Abstract
In Drosophila melanogaster, the Polycomb-group (PcG) and trithorax-group (trxG) genes have been identified as repressors and activators, respectively, of gene expression. Both groups of genes are required for the stable transmission of gene expression patterns to progeny cells throughout development. Several lines of evidence suggest a functional interaction between the PcG and trxG proteins. For example, genetic evidence indicates that the enhancer of zeste [E(z)] gene can be considered both a PcG and a trxG gene. To better understand the molecular interactions in which the E(z) protein is involved, we performed a two-hybrid screen with Enx1/EZH2, a mammalian homolog of E(z), as the target. We report the identification of the human EED protein, which interacts with Enx1/EZH2. EED is the human homolog of eed, a murine PcG gene which has extensive homology with the Drosophila PcG gene extra sex combs (esc). Enx1/EZH2 and EED coimmunoprecipitate, indicating that they also interact in vivo. However, Enx1/EZH2 and EED do not coimmunoprecipitate with other human PcG proteins, such as HPC2 and BMI1. Furthermore, unlike HPC2 and BMI1, which colocalize in nuclear domains of U-2 OS osteosarcoma cells, Enx1/EZH2 and EED do not colocalize with HPC2 or BMI1. Our findings indicate that Enx1/EZH2 and EED are members of a class of PcG proteins that is distinct from previously described human PcG proteins.
In Drosophila melanogaster, the genes of the Polycomb group (PcG) and trithorax group (trxG) are part of a cellular memory system, which is responsible for the stable inheritance of gene activity. The PcG and trxG genes have been identified in Drosophila as repressors (PcG) (18, 22, 27, 28, 38) and activators (trxG) (20, 21), respectively, of homeotic gene activity. PcG and trxG genes were originally found in Drosophila, but mammalian homologs have also been identified and appear to function like their Drosophila homologs (reviewed in reference 37). It has been proposed that PcG proteins repress gene expression through the formation of multimeric protein complexes. We have recently shown that the human PcG proteins HPH1 and HPH2 coimmunoprecipitate, cofractionate, and colocalize in nuclear domains with the human PcG proteins BMI1 (2, 12, 33) and HPC2, a recently identified, novel human Polycomb protein (33, 34). Furthermore, we have found that the human RING1 protein coimmunoprecipitates and colocalizes with HPC2 and other PcG proteins, indicating that RING1 is associated with, or is part of, the mammalian PcG complex (33, 35). These results indicate that mammalian PcG proteins form a multimeric protein complex. This observation is in agreement with observations that different PcG proteins, including Pc, bind in overlapping patterns on polytene chromosomes in Drosophila salivary gland cells (4, 10, 29).
Interestingly, also the trithorax gene product trx colocalizes with Drosophila PcG proteins at many sites on polytene chromosomes (6, 24). Even more strikingly, binding of the trx protein has been mapped to small DNA fragments that also contain binding sites for PcG proteins, the Polycomb response elements (5, 6). This finding is further substantiated by the observation that GAGA factor, the gene product of the trxG gene trithorax-like (Trl) (13), colocalizes with Pc protein within the close vicinity of a Polycomb response element (41). Furthermore, the PcG gene Enhancer of zeste [E(z)] contains a domain with sequence homology with the activator protein trx (17). This observation is in agreement with genetic data which indicate that E(z) can be considered both a PcG gene and a trxG gene (26). Double mutations of E(z) and trxG genes result in homeotic phenotypes which are similar to the homeotic phenotypes which are also observed in double mutants of trxG genes (26). Finally, polytene chromosome binding of the trx protein is strongly reduced in homozygous E(z) mutants (4), and vice versa, polytene chromosome binding of the E(z) protein is reduced in trx mutants (24). These data suggest functional interactions between activators (trxG proteins) and repressors (PcG proteins) that are important for their mode of action.
To start to investigate these puzzling features of the E(z) gene product, we used the two-hybrid system (8, 9) in order to identify proteins that interact with a mammalian homolog of E(z), the Enx1/EZH2 protein (15, 16). Here, we report the identification of the human EED protein, which interacts with Enx1/EZH2. EED is the human homolog of eed, a murine PcG gene (7, 36) which has extensive homology with the Drosophila PcG gene extra sex combs (esc) (14, 32, 39). Whereas Enx1/EZH2 and EED coimmunoprecipitate, they neither coimmunoprecipitate nor colocalize with other human PcG proteins, such as HPC2 and BMI1. Our findings indicate that both Enx1/EZH2 and EED form a class of mammalian PcG proteins that is distinct from previously described human PcG proteins.
MATERIALS AND METHODS
Yeast two-hybrid screen.
The full-length coding region of Enx1 (15, 16) was cloned into the pAS2 vector (8) (Clontech, Palo Alto, Calif.) and used as the target to screen for interacting proteins in a two-hybrid screen (8, 9). Plasmid pAS2-Enx1 was cotransformed with a human fetal brain Matchmaker two-hybrid library (Clontech) into Saccharomyces cerevisiae Y190. The transformants were plated on selective medium lacking the leucine, tryptophan, and histidine amino acid but containing 30 mM 3-amino-1,2,4-triazole (3-AT) (8, 12, 33). Approximately 5 × 105 independent clones were obtained; 50 growing colonies were obtained, of which 10 were β-galactosidase positive. After DNA isolation and rescreening, three colonies remained histidine and β-galactosidase positive. These clones were further characterized by sequencing and analyzed on gene homology by using the BLAST database. Two of these clones were the human homolog of the vertebrate PcG gene eed (36) and it was therefore named EED. The entire EED cDNA insert was used to screen a λACT human lymphocyte cDNA library (Clontech). Filters were hybridized overnight at 60°C in 0.25× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10× Denhardt’s solution–10% dextran sulfate–0.1% sodium dodecyl sulfate (SDS)–100 μg of denatured herring sperm DNA per ml–[α-32P]ATP-labeled probe (5 × 105 cpm/ml). After being washed three times for 45 min at 60°C in 0.25× SSC–0.1% SDS, the filters were autoradiographed with intensifying screens for 2 days at −70°C. This led to the isolation of the full-length EED cDNA. Potential interactions between Enx1 and EED and other vertebrate PcG proteins were tested. The transformants were plated on medium lacking the leucine, tryptophan, and histidine amino acids, with or without 30 mM 3-AT. Interactions that were scored negative failed to grow in the presence of 30 mM 3-AT. Due to residual HIS3 promoter activity they are able, however, to grow on medium that does not contain 3-AT (8, 12, 33). Under these nonselective conditions, negative interactions were β-galactosidase negative and the colony color was indicated as white (Table 1). Positive interactions are characterized by growth in the presence of 30 mM 3-AT and by being β-galactosidase positive. To exclude the possibility that the negative interactors did not produce either one of the fusion proteins, we Western blotted equal amounts of protein and incubated the blots with monoclonal antibodies that specifically recognize the GAL4 DNA-binding domain (GAL4-DBD) or the GAL4 transactivation domain (GAL4-TAD) protein (Clontech). All positive and negative interactors expressed both GAL4-DBD fusions and the GAL4-TAD fusions at approximately the same levels (data not shown).
