Abstract
The important roles played by branched polyubiquitin chains were recently uncovered in proteasomal protein degradation, mitotic regulation and NF-κB signaling. With the new realization of a wide presence of branched ubiquitin chains in mammalian cells, there is an urgent need of identifying the reader and eraser proteins of the various branched ubiquitin chains. In this work, we report the generation of non-cleavable branched triubiquitin probes with combinations of K11-, K48-, and K63-linkages. Through a pulldown approach using the branched triUb probes, we identified human proteins that recognize branched triubiquitin structures including ubiquitin-binding proteins and deubiquitinases (DUBs). Proteomics analysis of the identified proteins enriched by the branched triubiquitin probes points to possible roles of branched ubiquitin chains in cellular processes including DNA damage response, autophagy and receptor endocytosis. In-vitro characterization of several identified UIM-containing proteins demonstrated their binding to branch triubiquitin chains with moderate to high affinities. Availability of this new class of branched triubiquitin probes will enable future investigation into the roles of branched polyubiquitin chains through identification of specific reader and eraser proteins and the modes of branched ubiquitin chain recognition and processing using biochemical and biophysical methods.
Introduction
Ubiquitination, as a critical post-translational modification, signals protein degradation, DNA damage response, cell cycle progression and immune response1,2. Ubiquitin can be further modified via any of its seven lysine residues and N-terminal methionine residue to form polyubiquitin chains of different linkage, length and topology3. The ubiquitin pathway enzymes, including E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases, work in tandem to ubiquitinate a target protein with the consumption of ATP4. Deubiquitinases (DUBs) are proteases that hydrolyze the ubiquitin isopeptide bond thus antagonizing ubiquitination process5. Although the K48-linked polyubiquitin chain is a well-studied proteasomal degradation signal for unwanted cellular proteins, the cellular functions of other types of polyubiquitin chains are less well understood6.
More recently, branched ubiquitin chains were found to comprise a significant portion of total cellular ubiquitin pool and emerged as an essential polyubiquitin signal for a wide range of cellular processes7-10. Proteomics work has provided evidence of branched ubiquitin chains in human and yeast cells and the responsible ubiquitin ligases were identified11,12. An engineered viral protease, Lbpro, which cleaves ubiquitin chains at R74 and leaves a diglycine remnant on respective lysine residues of a largely intact ubiquitin, revealed that 10-20% of cellular ubiquitin participates in branched chains13. Although our knowledge of the cellular functions of branched ubiquitin chains is still very limited, earlier studies have revealed that different types of branched chains play indispensable roles in cell cycle progression14, NF-κB signaling15, and protein quality control16. Ube2s and the APC/C ubiquitin ligase complex were reported to assemble a K11 ubiquitin chain onto the K48 chain to form K11/K48 branched ubiquitin chains, which enhances the degradation of targeted proteins by proteasome14. The K48/K63 branched ubiquitin chain was found to be formed in response to interleukin-1b treatment15. It is believed that the formation of K48 branch on a K63 polyubiquitin chain prevents the DUB cleavage of the K63 chain, thus amplifying the downstream signal in the NF-κB pathway. It was also reported that misfolded proteins such as VHL is modified by a K11/K48 branched chain, which serves as a more stringent signal for cytoplasmic protein quality control as mediated by E3 ubiquitin ligases and chaperones16.
Branched ubiquitin chains are known to modulate cellular signals elicited by homotypic polyubiquitin chains7. For example, branched K11/K48-linked chains enhance proteasomal degradation and amplify the degradation signal by homotypic K48-linked polyubiquitin chains14,17. Conversely, M1/K63-linked branched chains reduce A20 deubiquitinase-catalyzed cleavage of K63-linked polyubiquitin chains in the case of inflammatory response18. Disassembly of branched ubiquitin chains by DUBs plays an important role in modulating the branched ubiquitin chain-mediated signalling process. In the proteasome, several DUBs function in conjunction with the regulatory particle proteins to remove ubiquitin chains while substrate proteins translocate to the main catalytic particle of the proteasome for degradation19. Activity of DUBs needs to be timed precisely with translocation of substrate protein into the catalytic particle of the proteasome for peptide cleavage. Recently the proteasome-associated DUB UCHL5/UCH37 was shown to process the K11/K48 branched chains through a debranching activity20,21.
More than 20 different families of ubiquitin-binding domains (UBDs) are estimated to exist in cells22 and most interaction between UBDs and ubiquitin occur through the hydrophobic patches on ubiquitin surface. Most UBDs contain an α-helix that that interact with the Ile44 patch on ubiquitin. Although the Ub-binding modes of many UBDs are similar, arrangement and number of UBDs in a reader protein have been shown to dictate UBD’s specificity for certain polyubiquitin chains. For example, ubiquitin-interacting motifs (UIMs) possess exquisite chain-linkage specificity for K63-diubiqitin when tandem UIMs (tUIMs) are separated by a linker region with a fixed length23. It is not clear whether proteins containing multiple UBDs recognize ubiquitin moieties specifically in branched polyubiquitin chains, leading to distinct signaling in a cellular context.
Previously di- and poly-ubiquitins containing non-cleavable linkages that mimic the native isopeptide linkage were prepared24-31. We reported a class of hybrid linear triubiquitin probes containing a Michael acceptor warhead as well as a non-cleavable linker, which allowed the interrogation of endo- and exo-cleavage modes of the human USP9X catalytic domain32. A similar strategy was also used to generate branched triubiquitin hybrid probes connected through a native isopeptide and a Michael acceptor-containing linkage with promise for identifying DUBs that process branched ubiquitin chains33. Several other methods were also used to generate branched ubiquitin chains through solid phase synthesis34, Lys(Boc) unnatural amino acid approach35, thiol-ene chemistry26, click chemistry36 and orthogonal sortylation37. Although DUBs are known to possess debranching activity in which a ubiquitin branch is removed to yield a linear polyubiquitin chain38, to date no DUBs have been reported capable of hydrolyzing the isopeptide at the C-terminus of the acceptor ubiquitin in the branched ubiquitin structure, which we termed ‘destemming’.
In this work, we report a new class of non-cleavable branched polyubiquitin probes. Using a semisynthetic strategy, we prepared branched triUb probes that resist cleavage of the branching isopeptide by DUBs while introducing a warhead at the C-terminus of the acceptor ubiquitin in the branched triUb. This class of probes allow not only the capturing of reader proteins that bind the branched triUb structure through noncovalent interactions, but also covalent trapping of DUBs that may not bind the triUb probe tightly enough for pulldown and identification. We generated K11/K48, K11/K63, and K48/K63 triubiquitin probes and used them in cell lysate-based pulldown and MS identification of both reader and eraser proteins from the human HEK-293T cell lysate. We identified a number of proteins that contain multiple ubiquitin-binding domains and ubiquitin-pathway enzymes particularly DUBs. Of particular interest are proteins in the DNA damage response, autophagy and receptor endocytosis pathways identified by the branched ubiquitin probes. Subsequent biochemical characterization was performed with recombinantly generated proteins to demonstrate the binding of several UIM-containing proteins toward branched ubiquitin chains.
