Abstract
Although Cks proteins were the first identified binding partners of cyclin-dependent protein kinases (cdks), their cell cycle functions have remained unclear. To help elucidate the function of Cks proteins, we examined whether their binding to p34cdc2 (the mitotic cdk) varies during the cell cycle in Xenopus egg extracts. We observed that binding of human CksHs2 to p34cdc2 was stimulated by cyclin B. This stimulation was dependent on the activating phosphorylation of p34cdc2 on Thr-161, which follows cyclin binding and is mediated by the cdk-activating kinase. Neither the inhibitory phosphorylations of p34cdc2 nor the catalytic activity of p34cdc2 was required for this stimulation. Stimulated binding of CksHs2 to another cdk, p33cdk2, required both cyclin A and activating phosphorylation. Our findings support recent models that suggest that Cks proteins target active forms of p34cdc2 to substrates.
Cell cycle transitions in eukaryotic cells are controlled by a family of cyclin-dependent protein kinases (cdks) and their cyclin-binding partners. The cyclin B-p34cdc2 complex is required for the G2-to-M phase transition. Regulation of p34cdc2 activity is crucial to the proper timing and execution of mitosis and is accomplished via regulation of cyclin B levels and by multiple phosphorylations of p34cdc2 (for reviews, see references 29, 33, 37, 44, and 51). Inactive, monomeric p34cdc2 binds to cyclin B, which accumulates during interphase, permitting phosphorylation of p34cdc2 on three sites: Thr-14, Tyr-15, and Thr-161 (50). Phosphorylation of Thr-161 is necessary for complete activation of the kinase and is carried out by the cdk-activating kinase (CAK) (for reviews, see references 48 and 49). Vertebrate and starfish CAKs consist of the cdk7-cyclin H-MAT1 complex (16, 18, 34, 45, 51, 56, 57), although budding yeast has a distinct CAK, Cak1p, that functions as a monomer (15, 27, 58). p34cdc2 also undergoes inhibitory phosphorylations on Thr-14 and Tyr-15 that keep it inactive until cdc25, a dual-specificity phosphatase, dephosphorylates these sites just prior to mitosis (for a review, see reference 8), thus producing an active, Thr-161-phosphorylated p34cdc2 responsible for entry into mitosis. Inactivation of p34cdc2 and exit from mitosis require the ubiquitin-dependent proteolysis of cyclin B (20, 28, 39). Ubiquitination of cyclin B is mediated by the ubiquitin ligase termed the anaphase-promoting complex, a multiprotein assembly that is activated by phosphorylation during mitosis (24, 30, 32, 53).
p13suc1 is the founding member of a family of p34cdc2-binding proteins whose functions are less clearly understood than those of the cyclins. suc1 was initially identified in fission yeast from a screen for high-copy-number suppressors of a temperature-sensitive allele of cdc2 (23). Homologs have since been identified in many organisms including budding yeast (CKS1), humans (CksHs1 and CksHs2), and frogs (Xe-p9) (21, 41, 46). In addition to extensive primary amino acid sequence conservation among these homologs, crystal structures of p13suc1, Cks1p, CksHs1, and CksHs2 reveal a common core motif (2, 3, 6, 13). Ternary complexes containing a cdk, a cyclin, and a Cks protein have been detected biochemically (1, 5), and the Cks proteins (in particular p13suc1) covalently coupled to Sepharose beads have become universal reagents for precipitating monomeric as well as active (cyclin-bound) forms of p34cdc2 from all eukaryotes (1, 5, 11, 46, 50).
Although both suc1 and CKS1 are essential for cell viability in fission yeast and budding yeast, respectively (21, 23, 25), the functions of the Cks proteins have remained unclear because no single model that can account for the conflicting observations has been proposed (for reviews, see references 14 and 43). While p13suc1 can stabilize the activity of some temperature-sensitive forms of fission yeast p34cdc2 in vitro (5, 36), there are also several lines of evidence that suggest that Cks proteins antagonize p34cdc2 activation. For example, overexpression of suc1 in fission yeast and of CKS1 in budding yeast delays entry into mitosis (22, 25, 46). In vitro, excess p13suc1 or Xe-p9 prevents the dephosphorylation of p34cdc2 on Tyr-15 in Xenopus interphase egg extracts, thereby keeping p34cdc2 inactive (12, 41). In fact, excess p13suc1 or Xe-p9 can directly inhibit the dephosphorylation of Tyr-15 by recombinant cdc25 (19, 41). These data are difficult to reconcile with the consequences of immunodepletion of Xe-p9 from Xenopus interphase egg extracts, which also prevents entry into mitosis (41). A role for Cks proteins in the exit from mitosis is suggested by the anaphase arrest of fission yeast suc1 deletion mutants (36). Inactivation of suc1 leads to elevated cyclin B levels and high p34cdc2 activity (4). In Xenopus, immunodepletion of Xe-p9 from mitotic egg extracts leads to a stabilization of cyclin B and high p34cdc2 activity, which prevents exit from mitosis (41). In budding yeast, conditional cks1 mutants display defects at both the G1/S and the G2/M transitions (55). Thus, Cks proteins present a complex picture from which their primary function is not easily deduced.
Recent X-ray crystallographic studies of CksHs1 bound to p33cdk2 led to the suggestion that Cks proteins might target cdks to specific substrates or to particular phosphoproteins (7). CksHs1 has a phosphate-binding pocket that is positioned along the putative substrate-binding surface of p33cdk2, thus possibly extending this surface and permitting more specific or tighter binding of substrates. A related model suggesting that Xe-p9 might serve as a “docking factor” has been proposed by Patra and Dunphy (41), based on a biochemical analysis using Xenopus egg extracts. These models, which we refer to as targeting models, have recently gained support from a study showing that p13suc1 binds to the phosphorylated (active) form of the anaphase-promoting complex (54).
Previous studies have not addressed whether Cks proteins might interact with p34cdc2 in a cell cycle-specific manner or preferentially with distinct subsets of p34cdc2 molecules. The existence of variations in binding would help guide proposed models of Cks protein function. We have addressed these questions by using Xenopus egg cytoplasmic extracts in which we could easily and synchronously manipulate the activation state of p34cdc2. Binding of Cks proteins to monomeric p34cdc2 in an interphase-arrested extract was compared to binding to p34cdc2 activated by addition of recombinant cyclin B. The role of p34cdc2 phosphorylation in binding of CksHs2 was assessed using p34cdc2 mutant proteins as well as p34cdc2 phosphorylated in vitro by Cak1p. We found that CksHs2 preferentially associated with the active form of p34cdc2. Finally, we discuss our observations in the context of current models of Cks function.
MATERIALS AND METHODS
Buffers.
EB contains 80 mM β-glycerophosphate (pH 7.3), 20 mM EGTA, 15 mM MgCl2, 10 mM dithiothreitol (DTT), 1 mg of ovalbumin per ml, and 1× protease inhibitors (10 μg each of leupeptin, chymostatin and pepstatin per ml). XB contains 10 mM K-HEPES (pH 7.7), 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 50 mM sucrose, 100 μg of ovalbumin per ml, and 0.1× protease inhibitors. 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (SB) contains 6.6% SDS, 26% (vol/vol) glycerol, 100 mM DTT, 262 mM Tris base, and bromophenol blue. Tris-buffered saline (TBS) contains 150 mM NaCl and 10 mM Tris-HCl (pH 8.0). TBST is TBS with 0.1% Tween 20.
Xenopus egg extracts.
Cytostatic factor-arrested extracts were released from cytostatic factor arrest with calcium and arrested in interphase by treatment with cycloheximide as described previously (50).
Protein purification.
Glutathione S-transferase (GST)–cyclin B was expressed from a pGEX vector encoding GST as an N-terminal fusion to residues 13 to 409 of sea urchin cyclin B (50). The protein was prepared from a 6-liter culture of Escherichia coli and purified by precipitation with glutathione-agarose followed by elution with free glutathione as described previously (50). The protein was desalted for use in glutathione-agarose precipitations by chromatography on a 1-ml Sephadex G-25 (Sigma) column equilibrated with 50 mM HEPES (pH 7.5)–3 mM DTT–1× protease inhibitors.
GST-cyclin A was expressed from a pGEX vector (42) and purified as for GST-cyclin B except that expression was induced at an A600 of 0.8 with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C overnight. The yield from 6 liters was 350 μg.