TABLE 1.
β-Galactosidase activities of Enx1 and EED interactions in the two-hybrid system
| GAL4-DBD fusion (aa) | GAL4-TAD fusion (aa) | Colony colora | Relative β-galacto- sidase activityb |
|---|---|---|---|
| Enx1 (1–746) | EED (1–535) | Blue | 100 |
| HPC2 (1–558) | White | <1 | |
| BMI1 (1–326) | White | <1 | |
| Xbm1 (1–326) | White | <1 | |
| RING1 (1–377) | White | <1 | |
| HPH1 (1–676) | White | <1 | |
| HPH1 (713–1013) | White | <1 | |
| HPH2 (137–432) | White | <1 | |
| EED (1–535) | Enx1 (1–746) | Blue | 100 |
| HPC2 (1–558) | White | <1 | |
| BMI1 (1–326) | White | <1 | |
| Xbm1 (1–326) | White | <1 | |
| RING1 (1–377) | White | <1 | |
| HPH1 (1–676) | White | <1 | |
| HPH1 (713–1013) | White | <1 | |
| HPH2 (137–432) | White | <1 |
White colonies were obtained on medium lacking both histidine and 3-AT. Blue colonies were obtained on medium lacking histidine but in the presence of 3-AT.
The average β-galactosidase activity in a triplicate experiment was 135 U. This activity was set at 100%.
RNA analysis.
Multitissue Northern blots containing approximately 2 μg of poly(A)+ RNA from different human tissues or human cell lines per lane were obtained commercially (Clontech). The U-2 OS osteosarcoma cell line was not present on the commercial Northern blot. Poly(A)+ RNA of U-2 OS was isolated and blotted, and the expression pattern of EED was analyzed. To allow a comparison with the commercial Northern blot, we blotted poly(A)+ RNA of SW480 cells, which is represented on the commercial blot and in which the EED gene is strongly expressed. The blots were hybridized with [α-32P]dATP-labeled DNA probes, and the blots were autoradiographed with intensifying screens at −70°C, using X-ray films.
Production of the Enx1/EZH2 and EED polyclonal rabbit antibodies.
Fusion proteins were made of the N-terminal region of Enx1 (amino acids [aa] 1 to 286) and EED (aa 95 to 283). cDNAs were cloned into pET-23 expression vectors (Novagen, Madison, Wis.). Fusion proteins were produced in Escherichia coli BL21(DE), and the purified fusion proteins were injected into a rabbit. Serum was affinity purified over an antigen-coupled CNBr-Sepharose column (Pharmacia, Uppsala, Sweden).
IPs and Western blotting.
U-2 OS osteosarcoma cells, which were grown to confluence, were lysed in ELB lysis buffer (250 mM NaCl, 0.1% Nonidet P-40, 50 mM HEPES [pH 7.0], 5 mM EDTA) containing 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and the protease inhibitors leupeptin, benzamidine, pepstatin and aprotinin. The cell lysate was sonicated three times with bursts of 15 s. The lysate was centrifuged at 14,000 × g at 4°C for 10 min, and the supernatant (500 μl) was aliquoted and stored at −70°C; 25 μl of the supernatant was subsequently incubated with the indicated antibodies for 2 h at 4°C. Goat anti-rabbit immunoglobulin G (IgG) antibodies (Jackson ImmunoResearch Laboratories) were added to the mixture and incubated for 1 h at 4°C. Protein A-Sepharose CL-4B (Pharmacia) and ELB buffer were added to enlarge the volume of the mixture to 300 μl. The mixture was incubated for 1 h at 4°C under continuous mixing. The mixture was centrifuged at 1,500 × g at 4°C for 1 min, washed with 1 ml of ice-cold ELB buffer without protease inhibitors, and centrifuged at 1,500 × g at 4°C for 1 min. This washing procedure was repeated five times. After heating and centrifugation to remove the protein A-Sepharose beads, the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose. The blots were probed with a 1:1,000 dilution of affinity-purified rabbit antibodies against EED and Enx1/EZH2 (Fig. 6A and B, respectively) or chicken anti-BMI1 antibody (Fig. 6C). The secondary alkaline phosphatase goat anti-rabbit or donkey anti-chicken IgG (heavy plus light chain) antibodies (Jackson ImmunoResearch Laboratories) were diluted 1:10,000, and nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Boehringer) was used as substrate for detection. The heavy chains of the rabbit immunoprecipitation (IP) antibodies (approximately 50 kDa) were recognized by the rabbit antibodies against EED and Enx1/EZH2. This lower molecular weight range is, however, not shown in Fig. 4A and B. To determine the relative molecular weight of the EED protein, T7-tagged EED cDNAs were transfected to U-2 OS cells. These cells were harvested, and the cell lysates were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with a 1:10,000 dilution of mouse monoclonal antibody against T7 (Novagen).
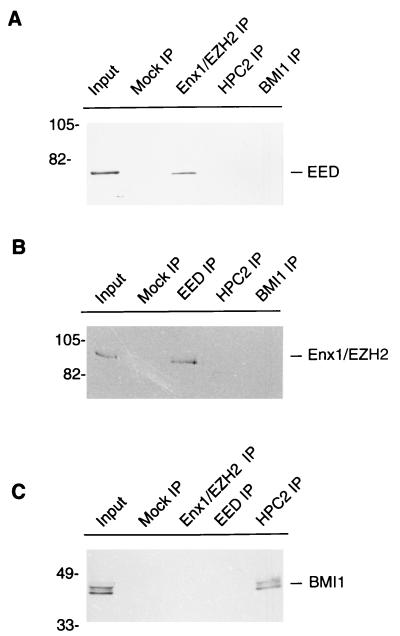
FIG. 6.