Results
Generation and characterization of branched triUb probes.
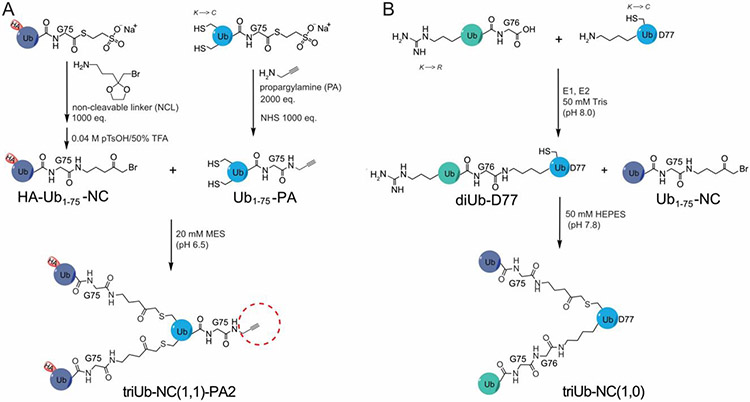
Branched triUb probes were generated using a semisynthetic approach as shown in Figure 1A. A recombinantly generated HA-tagged Ub1-75-MESNA was reacted with a linker molecule (NCL)39 and subsequently deprotected to generate the HA-Ub1-75-NC species. In parallel, a mutant Ub1-75-MESNA containing cysteines introduced at two selected lysine positions was reacted with propargylamine (PA) to generate Ub1-75-PA. Ligating HA-Ub1-75-NC with Ub1-75-PA yielded the desired branched triUb probes. The branched triUb probes (K11/K48, K11/K63, and K48/K63) were generated by using the acceptor ubiquitin containing any two of the three cysteine residues (K11C, K48C and K63C) in Ub1-75-PA (Figure 1A). The resulting branched triUb probes are named triUb-NC(1,1)-PA2 probes, abbreviated as branched triUb-PA, with (1,1) indicating branching of the ubiquitin chain. This follows a previously described nomenclature32, in which NC represents the non-cleavable linkage and the number indicates the position of the linkage with 1 indicating the position between two ubiquitins and 2 indicating the position at the C-terminus of the proximal Ub. The branched triUb-PA probes showed good purity in SDS-PAGE analysis (Figure S1A). Mass spectrometry analysis of intermediates and final triUb-PA probes showed correct molecular weight (Figure S1 B and C) and site of conjugation (Figure S2). Three diubiquitin probes, namely diUb-NC1-PA2, of different linkages (K11, K48 and K63) were also generated as previously described32.
Figure 1.
Generation of branched warhead-containing triUb probe for pulldown and branched triUb for binding assessment. A. Synthesis of triUb-NC(1,1)-PA2 probes via ligation of HA-Ub1-75-NC and Ub1-75-PA with a double-cysteine mutation introduced at requi-site lysine residues. B. Synthesis of triUb-NC(1,0) via ligation of enzymatically generated diUb-D77 with Ub1-75-NC.
Similarly, hybrid branched triUb probes without a warhead were generated by reacting Ub1-75-NC with an enzymatically generated diUb containing a cysteine introduced in the proximal ubiquitin at position 11 or 63 (Figure 1B). The resulting hybrid probe contains one non-cleavable linkage and one native isopeptide bond, thus named triUb-NC(1,0) following the above described nomenclature and abbreviated as triUb-(1,0), with 0 indicating a native bond in one of the two branches. The three branched triUbs were confirmed by SDS-PAGE and mass spectrometry (Figure S3).
We first assessed the branched triUb-PA probes in a DUB labeling assay. A successful labeling will demonstrate binding of the branched triUb-PA probes to DUBs and a normal reactivity of the warhead introduced at the C-terminus of the acceptor ubiquitin. Two OTU family DUBs, i.e. OTUD2 and OTUD3, with known linkage specificity40,41 were chosen to test labeling activity of triUb-PA probes. Recombinantly expressed and purified full-length OTUD2 (OTUD2-FL) and OTUD3 catalytic domain (OTUD3-CD52-275) were incubated with branched triUb-PA probes and the labeling was analyzed using SDS-PAGE and Coomassie blue staining (Figure S4). A comparison was made by using the corresponding diUb-PA probes and a monoUb-PA probe. OTUD2-FL showed labeling by the K11/K48- and K11/K63-triUb-PA probes but not the K48/K63-triUb-PA probe (Figure S4A). The labeling of OTUD2-FL by K11/K48-triUb-PA probe is the strongest among the three branched triUb probes. It is comparable to the K11-diUb-PA probe labeling but stronger than the K48-diUb-PA probe. A similar trend was observed for OTUD3-CD except that a weak but discernable labeling was detected for the K48/K63-triUb-PA probe (Figure S4B). These results support that the novel branched triUb-PA probes are functional in interacting with and covalently trapping DUBs, and the specific linkages in the branched triUb probes may influence the efficiency of DUB labeling. Notably, our branched triUb DUB probes allow the assessment of a new mode of DUB action in processing branched ubiquitin chains. Our results suggest that the removal of the whole branched ubiquitin chain moiety, which we termed “destemming”, is biochemically feasible. This DUB activity is distinct from the debranching activity previously known for several DUBs38.
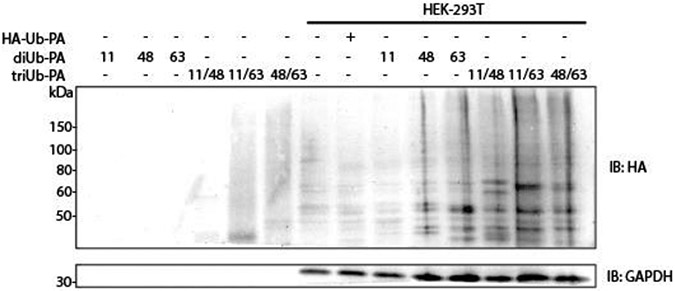
We further assessed the branched triUb-PA probes in labeling DUBs in HEK-293T cell lysate. Western blotting analyses using anti-HA antibody showed a number of discrete bands in all three triUb-PA probes (Figure 2). Despite some common bands being detected comparing the three branched triUb-PA probes, several uniquely labeled bands are discernable. A more drastic difference was observed when comparing the labeling bands by the branched triUb-PA and corresponding diUb-PA probes (Figure 2). These observations suggest that the branched tri-Ub-PA probe can effectively capture DUBs through its PA warhead in cell lysates.
Figure 2.
Western blot analysis of labeling of DUBs in HEK-293T cell lysates using HA-tagged branched triUb-PA probes in comparison to HA-tagged mono- and diUb-PA probes. The K11, K48 and K63 linkages in the branched triUb and diUb probes are denoted by the numerals.
Pulldown and proteomics analysis using branched triUb-PA probes.
