Abstract
p300 and the closely related CREB binding protein (CBP) are transcriptional adaptors that are present in intracellular complexes with TATA binding protein (TBP) and bind to upstream activators including p53 and nuclear hormone receptors. They have intrinsic and associated histone acetyltransferase activity, suggesting that chromatin modification is an essential part of their role in regulating transcription. Detailed characterization of a panel of antibodies raised against p300/CBP has revealed the existence of a 270-kDa cellular protein, p270, distinct from p300 and CBP but sharing at least two independent epitopes with p300. The subset of p300/CBP-derived antibodies that cross-reacts with p270 consistently coprecipitates a series a cellular proteins with relative molecular masses ranging from 44 to 190 kDa. Purification and analysis of various proteins in this group reveals that they are components of the human SWI/SNF complex and that p270 is an integral member of this complex.
The cellular protein p300 is a direct target of the transforming functions of the adenovirus E1A gene (19, 51) and as such is implicated in the regulation of both cell cycle-specific and tissue-specific gene expression (18, 27, 38, 43, 46, 52, 54). p300 is highly homologous (about 64% identical) to the cyclic AMP response element binding protein (CREB) coactivator, CBP (CREB binding protein) (5, 11, 17, 30, 34). Both p300 and CBP are present in intracellular complexes with the TATA binding protein (TBP) (1, 13). Both act as cofactors for p53 (6, 21, 33, 44) and nuclear hormone receptors (9, 24, 28). Both also contain intrinsic and associated histone acetyltransferase (HAT) activity (39, 53), suggesting that chromatin modification is an essential part of their role in regulating transcription.
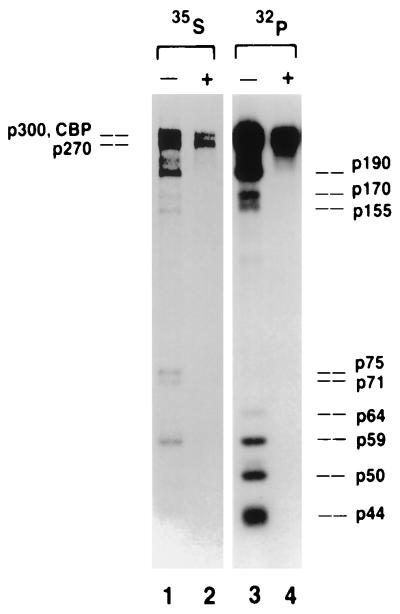
A recent detailed characterization of a panel of antibodies raised against a mixture of native p300 and CBP revealed the existence of a 270-kDa cellular protein, distinct from p300 and CBP but sharing at least two independent antigenic determinants with p300 (13). Four of the eleven antibodies in the panel recognize p270. The subset of p300/CBP-derived antibodies that recognizes p270 consistently coprecipitates a series of cellular proteins with relative molecular masses ranging from 44 to 190 kDa. Typical of these is the antibody designated NM1, whose immunoprecipitation pattern is shown in Fig. 1. TBP-specific antibodies coprecipitate a subset of these proteins including p300, CBP, and the phosphoprotein species indicated in Fig. 1 as p64 and p59 (1, 13). Because the TBP-specific antibodies do not coprecipitate all of the p300 family-associated proteins, it is likely that the array of proteins seen in Fig. 1 represents more than one intracellular complex.
FIG. 1.
Immune complexes precipitated by a p300/CBP/p270-reactive antibody. 35S- or 32P-labeled 293 cell lysates were immunoprecipitated with monoclonal antibody NM1 under nondenaturing conditions (−). To distinguish associated from cross-reactive species, half of the complex was denatured (+) by boiling in SDS and reprecipitated with fresh NM1 in conditions where only proteins directly recognized by the antibodies are recovered. The proteins from each immunoprecipitation were separated by electrophoresis and visualized by autoradiography. The positions of the associated protein species are shown on the right; the positions of p300, CBP, and p270 are shown on the left. The 35S panel is lightly exposed to enhance the resolution of p300, CBP, and p270. Only the bands corresponding to p300, CBP, and p270 are recovered by the antibody after the complex has been denatured.
We have now identified four of the remaining p300/CBP/p270-associated proteins as members of another important cellular complex: the mammalian SWI/SNF complex. The 190-kDa protein visible in the p300-related complex is BRG1, the human homolog of the yeast transcriptional activator, SWI2/SNF2. The 170- and 155-kDa species are the recently identified BRG1-associated factors, BAF-170 and BAF-155, both homologs of yeast SWI3 (50). A component of the 44-kDa band is the previously identified BRG1/hbrm-associated factor, hSNF5 (INI-1) (36). Reciprocal precipitation of the immune complex using antibodies raised against BAF-155 reveals p270 to be the p300-related protein in direct association with the mammalian SWI/SNF complex.
MATERIALS AND METHODS
Cell culture.
HeLa cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium containing 5% fetal bovine serum (Research Sera, Summit Biotechnology) and supplemented with 50 penicillin (IU/ml) and streptomycin (50 μg/ml). The cells were grown at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Protein purification and peptide sequencing.
