Abstract
X-ray mammography is currently considered the golden standard method for breast cancer screening, however, it has limitations in terms of sensitivity and specificity. With the rapid advancements in deep learning techniques, it is possible to customize mammography for each patient, providing more accurate information for risk assessment, prognosis, and treatment planning. This paper aims to study the recent achievements of deep learning-based mammography for breast cancer detection and classification. This review paper highlights the potential of deep learning-assisted X-ray mammography in improving the accuracy of breast cancer screening. While the potential benefits are clear, it is essential to address the challenges associated with implementing this technology in clinical settings. Future research should focus on refining deep learning algorithms, ensuring data privacy, improving model interpretability, and establishing generalizability to successfully integrate deep learning-assisted mammography into routine breast cancer screening programs. It is hoped that the research findings will assist investigators, engineers, and clinicians in developing more effective breast imaging tools that provide accurate diagnosis, sensitivity, and specificity for breast cancer.
Keywords: breast cancer, classification, X-ray mammography, artificial intelligence, machine learning, deep learning, medical imaging, radiology
1. Introduction
Breast cancer is one of the most prevalent cancers among females worldwide (1). Several factors, including gender, age, family history, obesity, and genetic mutations, contribute to the development of breast cancer (2). Early diagnosis with prompt treatment can significantly improve the 5-year survival rate of breast cancer (3). Medical imaging techniques like mammography and ultrasound are widely used for breast cancer detection (4, 5). Mammography utilizes low-dose X-rays to generate breast images that aid radiologists in identifying abnormalities like lumps, calcifications, and distortions (6). Mammography is recommended for women over 40, particularly those with a family history of breast cancer, as it effectively detects early-stage breast cancer (7). However, mammography has limitations, such as reduced sensitivity in women with dense breast tissue. To overcome these limitations, various imaging methods, such as digital breast tomosynthesis (DBT), ultrasound, magnetic resonance imaging (MRI), and positron emission tomography (PET), have been investigated as alternative tools for breast cancer screening.
DBT uses X-rays to generate three-dimensional breast images, which is particularly useful for detecting breast cancer in dense breasts (8). Compared to mammography, DBT provides higher accuracy and sensitivity in detecting breast cancer lesions. However, the interpretation of DBT images still faces inter-observer variability, which can affect its accuracy. Ultrasound imaging uses high-frequency sound waves to produce detailed images of breast tissue. Unlike mammography, ultrasound does not involve radiation, making it a safe method for detecting breast abnormalities, especially in women with dense breast tissue. Ultrasound helps evaluate abnormalities detected on a mammogram and can be used to monitor disease progression and assess treatment effectiveness (9). MRI has been recommended for women with high risks of breast cancer (10). PET utilizes a radioactive tracer to create breast images and is often used in conjunction with other imaging techniques, such as CT or MRI, to identify areas of cancer cells (11). Each of these imaging methods has its own set of advantages and disadvantages (12).
Artificial intelligence (AI) technologies have been extensively investigated to develop cancer prediction models (13, 14). AI-based models, such as machine learning (ML) algorithms, can analyze medical image datasets and patient characteristics to identify breast cancer or predict the risk of developing breast cancer. ML algorithms can extract quantitative features from medical images, such as mammograms or ultrasound images, through radiomics. AI-based prediction models can incorporate various cancer risk factors, including genetics, lifestyle, and environmental factors, to establish personalized imaging and treatment plans. In recent years, deep learning (DL) algorithms have emerged as promising AI tools to enhance the accuracy and efficiency of breast cancer detection (15). These data-driven techniques have the potential to revolutionize breast imaging by leveraging large amounts of data to automatically learn and identify complex patterns associated with malignancy.
This paper provides an overview of the recent developments in DL-based approaches and architectures used in mammography, along with their strengths and limitations. Additionally, the article highlights challenges and opportunities associated with integrating DL-based mammography to enhance breast cancer screening and diagnosis. The remaining sections of the paper are as follows: Section 2 describes the most popular medical imaging application for breast cancer detection. Section 3 discusses DL-based mammography techniques. Section 4 describes breast cancer prediction using DL techniques. Section 5 highlights the challenges and future research directions of DL approaches in mammography. Finally, Section 6 concludes the present study.
2. Medical imaging techniques for breast cancer detection
Medical imaging techniques have become essential in the diagnosis and management of breast cancer. This section provides an overview of several commonly used medical imaging techniques for breast cancer detection. Table 1 compares the most widely utilized medical imaging methods for breast cancer.
Table 1.
Comparison of medical imaging methods for breast cancer.
| Reference | Techniques | Sensitivity | Tumor size corresponding to sensitivity |
Advantages | Disadvantages |
|---|---|---|---|---|---|
| (16) | Mammography | 85% | ≤2 cm | Improved image resolution, widely available | Limited sensitivity in dense breast tissue, exposure to radiation |
| (17) | Ultrasound | 82% | 2 cm | No ionizing radiation, suitable for dense breasts and implant imaging | Operator-dependent, limited specificity |
| (18) | MRI | 95% | ≤2 cm | Images small details of soft tissues | Expensive |
| (19) | Diffused optical tomography |
92.35% | 1 cm | Non-invasive, safe | Illposed problem during reconstruction |
2.1. Mammography
This section presents the working principle, recent advancements, advantages, and disadvantages of mammography. Mammography is a well-established imaging modality used for breast cancer screening. It is a non-invasive technique that utilizes low-dose X-rays to generate high-resolution images of breast tissue. Mammography operates based on the principle of differential X-ray attenuation. The breast tissue is compressed between two plates, and a low-dose X-ray beam is directed through the breast to create an image. Different types of breast tissues, such as fatty, glandular, and cancerous tissue, attenuate X-rays differently. The X-rays that pass through the breast tissue are detected by a digital detector, and an image of the breast is formed. The resulting image is a two-dimensional projection of the breast tissue. In recent years, mammography has undergone significant advancements. Digital mammography has replaced film-screen mammography, leading to improved image quality and reduced radiation dose. Digital breast tomosynthesis (DBT), a 3D mammography technique, has enhanced breast cancer detection rates and reduced false positives. Automated breast ultrasound (ABUS) is another imaging modality used in conjunction with mammography for breast screening, particularly in women with dense breast tissue.
Numerous studies have investigated the effectiveness of mammography for breast cancer screening, demonstrating that it can reduce breast cancer mortality rates, especially for women aged 50-74 years. Additional screening with MRI or ultrasound may be recommended for women with higher risk of breast cancer, such as those with a family history or genetic predisposition. Several leading companies and research groups have achieved significant advancements in the past decade. For example, Hologic’s Genius 3D mammography technology provides higher-resolution 3D images, increasing detection rates while reducing false positives (20). However, it entails higher radiation exposure and higher costs compared to traditional mammography.
Other developments include GE Healthcare and Siemens Healthineers’ contrast-enhanced spectral mammography (CESM), which combines mammography with contrast-enhanced imaging to improve diagnostic accuracy (21). Artificial intelligence tools developed by companies like iCAD and ScreenPoint Medical have been utilized to enhance mammography interpretation, leading to earlier breast cancer detection (22). Gamma Medica and Dilon Technologies have introduced new breast imaging technologies, such as molecular breast imaging and breast-specific gamma imaging, which utilize different types of radiation to provide more detailed images of breast tissue (23).
The University of Chicago has made strides in contrast-enhanced mammography (CEM), which is more accurate in detecting invasive breast cancers than traditional mammography alone. CEM provides detailed images of breast tissue without ionizing radiation, though it is not widely available and may not be covered by insurance (24). The Karolinska Institute’s work on breast tomosynthesis has shown that it is more sensitive in detecting breast cancer than traditional mammography. Tomosynthesis provides a 3D image of the breast, facilitating the detection of small tumors and reducing the need for additional imaging tests. However, it exposes patients to slightly more radiation, takes longer to perform, and is more expensive (25).
Mammography has certain limitations, including limited sensitivity in women with dense breast tissue, false positives leading to unnecessary procedures, radiation exposure that accumulates over time, inability to distinguish between benign and malignant lesions, inaccuracy in detecting small cancers or cancers in certain breast regions, and limited utility in detecting specific types of breast cancer, such as inflammatory breast cancer. To address these limitations, various new imaging technologies, such as DBT, ultrasound elastography, and molecular breast imaging, have been proposed and investigated. These technologies aim to provide more accurate and reliable breast cancer detection, particularly in high-risk individuals. Future research directions for mammography include improving test accuracy, utilizing AI for image interpretation, and developing new techniques utilizing different radiation or contrast agents.
2.2. Digital breast tomosynthesis
DBT was first introduced in the early 2000s. Unlike traditional Mammography, DBT can generate three-dimensional images, leading to more accurate breast cancer detection by reducing tissue overlap. DBT is particularly effective in detecting small tumors and reducing false positive results compared to mammography (26). Additionally, it exposes patients to less radiation. However, DBT is more expensive and may not be covered by insurance for all patients. It also requires specialized equipment and training for interpretation, which may not be widely available in all areas.
2.3. Ultrasound
Ultrasound imaging is a non-invasive, relatively low-cost imaging technique that does not involve exposure to ionizing radiation. It can be used as an adjunct to mammography for breast cancer screening, especially in women with dense breast tissue. Nakano et al. (27) developed real-time virtual sonography (RVS) for breast lesion detection. RVS combines the advantages of ultrasound and MRI and can provide real-time, highly accurate images of breast lesions. However, RVS requires specialized equipment and software, and its diagnostic accuracy may depend on the operator. Standardization of RVS protocols and operator training may improve its accuracy and accessibility.
Zhang et al. (28) conducted a study on a computer-aided diagnosis (CAD) system called BIRADS-SDL for breast cancer detection using ultrasound images. BIRADS-SDL was compared with conventional stacked convolutional auto-encoder (SCAE) and semi-supervised deep learning (SDL) methods using original images as inputs, as well as an SCAE using BIRADS-oriented feature maps (BFMs) as inputs. The experimental results showed that BIRADS-SDL performed the best among the four networks, with classification accuracy of around 92.00 ± 2.38% and 83.90 ± 3.81% on two datasets. These findings suggest that BIRADS-SDL could be a promising method for effective breast ultrasound lesion CAD, particularly with small datasets. CAD systems can enhance the accuracy and efficiency of breast cancer detection while reducing inter-operator variability. However, CAD systems may produce false-positive or false-negative results, and their diagnostic accuracy may depend on the quality of the input images. Integrating CAD systems with other imaging modalities and developing algorithms to account for image quality variations may improve their accuracy and reliability (29).
GE Healthcare (USA) developed the Invenia Automated Breast Ultrasound (ABUS) 2.0, which improves breast cancer detection, especially in women with dense breasts, by providing high-resolution 3D ultrasound images (30). Siemens Healthineers (Germany) developed the ACUSON S2000 Automated Breast Volume Scanner (ABVS), which also provides high-resolution 3D ultrasound images for accurate breast cancer detection, particularly in women with dense breasts (31). These automated systems enhance breast cancer detection rates, improve workflow, and reduce operator variability.
