Abstract
The most widely recognized biochemical change associated with the majority of apoptotic systems is the degradation of genomic DNA. Among the enzymes that may participate in this cleavage, the acidic cation-independent DNase II is a likely candidate since it is activated in many apoptotic cells. To better understand its role, we purified and sequenced a DNase II extracted from porcine spleen. Protein sequencing of random peptides demonstrated that this enzyme is derived from a ubiquitous serpin, the leukocyte elastase inhibitor (LEI), by an acidic-dependent posttranslational modification or by digestion with elastase. We call this novel enzyme L-DNase II. In vitro experiments with purified recombinant LEI show that the native form has no effect on purified nuclei whereas its posttranslationally activated form induces pycnosis and DNA degradation. Antibodies directed against L-DNase II showed, in different cell lines, an increased expression and a nuclear translocation of this enzyme during apoptosis. Since the appearance of the endonuclease activity results in a loss of the anti-protease properties of LEI, the transformation from LEI to L-DNase II may act as a switch of protease and nuclease pathways, each of which is activated during apoptosis.
Apoptosis, or programmed cell death, is a mechanism of cell clearance in many physiological processes such as embryogenesis, metamorphosis, and tumor regression (2). Although the signals inducing apoptosis are very different, nuclear condensation, membrane blebbing, and formation of apoptotic bodies are morphological features common to all apoptotic cells. By far the most widely recognized biochemical change is the degradation of genomic DNA. The identity of the enzymes responsible for this cleavage is the subject of considerable debate. Several endonucleases have been proposed to be responsible for DNA fragmentation. Ca2+- plus Mg2+-dependent endonucleases in thymocytes, such as NUC 18/cyclophylin A (16), DNase I (21), DNase γ (29), and a new 97-kDa DNase (18), are examples. Mg2+-dependent, Ca2+-independent endonucleases have been implicated in human myeloid cell line apoptosis (7, 8). Barry and Eastman (1) implicated DNase II, a cation-independent acidic endonuclease, as the enzyme that degrades DNA in apoptosis associated with intracellular acidification. We have shown in our laboratory the involvement of DNase II in nuclear degradation in terminally differentiating lens fiber cells (32).
To date, our knowledge of the molecular structure of DNase II is very limited. The enzymatic properties of DNase II from different tissues and animals were found to be very similar, but its physical and chemical properties showed high variability. For instance, the molecular mass of mammalian DNase II ranges between 26 and 45 kDa. The reasons for this variability remain unknown (15).
To better understand the biology of DNase II, the knowledge of its protein sequence seemed to be a mandatory step. In this study, we showed that this ubiquitous L-DNase II arises from leukocyte elastase inhibitor (LEI) by a posttranslational modification that involves a shift in the molecular weight of LEI. This shift in the apparent molecular weight of LEI is followed by a loss of its elastase-inhibiting activity and the appearance of DNase II activity. We also investigated, using cultured cells or purified nuclei, its involvement in apoptosis.
MATERIALS AND METHODS
Protein sequence.
DNase II (200 μg) was purified by polyacrylamide gel electrophoresis (PAGE) from a commercial preparation (Worthington) and transferred to a polyvinylidene difluoride membrane (Millipore). The protein was visualized by staining in 0.001% amido black diluted in 40% methanol–10% acetic acid. The 27-kDa protein was cut out and digested with trypsin or lysine endopeptidase. The resulting peptides were separated by high-pressure liquid chromatography on a DEAE-C18 column. The N-terminal peptide sequence and the sequences of selected peptides were determined by Edman degradation (Laboratoire des Biotechnologies, Institut Pasteur, Paris, France).
LEI cloning and nucleotide sequence.
Total RNA from porcine spleen was purified with TriINSTAPUR (Eurogentech) and retrotranscribed from a poly(dT) primer by reverse transcriptase from Moloney murine leukemia virus (Bethesda Research Laboratories). The cDNA was amplified by PCR with horse primers. We therefore obtained a porcine sequence that was used to design specific porcine primers. The PCR products obtained with these primers and an anchoring poly(dT) primer (dT17-AGC TAC AGC TGA GCT CAG) were cloned in pGEM.T (Promega). Four partial clones were obtained and sequenced with the universal reverse and forward IRD-41-labeled primers. Sequencing reactions were performed with Thermo Sequenase cycle-sequencing kit (Amersham) and analyzed in a Licor automatic sequencer.
Construction of the recombinant vectors and expression of porcine LEI.
The complete coding sequence was reconstituted from two partial clones in pGEM by using the unique Eco0109I site. Isolation of plasmid DNA, conditions for digestion with restriction enzymes, and agarose gel electrophoresis were as described previously (27). Expression of the recombinant protein with the SP6 transcription promoter of pGEM vector was carried out in the presence of [35S]methionine in an in vitro transcription-translation system with rabbit reticulocyte lysate as indicated by the manufacturer (25).
A prokaryote expression system was constructed by inserting the cDNA of porcine LEI into NcoI and XhoI restriction sites of pET 23d(+) (Novagen). Escherichia coli BL21 was electroporated and grown in Luria-Bertani medium. The synthesis of LEI was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 10 mM. The resulting protein bears a polyhistidine tag at its N-terminal end, which allows purification with His-Bind resin in the presence of 6 M urea, as specified by the manufacturer (Novagen Inc.) (17).
Southern Blot analysis.
A 5-μg portion of porcine total genomic DNA was digested with PstI, EcoRI, and HindIII overnight at 37°C. The fragments obtained were separated on a 1% agarose–Tris-acetate-EDTA (TAE) gel and transferred by capillary blotting onto a Hybond N+ Amersham membrane. The membrane was prehybridized and hybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–0.5% sodium dodecyl sulfate (SDS) for several hours. The probe used was a 550-bp DNA fragment (from nucleotides [nt] 409 to 850 of the porcine DNA coding sequence), labeled by [32P]dCTP random priming as specified by the manufacturer (Amersham) and added to a final concentration of 106 cpm/ml. Filters were washed at low stringency and exposed for autoradiography at −80°C, using Kodak X-Omat S film, between two intensifying screens.
Northern blot analysis.
Total RNA was electrophoresed in a 1% agarose gel in a formaldehyde running buffer and transferred to a Hybond N+ membrane (Amersham). Transfer and hybridization were carried out as described previously (27). The blots were then washed twice for 15 min in 1× SSC–0.1% SDS at room temperature and twice for 15 min in 0.25× SSC–0.1% SDS at 50°C. The membranes were exposed for 1 week as above. For the probe, 25 ng of LEI complete coding sequence was prepared as above.
