Abstract
In yeast, the pheromone α-factor acts as an antiproliferative factor that induces G1 arrest and cellular differentiation. Previous data have indicated that Far1, a factor dedicated to pheromone-induced cell cycle arrest, is under positive and negative posttranslational regulation. Phosphorylation by the pheromone-stimulated mitogen-activated protein (MAP) kinase Fus3 has been thought to enhance the binding of Far1 to G1-specific cyclin-dependent kinase (Cdk) complexes, thereby inhibiting their catalytic activity. Cdk-dependent phosphorylation events were invoked to account for the high instability of Far1 outside early G1 phase. To confirm any functional role of Far1 phosphorylation, we undertook a systematic mutational analysis of potential MAP kinase and Cdk recognition motifs. Two putative phosphorylation sites that strongly affect Far1 behavior were identified. A change of serine 87 to alanine prevents the cell cycle-dependent degradation of Far1, causing enhanced sensitivity to pheromone. In contrast, threonine 306 seems to be an important recipient of an activating modification, as substitutions at this position abolish the G1 arrest function of Far1. Only the phosphorylated wild-type Far1 protein, not the T306-to-A substitution product, can be found in stable association with the Cdc28-Cln2 complex. Surprisingly, Far1-associated Cdc28-Cln2 complexes are at best moderately inhibited in immunoprecipitation kinase assays, suggesting unconventional inhibitory mechanisms of Far1.
In yeast, mating pheromone induces the competence of responsive cells to mate with cells of the opposite mating type by causing G1 cell cycle arrest and concomitant differentiation into mating-competent gametes (for a review, see reference 20). According to the current view, mating-factor-dependent signal transduction is initiated by the interaction of pheromone with an integral membrane-bound receptor which is associated with a heterotrimeric G protein. Upon stimulation, the G(β, γ) dimer dissociates from the G(α) subunit, which acts as an effector protein by causing the Ste5-dependent propagation of the signal to a tripartite mitogen-activated protein (MAP) kinase cascade (23). This signaling step, whose molecular details are only now emerging, involves many proteins and ultimately leads to the stimulation of the MAP kinase Fus3. The activity of Fus3, which can be compensated for by Kss1 in FUS3 deletion strains, serves as output of the pheromone response pathway (6, 13, 18, 27, 31). There is evidence that Fus3 and Kss1 phosphorylate the Ste12 transcription factor and repressors of Ste12, called Dig1/Rst1 and Dig2/Rst2 (7–9). Fus3 but not Kss1 is also believed to phosphorylate the putative cyclin-dependent kinase (Cdk) inhibitor Far1 (35, 47). However, the identity of the relevant phosphorylation sites has not been reported for any of these substrates. The G1 cyclins Cln1, Cln2, and Cln3 are regulators of the yeast Cdc28 kinase necessary for progression from G1 to S phase (17, 30, 37, 38, 52). The deletion of all three G1 cyclins (9, 38), a temperature-sensitive Cdc28 kinase under nonpermissive conditions (36), as well as pheromone action, prevents the induction of all late G1-specific cell cycle events and the subsequent entry into S phase. Therefore, the pheromone response apparently occurs by counteracting the activity of the G1 cyclin-Cdc28 kinase complex. G1 cyclins are unstable proteins whose phosphorylation by Cdc28 is believed to induce its Cdc34- and Grr1-dependent degradation (1, 25). Alternatively, Cln2 degradation might also be regulated indirectly via the upregulation of mitotic Clb kinases (2). G1 cyclins were initially considered to be functionally redundant, since the activity of any single G1 cyclin is sufficient to promote cell cycle progression. This simplistic view, however, is complicated by the fact that the transcriptional induction of many G1-specific transcripts (including Cln1 and Cln2) depends on the Cln3-Cdc28 kinase complex (10, 45, 48). Once Cln3-dependent transcription leads to a threshold activity of Cln1 and Cln2, these cyclins cause the phosphorylation-induced degradation of the p40sic1 Cdk inhibitor (15, 39, 42, 50, 51). Upon p40sic1 degradation, the S-phase-promoting cyclins Clb5 and Clb6 become active and induce DNA replication (40). In addition to the mechanisms mentioned above, Cln1 and Cln2 are further implicated in the regulation of polarized growth and in the cell cycle-specific downregulation of the responsiveness of the pheromone response pathway (26, 33).
FAR genes (designated FAR1 for their role in mating factor arrest) were initially cloned as genes involved specifically in the pheromone-dependent G1 cell cycle arrest (3, 21). Accordingly, mutation of FAR genes does not affect many other pheromone responses unrelated to cell cycle arrest, such as the induction of pheromone-responsive genes or the promotion of morphological changes which are needed for mating (3, 21). Among the two known FAR genes, FAR1 is most extensively characterized. Besides the classical role of FAR1 in cell cycle arrest, recent data suggest that FAR1 is required for efficient mating as it is needed for the directed morphogenesis toward a mating partner (11, 49). However, the domains needed for these morphogenetic events and the G1 arrest function can be separated (49). The G1 arrest-promoting activity of Far1 resides within the first 393 amino acids (aa), whereas the domain effecting morphogenesis is located more C terminally (49). Therefore, the amino-terminal portion of Far1 can be treated as the sole and independent entity containing the cell cycle-inhibitory function.
Several mechanisms ensure that Far1 can exert its function only in G1. Most importantly, Far1 is quickly proteolyzed in all phases of the cell cycle outside G1. It is presumed that Cdc28 kinase plays a crucial role in this phenomenon perhaps by modifying Far1 and thereby targeting the protein for destruction (28, 29). Furthermore, FAR1 transcription is maximal between mitosis and early G1 and is induced approximately fivefold by pheromone (28, 29, 32). This elevated level of FAR1 transcription in response to pheromone is necessary but not sufficient for the G1 arrest function, because FAR1 overexpression itself is not sufficient to cause cell cycle arrest (4, 17a). Therefore, it was proposed that Far1 must be activated posttranslationally by pheromone-induced Fus3-dependent phosphorylation (4, 12, 13, 35, 47). Indeed, Fus3 is able to phosphorylate Far1 in in vitro kinase assays, and in vivo phosphorylation of Far1 depends on functional Fus3. The correlation between phosphorylation and function can be extended by the observation that only phosphorylated Far1 seems to become associated with the Cln2-Cdc28 kinase complex (12, 35, 47). Since bacterially produced Far1 acts as a dose-dependent inhibitor of Cln2 kinase activity in vitro, it was proposed that Far1 might act as a Cdk inhibitor of the Cln2-Cdc28 kinase complex (19, 34).
In summary, Far1 is subjected to both positive and negative posttranslational regulatory events. Far1 activation is needed for pheromone-dependent cell cycle arrest, whereas its cell cycle-specific destabilization might be part of a recovery mechanism. Far1 destabilization could be a protective measure against inappropriate inhibition of Cdc28 activity. Fus3 kinase and Cdc28 kinases were suggested as the most likely direct mediators of these effects. However, neither Fus3-dependent phosphorylation sites nor Cdc28-dependent sites within Far1 have been identified. To determine whether Far1-specific regulatory mechanisms involve phosphorylation events, we systematically mutated potential MAP and Cdk consensus phosphorylation sites. Our results implicate two amino acids in the control of Far1 behavior. Changing serine 87 to alanine slows the degradation of Far1. Correspondingly, phosphorylation at this site might serve as a signal for the destruction machinery. In contrast, threonine 306 is a major Fus3-dependent phosphorylation site involved in the activation process. An alanine substitution at this site diminishes G1 arrest and prevents the stable interaction between Far1 and the Cdc28-Cln2 Cdk complex. In contrast to our expectations, we find that although pheromone-induced Far1 binding to Cdc28-Cln2 is essential for G1 arrest, Far1 does not significantly inhibit Cdc28-Cln2 kinase activity in immunoprecipitation kinase assays. Therefore, the phosphorylation-induced binding of Far1 to Cln2 (as opposed to the inhibition of Cln2 kinase activity by Far1) might be part of the mechanism mediating pheromone-dependent cell cycle arrest.
MATERIALS AND METHODS
Plasmid constructions.
