SUMMARY
The positioning of nucleosomes with respect to DNA plays an important role in regulating transcription. However, nucleosome mapping has been performed for only limited genomic regions in humans. We have generated genome-wide maps of nucleosome positions in both resting and activated human CD4+ T cells by direct sequencing of nucleosome ends using the Solexa high-throughput sequencing technique. We find that nucleosome phasing relative to the transcription start sites is directly correlated to RNA polymerase II (Pol II) binding. Furthermore, the first nucleosome downstream of a start site exhibits differential positioning in active and silent genes. TCR signaling induces extensive nucleosome reorganization in promoters and enhancers to allow transcriptional activation or repression. Our results suggest that H2A.Z-containing and modified nucleosomes are preferentially lost from the −1 nucleosome position. Our data provide a comprehensive view of the nucleosome landscape and its dynamic regulation in the human genome.
INTRODUCTION
Nucleosomes consisting of approximately 146 base pairs (bp) of DNA wrapped around a histone octamer are the fundamental structural units of chromatin (Kornberg and Lorch, 1999; Kornberg and Lorch, 2002). The covalent modifications of histones as well as the translational positioning of nucleosomes along the DNA have been implicated in profoundly influencing gene expression (reviewed in Berger, 2002; Henikoff et al., 2004; Kingston and Narlikar, 1999; Kouzarides, 2007; Li et al., 2007; Peterson and Laniel, 2004; Shahbazian and Grunstein, 2007; Zhang and Reinberg, 2001). During the past few years, large amounts of data pertaining to genome-wide profiles of histone modifications have been generated for a number of human cell types (Barski et al., 2007; Bernstein et al., 2005, 2006; Boyer et al., 2006; Guenther et al., 2007; Heintzman et al., 2007; Kim et al., 2005; Lee et al., 2006; Mikkelsen et al., 2007; Roh et al., 2004, 2006; Squazzo et al., 2006). These studies have provided novel insights into the mechanisms by which histone modifications regulate genome function. Specific histone modifications are enriched at important regulatory elements of transcription such as promoters and enhancers (Barski et al., 2007; Heintzman et al., 2007; Roh et al., 2005, 2007). Although nucleosome positions have been mapped in the S. cerevisiae (Lee et al., 2007; Yuan et al., 2005) and C. elegans genomes (Johnson et al., 2006), across human promoters and other human genomic regions (Dennis et al., 2007), a genome-wide map of nucleosomes in the human genome has not yet been produced.
Traditionally, nucleosome positioning at a specific locus has been determined by microccocal nuclease (MNase) digestion, followed by ligation-mediated PCR analysis. Recently, DNA microarrays with tiling oligonucleotide probes have been utilized to analyze MNase-digested chromatin to determine nucleosome positioning at a genome scale in yeast (Lee et al., 2007; Yuan et al., 2005). Similar strategies have been employed to analyze nucleosome positioning in limited genomic regions in the human genome (Dennis et al., 2007; Ozsolak et al., 2007). However, because the resolution of DNA microarray-based methods is dependent on the spacing between neighboring probes, it would be extremely expensive and tedious to generate a genome-wide map of nucleosome positioning at a high resolution for the entire human genome using this strategy.
We previously analyzed the genome-wide distribution patterns of histone methylation in human CD4+ T cells using ChIP-Seq by combining chromatin immunoprecipitation with Solexa high-throughput sequencing technology (Barski et al., 2007). We noticed that the H3K4me3 profiles in promoter regions displayed several subpeaks, each spanning approximately 150 bp, suggesting that these short sequence reads may elucidate nucleosome positions. To address this possibility directly, we sequenced the ends of the mononucleosome-sized DNA isolated from MNase-digested chromatin using the Solexa sequencing technique. Our data indicate that this strategy is an efficient and precise method to map genome-wide nucleosome positions in the human genome. Using this strategy, we have generated genome-wide maps of nucleosome positions in both resting and activated human T cells. We find that nucleosomes are highly phased relative to the transcription start sites (TSSs) of expressed genes, but this phasing disappears for unexpressed genes. Expressed genes and unexpressed genes exhibit differential positioning for the +1 nucleosome. We find that promoters with stalled Pol II exhibit a nucleosome phasing similar to promoters of transcriptionally active genes. Gene activation by T cell receptor (TCR) signaling is accompanied by nucleosome reorganization in promoters and enhancers. Furthermore, our data suggest that H2A.Z deposition and H3K4me3 modification may facilitate nucleosome eviction or repositioning in promoter regions of the human genome.
