholar]
- 171.Lin MH et al. B lymphocyte-induced maturation protein 1 (BLIMP-1) attenuates autoimmune diabetes in NOD mice by suppressing Th1 and Th17 cells. Diabetologia 56, 136–146 (2013). [DOI] [PubMed] [Google Scholar]
- 172.Hu M. et al. Infiltrating Foxp3(+) regulatory T cells from spontaneously tolerant kidney allografts demonstrate donor-specific tolerance. Am J Transplant 13, 2819–2830 (2013). [DOI] [PubMed] [Google Scholar]
- 173.Norton SE et al. High-Dimensional Mass Cytometric Analysis Reveals an Increase in Effector Regulatory T Cells as a Distinguishing Feature of Colorectal Tumors. J Immunol 202, 1871–1884 (2019). [DOI] [PubMed] [Google Scholar]
- 174.Betz BC et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med 207, 933–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Murphy TL, Tussiwand R & Murphy KM Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol 13, 499–509, doi: 10.1038/nri3470 (2013). [DOI] [PubMed] [Google Scholar]
- 176.Schraml BU et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Delacher M. et al. Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity 52, 295–312 e211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Delacher M. et al. Single-cell chromatin accessibility landscape identifies tissue repair program in human regulatory T cells. Immunity 54, 702–720 e717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang C. et al. BATF is required for normal expression of gut-homing receptors by T helper cells in response to retinoic acid. J Exp Med 210, 475–489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu C. et al. BATF Regulates T Regulatory Cell Functional Specification and Fitness of Triglyceride Metabolism in Restraining Allergic Responses. J Immunol 206, 2088–2100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hayatsu N. et al. Analyses of a Mutant Foxp3 Allele Reveal BATF as a Critical Transcription Factor in the Differentiation and Accumulation of Tissue Regulatory T Cells. Immunity 47, 268–283 e269 (2017). [DOI] [PubMed] [Google Scholar]
- 182. Itahashi K. et al. BATF epigenetically and transcriptionally controls the activation program of regulatory T cells in human tumors. Sci Immunol 7, eabk0957 (2022). This publication, together with ref 180 (Xu et al.), shows that BATF regulates a key gene signature in Treg cells and specific ablation of Batf results in an inflammatory disorder characterized by TH2-type dominant responses.
- 183.Cowell IG E4BP4/NFIL3, a PAR-related bZIP factor with many roles. Bioessays 24, 1023–1029 (2002). [DOI] [PubMed] [Google Scholar]
- 184.Cowell IG & Hurst HC Transcriptional repression by the human bZIP factor E4BP4: definition of a minimal repression domain. Nucleic Acids Res 22, 59–65 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang W. et al. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol 15, 6055–6063 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kashiwada M. et al. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci U S A 107, 821–826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gascoyne DM et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol 10, 1118–1124 (2009). [DOI] [PubMed] [Google Scholar]
- 188.Kamizono S. et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med 206, 2977–2986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kashiwada M, Pham NL, Pewe LL, Harty JT & Rothman PB NFIL3/E4BP4 is a key transcription factor for CD8alpha(+) dendritic cell development. Blood 117, 6193–6197 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kobayashi T. et al. NFIL3-deficient mice develop microbiota-dependent, IL-12/23-driven spontaneous colitis. J Immunol 192, 1918–1927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kashiwada M, Cassel SL, Colgan JD & Rothman PB NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO J 30, 2071–2082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Layland LE et al. Pronounced phenotype in activated regulatory T cells during a chronic helminth infection. J Immunol 184, 713–724 (2010). [DOI] [PubMed] [Google Scholar]
- 193. Kim HS, Sohn H, Jang SW & Lee GR The transcription factor NFIL3 controls regulatory T-cell function and stability. Exp Mol Med 51, 80 (2019). This study shows that NFIL3 directly binds to and negatively regulates the expression of FOXP3, in a mechanism that induces methylation at the FOXP3 locus CpG sites.
- 194.Motomura Y. et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol 12, 450–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bagadia P, Huang X, Liu TT & Murphy KM Shared Transcriptional Control of Innate Lymphoid Cell and Dendritic Cell Development. Annu Rev Cell Dev Biol 35, 381–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Frias AB Jr. et al. The Transcriptional Regulator Id2 Is Critical for Adipose-Resident Regulatory T Cell Differentiation, Survival, and Function. J Immunol 203, 658–664 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Miyazaki M. et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol 15, 767–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Masson F. et al. Id2 represses E2A-mediated activation of IL-10 expression in T cells. Blood 123, 3420–3428 (2014). [DOI] [PubMed] [Google Scholar]
- 199.Hwang SM et al. Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat Commun 9, 4736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Boland BS et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol 5, eabb4432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schlenner S. et al. NFIL3 mutations alter immune homeostasis and sensitise for arthritis pathology. Ann Rheum Dis 78, 342–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sakaguchi S, Vignali DA, Rudensky AY, Niec RE & Waldmann H The plasticity and stability of regulatory T cells. Nat Rev Immunol 13, 461–467 (2013). [DOI] [PubMed] [Google Scholar]
- 203.Polansky JK et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol 38, 1654–1663 (2008). [DOI] [PubMed] [Google Scholar]
- 204.Deknuydt F, Bioley G, Valmori D & Ayyoub M IL-1beta and IL-2 convert human Treg into T(H)17 cells. Clin Immunol 131, 298–307 (2009). [DOI] [PubMed] [Google Scholar]
- 205. Beriou G. et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood 113, 4240–4249 (2009). This paper demonstrates that IL-17-producing FOXP3+ Treg cells induced by pro-inflammatory cytokines maintain suppressive function and express CCR6.