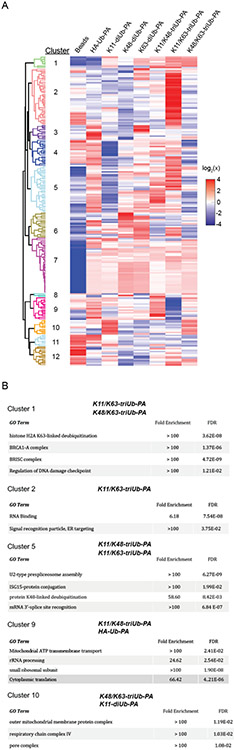
In order to identify cellular proteins that interact with the branched triUb-PA probes either through non-covalent binding or through activity-based covalent trapping, we developed a workflow for IP-MS/MS and label-free quantification (LFQ) analysis using the triUb-PA probes and HEK-293T cell lysates. Anti-HA magnetic beads were used for the pulldown and the captured proteins were subjected to trypsin digestion and subsequent nanoLC-MS/MS analysis on an Orbitrap mass spectrometer. A control pulldown was performed using empty beads in parallel. Mono- and di-Ub-PA probes were subjected to the pulldown and MS analysis as comparisons. The proteomics data were processed using MaxQuant (Figure S5) and subsequently analyzed using a multi-sample ANOVA test to group significantly enriched proteins. The results were visualized in a heatmap with hierarchical clustering of protein hits reflecting a Z-score normalization of log2(LFQ) value of a captured protein by the ubiquitin probes used in the parallel pulldown (Figures 3A and S6).
Figure 3.
Pulldown analysis of branched triUb probes in comparison to mono- and di-Ub probes. A. Analysis of HEK-293T cell lysate pulldown using triUb-NC(1,1)-PA2 probes. B. GO enrichment terms of selected clusters (includes biological pathway and cellular component).
As shown in Figure 3A, the pulled down proteins were clustered into twelve individual groups. Clusters present in branched triUb-PA probe pulldowns were further subjected to Gene Ontology (GO) analysis (Figures 3B and S6). Notably, proteins in cluster 1 bind all three branched triUb-PA probes, with stronger binding to the K11/K63- and K48/K63-triUb-PA probes as judged by the LFQ values. Interestingly, components of the BRCA1-A complex (Abraxas/F175A, BRCC36/BRCC3, BABAM2/BRE, UIMC1/Rap80 and BABA1/MERIT40) are among the cluster 1 proteins. Proteins in cluster 2 bind strongly to the K11/K63-triUb-PA probe, but not the K11/K48- and K48/K63-triUb-PA probes and the diUb-PA probes. The proteins in this cluster are found in the GO families of RNA binding and regulation of translation. Cluster 5 proteins were enriched in K11/K48 and K11/K63-triUb-PA probes. Proteins in clusters 8 and 9 were found to bind to the K11/K48-triUb-PA probe much stronger than the other two branched triUb-PA probes and the corresponding K11- and K48-diUb-PA probes. Cluster 10 contained outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM) proteins enriched in K48/K63-triUb-PA and K11-diUb-PA probe pulldowns. We also found proteins that bind more strongly to the diUb-PA probes than the branched triUb probes. For example, proteins in the cluster 6 bind K48- and K63-diUb-PA probes much stronger than any of the branched triUb-PA probes. Some pulled-down proteins, such as cluster 7, are rather promiscuous in binding to the mono-, di- and branched triUb probes.
Proteins contain different ubiquitin-binding domains or catalytic cores.
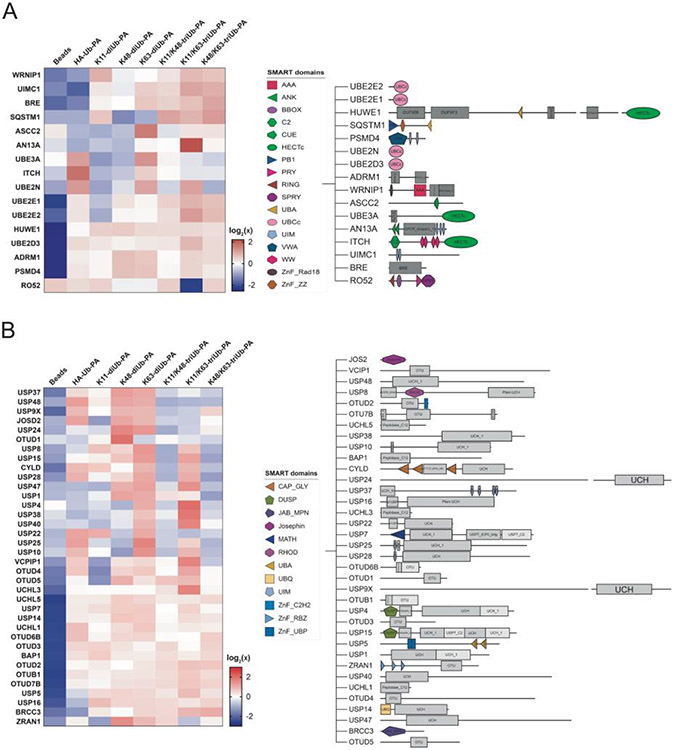
With the large number of proteins pulled down by the branched triUb-PA probes, we were particularly interested in proteins that contain ubiquitin-binding domain or catalytic core. These proteins most likely recognize and bind the ubiquitin moieties in the probes. We classified these proteins into three categories based on the nature of the ubiquitin-binding elements. First, we identified a number of proteins that contain at least one of the well characterized UBDs such as UIM and UBA (Figure 4A). Second, proteins in the ubiquitin-conjugating machineries including E2 ubiquitin-conjugating enzymes and HECT E3 ubiquitin ligases were also found among the pulled down proteins (Figure 4A). Third, we identified a number of DUBs that likely recognize the ubiquitin moiety and are trapped covalently by the PA warhead in the probes (Figure 4B). There are also many proteins among the pulled down proteins that do not contain any of the above three elements, which were likely pulled down indirectly in a protein complex, as exemplified by the several proteins in the BRCA1-A complex.
Figure 4.
Analysis of identified ubiquitin-binding proteins along with corresponding domains from SMART domain analysis. A. Enriched ubiquitin-binding proteins including ubiquitin-conjugating enzymes and ubiquitin ligases. B. Enriched DUBs from USP, UCH and OTU families.
We focused our attention to several proteins that contain multiple UIMs, which are known to recognize and bind mono and di-Ub structures. We reasoned that the presence of multiple UIMs in the same protein may provide an opportunity to identify proteins capable of binding to the more complex branched ubiquitin chain structures. Three proteins caught our attention, UIMC1/Rap80, AN13A and USP37. These proteins contain 2, 4, and 3 UIMs in their sequence respectively (Figure S7). Our LFQ analysis showed that UIMC1 was pulled down most efficiently by the K11/K63-triUb-PA probe followed by the K48/K63-triUb-PA and K63-diUb-PA probes. Notably, AN13A was pulled down most strongly by the K11/K63-triUb-PA probe, compared to other branched triUb-PA and corresponding di-Ub-PA probes. Because neither UIMC1 nor AN13A possess ubiquitin conjugating and deconjugating activities, it is possible that the UIM domains in their sequences contributed to the binding to the branched Ub probes.
