Abstract
Powdery mildew (PM) is one of the most destructive diseases that threaten cucumber production globally. Efficient breeding of novel PM-resistant cultivars will require a robust understanding of the molecular mechanisms of cucumber resistance against PM. Using a genome-wide association study, we detected a locus significantly correlated with PM resistance in cucumber stem, pm-s5.1. A 1449-bp insertion in the CsMLO8 coding region at the pm-s5.1 locus resulted in enhanced stem PM resistance. Knockout mutants of CsMLO8 and CsMLO11 generated by CRISPR/Cas9 both showed improved PM resistance in the stem, hypocotyl, and leaves, and the double mutant mlo8mlo11 displayed even stronger resistance. We found that reactive oxygen species (ROS) accumulation was higher in the stem of these mutants. Protein interaction assays suggested that CsMLO8 and CsMLO11 could physically interact with CsRbohD and CsCRK2, respectively. Further, we showed that CsMLO8 and CsCRK2 competitively interact with the C-terminus of CsRbohD to affect CsCRK2-CsRbohD module-mediated ROS production during PM defense. These findings provide new insights into the understanding of CsMLO proteins during PM defense responses.
Introduction
Powdery mildew (PM) is one of the most common and widespread diseases in cucumber (Cucumis sativus L.) [1]. PM is caused by the fungus Podosphaera fusca, and the symptoms are characterized by small, white, powdery fungal growth on stems and leaves, leading to leaf shrinkage, premature senescence, and death [2]. At present, the application of fungicides is the primary way to control PM in commercial agricultural production. However, PM pathogens often acquire resistance to fungicides, rendering them ineffective, while these chemicals remain harmful to human health and to the environment [3]. Breeding PM-resistant cucumber cultivars is the most efficient and environmentally friendly method to control PM.
PM-resistant cucumber lines have been reported since the 1940s, mostly from Asian germplasm. They include ‘Puerto Rico 37’ from Chinese germplasm [4], PI 197087 from India [5], ‘Yomaki’ (PI288238) and ‘Natsufushinari’ (PI 279465) from Japan [6], and ‘Bangalore’ and ‘Burma’ lines (PI 200815 and PI 200818) from India [4]. These germplasms and their derived lines have been used for subsequent cucumber PM resistance breeding. Resistance in these accessions was primarily due to the presence of multiple recessive genes.
Quantitative trait loci (QTLs) for PM resistance in cucumber leaves have been detected on all seven cucumber chromosomes. Sakata et al. [7] first reported QTLs for PM resistance using 95 cucumber accessions; six QTLs underlying PM resistance in the Indian accession PI197088-1 were detected, including one major QTL conferring resistance. Liu et al. [8] identified five QTLs responsible for PM resistance originating from a European greenhouse cucumber line, ‘S06’. Zhang et al. [9] detected four QTLs (pm5.1, pm5.2, pm5.3, and pm6.1) controlling PM resistance in North China cucumber line ‘K8’ and identified a major QTL on chromosome 5. He et al. [10] detected four QTLs (pm-tl1.1, pm-tl1.2, pm-tl5.1, and pm-tl5.2) for PM resistance on the true leaf of ‘WI 2757’. Nie et al. [11] detected a major QTL, pm5.1, in an East Asian line, ‘S1003’. Moreover, Zhang et al. [12] detected two major QTLs on chromosomes 1 and 6 using ‘BK2’ and ‘H136’. Wang et al. [13] identified four QTLs (pm1.1, pm2.1, pm5.1, pm 6.1) in PI197088. Liu et al. [14] performed a genome-wide association study (GWAS) on leaf PM resistance and detected 13 PM loci distributed across almost all chromosomes.
While many QTLs for leaf PM resistance have been identified, only a few candidate genes underlying these QTLs have been predicted. Xu et al. [15] identified Csa1M064780 and Csa1M064790, encoding cysteine-rich receptor-like kinases, as candidate genes. Zhang et al. [16] suggested that Csa5M622830, which encodes a GATA transcription factor, was the most likely candidate gene. Recently, Zhang et al. [17] suggested that CsGy5G015660, which encodes a putative leucine-rich repeat receptor-like serine/threonine-protein kinase, was the causal gene for PM resistance.
To date, very few studies have focused on PM resistance on cucumber stems or hypocotyls. He et al. [10] detected three QTLs (pm-hy3.1, pm-hy4.1, and pm-hy5.1) for hypocotyl resistance in ‘WI 2757’, with pm-hy5.1 playing a key role. Liu et al. [18] found a recessive nuclear gene (pm-s) controlling stem PM resistance in ‘NCG-122’, and predicted that the Mildew Resistance Locus O (MLO)-related protein-encoding gene Csa5G623470 as the most possible candidate gene. However, the function of these genes has not yet been verified via genetic complementation studies, and therefore the genetic and molecular mechanisms underlying PM resistance remain unclear.
S-genes have been the focus of many plant–pathogen interaction studies because they encode proteins that facilitate host recognition and defense suppression, and enable pathogen penetration of plant tissue [19]. Loss of S-gene function confers broad-spectrum resistance against a variety of diseases in plants [20]. Over 180 S-genes have been identified in various plant species [19]. Among them, Mildew Resistance Locus O (MLO), which encodes a plasma membrane-anchored protein, is the most widely known S-gene responsible for PM susceptibility in plants [21]. Resistance mediated by mlo was first discovered in barley [22], and since then, mlo-mediated resistance to PM has been reported in other plant species, e.g. AtMLO2, AtMLO6, and AtMLO12 in Arabidopsis [23], SlMLO1 in tomato [24], TaMLO_A1 and TaMLO_B1 in wheat [25], CaMLO2 in pepper [26], VvMLO3, VvMLO6, and VvMLO7 in grapevine [27–29], FaMLO10 and FaMLO20 in strawberry [30], and PsMLO1 in pea [31, 32]. In cucumber, 16 CsMLO genes have been identified, and three genes that belong to clade V, i.e. CsMLO1, CsMLO8, and CsMLO11, were associated with PM susceptibility [33]. CsMLO8 was co-located with a major-effect recessive QTL on chromosome 5 for leaf and hypocotyl PM resistance in several studies [10, 11, 18, 34], while CsMLO1 and CsMLO11 were co-localized with QTLs pm1.1 and pm6.1, respectively [35]. While some studies showed that mlo-mediated resistance by plants depends on enhanced callose deposition [36] and increased defense responses such as hydrogen peroxide bursts [37], the molecular mechanism underlying this defense response is still ambiguous, especially in cucumber.
In this study we performed a GWAS using 95 diverse cucumber accessions in two seasons, and detected a major locus significantly associated with stem PM resistance. The candidate gene CsMLO8 within the pm-s5.1 locus, and its homolog CsMLO11, were functionally characterized. Mutants of mlo8, mlo11, and mlo8mlo11 showed improved PM resistance. In addition, the accumulation of reactive oxygen species (ROS) increased in the stems of these mutants. We further demonstrated that CsMLO8 and CsMLO11 could physically interact with respiratory burst oxidase homolog D (CsRbohD) and cysteine-rich receptor-like kinase 2 (CsCRK2), respectively. Further, CsMLO8 and CsCRK2 competitively interact with the C-terminus of CsRbohD to regulate ROS production during PM defense. These findings provide new insights into the function of the CsMLO genes for PM defense responses.
Results
Genetic diversity of powdery mildew resistance among accessions of cucumber core germplasms
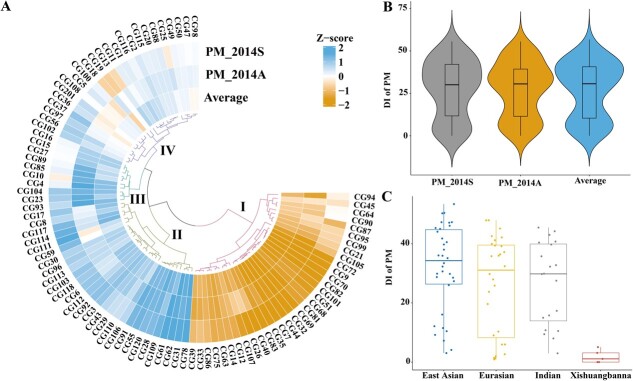
A collection of 95 accessions from cucumber core germplasm (CG), acquired from different regions of the world (Supplementary Data Table S1), was characterized for PM resistance in stems in spring 2014 and fall 2014. The average disease index (DI) of each accession was calculated in each experiment (Fig. 1a, Supplementary Data Table S1). PM resistance showed diverse phenotypic variations, ranging from 0 to 55.56 in both the spring and fall trials (Fig. 1b; Supplementary Data Table S2). The cucumber accessions were classified into four clusters based on the average DI: I, highly tolerant; II, tolerant; III, sensitive; and IV, highly sensitive (Fig. 1a; Supplementary Data Table S3). A total of 33 highly tolerant germplasms with DIs ranging from 0.00 to 20.99 and seven highly sensitive germplasms with DIs ranging from 46.28 to 53.09 were identified. The 95 accessions consisted of four ecotypes, including East Asian (n = 35 lines), Eurasian (n = 29), Indian (n = 21), and Xishuangbanna (n = 10) types (Fig. 1c; Supplementary Data Table S1). The mean DI of PM in the Xishuangbanna ecotype was lower than all others, suggesting that the Xishuangbanna ecotype is more resistant to PM infection (Fig. 1c).