Canon Medical Systems (Japan) developed the Aplio i-series ultrasound system with the iBreast package, which offers high-resolution breast imaging, leading to improved diagnostic performance for breast cancer detection. Invenia ABUS 2.0 and ACUSON S2000 ABVS are automated systems, while Aplio i-series with iBreast package requires manual scanning. The advantages of ABUS 2.0 and ACUSON S2000 ABVS include enhanced image quality, improved workflow, and reduced operator variability. However, they are more expensive than traditional mammography, and image interpretation may be time-consuming. Ultimately, the choice of system depends on the needs and preferences of healthcare providers and patients. Future research is likely to focus on improving the accuracy of ultrasound imaging techniques, developing new methods for detecting small calcifications, and reducing false-positive results.
2.4. Magnetic resonance imaging
MRI utilizes strong magnetic fields and radio waves to generate images of the body’s internal structures, making it one of the most important diagnostic tools. It has various applications, including the diagnosis and monitoring of neurological, musculoskeletal, cardiovascular, and oncological conditions. Its ability to image soft tissues makes it well-suited for breast imaging. Breast MRI is a non-invasive technique used for the detection and monitoring of breast cancer. It is often used in conjunction with mammography and ultrasound to provide a comprehensive evaluation of breast tissue.
Kuhl et al. (32) were the first to investigate post-contrast subtracted images and maximum-intensity projection for breast cancer screening with MRI. This approach offers advantages in terms of speed, cost-effectiveness, and patient accessibility. However, abbreviated MRI has limitations, including lower specificity and the potential for false positives. Mann et al. (33) studied ultrafast dynamic contrast-enhanced MRI for assessing lesion enhancement patterns. The use of new MRI sequences and image reconstruction techniques improved the specificity in distinguishing between malignant and benign lesions. Zhang et al. (34) explored a deep learning-based segmentation technique for breast MRI, which demonstrated accurate and consistent segmentation of breast regions. However, this method has limitations, such as its reliance on training data and potential misclassification.
MRI has several advantages, including the absence of ionizing radiation and increased accuracy in detecting small tumors within dense breast tissue. However, it is expensive, time-consuming, and associated with a higher false-positive rate. Future research directions involve developing faster and more efficient MRI techniques and utilizing AI techniques to enhance image analysis and interpretation.
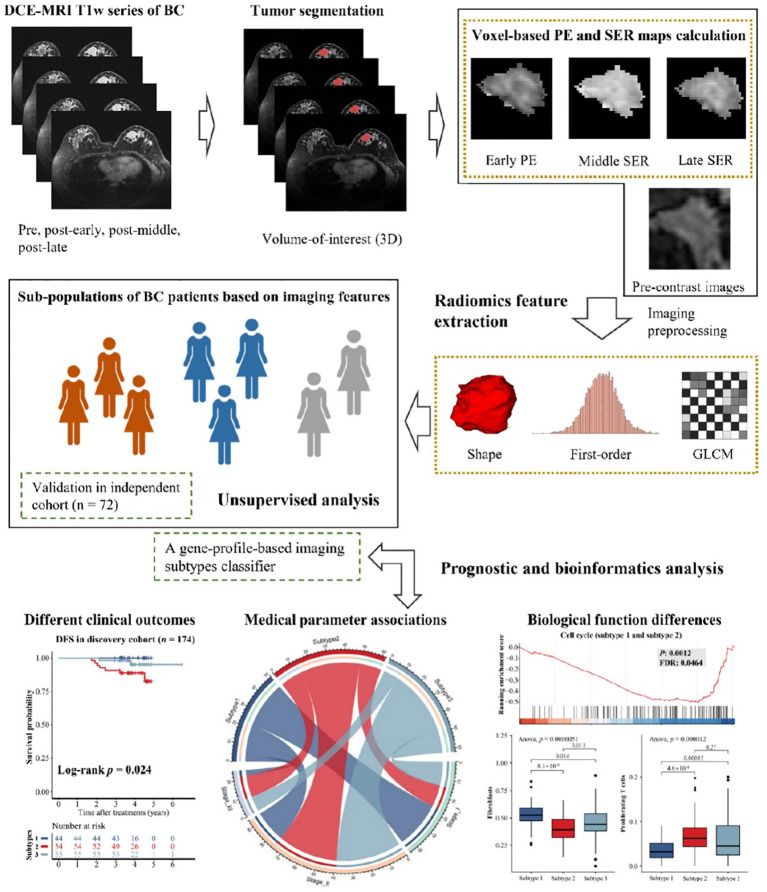
Contrast-enhanced MRI (DCE-MRI) has recently become a crucial method in clinical practice for the detection and evaluation of breast cancer. Figure 1 illustrates the workflow of unsupervised analysis based on DCE-MRI radiomics features in breast cancer patients (35). Ming et al. (35) utilized DCE-MRI to calculate voxel-based percentage enhancement (PE) and signal enhancement ratio (SER) maps of each breast. This study collected two independent radiogenomics cohorts (n = 246) to identify and validate imaging subtypes. The results demonstrated that these imaging subtypes, with distinct clinical and molecular characteristics, were reliable, reproducible, and valuable for non-invasive prediction of the outcome and biological functions of breast cancer.
Figure 1.
Workflow of unsupervised analysis based on DCE-MRI features in breast cancer patients (35).
2.5. Positron emission tomography
PET is an advanced imaging technique that has made significant contributions to the diagnosis and treatment of breast cancer. It is a non-invasive procedure that provides healthcare professionals with valuable information about the spread of cancer to other parts of the body, making it an essential tool in the fight against breast cancer. With ongoing technological advancements, PET plays a crucial role in the detection and treatment of breast cancer.
PET utilizes radiotracers to generate three-dimensional images of the interior of the body. It operates by detecting pairs of gamma rays emitted by the radiotracer as it decays within the body. PET imaging was first introduced in the early1950s, and the first PET scanner was developed in the 1970s. Since then, PET has become an indispensable tool for cancer detection. It has been commonly used to diagnose and stage cancer and assess the effectiveness of cancer treatments. It is also utilized in cardiology, neurology, breast, and psychiatry.
PET is more sensitive than mammography and ultrasound in detecting small breast tumors, and it can also distinguish between benign and malignant lesions with higher accuracy (36). The advantages of PET include its non-invasive and safety for repeated use. However, PET does have limitations, including limited availability, higher cost compared to mammography and ultrasound, a higher rate of false positives, and the requirement for radiotracer injection.
3. Deep learning-based mammography techniques
Several DL architectures, including convolutional neural networks (CNN), transfer learning (TL), ensemble learning (EL), and attention-based methods, have been developed for various applications in mammography. These applications include breast cancer detection, classification, segmentation, image restoration and enhancement, and computer-aided diagnosis (CAD) systems.
CNN is an artificial neural network with impressive results in image recognition tasks. CNN recognizes image patterns using convolutional layers that apply filters to the input image. The filters extract features from the input image, passing through fully connected layers to classify the image. Several CNN-based methods have been proposed in mammography for breast tumor detection. Wang et al. (37) applied CNN with transfer learning in ultrasound for breast cancer classification. The proposed method achieved an area under the curve (AUC) value of 0.9468 with five-folder cross-validation, for which the sensitivity and specificity were 0.886 and 0.876, respectively. Shen et al. (38) proposed a deep CNN in Mammography to classify benign and malignant and achieved an accuracy of 0.88, higher than radiologists (0.83). The study showed that CNN had a lower false-positive rate than radiologists. Yala et al. (39) developed CNN-based mammography to classify mammograms as low or high risk for breast cancer and achieved an AUC of 0.84, which was higher than that of radiologists (0.77). CNN had a lower false-positive rate than radiologists, which has shown promising results in improving the accuracy of mammography screening. CNN has several advantages over traditional mammography screening, including higher accuracy, faster processing, and the ability to identify subtle changes in mammograms. CNN requires large amounts of data to train the network and may not be able to detect all types of breast cancer. Further research is needed to investigate the use of CNN in Mammography.
CNN is an artificial neural network that has shown impressive results in image recognition tasks. It recognizes image patterns using convolutional layers that apply filters to the input image. These filters extract features from the input image, which then pass through fully connected layers to classify the image. In Mammography, several CNN-based methods, such as DenseNet, ResNet, and VGGNet, have been proposed for breast tumor detection. For example, Wang et al. (37) applied CNN with transfer learning in ultrasound for breast cancer classification, achieving an area under the curve (AUC) value of 0.9468 with five-fold cross-validation. The sensitivity and specificity were 0.886 and 0.876, respectively. Shen et al. (38) proposed a deep CNN in mammography to classify between benign and malignant tumors, achieving an accuracy of 0.88, higher than that of radiologists (0.83). Yala et al. (39) developed a CNN-based mammography system to classify mammograms as low or high risk for breast cancer, achieving an AUC of 0.84, higher than that of radiologists (0.77). These studies demonstrated that CNN had a lower false-positive rate than radiologists, showing promise in improving the accuracy of mammography screening. CNN offers advantages over traditional mammography screening, including higher accuracy, faster processing, and the ability to identify subtle changes in mammograms. However, CNN requires large amounts of data to train the network and may not be able to detect all types of breast cancer. Further research is needed to investigate the use of CNN in mammography.
TL utilizes pre-trained DL models to train on small datasets. TL-based methods have shown promising results in improving the accuracy of mammography for breast tumor detection. EL combines multiple DL models to improve the accuracy of predictions. EL-based approaches, such as stacking, boosting, and bagging, have been proposed in mammography for breast tumor detection.
Attention-based methods use attention mechanisms to focus on critical features of the image. Several attention-based methods, such as SE-Net and Channel Attention Networks (CAN), have been proposed for breast tumor detection in mammography. DL is a type of ML that uses neural networks to learn and make predictions. DL methods have gained popularity in recent years due to their ability to work with large datasets and extract meaningful patterns and insights.
DL methods have revolutionized the field of machine learning and are being used in an increasing number of applications, ranging from self-driving cars to medical imaging. As datasets and computing power continue to grow, these methods are expected to become even more powerful and prevalent in the future.
4. Breast cancer prediction using deep learning
This section presents the recent developments in DL methods for breast cancer prediction. The DL-based breast cancer prediction techniques involves the following steps:
Data Collection: Breast datasets are obtained from various sources such as medical institutions, public repositories, and research studies. These datasets consist of mammogram images, gene expression profiles, and clinical data.
Data Preprocessing: The collected datasets are preprocessed to eliminate noise, normalize, and standardize the data. This step involves data cleaning, feature extraction, and data augmentation.
Model Building: DL models, such as CNNs, RNNs, DBNs, and autoencoders, are developed using the preprocessed breast cancer datasets. These models are trained and optimized using training and validation datasets.
Model Evaluation: The trained DL models are assessed using a separate test dataset to determine their performance. Performance metrics, including sensitivity, specificity, accuracy, precision, F1 score, and AUC, are used for evaluation.
Model Interpretation: The interpretability of the DL models is evaluated using techniques such as Grad-CAM, saliency maps, and feature visualization. These techniques help identify which features of the input data are utilized by the DL models for making predictions.
Deployment: The DL model is deployed in a clinical setting to predict breast cancer in patients. The performance of the model is regularly monitored and updated to enhance accuracy and efficiency.
By utilizing DL techniques, breast cancer prediction can be significantly improved, leading to better detection and treatment outcomes.