Enx1/EZH2 and EED coimmunoprecipitate from extracts of U-2 OS cells. IP experiments were performed with extracts of U-2 OS human osteosarcoma cells. (A) IP performed with polyclonal rabbit antibodies against Enx1/EZH2 (Enx1/EZH2 IP), HPC2 (HPC2 IP), and BMI1 (BMI1 IP) or with preimmune serum (Mock IP). The resulting IPs were Western blotted and incubated with rabbit anti-EED antibody. The 68-kDa EED protein was detected in the U-2 OS cell extract (Input) and in the Enx1/EZH2 IP but not in the HPC2 IP and BMI1 IP. (B) IP performed with polyclonal rabbit antibodies against EED (EED IP), HPC2 (HPC2 IP), and BMI1 (BMI1 IP) or with preimmune serum (Mock IP). The resulting IPs were Western blotted and incubated with rabbit anti-Enx1 antibody. The approximately 90-kDa Enx1/EZH2 protein was detected in the U-2 OS cell extract (Input) and in the EED IP, but not in the HPC2 IP and BMI1 IP. (C) IP performed using polyclonal rabbit antibodies against Enx1/EZH2 (Enx1/EZH2 IP), EED (EED IP), and HPC2 (HPC2 IP) or with preimmune serum (Mock IP). The resulting IPs were Western blotted and incubated with chicken anti-BMI1 antibody. The approximately 44- to 47-kDa BMI1 protein was detected in the U-2 OS cell extract (Input) and in the HPC2 IP, but not in the Enx1/EZH2 IP and the EED IP. Molecular weights are indicated in thousands.
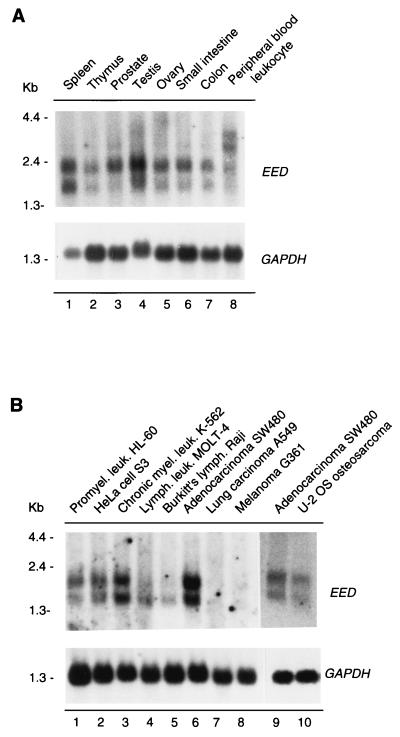
FIG. 4.
Expression patterns of EED in human tissues (A) and in human cancer cell lines (B). (A) Expression levels in spleen (lane 1), thymus (lane 2), prostate (lane 3), testis (lane 4), ovary (lane 5), small intestine (lane 6), colon (lane 7), and peripheral blood leukocytes (lane 8). (B) Expression levels in promyelocytic leukemia HL-60 (lane 1), HeLa cell S3 (lane 2), chronic myelogenous leukemia K-562 (lane 3), lymphoblastic leukemia MOLT-4 (lane 4), Burkitt’s lymphoma Raji (lane 5), colorectal adenocarcinoma SW480 (lane 6), lung carcinoma A549 (lane 7), and melanoma G361 (lane 8) cell lines. Lanes 1 to 8 represent a commercially obtained Northern blot. We also isolated and blotted poly(A)+ RNA from U-2 OS (lane 10). To allow comparison with the commercial multiple-tissue Northern blot, we isolated and blotted poly(A)+ RNA from SW480 cells (lane 9). The filters were rehybridized with a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to verify the loading of RNA in each lane.
Immunofluorescence labeling of tissue culture cells.
Coverslips with attached U-2 OS cells were rinsed once with phosphate-buffered saline (PBS) and incubated with freshly prepared 2% (wt/vol) paraformaldehyde in PBS for 15 min at room temperature. After fixation, cells were rinsed twice with PBS and permeabilized with 0.5% (wt/vol) Triton X-100 (Sigma) for 5 min at room temperature. Cells were subsequently rinsed twice with PBS, incubated in PBS containing 100 mM glycine for 10 min, and incubated for 10 min in PBG (PBS containing 0.5% bovine serum albumin and 0.05% gelatin from cold-water fish skin [Sigma]). Fixed cells were incubated for 2 h at room temperature with primary antibodies diluted in PBG. Subsequently, cells were washed four times for 5 min in PBG and incubated with secondary antibodies diluted in PBG for 1.5 h at room temperature. Secondary antibodies used were donkey anti-rabbit IgG coupled to fluorescein isothiocyanate and donkey anti-chicken IgG-coupled Cy3 (both from Jackson ImmunoResearch Laboratories). After labeling, cells were washed four times for 5 min in PBG and twice for 5 min in PBS. Images of labeled cells were produced on a Leica confocal laser scanning microscope with a 100×/1.35 oil immersion lens. Pairs of images were collected simultaneously in the green and red channels. Single optical sections are shown. The first two pictures of each row (Fig. 7) represent the two different scanned channels for imaging the double labeling, whereas the last picture represents the reconstituted image. To determine potential colocalization between the Enx1/EZH2 and EED proteins, we transiently transfected U-2 OS cells with T7-tagged EED protein. Double labeling was performed with a mouse monoclonal antibody against T7 (Novagen) and affinity-purified rabbit antibody against Enx1/EZH2.
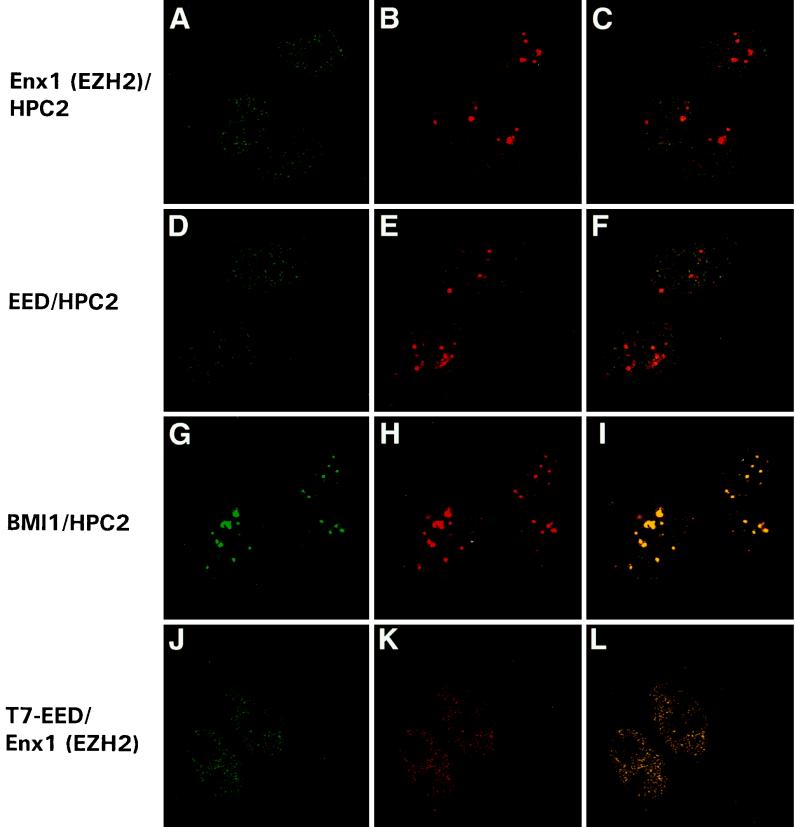
FIG. 7.