A preparative NM1 immune precipitation from a lysate of approximately 2 × 109 HeLa cells was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the proteins in the immune complex were visualized by staining with Coomassie Blue-G (Sigma). The gel fragments containing the p270, p190, p170, and p155 bands were excised and digested with lysylendopeptidase (Achromobacter protease I; Wako). Peptide fragments were extracted from the gel, separated by high-pressure liquid chromatography using a Vydac C18 column and were sequenced by automated sequencers (Applied Biosystems models 470, 473, and 477).
Antibodies.
The generation of monoclonal antibodies to p300/CBP (NM1 and NM11) and the reactivity of NM1 with p270 have been described elsewhere (13). A peptide corresponding to residues 2 to 15 from the amino-terminal sequence of BRG1 reported by Khavari et al. (29) was synthesized and used to raise rabbit immune sera specific for BRG1. A similar antibody to BRG1 (residues 2 through 15) was also generated in BALB/c mice. Rabbit antipeptide antibodies to BAF-170 and BAF-155 were developed by using keyhole limpet hemocyanin-conjugated peptides corresponding to residue positions 15 to 27 of the p170.K47 peptide and residues 592 to 605 of the p155.K31 sequences shown in Fig. 3. An antipeptide antibody specific for p270, showing no cross-reactivity with p300 or CBP, was developed in a similar manner from p270-specific peptide sequence (peptide sequence, RITATMDDMLL). All rabbit polyclonal sera were produced by CoCalico Biologicals (Reamstown, Pa.). The simian virus 40 T-antigen-specific antibody 419 (22) and the TBP-specific monoclonal antibody SL8 (42) were provided by Ed Harlow and Nouria Hernandez, respectively. The BRG1- and INI-1-specific polyclonal antibodies (50) used for Fig. 5 were provided by G. Crabtree.
FIG. 3.
The NM1 complex proteins p155 and p170 are BAF-155 and BAF-170. The sequences of two peptides derived from micropeptide sequencing of gel-purified p155 (K31 and K26) and two peptides derived from p170 (K43 and K47) are represented on the top line of each pair. The sequences show near-perfect identity with the deduced amino acid sequences of BAF-155 and BAF-170, respectively, as reported by Wang et al. (50). The R value indicates the amino acid position of the first residue in the deduced BAF sequence. An uncertain reading of the peptide sequencing cycle is indicated by underlining; an unreadable cycle is indicated by an X. Identity is indicated by a dot between residues. An initial K residue, not written in the sequenced peptides, is inferred from the nature of the enzyme used in the digest (Fig. 2). A K occurs at each corresponding predicted position in the BAF proteins. Peptide sequence was obtained as described in the legend to Fig. 2.
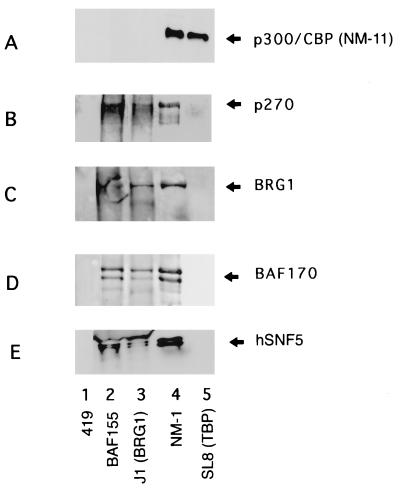
FIG. 5.
Immune complexes probed with antibodies specific for p300 family members and mammalian SWI/SNF complex components. The immune complexes precipitated by the series of antibodies indicated below lanes 1 through 5 were transferred to nitrocellulose and probed with a panel of antibodies individually specific for p300 family members and mammalian SWI/SNF complex components. The 419 antibody (lane 1) recognizes the simian virus 40 virus T antigen and serves as a negative control. The BAF-155 antibody (lane 2) is the antipeptide antibody raised against the K31 peptide sequence from BAF-155, also used for Fig. 4. J1 (lane 3) is a rabbit polyclonal antibody raised against the C-terminal portion of BRG1 (50). NM1 (lane 4) is a monoclonal antibody raised against human p300 that cross-reacts with CBP and p270 (13). SL8 (lane 5) is a monoclonal antibody directed against human TBP (42). The specifities of the antibodies used to probe the nitrocellulose blot are indicated at the right. The p300/CBP-specific antibody (A) is monoclonal antibody NM11, which does not cross-react with p270 (13, 14). The p270 antibody (B) is an antipeptide antibody raised against peptide sequence obtained from purified p270. The BRG1 antibody (C) is the antipeptide antibody described in the legend to Fig. 2. The BAF-170 antibody (D) is an antipeptide antibody raised against a portion of the BAF-170 K47 sequence. (Although BAF-170 appears as a single band in 35S- or 32P-labeled images, it consistently shows as a doublet in silver stains or Western blots such as in panel D.) The hSNF5 (INI-1) antibody is a rabbit polyclonal antibody described by Wang et al. (50). Membranes used for these Western blots were checked by autoradiography to verify the efficient transfer of the proteins prior to antibody probing.
Immunoprecipitations.
32P- or 35S-labeled or unlabeled total cell lysates were immunoprecipitated with antibody and protein A-Sepharose CL-4B beads (Pharmacia) for 1.5 h at 4°C as described previously (49, 52). Immune complexes were washed four times with E1A lysis buffer (49) and then either boiled off the beads in SDS in preparation for SDS-PAGE or used in further enzymatic assays. Sequential immunoprecipitations were performed as described by Dallas et al. (13).