4.1. Data preprocessing techniques and evaluation
4.1.1. Preprocessing techniques
When applying DL algorithms to analyze breast images, noise can have a negative impact on the accuracy of the image classifier. To address this issue, several image denoising techniques have been developed. These techniques, including the Median filter, Wiener filter, Non-local means filter, Total variation (TV) denoising, Wavelet-based denoising, Gaussian filter, anisotropic diffusion, BM3D denoising, CNN, and autoencoder, aim to reduce image noise while preserving important features and structures that are relevant for breast cancer diagnosis.
After denoising, a normalization method, such as min-max normalization, is typically employed to rescale the images and reduce the complexity of the image datasets before feeding them into the DL model. This normalization process ensures that the model can effectively learn meaningful patterns from the images and improve its ability to accurately classify them.
4.1.2. Performance metrics
Several performance metrics are utilized to evaluate DL algorithms for breast screening. The selection of a specific metric depends on the task at hand and the objectives of the model. Some of the most commonly employed metrics include:
Accuracy: measures the proportion of correct predictions made by the model.
Precision: measures the proportion of true positive predictions out of all positive predictions made by the model.
Sensitivity: measures the proportion of true positive predictions out of all actual positive cases in the dataset.
F1 score: a composite metric that balances precision and sensitivity.
Area under the curve (AUC): distinguishes between positive and negative points across a range of threshold values.
Mean Squared Error (MSE): measures the average squared difference between predicted and actual values in a regression task.
Mean Absolute Error (MAE): measures the average absolute difference between the predicted and actual values in a regression task.
The commonly used equation for calculating accuracy, as stated in reference (40), is:
| (1) |
Where TP and TN are the numbers of true positives and true negatives, FP and FN are the numbers of false positives and false negatives, respectively.
| (2) |
| (3) |
| (4) |
AUC is typically computed by plotting the true positive rate against the false positive rate at different threshold values and then calculating the area under this curve.
| (5) |
| (6) |
Where ytru is the true value and ypred is the predicted value, and n is the number of samples.
Equations 1 – 6 provide a general idea of how performance metrics are computed, but the actual implementation may vary depending on the specific task and the software.
4.2. Datasets
Breast datasets play a crucial role in evaluating DL approaches. These datasets offer a comprehensive collection of high-quality and labelled breast images that can be utilized for training and testing DL algorithms. Table 2 presents commonly utilized publicly available breast datasets in mammography for breast screening.
Table 2.
Breast image dataset.
| Year | Country | Dataset | Sample Number | Human Number | Task |
|---|---|---|---|---|---|
| 1998 (41) | US | Digital Database for Screening Mammography (DDSM) | 2,620 | N/A | Breast cancer detection |
| 1998 (42) | US | Mammographic Image Analysis Society (Mini-MIAS) | 322 | N/A | Breast cancer classification |
| 2012 (43) | US | INbreast | 410 | N/A | Breast cancer detection |
| 2017 (44) | US | Breast Cancer Digital Repository (BCDR) | 1,224 | N/A | Breast cancer classification |
| 2017 (45) | US | Curated Breast Imaging Subset of DDSM (CBIS-DDSM) | 753 | N/A | Breast cancer detection |
| 2011 (46) | US | BCDR-F01 | 362 | N/A | Breast cancer classification |
| 2018 (47) | USA | DDSM | 2,620 | N/A | Classification, segmentation |
| 2016 (48) | Netherlands | Mammographic Image Analysis Society (MIAS) | 322 | N/A | Classification, segmentation |
| 2012 (49) | Multi-country | BCDR | 1,875 | N/A | Classification, density |
N/A, Not Applicate.
4.3. Breast lesion segmentation
The Nottingham Histological Grading (NHG) system is currently the most commonly utilized tool for assessing the aggressiveness of breast cancer (50). According to this system, breast cancer scores are determined based on three significant factors: tubule formation (51), nuclear pleomorphism (52), and mitotic count (53). Tubule formation is an essential assessment factor in the NHG grading system for understanding the level of cancer. Before identifying tubule formation, detection or segmentation tasks need to be performed. Pathologists typically conduct these tasks visually by examining whole slide images (WSIs). Medical image segmentation assists pathologists in focusing on specific regions of interest in WSIs and extracting detailed information for diagnosis. Conventional and AI methods have been applied in medical image segmentation, utilizing handcrafted features such as color, shapes, and texture (54–56). Traditional manual tubule detection and segmentation techniques have been employed in medical images. However, these methods are challenging, prone to errors, exhaustive, and time-consuming (57, 58).
Table 3 provides a comparison of recently developed DL methods in mammography for breast lesion segmentation. These methods include the Conditional Random Field model (CRF) (59), Adversarial Deep Structured Net (60), Deep Learning using You-Only-Look-Once (61), Conditional Residual U-Net (CRU-Net) (62), Mixed-Supervision-Guided (MS-ResCU-Net) and Residual-Aided Classification U-Net Model (ResCU-Net) (63), Dense U-Net with Attention Gates (AGs) (64), Residual Attention U-Net Model (RU-Net) (65), Modified U-Net (66), Mask RCNN (67), Full-Resolution Convolutional Network (FrCN) (68), U-Net (69), Conditional Generative Adversarial Networks (cGAN) (70, 71), DeepLab (72), Attention-Guided Dense-Upsampling Network (AUNet) (73), FPN (74), modified CNN based on U-Net Model (76), deeply supervised U-Net (77), modified U-Net (78), and Tubule-U-Net (79). Among these DL methods, U-Net is the most commonly employed segmentation method.
Table 3.
Deep learning approaches in mammography for breast lesion segmentation.
| Year | Model | Evaluation Dataset | Noise remove method | Performance Metrics (results) |
|---|---|---|---|---|
| 2015 (59) | CRF | INbreast and DDSM-BCRP | NA | The method achieved an 89.0% Dice index in 0.1. |
| 2018 (60) | adversarial deep structured net | INbreast and DDSM-BCRP | NA | The method achieved a segmentation rate of 97.0%. |
| 2018 (61) | deep learning using You-Only-Look-Once | INbreast | NA | The method achieved detection rate of 98.96%, Matthews correlation coefficient (MCC) of 97.62%, and F1 score of 99.24%. |
| 2018 (62) | CRU-Net | INbreast and DDSM-BCRP | NA | The CRU-Net achieved a Dice Index DI of 93.66% for INbreast and a DI of 93.32% for DDSM-BCRP. |
| 2019 (63) | MS-ResCU-Net and ResCU-Net | INbreast | NA | The MS-ResCU-Net achieved an accuracy of 94.16%, sensitivity of 93.11%, specificity of 95.02%, DI of 91.78%, Jac of 85.13%, and MCC of 87.22%, while ResCU-Net correspondingly achieved 92.91%, 91.51%, 94.64%, 90.50%, 83.02%, and 84.99%. |
| 2019 (64) | dense U-Net with AGs | DDSM | NA | The method achieved 82.24% F1 score, 77.89% sensitivity, and overall accuracy of 78.38%. |
| 2019 (65) | RU-Net | DDSM, BCDR-01, and INbreast | cLare filter | The proposed model achieved a mean test pixel accuracy of 98.00%, a mean Dice coefficient index (DI) of 98.00%, and mean IOU of 94.00%. |
| 2019 (66) | modified U-Net | DDSM | Laplacian filter | The method produced 98.50% of the F-measure and a 97.80% Dice score, Jaccard index of 97.40%, and average accuracy of 98.20%. |
| 2020 (67) | mammographic CAD based on pseudocolour mammograms and mask RCNN | INbreast | morphological filters | The DSI achieved for mass segmentation was 0.88Â ± 0.10, and GMs and mask RCNN yielded an average TPR of 0.90Â ± 0.05. |
| 2020 (68) | FrCN | INbreast | NA | FrCN achieved an overall accuracy of 92.97%, 85.93% for MCC, 92.69% for Dice, and 86.37% for the Jaccard similarity coefficient. |
| 2020 (69) | U-Net | CBIS-DDSM, INbreast, UCHCDM, and BCDR-01 | adaptive median filter | The U-Net model achieved a mean Dice coefficient index of 95.10% and a mean IOU of 90.90%. |
| 2020 (70) | cGAN | INbreast | median filter | The cGAN achieved an accuracy of 98.0%, Dice coefficient of 88.0%, and Jaccard index of 78.0%. |
| 2020 (71) | cGAN | DDSM and INbreast | Morphological operations | The proposed cGAN model achieved a Dice coefficient of 94.0% and an intersection over union (IoU) of 87.0% |
| 2020 (72) | mask RCNN and DeepLab | MIAS and DDSM | Savitzky Golay filter | The mask RCNN achieved an AUC of 98.00%, DeepLab achieved an AUC of 95.00%. |
| 2020 (73) | AUNet | CBIS-DDSM and INbreast | NA | produced an average Dice similarity coefficient of 81.80% for CBIS-DDSM and 79.10% for INbreast |
| 2020 (74) | mask RCNN-FPN | training on DDSM and testing on the INbreast database | NA | The model achieved a mean average precision of 84.0% for multidetection and 91.0% segmentation accuracy. |
| 2020 (75) | U-Net | DDSM | NA | The model achieved a sensitivity of 92.32%, specificity of 80.47%, accuracy of 85.95%, Dice coefficient index of 79.39%, and AUC of 86.40%. |
| 2021 (76) | modified CNN based on U-Net model | DDSM-400 and CBIS-DDSM | NA | The method achieved a diagnostic performance of 89.8% and AUC of 86.20% based on ground-truth segmentation maps and a maximum of 88.0% and 86.0% for U-Net-based segmentation for DDSM-400 and CBIS-DDSM, respectively. |
| 2021 (77) | deeply supervised U-Net | DDSM and INbreast | cLare filter | The method achieved 82.70% of Dice, 85.70% of Jaccard coefficient, 99.70% accuracy, 83.10% sensitivity, and 99.80% specificity. |
| 2021 (78) | modified U-Net | MIAS, DDSM, and CBIS-DDSM | NA | The method achieved accuracy of 98.87%, AUC of 98.88%, sensitivity of 98.98%, precision of 98.79%, and F1 score of 97.99% on the DDSM datasets |
| 2023 (79) | Tubule-U-Net | 30820 polygonal annotated tubules in 8225 patches | NA | achieved 95.33%, 93.74%, and 90.02%, dice, sensitivity, and specificity scores, respectively |
N/A, Not Applicate.