Enx1/EZH2 and EED do not colocalize with the PcG protein HPC2 in nuclear domains of U-2 OS cells. Confocal single optical sections of double-labeled cells are presented. (A to C) Rabbit anti-Enx1/EZH2 and chicken anti-HPC2 double labeling. Enx1/EZH2 (A), like HPC2 (B), is homogeneously distributed in the nucleus, but unlike HPC2, Enx1/EZH2 is not concentrated in large, brightly labeled domains (B and C). (D to F) Rabbit anti-EED and chicken anti-HPC2 double labeling. EED (D), like HPC2 (E), is homogeneously distributed in the nucleus, but unlike HPC2, EED is not concentrated in large, brightly labeled domains (E and F). Rabbit anti-BMI1 (G) and chicken anti-HPC2 (H) double labeling demonstrates colocalization of HPC2 and BMI1 in the large bright domains (I) (indicated by yellow). We transiently transfected U-2 OS cells with the T7-tagged EED535 protein. Double labeling was performed with a mouse monoclonal antibody against T7 (J) and the affinity-purified rabbit antibody against Enx1/EZH2 (K). We observed complete colocalization between T7-EED535 and Enx1/EZH2 (L).
LexA fusion reporter gene-targeted repression assay.
The LexA repression assay was performed as described previously (3, 33, 34). U-2 OS cells were cultured in a 25-cm2 flask and cotransfected with 2 μg of the heat shock factor (HSF)-inducible luciferase (LUC) reporter plasmid (33, 34), 4 μg of the LexA-fusion constructs, and 2 μg of the pSV/β-Gal construct (Promega), using the calcium phosphate transfection method. The HSF-inducible LUC reporter plasmid was activated by exposure of the cells at 43°C for 1 h, followed by a 6-h recovery at 37°C. LUC activity was normalized to β-galactosidase activity. The LUC activity in cells transfected with the LUC reporter plasmid only was therefore set at 100%, and LUC activities in cells cotransfected with the indicated plasmids were expressed as percentage of this control. The degree of repression by LexA-fusion proteins is expressed as mean ± standard error of the mean.
RESULTS
Identification of the human EED protein which interacts with Enx1.
To identify genes encoding proteins that interact with Enx1, the vertebrate homolog of the Drosophila PcG protein E(z), we performed a two-hybrid screen (8). The full-length coding region of Enx1 (16) was cloned into the pAS2 vector (8). Plasmid pAS2-Enx1 was cotransformed with a human fetal brain Matchmaker two-hybrid library (Clontech) into the yeast strain Y190. The transformants were plated on selective medium lacking histidine, tryptophan, and leucine (8, 12, 33). Of approximately 5 × 105 independent clones, 50 colonies were His+; of these, 10 were β-galactosidase positive. After DNA isolation and rescreening, three colonies remained histidine and β-galactosidase positive. Two 1,628-bp-long cDNA clones were identical. The entire EED cDNA insert was used to screen a λACT human fetal brain cDNA library (Clontech). We isolated a 1,837-bp-long cDNA (Fig. 1). The predicted 535-aa-long protein is identical to the mouse eed protein (7) and we therefore name the novel human protein EED. The eed (for embryonic ectoderm development) gene (7, 36) has been identified as being a vertebrate homolog of the Drosophila PcG gene esc (14, 32, 39). Within the 1,605-bp-long coding region, EED is 93% identical with eed at the nucleotide level. The N-terminal region of the protein (from aa 115 to 147) is rich in proline (P), glutamic acid (E), serine (S), and threonine (T), a potential PEST sequence, which has been implicated in protein degradation (31). Most important is the presence of five WD-40 domains throughout the protein. In these domains, the homology between EED and esc is highest, ranging from 54% identity in WD-40 repeat 1 to 83% identity in WD-40 repeat 4 (36) (Fig. 2).
FIG. 1.
Nucleotide sequence of EED and its predicted amino acid sequence. The point mutations (bp 872 and 881) in eed that are found in the mutant eed mice are boxed. The stop codon of the EED gene is indicated with an asterisk.
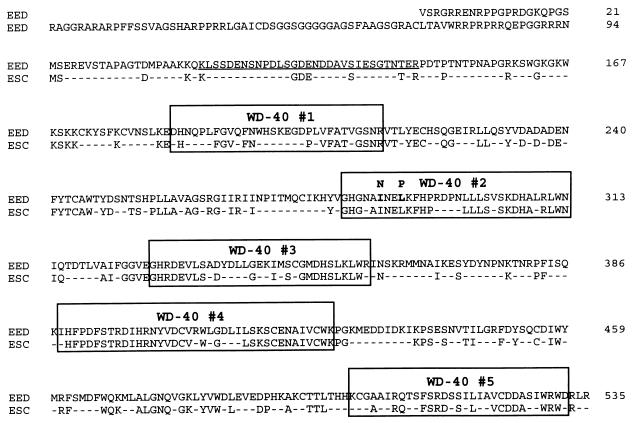
FIG. 2.
Comparison of the EED/eed protein with the Drosophila PcG protein esc. Identical amino acids are shown; nonidentical amino acids are indicated with a dash. The five WD-40 repeats are indicated with boxes. A putative PEST sequence is underlined. The point mutations (aa 287 and 290) in eed that are found in the mutant eed mice are shaded in the boxed WD-40 domain 2.
In conclusion, a two-hybrid screen with the Enx1 protein as the target resulted in the isolation of a human cDNA which encodes a protein that is identical with the mouse PcG protein eed. We name this human protein EED.
Mapping of the domains of interaction between Enx1 and EED.