Figure 1.
Genetic diversity of stem PM resistance among accessions of cucumber core germplasms. a Heat map indicating the DI of stem PM resistance in spring 2014 (PM_2014S) and fall 2014 (PM_2014A). The four clusters of the CG accessions are numbered I–IV. Yellow means resistant and blue susceptible. b Violin plots depicting the distribution of DI in two seasons. c Box plots of DI of PM among different ecotypes.
Genome-wide association studies reveal CsMLO8 as a candidate gene for stem powdery mildew resistance
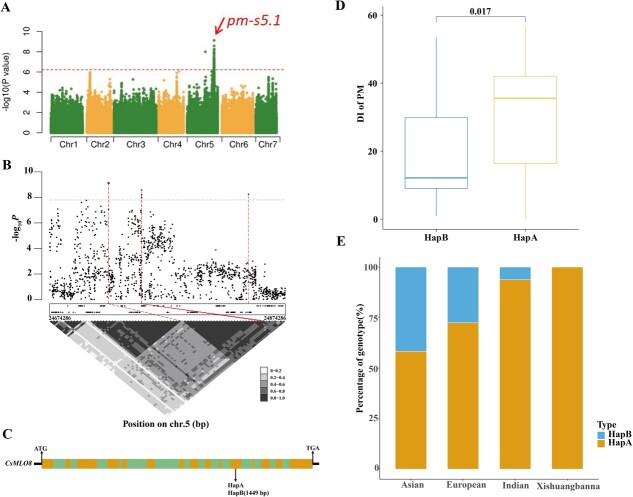
To identify the key genes for PM resistance on cucumber stem, the DI of each experiment was used for a GWAS using a mixed linear model. A stable association signal in a 200-kb region on chromosome 5 was detected across the two seasons and was named pm-s5.1 (Fig. 2a; Supplementary Data Fig. S1; Supplementary Data Table S4). To identify potential candidate genes within this locus, SNPs of chromosome 5 between nucleotides 24 674 286 and 24 874 286 were analyzed by pairwise linkage disequilibrium (LD) correlations (Fig. 2b). We then focused on the interval from 24 724 286 to 24 842 958 nucleotides (r2 ≥ 0.6). There are 19 annotated ‘protein-coding’ genes in this region, using the cucumber ‘9930’ reference genome (Supplementary Data Table S5). Among these genes, Csa5G623470 was of interest; it encoded a seven-transmembrane-domain protein and there was a 1449-bp insertion (HapB) in the 11th exon (Fig. 2c). The HapB accessions all had significantly (P = 0.017) lower DIs than those without the insertion (HapA types) (Fig. 2d), indicating that Csa5G623470 is a candidate gene for PM resistance in cucumber stem. Interestingly, the HapB allele existed in the Eurasian, East Asian, and Indian types, but was absent in the Xishuangbanna-type cucumbers (Fig. 2e), suggesting that the strong resistance seen on the stems of the 10 Xishuangbanna-type accessions is not caused by the loss of CsMLO8.
Figure 2.
GWASs reveal that natural variation of CsMLO8 confers PM resistance in cucumber stem. a GWAS Manhattan plot for stem PM resistance in spring of 2014. The genome-wide significance threshold was P = 5.9 × 10−7. The red arrow indicates the position of a strong peak (pm-s5.1). b The LD block surrounding the pm-s5.1 peak. The red dotted lines indicate the candidate region. c Gene structure of Csa5G623470 in the interval. Filled yellow box, filled green box, and black line represent exons, introns, and the UTR respectively. d Box plots of DI, based on the haplotypes of Csa5G623470: HapA (WT) and HapB (with a 1449-bp insertion). e Proportion of HapA and HapB alleles within each ecotype.
Mutations in CsMLO8 and CsMLO11 confer resistance of the relevant mutants to powdery mildew
To validate the function of CsMLO8 in PM resistance, we knocked out CsMLO8 in ‘CU2’, a variety harboring the HapA allele, which shows PM susceptibility on the stem, using the CRISPR/Cas9 system. Two independent homozygous knockout lines, named mlo8CR1 and mlo8CR2, respectively, were obtained. mlo8CR1 had a 1-bp deletion which led to an early stop codon, while mlo8CR2 had a 6-bp deletion on the third exon. These deletions are predicted to produce polypeptides with a 446- and 2-amino acid deletion in mlo8CR1 and mlo8CR2, respectively (Fig. 3a). Since CsMLO11 has been previously reported as a PM-susceptible gene, we also knocked out CsMLO11 in ‘CU2’. Two independent homozygous knockout lines, named mlo11CR1 and mlo11CR2, respectively, were obtained; mlo11CR1 had a 1-bp insertion while mlo11CR2 had a 16-bp deletion in the third exon, and both polymorphisms led to the creation of an early stop codon (Fig. 3b). A double mutant of CsMLO8 and CsMLO11 (mlo8CR1mlo11CR1) was also generated by crossing the single mutants.
Figure 3.
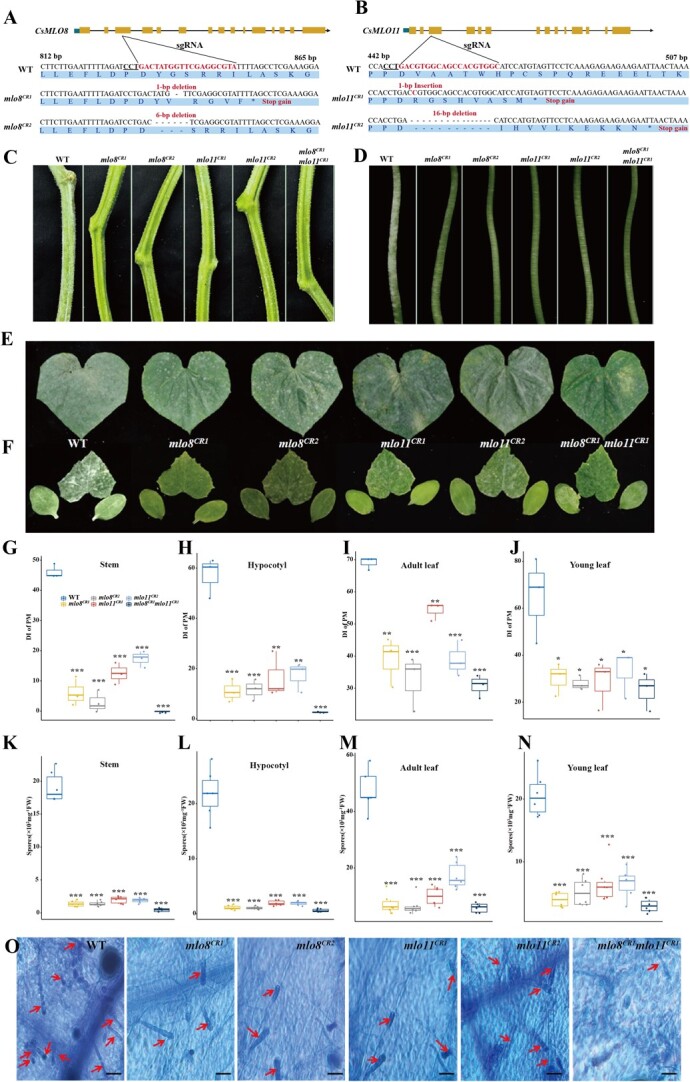
PM resistance in CRISPR/Cas9 mutants of CsMLO8/11. a, b Schematic illustrating the gene structure of (a) CsMLO8 and (b) CsMLO11; filled blue, yellow boxes, and black lines represent the UTR, exons, and introns respectively. mlo8CR1 and mlo8CR2 are homozygous mutants for CsMLO8. mlo11CR1 and mlo11CR2 are homozygous mutants for CsMLO11. c–f PM infection phenotype of mutants (mlo8CR1, mlo8CR2, mlo11CR1, mlo11CR2, mlo8CR1mlo11CR1) and WT plants in (c) stem, (d) hypocotyl, (e) adult leaf, and (f) young leaf. Photographs were taken at 62 dps for the stem, 8 dpi for the hypocotyl, 55 dps for the adult leaves, and 6 dpi for the young leaves. g–j Statistical analyses of DI for (g) stem, (h) hypocotyl, (i) adult leaf, and (j) young leaf. ***P < 0.001; **P < 0.01; *P < 0.05, Student’s t-test, n = 3. k–n Number of spores in (k) stem, (l) hypocotyl, (m) adult leaf, and (n) young leaf. ***P < 0.001, Student’s t-test, n = 6. o Trypan blue staining of young leaves of the WT and mlo mutants at 6 dpi. Red arrows indicate spore hyphae. Scale bars, 50 μm.
