Abstract
Chromosomal translocations in acute leukemia that affect the AML-1/CBFβ transcription factor complex create dominant inhibitory proteins. However, the mechanisms by which these proteins act remain obscure. Here we demonstrate that the multidrug resistance 1 (MDR-1) promoter is a target for AML/ETO transcriptional repression. This repression is of basal, not activated, expression from the MDR-1 promoter and thus represents a new mechanism for AML/ETO function. We have defined two domains in AML/ETO that are required for repression of basal transcription from the MDR-1 promoter: a hydrophobic heptad repeat (HHR) motif and a conserved zinc finger (ZnF) domain termed the MYND domain. The HHR mediates formation of AML/ETO homodimers and AML/ETO-ETO heterodimers. Single serine substitutions at conserved cysteine residues within the predicted ZnFs also abrogate transcriptional repression. Finally, we observe that AML/ETO can also inhibit Ets-1 activation of the MDR-1 promoter, indicating that AML/ETO can disrupt both basal and Ets-1-dependent transcription. The fortuitous inhibition of MDR-1 expression in t(8;21)-containing leukemias may contribute to the favorable response of these patients to chemotherapeutic drugs.
AML-1 is the direct or indirect target of multiple chromosomal translocations in acute B-cell and myeloid leukemia. t(8;21) and inv(16) disrupt AML-1 and its heterodimeric partner, CBFβ, respectively, and are the most frequent translocations in acute myeloid leukemia (AML). These translocations are found in the leukemic blasts of up to 30% of patients with AML with discernable translocations (28, 37). t(12;21) also disrupts AML-1 in B-cell acute lymphocytic leukemias of children (43). Thus, AML-1 is one of the most frequently mutated genes in human leukemia. Interestingly, patients containing these translocations uniformly respond better to chemotherapy, with an increased 5-year survival rate (3, 9, 21, 42).
t(8;21)(q22;q22) fuses the N-terminal 177 amino acids (aa) of AML-1 to the C-terminal 575 aa of ETO to form the chimeric AML/ETO protein (36). An analysis of the structure of AML/ETO reveals that the DNA binding runt domain of AML-1 is not altered but that the transactivation domain of AML-1 has been replaced by ETO. This led to the hypothesis that the fusion protein acts as a dominant inhibitor of AML-1B function (32, 34). AML/ETO interfered with AML-1B-activated transcription of the T-cell receptor β (TCRβ) enhancer, and the interleukin 3 and granulocyte-macrophage colony-stimulating factor (GM-CSF) promoters, but did not affect the basal expression of these promoters (13, 32, 45). The dominant inhibitory action of the t(8;21) and the inv(16) fusion proteins has been confirmed biologically by expressing these fusion proteins during murine development (4, 48). These mice display the same phenotype as that displayed by AML-1 (and CBFβ-)-deficient mice (39, 46).
Multiple mechanisms have been proposed for transcriptional repression, including competition for binding sites and interaction with surrounding factors, with corepressors, or with the basal transcriptional machinery (7, 18). Because AML/ETO acts at substoichiometric levels, AML/ETO interference with AML-1B-mediated transactivation is unlikely to be due to competition for DNA binding sites (34). Moreover, the fusion protein failed to repress basal expression from the TCRβ enhancer-simian virus 40 early chimeric promoter, suggesting that the fusion protein does not interact with the basal machinery to globally repress transcription (34). C-terminal ETO sequences are required for function, suggesting that the fusion protein may contact other factors that mediate transcriptional interference (26, 34).
ETO contains four domains that have homology to the Drosophila protein nervy (12). Overall, ETO and nervy are 30% identical, but these four regions display 50 to 55% identity. Two of these domains are putative protein interaction domains, an amphipathic helix that contains a hydrophobic heptad repeat (HHR) (33) and the MYND domain that contains two putative zinc fingers (ZnFs) (17). In AML/ETO, deletion of the C-terminal 283 aa, including the ZnFs, the HHR, and a third domain of unknown function (the nervy domain), inactivates the protein’s ability to inhibit AML-1B-dependent transcription (26).
The development of drug-resistant neoplastic cells during chemotherapeutic regimens is a major determinant in treating many types of cancer, including acute leukemia (1). MDR-1 encodes a transmembrane “pump,” P-glycoprotein, that extrudes anthracyclines, epipodophyllotoxins, and vinca alkaloids, drugs which are commonly used to treat AML (1). A subset of de novo AMLs are MDR-1 negative (40). These MDR-1-negative AMLs have recently been linked to the t(8;21) translocation found primarily in adult cases of AML (25), and for these cases, MDR-1 could not be detected (40). Thus, the great majority of t(8;21) cases fail to express MDR-1, and this correlates with a better response to therapy.
Previously, we identified an AML-1 binding site adjacent to an Ets-1 binding site within the first 137 bp upstream of the MDR-1 transcriptional start site. AML-1B can bind this site and stimulate expression of MDR-1 three- to fourfold (42a). In this report, we identified MDR-1, a gene that is expressed in many cell types (including myeloid cells), as a target for AML/ETO repression. Our results also indicate that this repression is not simply the result of AML/ETO interfering with AML-1B function. We analyzed in detail the domains of ETO that mediate repression and found that both the HHR and the ZnF domains are required. Individual serine substitutions at either of two conserved cysteine residues in the putative ZnFs of the MYND domain resulted in similar abrogations of transcriptional repression. We found that AML/ETO can form homodimers as well as heterodimers with ETO and that the HHR motif mediates this dimerization. Finally, we observed that AML/ETO can block Ets-1-mediated transactivation of the MDR-1 promoter. Thus, MDR-1 may represent a fortuitous physiological target for AML/ETO because myeloid leukemias with t(8;21) rarely express MDR-1, a characteristic which is correlated with an improved response to therapy (40).
MATERIALS AND METHODS
Cell culture.