For cloning of FAR1, the region of FAR1 encoding aa 1 to 393 was PCR amplified by the primer pair FAR1-PCR11 (5′-AAA AGA ATT CAT ATG AAG ACA CCA ACA AG-3′) and FAR1-PCR10 (5′-TTC TGG ATC CTA CAC ACT GAC CAT AAC-3′), generating a fragment with an NdeI site at the start codon, an EcoRI site before the start codon, and a BamHI site after the introduced stop codon. This fragment was then cut with EcoRI and BamHI and cloned behind the triosephosphate isomerase promoter of pGA1840 (CEN plasmid with TRP1 selection marker [18]). For mutagenesis, this EcoRI-BamHI fragment was then transferred into M13mp18 and into M13mp19. The mutagenesis reactions were performed by single-strand mutagenesis according to a commercial protocol (Amersham). The mutants were confirmed by sequencing, and in case of the T306A mutant it was confirmed by sequencing that no other mutation was present. The mutant clones were then shuffled back to pGA1840, transformed into yeast, and assayed for biological activity. For the mutagenesis reactions, the following oligonucleotides were used: FAR1-Mut1 (S87A) (5′-GGT GGG GGA GCT ATA GGC TTG G-3′), FAR1-Mut2 (S114A) (5′-ATT TAC TGG CGC CTG TAA AGC GC-3′), FAR1-Mut5 (S341V,S346A) (5′-AGC TAT CTT GAC GTC CCA TTT TTA AAT GCA CCA TTC GTT AAT-3′), FAR1-Mut8 (S194A) (5′-GAA AGT TGG TGC ATC TTC TTC G-3′), FAR1-Mut9 (S213A) (5′-GTA CCT TTT TGG CGC GAA TTG TGA ATT G-3′), FAR1-Mut12 (T324A) (5′-GAT GGG TTC AGA GCT CCA CGC TTA TCG-3′), FAR1-Mut13 (T306A) (5′-CCT CAG TTT GCA CCA CAG GAG CAG-3′), FAR1-Mut14 (T63A) (5′-GAG AGT CAA TTT GCG CCA AAT CTT GG-3′), FAR1-Mut15 (T306Q) (5′-TTC CTC AGT TCG AAC CAC AGG AGC AG-3′), and FAR1-Mut16 (T306S) (5′-TTC CTC AGT TTA GCC CAC AGG AGC AG-3′).
For tagging FAR1 N terminally with hemagglutinin (HA) or Myc, a NotI site was introduced right after the start codon of FAR1 by PCR using the primer pair FAR1-PCR-T1 (5′-GCG GCG AAT TCC ATA TGG GCG GCC GCA AGA CAC CAA CAA GAG TTT CG-3′) and FAR1-PCR10. The resulting EcoRI-BamHI fragment was then transferred into pGA1840 as described above, and HA6 or Myc6 tags were cloned into the NotI site. Furthermore, to generate specific mutant combinations or to add either the HA6 or Myc6 tag, fragments were shuffled together via cloning employing the DraIII site right after the start codon, the BstBI site between the S87 and S114 substitutions, and the HindIII situated further 3′ terminally. For simplicity, only those FAR1-expressing plasmids needed in this study are listed: pGA2046 (FAR1N), pGA2084 (FAR1N-T306A), pGA2209 (FAR1N-Myc), pGA2210 (FAR1N-Myc-T306A), pGA2214 (FAR1N-HA), pGA2215 (FAR1N-HA-T306A), pGA2216 (FAR1N-HA-S87A,S114A,S341V,S346A), pGA2217 (FAR1N-HA6-S87A,S114A,T306A,S341V,S346A), pGA2218 (FAR1N-HA-S87A), pGA2219 (FAR1N-HA-S87A,T306A), pGA2220 (FAR1N-HA-stop codon at position 338), pGA2221 (FAR1N-HA-T63A), and pGA2222 (FAR1N-HA-T306S).
A plasmid containing the full-length FAR1 gene was constructed by amplifying the gene by PCR (relevant primer sequences, CTGCAGCGATGCAATTTTCAACATGC and GAGCTCTAGTGCATATATGACGAGAT) and subcloning the cut fragment into a PstI-SacI-cut YCplac22 derivative (which had its SpeI site destroyed). The resulting plasmid (pGA2273), which fully restored pheromone sensitivity to a far1Δ strain, was cut with BamHI and SpeI. Using FAR1 and far1N-T306A plasmids as the template, we generated and ligated PCR fragments that introduced an HpaI site at position 1045 of FAR1. They covered the region from SpeI to position 1048 or from position 1049 to the BamHI site. Both fragments were introduced into cut pGA2273 vector to give plasmids pGA2274 (FAR1) and pGA2275 (far1-T306A), respectively.
Plasmid pTP411 (pRS314 derivative, CEN plasmid with a TRP1 selection marker), allowing the expression of C-terminally HA3-tagged CLN2 from the CLN3 promoter, was obtained via Fred Cross. The CLN2 expression cassette was transferred to pRS315 (LEU selection) via restriction with SalI and SacII, generating pGA2238. To obtain Myc9-tagged CLN2, pGA2238 was cut with NotI to replace the HA tag with the Myc tag, generating pGA2236. A description of the disruption plasmids pGA1832, pGA1844, and pGA2002 can be obtained from G. Ammerer upon request. Plasmids used for the GAL promoter-driven expression of full-length FAR1 were JM306 (FAR1) and FC361 (FAR1-S87A). Details of their construction can be obtained from F. Cross upon request.
Immunoprecipitations, Western blotting, and kinase assays.
Cells from overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.2 and further grown in the appropriate medium to an OD600 of 0.8 to 1, and α-factor (αF) induction (1 μg/ml of culture) was done as indicated. For harvesting, 35-ml cell cultures were immediately chilled with crushed ice, centrifuged, and washed twice with 5 ml and then 1 ml of stop buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, 50 mM NaF). Cell pellets were dissolved in 300-μl of ice-cold lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1% Nonidet P-40 [NP-40], containing the following protease and phosphatase inhibitors: 60 mM β-glycerol phosphate, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 400 μM tosylsulfonyl phenylalanyl chloromethyl ketone 200 μM Nα-p-tosyl-l-lysine chloromethyl ketone, 40 μg of aprotinin per ml, 20 μg of leupeptin per ml, 5 mM EDTA, 5 mM EGTA, 1 mM dithothreitol, and 0.1 mM orthovanadate). Subsequently, cell lysis was effected by the addition of 2/3 volume of glass beads and two 5-min pulses of shaking. The extract was then clarified by centrifugation (5, 15, and 5 min) in a microcentrifuge, and protein concentrations were determined by the commercial Bio-Rad protein assay.
For immunoprecipitation, 400 μg of extract was diluted to 10 μg/μl, and 20 μl of hybridoma supernatants containing either anti-Myc 9E10 or anti-HA monoclonal antibodies was added and incubated on ice for 2 h. Subsequently, 20 μl of a 1/1 suspension of protein A beads was added, and the mixture was incubated on ice for an additional hour with occasional shaking. The absorbed immunocomplexes were then washed five times with lysis buffer containing 1/10 of the above inhibitors and solubilized by boiling for 3 min after the addition of 40 μl of 5× sample buffer. For kinase assays, washing was performed three times with lysis buffer and twice with kinase assay buffer (10 mM Tris [pH 7.5], 10 mM MgCl2). Kinase reactions were then performed for 20 min at 30°C in 15 μl of kinase assay buffer containing 1 μl of [γ-32P]ATP (6,000 Ci/mmol) and 10 μg of H1 (Boehringer Mannheim) or 1 μg of Far1 peptide (aa 51 to 301, purified as described by Peter et al. [35]) as the substrate; the reactions were stopped by boiling for 3 min in sample buffer. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% polyacrylamide gel containing acrylamide and N,N-bisbisacrylamide in a ratio of 40:1. For kinase assays gels were dried for 2 h and subsequently exposed to autoradiography for 4 to 24 h. Western blotting was done by transfer to nitrocellulose filters (Boehringer Mannheim) and according to the instructions for the enhanced chemiluminescence (ECL) blotting system (Amersham) with the exception of blocking with PBST–10% milk and antibody incubation and washing with PBST–4% milk. Primary antibodies were diluted 1:1,000 for anti-Far1 polyclonal antibodies and 1:100 and 1:50 for hybridoma supernatants containing HA and Myc antibodies, respectively.
(i) Kinase assay with full-length Far1.
Cells were in the BF264-15Dau (MATa bar1 ade1 his2 leu2 ura3 trp1) background. They contained a GAL1::FAR1 cassette integrated at LEU2 and were disrupted at the chromosomal far1 gene with URA3. The cells were transformed with vector or with plasmid KH100 (TRP1/ARS-CEN/GAL1::CLN2-HA). The expression level of Cln2 induced by this plasmid is approximately 5- to 10-fold above normal levels of Cln2. This level is insufficient to cause mating factor resistance, especially when FAR1 is overproduced from the GAL1 promoter. These strains are fully mating factor sensitive by replica plating assay. The cultures were grown in Trp-deficient synthetic complete medium containing galactose (SCGal-Trp) at 30°C. Mating factor was added to a final concentration of 0.5 μM when cultures were at an OD600 of approximately 0.5. Mating factor induction was done for 2.5 h.