These mutants (mlo8CR1, mlo8CR2, mlo11CR1, mlo11CR2, and mlo8CR1mlo11CR1) together with the wild-type control (WT) were evaluated for PM resistance at the seedling and adult stages. For adult stem, at 62 days after sowing (dps), the WT showed severe PM infection with an average DI of 46.2. The mlo8CR1 and mlo8CR2 genotypes both showed significantly increased stem PM resistance, with a DI of 6.0 and 3.0, respectively. The mlo11CR1 and mlo11CR2 genotypes also showed significantly reduced stem PM infection, with a DI of 12.6 and 17.4, respectively. For the double mutant mlo8CR1mlo11CR1, no PM infection was detected on the stem (Fig. 3c and g). For hypocotyls, the WT showed severe PM infection at 8 days post-inoculation (dpi), with an average DI of 57.1. However, mlo8CR1 and mlo8CR2 both showed significantly increased PM resistance, with a DI of 11.0 and 11.7, respectively. The mlo11CR1 and mlo11CR2 genotypes also showed significantly reduced PM infection, with a DI of 16.50 and 17.3, respectively. The mlo8CR1mlo11CR1 double mutant exhibited the strongest resistance, with a DI of 2.7 (Fig. 3d and h). We also evaluated PM resistance in young and adult leaves. At 55 dps, the adult leaf of the WT showed severe PM symptoms, with an average DI of 69, while the mlo mutants were less affected, with DIs of 32.5–39 in mlo8, 39–54.2 in mlo11, and 30.9 in mlo8CR1mlo11CR1 (Fig. 3e and i). At 6 dpi, the first true leaf of the WT showed severe disease symptoms, while symptoms were mild in the mutants. The average DI of the WT was 65, and was significantly higher than those of mlo8 (30.2, 28), mlo11 (28.5, 33.2), and mlo8CR1mlo11CR1 (25.1) mutants (Fig. 3f and j). Consistent with this observation, compared with the WT control, mlo8, mlo11, and mlo8CR1mlo11CR1 had 13-, 9-, and 42-fold fewer spores on the infected stems at 62 dps (Fig. 3k), and 20-, 10-, and 35-fold fewer spores on hypocotyls at 8 dpi, respectively (Fig. 3l). In addition, mlo8, mlo11, and mlo8CR1mlo11CR1 had 6-, 4-, and 7-fold fewer spores on infected adult leaves at 55 dps (Fig. 3m), and 4-, 2-, and 6-fold fewer spores on young leaves at 6 dpi, respectively (Fig. 3n). Moreover, Trypan blue staining revealed that there were fewer fungal hyphae and conidia in the mutants compared with the WT at 6 dpi (Fig. 3o). These results indicate that disruption of CsMLO8 and/or CsMLO11 results in enhanced resistance to PM, and the resistance is more obvious in the stem and hypocotyl.
Multiple metabolism- and defense-related genes were upregulated in the CsMLO8/11 mutants
To further investigate the molecular mechanism by which CsMLO8 and CsMLO11 regulate PM resistance in cucumber, we compared the transcriptomic profiles of mutant (mlo8CR1, mlo11CR1, mlo8CR1mlo11CR1) and WT hypocotyls before inoculation (0 hpi) and 24 h post-inoculation (24 hpi).
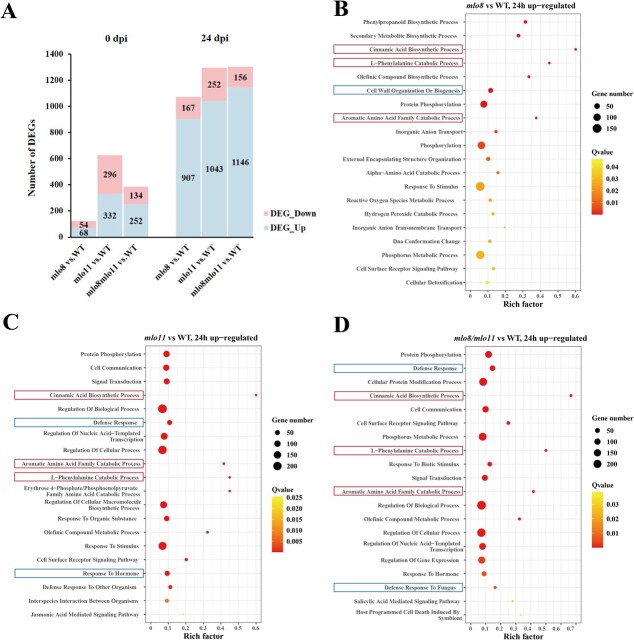
Before inoculation, a total of 122 differentially expressed genes (DEGs) were identified between mlo8CR1 and the WT, and 628 and 386 DEGs between the WT and mlo11CR1 and mlo8CR1mlo11CR1, respectively (Fig. 4a). Interestingly, at 24 hpi the number of DEGs was significantly increased compared with 0 hpi: 1074 in mlo8CR1, 1295 in mlo11CR1, and 1302 in mlo8CR1mlo11CR1 compared with the WT. Among them, 84.5, 80.5, and 88.0% of the DEGs were upregulated in mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1, respectively (Fig. 4a).
Figure 4.
DEGs between mlo8CR1 and mlo11CR1 CRISPR-Cas9 mutant lines and controls. a Up- and downregulated DEGs in mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 relative to WT control at 0 and 24 hpi. GO pathways enriched among upregulated genes of (b) mlo8CR1 vs WT, (c) mlo11CR1 vs WT, and (d) mlo8CR1mlo11CR1 vs WT. The red boxes indicate the common pathways detected in the three tested mutants, while the blue boxes represent the unique pathways found in the different mutants.
To better understand the DEGs in the aforementioned mutants at 24 hpi, Gene Ontology (GO) enrichment was performed. Several significantly upregulated pathways (q value <0.05) related to plant disease resistance were enriched. Primary and secondary metabolites, including l-phenylalanine, aromatic amino acid, and cinnamic acid, were also upregulated in mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 (Fig. 4b–d; Supplementary Data Table S6). Interestingly, pathways related to cell wall organization or biogenesis were uniquely enhanced in mlo8CR1 (Fig. 4b), while pathways related to defense response were significantly upregulated in mlo11CR1 and mlo8CR1mlo11CR1 (Fig. 4c and d). These results indicated that CsMLO8 and CsMLO11 may function in overlapping cellular signaling pathways.
More reactive oxygen species accumulation in mlo8/11 stem after powdery mildew inoculation
ROS production is important in plant pathogen-defense responses. NADPH oxidase/respiratory burst oxidase homolog (Rboh) proteins function in localized ROS ‘bursts’ [38]. RbohD is required for ROS production during the immune response in Arabidopsis [39]. CYSTEINE-RICH RLK2 (CRK2) kinase could interact with RbohD and is required for the full elicitor-induced ROS burst [40]. Interestingly, we found that CsaV3_3G043480 (CsRbohD) was significantly upregulated by PM inoculation in mlo8CR1, and mlo8CR1mlo11CR1, but not in the WT controls. Moreover, the expression level of CsaV3_2G024820 (CsCRK2) increased in WT, mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 after PM inoculation, and the upregulation was more significant in mlo8CR1 and mlo8CR1mlo11CR1 (Fig. 5a; Supplementary Data Table S7). Moreover, from the RNA-seq data, we found that the transcripts of genes encoding key antioxidant enzymes involved in ROS scavenging, e.g. superoxide dismutase (SOD), glutathione peroxidase (GPX), ascorbate peroxidase (APX), and glutathione S-transferase (GST), were decreased in WT, mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 at 24 hpi compared with 0 hpi (Supplementary Data Table S7).