C33A cells and Cos-7 cells were maintained in Dulbecco modified Eagle medium (BioWhittaker Inc, Walkersville, Md.) containing 10% fetal calf serum, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 2 mM l-glutamine (all from BioWhittaker). NIH 3T3 cells were maintained in DMEM with 10% calf serum, antibiotics, and l-glutamine, and HEL cells were cultured in RPMI 1640 (BioWhittaker) containing 10% fetal calf serum, antibiotics, and l-glutamine.
Plasmid constructions.
Deletions and point mutations in AML/ETO were generated in pBluescript ETO by oligonucleotide-mediated mutagenesis (24). Deletions include the TAF homology domain (residues 277 to 344 in AML/ETO), the HHR (residues 500 to 520), the nervy domain (residues 594 to 636), and the ZnF domain (residues 663 to 700). After sequence analysis to confirm that the mutations were present, the HpaII-BglII fragment (for TAF110 and HHR deletions) or the BglII-XbaI fragment (for nervy and ZnF deletions and point mutations) was then subcloned into pCMV5 AML/ETO. The pMLV Ets-1 expression plasmid was a gift from J. Ghysdael.
Transcriptional analysis.
The −137 MDR-1 chloramphenicol acetyltransferase (CAT) plasmid has been described previously (44). Transfection of C33A cells (2 × 106 cells in 60-mm-diameter dishes) by calcium phosphate coprecipitation was performed as previously described (16). HEL cells (2 × 106 cells in 60-mm-diameter dishes) were transfected with 10 μl of Lipofectamine (Gibco-BRL, Gaithersburg, Md.) per transfection, and NIH 3T3 cells (2 × 105 cells in 60-mm-diameter dishes) were transfected with 15 μl of Superfect reagent (Qiagen) per transfection. Cytomegalovirus (CMV) β-galactosidase or Rous sarcoma virus secreted alkaline phosphatase (SEAP) was included as an internal control for transfection efficiency. Measurement of β-galactosidase activity and SEAP activity followed standard procedures (2, 38). CAT activity was measured as previously described (16) and was quantitated on a Molecular Dynamics PhosphorImager with Image-Quant software and normalized with respect to β-galactosidase activity. Experiments were repeated a minimum of three times, and results are indicated as the means with standard deviations.
Immunoprecipitation.
For coprecipitation experiments to detect heterodimers, Cos-7 cells or NIH 3T3 cells (106 cells in 60-mm-diameter dishes) were cotransfected with 2 μg of CMV5 AML/ETO or AML/ETO deletion mutants and 2 μg of CMV5 ETO. For detecting homodimers, we cotransfected 2 μg of hemagglutinin (HA)-tagged CMV5 AML/ETO (HA-AML/ETO) with 2 μg of CMV5 AML/ETO deletion mutants, and 40 h after transfection the cells were labeled with 100 μCi of [35S]methionine for 3 h in methionine- and cysteine-free Dulbecco modified Eagle medium (PROMIX; Amersham) containing 2% dialyzed calf serum. Cells were lysed in antibody buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 0.5% sodium dodecyl sulfate (SDS), 1.5 mg of iodoacetamide per ml, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1 trypsin-inhibiting units (TIU) of aprotinin per ml), followed by incubation with 100 μl of formalin-fixed Staphylococcus aureus membranes (Immunoprecipitin; GIBCO-BRL) for 30 min to eliminate nonspecific protein binding. After centrifugation for 5 min at 4°C, the supernatants were collected and immunoprecipitated for 1 h with affinity-purified primary antibody. Fifteen microliters of a 50% slurry of protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden) was then added for 1 h to collect the immune complexes. The immune complexes were then washed three times with lysis buffer, and proteins were eluted by boiling the immune complexes for 2 min in 1× Laemmli buffer. Samples were then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) in an 8% gel, and the gel was then fixed in 45% methanol–10% acetic acid for 30 min and incubated with Amplify (Amersham) for 20 min. The gel was then dried and subjected to autoradiography.
Western blotting.
Western blotting was performed on cell lysates from calcium phosphate-transfected Cos cells (106 cells in 60-mm-diameter dishes) at 48 h posttransfection. Cells were lysed in antibody buffer and sonicated, followed by protein quantitation with the Bio-Rad DC protein assay. Then, 150 μg of protein was boiled in Laemmli buffer for 2 min, fractionated by SDS-PAGE, and transferred to nitrocellulose. Blots were blocked for 1 h with 5% milk, and primary antibody incubation took place overnight at 4°C. After being washed and incubated with secondary antibody, followed by a further washing, proteins were visualized by enhanced chemiluminescence (Pierce). For Western blotting with C33A and HEL cells transiently transfected for CAT assays, identical procedures were followed, except that cell lysates were prepared in 250 mM Tris, pH 7.5, containing 0.1 TIU of aprotinin, 0.1 mM phenylmethysulfonyl fluoride, and 1 μg of iodoacetamide per ml.
The antibodies used in these experiments included the AML N-terminal antibody that has been described previously (35), and the α-HA (12CA5) antibody was purchased from Babco (Berkeley, Calif.). The ETO antibody was generated against a glutathione S-transferase (GST)–ETO fusion protein and was affinity purified against the GST-ETO protein. This affinity-purified antibody is highly specific for ETO (28a).
RESULTS
AML/ETO represses transcription of the MDR-1 promoter.