(ii) Immunoprecipitation, histone H1 kinase activity assay, and immunoblot analysis.
Cells from 100 ml of culture were collected by filtration on a Millipore filter, rinsed off the filter in LSHN buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 10% glycerol), and centrifuged at 1,000 rpm. Before centrifugation, 1 ml from each 100-ml culture was saved for fixing. Fixing solution contained 0.74% formaldehyde in 1× PBS buffer. Cell pellets were resuspended in 1 ml of LSHN, transferred to a microcentrifuge tube, and pelleted. Washed cell pellets were resuspended in buffer N (39) (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 60 mM β-glycerophosphate, 0.1% NP-40) with protease inhibitors (10 μg of aprotinin per ml, 10 μg of pepstatin per ml, and 0.5 μM phenylmethylsulfonyl fluoride. Glass beads (425 to 600 μm in diameter) were added, and samples were vortexed in a Vortex Genie Sleeve. Vortexing was done twice for 3 min at 4°C. Cell lysates were centrifuged at 15,000 rpm for 2 min to remove cell debris. Cell lysates were immunoprecipitated with anti-HA monoclonal antibody 12CA5 (Babco) (5 μg in each reaction) for 1 h on ice. After centrifugation at 15,000 rpm for 2 min, immune complexes were adsorbed onto protein A-agarose (Repligen) by addition of 30 μl of slurry followed by 1-h rotation at 4°C. Immune complexes were washed with LSHNN buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 10% glycerol, 0.1% NP-40) three times, then washed with kinase buffer twice, and resuspended in kinase buffer (10 mM HEPES [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol). Immune complexes were incubated with 5 μM ATP, 2 μg of histone H1 (Boehringer), and [32P]ATP (10 μCi). Incubation was carried out at 30°C for 10 min. The reaction was stopped by adding sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 0.0025% bromophenol blue, 2% β-mercaptoethanol). Samples were denatured at 95°C for 5 min. Denatured samples were subjected to SDS-PAGE (12% gel). Gels were transferred to Hybond membranes by semidry blot transfer (Hoefer), and membranes were exposed to film for 30 to 60 min to detect kinase activity. Kinase assays were quantitated with a PhosphorImager.
Antibodies.
Affinity-purified rabbit antibodies against Far1 (Escherichia coli-purified peptide containing aa 50 to 301 [11]) were generated by using standard techniques (18). The anti-Myc (9E10) and anti-HA (12CA5) mouse monoclonal antibody-producing hybridomas are described elsewhere (14, 16).
PP2A treatment.
One hundred micrograms of the indicated extracts was diluted in lysis buffer to 100 μl, SDS was added to a final concentration of 1% and the extracts were heated to 95°C for 5 min. Then the SDS concentration was diluted to 0.1% by the addition of 1 ml of lysis buffer, and immunoprecipitation was carried out as described above except that the reaction mixtures were constantly mixed by rolling at 4°C. Washing was done twice with lysis buffer containing 1/10 of the standard inhibitor concentration and three times with protein phosphatase 2A (PP2A) buffer (50 mM Tris [pH 7.5], 0.1 mM CaCl2). The phosphatase reactions proceeded for 15 min at 30°C in 15 μl of PP2A buffer containing either 2 U of PP2A or 2 U of PP2A with 10 μM okadaic acid, as indicated. Reactions were stopped by adding okadaic acid to a concentration of 10 μM and boiling after the addition of sample buffer.
Pheromone response assays.
Halo plate assays were carried out as described previously (44). Briefly, 5 μl of exponentially growing cells at an OD600 of 0.5 was poured with 6 ml of 0.5% agarose (cooled to 42°C) onto Trp-deficient plates. Filter discs containing 2 and 0.4 μg of αF were placed on top of the solidified agarose, plates were incubated at 30°C, and photographs were taken after 2 days of incubation. For plate assays, 5-μl aliquots of yeast cultures at OD600 0.1, 0.02, and 0.005 were spotted on Trp-deficient plates containing the indicated αF concentrations. Photographs were taken after 2 days of incubation at 30°C.
Yeast strains.
Yeast strains used were GA250 (MATα cdc15 bar::URA3 trp1-1 leu2-3), GA341 (MATa bar1::HISG fus3-A kss1::URA3), GA289 (MATa bar1::HISG), GA299 (MATa bar1::HISG far1::URA3), GA305 (MATα bar1::HISG far1::HISG), GA314 (MATa bar1::HISG far1::HISG cln2::URA3), GA343 (MATa bar1::HISG kss1::URA3), GA161 (MATa bar1::HISG ste12::URA3), GA153 (MATa bar1::HISG fus3::URA3), GA224 (MATa bar1::HISG ste7::URA3), GA367 (MATa bar1::HISG, far1::HISG cln2::URA3), and K3446 (Matα cdc28-13 URA3). All GA strains except GA250 are derivatives from K1107, whose full genotype is MATa HMLa HMRa ho::lacZ ura3-52 ade2-1 can1-100 his3 leu2-3,112 trp1-1 met. GA250 and K3446 are K699 (W303) derivatives (MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL psi+).
RESULTS
The amino-terminal domain of Far1 (aa 1 to 393) is phosphorylated in response to pheromone.
In this study, we aimed to address the question of whether posttranslational modifications of Far1 are functionally relevant. To facilitate our analysis and to ensure that we measure only the effects of posttranslational regulatory mechanisms, we manipulated the experimental system as follows. (i) To exclude transcriptional effects (the FAR1 promoter is induced about fivefold in response to pheromone [4]), we cloned FAR1 behind the constitutive triosephosphate isomerase promoter, which drives expression as strongly as the pheromone-induced FAR1 promoter (reference 18 and data not shown). (ii) To limit our studies to a minimal number of potential phosphorylation sites, we concentrated on the smallest possible fragment of Far1 (encoding aa 1 to 393) which had previously been reported to be necessary and sufficient G1 arrest function as well as for cell cycle-specific degradation of Far1 (28, 35). We will refer to this N-terminal fragment of Far1 as Far1N throughout the text. (iii) To facilitate the detection and immunoprecipitation of Far1N, the protein was tagged at its N terminus with either a Myc6 or HA6 tag, generating Far1N-Myc or Far1N-HA, respectively (see Materials and Methods).
To ascertain that the different FAR1N constructs are functional for G1 arrest, the corresponding plasmids were transformed into a far1Δ strain and pheromone sensitivity was estimated by a halo plate assay. For a FAR1N-HA plasmid, the result is shown in Fig. 1A. Even though its sensitivity to pheromone does not quite match the sensitivity of a wild-type strain, the FAR1N-HA transformant is highly sensitive compared to a FAR1-deficient strain. To confirm that Far1N is modified in response to pheromone, Western blotting was performed with extracts from pheromone-induced and uninduced cells. As shown in Fig. 1B, Far1N migrates mostly as one homogeneous band under uninduced conditions. Shortly after pheromone exposure, a dramatic shift to a more complex pattern occurs. After 15 min, the slowest-migrating species has become the predominant form of Far1. At least four to five slower-migrating bands can now be resolved, suggesting that the protein becomes phosphorylated at multiple sites during pheromone induction. The mobility shifts must be the consequence of phosphorylation because they can be reversed by PP2A treatment in vitro (Fig. 1C). We therefore believe that the phosphorylation status of Far1N can be inferred to some degree by the migration pattern on SDS-acrylamide gels. Overall, the data are consistent with previous reports on the full-length protein (4, 12, 34). Phosphorylation is highly dependent on the activity of the pathway and on the function of Fus3 kinase (Fig. 1D). Phosphorylation of Far1N also is necessary for interaction with Cln2. Far1N from ste7 or kss1 fus3 mutants does not exhibit the pheromone-induced mobility shift and does not recruit Cln2 into a complex (Fig. 1D). On the other hand, the kinetics of phosphorylation are fast enough to be causally linked with cell cycle arrest. The data also emphasize that Far1 phosphorylation events are prevalent within the G1 arrest-promoting domain as would be expected if they are functionally important.