CksHs2 was expressed from pRK171 in E. coli BL21(DE3)pLysS cells (46). A 3-liter culture was induced at an A600 of 0.4 with 0.4 mM IPTG. After 3.5 h at 37°C, the cells were pelleted at 4,200 × g for 5 min in a Sorvall GS-3 rotor. Cells were washed twice with 0.9% NaCl, resuspended in 30 ml of 50 mM Tris-HCl (pH 8.0)–2 mM EDTA–10% glycerol, and sonicated on ice (four 30-s continuous pulses on output 5, using a Heat Systems-Ultrasonics, Inc. model W-375 apparatus with a microtip). The extract was clarified by ultracentrifugation for 40 min at 1.78 × 105 × g (50,000 rpm) in a Beckman 60 Ti rotor. A 50 to 70% ammonium sulfate fraction was resuspended in 2.5 ml of 50 mM Tris-HCl (pH 8.0)–2 mM EDTA–100 mM NaCl. The protein was applied to a 100-ml Bio-Gel P-30 column (1.5-cm diameter; Bio-Rad) that had been equilibrated with the same buffer. Fractions containing CksHs2 were pooled and either dialyzed into EB or coupled to CNBr-activated Sepharose (see below).
Recombinant human p33cdk2 was purified from E. coli as described by Connell-Crowley et al. (9); the p33cdk2 used for Fig. 2D was a gift of P. Kaldis. Purified recombinant Cak1p was a gift of P. Kaldis.
FIG. 2.
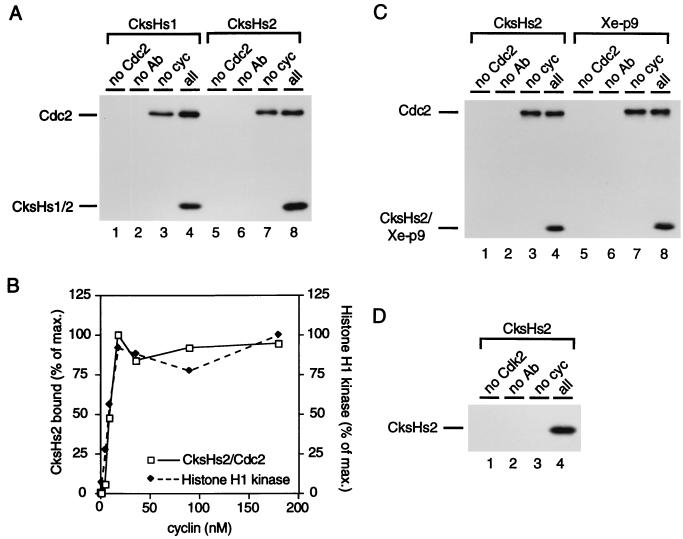
Cyclin stimulates binding of Cks proteins to cdks. (A) CksHs1 and CksHs2 exhibit cyclin-stimulated binding to p34cdc2. In vitro-translated, 35S-labeled CksHs1 (lanes 1 to 4) or CksHs2 (lanes 5 to 8) and in vitro-translated, 35S-labeled p34cdc2-HA (lanes 2 to 4 and 6 to 8) were incubated in Xenopus interphase egg extract with (lanes 1, 2, 4, 5, 6, and 8) or without (lanes 3 and 7) 36 nM GST-cyclin B. p34cdc2-HA and associated CksHs1 or CksHs2 were immunoprecipitated with 12CA5 monoclonal antibody (Ab) against the HA epitope and were analyzed by SDS-PAGE and fluorography. Controls for background precipitation of CksHs1 or CksHs2 were performed by substituting unprogrammed reticulocyte lysate for p34cdc2 (lanes 1 and 5) or using protein A-agarose beads without antibody (lanes 2 and 6). (B) Effect of cyclin concentration on association of CksHs2 with p34cdc2 and on p34cdc2 activation. CksHs2 and p34cdc2 were incubated in extract (as for panel A) along with 0.05 to 120 nM cyclin B. Aliquots were analyzed for histone H1 kinase activity and for the amount of CksHs2 bound to p34cdc2 (as for panel A). (C) Xe-p9 exhibits cyclin-stimulated binding to p34cdc2. In vitro-translated, 35S-labeled CksHs2 (lanes 1 to 4) or Xe-p9 (lanes 5 to 8) and in vitro-translated, 35S-labeled p34cdc2 (lanes 2 to 4 and 6 to 8) were incubated in Xenopus egg interphase extract with (lanes 1, 2, 4, 5, 6, and 8) or without (lanes 3 and 7) cyclin B. p34cdc2 and associated CksHs2 or Xe-p9 were immunoprecipitated and analyzed by SDS-PAGE and fluorography. The amounts of CksHs2 and Xe-p9 and their specific activities were equal. Controls for background precipitation of CksHs2 or Xe-p9 were as described above. (D) CksHs2 exhibits cyclin-stimulated binding to p33cdk2. Purified bacterially expressed HA-p33cdk2 was incubated with in vitro-translated, 35S-labeled CksHs2 in Xenopus egg interphase extract with (lanes 1, 2, and 4) or without (lane 3) cyclin B. p33cdk2 and associated CksHs2 were immunoprecipitated and analyzed by SDS-PAGE and fluorography. Controls for background precipitation of CksHs2 were performed by substituting buffer for p33cdk2 (lane 1) or using protein A-agarose beads without antibody (lane 2); 0.25 to 0.5% of the added CksHs2 bound to p33cdk2.
In vitro transcription and translation.
PCR was used to introduce an NcoI site at the start codon and a BamHI site downstream of the stop codon of CksHs1, CksHs2, and Xe-p9. NcoI-BamHI fragments were subcloned into NcoI-BamHI-cut cycΔ13Tb. This vector contains a T7 promoter and the 5′ and 3′ untranslated regions of β-globin to facilitate in vitro transcription and translation (19). The 5′ and 3′ primers used for PCR were as follows (start and stop codons are underlined): for CksHs1, GGGGAATTCCATGGCGCACAAACAAATTTACTATTCG and GGGGGATCCTCATTTCTTTGGTTTCTTGG; for CksHs2, GGGGAATTCCATGGCCCACAAGCAGATCTAC and GGGGGATCCTCATTTTTGTTGATCTTTTGG; and for Xe-p9, GGGGAATTCCATGGCATACAAGAACATTTACTACTCGG and GGGGGATCCTCATTTCTGTTGATCTTTTGG. The complete sequence of each gene was verified. Hemagglutinin epitope (HA)-tagged versions of wild-type and mutant forms of Xenopus p34cdc2 in the same vector have been described elsewhere (19, 52).
In vitro transcription reactions were performed as described previously (19) except that rRNasin (1.6 U/μl; Promega) was included and the products were treated with RNase-free DNase (0.9 U/μl) for 15 min at 37°C. The RNA was translated in reticulocyte lysates (Promega) according to the manufacturer’s instructions. p34cdc2 translations used 0.15 mCi of [35S]methionine per ml and 40 μM unlabeled methionine. The luciferase translation used 0.3 mCi of [35S]methionine per ml and included 40 μM unlabeled methionine. (Luciferase mRNA was obtained from Promega.) The Cks protein translations used 0.3 mCi of [35S]methionine per ml without any additional unlabeled methionine. The concentration of the p34cdc2 was determined by immunoblotting compared to a known concentration of Xenopus p33cdk2, using an anti-PSTAIR antiserum (50). The concentration of CksHs2 was calculated by comparison to the p34cdc2 translation, using SDS-PAGE and phosphorimage quantitation and adjusting for the respective specific activities of methionine and the number of methionines in each protein. The concentration of wild-type and mutant p34cdc2 was 70 nM, and the concentration of the Cks proteins was ∼100 nM.
Glutathione-agarose precipitation of cyclin-bound Cks proteins.