Reverse transcription-PCR (RT-PCR) and nested PCR.
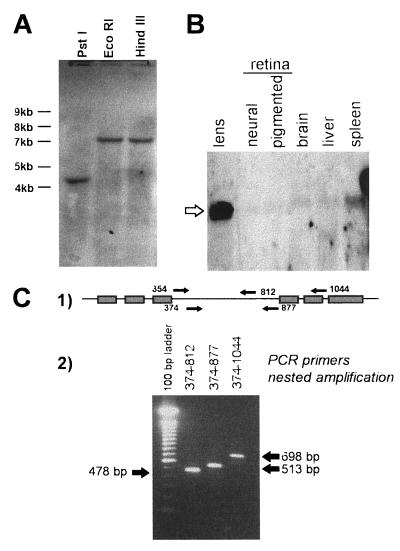
mRNA was purified from 150 μg of porcine spleen total RNA with poly(dT)-containing magnetic beads (Dynal). Retrotranscription was performed as above. mRNA was eliminated by incubation for 1 h at 65°C in 0.2 M NaOH, and free nucleotides were eliminated on a spun column (Pharmacia Biotech). Poly(dG) was added at the 5′ end of the cDNA with a polynucleotide terminal transferase (Boehringer Mannheim). The first PCR step was performed with primers oligo(dT) and 354; nested PCR was performed with primers 374 plus 812, 374 plus 877, and 374 plus 1044. The primer sequences are as follows: 354, 5′-AAA CCT ACG GGG CTG AAC TG-3′; 374, 5′-AGC GTG GAC TTC CTG CGG-3′; 812, 5′-ACC TCG GGC AGT GGA CAT TG-3′; 877, 5′-AAA GAG ATC CTG CAC GCC CA-3′; 1044, 5′-CAT TGA AAT TTT CCT CTG GCA-3′.
Posttranslational modification of the recombinant protein.
The recombinant protein produced by the reticulocyte lysate was incubated at different pHs overnight. The different pHs were obtained with the following reagents: pH 1.0, 50 mM H2SO4; pH 2.0, 50 mM H2SO4 (adjusted with NaOH); pH 4.0, 50 mM acetic acid; pH 6.0, 50 mM H3PO4. Alternatively, the mixture was diluted in phosphate-buffered saline (PBS) and digested overnight with 1 μg of bovine elastase (Sigma). A 3-μl volume of each sample was mixed with 30 μl of Laemmli sample buffer and loaded on a 12% polyacrylamide gel. The gel was subsequently fixed, treated with En3Hance enhancer (NEN Bioproducts), and exposed for autoradiography as above.
Porcine spleen crude extract.
A 2-g portion of porcine spleen was homogenized in 20 ml of 25 mM Tris–1 mM EDTA (pH 7.4) containing 1 M NaCl and centrifuged for 15 min at 13,000 × g, and the supernatant was dialyzed overnight against the same buffer without NaCl. The protein concentration, as determined by the bicinchoninic acid method (Pierce), was 4.5 mg/ml.
DNase II activity assays.
A 50-μl volume of the pH 2-treated protein (obtained from reticulocyte lysate or from E. coli) was incubated in a final volume of 600 μl containing 10 mM Tris and 10 mM EDTA (pH 5.5) with 25 μg of genomic DNA. Aliquots (100 μl) were ethanol precipitated at different incubation times, resuspended, and loaded on a 1% agarose gel. DNase activity on plasmid DNA was assayed under the same conditions with 4.5 μg of pGEM and 5 μl of the acid-treated protein.
Anti-elastase activity.
The anti-elastase activities of the 42-kDa protein and of its posttranslational product were evaluated by the capacity of these polypeptides to bind to elastase (5). A 6-μl volume of the posttranslational reaction mixture was incubated in PBS containing 414 ng of elastase. The reaction was allowed to proceed for 5 min at 20°C and then stopped by the addition of twofold-concentrated Laemmli sample buffer without S-S reducing agent. The samples were then loaded on a 12% polyacrylamide gel and subjected to autoradiography.
Western blot analysis.
Polyclonal antibody against DNase II and immunoblot analyses were performed as described previously (32).
Effect of LEI and pH 2-treated LEI on purified nuclei.
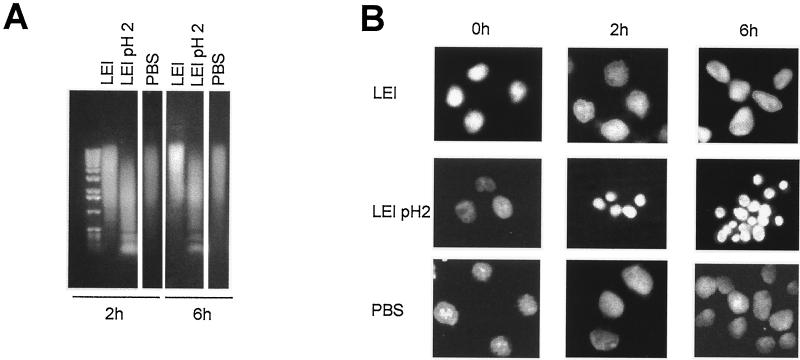
Baby hamster kidney (BHK) cells were grown as a monolayer in Dulbecco’s modified Eagle’s medium (GIBCO-BRL) supplemented with 10% fetal calf serum, 4 mM glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml (all from GIBCO-BRL) at 37°C in a humidified atmosphere containing 5% CO2. The cells were detached with trypsin, rinsed in PBS, and extracted by incubation in 1.5 mM MgCl2 for 15 min at 4°C. They were then subjected to five strokes in a Dounce homogenizer and washed by centrifugation three times in 1.5 mM MgCl2. The obtained pellet containing nuclei was stored in 10 mM Tris (pH 7.4)–200 mM sucrose–60 mM NaCl at a concentration of 108 nuclei/ml. To test the effects of LEI, 106 nuclei were incubated for different periods with 280 ng of E. coli-produced LEI, treated overnight at pH 2.0 or untreated. Negative control experiments were done with PBS. The reaction mixture was adjusted to DNase II ionic conditions by adding 2 μl of a 10-fold appropriate buffer (see above). After incubation of these mixtures at 37°C for 0, 2, or 6 h, nuclei were prepared for morphological or DNA analysis.