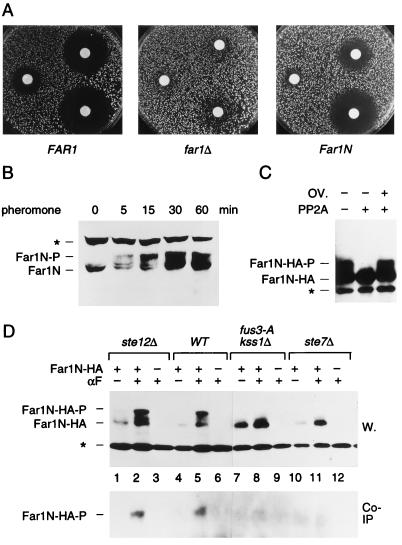
FIG. 1.
Far1N is phosphorylated in response to pheromone. (A) To measure pheromone sensitivity, halo assays were performed with FAR1 cells (K2149; left panel) and far1Δ cells (K2166) transformed with YCplac122 (middle panel) or pGA2046 (FAR1N; right panel). Pheromone concentrations on the filter discs were 1, 0.2, and 0.04 μg, respectively. Photographs were taken after incubation of plates at 30°C for 2 days. (B) To measure Far1N phosphorylation (as revealed by altered mobility), Western blotting was performed with Far1N-containing extracts. Cells were treated with αF for the specified times. Far1N-P indicates the phosphorylated form of Far1N. ∗ marks an unspecific band recognized by the polyclonal anti-Far1 antiserum. Cells used were far1Δ (K2166) transformed with FAR1N expressing pGA2042. (C) To measure the phosphatase sensitivity of Far1N-HA (pGA2214) from αF-treated cells, Far1N-HA was immunoprecipitated. The immunoprecipitate was then treated either with PP2A alone (lane 2) or with both PP2A and the phosphatase inhibitor okadaic acid (OV.). ∗ marks an unspecific band. (D) Far1N-HA-T306 phosphorylation depends on upstream components of the pheromone response pathway. Far1N-HA and Cln2-Myc containing extracts were prepared, and coprecipitation experiments were performed as described for Fig. 4A. The upper panel shows the detection of Far1N-HA in a Western blot (W.) of the total cell extract. In the lower panel, coprecipitated (Co-IP) Far1N-HA is shown. ∗ indicates an unspecific band. Strains used were ste12Δ (GA161; lanes 1 to 3), WT (GA314; lanes 4 to 6), fus3-A (a loss-of-function mutation of FUS3 that behaves like fus3Δ) kss1Δ (GA341; lanes 7 to 9), and ste7Δ (GA224; lanes 10 to 12).
Generation and analysis of putative phosphorylation site mutants.
It seemed reasonable to start with the assumption that Fus3 recognizes the generic MAP kinase consensus phosphorylation site (L/P)X(T/S)P (5). There are three SP sites between aa 1 and 393 that satisfy this sequence criterion. However, since even classical MAP kinase modifies S/T residues that do not contain such a defined sequence context, we decided to include all potential (S/T)P phosphorylation sites. Note that this includes both potential MAP kinase and Cdk phosphorylation sites. Because neither an N-terminal deletion of aa 1 to 50 nor the mutation of all four (S/T)P motifs within aa 1 to 50 affects the pheromone response, we mutated to APs only those nine (S/T)Ps that are situated between aa 51 and 393 (9a, 28). The position of the consensus sites is shown in Fig. 2A.
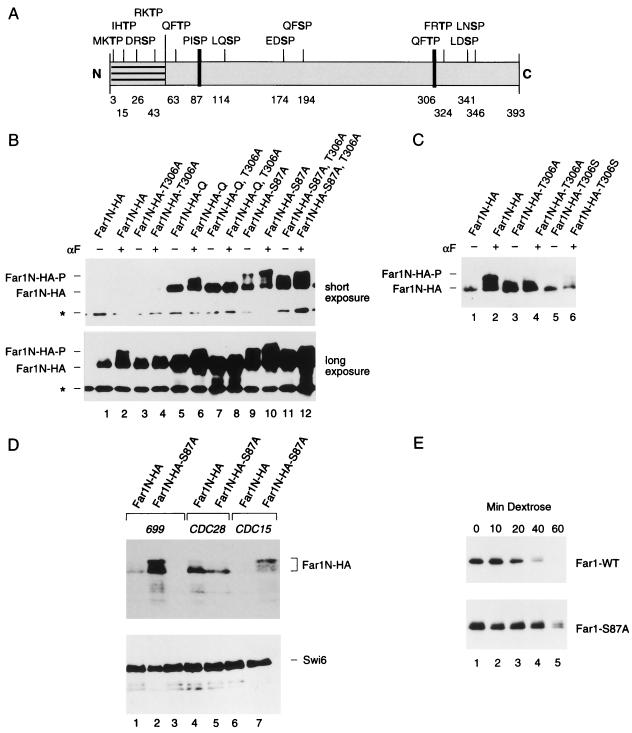
FIG. 2.
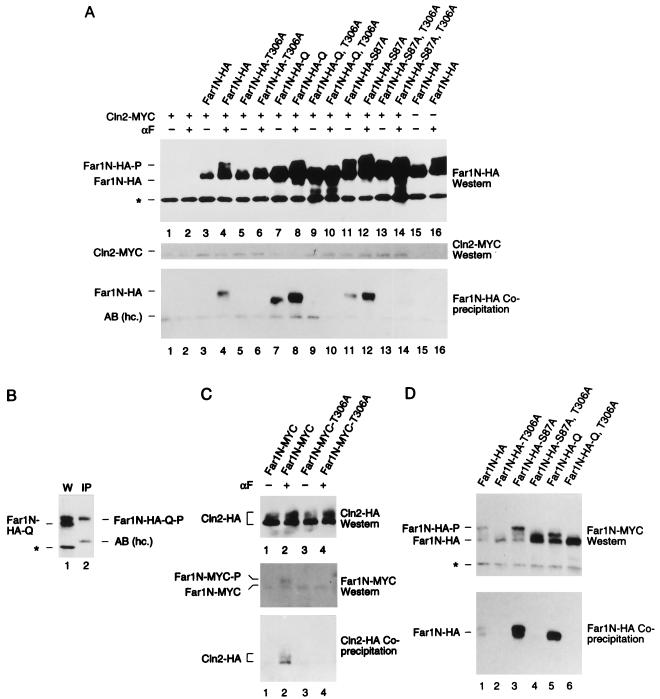
Mutational determination of potential Far1N phosphorylation sites. (A) Diagram showing all SP and TP sites within Far1N. Sites whose mutation leads to a biological phenotype are indicated by bold lines. (B) Western analysis of Far1N-HA substitution mutants. Cells exposed to αF (30 min) are indicated by +. Far1N-HA indicates the phosphorylated version of Far1N-HA. * marks an unspecific band which is recognized by HA antibodies and which was used as internal control to ensure equal loading. The upper and lower panels show short (1-min) and long (5-min) exposures of the Western blot. The following cells were analyzed. GA314 (Mata far1Δ cln2Δ) was transformed with a Cln2-Myc expression plasmid (pGA2236). Furthermore, this strain contained plasmids expressing FAR1N-HA (pGA2214; lanes 1 and 2), FAR1N-HA-T306A (pGA2215; lanes 3 and 4), FAR1N-HA-Q (pGA2216; lanes 5 and 6), FAR1N-HA-Q-T306A (pGA2217; lanes 7 and 8), FAR1N-HA-S87A (pGA2218; lanes 9 and 10), and FAR1N-HA-S87A-T306A (pGA2219) (lanes 11 and 12). “Q” in Far1N-HAQ and Far1N-HA-Q-T306A stands for quadruple substitution, which includes the substitutions S87A, S114A, S341V, and S346A. (C) Far1N-HA-T306S is phosphorylated in response to pheromone. Mata far1Δ cells (K2166) transformed with FAR1N-HA (pGA2214; lanes 1, 2, and 7), FAR1N-HA-T306A (pGA2215; lanes 3 and 4), and FAR1N-HA-T306S (lanes 5 and 6) were analyzed by Western blotting. αF induction is indicated by +. (D) S87 phosphorylation is CDC28 dependent and results in cell cycle-specific Far1 degradation. Extracts were prepared from cycling cells (lanes 1 and 2) and from G1 and M-arrested cells (lanes 4 and 5 and lanes 6 and 7, respectively). Strains used for the experiment were K699 (WT), GA250 (cdc15), and K3446 (cdc28-13). Cell cycle arrest was achieved via arresting cells by incubation at the restrictive temperature for 3 h, and uniform cell cycle arrest was confirmed by microscopic examination of cells. A Western blot to detect Far1-HA is shown in the upper panel. Equal loading was confirmed by Ponceau staining of the blot (data not shown). To assess the quality of the extracts, a parallel Western blot was prepared and probed with Swi6 antibodies. (E) Far1 S87A has a decreased turnover. Full-length FAR1 and FAR1-S87A were expressed in a cycling cell population from the GAL promoter by continuous galactose induction. Far1 (detected via anti-Far1 antibodies) is shown after inactivation of the GAL promoter as indicated. Plasmids used were JM306 (GAL-FAR1) and FC361 (GAL-FAR1-S87A).