Six microliters of in vitro-translated CksHs1 or CksHs2, 20 μl of Xenopus egg extract or XB, and 6 μl of XB were incubated for 10 min at 23°C; 8 μl of GST-cyclin B was added to a final concentration of either 36 or 360 nM, and reaction mixtures were incubated for 20 min at 23°C. As a control, 8 μl of GST was added to a final concentration of 3.5 μM. Reaction mixtures were diluted to 200 μl with XB containing 0.5% Nonidet P-40 (NP-40) and rotated for 1 h at 4°C with 30 μl of glutathione-agarose beads (Sigma). The beads were washed five times with 400 μl of XB containing 0.5% NP-40 then once with XB. The beads were resuspended in 25 μl of 2× SB, and 10 μl was analyzed by using a 10% acrylamide stacking gel and a 16.5% acrylamide separating gel containing 13% glycerol and 3.6 M urea (35). A set of dilution standards of CksHs1 or CksHs2 (representing from 24 to 0.75% of the total in each reaction) was run simultaneously for comparison. Gels were analyzed by phosphorimaging (Molecular Imager GS-250; Bio-Rad) or by fluorography.
Immunoprecipitation of p34cdc2-bound Cks proteins.
Six microliters of a p34cdc2 in vitro translation, 6 μl of a CksHs1, CksHs2, or Xe-p9 in vitro translation, and 20 μl of Xenopus egg extract were incubated for 10 min at 23°C. (Control incubations contained translations performed without RNA or XB in place of Xenopus extract.) Eight microliters of 180 nM GST-cyclin B in XB (or 8 μl of XB alone) was added, and reaction mixtures were incubated for 20 min at 23°C. The reaction mixtures were then diluted with 160 μl of XB containing 0.5% NP-40, and the in vitro-translated p34cdc2 was immunoprecipitated by its C-terminal HA tag. Immunoprecipitation was performed with 10 μl of protein A-agarose beads bound to 1 μl of ascites fluid containing the 12CA5 antibody (17, 59). After rotating at 4°C for 1 h, the beads were washed as for glutathione-agarose precipitations (see above) (0.5 M NaCl was included in the third and fourth washes for Fig. 5 and 6). The beads were resuspended in 20 μl of 1× SB or in EB (10-μl total volume including the beads) for histone H1 kinase assays; 10 μl was analyzed by SDS-PAGE and phosphorimage analysis or fluorography as above for glutathione-agarose precipitations. The amount of bound CksHs2 was calculated as the ratio of the integrated intensity of the CksHs2 band to that of the p34cdc2 band. The gel background was subtracted from the p34cdc2 band intensity, and the amount of CksHs2 precipitated in the absence of p34cdc2-HA was subtracted from the CksHs2 value.
FIG. 5.
Phosphorylation of p34cdc2 plays an important role in the cyclin-stimulated binding of CksHs2 to p34cdc2. In vitro-translated, 35S-labeled CksHs2 and in vitro-translated, 35S-labeled wild-type (wt) p34cdc2-HA (lanes 1 to 4, 7, and 8) or an unphosphorylatable mutant of p34cdc2-HA (AF/ala; lanes 5 and 6) were incubated in the presence (lanes 1 to 6) or absence (lanes 7 and 8) of Xenopus egg interphase extract (EXT) with (lanes 1, 2, 4, 6, and 8) or without (lanes 3, 5, and 7) cyclin B. (AF/ala is mutated at all three phosphorylation sites on p34cdc2: T14A, Y15F, and T161A.) p34cdc2-HA and associated CksHs2 were immunoprecipitated and analyzed by SDS-PAGE and fluorography. Controls for background precipitation of CksHs2 were performed as described for Fig. 2. Equal amounts of each form of p34cdc2 were used in each reaction, and their specific activities were the same. Ab, antibody.
FIG. 6.
Phosphorylation of Thr-161 of p34cdc2 is necessary for the cyclin-stimulated binding of CksHs2. In vitro-translated, 35S-labeled CksHs2 and in vitro-translated, 35S-labeled p34cdc2-HA mutants were incubated in Xenopus egg interphase extract with (lane 1 and even-numbered lanes) or without (odd-numbered lanes except lane 1) cyclin B. The binding reactions contained the following forms of p34cdc2-HA: wild type (wt; lanes 2 to 4), AF (lanes 5 and 6), T161A (lanes 7 and 8), AF/ala (lanes 9 and 10), and K33R (lanes 11 and 12). (AF is mutated at the two inhibitory phosphorylation sites; T161A is mutated at the activating phosphorylation site; AF/ala is mutated at all three sites; K33R is mutated in the ATP-binding site.) p34cdc2-HA and associated CksHs2 were immunoprecipitated and analyzed by SDS-PAGE and fluorography. Controls for background precipitation of CksHs2 were performed as described for Fig. 2. Equal amounts of each form of p34cdc2 were used in the reactions, and the specific activities of these proteins were equal. Ab, antibody.
Histone H1 kinase assays.
Histone H1 kinase assays were performed as described previously (50), using either 10 μl of immunoprecipitated p34cdc2 or 10 μl of 50-fold-diluted Xenopus egg extract in EB. Quantitation was performed by phosphorimage analysis.
Cak1p activation of p34cdc2.
Three microliters of in vitro-translated p34cdc2 and 0.8 μl of 0.9 μM GST-cyclin B were incubated with 8.5 ng of budding yeast Cak1p in EB containing 6 mM MgCl2 and 0.6 mM ATP in a total volume of 17 μl for 30 min at 23°C. Pilot experiments indicated that this amount of Cak1p maximally activated p34cdc2 under these conditions. For coimmunoprecipitation with CksHs2, 3 μl of in vitro-translated CksHs2 was subsequently added for 5 min at 23°C, and then p34cdc2 was immunoprecipitated. Immunoprecipitation was carried out as described above with substitution of EB for XB. For assessment of histone H1 kinase activity, CksHs2 was omitted and the beads were resuspended in 10 μl of EB after immunoprecipitation.
To determine the effect of CksHs2 on Cak1p activation of p34cdc2, 3 μl of in vitro-translated p34cdc2 and 0.8 μl of 0.9 μM GST-cyclin B were incubated with bacterially produced and purified CksHs2 in EB in a 16-μl total volume for 10 min at 23°C. Activation with Cak1p, immunoprecipitation, and histone H1 kinase assay were then performed as described above except that 18 ng of Cak1p was used in a final volume of 20 μl.
To determine the effect of CksHs2 on the phosphorylation of p34cdc2 by Cak1p, 30 μl of in vitro-translated p34cdc2 was immunoprecipitated as described above; 0.8 μl of 0.9 μM GST-cyclin B and various amounts of purified CksHs2 were added in a final volume of 16 μl, and the reaction mixtures were incubated for 25 min at 23°C. Phosphorylation with 18 ng of GST-Cak1p was performed as described above in a total reaction volume of 20 μl except that 10 μCi of [γ-32P]ATP was included. The reaction was stopped with 2× SB and analyzed by SDS-PAGE and autoradiography.
Cak1p phosphorylation of p33cdk2.
Five microliters of 600 nM HA-p33cdk2 with or without 5 μl of 1.8 μM GST-cyclin A was incubated with or without 17.5 ng of purified GST-Cak1p in a total volume of 15 μl of EB containing 165 μM ATP and an extra 1.7 mM MgCl2 for 20 min at 23°C; 15 μl of 35S-labeled, in vitro-translated CksHs2 was then added to each reaction for 30 min at 23°C. The HA-p33cdk2 was immunoprecipitated and analyzed by SDS-PAGE and fluorography as described above except that the EB used for the immunoprecipitation and washes lacked NP-40 in all but the two middle washes.
To confirm the phosphorylation of HA-p33cdk2 by Cak1p, reactions of the same composition as above were performed with 5 μCi of [γ-32P]ATP per reaction. After the first incubation, these reactions were analyzed by SDS-PAGE and autoradiography and quantitated by phosphorimage analysis.
Binding of purified CksHs2 to monomeric HA-p33cdk2.