Immunoblotting.
Total cell lysates were immunoprecipitated as described above and then separated through an SDS–7.5% polyacrylamide gel. After electrophoresis, proteins were blotted onto Immobilon P membranes (Millipore) by using standard procedures (23). The membrane was blocked with 5% nonfat milk in either Tris-buffered saline (20 mM Tris [pH 7.5], 150 mM NaCl) for Western blots using alkaline phosphatase-conjugated secondary antibody or PBST (0.2% Tween in phosphate-buffered saline) for enhanced chemiluminescence detection using horseradish peroxidase-conjugated secondary antibody. Chemiluminescence detection was performed with enhanced chemiluminescence reagents (Amersham) according to the manufacturer’s guidelines.
HAT assays.
Immune complexes were prepared as described above from 2.5 mg of total cell lysate but were treated additionally with ethidium bromide (12.5 μg/ml, final concentration) for 15 min at 4°C and then cleared by centrifugation at 3,500 × g prior to the immunoprecipitation. The ethidium bromide treatment was used to ensure that the immune complex represents only specific protein-protein interactions, not nonspecific protein associations mediated through DNA binding. In most experiments, an aliquot of the immune complex was reserved for parallel ATPase assays. The complexes were washed four times in a buffer consisting of 20 mM sodium phosphate, 250 mM NaCl, 0.1% Nonidet P-40 and 5 mM EDTA (pH 7.0) and were resuspended in HAT buffer (7) with 1.0 mg of calf thymus histones (Sigma type IIA). HAT assays were performed as outlined by Brownell and Allis (7) in the presence of 3H-acetyl coenzyme A and scored by detecting acetylation of histone substrates in a filter binding assay. 3H incorporation into histone substrate was determined by liquid scintillation counting.
ATPase assays.
Protein extracts were prepared for ATPase assays as described for the HAT assay. Equal amounts of total cellular lysate were used for corresponding samples in these two assay systems. ATPase activity was measured in a polyethyleneimine-cellulose thin-layer chromatography system as the ability to hydrolyze the terminal phosphate from [γ-32P]ATP. Reactions were performed essentially as described by Laurent et al. (32) except that the running buffer was modified to 0.75 M KH2PO4 (pH 3.5). Reaction conditions consisted of the immune complex in a 20-μl reaction volume containing 25 mM Tris (pH 6.9), 10 mM MgCl2, 1 mM dithiothreitol, 0.2 mg of salmon sperm DNA per ml, and 5 nM [γ-32P]ATP (3,000 Ci/mmol; NEN). Reactions were incubated at 37°C for the times indicated and stopped by the addition of EDTA to a final concentration of 0.025 M. The percent conversion from ATP-bound 32P to free 32Pi was quantified on a Fuji phosphoimaging system. Release of 32P-labeled Pi from the ATP substrate was also detected by autoradiography.
RESULTS
Identification of multiple members of the NM1 immune complex as BRG1 and BRG1-associated factors.
Figure 1 shows the immune precipitation pattern obtained with NM1, 1 of 4 monoclonal antibodies in our panel of 11 raised against p300/CBP that also recognizes p270. A combination of immunoprecipitation and Western blotting with p300-specific versus CBP-specific antibodies has confirmed that the 300-kDa band in these immunoprecipitations contains both p300 and CBP, while the 270-kDa band is distinct from both (13). p270 is related to p300 at a minimum of two epitopes because the four p300/CBP-derived antibodies that recognize p270 include ones that are specific for p300 as well as ones that recognize an epitope shared by p300 and CBP (13).
Each of the antibodies that recognizes p270 coprecipitates a series of cellular products. Analysis of 35S-labeled lysates (Fig. 1, lane 1) reveals coprecipitated species of 190, 170, 155, 75, 71, 64, 59, 50, and 44 kDa. These are relatively low percentage gels, designed to improve the resolution of the high-molecular-weight species, and thus do not show products migrating at less than 40 kDa. We have examined the complexes at lower-molecular-weight ranges and do not find proteins of lower molecular weight consistently part of the complex. The only prominent species in the 35S-labeled lysate that do not have phosphoprotein counterparts are the products migrating at 71 and 75 kDa (lane 3 compared with lane 1). Denaturation of the complexes and reprecipitation with NM1 shows that only p300, CBP, and p270 are recognized directly by the antibody; the remaining species are associated with the antibody targets (Fig. 1, lanes 2 and 4).