RESULTS
Mapping Nucleosome Positions by the Solexa Sequencing Technique
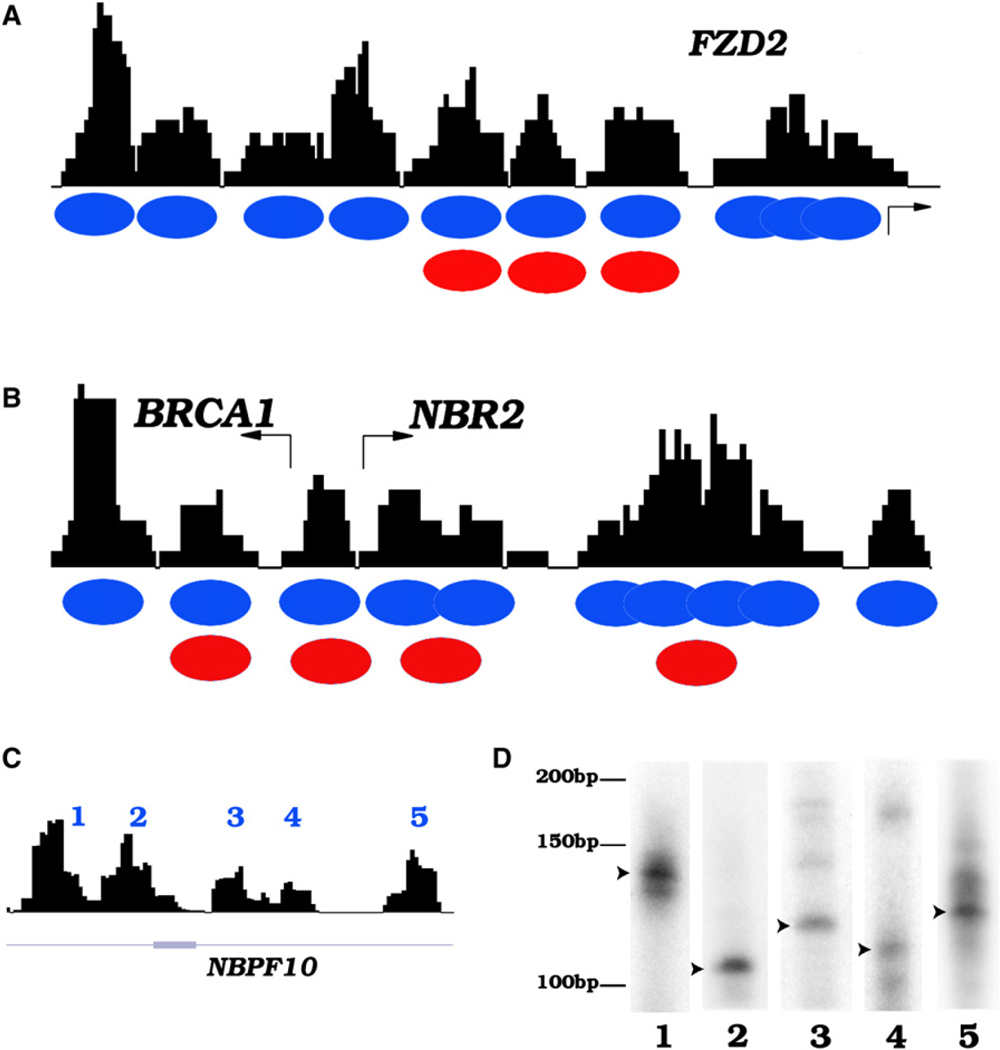
To analyze nucleosome positioning across the genome in resting and activated human CD4+ T cells, we isolated mononucleosome-sized DNA from MNase-digested chromatin and sequenced the DNA ends using the Solexa sequencing technology as described previously (Barski et al., 2007). We achieved approximately 10-fold coverage of all nucleosomes of each cell state assuming one nucleosome every 200 bp of DNA throughout the genome. The short reads obtained from sequencing were mapped to the human genome (hg18), and a simple scoring function (see Experimental Procedures) was applied to create a profile of nucleosomes across the genome. A previous study reported the mapping of nucleosome positions across 3692 human promoters using high-density tiling DNA microarrays (Ozsolak et al., 2007). Comparison of our data with the DNA microarray results revealed similar nucleosome positioning at the FZD2, BRCA1, and NBR2 promoters (Figures 1A and 1B). In addition, we detected several overlapping nucleosomes in the region downstream of the NBR2 TSS whereas only one single nucleosome was detected by the DNA microarray method (Figure 1B). These results indicate that the short sequence reads obtained from sequencing of MNase-digested chromatin using the Solexa technique can reliably define the nucleosome boundaries in the human genome at high resolution. In addition to promoter regions, well-positioned nucleosomes were detected in intergenic and transcribed regions, as exemplified in Figure 1C for a transcribed region of the NBPF10 gene on chromosome 1. To confirm the nucleosome positions revealed by the sequencing strategy, we used LM-PCR assays to determine the boundaries of the nucleosomes indicated in Figure 1C. As shown in Figure 1D, one major nucleosome position was detected for each of these nucleosomes.
Figure 1. The Solexa Sequencing Tags Define Nucleosome Boundaries in the Human Genome.
(A) Nucleosome profile at the FZD2 promoter. The mononucleosome-sized DNA was isolated from MNase-digested chromatin of human T cells and sequenced to read 25 bp from the end using the Solexa sequencing technique. The sequence reads mapped to a given region were used to generate the nucleosome profiles using a scoring function (see Experimental Procedures) as shown by the black track. Blue ovals indicate inferred nucleosome positions in this study and red ovals indicate nucleosome positions identified previously (Ozsolak et al., 2007).
(B) Nucleosome profile at the BRCA1 and NBR2 promoters.
(C) Nucleosomes identified in the gene body of NBPF10 (region shown is chr1:16763501–16764673).
(D) Confirmation of nucleosome boundaries using LM-PCR. The mononucleosome DNA was ligated to a pair of Solexa adaptors, followed by amplification using one Solexa primer and one sequence-specific primer recognizing one of the nucleosomes indicated in Figure 1C. The product was labeled using a 32P-labeled nested primer, resolved by polyacrylamide gel electrophoresis, and visualized by exposing to X-ray films. An arrowhead indicates the major nucleosome boundaries.
Nucleosome Phasing surrounding TSSs
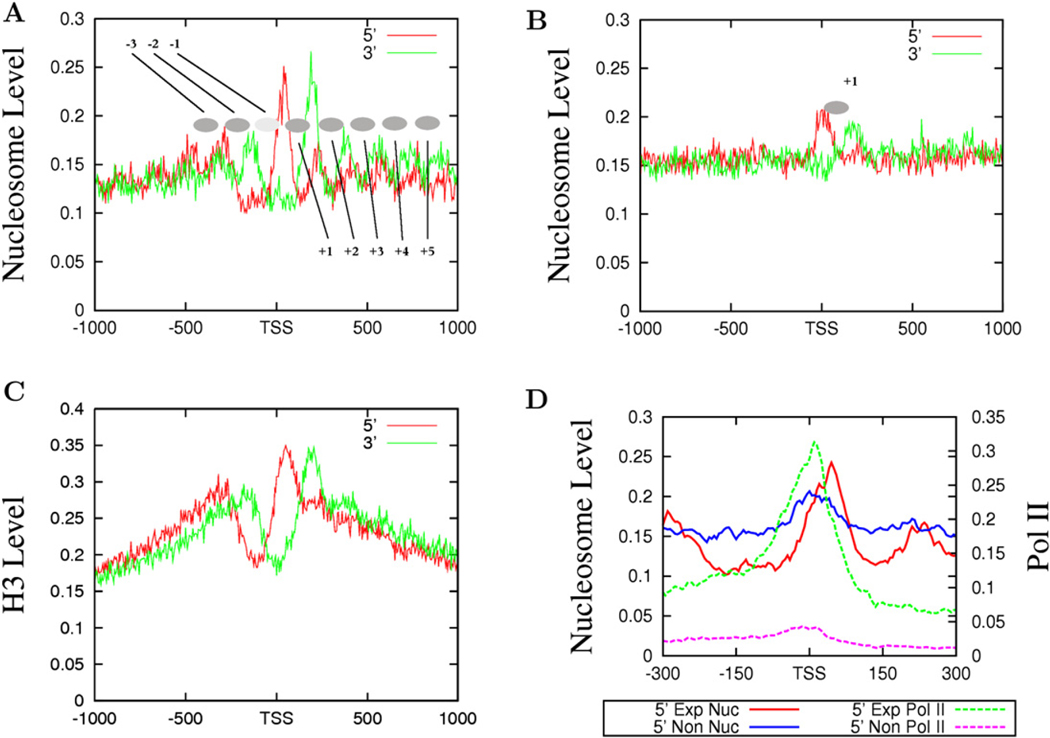
Previous analyses of nucleosome positioning in yeast have suggested that nucleosomes in promoter regions are highly phased (Albert et al., 2007; Yuan et al., 2005). To determine if nucleosomes are phased relative to the TSS in the human genome, we counted the total number of sequence reads (tags) from both the sense and antisense strands of DNA in 5 bp windows surrounding TSSs for expressed and unexpressed genes (see Experimental Procedures for definitions). The reads from the sense and antisense strands represent the 5′ and 3′ nucleosome boundaries, respectively. As shown in Figure 2A, we detected eight phased nucleosomes as indicated by the filled ovals (from −3 to +5), three upstream and five downstream of the TSSs in the promoter regions of expressed genes. Interestingly, we detected only one well-positioned nucleosome, the +1 nucleosome, in the promoter regions of unexpressed genes (Figure 2B). To validate the nucleosome distribution revealed by MNase digestion, we examined the distribution of histone H3 by sequencing the ChIP DNA using an H3 antibody and chromatin fragmented by sonication of formaldehyde-crosslinked CD4+ T cells. Our data indicate that the two methods revealed generally similar nucleosome levels in most regions (Figure S1 available online). However, the fine resolution in nucleosome positioning surrounding the TSS detected by the direct sequencing of mononucleosomes from MNase digestion was not achieved by the H3-ChIP procedure (compare Figure 2A with Figure 2C), which is likely due to the heterogeneous sizes of chromatin fragments used for ChIP.