Figure 5.
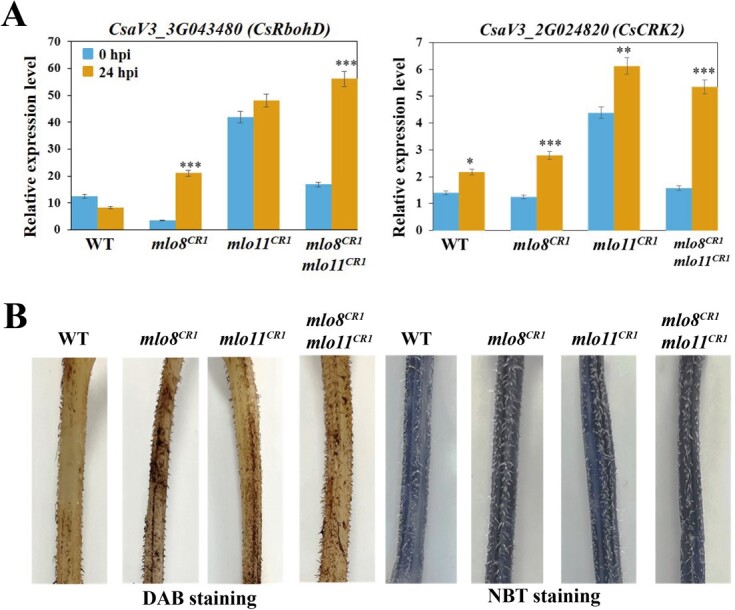
Changes in ROS accumulation in stems of CsMLO mutants and WT after PM infection. a Relative expression levels of CsRbohD and CsCRK2 at 0 and 24 hpi *P < 0.05, **P < 0.01, ***P < 0.001, ANOVA, Student’s t-test. b Stems harvested at 6 dpi were stained. Blue and brown precipitate indicates O2− and H2O2 location and content.
To investigate whether the enhanced stem PM resistance of CsMLO mutants was related to ROS accumulation, we examined hydrogen peroxide (H2O2) and superoxide anions (O2−) in PM-inoculated stems. Diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining revealed a higher increase in H2O2 and O2− accumulation, respectively, in the stem of the three mutants compared with control plants, suggesting that mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 conferred increased ROS production (Fig. 5b). These results suggested that dysfunction of the CsMLO8 and CsMLO11 genes facilitates a burst of ROS production which contributes to the PM immune response.
CsMLO8 and CsMLO11 could physically interact with CsRbohD and CsCRK2
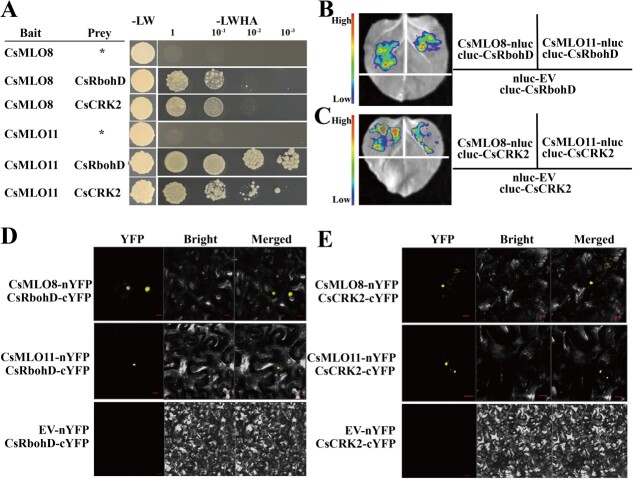
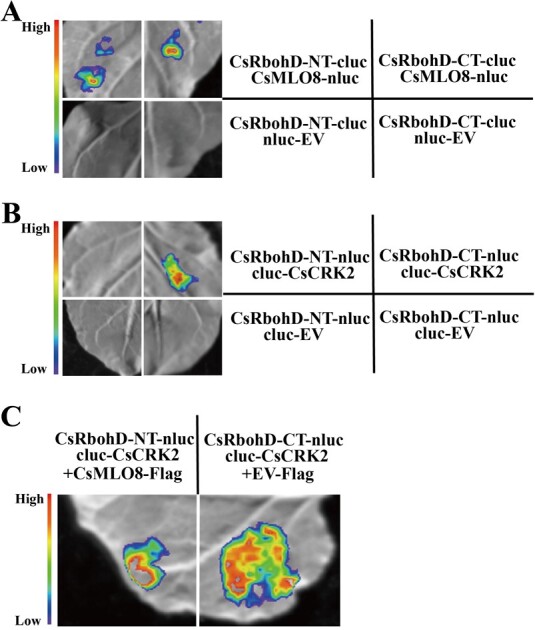
To further investigate whether CsMLO8 or CsMLO11 could physically interact with CsRbohD and CsCRK2, we carried out diverse biochemical assays. First, yeast two-hybrid (Y2H) assays demonstrated that CsMLO8 or CsMLO11 indeed interacts with CsRbohD and CsCRK2 (Fig. 6a). To further confirm the protein interactions in plant cells, we performed luciferase complementation imaging assays (LCIs) in Nicotiana benthamiana leaves. CsMLO8 and CsMLO11 were fused to the N-terminus of LUC (nLUC) to generate CsMLO8-nLUC and CsMLO11-nLUC, respectively; meanwhile, CsRbohD and CsCRK2 were fused to the C-terminus of LUC (cLUC) to generate cLUC-CsRbohD and cLUC-CsCRK2. As revealed in Fig. 6b, strong LUC activities were detected in the samples co-expressing CsMLO8-nLUC/cLUC-CsRbohD and CsMLO11-nLUC/cLUC-CsRbohD. Similarly in Fig. 6c, strong LUC activities were detected in the samples co-expressing CsMLO8-nLUC/cLUC-CsCRK2 and CsMLO11-nLUC/cLUC-CsCRK2. We then performed transient bimolecular fluorescence complementation (BiFC) assays in N. benthamiana leaves. CsRbohD and CsCRK2 were fused to the C-terminus of yellow fluorescent protein (cYFP). CsMLO8 and CsMLO11 were fused to the N-terminus of YFP (nYFP). Strong interaction signals were detected in the CsMLO8-nYFP/CsRbohD-cYFP, CsMLO11-nYFP/CsRbohD-cYFP, CsMLO8-nYFP/CsCRK2-cYFP, and CsMLO11-nYFP/CsCRK2-cYFP co-expressing samples (Fig. 6d and e). Taking these results together, we concluded that CsMLO8/11 interacts with CsRbohD and CsCRK2.
Figure 6.
Physical interaction assays for CsMLO8/11, CsRbohD, and CsCRK2. a Y2H analyses showing the interaction between CsMLO8/11 and CsRbohD and between CsMLO8/11 and CsCRK2. Asterisks indicate empty vectors. Positive interactions were tested using yeast cells grown on synthetic defined medium lacking Leu, Trp, His, and adenine (−LWHA). b, c LCI assays showing the interaction between (b) CsMLO8/11 and CsRbohD, and (c) CsMLO8/11 and CsCRK2. d, e BiFC assays showing the interaction between (d) CsMLO8/11 and CsRbohD, and (e) CsMLO8/11 and CsCRK2. Scale bars, 10 μm.
CsMLO8 and CsCRK2 competitively interact with the C-terminus of CsRbohD
To determine which region of CsRbohD mediates the CsRbohD–CsMLO8 interaction, the N- and C-termini of CsRbohD were fused with cLUC, respectively, for LCI assays in N. benthamiana leaves. Obvious LUC activities were observed in both the CsMLO8-nLUC/cLUC-CsRbohD-CT and CsMLO8-nLUC/cLUC-CsRbohD-NT co-expression samples, but no LUC activity was observed in the control samples (Fig. 7a). These results suggest that CsMLO8 interacts with both the N- and C-termini of CsRbohD. Furthermore, it has been reported that CRK2 contributes to the activation of RbohD via phosphorylation of its C-terminus in Arabidopsis [40]. Our experiment (Fig. 7b) also showed LUC activities in the CsRbohD-CT-nLUC/cLUC-CsCRK2, but not in the CsRbohD-NT- nLUC/cLUC-CsCRK2 co-expression samples, demonstrating that CsCRK2 interacts with the C-terminus of CsRbohD. As both CsMLO8 and CsCRK2 interact with the C-terminus of CsRbohD, we wondered if CsMLO8 competitively interferes with the interaction between CsRbohD and CsCRK2. We therefore performed LCI assays in N. benthamiana leaves. As shown in Fig. 7c, the luminescence signals of the samples co-expressing CsRbohD-CT-nLUC/cLUC-CsCRK2 and CsMLO8-flag were significantly inhibited compared with those of samples co-expressing CsRbohD-CT-nLUC/cLUC-CsCRK2 and control vector. Taken together, these results indicate that CsMLO8 and CsCRK2 competitively interact with the C-terminus of CsRbohD.
