Abstract
We explore the role of histone H1 as a DNA sequence-dependent architectural determinant of chromatin structure and of transcriptional activity in chromatin. The Xenopus laevis oocyte- and somatic-type 5S rRNA genes are differentially transcribed in embryonic chromosomes in vivo depending on the incorporation of somatic histone H1 into chromatin. We establish that this effect can be reconstructed at the level of a single nucleosome. H1 selectively represses oocyte-type 5S rRNA genes by directing the stable positioning of a nucleosome such that transcription factors cannot bind to the gene. This effect does not occur on the somatic-type genes. Histone H1 binds to the 5′ end of the nucleosome core on the somatic 5S rRNA gene, leaving key regulatory elements in the promoter accessible, while histone H1 binds to the 3′ end of the nucleosome core on the oocyte 5S rRNA genes, specifically blocking access to a key promoter element (the C box). TFIIIA can bind to the somatic 5S rRNA gene assembled into a nucleosome in the presence of H1. Because H1 binds with equivalent affinities to nucleosomes containing either gene, we establish that it is the sequence-selective assembly of a specific repressive chromatin structure on the oocyte 5S rRNA genes that accounts for differential transcriptional repression. Thus, general components of chromatin can determine the assembly of specific regulatory nucleoprotein complexes.
Determination of the structure of eukaryotic transcription factors has led to the recognition that several DNA recognition motifs are shared with proteins that are conventionally viewed as having the general function of packaging the vast majority of DNA within the chromosome (61, 71, 105). These similarities occur between the core histones and particular TATA binding protein-associated factors within transcription factor TFIID (3, 9, 118), between linker histones and sequence-specific DNA binding proteins such as hepatocyte nuclear factor 3 (HNF3) (15, 72), and between high-mobility-group HMG1 and DNA binding proteins such as the human sex-determining factor SRY (73, 103, 105). The proteins that package DNA into the chromosome do so by bending or wrapping the double helix. Transcription factors that share structural features with histones or HMG proteins might function as architectural determinants that remodel DNA to facilitate the assembly of higher-order nucleoprotein structures that activate or repress transcription (4, 24, 26, 30, 31, 51, 109). HNF3, which contains a winged-helix motif similar to that found in histone H1, can replace H1 in the chromatin of the mouse serum albumin enhancer (53, 54). HNF3 contributes to the positioning of nucleosomes on this regulatory DNA (14, 81, 111).
The core histones, linker histones, and HMG proteins could also contribute to the assembly of specific regulatory nucleoprotein architectures through structure- and sequence-selective interactions with DNA (87, 109, 112). Core histones may selectively recognize both DNA structure such as curvature (83, 112) and sequence (25, 101). Sequence-specific DNA binding proteins and transcriptional repressors can target the association of nucleosomes with specific DNA sequences (16, 20). The wrapping of DNA around a core histone octamer can stimulate transcription (75, 92) or contribute to the repression of biological function (20, 86, 91). In model systems using the Xenopus borealis 5S rRNA gene, histone H1 and HMG1 can be incorporated into specific nucleosomes and repress transcription (95–97). Histone H1 and HMG1 can also facilitate the binding of transcription factors to DNA (59, 77, 110). HMGI/Y contributes to the assembly of a specific regulatory nucleoprotein complex on the enhancer of the human beta interferon gene (93). These observations illustrate the potential for the structural proteins of the chromosome to be co-opted for specific regulatory functions.
In Xenopus laevis, 5S rRNA gene expression is developmentally regulated (100, 116). The oocyte 5S rRNA genes (20,000 per haploid) are active in growing oocytes and transiently active at the mid-blastula transition (MBT) yet are repressed in somatic cells. In contrast, the somatic 5S rRNA genes (400 per haploid) are active in oocytes, at the MBT, and in somatic cells. These genes share the same transcription factors, TFIIIA, TFIIIB, and TFIIIC (78), yet assemble transcription complexes with differential stabilities (107, 111). Transcription factors such as TFIIIA are abundant in growing oocytes yet are limiting for transcription in eggs and embryos (2, 111). This limitation in transcription factor abundance coupled to the relative instability of the oocyte 5S rRNA gene transcription complex contributes to the inactivation of the oocyte 5S rRNA gene during embryogenesis (reviewed in references 106 and 111). However, even if transcription factors are maintained at high concentrations in vivo, other mechanisms can direct the silencing of the oocyte 5S rRNA genes (2, 8). A second contributory factor to differential 5S rRNA gene expression is an alteration in chromatin structure as Xenopus embryogenesis proceeds. Histone H1 functions as a specific repressor for the oocyte 5S rRNA genes (2, 45). Removal of histone H1 from the chromosomal chromatin of somatic cells selectively allows transcription factor access to the oocyte 5S rRNA genes (76). Reconstitution of histone H1 into chromosomal chromatin in vitro reestablishes a state of repression for the oocyte 5S rRNA genes (76). Moreover, histone H1 can direct the dominant and specific repression of the oocyte 5S rRNA genes in the presence of H1-deficient nucleosomal arrays and the transcriptional machinery (12, 108). These results have direct physiological relevance since in X. laevis during embryogenesis there is a transition from a cleavage (B4) to an adult (H1) linker histone that occurs prior to gastrulation and that is partially complete at the MBT (reviewed in reference 8). This transition from histone B4 to histone H1 can be experimentally manipulated to drive the selective repression of the oocyte 5S rRNA genes even in the continued presence of excess transcription factors (8, 45). However, the molecular mechanisms by which histone H1 acts as a highly selective transcriptional repressor have not yet been determined.
The nature of linker histone association with nucleosomal DNA is potentially variable (17). One model for the interaction of the central winged-helix domain of the linker histone (15, 72) in the nucleosome has this domain interacting with DNA like a transcription factor through contacts made in the major groove inside the gyres of DNA (68, 69). This type of interaction could explain the sequence-selective interactions of linker histones with nucleosomal DNA and their capacity to influence the positioning of nucleosomes with respect to DNA sequence (55). Low-resolution nucleosome mapping suggests that histone H1 selectively contributes to the organization of nucleosomes over the X. laevis oocyte 5S rRNA genes (12, 94). This selectivity could manifest itself in two ways: either histone H1 might much prefer to interact with nucleosomal DNA containing the oocyte 5S rRNA genes, or histone H1 might have a much more dramatic organizational role on the oocyte 5S rRNA genes compared to the somatic 5S rRNA genes. In both respects, histone H1 is known to constrain nucleosome positioning much more effectively than histone B4 and to interact with nucleosomal DNA much more tightly than histone B4 (96). In this study, we first establish that histone H1 interacts with nucleosomes assembled on oocyte and somatic 5S rRNA genes with equivalent affinities. We then examine the capacity of histone H1 to direct nucleosome positioning on X. laevis oocyte and somatic 5S rRNA genes by using a high-resolution micrococcal nuclease mapping method. We find that histone H1 positions a nucleosome so as to occlude the oocyte 5S rRNA gene from the transcriptional machinery, whereas on the somatic 5S rRNA genes nucleosome positioning occurs so as to leave the essential promoter elements accessible and the genes transcriptionally competent. Thus, histone H1 can determine differential transcriptional activity in a nucleosomal context by acting as a chromatin-organizing factor rather than having a differential affinity for oocyte versus somatic 5S rRNA genes.
MATERIALS AND METHODS
Preparation of X. laevis 5S rRNA gene fragments.
The Xlo/270 fragment was prepared from pXlo31 (115) in two steps. First, a 660-bp DNA fragment was isolated from a HindIII digest of the plasmid. Second, this fragment was recut with SfaNI and MaeII to produce the Xlo/270 fragment, followed by purification by 6% nondenaturing polyacrylamide gel electrophoresis (PAGE). Xls/240 was prepared by DdeI digestion of the PCR products of a 383-bp fragment of pXls 11 (115) from −105 to +277. Other oocyte and somatic 5S RNA gene fragments used were amplified from pXlo31 and pXls11, respectively, by PCR (Pfu polymerase; Stratagene). All PCR products were purified by PAGE (6% nondenaturing polyacrylamide gel).
Preparation of histone octamers, linker histones, TFIIIA, and GV extract.
Histone octamers were prepared from chicken erythrocytes by the method of Simon and Felsenfeld (84). Linker histone H1 was prepared from X. laevis erythrocyte nuclei. After separation of crude H1 from the chromatin with a hydroxyapatite column, H1 was purified by using a Bio-Rex 70 column (eluted with 1 M NaCl). TFIIIA was isolated from the ovaries of several young X. laevis frogs as described by Zwieb and Brown (123). These proteins are >95% homologous as judged by sodium dodecyl sulfate (SDS)-PAGE. Germinal vesicle (GV) extract was prepared from X. laevis oocytes by the method of Birkenmeier et al. (6).
Reconstitution of nucleosome cores.
Nucleosome cores were reconstituted onto radiolabeled DNA fragments by a salt dialysis method with purified chicken erythrocyte histone octamers (10). Solutions of 100 μl containing 5 μg of 32P-labeled DNA fragments, 3 μg of histone octamers, and 2.0 M NaCl were dialyzed overnight at 4°C against 10 mM Tris-HCl (pH 7.4)–1 mM EDTA–10 mM 2-mercaptoethanol–0.1 mM phenylmethylsulfonyl fluoride–2.0 M NaCl. The salt concentration was then lowered to 0.1 M by stepwise dialysis: 1.5 M for 4 h, then 1.0 M for 4 h, then 0.75 M for 4 h, and finally 0.1 M for overnight with 0.1 mM EDTA in the final dialysis step. The resulting reconstituted nucleosome cores were purified by 5 to 20% sucrose gradient sedimentation as described by Ura et al. (95).
H1 binding experiments.