Figure 2. Nucleosomes near the TSS of Actively Transcribed Genes Are Strongly Phased.
(A) The nucleosomes near the TSS of expressed genes are phased with respect to the TSS. The y axis shows the normalized number of sequence tags from the sense strand (red) and antisense strand (green) of DNA at each position. The inferred nucleosomes are shown by the filled ovals that are numbered as indicated.
(B) Only one well-positioned nucleosome exists near the TSS of unexpressed genes (see panel A for details).
(C) Histone distribution surrounding the TSS of expressed genes analyzed by ChIP-Seq using an H3 antibody and crosslinked and sonicated chromatin. The y axis shows the normalized number of sequence tags from the sense strand (red) and antisense strand (green) of DNA at each position.
(D) The +1 nucleosomes are differentially positioned in expressed and unexpressed genes. The nucleosome tags from the sense strand of DNA of expressed (indicated as 5′ Exp Nuc, red) and unexpressed (indicated as 5′ Non Nuc, blue) genes are shown. The Pol II tags obtained from the ChIP-Seq analysis (Barski et al., 2007) are also shown for the expressed and unexpressed genes.
Nucleosome Positioning surrounding TSSs
To examine the relationship between the positioning of nucleosomes and transcriptional activity, we plotted the nucleosome and Pol II tags from the sense strand for both the expressed and unexpressed promoters (Figure 2D). It is interesting to note that the 5′ end of the +1 nucleosome in the active promoters peaked at +40 bp, whereas the 5′ end of the +1 nucleosome in the inactive promoters peaked at +10 bp. A similar shift of the 3′ end was also observed (data not shown). Examination of Pol II binding in the promoter region of active genes indicates that it peaked at around +10 bp, overlapping with the nucleosome peak in the inactive promoters.
Nucleosome Phasing near TSSs Is Correlated with Pol II Binding
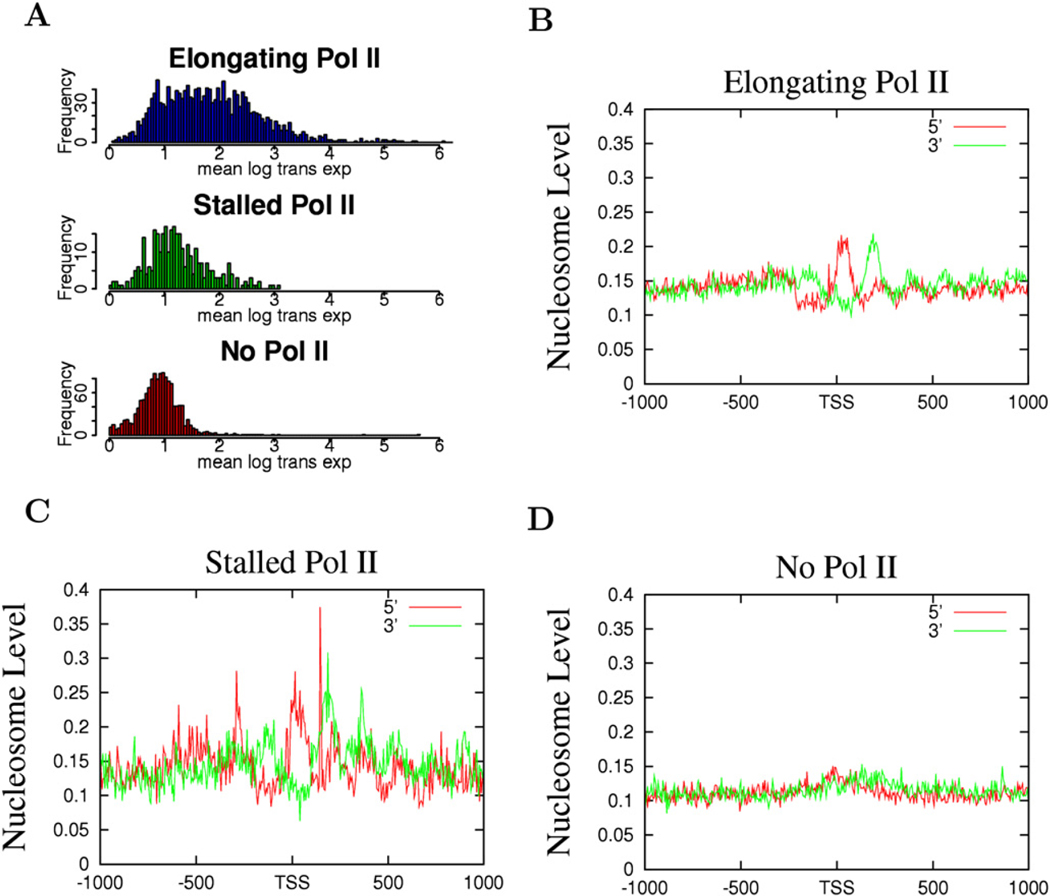
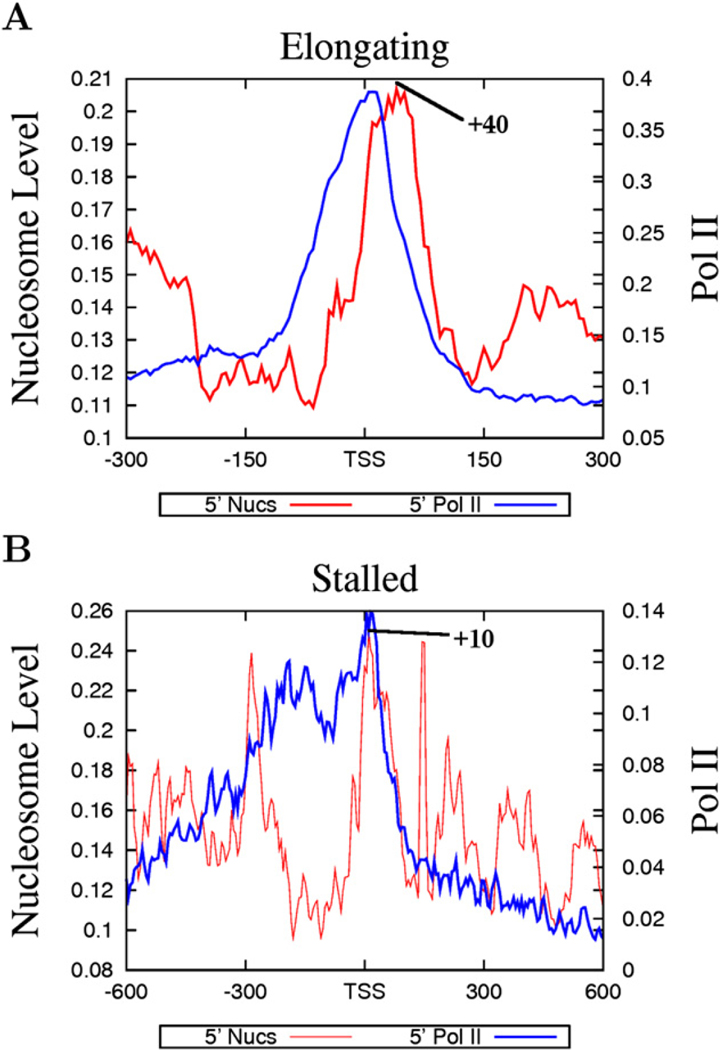
The above data suggest that nucleosome phasing and positioning surrounding the TSSs may be related with Pol II binding. Indeed, we find that the nucleosome phasing patterns appear to correlate with the levels of Pol II in the promoter region: better phasing was observed with higher levels of Pol II and less phasing with lower levels of Pol II (Figure S2). Recent data suggest that a significant fraction of Drosophila genes are associated with poised or stalled Pol II (Muse et al., 2007; Zeitlinger et al., 2007). To examine the nucleosome organization at promoters with poised Pol II, we selected genes with stalled Pol II, elongating Pol II, or no Pol II, adopting a strategy previously employed in Drosophila (Zeitlinger et al., 2007) (see Experimental Procedures for details). The genes without any Pol II binding exhibited very low levels of expression (Figure 3A, bottom panel), whereas the genes with elongating Pol II showed a broad range of expression (Figure 3A, top panel). The majority of the genes with poised Pol II exhibited similar expression levels to those without any Pol II binding (Figure 3A, middle panel). As expected, the genes with elongating Pol II demonstrated a similar nucleosome phasing pattern to the expressed genes shown in Figure 2A (Figure 3B). Interestingly, the genes with poised Pol II also exhibited a similar pattern of nucleosome phasing to the expressed genes (Figure 3C), whereas the genes without any Pol II did not show any significant nucleosome phasing surrounding the TSS (Figure 3D). Despite the similar patterns of