Abstract
Suppression of tumor cell growth by p53 results from the activation of both apoptosis and cell cycle arrest, functions which have been shown to be separable activities of p53. We have characterized a series of p53 mutants with amino acid substitutions at residue 175 and show that these mutants fall into one of three classes: class I, which is essentially wild type for apoptotic and cell cycle arrest functions; class II, which retains cell cycle arrest activity but is impaired in the induction of apoptosis; and class III, which is defective in both activities. Several residue 175 mutants which retain cell cycle arrest function have been detected in cancers, and we show that these have lost apoptotic function. Furthermore, several class II mutants have been found to be temperature sensitive for apoptotic activity while showing constitutive cell cycle arrest function. Taken together, these mutants comprise an excellent system with which to investigate the biochemical nature of p53-mediated apoptosis, the function of principal importance in tumor suppression. All of the mutants that showed loss of apoptotic function also showed defects in the activation of promoters from the potential apoptotic targets Bax and the insulin-like growth factor-binding protein 3 gene (IGF-BP3), and a correlation between full apoptotic activity and activation of both of these promoters was also seen with the temperature-sensitive mutants. However, a role for additional apoptotic activities of p53 was suggested by the observation that some mutants retained significant apoptotic function despite being impaired in the activation of Bax- and IGF-BP3-derived promoters. In contrast to the case of transcriptional activation, a perfect correlation between transcriptional repression of the c-fos promoter and the ability to induce apoptosis was seen, although the observation that Bax expression induced a similar repression of transcription from this promoter suggests that this may be a consequence, rather than a cause, of apoptotic death.
p53 is the most commonly mutated gene in human cancers, and the wild-type p53 protein plays an important role in preventing malignant progression (17). p53 expression is elevated in response to stress, such as DNA damage or abnormal proliferative signals, and activation of p53 function is thought to prevent the outgrowth of cells which harbor potentially oncogenic alterations (23). Several activities of p53 which may contribute to its tumor suppressive functions have been described, most notably the ability to block cell cycle progression and the induction of programmed cell death or apoptosis (2). Although either of these activities results in inhibition of cell growth, there is some evidence that the apoptotic function of p53 is the principal tumor-suppressive function (35, 39). Reintroduction of wild-type p53 into many tumor cell lines results in the activation of both the cell cycle arrest and apoptotic responses, although normal fibroblasts appear to respond to p53 by showing only cell cycle arrest (12, 25). This potential differential between normal and tumor cells, where normal cells undergo a reversible cell cycle arrest and tumor cells undergo irreversible apoptotic cell death in response to p53, offers great potential for the use of p53 as a therapeutic agent.
One of the most important activities of p53 is the ability to function as a sequence-specific DNA binding protein and transcription factor (9, 33). The expression of many cellular genes has been shown to be regulated by p53, and several of these genes are clearly involved in mediating the p53 response. Among the reported p53-responsive genes with potential as downstream mediators of the p53 response are p21Waf1/Cip1 (encoding a cyclin-dependent kinase inhibitor), Bax, the insulin-like growth factor-binding protein 3 gene (IGF-BP3), the cyclin G1 gene, and Gadd45 (24). In contrast, Mdm2, one of the first p53-inducible genes to be described, is not thought to mediate p53 effects but plays an important role in regulating p53 function by binding to the trans-activation domain of p53 (30) and targeting it for degradation (15, 21). Although transcriptional activation by p53 plays a role in both cell cycle arrest and apoptosis, poorly defined transcriptionally independent activities of p53 are also important, particularly to the apoptotic response (5, 16, 41). p53 is also able to repress transcription from several cellular and viral promoters (14, 27, 38) and to negatively regulate expression of some cellular genes (31), and there is evidence that this transcriptional repression activity may also be important for apoptosis (36, 37). Several lines of evidence show that the cell cycle arrest and apoptotic activities of p53 are separable functions (34), and p53 mutants which retain the ability to induce cell cycle arrest but fail to activate apoptosis have been described previously (10, 35). These mutants demonstrate that apoptosis is not simply a consequence of cell cycle arrest in the context of opposing proliferative signals (the conflict of signals model); they also suggest that the response to p53 can follow alternative pathways, depending on cell type, cell environment, and the presence of other genetic alterations (2).