Aliquots of 33 ng of reconstituted nucleosome cores of Xlo/200 or Xls/200 and small amounts of naked DNA were incubated with various amounts of linker histone H1 in 10 μl of TFIIIA binding buffer (20 mM Tris-HCl [pH 7.4], 70 mM KCl, 2 mM MgCl2, 10 μM ZnSO4, 1 mM dithiothreitol, 0.1% Nonidet P-40, bovine serum albumin [0.3 mg/ml], 10% glycerol) with 1 μg of poly(dI-dC) · poly(dI-dC) at room temperature for 30 min. The samples were loaded directly onto a 0.7% agarose gel in 0.5× Tris-borate-EDTA (TBE). After electrophoresis in small gels (10 by 10 by 0.5 cm) at 100 V for 3 h, the gel was dried and subjected to autoradiography.
Mapping of positions of nucleosome cores and chromatosomes.
Reconstituted nucleosome cores (50 ng, DNA content) in the absence or presence of 1 ng of linker histone H1 were incubated with 0.019 to 0.15 U of micrococcal nuclease in 50 μl of 10 mM Tris-HCl (pH 8.0)–1 mM CaCl2 for 5 min at room temperature. After addition of 1 μl of 0.5 M EDTA, the DNA fragments were extracted with phenol and labeled with [γ-32P]ATP and T4 polynucleotide kinase. After the incubation at 37°C for 30 min, the resulting samples were immediately loaded onto a 6% nondenaturing polyacrylamide gel. After electrophoresis, the DNA fragments corresponding to nucleosome cores or chromatosomes were recovered and then digested with two kinds of restriction endonucleases to determine the boundaries of micrococcal nuclease cleavage (39). In the mapping of mutant Xlo, Sau96 I was additionally used to determine the chromatosome position because no digestion product by EaeI was observed.
Competition experiments for TFIIIA binding to nucleosome cores in the presence of H1.
After the reconstitution of nucleosome cores (33 ng, DNA content) containing either Xlo/200 or Xls/200 5S DNA, these nucleosome cores were mixed with small amounts of the naked DNA and incubated with H1 (41 ng) at room temperature for 30 min in 10 μl of the TFIIIA binding buffer with 1 μg of poly(dI-dC) · poly(dI-dC) to form a complete 1:1 complex of the nucleosome core and H1. Various amounts of TFIIIA were then added to the samples, which were incubated at room temperature for additional 30 min and then loaded directly onto a 0.7% large agarose gel (20 by 25 by 0.5 cm) in 0.5× Tris-borate (TB). After electrophoresis at 30 mM for 3.5 h, the gel was dried and subjected to autoradiography.
Immunoblotting of H1 complexes.
After the binding experiments were carried out as described above, using unradiolabeled Xls/200 cores, the agarose gels containing nucleosome core complexes were stained with ethidium bromide (0.5 μg/ml in 0.5× TB) for 15 min, immediately photographed under UV illumination, and soaked in 0.01% SDS–0.5× TB buffer for 20 min. The gel was then electroblotted onto a polyvinylidene difluoride membrane at 250 mA for 30 min with a semidry transfer unit, probed with anti-H1 antiserum, and visualized by standard methods. For the chemiluminescence detection, ECL Plus (Amersham) was used.
DNase I footprinting.
Reconstituted nucleosome cores (0.13 μg, DNA content) of 3′-end-labeled Xls/240 were incubated with H1 (0.17 μg) at room temperature for 30 min in 40 μl of the TFIIIA binding buffer with 1 μg of poly(dI-dC) · poly(dI-dC) (it was confirmed by 0.7% agarose gel electrophoresis that under this condition, the nucleosome cores completely formed the 1:1 complex with H1 and that no free nucleosome core and no aggregated complex were observed). Then, 0.4 μg of TFIIIA was added to the samples and the samples were incubated at room temperature. After 30 min, 5 μl of 20 mM MgCl2 and 5 μl of diluted DNase I solution (0.2 U/μl) were added and the samples were incubated at room temperature for 1 min. The reaction was quenched by the addition of 6 μl of a solution of 0.2 M EDTA–50% glycerol, and the samples were immediately loaded onto a 0.7% large agarose gel (20 by 25 by 0.5 cm) in 0.5× TB. After electrophoresis at 4°C and 140 V for 5 h, the wet gel was subjected to autoradiography, the complexes corresponding to the core-TFIIIA complex (Fig. 5B, lanes 7 to 9) were excised, and the DNA fragments were extracted and analyzed by PAGE (8% denaturing polyacrylamide gel). In parallel, the naked DNA, the DNA-TFIIIA complex, the nucleosome core, the core-H1 complex, and the core-TFIIIA complex were digested with DNase I, and the resulting samples were treated as described above. Amounts of DNase I used were 0.05, 0.1, 0.13, 0.17, and 1 U, respectively.
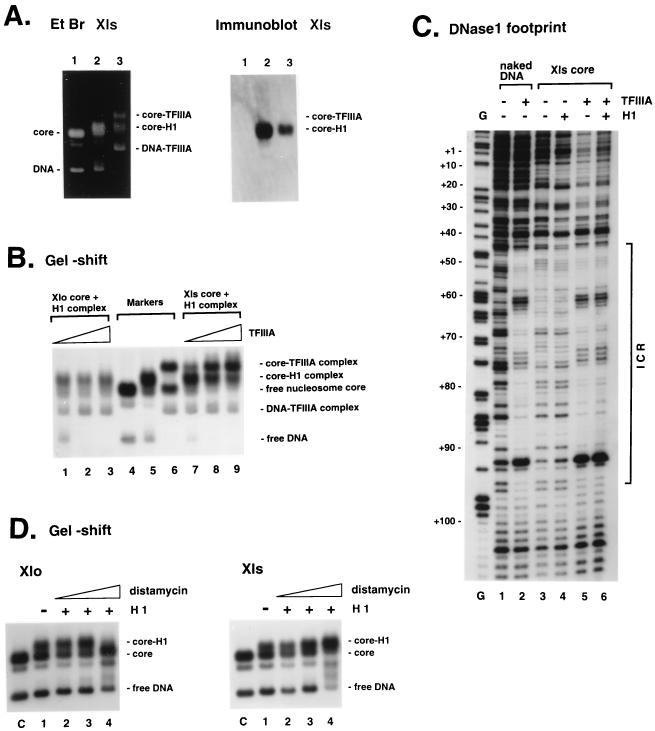
FIG. 5.
(A) Elimination of H1 bound to Xls core by TFIIIA. Nucleosome core-H1 complexes of Xls/200 were mixed with 200 ng of TFIIIA and analyzed by agarose gel electrophoresis (Materials and Methods) before ethidium bromide (Et Br) staining (left) and Western blotting with an anti-H1 antiserum (right). Lanes 1 and 2 show the nucleosome core and core-H1 complex used, respectively; lane 3 shows core complexes formed after incubation of TFIIIA with the core-H1 complex. (B) Specific binding of TFIIIA to the somatic 5S RNA gene incorporated into a nucleosome containing histone H1. Nucleosome core-H1 complexes of Xlo/200 (lanes 1 to 3) or Xls/200 (lanes 7 to 9) were mixed with various amounts of TFIIIA and analyzed by 0.7% agarose gel electrophoresis. After the reconstituted nucleosome core was incubated with linker histone H1 for 30 min at room temperature to form a complete 1:1 complex of the nucleosome core and H1 (see Materials and Methods), TFIIIA was added to the reaction mixture and the mixture was incubated for additional 30 min. Amounts of TFIIIA used were 1 ng (lanes 1 and 7), 10 ng (lanes 2 and 8), and 100 ng (lanes 3 and 9). Each reaction mixture also contained small amounts of the naked DNA as internal control for TFIIIA binding. Positions of the nucleosome core, nucleosome core-H1 complex, and nucleosome core-TFIIIA complex are shown as markers in lanes 4, 5, and 6, respectively. (C) DNase I digestion of the TFIIIA-nucleosome core complexes assembled on the somatic 5S RNA gene. Autoradiographs show digestion patterns of the coding strand: naked DNA (lane 1), DNA-TFIIIA complex (lane 2), nucleosome core (lane 3), core-H1 complex (lane 4), and core-TFIIIA complex (lane 5). Lane 6 shows the DNase I digestion pattern of the complex observed in panel B, lanes 7 to 9, corresponding to the core-TFIIIA complex. Conditions for binding of TFIIIA and H1 and subsequent DNase I digestion are described in Materials and Methods. Lane G shows positions of guanines in the sequence, cut by dimethylsulfate-piperidine. Numbers on the left correspond to positions in the sequence of the somatic 5S RNA gene. The location of the internal control region (ICR) is shown on the right. (D) Preferential inhibition of H1 binding to Xlo core by distamycin. After incubation of Xlo/200 (left) or Xls/200 (right) with distamycin (lane 1, 0 M; lane 2, 8.6 × 10−5 M; lane 3, 1.7 × 10−4 M; lane 4, 3.4 × 10−4 M), binding abilities of H1 to the cores were analyzed in a 0.7% nondenaturing agarose gel. Conditions for binding of distamycin and H1 are described in Materials and Methods.
Inhibition of H1 binding to nucleosome cores by distamycin.
After reconstitution, nucleosome cores (33 ng, DNA content) containing either Xlo/200 or Xls/200 5S DNA, and small amounts of the naked DNA, were incubated with various amounts of distamycin A (Sigma) in 10 μl of TFIIIA binding buffer with 1 μg of poly(dI-dC) · poly(dI-dC); H1 (8 ng) was added to the samples, which were incubated at room temperature for an additional 30 min and then loaded directly onto a 0.7% large agarose gel. After electrophoresis, the gel was dried and subjected to autoradiography.
Transcription reactions.