nucleosome phasing in the promoters with elongating and stalled Pol II, the +1 nucleosome in these two sets of promoters exhibited different positions, peaking at +10 bp for promoters with stalled Pol II and +40 bp for promoters with elongating Pol II (Figure 4). In addition, the Pol II signal was localized within a narrow region immediately upstream of the TSS at the elongating promoters, whereas it was distributed broadly at the poised promoters (Figure 4). These data suggest that the nucleosome phasing surrounding the TSS is correlated with Pol II binding but not necessarily with the activity of the bound Pol II. Furthermore, the single positioned +1 nucleosome observed in the unexpressed genes (Figure 2B) may be attributed to the fraction of the unexpressed genes that are associated with stalled Pol II.
Figure 3. Promoters with Stalled Pol II Exhibit Similar Patterns of Nucleosome Phasing to the Promoters with Elongating Pol II.
(A) Expression patterns of the genes with elongating, stalled, or no Pol II (see Experimental Procedures for details). The y axis indicates the number of genes exhibiting the expression level shown by the x axis.
(B) The nucleosome pattern near the TSS of the genes with elongating Pol II. The y axis shows the normalized number of sequence tags from the sense strand (red) and antisense strand (green) of DNA at each position.
(C) The nucleosome pattern near the TSS of the genes with stalled Pol II in the promoter region.
(D) The nucleosome pattern near the TSS of the genes without any Pol II binding in the promoter region.
Figure 4. The Distinct Nucleosome Positioning Patterns at Poised and Elongating Promoters.
(A) Nucleosome positioning and Pol II binding at elongating promoters.
(B) Nucleosome positioning and Pol II binding at poised promoters. The peak of the +1 nucleosome position is indicated in both panels.
To ensure that the methods we used to identify genes with stalled, elongated, and no Pol II were appropriate, we analyzed the genes that are associated with initiating Pol II by ChIP-Seq using an antibody specifically recognizing the unphosphorylated form of Pol II. We find that 41% of all promoters are associated with significant levels of initiating Pol II (p < 0.05; see Experimental Procedures for details) in resting human CD4+ T cells. The distribution of unphosphorylated Pol II in the promoters of active and inactive genes (Figure S3A) was similar to what has been shown previously (Guenther et al., 2007). Furthermore, we used the H3K36me3 level in gene bodies as a mark of active transcription (Barski et al., 2007; Guenther et al., 2007) and compared the H3K36me3 profile across the genes we defined as elongated, silent, and stalled with the profiles of genes that are expressed and unexpressed and those containing significant unphosphorylated Pol II signals in the promoter regions but no detectable mRNAs. The genes classified as elongated had similar H3K36me3 profiles to those classified as expressed, the silent genes were similar to those classified as unexpressed, and the genes classified as stalled were similar to those with an unphosphorylated Pol II signal in the promoter but no mRNAs (Figures S3B and S3C). Finally, our data indicate that the inactive genes associated with initiating Pol II identified using the unphospohorylated Pol II antibody exhibited a similar nucleosome phasing pattern to those identified as stalled genes (Figure S3D). These results indicate that the methods we employed to identify genes with stalled, elongated, and no Pol II were indeed valid.
