Abstract
E-cadherin plays a pivotal role in the biogenesis of the first epithelium during development, and its down-regulation is associated with metastasis of carcinomas. We recently reported that inactivation of RB family proteins by simian virus 40 large T antigen (LT) in MDCK epithelial cells results in a mesenchymal conversion associated with invasiveness and a down-regulation of c-Myc. Reexpression of RB or c-Myc in such cells allows the reexpression of epithelial markers including E-cadherin. Here we show that both RB and c-Myc specifically activate transcription of the E-cadherin promoter in epithelial cells but not in NIH 3T3 mesenchymal cells. This transcriptional activity is mediated in both cases by the transcription factor AP-2. In vitro AP-2 and RB interaction involves the N-terminal domain of AP-2 and the oncoprotein binding domain and C-terminal domain of RB. In vivo physical interaction between RB and AP-2 was demonstrated in MDCK and HaCat cells. In LT-transformed MDCK cells, LT, RB, and AP-2 were all coimmunoprecipitated by each of the corresponding antibodies, and a mutation of the RB binding domain of the oncoprotein inhibited its binding to both RB and AP-2. Taken together, our results suggest that there is a tripartite complex between LT, RB, and AP-2 and that the physical and functional interactions between LT and AP-2 are mediated by RB. Moreover, they define RB and c-Myc as coactivators of AP-2 in epithelial cells and shed new light on the significance of the LT-RB complex, linking it to the dedifferentiation processes occurring during tumor progression. These data confirm the important role for RB and c-Myc in the maintenance of the epithelial phenotype and reveal a novel mechanism of gene activation by c-Myc.
The retinoblastoma gene product (RB) was first identified as a suppressor of tumor formation because it was absent or mutated in many human tumors (54). RB is thought to function as a tumor suppressor by controlling the cell cycle progression at the G1/S boundary by inactivating the E2F transcription factor (55). Indeed, RB regulates the activity of several transcription factors in either a negative manner (for E2F and Elf-1) or a positive manner (for SP1, SP3, ATF-1, ATF-2, MyoD, TAF-II 250, NF-IL6, and C/EBPs) (reviewed in reference 53; 10, 11). Therefore, several genes including those encoding c-Fos, c-Myc, transforming growth factors β1 and β2, insulin-like growth factor II, interleukin-6, c-Jun, and Her2/Neu, in addition to differentiation-inducing genes, have been shown to be regulated negatively and/or positively by RB (53). Besides these observations, several studies of transgenic and null mice have demonstrated a role for RB in the proper timing and execution of cellular differentiation during development, more specifically during neuronal and hematopoietic differentiation. In these cases, when RB function is inactivated, apoptosis occurred with aberrant terminal differentiation (see reference 53 for a review). Developmental studies of RB have correlated its expression with the more differentiated epithelial tissues (49). More recently, RB has also been described as the product of a survival gene (15, 19, 31), and in one case this property was linked to its role in maintaining epithelial differentiation (32).
The c-myc proto-oncogene, which encodes two amino-terminally distinct Myc proteins (17), acts as a transcription factor (22). Its expression results in the activation and the repression of several genes involved in growth regulation and differentiation (22). However, the Myc target genes do not form a homogeneous group related only to cell proliferation. Myc also alters the expression of genes involved in cytoskeleton organization (39), extracellular matrix structure and stability (41, 58), and cell adhesion (6, 25, 52) and was also shown to reverse a transformed phenotype (48). Each Myc protein dimerizes with the Max protein, and the Myc-Max heterodimer binds to the E-box sequence, CACGTG (22). Myc also interacts with several transcription factors (reviewed in reference 22). The non-AUG-initiated form of Myc, Myc1, strongly and specifically activates transcription through a noncanonical DNA-binding site (16). Therefore, the molecular mechanisms by which Myc regulates transcriptional activity appear to be quite complex and are not yet fully elucidated.
The cell adhesion molecule E-cadherin, specifically expressed in epithelial tissues, belongs to a large family of transmembrane glycoproteins. E-cadherin is essential for the maintenance and function of epithelial cell layers and also plays a pivotal role very early in development, during the compaction process of the preimplantation embryo, i.e., in the biogenesis of epithelium (29, 43). E-cadherin expression is down-regulated in tumor progression (5). In carcinomas, this down-regulation is associated with invasiveness and with dedifferentiation and metastasis of carcinoma cells in vivo. The reexpression of E-cadherin in these cells decreases their invasiveness. E-cadherin is therefore considered a tumor suppressor (7). The molecular mechanism responsible for E-cadherin down-regulation in dedifferentiated carcinoma cells is not yet understood. Knowledge about the regulation of E-cadherin should provide further insight into the processes occurring during developmental morphogenesis and tumor invasion.
We have recently shown that RB inactivation by simian virus 40 (SV40) large T antigen (LT) specifically induces in differentiated epithelial MDCK cells a massive apoptosis and a mesenchyme-like conversion, i.e., a loss of expression of epithelial markers including E-cadherin and cytokeratin and simultaneously an important invasiveness of the cells and a strong down-regulation of c-myc (31, 32). The reexpression of RB and Myc allow a partial reexpression of epithelial markers such as E-cadherin, cytokeratin, and desmoplakin (32).
These results raised the possibility that E-cadherin, considered a master gene of the epithelial phenotype, might be directly regulated by RB and Myc.
These data prompted us to test whether RB and Myc directly regulate the transcription of E-cadherin. We report here that RB and Myc transcriptionally activate the expression of the E-cadherin promoter in epithelial MDCK cells through interaction with the AP-2 transcription factor. When RB family proteins are inactivated by SV40 LT, this property is lost. In fibroblasts, RB and Myc are unable to activate the E-cadherin promoter, showing that these interactions are likely to be particularly important and specific in epithelial lineages.
MATERIALS AND METHODS
Cell culture and transfection.
NIH 3T3 and HepG2 cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS); MDCK, LT-transformed MDCK [MDCK(LT)], and HaCat cells were grown in 5% serum. MDCK(1-6) and MDCK(2a5) (31) are clonal derivatives of MDCK which were transformed either with wild-type SV40 LT or with the pAT-D2H LT mutant having a deletion between positions 101 and 118 and unable to bind RB (40). Transfection assays were performed as previously described (2). The amount of transfected DNA was kept constant by adding pUC18 plasmid DNA. As internal control, 1 μg of the β-actin–β-galactosidase construct pHβALacZ was cotransfected with the other plasmids in each sample. Chloramphenicol acetyltransferase (CAT) activity was calculated as the percentage of chloramphenicol converted to acetylated forms. The basal level of the reporter plasmid without cotransfection of an expression plasmid was set at unity. Each transfection was performed in duplicate and was reproduced at least three times with different plasmid preparations; the histograms shown in the figures are representative of these experiments. Each cotransfection experiment was also independently performed with a plasmid control containing only the promoter region of the expression plasmid pSV2Δ promoter. When necessary, the final CAT activity was corrected, depending on these controls. In addition, β-actin and SV40 promoters linked to the CAT gene were used in some experiments (Fig. 2) as promoter controls.
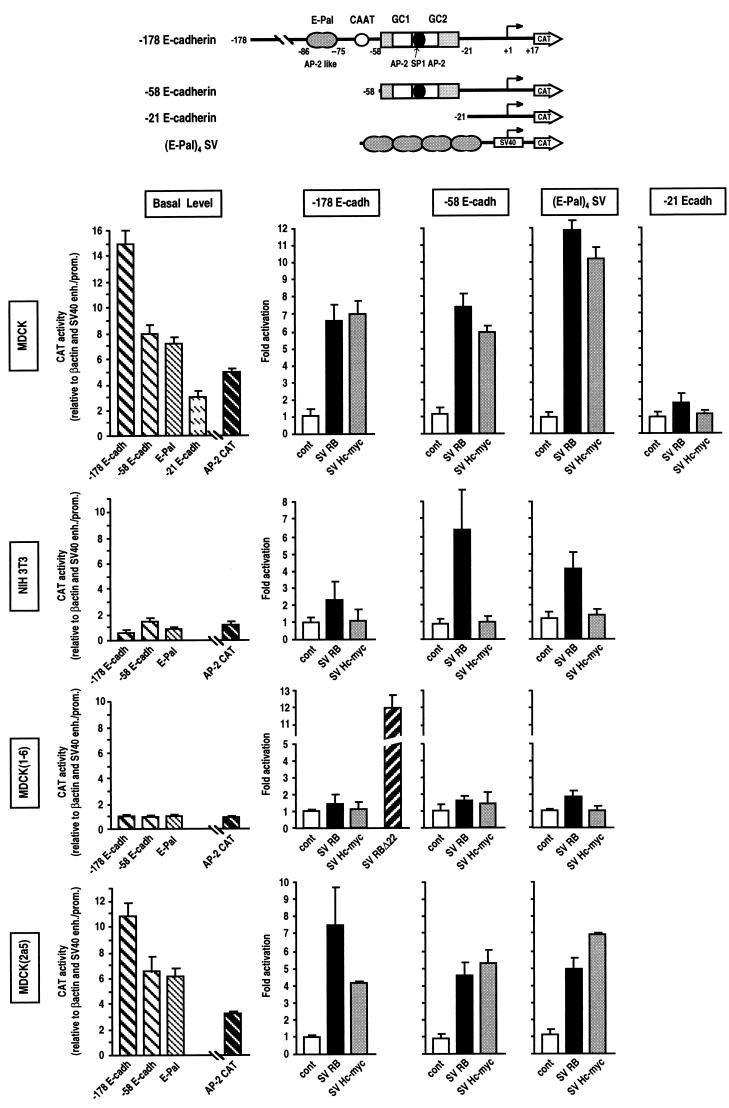
FIG. 2.