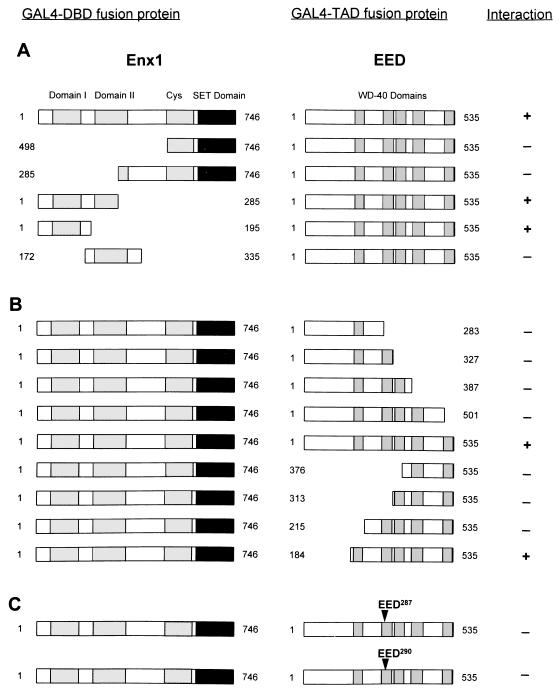
To define the domains that are responsible for the interaction between Enx1 and EED, we subcloned different parts of Enx1 and EED in frame with the GAL4-DBD and tested whether these proteins could still interact with full-length EED or full-length Enx1. Enx1 comprises two N-terminal domains which show strong homology between Drosophila E(z) and its mammalian homologs. These domains have been designated domains I and II (25). Furthermore, Enx1 contains a C-terminal cysteine-rich domain and a SET domain. This last domain is found in a number of different proteins such as the trithorax protein (17). We found that the region encompassing both the cysteine-rich domain and the SET domain (aa 498 to 746) did not interact with EED (Fig. 3A). Also a region extended toward the N terminus (aa 285 to 746) did not interact with EED. In contrast, the region encompassing domain I and a part of domain II (aa 1 to 285) did interact with EED. To analyze this region in more detail, we made two constructs, one containing homology domain I (aa 1 to 195) and the other containing homology domain II (aa 172 to 335). Only the region of Enx1 which contains homology domain I interacted with the EED protein (Fig. 3A). We conclude that EED binds to the N-terminal region of Enx1 which encompasses homology domain I.
FIG. 3.
Mapping of interaction domains between Enx1 and EED. (A) Indicated portions of Enx1 were fused to the GAL4-DBD (GAL4-DBD fusion protein). These Enx1 regions include homology domains I (aa 94 to 159), homology domain II (aa 218 to 329), a cysteine-rich domain (aa 498 to 612), and the SET domain (aa 613 to 742). Constructs that encompass different portions of the Enx1 protein are indicated. The plasmids were cotransformed with full-length EED (aa 1 to 535), which is fused to the GAL4-TAD (GAL4-TAD fusion protein). Interactions were positive (+) when the transformants were able to grow on selective medium lacking histidine and when they were also β-galactose positive. (B) Full-length Enx1 (aa 1 to 746) fused to the GAL4-DBD was tested for interaction against indicated portions of EED fused to the GAL4-TAD. (C) Indicated point mutations in the second WD-40 domain of EED were made and tested against the full-length Enx1 protein.
EED contains five WD-40 domains which are thought to be involved in protein-protein interactions (14). We tested the importance of these WD-40 domains for the interaction between Enx1 and EED. We made truncated EED protein constructs that contain an increasing number of WD-40 domains. We found that none of the truncated EED proteins that contain up to four WD-40 domains interacted with Enx1 (Fig. 3B). Only when all five WD-40 domains were present was this truncated EED protein (aa 184 to 535) able to interact with Enx1 (Fig. 3B). The most N-terminal region of EED (aa 1 to 184), which does not contain WD-40 domains, was not important for mediating the interaction between Enx1 and EED (Fig. 3B).
This last result implies that all WD-40 domains of EED are necessary for interaction with Enx1. We tested this notion further by creating mutant EED proteins that contain point mutations in the second WD-40 domain. A recessive embryonic-lethal eed mutation has been shown to be due to point mutations that result in altered aa 287 or 290 (36). These mutations represent a null or hypomorphic allele, respectively. In the null mutant, aa 287 is changed from isoleucine (I) to asparagine (N) by a change in the codon ATC to AAC (Fig. 1 and 2). In the hypomorphic mutant, aa 290 is changed from leucine (L) to proline (P) by a change in the codon CTG to CCG. We created these mutations by using PCR primers which contained the respective mutations and tested whether the mutant EED proteins are still able to interact with Enx1 in the two-hybrid system. We found that both point mutations abolished the interaction with Enx1 in the two-hybrid system (Fig. 3C). This result underlines the importance of intact WD-40 domains of EED for the interaction with Enx1.
In conclusion, we find that the N-terminal region of Enx1 that encompasses homology domain I mediates the interaction between Enx1 and EED. For this interaction, all five WD-40 domains of EED are necessary. Furthermore, the WD-40 domains need to be intact since point mutations in the second WD-40 domain abolish the interaction between Enx1 and EED.
No interactions between Enx1 and EED and other, previously identified PcG proteins.
We next tested whether we could identify interactions between Enx1 or EED with other, previously identified PcG proteins. This was done by cloning Enx1 and EED in frame with either the GAL4-DBD or the GAL4-TAD. Enx1 and EED interacted equally strongly with each other, no matter whether they were fused to GAL4-DBD or GAL4-TAD (Table 1). However, we found no interactions between Enx1 or EED and the vertebrate PcG proteins HPC2 (34), BMI1 (1), Xbmi1 (30), RING1 (33), or HPH1 or HPH2 (12) (Table 1). We conclude that there are no interactions between Enx1 and EED and other human PcG proteins in the two-hybrid system.
Distribution of EED transcripts in human tissues and cancer cell lines.
We next studied the expression level of the EED gene by analyzing multiple-tissue Northern blots containing poly(A)+ mRNA from different human tissues or human cell lines (Clontech). As the probe we used the entire EED cDNA. We detected two transcripts of approximately 1.5 and 2 kb in all the tissues and cell lines tested (Fig. 4). In normal tissue also higher transcripts were detected, but at a much weaker level (Fig. 4A). Only in peripheral blood leukocytes were these higher transcripts of approximately 3 and 3.5 kb expressed at a higher level (Fig. 4A, lane 8). The significance of this observation is not clear. One possibility is that these transcripts selectively arise from different cell types such as granulocytes or lymphocytes within the heterogeneous peripheral blood leukocytes. In normal human tissues, the highest level of EED expression is found in the testis (Fig. 4A, lane 4). Expression levels are still well pronounced in the spleen (lane 1), prostate (lane 3), ovary (lane 5), and small intestine (lane 6). The expression levels of EED are somewhat lower in the thymus (lane 2), colon (lane 7), and peripheral blood leukocytes (lane 8). The differences in abundance of EED transcripts are more pronounced in human cell lines than in normal human tissues (Fig. 4B). Highest expression levels are observed in the colorectal adenocarcinoma SW480 (lane 6 and 9), K-562 (lane 3), and U-2 OS osteosarcoma (lane 10) cells. In Burkitt’s lymphoma Raji (Fig. 4B, lane 5), lung carcinoma (lane 7), and melanoma G361 (lane 8) cells, EED is expressed at a lower level.
Comparison between different EED proteins.