Nucleosome Levels in the Promoter Region
Previous studies have suggested that a nucleosome-free region (NFR) exists upstream of the TSS of expressed genes but not unexpressed genes in the human genome (Ozsolak et al., 2007). Comparison of the nucleosome peaks in Figures 2A and 2B indicates that the −1 nucleosome level was lower than either the −2 or +1 nucleosome level in both the active and inactive promoters, suggesting some nucleosome depletion in both cases. To better quantify the nucleosome level in this region, we counted the total number of tags detected in each of these regions from the −2 to the +5 nucleosome. Interestingly, the level of the +1 nucleosome was higher than other nucleosomes, being 11% and 14% higher than the −2 nucleosome level in expressed and unexpressed genes, respectively (see Figures 7C and 7D). Compared with the −2 nucleosome, the level of the −1 nucleosome decreased 40% in expressed genes and only 16% in unexpressed genes (Figure S4A; Figures 7C and 7D). These results are consistent with the previous observation that active transcription is detected at only a fraction of cells at any one moment (Chubb et al., 2006). We further separated the expressed genes according to their expression levels and analyzed the depletion of the −1 nucleosome. We did not find significant differences in the −1 nucleosome depletion between the different expression groups (data not shown). Even the group of the 100 most active genes exhibited similar −1 nucleosome depletion to the set of all expressed genes (Figure S4A). However, our analysis of the Pol II levels in promoter regions indicates that the −1 nucleosome depletion appears to correlate with promoter Pol II binding levels (Figure S4B).
Figure 7. Modification Profiles and Nucleosome Loss of Individual Nucleosomes near the TSS.
(A) Methylation of H3K4 near the TSSs of actively transcribed genes (data from Barski et al., 2007) overlaid with nucleosome positions. The sequence tags from the sense strand are shown for nucleosomes (black), H3K4me3 (red), H3K4me2 (green), and H3K4me1 (blue). H3K4me3 marks the −2, +1, +2, and +3 nucleosomes, H3K4me2 marks the +3 and +4 nucleosomes, and H3K4me1 marks the +5 and +6 nucleosomes.
(B) Histone variant H2A.Z marks the −3, −2, +1, +2, and +3 nucleosomes in actively transcribed genes.
(C) Quantification of the nucleosome levels by counting the sequence tags at the nucleosome positions (indicated in Figure 2A) for actively transcribed genes in resting T cells.
(D) Quantification of the nucleosome levels by counting the sequence tags at the nucleosome positions (indicated in Figure 2A) for silent genes in resting T cells.
(E) Quantification of the H2A.Z nucleosome levels by counting the sequence tags obtained from the ChIP-Seq analysis (Barski et al., 2007) at the nucleosome positions (indicated in Figure 2A) for actively transcribed genes in resting T cells.
(F) Quantification of the H2A.Z nucleosome levels by counting the sequence tags obtained from ChIP-Seq analysis (Barski et al., 2007) at the nucleosome positions (indicated in Figure 2A) for silent genes in resting T cells.
(G) Total nucleosomes and H2A.Z-containing nucleosomes near the TSS of the CDC23 gene in resting T cells. The −1 nucleosome region is highlighted.
(H) Total nucleosomes and H2A.Z-containing nucleosomes near the TSS of the ADIPOR2 gene in resting T cells. The −1 nucleosome region is highlighted.
T Cell Activation Induces Nucleosome Reorganization
Activation of human CD4+ T cells by TCR signaling activated 417 and repressed 580 genes (see Experimental Procedures for definitions). To determine if the nucleosome structure is dynamically regulated by TCR signaling, we compared the nucleosome distribution across the promoter regions of the repressed and induced genes (Figures 5A, 5B, and 5E). Our data indicate that the nucleosome level did not show an apparent change in the region of the −1 nucleosome (highlighted region) for induced genes. Instead, we observed an increase in the +1 and +2 nucleosome levels downstream of the TSS (Figure 5A). This suggests that the −1 nucleosome may already be depleted and prepared for gene activation before TCR signaling. Indeed, comparison of these genes with actively expressed genes indicates that they had similar levels of the −1 nucleosome in resting T cells (Figure 5B). Pol II may have already bound to the promoters in the resting state and TCR signaling may have induced a switch of Pol II from a stalled form to an elongating form. Indeed, more hypophosphorylated Pol II than ser5-phosphorylated Pol II was detected at the inducible promoters before TCR signaling (Figure 5C), whereas more ser5-phosphorylated Pol II was detected after TCR signaling (Figure 5D). Analysis of the promoters repressed by TCR signaling revealed a significant increase of nucleosome level at the −1 nucleosome position upon T cell activation (highlighted region), although the +1 nucleosome level also increased (Figure 5E). The increase of the −1 nucleosome level may help to repress these genes. Indeed, our examination of Pol II levels indicates that both the ser5-phosphorylated and hypophosphorylated forms of Pol II were decreased significantly at the repressed promoters after TCR signaling (compare Figures 5F and 5G).
Figure 5. Nucleosome Reorganization at Genes Induced and Repressed by TCR Signaling.
(A) Nucleosome profiles in resting and activated T cells for genes induced by TCR signaling. The y axis shows the normalized number of sequence tags from the sense strand of DNA of induced genes in resting (blue) and activated (red) T cells in 5 bp windows. Highlighted region indicates the −1 nucleosome position.
(B) Comparison of nucleosome levels between the expressed and inducible genes in resting T cells.
(C) Pol II density as determined by performing ChIP-Seq for Pol II using antibodies against Ser5 phosphorylated and unphosphorylated Pol II in resting T cells across genes induced by TCR signaling.
(D) Pol II density for Ser5 phosphorylated and unphosphorylated Pol II in activated T cells across genes induced by TCR signaling.
(E) Nucleosome profiles in resting and activated T cells for genes repressed by TCR signaling. The y axis shows the number of sequence tags from the sense strand of DNA of repressed genes in resting (blue) and activated (red) T cells. Highlighted region indicates the −1 nucleosome position.
(F) Pol II density for Ser5 phosphorylated and unphosphorylated Pol II in resting T cells across genes repressed by TCR signaling.
(G) Pol II density for Ser5 phosphorylated and unphosphorylated Pol II in activated T cells across genes repressed by TCR signaling.
Nucleosome Reorganization at Functional Enhancers
Conserved noncoding sequence 1 (CNS1) downstream of the IL-13 gene is required for the coordinated expression of the Th2 cytokine genes, including IL-13 and IL-4 (Loots et al., 2000). We have previously reported that a DNA element immediately next to CNS1, which was identified as an acetylation island, termed as AI1, and was not as conserved as CNS1, can activate a reporter promoter in response to Ionomycin and PMA stimulation that mimics TCR signaling (Roh et al., 2005). Interestingly, AI1 constitutively activates the reporter promoter in a nonchromatin vector, suggesting that the factors required for activating transcription via AI1 are present in the cells and can access the target sites to activate the transcription. However, AI1 fails to activate the reporter promoter in a chromatin vector without stimulation (Roh et al., 2005). This suggests that the transcription activators cannot access their target sites in a chromatin environment without TCR signaling. Therefore, TCR signaling may induce chromatin remodeling and make the target sites more accessible to the transcription activators. To determine if changes to the chromatin structure occur at the endogenous locus in response to TCR signaling, we examined the nucleosome organization at the AI1 and CNS1 regions in both resting and activated T cells. As shown in Figure 6A, this region contains several smaller CNSs (labeled a to i). Examination of the nucleosome profile revealed that there are multiple nucleosomes, both well-positioned and delocalized, in this region in resting T cells. All of these CNSs, except CNSi, are located within nucleosomes. Interestingly, after T cell activation, we observed a significant reorganization of the nucleosome structure such that the nucleosomes became more localized. Furthermore, several nucleosomes were either removed or shifted so that CNSb, c, d, e, and f and half of the CNSg sequence were now located in linker regions (Figure 6A), suggesting that they had become accessible to regulatory factors. These results also suggest that localization of the regulatory sequences in linker regions may be important for their function in regulating transcription. Indeed, we observed that many CNSs, which are potential regulatory elements, are located in linker regions, as exemplified for a region in the RAD50 gene (Figure 6B).