Cell-type-specific activation of E-cadherin promoter by RB and Myc. The reporters used in this study are depicted at the top. For graphs of basal level, the −178, −58, and −21 E-cadherin CAT, (E-Pal)4 SV CAT, and AP-2 CAT constructs (3 μg of each) were transfected into MDCK, NIH 3T3, MDCK(1-6), and MDCK(2a5) cells. Basal CAT activities are expressed after normalization with the β-actin promoter and SV40 promoter/enhancer activities and are presented, except for AP-2 CAT, relative to the −178 E-cadherin promoter in MDCK(1-6) cells, which is set at 1. AP-2 CAT activities are presented relative to its activity in MDCK(1-6) cells, which is set at 1. For graphs related to cell type specificity, 2 μg [1 μg for (E-Pal)4 SV CAT] of each E-cadherin promoter construct was cotransfected with 7 μg [or 10 μg for (E-Pal)4 SV CAT] of RB, RBΔ22, and Myc expression vectors or the control pSV2Δ (cont) in the indicated cell lines. The values indicated are averages expressed as fold activation of CAT activity relative to the baseline value obtained by cotransfecting each E-cadherin CAT construct with pUC DNA.
Plasmid constructs.
The E-cadherin promoter CAT constructs (−178, −58, and −21 E-cadherin CAT), (E-Pal)4 SV CAT, and constructs having point mutations in E-Pal (palindromic element), CCAAT box, GC1 box, and GC2 box were previously described (4, 20, 21). The human genomic pHc-myc1-2-3 (Hc-myc), Hc-mycΔ, and pSV Hc-myc1-2-3 (SV Hc-myc) constructs were also described previously (28, 39). The human RB (SV RB) and RBΔ22 (SV RBΔ22) cDNA expression plasmids were gifts of B. Weinberg and P. Hinds (50). SV RBΔ was derived from SV RB by removing the EcoRI fragment carrying the RB cDNA nucleotides 900 to 4600. The SV N-myc expression vector contains the murine genomic N-myc under the control of the SV40 promoter and was provided by G. Fourel. The reporter plasmid endoA CAT was previously described (39). TK CAT is the pBLCAT2 plasmid containing the herpesvirus thymidine kinase (TK) promoter (−109 to +55) fused to the CAT gene and was provided by G. Schütz. AP-2 cona CAT, a kind gift of A. Israël, contains three copies of the metallothionein AP-2 binding site linked to the conaβ2-microglobulin promoter and to the CAT gene. pPADH-AP2, which is a human cDNA under alcohol dehydrogenase promoter control (a gift of T. Williams) (57), was used in transfection assays. AP-2 ΔTA, the dominant-negative mutant, results from a deletion (amino acids [aa] 51 to 138) of the N-terminal domain of AP-2 (21). The vectors used for in vitro translation of AP-2 and AP-2Δ2 (deletion of aa 1 to 163) were previously described (36, 37), as were the reporters HTLV-1 6×ABC and HTLV-1 6×NBC. The latter contain six copies of either the wild-type 21-bp repeat (6×ABC) or a mutation in the A motif (AP-2 binding site) of the 21-bp repeat (6×NBC) upstream of the human T-cell leukemia virus type 1 (HTLV-1) TATA element and fused to the CAT gene (37). The glutathione S-transferase (GST)–RB(379-928), -(379-928; 706C-to-F mutation), -(379-792), and -(763-928) fusions were previously described (26). A GST-HP1 construct (a gift of J.-S. Seelers), in which HP1 is a chromatin protein, was used as a negative control in the in vitro binding assays.
In vitro binding assay with GST-RB.
GST-RB fusion proteins were expressed in Escherichia coli BL21(DE3)(pLysS) and purified as described by Smith and Johnson (46) by adsorption onto glutathione-agarose (Sigma). Equal amounts of immobilized proteins were used in the protein-protein interaction studies. For in vitro translation, AP-2 and AP-2Δ (37) were labeled with [35S]methionine and generated in a coupled in vitro transcription-translation rabbit reticulocyte lysate system (Promega). Glutathione-agarose beads bearing the GST fusion proteins were rocked for 1 h at 4°C with 10% FCS and 1 μl of in vitro-synthesized AP-2 or AP-2Δ in a final volume of 200 μl in 50 mM TNENI (50 mM NaCl, 0.5% Nonidet P-40 [NP-40], 50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). The glutathione-agarose beads were then washed four times in 10 ml of TKENI buffer (300 mM KCl), and bound proteins were eluted with 25 μl of elution buffer (8 M urea, 0.1 M NaH2PO4, 10 mM Tris-HCl [pH 8.0]) and resolved on sodium dodecyl sulfate (SDS)-containing 9% and, for AP-2Δ, 12% polyacrylamide gels. The gel was first stained by Coomassie blue to ensure that similar amounts of fusion proteins were recovered in each sample and then dried and autoradiographed. Nonsaturating autoradiographs were quantified by densitometry scanning, and the percentage of bound proteins was calculated.
Cells lysates, immunoprecipitation, and Western blotting.
Cells were washed three times with cold phosphate-buffered saline and disrupted as described previously (11) in lysis 250 buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 0.1% NP-40, 5 mM EDTA, 1 mM PMSF, and 10 μg each of leupeptin, aprotinin, and pepstatin per ml) by subjecting them to five freeze-thaw cycles (liquid nitrogen, 37°C) and clearing by centrifugation (14,000 rpm, 10 min at 4°C). The supernatant was then used for coimmunoprecipitation. Two human RB antibodies (αRB) were used: mouse monoclonal antibody PMG3-245 (Pharmingen), raised against a peptide corresponding to aa 300 to 380; and polyclonal rabbit antibody C-15, generated against aa 914 to 928 (Santa Cruz Biotechnology Inc.). Three distinct anti-AP-2 antibodies (αAP-2) were used: polyclonal rabbit antibody C-18, made against a peptide corresponding to amino acids 420 to 437 of the C-terminal part of the human AP-2 protein (Santa Cruz Biotechnology); mouse polyclonal antiserum OB2-1, directed against human AP-2 protein (9); and affinity-purified rabbit polyclonal antiserum HCH16, against the N-terminal peptide of AP-2 (9). The mouse monoclonal anti-LT antibody (αLT) 419 was used for LT. All antibodies except OB2-1 and HCH16 were coupled to protein A/G-agarose beads as described previously (18). For immunoprecipitation experiments, 500 μl of cell extract (107 cells) and 25 μl of coupled protein A/G-agarose beads were incubated for 4 h at 4°C. Competition to test the specificity of the αAP-2 (C-18) was done by preincubation of the antibody reagents with the appropriate cognate peptide (threefold excess) for 1 h at 26°C prior to addition of the cell lysate. For OB2-1 and HCH16 immunoprecipitations, cell extracts were incubated with 5 μl of OB2-1 or 30 μl of HCH16 for 3 h at 4°C, then 20 μl of protein A/G-agarose beads was added, and the mixture was incubated for 1 h at 4°C. Immune complexes with protein A/G-agarose beads were then washed five times with lysis 125 buffer (containing 125 mM NaCl). Beads were then boiled in SDS loading buffer, and the proteins were separated by SDS-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Western blots and SDS whole-cell extracts were prepared as described previously (33). Immunoprecipitations of radiolabeled MDCK(1-6) cells were done as follows. Cells (7 × 107) plated into 100-diameter dishes were starved in methionine-free medium supplemented with 2% FCS for 1 h and labeled for 4 h with 0.5 mCi of [35S]methionine in 3 ml of methionine-free medium. The cells were washed four times with cold phosphate-buffered saline, lysed in lysis 250 buffer, and subjected to precipitation with rabbit preimmune serum, C-15 αRB, or C-18 αAP-2. αRB immunoprecipitates were washed five times in lysis 125 buffer. The nonimmunoreactive component of the complex was dissociated with 250 μl of 1% SDS-containing radioimmunoprecipitation (RIPA) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% deoxycholate, 0.5% NP-40, 1 mM PMSF, 10 μg each of leupeptin, aprotinin, and pepstatin per ml) and incubated for 30 min at room temperature. The agarose bead solution was centrifuged, and the supernatant was transferred into a new tube. An equal volume of RIPA buffer without SDS was added, and proteins were reimmunoprecipitated with C-18 αAP-2 for 2 h at 4°C. Then the immune complexes were washed five times with 0.1% SDS-containing RIPA buffer. The proteins were resuspended in Laemmli SDS loading buffer. Samples were electrophoresed on SDS-polyacrylamide gels, fluorographed, dried, and autoradiographed.
RESULTS
RB and Myc transactivate the E-cadherin promoter in epithelial cells.