To study DNA degradation, the reaction was stopped by the addition of 20 μl of 10 mM Tris (pH 7.4)–100 mM NaCl–25 mM EDTA–1% Sarkosyl–5 μl of proteinase K (10 μg/μl). The mixture was incubated overnight at 37°C, 30 μg of DNase-free RNase was added, and the mixture was incubated for an additional 1 h. Samples were then loaded on a 1% agarose gel as above.
To analyze nuclear morphology, nuclei were washed in PBS at the end of the incubation time, stained with 4′,6-diamidino-2-phenylindole (DAPI), spread on a microscope mounting plate, covered with a coverslip, and analyzed under a Leitz Aristoplan microscope.
The same experiments were performed with 10 μl of LEI produced in reticulocyte lysate treated at pH 2.0 or left untreated. Control experiments with PBS and reticulocyte lysate crude extracts treated at pH 2 were also performed. Nuclear incubation, DNA analysis, and nuclear morphology investigation were done as described above.
Induction of apoptosis in cultured cells.
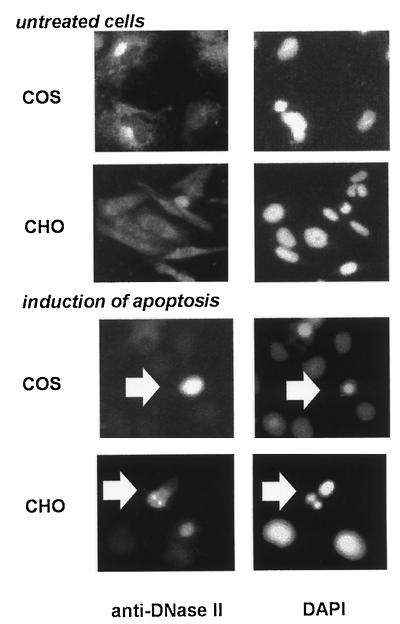
Chinese hamster ovary (CHO) cells and COS-7 cells were grown as a monolayer in Dulbecco’s modified Eagle’s medium Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 4 mM glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml at 37°C in a humidified atmosphere containing 5% CO2. The cells were seeded on coverslips (5,000 cells/cm2), maintained in culture for 2 days, and then treated for 1 h with 1 nM Thapsigargin (COS-7 cells) or incubated in the same medium for 6 days (CHO cells).
Immunocytochemistry.
Cells were fixed in 4% paraformaldehyde for 15 min, washed in PBS, and permeabilized with 0.3% Triton X-100 in PBS for 30 min. After being washed with PBS, the cells were saturated for 30 min at room temperature with PBS containing 1% skim milk and then incubated for 1 h at room temperature with anti-DNase II (1/100) in PBS containing 0.1% skim milk (32). This incubation was followed by five washes for 5 min per wash in PBS–0.1% skim milk. The antibody was localized with tetramethylrhodamine-5-isothiocyanate (TRITC) goat anti-rabbit immunoglobulin G (1/500 dilution) in PBS–0.1% skim milk (1 h room temperature). The cells were then washed five times for 5 min in PBS. During the last wash in PBS, nuclei were stained with the fluorescent nuclear stain DAPI and then mounted with 50% glycerol in PBS. The cells were evaluated under a Leitz Aristoplan microscope equipped with an epi-illuminator HBO and filters for rhodamine and DAPI fluorescence. They were photographed with Ilford HP5 film (400 ASA). Control experiments with anti-DNase II preimmune serum and PBS–0.1% skim milk instead of anti-DNase II were also performed.
RESULTS
Nucleotide sequence of DNase II.
DNase II obtained from porcine spleen (Worthington) was purified by PAGE and digested by lysine-endopeptidase and trypsin. The resulting peptides were separated by high-pressure liquid chromatography and sequenced by Edman degradation. The N-terminal sequence was also determined. We obtained the sequence of seven peptides, representing 78 amino acids and accounting for about one-third of the 27 kDa estimated for DNase II. An identity search in data bank showed that the sequence of these peptides all corresponded to the LEI (31), an antiprotease of the serpin superfamily. The LEI coding sequence is known for humans and horses but unknown for pigs (9, 26). By using PCR primers from the horse (Fig. 1), we amplified pig spleen cDNA and obtained the complete nucleotide coding sequence of porcine LEI. At the nucleotide level, the open reading frame showed 81% homology between pigs and horses. The mRNA from porcine spleen showed three putative polyadenylation signals (Fig. 1). At the protein level, our deduced sequence showed a difference of two amino acid substitutions with respect to the previously published protein (31).
FIG. 1.
Nucleotide and deduced protein sequence of porcine LEI. The sequences of internal peptides obtained by Edman degradation are indicated by a dotted underline. The obtained N-terminal sequence of DNase II is indicated by a continuous underline. Grey boxes represent the localization of the nucleotide sequences from horse LEI used to clone porcine LEI. Open boxes represent the difference between our deduced sequence and the already published sequence for porcine LEI. The arrow indicates the Eco0109I site. ∗∗, the P1-P1′ site. Presumptive polyadenylation sites are indicated by a double underline. The most frequently used site is located at the 3′ end.
Since DNase II and LEI had different biological activities and molecular weights, the existence of different genes carrying related sequences was verified by genomic Southern blot analysis (Fig. 2A). Total pig genomic DNA was digested with three restriction enzymes, transferred, and probed with a 550-bp DNA fragment from the middle region of the LEI coding sequence (from nt 409 to 850). After washing at low stringency, only one band was seen in each digestion.
FIG. 2.
Southern blot, Northern blot, and nested-PCR analyses. (A) A 25-μg portion of porcine DNA was digested with PstI, EcoRI, and HindIII, loaded onto a 1% agarose gel, and transferred to an N+ membrane. The membrane was hybridized with a 32P-labeled probe of porcine LEI. A single band is detectable in each lane. (B) Total mRNAs from porcine lens, neural retina, pigmented retina, brain, liver, and spleen were separated on a 1% agarose gel, transferred to a N+ membrane, and labeled with a 32P-labeled probe of porcine LEI. The membrane was then subjected to autoradiography. The open arrow indicates the band of 2,200 bases labeled in each tissue. (C) poly(A)+ mRNA from porcine spleen was retrotranscribed and amplified with primers 354 and poly(dT). A second amplification was performed with three pairs of internal primers (labeled 1), where the line represents LEI mRNA and the boxes indicate the positions of the peptides sequenced by Edman degradation of DNase II. The analysis of the second amplifications is also shown (labeled 2).