To determine whether any of the mutations affect the phosphorylation pattern of Far1N-HA, we analyzed the migration patterns of the mutant products on SDS-gels. Of the single mutants which included substitutions at T63, S87, S114, S174, S194, T306, T324, S341, and S346, only the conversion of T306 to alanine caused a distinct reduction in the pheromone-induced mobility shift (Fig. 2B, lanes 3 and 4, and data not shown). Loss of pheromone-dependent mobility is observed both in the single T306A mutant and in the mutant containing all other substitutions analyzed (Fig. 2B, lanes 4, 8, and 12). To provide further evidence that mutating T306 to A affects a genuine pheromone-dependent phosphorylation site, T306 was changed to serine, which revealed that the pheromone-dependent mobility shift caused by T306 phosphorylation (which is lost in the T306-to-A substitution) is restored in the T306S substitution (Fig. 2C). Note that the stability of Far1 T306S is greatly reduced (Fig. 2C). This vast reduction of the steady-state level of Far1-306S is consistent with the finding that this version of Far1 is inactive (data not shown).
S87 is required for the cell cycle-specific regulation of Far1 turnover.
The Far1 S87A substitution leads to an enhanced steady-state level of Far1N-HA (Fig. 2B). This effect is solely dependent on the S87A mutant and is not further enhanced either in the S87A,S114A,S341V,S346A quadruple mutant or in a sextuple mutant that also includes S174A and S194A substitutions (Fig. 2B and data not shown). The enhanced S87A substitution-dependent steady-state level of Far1N-HA does not alleviate pheromone-dependent, T306 phosphorylation-mediated mobility shifts either as a single mutant or as part of the quadruple mutant (for details of the S87A single substitution, see below).
Since we observe that the mobility of the S87A,S114A,S341V,S346A quadruple mutant is reduced in response to pheromone (compared to the wild-type [WT] protein), either S114, S341, or S346, alone or in combination, might be phosphorylated in response to pheromone (Fig. 2B). In addition to the consensus phosphorylation site mutants described above, we also analyzed the T63A and the T324A substitutions, which did not have a discernible effect on Far1N-HA mobility (data not shown). We noted that a large proportion of Far1N-HA-S87A has a reduced mobility, even in the absence of pheromone (Fig. 2B; compare lane 1 in the lower panel to lane 9 in the upper panel). It was previously reported that Far1 becomes phosphorylated, in a CDC28-dependent way, immediately prior to its degradation at the end of G1 (36, 37). Consistently, we find hyperphosphorylated Far1N-HA (from cycling cells) in the absence of pheromone after prolonged exposures of Western blots (data not shown). Since the prevalence of pheromone-independent hyperphosphorylation of Far1N-HA-S87A could be explained by its failure to become degraded at the end of G1, we wondered whether S87 phosphorylation might be part of the mechanism that ensures cell cycle-specific degradation of Far1. To address this hypothesis, we transformed Far1N-HA and Far1N-HA-S87A expression plasmids into strains containing temperature-sensitive mutations of CDC28 and CDC15, to arrest cells in G1 and mitosis, respectively. Consistent with the data shown in Fig. 2B, the S87A substitution leads to an increased steady-state level and to an enhanced level of the low mobility of Far1 in cycling cells (Fig. 2D; compare lanes 1 and 2). In G1-arrested cells (CDC28-13 arrest), however, both Far1N-HA and Far1N-HA-S87A are about equally abundant and neither protein is phosphorylated. However, in M-phase arrest (CDC15-1), the steady-state level of Far1N-HA is much lower than that of Far1N-HA-S87A, which accumulates in its hyperphosphorylated form (Fig. 2D, lanes 6 and 7). The above findings are consistent with the requirement of CDC28 for Far1 phosphorylation at the end of G1 (28, 29) and with the notion that S87 phosphorylation might be needed for Far1 degradation (Fig. 2D, lanes 4 and 5). To determine whether the increased steady-state level of Far1N-HA-S87A that we observed is the result of a decreased turnover of the protein, we determined its biological half-life by measuring the time course of the depletion of Far1 after shutting off its expression from the GAL promoter. These experiments reveal that the half-life of Far1N-S87A is about twice as long as that of the WT protein (Fig. 2E).
In short, we have shown that two substitutions, S87A and T306A, cause major changes in the behavior of Far1N. We could not confirm these phosphorylation sites by conventional peptide mapping techniques since only a very small, almost undetectable, portion of Far1N is phosphorylated in response to pheromone if cells are grown in phosphate-depleted medium needed for 32Pi labeling (data not shown). T306 is likely to be phosphorylated because we observe an increased mobility of Far1N-HA-T306A, which is reversed in the T306A substitution. Evidence of S87 phosphorylation is only implied by the phenotype that we observe in the S87A substitution mutant. Since the S87A substitution does not directly cause a mobility shift of the protein, we assume that S87 modification-dependent phosphorylation events might cause the cell cycle-specific phosphorylation shift of Far1 that precedes its degradation. Since the stabilized quadruple mutant that contains besides S87A the S114A, S341V, and S345A substitutions is not hyperphosphorylated in the absence of pheromone, in contrast to the S87A single mutant, at least one of the three additional sites of the quadruple mutant seems to be phosphorylated concomitantly with the degradation of Far1 (Fig. 2B). Our data concerning S87 phosphorylation are in accordance with a recent publication (19).
Effects of the mutants on pheromone sensitivity.
To analyze the physiological consequences of the mutations, we used a far1Δ strain transformed with the different FAR1N-HA constructs. Pheromone sensitivity was compared by the halo plate assay (data not shown) and by spotting cells directly onto αF-containing plates (Fig. 3A). In addition, we determined changes in budding index (Fig. 3B) and DNA content by fluorescence-activated cell sorting analysis (data not shown). In all assays, we observed that the T306A substitution causes pheromone resistance to an extent which is comparable to a complete loss of FAR1 function (Fig. 3A, lanes 2 and 7). The phenotypic analysis also reveals that the S87A substitution shows an enhanced sensitivity to pheromone compared to WT (Fig. 3A, lane 3 and 4). This enhanced pheromone sensitivity, which can be explained by the increased steady-state level of the S87A single mutant, is alleviated by the additional S114A, S341V, and S346A substitutions. Accordingly, phosphorylation at one or more of these sites might downregulate pheromone sensitivity. Concerning the S87A substitution, it should be noted that an S87P substitution leads, if overexpressed from the GAL promoter, to a dominant pheromone-independent activation of Far1, resulting in cell cycle arrest (19, 34). Since we observe that the pheromone sensitivity of the S87A,T306A double substitution is considerably less than that of the S87A substitution, we conclude that T306 phosphorylation is still needed for Far1 activation (Fig. 3). The reported pheromone-independent activation of S87P could therefore be the consequence of the vast overexpression of an already stabilized protein. Alternatively, the S87P substitution might cause an unphysiological conformational change within Far1 that locks the protein in a “pseudoactive” conformation. Since T306, the mutation of which causes pheromone resistance, is embedded within a QFTP motif also found around T63 and S194, we wondered whether substitutions at these sites would cause pheromone resistance. Since Far1N-HA-T63A is partially pheromone resistant, this site might also be phosphorylated in response to pheromone (data not shown). Unfortunately, we could not assess phosphorylation at these sites directly, as the single T63A substitution does not cause any discernible change in the mobility of Far1N-HA-T63A in pheromone-treated cells (data not shown).
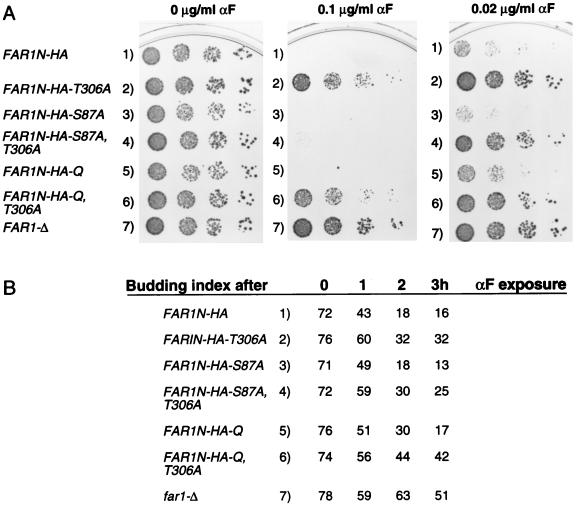
FIG. 3.