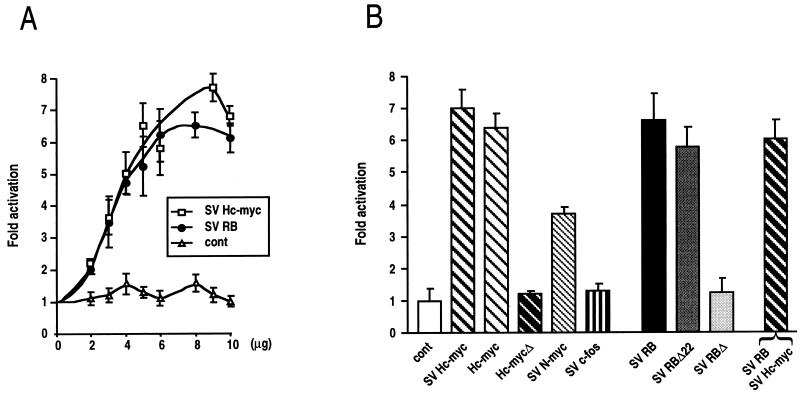
To determine whether E-cadherin is a target for RB and Myc regulation, we examined the ability of human RB and Myc expression vectors to transactivate a murine E-cadherin CAT gene fusion construct in transient cotransfection assays. Previous analysis of the E-cadherin promoter showed that a region of 178 bp upstream from the start site is necessary and sufficient to confer epithelial cell type specificity (4, 44). Therefore, the −178 E-cadherin CAT construct was cotransfected into MDCK epithelial cells with increasing amounts of RB expression plasmid or SV Hc-myc (Fig. 1A). SV Hc-myc is a human c-myc genomic clone derived from normal cells and containing all three exons and is thus able to synthesize the two Myc proteins Myc1 and Myc2 (28). Figure 1A shows that the E-cadherin CAT construct was strongly activated by both RB and Myc in a concentration-dependent fashion. E-cadherin CAT was also cotransfected with increasing amounts of control vectors to exclude the possibility that the increasing CAT activity was due to competition of a negative trans-acting factor(s) with the SV40 early promoter. To confirm the specificity of Myc activation, we used a second c-myc genomic clone, Hc-myc, also derived from normal cells and isolated by another group (14). The pHc-myc vector, like the previous construct, stimulates the expression of the E-cadherin promoter six- to sevenfold (Fig. 1B). Large deletions in the coding sequences of Myc and RB resulted in loss of their transactivating capacity, attesting to the need for functional proteins and to the absence of quenching (Fig. 1B, Hc-mycΔ and SV RBΔ). However, RB mutant RBΔ22, having a deletion of exon 22 inside the B region of the pocket, transactivated the E-cadherin promoter as efficiently as RB (Fig. 1B). RBΔ22 also displayed defective binding to oncoproteins (53). The specificity of RB and Myc activation was also demonstrated by the fact that another nuclear oncogene, c-fos, had no effect on E-cadherin promoter expression, in contrast to N-myc, which was found to also transactivate the E-cadherin promoter about fourfold (Fig. 1B). Cotransfection of both RB and Myc showed that their transactivating effect was not additive (Fig. 1B). The increase in CAT activity was similar to that observed with RB or Myc alone. These results (Fig. 1) indicate that RB and Myc can transactivate the E-cadherin promoter in MDCK epithelial cells to similar extents.
FIG. 1.
Activation of the murine E-cadherin promoter by RB and Myc in MDCK epithelial cells. (A) Dose-dependent effects of SV RB and SV Hc-myc on transcriptional activity of the −178 E-cadherin promoter. Three micrograms of −178 E-cadherin CAT was cotransfected with pUC DNA (baseline value) or with increasing amounts of SV RB and SV Hc-myc expression vectors. Total DNA was always kept constant by adding pUC DNA. As a control, −178 E-cadherin CAT was cotransfected with an expression plasmid without insert, pSV2Δ (cont). The values indicated are averages expressed as fold activation of CAT activity relative to the baseline value obtained by cotransfecting −178 E-cadherin promoter CAT with pUC DNA. Each value is expressed after normalization for transfection efficiency, using pHβALacZ as an internal control. The results are from at least three experiments performed in duplicate, and standard deviation bars are shown. (B) Activation of E-cadherin transcriptional activity by different nuclear regulators. Three micrograms of −178 E-cadherin CAT was cotransfected into MDCK cells with 7 μg of the indicated vectors.
We previously demonstrated that c-myc positively regulates the murine cytokeratin endoA and endoB genes (39). Since RB and Myc had similar effects on the E-cadherin promoter, we therefore tested whether RB also could transactivate the endoA promoter. We found that RB and Myc stimulated endoA CAT activity 4.6- and 5.5-fold, respectively (data not shown).
RB and Myc transactivation of the E-cadherin promoter is cell type specific.
To determine whether the positive regulation of E-cadherin expression by RB and Myc was specific to epithelial cells, we studied the expression of the −178 E-cadherin promoter and its regulation by RB and Myc in mesenchymal NIH 3T3 cells. Figure 2 shows that in fibroblasts, basal expression of the E-cadherin promoter, compared to that of two other control promoters, β-actin and SV40, was about 12 to 15 times lower than in MDCK cells, in agreement with previous results (4). Furthermore RB and Myc failed to significantly transactivate this promoter in fibroblasts. We also analyzed ectopic E-cadherin regulation in MDCK cells transformed by SV40 large T antigen. In these MDCK(1-6) fibroblast-like cells, RB is inactivated, c-myc is repressed, and endogenous E-cadherin is down-regulated (31, 32). Figure 2 shows that in MDCK(1-6) cells, basal expression of the E-cadherin promoter was greatly reduced and the RB- and Myc-mediated activation was abolished, thus mimicking the results for NIH 3T3 fibroblasts.
To confirm the involvement of the RB protein family, we repeated these experiments with MDCK cells transformed by an LT mutant, pATD-2H, having a deletion between positions 101 and 118 and unable to bind to RB but still inactivating p53 (40). This clone, called MDCK(2a5), has an epithelial phenotype and expresses high levels of Myc and E-cadherin, like the MDCK parental cells (31, 32). In these cells, the −178 E-cadherin promoter behaved as in MDCK cells: its basal expression was high, and it could be similarly activated by the two nuclear regulators RB and Myc (Fig. 2). All of these results indicate that RB and c-Myc transactivation of E-cadherin expression is specific to epithelial cells and requires an active RB protein family.
Characterization of RB- and Myc-responsive elements.
Several important elements (Fig. 2) on the E-cadherin promoter have been previously characterized; these include a palindromic element called E-Pal (−98 to −78), which is highly homologous to KER elements found in keratin genes (4). E-Pal is responsible for the cell type specificity of the promoter, acting positively in epithelial cells and negatively in mesenchymal cells (21). Basal expression is conferred by a CCAAT box (−69 to −58) and a GC region (−58 to −25) (4). The AP-2 transcription factor and/or AP-2-related protein has been shown by several complementary experimental approaches, including in vivo footprinting, gel retardation, point mutations, and transient transfection assay, to be crucial for the activities of both E-Pal and the GC-rich region (4, 20, 21).
Figure 2 shows that after deletion of E-Pal and the CCAAT box, basal expression of −58 E-cadherin in MDCK cells decreased by 50%, in agreement with previous results (4). Nevertheless, the GC region was still sufficient to be specifically activated by RB and Myc. RB activation was 7.5-fold, and Myc activation was 6-fold (Fig. 2). When the GC boxes were deleted, basal expression of −21 E-cadherin dropped an additional twofold and could no longer be significantly activated by RB and Myc (Fig. 2). Since the E-Pal element plays a crucial role in specific E-cadherin expression in epithelial cells (4, 21), we tested whether a chimeric reporter containing four E-Pal elements linked to the SV40 promoter would be activated by RB and Myc. This construct, (E-Pal)4 SV, was activated to a similar high extent (more than 10-fold) by RB and Myc in MDCK epithelial cells (Fig. 2). All of these results show that the E-Pal element and the GC region are similarly activated by both RB and Myc.
In MDCK(1-6) cells, neither the −178 E-cadherin, −58 E-cadherin, nor (E-Pal)4 SV reporter construct could be transactivated by RB or Myc. Likewise, in NIH 3T3 cells, the three reporter constructs could not be activated by Myc, and the RB-mediated activation of −178 E-cadherin and (E-Pal)4 SV expression was 66% lower than in MDCK cells. However, in these cells, the −58 cadherin construct was stimulated by RB six- to sevenfold (Fig. 2).
RB and c-Myc transcriptional activation is mediated by the transcription factor AP-2.