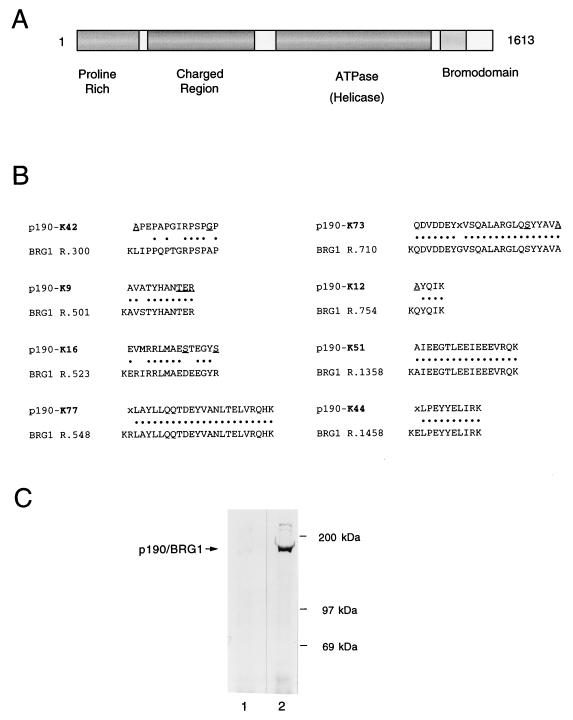
Because the products in these complexes are likely to represent important cellular partners of the p300-related proteins, we have begun to purify and obtain micropeptide sequence on each of the components. The sequence from the protein designated p190 in Fig. 1 reveals that this species is identical or highly related to the transcriptional coactivator BRG1. Eight peptides were obtained (Fig. 2), and all show a very high degree of identity to the cDNA-derived sequence of BRG1 (29). The relationship is not limited to any one domain, as peptides were recovered corresponding to regions throughout the BRG1 sequence (Fig. 2A and B). Three of the peptides (K9, K16, and K42) were obtained from very limited amounts of material and show some mismatches relative to the published BRG1 sequence. We do not think that these represent alternative forms of BRG1 because attempts to isolate products with these sequences were negative. The presence of BRG1 specifically is supported by the identity to BRG1 in the majority of the peptides, even in areas where BRG1 sequence differs from that of other very closely related human forms such as hbrm which is 80 to 90% identical to BRG1 over most of its length (10, 37; reviewed in reference 41). The presence of BRG1 in the NM1 complex is confirmed by reactivity with antiserum raised against the BRG1-specific amino-terminal sequence (Fig. 2C). The BRG1-specific serum (lane 2), but not preimmune serum (lane 1), reacts strongly and specifically in Western blots with the 190-kDa band immobilized by electrophoretic transfer of the NM1 immune complex.
FIG. 2.
Identification of the 190-kDa protein in NM1 complexes as BRG1. (A) Physical map of identified or predicted domains of BRG1, adapted from reference 29. BRG1 has a 1,613-amino-acid open reading frame. BRG1, like other members of the Swi2 family, contains multiple motifs characteristic of helicases and ATPases, although helicase activity has not been detected in these proteins. The ATPase/helicase domain spans amino acid residues 774 to 1237, and the bromodomain spans amino acid residues 1448 to 1523. BRG1 also contains a functional retinoblastoma protein binding motif (LXCXE) encompassing residues 1322 to 1326 (16). (B) The sequences of eight peptides derived from micropeptide sequencing of gel-purified p190 isolated from human cells are represented on the top line of each pair. Dots between residues indicate identity with the cDNA-derived human BRG1 sequence. Uncertainties in the reading of the sequencing cycles are indicated by underlining; X indicates an unreadable cycle. Uncertainties and unreadable cycles were not counted as identities, although they may be matches. The enzyme used to digest p190 prior to sequencing (lysylendopeptidase) cuts almost exclusively at lysine residues, so that each peptide sequence is expected to be immediately preceded by a lysine (K). As shown in the BRG1 sequence, a lysine occurs in BRG1 at each position where it is predicted to occur in p190, increasing the percent identity in the match. R represents the amino acid number of the initial lysine residue in BRG1 as reported in Khavari et al. (29). Peptides were recovered from regions throughout BRG1. (C) NM1-precipitated proteins from HeLa cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and probed by Western blotting with a BRG1-specific amino-terminal antipeptide antibody (lane 2) or preimmune serum (lane 1).
BRG1 (brm-related gene) (29) is a mammalian homolog of the Drosophila brm gene, which is required during development for activation of multiple homeotic genes (48). Both BRG1 and brm are close homologs of the yeast SWI2/SNF2 gene (2, 29, 32), which regulates the yeast mating-type switch and the switch to different carbohydrate utilization pathways (8, 47). In each system, the BRG1-related product exists in complex with a series of associated proteins (12, 15, 31, 40, 50). A recent detailed characterization of mammalian SWI/SNF complexes (50) indicates that they are present in multiple forms made up of 9 to 12 proteins now designated BRG1-associated factors (BAFs), ranging in estimated size from 47 to 250 kDa. These complexes include species of 155 and 170 kDa (BAF-155 and BAF-170) whose sequence has been determined by these investigators. A comparison of the peptide sequence that we have obtained for the 155- and 170-kDa species in the NM1 complex with the deduced amino acid sequences reported for BAF-155 and BAF-170 indicates that these are the same products (Fig. 3).
p270 is a p300-related protein associated with the mammalian SWI/SNF complex.