Pheromone sensitivity of Far1N-HA substitution mutants. (A) Plate assay. Various FAR1N-HA-expressing plasmids were transformed into a far1Δ strain (K21669), and pheromone sensitivity was measured by spotting a serial dilution of cells onto plates containing the indicated concentrations of αF. Plasmids used were pGA2214 (FAR1N-HA), pGA2215 (FAR1N-HA-T306A), pGA2218 (FAR1N-HA-S87A), pGA2219 (FAR1N-HA-S87A,T306A), pGA2216 2216 (FAR1N-HA-Q), and pGA2217 (FAR1N-HA-Q-T306A). Q in FAR1N-HA-Q indicates quadruple substitution, which encompasses S87A, S114A, S341V, and S346A. Photographs were taken after 2 days of incubation at 30°C. (B) Budding index of far1Δ cells transformed with various FAR1N-HA-expressing plasmids (see above) after 0, 1, 2, and 3 h of αF treatment.
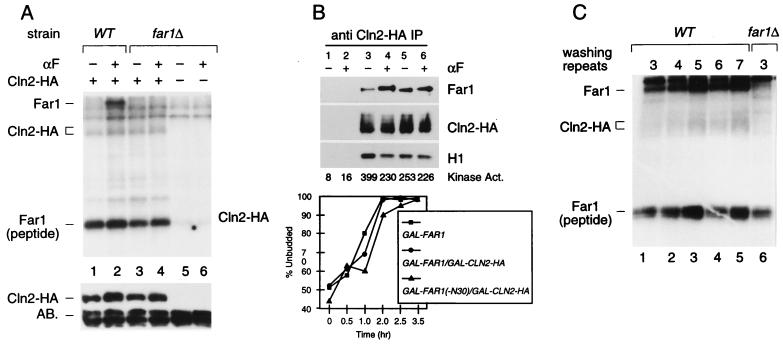
Since we analyzed pheromone resistance caused by the T306A mutation only in the N-terminal truncation version of Far1, we wished to confirm that the T306 mutation also results in a loss of function phenotype if the mutation is introduced into full-length Far1 driven by its own promoter. As shown in Fig. 4B, Far1-T306A indeed causes pheromone resistance. Moreover, a Western blot indicates that Far1-T306A accumulates to steady-state levels comparable to WT levels and that the pheromone-induced phosphorylation (as indicated by a mobility shift) of Far1-T306A is reduced compared to WT (Fig. 4A).
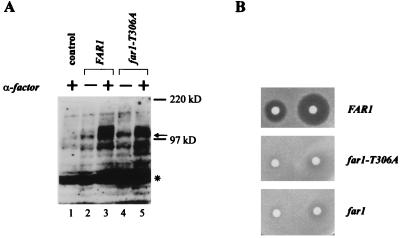
FIG. 4.
The cell cycle arrest function of Far1 is abolished in full-length Far1-T306A as in Far1N-HA-T306A. (A) Far1 and Far1-T306A phosphorylation was detected by Western blotting of total-cell extract. Cells used were far1Δ (GA305) transformed with YCPlac22 (lane 1) or FAR1 (pGA2274; lanes 2 and 3) and far1-T306A (pGA2275; lanes 4 and 5) plasmids. Cells were treated with αF (1 μg/ml) for 15 min (lanes 1, 3, and 5). The arrow indicates Far1 protein. Note that the pheromone-dependent shift (as indicated by the broad signal) of Far1 is much more prominent than in Far1-T306A. * marks an unspecific band recognized by the polyclonal Far1 antiserum. The band seen below the 97-kDa marker seems to be a degradation product of Far1. (B) Pheromone sensitivity was measured by halo assay with a far1Δ strain (GA305) transformed with YCPlac22 (lower panel), far1-T306A (pGA2275; middle panel), and FAR1 (pGA2274; upper panel) plasmids. Pheromone concentrations on filter discs were 0.3 and 1 μg. Photos were taken after incubation of the plates at 30°C for 2 days.
Pheromone-induced T306 phosphorylation is required for the pheromone-dependent interaction of Far1N-HA with Cln2-Myc.
Previous studies indicated that Far1 stabilizes its association with the Cln2-Cdc28 kinase in response to pheromone (12, 35, 47). We therefore wanted to determine whether T306 substitutions influence this interaction. To exclude any effects related to the transcriptional regulation of CLN2, we used in all subsequent experiments a construct allowing the weak constitutive expression of Myc epitope-tagged CLN2 from the pheromone-unaffected CLN3 promoter. Expression of CLN2 from the CLN3 promoter is approximately as strong as from its own promoter in cycling cells and does not affect pheromone sensitivity (data not shown). To perform coprecipitation experiments, Cln2-Myc and Far1N-HA were expressed in a far1Δ cln2Δ strain. The precipitation of Cln2-Myc was effected with anti-Myc antibodies, and the amount of coprecipitated Far1N-HA was measured by Western blotting with HA antibodies (Fig. 5A). From this experiment, we concluded that pheromone leads to the specific coprecipitation of the T306-phosphorylated form of Far1N-HA (Fig. 5A, lanes 3, 4, 7, 8, 11, and 12; Fig. 5B). The amount of Far1N-HA coprecipitated is elevated if the steady-state level of Far1N-HA is increased due to the S87A substitution (Fig. 5A). Importantly, however, we do not observe coimmunoprecipitation of Far1N-HA-T306A substitution mutants, either as single mutant or in combination with other substitutions. The finding that the Far1N-HA-T306A substitution mutant which does not stably bind Cdc28 in response to pheromone does not accumulate to higher steady-state levels indicates that it can still be modified on the S87A site to induce its degradation. The observed coprecipitation of residual amounts of Far1N-HA-S87A or Far1N-HA-Q in the absence of pheromone can be explained by their increased steady-state levels and by the basal level of signaling that occurs even in the absence of pheromone (Fig. 5A, lanes 7 and 11). To confirm these above interactions, we performed the reciprocal experiment and precipitated Far1N-Myc and measuring coprecipitated Cln2-HA by Western blotting (Fig. 5C).
FIG. 5.
Pheromone-induced Far1N-HA-T306 phosphorylation is required for the pheromone-dependent interaction of Far1N-HA with Cln2-Myc. (A) Coprecipitation of Far1N-HA derivatives with Cln2-Myc. Total extracts from the indicated cells (described in Fig. 2) were generated. Far1N-HA derivatives (from the total-cell extracts) were analyzed by Western blotting (upper panel). After immunoprecipitation of Cln2-Myc by monoclonal anti-Myc antibodies, Cln2-Myc (middle panel) and coprecipitated Far1-HA derivatives (lower panel) were detected by Western blotting. αF induction and the presence of Cln2-Myc are indicated by +. AB (hc.), antibody (heavy chain). (B) Only phosphorylated Far1-HA coprecipitates with Cln2-Myc. Western blotting and immunoprecipitation were performed as for panel A, and Western extract and an immunoprecipitate were loaded on a single gel next to each other as indicated. (C) Coprecipitation of Cln2-HA with Far1N-Myc. Cln2-HA from the total extract was analyzed by Western blotting (top panel) after immunoprecipitation of Far1N-Myc. Both Far1N-Myc (middle panel) and coprecipitated Cln2-HA were detected by Western blotting. For the experiment, we used GA314 (far1Δ cln2Δ) cells which were transformed with pGA2238 (expressing Cln2-HA) and with pGA2209 and pGA2210 (expressing Far1N-Myc [lanes 1 and 2] and Far1N-Myc-T306A [lanes 3 and 4], respectively). (D) Coprecipitation of Far1N-HA derivatives in a FAR1 (GA367) background. The precipitation was performed as for panel A except that all cells were treated with αF for 1 h. Uniform, αF-dependent cell cycle arrest was monitored after further incubating cells for 1.5 h with αF, and cells were fixed for documentation. The upper panel shows the detection of Far1N-HA derivatives from total-cell extracts. In the lower panel, coprecipitated Far1N-HA derivatives were detected after immunoprecipitation of Cln2-Myc.
To ascertain that the failure of Far1N to bind to Cln2 is the cause rather than the consequence of the failure to arrest in G1, we expressed the indicated Far1N-HA alleles in a strain containing a functional chromosomal copy of FAR1. This arrangement allows pheromone-dependent cell cycle arrest irrespective of whether a cell contains functionally compromised Far1N molecules. As expected, Far1N-HA but not Far1N-HA-T306A (which is not phosphorylated in response to pheromone) interacts with Cln2-Myc (Fig. 5D).