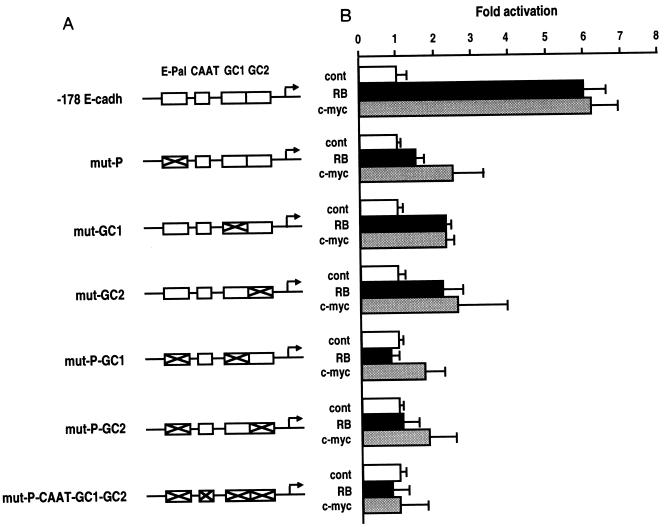
The cell-type-specific expression of the E-cadherin promoter is mediated by the E-Pal element and AP-2-related proteins (4, 20, 21). To determine if RB- and Myc-mediated activation required the AP-2 transcription factor, we used several specific point mutations within each of the AP-2 binding sites of the E-cadherin promoter: E-Pal, GC1, and GC2 elements (20). The loss of the functionality of these AP-2 sites resulted in 60 to 100% inhibition of RB- and Myc-mediated activation (Fig. 3). The use of a dominant-negative mutant of AP-2, AP-2 ΔTA, which lacks the transactivation domain (21), inhibited in a dose-dependent manner RB- and Myc-mediated activation of the three reporters, −178 E-cadherin, −58 E-cadherin, and E-Pal (Table 1). Two controls were performed to ensure that the effect of the dominant-negative mutant of AP-2 was specific and not due to a general effect on cells. We used the −21 E-cadherin promoter, which does not contain any AP-2 binding site, and TK CAT. In both cases, the cotransfection of these constructs with AP-2 ΔTA mutant did not affect their basal level (Table 1). We next performed cotransfection assays with an AP-2 expression vector, pPADH-AP2. To exclude self-interference phenomena which occur between endogenous and exogenous transfected AP-2 (27), we performed these assays in HepG2 cells that lack endogenous AP-2 activity (24). Whereas RB and Myc alone showed very modest stimulation of E-cadherin promoter activity, they significantly enhanced its transcriptional activity eight- to ninefold when cotransfected with the pPADH-AP2 expression vector (Table 2).
FIG. 3.
Specific point mutations of the AP-2 binding sites of the E-cadherin promoter result in the loss of RB- and Myc-mediated activation. (A) Diagrams of mutant constructs of the −178 E-cadherin promoter. Point mutations abolishing AP-2-like and AP-2 binding within the E-Pal and GC boxes are indicated by crosses (21). (B) The E-cadherin constructs (2 μg of each) were cotransfected with 7 μg of RB and Myc expression vectors (SV RB and SV Hc-myc, respectively) or the control pSV2Δ (cont) in MDCK epithelial cells. The values are averages expressed as fold activation of CAT activities relative to the baseline obtained by cotransfecting each E-cadherin CAT construct with 7 μg of pUC DNA.
TABLE 1.
Dominant-negative AP-2 suppresses the RB or c-Myc-mediated activation of the E-cadherin promotera
| Activation by: | Amt of dominant-negative AP-2 (μg) | Promoter activity (%)
|
||||
|---|---|---|---|---|---|---|
| −178 | −58 | E-Pal | −21 | TK CAT | ||
| SV Hc-myc | 100 | 100 | 100 | |||
| 1 | 11 | 20 | 16 | |||
| 4 | 2 | 3 | 11 | |||
| SV RB | 100 | 100 | 100 | |||
| 1 | 15 | 6 | 2 | |||
| 4 | 0.7 | 1.3 | 0.6 | |||
| Basal levelb | 100 | 100 | ||||
| 1 | 98 | 96 | ||||
| 4 | 99 | 92 | ||||
Three micrograms of −178 or −58 E-cadherin CAT and 1 μg of (E-Pal)4 SV CAT were transfected into MDCK cells either with 5 μg of pUC DNA or with RB and Myc expression vectors as indicated, plus increasing amounts of dominant-negative AP-2 in which the N-terminal region has been deleted. Activities of the −178 and −58 E-cadherin CAT and (E-Pal)4 SV CAT reporters in the presence of control vector without the AP-2 sequence were set at 100%.
Two negative controls, −21 E-cadherin CAT, which does not contain AP-2 binding sites, and TK CAT, were used; their basal levels were determined in a cotransfection assay with increasing amounts of dominant-negative AP-2.
TABLE 2.
Transcriptional activation of the E-cadherin promoter in HepG2 cellsa
| Vector addition | Fold activation
|
|
|---|---|---|
| −pPADH-AP-2 | +pPADH-AP-2 | |
| Control | 1.0 | 2.3 |
| SV RB | 1.5 | 8.5 |
| SV Hc-myc | 3.5 | 9.3 |
One microgram of −178 E-cadherin CAT was transfected into HepG2 cells with 4 μg of AP-2 expression vector (pPADH-AP2; gift of T. Williams) and 6 μg of pSV2Δ (control), pSV Hc-myc, or SV RB. The values are averages expressed as fold activation of CAT activities relative to the baseline obtained by cotransfecting the −178 E-cadherin CAT construct with 10 μg of pUC DNA.
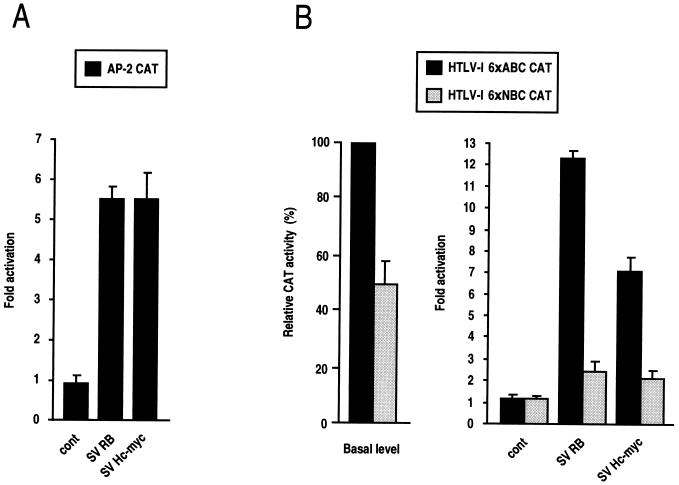
To confirm these results in other systems and to ensure that the functional interaction of RB and Myc with AP-2 activity is a general mechanism, we used two chimeric reporter constructs, one containing three AP-2 binding sites from the metallothionein (MTIIA) promoter (AP-2 CAT) and the other including part of the HTLV-1 long terminal repeat (LTR) 21-bp repeats (HTLV-1 6×ABC) known to be transcriptionally activated by AP-2 protein (37). Figure 4 shows that these constructs are transactivated 12-fold by RB and 7-fold by Myc. When the AP-2 binding site was functionally inactivated by point mutation (HTLV-1 6×NBC) (37), RB and Myc lost their capacity to induce transcriptional activity. Interestingly, in MDCK cells the basal level of the mutated HTLV-1 6×NBC reporter was reduced by 50% compared to the wild-type HTLV-1 6×ABC, in contrast to the situation in NIH 3T3 cells, where it had a comparable low level (data not shown). This finding indicates that endogenous AP-2 activity is greatly reduced in NIH 3T3 mesenchymal cells. Similarly, endogenous AP-2 activity, as monitored by transfecting the chimeric AP-2 CAT construct into the different cell lines and by relating its basal level expression to the two control promoter activities, SV40 and β-actin, perfectly reflects the differential expression of the E-cadherin promoter and E-Pal (Fig. 2). Thus, AP-2 CAT activity was high in MDCK and MDCK(2a5) cells and low in MDCK(1-6) and NIH 3T3 cells (Fig. 2). These results show that endogenous AP-2 activity is linked in MDCK cells to the functionality of RB. When RB is inactivated by LT as in MDCK(1-6) cells, AP-2 activity is down-regulated; when RB is still active, even in the presence of inactive p53 as in MDCK(2a5) cells, AP-2 activity is preserved. AP-2 activity is also greatly reduced in NIH 3T3 mesenchymal cells. These findings, together with those of previous studies (4, 20, 21), indicate that RB- and Myc-mediated activation of the cadherin gene requires the AP-2 transcription factor.
FIG. 4.
RB and Myc activate expression of the HTLV-1 LTR through AP-2. (A) MDCK cells were cotransfected with 3 μg of AP-2 cona CAT (AP-2 CAT) reporter containing three metallothionein AP-2 binding sites linked to the conaβ2 promoter and either 5 μg of RB and Myc expression vectors or pSV2Δ (cont). Controls with only the conaβ2 promoter were also performed (not shown). (B) MDCK cells were cotransfected with 3 μg of either HTLV-1 6×ABC or HTLV-1 6×NBC (see Materials and Methods) and with 5 μg of RB and Myc expression vectors or pSV2Δ (cont). The panel on the left shows the basal level of the wild-type and mutated HTLV-1 reporters. All values indicated are averages expressed as fold activation of CAT activity relative to the baseline value obtained by cotransfecting each E-cadherin CAT construct with pUC DNA.
RB and AP-2 interact in vitro.