Figure 7.
CsMLO8 interferes with the interaction of CsCRK2 and CsRbohD. a, b LCI assays showing the interaction between the truncated versions of CsRbohD and (a) CsMLO8 and (b) CsCRK2. c CsMLO8 inhibits the interaction of CsCRK2 and the C-terminus of CsRbohD. CsRbohD-NT, amino terminal domain of CsRbohD; CsRbohD-CT, carboxyl terminal domain of CsRbohD. pCAMBIA1300-flag was used as a control (EV-Flag) instead of CsMLO8-Flag. Agrobacterium tumefaciens strain GV3101 transformed with equal quantities of these constructs was injected into N. benthamiana leaves.
Discussion
PM is one of the most severe diseases that threaten cucumber production globally; breeding PM-resistant cultivars enables sustainable cucumber cultivation. However, the genes responsible for PM resistance and their underlying molecular mechanism in cucumber are relatively unknown. Here, we characterized PM resistance on the stems of 95 accessions of the core collection, which is a subset of the 3000 globally acquired cucumber accessions. Using 1 702 257 SNPs, we detected one locus, pm-s5.1, significantly associated with stem PM resistance on chromosome 5 in two seasons. The stem resistance conferred by pm-s5.1 in this study is co-localized with a QTL, pm-s, for stem PM resistance we previously identified [18] and a QTL, pm5.2, for hypocotyl resistance detected by He et al. [10] . In addition, we had previously characterized PM resistance in the leaves of individuals from the core accessions [14], and identified 13 loci associated with leaf PM resistance. Among them, the pmG5.4 locus co-localized with pm-s5.1, indicating that the major locus pm-s5.1 confers both stem and leaf PM resistance. Compared with the loci for stem resistance, more QTLs for leaf resistance were detected across all seven chromosomes [7, 8, 10–14], suggesting that there are more genes involved in leaf PM resistance compared with stem PM resistance in cucumber.
Within the pm-s5.1 locus, we found a 1449-bp insertion in the coding region of CsMLO8 that resulted in significantly higher PM resistance on the stem. The result is consistent with the study by Berg et al. [34], who showed that a non-autonomous Class LTR retrotransposable element in CsMLO8 was present frequently in cultivated cucumber germplasm, and caused hypocotyl PM resistance. Moreover, in our study, the Xishuangbanna ecotype generally had higher PM resistance in the stem compared with the other ecotypes. However, the geographical distribution of the HapA and HapB alleles, associated with susceptibility and resistance, respectively, showed that the HapB allele is absent among Xishuangbanna ecotypes, suggesting that it is not CsMLO8 but another gene that is responsible for the stem PM resistance of Xishuangbanna-type cucumbers.
Among the 16 CsMLO genes in cucumber, CsMLO1, CsMLO8, and CsMLO11 were associated with PM susceptibility [33]. The heterologous overexpression of CsMLO8 in a tomato mlo mutant restored PM susceptibility [34]. Later studies showed that the overexpression of CsMLO1 in the tomato mlo mutant completely restored PM susceptibility, while overexpression of CsMLO11 only led to partial restoration [33]. CsMLO1, CsMLO8, and CsMLO11 mutants were recently generated in cucumber plants [41]; the PM symptoms on mlo8mlo11 and mlo1mlo11 leaves were significantly decreased compared with mlo1 plants, and no PM symptoms appeared on mlo1mlo8mlo11 leaves [41]. However, PM resistance on stem or hypocotyl was not characterized in these studies. To verify the role of CsMLO8 in stem resistance, we generated two independent CsMLO8 knockout lines (named mlo8CR1 and mlo8CR2) via CRISPR/Cas9. Mlo8CR1 and mlo8CR2 both showed significantly increased PM resistance on the stem and hypocotyl compared with non-transformed controls (Fig. 3). We also knocked out CsMLO11 to obtain mlo11CR1 and mlo11CR2, and generated a double mutant of CsMLO8 and CsMLO11 (mlo8CR1mlo11CR1). The mlo11CR1 and mlo11CR2 mutants showed significantly reduced PM infection on both stem and hypocotyl, but the resistance was comparatively weak compared with that of mlo8CR1 and mlo8CR2. In contrast, the double mutant mlo8CR1mlo11CR1 exhibited stronger resistance on the stem and hypocotyl compared with the single mutant mol8 or mlo11 (Fig. 3). This revealed that CsMLO8 has a higher impact on stem PM resistance than CsMLO11. PM resistance in the adult and young leaf was also evaluated. Consistent with data from Tek et al. [41], the PM infection on mlo8 and mlo11 leaves was markedly decreased compared with the WT. Moreover, the PM symptoms and spore density decreased more noticeably on the stem and hypocotyls compared with the leaves, indicating that disruption of CsMLO8 and/or CsMLO11 results in greater reduction of PM infection on stems than on leaves.
To further illustrate the molecular mechanism of mlo8- and mlo11-mediated PM resistance, we compared the hypocotyl transcriptome of the mutants, i.e. mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1, with that of the WT control. The results suggested that the transcript levels of a number of genes in primary and secondary metabolic pathways were higher in mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 plants relative to control after pathogen inoculation; these metabolites included the aromatic amino acid l-phenylalanine, cinnamic acid, and phenylpropanoid. A diversity of secondary metabolites function in defense against biotic stress in plants [42, 43]. Aromatic amino acids (tryptophan, phenylalanine, and tyrosine) are the branch points where primary metabolites may enter secondary metabolic pathways that function in protecting plants from biotic stresses [44, 45]. Phenylpropanoids are associated with plant biotic stress tolerance [46]. In the phenylpropanoid biosynthesis pathway, l-phenylalanine is converted to trans-cinnamic acid to produce the precursor for the biosynthesis of salicylic acid, a crucial hormone regulating pathogen response and disease resistance [47, 48]. Moreover, several different pathways were enhanced in mlo8CR1 and mlo11CR1. In mlo8CR1, cell wall organization or biogenesis pathways were enriched. Sun et al. [49] recently screened a PM-susceptible mutant from an EMS-mutagenized population of a natural mlo8 mutant, and also found that PM resistance of mlo8 was associated with increased cell wall deposition, while in mlo11CR1 and mlo8CR1mlo11CR1 defense response pathways were significantly upregulated, suggesting that CsMLO8 and CsMLO11 might have different defense mechanisms. Our findings are supported by the study of Tek et al. [41], in which CsMLO8 was proposed as a negative regulator in the pre-invasive response, while CsMLO11 could be associated with the post-invasive defense response.
ROS can induce damage to cellular molecules, and contribute to programmed cell death and defense response [50, 51]. The enhanced PM resistance in transgenic Vitis vinifera plants expressing VqWRKY31 and VqWRKY56 was correlated with increased levels of ROS [52, 53]. In cucumber, it was reported that PM resistance involved ethylene-mediated ROS metabolism [3, 54]. In our study, compared with the WT, a burst of ROS production was also detected in the stem of mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1. The ROS accumulation induced by pathogen infection was linked to calcium signaling and callose deposition at the plasmodesmata [55]. Apoplastic ROS are produced by cell wall peroxidases and Rbohs in plants [56, 57]. During pathogen invasion, the activation of pattern recognition receptors leads to apoplastic ROS production through Rbohs and peroxidase activity [57–59], and RbohD is the highest expressed Rbohs in Arabidopsis [58, 60, 61]. CYSTEINE-RICH RLK2 (CRK2) kinase phosphorylates the C-terminal region of RBOHD and is required for the full elicitor-induced ROS burst [40]. Here, we found that both CsRbohD and CsCRK2 were more highly expressed in mlo8CR1, mlo11CR1, and mlo8CR1mlo11CR1 (Fig. 5a). These results suggest that the loss of function of CsMLO8 and CsMLO11 promoted the ROS burst, contributing to PM resistance.