The p53 protein can be divided into three principal domains: the N-terminal transcriptional activation domain, the C-terminal regulatory domain, and the central portion of the protein, which is responsible for sequence-specific DNA binding (20). Most of the tumor-derived p53 mutants harbor single amino acid substitutions within the central region which either prevent normal DNA contacting or alter the conformation of this domain (7). In both cases, the result of the mutation is a defect in the ability to bind DNA, activate transcription, and carry out normal tumor-suppressive functions. Most of these tumor-derived mutants are defective for both apoptosis and cell cycle arrest; however, tumors harboring a p53 point mutant and showing a loss only of apoptotic function, while retaining a normal ability to induce cell cycle arrest, have also arisen, albeit rarely (35). Closer examination of this mutant revealed a selective loss in transcriptional trans-activation activity, with the retention of essentially a wild-type ability to activate expression of p21Waf1/Cip1but defects in the activation of promoters containing p53 binding sites from the Bax and IGF-BP3 genes (26). Analysis of p21Waf1/Cip1-null mouse and human cells clearly indicated a role for this protein in p53-induced cell cycle arrest, although there is no clear contribution to apoptosis (3, 11, 42). Bax and IGF-BP3 both show apoptotic activities, although IGF-BP3 is also likely to contribute to cell cycle arrest by interfering with both the mitogenic and survival activities of IGF (4). The differential transcriptional activity shown by the p53 mutants strongly supported a role for p21Waf1/Cip1 in cell cycle arrest and suggested that Bax and IGF-BP3 may be the apoptotic transcriptional targets of p53.
Codon 175 in human p53 is one of the hot spots for mutation in human cancers, and crystallographic analysis has shown that while it does not contact the DNA directly, this residue plays an important role in maintaining the structure of the DNA binding domain (7). Mutation of this residue to histidine, the most common substitution detected in cancer cells, results in loss of all known p53 functions. We previously reported that mutation of this residue to proline, a much rarer event in cancers, ablates apoptotic activity but has no effect on the ability to induce cell cycle arrest. This phenotype was shown to correlate with retention of the ability to activate expression of p21Waf1/Cip1 but not Bax or IGF-BP3 (26, 35). In this study we describe the phenotype of a series of residue 175 mutants (32) and examine the correlation of apoptotic activity with transcriptional activation and repression.
MATERIALS AND METHODS
Plasmids.
p53 sequences were expressed from the cytomegalovirus (CMV) early promoter and have been described previously by Ory et al. (32). Transcriptional activation and repression by p53 was assessed by using the luciferase reporter gene under the control of promoter fragments from p53-responsive genes. The promoter fragments used in this study were the SmaI-SacI fragment of the human bax promoter derived from the plasmid Bax-CAT (29) and cloned into pGL3 in the plasmid Bax-luc (13), the Box B responsive element from the IGF-BP3 promoter in the plasmid boxB-luc (4), and a fragment of the c-fos promoter derived from plasmid pF711/CAT (40) and described here as fos-luc. As an assessment of transfection efficiency, transfections also included pJ4Ωβgal, in which the β-galactosidase gene is regulated by the long terminal repeat of the Moloney murine leukemia virus, which is both constitutive and nonresponsive to p53 (8). Full-length Bax protein was also expressed from the CMV promoter in plasmid pCB6+Bax (26).
Cell culture and transfection.
p53-null human tumor Saos-2 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum at 37°C in an atmosphere of 10% CO2 in air. For transient transfections, 5 × 105 to 8 × 105 cells were plated in 10-cm-diameter dishes and transfected with calcium phosphate coprecipitate the following day. Where indicated, cells were transfected with 50 ng to 5 μg of either p53 or Bax expression plasmids or empty vector and 5 μg of each reporter construct. Cells were washed the day after transfection and then harvested 24 to 48 h later and prepared for flow cytometry, Western blotting (immunoblotting), and luciferase and β-galactosidase assays. For temperature shift experiments, all cells were transfected at 37°C and then moved to different temperatures after washing.
Transcriptional activity and protein analysis.
Cell lysates were prepared 24 h posttransfection for luciferase and β-galactosidase reporter assays, as previously described (22). For Western blot analysis, cell lysates were normalized either by equal volume or by expression of cotransfected β-galactosidase, both giving identical results. Lysates were subjected to polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membranes. As an assessment of transfer, membranes were stained with Ponceau S prior to protein detection with the p53-specific monoclonal antibody DO-1 (Ab-6; Oncogene Science) and a monoclonal antibody specific to p21Waf1/Cip1 (Pharmingen).