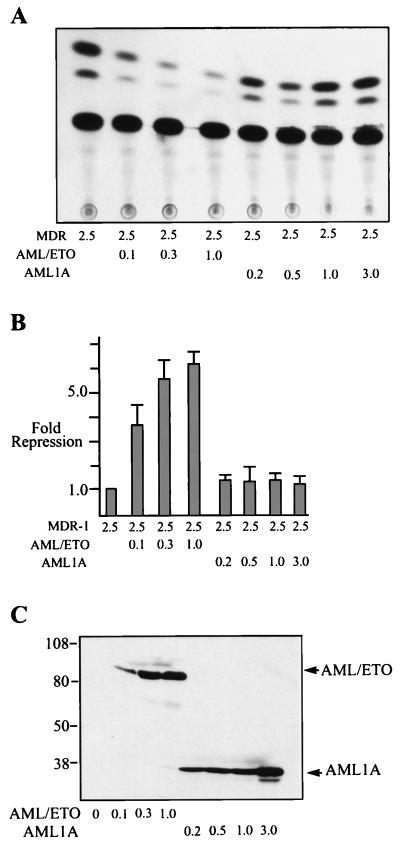
An AML-1 binding site is located at nucleotide position −94 to −89 in the MDR-1 promoter and in some cell types contributes to the basal expression of MDR-1 (42a). To test whether AML/ETO could affect the transcription of MDR-1 we transfected MDR-1(−137)–CAT together with increasing amounts of AML/ETO. Because previous results revealed that AML-1A (an AML protein missing the transactivation domain) interferes with AML-1B-activated transcription (34), we also tested AML-1A in this assay. We observed repression of MDR-1 with increasing amounts of AML/ETO, but not with increasing amounts of AML1A (Fig. 1A and B). Western blotting of the same cell extracts used to measure CAT activity with antibodies directed to the N terminus of AML-1 revealed that these proteins were synthesized at similar levels (Fig. 1C). These results indicate that ETO sequences in AML/ETO are required for transcriptional repression. Also, because high-level expression of AML1A did not interfere with MDR-1 transcription, we conclude that inhibition of AML-1B activity does not significantly affect MDR-1 basal transcription in C33A cells (confirmed by using constructs in which the AML-1B binding sites were deleted; data not shown).
FIG. 1.
AML/ETO, but not AML1A, represses basal transcription of the MDR-1 promoter. (A) C33A cells were transfected with 2.5 μg of MDR-CAT (−137), 100 ng of CMV β-galactosidase as an internal control, and increasing amounts (in micrograms) of CMV5 AML1A or CMV5 AML/ETO plasmids as indicated. CAT activity was measured from whole-cell extracts as described in Materials and Methods. (B) Quantitation of the results in panel A. CAT activity was normalized with respect to β-galactosidase activity. Fold repression represents the normalized promoter activities from cells transfected with expression plasmids compared to that from cells transfected with MDR-1 alone. (C) Western blot of cell extracts used in panel A blotted with our anti-AML N-terminal antibody and detected by enhanced chemiluminescence.
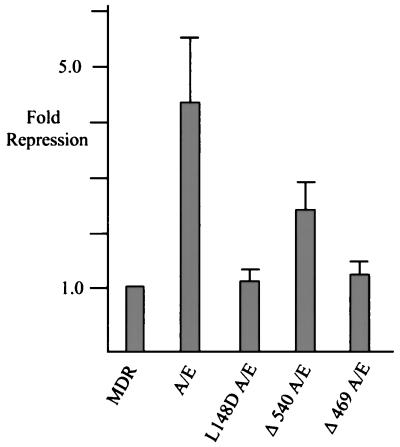
To further test the requirement for ETO sequences in MDR repression we tested two C-terminal deletion mutants of AML/ETO. Deletion of residues 540 to 752 (the nervy domain and the putative ZnF domain; Δ540 AML/ETO) significantly impaired AML/ETO function (Fig. 2). Further deletion to residue 469 (Δ469 AML/ETO), which removes the amphipathic helix that contains a hydrophobic heptad repeat, completely ablated repression (Fig. 2). We also tested whether the DNA binding ability of AML/ETO was necessary for repression. The AML/ETO protein with the L148D substitution (L148D AML/ETO) is unable to bind DNA (26), and this protein was also unable to repress MDR-1 transcription (Fig. 2).
FIG. 2.
AML/ETO repression of MDR-1 requires DNA binding and fusion with ETO sequences. C33A cells were transfected with 5 μg of MDR-1 CAT and 1 μg of Rous sarcoma virus SEAP plasmids and 2 μg of the indicated CMV5 AML/ETO (A/E) fusion proteins. CAT activity was quantitated with a Molecular Dynamics PhosphorImager and normalized relative to SEAP activity.
HHR and ZnF motifs are required for AML/ETO transcriptional repression.
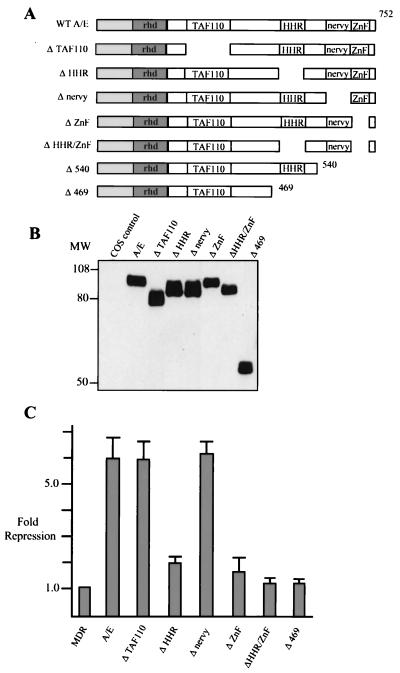
The impaired function of Δ540 AML/ETO for the first time suggested a function for the putative ZnF domain of AML/ETO. To precisely determine the C-terminal sequences of ETO required for repression, we constructed internal deletions in each of the conserved domains of ETO (Fig. 3A). These deletions included the region of ETO that has homology to the TAF110 coactivator (20) that has not been previously tested. We confirmed that each of these mutant proteins was appropriately expressed by transfecting Cos cells, followed by Western blot analysis with our anti-AML antibody (Fig. 3B). These proteins were expressed at levels similar to those of wild-type AML/ETO, although we consistently observed that AML/ETO with the ZnF domain deleted (ΔZnF AML/ETO) migrated more slowly than predicted. This was also true when the ZnF was independently deleted by using restriction enzymes flanking this domain (data not shown). We also determined that the AML/ETO deletion mutants were not significantly altered in DNA binding or subcellular localization relative to wild-type AML/ETO (data not shown).