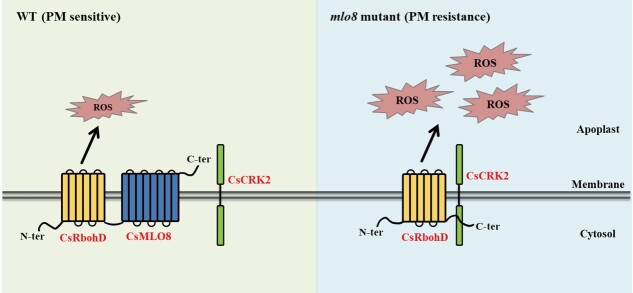
MLOs are involved in several biological processes, including root thigmomorphogenesis [62], fertilization [63], and stress response [64, 65]. The function of MLO proteins in multiple plant processes has been summarized [66], and it was proposed that MLO proteins function in responses to mechanophysical stimuli. With respect to pathogen response, barley HvMLO1 promotes arbuscular mycorrhizal colonization by Rhizophagus. irregularis [64] and Serendipita. indica infection of barley roots [65], and the development of the pathogen is terminated upon cell wall penetration in barley mlo mutants [67]. In Arabidopsis, three MLO proteins are associated with PM susceptibility: the single mutant Atmlo2 is partially resistant and the triple mutant mlo2mlo6mlo12 showed complete PM resistance [23]. Protein interactome analyses found that MLOs could interact with cyclic nucleotide-gated channels (CNGCs) to regulate polarized tip growth of the pollen tube [68, 69], with the vesicle-associated membrane protein VAMP72 clade to regulate calcium-dependent root thigmomorphogenesis [69], and with a cytoplasmic calcium sensor (CaM) to modulate HvMLO1-mediated PM susceptibility [70] and AtMLO10-mediated pollination [71]. Although it has been reported that MLOs regulate ROS [72, 73], the underlying molecular mechanism of how MLOs regulate the ROS signaling pathway remains unclear. In this study, our experiments demonstrated that CsMLO8 and CsMLO11 could physically interact with CsRbohD and CsCRK2. Moreover, CsMLO8 and CsCRK2 competitively interact with the C-terminus of CsRbohD. Therefore, we propose a working model to explain the possible mechanism of CsMLO8-mediated susceptibility to PM in cucumber stem (Fig. 8). In WT cucumber, CsMLO8 inhibits the interaction between CsCRK2 and the C-terminus of CsRbohD, thus less ROS is produced, resulting in PM susceptibility. In the mlo8 mutant, the inhibition of CsCRK2 by CsMLO8 is relieved, and thus CsCRK2 is able to phosphorylate the C-terminal region of CsRbohD and induce a ROS burst, resulting in a PM-resistant phenotype. Taken together, our results revealed that CsMLO8/11 interact with CsCRK2 and CsRbohD to regulate PM resistance in cucumber stem. CsMLO8 and CsMLO11 are therefore pivotal target genes that could be manipulated to breed new cultivars with PM resistance.
Figure 8.
A schematic model explaining the possible mechanism of CsMLO8-triggered PM susceptibility in cucumber. CsCRK2 contributes to the activation of CsRbohD via phosphorylation of its C-terminus. In WT cucumber, CsMLO8 binds to the C-terminus of CsRbohD, resulting in inhibition of the interaction between CsCRK2 and CsRbohD; thus less ROS is produced, resulting in PM susceptibility. In an mlo8 mutant, CsCRK2 phosphorylates the C-terminal region of CsRbohD and induces an ROS burst, resulting in the PM-resistant phenotype.
Materials and methods
Investigation of stem powdery mildew resistance in 95 cucumber accessions
PM resistance of the adult stem was evaluated in 95 cucumber accessions collected from different geographical origins (Supplementary Data Table S1). The PM resistance of each accession was evaluated in spring 2014 and fall 2014. The experiments were conducted in the greenhouse at Changping (40°13′ N, 116°05′ E) and Shunyi farms (40°15′ N, 116°83′ E) in Beijing, China, in a randomized complete block design. Three replicates were set for each experiment, and five plants per accession were used for each replicate.
PM infection naturally occurred on the stems of adult plants 7 weeks after sowing in the greenhouse. Ten weeks after sowing, PM symptoms on the stem were ranked by disease severity of each plant on a scale of 0, 1, 3, 5, 7, or 9, based on the proportion of the stem surface with the characteristic PM spots, as follows: 0, no symptoms; 1, ≤25%; 3, 25–50%; 5, 50–75%; 7, ≥ 75%; 9, PM spots covered the entire stem. For each line, a disease index (DI) was used as an indicator to represent PM resistance in each plant. DI = 100 × ∑(number of plants with disease rating × disease rating)/(total number of plants × highest disease rating) [9].
To identify similarities and differences among the core accessions in their response to PM infection, a phylogenetic tree was constructed using SAS 9.0 based on the average DI of each accession in two seasons.
Genome-wide association analysis
GWAS was performed with 1 702 257 SNPs using the standard mixed linear model of the software TASSEL 5.0 [74]. Manhattan plots and LD heat maps were drawn using the qqman package in the R environment [75]. The strong QTL was analyzed [−log10 (P-value) = 6.23]. SNPs in candidate genes within 200 kb of the linkage peak were identified.
Plasmid construction and plant transformation
To generate CRISPR/Cas9 mutations in the CsMLO8 and CsMLO11 genes, single guide RNA (sgRNA) target sites were designed to insert into pKSE402 with BsaI and T4 ligase [76]. The construct was transformed into Agrobacterium tumefaciens strain EHA105. PM-susceptible cucumber cultivar ‘CU2’, was used for transformation, referring to the previously published method [76]. Mutated plants were verified by sequencing the target sequence. T2 homozygous mutants of CsMLO8 and CsMLO11 were crossed and then selfed to screen for homozygous double mutants of CsMLO8CsMLO11. The oligos used are listed in Supplementary Data Table S8.
Powdery mildew inoculation, disease index, and spore density
PM resistance of the CsMLO mutants (mlo8CR1, mlo8CR2, mlo11CR1, mlo11CR2, mlo8CR1mlo11CR1) and WT plants was evaluated at the seedling (hypocotyl, young leaf) and adult (stem, adult leaf) stages.
Experiments evaluating PM resistance at the adult stage were conducted in the greenhouse at a Changping farm (40°13′ N, 116°05′ E) in Beijing, China, in a randomized complete block design. Three replicates were set for each line, and five plants were used for each replicate. The seeds were sown on 27 July 2022. PM infection naturally occurred on the leaves ~6 weeks after sowing. PM symptoms on leaves were scored on 21 September (55 dps). PM on stems appeared ~7 weeks after sowing and was evaluated on 28 September (62 dps).
For phenotyping PM resistance at seedling stage, all lines were cultivated in an environmentally controlled growth chamber under normal conditions, with the temperature of 28/18°C (14 h day/10 h night) and 60–80% relative humidity, in a randomized complete block design. Three replicates were set for each line, and five plants were used for each replicate. The PM pathogen was collected from infected cucumber leaves in the greenhouse. When the first true leaf was fully expanded, a pathogen suspension with a concentration of 5 × 105 spores/ml was sprayed evenly on the leaf and hypocotyl. PM symptoms on the first true leaf and hypocotyl were scored separately at 6 and 8 dpi.
PM symptoms on the stem, adult leaf, hypocotyl, and young leaf were ranked by disease severity based on the proportion of the surface area covered with PM spores, which was classified using the method described above for the stem. Likewise, the calculation of the DI was done in the same way [9]. At the same time, infected stem (1.0 g), hypocotyl (0.8 g), adult leaf (0.4 g), and young leaf (0.1 g) were soaked in 2 ml of sterile water with gentle agitation. Spores were counted with an automated fluorescence microscope (BX63, Olympus, Japan).
RNA-seq analysis of transgenic cucumber plants
For RNA-seq analysis, the hypocotyl of the WT and mutants (mlo8CR1, mlo11CR1, mlo8CR1mlo11CR1) were sampled at 0 and 24 h after pathogen inoculation. Three independent biological replicates were used for RNA-seq experiments. Raw reads were filtered using fastp software [77]. The clean reads were then aligned to the reference genome of ChineseLong v3 (http://cucurbitgenomics.org/v2/ftp/genome/cucumber/Chinese_long/v3/) using the STAR program [78] and unique alignments were retained. DEGs between different genotypes and time-points were identified using the R DESeq2 package [79]. Genes with an absolute value of log2 fold change ≥2 were considered reliable DEGs. GO enrichment analysis was performed using the R clusterProfiler package [80].
Quantitative reverse transcription PCR
Total RNA was extracted from hypocotyls using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). qRT–PCR was performed using SYBR Premix Ex Taq™ II (TaKaRa, Kyoto, Japan). Actin1 (CsaV3_2G018090) was used as the reference gene for normalization [81]. Gene relative expression level was analyzed using the 2−ΔΔCt method [82]. Three biological replicates were applied. Primers used are listed in Supplementary Data Table S8.