First, reconstituted nucleosome cores purified by 5 to 20% sucrose gradient sedimentation (95) such that no free DNA contaminated the sample (40 ng, DNA content) of Xlo/270 or Xls/265 were incubated with or without 11.3 ng of linker histone H1 in 10 mM HEPES (pH 7.4)–50 mM KCl–7 mM MgCl2–2.5 mM dithiothreitol–2.5 U of RNasin–0.1 mM EDTA–1 μg of poly(dI-dC) · poly(dI-dC) for 30 min at room temperature. It was confirmed by 0.7% agarose gel electrophoresis that the nucleosome cores completely formed the 1:1 complex with H1 under this condition. No free nucleosome core and no aggregated complex were observed. Then transcription reactions using Xenopus GV extract were carried out as described by Ura et al. (95) except that labeling reactions were continued for 1 h after addition of exogenous nucleotides. In the experiment shown in Fig. 2B, mixtures of nucleosome cores of Xlo/270 and Xls/270 (40 ng of each) were used as templates for transcription. The transcripts were analyzed in a semidenaturing gel (15% polyacrylamide, 4 M urea, 1× TBE) to quantitate the oocyte and somatic 5S RNA products (62).
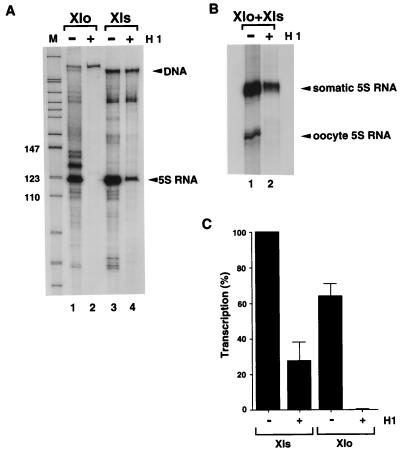
FIG. 2.
Specific repression of the oocyte 5S RNA gene transcription by linker histone H1. For conditions for binding of H1 and subsequent transcription by Xenopus GV extract, see Materials and Methods. (A) Transcription using Xlo/270 core alone (lane 1), Xlo/270 core-H1 complex (lane 2), Xls/265 core alone (lane 3), or Xls/265 core-H1 complex (lane 4) as the template for transcription. The transcripts were analyzed by PAGE (8% denaturing polyacrylamide gel). Sizes are indicated in nucleotides. (B) Transcription using mixtures of Xlo/270 and Xls/270 nucleosome cores in the absence (lane 1) or presence (lane 2) of H1. The oocyte and somatic 5S RNA products were analyzed by semidenaturing PAGE.
RESULTS
Histone H1 binds to nucleosomes containing the X. laevis oocyte and somatic 5S rRNA genes with equivalent affinities yet directs differential transcriptional repression.
The 5S rRNA genes have been useful for the reconstitution of specific chromatin structures in vitro. Nucleosomes assembled by using 5S rRNA genes from Lytichinus variegatus (19, 55, 88, 89) and the somatic-type 5S DNA of X. borealis (37, 68, 95) have been extensively studied. X. borealis 5S rRNA genes have major differences in sequence and organization compared to X. laevis 5S rRNA genes (11, 22, 49, 65). The X. laevis oocyte and somatic 5S rRNA genes have provided a paradigm for investigating the role of chromatin structure in the developmental regulation of differential gene expression (111). The oocyte-type 5S rRNA genes of X. laevis are repressed during embryogenesis, while the somatic-type genes remain active (100, 116). Early work by Gottesfeld and Bloomer established that the X. laevis oocyte 5S rRNA genes were assembled into a nonrandomly organized nucleosomal array (27, 29; see also references 12 and 119). This nucleosomal organization was absent when the X. laevis oocyte 5S rRNA genes were active (21). Because the primary sequence-specific transcription factor TFIIIA binds to oocyte-type and somatic-type genes with equivalent affinities (52), it is clear that the assembly of higher-order nucleoprotein complexes must contribute to gene regulation (47, 106, 107). Histone H1 was proposed to be a gene-specific repressor for the oocyte-type 5S rRNA genes based on the functional consequences of the removal or addition of the protein to chromatin isolated from somatic cells or reconstituted on genomic DNA (12, 76, 108). In vivo manipulation of histone H1 levels confirmed this hypothesis (8, 45). However, because structural studies of chromatin focused on the X. borealis 5S rRNA genes with sequences very different from those of the X. laevis genes, the molecular basis for the specific repression by histone H1 of the X. laevis oocyte-type 5S rRNA genes compared to the X. laevis somatic-type genes was not resolved. X. laevis oocyte-type and somatic-type 5S rRNA genes and their flanking sequences also contain significant differences in sequence, especially with respect to AT-rich versus GC-rich content (65). Our first experiments therefore examined the capacity of histone H1 to direct the assembly of specific chromatin structures on X. laevis oocyte-type and somatic-type 5S rRNA genes.
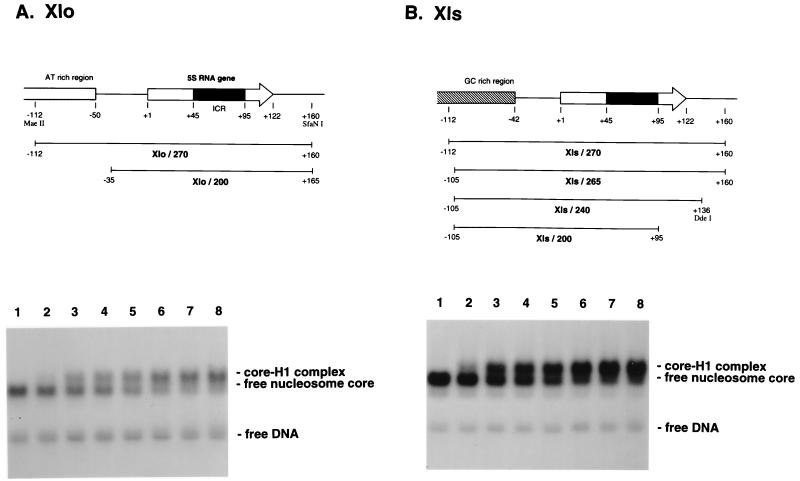
In our experiments, we make use of DNA fragments that are at least 200 bp in length containing oocyte (Xlo) or somatic (Xls) genes (Fig. 1). This is because we wish to provide the histone octamer with or without histone H1 the full opportunity to choose a position that would include the entire nucleosome core (146 bp) plus a linker segment of ∼50 bp. We monitored the assembly of nucleosomes onto DNA fragments (270 bp in length) by using gel retardation assays (39). Titration of Xenopus histone H1 into these nucleosomes revealed that the linker histone interacts preferentially with DNA wrapped around the histone octamer compared to naked DNA (Fig. 1) and that equivalent association of H1 with nucleosomes containing either oocyte or somatic 5S rRNA genes occurred. Quantitation of the binding affinities revealed equivalent dissociation constants of ∼10 nM (data not shown; see also reference 57).
FIG. 1.
Upper panels, DNA structures of the X. laevis oocyte (A) and somatic 5S RNA genes (B) used. Arrows show the location and orientation of the 120-bp 5S RNA gene. Black boxes indicate the internal control region (ICR). Numbers represent the positions relative to the start site of transcription, +1. Lower panels, lack of significant differences in the affinities of linker histone H1 toward nucleosome cores of Xlo/200 (A) and Xls/200 (B). Reconstituted nucleosome cores were mixed with various amounts of H1 and analyzed by 0.7% agarose gel electrophoresis. Concentrations of H1 added: 0 M (lane 1), 3.5 nM (lane 2), 8.7 nM (lane 3), 17 nM (lane 4), 35 nM (lane 5), 67 nM (lane 6), 106 nM (lane 7), and 174 nM (lane 8). Small amounts of the naked DNA were included to show no affinity of H1 toward the naked DNA under the experimental conditions.
To establish the regulatory significance of histone H1 incorporation into nucleosomes containing X. laevis oocyte- and somatic-type 5S rRNA genes, we next examined 5S rRNA gene transcription from nucleosomal templates purified on sucrose gradients in the absence and presence of histone H1 (Fig. 2A). The addition of a single molecule of histone H1 per histone octamer assembled on the X. laevis oocyte 5S rRNA gene completely inhibits transcription of the oocyte 5S rRNA gene (Fig. 2A; compare lanes 1 and 2; quantitation is shown in Fig. 2C). In contrast, substantial transcriptional activity from the somatic 5S rRNA gene remains (Fig. 2A; compare lanes 3 and 4; quantitation is shown in Fig. 2C). We repeated this transcription experiment by using an equimolar mixture of oocyte and somatic 5S rRNA genes. The somatic 5S rRNA gene is transcribed approximately fivefold more effectively than the oocyte 5S rRNA gene under these conditions (Fig. 2B, lane 1) (115). Addition of histone H1 again selectively represses oocyte 5S rRNA gene transcription (Fig. 2B, lane 2). We conclude that the addition of histone H1 to a mixture of transcriptionally competent X. laevis oocyte and somatic 5S rRNA genes assembled into nucleosomes recapitulates the selective repression of oocyte 5S rRNA genes dependent on histone H1 accumulation that is seen in vivo (8, 45). Note that in this and subsequent experiments, the stoichiometry of histone H1 addition is always monitored empirically by gel shift and is in excellent agreement with predictions for binding stoichiometry estimated from mass measurements (data not shown). This level of histone H1 has no effect on the transcription of naked DNA templates because it does not bind stably to them (data not shown and Fig. 1). Thus, we establish that our in vitro system gives differential 5S rRNA gene transcription, despite the equivalent binding of H1 to nucleosomes containing oocyte or somatic type 5S DNA (Fig. 1). We next explored the possibility that histone H1 might contribute to the assembly of distinct nucleosomal structures on the oocyte- and somatic-type 5S rRNA genes.
Histone H1 has differential effects on nucleosome positioning on X. laevis oocyte and somatic 5S rRNA genes.