Naik et al. (80) developed a likelihood method for the segmentation of lumen, cytoplasm, and nuclei based on a constraint: a lumen area must be surrounded by cytoplasm and a ring of nuclei to form a tubule. Tutac et al. (81) introduced a knowledge-guided semantic indexing technique and symbolic rules for the segmentation of tubules based on lumen and nuclei. Basavanhally et al. (82) developed the O’Callaghan neighborhood method for tubule detection, allowing for the characterization of tubules with multiple attributes. The process was tested on 1226 potential lumen areas from 14 patients and achieved an accuracy of 89% for tubule detection. In reference (83), the authors applied a k-means clustering algorithm to cluster pixels of nuclei and lumens. They employed a level-set method to segment the boundaries of the nuclei surrounding the lumen, achieving an accuracy of 90% for tubule detection. Romo-Bucheli et al. (84) developed a Convolutional Neural Network (CNN) based detection and classification method to improve the accuracy of nuclei detection in tubules, achieving an accuracy of 90% for tubule nuclei detection. Hu et al. (85) proposed a breast mass segmentation technique using a full CNN (FCNN), which showed promising results with high accuracy and speed. Abdelhafiz et al. (86) studied the application of deep CNN for mass segmentation in mammograms and found increased performance in terms of accuracy. Tan et al. (87) recently developed a tubule segmentation method that investigates geometrical patterns and regularity measurements in tubule and non-tubule regions. This method is based on handcrafted features and conventional segmentation techniques, which are not effective and efficient for tubule structures due to their complex, irregular shapes and orientations with weak boundaries.
4.4. Deep learning approaches in mammography for breast lesion detection and classification
DL approaches have garnered considerable attention in mammography for the detection and classification of breast lesions, primarily due to their ability to automatically extract high-level features from medical images. Numerous popular DL algorithms have been employed in mammography for breast screening, including convolutional neural networks (CNN), deep belief networks (DBN), recurrent neural networks (RNN), autoencoders, generative adversarial networks (GAN), capsule networks (CN), convolutional recurrent neural networks (CRNN), attention mechanisms, multiscale CNN, and ensemble learning (EL).
CNN proves highly effective in extracting and classifying image features into distinct categories. DBN is particularly advantageous in identifying subtle changes in images that may be challenging for human observers to discern. RNN utilizes feedback loops to facilitate predictions, thereby aiding in the analysis of sequential data. Autoencoders are utilized for unsupervised feature learning, which aids in the detection and classification of mammography images. GAN is exceptionally effective in generating synthetic mammography images for training DL models. CN is highly proficient in detecting and classifying mammography images. CRNN combines CNN and RNN, making it particularly useful in analyzing sequential data. Attention mechanisms focus on specific areas of mammography images, proving beneficial in detecting and classifying images that encompass intricate structures and patterns. Multiscale CNN analyzes images at multiple scales, proving invaluable in detecting and classifying images with complex structures and patterns at varying scales. EL combines multiple DL models to enhance accuracy and reduce false positives.
Table 4 analyzes the recently developed DL methods for breast lesion detection using mammography. These methods have the potential to greatly enhance the accuracy and efficiency of breast cancer diagnosis. However, it is important to note that most DL methods for biomedical imaging applications come with certain limitations. These limitations include the need for large training datasets, being limited to mass spectrometry images, and being computationally expensive.
Table 4.
DL-based mammography for breast tumor detection.
| Reference | Year | Method | Database | Number of images | Accuracy | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| (88) | 2016 | Deep CNN | DDSM | 600 | 96.7% | NA | NA | NA |
| (89) | 2016 | AlexNet | FFDM | 607 | NA | 86% | NA | NA |
| (90) | 2016 | CNN | BCDR-F03 | 736 | NA | 82% | NA | NA |
| (91) | 2016 | SNN | UCI, DDSM | NA | 89.175%, 86% | NA | NA | NA |
| (92) | 2016 | ML-NN | ED(US) | NA | 98.98% | 98% | NA | NA |
| (93) | 2016 | DBN | ED(US-SW E) | NA | 93.4% | 94.7% | 88.6% | 97.1% |
| (94) | 2017 | Deep CNN | FFDM | 3185 | 82% | 88% | 81% | 72% |
| (95) | 2017 | CNN (COM) | INbreast | 115 | 95% | 91% | NA | NA |
| (96) | 2017 | Deep CNN | SFM, DM | 2242 | NA | 82% | NA | NA |
| (97) | 2017 | CNN-CT | IRMA | 2796 | 83.74% | 83.9% | 79.7% | 85.4% |
| (97) | 2017 | CNN-WT | IRMA | 2796 | 81.83% | 83.9% | 78.2% | 83.3% |
| (98) | 2017 | VGG19 | FFDM | 245 | NA | 86% | NA | NA |
| (99) | 2017 | Custom CNN | FFDM | 560 | NA | 79% | NA | NA |
| (100) | 2017 | VGG16 | IRMA | 2795 | 100% | 100% | NA | NA |
| (101) | 2017 | SNN | DDSM | 480 | 79.5% | NA | NA | NA |
| (101) | 2017 | CNN (COM) | MIAS, CBIS-INBreast | NA | 57% | 77% | NA | NA |
| (96) | 2017 | Multitask DNN | ED(Mg),DD SM | 1057 malignant, 1397 benign | 82% | NA | NA | NA |
| (102) | 2017 | CNN (COM) | ED (HP) | NA | 95.9% (2 classes), 96.4% (15 classes) | NA | NA | NA |
| (103) | 2017 | ImageNet | BreakHis | NA | 93.2% | NA | NA | NA |
| (104) | 2018 | GoogLeNet | BCDR-F03 | 736 | 81% | 88% | NA | NA |
| (104) | 2018 | AlexNet | BCDR-F03 | 736 | 83% | 79% | NA | NA |
| (104) | 2018 | Shallow CNN | BCDR-F03 | 736 | 73% | 82% | NA | NA |
| (105) | 2018 | Faster R-CNN | INbreast | 115 | NA | 95% | NA | NA |
| (105) | 2018 | Faster R-CNN | DREAM | 82,000 | NA | 85% | NA | NA |
| (106) | 2018 | ROI based CNN | DDSM | 600 | 97% | NA | NA | NA |
| (107) | 2018 | Inception V3 | DDSM | 5316 | 97.35% ( ± 0.80) | 98% | NA | NA |
| (107) | 2018 | Inception V3 | INbreast | 200 | 95.50% ( ± 2.00) | 97% | NA | NA |
| (107) | 2018 | Inception V3 | BCDR-F03 | 600 | 96.67% ( ± 0.85) | 96% | NA | NA |
| (107) | 2018 | VGG16 | DDSM | 5316 | 97.12% ( ± 0.30) | NA | NA | NA |
| (107) | 2018 | ResNet50 | DDSM | 5316 | 97.27% ( ± 0.34) | NA | NA | NA |
| (108) | 2018 | Deep CNN | MIAS | 120 | 96.7% | NA | NA | NA |
| (109) | 2018 | AlexNet, Transfer Learning | University of Pittsburgh | 20,000 | NA | 98.82% | NA | NA |
| (110) | 2018 | Faster R-CNN | DDSM, INbreast & Semmelweis University data | 2620,115, 847 | NA | 95% | NA | NA |
| (111) | 2018 | CNN | FFDM | 78 | NA | 81% | NA | NA |
| (112) | 2018 | MV-DNN | BCDR-F03 | 736 | 85.2% | 89.1% | NA | NA |
| (113) | 2018 | Deep CNN | MIAS | 322 | 65% | NA | NA | NA |
| (114) | 2018 | SDAE | ED (HP) | 58 | 98.27% (Benign), 90.54% (Malignant) | NA | 97.92% (Benign), 90.17% (Malignant) | NA |
| (115) | 2018 | CNN (UDM) | BreakHis | NA | 96.15%, 98.33% (2 Classes), 83.31-88.23% (8 Classes) | NA | NA | NA |
| (116) | 2018 | CNN-CH | BreakHis | 400× (× represents magnificati on factor) | 96% | NA | 97.79% | 90.16% |
| (116) | 2018 | CNN-CH | BreakHis | 400× (× represents magnificati on factor) | 97.19% | NA | 98.20% | 94.94% |
| (117) | 2019 | CNN | DDSM | 190 | 93.24% | NA | 91.92% | 91.92% |
| (117) | 2019 | CNN based LBP | DDSM | 190 | 96.32% | 97% | 96.81% | 95.83% |
| (118) | 2020 | InceptionV3 | DDSM | 2620 | 79.6% | NA | 89.1% | NA |
| (118) | 2020 | ResNet 50 | DDSM | 2620 | 85.7% | NA | 87.3% | NA |
| (119) | 2020 | ResNet50 | DDSM patch | 10713 | 75.1% | NA | NA | NA |
| (119) | 2020 | Mobile Net | DDSM patch | 10713 | 77.2% | NA | NA | NA |
| (119) | 2020 | MVGG16 | DDSM patch | 10713 | 80.8% | NA | NA | NA |
| (119) | 2020 | MVGG16 + ImageNet | DDSM patch | 10713 | 88.3% | 93.3% | NA | NA |
| (71) | 2020 | GAN and CNN | DDSM | 292 | 80% | 80% | NA | NA |
| (120) | 2021 | Optimal Multi-Level Thresholding-based Segmentation with DL enabled Capsule Network (OMLTS-DLCN) | Mini-MIAS dataset and DDSM dataset | NA | 98.5% for Mini-MIAS, 97.55% for DDSM | NA | NA | NA |
| (121) | 2021 | Inception-ResNet-V2 | BreastScreen Victoria dataset | 28,694 | 0.8178 | 0.8979 | NA | NA |
| (122) | 2021 | AI-powered imaging biomarker | 2,058 | NA | 0.852 | NA | NA | |
| (123) | 2022 | DualCoreNet | DDSM | NA | NA | 0.85 | NA | NA |
| (123) | 2022 | DualCoreNet | INbreast | NA | NA | 0.93 | NA | NA |
N/A, Not Applicate.
Table 5 presents a comprehensive list of the latest DL-based mammogram models developed for breast lesion classification. DL models offer numerous benefits, including exceptional accuracy and optimal performance achieved with fewer parameters. However, it is important to acknowledge certain limitations associated with existing DL methods for breast tumor classification using mammographies. These limitations include the substantial computational power and extensive datasets required for training the models, which can be computationally expensive, intricate, and time-consuming.
Table 5.