We also noted that sequestosome-1 (SQSTM1 or p62) was among the pulled down proteins. SQSTM1 was pulled down more strongly by the branched triUb-PA probes and the K11-diUb-PA probes. Although SQSTM1 only contains a single UBA at its C-terminal region42, it is known to form an oligomeric structure43, which may contribute to the overall preference for the branched triUb structure. Werner helicase-interacting protein 1 (WRNIP1/WRIP1) share a similar profile with a preference in binding to the branched triUb-PA probes. Interesting, WRNIP1 also contains a single ubiquitin-binding zinc finger domain (UBZ) and likely form an oligomeric structure44,45.
Biochemical characterization of binding of UIM-containing proteins to branched triUbs.
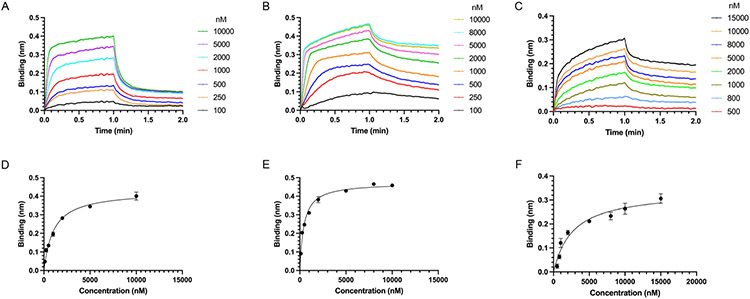
We expressed and purified UIMC176-128 and AN13A479-594 that contain the UIM domains. To assess their binding to the various branched triUb structures, Bio-layer Interferometry (BLI) was used to compare the three branched triUbs as well as mono- and di-Ubs of different linkages. We also expressed and purified the UIM-containing segment of USP37700-851 given the observation that it contains three UIMs in its C-terminal region despite a weaker pulldown by the branched triUb-PA probes. For BLI measurement, the UIM-containing recombinant proteins were immobilized onto the biosensor through an anti-His-tag antibody with the various Ub species as analytes. The measurement was done in duplicates. We first compared the binding of the K63-diUb containing a native linkage versus the chemically linked K63-diUb (K63 diUb-NC1) to UIMC176-128. Our data showed a similar binding affinity indicating that the thioether linkage introduced between the ubiquitin moieties has no adverse effect on their binding to UIMs (Figure S8).
Next, we determined the Kd of UIMC176-128, AN13A479-594 and USP37700-851 binding to the K11/K48-, K11/K63- and K63/K48-triUb-(1,0) in comparison to the K11-, K48- and K63-diUbs respectively. UIMC176-128 showed the highest binding affinity for the K11/63-triUb-(1,0) with a Kd of 1.04 μM, followed by K63-diUb (Kd of 2.17 μM), K11-diUb (Kd of 2.92 μM) and K63/48-triUb (Kd of 2.43 μM) (Table 1; Figures 5 and S9). No significant binding was observed for K11/48-triUb-(1,0) in a single analyte concentration binding assessment at 2 μM. AN13A479-594 showed preference for K11/63-triUb(1,0) and K63 diUb with a Kd of 0.43 μM and 0.25 μM respectively, followed by K63/K48-triUb-(1,0) and K11-diUb (Kd of 0.74 μM and 1.28 μM respectively) and K11/48-triUb-(1,0) (Kd of 7.83 μM) (Table 1; Figures 5 and S10). USP37700-851 binds branched triUb with a Kd of 2.71 μM for K63/K48-triUb-(1,0) and 3.36 μM for K11/K63-triUb-(1,0) in comparison to a Kd of 4.97 μM and 8.89 μM for K63-diUb and K11-diUb respectively (Table 1; Figures 5 and S11). No significant binding to USP37700-851 was observed for K11/48-triUb-(1,0) in a single analyte concentration binding assessment at 2 μM. For all three UIM-containing proteins, no significant binding was observed for K48-diUb in a single analyte concentration binding assessment at 2 μM.
Table 1.
Binding constant of UIMC1, AN13A and USP37 UIMs interacting with diUb and branched triUb determined by BLI.
| Probe | UIMC176-128 Kd (μM) |
AN13A479-594 Kd (μM) |
USP37700-851 Kd (μM) |
|---|---|---|---|
| K11-diUb | 2.92 ± 0.46 | 1.28 ± 0.21 | 8.89 ± 0.83 |
| K63-diUb | 2.17 ± 0.23 | 0.25 ± 0.03 | 4.97 ± 1.07 |
| K11/K48-triUb-(1,0) | --- | 7.83 ± 1.89 | --- |
| K63/K48-triUb-(1,0) | 2.43 ± 0.62 | 0.74 ± 0.69 | 2.71 ± 0.52 |
| K11/K63-triUb-(1,0) | 1.04 ± 0.10 | 0.43 ± 0.03 | 3.36 ± 0.89 |
--- indicates no significant binding observed.
Figure 5.
BLI traces of branched triUb binding to UIM domains of UIMC1, AN13A and USP37 immobilized onto the BLI sensor tip surface. BLI traces averaged from two repeats are shown for concentration series of (A) UIMC176-128 binding K11/K63 triUb(1,0); (B) AN13A479-594 binding K11/K63 triUb(1,0); (C) USP37700-851 binding K63/K48 triUb(1,0). The Kd of binding was determined by fitting the maximum BLI binding observed versus the concentration of triUb species used in the BLI binding experiment for (D) UIMC176-128, (E) AN13A479-594 and (F) USP37700-851.
Deubiquitinases and ubiquitination pathway enzymes enriched in branched triUb-PA probe pulldowns.
We observed a number of DUBs in the pulldown and most DUBs except BRCC3 were only identified in pulldown using Ub probes but not in the control pulldown (Figures 4B and S12). Spiderplot analysis of captured DUBs showed that several DUBs, USP4, USP38, USP40, OTUD4, OTUD5, VCPIP1 and UCHL3, were captured more efficiently by branched K11/K63 triUb-PA probe than the corresponding K11-diUb and K63-diUb probes and the monoUb probe (Figure S12). No DUBs were determined to be particularly specific for K11/K48-triUb-PA or K48/K63-triUb-PA. Although numerous DUBs were enriched by triUb-PA probes, most also showed comparable enrichment with diUb-PA probes. We cannot rule out that some pulled-down DUBs may noncovalently bind to the triUb probes given that a mild wash condition was used in the pulldown. Notably, several ubiquitin conjugating enzymes, particularly UBE2N, UBE2E2 and UBE2D3, were identified to be enriched by K11/K63-triUb-PA probes (Figure 4A). In addition, a HECT E3 ligase HUWE1 was also found enriched in diUb and triUb-PA probe pulldowns (Figure 4A).