DNase II displayed the N- and C-terminal regions, of LEI but they had different molecular masses: 27 and 42 kDa, respectively. An alternative splicing of LEI mRNA might therefore be responsible for the synthesis of DNase II. To verify this hypothesis, we studied the mRNA from different porcine tissues (lens, neural retina, retinal pigmented epithelium, brain, liver, and spleen) by Northern blot hybridization with a radioactively labeled cDNA probe of LEI (Fig. 2B). In all the tissues, a single band of 2,200 nt was labeled, corresponding to the size expected for LEI transcript polyadenylated at the 3′ foremost site. No smaller mRNA was recorded. Since the presence of a rare alternatively spliced transcript would not be detected by this technique, we studied porcine spleen mRNA by RT-PCR. Figure 2C shows the result of a representative experiment: poly(A)+ mRNA was retrotranscribed and then amplified with primers 354 and poly(dT). The product of this amplification was amplified a second time with internal primers (primers 374 plus 812, 374 plus 877, and 374 plus 1044). Only one band, corresponding to the mRNA size expected for full-length LEI, was observed. Similar experiments performed in the 5′ region (nt 35 to 392) gave identical results (not shown).
In vitro and in vivo expression of DNase II.
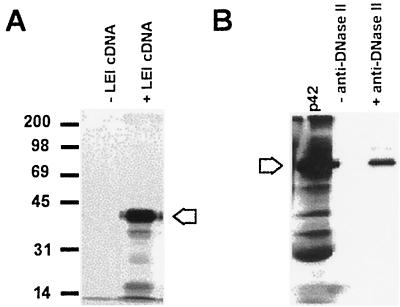
These results raised the possibility that DNase II was derived from LEI by a posttranslational modification, leading to a size shift. To verify this hypothesis, the complete coding sequence was reconstructed in a pGEM plasmid. The protein was then expressed in an in vitro transcription-translation system with a TNT rabbit reticulocyte lysate (Promega). The reaction was performed in the presence of [35S]methionine. The major protein band obtained (42 kDa) (Fig. 3A), was not recognized in Western blots but could be immunoprecipitated by an antibody raised against the 27-kDa DNase II (32) (Fig. 3B).
FIG. 3.
Expression of porcine LEI in vitro. (A) The cDNA of porcine LEI was inserted in the pGEM vector and expressed with Promega reticulocyte lysate. The reaction was allowed to proceed for 1.5 h at 30°C in the presence or absence of plasmid DNA. A 3-μl volume of reaction mixture was mixed with the same volume of 2× Laemmli sample buffer. The samples were separated on a 12% acrylamide gel and treated for autoradiography. The arrow indicates the 42-kDa main band. (B) A 5-μl volume of reticulocyte lysate containing [35S]methionine-labelled LEI was immunoprecipitated in the presence or absence of anti-DNase II and then analyzed by PAGE and autoradiography. The arrow indicates the 42-kDa band.
An aliquot of this protein was incubated at 37°C under DNase II-activating conditions (10 mM Tris, 10 mM EDTA [pH 5.5]), with either a plasmid or genomic DNA. No degradation of the plasmid or the genomic DNA was recorded even after overnight incubation (data not shown).
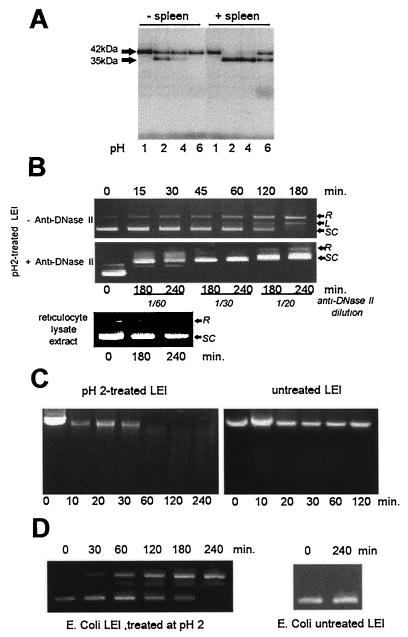
Since the DNase II purification method of Bernardi et al. (3) included the exposure of spleen extracts to an acidic pH, we incubated the 35S-labeled protein at 37°C overnight at pH 1, 2, 4, and 6 (Fig. 4A, lanes − spleen). A band shift from 42 to 35 kDa was observed mainly at pH 2 but also at pH 4. Although we obtained the band shift with strong-acid treatment in vitro, some soluble factors might permit this maturation under physiological pH conditions in the cell. Indeed, the incubation of labeled LEI in the presence of a crude spleen extract enhanced the production of the 35-kDa band and made the maturation possible at pH 6 (Fig. 4A, +spleen).
FIG. 4.
Posttranslation modification of porcine LEI. (A) Porcine LEI was expressed and [35S]methionine labeled with the TNT Promega reticulocyte lysate system. Aliquots of 3 μl were treated at 37°C overnight at different pHs in the presence or absence of crude porcine spleen extract. The reaction was stopped by adding the same volume of 2× Laemmli sample buffer. The samples were then separated on a 12% acrylamide gel and subjected to autoradiography. (B) A 4.5-μg sample of a supercoiled plasmid (pGEM) was incubated in 10 mM Tris–10 mM EDTA (pH 5.5) with 5 μl of the acid-treated protein; aliquots were ethanol precipitated at different incubation times, resuspended, and loaded on a 1% agarose gel (SC, supercoiled; L, linear; R, relaxed). The same experiment was performed, in the presence of anti-DNase II or with the TNT reticulocyte lysate alone, after treatment at pH 2.0. (C) DNase activity of pH 2-treated or untreated LEI was measured as in panel B, with genomic DNA as the substrate. (D) DNase activity was tested as in panel B, with a pH 2-treated LEI produced by E. coli and purified by the His-Bind system (Novagen).
Since the 42-kDa protein did not have endonuclease activity, we verified whether the appearance of the 35-kDa band was related to a DNase II activity. An aliquot of LEI incubated at pH 2 overnight was tested for DNase II activity on plasmid (Fig. 4B) and genomic (Fig. 4C) DNA. Both plasmid and genomic DNA were digested. Control samples containing untreated LEI, as well as the reticulocyte lysate extract treated at pH 2.0 (Fig. 4B and C), showed no DNA-cleaving activity. In addition, polyclonal anti-DNase II was able to inhibit this enzymatic activity (Fig. 4B, +Anti-DNase II). In the presence of the antiserum, a complex was formed with DNA, leading to a slower migration of plasmid DNA in the gel. The same results were obtained with the recombinant protein produced and purified from E. coli (Fig. 4D). No activity was found in bacterial extracts not expressing LEI treated at pH 2. Therefore, we had identified a protein with DNase II activity, derived from LEI. We called this protein LEI-derived DNase II (L-DNase II).