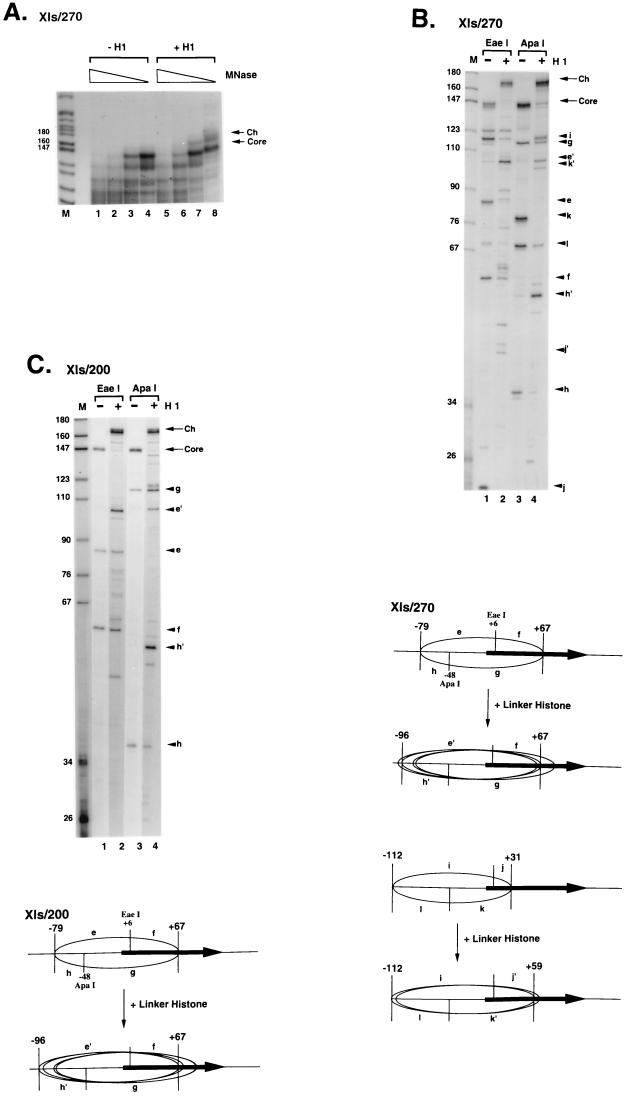
Our next experiments examined the positioning of histone-DNA contacts on X. laevis oocyte and somatic 5S rRNA genes in the presence or absence of histone H1. The core histones H2A, H2B, H3, and H4 assemble an octamer (H2A, H2B, H3, H4)2 that protects about 146 bp of DNA from digestion from micrococcal nuclease (58); incorporation of histone H1 into the nucleosome protects an additional 20 bp from digestion (1, 85). Inclusion of histone H1 into nucleosomes assembled on the X. laevis oocyte 5S rRNA gene leads to the accumulation of a kinetic intermediate (chromatosome stop) on micrococcal nuclease digestion that is about 20 bp longer than the 146 bp protected by the core histones (Fig. 3A; compare lanes 1 to 4 with lanes 5 to 8). This result indicates that histone H1 is stably and appropriately incorporated into nucleosomes containing the oocyte 5S rRNA gene. The accumulation of kinetic intermediates of about 146 and 167 bp in length during micrococcal nuclease digestion (Fig. 3A) allows the boundaries of the nucleosomes to be determined at base pair resolution. This is accomplished by gel isolation of the DNA fragments and end labeling of these fragments with polynucleotide kinase and [γ32P]ATP, followed by restriction endonuclease cleavage and resolution on a denaturing polyacrylamide gel (19, 55). If a single position exists, then two DNA fragments should be obtained whose lengths add up to 146 bp for nucleosome cores and to 166 bp for chromatosomes. For the oocyte 5S rRNA gene reconstituted with a histone octamer by using DNA fragments of either 270 or 200 bp multiple DNA fragments are obtained (Fig. 3B, lanes 1 and 3), demonstrating that many translational positions can be occupied by the histone octamer along the DNA sequence. The addition of one molecule of histone H1 per histone octamer greatly reduces this complexity such that two major DNA fragments are recovered (Fig. 3B, lanes 2 and 4). Other minor positions remain; however, these are significantly reduced compared to the complexity existing before the addition of histone H1. This result indicates that the incorporation of histone H1 into a nucleosome containing the oocyte 5S rRNA gene acts as a major determinant of nucleosome positioning such that a single predominant position is attained. To determine that the boundaries we are analyzing are bona fide, we performed a kinetic analysis of micrococcal nuclease digestion for each construct with or without histone H1. A typical result is shown in Fig. 3C for the Xlo/200 construct: only two stable DNA fragments are obtained following digestion with DdeI (Fig. 3C, labeled a and b) indicating that these are the correct boundaries. Our interpretation of the nucleosome positioning data for the oocyte 5S rRNA gene is shown in Fig. 3D. Although the histone octamer occupies many potential translational positions, surprisingly the inclusion of histone H1 leads to the appearance of a single predominant position from nucleotides (nt) −24 to +148 in which the entire oocyte-type 5S rRNA gene including all known promoter elements is contained within the nucleosome. Our result immediately suggests that the binding of H1 to the nucleosome containing the oocyte 5S rRNA gene may prevent TFIIIA and other components of the RNA polymerase III transcriptional machinery from associating with a chromatin template (see Fig. 5).
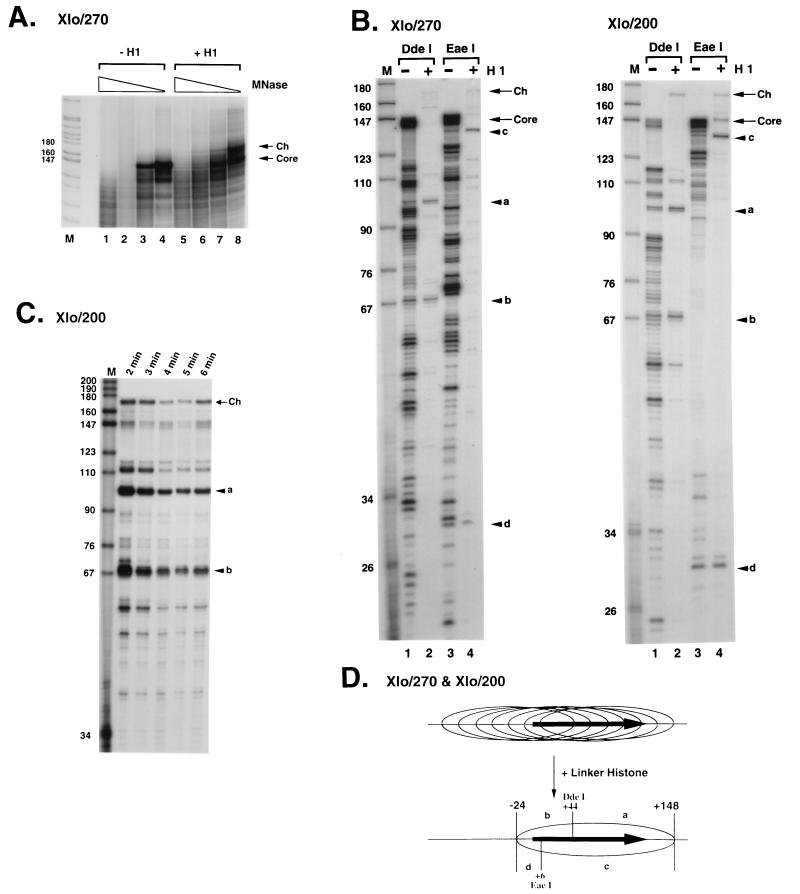
FIG. 3.
Nucleosome positioning on the X. laevis oocyte 5S rRNA gene. (A) Micrococcal nuclease digestion of reconstituted nucleosome cores of Xlo/270. Reconstituted nucleosome cores (50 ng of DNA) in the absence (lanes 1 to 4) or presence (lanes 5 to 8) of 1 ng of linker histone H1 were digested with 0.15, 0.075, 0.038, and 0.019 U of micrococcal nuclease (MNase; 5 min, room temperature). Products of digestion were labeled with [γ-32P]ATP and analyzed by PAGE (6% nondenaturing polyacrylamide gel). Lane M shows fragments of pBR322 digested by MspI. The positions of digestion products corresponding to the nucleosome core DNA fragments and chromatosome DNA fragment are indicated on the right as arrows labeled Core and Ch, respectively. Sizes are indicated in nucleotides. (B) Mapping of nucleosomes cores and chromatosomes by the combination of micrococcal nuclease and restriction endonuclease digestion. DNA from nucleosome cores and chromatosomes protected from micrococcal nuclease digestion (A) was recovered and digested with two kinds of restriction endonucleases to determine DNA regions contacting with histones at the nucleotide level. Although many digestion fragments were observed in the experiment with the Xlo/270 nucleosome core (lane 1 and 3), only one predominant set of digestion products was observed in each digestion using the chromatosome: fragments of 101 nt (a) and 68 nt (b) in DdeI digestion (lane 2) and fragments of 138 nt (c) and 30 nt (d) in EaeI digestion (lane 4). For the Xlo/200 nucleosome core, the mapping experiment yielded the same result as that in panel A. Lane M, MspI digestion products of pBR322 as size markers. Arrows labeled Core and Ch show DNA fragments recovered from nucleosome cores and chromatosomes, respectively. (C) Time independence of micrococcal nuclease digestion pattern of Xlo/200 cores. After micrococcal nuclease digestion, 170-bp DNA fragments corresponding to the chromatosomes were isolated and digested with DdeI to determine the positioning. Incubation times for micrococcal nuclease digestion are shown at the top. (D) Summary of mapping data shown in panel B. Positions of restriction fragments shown in panel B and restriction sites of DdeI and EaeI are indicated. Positions of nucleosome cores and the chromatosome composed of Xlo/270 and Xlo/200 are indicated by ellipsoids. The horizontal closed arrows are the 5S RNA genes.