Figure 6. Nucleosome Reorganization at Functional Enhancers.
(A) Nucleosome profiles in resting and activated T cells near the functional enhancer elements CNS1 and AI1. CNS1 and AI1 regions are indicated below the nucleosome profiles. The CNSs are alphabetically labeled and their correspondence to nucleosome structure is highlighted in yellow. Region shown is chr5:132,026,345–132,028,199.
(B) Nucleosome profiles in resting and activated T cells in an intron of the RAD50 gene. The correspondence of CNSs in this locus to nucleosome linker regions is highlighted in yellow. Region shown is chr5:131,960,529–131,964,566.
Modification of the Promoter-Proximal Nucleosomes
Previous studies have suggested that promoter regions are often associated with H3K4 methylation, with nucleosomes near the TSS being modified with H3K4me3 and nucleosomes further away being modified with H3K4me2 and H3K4me1 (Barski et al., 2007; Heintzman et al., 2007; Pokholok et al., 2005). To pinpoint the specific modifications associated with each individual nucleosome in the promoter region, we examined the H3K4 modification tags (Barski et al., 2007) along with the nucleosome tags near the TSSs of active genes (Figure 7A). The H3K4me3 modification was mainly associated with the −2, +1, +2, and +3 nucleosomes; H3K4me2 peaks with the +3 and +4 nucleosomes; and H3K4me1 peaks with the +5 and +6 nucleosomes.
Histone variant H2A.Z has been reported to decorate two nucleosomes, one upstream and one downstream of TSSs in yeast (Raisner et al., 2005). Our data indicate that it is associated with the −3, −2, +1, +2, and +3 nucleosomes of actively transcribed genes in the human genome (Figure 7B). The deeper valley at the −1 nucleosome region of the H2A.Z distribution versus the total nucleosome distribution suggests that H2A.Z nucleosomes are preferentially lost in this region compared to all nucleosomes. To quantify this, we counted all the nucleosome and H2A.Z nucleosome tags corresponding to each nucleosome position (see Experimental Procedures) (Figures 7C, 7D, 7E, and 7F). Even though the total nucleosome loss at the −1 nucleosome position was only 40% in active genes, the H2A.Z nucleosome was depleted 74% compared to the −2 nucleosome (Figures 7C and 7E). A remarkable 52% loss was observed for the −1 H2A.Z nucleosome in the silent genes, even though there was only 16% total nucleosome loss at the same position (Figures 7D and 7F). Examination of specific promoters indeed revealed that H2A.Z-containing nucleosomes are preferentially lost from the −1 nucleosome position even when there is still nucleosome structure in the region (Figures 7G and 7H). It is also interesting to note that the loss of the H2A.Z-containing nucleosome from the −1 nucleosome position in the TCR-inducible genes in resting T cells was similar to the loss in expressed genes (Figure S5). We also observed a preferential loss of H3K4me3-modified nucleosomes from the −1 nucleosome position as compared to the total nucleosomes (Figure 7A). These data suggest that deposition of histone variant H2A.Z or modification by H3K4me3 may facilitate nucleosome eviction or repositioning in the −1 nucleosome region.
DISCUSSION
We have demonstrated that the short sequence reads generated by sequencing mononucleosome DNA ends using the Solexa sequencing technology can precisely define nucleosome positions in the human genome. While the resolution of DNA microarray-based techniques is dependent on the spacing of probes and mononucleosome preparation, the resolution of the sequencing strategy is only dependent on the size distribution of the mononucleosome templates. Different methods of chromatin preparation, including sonication or MNase digestion of crosslinked or native chromatin, have been used for analysis of histone modification and histone/nucleosome positioning. One concern using the MNase digestion strategy is that it may preferentially degrade open chromatin in regions such as promoters of active genes. Previous results suggest that this is not the case. For example, a similar nucleosome depletion in the promoter region has been observed using both the MNase digestion strategy and sonication followed by H3-ChIP (Bernstein et al., 2004; Lee et al., 2004; Yuan et al., 2005). Furthermore, our sequencing analysis of the H3-ChIP DNA reveals a similar depletion of the −1 nucleosome as detected by the direct sequencing of MNase-generated mononucleosomes (Figure 2). Another concern is that MNase digestion may preferentially release more active chromatin. Indeed, when very limited digestion is used, MNase can be used to probe nuclease hypersensitive sites (Langst et al., 1997). However, when the chromatin is digested to 80% of mononucleosomes, the active and inactive genes are equally accessed. For example, the fractions of the inactive β-globin and active β-actin gene regions in the mononucleosome DNA were similar (data not shown). The third concern is that nucleosomes prepared from native noncrosslinked chromatin may freely slide and lose their positioning information. Published results suggest that nucleosomes are stable under these in vitro conditions; both crosslinked (Albert et al., 2007) and noncrosslinked nucleosomes (Segal et al., 2006) show similar preference to the dinucleotide repeat patterns.