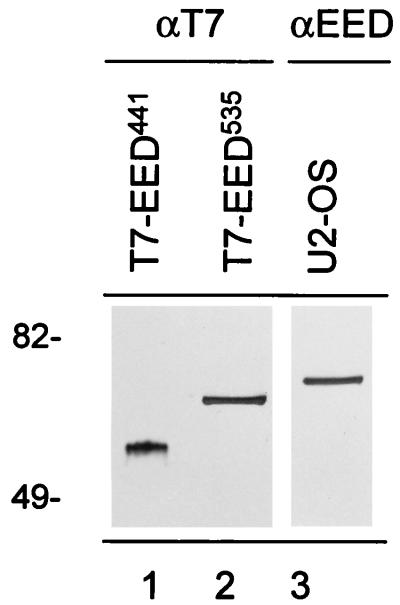
The first published eed cDNA encodes a 441-aa-long protein (36). However, a more recent report describes a longer version of the eed protein, which contains an additional 94 N-terminal amino acids (7). This longer protein starts from a codon that encodes valine (7). For convenience, we will call the smaller eed protein eed441 and the larger protein eed535. The cDNA clone that we isolated potentially encodes the EED535 protein (Fig. 1 and 2). To test this notion, we created two EED constructs which contain a T7 tag at the N terminus. One construct contains the EED cDNA starting from the GTG, thus potentially encoding the EED535 protein. The other construct contains the EED cDNA starting from the ATG (at putative aa position 94), thus encoding the EED441 protein. Both constructs were transiently transfected to U-2 OS cells to determine the relative molecular weights of the T7-tagged EED proteins. The cells were harvested, and the cell lysates were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with a mouse monoclonal antibody against T7 (Fig. 5, lane 1 and 2). A 53-kDa protein was recognized in extracts from T7-EED441-transfected U-2 OS cells (Fig. 5, lane 1), and a 65-kDa protein was recognized in extracts from T7-EED535-transfected cells (Fig. 5, lane 2). It being taken into account that the T7 tag encodes an approximately 2-kDa protein fragment, this finding indicates molecular masses of 51 kDa for the EED441 protein and 63 kDa for the EED535 protein. On Western blots, an affinity-purified antibody against EED recognizes a protein of 68 kDa (Fig. 5, lane 3). This result suggests that the endogenous 68-kDa EED protein is encoded by the EED535 cDNA.
FIG. 5.
Comparison between different EED proteins. Two T7-tagged EED constructs which potentially encode the EED441 protein and EED535 protein were made. U-2 OS cells were transfected with T7-EED441 (lane 1) and T7-EED535 (lane 2) cDNAs, and the respective cell lysates were analyzed by Western blotting. Blots were probed with a mouse monoclonal antibody against T7 (αT7; lanes 1 and 2). The endogenous EED protein was detected in a cell lysate of U-2 OS cells (lane 3), using an affinity-purified antibody against EED (αEED). Molecular weights are indicated in thousands.
Enx1/EZH2 and EED coimmunoprecipitate from extracts of U-2 OS cells.
To test whether Enx1 and EED exist in vivo as part of a protein complex, we performed IP experiments using antibodies raised against the EED and Enx1 proteins. We used extracts from U-2 OS cells in which PcG proteins are expressed at a high level (2, 12, 33, 34) and in which a high expression level of the EED gene is found (Fig. 3). On Western blots, the affinity-purified antibody against EED recognizes a protein of 68 kDa (Fig. 6A).
Different splicing variants of the human Enx1 homolog result in proteins of different sizes (15, 25), and there is confusion in the literature concerning the nomenclature of these genes and their corresponding proteins. The human homolog of Enx1 has been named ENX1, but it encodes a protein that is 133 aa shorter than the Enx1 protein (15). Recently another Enx1 homolog, called EZH2, has been isolated (25). The EZH2 cDNA encodes a protein that is 98.3% identical with the Enx1 protein and contains the 133 N-terminal aa which are lacking from the ENX1 protein (25). The C-terminal part of the EZH2 protein from aa 133 to 746 is 100% identical with the ENX1 protein, supporting the idea that the two proteins arise from differential splicing (25). We raised antibodies against the first 286 N-terminal aa of the Enx1 protein, which includes the 133 N-terminal aa that are missing from the ENX1 protein. Within these 133 N-terminal aa, the Enx1 and EZH2 proteins are 97% identical. In human cells, the antibodies raised against Enx1 (aa 1 to 286) are therefore more likely to recognize the larger EZH2 protein than the smaller ENX1 protein. We found that the affinity-purified antibody recognizes a protein of approximately 90 kDa (Fig. 6A), which is close to the predicted 85 kDa of the EZH2 protein (7, 25). We will therefore refer to this human protein, which is recognized by the antibody against the mouse Enx1 protein, as Enx1/EZH2.
We found that EED was present in the Enx1/EZH2 IP (Fig. 6A) and that Enx1/EZH2 was present in the EED IP (Fig. 6B). In contrast, in IPs with the HPC2 and BMI1 antibodies, we did not detect the presence of EED (Fig. 6A) or Enx1/EZH2 (Fig. 6B). In the reciprocal experiment, we did not detect BMI1 in the Enx1/EZH2 and EED IPs (Fig. 6C). In this experiment we did, as observed previously (33), detect BMI1 in the IP with the HPC2 antibody (Fig. 6C). Finally, no antigens were detected when the specific IP antibodies were replaced by preimmune sera (Fig. 6, Mock IP) or unrelated antibodies or when the first antibody was merely omitted from the IPs (data not shown). This result underlines the specificity of the IPs.
In conclusion, we show that Enx1/EZH2 and EED coimmunoprecipitate from extracts of U-2 OS human osteosarcoma cells. The results indicate that Enx1/EZH2 and EED are in vivo part of a protein complex, but that they are not included in complexes which contain the human PcG proteins HPC2 and BMI1.
Enx1/EZH2 and EED do not colocalize with HPC2 in nuclei of U-2 OS cells.
We next analyzed the subcellular localization of the Enx1/EZH2 and EED proteins in relation to the PcG protein HPC2 by performing immunofluorescence labeling experiments. We used U-2 OS cells, in which we found that Enx1/EZH2 and EED coimmunoprecipitate (Fig. 6). The use of chicken anti-HPC2 (33) and rabbit anti-Enx1/EZH2 and anti-EED allows double-labeling experiments. Both Enx1/EZH2 and EED proteins were found in the nuclei of U-2 OS, throughout the nucleoplasm in a fine granular pattern (Fig. 7A and D, respectively). In striking contrast, the HPC2 protein is found in a punctate, fine granular pattern, but also in large, brightly labeled domains (Fig. 7B, E, and H). In these large domains, we and others observed complete colocalization between the human PcG proteins HPC2 and BMI1 (Fig. 7G to I) as well as between HPC2, BMI1, RING1, HPH1, and HPH2 (2, 12, 33–35). No colocalization between Enx1/EZH2 or EED and HPC2 or other human PcG proteins (data not shown) in these large domains was found, since neither Enx1/EZH2 nor EED is localized in large domains.