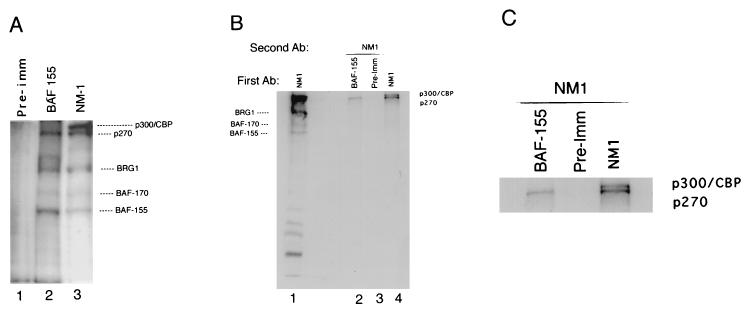
A comparison between the higher-molecular-weight proteins precipitated by BAF-155-specific antibodies and those in the NM1 immune complex suggests that p270 is the major p300-related protein in the complex (Fig. 4A). The two antibodies precipitate similar arrays of proteins. However, while a band comigrating with p270 is present in the BAF-155 complex, there is no evidence of species corresponding to p300 or CBP. Sequential immunoprecipitation (Fig. 4B and C) using the BAF-155 antibody, followed by denaturation and reimmunoprecipitation with the NM1 antibody, shows that the 270-kDa species from the BAF-155-specific complex is recovered by NM1 (lane 2), confirming that this species is the p300-related protein p270. Neither p300 nor CBP was found in the BAF-155 complex although p300, CBP, and p270 were all successfully reimmunoprecipitated from the original NM1 immune complexes (lane 4).
FIG. 4.
p270 is the p300 family member present in BAF155 complexes. (A) The high-molecular-weight proteins in the 32P-labeled immune complex precipitated with BAF-155-specific antibodies (lane 2) are compared with the complex precipitated by the NM1 antibodies (lane 3). The preimmune (Pre-imm) serum from the BAF-155 rabbit is used as a background control (lane 1). The positions of p300/CBP, p270, BRG1, BAF-170, and BAF155 in the NM1 complex are indicated. A species comigrating with p270 is seen in the BAF-155 complex, but there is no detectable appearance of species at the p300/CBP position. (B) 32P-labeled HeLa cell lysates were immunoprecipitated with either the BAF-155-specific antipeptide antibody (Ab), a preimmune control, or the NM1 antibodies and then denatured and reprecipitated with NM1 to identify the NM1-reactive species present in each complex (lanes 2 through 4). The standard NM1 immune complex is shown in lane 1. The NM1 antibody recovers p300, CBP, and p270 in the NM1 complex (lane 4) but detects only p270 in the BAF-155 complex (lane 2). (C) Enlargement of the high-molecular-weight region of lanes 2 through 4 of the gel shown in panel B to provide better resolution of the p300 and p270 bands.
We have also generated antipeptide antibodies specific for p270 which are suitable for Western blots. These antibodies do not cross-react with p300 or CBP. Electroblotting the BAF-155 immune complex (Fig. 5) and probing with the p270-specific antibodies shows again that p270 is a component of the mammalian SWI/SNF complex (Fig. 5B, lane 2). Analysis of the immune complex precipitated by the BRG1-specific antibodies (J1) that were used to define BAF-155 and BAF-170 (50) likewise shows a positive reaction with p270 antibodies (Fig. 5B, lane 3). When the immune complexes are probed with NM-11 (Fig. 5A), a monoclonal antibody that, in contrast to NM-1, recognizes p300 and CBP but not p270 (13, 14), a positive reaction is seen with the NM1 complex (lane 4) but not with the BAF-155 or BRG1 complexes (lanes 2, 3). Antibodies to BRG1 and BAF-170 indicate the presence of their respective proteins in all three complexes, NM1, BRG1, and BAF-155 (Fig. 5C and D, lanes 2 to 4). (Although BAF-170 appears as a single band in 35S- or 32P-labeled images, it consistently shows as a doublet in silver stains or Western blots such as in Fig. 5D.) Antibodies to the BRG1-associated protein hSNF5 (INI-1) also react positively with all three complexes (Fig. 5E), indicating that hSNF5 is also present in the NM1 complex. The hSNF5-reactive band runs just below the immunoglobulin heavy chain, which can be seen in lanes 2 and 3 of Fig. 5E, and coincides with the band designated p44 in the NM1 immune complex. One additional immune complex, the TBP-specific complex brought down by the SL8 monoclonal antibody, was probed in these experiments. The TBP complex shows the presence of p300/CBP (Fig. 5A, lane 5) as reported previously (13) but shows no reaction with p270-specific antibodies (Fig. 5B, lane 5).
The NM1 complexes contain HAT and ATPase activities that segregate with individual complex components.
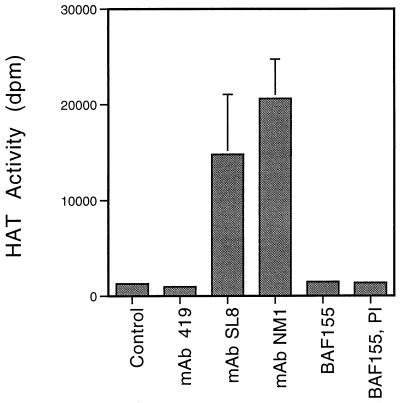
BRG1 has multiple motifs characteristic of ATPases (indicated schematically in Fig. 2), and SWI/SNF complexes contain a BRG1-associated ATPase activity (31). p300 and CBP have intrinsic and associated HAT activity (39, 53). The presence of these activities in NM1 complexes was assayed as shown in Fig. 6 and 7. HAT assays were performed on immunoprecipitated complexes and scored by detecting acetylation of histone substrates in a filter binding assay. NM1 immune complexes contain p300 and CBP and show readily detectable HAT activity (Fig. 6). Also positive in this assay were SL8 (TBP-specific) immune complexes, which contain p300, CBP, and TAFII250, which has also been found to contain HAT activity (35). However, the BRG1 complex (BAF-155 antibodies) showed no HAT activity above the background levels represented by the 419 and preimmune antibodies.