Inclusion of histone H1 into nucleosomes assembled on the X. laevis somatic 5S rRNA gene again leads to the accumulation of a kinetic intermediate (chromatosome stop) in micrococcal nuclease digestion that is about 20 bp longer than the 146 bp protected by the core histones (Fig. 4A; compare lanes 1 to 4 with lanes 5 to 8). However, mapping of the boundaries of the nucleosomes leads to a result very different from that obtained with the oocyte-type 5S rRNA genes. Various positions of the histone octamer exist on the somatic 5S rRNA gene in the absence of histone H1, depending on DNA length (Fig. 4B and C). For the Xls/270 construct, two predominant positions exist, with the first nucleosome core having a 5′ boundary at −79 and a 3′ boundary at +67 (relative to the start site of transcription at +1; fragments marked e and f in Fig. 4B, lane 1) and the second with a 5′ boundary at −112 and a 3′ boundary at +31 (fragments marked i and j in Fig. 4B, lane 1, and lower panels). For the Xls/200 construct, a single predominant position exists, with a 5′ boundary at −79 and a 3′ boundary at +67 (Fig. 4C, lanes 1 and 3). On the addition of a single molecule of histone H1 per histone octamer, micrococcal nuclease cleavage patterns become more heterogeneous (Fig. 4B and C, lanes 2 and 4). This result suggests that the inclusion of histone H1 into the X. laevis somatic-type 5S rRNA gene destabilizes nucleosome positioning signals that determine the selective association of the core histone octamer with particular sequences. This result is in marked contrast to the properties of the X. laevis oocyte-type gene (Fig. 3) and earlier work with model systems (56, 95–97). In terms of the implications for differential oocyte and somatic 5S rRNA gene transcription, the nucleosomal structures assembled on the two genes incorporate histone H1 with equivalent affinities (Fig. 1) but differ significantly in the consequences of histone H1 incorporation for nucleosome positioning.
FIG. 4.
Nucleosome positioning on the X. laevis somatic 5S rRNA gene. (A) Micrococcal nuclease digestion of reconstituted nucleosome cores of Xls/270. Reconstituted nucleosome cores (50 ng of DNA) in the absence (lanes 1 to 4) or presence (lanes 5 to 8) of 1 ng of linker histone H1 were digested with 0.15, 0.075, 0.038, and 0.019 U of micrococcal nuclease (MNase; 5 min, room temperature). Products of digestion were labeled with [γ-32P]ATP and analyzed by PAGE (6% nondenaturing polyacrylamide gel). Lane M shows fragments of pBR322 digested by MspI. The positions of digestion products corresponding to the nucleosome core DNA fragments and chromatosome DNA fragment are indicated on the right as arrows labeled Core and Ch, respectively. Sizes are indicated in nucleotides. (B and C) Upper panels, mapping of nucleosome cores and chromatosomes by the combination of micrococcal nuclease and restriction endonuclease digestion. DNA from nucleosome cores and chromatosomes protected from micrococcal nuclease digestion (A) was recovered and digested with two kinds of restriction endonucleases to determine DNA regions contacting histones at the nucleotide level. In the mapping of the Xls/270 nucleosome core, two sets of digestion products were observed in each digestion: fragments of 85 nt (e) and 57 nt (f) and fragments of 118 nt (i) and 21 nt (j) in EaeI digestion (lane 1); fragments of 114 nt (g) and 36 nt (h) and fragments of 79 nt (k) and 69 nt (l) in ApaI digestion (lane 3). In the same experiment using the chromatosome, two sets of major digestion products were observed: fragments e′ (104 nt) and f and fragments i and j′ (42 nt) in EaeI digestion (lane 2); fragments g and h′ (53 nt) and fragments k′ (101 nt) and l in ApaI digestion (lane 4). For Xls/200 nucleosome core, only one set of major digestion products was observed in each digestion. Nucleosome core, fragments e and f in EaeI digestion (lane 1) and fragments g and h in ApaI digestion (lane 3); chromatosome, fragments e′ and f in EaeI digestion (lane 2) and fragments g and h′ in ApaI digestion (lane 4); lane M, MspI digestion products of pBR322 as size markers. Arrows labeled Core and Ch show DNA fragments recovered from nucleosome cores and chromatosomes, respectively. Lower panels, summary of mapping data shown in the upper panels. Positions of restriction fragments shown in upper panels and restriction sites of EaeI and ApaI are indicated. In Xls/200, one nucleosome core positioning from −79 to +67 and multiple chromatosomes positioning from −96 to +67 are observed. Ellipses represent the positions of major nucleosome cores and chromatosomes. The horizontal closed arrows are the 5S RNA genes.
In summary, histone H1 binds predominantly at the 5′ end of the nucleosome core on the somatic 5S rRNA gene and at the 3′ end of the core of the oocyte 5S rRNA gene. The binding of histone H1 will specifically block key regulatory elements on the oocyte 5S rRNA gene but not on the somatic gene. Importantly for the somatic 5S rRNA gene, assembly into a nucleosome containing histone H1, in spite of the heterogeneity of position, still leaves key recognition elements for the RNA polymerase III transcriptional machinery exposed, especially the C box (+81 to +91) (66). These elements lie toward the 3′ end of the internal control region, which represents the binding site for transcription factor TFIIIA. TFIIIA is the key specificity factor in the assembly of a transcription complex on a 5S rRNA gene (7, 36, 74). Interaction of TFIIIA with the C box is sufficient to allow the assembly of a specific transcription complex on the 5S rRNA gene (66). Therefore, the positioning of nucleosomes directed by histone H1 on the oocyte and somatic 5S rRNA genes could explain the restriction of transcription by H1 of the oocyte 5S rRNA genes in a nucleosome (Fig. 2) and why the somatic 5S rRNA genes are affected to a much smaller extent. In this way, histone H1 could function as a determinant of differential 5S rRNA gene transcription. Our next experiments tested this hypothesis directly.
A nucleosome containing histone H1 blocks TFIIIA from access to the X. laevis oocyte 5S rRNA gene but not to the somatic 5S rRNA gene.
We and others have investigated the influence of nucleosome assembly on transcription factor access to their cognate sequences (reviewed in references 38 and 60). In general, the incorporation of histone H1 into nucleosomes renders DNA less accessible to the transcriptional machinery (42, 43, 95–97). However, certain transcription factors can bind to positioned nucleosomes even in the presence of histone H1 (114). We wished to explore this issue by using the positioned nucleosomes on the X. laevis oocyte and somatic 5S rRNA genes because in this system the association of these genes with histone H1 has in vivo regulatory significance (8, 45).
A nucleosome containing histone H1 assembled on the somatic 5S rRNA gene does not prevent TFIIIA binding (Fig. 5A, left panel; Fig. 5B, lanes 7 to 9). Immunoblotting of nucleosomes assembled on the somatic 5S rRNA gene in the presence of histone H1 and TFIIIA, using antibodies against histone H1 (Fig. 5A, right panel), shows that the addition of TFIIIA displaces histone H1 from the tertiary complex with the histone octamer. We attribute differences between our result and that of Gottesfeld (28) to our use of substantially longer DNA fragments in nucleosome reconstitution that may promote nucleosome mobility (95). In contrast, we find that the nucleosome assembled on the oocyte 5S rRNA gene prevents the association of TFIIIA (Fig. 5B, lanes 1 to 3). We next tested the specificity of association of TFIIIA with the X. laevis somatic gene by DNase I treatment of the reaction mixture before gel resolution of nucleoprotein complexes, isolation of DNA contained within defined complexes, and resolution of DNA fragments on a sequencing gel (113). TFIIIA binds specifically to the somatic 5S rRNA gene in a nucleosomal context in the presence of H1 (Fig. 5C, lanes 5 and 6). This result indicates that the association of histone H1 in a nucleosome having a 3′ boundary that extends at least 69 bp into the somatic 5S rRNA gene does not significantly impede TFIIIA binding. Importantly, it is clear from the DNase I footprints shown in Fig. 5C that TFIIIA occupies the full internal control region (promoter) of the somatic 5S rRNA gene after binding to the nucleosome on the somatic gene. This suggests that access of TFIIIA to the key binding site within the internal control region (C box [66]) allows TFIIIA access to the entire internal promoter, resulting in a rearrangement of the nucleosome. Note that in this experiment, the Xls/240 DNA fragment is used because only a single nucleosome position can then be reconstituted (−79 to +69). Truncation of the DNA fragment at the 5′ end removes the contribution of the second nucleosome position (−112 to +59) (Fig. 4, Xls/200).
We next examined the determinants of differential H1 association with oocyte and somatic 5S rRNA genes by using the drug distamycin A, which specifically recognizes oligo(dA) · oligo(dT) runs (23, 98). Distamycin interacts with the minor groove of adjacent adenine residues and the thymine residues on the opposite strand (48). We find that distamycin will selectively displace histone H1 from the nucleosome containing the Xlo/200 compared to the Xls/200 DNA fragment (Fig. 5D, left and right panels; compare lanes 1 to 4). This result is consistent with their being a different quality of interaction of histone H1 with the nucleosome containing the oocyte-type gene compared to the somatic-type gene in spite of identical binding affinities (Fig. 1), which would be explained by the differential positioning of nucleosomes on the oocyte and somatic 5S rRNA gene segments in response to the addition of H1 (Fig. 3 and 4). Since the Xlo/200 fragment lacks the major AT-rich segment of oocyte 5S DNA (Fig. 1), our results demonstrate that sequences within 40 bp to either side of the oocyte 5S rRNA gene itself will control nucleosome positioning (based on a gene sequence of 120 bp flanked by two sequences of 40 bp each in the Xlo/200 DNA fragment).