FIG. 3.
HHR and ZnF motifs are required for transcriptional repression. (A) Schematic diagram of AML/ETO (A/E) and AML/ETO deletion constructs. (B) Cos cells were transfected with 3 μg of the indicated deletion constructs, and at 48 h posttransfection cells were collected for Western analysis as described in Materials and Methods. Blotting was performed with the anti-AML N-terminal antibody. (C) C33A cells were transfected with 2.5 μg of MDR-1 CAT, 100 ng of CMV β-galactosidase expression plasmid, and 0.5 μg of CMV5 AML/ETO or the indicated CMV5 AML/ETO deletion constructs. CAT activity was quantitated and normalized to β-galactosidase activity.
These deletion constructs were tested for repression of MDR-1 by transfecting C33A cells (Fig. 3C). Deletion of either the TAF homology domain or the nervy homology region did not significantly alter transcriptional repression. By contrast, deletion of either the HHR or the ZnF domains reduced AML/ETO-mediated repression from six- to twofold (Fig. 3C). Deletion of the HHR and ZnF domains together completely abrogated transcriptional repression, similar to what was seen for Δ469 AML/ETO, indicating that both motifs contribute to the repression. Western blotting of representative cell extracts used in the CAT assays revealed that proteins were synthesized at similar levels in these same transfections (data not shown). To confirm that this result was not cell type specific we used the hematopoietic cell line HEL (30) and obtained similar results (data not shown, but see Fig. 4 below). In fact, these cell lines were used interchangeably in these assays.
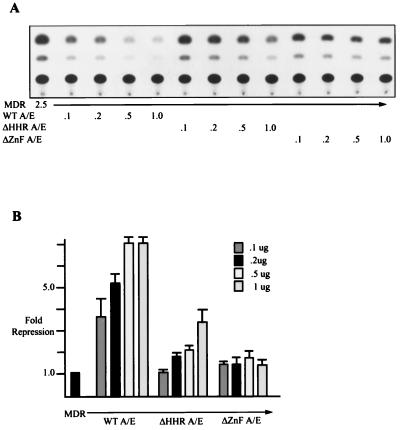
FIG. 4.
ΔZnF AML/ETO does not repress efficiently even at high expression levels. (A) Two micrograms of MDR-1 CAT, 100 ng of CMV β-galactosidase expression plasmid, and 0.1, 0.2, 0.5, or 1 μg of the indicated AML/ETO (A/E) expression plasmids were transfected into HEL cells. At 48 h posttransfection CAT assays were performed as described in Materials and Methods. (B) CAT activity was quantitated and normalized to β-galactosidase activity.
Because AML/ETO with the HHR deleted (ΔHHR AML/ETO) and ΔZnF AML/ETO still maintained some ability to repress transcription, we performed titration experiments (Fig. 4). As the amount of input DNA increased, we observed an increase in the ability of the ΔHHR AML/ETO mutant to repress transcription, but the levels of repression did not reach those of the wild-type protein. However, an increase in the amount of the ΔZnF AML/ETO did not result in a further increase in the amount of repression (Fig. 4). Similar results were obtained with C33A cells (data not shown). Western blotting of the cell extracts confirmed that the levels of proteins increased with increasing levels of input DNA (data not shown). Thus, high levels of the ΔHHR AML/ETO protein can overcome the defect to some degree, whereas the modest ΔZnF AML/ETO repression activity cannot be augmented with increased levels of protein.
Point mutations in the putative ZnFs of AML/ETO impair transcriptional repression.
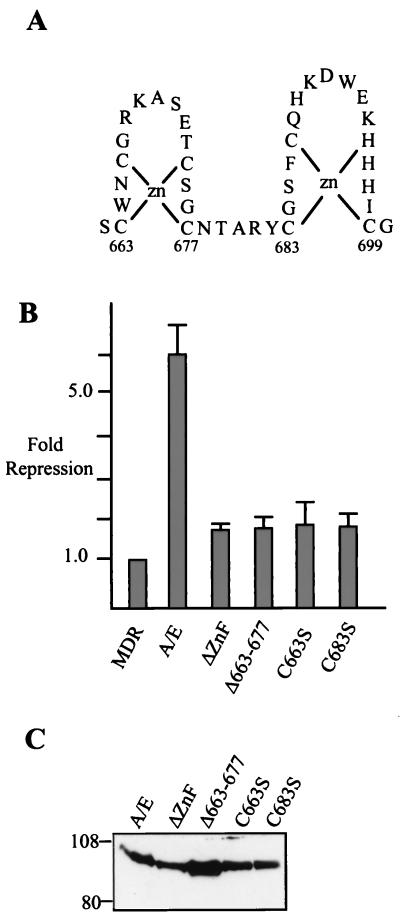
Because ΔZnF AML/ETO is impaired for transcriptional repression even at high protein levels, we constructed more subtle alterations in this domain. The predicted structure of the two ZnFs is shown in Fig. 5A. To determine whether both putative ZnFs were required for function, we deleted the predicted N-terminal ZnF (residues 663 to 673) and we also introduced serine substitutions at either C663 or C683. If this structure forms the predicted ZnFs, serine substitutions at either C663 or C683 would disrupt these structures (Fig. 5A). Moreover, these residues are invariant among the proteins with a homologous MYND motif (17). Each of these mutants was impaired for MDR-1 repression relative to wild-type AML/ETO, and each was similar in activity to ΔZnF AML/ETO (Fig. 5B). Western blotting of the CAT assay cell extracts indicated that these proteins were expressed at similar levels (Fig. 5C). Thus, the serine substitutions at either C663 and C683 provide indirect evidence that these motifs do form ZnF structures that are required for AML/ETO transcriptional repression.
FIG. 5.