Since both antibodies against Enx1/EZH2 and EED are rabbit derived, it is not possible to directly test for potential colocalization between those two proteins. However, to determine potential colocalization between the Enx1/EZH2 and EED proteins, we transiently transfected U-2 OS cells with the T7-tagged EED535 protein. Double labeling was performed with a mouse monoclonal antibody against T7 (Fig. 7J) and the affinity-purified rabbit antibody against Enx1/EZH2 (Fig. 7K). We observed complete colocalization between T7-EED535 and Enx1/EZH2 (Fig. 7L), which indicates that endogenous EED and Enx1/EZH2 also colocalize with each other.
We conclude that Enx1/EZH2 and EED do not colocalize with known human PcG proteins in large nuclear domains of U-2 OS cells. This is in striking contrast with previous observations which showed that the human PcG proteins HPC2, BMI1, RING1, HPH1, and HPH2 all colocalize in these domains. These results are in agreement with the observation that Enx1/EZH2 and EED do not coimmunoprecipitate with the other human PcG proteins, and they strengthen the notion that Enx1/EZH2 and EED form a distinct protein complex.
Repression of HSF-induced LUC gene activity by Enx1 and EED.
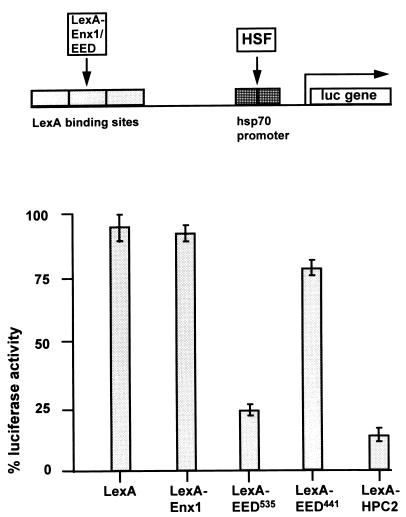
In Drosophila, the PcG proteins have been identified as repressors of gene activity. So far, no PcG protein has been found to bind directly to DNA. Nevertheless, the ability of PcG proteins to repress gene activity can be tested by targeting LexA fusion proteins to reporter genes (3). Previously, we have found that a LexA-HPC2 as well as LexA-RING1 fusion proteins repress gene activity when targeted to a reporter gene (33, 34). We therefore analyzed the ability of LexA-Enx1, LexA-EED441, and LexA-EED535 fusion proteins to repress gene activity when targeted to a reporter gene.
U-2 OS human osteosarcoma cells were transfected with a construct containing a tandem of four LexA operators, binding sites for the HSF transcriptional activator, and the hsp70 TATA promoter region, immediately upstream of the LUC reporter gene (3, 33, 34). As a transcriptional activator, the endogenous HSF was used. In the absence of HSF, no LUC activity was observed (data not shown). Maximum LUC activity in the presence of HSF was set at 100%. Cotransfection of LexA alone had no significant influence on HSF-induced LUC activity (Fig. 8; 96% ± 6% [n = 3]). We found that LexA-Enx1 was not able to repress LUC expression significantly (Fig. 8; 93% ± 3% [n = 3]). We also found that whereas LexA-EED535 was able to significantly repress LUC expression (Fig. 8; 25% ± 4% [n = 3]), LexA-EED441 was not able to repress LUC expression (78% ± 5% [n = 3]). In the same experiments, LexA-HPC2 repressed LUC expression most efficiently (Fig. 6; 10% ± 5% [n = 3]). This degree of repression has been observed previously (3, 33, 34).
FIG. 8.
Repression of HSF-induced LUC gene activity by Enx1 and EED. Activation of LUC reporter expression is maximally induced by endogenous HSF in the absence of any LexA fusion protein and was set at 100%. LUC activities in cells cotransfected with other plasmids were expressed as a percentage of this control value. Bars represent the average degree of repression by LexA, LexA-Enx1, LexA-EED535, LexA-EED441, or LexA-HPC2 in three independent experiments (mean ± standard error of the mean).
These results are in agreement with the previous report in which the eed535 protein but not the eed441 protein was able to repress gene activity when targeted to a reporter gene (7). We conclude that the EED535 protein but not the Enx1 protein is able to repress gene activity when targeted to a reporter gene.
DISCUSSION
Identification of an interaction between Enx1/EZH2 and EED.
In this report, we describe the identification of an interaction between Enx1/EZH2 and EED, mammalian homologs of, respectively, the Drosophila PcG proteins E(z) and esc. Our interest in searching for proteins that interact with Enx1/EZH2 is inspired by observations that in Drosophila, E(z) can be considered to be a PcG and a trxG gene. Double mutations of E(z) and a trxG gene result in homeotic phenotypes which are similar to the homeotic phenotypes which are also observed in double mutants of trxG genes (26). Also, within imaginal discs of larvae hemizygous for certain mutant alleles of E(z) there is no accumulation of homeotic proteins such as Antennapedia and Ultrabithorax (26). Lack of accumulation of these homeotic proteins is a hallmark for trxG mutations. Another line of evidence that points toward a functional convergence between PcG and trxG proteins is the observation that E(z) contains the SET domain, a stretch of 114 aa in the C-terminal region of the E(z) protein, which is 48% identical and 68% similar with the corresponding region in the trx protein (17). Finally, polytene chromosome binding of the trx protein is strongly reduced in homozygous E(z) mutants (4), and vice versa, polytene chromosome binding of the E(z) protein is reduced in trx mutants (24). These data suggest a role for the E(z) which may differ considerably from other PcG proteins, providing an important rationale to perform a two-hybrid screen with a mouse homolog of E(z) as the target.
Here, we report the identification of EED, the human homolog of the murine eed protein, a homolog of the Drosophila esc protein. EED interacts with the Enx1 protein, both in the two-hybrid assay and in vivo. esc is a PcG protein which also stands apart from other PcG proteins. PcG genes play a crucial role in the maintenance of homeotic gene activity during later phases of embryonic development, but for esc an earlier role has been proposed (14, 32, 39, 40). First, the esc gene is expressed only during a limited, very early developmental phase of Drosophila development (39). Absence of the esc gene product during this short period results in homeotic transformations which are very similar to those of other PcG mutations. However, when the esc protein is missing during earlier or, most importantly, during later phases in development, no phenotypical defects are observed (39, 40). This finding indicates a role for esc which is different from these other PcG genes. It may be significant that the two atypical PcG proteins Enx1 and EED interact with each other and not with other PcG proteins.
Do Enx1/EZH2 and EED form a class of PcG proteins that differ from other, previously identified mammalian PcG proteins?
We found that Enx1/EZH2 and EED interact in vivo but not with other, previously identified human PcG proteins. We base this conclusion on the observations that Enx1/EZH2 and EED do not interact with other human PcG proteins in the two-hybrid system and that they do not coimmunoprecipitate or colocalize in interphase nuclei with any of these other human proteins. This is in striking contrast with the other, previously identified human PcG proteins HPC2, BMI1, RING1, HPH1, and HPH2, which all coimmunoprecipitate with each other and colocalize in large nuclear domains of several human cell lines (2, 12, 33–35).