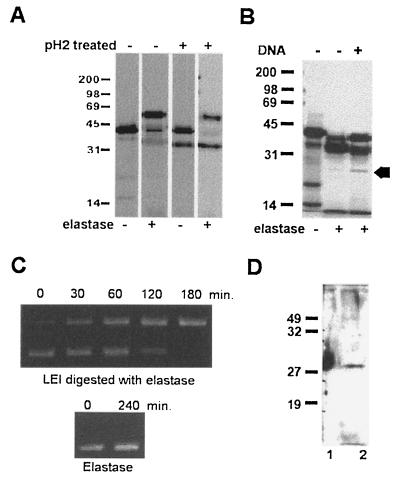
Since the sequences responsible for the elastase inhibitory action (10) were present in L-DNase II, we investigated if the anti-elastase activity remained in the 35-kDa protein (Fig. 5A). The formation of an SDS-resistant complex between the protease and its inhibitor is a mandatory condition for the anti-elastase activity (24). Therefore, we incubated either the 42-kDa labeled LEI or the acid-treated labeled protein with elastase and used PAGE to separate the complexes formed. We observed a shift of the 42-kDa band (corresponding to the native LEI protein) to about 66 kDa, the expected size for the LEI-elastase complex (42 kDa for LEI and 24 kDa for elastase). In contrast, no shift of the 35-kDa band was observed.
FIG. 5.
Posttranslational modification induced by elastase and DNA. (A) pH 2-treated and untreated LEI were incubated in PBS in the presence or absence of 414 ng of elastase. The reaction was allowed to proceed for 5 min at 20°C and then stopped by the addition of 2× Laemmli sample buffer without S-S reducing agent. The samples were then loaded on a 12% polyacrylamide gel and subjected to autoradiography. (B) Porcine LEI was expressed and [35S]methionine labeled with the TNT Promega reticulocyte lysate system. Aliquots of 3 μl were treated at 37°C overnight with elastase alone or with elastase followed by a second incubation overnight with DNA. The arrow indicates the 27-kDa band. The reaction was stopped by adding the same volume of 2× Laemmli sample buffer. The samples were then separated on a 12% acrylamide gel and treated for autoradiography. (C) A 4.5-μg portion of a supercoiled plasmid (pGEM) was incubated in 10 mM Tris–10 mM EDTA (pH 5.5) with 5 μl of LEI produced by E. coli previously digested overnight with elastase. Aliquots were ethanol precipitated for different incubation times, resuspended, and loaded on a 1% agarose gel. Similar results were obtained with LEI produced by the TNT system (not shown). (D) Porcine LEI treated with elastase and DNA as above (right-hand lane of panel B) was transferred to a membrane and visualized with polyclonal anti-DNase II. Lanes: 1, commercial DNase II; 2, LEI after treatment with elastase and DNA.
These results indicated that the acid treatment of LEI induced a band shift from 42 to 35 kDa and that this determined the appearance of DNase II activity and the loss of anti-elastase activity.
Because the acid treatment is unphysiological, we submitted the 42-kDa protein to different treatments at neutral pH. Among these, we found that the incubation of LEI with elastase (its preferred substrate) led to a modification similar to the acid treatment, i.e., a shift from 42 to 35 kDa (Fig. 5B). The protein treated with elastase also had DNase II activity (Fig. 5C).
It is worthwhile noting that the major protein appearing after acid and elastase treatment was the 35-kDa protein, while the originally sequenced DNase II was 27 kDa. Since measurement of the DNase activity involved incubation of the 35-kDa protein with DNA, we studied the influence of DNA on p35. Figure 5B shows that this treatment induced the appearance of a band at 27 kDa, which was recognized in Western blots by polyclonal anti-DNase II (Fig. 5D). Note that the p42 and p35 bands, although present at larger amounts, were not recognized by anti-DNase II. Enzyme-linked immunosorbent assays (data not shown) indicated that this antibody had higher affinity for DNase II than for LEI.
Induction of nuclear modification by L-DNase II.
To verify if L-DNase II could be responsible for nuclear changes related to apoptosis, we incubated L-DNase II with purified nuclei from normal BHK cells. These experiments were performed with both the protein produced by the reticulocyte lysate system and the recombinant LEI purified from bacteria (Fig. 6). Nuclei (106) were incubated for 0, 2, or 6 h at 37°C in the presence of LEI produced in bacteria, LEI treated at pH 2, and PBS. All the samples were loaded onto an agarose gel (Fig. 6A) or DAPI stained for morphological studies (Fig. 6B). As the time of incubation increased, a mild degradation of DNA was seen in nuclei incubated with either PBS or untreated LEI, but a nucleosomal ladder was seen in LEI acid-activated samples. No major modification in the shape of the nuclei was seen in the presence of PBS or native LEI (Fig. 6B), but a nuclear condensation similar to apoptotic nuclei (4) was observed in the sample incubated with pH 2-treated LEI. The same modifications were seen in nuclei incubated with purified DNase II used as a positive control (data not shown). Similar results were obtained with LEI synthesized by the reticulocyte lysate system (data not shown).
FIG. 6.
Induction of apoptotic morphology by posttranslationally modified LEI. Purified nuclei from BHK cells were incubated for 0, 2, or 6 h in the presence of LEI produced by E. coli without further treatment (LEI), LEI treated overnight at pH 2.0 (LEI pH 2), or PBS. (A) Neutral agarose gel of BHK purified nuclei after incubation for 2 or 6 h in the presence of the different proteins. (B) The purified nuclei were treated as above, stained with DAPI, and observed with a fluorescence microscope.
L-DNase II in apoptotic cells.
We further investigated the implication of L-DNase II in apoptotic cultured cells by using two cell lines and two different inductors of apoptosis. After induction of apoptosis, the cultured cells were fixed and treated for immunofluorescence with anti-DNase II (fully characterized in reference 32). Representative results with COS and CHO cells are shown in Fig. 7. L-DNase II has a cytoplasmic location in control cells. After induction of apoptosis in COS cells by the Ca2+-ATPase inhibitor Thapsigargin, we observed a strong increase in immunoreactivity in the apoptotic cells (seen by nuclear staining with DAPI). In CHO cells, apoptosis was induced by long-term culture. Although the increase of immunoreactivity in this cell line is questionable, a nuclear translocation of L-DNase II is clearly seen in apoptotic cells.