DL-based mammography for breast tumor classification.
| Reference | Year | Method | Database | Number of images | Accuracy | AUC | Sensitivity | Precision | F1-Score |
|---|---|---|---|---|---|---|---|---|---|
| (95) | 2017 | Transfer learning, Random Forest | INbreast | 108 | 90% | NA | 98% | 70% | NA |
| (124) | 2018 | Deep GeneRAtive Multitask | CBIS-DDSM | NA | 89% | 0.884 | NA | NA | NA |
| (125) | 2019 | VGG, Residual Network | CBIS-DDSM | NA | NA | NA | 86.10% | 80.10% | NA |
| (126) | 2019 | DCNN, Alexnet | CBIS-DDSM | 1696 | 75.0% | 0.80 | NA | NA | NA |
| (127) | 2019 | MA-CNN | MIAS | 322 | 96.47% | 0.99 | 96.00% | NA | NA |
| (128) | 2019 | DCNN, MSVM | MIAS | 322 | 96.90% | 0.99 | NA | NA | NA |
| (129) | 2019 | CNN Improvement (CNNI-BCC) |
MIAS | NA | 90.50% | 0.90 | 89.47% | 90.71% | NA |
| (130) | 2020 | MobileNet, VGG, Resnet, Xception |
CBIS-DDSM | 1696 | 84.4% | 0.84 | NA | NA | 85.0% |
| (131) | 2020 | MobilenetV1, MobilenetV2 | CBIS-DDSM | 1696 | 74.5% | NA | NA | 70.00% | 76.00% |
| (132) | 2020 | DE-Ada* | CBIS-DDSM | NA | 87.05% | 0.9219 | NA | NA | NA |
| (133) | 2020 | AlexNet | MIAS | 68 | 98.53% | 0.98 | 100% | 97.37% | 98.3% |
| (133) | 2020 | GoogleNet | MIAS | 68 | 88.24% | 0.94 | 80% | 94.74% | 85.71% |
| (134) | 2020 | Inception ResNet V2 | INbreast | 107 | 95.32% | 0.95 | NA | NA | NA |
| (132) | 2020 | De-ada* | INbreast | NA | 87.93% | 0.9265 | NA | NA | NA |
| (135) | 2021 | CNN | CBIS-DDSM | 1592 | 91.2% | 0.92 | 92.31% | 90.00% | 91.76% |
| (136) | 2021 | MobilenetV2, Nasnet Mobile, MEWOM | CBIS-DDSM | 1696 | 93.8% | 0.98 | 93.75% | 93.80% | 93.77% |
| (137) | 2021 | ResNet-18, (ICS-ELM) | MIAS | 322 | 98.13% | NA | NA | NA | NA |
| (135) | 2021 | CNN | MIAS | 322 | 93.39% | 0.94 | 92.72% | 94.12% | 93.58% |
| (136) | 2021 | Mobilenet V2 & NasNet Mobile, MEWOA |
MIAS | 300 | 99.80% | 1.00 | 99.00% | 99.33% | 99.16% |
| (137) | 2021 | ResNet-18, (ICS-ELM) | INbreast | 179 | 98.26% | NA | NA | NA | NA |
| (135) | 2021 | CNN | INbreast | 387 | 93.04% | 0.94 | 94.83% | 91.23% | 93.22% |
| (136) | 2021 | Fine-tuned MobilenetV2, Nasnet, MEWOM | INbreast | 108 | 99.7% | 1.00 | 99.0% | 99.0% | 99.0% |
| (123) | 2022 | DualCoreNet | CBIS-DDSM | NA | NA | 0.85± 0.021 | NA | NA | NA |
| (138) | 2022 | CNN classifier with different fine-tuning |
DDSM | 13128 | 99.96% | 1.00 | 100% | 99.92% | 99.96% |
N/A, Not Applicate.
5. Challenges and future research directions
The emergence of DL techniques has revolutionized medical imaging, offering immense potential to enhance the diagnosis and treatment of various diseases. DL algorithms present several advantages compared to traditional ML methods. For instance, DL algorithms can be trained using robust hardware such as graphical processing units (GPU) and tensor processing units (TPU), greatly accelerating the training process. This has enabled researchers to train large DL models with billions of parameters, yielding impressive results in diverse language tasks. However, to fully leverage the potential of DL in medical imaging, several challenges must be addressed. One of the primary challenges is the scarcity of data. DL algorithms require abundant, high-quality data for effective training. Yet, acquiring medical imaging data is often challenging, particularly for rare diseases or cases requiring long-term follow-up. Furthermore, data privacy regulations and concerns can further complicate the availability of medical imaging data. Another challenge lies in the quality of annotations. DL algorithms typically demand substantial amounts of annotated data for effective training. However, annotating medical imaging data can be subjective and time-consuming, leading to issues with annotation quality and consistency. This can significantly impact the performance of deep learning algorithms, particularly when accurate annotations are vital for diagnosing or treating specific conditions. Additionally, imbalanced classes pose another challenge in medical imaging.
In numerous instances, the occurrence of certain states may be relatively low, which can result in imbalanced datasets that have a detrimental effect on the performance of DL algorithms. This situation can pose a significant challenge, especially for rare diseases or conditions with limited data availability. Another crucial concern in medical imaging is the interpretability of models. Although DL algorithms have showcased remarkable performance across various medical imaging tasks, the lack of interpretability in these models can hinder their adoption. Clinicians frequently necessitate explanations for the predictions made by these algorithms in order to make informed decisions, but the opacity of DL models can make this task arduous.
Data privacy is a paramount concern in medical imaging. Medical images encompass confidential patient information, stringent regulations dictate the utilization and dissemination of such data. The effective training of DL necessitates substantial access to extensive medical imaging data, thereby introducing challenges concerning data privacy and security. Additionally, computational resources pose another challenge in the realm of medical imaging. DL algorithms mandate substantial computational resources for the effective training and of models. This predicament can prove particularly troublesome in medical imaging, given the size and intricacy of medical images, which can strain computing resources. DL algorithms can be vulnerable to adversarial attacks, where small perturbations to input data can cause significant changes in the model’s output. This can be particularly problematic for medical imaging, where even small changes to an image can have substantial implications for diagnosis and treatment.
Several potential strategies can be employed to address these challenges effectively. One approach involves the development of transfer learning techniques, enabling DL models to be trained on smaller datasets by leveraging information from related tasks or domains. This approach holds particular promise in medical imaging, where data scarcity poses a significant obstacle. Another approach involves placing emphasis on the development of annotation tools and frameworks that enhance the quality and consistency of annotations. This becomes important in cases where annotations play a critical role in diagnosing or treating specific conditions. Furthermore, improved data sharing and collaboration between institutions can help alleviate both data scarcity and privacy concerns. By pooling resources and sharing data, it becomes feasible to construct more extensive and diverse datasets that can be employed to train DL models with greater effectiveness. Additionally, enhancing the interpretability of DL models in medical imaging techniques stands as another critical area of research. The development of explainable AI techniques can provide clinicians with valuable insights into the underlying factors contributing to a model’s predictions. Lastly, bolstering the robustness of DL models constitutes a crucial focal point. This entails exploring adversarial training techniques, as well as leveraging ensemble methods and other strategies to enhance the overall robustness and generalizability of DL models.
DL techniques have the potential to revolutionize medical imaging. However, to fully leverage this potential, it is crucial to address several challenges. These challenges encompass data scarcity, annotation quality, imbalanced classes, model interpretability, data privacy, computational resources, and algorithm robustness. By prioritizing strategies to tackle these challenges, it becomes possible to develop DL models that are more effective and reliable for various medical imaging applications.
6. Conclusion
This paper examines the recent advancements in DL-based mammography for breast cancer screening. The authors have investigated the potential of DL techniques in enhancing the accuracy and efficiency of mammography. Additionally, they address the challenges that need to be overcome for the successful adoption of DL techniques in clinical practice.
Author contributions
LW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the International Science and Technology Cooperation Project of the Shenzhen Science and Technology Innovation Committee (GJHZ20200731095804014).
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Chon JW, Jo YY, Lee KG, Lee HS, Kweon HY. Effect of silk fibroin hydrolysate on the apoptosis of mcf-7 human breast cancer cells. Int J Ind Entomol (2013) 27(2):228–36. doi: 10.7852/ijie.2013.27.2.228 [DOI] [Google Scholar]
- 2. Habtegiorgis SD, Getahun DS, Telayneh AT, Birhanu MY, Feleke TM, Mingude AB, et al. Ethiopian women's breast cancer self-examination practices and associated factors a systematic review and meta-analysis. Cancer Epidemiol (2022) 78:102128. doi: 10.1016/j.canep.2022.102128 [DOI] [PubMed] [Google Scholar]
- 3. Ginsburg O, Yip CH, Brooks A, Cabanes A, Caleffi M, Dunstan Yataco JA, et al. Breast cancer early detection: A phased approach to implementation. Cancer (2020) 126:2379–93. doi: 10.1002/cncr.32887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jalalian A, Mashohor SB, Mahmud HR, Saripan MIB, Ramli ARB, Karasfi B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clin Imaging (2013) 37(3):420–6. doi: 10.1016/j.clinimag.2012.09.024 [DOI] [PubMed] [Google Scholar]
- 5. Gilbert FJ, Pinker-Domenig K. (2019). Diagnosis and staging of breast cancer: when and how to use mammography, tomosynthesis, ultrasound, contrast-enhanced mammography, and magnetic resonance imaging. In: Diseases of the chest, breast, heart and vessels 2019-2022. Springer, Cham: (2019). pp. 155–66. doi: 10.1007/978-3-030-11149-6_13 [DOI] [PubMed] [Google Scholar]
- 6. Alghaib HA, Scott M, Adhami RR. An overview of mammogram analysis. IEEE Potentials (2016) 35(6):21–8. doi: 10.1109/MPOT.2015.2396533 [DOI] [Google Scholar]
- 7. Monticciolo DL, Newell MS, Moy L, Niell B, Monsees B, Sickles EA. Breast cancer screening in women at higher-than-average risk: recommendations from the ACR. J Am Coll Radiol (2018) 15(3):408–14. doi: 10.1016/j.jacr.2017.11.034 [DOI] [PubMed] [Google Scholar]
- 8. Alabousi M, Zha N, Salameh JP, Samoilov L, Sharifabadi AD, Pozdnyakov A, et al. Digital breast tomosynthesis for breast cancer detection: a diagnostic test accuracy systematic review and meta-analysis. Eur Radiol (2020) 30:2058–71. doi: 10.1007/s00330-019-06549-2 [DOI] [PubMed] [Google Scholar]
- 9. Brem RF, Lenihan MJ, Lieberman J, Torrente J. Screening breast ultrasound: past, present, and future. Am J Roentgenol (2015) 204(2):234–40. doi: 10.2214/AJR.13.12072 [DOI] [PubMed] [Google Scholar]
- 10. Heller SL, Moy L. MRI breast screening revisited. J Magnetic Resonance Imaging (2019) 49(5):1212–21. doi: 10.1002/jmri.26547 [DOI] [PubMed] [Google Scholar]
- 11. Schöder H, Gönen M. Screening for cancer with PET and PET/CT: potential and limitations. J Nucl Med (2007) 48(1):4S–18S. doi: 10.1016/S0148-2963(03)00075-4 [DOI] [PubMed] [Google Scholar]
- 12. Narayanan D, Berg WA. Dedicated breast gamma camera imaging and breast PET: current status and future directions. PET Clinics (2018) 13(3):363–81. doi: 10.1016/j.cpet.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med (2019) 25:30–6. doi: 10.1038/s41591-018-0307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim KH, Lee SH. Applications of artificial intelligence in mammography from a development and validation perspective. J Korean Soc Radiol (2021) 82(1):12. doi: 10.3348/jksr.2020.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamed G, Marey MAER, Amin SES, Tolba MF. (2020). Deep learning in breast cancer detection and classification. In: Proceedings of the international conference on artificial intelligence and computer vision, advances in intelligent systems and computing. Springer, Cham: (2020) 1153:322–33. doi: 10.1007/978-3-030-44289-7_30 [DOI] [Google Scholar]
- 16. Wang J, Gottschal P, Ding L, Veldhuizen DAV, Lu W, Houssami N, et al. Mammographic sensitivity as a function of tumor size: a novel estimation based on population-based screening data. Breast (2021) 55:69–74. doi: 10.1016/j.breast.2020.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stein RG, Wollschläger D, Kreienberg R, Janni W, Wischnewsky M, Diessner J, et al. The impact of breast cancer biological subtyping on tumor size assessment by ultrasound and mammography-a retrospective multicenter cohort study of 6543 primary breast cancer patients. BMC Cancer (2016) 16(1):1–8. doi: 10.1186/s12885-016-2426-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen HL, Zhou JQ, Chen Q, Deng YC. Comparison of the sensitivity of mammography, ultrasound, magnetic resonance imaging and combinations of these imaging modalities for the detection of small (≤2 cm) breast cancer. Medicine (2021) 100(26):e26531. doi: 10.1097/MD.0000000000026531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gunther JE, Lim EA, Kim HK, Flexman M, Altoé M, Campbell JA, et al. Dynamic diffuse optical tomography for monitoring neoadjuvant chemotherapy in patients with breast cancer. Radiology (2018) 287(3):778–86. doi: 10.1148/radiol.2018161041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Movik E, Dalsbø TK, Fagerlund BC, Friberg EG, Håheim LL, Skår Å. (2017). Digital breast tomosynthesis with hologic 3d mammography selenia dimensions system for use in breast cancer screening: a single technology assessment. Oslo, Norway: Knowledge Centre for the Health Services at The Norwegian Institute of Public Health. [PubMed] [Google Scholar]
- 21. Marion W. Siemens Healthineers, GE HealthCare Race To Develop Next-Gen AI Solutions For Personalized Care (2023). Available at: https://medtech.pharmaintelligence.informa.com/MT147893/Siemens-Healthineers-GE-HealthCare-Race-To-Develop-NextGen-AI-Solutions-For-Personalized-Care.