FIG. 6.
HAT activity. HAT assays were performed on immunoprecipitated HeLa cell complexes in the presence of 3H-acetyl coenzyme A and scored by detecting acetylation of histone substrates in a filter binding assay. Results are represented as disintegrations/minute per filter.
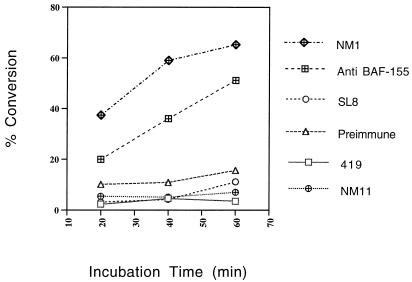
FIG. 7.
ATPase activity. Antibody complexes isolated from HeLa cells in parallel with those used in the HAT assay (Fig. 6) were assayed for associated ATPase activity. Equal amounts of total cell protein were immunoprecipitated with the appropriate antibody. ATPase activity was measured in a thin-layer chromatography assay as the ability to hydrolyze the terminal phosphate from [γ-32P]ATP. The percent conversion from ATP-bound 32P to free 32Pi was quantified on a Fuji phosphoimaging system. The results shown are the averages of six independent experiments.
ATPase activity was measured in parallel immunoprecipitations as percent conversion from ATP-bound 32P to free 32Pi in a thin-layer chromatography assay (Fig. 7). The NM1- and BAF155-derived complexes both show ATPase activity. The TBP-specific complex (SL8) does not show ATPase activity above background levels. Similarly, the NM11 complex, which contains p300 and CBP but not p270 or the associated SWI/SNF complex (13, 14), does not show ATPase activity.
The presence of these activities in NM1 immune complexes and their segregation into TBP-specific and BAF-155-specific complexes are consistent with the interpretation that NM1 immune complexes represent multiple distinct complexes involving the direct NM1 targets, p300, CBP, and p270. The lack of detectable HAT activity in the SWI/SNF (BAF-155) complex implies that p270 is not a HAT like the antigenically related p300 and CBP.
p270 association with BAF-155 and BAF-170 is not dependent on BRG1.
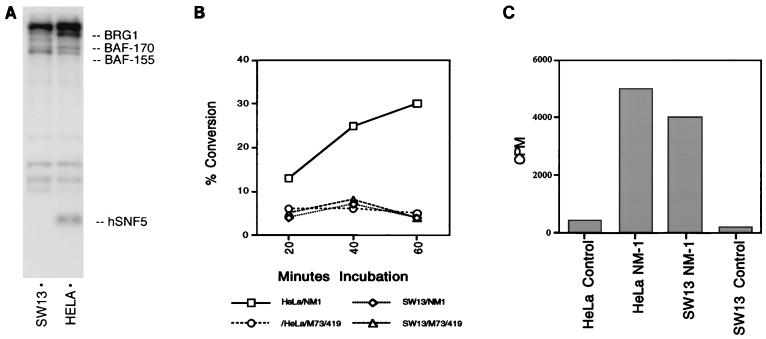
Analysis of mammalian SWI/SNF complexes is facilitated by the availability of the human carcinoma cell line SW13, which is severely deficient in BRG1 expression (16, 37). As expected from the identification of the p190 band in NM1 complexes as BRG1, this band is greatly reduced in NM1 complexes precipitated from SW13 cells (Fig. 8A). The lowest-molecular-weight species, p44, now identified as hSNF5 (INI-1), is also absent from the SW13 NM1 complex. The BAF-170 and BAF-155 species are still present when the complex is isolated from SW13 cells. The stability of the in vivo association between p270 and BAF-155 and BAF-170 in the absence of BRG1 supports the interpretation that p270 is an integral member of this complex. ATPase activity was compared in NM1 complexes isolated from HeLa and SW13 cells (Fig. 8B). The background level of ATPase activity in the reaction is indicated by immunoprecipitation of HeLa or SW13 cells with the 419 control antibody. NM1 complexes precipitated from HeLa cells show readily detectable ATPase activity, while NM1 complexes precipitated from SW13 cells show no ATPase activity above background levels. In contrast, the loss of SWI/SNF complex components in SW13 cells has little effect on HAT activity in the NM1 complexes (Fig. 8C). These results support the conclusion that BRG1 is in stable intracellular association with p270 and is the source of the ATPase activity in the NM1 complexes.
FIG. 8.
Analysis of NM1 complexes in BRG1-deficient cells. (A) Immune complexes. NM1 complexes were precipitated from the BRG1-deficient cell line SW13 (lane 1) or from HeLa cells (lane 2). The position of the p190/BRG1 band in the HeLa cell complex is indicated at the right. In all comparisons between HeLa and SW13 cells, protein concentrations from each cell lysate were normalized before immune precipitation. (B) ATPase assay. NM1 complexes isolated from SW13 cells were assayed for associated ATPase activity in comparison with HeLa cell-derived complexes. Assays were performed as described for Fig. 7, and results are expressed as percent conversion from ATP-bound 32P to free 32Pi. The results shown are the averages of four independent experiments (standard deviation = 13.7%). Baseline activities in HeLa and SW13 lysates precipitated with control monoclonal antibody 419 are represented by circles and triangles, respectively; activity in HeLa-derived NM1 complexes is represented by squares; activity in SW13-derived NM1 complexes is represented by diamonds. (C) HAT assay. HAT activity was determined as described for Fig. 6, using aliquots of the same lysates shown in panel B. Antibody 419 was again used as a background control.