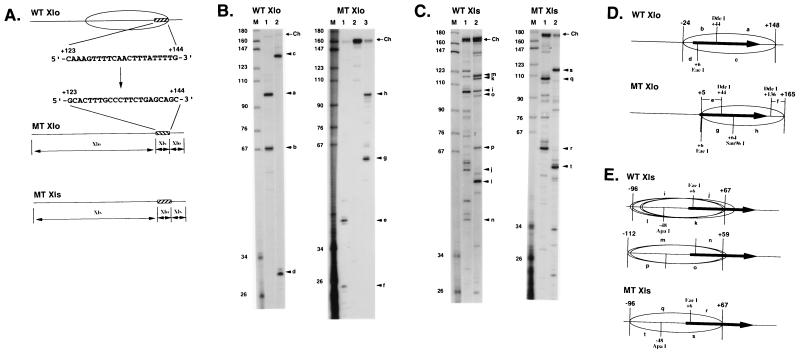
A major sequence determinant of nucleosome positioning on the X. laevis oocyte-type 5S rRNA gene lies between +123 and +144.
Our results with distamycin (Fig. 5D) suggest that oligo(dA) · oligo(dT) tracts contribute to nucleosome positioning on the oocyte-type 5S rRNA genes. The only such tracts in the Xlo/200 DNA fragment lie between +123 and +144 to the 3′ of the gene sequence (Fig. 6A). The positioning of nucleosomes on both the Xlo/200 and Xlo/270 DNA fragments is identical in the presence of histone H1 (Fig. 3). The sequence from +123 to +144 is at the very edge of nucleosomal DNA in a region that we would predict to interact with the globular domain of H1 (17, 68). Replacement of this portion of the Xlo sequence with the comparable sequence from Xls (Fig. 6A) leads to a major change in the translational position of a nucleosome on the DNA fragment (Fig. 6B and D). The nucleosome moves to the 3′ of the DNA fragment (Fig. 6D; compare WT and MT Xlo). This result suggests that the AT-rich sequence from +123 to +144 helps delimit the 3′ boundary of the nucleosome assembled on the oocyte-type gene. We next examined whether this sequence was a dominant determinant of nucleosome position by inserting it 3′ of the somatic-type gene (Fig. 6A, MT Xls). Mapping of nucleosome positioning on wild-type and mutant Xls sequences (Fig. 6C and D) indicates that the tendency of nucleosomes to occupy the 5′ end of the Xls gene remains intact. Thus, the oocyte-type 5S DNA sequence from +123 to +144 is important in the context of the oocyte-type 5S rRNA gene for nucleosome positioning but cannot dominate positioning signals intrinsic to the somatic-type 5S rRNA gene.
FIG. 6.
Mapping of nucleosome positioning on wild-type and mutant X. laevis oocyte- and somatic-type 5S rRNA genes. (A) DNA sequence 3′ to the wild-type oocyte-type 5S rRNA (WT Xlo) gene from +123 to +144 is shown by a hatched box relative to the wild-type chromatosome position (ellipsoid). This is replaced by the sequence 3′ to the somatic-type 5S rRNA (Xls) gene from +123 to +144 to create the MT Xlo DNA sequence. Replacement of the sequence from +123 to +144 3′ to the wild-type somatic gene (WT Xls) with the oocyte sequence generates the MT Xls DNA sequence. (B and C) Mapping of chromatosome boundaries on WT Xlo, MT Xlo, WT Xls, and MT Xls sequences by using DNA fragments 270 bp in length. Procedures for WT Xlo and WT Xls were as for Fig. 3 and 4. The digestion fragments in the MT Xlo chromatosome are completely different from those observed in the WT Xlo chromosome: fragments of 41 nt (e) and 25 nt (t) in DdeI digestion (lane 1); fragments of 60 nt (g) and 98 nt (h) in Sau96I digestion (lane 3). In the mapping, Sau96I (lane 3) was additionally used to determine the chromatosome position because of no digestion by EaeI (lane 2). In the MT Xls chromatosome, the same fragments as those derived from one chromatosome position from −96 to +67 in WT Xls were observed in digestion by EaeI (lane 1) and ApaI (lane 2). (D and E) Chromatosome positions. DNA fragments are indicated as resolved in panels B and C.
DISCUSSION
We have examined the molecular mechanism by which histone H1 regulates differential transcription of the X. laevis oocyte and somatic 5S rRNA genes in a nucleosomal context. The major conclusion from our work is that histone H1 acts to a remarkable degree as an architectural determinant of nucleosome positioning on the oocyte 5S rRNA gene (Fig. 3). While histone octamers do not repress oocyte 5S rRNA gene transcription, the addition of histone H1 to complete the assembly of the nucleosome prevents TFIIIA binding and directs transcriptional repression (Fig. 2 and 5). Prevention of TFIIIA binding is due to the positioning of histone-DNA contacts in the nucleosome containing histone H1 such that all promoter sequences are occluded (Fig. 3). Histone H1 also influences histone-DNA interactions on the somatic 5S rRNA genes (Fig. 4). However, on these genes histone-DNA contacts are directed away from key regulatory elements, and thus both TFIIIA binding to nucleosomal DNA and transcription are retained (Fig. 2 and 5). We conclude that the capacity of nucleosomes to be differentially positioned on the X. laevis oocyte and somatic 5S rRNA genes can account for the differential effect of histone H1 on transcription in vivo (8, 45). The in vitro positioning of nucleosomes that we obtain dependent on histone H1 on the oocyte 5S rRNA genes is consistent with our earlier work on chromatin organization in vivo (12). The consequences of histone H1 addition for the transcription of chromatin templates in vitro recapitulates our earlier work using endogenous chromosomes (12), reconstituted nuclei (108), and developing embryos (8).
Requirements for the assembly of a specific repressive nucleoprotein complex on the oocyte-type 5S RNA gene.
We find that an oligo(dA) · oligo(dT)-rich sequence 3′ of the oocyte 5S rRNA gene is important in determining nucleosomal positioning on these genes in the presence of histone H1 (Fig. 6). A selective role for AT-rich DNA in the incorporation of histone H1 into nucleosomes containing oocyte 5S DNA is suggested by the selective dissociation of H1 from nucleosomes containing these genes in the presence of distamycin (Fig. 5D). This minor groove binding drug (23, 48, 98) has previously been shown to displace H1 from DNA (46). This displacement might be through competition for association with the minor groove (13), or it might be due a straightening of any DNA curve imposed by the two runs of four A · T base pairs separated by a helical turn of DNA (117). The globular domain of histone H1 is very similar in structure to the winged-helical domain of HNF3 (15, 72). Since HNF3 will contribute to nucleosome positioning (14, 53, 54, 81), one possibility is that the globular domain of histone H1 interacts with DNA in a nucleosome in a similar manner to the interaction of HNF3 with DNA (15, 67). HNF3 interacts with DNA via the contacts of a recognition α-helix with the major groove of DNA, bending the double helix toward it. We have proposed that the globular domain of H1 might bind to a nucleosome core inside the gyres of DNA (33, 35, 40, 68, 69). An interaction of the winged-helix domain with the major groove of DNA in the same way as that by HNF3 could explain the sequence selectivity of histone H1 in determining nucleosome position on the oocyte 5S rRNA genes (Fig. 3) (99a). A local effect on nucleosome positioning mediated by histone H1 would explain why higher-order chromatin structures are not required to maintain the oocyte 5S rRNA genes in a repressed state (32). It should be noted that the X. laevis somatic- and oocyte-type 5S rRNA genes differ from each other substantially over the DNA sequence outside the 120-bp-long coding sequence (65). Our mutational analysis of oocyte- and somatic-type 5S rRNA genes suggests that multiple independent sequence features contribute to the final positions of histone-DNA contacts on both genes.
Bradbury and colleagues demonstrated that histone octamers are mobile on long DNA (>200 bp) under conditions of physiological ionic strength (56, 63). Nucleosome mobility has been suggested to have a major role in allowing access of the transcriptional machinery to regulatory DNA in chromatin (5, 95, 99). The histone octamer can occupy many alternate translational positions on oocyte 5S DNA, reflective of mobility along the double helix (Fig. 3); this variation in histone-DNA contacts most probably accounts for the transcriptional competence of this template (Fig. 2). Addition of histone H1 restricts nucleosome position on the oocyte 5S rRNA gene to one that occludes TFIIIA access (Fig. 3 and 5) and represses transcription (Fig. 2). Earlier work had demonstrated that removal of histone H1 was all that was required for transcription factors to activate the oocyte 5S rRNA genes in somatic nuclei (12, 76). The in vitro positioning that we obtain in the presence of histone H1 on the oocyte 5S rRNA genes is consistent with the low-resolution in vivo mapping on nucleosomes on these repeats in the chromosome of somatic cells (12, 29, 119). In our earlier studies, we concluded that the addition of histone H1 directed the positioning of a nucleosome over the oocyte 5S rRNA gene (12). This nucleosome has micrococcal nuclease boundaries very similar to those delineated in the present work (12) (Fig. 1) in which the entire oocyte type 5S rRNA gene is incorporated into a positioned nucleosome. Our results also indicate that nucleosome positioning on the oocyte-type 5S rRNA genes can be determined by DNA sequences within 40 bp of the gene (Fig. 3) and that AT-rich flanking sequences 5′ of the gene (22) are not essential for either nucleosome positioning (Fig. 3) or selective transcriptional repression (Fig. 2). Tomaszewski and Jerzmanowski (94) have suggested that these upstream AT-rich flanking sequences might also have regulatory significance by contributing to nucleosome positioning. This possibility remains to be tested at a functional level. Our data show that the presence of these sequences is not essential in order to assemble a specific repressive chromatin structure dependent on histone H1.