Point mutations in the predicted ZnFs abrogate AML/ETO function. (A) predicted structure of the ZnF domain in the ETO portion of AML/ETO. (B) C33A cells were transfected with 2 μg of MDR-1 CAT, 100 ng of CMV β-galactosidase expression plasmid, and 0.5 μg of the indicated AML/ETO (A/E) expression plasmids. CAT assays were quantitated and normalized to β-galactosidase activity. (C) Western blotting (as described in Materials and Methods) was performed on representative cell lysates used in panel B with the anti-AML N-terminal antibody.
AML/ETO can homodimerize and form heterodimers with ETO.
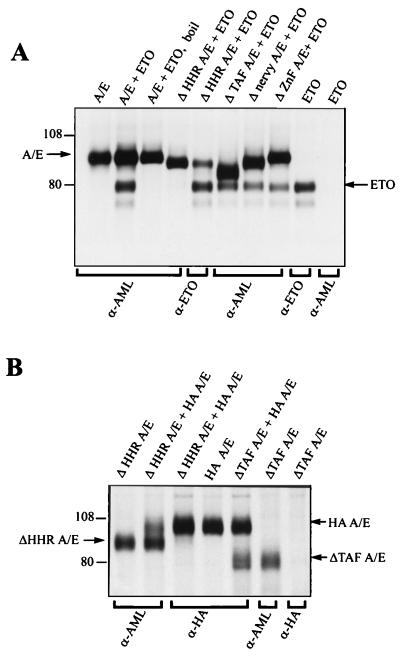
The t(12;21) fusion protein can also interfere with AML-1B-dependent activation of the TCRβ enhancer and the M-CSF-1 receptor (11, 19). This activity requires a putative repression domain that can also mediate homodimer formation (19). Because the HHR and ZnF domains of ETO are putative protein interaction domains, we asked whether AML/ETO and ETO could form heterodimers. Cos-7 cells were metabolically labeled 48 h posttransfection, and cell lysates were prepared for immunoprecipitation with the N-terminal AML-1 antibody (32). We found that ETO could be coprecipitated with AML/ETO and that this interaction was stable even when the immunoprecipitations were performed in the presence of 0.5% SDS (Fig. 6A). However, we did find that boiling the cell lysate disrupted the interaction (Fig. 6A, third lane from left). We tested our panel of AML/ETO deletion mutants and found that the HHR was the only conserved domain required for ETO to coprecipitate with AML/ETO (Fig. 6A). Control immunoprecipitation with our anti-ETO antibody revealed that ETO was expressed appropriately with ΔHHR AML/ETO but was unable to coprecipitate (Fig. 6A, fifth lane from left). Similar results were obtained upon transfecting NIH 3T3 cells (data not shown).
FIG. 6.
The HHR motif is required for AML/ETO/ETO heterodimers and AML/ETO homodimers. (A) Cos cells were transfected with 2 μg of the indicated CMV5 AML/ETO (A/E) and CMV5 ETO plasmids, and 48 h posttransfection cells were labeled with [35S]methionine as described in Materials and Methods. Equal trichloroacetic acid-precipitable counts were immunoprecipitated with the antibodies listed on the bottom of the diagram, as described in Materials and Methods. “Boil” indicates that the sample was heated to 100°C for 1 min prior to the immunoprecipitation. Protein A-Sepharose was used to collect immunocomplexes, which were then analyzed by SDS-PAGE in an 8% gel. The gel was then dried and subjected to autoradiography. (B) Cos cells were transfected with HA-AML/ETO (HA A/E) and the indicated AML/ETO deletion constructs and were labeled with [35S]methionine, immunoprecipitated, and analyzed by SDS-PAGE as described for panel A.
To test whether AML/ETO can form homodimers through the HHR motif, we tested the ability of HA-AML/ETO to interact with either AML/ETO with the TAF domain deleted (ΔTAF AML/ETO) or ΔHHR AML/ETO. ΔTAF AML/ETO was efficiently coprecipitated with HA-AML/ETO (Fig. 6B), whereas ΔHHR AML/ETO did not coprecipitate with HA-AML/ETO. Control experiments indicated that the HA antibody did not precipitate ΔTAF AML/ETO alone and that ΔHHR AML/ETO was efficiently expressed in the cell lysate (Fig. 6B). These precipitations were performed in the presence of 0.2% SDS (0.5% SDS interfered with the anti-HA antibody immunoprecipitations), again indicating the stability of interactions with the HHR motif. Taken together, these experiments reveal that the HHR mediates AML/ETO homodimer and AML/ETO-ETO heterodimer formation.
Functional analysis of AML/ETO-ETO heterodimers.
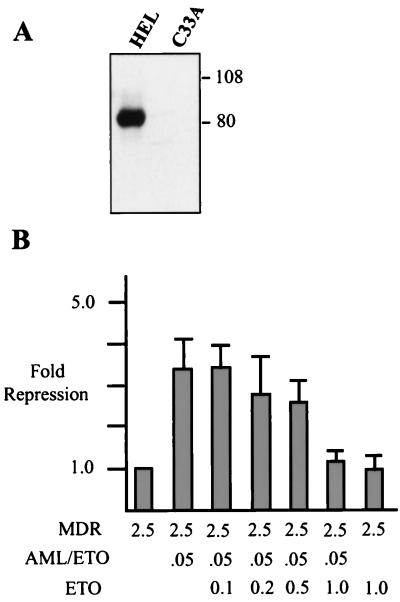
Because AML/ETO can interact with ETO, we determined whether this interaction affects AML/ETO function. This interaction is significant given that cells carrying t(8;21) also express ETO protein (10). We observed levels of AML/ETO-mediated repression of MDR-1 in HEL cells similar to those in C33A cells (Fig. 3 and 4). Therefore, we determined the levels of endogenous ETO protein in these cells by Western blot analysis using anti-ETO antibodies directed against the C-terminal domain. HEL cells express readily detectable levels of ETO, whereas C33A cells do not express detectable protein (Fig. 7A). These results suggest that high levels of ETO are not required for AML/ETO function. In fact, by comparison to the levels of AML/ETO expressed in transient transfection/repression assays, it appears that ETO may be dispensable for this activity. To directly test ETO involvement in AML/ETO function we cotransfected increasing amounts of ETO with AML/ETO and MDR-1–CAT into C33A cells. We chose C33A cells because they do not appear to express endogenous ETO. In these experiments we also transfected less AML/ETO (100 ng) so that potential increases in repression due to the addition of ETO could be observed. However, we found that cotransfection of ETO did not stimulate the ability of AML/ETO to repress, but rather at high levels inhibited AML/ETO function (Fig. 7B).