DISCUSSION
The monoclonal antibodies that we have raised against p300 identify an immunologically related cell protein, p270, that shares at least two distinct antigenic determinants with p300 (13). The subset of antibodies that cross-reacts with p270, exemplified by NM1, recognizes p300, CBP, and p270 directly and also indirectly coprecipitates a series of at least nine cellular proteins stably associated with p300, CBP, or p270 in physiological conditions. These proteins resolve into at least two complexes involving the p300/CBP/p270 family. One complex contains TBP along with p300 or CBP (apparently as alternates to TAFII250) in association with at least two other TBP-associated proteins, p64 and p59 (1, 13). The SL8 (TBP-specific) complex, consistent with the presence of p300, CBP, and TAFII250, contains HAT activity. This complex does not contain detectable p270 or ATPase activity.
A second group of proteins brought down with the NM1 antibodies contains p270 as an integral component of SWI/SNF complexes. We have confirmed the presence in the p270-containing complexes of BRG1, the BRG1-associated factors BAF-155 and BAF-170 (50), and the BRG1/hbrm-associated factor hSNF5 (INI-1) (36). Conversely, probes of SWI/SNF complexes isolated through either BAF-155-specific antibodies or BRG1-specific antibodies confirm the presence of p270. The identification of p270 as an integral member of human SWI/SNF complexes is consistent with the presence of a 250-kDa band noted by Wang et al. (50) and designated BAF-250. It is likely that p270 is equivalent to BAF-250.
Both yeast and human SWI/SNF complexes contain a nucleosome remodelling activity (reviewed in reference 41). Taken together with the HAT activities associated with p300/CBP (39, 53), the presence of p270 in SWI/SNF complexes suggests that multiple aspects of chromatin remodelling may be mediated through a series of proteins with structural homology to p300.
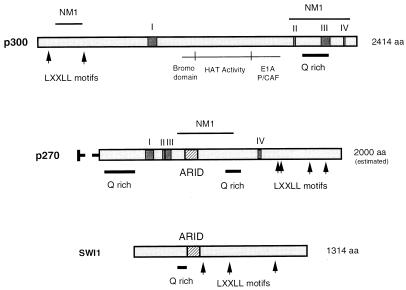
Identification of the precise structural determinants shared by p270 and p300 will likely require a better understanding of the three-dimensional structure of p270 and p300 than is available now. The relationship is likely to be limited, as the majority of antibodies developed against p300 do not cross-react with p270 and because p270 and p300 show distinguishable activities. The p270-containing complexes do not show HAT activity if p300 and CBP are not present (Fig. 6). p270 is also not targeted by E1A (not shown) and is not present in the TBP-specific complexes that contain p300 and CBP (Fig. 5). We are currently sequencing p270 (12a). The sequence (represented schematically in Fig. 9) reveals some unique features of p270, in particular, the presence of a newly recognized DNA binding domain, termed AT-rich interactive domain (ARID), first identified in the mouse Bright and Drosophila Dead-Ringer proteins (20, 26). This feature is also present in yeast Swi1, suggesting that p270 is the human homolog of this member of the yeast SWI/SNF complex. p300 does not contain an ARID region. p270 shows some specific features in common with p300: both contain Q-rich regions spanning about 200 residues (implicated in transactivation functions [45]), and both contain potential nuclear hormone receptor binding motifs, LXXLL (25). These two features are present in SWI1 as well and support the interpretation that p270 is a SWI1 homolog. The LXXLL motifs have not previously been noted in SWI1, as their potential significance has only recently become apparent. p270 does not contain long stretches of sequence homology to p300 like those seen in comparison of p300 and CBP. However, a filtered BLAST search program (3, 4) identifies at least four homology regions (I to IV) distinct from the Q-rich regions or LXXLL motifs. These are amino acid stretches of 22 to 66 residues showing 27 to 46% identity between p300 and p270. Interestingly, all but one of these regions (IV) are in areas that are not highly conserved between p300 and CBP. Also interestingly, mapping of the NM1 epitope in p270 indicates that it is distinct from all of these regions, implying that the extent of the relationship is greater than indicated by the degree of linear sequence homology. The NM1 epitope appears to identify a structural epitope shared by p270 and p300. This antibody reacts with native protein but not with denatured proteins such as that present in Western blots (13); its reaction in sequential immunoprecipitations depends on at least partial renaturing of the proteins. There are at least two NM1-reactive sites in p300, near the amino and carboxy ends. The NM1 epitope in p270 lies in the middle of the protein. There is no recognizable linear sequence that is shared by the regions containing the NM1 epitopes, supporting the idea that this is a structural determinant. Because the p300/CBP population that we used as antigen was native, enzymatically active protein, as determined from the presence of HAT activity in stored immunogen samples, it is likely that the surface epitopes that served as antigenic determinants correlate with functional regions, perhaps with cofactor binding sites. p270 may recruit SWI/SNF complexes to transcription sites in a manner analogous to p300, bringing histone-modifying activity to such sites.