One hundred micrograms of HA-p33cdk2 in 200 μl of EB was immunoprecipitated with 10 μl of protein A-agarose beads bound to 1 μl of ascites fluid containing the 12CA5 antibody. The beads were washed five times with 400 μl of EB. The immunoprecipitated HA-p33cdk2 was incubated with rotation in a final volume of 100 μl with 11 μM CksHs2 for 45 min at 4°C and was then washed once with 400 μl of XB containing 0.5% NP-40 and once with XB alone. The beads were resuspended in 20 μl of 2× SB and were analyzed by SDS-PAGE as described above for glutathione-agarose precipitations. The bound CksHs2 protein was detected by immunoblotting. The Immobilon-P (Millipore) membrane was incubated for 1 h at 23°C with a 1:1,000 dilution of an anti-Cks1 antiserum (Santa Cruz Biotechnology) in TBST containing 5% powdered milk (TBST-BLOTTO) followed by one 15-min wash in TBST-BLOTTO and three washes in TBST for 15 min. Successive 30-min incubations were performed with 1:5,000 dilutions of mouse anti-rabbit and rabbit anti-mouse antibodies in TBST-BLOTTO. The membrane was then incubated with a mixture of 1:10,000 dilutions of horseradish peroxidase-coupled goat anti-rabbit and goat anti-mouse antibodies in TBST-BLOTTO for 1 h at 23°C followed by washes as above and an additional wash in TBS. Detection was performed with Pierce Supersignal substrate and fluorography.
Preparation and use of CksHs2-Sepharose and ovalbumin-Sepharose.
Ovalbumin (30 mg) and CksHs2 (5 mg) were each dialyzed against 0.1 M NaHCO3 (pH 8.3)–0.5 M NaCl and coupled to CNBr-activated Sepharose (Pharmacia) according to the manufacturer’s instructions. Unreacted groups were quenched with 0.1 M Tris-HCl (pH 8.0), and the beads were stored at 4°C in 50 mM Tris-HCl (pH 8.0)–2 mM EDTA–100 mM NaCl. The final concentration of ovalbumin-Sepharose was 10 mg/ml. The final concentration of CksHs2-Sepharose was 5 mg/ml.
For use in precipitation experiments, 10 μl of beads was washed into XB containing 0.5% NP-40 and 0.5 M NaCl and incubated with 2 to 10 μl of in vitro-translated protein in a 200-μl total volume. Alternatively, these beads were incubated with 35 nM GST-cyclin B or 3.5 μM GST (a gift of J. Holmes) in a 100-μl total volume. After incubation for 1 h at 4°C with rotation, the beads were washed once with XB containing 0.5% NP-40 and 0.5 M NaCl and once with XB. The beads were resuspended in 2× SB, and the bound 35S-labeled, in vitro-translated proteins were detected by SDS-PAGE and quantitated by phosphorimage analysis as described above for glutathione-agarose precipitations. Protein standards were used to quantitate the percentage of total protein bound. Precipitation of GST-cyclin B and GST was assessed by immunoblotting with affinity-purified anti-GST polyclonal antibodies (a gift of Z. Pitluk) and was quantitated by phosphorimage analysis and comparison to standards on the same blots.
RESULTS
CksHs1 and CksHs2 exhibit cyclin-stimulated binding to p34cdc2.
We used Xenopus egg extracts to study the binding of in vitro-translated Cks proteins to p34cdc2. This system allowed us to manipulate the activation state of p34cdc2. Xenopus egg extracts undergo successive cell cycles dependent on the synthesis of only one protein, cyclin B (38). Extracts treated with cycloheximide to inhibit protein synthesis arrest in interphase; subsequent addition of exogenous cyclin B induces mitosis (50). We first wished to verify that exogenously supplied human Cks proteins would interact with Xenopus p34cdc2 under physiological conditions and to determine the efficiency of their binding. We added trace amounts of 35S-labeled, in vitro-translated CksHs1 or CksHs2 proteins to a Xenopus egg extract in interphase. GST-cyclin B was added to activate the p34cdc2, thus driving the extract into mitosis (50). GST-cyclin-p34cdc2 complexes and any associated proteins were then precipitated by using glutathione-agarose beads. We observed that 25 to 50% of the added CksHs1 or CksHs2 was precipitated under these conditions (Fig. 1, lanes 2 and 4), indicating that in vitro-translated CksHs1 and CksHs2 can bind efficiently to Xenopus p34cdc2.
FIG. 1.
Association of in vitro-translated CksHs1 and CksHs2 with cyclin B-p34cdc2 in a Xenopus egg extract. In vitro-translated, 35S-labeled CksHs1 (lanes 1 and 2) or CksHs2 (lanes 3 and 4) was incubated in Xenopus egg interphase extracts with (lanes 2 and 4) or without (lanes 1 and 3) 36 nM GST-cyclin B. Cyclin-p34cdc2 complexes and any associated CksHs1 or CksHs2 were precipitated by using glutathione-agarose beads. The bound Cks proteins were detected by fluorography following SDS-PAGE.
As a first step toward determining whether the association of CksHs1 or CksHs2 with p34cdc2 varies at any point in the stepwise activation of p34cdc2, we examined whether the presence of cyclin B affected the binding of CksHs1 or CksHs2 to p34cdc2. We used exogenously supplied p34cdc2 for this and many subsequent experiments. This in vitro-translated p34cdc2 contained a C-terminal HA tag to facilitate its selective immunoprecipitation using monoclonal antibody 12CA5. We incubated 35S-labeled, in vitro-translated CksHs1 or CksHs2 with 35S-labeled, in vitro-translated p34cdc2 in a Xenopus interphase egg extract. When added, the concentration of GST-cyclin B (36 nM) was just sufficient to maximally activate p34cdc2 (Fig. 2B). The p34cdc2-HA was immunoprecipitated, and the amount of bound CksHs1 and CksHs2 was determined following SDS-PAGE. Because the added in vitro-translated p34cdc2-HA represented only a small fraction (∼10%) of the total p34cdc2 in the extracts (see Materials and Methods and reference 31) and because the immunoprecipitations were ∼20% efficient, we expected at most only a small percentage of the added Cks protein to coimmunoprecipitate with p34cdc2-HA even if binding was as efficient as seen in Fig. 1. Assuming that glutathione-agarose precipitated all of the GST-cyclin B-p34cdc2 and associated Cks proteins (Fig. 1), we expected to precipitate (0.10)(0.20) or 2% as much CksHs1 and CksHs2 as in Fig. 1. In the absence of cyclin, we detected little or no specific association of CksHs1 or CksHs2 with monomeric p34cdc2-HA (Fig. 2A, lanes 3 and 7). In contrast, the presence of cyclin B stimulated binding of CksHs1 or CksHs2 to p34cdc2-HA (Fig. 2A, lanes 4 and 8). The proportion of input Cks protein bound was 0.4 to 1.0%, which was almost exactly the anticipated 2% of the level of Cks proteins that precipitated with GST-cyclin-p34cdc2 above (Fig. 1), indicating that binding is quite efficient under these conditions. Control precipitations from reactions containing cyclin B confirmed that both antibody and p34cdc2 were required for coprecipitation of CksHs1 or CksHs2 (Fig. 2A, lanes 1, 2, 5, and 6). Because CksHs1 and CksHs2 consistently bound to p34cdc2 equally well (Fig. 1 and 2 and data not shown), we arbitrarily selected CksHs2 for most of the in-depth analysis that follows.
To evaluate the physiological relevance of cyclin-stimulated binding of CksHs1 and CksHs2 to p34cdc2, we performed a titration of cyclin B to determine whether stimulation of CksHs2 binding to p34cdc2 paralleled the activation of p34cdc2. In vitro-translated, 35S-labeled CksHs2 was incubated with p34cdc2-HA in a Xenopus egg extract to which various amounts of GST-cyclin B were added. The association of CksHs2 with p34cdc2-HA was determined by coimmunoprecipitation (as described above), and the activity of the p34cdc2-HA was determined by a histone H1 kinase assay. The binding of CksHs2 to p34cdc2-HA at a given cyclin concentration closely followed the activation of p34cdc2-HA (Fig. 2B). Maximal activation and maximal stimulation of CksHs2 binding both occurred at 18 nM GST-cyclin B, confirming that the concentrations of cyclin B that stimulate binding are reasonable and suggesting that there is a correlation between the activation of p34cdc2 and its association with CksHs2. We chose to perform subsequent experiments with 36 nM GST-cyclin B to be firmly within the range of maximal CksHs2 association with p34cdc2-HA.
Cyclin-stimulated binding of Cks proteins to cdks is conserved.