Reactive oxygen species and trypan blue staining of O2− and H2O2
When the first true leaf was fully expanded, mlo8CR1, mlo11CR1, mlo8CR1mlo11CR1, and WT seedlings were inoculated with PM pathogen, and the stem was sampled for histochemical staining at 6 days after PM inoculation. Histochemical staining of H2O2 and O2− was performed following our previously published method [83]. Young leaf was used for trypan blue staining as outlined in [52]. Six days after pathogen inoculation the first true leaf was submerged in lactophenol–trypan blue solution (30 ml ethanol, 10 ml phenol, 10 ml glycerin, 10 ml lactic acid, and 20 mg trypan blue) in glass containers, and then boiled for 1 min. Leaves were then incubated in 2.5 g ml−1 chloral hydrate solution for 24 h. PM incidence was observed with a microscope (BX63, Olympus, Japan) under 10× magnification.
Yeast two-hybrid assays
Y2H assays were performed using a DUALmembrane starter kit STE (Dualsystem Biotech AG, catalog no. P01401). The coding sequence of CsMLO8 and CsMLO11 were separately fused to pBT3-STE as bait, and the full lengths of CsRbohD and CsCRK2 were individually fused to pPR3-N as prey. Each bait and prey construct and empty vectors were paired, and transformed into yeast strain NMY51. The yeast cells were first selected on synthetic defined medium lacking leucine and tryptophan (SD−L/W), and then transferred to SD medium lacking Leu, Trp, His, and adenine (SD−L/W/H/A) to test protein interactions.
Firefly luciferase complementation imaging assays
The LCI assay was performed following a previously described method [84]. Firstly, the coding sequences (CDSs) of CsMLO8, CsMLO11, and full-length or truncated CsRbohD were separately fused to pCAMBIA1300-nLUC, and full-length or truncated CsRbohD and CsCRK2 were separately fused to pCAMBIA1300-cLUC vector, and the CDS of CsMLO8 was also fused to pCAMBIA1300-Flag. Primers are listed in Supplementary Data Table S8. Agrobacterium strain GV3101 cells transformed with the nLUC and cLUC expression vectors were equally mixed and were injected into young N. benthamiana leaves. The LUC image was captured at 48 h after infiltration using imaging apparatus (NightSHADE LB 985, Berthold). Five independent N. benthamiana leaves were analyzed.
Bimolecular fluorescence complementation assays
For BiFC analysis, the full-length CDSs of CsMLO8, CsMLO11, and CsCRK2 were separately cloned into the pEarleyGate201-nYFP vector, and full-length CDSs of CsCRK and CsRbohD were separately cloned into the pEarleyGate202-cYFP vector. Primers are summarized in Supplementary Data Table S8. Agrobacterium tumefaciens strain GV3101 cells transformed with the above nYFP and cYFP constructs were equally mixed and injected into N. benthamiana leaves. The YFP signals were observed with a confocal microscope (Carl Zeiss, LSM880). Five independent N. benthamiana leaves were analyzed.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (32202487), the Beijing Municipal Science and Technology Commission Program (Z231100003723005), the Key-Area Research and Development Program of Guangdong Province (2020B020220001), the Key-Area Research and Development Program of Shandong Province (2021LZGC016), the Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding (G20220628003-03), the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-23), and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-IVFCAAS).
Author contributions
S.D. drafted the manuscript; S.D., X.L., J.H., and H.M. conducted the experiments and analyzed the data; D.M.B., Y.B., J.G., X.P.L., R.Y., and X.G. helped collect the data; J.S., X.Y., and S.Z. conceived and designed the study.
Data availability
All data that support the findings of this study are available in the article and in the supplementary materials published online.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Shaoyun Dong, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Xin Liu, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Jianan Han, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Han Miao, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Diane M Beckles, Department of Plant Sciences, University of California Davis, One Shield Avenue, Davis, CA 95616, USA.
Yuling Bai, Plant Breeding, Wageningen University & Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands.
Xiaoping Liu, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Jiantao Guan, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Ruizhen Yang, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Xingfang Gu, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Jiaqiang Sun, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, 100081 Beijing, China.
Xueyong Yang, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
Shengping Zhang, State Key Laboratory of Vegetable Biobreeding, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 100081, Beijing China.
References
- 1. Perez-Garcia A, Romero D, Fernandez-Ortuno D. et al. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol. 2009;10:153–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zitter TA, Hopkins DL, Thomas CE. Compendium of Cucurbit Diseases. Saint Paul: APS Press; 1996: [Google Scholar]
- 3. Xu XW, Liu XL, Yan YL. et al. Comparative proteomic analysis of cucumber powdery mildew resistance between a single-segment substitution line and its recurrent parent. Hortic Res. 2019;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kooistra E. Powdery mildew resistance in cucumber. Euphytica. 1968;17:236–44 [Google Scholar]
- 5. Barnes W, Epps W. Powdery mildew resistance in South Carolina cucumbers. Plant Dis Rep. 1956;40:1093 [Google Scholar]
- 6. Fugieda KAR. Genetic study of powdery mildew resistance and spine color on fruit in cucumber. Jpn Soc Hortic Sci. 1962;31:30–2 [Google Scholar]
- 7. Sakata Y, Kubo N, Morishita M. et al. QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus l.). Theor Appl Genet. 2006;112:986–6 [DOI] [PubMed] [Google Scholar]
- 8. Liu LZ, Yuan XJ, Cai R. et al. Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J Phytopathol. 2008;156:691–7 [Google Scholar]
- 9. Zhang SP, Liu MM, Miao H. et al. QTL mapping of resistance genes to powdery mildew in cucumber. Sci Agr Sin. 2011;44:3584–93 [Google Scholar]
- 10. He XM, Li YH, Pandey S. et al. QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor Appl Genet. 2013;126:2149–61 [DOI] [PubMed] [Google Scholar]
- 11. Nie JT, He HL, Peng JL. et al. Identification and fine mapping of pm5.1: a recessive gene for powdery mildew resistance in cucumber (Cucumis sativus L.). Mol Breeding. 2015;35:7 [Google Scholar]
- 12. Zhang P, Zhu YQ, Wang LL. et al. Mining candidate genes associated with powdery mildew resistance in cucumber via super-BSA by specific length amplified fragment (SLAF) sequencing. BMC Genomics. 2015;16:1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang YH, VandenLangenberg K, Wen CL. et al. QTL mapping of downy and powdery mildew resistances in PI 197088 cucumber with genotyping-by-sequencing in RIL population. Theor Appl Genet. 2018;131:597–611 [DOI] [PubMed] [Google Scholar]
- 14. Liu XP, Gu XF, Lu HW. et al. Identification of novel loci and candidate genes for resistance to powdery mildew in a resequenced cucumber germplasm. Genes (Basel). 2021;12:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu X, Yu T, Xu R. et al. Fine mapping of a dominantly inherited powdery mildew resistance major-effect QTL, Pm1.1, in cucumber identifies a 41.1 kb region containing two tandemly arrayed cysteine-rich receptor-like protein kinase genes. Theor Appl Genet. 2016;129:507–16 [DOI] [PubMed] [Google Scholar]
- 16. Zhang KJ, Wang X, Zhu WW. et al. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor Appl Genet. 2018;131:2229–43 [DOI] [PubMed] [Google Scholar]
- 17. Zhang CY, Anarjan MB, Win KT. et al. QTL-seq analysis of powdery mildew resistance in a Korean cucumber inbred line. Theor Appl Genet. 2021;134:435–51 [DOI] [PubMed] [Google Scholar]
- 18. Liu PN, Miao H, Lu HW. et al. Molecular mapping and candidate gene analysis for resistance to powdery mildew in Cucumis sativus stem. Genet Mol Res. 2017;16:gmr16039680. [DOI] [PubMed] [Google Scholar]
- 19. Van Schie CCN, Takken FLW. Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol. 2014;52:551–81 [DOI] [PubMed] [Google Scholar]
- 20. Engler JD, Favery B, Engler G. et al. Loss of susceptibility as an alternative for nematode resistance. Curr Opin Biotechnol. 2005;16:112–7 [DOI] [PubMed] [Google Scholar]
- 21. Devoto A, Piffanelli P, Nilsson I. et al. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–5004 [DOI] [PubMed] [Google Scholar]
- 22. Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–52 [Google Scholar]
- 23. Consonni C, Humphry ME, Hartmann HA. et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–20 [DOI] [PubMed] [Google Scholar]
- 24. Bai YL, Pavan S, Zheng Z. et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol Plant Microbe Interact. 2008;21:30–9 [DOI] [PubMed] [Google Scholar]
- 25. Varallyay E, Giczey G, Burgyan J. Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol. 2012;157:1345–50 [DOI] [PubMed] [Google Scholar]