It is important to note that the distribution of nucleosome positions including histone H1 on the somatic 5S rRNA gene (Fig. 4) does not prevent TFIIIA from binding to the internal control region (Fig. 5) or transcription from occurring (Fig. 2). This is because key regulatory elements on the nucleosome that have been defined by using naked DNA within the internal control region remain accessible to TFIIIA (Fig. 4) (34, 36, 66). Even though our experiments employ a single nucleosome, the key binding sites (+81 to +91) for TFIIIA would be predicted to remain accessible within the 40- to 50-bp linker DNA present in the somatic 5S rRNA gene nucleosomal repeat (119). A final point is histone H1 binds selectively to nucleosomal DNA compared to naked DNA (Fig. 1) and binds with equivalent affinities to nucleosomes containing oocyte and somatic 5S rRNA genes. This observation excludes the possibility that differential association of H1 with either gene (41) accounts for the selective regulatory effect.
Histone H1 as a gene-specific repressor.
Linker histones have been proposed to act as general repressors of transcription (102). In vitro reconstitution experiments support this repressive role (18, 44, 50, 82). More recent in vivo experiments suggest that a more specific role exists for linker histones in gene regulation (8, 45, 70, 79, 80, 90). However, the molecular mechanisms by which linker histones might contribute to specific gene regulation have remained obscure. Here we show that histone H1 contributes to the organization of a specific repressive nucleosomal architecture on the oocyte 5S rRNA genes. This can account for the specificity of transcriptional repression observed in vivo (8, 45, 99a). Thus, histone H1 provides a clear example of how a general constituent of chromatin can assume a specific regulatory role dependent on the assembly of a particular nucleoprotein architecture. Other examples of this phenomenon include the assembly of HMG1 (110), histones H3 and H4 (20), and HMGI/Y (93) into specific regulatory complexes. Future experiments will explore the role of histone H1 in the activation and repression of other genes that are developmentally regulated (90, 99a).
REFERENCES
- 1.Allan J, Hartman P G, Crane-Robinson C, Aviles F X. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 2.Andrews M T, Brown D D. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell. 1987;51:445–453. doi: 10.1016/0092-8674(87)90640-4. [DOI] [PubMed] [Google Scholar]
- 3.Arents G, Burlingame R W, Wang B W, Love W E, Moudrianakis E N. The nucleosomal core histone octamer at 3.1Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazett-Jones D P, LeBlanc B, Herfort M, Moss T. Short range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1136. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 5.Becker P B. The establishment of active promoters in chromatin. Bioessays. 1994;16:541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- 6.Birkenmeier E H, Brown D D, Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978;15:1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- 7.Bogenhagen D F, Sakonju S, Brown D D. A control region in the center of the 5S RNA gene directs specific initiation of transcription II. The 3′ border of the region. Cell. 1980;19:27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- 8.Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 9.Burley S K, Xie X, Clark K L, Shu F. Histone-like transcription factors in eukaryotes. Curr Opin Struct Biol. 1997;7:94–102. doi: 10.1016/s0959-440x(97)80012-7. [DOI] [PubMed] [Google Scholar]
- 10.Camerini-Otero R D, Sollner-Webb B, Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976;8:333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- 11.Carroll D, Brown D D. Repeating units of Xenopus laevis oocyte type 5S RNA genes are heterogeneous in length. Cell. 1976;7:467–475. doi: 10.1016/0092-8674(76)90198-7. [DOI] [PubMed] [Google Scholar]
- 12.Chipev C C, Wolffe A P. Chromosomal organization of Xenopus laevis oocyte and somatic 5S rRNA genes in vivo. Mol Cell Biol. 1992;12:45–55. doi: 10.1128/mcb.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churchill M E, Suzuki M. SPKK motifs prefer to bind to DNA at A-T rich sites. EMBO J. 1989;8:4189–4195. doi: 10.1002/j.1460-2075.1989.tb08604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirillo L A, McPherson C E, Bossard P, Stevens K, Cherian S, Shim E Y, Clark K L, Burley S K, Zaret K S. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark K L, Halay E D, Lai E, Burley S K. Co crystal structure of the HNF-3/fork head DNA recognition motif resembles histone H5. Nature. 1993;364:412–417. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 16.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators SSN6 and Tup1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 17.Crane-Robinson C. Where is the globular domain of linker histone located on the nucleosome? Trends Biochem Sci. 1997;22:75–77. doi: 10.1016/s0968-0004(97)01013-x. [DOI] [PubMed] [Google Scholar]
- 18.Croston G E, Kenigan K A, Lira L M, Marshak D R, Kadonaga J T. Sequence specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 19.Dong F, Hansen J C, van Holde K E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 21.Engelke D R, Gottesfeld J M. Chromosomal footprinting of transcriptionally active and inactive oocyte-type 5S RNA genes of Xenopus laevis. Nucleic Acids Res. 1990;18:6031–6037. doi: 10.1093/nar/18.20.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedoroff N V, Brown D D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. I. The AT-rich spacer. Cell. 1978;13:701–716. doi: 10.1016/0092-8674(78)90220-9. [DOI] [PubMed] [Google Scholar]
- 23.Fox K R, Waring M J. DNA structural variations produced by actinomycin and distamycin as revealed by DNase I footprinting. Nucleic Acids Res. 1984;12:9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interaction. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 25.Godde J S, Wolffe A P. Nucleosome assembly of CTG triplet repeats. J Biol Chem. 1996;271:15222–15229. doi: 10.1074/jbc.271.25.15222. [DOI] [PubMed] [Google Scholar]
- 26.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3156. [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesfeld J M. Organization of 5S genes in chromatin of Xenopus laevis. Nucleic Acids Res. 1980;8:905–922. [PMC free article] [PubMed] [Google Scholar]
- 28.Gottesfeld J M. DNA sequence-directed nucleosome reconstitution on 5S RNA genes of Xenopus laevis. Mol Cell Biol. 1987;7:1612–1622. doi: 10.1128/mcb.7.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottesfeld J M, Bloomer L S. Non random alignment of nucleosomes on 5S RNA genes of Xenopus laevis. Cell. 1980;21:751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- 30.Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 31.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 32.Gurdon J B, Dingwall C, Laskey R A, Korn L J. Developmental inactivity of 5S RNA genes persists when chromosomes are cut between genes. Nature. 1982;299:652–653. doi: 10.1038/299652a0. [DOI] [PubMed] [Google Scholar]
- 33.Hayes J J. Site-directed cleavage of DNA by a linker histone-Fe(II) EDTA conjugate: localization of a globular domain binding site within a nucleosome. Biochemistry. 1996;35:11931–11937. doi: 10.1021/bi961590+. [DOI] [PubMed] [Google Scholar]
- 34.Hayes J J, Clemens K R. Location of contacts between individual zinc fingers of Xenopus laevis transcription factor IIIA and the internal control region of a 5S RNA gene. Biochemistry. 1992;31:11600–11605. doi: 10.1021/bi00161a045. [DOI] [PubMed] [Google Scholar]
- 35.Hayes J J, Kaplan R, Ura K, Pruss D, Wolffe A P. A putative DNA binding surface in the globular domain of a linker histone is not essential for specific binding to the nucleosome. J Biol Chem. 1996;271:25817–25822. doi: 10.1074/jbc.271.42.25817. [DOI] [PubMed] [Google Scholar]
- 36.Hayes J J, Tullius T D. Structure of the TFIIIA/5S DNA complex. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 37.Hayes J J, Tullius T D, Wolffe A P. The structure of DNA in a nucleosome. Proc Natl Acad Sci USA. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayes J J, Wolffe A P. The interaction of transcription factors with nucleosomal DNA. Bioessays. 1992;14:597–603. doi: 10.1002/bies.950140905. [DOI] [PubMed] [Google Scholar]
- 39.Hayes J J, Wolffe A P. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc Natl Acad Sci USA. 1993;90:6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayes J J, Pruss D, Wolffe A P. Histone domains required to assemble a chromatosome including the Xenopus borealis somatic 5S rRNA gene. Proc Natl Acad Sci USA. 1994;91:7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jerzmanowski A, Cole R D. Flanking sequences of Xenopus 5S RNA genes determine differential inhibition by H1 histone in vitro. J Biol Chem. 1990;265:10726–10732. [PubMed] [Google Scholar]
- 42.Juan L J, Utley R, Adams C C, Vettese-Dadey M, Workman J L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994;13:6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juan L J, Utley R T, Vignali V, Bohm L, Workman J L. H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J Biol Chem. 1997;272:3635–3640. doi: 10.1074/jbc.272.6.3635. [DOI] [PubMed] [Google Scholar]
- 44.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 45.Kandolf H. The H1A histone variant is the in vivo repressor of oocyte-type 5S gene transcription in Xenopus laevis embryos. Proc Natl Acad Sci USA. 1994;91:7257–7260. doi: 10.1073/pnas.91.15.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kas E, Izaurralde E, Laemmli U K. Specific inhibition of DNA binding to nuclear scaffolds and H1 by distamycin. The role of oligo(dA) · oligo(dT) tracts. J Mol Biol. 1989;210:587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- 47.Keller H J, Romaniuk P J, Gottesfeld J M. Interaction of Xenopus TFIIIC with the TFIIIA.5S RNA gene complex. J Biol Chem. 1992;267:18190–18198. [PubMed] [Google Scholar]
- 48.Kopka M L, Yoon C, Goodsell D, Pjura P, Dickerson R E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci USA. 1985;82:1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korn L J, Brown D D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978;15:1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- 50.Laybourn P J, Kadonaga J T. Role of nucleosome cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 51.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 52.McConkey G A, Bogenhagen D F. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 1988;2:205–214. doi: 10.1101/gad.2.2.205. [DOI] [PubMed] [Google Scholar]
- 53.McPherson C E, Horowitz R, Woodcock C L, Jiang C, Zaret K S. Nucleosome positioning properties of the albumin transcriptional enhancer. Nucleic Acids Res. 1996;24:397–404. doi: 10.1093/nar/24.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McPherson C E, Shim E Y, Friedman D S, Zaret K S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 55.Meersseman G, Pennings S, Bradbury E M. Chromatosome positioning on assembled long chromatin: linker histones affect nucleosome placement on 5S DNA. J Mol Biol. 1991;220:89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- 56.Meersseman G, Pennings S, Bradbury E M. Mobile nucleosomes—a general behavior. EMBO J. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nightingale K P, Pruss D, Wolffe A P. A single high affinity binding site for histone H1 in a nucleosome containing the Xenopus borealis 5S ribosomal RNA gene. J Biol Chem. 1996;271:7090–7094. doi: 10.1074/jbc.271.12.7090. [DOI] [PubMed] [Google Scholar]