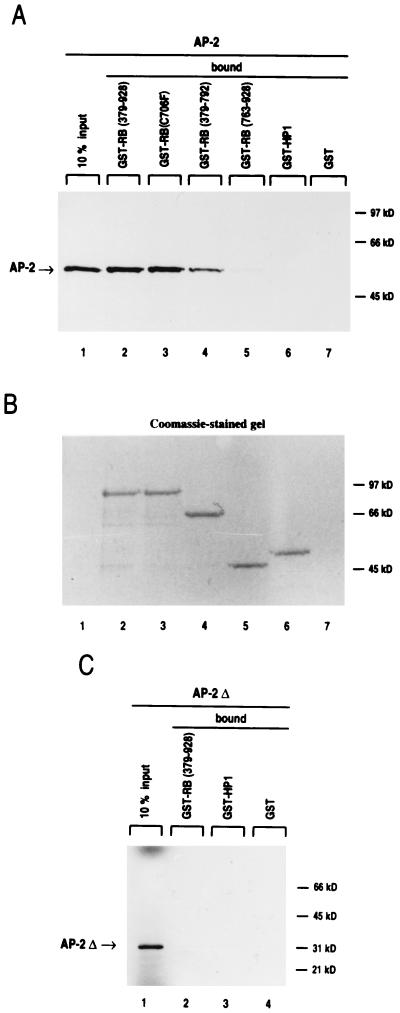
To address how RB activates AP-2-mediated transcription, their in vitro interaction was examined. The cDNAs encoding RB(379-928) and various RB deletion mutations, RB(379-928; 706C-to-F mutation), RB(379-792), and RB(763-928), inserted into a GST expression plasmid were expressed as GST fusion proteins in E. coli. The AP-2 expression vector was used to synthesize 35S-radiolabeled AP-2 protein in a reticulocyte lysate. Unlike GST alone or a GST-HP1 chromatin protein (unrelated to RB), GST-RB(379-928) bound specifically to 9 to 10% of the input AP-2 protein (Fig. 5A, lane 2). The GST-RB fusion protein mutated in the B region of the small pocket (C706F), abolishing its interaction with the oncoproteins, still binds to the AP-2 protein with as much affinity as the wild-type form of RB (Fig. 5A, lane 3). This is consistent with our earlier result with the RBΔ22 mutant, also affected in the B region, which transactivated the E-cadherin promoter as efficiently as wild-type RB (Fig. 1B). In contrast, RB(379-792), which lacks the C-terminal domain, shows binding to AP-2 reduced by 50% (Fig. 5A, lane 4). However the C-terminal domain alone did not bind significantly to the transcription factor, with only 1 to 2% of input protein retained (Fig. 5A, lane 5). These results indicate that two regions of RB are required for interaction with AP-2, the carboxy-terminal domain and part of the oncoprotein binding domain. To determine the region of AP-2 required for binding to RB, an AP-2 N-terminal deletion protein lacking aa 1 to 165 was assayed for in vitro binding (37). The results show that the N-terminal region of AP-2 is required for interaction with RB (Fig. 5C). It is interesting that deletion of the same N-terminal region of AP-2 also suppresses RB- and Myc-mediated activation (Table 1), strongly suggesting that the physical interaction with AP-2 is required for transcriptional activation in vivo.
FIG. 5.
AP-2 and RB proteins interact in vitro. (A and B) Both the RB small pocket and the C-terminal domain are required for interaction with AP-2. GST, GST-RB(379-928), GST-RB(379-928; 706C-to-F mutation) [GST(C706F)], GST-RB(379-792), GST-RB(763-928), and GST-HP1 fusion proteins immobilized on glutathione-agarose beads were used to bind radiolabeled AP-2 protein. Lane 1 shows 10% of the input of in vitro-translated [35S]AP-2. In lanes 2 to 7, the same amounts of [35S]AP-2 were incubated with glutathione-agarose beads containing similar amounts of the various GST fusion proteins. GST-HP1, a chromatin protein unrelated to RB, was used as negative control. Nine to 10% of AP-2 bound to GST-RB(379-928) and to GST-RB(C706F), 4.5 to 5% bound to GST-RB(379-792), and 1 to 2% bound to GST-RB(763-928). (B) Before drying and autoradiography, the gel was stained with Coomassie blue to ensure that all fusion proteins were correctly recovered. (C) N-terminally deleted AP-2 protein does not bind to RB. A 35S-labeled N-terminal deletion AP-2 protein (AP-2Δ) was incubated with various GST fusion proteins (lanes 2 to 4). Similar amounts of radiolabeled AP-2 and AP-2Δ were used in panels A and C, and gels were exposed for the same length of time.
Interactions between RB and AP-2 in vivo.
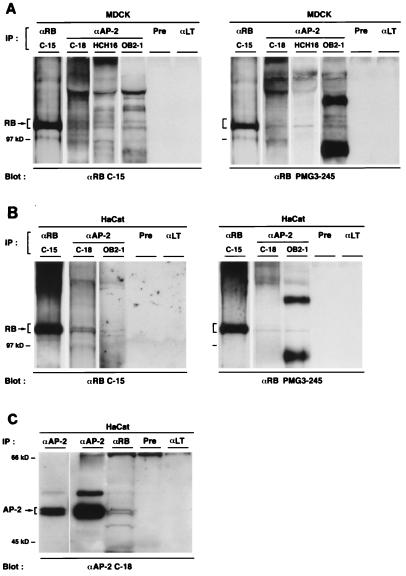
Having established an interaction between RB and AP-2 in vitro, we next explored the interaction in vivo. Evidence of RB–AP-2 complexes was detected in untransfected, asynchronous MDCK cell lysates by coimmunoprecipitation with each of three αAP-2, and the presence of RB in each immunoprecipitate was revealed by Western blotting using the C-15 and PMG3-245 αRB (Fig. 6A). The αAP-2 used were (i) a rabbit polyclonal antibody (C-18) raised against the C-terminal part of the protein, (ii) a rabbit polyclonal antiserum (HCH16) against the N-terminal peptide of AP-2 (9), and (iii) a mouse polyclonal antiserum (OB2-1) raised against purified human AP-2 protein (9). These results, with three distinct αAP-2, clearly demonstrate that RB and AP-2 form stable complexes in vivo. To establish that the interaction between the two proteins was a general phenomenon in epithelial cells, and to more easily detect AP-2 in the reverse coimmunoprecipitation with human αRB, we also used human immortalized HaCat keratinocytes. As shown in Fig. 6B and C, both RB and AP-2 were detected in the immunoprecipitations with αAP-2 and αRB. Therefore, RB and AP-2 form stable complexes in HaCat cells as well.
FIG. 6.
RB and AP-2 interact in vivo. (A) RB–AP-2 complexes in MDCK cells. Lysates of MDCK cells were immunoprecipitated (IP) with the three αAP-2, C-18, HCH16, and OB2-1, αRB (C-15) as a positive control, or rabbit preimmune serum (Pre) and αLT as negative controls. Immune complexes were separated on a 7.5% polyacrylamide gel and transferred to a PVDF membrane, which was then probed with αRB (C-15 and PMG3-245). (B) Stable RB–AP-2 complexes in HaCat cells. Coimmunoprecipitations similar to those described for panel A were performed with HaCat cells with αAP-2 (C-18 and OB2-1). (C) HaCat lysates were also subjected to the reverse immunoprecipitation with αRB (PMG3-245) or αAP-2 (C-18) as a positive control. As negative controls, mouse preimmune serum (Pre) and αLT were used. After transfer, the PVDF membrane was probed with αAP-2 (C-18). A short exposure of the positive control is also shown.
RB mediates an interaction between LT and AP-2 in epithelial cells in vivo.