Far1N phosphorylation-induced binding to Cdc28-Cln2 leads to pheromone arrest but does not significantly inhibit Cdc28-Cln2 kinase activity.
Previous reports indicated that Cdc28 kinase activity is reduced in pheromone-treated cells. This reduction was attributed to the direct inhibition of kinase activity due to the binding of Far1 to the Cln2-Cdc28 kinase. Previous measurements of Cln2 kinase activity, however, were hampered by the fact that they were based on assays in which CLN2 was expressed either from its own pheromone-repressed promoter or from the ADH promoter, which leads to vast overexpression (34, 47). To overcome these constraints, we expressed HA-tagged Cln2 from the weak pheromone-unaffected CLN3 promoter and assayed for kinase activity by using either a small N-terminal fragment of Far1 (aa 51 to 297) or H1 as the substrate (Fig. 6A [compare lanes 1 and 5] and data not shown). Surprisingly, we do not detect any reduction of Cln2-HA kinase activity when Cln2-HA is immunoprecipitated from pheromone-induced cells, although in response to pheromone, Far1 is coimmunoprecipitated with Cln2-HA–Cdc28 and is phosphorylated in these immune complexes, presumably by Cln2-HA–Cdc28. Furthermore, kinase activity is not affected when Cln2-HA is precipitated from far1Δ cells.
FIG. 6.
Specific Cln2-HA kinase activity is not significantly affected by pheromone-dependent Far1 association. (A) Cln2-HA kinase activity was measured after immunoprecipitation with anti-HA antibodies (upper panel). Far1 indicates coprecipitated Far1 which is phosphorylated by Cln2-HA. The substrate for the kinase reaction was a bacterially produced Far1 peptide encoding aa 51 to 301 (5). The same results were obtained if H1 was used as a substrate or if the extraction buffer described by Peter and Herskowitz (34) was used (data not shown). Equal precipitation of Cln2-HA was ensured by Western blotting of the immunoprecipitate. AB., antibody (heavy chain). The strains used for the experiment were K2149 (WT) and K2166 (far1Δ). Both strains were transformed with pTP411, which allows the expression of HA-tagged CLN2 from the CLN3 promoter. (B) GAL1::FAR1 (lanes 1 and 2), GAL1::FAR1 GAL1::CLN2-HA (lanes 3 and 4), and GAL1::FAR1Δ30 GAL1::CLN2-HA (lanes 5 and 6) strains were grown in SCGal-Trp medium and incubated in mating factor or left untreated for 2.5 h. Cln2-HA was immunoprecipitated, and immunoprecipitates assayed for histone H1 kinase activity. In parallel, precipitated Cln2-HA protein and associated Far1 protein were determined by Western blotting. The percentage of unbudded cells in mating factor-treated cultures is indicated in the lower panel. Note that effective cell cycle arrest continued for at least 1 h after the time at which Cln2-associated kinase and Far1 binding were determined. (C) The association between Far1 and Cln2-HA is unaffected by repeated washing of the immunoprecipitate. Kinase assays were performed as described for panel A except that washing of the immunoprecipitate was repeated as indicated.
The finding that Cln2-HA–Cdc28 kinase activity is unaffected by pheromone treatment was unexpected; we therefore carried out a second independent experiment in a different lab (that of F. Cross). We analyzed the interaction of Cln2-HA with untagged Far1. Both proteins were expressed from a galactose-inducible promoter which is not affected by pheromone treatment. Pheromone response was checked by counting the budding index in response to pheromone (Fig. 6B, lower panel). As expected, pheromone leads to an increased coprecipitation of Far1 whereas the Cln2 kinase activity toward H1 is only slightly affected by pheromone treatment (Fig. 6B). Similar results were obtained by analyzing Far1Δ30, a stabilized version of Far1 due to the deletion of the N-terminal 30 amino acids (Fig. 6B, lanes 5 and 6) (36). In accordance with Fig. 4A, an increased steady-state level of Far1 as caused by the Far1Δ30 substitution in addition to its overexpression from the GAL1-10 promoter leads to a considerable amount of Far1 coprecipitation in the absence of pheromone (Fig. 6B, lanes 5 and 6).
To further corroborate the finding that specific Cln2-HA kinase activity is not significantly affected by pheromone, we confirmed that the kinase activity that we observe in pheromone-induced cells is not an artifact resulting from the partial loss of Far1 binding to the Cln2-HA–Cdc28 kinase during the immunoprecipitation procedure. Therefore, we performed a standard immunoprecipitation of Cln2-HA from pheromone-induced cells and compared both Cln2-HA kinase activity and coprecipitated Far1 after repeating washing the immunocomplexes as indicated (Fig. 6C). Since we always observe a constant amount of coprecipitated Far1- and Cln2-HA-dependent kinase activity, we conclude that our washing conditions do not lead to a dissociation of Far1 from the Cln2-HA–Cdc28 complex. As a result, this experiment is in accordance with the notion that our kinase measurements reflect the true physiological situation. In summary, the Cln2-HA kinase assays indicate that the specific Cln2-HA kinase activity does not change significantly in response to pheromone, although Far1 is specifically bound to Cln2-HA–Cdc28 in pheromone-induced cells.
DISCUSSION
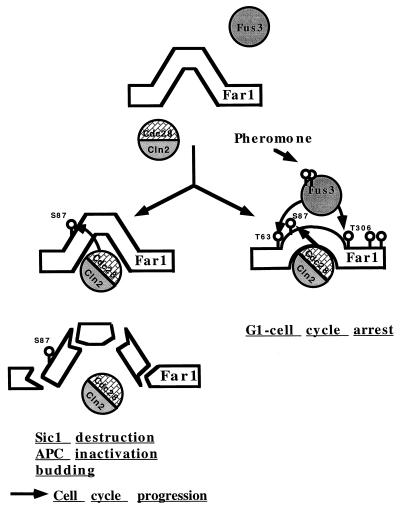
In pheromone-responding yeast cells, Far1 is needed to prevent the initiation of cell division. This halt of G1/S progression has been proposed to rely on the direct interaction between Far1 and G1 cyclin kinase complexes for which the accumulation of Far1 coupled with an increased affinity for the kinase seem to be a prerequisite. The pheromone-induced MAP kinase Fus3 seems to play a crucial role in the Far1 activation step. In contrast, to allow reentry into the cell cycle after pheromone arrest or to protect against inappropriate cell cycle attenuation, the interaction between Far1 and Cdc28 kinases needs to be broken or prevented. It has been speculated that a cell might accomplish this via Cdc28-induced degradation of Far1. Considering these two antagonistic influences on Far1 function, the system can be imagined to resemble a tug of war between a pheromone-induced event opposing Cdc28-Cln function and a Cdc28-induced event counteracting Far1 function (Fig. 7). From many correlative observations, it has been speculated that direct phosphorylation of Far1 might be at the heart of both phenomena. Even though our studies do not provide a final answer, they clearly support such a view by highlighting the existence of putative phosphorylation sites within Far1. We suggest that Far1 activation follows from the modification of position 306 whereas position 87 provides the key for its destruction (Fig. 7). It is not a trivial undertaking to provide a definitive biochemical proof for a protein phosphorylation event. With respect to Far1, the problem is compounded by the fact that Far1 interacts with several protein kinases which exhibit closely related specificities for the phosphorylation motif and whose substrate interactions might be interdependent. This situation made in vitro observations almost worthless because many phosphorylation sites (e.g., S87) seemed to be recognized by both Fus3 and Cdc28 kinases (17a). A biochemical characterization of in vivo-modified Far1 also proved to be an unattractive option. First, the protein is highly unstable and easily degraded during prolonged extraction procedures. Second, under closer scrutiny, we found that the procedures commonly used for radioactive labeling in yeast influence the modification event under study (data not shown). Therefore, a genetic analysis seemed to be the best way to test the current models since they predicted that Far1 should be modified at SP and/or TP motifs. If any of the projected phosphorylation events were causally involved in either activation or degradation, then certain alanine substitutions would have to exhibit an appropriate phenotype. This is exactly what we found. Of the consensus phosphorylation site mutants that we generated and analyzed, three (S63A, S87A, and T306A) clearly supported the notion that Far1 phosphorylation could have a direct role in activation as well as destruction. Of these, T306 seems to be the prime site involved in the activation event, as even the single mutant has a remarkably strong loss of function phenotype.
FIG. 7.