FIG. 7.
Involvement of L-DNase II in apoptosis. L-DNase II was located in the cytoplasm of COS and CHO control cells (upper panel, left), compared to the nuclear staining DAPI (right). Apoptosis was induced in COS cells with the Ca2+-ATPase inhibitor, Thapsigargin (1 nM) for 1 h or by long-term culture in CHO cells (lower panel) Apoptotic COS cells show an increased reactivity of L-DNase II; apoptotic CHO cells show a nuclear translocation of the enzyme.
DISCUSSION
L-DNase II is encoded by LEI mRNA.
DNase II was studied from an enzymatic point of view in the early 1970s. Later, interest in this nuclease was lost. Recently, its involvement in apoptosis (1, 32) brought it to our attention. To clone the cDNA, we determined the amino acid sequence of seven peptides derived by proteolysis from DNase II. This allowed us to determine the sequence of about one-third of the 27-kDa DNase II. Astonishingly, the sequences obtained belong to porcine LEI, a protein from the serpin superfamily (for this reason, we call the enzyme L-DNase II). The attempt to sequence the N-terminal part of DNase II resulted in a sequence belonging to the C-terminal region of LEI (Fig. 1). This result may have two explanations: (i) this sequence actually represents the N-terminal sequence of L-DNase II (in this case, L-DNase II could result from a novel organization of LEI), or (ii) this result is an artifact. Of these two hypothesis, we favor the second for two reasons. (i) Teschauer et al. (31) have reported that LEI is blocked at its N-terminal end. They show that only the peptide resulting from the natural cleavage of LEI (P1-P1′) may be directly sequenced by Edman degradation. This sequence corresponds to our N-terminal sequence. (ii) DNase II may arise from gene duplication and/or reorganization of LEI at the chromosomal level (frequently described in the serpin superfamily [6]). Southern blotting of genomic DNA probed with an LEI cDNA shows the presence of one band in all the digestions, suggesting the presence of only one gene coding for this sequence. In addition, the labeled bands do not display very high molecular weights. This makes the possibility of the existence of two genes next to each other very low.
Since L-DNase II is smaller than LEI and displays the N- and C-terminal regions of LEI, we raised the hypothesis of the existence of alternative splicing of LEI mRNA leading to the production of L-DNase II. The Northern blotting and RT-PCR (which allows the detection of very poorly expressed mRNA) studies demonstrated the presence of only one mRNA with the size expected for the full-length LEI.
L-DNase II is derived from LEI by a posttranslational modification.
The results described above suggest that L-DNase II might be derived from LEI by a posttranslational modification. Therefore, we expressed a recombinant LEI by using an in vitro transcription-translation system. We obtained a single protein showing the molecular weight and properties of LEI (i.e., formation of a complex with elastase, a mandatory condition for the elastase inhibitory activity) (24). Since the DNase II purification procedure of Bernardi et al. (3) includes an acidic extraction, we have incubated LEI at different acidic pHs. This treatment produced a 35-kDa protein that is correlated with the appearance of DNase II activity. In addition, this protein has lost its capacity to bind elastase. This indicates that L-DNase II is derived from LEI by a posttranslational modification. It is important to note that soluble factors present in spleen extract facilitate this transition. Indeed, in the presence of this extract, the band shift was observed at pH 6, making this process likely to occur in apoptotic cells, which are subjected to a decrease in intracellular pH. This may explain why cytoplasmic alkalinization inhibits apoptosis in some apoptotic models (19, 23, 33).
Three hypotheses can be put forward to explain the nature of the posttranslational modification leading to production of L-DNase II from LEI. (i) LEI belongs to the family of serpin proteins. Several proteins of this family are known to have “relaxed” and “stressed” states which modulate their activity (12, 30) and are correlated with different apparent molecular weights. This may be also the case for LEI. However, the molecular weight shift is too large to be explained by conformational changes alone, and we see this band shift under denaturing conditions (SDS-PAGE). (ii) The transition from LEI to L-DNase II may be the result of a proteolytic cleavage of N- or C-terminal regions. In fact, the elastase releases a peptide at the N-terminal end of the protein (31) and mimics the exposure of p42 to acidic pH (i.e., a band shift from p42 to p35). This result favors the hypothesis of proteolytic cleavage as the most likely mechanism. Nevertheless, it must be noted that the peptide (4 kDa) liberated from LEI by elastase is found in L-DNase II, and it alone cannot account for the loss in molecular mass. This suggests that the elastase cleavage does not induce the complete transition. We believe that the transition from p42 to p27 takes place in two steps. The first step, induced by acidification (and similar to the elastase cleavage), determines the formation of an intermediate form, p35, that has lost its antiprotease activity. The second step, induced by DNA, completes the modification to p27 L-DNase II. In this model, L-DNase II may be an element of the proteolytic cascade activated in apoptosis (20). (iii) Since we were unable to sequence any polypeptide from the central region of LEI, suggesting the loss of this region in the mature protein, the existence of splicing of the protein might also be hypothesized. Protein splicing is a posttranslational modification in which, in a precursor protein, an internal segment is excised and the external domains are joined by a peptide bond. This process has already been described in prokaryotes and lower eukaryotes and concerns proteins that are often related to DNA metabolism (35).
L-DNase II is involved in apoptosis.
DNase II has been partially purified from human gastric mucosa, cervix, and urine (36, 37). In 1985, Liao (14) purified a different enzyme displaying DNase II activity from porcine spleen and established the sequence of its active site. We presume that two or more enzymes with similar activities may exist, as is the case for DNase I (28, 38). The involvement of this DNase in apoptosis was investigated in purified nuclei (a method previously used to identify nucleases involved in nuclear degradation during apoptosis [18]) and in several cell lineages by using different inductors of apoptosis. We found that L-DNase II induces the cleavage of DNA into an oligonucleosomal ladder, a hallmark of apoptosis, and the appearance of nuclear pycnosis, resembling morphological changes seen in apoptosis. In cellular models, an increase of the immunoreactivity for L-DNase II and a nuclear translocation of the enzyme are seen during apoptosis. These facts further support the role of L-DNase II in this process and are in agreement with previous results in differentiating lens cells (32).
L-DNase II activation and the current models of apoptosis.