Discussion
Prior proteomics work using cleavage-resistant, triazole-linked diubiquitin probes without an activity-based warhead was successful in identifying DUBs and ubiquitin-binding proteins specific to individual ubiquitin chain linkages30,46. Several DUBs were pulled down by diUb probes of specific linkage types and a number of Ub-binding proteins known to bind to K6, K48 and K63 linkages were also identified. Although previous studies have shown the presence of branched ubiquitin chains in cells and demonstrated their specific roles in cellular processes including proteasomal protein degradation, mitotic regulation and NF-κB signaling11,15,47, our understanding of how reader and eraser proteins recognize and process branched polyubiquitin chains of different linkages and topologies is still very limited. In this study we prepared triUb probes that resist DUB cleavage and allow identification of ubiquitin-binding proteins that potentially recognize branched triUb chains (K11/K63, K11/K48, and K48/K63). The presence of a terminal alkyne warhead in the branched triUb probes allows for covalent trapping of DUBs capable of processing the branched polyUb chain through “destemming”, in which the branched polyUb chain is removed en bloc by DUBs.
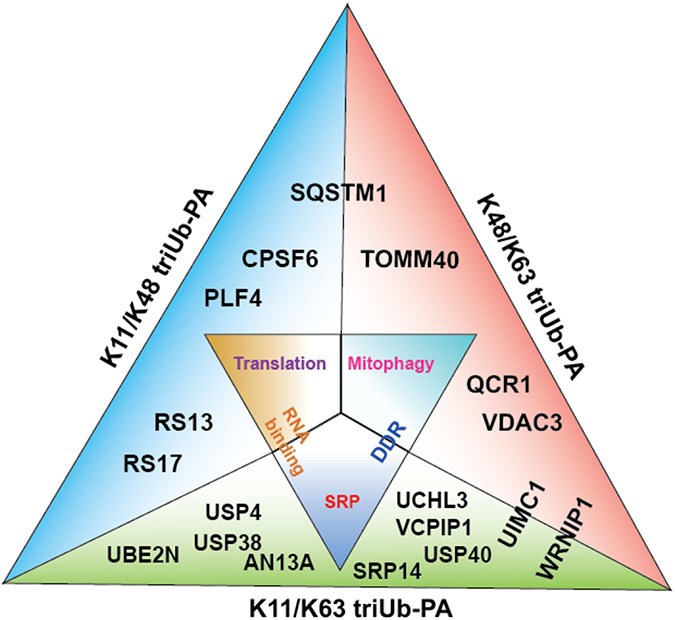
In a cell lysate-based pulldown experiment using the branched triUb probes of different linkages, we captured various ubiquitin-binding proteins, DUBs and ubiquitin conjugating enzymes. Notably, a cluster of proteins pulled down by the K11/K63 and K48/K63 branched triUb probes are involved in DNA damage response (Figure 6). Particularly, proteins from the BRCA1-A complex are among the proteins captured by the K11/K63-triUb probes. Quantitative analysis of the pulled-down proteins from cell lysate using MaxQuant LFQ revealed stronger pulldown of these proteins by the branched triUb probes than the corresponding diUb probes. This raised an interesting possibility of the involvement of branched ubiquitin chains in cellular DNA damage response. To date there has been no direct evidence of branched ubiquitin chains functioning in DNA repair or damage response. Nonetheless, it was suggested that mixed K11/K63-Ub chains may exist in DNA double-strand break repair and the DUB activity of Cezanne counteracts its formation48. Our finding points to a potential role of the branched polyUb chain in DNA damage response that requires future investigation.
Figure 6.
Illustration of representative proteins identified by each of the three triUb-PA probes and the associated biological pathways. Each side of the triangle reflects enrichment by each of the triUb probes. Inner triangle illustrates enriched protein complexes and associated biological pathways. Outer triangle contains selected proteins enriched by each of the probe. SRP and DDR refer to signal recognition particle and DNA damage response respectively.
The role of branched ubiquitin chains has been found to more efficiently target substrate for proteasomal degradation compared to homotypic polyUb chains14,47. The proteasome function is fine-tuned by the opposing activities of ubiquitin ligases and DUBs. The DUBs act to limit substrate’s dwell time and activate substrate entry into the 20S core particle. Notably proteasome-associated DUBs, USP14 and UCHL5, were enriched by branched probes in our pulldown-mass spectrometry experiments (Figure 4B). Recent studies have shown that UCHL5 in association with RPN13 plays a significant role in debranching K48 chains from K11/K48 branched chains and promoting substrate clearance through the proteasome20,21. The rate of UCHL5 cleavage of K48 branchpoints was shown to be equivalent to the commitment step in proteasomal degradation and introducing a UCHL5 active site mutant slowed down the degradation process20,21. We reason that the cleavage of branched polyubiquitin chains en bloc may provide an efficient way of removing branched polyubiquitin chains from the target protein. DUBs capable of destemming branched ubiquitin chains can quickly remove ubiquitin chains from substrate proteins in order to recycle ubiquitin and shuttle substrate into the core particle in the 20S proteasome49.
Compared to proteasome-mediated protein degradation, cellular degradation through the autophagy lysosomal pathway (ALP) and the roles of linear and branched ubiquitin chains in this process are less well understood. Recently, K29/K48-branched ubiquitination of Class III PI3-kinase complex (VPS34) was shown to modulate autophagosome formation and maturation50. Interestingly, in our study, the branched triUb probes pulled down autophagosome receptor SQSTM1/p62 and OMM proteins more efficiently than the corresponding diUb probes (Figure 6). SQSTM1/p62 is an autophagy receptor and has been shown to bind and aggregate on K63-linked polyubiquitin chain through its UBA domain51. SQSTM1 was preferentially pulled down by K48/K63-triUb and K11/K48-triUb probes as revealed by quantitative LFQ analysis (Figure 4A). Although the involvement of branched ubiquitin chains in mitophagy, a specific type of selective autophagy, has been suggested52, direct evidence is still lacking. OMM proteins are known to be polyubiquitinated and targeted to phagophore via ubiquitin binding receptor proteins like SQSTM1 in response to mitochondrial depolarization in a PINK1/Parkin-dependent ubiquitination cascade53. Our findings suggest the possible participation of branched ubiquitin chains in mitophagy through the involvement of SQSTM1 recognizing branched polyubiquitin chains and facilitating the degradation of mitochondria by autophagy lysosomal process.