FIG. 9.
Alignment of p300, p270, and SWI1. The linear amino acid sequences of p300, p270, and SWI1 are represented schematically. Regions highly conserved between p300 and CBP include the bromodomain, the HAT domain, and the E1A and P/CAF binding region. The sequence of p270 is over 90% complete (assuming an estimated size of 2,000 amino acids [aa]; the uncertain sequence is represented by a dashed line). p270 does not show HAT activity of E1A binding activity. There is also no evidence thus far of a bromodomain consensus sequence. p270 contains an ARID DNA binding region (hatched box), a feature not present in p300. p270 does show several specific features in common with p300. Regions of conserved sequence between p270 and p300 (homology regions I to IV) are indicated by solid boxes within the bars. In addition to the four regions of conserved sequence, p270 and p300 have in common the presence of Q-rich regions (implicated in transactivation functions) and LXXLL motifs (implicated in nuclear receptor binding). p270 and p300 also have in common the structural epitope recognized by NM1. p270 and SWI1 have an overall similarity of structure, in that they both contain ARID regions, and they both have multiple LXXLL motifs as well as Q-rich regions. SWI1 has an unusual asparagine/threonine-rich stretch near the N terminus; we do not yet know if this feature also occurs in p270. Other than these common features, p270 and SWI1 do not show direct sequence homology.
ACKNOWLEDGMENTS
The first three authors contributed equally to this work.
We thank Gerry Crabtree, Nouria Hernandez, and Ed Harlow for generous gifts of antibodies, and we thank Annette Heagy, Patty Baxter, Steve Dorfman, and John Gibas for expert technical assistance.
This work was supported by PHS grants CA53592, CA55330 (E.M.), and CA68066 (P.Y.) from the NIH.
REFERENCES
- 1.Abraham S E, Corrigan Carter M, Moran E. Transforming growth factor β1 (TGF β1) reduces cellular levels of p34cdc2, and this effect is abrogated by adenovirus independently of the E1A-associated pRB binding activity. Mol Biol Cell. 1992;3:655–665. doi: 10.1091/mbc.3.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3643–3651. doi: 10.1128/mcb.6.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Boguski M S, Gish W, Wootton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lippman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of Coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 6.Avantagiatti M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53 dependent pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson M, Osmond B C, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti D, La Morte V, Nelson M, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 10.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 12.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 12a.Dallas, P., and E. Moran. Unpublished data.
- 13.Dallas P B, Yaciuk P, Moran E. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J Virol. 1997;71:1726–1731. doi: 10.1128/jvi.71.2.1726-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dallas P B, Yaciuk P, Moran E. The monoclonal antibody NM11 recognizes a C-terminal epitope shared by p300 and CBP. Hybridoma. 1997;16:273–275. doi: 10.1089/hyb.1997.16.273. [DOI] [PubMed] [Google Scholar]
- 15.Dingwall A K, Beck S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG-1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 17.Eckner R, Ewen M E, Newsome D, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kd protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and BHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 19.Egan C, Jelsma T N, Howe J A, Bayley S T, Ferguson B, Branton P E. Mapping of cellular protein-binding sites on the products of early region 1A of human adenovirus type 5. Mol Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory S L, Kortschak R D, Kalionis B, Saint R. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA binding proteins. Mol Cell Biol. 1996;16:792–799. doi: 10.1128/mcb.16.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–822. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 22.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for SV40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 24.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 26.Herrscher R F, Kaplan M H, Leisz D L, Das C, Scheuermann R, Tucker P W. The immunoglobulin heavy chain MARs are bound by bright: a B cell specific transactivator that describes a new DNA binding protein family. Genes Dev. 1995;9:3067–3082. doi: 10.1101/gad.9.24.3067. [DOI] [PubMed] [Google Scholar]
- 27.Howe J A, Mymryk J S, Egan C, Branton P E, Bayley S T. Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci USA. 1990;87:5883–5887. doi: 10.1073/pnas.87.15.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 30.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 31.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 32.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lill N L, Grossman S R, Ginsburg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 34.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 35.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 36.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mymryk J S, Lee R W, Bayley S T. Ability of adenovirus 5 E1A proteins to suppress differentiation of BC3H1 myoblasts correlates with their binding to a 300 kDa cellular protein. Mol Biol Cell. 1992;3:1107–1115. doi: 10.1091/mbc.3.10.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 40.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodelling machine. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 42.Ruppert S M L, McCulloch V, Meyer M, Bautista C, Falkowski M, Stunnenberg H G, Hernandez N. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-react with TBP from other species. Hybridoma. 1996;15:55–68. doi: 10.1089/hyb.1996.15.55. [DOI] [PubMed] [Google Scholar]
- 43.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300—direct interaction with the activation domain of MyoD and with the MADS Box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scolnick D M, Chehab N, Stavridi E S, Lien M, Caruso L, Moran E, Berger S L, Halazonetis T D. CBP and P/CAF are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 45.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 48.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 49.Wang H-G H, Moran E, Yaciuk P. E1A promotes association between p300 and pRB in multimeric complexes that are required for normal biological activity. J Virol. 1995;69:7917–7924. doi: 10.1128/jvi.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 51.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 52.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300 kilodalton product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 54.Yuan W, Condorelli G, Caruso M, Falsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]