- 22. NEWS BREAST IMAGING, ScreenPoint Medical: Transpara Breast AI Demonstrates Value in Real-world Clinical Usage (2023). Available at: https://www.itnonline.com/content/screenpoint-medical-transpara-breast-ai-demonstrates-value-real-world-clinical-usage.
- 23. Gong Z, Williams MB. Comparison of breast specific gamma imaging and molecular breast tomosynthesis in breast cancer detection: evaluation in phantoms. Med Phys (2015) 42(7):4250. doi: 10.1118/1.4922398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Łuczyńska E, Heinze-Paluchowska S, Hendrick E, Dyczek S, Ryś J, Herman K, et al. Comparison between breast MRI and contrast-enhanced spectral mammography. Med Sci Monit (2015) 21:1358–67. doi: 10.12659/MSM.893018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen EO, Perry RE, Tso HH, Phalak KA, Leung JWT. Breast cancer screening in women with and without implants: retrospective study comparing digital mammography to digital mammography combined with digital breast tomosynthesis. Eur Radiol (2021) 31(12):9499–510. doi: 10.1007/s00330-021-08040-3 [DOI] [PubMed] [Google Scholar]
- 26. Gilbert FJ, Tucker L, Young KC. Digital breast tomosynthesis (DBT): a review of the evidence for use as a screening tool. Clin Radiol (2016) 71(2):141–50. doi: 10.1016/j.crad.2015.11.008 [DOI] [PubMed] [Google Scholar]
- 27. Nakano S, Fujii K, Yorozuya K, Yoshida M, Fukutomi T, Arai O, et al. P2-10-10: a precision comparison of breast ultrasound images between different time phases by imaging fusion technique using magnetic position tracking system. Cancer Res (2011) 71(24):P2–10-10-P2-10-10. doi: 10.1158/0008-5472.SABCS11-P2-10-10 [DOI] [Google Scholar]
- 28. Zhang E, Seiler S, Chen M, Lu W, Gu X. Birads features-oriented semi-supervised deep learning for breast ultrasound computer-aided diagnosis. Phys Med Biol (2020) 65(12):125005. doi: 10.1088/1361-6560/ab7e7d [DOI] [PubMed] [Google Scholar]
- 29. Santos MK, Ferreira Júnior JR, Wada DT, Tenório APM, Barbosa MHN, Marques PMA. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: advances in imaging towards to precision medicine. Radiol Bras (2019) 52(6):387–96. doi: 10.1590/0100-3984.2019.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. GE Healthcare . Invenia ABUS 2.0. Available at: https://www.gehealthcare.com/en-ph/products/ultrasound/abus-breast-imaging/invenia-abus.
- 31. Siemens Healthineers . ACUSON S2000 ABVS System HELX Evolution with Touch Control. Available at: https://shop.medicalimaging.healthcare.siemens.com.sg/acuson-s2000-abvs-system-helx-evolution-with-touch-control/.
- 32. Kuhl CK, Schrading S, Strobel K, Schild HH, Hilgers RD, Bieling HB. Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection-a novel approach to breast cancer screening with MRI. J Clin Oncol (2014) 32(22):2304–10. doi: 10.1200/JCO.2013.52.5386 [DOI] [PubMed] [Google Scholar]
- 33. Mann RM, Mus RD, van Zelst J, Geppert C, Karssemeijer NA. Novel approach to contrast-enhanced breast magnetic resonance imaging for screening: high-resolution ultrafast dynamic imaging. Invest Radiol (2014) 49(9):579–85. doi: 10.1097/RLI.0000000000000057 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y, Chen JH, Chang KT, Park VY, Kim MJ, Chan S, et al. Automatic breast and fibroglandular tissue segmentation in breast MRI using deep learning by a fully-convolutional residual neural network U-net. Acad Radiol (2019) 26(11):1526–35. doi: 10.1016/j.acra.2019.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ming W, Li F, Zhu Y, Bai Y, Gu W, Liu Y, et al. Unsupervised analysis based on DCE-MRI radiomics features revealed three novel breast cancer subtypes with distinct clinical outcomes and biological characteristics. Cancers (2022) 14(22):5507. doi: 10.3390/cancers14225507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcus C, Jeyarajasingham K, Hirsch A, Subramaniam RM. PET/CT in the management of thyroid cancers. Am Roentgen Ray Soc Annu Meeting (2014) 202:1316–29. doi: 10.2214/AJR.13.11673 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y, Choi EJ, Choi Y, Zhang H, Jin GY, Ko SB. Breast cancer classification in automated breast ultrasound using multiview convolutional neural network with transfer learning - ScienceDirect. Ultrasound Med Biol (2020) 46(5):1119–32. doi: 10.1016/j.ultrasmedbio.2020.01.001 [DOI] [PubMed] [Google Scholar]
- 38. Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci Rep (2019) 9:12495. doi: 10.1038/s41598-019-48995-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology (2019) 292(1):182716. doi: 10.1148/radiol.2019182716 [DOI] [PubMed] [Google Scholar]
- 40. Wang L. Deep learning techniques to diagnose lung cancer. Cancers (2022) 14:5569. doi: 10.3390/cancers14225569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heath M, Bowyer K, Kopans D, Kegelmeyer P, Jr, Moore R, Chang K. Current Status of the digital database for screening mammography. In: Digital mammography. computational imaging and vision. (Springer, Dordrecht: ) (1998) 11:457–60. doi: 10.1007/978-94-011-5318-8_75 [DOI] [Google Scholar]
- 42. Li B, Ge Y, Zhao Y, Guan E, Yan W. Benign and malignant mammographic image classification based on convolutional neural networks. In: 10th international conference on machine learning and computing. (Beijing, China: Association for Computing Machinery; ) (2018). pp. 11–15. doi: 10.1145/3195106.3195163 [DOI] [Google Scholar]
- 43. Inês CM, Igor A, Inês D, António C, Maria JC, Jaime SC. Inbreast: toward a full-field digital mammographic database. Acad Radiol (2012) 19:236–48. doi: 10.1016/j.acra.2011.09.014 [DOI] [PubMed] [Google Scholar]
- 44. Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, et al. Classification of breast cancer histology images using convolutional neural networks. PloS One (2017) 12(6):e0177544. doi: 10.1371/journal.pone.0177544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee R, Gimenez F, Hoogi A, Miyake K, Gorovoy M, Rubin D. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data (2017) 4:170177. doi: 10.1038/sdata.2017.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramos-Pollán R, Guevara-López M, Suárez-Ortega C, Díaz-Herrero G, Franco-Valiente J, Rubio-del-Solar M, et al. Discovering mammography-based machine learning classifiers for breast cancer diagnosis. J Med Syst (2011) 1:11. doi: 10.1007/s10916-011-9693- [DOI] [PubMed] [Google Scholar]
- 47. Heath M, Bowyer K, Kopans D, Moore R, Kegelmeyer WP. (2001). Current status of the digital database for screening mammography. In: Digital mammography. computational imaging and vision. Springer Netherlands: (1998) 13:457–60. doi: 10.1007/978-94-011-5318-8_75 [DOI] [Google Scholar]
- 48. Yoon WB, Oh JE, Chae EY, Kim HH, Lee SY, Kim KG. Automatic detection of pectoral muscle region for computer-aided diagnosis using MIAS mammograms. BioMed Res Int (2016) 2016:5967580. doi: 10.1155/2016/5967580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lopez MG, Posada N, Moura DC, Pollán RR, Valiente JMF, Ortega CS, et al. (2012). BCDR: a breast cancer digital repository. In: International conference on experimental mechanics. Porto, Portugal: (2012). pp. 113–20. [Google Scholar]
- 50. Dalle J-R, Leow WK, Racoceanu D, Tutac AE, Putti TC. (2008). Automatic breast cancer grading of histopathological images. In: Annu Int Conf IEEE Eng Med Biol Soc. Vancouver, BC, Canada: IEEE; (2008). pp. 3052–5. doi: 10.1109/IEMBS.2008.4649847 [DOI] [PubMed] [Google Scholar]
- 51. Lee S, Fu C, Salama P, Dunn K, Delp E. Tubule segmentation of fluorescence microscopy images based on convolutional neural networks with inhomogeneity correction. Int Symp Electr Imaging (2018) 30:199–1–199–8. doi: 10.2352/ISSN.2470-1173.2018.15.COIMG-199 [DOI] [Google Scholar]
- 52. Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi AA. Dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging (2017) 36(7):1550–60. doi: 10.1109/TMI.2017.2677499 [DOI] [PubMed] [Google Scholar]
- 53. Saha M, Chakraborty C, Racoceanu D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput Med Imaging Graph (2018) 64:29–40. doi: 10.1016/j.compmedimag.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 54. Ronneberger O, Fischer P. Brox, T. U-net: Convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention. Springer Switzerland: (2015). p. 234–41. doi: 10.1007/978-3-319-24574-4_28 [DOI] [Google Scholar]
- 55. Borgli H, Thambawita V, Pia Helén S, Hicks SA, Lange TD. A comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data (2020) 7:1–14. doi: 10.1038/s41597-020-00622-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mamonov AV, Figueiredo IN, Figueiredo PN, Tsai YH. Automated polyp detection in colon capsule endoscopy. IEEE Trans Med Imaging (2013) 33:1488–502. doi: 10.1109/TMI.2014.2314959 [DOI] [PubMed] [Google Scholar]
- 57. Bellens S, Probst GM, Janssens M, Vandewalle P, Dewulf W. Evaluating conventional and deep learning segmentation for fast X-ray CT porosity measurements of polymer laser sintered am parts. Polym Test (2022) 110:107540. doi: 10.1016/j.polymertesting.2022.107540 [DOI] [Google Scholar]
- 58. Yuan X, Yuxin W, Jie Y, Qian C, Xueding W, Carson PL. Medical breast ultrasound image segmentation by machine learning. Ultrasonics (2018) 91:1–9. doi: 10.1016/j.ultras.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 59. Dhungel N, Carneiro G, Bradley AP. Tree RE-weighted belief propagation using deep learning potentials for mass segmentation from mammograms. In: 2015 IEEE 12th international symposium on biomedical imaging Brooklyn, NY, USA: IEEE; (2015). pp. 760–3. doi: 10.1109/ISBI.2015.7163983 [DOI] [Google Scholar]
- 60. Zhu W, Xiang X, Tran TD, Hager GD, Xie X. Adversarial deep structured nets for mass segmentation from mammograms. In: International symposium on biomedical imaging. Washington, DC, USA: IEEE; (2018). 847–50. doi: 10.1109/ISBI.2018.8363704 [DOI] [Google Scholar]