- 206.Hoffmann P. et al. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur J Immunol 39, 1088–1097 (2009). [DOI] [PubMed] [Google Scholar]
- 207.Xu L, Kitani A, Fuss I & Strober W Cutting edge: regulatory T cells induce CD4+CD25−Foxp3− T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol 178, 6725–6729 (2007). [DOI] [PubMed] [Google Scholar]
- 208.Komatsu N. et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A 106, 1903–1908 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zhou X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol 10, 1000–1007 (2009). This publication indicates that Treg cells lose expression of FOXP3 in inflamed microenvironments and acquire an effector-memory phenotype.
- 210.Noval Rivas M. et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity 42, 512–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rubtsov YP et al. Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hori S. Regulatory T cell plasticity: beyond the controversies. Trends Immunol 32, 295–300 (2011). [DOI] [PubMed] [Google Scholar]
- 213.Hori S. Developmental plasticity of Foxp3+ regulatory T cells. Curr Opin Immunol 22, 575–582 (2010). [DOI] [PubMed] [Google Scholar]
- 214.Hori S. Lineage stability and phenotypic plasticity of Foxp3(+) regulatory T cells. Immunol Rev 259, 159–172 (2014). [DOI] [PubMed] [Google Scholar]
- 215.Szabo SJ et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). [DOI] [PubMed] [Google Scholar]
- 216.Hwang ES, Szabo SJ, Schwartzberg PL & Glimcher LH T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307, 430–433 (2005). [DOI] [PubMed] [Google Scholar]
- 217.Lazarevic V et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol 12, 96–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Djuretic IM et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol 8, 145–153 (2007). [DOI] [PubMed] [Google Scholar]
- 219.Amarnath S. et al. Tbet is a critical modulator of FoxP3 expression in autoimmune graft-versus-host disease. Haematologica 102, 1446–1456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Levine AG et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 546, 421–425 (2017). This report shows that T-bet expression is essential for maintaining Treg cell suppressive function for controlling TH1 and CD8+ T cell responses.
- 221. Dominguez-Villar M, Baecher-Allan CM & Hafler DA Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med 17, 673–675 (2011). This paper identifies IFNγ+T-bet+ Treg cells in patients with multiple sclerosis and suggests that they may have reduced in vitro suppressive function.
- 222.Wang Z. et al. Role of IFN-gamma in induction of Foxp3 and conversion of CD4+ CD25− T cells to CD4+ Tregs. J Clin Invest 116, 2434–2441 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Di Giovangiulio M. et al. Tbet Expression in Regulatory T Cells Is Required to Initiate Th1-Mediated Colitis. Front Immunol 10, 2158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Afkarian M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol 3, 549–557 (2002). [DOI] [PubMed] [Google Scholar]
- 225. Koch MA et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 10, 595–602 (2009). This report shows that T-bet expression is essential for Treg cells to access sites of TH1-type inflammation.
- 226.Tan TG, Mathis D & Benoist C Singular role for T-BET+CXCR3+ regulatory T cells in protection from autoimmune diabetes. Proc Natl Acad Sci U S A 113, 14103–14108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Warunek J. et al. Tbet Expression by Regulatory T Cells Is Needed to Protect against Th1-Mediated Immunopathology during Toxoplasma Infection in Mice. Immunohorizons 5, 931–943 (2021). [DOI] [PubMed] [Google Scholar]
- 228.Kachler K, Holzinger C, Trufa DI, Sirbu H & Finotto S The role of Foxp3 and Tbet co-expressing Treg cells in lung carcinoma. Oncoimmunology 7, e1456612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Szabo SJ et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295, 338–342 (2002). [DOI] [PubMed] [Google Scholar]
- 230.Zheng W & Flavell RA The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997). [DOI] [PubMed] [Google Scholar]
- 231.Ho IC et al. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J 10, 1187–1192 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chapoval S, Dasgupta P, Dorsey NJ & Keegan AD Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by IL-4 and STAT6. J Leukoc Biol 87, 1011–1018 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wohlfert EA et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest 121, 4503–4515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang Y, Su MA & Wan YY An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Siede J. et al. IL-33 Receptor-Expressing Regulatory T Cells Are Highly Activated, Th2 Biased and Suppress CD4 T Cell Proliferation through IL-10 and TGFbeta Release. PLoS One 11, e0161507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hayakawa M. et al. T-helper type 2 cell-specific expression of the ST2 gene is regulated by transcription factor GATA-3. Biochim Biophys Acta 1728, 53–64 (2005). [DOI] [PubMed] [Google Scholar]
- 237.Sawant DV et al. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol 189, 4759–4769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sakai R. et al. Kidney GATA3(+) regulatory T cells play roles in the convalescence stage after antibody-mediated renal injury. Cell Mol Immunol 18, 1249–1261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Kalekar LA et al. Regulatory T cells in skin are uniquely poised to suppress profibrotic immune responses. Sci Immunol 4, aaw2910 (2019). This paper shows that skin Treg cells express high levels of GATA3 and have a key role during dermal fibrosis.