While these studies were near completion, Patra and Dunphy reported the cloning and characterization of the Xenopus homolog of CksHs2, Xe-p9 (41). Although their experimental approach was different, their data could be interpreted as indicating that Xe-p9 binding to p34cdc2 is not stimulated by cyclin. We therefore tested 35S-labeled, in vitro-translated Xe-p9 for cyclin-stimulated binding to 35S-labeled, in vitro-translated p34cdc2-HA in a Xenopus egg extract. Virtually no Xe-p9 coimmunoprecipitated with p34cdc2-HA in the absence of cyclin (Fig. 2C, lane 7); however, 1% of the added Xe-p9 bound to p34cdc2-HA in the presence of GST-cyclin B (Fig. 2C, lane 8), comparable to the level of cyclin-stimulated binding of CksHs2 to p34cdc2-HA (Fig. 2C; compare lanes 4 and 8).
To test whether CksHs2 would exhibit cyclin-stimulated binding to a cdk other than p34cdc2, we substituted purified human HA-p33cdk2 for p34cdc2-HA in the coimmunoprecipitation assay. Although binding of CksHs2 to monomeric p33cdk2 was undetectable (Fig. 2D, lane 3), significant binding (0.25 to 0.5%) of CksHs2 to p33cdk2 was observed in the presence of cyclin B (Fig. 2D, lane 4). This percentage of total bound CksHs2 was comparable to that seen in Fig. 2. However, the concentration of CksHs2 in this experiment was fourfold higher than that in Fig. 2, and the concentration of HA-p33cdk2 (produced in E. coli rather than by in vitro translation) was ∼15-fold higher than the concentration of p34cdc2-HA used for Fig. 2. Thus, while this experiment indicates that cyclin-stimulated binding of CksHs2 is not limited to p34cdc2, it also suggests that the affinity of CksHs2 for cyclin B-p33cdk2 is much lower than for cyclin B-p34cdc2.
Binding of monomeric cdks to CksHs2.
Although our experiments indicate that the presence of cyclin increases the amount of Cks protein that coimmunoprecipitates with p34cdc2, they do not preclude a weak interaction of Cks proteins with p34cdc2 in the absence of cyclin. In fact, many studies have used Sepharose-coupled CksHs2 homologs to purify both monomeric and cyclin-bound forms of p34cdc2 (5, 10, 50). It is possible that Cks proteins and p34cdc2 interact weakly in the absence of cyclin, resulting in a high dissociation rate and making detection of physiological concentrations of the complex difficult on the time scale of immunoprecipitations. High concentrations of Cks protein coupled to beads might drive a low-affinity interaction between Cks and monomeric p34cdc2 and facilitate rebinding of any p34cdc2 molecules that dissociate, thereby allowing detection of the complex. To test this prediction using our reagents, we used CNBr-activated Sepharose beads to which CksHs2 was coupled at a concentration of 5 mg/ml to precipitate 35S-labeled, in vitro-translated p34cdc2-HA. We found that 33% of the input p34cdc2-HA bound to the CksHs2-Sepharose beads, which was much greater than the association of p34cdc2 with control beads (coupled to ovalbumin) or the association of an unrelated protein (luciferase) with the CksHs2-Sepharose beads (Fig. 3A). These results indicate that the presence of a vast excess of Cks protein can drive a low-affinity interaction with monomeric p34cdc2.
FIG. 3.
Binding of Cks proteins to monomeric cdks. (A) Binding of p34cdc2 and cyclin B to CksHs2-Sepharose beads. Binding of in vitro-translated, 35S-labeled p34cdc2 or luciferase (as a control) to either ovalbumin (ova)-Sepharose beads or CksHs2-Sepharose beads was assessed by SDS-PAGE and phosphorimage quantitation. Values represent the means of five experiments with the indicated standard errors. Binding of GST-cyclin B or GST to either ovalbumin-Sepharose beads or CksHs2-Sepharose beads was assessed by immunoblotting with antibody specific for GST and quantitation by phosphorimage analysis. The GST was added at 100-fold higher concentration (3.5 μM) than the GST-cyclin B. Values for GST-cyclin B binding represent the means of three experiments. The standard errors were too small to show on this graph. (B) Binding of CksHs2 to monomeric p33cdk2. A high concentration of immunoprecipitated HA-p33cdk2 was incubated with 11 μM partially purified CksHs2 and then pelleted and washed. Immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting with an anti-Cks antibody. A control for background precipitation of CksHs2 was performed by substituting buffer for p33cdk2 (lane 1).
To further confirm that high protein concentrations can stabilize the weak interaction between CksHs2 and monomeric cdk, we incubated 11 μM (0.1 mg/ml) CksHs2 with a high concentration of immunoprecipitated HA-p33cdk2 and then pelleted the HA-p33cdk2. Coprecipitating CksHs2 was detected by SDS-PAGE followed by immunoblotting with an antibody that recognizes CksHs2. We were able to detect a low level of binding of CksHs2 to monomeric HA-p33cdk2 (∼0.2% of input) in this assay (Fig. 3B).
Phosphorylation of p34cdc2 is critical for the cyclin-stimulated binding of CksHs1 and CksHs2.
In principle, cyclin could stimulate Cks binding to p34cdc2 by a number of mechanisms, including direct interaction with the Cks protein, induction of a conformational change in p34cdc2, or induction of phosphorylation of p34cdc2, or indirectly through production of the mitotic state. We first examined whether CksHs2 could bind directly to cyclin B by attempting to precipitate GST-cyclin B with CksHs2-Sepharose beads in the absence of p34cdc2. A low level of binding of GST-cyclin B to the CksHs2 beads was detected in this assay (Fig. 3A). Although this binding was 18-fold higher than that to control ovalbumin-coupled Sepharose beads, it was relatively inefficient (3.3%) compared to the binding of p34cdc2 (33%) to the same beads, indicating that it reflects a very weak interaction between cyclin and CksHs2. A control protein, GST, bound to CksHs2-Sepharose beads at background levels.
To assess the contribution this interaction may play to the cyclin-stimulated binding that we observed in Fig. 2, we incubated 35S-labeled, in vitro-translated CksHs1 or CksHs2 with GST-cyclin B and precipitated the GST-cyclin B by using glutathione-agarose beads. There was no detectable association of CksHs2 with GST-cyclin B over background with 36 nM cyclin B (Fig. 4, lanes 5 to 8), the standard cyclin concentration for CksHs1 and CksHs2 binding experiments (Fig. 2B). A low level of CksHs1 and CksHs2 binding could be detected when the concentration of GST-cyclin B was raised 10-fold to 360 nM (Fig. 4, lanes 9 to 12). However, even this level of binding of CksHs2 was 30-fold less than the amount of CksHs2 that bound to GST-cyclin B-p34cdc2 in an egg extract containing only 36 nM cyclin B (Fig. 4, lanes 1 to 4). Thus, although CksHs2 appears to bind weakly to cyclin B, this interaction plays at most a minor role in the cyclin-stimulated binding to p34cdc2. The physiological significance of this binding, if any, remains unclear.
FIG. 4.
Binding of CksHs1 and CksHs2 to GST-cyclin B. In vitro-translated, 35S-labeled CksHs1 (lanes 1, 2, 5, 6, 9, and 10) or CksHs2 (lanes 3, 4, 7, 8, 11, and 12) was incubated with (even-numbered lanes) or without (odd-numbered lanes) GST-cyclin B in the presence (lanes 1 to 4) or absence (lanes 5 to 12) of Xenopus egg interphase extract; 36 nM GST-cyclin B was used in lanes 1 to 8, and 360 nM was used in lanes 9 to 12. GST-cyclin B and associated proteins were precipitated with glutathione-agarose beads and analyzed by SDS-PAGE and fluorography.