Transcriptional activation requires assembly of bulky transcription machinery immediately upstream of the TSS. The nucleosome structure is apparently incompatible with the process and needs to be removed for active transcription (Lorch et al., 1987). Previous studies have provided evidence that nucleosomes are lost in the Pho5 promoter region during transcriptional activation in yeast (Boeger et al., 2003; Reinke and Horz, 2003). A similar nucleosome loss has been observed for the mouse mammary tumor virus promoter upon its activation by hormone (Richard-Foy and Hager, 1987). Large-scale analyses have also revealed nucleosome loss surrounding TSSs in both yeast and humans (Bernstein et al., 2004; Lee et al., 2004; Ozsolak et al., 2007; Yuan et al., 2005). Consistent with these observations, we find that the nucleosome level immediately upstream of TSSs is decreased in a Pol II-dependent manner in the human genome. We can imagine that there are at least two mechanisms involved in the nucleosome loss: nucleosome eviction and nucleosome sliding. As mentioned above, nucleosome loss has been shown to accompany gene activation (Boeger et al., 2003; Reinke and Horz, 2003). Similarly, it has been reported that nucleosome sliding is associated with activation of the interferon-β promoter by viral infection (Lomvardas and Thanos, 2002). Whichever process takes place probably depends on what is energetically more favorable in each situation. Our data suggest that the binding of Pol II may prevent nucleosomes from occupying the −1 nucleosome position. This could also explain why nucleosomes are phased in active promoters. If the +1 nucleosome is positioned by the transcriptional machinery assembly, the other nucleosomes downstream are constrained by the positioning of the first nucleosome and the result is phasing (Kornberg and Stryer, 1988). However, nucleosomes might also assume energetically favorable positions based on DNA sequence features (Albert et al., 2007; Ioshikhes et al., 2006; Satchwell et al., 1986; Segal et al., 2006) and therefore gradually lose the phasing further away from the TSS. This interpretation is also consistent with the observation that the promoters with stalled Pol II assume similar nucleosome organization to the promoters with actively elongating Pol II. We speculate that the nucleosome positioning at regulatory regions such as promoters may be maintained by multiple factors including ATP-dependent chromatin-remodeling enzymes, DNA/chromatin-binding factors, and Pol II machinery assembly, whereas the nucleosome positioning at nonregulatory regions may be mainly controlled by the underlying DNA sequence features, as described previously.
How is the −1 nucleosome eviction or sliding regulated? Previous studies suggest that an ATP-dependent chromatin-remodeling enzyme, the SWI/SNF complex, is involved in nucleosome eviction (Gutierrez et al., 2007; Owen-Hughes et al., 1996; Schwabish and Struhl, 2007) and the Iswi2 enzyme is involved in shifting nucleosomes to a position with an energetically unfavorable DNA sequence (Whitehouse and Tsukiyama, 2006). However, histone modifications may also be involved in the process of nucleosome eviction. In the yeast Pho5 promoter, nucleosomes are first hyperacetylated and then lost upon activation (Boeger et al., 2003; Reinke and Horz, 2003). In addition, mutation of the histone acetyltransferase GCN5 has been found to decrease nucleosome eviction from GAL1 promoter and transcribed regions (Govind et al., 2007). Deposition of the histone variant H2A.Z by the ATP-dependent Ino80 into nucleosomes (Kobor et al., 2004; Krogan et al., 2003; Mizuguchi et al., 2004) may also facilitate nucleosome eviction or repositioning by destabilizing the nucleosome structure (Luger et al., 1997). Indeed, H2A.Z appears to be preferentially lost upon induction of the Pho5 gene (Santisteban et al., 2000). H2A.Z is highly enriched at yeast promoter regions (Guillemette et al., 2005; Zhang et al., 2005) and has been found to flank an NFR near the TSS (Raisner et al., 2005), suggesting a potential role for H2A.Z in maintaining the NFR. Our data that H2A.Z-containing nucleosomes are more depleted at the −1 nucleosome position than non-H2A.Z nucleosomes are consistent with this notion. Furthermore, H2A.Z-containing nucleosomes are located farther away from TSSs than non-H2A.Z nucleosomes (Figure 7). This suggests that they might be preferentially shifted and that it might be energetically more favorable to shift an H2A.Z-containing nucleosome than a regular nucleosome. It is important to point out that we cannot rule out the possibility that what we are observing in these regions is histone turnover from H2A.Z nucleosomes to nucleosomes containing the canonical H2A histone. Another histone variant, H3.3, is also found at transcriptional regulatory elements (Mito et al., 2007), and its incorporation together with H2A.Z into a nucleosome further destabilizes the nucleosome structure (Jin and Felsenfeld, 2007). Therefore, we expect that H3.3 may be also involved in the nucleosome reorganization at promoter regions upon gene activation.
EXPERIMENTAL PROCEDURES
Mononucleosome Preparation and Sequencing using the Solexa Technique
CD4+ T cells were purified from human blood as described (Barski et al., 2007). Incubating with anti-CD3 and anti-CD28 antibodies for 18 hr activated the cells. To prepare mononucleosomes, the resting and activated T cells were treated with MNase to generate approximately 80% mononucleosomes and 20% dinucleosomes. The DNA fragments of approximately 150 bp were isolated from agarose gel, blunt-ended, ligated to the Solexa adaptors, and sequenced using the Illumina 1G Genome Analyzer as described previously (Barski et al., 2007).
ChIP-Seq experiments were performed as described previously (Barski et al., 2007) using antibodies directed against histone H3 (Abcam, AB1791), Ser5 phosphorylated Pol II (Abcam, AB5131), and unphosphorylated Pol II (Abcam, AB817).
Data Analysis
Solexa Pipeline Analysis
Sequenced reads of mostly 25 bp were obtained using the Solexa Analysis Pipeline. All reads were mapped to the human genome (hg18) and all uniquely matching reads were retained. Unique read numbers for each library are listed in Table S1.
Nucleosome Scoring
Nucleosome profiles were obtained by applying a simple scoring function to the sequenced reads. A sliding window of 10 bp was applied across all chromosomes and at each window all reads mapping to the sense strand 80 bp upstream of the window and reads mapping to the antisense strand 80 bp downstream of the window contributed equally to the score of the window.
Gene Sets
Expression microarray experiments were performed for both resting and activated T cells using Affymetrix Human Genome U133 Plus 2.0 GeneChip array. The Affymetrix Microaray Suite 5 (MAS5.0) algorithm (Affymetrix, 2002) was applied to the expression data to make Absent/Present calls. Expressed (nonexpressed) probes were selected for both resting and activated T cells by selecting all probes that were unambiguously called Present (Absent) across all replicates. The Affymetrix probe ids were then mapped to UCSC genes (Hsu et al., 2006) using the mapping table provided by the UCSC Genome Table Browser (Karolchik et al., 2004). All probes not mapping uniquely to a UCSC gene were removed from analysis. Induced genes were defined as genes unambiguously called Absent in resting T cells and Present in activated T cells. Alternatively, repressed genes were defined as genes unambiguously called Present in resting T cells and Absent in activated T cells. For identifying induced and repressed genes, in an effort to get larger gene sets, no filter for nonunique mapping of Affymetrix probe ids to UCSC genes was applied.
Polymerase II Stalling Analysis
To classify genes as containing stalled or elongating polymerase II (Pol II), we utilized a stalling index similar to that employed in Zeitlinger et al. (2007). We calculated the promoter Pol II level as the sum of all Pol II signals in a 1 kilobase (kb) region surrounding the TSS and calculated the average Pol II signal for 1 kb windows through the gene body, starting 1 kb downstream of the TSS. A stalling index was then defined as the ratio of the promoter Pol II level over the average gene body level. Genes with stalled Pol II were defined as those with a stalling index of at least 10 and no detectable Pol II signal within the gene body. Genes with elongating Pol II were defined as those with a stalling index between 1 and 3 and an average gene body Pol II signal of at least 5. Genes with no promoter Pol II were defined as those with no Pol II signal in the 1 kb region surrounding the TSS.