- 240.Sun Z. et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000). [DOI] [PubMed] [Google Scholar]
- 241.Kurebayashi S et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A 97, 10132–10137 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Eberl G. et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol 5, 64–73 (2004). [DOI] [PubMed] [Google Scholar]
- 243.Croft CA et al. Notch, RORC and IL-23 signals cooperate to promote multi-lineage human innate lymphoid cell differentiation. Nat Commun 13, 4344 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ivanov II et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006). [DOI] [PubMed] [Google Scholar]
- 245.Lochner M. et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med 205, 1381–1393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Sefik E. et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997, aaa9420 (2015). This article shows that RORγt is preferentially expressed in colonic Treg cells and induced by gut microbiota.
- 247.Bhaumik S, Mickael ME, Moran M, Spell M & Basu R RORgammat Promotes Foxp3 Expression by Antagonizing the Effector Program in Colonic Regulatory T Cells. J Immunol 207, 2027–2038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Langrish CL et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 201, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.McGeachy MJ et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol 8, 1390–1397 (2007). [DOI] [PubMed] [Google Scholar]
- 250.Ghoreschi K. et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wei G. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ohnmacht C. et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 349, 989–993 (2015). [DOI] [PubMed] [Google Scholar]
- 253.Yang BH et al. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol 9, 444–457 (2016). [DOI] [PubMed] [Google Scholar]
- 254.Zhou L. et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 8, 967–974 (2007). [DOI] [PubMed] [Google Scholar]
- 255. Kedmi R. et al. A RORgammat(+) cell instructs gut microbiota-specific Treg cell differentiation. Nature 610, 737–743 (2022). This paper indicates that RORγt is preferentially expressed in colonic Treg cells and induced by gut microbiota.
- 256.Akagbosu B. et al. Novel antigen-presenting cell imparts T(reg)-dependent tolerance to gut microbiota. Nature 610, 752–760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lyu M. et al. ILC3s select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature 610, 744–751 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chaudhry A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Afzali B. et al. CD161 expression characterizes a subpopulation of human regulatory T cells that produces IL-17 in a STAT3-dependent manner. Eur J Immunol 43, 2043–2054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ichiyama K. et al. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem 283, 17003–17008 (2008). [DOI] [PubMed] [Google Scholar]
- 261.Neumann C. et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat Immunol 20, 471–481 (2019). [DOI] [PubMed] [Google Scholar]
- 262.Imbratta C, Hussein H, Andris F & Verdeil G c-MAF, a Swiss Army Knife for Tolerance in Lymphocytes. Front Immunol 11, 206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gabrysova L. et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4(+) T cells. Nat Immunol 19, 497–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Xu J. et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol 182, 6226–6236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Xu M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kullberg MC et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun 66, 5157–5166 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fukuda T. et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med 186, 439–448 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ye BH et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 16, 161–170 (1997). [DOI] [PubMed] [Google Scholar]
- 269.Yu D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009). [DOI] [PubMed] [Google Scholar]
- 270.Nurieva RI et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Koenig A. et al. NFATc1/alphaA and Blimp-1 Support the Follicular and Effector Phenotype of Tregs. Front Immunol 12, 791100 (2021). This paper shows that BLIMP1 cooperates with NFATc1 to mediate CXCR5 transactivation for promoting the migration of TFR cells into B cell follicles.
- 272.Linterman MA et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med 17, 975–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Chung Y. et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med 17, 983–988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Aloulou M. et al. Follicular regulatory T cells can be specific for the immunizing antigen and derive from naive T cells. Nat Commun 7, 10579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kumar S. et al. Developmental bifurcation of human T follicular regulatory cells. Sci Immunol 6, eabd8411 (2021). [DOI] [PubMed] [Google Scholar]
- 276.Fu W. et al. Deficiency in T follicular regulatory cells promotes autoimmunity. J Exp Med 215, 815–825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wen Y. et al. Imbalance of circulating CD4(+)CXCR5(+)FOXP3(+) Tfr-like cells and CD4(+)CXCR5(+)FOXP3(−) Tfh-like cells in myasthenia gravis. Neurosci Lett 630, 176–182 (2016). [DOI] [PubMed] [Google Scholar]
- 278.Fonseca VR et al. Human blood T(fr) cells are indicators of ongoing humoral activity not fully licensed with suppressive function. Sci Immunol 2, eaan1487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]