- 58.Noll M, Kornberg R D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 59.Onate S E, Prendergast P, Wagner J P, Nissen M, Reeves R, Pettijohn D E, Edwards D P. The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcriptional control. Crit Rev Eukaryot Gene Expr. 1994;4:1–39. [PubMed] [Google Scholar]
- 61.Patikoglou G, Burley S K. Eukaryotic transcription factor-DNA complexes. Annu Rev Biophys Biomol Struct. 1997;26:289–325. doi: 10.1146/annurev.biophys.26.1.289. [DOI] [PubMed] [Google Scholar]
- 62.Peck L J, Millstein L, Eversole-Cire P, Gottesfeld J M, Varshavsky A. Transcriptionally inactive oocyte-type 5S RNA genes of Xenopus laevis are complexed with TFIIIA in vitro. Mol Cell Biol. 1987;7:3503–3510. doi: 10.1128/mcb.7.10.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pennings S, Meersseman G, Bradbury E M. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci USA. 1994;91:10275–10279. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perry M, Thomson G H, Roeder R G. Genomic organization and nucleotide sequence of two distinct histone gene clusters from Xenopus laevis: identification of novel conserved upstream sequence elements. J Mol Biol. 1985;185:479–499. doi: 10.1016/0022-2836(85)90065-8. [DOI] [PubMed] [Google Scholar]
- 65.Peterson R C, Doering J L, Brown D D. Characterization of two Xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980;20:131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- 66.Pieler T, Hamm J, Roeder R G. The 5S internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987;48:91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- 67.Pierrou S, Hellquist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5014. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis E N, Wolffe A P. An asymmetric model for the nucleosome: a binding site for linker histones inside the DNA gyres. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 69.Pruss D, Hayes J J, Wolffe A P. Nucleosomal anatomy—where are the histones? Bioessays. 1995;17:161–170. doi: 10.1002/bies.950170211. [DOI] [PubMed] [Google Scholar]
- 70.Prymakowska-Boasak M, Przewloka M R, Iwkiewicz J, Egierszdorff S, Kuras M, Chaubet N, Gigot C, Spiker S, Jerzmanowski A. Histone H1 overexpressed to high level in tobacco affects certain developmental programs but has limited effect on basal cellular functions. Proc Natl Acad Sci USA. 1996;93:10250–10255. doi: 10.1073/pnas.93.19.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- 72.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–224. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 73.Read C M, Cary P D, Crane-Robinson C, Driscoll P C, Norman D G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21:3427–3437. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakonju S, Bogenhagen D F, Brown D D. A control region in the center of the 5S RNA gene directs specific initiation of transcription 1. The 5′ border of the region. Cell. 1980;19:13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- 75.Schild C, Claret F-X, Wahli W, Wolffe A P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlissel M S, Brown D D. The transcriptional regulation of Xenopus 5S RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984;37:903–911. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- 77.Schultz T F, Spiker S, Quatrano R S. Histone H1 enhances the DNA binding activity of the transcription factor EmBP-1. J Biol Chem. 1996;271:25742–25745. doi: 10.1074/jbc.271.42.25742. [DOI] [PubMed] [Google Scholar]
- 78.Segall J, Matsui T, Roeder R G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–11991. [PubMed] [Google Scholar]
- 79.Seguchi K, Takami Y, Nakayama T. Targeted disruption of 01H1 encoding a particular H1 histone variant causes changes in protein patterns in the DT40 chicken B cell line. J Mol Biol. 1995;254:869–880. doi: 10.1006/jmbi.1995.0662. [DOI] [PubMed] [Google Scholar]
- 80.Shen X, Gorovsky M A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 81.Shim E Y, Woodcock C, Zaret K S. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimamura A, Sapp M, Rodriguez-Canmpos A, Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989;9:5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shrader T E, Crothers D M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simon R H, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simpson R T. Structure of the chromatosome, a chromatin core particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 86.Simpson R T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 87.Simpson R T. Nucleosome positioning: occurrence, mechanisms and functional consequences. Prog Nucleic Acid Res Mol Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 88.Simpson R T, Stafford D W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci USA. 1983;80:51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simpson R T, Thoma F, Brubaker J M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 90.Steinbach O, Wolffe A P, Rupp R. Accumulation of somatic linker histones causes loss of mesodermal competence in Xenopus. Nature. 1997;389:406–412. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 91.Straka C, Horz W. A functional role for nucleosomes in the repression of a yeast promoter. EMBO J. 1991;10:361–368. doi: 10.1002/j.1460-2075.1991.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stunkel W, Kober I, Seifart K H. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol. 1997;17:4397–4405. doi: 10.1128/mcb.17.8.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thanos D, Maniatis T. The high mobility group protein HMGI (Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 94.Tomaszewski R, Jerzmanowski A. The AT-rich flanks of the oocyte-type 5S RNA gene of Xenopus laevis act as a strong local signal for histone H1-mediated chromatin reorganization in vitro. Nucleic Acids Res. 1997;25:458–465. doi: 10.1093/nar/25.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ura K, Hayes J J, Wolffe A P. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 1995;14:3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ura K, Nightingale K, Wolffe A P. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J. 1996;15:4959–4969. [PMC free article] [PubMed] [Google Scholar]
- 97.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription by RNA polymerase, nucleosome mobility and positioning, and linker histone dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Dyke M W, Hertzberg R P, Dervan P B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II) Proc Natl Acad Sci USA. 1982;79:5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Varga-Weisz P D, Blank T A, Becker P B. Energy-dependent chromatin accessibility and nucleosome mobility in a cell free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99a.Vermaak D, Steinbach O C, Dimitrev S, Rupp R A W, Wolffe A P. The globular domain of histone H1 is sufficient to direct specific gene repression in early Xenopus embryos. Curr Biol. 1998;8:533–536. doi: 10.1016/s0960-9822(98)70206-4. [DOI] [PubMed] [Google Scholar]
- 100.Wakefield L, Gurdon J B. Cytoplasmic regulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 1983;2:1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y-H, Griffith J. Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics. 1995;25:570–573. doi: 10.1016/0888-7543(95)80061-p. [DOI] [PubMed] [Google Scholar]
- 102.Weintraub H. Histone H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984;38:17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- 103.Weir H M, Kraulis P J, Hill C S, Raine A R C, Laue E D, Thomas J O. Structure of the HMG box motif in the B domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Werner M H, Huth J R, Gronenborn A M, Clore G M. Molecular basis of human 46X,Y sex reversal revealed from the three dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–717. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 105.Werner M H, Burley S K. Architectural transcription factors: proteins that remodel DNA. Cell. 1997;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- 106.White R. RNA polymerase III transcription. R. G. Austin, Tex: Landes; 1994. [Google Scholar]
- 107.Wolffe A P. Transcription fraction TFIIIC can regulate differential Xenopus 5S RNA gene transcription in vitro. EMBO J. 1988;7:1071–1079. doi: 10.1002/j.1460-2075.1988.tb02915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolffe A P. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 1989;8:527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolffe A P. Architectural transcription factors. Science. 1994;264:1100–1101. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- 110.Wolffe A P. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994;19:240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 111.Wolffe A P, Brown D D. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988;241:1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]
- 112.Wolffe A P, Drew H R. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci USA. 1989;86:9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolffe A P, Hayes J J. The analysis of transcription factor interactions with model nucleosomal templates. Methods Mol Genet. 1993;2:314–330. [Google Scholar]
- 114.Wong J, Li Q, Levi B-Z, Shi Y-B, Wolffe A P. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 1997;16:7130–7145. doi: 10.1093/emboj/16.23.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wormington W M, Bogenhagen D F, Jordan E, Brown D D. A quantitative assay for Xenopus 5S rRNA gene transcription in vitro. Cell. 1981;24:809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]
- 116.Wormington W M, Brown D D. Onset of 5S RNA gene regulation during Xenopus embryogenesis. Dev Biol. 1983;99:248–257. doi: 10.1016/0012-1606(83)90273-7. [DOI] [PubMed] [Google Scholar]
- 117.Wu H M, Crothers D M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 118.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 119.Young D, Carroll D. Regular arrangement of nucleosomes on 5S rRNA genes in Xenopus laevis. Mol Cell Biol. 1983;3:720–730. doi: 10.1128/mcb.3.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi M E. HMG 1 interacts with Hox proteins and enhances their DNA binding and transcription activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 121.Zaret K S. Nucleoprotein architecture of the albumin transcriptional enhancer. Semin Cell Biol. 1995;6:209–218. doi: 10.1006/scel.1995.0029. [DOI] [PubMed] [Google Scholar]
- 122.Zhang X Y, Fitter F, Horz W. Eight different highly specific nucleosome phases on alpha-satellite DNA in the African green monkey. Nucleic Acids Res. 1983;11:4287–4306. doi: 10.1093/nar/11.13.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zwieb C, Brown R S. Absence of substantial bending in Xenopus laevis transcription factor IIIA-DNA complexes. Nucleic Acids Res. 1990;18:583–587. doi: 10.1093/nar/18.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]