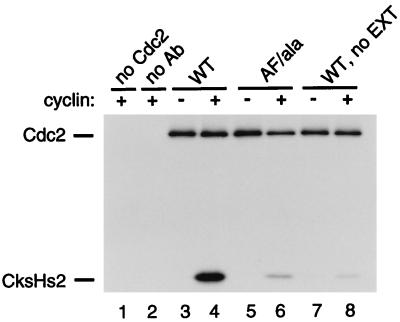
We next examined whether any of the three phosphorylations of p34cdc2 that occur subsequent to cyclin-binding in Xenopus egg extracts (50, 52) affected the cyclin-stimulated binding of CksHs2 to p34cdc2. The levels of cyclin-stimulated binding of 35S-labeled CksHs2 to wild-type p34cdc2-HA and to a nonphosphorylatable mutant form of p34cdc2-HA in a Xenopus egg extract were compared following immunoprecipitation of the epitope-tagged p34cdc2. The nonphosphorylatable p34cdc2, called AF/ala, contained conservative mutations of all three phosphorylation sites: Thr-14 to Ala, Tyr-15 to Phe, and Thr-161 to Ala. Although cyclin strongly stimulated the binding of CksHs2 to wild-type p34cdc2, binding to the nonphosphorylatable mutant was consistently reduced by 80 to 90% (Fig. 5; compare lanes 4 and 6). Because cyclin can bind to the p34cdc2 AF/ala mutant (52), we conclude that the conformational change of p34cdc2 induced by binding to cyclin is insufficient for cyclin-stimulated binding. Additionally, since the addition of cyclin to this extract activates the endogenous p34cdc2 and drives the Xenopus egg extract into mitosis, this experiment also indicates that the simple induction of mitosis does not stimulate CksHs2 binding.
In a complementary experiment, we measured the association of 35S-labeled CksHs2 with 35S-labeled p34cdc2-HA in the absence of Xenopus egg extract (and hence in the absence of any protein kinases acting on p34cdc2). The cyclin-stimulated binding of CksHs2 to p34cdc2 in the absence of extract was only 5 to 10% of the level of binding to p34cdc2 in an egg extract (Fig. 5; compare lanes 4 and 8). The comparable binding of CksHs2 to wild-type p34cdc2 in the absence of egg extract and to nonphosphorylatable p34cdc2 in the presence of egg extract supports the conclusion that phosphorylation of p34cdc2 plays a major role in binding to CksHs2.
Phosphorylation of Thr-161 stimulates binding of CksHs2 to p34cdc2.
We used additional p34cdc2 mutants to determine which specific phosphorylations were important for the cyclin-stimulated binding to CksHs2. Coimmunoprecipitation of 35S-labeled CksHs2 with 35S-labeled p34cdc2-HA from a Xenopus egg extract was performed as described above, substituting p34cdc2-HA phosphorylation site mutants for wild-type p34cdc2-HA. Conservative mutation of the inhibitory phosphorylation sites (T14A and Y15F) either individually (data not shown) or in combination (AF) did not affect the cyclin-stimulated binding of CksHs2 to p34cdc2 (Fig. 6; compare lanes 4 and 6). In contrast, mutation of the activating phosphorylation site (T161A) consistently reduced binding by 80 to 90% (Fig. 6; compare lanes 4 and 8), comparable to the reduction in binding of CksHs2 to the triple phosphorylation site mutant of p34cdc2 (AF/ala) (Fig. 6, lane 10). Similar results were obtained with CksHs1 (data not shown).
Since phosphorylation of Thr-161 is necessary for catalytic activity, we tested whether the kinase activity of p34cdc2 was important for CksHs2 binding. We found that CksHs2 bound comparably to wild-type p34cdc2 and to a mutant p34cdc2 (K33R) that is inactive as a protein kinase due to a conservative mutation of a lysine required for catalysis (Fig. 6; compare lanes 4 and 12). Thus, cyclin-stimulated binding of CksHs2 to p34cdc2 depends on the phosphorylation of Thr-161 and/or the active conformation that this phosphorylation causes p34cdc2 to adopt, but not on the catalytic activity of p34cdc2.
We then tested whether activating phosphorylation of p34cdc2 by purified CAK could stimulate binding to CksHs2 in the complete absence of Xenopus egg extract. We first confirmed that purified budding yeast Cak1p could activate GST-cyclin B-p34cdc2-HA. As expected, wild-type p34cdc2 showed significant cyclin- and Cak1p-dependent histone H1 kinase activity, but the T161A activation site mutant was inactive (Fig. 7A). This phosphorylation was accompanied by a 13-fold increase in the binding of added CksHs2 (Fig. 7B; compare lanes 3 and 4), whereas incubation with Cak1p did not affect the binding of CksHs2 to the T161A mutant (Fig. 7B, lanes 5 and 6).
FIG. 7.
In vitro phosphorylation of p34cdc2 by CAK stimulates binding of CksHs2. (A) Histone H1 kinase assays to assess Cak1p activation of p34cdc2. Purified yeast Cak1p was incubated in the presence of cyclin B with in vitro-translated, 35S-labeled wild-type (wt) p34cdc2-HA (lanes 3 and 4) or p34cdc2-HA (T161A) (lanes 5 and 6). p34cdc2-HA was immunoprecipitated, and histone H1 kinase assays were performed on the immunoprecipitates. Controls for background histone H1 kinase activity include substituting unprogrammed reticulocyte lysate for the p34cdc2 translation (lane 1) and substituting buffer for cyclin B (lane 2). The histone H1 kinase activity seen in lane 3 is 6% of that in lane 4 (by phosphorimage quantitation) and is due to a low level of CAK activity in the reticulocyte lysate. (B) Cak1p phosphorylation of p34cdc2 stimulates association with CksHs2. Cak1p was incubated in the presence of cyclin B with in vitro-translated, 35S-labeled wild-type p34cdc2-HA (lanes 2 to 4) or p34cdc2-HA (T161A) (lanes 5 and 6). In vitro-translated, 35S-labeled CksHs2 was then added, and the p34cdc2 and associated CksHs2 were immunoprecipitated and examined by SDS-PAGE and fluorography. Controls include wild-type cyclin B-p34cdc2 and cyclin B-p34cdc2 (T161A) that were not incubated with Cak1p (lanes 3 and 5) and controls for background association of CksHs2 with the protein A-agarose beads, substituting unprogrammed reticulocyte lysate for the p34cdc2 translation (lane 1) or performing the experiment as for lane 4 but precipitating with protein A-agarose beads in the absence of antibody (Ab) (lane 2). (C) Both cyclin binding and Cak1p phosphorylation of p33cdk2 are required to stimulate association with CksHs2. Monomeric HA-p33cdk2 (lanes 3 and 4) or GST-cyclin A-p33cdk2 (lanes 5 and 6) was incubated with GST-Cak1p (lanes 4 and 6) or a buffer control (lanes 3 and 5) and then incubated with in vitro-translated, 35S-labeled CksHs2. The p33cdk2 and associated CksHs2 were immunoprecipitated and analyzed by SDS-PAGE and fluorography. Buffer was substituted for p33cdk2 to control for background immunoprecipitation of CksHs2 (lanes 1 and 2). (D) Cak1p phosphorylation of monomeric p33cdk2 and GST-cyclin A-p33cdk2. Phosphorylation of HA-p33cdk2 (lane 2) or GST-cyclin A-p33cdk2 (lane 3) was performed as for panel C in the presence of [γ-32P]ATP. The incorporation of 32P into p33cdk2 was assessed by SDS-PAGE and autoradiography. A control reaction contained only GST-Cak1p (lane 1).
We wondered whether activating phosphorylation alone (in the absence of cyclin) would be sufficient to stimulate association with CksHs2. We tested this possibility in an assay using p33cdk2 rather than p34cdc2. Although p34cdc2 phosphorylation by CAK is dependent on its association with cyclin B, cyclin-free p33cdk2 can be phosphorylated efficiently by budding yeast Cak1p (Fig. 7D; see also reference 27). Both phosphorylation by Cak1p and binding to cyclin A were required to stimulate association of CksHs2 with p33cdk2 (Fig. 7C, lane 6). Neither phosphorylation nor cyclin binding alone had a significant effect on binding (Fig. 7C, lanes 4 and 5). Thus, although cyclin-stimulated binding of Cks proteins to cdks is largely dependent on the activating phosphorylation of the cdk, it is the phosphorylated, cyclin-bound cdk complex that preferentially associates with Cks proteins.
CksHs2 blocks Cak1p phosphorylation and activation of p34cdc2.