- 61. Al-antari MA, Al-masni MA, Choi MT, Han SM, Kim TS. A fully integrated computer-aided diagnosis system for digital x-ray mammograms via deep learning detection, segmentation, and classification. Int J Med Inform (2018) 117:44–54. doi: 10.1016/j.ijmedinf.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 62. Li H, Chen D, Nailon WH, Davies ME, Laurenson D. Image analysis for moving organ, breast, and thoracic images. In: Improved breast mass segmentation in mammograms with conditional residual u-net. Cham: Springer; (2018). p. 81–9. [Google Scholar]
- 63. Shen T, Gou C, Wang J, Wang FY. Simultaneous segmentation and classification of mass region from mammograms using a mixed-supervision guided deep model. IEEE Signal Process Lett (2019) 27:196–200. doi: 10.1109/LSP.2019.2963151 [DOI] [Google Scholar]
- 64. Li GDS, Dong M, Xiaomin M. Attention dense-u-net for automatic breast mass segmentation in digital mammogram. IEEE Access (2019) 7:59037–47. doi: 10.1109/ACCESS.2019.2914873 [DOI] [Google Scholar]
- 65. Abdelhafiz D, Nabavi S, Ammar R, Yang C, Bi J. Residual deep learning system for mass segmentation and classification in mammography. In: Proceedings of the 10th ACM international conference on bioinformatics. Association for Computing Machinery, New York, NY, USA: Computational Biology and Health Informatics (2019). pp. 475–84. doi: 10.1145/3307339.3342157 [DOI] [Google Scholar]
- 66. Hossain MS. Microcalcification segmentation using modified u-net segmentation network from mammogram images. J King Saud University Comput Inf Sci (2022) 34(2):86–94. doi: 10.1016/j.jksuci.2019.10.014 [DOI] [Google Scholar]
- 67. Min H, Wilson D, Huang Y, Liu S, Crozier S, Bradley AP, et al. (2020). Fully automatic computer-aided mass detection and segmentation via pseudo-color mammograms and mask R-CNN. In: 17th international symposium on biomedical imaging (ISBI), Iowa City, IA, USA: IEEE; (2000). pp. 1111–5. doi: 10.1109/ISBI45749.2020.9098732 [DOI] [Google Scholar]
- 68. Al-antari MA, Al-masni MA, Kim TS. Advances in experimental medicine and biology. In: Deep learning computer-aided diagnosis for breast lesion in digital mammogram. Cham: Springer; (2020). p. 59–72. [DOI] [PubMed] [Google Scholar]
- 69. Abdelhafiz D, Bi J, Ammar R, Yang C, Nabavi S. Convolutional neural network for automated mass segmentation in mammography. BMC Bioinf (2020) 21(S1):1–19. doi: 10.1186/s12859-020-3521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Saffari N, Rashwan HA, Abdel-Nasser M, Singh VK, Puig D. Fully automated breast density segmentation and classification using deep learning. Diagnostics (2020) 10(11):988. doi: 10.3390/diagnostics10110988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kumar Singh V, Rashwan HA, Romani S, Akram F, Pandey N, Kamal Sarker MM, et al. Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst Appl (2020) 139:112855. doi: 10.1016/j.eswa.2019.112855 [DOI] [Google Scholar]
- 72. Ahmed L, Iqbal MM, Aldabbas H, Khalid S, Saleem Y, Saeed S. Images data practices for semantic segmentation of breast cancer using deep neural network. J Ambient Intell Humanized Comput (2020) 14:15227–243. doi: 10.1007/s12652-020-01680-1 [DOI] [Google Scholar]
- 73. Sun H, Cheng L, Liu B, Zheng H, Feng DD, Wang S. AUNet: attention-guided dense-upsampling networks for breast mass segmentation in whole mammograms. Phys Med Biol (2020) 65(5):55005. doi: 10.1088/1361-6560/ab5745 [DOI] [PubMed] [Google Scholar]
- 74. Bhatti HMA, Li J, Siddeeq S, Rehman A, Manzoor A. (2020). Multi-detection and segmentation of breast lesions based on mask RCNN-FPN. In: International conference on bioinformatics and biomedicine (BIBM). Seoul, Korea (South): IEEE; (2000). pp. 2698–704. doi: 10.1109/BIBM49941.2020.9313170 [DOI] [Google Scholar]
- 75. Zeiser FA, da Costa CA, Zonta T, Marques NMC, Roehe AV, Moreno M, et al. Segmentation of masses on mammograms using data augmentation and deep learning. J Digital Imaging (2020) 33(4):858–68. doi: 10.1007/s10278-020-00330-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsochatzidis L, Koutla P, Costaridou L, Pratikakis I. Integrating segmentation information into cnn for breast cancer diagnosis of mammographic masses. Comput Methods Programs Biomed (2021) 200:105913. doi: 10.1016/j.cmpb.2020.105913 [DOI] [PubMed] [Google Scholar]
- 77. Ravitha Rajalakshmi N, Vidhyapriya R, Elango N, Ramesh N. Deeply supervised u-net for mass segmentation in digital mammograms. Int J Imaging Syst Technol (2021) 31(1):59–71. doi: 10.1002/ima.22516 [DOI] [Google Scholar]
- 78. Salama WM, Aly MH. Deep learning in mammography images segmentation and classification: automated cnn approach. Alexandria Eng J (2021) 60(5):4701–9. doi: 10.1016/j.aej.2021.03.048 [DOI] [Google Scholar]
- 79. Tekin E, Yazıcı Ç, Kusetogullari H, Tokat F, Yavariabdi A, Iheme LO, et al. Tubule-U-Net: a novel dataset and deep learning-based tubule segmentation framework in whole slide images of breast cancer. Sci Rep (2023) 13:128. doi: 10.1038/s41598-022-27331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Naik S, Doyle S, Agner S, Madabhushi A, Tomaszewski J. (2008). Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In: International Symposium on Biomedical Imaging: From Nano to Macro. Paris, France: IEEE; (2008) pp. 284–7. doi: 10.1109/ISBI.2008.4540988 [DOI] [Google Scholar]
- 81. Tutac AE, Racoceanu D, Putti T, Xiong W, Cretu V. (2008). Knowledge-guided semantic indexing of breast cancer histopathology images. In: International conference on biomedical engineering and informatics. China: IEEE; (2008). pp. 107–12. doi: 10.1109/BMEI.2008.166 [DOI] [Google Scholar]
- 82. Basavanhally A, Summers RM, Van Ginneken B, Yu E, Xu J, Ganesan S, et al. Incorporating domain knowledge for tubule detection in breast histopathology using o'callaghan neighborhoods. Proc SPIE Int Soc Optical Eng (2011) 7963:796310–796310-15. doi: 10.1117/12.878092 [DOI] [Google Scholar]
- 83. Maqlin P, Thamburaj R, Mammen JJ, Nagar AK. Automatic detection of tubules in breast histopathological images. In: Proceedings of Seventh International Conference on Bio-Inspired Computing: Theories and Applications. Springer, India: Adv Intelligent Syst Comput. (2013) 202:311–21. doi: 10.1007/978-81-322-1041-2_27 [DOI] [Google Scholar]
- 84. Romo-Bucheli D, Janowczyk A, Gilmore H, Romero E, Madabhushi A. Automated tubule nuclei quantification and correlation with oncotype dx risk categories in er+ breast cancer whole slide images. Sci Rep (2016) 6:32706. doi: 10.1038/srep32706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hu Y, Guo Y, Wang Y, Yu J, Li J, Zhou S, et al. Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model. Med Phys (2019) 46(1):215–28. doi: 10.1002/mp.13268 [DOI] [PubMed] [Google Scholar]
- 86. Abdelhafiz D, Yang C, Ammar R, Nabavi S. Deep convolutional neural networks for mammography: advances, challenges and applications. BMC Bioinf (2019) 20:1–20. doi: 10.1186/s12859-019-2823-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tan XJ, Mustafa N, Mashor MY, Rahman KSA. A novel quantitative measurement method for irregular tubules in breast carcinoma. Eng Sci Technol an Int J (2022) 31:101051. doi: 10.1016/j.jestch.2021.08.008 [DOI] [Google Scholar]
- 88. Jiao Z, Gao X, Wang Y, Li J. A deep feature based framework for breast masses classification. Neurocomputing (2016) 197:221–31. doi: 10.1016/j.neucom.2016.02.060 [DOI] [Google Scholar]
- 89. Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging (2016) 3(3):034501. doi: 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Arevalo J, Gonzalez FA, Ramos-Pollan R, Oliveira JL, Guevara Lopez MA. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed (2016) 127:248–57. doi: 10.1016/j.cmpb.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 91. Leod PM, Verma B. (2016). Polynomial prediction of neurons in neural network classifier for breast cancer diagnosis. In: Proceedings of the international conference on natural computation (ICNC). Zhangjiajie, China: IEEE; (2015). pp. 775–80. doi: 10.1109/ICNC.2015.7378089 [DOI] [Google Scholar]
- 92. Nascimento CDL, Silva SDS, da Silva TA, Pereira WCA, Costa MGF, Costa Filho CFF. Breast tumor classification in ultrasound images using support vector machines and neural networks. Rev Bras Engenharia Biomedica (2016) 32(3):283–92. doi: 10.1590/2446-4740.04915 [DOI] [Google Scholar]
- 93. Zhang Q, Xiao Y, Dai W, Suo JF, Wang CZ, Shi J, et al. Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics (2016) 72:150–7. doi: 10.1016/j.ultras.2016.08.004 [DOI] [PubMed] [Google Scholar]
- 94. Sun W, Tseng TL, Zhang J, Qian W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graphics (2017) 57:4–9. doi: 10.1016/j.compmedimag.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 95. Dhungel N, Carneiro G, Bradley AP. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med Image Anal (2017) 37:114–28. doi: 10.1016/j.media.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 96. Samala RK, Chan HP, Hadjiiski LM, Helvie MA, Cha K, Richter C. Multi-task transfer learning deep convolutional neural network: application to computer aided diagnosis of breast cancer on mammograms. Phys Med Biol (2017) 62(23):8894–908. doi: 10.1088/1361-6560/aa93d4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jadoon MM, Zhang Q, Ul Haq I, Butt S, Jadoon A. Three-class mammogram classification based on descriptive CNN features. BioMed Res Int (2017) 2017:3640901. doi: 10.1155/2017/3640901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys (2017) 44(10):5162–71. doi: 10.1002/mp.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Qiu Y, Yan S, Gundreddy RR, Wang Y, Zheng B. A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J X-Ray Sci Technol (2017) 25(5):751–63. doi: 10.3233/XST-16226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gardezi SJS, Awais M, Faye I, Meriaudeau F. (2017). Mammogram classification using deep learning features. In: Proceedings of the 2017 IEEE international conference on signal and image processing applications (ICSIPA). Kuching, Malaysia: IEEE; (2017). pp. 485–8. doi: 10.1109/ICSIPA.2017.8120660 [DOI] [Google Scholar]