- 26. Zheng Z, Nonomura T, Appiano M. et al. Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS One. 2013;8:e70723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pessina S, Lenzi L, Perazzolli M. et al. Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic Res. 2016;3:16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wan DY, Guo Y, Cheng Y. et al. CRISPR/Cas9-mediated mutagenesis of vvmlo3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res. 2020;7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiu W, Feechan A, Dry I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic Res. 2015;2:15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tapia RR, Barbey CR, Chandra S. et al. Evolution of the MLO gene families in octoploid strawberry (Fragaria × ananassa) and progenitor diploid species identified potential genes for strawberry powdery mildew resistance. Hortic Res. 2021;8:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santo T, Rashkova M, Alabaca C. et al. The ENU-induced powdery mildew resistant mutant pea (Pisum sativum l.) lines S(er1mut1) and F(er1mut2) harbour early stop codons in the PsMLo1 gene. Mol Breeding. 2013;32:723–7 [Google Scholar]
- 32. Humphry M, Reinstadler A, Ivanov S. et al. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLo1. Mol Plant Pathol. 2011;12:866–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berg JA, Appiano M, Bijsterbosch G. et al. Functional characterization of cucumber (Cucumis sativus L.) Clade V MLO genes. BMC Plant Biol. 2017;17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berg JA, Appiano M, Martinez MS. et al. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015;15:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fukino N, Yoshioka Y, Sugiyama M. et al. Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol Breeding. 2013;32:267–77 [Google Scholar]
- 36. Skou JP. Callose formation responsible for the powdery mildew resistance in barley with genes in the ml-o locus. J Phytopathol. 1982;104:90–5 [Google Scholar]
- 37. Piffanelli P, Zhou FS, Casais C. et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta. 2014;1840:1596–604 [DOI] [PubMed] [Google Scholar]
- 39. Castro B, Citterico M, Kimura S. et al. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat Plants. 2021;7:403–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kimura S, Hunter K, Vaahtera L. et al. CRK2 and C-terminal phosphorylation of NADPH oxidase RBOHD regulate reactive oxygen species production in Arabidopsis. Plant Cell. 2020;32:1063–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tek MI, Calis O, Fidan H. et al. CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front Plant Sci. 2022, 13:1081506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jha Y, Mohamed HI. Plant secondary metabolites as a tool to investigate biotic stress tolerance in plants: a review. Gesunde Pflanzen. 2022;74:771–90 [Google Scholar]
- 43. Liu W, Yan C, Li R. et al. VQMAPK3/VQMAPK6, VQWRKY33, and VQNSTS3 constitute a regulatory node in enhancing resistance to powdery mildew in grapevine. Hortic Res. 2023;10:uhad116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliva M, Guy A, Galili G. et al. Enhanced production of aromatic amino acids in tobacco plants leads to increased phenylpropanoid metabolites and tolerance to stresses. Front. Plant Sci. 2021;11:604349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoo H, Widhalm JR, Qian YC. et al. An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase. Nat Commun. 2013;4:2833. [DOI] [PubMed] [Google Scholar]
- 46. Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105 [DOI] [PubMed] [Google Scholar]
- 47. Dixon RA, Achnine L, Kota P. et al. The phenylpropanoid pathway and plant defence – a genomics perspective. Mol Plant Pathol. 2002;3:371–90 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y, Li X. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol. 2019;50:29–36 [DOI] [PubMed] [Google Scholar]
- 49. Sun YH, Li X, Zhang QZ. et al. The reverse mutation of CsMLO8 results in susceptibility to powdery mildew via inhibiting cell wall apposition formation and cell death in cucumber (Cucumis sativus L.). Sci Hortic. 2023;313:111894 [Google Scholar]
- 50. El-Shetehy M, Wang CX, Shine MB. et al. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal Behav. 2015;10:e998544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tuteja N, Sopory SK. Plant signaling in stress G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yin W, Wang X, Liu H. et al. Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic Res. 2022;9:uhab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Y, Wang X, Fang J. et al. Vqwrky56 interacts with vqbzipc22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 2023;237:1856–75 [DOI] [PubMed] [Google Scholar]
- 54. Zhang D, Wu S, Li N. et al. Chemical induction of leaf senescence and powdery mildew resistance involves ethylene-mediated chlorophyll degradation and ROS metabolism in cucumber. Hortic Res. 2022;9:uhac101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheval C, Samwald S, Johnston MG. et al. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc Natl Acad Sci USA. 2020;117:9621–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang HL, Ullah F, Zhou DX. et al. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019;10:800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daudi A, Cheng ZY, O'Brien JA. et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56:1472–80 [DOI] [PubMed] [Google Scholar]
- 59. O'Brien JA, Daudi A, Finch P. et al. A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 2012;158:2013–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Waszczak C, Carmody M, Kangasjarvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018;69:209–36 [DOI] [PubMed] [Google Scholar]
- 61. Wang CX, Liu RY, Lim GH. et al. Pipecolic acid confers systemic immunity by regulating free radicals. Sci Adv. 2018;4. 10.1126/sciadv.aar4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Z, Noir S, Kwaaitaal M. et al. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell. 2009;21:1972–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kessler SA, Shimosato-Asano H, Keinath NF. et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968–71 [DOI] [PubMed] [Google Scholar]
- 64. Jacott CN, Charpentier M, Murray JD. et al. Mildew Locus O facilitates colonization by arbuscular mycorrhizal fungi in angiosperms. New Phytol. 2020;227:343–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hilbert M, Novero M, Rovenich H. et al. MLO differentially regulates barley root colonization by beneficial endophytic and mycorrhizal fungi. Front. Plant Sci. 2020;10:1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jacott CN, Ridout CJ, Murray JD. Unmasking Mildew Resistance Locus O. Trends Plant Sci. 2021;26:1006–13 [DOI] [PubMed] [Google Scholar]
- 67. Aist JR, Gold RE, Bayles CJ. et al. Evidence for the involvement of molecular components of papillae in ml-o resistance to barley powdery mildew. Physiol Mol Plant Pathol. 1988;33:17–32 [Google Scholar]
- 68. Frietsch S, Wang YF, Sladek C. et al. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc Natl Acad Sci USA. 2007;104:14531–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen J, Lalonde S, Obrdlik P. et al. Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front. Plant Sci. 2012;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim MC, Panstruga R, Elliott C. et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416:447–51 [DOI] [PubMed] [Google Scholar]
- 71. Zhang Q, Hou CC, Tian YD. et al. Interaction between AtCML9 and AtMLO10 regulates pollen tube development and seed setting. Front. Plant Sci. 2020;11:1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Morandini P, Costa A. et al. cROStalk for life: uncovering ROS signaling in plants and animal systems, from gametogenesis to early embryonic development. Genes (Basel). 2021;12:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cui FQ, Wu HP, Safronov O. et al. Arabidopsis MLO2 is a negative regulator of sensitivity to extracellular reactive oxygen species. Plant Cell Environ. 2018;41:782–96 [DOI] [PubMed] [Google Scholar]
- 74. Bradbury PJ, Zhang Z, Kroon DE. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–5 [DOI] [PubMed] [Google Scholar]
- 75. Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. J Open Source Softw. 2014;3:731 [Google Scholar]
- 76. Hu BW, Li DW, Liu X. et al. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol Plant. 2017;10:1575–8 [DOI] [PubMed] [Google Scholar]
- 77. Chen SF, Zhou YQ, Chen YR. et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dobin A, Davis CA, Schlesinger F. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yu GC, Wang LG, Han YY. et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xie Q, Liu P, Shi L. et al. Combined fine mapping, genetic diversity, and transcriptome profiling reveals that the auxin transporter gene ns plays an important role in cucumber fruit spine development. Theor Appl Genet. 2018;131:1239–52 [DOI] [PubMed] [Google Scholar]
- 82. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 83. Dong S, Li C, Tian H. et al. Natural variation in STAYGREEN contributes to low temperature tolerance in cucumber. J Integr Plant Biol. 2023;65:2552–68 [DOI] [PubMed] [Google Scholar]
- 84. He GH, Liu J, Dong HX. et al. The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol Plant. 2019;12:689–703 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available in the article and in the supplementary materials published online.