FIG. 7.
High expression levels of ETO can inhibit AML/ETO function. (A) Western blot analysis of ETO expression in HEL and C33A cells. Cell lysates (150 μg) from HEL or C33A cells were fractionated by SDS-PAGE in an 8% gel, transferred to nitrocellulose, and blotted with ETO antiserum as described in Materials and Methods. (B) C33A cells were transfected with 2 μg of MDR-1, 0.1 μg of CMV5 AML/ETO, 100 ng of CMV β-galactosidase, and increasing amounts of CMV5 ETO as indicated. CAT assays were quantitated and normalized to β-galactosidase activity.
AML/ETO can repress Ets-1 activation of the MDR-1 promoter.
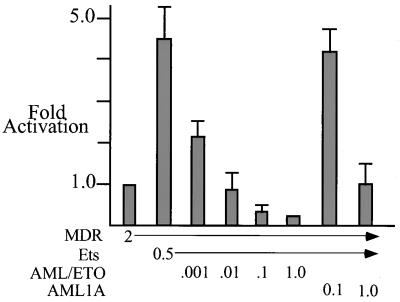
Recent work indicates that MDR1 transcription can be stimulated by Ets-1 (42a). Although MDR-1 is only modestly activated by AML-1B (approximately twofold in C33A or NIH 3T3 cells), Ets-1 can activate the promoter three- to fivefold in NIH 3T3 cells. This activation is dependent on an Ets binding site (42a) and is not observed in C33A cells, likely due to higher levels of basal activity (data not shown). Therefore, we tested the ability of AML/ETO to block Ets-1-activated MDR-1 transcription and found that AML/ETO could efficiently inhibit Ets-1-dependent activation (Fig. 8). A titration experiment revealed that higher levels of AML/ETO could repress basal transcription of MDR-1 in NIH 3T3 cells, as we observed for C33A cells (data not shown, but see the results for AML/ETO [0.1 μg] in Fig. 8). AML1A was unable to repress Ets-1 activation except at a 100-fold excess relative to AML/ETO. This repression may be due to direct physical interaction (titration) between the runt domain of AML1A and Ets-1 (15).
FIG. 8.
AML/ETO can inhibit Ets-1-mediated activation of MDR-1. NIH 3T3 cells were transfected with 2 μg of MDR-1 and 20 ng of CMV β-galactosidase. Where indicated Ets-1 (Ets; 0.5 μg) was added, along with the indicated increasing amounts of CMV5 AML/ETO or CMV5 AML1A. CAT activity was quantitated and normalized to β-galactosidase activity.
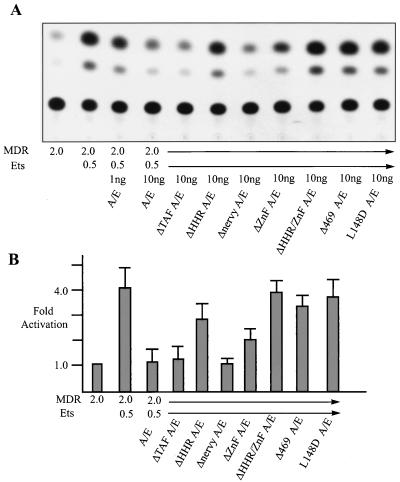
To further characterize the inhibition of Ets-1 activation, we tested a series of AML/ETO mutants. The L148D AML/ETO protein was unable to inhibit Ets-1, indicating that the DNA binding ability of the fusion protein is required for repression (Fig. 9). Δ469 AML/ETO was deficient for blocking Ets-1 function, as was ΔHHR/ΔZnF (double mutant) AML/ETO, revealing that the C-terminal ETO sequences are again required for repression. ΔHHR AML/ETO was impaired threefold relative to wild-type AML/ETO, while ΔZnF AML/ETO showed a twofold decrease relative to the wild type. Western blotting with anti-Ets-1 indicated that Ets-1 protein levels were not altered by cotransfection with AML/ETO or AML/ETO deletion mutants (data not shown). Thus, both the HHR and ZnF domains contribute to the inhibition of Ets-1 activation.
FIG. 9.
AML/ETO inhibition of Ets-1 activation requires AML/ETO DNA binding and HHR and ZnF domains. (A) NIH 3T3 cells were transfected with 2 μg of MDR-1 CAT, 20 ng of CMV β-galactosidase, 0.5 μg of Ets-1, and 1 ng of AML/ETO (A/E) or 10 ng of AML/ETO or AML/ETO deletion constructs. (B) CAT assays were quantitated and normalized to β-galactosidase activity. Because Ets-1 consistently activated β-galactosidase activity twofold, the apparent levels of activated transcription shown in panel A are reduced in panel B.
DISCUSSION
AML/ETO interferes with AML-1B-dependent transactivation of the TCRβ, interleukin 3, neutrophil protein 3, and GM-CSF promoters (13, 34, 45, 47) and AML-2 and PEBP2A1 (AML-3) activation of TCRβ (35). However, the fusion protein fails to inhibit basal transcription from these promoters, suggesting that it acts as a dominant inhibitory protein to interfere with AML family member functions. As well, AML/ETO can interfere with AML-1B and C/EBPα synergistic activation of the NP-3 promoter, but not with basal transcription (47). Thus, the results for the MDR-1 promoter represent the first example of AML/ETO repression of basal transcription.