- 101. Kumar I, Bhadauria HS, Virmani J, Thakur S. A classification framework for prediction of breast density using an ensemble of neural network classifiers. Biocybernetics Biomed Eng (2017) 37(1):217–28. doi: 10.1016/j.bbe.2017.01.001 [DOI] [Google Scholar]
- 102. Zheng Y, Jiang Z, Xie F, Zhang H, Ma Y, Shi H, et al. Feature extraction from histopathological images based on nucleus-guided convolutional neural network for breast lesion classification. Pattern Recognit (2017) 71:14–25. doi: 10.1016/j.patcog.2017.05.010 [DOI] [Google Scholar]
- 103. Han S, Kang HK, Jeong JY, Park MH, Kim W, Bang WC, et al. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol (2017) 62(19):7714–28. doi: 10.1088/1361-6560/aa82ec [DOI] [PubMed] [Google Scholar]
- 104. Yu S, Liu LL, Wang ZY, Dai GZ, Xie YQ. Transferring deep neural networks for the differentiation of mammographic breast lesions. Sci China Technol Sci (2018) 62(3):441–7. doi: 10.1007/s11431-017-9317-3 [DOI] [Google Scholar]
- 105. Ribli D, Horv'ath A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep (2018) 8(1):85–94. doi: 10.1038/s41598-018-22437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. A l-masni MA, Al-antari MA, Park JM, Gi G, Kim TY, Rivera P, et al. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput Methods Programs Biomed (2018) 157:85–94. doi: 10.1016/j.cmpb.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 107. Chougrad H, Zouaki H, Alheyane Q. Deep convolutional neural networks for breast cancer screening. Comput Methods Programs Biomed: An International Journal Devoted to the Development, Implementation and Exchange of Computing Methodology and Software Systems in Biomedical Research and Medical Practice (2018) 157:19–30. doi: 10.1016/j.cmpb.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 108. Jiao Z, Gao X, Wang Y, Li J. A parasitic metric learning net for breast mass classification based on mammography. Pattern Recognit (20118) 75:292–301. doi: 10.1016/j.patcog.2017.07.008 [DOI] [Google Scholar]
- 109. Mohamed AA, Berg WA, Peng H, Luo Y, Jankowitz RC, Wu S. A deep learning method for classifying mammographic breast density categories. Med Phys (2018) 45(1):314–21. doi: 10.1002/mp.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep (2018) 8(1):4165. doi: 10.1038/s41598-018-22437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mendel K, Li H, Sheth D, Giger M. Transfer learning from convolutional neural networks for computer-aided diagnosis: a comparison of digital breast tomosynthesis and full-field digital mammography. Acad Radiol (2019) 26(16):735–43. doi: 10.1016/j.acra.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang H, Feng J, Zhang Z, Su H, Cui L, He H, et al. Breast mass classification via deeply integrating the contextual information from multi-view data. Pattern Recognit (2018) 80:42–52. doi: 10.1016/j.patcog.2018.02.026 [DOI] [Google Scholar]
- 113. Charan S, Khan MJ, Khurshid K. (2018). Breast cancer detection in mammograms using convolutional neural network. In: Proceedings of the 2018 international conference on computing, mathematics and engineering technologies (iCoMET). Sukkur, Pakistan: IEEE; (2018). pp. 1–5. doi: 10.1109/ICOMET.2018.8346384 [DOI] [Google Scholar]
- 114. Feng Y, Zhang L, Yi Z. Breast cancer cell nuclei classification in histopathology images using deep neural networks. Int J Comput Assist Radiol Surg (2018) 13(2):179–91. doi: 10.1007/s11548-017-1663-9 [DOI] [PubMed] [Google Scholar]
- 115. Bardou D, Zhang K, Ahmad SM. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access (2018) 6: 24680–93. doi: 10.1109/access.2018.2831280 [DOI] [Google Scholar]
- 116. Nahid AA, Kong Y. Histopathological breast-image classification using local and frequency domains by convolutional neural network. Inf (Switzerland) (2018) 9(1):19. doi: 10.3390/info9010019 [DOI] [Google Scholar]
- 117. Touahri R, AzizI N, Hammami NE, Aldwairi M, Benaida F. (2019). Automated breast tumor diagnosis using local binary patterns (LBP) based on deep learning classification. In: Proceedings of the 2019 international conference on computer and information sciences (ICCIS). Sakaka, Saudi Arabia: IEEE; (2019). pp. 1–5. doi: 10.1109/ICCISci.2019.8716428 [DOI] [Google Scholar]
- 118. Abdel Rahman AS, Belhaouari SB, Bouzerdoum A, Baali H, Alam T, Eldaraa AM. (2020). Breast mass tumor classification using deep learning. In: Proceedings of the 2020 IEEE international conference on informatics, iot, and enabling technologies (ICIoT). Doha, Qatar: IEEE; (2020). pp. 271–6. doi: 10.1109/ICIoT48696.2020.9089535 [DOI] [Google Scholar]
- 119. Khamparia A, Khanna A, Thanh DNH, Gupta D, Podder P, Bharati S, et al. Diagnosis of breast cancer based on modern mammography using hybrid transfer learning. Multidim Syst Sign Process (2021) 32:747–65. doi: 10.1007/s11045-020-00756-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kavitha T, Mathai PP, Karthikeyan C, Ashok M, Kohar R, Avanija J, et al. Deep learning based capsule neural network model for breast cancer diagnosis using mammogram images. Interdiscip Sci Comput Life Sci (2022) 14:113–29. doi: 10.1007/s12539-021-00467-y [DOI] [PubMed] [Google Scholar]
- 121. Frazer HM, Qin AK, Pan H, Brotchie P. Evaluation of deep learning-based artificial intelligence techniques for breast cancer detection on mammograms: results from a retrospective study using a breastscreen victoria dataset. J Med Imaging Radiat Oncol (2021) 65(5):529–37. doi: 10.1111/1754-9485.13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kim KH, Nam H, Lim E, Ock CY. Development of AI-powered imaging biomarker for breast cancer risk assessment on the basis of mammography alone. J Clin Oncol (2021) 39(15):10519–9. doi: 10.1200/JCO.2021.39.15_suppl.10519 [DOI] [Google Scholar]
- 123. Li H, Chen D, Nailon WH, Davies ME, Laurenson DI. Dual convolutional neural networks for breast mass segmentation and diagnosis in mammography. IEEE Trans Med Imaging (2022) 41(1):3–13. doi: 10.1109/TMI.2021.3102622 [DOI] [PubMed] [Google Scholar]
- 124. Shams S, Platania R, Zhang J, Kim J, Lee K, Park SJ. (2018). Deep generative breast cancer screening and diagnosis. In: Proceedings of the international conference on medical image computing and computer-assisted intervention, Granada, Spain. Granada, Spain: Springer; (2018). 11071:859–67. doi: 10.1007/978-3-030-00934-2_95 [DOI] [Google Scholar]
- 125. Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci Rep (2019) 9:1–12. doi: 10.1038/s41598-019-48995-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tsochatzidis L, Costaridou L, Pratikakis I. Deep learning for breast cancer diagnosis from mammograms—A comparative study. J Imaging (2019) 5:37. doi: 10.3390/jimaging5030037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Agnes SA, Anitha J, Pandian SIA, Peter JD. Classification of mammogram images using multiscale all convolutional neural network (MA-CNN) J. Med Syst (2020) 44:1–9. doi: 10.1007/s10916-019-1494-z [DOI] [PubMed] [Google Scholar]
- 128. Kaur P, Singh G, Kaur P. Intellectual detection and validation of automated mammogram breast cancer images by multi-class SVM using deep learning classification. Inform Med Unlocked (2019) 16:100151. doi: 10.1016/j.imu.2019.01.001 [DOI] [Google Scholar]
- 129. Ting FF, Tan YJ, Sim KS. Convolutional neural network improvement for breast cancer classification. Expert Syst Appl (2019) 120:103–15. doi: 10.1016/j.eswa.2018.11.008 [DOI] [Google Scholar]
- 130. Falconi LG, Perez M, Aguilar WG, Conci A. Transfer learning and fine tuning in breast mammogram abnormalities classification on CBIS-DDSM database. Adv Sci Technol Eng Syst (2020) 5:154–65. doi: 10.25046/aj050220 [DOI] [Google Scholar]
- 131. Ansar W, Shahid AR, Raza B, Dar AH. Breast cancer detection and localization using mobilenet based transfer learning for mammograms. In: International symposium on intelligent computing systems. Sharjah, United Arab Emirates: Springer, Cham; (2020) 1187:11–21. doi: 10.1007/978-3-030-43364-2_2 [DOI] [Google Scholar]
- 132. Zhang H, Wu R, Yuan T, Jiang Z, Huang S, Wu J, et al. DE-Ada*: A novel model for breast mass classification using cross-modal pathological semantic mining and organic integration of multi-feature fusions. Inf Sci (2020) 539:461–86. doi: 10.1016/j.ins.2020.05.080 [DOI] [Google Scholar]
- 133. Shayma'a AH, Sayed MS, Abdalla MI, Rashwan MA. Breast cancer masses classification using deep convolutional neural networks and transfer learning. Multimed Tools Appl (2020) 79:30735–68. doi: 10.1007/s11042-020-09518-w [DOI] [Google Scholar]
- 134. Al-Antari MA, Han SM, Kim TS. Evaluation of deep learning detection and classification towards a computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput Methods Programs Biomed (2020) 196:105584. doi: 10.1016/j.cmpb.2020.105584 [DOI] [PubMed] [Google Scholar]
- 135. El Houby EM, Yassin NI. Malignant and nonmalignant classification of breast lesions in mammograms using convolutional neural networks. Biomed Signal Process Control (2021) 70:102954. doi: 10.1016/j.bspc.2021.102954 [DOI] [Google Scholar]
- 136. Zahoor S, Shoaib U, Lali IU. Breast cancer mammograms classification using deep neural network and entropy-controlled whale optimization algorithm. Diagnostics (2022) 12(2):557. doi: 10.3390/diagnostics12020557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chakravarthy SS, Rajaguru H. Automatic detection and classification of mammograms using improved extreme learning machine with deep learning. IRBM (2021) 43:49–61. doi: 10.1016/j.irbm.2020.12.004 [DOI] [Google Scholar]
- 138. Mudeng V, Jeong JW, Choe SW. Simply fine-tuned deep learning-based classification for breast cancer with mammograms. Comput Mater Contin (2022) 73(3):4677–93. doi: 10.32604/cmc.2022.031046 [DOI] [Google Scholar]