Of the ubiquitin binding proteins enriched in branched triUb probe pulldowns, three of the tUIM-containing proteins, UIMC1, AN13A and USP37, were found with varying affinities for branched ubiquitin chains. BLI results indicate that the UIMC176-128 binds K11/K63-triUb with two-fold higher affinity than the K63- and K11-diUb. UIMC1 contains two UIMs in an extended α-helix, thus it is likely that the binding of UIMC1 to K11/K63-triUb and K63-diUb share a common binding mode. The slight higher affinity for K11/K63-triUb could be due to the UIMC1 tUIM’s ability to bind K11-diUb. Alternatively, the addition of K11 ubiquitin branch stabilizes the K63-diUb conformation that favors its binding by UIMC1’s tUIM. Given the known role of UIMC1/Rap80 in binding polyubiquitin chains at DNA damage sites to recruit BRCA1 for DNA damage repair response54, our results suggest branched polyubiquitin chains may enhance the binding of UIMC1/Rap80 to the polyubiquitin structure in DNA damage response.
Notably, ankyrin repeat domain-containing protein 13A (AN13A) showed a preference for the K11/K63 branched triUb over the other two types of branched triUbs and over the corresponding K11- and K63-diUbs in our cell lysate pulldown LFQ analysis. Relatively little is known about AN13A’s function, except one report of AN13A promoting ubiquitination-mediated EGF receptor endocytosis, possibly through K63-linked polyubiquitination55. There was no report of AN13A binding to branched ubiquitin chains. Strikingly, AN13A contains four UIMs, which are located at its C-terminal region (a.a. 483-590). Although the information of branched Ub chains’ role in endocytosis of cell surface receptors is very limited, mass spectrometry analysis, combined with Ub Lys mutant analysis, suggested the existence of K11/K63 branched ubiquitin chains on the ubiquitinated MHC I complex in endocytosis and an important role of this chain type in MHC I receptor internalization56. When testing the binding of the UIM-containing region of AN13A (AN13A479-594) to the branched triUb and diUb, we observed more promiscuous binding to the various Ub structures. This may reflect the difference of the relative positioning and orientation of UIMs in isolated UIM-containing sequence from that in the full-length protein. As a result, a truncated protein sequence containing UIMs may not fully recapitulate the ubiquitin chain-binding preference of the full-length protein. Further investigation will be needed to understand the ubiquitin structure preference of the full-length AN13A in receptor endocytosis. Availability of the branched polyubiquitin probes will facilitate future investigation into the roles of the poorly understood branched ubiquitin chains through identification of specific reader and eraser proteins and characterization of their mode of recognition and processing of branched ubiquitin chains using biochemical and biophysical methods.
Conclusions
Using a semisynthetic approach, we generated a new class of branched triubiquitin probes with a C-terminal alkyne warhead installed on the acceptor ubiquitin. The thioether linkage in the branched ubiquitin chain mimics the native isopeptide linkage as judged by comparable binding affinity to a known UIM-containing protein UIMC1/Rap80. Pulldown using these branched triubiquitin probes coupled with quantitative mass spectrometry analysis identified proteins and complexes in several biological pathways, including DNA damage response, autophagy and receptor endocytosis. BLI binding experiment using recombinant UIM domain-containing segment of identified ubiquitin reader proteins confirmed binding to branched triubiquitin chains with moderate to high affinities. The availability of this new class of branched triUb probes will enable future studies into the ubiquitin-binding domains and catalytic cores potentially responsible for branched ubiquitin chain recognition and the functional significance of branched ubiquitin chains in biological processes.
Methods
Generation of triUb-NC(1,1)-PA2
Generation of branched triUb probes followed a similar method to prepare diUb-NC1-PA2 probes (see Supplemental Methods) with a different stoichiometry of 2 eq. of HA-Ub1-75-NC incubated with 1 eq. of K11C/K48C-Ub1-75-PA, K11C/K63C-Ub1-75-PA, or K48C/K63C-Ub1-75-PA. The reaction mixture was incubated with 1 mM TCEP and purified in a HiTrap SP FF column to remove excess monoubiquitin. The di-and tri-ubiquitin fractions were combined and purified using a Superdex 75 10/300 size exclusion column.
Generation of triUb-NC(1,0)
Enzymatically generated diubiquitin following a reported method57 (see Supplementary Method) containing a free Cys at the desired branch point was reacted with 3 molar equivalents of HA-Ub1-75-NC species at room temperature for 16 hrs in a ligation buffer containing 50 mM HEPES (pH 7.8), 100 mM NaCl, 1 mM EDTA to generate hybrid triubiquitin branched chains. The reaction mixture was purified using a Superdex 75 10/300 column in 50 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM DTT.
Pulldown using HEK-293T cell lysates
Design of pulldown experiment was based on prior literature reports using non-cleavable diubiquitin probes30,46. For each pulldown, 5 μg HA-Ub-PA, 9.4 μg diUb-NC1-PA2 or 14.4 μg triUb-NC(1,1)-PA2 probes were incubated with 2 mg HEK-293T cell lysate for 2 hrs at room temperature. The probe-treated lysate was either processed immediately or stored in −80 °C with 1X protease inhibitor cocktail from Millipore Sigma. The probe-treated lysate was diluted 10-fold with a buffer containing 50mM Tris-HCl, pH 7.5, 150mM NaCl, 5mM EDTA supplemented with 0.1% Tween-20 and protease inhibitor cocktail. Then, 100 μL of anti-HA magnetic beads were added to the solution and rotated at 4 °C overnight. Unbound proteins were aspirated and anti-HA beads were washed three times with ice-cold phosphate-buffered saline (PBS) buffer (pH 7.4) with 0.05% Tween-20 before a one-time wash with ice-cold PBS buffer. Bead-bound samples were incubated with 100 μL of 50 mM NaOH solution for 10 min at 37 °C and eluted, followed by two additional 50 μL washes. The elution and wash solutions were combined.
Proteomics using nanoLC-MS/MS Orbitrap
The eluate was adjusted to pH 8.0 using a 1M Tris (pH 8.0) solution and reduced with 10 mM DTT at 60 °C for 1 hr. The sample was then subsequently alkylated with 40 mM iodoacetamide at room temperature for 30 min (protected from light). The alkylation reaction was quenched by addition of 10 mM DTT prior to digestion with 1:50 mass ratio of trypsin protease to total protein at 37 °C for 18 hrs. Tryptic digestion was quenched and acidified by 0.1% trifluoroacetic acid. Digested peptide samples were then desalted using Thermo Fisher 100 μL precast C18 resin Zip-tip and lyophilized prior to liquid chromatography-mass spectrometry (LC-MS/MS) analysis. The lyophilized tryptic digests were dissolved in 0.1% formic acid in diH2O prior to LC-MS/MS (15 cm x 75 μm) reverse phase nano-LC (Thermo Fisher Scientific, Waltham, MA) analysis using Orbitrap Q-Exactive (positive polarity mode, collision induced dissociation) (Thermo Fisher Scientific, Waltham, MA) with a nano-electrospray ion source. A linear gradient from 5% to 60% acetonitrile in 0.1% formic acid water solution was used to separate peptides for 150 min at a constant flow of 200 nL/min. The system was set to operate in a data-dependent mode with MS/MS scan of the six most abundant peaks from a full MS scan. Full scans were acquired between 300 to 1800 m/z with a resolution of 60,000.