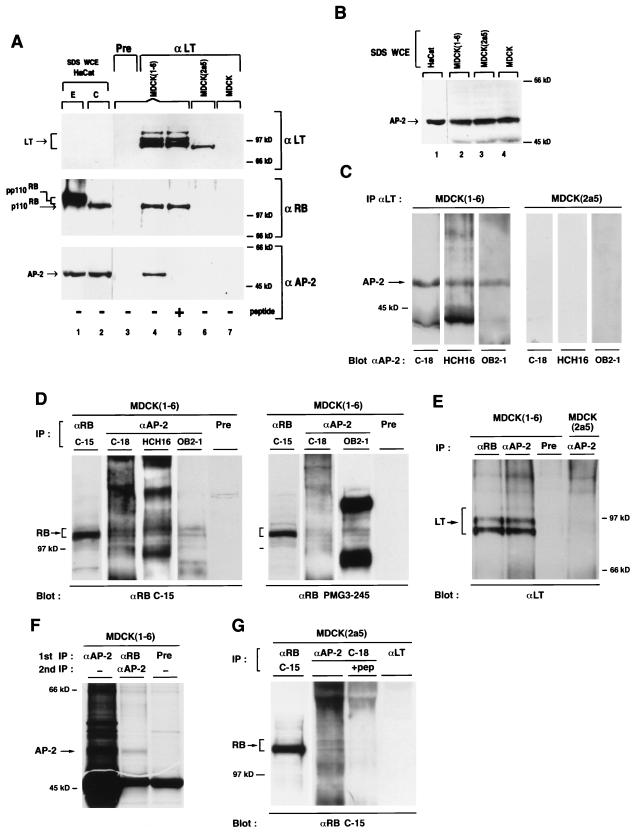
We have shown that LT is able to disrupt the activation of AP-2 by RB. In an effort to understand how this may be achieved and to further examine RB–AP-2 interactions, we went on to look for in vivo molecular complexes between LT, RB, and AP-2 proteins. In these experiments, we took advantage of our different MDCK cell lines transformed by wild-type and mutant LT. It seemed likely that the interaction between LT and AP-2 is dependent on the RB-binding site of the oncoprotein, since endogenous AP-2 activity is greatly inhibited in MDCK(1-6) cells and preserved in MDCK(2a5) cells, which express a non-RB-binding form of LT (Fig. 2). Consequently, immunoprecipitation of wild-type LT in MDCK(1-6) cells should coimmunoprecipitate RB and AP-2, whereas in MDCK(2a5) cells, an LT antibody should be unable to coprecipitate either RB or AP-2. Figure 7A shows that indeed αLT coprecipitated hypophosphorylated RB plus AP-2 only in MDCK(1-6) cells. Neither of these proteins was immunoprecipitated in MDCK(2a5) or nontransformed MDCK cells. The inability of αLT to detect AP-2 protein in MDCK(2a5) cells could not be attributed to the absence of LT and/or AP-2 proteins since both proteins could be detected in these cells with the appropriate antibodies (Fig. 7B). The specificity of αLT was also demonstrated by the use of a preimmune serum. AP-2 protein was detected with the C-18 rabbit polyclonal antibody, and we could also show that incubation of αAP-2 with the cognate peptide resulted in a loss of the detection of the 52-kDa protein on the Western blot (Fig. 7A, lane 5). To confirm the identity of this protein, further coimmunoprecipitations with αLT were performed and the Western blots were probed with the two additional αAP-2, HCH16 and OB2-1 (9). Both antisera specifically recognized the 52-kDa protein coprecipitating with αLT from MDCK(1-6) cells but not from MDCK(2a5) cells (Fig. 7C). Taken together, these results again demonstrate that the 52-kDa protein coimmunoprecipitating with RB is indeed AP-2. Furthermore, immunoprecipitation of AP-2 from MDCK(1-6) cells with either the C-18, HCH16, or OB2-1 antibody coimmunoprecipitated both RB (Fig. 7D) and LT (Fig. 7E). The reverse coimmunoprecipitation by αRB also led to detection of AP-2 (Fig. 7F) and LT (Fig. 7E). In addition, in MDCK(2a5) cells an immunoprecipitation using αAP-2 coimmunoprecipitated RB only in the absence of the cognate peptide (Fig. 7G), not LT (Fig. 7E). Table 3 summarizes the coimmunoprecipitation results.
FIG. 7.
In vivo interaction between LT, RB, and AP-2. (A) αLT coimmunoprecipitates RB and AP-2 in LT-transformed MDCK cells when the RB oncoprotein binding domain is not mutated. Total lysates of MDCK(1-6), MDCK(2a5), and MDCK cells were immunoprecipitated (IP) with mouse preimmune serum (Pre) or the mouse monoclonal 419 αLT coupled to protein A/G-agarose beads. Immune complexes were separated on an SDS–7.5% polyacrylamide gel, and proteins were transferred onto a PVDF membrane. After transfer, the membrane was cut into two pieces. The upper part was successively probed with the monoclonal PMG3-245 αRB and then αLT. In lanes 1 and 2, 30-μg aliquots of 1% SDS whole-cell HaCat extract (SDS WCE) of exponential (E) and confluent (C) cells were loaded on the gel as positive controls for hyper- and hypophosphorylated RB. Lane 5 was isolated from the lower part; then lanes 1 to 4, 6, and 7 were probed with the rabbit polyclonal αAP-2 (C-18) raised against the C-terminal peptide of AP-2 (Santa Cruz). Lane 5 alone was incubated with αAP-2 and a 10-fold excess of the cognate peptide. (B) The 52-kDa AP-2 protein is expressed in all cells tested. Aliquots (30 μg) of 1% SDS whole-cell extracts (SDS WCE) of HaCat, MDCK(1-6), MDCK(2a5), and MDCK cells were separated on a 7.5% polyacrylamide gel, transferred to a PVDF membrane, and probed with the polyclonal αAP-2 (C-18). (C) The LT-immunoprecipitated 52-kDa protein is recognized by three different αAP-2. Total lysates of MDCK(1-6) and MDCK(2a5) cells were immunoprecipitated with αLT as described for panel A, and the different Western blots were probed with the rabbit polyclonal C-18, the rabbit polyclonal antiserum HCH16 against the N-terminal peptide of AP-2 (9), and the mouse polyclonal OB2-1 antiserum directed against the whole human AP-2 protein (9). (D) RB is coimmunoprecipitated by several distinct αAP-2 in MDCK(1-6) cells. Total lysates of MDCK(1-6) cells were immunoprecipitated with αAP-2 (C-18, HCH16, and OB2-1), αRB (C-15) as positive control, or rabbit preimmune serum (Pre) as a negative control. After separation on a 7.5% polyacrylamide gel and transfer, the PVDF membrane was probed with αRB (C-15 and PMG3-245). (E) LT is coimmunoprecipitated with αAP-2 only in MDCK(1-6) and not in MDCK(2a5) cells. Total lysates of MDCK(1-6) and MDCK(2a5) cells were subjected to precipitation with rabbit preimmune serum (Pre), αRB (C-15), or αAP-2 (C-18) as indicated. After separation on 7.5% polyacrylamide gels and transfer, the PVDF membrane was probed with αLT. (F) AP-2 is coimmunoprecipitated by αRB in MDCK(1-6) cells. To allow easier detection of AP-2, cell lysates of MDCK(1-6) cells (7 × 107) labeled with [35S]methionine were subjected to precipitation with rabbit preimmune serum (Pre), αRB (C-15), or αAP-2 (C-18) as a positive control. The washed RB immunoprecipitate was then dissociated with 1% SDS and reprecipitated with αAP-2 (C-18). Immune complexes were separated on a 10% polyacrylamide gel. The gel was fluorographed and dried, and proteins were detected by autoradiography. (G) RB coprecipitates with αAP-2 in MDCK(2a5) cells. Lysates of MDCK(2a5) cells were immunoprecipitated with αAP-2 (C-18) in the absence or in the presence of the cognate peptide (pep) in threefold excess, αRB (C-15) as a positive control, or αLT as a negative control. Immunoprecipitates were analyzed on a 7.5% polyacrylamide gel; proteins were transferred onto a PVDF membrane and probed with αRB (C-15).
TABLE 3.
Summary of in vivo interactions between RB, AP-2, and LT
| IPa | Protein detected in coimmunoprecipitates | Coimmunoprecipitation result in:
|
|||
|---|---|---|---|---|---|
| Immortalized epithelial cells
|
LT-transformed cells
|
||||
| MDCK | HaCat | MDCK(LT)b | MDCK(LTm)c | ||
| RB | AP-2 | NDd | + | + | ND |
| LT | + | − | |||
| AP-2 | RB | + | + | + | + |
| LT | + | − | |||
| LT | RB | + | − | ||
| AP-2 | + | − | |||
All αAP-2 immunoprecipitations (IP) were done with two or three distinct αAP-2 except with MDCK(LTm) lysates. Three different αAP-2 were used to identify AP-2 in the LT immunoprecipitations. Two αRB were always used to reveal RB except for the MDCK(LTm) cells.
MDCK(1-6) cells transformed by wild-type LT.
MDCK(2a5) cells transformed by an LT mutant unable to bind RB.
ND, not determined.
Overall, these results demonstrate that in MDCK(LT) cells there is a protein complex between LT, RB, and AP-2 and that a mutation in LT, leading to inhibition of its interaction with RB, simultaneously inhibits its interaction with AP-2, whereas RB and AP-2 still interact stably (Fig. 7G). Consequently, the interaction between LT and AP-2 is indeed mediated by RB. Consistent with these findings, as mentioned earlier, both of the RB mutants with defective oncoprotein binding, RBC706F and RBΔ22, still showed wild-type levels of binding to AP-2 (Fig. 5A, lane 3) and wild-type effects on E-cadherin activation (Fig. 1B). To confirm further that LT requires RB to interact with AP-2, we cotransfected RBΔ22 with the E-cadherin promoter in MDCK(1-6) cells. Whereas wild-type RB was unable to activate the E-cadherin promoter in these cells (Fig. 2), RBΔ22, which is not inactivated by LT, strongly activated the E-cadherin promoter (Fig. 2). Thus, AP-2 inactivation by LT is mediated by RB.
DISCUSSION
We demonstrate here that both RB and Myc are specific transactivators of the E-cadherin promoter in epithelial cells. The use of different 5′ deletions of this promoter showed that several target sequences are similarly and significantly activated by both RB and Myc. These sequence elements are E-Pal and the GC boxes, which were previously shown to play a crucial role in vivo in cell-type-specific and basal expression, respectively, with both elements absolutely requiring AP-2 or an AP-2-like transcription factor to be functional (4, 20, 21). Here we definitively show, using reporter constructs with point mutations within the AP-2 binding sites (Fig. 3), that transcriptional stimulation of the E-cadherin promoter by RB and Myc is mediated by AP-2 proteins. Furthermore, the RB and Myc AP-2-mediated transcriptional activation is not restricted to the E-cadherin promoter but is also observed with the HTLV-1 LTR, also previously shown to be stimulated by AP-2 (37). The implication therefore is that interaction with AP-2 is a general mechanism by which RB and Myc can function. We also demonstrate that at least part of the mechanism behind this interaction is a stable complex that can form between AP-2 and RB proteins both in vitro (Fig. 5) and in vivo (Fig. 6 and 7). The in vitro interaction requires the N-terminal region of AP-2 and both the small pocket and the C-terminal domain of RB. The intracellular interaction between RB and AP-2 could be shown in epithelial MDCK cells as well as in immortalized keratinocyte HaCat cells.