While we have shown that binding of Cks proteins to cdks is stabilized by the presence of cyclin and activating phosphorylation of the cdk, we also wished to determine whether a weaker interaction between CksHs2 and an unphosphorylated cyclin-cdk complex might exist at roughly physiological concentrations of these proteins. We developed an assay for interaction between Cks proteins and cdks free in solution at near physiological concentrations, based on a suggestion by Bourne et al. (7). Using a cocrystal structure of CksHs1 and p33cdk2, they hypothesized that CksHs1 binding to cyclin A-p33cdk2 might sterically block CAK from phosphorylating Thr-160 of p33cdk2. Studying the effect of a Cks protein on cdk activation and phosphorylation by CAK might therefore provide an assay to detect an interaction without the time delay and dilution inherent in immunoprecipitations. Thus, to look for an interaction between Cks proteins and unphosphorylated cyclin-cdk while at the same time testing the prediction of Bourne et al., we incubated complexes of p34cdc2-HA and GST-cyclin B with various concentrations of bacterially expressed and purified CksHs2. Yeast Cak1p was added, and the p34cdc2-HA was subsequently assayed for its histone H1 kinase activity and for its level of phosphorylation by Cak1p (Fig. 8). The histone H1 kinase assays showed that high concentrations of CksHs2 virtually completely inhibited both p34cdc2 activation and its phosphorylation by Cak1p (Fig. 8). Maximal inhibition occurred at supraphysiological concentrations of CksHs2 (Fig. 8, lanes 4 to 6), although partial effects were observed at near physiological concentrations of CksHs2 (Fig. 8, lanes 7 and 8; see reference 41). In contrast, up to 26 μM CksHs2 had no effect on the activity of p34cdc2 that had already been activated by Cak1p (data not shown). We conclude that binding of CksHs2 can interfere with the phosphorylation of p34cdc2 by yeast Cak1p. This solution assay also indicates that CksHs2 is able to interact with a cyclin B-p34cdc2 complex prior to phosphorylation of Thr-161, even though this interaction is too weak to detect by the coimmunoprecipitation assay.
FIG. 8.
CksHs2 blocks the activation and phosphorylation of p34cdc2 by Cak1p. In vitro-translated, 35S-labeled p34cdc2 was incubated with cyclin B and partially purified CksHs2 (lanes 4 to 10). Following incubation with added Cak1p, p34cdc2-HA was immunoprecipitated and assayed for histone H1 kinase activity (top panel). In the bottom panel, in vitro-translated p34cdc2-HA was immunoprecipitated and incubated with GST-cyclin B and partially purified CksHs2 (lanes 4 to 10) prior to phosphorylation by GST-Cak1p in the presence of [γ-32P]ATP. The incorporation of 32P into p34cdc2 was assessed by SDS-PAGE and autoradiography. Control reactions were performed in the absence of p34cdc2 (lane 1) or of Cak1p (lane 2). The final concentrations of CksHs2 after addition of Cak1p were as follows: lane 3, no CksHs2; lane 4, 26 μM; lane 5, 7 μM; lane 6, 1.6 μM; lane 7, 410 nM; lane 8, 100 nM; lane 9, 26 nM; lane 10, 6 nM.
DISCUSSION
Cyclin-stimulated binding of Cks proteins to cdks.
This study was designed to determine whether human Cks proteins bound preferentially to different forms of p34cdc2 or to p34cdc2 at different stages of the cell cycle in Xenopus egg extracts. We observed that binding of CksHs2 was stimulated in the presence of cyclin B. We also observed cyclin-stimulated binding of a Xenopus Cks protein (Xe-p9) to Xenopus p34cdc2 and of human CksHs2 to human p33cdk2, indicating that this effect is conserved between species and between different Cks and cdk proteins. The major determinant of this stimulation was the activating phosphorylation of p34cdc2 on Thr-161. Binding was reduced by ∼90% in the absence of this phosphorylation, either in the absence of Xenopus egg extract, which provides the CAK to phosphorylate this site, or when a nonphosphorylatable T161A mutant of p34cdc2 was used. Furthermore, binding to wild-type p34cdc2 in the absence of Xenopus egg extract was stimulated when p34cdc2 was phosphorylated by purified budding yeast CAK. Activating phosphorylation of monomeric p33cdk2 was not, however, sufficient to stimulate binding of CksHs2; both phosphorylation and the presence of cyclin were required.
At first glance, these results seem to conflict with previous indications that Cks proteins can bind to all forms of p34cdc2, regardless of phosphorylation state or the presence of cyclin. For example, p13suc1-coupled Sepharose beads are a universal reagent for binding monomeric and cyclin-bound, active and inactive forms of p34cdc2 from all eukaryotic cells (1, 5, 10, 11). We believe that the stimulated binding of Cks proteins to p34cdc2 was not appreciated previously because the high concentrations of the Cks proteins that were coupled to the beads would have favored association of even weakly bound proteins and would have masked any differences between low- and high-affinity interactions. Whereas the concentration of a Cks protein coupled to beads is typically at least 100 μM (5, 10, 11), our coimmunoprecipitation experiments used approximately physiological (41) or even subphysiological levels. It seems unlikely that the bead experiments could have distinguished weak from strong binding. In fact, we found that the same monomeric, unphosphorylated p34cdc2 that did not bind detectably to a physiological concentration of CksHs2 nevertheless bound strongly to CksHs2-Sepharose beads (Fig. 3). We also infer from the ability of supraphysiological concentrations of CksHs2 to block phosphorylation of p34cdc2 by Cak1p that CksHs2 is able to interact transiently with unphosphorylated cyclin B-p34cdc2 complexes. (It is unclear whether this inhibition of Cak1p plays any role in vivo.) Thus, the presence of cyclin and of activating phosphorylation of the cdk stabilizes an otherwise weak and transient interaction, thereby permitting isolation of an intact Cks-cdk complex.
A few earlier studies suggested that Cks proteins may exhibit cell cycle-specific association with p34cdc2, observations that might have been due to the effect that we report here. Using synchronized fission yeast cells, Booher et al. (5) observed that the peak in p34cdc2 binding to p13suc1-Sepharose beads paralleled the peak in the levels of the p63cdc13 cyclin and the peak in p34cdc2- and p63cdc13-associated histone H1 kinase activity. Nigg et al. (40) detected no p13suc1 precipitation of p34cdc2 from G1 lysates of synchronized chicken cells but found equivalent precipitation of p34cdc2 from lysates of cells in S, G2, or M phase. In both of these cases, binding correlated with increased levels of cyclins, and hence also with activating phosphorylation of p34cdc2, although it is unclear to what extent binding of an endogenous Cks protein may have prevented binding of p34cdc2 to the p13suc1-Sepharose beads.
How does cdk phosphorylation stimulate Cks binding?
Some structural hints as to how activating phosphorylation of a cdk might stimulate binding of a Cks protein come from X-ray diffraction studies of a CksHs1-p33cdk2 complex. In this structure, the T-loop of p33cdk2 (which contains Thr-160) lies at least 7.5 Å from CksHs1 and even further from a phosphate-binding site on CksHs1 (7). However, the p33cdk2 in this structure was unphosphorylated and not bound to a cyclin. Binding to a fragment of cyclin A induces large movements of T-loop residues (26). Subsequent phosphorylation of Thr-160 induces further movement of T-loop residues by up to 7 Å (47). Whether these extensive changes would be sufficient to place the phosphate on Thr-160 in the phosphate-binding site of CksHs1 is unclear, although such an interaction would nicely explain the stimulated binding upon Cdk phosphorylation and cyclin binding. An alternative possibility, proposed by Bourne et al. (7), is that binding to cyclin and Thr-160 phosphorylation may move other T-loop residues into contact with CksHs1. Stabilization of binding following Thr-160 phosphorylation could follow from general contacts between T-loop residues and CksHs1, or possibly from ionic interactions between the phosphate on Thr-160 and a positively charged surface on CksHs1 formed by Lys-24, Lys-30, and Lys-34 (7). Finally, it is possible that Thr-160 phosphorylation of cyclin-bound p33cdk2 communicates a more subtle conformational change to the CksHs1-p33cdk2 interface to stabilize the interaction.
Implications for Cks protein function.
The preferential association of CksHs2 with fully active, cyclin-bound, Thr-161 phosphorylated p34cdc2 is consistent with current models that suggest a p34cdc2 targeting function for Cks proteins (7, 41). Bourne et al. (7) observed that the surface of CksHs1 that faces the active site cleft of p33cdk2 may extend the substrate recognition surface of p33cdk2. The putative phosphate-binding site of CksHs1, located on this surface, may guide the protein kinase to specific phosphoproteins. Thus, a Cks protein could serve to stimulate the phosphorylation of certain substrates or to direct the subcellular localization of an active cdk. Based on biochemical experiments in Xenopus egg extracts, Patra and Dunphy (41) suggested that Xe-p9 might serve as a docking factor to target p34cdc2 to potential substrates such as cdc25 or the anaphase-promoting complex. Such targeting functions could go a long way toward rationalizing the often conflicting evidence concerning whether Cks proteins are activators or inhibitors of p34cdc2 function.