The finding that L-DNase II is derived from LEI is very interesting for the understanding of the molecular mechanisms involved in apoptosis. Two main conclusions can be highlighted. (i) It expands the participation of the serpin superfamily in apoptosis. It has been shown that several human and viral serpins can regulate apoptosis. The best characterized are the cytokine response modifier gene A (crmA), which inhibits interleukin-1β converting enzyme, and the plasminogen activator inhibitor type 2, which inhibits apoptosis induced by tumor necrosis factor (22). (ii) Wright et al. (34) showed the implication of an elastase-like protease activated during apoptosis of U937 cells. Several proteases, such as caspases and other noncaspase proteases, have also been implicated in this phenomenon (39).
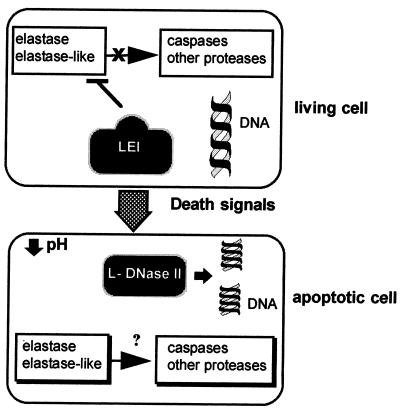
We propose that LEI plays a critical role in apoptosis (Fig. 8), acting as a molecular switch between living cells and apoptotic cells. Its double function is to prevent the proteolytic cascade of apoptosis in living cells while, via the transition to DNase, releasing this proteolytic inhibition and inducing nuclear degradation in apoptotic cells. We hypothesize that the major control element operating this switch is the intracellular pH. When the intracellular pH decreases in apoptotic cells, the transition from LEI to L-DNase II is made possible. The presence of cofactors (probably acting as other control elements) completes the process. In forms of apoptosis lacking acidification (13), LEI may be a simple substrate for elastase-like activity. This may induce the activation of L-DNase II by an alternative pathway.
FIG. 8.
Hypothetical role of L-DNase II in the apoptotic pathway. In a living cell, LEI is in its native (p42) form. No L-DNase II activity is present in the cell, and protease activities are inhibited by the LEI anti-protease action. During apoptosis, cytoplasm acidification induces the posttranslational modification of LEI, leading to the loss of anti-protease activity and the appearance of L-DNase II. Two degradation pathways are then activated: the endonuclease pathway, by generating L-DNase II, and the protease pathway, by releasing its inhibition.
These findings establish for the first time a possible link between the activation of endonucleases and proteases involved in apoptosis. In addition, we show here that this link between these systems may not be linear (i.e., activation of proteases leads to activation of endonucleases), as currently believed (4, 11), but that both pathways might be activated at the same time. The discovery of this pathway gives a new insight into apoptosis research and may lead to the development of new tools to regulate cell death.
ACKNOWLEDGMENTS
We acknowledge Pascal Egger and Delphine Goux for their help with DNA sequencing and David McDevitt for correcting the English in the manuscript.
A.T. was supported by Retina France-AFRP and Rhône-Poulenc Rorer, and P.P. was supported by Association Claude Bernard and Association pour Recherche sur le Cancer-ARC.
REFERENCES
- 1.Barry M A, Eastman A. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch Biochem Biophys. 1993;300:440–450. doi: 10.1006/abbi.1993.1060. [DOI] [PubMed] [Google Scholar]
- 2.Bellamy C O C, Malcomson R D G, Harrison D J, Wyllie A H. Cell death in health and disease: the biology and regulation of apoptosis. Semin Cancer Biol. 1995;6:3–16. doi: 10.1006/scbi.1995.0002. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi G, Bernardi A, Chersi A. Studies on acid hydrolases. I. A procedure for the preparation of acid deoxyribonuclease and other acid hydrolases. Biochim Biophys Acta. 1966;129:1–11. [PubMed] [Google Scholar]
- 4.Chinnaiyan A M, Dixit V M. The cell death machine. Curr Biol. 1996;6:555–562. doi: 10.1016/s0960-9822(02)00541-9. [DOI] [PubMed] [Google Scholar]
- 5.Christensen S, Valnickova Z, Thögersen I B, Pizzo S V, Nielsen H R, Roepstorff P, Enghild J J. Sodium dodecyl sulfate-stable complexes between serpins and active or inactive proteinases contain the region COOH-terminal to the reactive site loop. J Biol Chem. 1995;270:14859–14862. doi: 10.1074/jbc.270.25.14859. [DOI] [PubMed] [Google Scholar]
- 6.Gettins P, Patston P, Olson S. Serpins: structure, function and biology. New York, N.Y: Springer Editor; 1996. [Google Scholar]
- 7.Kawabata H, Anzai N, Masutani H, Hirama T, Hishita T, Dodo M, Masuda T, Yoshida Y, Okuma M. Mg2+- or Mn2+-dependent endonuclease activities of human myeloid leukemia cells capable of producing nucleosomal-size DNA fragmentation. Biochem Biophys Res Commun. 1997;233:133–138. doi: 10.1006/bbrc.1997.6362. [DOI] [PubMed] [Google Scholar]
- 8.Kawabata H, Anzai N, Masutani H, Hirama T, Yoshida Y, Okuma M. Detection of Mg2+-dependent endonuclease activity in myeloid leukemia cell nuclei capable of producing internucleosomal DNA cleavage. Biochem Biophys Res Commun. 1993;191:247–254. doi: 10.1006/bbrc.1993.1209. [DOI] [PubMed] [Google Scholar]
- 9.Kordula T, Dubin A, Schooltink H, Koj A, Heinrich P C, Rose-John S. Molecular cloning and expression of an intracellular serpin: an elastase inhibitor from horse leukocytes. Biochem J. 1993;293:187–193. doi: 10.1042/bj2930187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korpula-Mastalerz R, Dubin A. The intracellular serpin family. Acta Biochim Pol. 1996;43:419–430. [PubMed] [Google Scholar]
- 11.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence D A. The serpin-proteinase complex revealed. Nat Struct Biol. 1997;4:339–341. doi: 10.1038/nsb0597-339. [DOI] [PubMed] [Google Scholar]