Far1 is regulated by positive and negative phosphorylation events. Pheromone treatment results in the activation of the Fus3 kinase, which in turn causes the phosphorylation of Far1 at T306 and possibly at T63 (left diagram). This phosphorylation event is necessary for the binding of Far1 to the Cln2-Cdc28 kinase. This interaction results into the functional inactivation of the Cln2-Cdc28 cell cycle-promoting activity and results into a G1 cell cycle arrest (right diagram). The detailed molecular consequences of the Far1 Cln2-Cdc28 interaction will be the subject of further studies. Cdc28-dependent S87 phosphorylation counteracts Far1 activation by targeting the protein for destruction (left diagram). As a consequence, negative regulation on the Cln2-Cdc28 complex is alleviated and critical phosphorylation events needed for cell cycle progression can be executed.
The observed stabilization of the Far1 S87A mutant raises the issue of whether phosphorylation at this site by Cdc28 precipitates Far1 degradation. Previous reports indicated that the first 50 aa are important for the cell cycle-specific degradation of Far1; however, mutating all possible consensus phosphorylation sites within the amino-terminal 50 aa had no discernible effect (9b, 29). Since an extensive deletion analysis within the N-terminal 50 aa also failed to delineate unique sequences specifically required for stabilization (9a), modifications within this region may not be what is important for its function. The effects of deletions in this region could be entirely indirect, perhaps due to affecting protein context or localization. In summary, S87 is the only serine/threonine within any putative Cdc28 recognition motif that leads to the expected stabilization phenotype.
It is generally assumed that antiproliferative factors cause cell cycle arrest via the inhibition of Cdk activity. Even though Far1 does not show significant sequence similarity to any of the mammalian inhibitors or to the bona fide yeast Cdk inhibitors such as Sic1 or Rum1 (8, 40), it was not unreasonable to expect that Far1 also functions as a stoichiometric Cdk inhibitor. Indeed, the finding that the addition of Far1 to Cln2-Cdc28 kinase complexes in vitro causes a dose-dependent reduction of kinase activity supported this proposal (34). Moreover, recent evidence suggests that Cdc28-Cln3 kinase activity is downregulated by pheromone (24). The data indicating Cln2 inhibition were further corroborated by showing that Cln2-Cdc28 kinase when highly overexpressed from the ADH promoter is inhibited about four- to fivefold in response to pheromone. Therefore, it has been a surprise to us that the specific kinase activity of Cln2 as measured in our in vitro kinase assays is not significantly reduced in response to pheromone. How could these apparently conflicting data be reconciled? It is certainly difficult to find conditions where pheromone-responding and nonresponding cells can be properly compared and where posttranscriptional and transcriptional effects can be dissected. Of course, the opportunity for obtaining artifacts due to the necessary ectopic or artificially high expression is not negligible. For example, even in the mammalian system it was shown that the CDK-p21 complex can exist in both active and inactive states, depending on the relative concentrations of the proteins (22, 41). Also, since it is evident that Far1 is able to associate with the Cln2 kinase, it is not surprising that the addition of a high excess of purified Far1 to Cln2-Cdc28 kinase is able to lower kinase activity toward a second substrate. Differences in the results may therefore just reflect differences in the experimental setup. In any case, even if the Cln2-Cdc28 kinase was repressed in cells that overexpressed CLN2 from the ADH promoter, the absolute levels of kinase activity were well within the levels found in normal unchallenged WT cells (34). Thus, we believe that the inhibition of Cln2 kinase alone is unlikely to provide a functional explanation for the arrest in vivo. In this regard, our observations are not without precedent. It has also been shown by others that immunoprecipitated Cln2 complexes from pheromone-induced cells indeed show some level of kinase activity toward histone H1 (47). Even more convincing evidence that Far1 binding may not cause inhibition of catalytic activity per se stems from immunoprecipitation kinase assays with Far1 (35). These assays revealed that Far1 immunoprecipitates from pheromone-induced cells contain Cdc28-dependent kinase activity. If not by inhibition of catalytic activity, how then might Far1 prevent the function of Cdc28-Cln2? Far1 could obscure the interaction of Cln2 with some selected target sites or substrates. Alternatively, Far1 might restrict Cln2 to subcellular compartments where the kinase cannot promote its cell cycle-specific function. Testing of these ideas might be possible in studies oriented more cell biologically.
ACKNOWLEDGMENTS
We are grateful to members of the Nasmyth and Ammerer labs for helpful discussions. Martin Tögel’s help was instrumental for culturing hybridoma cells. We thank Knud Naiz and Alexander Schleiffer for comments. We are especially grateful to Brandt Schneider and Bruce Futcher for advice and for instrumental help in writing the manuscript and to the members of the CSHL graphics facility for helping with the artwork.
We thank Michael Hengartner for paying for the artwork. This study was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung, by the Austrian Jubiläumsfonds der Nationalbank (grants 5760 and 6431), by TMR Network grant ERB 4061 PL95-0662, and by NIH grant GM 49716 to F.C.
REFERENCES
- 1.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 2.Blondel M, Mann C. G2 cyclins are required for the degradation of G1 cyclins in yeast. Nature. 1996;384:279–281. doi: 10.1038/384279a0. [DOI] [PubMed] [Google Scholar]
- 3.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 4.Chang F, Herskowitz I. Phosphorylation of Far1 in response to α-factor: a possible requirement for cell cycle arrest. Mol Biol Cell. 1992;3:445–450. doi: 10.1091/mbc.3.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark-Lewis I, Sanghera J S, Pelech S L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 6.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signaling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 7.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 8.Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- 9.Cross F. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START I arrest and is independent of the mating pheromone signal. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cross, F. Unpublished data.
- 9b.Cross, F., and J. McKinney. Unpublished data.
- 10.Dirick L, Böhm T, Nasmyth K. Roles and regulation of the Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorer R, Pryciak P M, Hartwell L H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J Cell Biol. 1996;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elion E A, Satterberg B, Kranz J R. Fus3 phosphorylates multiple components of the mating signal transduction cascade: evidence for Ste12 and Far1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Errede B, Gartner A, Zhaoqing Z, Nasmyth K, Ammerer G. A pheromone induced kinase cascade in S. cerevisiae: activation of the Fus3 kinase by the Ste7 kinase in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 14.Evan G L, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman R M, Correll C C, Kaplan K B, Deshais R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 16.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I, Lerner I, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2149–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futcher B A. Saccharomyces cerevisiae cell cycle: cdc28 and the G1 cyclins. Semin Cell Biol. 1991;2:205–212. [PubMed] [Google Scholar]
- 17a.Gartner, A. Unpublished data.
- 17b.Gartner, A., and G. Ammerer. Unpublished data.
- 18.Gartner A, Nasmyth K, Ammerer G. Signal transduction in S. cerevisiae requires threonine and tyrosine phosphorylation of Fus3 and Kss1. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 19.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 21.Horecka J, Sprague G F., Jr Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics. 1996;144:905–921. doi: 10.1093/genetics/144.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui Z, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 23.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 24.Jeoung D-I, Oehlen L J W M, Cross F R. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 26.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 28.McKinney J D, Cross F R. FAR1 and the G1 phase specificity of cell cycle arrest by mating factor in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2509–2516. doi: 10.1128/mcb.15.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinney J D, Chang F, Heintz N, Cross F R. Negative regulation of FAR1 at the start of the yeast cell cycle. Genes Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- 30.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 31.Neiman A, Herskowitz I. Regulation of a yeast protein kinase cascade, in vitro: activation of the yeast MEK homologue Ste7 by Ste11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oehlen L J, McKinney J R, Cross F R. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oehlen L J, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at START in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 34.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 35.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. Far1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 36.Reed S I. The selection of S. cerevisiae mutants defective in the START event of cell division. Genetics. 1980;95:561–567. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed S I. The role of p34 kinases in the G1 to S-phase transition. Annu Rev Cell Biol. 1992;8:529–561. doi: 10.1146/annurev.cb.08.110192.002525. [DOI] [PubMed] [Google Scholar]
- 38.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 39.Schneider B L, Yang Q F, Futcher A B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 40.Schwob E, Böhm T, Mendelhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40sic1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 41.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 42.Skowyra D, Craig K I, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 43.Song O K, Dolan J W, Yuan Y I, Fiels S. Pheromone dependent phosphorylation of the yeast Ste12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 44.Sprague G F. Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- 45.Stuart D, Wittenberg C. CLN3, not positive feedback determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 46.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by the two MAP kinase substrates Rst1 and Rst2. Curr Biol. 1997;7:22–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 47.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces G1 cyclins: Cln3 may be an upstream activator Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of sic1p by G1 cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 51.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 52.Wittenberg C, Sugimoto K, Reed S I. G1 specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34cdc28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]