Biolayer Interferometry binding assay
Anti-Penta-His sensors were utilized on a ForteBio BLItz system. The baseline was established for 30 s using an assay buffer containing 50 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM DTT, 0.25% Tween20, and 0.16 mg/mL bovine serum albumin (BSA). The ligand, 5 μM His-tagged UIM- containing proteins, was loaded to the sensor for 60 seconds, then fresh buffer was used for 30 seconds to remove excess ligand that did not bind. The initial baseline for the binding measurement was performed with fresh buffer for 60 seconds, then the ubiquitin species as analyte on the drop holder was used for the association step for 60 seconds (120 s was used for slow binding analytes). The dissociation step with fresh buffer, proceeded for 60 seconds before the regeneration procedure, which consisted of three iterations of 5 second incubation with 100 mM glycine (pH 3.0), then 5 second incubation with fresh assay buffer.
Using the ForteBio software, sensorgram was subtracted with a blank run obtained using the assay buffer and step-corrected. The BLI binding response at the end of the association phase (mean ± SD) was plotted against the analyte concentration. Kd was calculated using the binding-saturation/one-site binding model on GraphPad Prism 7.
Supplementary Material
ACKNOWLEDGMENT
This work is supported in part by National Institutes of Health (NIH) R01 GM129468 to Z.Z. and National Institute of General Medical Sciences P30 GM110758 and P20 GM104316.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
Purification of various ubiquitin species and DUBs; generation of diUb species; cell culturing and pulldown experiments; BLI analysis of triUb binding; LFQ proteomics analysis.
REFERENCES
- (1).Yau R; Rape M The increasing complexity of the ubiquitin code. Nature cell biology. 2016, 18, 579–86. [DOI] [PubMed] [Google Scholar]
- (2).Ciechanover A; Orian A; Schwartz AL The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J. Cell Biochem. Suppl 2000, 34, 40–51. [DOI] [PubMed] [Google Scholar]
- (3).Komander D; Rape M The ubiquitin code. Annu. Rev. Biochem 2012, 81, 203–29. [DOI] [PubMed] [Google Scholar]
- (4).Pickart CM Mechanisms underlying ubiquitination. Annu. Rev. Biochem 2001, 70, 503–33. [DOI] [PubMed] [Google Scholar]
- (5).Reyes-Turcu FE; Ventii KH; Wilkinson KD Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem 2009, 78, 363–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Swatek KN; Komander D Ubiquitin modifications. Cell Res. 2016, 26, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Haakonsen DL; Rape M Branching Out: Improved Signaling by Heterotypic Ubiquitin Chains. Trends Cell Biol. 2019, 29, 704–716. [DOI] [PubMed] [Google Scholar]
- (8).French ME; Koehler CF; Hunter T Emerging functions of branched ubiquitin chains. Cell Discov. 2021, 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Akutsu M; Dikic I; Bremm A Ubiquitin chain diversity at a glance. J. Cell Sci 2016, 129, 875–80. [DOI] [PubMed] [Google Scholar]
- (10).Ohtake F; Tsuchiya H The emerging complexity of ubiquitin architecture. J. Biochem 2017, 161, 125–133. [DOI] [PubMed] [Google Scholar]
- (11).Yau RG; Doerner K; Castellanos ER; Haakonsen DL; Werner A; Wang N; Yang XW; Martinez-Martin N; Matsumoto ML; Dixit VM; Rape M Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Valkevich EM; Sanchez NA; Ge Y; Strieter ER Middle-down mass spectrometry enables characterization of branched ubiquitin chains. Biochemistry 2014, 53, 4979–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Swatek KN; Usher JL; Kueck AF; Gladkova C; Mevissen TET; Pruneda JN; Skern T; Komander D Insights into ubiquitin chain architecture using Ub-clipping. Nature 2019, 572, 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Meyer HJ; Rape M Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ohtake F; Saeki Y; Ishido S; Kanno J; Tanaka K The K48-K63 Branched Ubiquitin Chain Regulates NF-kappaB Signaling. Mol Cell 2016, 64, 251–266. [DOI] [PubMed] [Google Scholar]
- (16).Samant RS; Livingston CM; Sontag EM; Frydman J Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 2018, 563, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Oh E; Mark KG; Mocciaro A; Watson ER; Prabu JR; Cha DD; Kampmann M; Gamarra N; Zhou CY; Rape M Gene expression and cell identity controlled by anaphase-promoting complex. Nature 2020, 579, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wertz IE; Newton K; Seshasayee D; Kusam S; Lam C; Zhang J; Popovych N; Helgason E; Schoeffler A; Jeet S; Ramamoorthi N; Kategaya L; Newman RJ; Horikawa K; Dugger D; Sandoval W; Mukund S; Zindal A; Martin F; Quan C; Tom J; Fairbrother WJ; Townsend M; Warming S; DeVoss J; Liu J; Dueber E; Caplazi P; Lee WP; Goodnow CC; Balazs M; Yu K; Kolumam G; Dixit VM Phosphorylation and Linear Ubiquitin Direct A20 Inhibition of Inflammation. Nature 2015, 528, 370–375. [DOI] [PubMed] [Google Scholar]
- (19).Finley D; Chen X; Walters KJ Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci 2016, 41, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Deol KK; Crowe SO; Du J; Bisbee HA; Guenette RG; Strieter ER Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation. Mol. Cell 2020, 80, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Song A; Hazlett Z; Abeykoon D; Dortch J; Dillon A; Curtiss J; Martinez SB; Hill CP; Yu C; Huang L; Fushman D; Cohen RE; Yao T Branched ubiquitin chain binding and deubiquitination by UCH37 facilitate proteasome clearance of stress-induced inclusions. eLife 2021, 10, e72798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dikic I; Wakatsuki S; Walters KJ Ubiquitin-binding domains — from structures to functions. Nat. Rev. Mol. Cell Bio 2009, 10, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sims JJ; Cohen RE Linkage-specific avidity defines the lysine 63-linked polyubiquitin binding preference of Rap80. Mol. Cell 2009, 33, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yin L; Krantz B; Russell NS; Deshpande S; Wilkinson KD Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochem. 2000, 39, 10001–10010. [DOI] [PubMed] [Google Scholar]
- (25).Eger S; Scheffner M; Marx A; Rubini M Synthesis of Defined Ubiquitin Dimer. J. Am. Chem. Soc 2010, 132, 16337–16339. [DOI] [PubMed] [Google Scholar]
- (26).Valkevich EM; Guenette RG; Sanchez NA; Chen YC; Ge Y; Strieter ER Forging isopeptide bonds using thiol-ene chemistry: site-specific coupling of ubiquitin molecules for studying the activity of isopeptidases. J. Am. Chem. Soc 2012, 134, 6916–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hemantha HP; Bavikar SN; Herman-Bachinsky Y; Haj-Yahya N; Bondalapati S; Ciechanover A; Brik A Nonenzymatic Polyubiquitination of Expressed Proteins. J. Am. Chem. Soc 2014, 136, 2665–2673. [DOI] [PubMed] [Google Scholar]