- 13.Lei H Y, Tang M J, Tsao N. Intracellular alkalinization in dexamethasone-induced thymocyte apoptosis. Apoptosis. 1997;2:304–312. doi: 10.1023/a:1026441221027. [DOI] [PubMed] [Google Scholar]
- 14.Liao T-H. The subunit structure and active site sequence of porcine spleen deoxyribonuclease. J Biol Chem. 1985;260:10708–10713. [PubMed] [Google Scholar]
- 15.Liao T-H, Liao W-C, Chang H-C, Lu K-S. Deoxyribonuclease II purified from the isolated lysosomes of porcine spleen and from porcine liver homogenates. Comparison with deoxyribonuclease II purified from porcine spleen homogenates. Biochim Biophys Acta. 1989;1007:15–22. doi: 10.1016/0167-4781(89)90124-3. [DOI] [PubMed] [Google Scholar]
- 16.Montague J W, Gaido M L, Frye C, Cidlowski J A. A calcium-dependent nuclease from apoptotic rat thymocytes is homologous with cyclophilin. Recombinant cyclophilins A, B, C have nuclease activity. J Biol Chem. 1994;269:18877–18880. [PubMed] [Google Scholar]
- 17.Novagen Inc. pET system manual. 6th ed. Madison, Wis: Novagen Inc.; 1995. [Google Scholar]
- 18.Pandey S, Walker P R, Sikorska M. Identification of a novel 97 kDa endonuclease capable of internucleosomal DNA cleavage. Biochemistry. 1997;36:711–720. doi: 10.1021/bi962387h. [DOI] [PubMed] [Google Scholar]
- 19.Pardhasaradhi B V V, Khar A, Srinivas U K. Effect of anti-apoptotic genes and peptide inhibitors on cytoplasmic acidification during apoptosis. FEBS Lett. 1997;411:67–70. doi: 10.1016/s0014-5793(97)00665-0. [DOI] [PubMed] [Google Scholar]
- 20.Patel T, Gores G J, Kaufmann S H. The role of proteases during apoptosis. FASEB J. 1996;10:587–597. doi: 10.1096/fasebj.10.5.8621058. [DOI] [PubMed] [Google Scholar]
- 21.Peitsch M C, Polzar B, Stephan H, Crompton T, MacDonald H R, Mannherz H G, Tschopp J. Characterization of the endogenous deoxyribonuclease involved in nuclear DNA degradation during apoptosis (programmed cell death) EMBO J. 1993;12:371–377. doi: 10.1002/j.1460-2075.1993.tb05666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pemberton P A. The role of serpin superfamily members in cancer. Cancer J. 1997;10:24–30. [Google Scholar]
- 23.Pérez-Sala D, Collado-Escobar D, Mollinedo F. Intracellular alkalinization suppresses Lovastatin-induced apoptosis in HL-60 cells through the inactivation of a pH-dependent endonuclease. J Biol Chem. 1995;270:6235–6242. doi: 10.1074/jbc.270.11.6235. [DOI] [PubMed] [Google Scholar]
- 24.Potempa J, Dubin A, Watorek W, Travis J. An elastase inhibitor from equine leukocyte cytosol belongs to serpin superfamily. Further characterization and amino acid sequence of the reactive center. J Biol Chem. 1988;263:7364–7369. [PubMed] [Google Scholar]
- 25.Promega Corp. TNT coupled retyculocyte lysate system. Technical bulletin. Madison, Wis: Promega Corp.; 1996. [Google Scholar]
- 26.Remold-O’Donnell E, Chin J, Alberts M. Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc Natl Acad Sci USA. 1992;89:5635–5639. doi: 10.1073/pnas.89.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Shak S, Capon D J, Hellmiss R, Marsters S A, Baker C L. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci USA. 1990;87:9188–9192. doi: 10.1073/pnas.87.23.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiokawa D, Ohyama H, Yamada T, Takahashi K, Tanuma S-I. Identification of an endonuclease responsible for apoptosis in rat thymocytes. Eur J Biochem. 1994;226:23–30. doi: 10.1111/j.1432-1033.1994.tb20022.x. [DOI] [PubMed] [Google Scholar]
- 30.Stein P E, Leslie A G W, Finch J T, Turnell W G, McLaughlin P J, Carrell R W. Cristal structure of ovalbumin as a model for reactive centre of serpins. Nature. 1990;347:99–102. doi: 10.1038/347099a0. [DOI] [PubMed] [Google Scholar]
- 31.Teschauer W F, Mentele R, Sommerhoff C P. Primary structure of a porcine leukocyte serpin. Eur J Biochem. 1993;217:519–526. doi: 10.1111/j.1432-1033.1993.tb18272.x. [DOI] [PubMed] [Google Scholar]
- 32.Torriglia A, Chaudun E, Chany-Fournier F, Jeanny J C, Courtois Y, Counis M F. Involvement of DNase II in nuclear degeneration during lens cell differentiation. J Biol Chem. 1995;270:28579–28585. doi: 10.1074/jbc.270.48.28579. [DOI] [PubMed] [Google Scholar]
- 33.Wolf C M, Morana S J, Eastman A. Zinc inhibits apoptosis upstream of ICE/CED-3 proteases rather than at the level of an endonuclease. Cell Death Differ. 1997;4:125–129. doi: 10.1038/sj.cdd.4400218. [DOI] [PubMed] [Google Scholar]
- 34.Wright S C, Wei Q S, Zhong J, Zheng H, Kinder D H, Larrick J W. Purification of a 24kDa protease from apoptotic tumor cells that activates DNA fragmentation. J Exp Med. 1994;180:2113–2123. doi: 10.1084/jem.180.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu M-Q, Perler F B. The mechanism of protein splicing and its modulation by maturation. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanaka M, Tsubota Y, Anai M, Ishimatsu K, Okumura M, Katsuki S, Takagi Y. Thermal inactivation of shrimp deoxyribonuclease with and without sodium dodecyl sulfate. J Biol Chem. 1974;249:3884–3889. [PubMed] [Google Scholar]
- 37.Yasuda T, Nadano D, Awazu S, Kishi K. Human urine deoxyribonuclease II (DNase II) isoenzymes: a novel immunoaffinity purification, biochemical multiplicity, genetic heterogeneity and broad distribution among tissues and body fluids. Biochim Biophys Acta. 1992;1119:185–193. doi: 10.1016/0167-4838(92)90390-y. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Z, Parmelee D, Hyaw H, Coleman T A, Su K, Zhang J, Gentz R, Ruben S, Rose C, Li Y. Cloning and characterization of a novel human DNase. Biochem Biophys Res Commun. 1997;231:499–504. doi: 10.1006/bbrc.1996.5923. [DOI] [PubMed] [Google Scholar]
- 39.Zhivotovsky B, Burgess D H, Orrenius S. Proteases in apoptosis. Experientia. 1996;52:968–978. doi: 10.1007/BF01920106. [DOI] [PubMed] [Google Scholar]