Our studies of MDCK(1-6) cells transformed by a wild-type LT and MDCK(2a5) cells transformed by an LT mutant inactivating only p53 and leaving RB active indicate that the inactivation of AP-2 activity by LT is linked to the integrity of its RB binding site. Moreover, αLT coimmunoprecipitated RB and AP-2 only when the RB binding site was intact (Fig. 7A), and αAP-2 coimmunoprecipitated both RB and LT in MDCK(LT) (Fig. 7D and E) and only RB in MDCK(2a5) cells (Fig. 7G). Mutations of the B domain of the RB pocket (RBΔ22 and RBC706F), which result in a loss of interaction with the oncoproteins, did not affect the transcriptional stimulation of E-cadherin (Fig. 1) or the in vitro binding to AP-2 (Fig. 5). In addition, RBΔ22, in contrast to RB, transactivated the E-cadherin promoter in MDCK(LT) cells. These results strongly suggest that there is a complex between LT, RB, and AP-2 in MDCK(LT) cells and that the physical and functional interaction between LT and AP-2 is mediated by RB. As RB appears to stimulate AP-2 activity, as has been shown for several other transcription factors (10, 11, 53), by binding to RB, LT would inhibit this activation possibly by preventing AP-2 from binding to DNA as previously suggested (35).
Overall, our observations are compatible with the hypothesis that RB functions as a molecular matchmaker assembling different protein complexes in different cell types (53, 56). The binding of AP-2 is distinct from all previously known RB binding mechanisms. As also shown for E2F (23, 42), AP-2 binding requires the small pocket and the C-terminal domain, but in contrast to E2F, the B region does not seem to be involved, which leads us to suggest that the A region is required, unless other sequences within the B region are involved. This difference between E2F and AP-2 binding might be relevant to the functioning of RB, as it acts in one case as an inhibitor and in the other as an activator.
AP-2 can also be activated by Myc. Classically, promoter activation by Myc has been associated with direct binding to an E box (22), but the E-cadherin promoter contains only a noncanonical E box, within the E-Pal element (21). However, Myc-mediated activation observed here involves AP-2 sites without E-box sequences (−58 E-cadherin [Fig. 2]; AP-2 CAT and HTLV LTR [Fig. 4]) which have been shown to bind AP-2 in vivo (20). Moreover, we have also shown that point mutations within these sites that destroy AP-2 binding strongly inhibited Myc-mediated activation (Fig. 3 and 4). However, AP-2 and Myc have previously been shown to form a complex in vivo via their C-terminal domains (13). This finding, together with our results, implies that a novel Myc interaction with promoter region DNA at the level of AP-2 binding sites can also lead to Myc activation of a promoter. Work is in progress to detect such complexes in vivo.
The protein interaction between Myc and AP-2 reported by Gaubatz et al. (13) used HeLa cells, which synthesize large amounts of AP-2 protein (24, 35). They examined the regulation by exogenous AP-2 and Myc of a chimeric reporter construct, containing adjacent E-box and AP-2 binding sites isolated from an enhancer in the first intron of the α-prothymosin gene. Endogenous AP-2 has been reported to cause self-interference in cotransfection studies using certain cell lines transfected by exogenous AP-2 (27). The reporter construct used in the study of Gaubatz et al. (13) was not stimulated by exogenous AP-2 alone but, in contrast, was repressed. However, mutation of the AP-2 binding site decreased Myc-mediated transactivation by 50%, and a further decrease was observed when the mutation was more proximal to the E box (13). Thus, reexamination of these results also highlights the importance of the AP-2 binding site in Myc-mediated activation. The conclusion by Gaubatz et al. (13) that AP-2 is a negative regulator of Myc function may be due to the self-interference phenomena previously described (27) or may be peculiar to the E-box and AP-2 binding sites used. Certainly in several experiments presented here, on both natural and chimeric promoters it is clear that Myc acts as an activator at AP-2 sites.
To date no in vivo interaction between RB and Myc has been demonstrated, although RB has been shown to stimulate Gal4-Myc-mediated transcription through protein-protein interactions (1). Both RB and Myc interact with several transcription factors involved in the basal transcription machinery (22, 53). It is therefore possible that they both participate in a large transcription complex involving many other proteins. However, the precise molecular mechanisms by which RB, Myc, and AP-2 cooperate to effect transcriptional activation of E-cadherin requires further study. Interestingly, the positive effects of RB and c-Myc were not additive (Fig. 1). This might imply that there are opposing effects which are counterbalanced or that they function on the same target.
What we have shown is that the transactivation of the E-cadherin promoter by RB and Myc occurs specifically in epithelial cells. In mesenchymal cells (NIH 3T3 fibroblasts), RB and Myc were totally inefficient in stimulating the E-cadherin promoter (Fig. 2). Several possibilities could explain these results. First, very low levels of AP-2 proteins (data not shown) and activity were detected in these cells (Fig. 2). Additional factors might also intervene. A cofactor and/or posttranslational modification may be missing in fibroblasts, or alternatively an inhibitor may be present. Max protein levels are probably a determining factor in Myc transcriptional activity. Max overexpression was found to repress Myc-mediated activation of the −178 E-cadherin promoter, as well as −58 E-cadherin and (E-Pal)4 SV constructs, whereas transfected alone it had no effect (unpublished results). In the present case, Max might compete with AP-2 for Myc binding as has been suggested previously (13). Work is in progress to test this possibility.
In tumor cells, the loss of E-cadherin expression parallels tumor progression toward a malignant invasive state and is correlated with a loss of the epithelial phenotype and acquisition of mesenchyme properties (5, 7). The positive regulation of E-cadherin by RB and Myc reported in the present study is perfectly consistent with our previous data (31, 32) and confirms that RB and Myc play an important role in the maintenance of the epithelial phenotype (32) by ensuring high expression of the E-cadherin gene, which is considered a master gene of the epithelial phenotype (7). Moreover, RB and Myc can transcriptionally activate other specific epithelial markers, such as cytokeratins endoA and endoB (39). We infer that similar mechanisms and transcription factors mediate endoA and endoB activation by RB and Myc.
Down-regulation of E-cadherin not only is observed during tumor progression but also is an important regulatory process in development, especially during gastrulation (51). Remarkably, Myc, like E-cadherin, is selectively down-regulated in the most highly proliferative tissue of the embryo, the primitive ectoderm, during gastrulation (47), which suggests that during embryogenesis as well, down-regulation of Myc activity contributes to invasiveness (12). Interestingly, RB and AP-2 also exhibit highly specific expression patterns during embryogenesis (49). For example, RB expression is high in the nervous system and in various epithelial cells, such as kidney collecting tubules and skin, where it is confined in the more differentiated layer; these are also the tissues where AP-2 expression is high (34). Moreover, Myc expression is also found during tubulogenesis in kidney (38, 45). Thus, all three proteins may cooperate during the developmental differentiation of particular epithelial tissues.
In summary, therefore, our results show that Myc and RB act as activators of AP-2 specifically in epithelial cells and indicate a novel mechanism of gene activation by Myc in addition to the activation through the E box (22). These results open new pathways for identifying RB and Myc target genes and exploring RB and Myc function. The inhibition by LT of RB-mediated activation of AP-2 activity might constitute an important mechanism through which the oncoprotein establishes its oncogenic property. Indeed, the AP-2 transcription factor is specially involved in epithelial gene expression (21, 30, 34), and the small DNA tumor viruses producing these oncoprotein all show various degrees of epithelial tropism. Moreover, epithelial cells are at the origin of 90% of all human tumors. Therefore, we hypothesize that the LT-RB complex inactivating AP-2 may play an important role during the dedifferentiation processes occurring during tumor progression. Furthermore, our results suggest that the overall transcription of genes containing an AP-2 sequence may be regulated by RB and Myc. One candidate is the AP-2 gene itself, which is positively autoregulated by its own product (3). In addition, since AP-2 is also involved in Myc expression (24), it may play a role in Myc autoregulation. Our present and previous results (32, 39) raise the possibilities that one of the primary biological effects of RB and Myc is to positively regulate cellular genes involved in epithelial differentiation and that inactivation of this function plays a major role in tumor progression.
ACKNOWLEDGMENTS
We thank T. Williams, A. Israël, J.-S. Seeler, and R. White for pPADH-AP2, AP-2 conaβ2 CAT, GST-HP1, and some GST-RB constructs. We are very grateful to M. Buckingham, M. Yaniv, J. C. Reyes, and G. Butler Brown for critical reading of the manuscript.
This work was supported by grants from Association pour la Recherche sur le Cancer (6256), the Fondation pour la Recherche Médicale, and the GEFLUX. E.B. was supported by predoctoral fellowships from the Ministère de l’enseignement Supérieur et de la Recherche and from La Ligue contre le Cancer.