ACKNOWLEDGMENTS
We thank Philipp Kaldis for providing purified recombinant Cak1p and HA-p33cdk2, Zach Pitluk for providing affinity-purified polyclonal anti-GST antibodies, Jennifer Holmes for providing purified GST protein, Steve Reed for providing CksHs1 and CksHs2 expression clones, Helen Piwnica-Worms for providing the GST-cyclin A expression clone, and Debra Patra and Bill Dunphy for providing the Xe-p9-containing vector from which Xe-p9 was cloned. We also thank Janet Burton, Deborah Enke, Philipp Kaldis, Jonathan Kimmelman, Jeffrey Peterson, and Karen Ross for critical reading of the manuscript and the entire Solomon lab for experimental advice.
This work was supported by MSTP grant GM07205 from NIH, grant GM47830 from NIH, and the Searle Scholars Program/The Chicago Community Trust. M.J.S. is a Leukemia Society of America Scholar.
REFERENCES
- 1.Arion D, Meijer L, Brizuela L, Beach D. cdc2 is a component of the M phase-specific histone H1 kinase: evidence for identity with MPF. Cell. 1988;55:371–378. doi: 10.1016/0092-8674(88)90060-8. [DOI] [PubMed] [Google Scholar]
- 2.Arvai A S, Bourne Y, Hickey M J, Tainer J A. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J Mol Biol. 1995;249:835–842. doi: 10.1006/jmbi.1995.0341. [DOI] [PubMed] [Google Scholar]
- 3.Arvai A S, Bourne Y, Williams D, Reed S I, Tainer J A. Crystallization and preliminary crystallographic study of human CksHs1: a cell cycle regulatory protein. Proteins. 1995;21:70–73. doi: 10.1002/prot.340210109. [DOI] [PubMed] [Google Scholar]
- 4.Basi G, Draetta G. p13suc1 of Schizosaccharomyces pombe regulates two distinct forms of the mitotic cdc2 kinase. Mol Cell Biol. 1995;15:2028–2036. doi: 10.1128/mcb.15.4.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booher R N, Alfa C E, Hyams J S, Beach D H. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 6.Bourne Y, Arvai A S, Bernstein S L, Watson M H, Reed S I, Endicott J E, Noble M E, Johnson L N, Tainer J A. Crystal structure of the cell cycle-regulatory protein suc1 reveals a beta-hinge conformational switch. Proc Natl Acad Sci USA. 1995;92:10232–10236. doi: 10.1073/pnas.92.22.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne Y, Watson M H, Hickey M J, Holmes W, Rocque W, Reed S I, Tainer J A. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 8.Coleman T R, Dunphy W G. Cdc2 regulatory factors. Curr Opin Cell Biol. 1994;6:877–882. doi: 10.1016/0955-0674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 9.Connell-Crowley L, Solomon M J, Wei N, Harper J W. Phosphorylation independent activation of human cyclin dependent kinase 2 by cyclin A in vitro. Mol Biol Cell. 1993;4:79–92. doi: 10.1091/mbc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducommun B, Brambilla P, Draetta G. Mutations at sites involved in Suc1 binding inactivate Cdc2. Mol Cell Biol. 1991;11:6177–6184. doi: 10.1128/mcb.11.12.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunphy W G, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 12.Dunphy W G, Newport J W. Fission yeast p13 blocks mitotic activation and tyrosine dephosphorylation of the Xenopus cdc2 protein kinase. Cell. 1989;58:181–191. doi: 10.1016/0092-8674(89)90414-5. [DOI] [PubMed] [Google Scholar]
- 13.Endicott J A, Noble M E, Garman E F, Brown N, Rasmussen B, Nurse P, Johnson L N. The crystal structure of p13suc1, a p34cdc2-interacting cell cycle control protein. EMBO J. 1995;14:1004–1014. doi: 10.1002/j.1460-2075.1995.tb07081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endicott J A, Nurse P. The cell cycle and suc1: from structure to function? Structure. 1995;3:321–325. doi: 10.1016/s0969-2126(01)00162-9. [DOI] [PubMed] [Google Scholar]
- 15.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 16.Fesquet D, Labbé J C, Derancourt J, Capony J P, Galas S, Girard F, Lorca T, Shuttleworth J, Dorée M, Cavadore J C. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 19.Gautier J, Solomon M J, Booher R N, Bazan J F, Kirschner M W. cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell. 1991;67:197–211. doi: 10.1016/0092-8674(91)90583-k. [DOI] [PubMed] [Google Scholar]
- 20.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 21.Hadwiger J A, Wittenberg C, Mendenhall M D, Reed S I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989;9:2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayles J, Aves S, Nurse P. Suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J. 1986;5:3373–3379. doi: 10.1002/j.1460-2075.1986.tb04653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayles J, Beach D, Durkacz B, Nurse P. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet. 1986;202:291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- 24.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen L H, Luca F C, Ruderman J V, Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 25.Hindley J, Phear G, Stein M, Beach D. suc1+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol Cell Biol. 1987;7:504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 27.Kaldis P, Sutton A, Solomon M J. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 28.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 29.King R W, Jackson P K, Kirschner M W. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 30.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 31.Kumagai A, Dunphy W G. The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell. 1991;64:903–914. doi: 10.1016/0092-8674(91)90315-p. [DOI] [PubMed] [Google Scholar]
- 32.Lahav-Baratz S, Sudakin V, Ruderman J V, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lew D J, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 34.Mäkelä T P, Tassan J-P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associated with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 35.Merle P, Kadenbach B. The subunit composition of mammalian cytochrome c oxidase. Eur J Biochem. 1980;105:499–507. doi: 10.1111/j.1432-1033.1980.tb04525.x. [DOI] [PubMed] [Google Scholar]
- 36.Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 37.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 38.Murray A W, Kirschner M W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 39.Murray A W, Solomon M J, Kirschner M W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 40.Nigg E A, Krek W, Peter M. Vertebrate cdc2 kinase: its regulation by phosphorylation and its mitotic targets. Cold Spring Harbor Symp Quant Biol. 1991;56:539–547. doi: 10.1101/sqb.1991.056.01.061. [DOI] [PubMed] [Google Scholar]
- 41.Patra D, Dunphy W G. Xe-p9, a Xenopus Suc1/Cks homolog, has multiple essential roles in cell cycle control. Genes Dev. 1996;10:1503–1515. doi: 10.1101/gad.10.12.1503. [DOI] [PubMed] [Google Scholar]
- 42.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pines J. Cell cycle: reaching for a role for the Cks proteins. Curr Biol. 1996;6:1399–1402. doi: 10.1016/s0960-9822(96)00741-5. [DOI] [PubMed] [Google Scholar]
- 44.Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon R Y, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson H E, Stueland C S, Thomas J, Russell P, Reed S I. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990;4:1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- 47.Russo A A, Jeffrey P D, Pavletich N P. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 48.Sclafani R A. Cyclin dependent kinase activating kinases. Curr Opin Cell Biol. 1996;8:788–794. doi: 10.1016/s0955-0674(96)80079-2. [DOI] [PubMed] [Google Scholar]
- 49.Solomon M J. The function(s) of CAK, the p34cdc2-activating kinase. Trends Biochem Sci. 1994;19:496–500. doi: 10.1016/0968-0004(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 50.Solomon M J, Glotzer M, Lee T H, Philippe M, Kirschner M W. Cyclin activation of p34cdc2. Cell. 1990;63:1013–1024. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 51.Solomon M J, Harper J W, Shuttleworth J. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sudakin V, Shteinberg M, Ganoth D, Hershko J, Hershko A. Binding of activated cyclosome to p13suc1: use for affinity purification. J Biol Chem. 1997;272:18051–18059. doi: 10.1074/jbc.272.29.18051. [DOI] [PubMed] [Google Scholar]
- 55.Tang Y, Reed S I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 56.Tassan J-P, Jaquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tassan J-P, Schultz S J, Bartek J, Nigg E A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thuret J-Y, Valay J-G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 59.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]