Basal expression from the MDR-1 promoter has not been completely characterized, but in C33A cells removal of both the canonical AML-1 binding site and an Ets binding site by deleting nucleotides −58 to −137 did not significantly alter basal transcription (data not shown). Moreover, these cells have low levels of AML-1B, and overexpression of a competitive inhibitor of AML-1B failed to inhibit basal expression (Fig. 1), indicating that loss of AML-1B function is not sufficient to alter this basal transcription. While C/EBPβ has been shown to regulate the MDR-1 promoter, its DNA binding site is deleted in the promoter constructs used here (6). Mutant forms of the p53 tumor suppressor protein have been shown to activate MDR-1 (5), and C33A cells lack functional p53 (8, 44). Therefore, at least part of the high level of basal activity observed in C33A cells could be due to derepression or activation by mutant p53.
The AML/ETO HHR dimerization motif contributes to the repression of the MDR-1 promoter. However, because repression occurs in cells that contain undetectable levels of ETO, the formation of AML-1/ETO-ETO heterodimers is not required for activity. Moreover, our anti-ETO serum is directed to the MYND domain, which is nearly identical to those in other ETO family proteins, and this antiserum cross-reacts with a second ETO family member (data not shown). Therefore, it is unlikely that C33A cells express high levels of other ETO family members. While coexpression of another ETO family member enhanced AML-1/ETO repression of the TCRβ enhancer approximately twofold (23), it is unlikely that heterodimerization with other family members is required for AML-1/ETO functions.
To address the role of AML/ETO homodimers in repression, we replaced the HHR of AML/ETO with the GCN4 leucine zipper. This motif mediates homodimeric interactions of GCN4 and has been used to investigate the functional role of p53 homodimerization (41). Thus, this chimeric AML/ETO protein should only form homodimers and not associate with other heterodimeric partners, including ETO. The GCN4-modified AML/ETO was impaired for transcriptional repression to a level similar to that of the HHR deletion mutant (three- to fourfold relative to the wild type; data not shown). Although these results suggest that the HHR may function to recruit a heterologous protein, the GCN4-AML/ETO protein formed relatively weak homodimers (barely detectable by immunoprecipitation from transfected cells in buffer lacking SDS) compared to the wild-type protein (easily detectable in buffer containing SDS). Therefore, we cannot rule out the possibility that homodimers do play a role in repression.
Our results have defined a second domain in AML/ETO that contributes to transcriptional repression. Even subtle mutations that would affect the structure of the predicted ZnFs impair repression. This domain has been termed the MYND motif due to the conservation of its general structure in ETO (also known as myeloid tumor gene 8 [MTG8]) and in the Drosophila proteins nervy and DEAF-1 (17). This motif is also found in numerous other proteins and predicted proteins in mammals, yeast, and Caenorhabditis elegans (17). Only one of these proteins has been assigned a function. DEAF-1 is a transcription factor that specifically binds DNA and cooperates with Deformed to regulate transcription, although the MYND domain does not participate in DNA binding. Our results strongly suggest that this motif interacts with other proteins to negatively regulate transcription. In support of this hypothesis, we have used the yeast two-hybrid assay and coimmunoprecipitation assays to identify a specific interaction between the MYND motif and N-CoR, a corepressor that recruits histone deacetylases to repress transcription (29). Thus, the interaction with N-CoR cosegregates with the function of the MYND domain in transcriptional repression of basal and Ets-1-dependent activation of the MDR-1 promoter.
Ets family transcription factors such as Ets-1 and PU.1 are involved in proliferation and differentiation of hematopoietic cells (27). C/EBPα is also a critical regulator of granulocytic differentiation (14). In addition to AML-1B and Ets-1, AML/ETO can inhibit CEBPα transactivation and AML-1B–C/EBPα synergistic transcriptional activation (47). Therefore, the block in differentiation observed in myeloid blasts containing t(8;21) and in 32D cells overexpressing AML/ETO (47) may result from AML/ETO inhibition of genes that are normally activated by differentiation-promoting factors such as AML-1B, Ets family proteins, and C/EBPα.
The expression of the MDR-1 gene in de novo AML is a poor prognostic factor, likely due to the role of MDR in chemotherapeutic insensitivity. However, recent studies indicate that in a subset of AML [those with t(8;21)] the expression of MDR is undetectable and clearly indicative of a good prognosis. Although childhood cases that carry the t(8;21) translocation did express MDR-1 (75%), the majority of the adult cases fail to express P-glycoprotein (40). Interestingly, this correlates with therapeutic outcome, as childhood t(8;21) cases respond poorly to chemotherapy (22, 31). Whether MDR-1 is regulated differently in children versus adults is a difficult question to address; however, it appears that a fortuitous outcome of t(8;21), at least in adults, is the inhibition of MDR-1 expression, perhaps through direct repression of transcription.
ACKNOWLEDGMENTS
We thank Dana King and Yue Hou for technical assistance and Jennifer Westendorf, Randy Fenrick, Shari Meyers, and Noel Lenny for plasmids, insightful discussions, and critical evaluation of data.
This work was supported by NIH/NCI grants RO1-CA64140 and RO1-CA77274, by American Cancer Society grant JFRA-591 (to S.W.H.), by the American Lebanese and Syrian Associated Charities, by the Vanderbilt Cancer Center, and by a center grant from NCI (CA68485).