It is important to point out that the E(z) protein colocalizes with other PcG proteins on only a subset of PcG binding sites on polytene chromosomes. Whereas the Drosophila PcG proteins Pc, Psc, Su(z)2, and Ph are found at 80 to 90 specific cytological sites, E(z) is found at only 42 of these sites (4). Only two additional E(z) binding sites do not overlap with PcG sites. The localization of the esc protein has not been reported as yet. Although E(z) and other PcG proteins bind to 42 common cytological sites, this does not automatically imply that E(z) is part of a common PcG protein complex. Also the trx protein has cytological binding sites in common with PcG proteins (6), but a direct, physical interaction between trx and PcG protein has not been established. Even within the resolution of Polycomb response elements (24, 41), there is probably still room for distinct protein complexes that have no physical interactions. Taken together, our current data and those from previous reports suggest that both the E(z) homolog Enx1/EZH2 and the esc homologs eed/EED behave differently from other PcG proteins.
Functional significance of the Enx1/EZH2 and EED interaction.
It has been proposed that esc interacts with the transcriptional machinery through the WD-40 domains (14). This model is based on the homology that is found between esc and Tup1, a yeast protein which also contains seven WD-40 domains. These WD-40 domains are important for the involvement of the Tup1 protein in the repression of gene activity and in its binding to the DNA-binding homeodomain protein α2 (19, 23). Also, in several esc mutants point mutations in the WD-40 domains have been found (32). We find that either one of two point mutations in the second WD-40 domain completely abolishes the interaction in the two-hybrid system between Enx1 and EED. Precisely these two point mutations are responsible for the severe developmental defects in eed mutant mice (36). It is significant that the ability of the eed535 protein to repress gene activity is also completely abolished by these point mutations (7). It is therefore tempting to speculate that both the interference with the binding capacity and the repressing abilities of the eed/EED protein through these point mutations contribute to the developmental defects in eed mutant mice. One immediate consequence of these point mutations can be that the Enx1 protein is no longer able to bind to eed with the subsequent loss of integrity of the protein complex of which Enx1/EZH2 and eed are part.
ACKNOWLEDGMENTS
R.G.A.B.S. and J.V. contributed equally to this work.
We thank A. Ullrich for the Enx1 clone.
REFERENCES
- 1.Alkema M J, Wiegant J, Raap A K, Berns A, van Lohuizen M. Characterization and chromosomal localization of the human proto-oncogene BMI-1. Hum Mol Genet. 1993;2:1597–1603. doi: 10.1093/hmg/2.10.1597. [DOI] [PubMed] [Google Scholar]
- 2.Alkema M J, Bronk M, Verhoeven E, Otte A P, van’t Veer L J, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 3.Bunker C A, Kingston R E. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington E A, Jones R S. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–4083. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y-L, King B O, O’Connor M, Mazo A, Huang D-H. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol Cell Biol. 1995;15:6601–6612. doi: 10.1128/mcb.15.12.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denisenko O N, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol Cell Biol. 1997;17:4707–4717. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type I catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 9.Fields S, Song O K. A novel genetic system to detect protein-protein interactions. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Franke A, DeCamillis M, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz E M, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature (London) 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 12.Gunster M J, Satijn D P E, Hamer C M, den Blaauwen J L, de Bruijn D, Alkema M J, van Lohuizen M, van Driel R, Otte A P. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and human homologs of Polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 14.Gutjahr T, Frei E, Spicer C, Baumgarter S, White R A H, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 1995;14:4296–4306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobert O, Jalla B, Ullrich A. Interaction of Vav with ENX-1, a putative transcriptional regulator of homeobox gene expression. Mol Cell Biol. 1996;16:3066–3073. doi: 10.1128/mcb.16.6.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobert O, Sures I, Ciossek T, Fuchs M, Ullrich A. Isolation and developmental expression of Enx-1, a novel mouse Polycomb group gene. Mech Dev. 1996;55:171–184. doi: 10.1016/0925-4773(96)00499-6. [DOI] [PubMed] [Google Scholar]
- 17.Jones R S, Gelbart W M. The Drosophila Polycomb-group gene enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature (London) 1985;316:153–155. [Google Scholar]
- 19.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 20.Kennison J A. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Gen. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 21.Kennison J A, Tamkun J W. Trans-regulation of homeotic genes in Drosophila. New Biol. 1992;4:91–96. [PubMed] [Google Scholar]
- 22.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 23.Komachi K, Redd M J, Johnson A D. The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev. 1994;8:2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- 24.Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- 25.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Junuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaJeunesse D, Shearn A. E(z): a polycomb group gene or a trithorax group gene? Development. 1996;122:2189–2197. doi: 10.1242/dev.122.7.2189. [DOI] [PubMed] [Google Scholar]
- 27.Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- 28.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 29.Rastelli L, Chan C S, Pirotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reijnen M J, Hamer K M, den Blaauwen J L, Lambrechts C, Schoneveld I, van Driel R, Otte A P. Polycomb and bmi-1 homologs are expressed in overlapping patterns in Xenopus embryos and are able to interact with each other. Mech Dev. 1995;53:35–46. doi: 10.1016/0925-4773(95)00422-x. [DOI] [PubMed] [Google Scholar]
- 31.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 32.Sathe S, Harte P J. The Drosophila extra sex combs protein contains WD motifs essential for its function as repressor of homeotic genes. Mech Dev. 1995;52:77–87. doi: 10.1016/0925-4773(95)00392-e. [DOI] [PubMed] [Google Scholar]
- 33.Satijn D P E, Gunster M J, van der Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the Polycomb-group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satijn D P E, Olson D J, van der Vlag J, Hamer C M, Lambrechts A C, Masselink H, Gunster M J, Sewalt R G A B, van Driel R, Otte A P. Interference with the expression of a novel human Polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoorlemmer J, Marcos C, Were F, Martinez R, Garcia E, Satijn D P E, Otte A P, Vidal M. RING1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature (London) 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 38.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 39.Simon J, Bornemann D, Lunde K, Schwartz C. The extra sex combs product contains WD40 repeats and its time of action implies a role distinct from other Polycomb group products. Mech Dev. 1995;53:197–208. doi: 10.1016/0925-4773(95)00434-3. [DOI] [PubMed] [Google Scholar]
- 40.Struhl G, Brower D. Early role of the esc+ gene product in the determination of segments in Drosophila. Cell. 1982;31:285–292. doi: 10.1016/0092-8674(82)90428-7. [DOI] [PubMed] [Google Scholar]
- 41.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]