- (28).Lewis YE; Abeywardana T; Lin YH; Galesic A; Pratt M Synthesis of a Bis-thio-acetone (BTA) Analogue of the Lysine Isopeptide Bond and its Application to Investigate the Effects of Ubiquitination and SUMOylation on α-Synuclein Aggregation and Toxicity. R. ACS Chem. Biol 2016, 11, 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Singh SK; Sahu I; Mali SM; Hemantha HP; Kleifeld O; Glickman MH; Brik A Synthetic Uncleavable Ubiquitinated Proteins Dissect Proteasome Deubiquitination and Degradation, and Highlight Distinctive Fate of Tetraubiquitin. J. Am. Chem. Soc 2016, 138, 16004–16015. [DOI] [PubMed] [Google Scholar]
- (30).Zhang X; Smits AH; van Tilburg GB; Jansen PW; Makowski MM; Ovaa H; Vermeulen M An Interaction Landscape of Ubiquitin Signaling. Mol. Cell 2017, 65, 941–955. [DOI] [PubMed] [Google Scholar]
- (31).Zheng Q, Wang T, Chu GC, Zuo C, Zhao R, Sui X, Ye L, Yu Y, Chen J, Wu X, Zhang W, Deng H, Shi J, Pan M, Li YM and Liu L. An E1-Catalyzed Chemoenzymatic Strategy to Isopeptide-N-Ethylated Deubiquitylase-Resistant Ubiquitin Probes. Angew. Chem., Int. Ed 2020, 59, 13496–13501. [DOI] [PubMed] [Google Scholar]
- (32).Paudel P; Zhang Q; Leung C; Greenberg HC; Guo Y; Chern YH; Dong A; Li Y; Vedadi M; Zhuang Z; Tong Y Crystal Structure and Activity-Based Labeling Reveal the Mechanisms for Linkage-Specific Substrate Recognition by Deubiquitinase USP9X. P. Natl. Acad. Sci. USA 2019, 116, 7288–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Liu J; Li Y; Deol K; Strieter E Synthesis of Branched Triubiquitin Active-Site Directed Probes. Org. Lett 2019, 21, 6790–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tang S; Liang LJ; Si YY; Gao S; Wang JX; Liang J; Mei Z; Zheng JS; Liu L Practical Chemical Synthesis of Atypical Ubiquitin Chains by Using an Isopeptide-Linked Ub Isomer. Angew. Chem. Int. Edit 2017, 56, 13333–13337. [DOI] [PubMed] [Google Scholar]
- (35).Dixon EK; Castañeda CA; Kashyap T; Wang Y; Fushman D Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies. Bioorg. Med. Chem 2013, 21, 3421–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zhang X; Scheffner M; Marx A Assembly of branched ubiquitin oligomers by click chemistry. Chem. Commun 2019, 55, 13903–13905. [DOI] [PubMed] [Google Scholar]
- (37).Fottner M; Weyh M; Gaussmann S; Schwarz D; Sattler M; Lang K A modular toolbox to generate complex polymeric ubiquitin architectures using orthogonal sortase enzymes. Nat. Commun 2021, 12, 6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Gopinath P; Ohayon S; Nawatha M; Brik A Chemical and semisynthetic approaches to study and target deubiquitinases. Chem. Soc. Rev 2016, 45, 4171–98. [DOI] [PubMed] [Google Scholar]
- (39).Yang K; Li G; Gong P; Gui W; Yuan L; Zhuang Z Chemical Protein Ubiquitylation with Preservation of the Native Cysteine Residues. Chembiochem 2016, 17, 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mevissen TE; Hospenthal MK; Geurink PP; Elliott PR; Akutsu M; Arnaudo N; Ekkebus R; Kulathu Y; Wauer T; El Oualid F; Freund SM; Ovaa H; Komander D OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013, 154, 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Flierman D; van der Heden van Noort GJ; Ekkebus R; Geurink PP; Mevissen TE; Hospenthal MK; Komander D; Ovaa H Non-hydrolyzable Diubiquitin Probes Reveal Linkage-Specific Reactivity of Deubiquitylating Enzymes Mediated by S2 Pockets. Cell Chem. Biol 2016, 23, 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bjørkøy G; Lamark T; Brech A; Outzen H; Perander M; Overvatn A; Stenmark H; Johansen T p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol 2005, 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wurzer B; Zaffagnini G; Fracchiolla D; Turco E; Abert C; Romanov J; Martens S Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife 2015, 4, e08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tsurimoto T; Shinozaki A; Yano M; Seki M; Enomoto T Human Werner helicase interacting protein 1 (WRNIP1) functions as a novel modulator for DNA polymerase delta. Genes Cells 2005, 10, 13–22. [DOI] [PubMed] [Google Scholar]
- (45).Crosetto N; Bienko M; Hibbert RG; Perica T; Ambrogio C; Kensche T; Hofmann K; Sixma TK; Dikic I Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J. Biol. Chem 2008, 283, 35173–35185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang X; Smits AH; van Tilburg G B; Ovaa H; Huber W; Vermeulen M Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc 2018, 13, 530–550. [DOI] [PubMed] [Google Scholar]
- (47).Liu C; Liu W; Ye Y; Li W Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun 2017, 8, 14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wu X; Liu S; Sagum C; Chen J; Singh R; Chaturvedi A; Horton JR; Kashyap TR; Fushman D; Cheng X; Bedford MT; Wang B Crosstalk between Lys63- and Lys11-polyubiquitin signaling at DNA damage sites is driven by Cezanne. Genes Dev. 2019, 33, 1702–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Collins GA; Goldberg AL The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chen YH; Huang TY; Lin YT; Lin SY; Li WH; Hsiao HJ; Yan RL; Tang HW; Shen ZQ; Chen GC; Wu KP; Tsai TF; Chen RH VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. Nat. Commun 2021, 12, 1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Seibenhener ML; Babu JR; Geetha T; Wong HC; Krishna NR; Wooten MW Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell Biol 2004, 24, 8055–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Deol KK; Eyles SJ; Strieter ER Quantitative Middle-Down MS Analysis of Parkin-Mediated Ubiquitin Chain Assembly. J. Am. Soc Mass Spectr 2020, 31, 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Harper JW; Ordureau A; Heo J-M Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Bio 2018, 19, 93–108. [DOI] [PubMed] [Google Scholar]
- (54).Kim H; Chen J; Yu X Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007, 316, 1202–1205. [DOI] [PubMed] [Google Scholar]
- (55).Tanno H; Yamaguchi T; Goto E; Ishido S; Komada M The Ankrd 13 family of UIM-bearing proteins regulates EGF receptor endocytosis from the plasma membrane. Mol. Biol. Cell 2012, 23, 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Boname JM; Thomas M; Stagg HR; Xu P; Peng J; Lehner PJ Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic 2010, 11, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Raasi S; Pickart CM Ubiquitin chain synthesis. Methods Mol. Biol 2005, 301, 47–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.