REFERENCES
- 1.Arceci R J. Clinical significance of P-glycoprotein in multidrug resistance malignancies. Blood. 1993;81:2215–2222. [PubMed] [Google Scholar]
- 2.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phophatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 3.Bloomfield C D. Prognostic factors for selecting curative therapy for adult acute myeloid leukemia. Leukemia. 1992;6:65–67. [PubMed] [Google Scholar]
- 4.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N, Eckhaus M, Marin-Padilla M, Collins F S, Wynashaw-Boris A, Liu P P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MY11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 5.Chin K-V, Ueda K, Pastan I, Gottesman M. Modulation of activity of the promoter of the human MDR1 gene by ras and p53. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 6.Combates N J, Rzepka R W, Chen Y N, Cohen D. NF-IL6, a member of the C/EBP family of transcription factors, binds and trans-activates the human MDR-1 promoter. J Biol Chem. 1994;269:29715–29719. [PubMed] [Google Scholar]
- 7.Cowell I. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 8.Crook T, Wrede D, Vousden K H. p53 point mutations in HPV negative human cervical carcinoma cell lines. Oncogene. 1991;6:873–875. [PubMed] [Google Scholar]
- 9.Dastague N, Payen C, Lafage-Pochitaloff M, Bernard P, Leroux D, Huguet-Rigal F, Stoppa A M, Marit G, Molina L, Michallet M. Prognostic significance of karyotype in de novo adult acute myeloid leukemia. The BGMT group. Leukemia. 1995;9:1491–1498. [PubMed] [Google Scholar]
- 10.Erickson P F, Dessev G, Lasher R S, Philips G, Robinson M, Drabkin H A. ETO and AML1 phosphoproteins are expressed in CD34+ hematopoietic progenitors: implications for t(8;21) leukemogenesis and monitoring residual disease. Blood. 1996;88:1813–1823. [PubMed] [Google Scholar]
- 11.Fears S, Gavin M, Zhang D E, Hetherington C, Ben-David Y, Rowley J D, Nucifora G. Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc Natl Acad Sci USA. 1997;94:1949–1954. doi: 10.1073/pnas.94.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 14.Friedman A D. Regulation of immature myeloid cell differentiation by PEBP2/CBF, Myb, C/EBP, and Ets family members. Curr Top Microbiol Immunol. 1996;211:149–157. doi: 10.1007/978-3-642-85232-9_15. [DOI] [PubMed] [Google Scholar]
- 15.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 16.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross C T, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 18.Herschbach B M, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 19.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 21.Joventino L P, Stock W, Lane N J, Daly K M, Mick R, Le Beau M M, Larson R A. Certain HLA antigens are associated with specific morphologic and cytogenetic subsets of acute myeloid leukemia. Leukemia. 1995;9:433–439. [PubMed] [Google Scholar]
- 22.Kalwinski D K, Raimondi S C, Schell M J, Mirro J, Santana V M, Behm F, Dahl G V, Williams D. Prognostic importance of cytogenetic subgroups in de novo pediatric acute nonlymphocytic leukemia. J Clin Oncol. 1990;8:75–83. doi: 10.1200/JCO.1990.8.1.75. [DOI] [PubMed] [Google Scholar]
- 23.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–494. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leith C P, Kopecky K J, Godwin J, McConnel T, Slovak M L, Chen I-M, Head D R, Appelbaum F R, Willman C L. Acute myeloid leukemia in the elderly: assessment of multidrug resistance (MDR1) and cytogenetics distinguishes biologic subgroups with remarkably distinct responses to standard chemotherapy. A Southwest Oncology Group study. Blood. 1997;9:3323–3329. [PubMed] [Google Scholar]
- 26.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 27.Lenny N, Westendorf J J, Hiebert S W. Transcriptional regulation during myelopoiesis. Mol Biol Rep. 1997;24:157–168. doi: 10.1023/a:1006859700409. [DOI] [PubMed] [Google Scholar]
- 28.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 28a.Lutterbach, B., and S. W. Hiebert. Unpublished data.
- 29.Lutterbach, B., J. J. Westendorf, A. Patten, K. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, E. Seto, and S. W. Hiebert. Mechanism of transcriptional repression by the t(8;21) fusion protein: ETO interacts with the N-CoR and mSin3 corepressors. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 30.Martin P, Papayannopoulou T. A new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 1982;216:1233–1237. doi: 10.1126/science.6177045. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Climent J A, Lane N J, Rubin C M, Morgan E, Johnstone H S, Mick R, Murphy S B, Vardiman J W, Larson R A, Le Beau M M, Rowley J D. Clinical and prognostic significance of chromosomal abnormalities in childhood acute myeloid leukemia de novo. Leukemia. 1995;9:95–101. [PubMed] [Google Scholar]
- 32.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers S, Hiebert S W. Indirect and direct disruption of transcriptional regulation in cancer: E2F and AML-1. Crit Rev Eukaryot Gene Expr. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 34.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers S, Lenny N, Sun W, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 36.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyoshi H, Shimizu K, Kou T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norton P A, Coffin J M. Bacterial B-galactosidase as a marker of Rous sacoma virus gene expression and replication. Mol Cell Biol. 1985;5:281–290. doi: 10.1128/mcb.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 40.Pearson L, Leith C P, Duncan M H, Chen I M, McConnell T, Trinkaus K, Foucar K, Willman C L. Multidrug resistance-1 (MDR1) expression and functional dye/drug efflux is highly correlated with the t(8;21) chromosomal translocation in pediatric acute myeloid leukemia. Leukemia. 1996;10:1274–1282. [PubMed] [Google Scholar]
- 41.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porwit-MacDonald A, Janossey G, Ivory K, Swirsky D, Peters R, Wheatley K, Walker H, Turker A, Goldstone A H, Burnett A. Leukemia-associated changes identified by quantitative flow cytometry. IV. CD34 overexpression in acute myelogenous leukemia M2 with t(8;21) Blood. 1996;87:1162–1169. [PubMed] [Google Scholar]
- 42a.Schuetz, J. Unpublished data.
- 43.Shurtleff S A, Buijs A, Behm F G, Rubnitz J E, Raimondi S C, Hancock M L, Chan G C, Pui C H, Grosveld G, Downing J R. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 44.Thottassery J V, Zambetti G P, Arimori D, Schuetz E G, Schuetz J D. p53-dependent regulation of MDR1 gene expression causes selective resistance to chemotherapeutic agents. Proc Natl Acad Sci USA. 1997;94:11037–11042. doi: 10.1073/pnas.94.20.11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchida H, Zhang J, Nimer S D. Am11a and Am11b can transactivate the human Il-3 promoter. J Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- 46.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T-L, Huang X, Bushweller J H, Bories J-C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 47.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1–ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]