For evaluation of these methods, we identified genes with stalled, elongated, and no Pol II in a complementary, mRNA-level-based approach. We identified genes that were unambiguously classified as Absent and Present by the Affymetrix MAS5.0 algorithm as analogous to genes with no Pol II and elongated Pol II, respectively. We then identified all genes with significant amounts of unphosphorylated Pol II in the promoter regions (see below) that were still classified as Absent as analogous to the genes with stalled Pol II.
Identification of Significant Windows of Pol II
To identify regions with significant amounts of Pol II, we modeled the distribution of sequenced reads throughout the genome as a Poisson process and calculated the number of reads necessary in 1 kb windows for a p value threshold of 0.05.
TSS Alignments
To examine nucleosomes near the TSSs of each gene set, we aligned all genes in each set using TSS coordinates of UCSC genes (Hsu et al., 2006). Sliding windows of 5 bp were applied in the regions of TSSs and all reads originating in these windows were tallied. Total counts were normalized by the numbers of genes in each set. Reads on the sense and antisense strands were treated separately. Similar methods were applied to the Pol II, H3K4me3, H3K4me2, H3K4me1, and H2A.Z libraries using data from Barski et al. (2007).
Gene Body Density of Pol II Signals
The densities for Ser5 phosphorylated and unphosphorylated Pol II (Figures 5C, 5D, 5F and 5G) were calculated by starting 5 kb upstream of the TSS of each gene and summing the Pol II signal in 250 bp windows up to the TSS. Within the gene bodies, the Pol II level was calculated in 5 kb windows. The density downstream of the gene was calculated similar to the upstream region. Densities were obtained by dividing read numbers by the total number of base pairs in each window.
Quantification of Nucleosome Levels
Nucleosome levels at individual nucleosome positions were calculated by defining the following positions to individual nucleosomes: −2 [−370: −196], −1 [−195: −46], +1 [−45:134], +2 [135:314], +3 [315:494], +4 [495:674], +5 [675:859]. Nucleosome levels corresponding to each nucleosome were calculated by summing the reads aligning to the sense strand in the window associated with each nucleosome.
ACCESSION NUMBERS
The raw sequence tags from the resting and activated human CD4+ T cells as well as from the H3 and Pol II ChIP-Seq analysis have been deposited in the Short Read Archive (SRA) (Wheeler et al., 2008) under accession number SRA000234. The microarray data have been deposited to the GEO repository under accession number GSE10437.
Supplementary Material
ACKNOWLEDGMENTS
The gene expression analysis using the Affymetrix microarrays was performed by the Gene Expression Core Facility of NHLBI. This work was supported by the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute. We thank Weiqun Peng, Chongzhi Zang, and Raja Jothi for helpful comments and discussions.
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include five figures and one table and can be found with this article online at http://www.cell.com/cgi/content/full/132/5/887/DC1/.
REFERENCES
- Affymetrix N (2002). Statistical Algorithms Description Document. http://www.affymetrix.com/support/technical/whitepapers/sadd_whitepaper.pdf.
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, and Pugh BF (2007). Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao N (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Berger SL (2002). Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, and Schreiber SL (2004). Global nucleosome occupancy in yeast. Genome Biol. 5, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, and Kornberg RD (2003). Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11, 1587–1598. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, and Singer RH (2006). Transcriptional pulsing of a developmental gene. Curr. Biol 16, 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JH, Fan HY, Reynolds SM, Yuan G, Meldrim JC, Richter DJ, Peterson DG, Rando OJ, Noble WS, and Kingston RE (2007). Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome Res. 17, 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, and Hinnebusch AG (2007). Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, and Young RA (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, and Gaudreau N (2005). Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384. 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, and Workman JL (2007). Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26, 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 39, 311–318. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T, and Ahmad N (2004). Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 20, 320–326. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, and Haussler N (2006). The UCSC known genes. Bioinformatics 22, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, and Pugh BF (2006). Nucleosome positions predicted through comparative genomics. Nat. Genet 38, 1210–1215. [DOI] [PubMed] [Google Scholar]
- Jin C, and Felsenfeld N (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Tan FJ, McCullough HL, Riordan DP, and Fire AZ (2006). Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 16, 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, and Ren N (2005). A high-resolution map of active promoters in the human genome. Nature 436, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, and Narlikar GJ (1999). ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, and Rine N (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2, E131. 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, and Lorch N (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, and Lorch N (2002). Chromatin and transcription: where do we go from here. Curr. Opin. Genet. Dev 12, 249–251. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, and Stryer N (1988). Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16, 6677–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides N (2007). Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Langst G, Schatz T, Langowski J, and Grummt N (1997). Structural analysis of mouse rDNA: coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res. 25, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, and Lieb JD (2004). Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet 36, 900–905. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. (2006). Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, and Nislow N (2007). A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet 39, 1235–1244. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, and Workman JL (2007). The role of chromatin during transcription. Cell 128, 707–719. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, and Thanos N (2002). Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110, 261–271. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, and Frazer KA (2000). Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- Lorch Y, LaPointe JW, and Kornberg RD (1987). Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, and Henikoff N (2007). Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, and Wu N (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, and Adelman N (2007). RNA polymerase is poised for activation across the genome. Nat. Genet 39, 1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T, Utley RT, Cote J, Peterson CL, and Workman JL (1996). Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273, 513–516. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, and Fisher DE (2007). High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol 25, 244–248. [DOI] [PubMed] [Google Scholar]
- Peterson CL, and Laniel MA (2004). Histones and histone modifications. Curr. Biol 14, R546–R551. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527. [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, and Madhani HD (2005). Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, and Horz N (2003). Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11, 1599–1607. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H, and Hager GL (1987). Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 6, 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, and Zhao N (2004). High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol 22, 1013–1016. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, and Zhao N (2005). Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, and Zhao N (2006). The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103, 15782–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Wei G, Farrell CM, and Zhao N (2007). Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 17, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, and Smith MM (2000). Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103, 411–422. [DOI] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, and Travers AA (1986). Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol 191, 659–675. [DOI] [PubMed] [Google Scholar]
- Schwabish MA, and Struhl N (2007). The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol 27, 6987–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, and Widom N (2006). A genomic code for nucleosome positioning. Nature 442, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, and Grunstein N (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem 76, 75–100. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, and Farnham PJ (2006). Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16, 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. (2008). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36, D13–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, and Tsukiyama N (2006). Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol 13, 633–640. [DOI] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, and Rando OJ (2005). Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, and Young RA (2007). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet 39, 1512–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, and Cairns BR (2005). Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Reinberg N (2001). Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.