REFERENCES
- 1.Adnane J P, Robbins D. The retinoblastoma susceptibility gene product regulates Myc-mediated transcription. Oncogene. 1995;10:381–387. [PubMed] [Google Scholar]
- 2.Batsché E, Lipp M, Crémisi C. Transcriptional repression and activation in the same cell type of the human c-myc promoter by the retinoblastoma gene protein: antagonisation of both effects by SV40 T antigen. Oncogene. 1994;9:2235–2243. [PubMed] [Google Scholar]
- 3.Bauer R, Imhof A, Pscherer A, Kopp H, Moser M, Seegers S, Kerscher M, Tainsky M, Hofstaedter F, Buettner R. The genomic structure of the human AP-2 transcription factor. Nucleic Acids Res. 1994;22:1413–1420. doi: 10.1093/nar/22.8.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens J, Löwrick O, Klein-Hitpass L, Birchmeier W. E-cadherin promoter: functional analysis of a G-C-rich region and an epithelial cell-specific palindromic regulatory element. Proc Natl Acad Sci USA. 1991;88:11495–11499. doi: 10.1073/pnas.88.24.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens J, Mareel M M, Van Roy F F, Birchmeier W. Dissecting tumor cell invasion: epithelial cells acquire invasive properties after the loss of uvomorulin-mediated cell-cell adhesion. J Cell Biol. 1989;108:2435–2447. doi: 10.1083/jcb.108.6.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernards R, Dessain S, Weinberg R. N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell. 1986;47:667–674. doi: 10.1016/0092-8674(86)90509-x. [DOI] [PubMed] [Google Scholar]
- 7.Birchmeier W, Behrens J, Weidner M, Frixen U, Schipper J. Dominant and recessive genes involved in tumor cell invasion. Curr Opin Cell Biol. 1991;3:832–840. doi: 10.1016/0955-0674(91)90057-6. [DOI] [PubMed] [Google Scholar]
- 8.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 9.Bosher J M, Williams T, Hurst H C. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P-L, Riley G J, Chen-Kiang S, Lee W-H. The retinoblastoma protein interacts with and activates NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P-L, Riley D J, Chen Y, Lee W-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPα. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 12.Downs K M, Martin G R, Bishop M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 13.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcriptional factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazin C, Dupont de Dinechin S, Hampe A, Masson J M, Martin P, Sthelin D, Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984;3:383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas-Kogan D A, Kogan C S, Levi D, Dazin P, T’Ang A, Fung T Y, Israel M A. Inhibition of apoptosis by the retinoblastoma gene product. EMBO J. 1995;14:461–472. doi: 10.1002/j.1460-2075.1995.tb07022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hann S R, Dixit M, Sears C R, Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a non- canonical DNA-binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 17.Hann S R, King M W, Bentley C W, Anderson C W, Eisenman R N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 521–523. [Google Scholar]
- 19.Haupt Y, Rowan S, Oren M. p53-mediated apoptosis in Hela cells can be overcome by excess pRB. Oncogene. 1995;10:1563–1571. [PubMed] [Google Scholar]
- 20.Hennig G, Behrens J, Truss M, Frisch S, Reichmann E, Birchmeier W. Progression of carcinoma cells is associated with alterations in chromatin structure and factor binding at the E-cadherin promoter in vivo. Oncogene. 1995;11:475–484. [PubMed] [Google Scholar]
- 21.Hennig G, Löwrick O, Birchmeier W, Behrens J. Mechanisms identified in the transcriptional control of epithelial gene expression. J Biol Chem. 1996;271:595–602. doi: 10.1074/jbc.271.1.595. [DOI] [PubMed] [Google Scholar]
- 22.Henriksson M, Lüscher B. Proteins of the myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 23.Hiebert W S. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol Cell Biol. 1993;13:3384–3391. doi: 10.1128/mcb.13.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 25.Inghirami G, Grignani F, Sternas L, Lombardi L, Knowles D M, Dalla-Favera R. Down-regulation of LFA-1 adhesion receptors by c-myc oncogene in human B lymphoblastoid cells. Science. 1990;250:682–686. doi: 10.1126/science.2237417. [DOI] [PubMed] [Google Scholar]
- 26.Kaelin W G, Krek W, Sellers W R, DeCaprio I A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 27.Kannan P, Buettner R, Chiao P J, Yim S D, Sarkiss M, Tainsky M A. N-ras oncogene causes AP-2 transcriptional self-interference, which leads to transformation. Genes Dev. 1994;8:1258–1269. doi: 10.1101/gad.8.11.1258. [DOI] [PubMed] [Google Scholar]
- 28.Land H, Chen A C, Morgenstern J P, Parada L F, Weinberg R. Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol Cell Biol. 1986;6:1917–1925. doi: 10.1128/mcb.6.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leask A, Byrne C, Fuchs E. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc Natl Acad Sci USA. 1991;88:7948–7952. doi: 10.1073/pnas.88.18.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martel C, Batsché E, Harper F, Crémisi C. Inactivation of the retinoblastoma gene product or an RB-related protein by SV40 T antigen in MDCK epithelial cells results in massive apoptosis. Cell Death Differ. 1996;3:61–74. [PubMed] [Google Scholar]
- 32.Martel C, Harper F, Cereghini S, Veerlé N, Mareel M, Crémisi C. Inactivation of retinoblastoma family proteins by SV40 T antigen results in creation of a hepatocyte growth factor/scatter factor autocrine loop associated with an epithelial-fibroblastoid conversion and invasiveness. Cell Growth Differ. 1997;8:165–178. [PubMed] [Google Scholar]
- 33.Martel C, Lallemand D, Crémisi C. Specific c-myc and max regulation in epithelial cells. Oncogene. 1995;10:2195–2205. [PubMed] [Google Scholar]
- 34.Mitchell P J, Timmons P M, Hebert J M, Rigby P W, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell P J, Wang C, Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 36.Muchardt C, Seeler J-S, Nirula A, Gong S, Gaynor R. Transcription factor AP-2 activates gene expression of HTLV-1. EMBO J. 1992;11:2573–2581. doi: 10.1002/j.1460-2075.1992.tb05322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muchardt C, Seeler J S, Gaynor R B. Regulation of HTLV-1 gene expression by tax and AP-2. New Biol. 1992;4:541–550. [PubMed] [Google Scholar]
- 38.Mugrauer G, Ekblom P. Contrasting expression patterns of three members of the myc family of protooncogenes in the developing and adult mouse kidney. J Cell Biol. 1991;112:13–25. doi: 10.1083/jcb.112.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onclercq R, Lavenu A, Crémisi C. Pleiotropic derepression of developmentally regulated cellular and viral genes by the product of c-myc protooncogene in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 1989;17:735–753. doi: 10.1093/nar/17.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietenpol J A, Stein R W, Moran E, Yaciuk P, Schlegel R, Lyons R M, Pittelkow M R, Munger K, Howley P M, Moses H L. TGFβ1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming pRB binding domains. Cell. 1990;61:777–785. doi: 10.1016/0092-8674(90)90188-k. [DOI] [PubMed] [Google Scholar]
- 41.Prendergast G C, Diamond L E, Dahl D, Cole M D. The c-Myc-regulated gene mr1 encodes plasminogen activator inhibitor 1. Mol Cell Biol. 1990;10:1265–1269. doi: 10.1128/mcb.10.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin X-Q, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 43.Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci USA. 1995;92:855–859. doi: 10.1073/pnas.92.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ringwald M, Baribault H, Schmidt C, Kemler R. The structure of the gene coding for the mouse cell adhesion molecule uvomorulin. Nucleic Acids Res. 1991;19:6533–6539. doi: 10.1093/nar/19.23.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmid P, Schulz W, Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989;243:226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- 46.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 47.Snow M H L. Gastrulation in the mouse: growth and regionalization of the epiblast. J Embryol Exp Morphol. 1977;42:293–303. [Google Scholar]
- 48.Suen T-C, Hung M-C. c-myc reverses neu-induced transformed morphology by transcriptional repression. Mol Cell Biol. 1991;11:354–362. doi: 10.1128/mcb.11.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szekely L, Jiang W, Bulic-Jakus F, Rosen A, Ringertz N, Klein G, Wiman K G. Cell type and differentiation dependent heterogeneity in retinoblastoma protein expression in SCID mouse fetuses. Cell Growth Differ. 1992;3:149–156. [PubMed] [Google Scholar]
- 50.Templeton D J, Park S H, Lanier L, Weinberg R A. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiery J-P, Duband J-L, Rutishauser U, Edelman G M. Cell adhesion molecules in early chicken embryogenesis. Proc Natl Acad Sci USA. 1982;79:6731–6741. doi: 10.1073/pnas.79.21.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Versteeg R, Noordermeer A, Krüse-Wolters M, Ruiter D J, Schrier P I. c-myc down-regulates class 1 HLA expression in human melanomas. EMBO J. 1988;7:1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J Y J, Knudsen E S, Welch P J. The retinoblastoma tumor-suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg R. Tumor suppressor genes. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 55.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 56.Welch P J, Wang J Y. Disruption of retinoblastoma protein function by coexpression of its C pocket fragment. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 57.Williams T, Admon A, Lüscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 58.Yang B-S, Geddes T J, Pogulis R J, de Crombrugghe B, Freytag S O. Transcriptional suppression of cellular gene expression by c-Myc. Mol Cell Biol. 1991;11:2291–2295. doi: 10.1128/mcb.11.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]