Flow cytometry.
Total populations of transfected cells, including floating and adherent cells, were harvested 24 to 48 h posttransfection, fixed, and stained for p53 with monoclonal antibody DO-1 and for DNA content with propidium iodide, as previously described (35). Samples were analyzed in a flow cytometer (FACScalibur; Becton Dickinson), and cells were measured for fluorescein isothiocyanate fluorescence (green channel) and propidium iodide fluorescence (red channel). Total populations were gated to remove doublets and very small particles. Background levels of fluorescence were established by using cells transfected with vector alone, and then cells in p53-transfected populations with high fluorescence were gated and analyzed as a separate p53-containing fraction. Cell cycle analysis of p53-expressing and nonexpressing cells from each transfection was performed with CellQuest analysis software (Becton Dickinson), and apoptosis induced by each p53 protein was expressed as the percentage of cells with a sub-G1 DNA content relative to the rest of the population of p53-positive cells. The specific amount of apoptosis induced as a result of the transfection was taken as the percentage of sub-G1 cells in excess of that observed in the untransfected population.
RESULTS
Cell cycle arrest and apoptosis by residue 175 mutants.
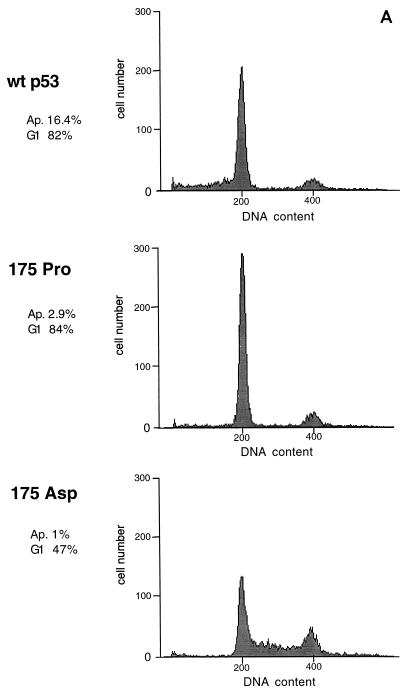
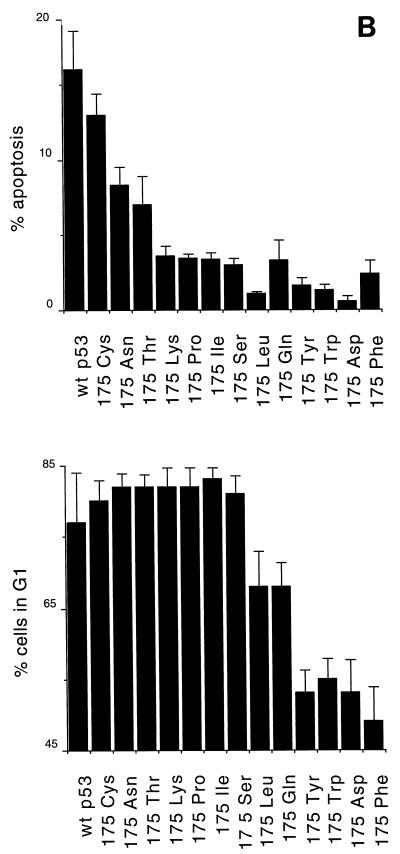
We have shown previously that mutation of arginine to proline at residue 175 of human p53 gives rise to a protein with impaired apoptotic but wild-type cell cycle arrest functions (35). In order to determine whether this is a general phenotype of mutations at this residue, we tested a series of previously described residue 175 substitutions (32) for the ability to induce cell cycle arrest and apoptosis following transient transfection of the p53 clones into the p53-null human osteosarcoma line Saos-2. Cell cycle distribution and apoptosis were determined by fluorescence-activated cell sorting analysis of the p53-expressing population of cells, as previously described (35). Wild-type p53 activates both cell cycle arrest, as characterized by an increase in cells in the G1 population, and enhanced apoptosis, as measured by the proportion of cells with a sub-G1 DNA content. It is important to note that this assay measures the apoptotic rate rather than cumulative apoptosis. In Saos-2 cells expressing inducible p53, an apoptotic rate of 10% after 24 h correlated with death of all cells within 5 days of p53 expression (data not shown). Figure 1A shows representative profiles of cells expressing wild-type p53 (G1 arrest and apoptosis), p53 175Pro (G1 arrest only), or p53 175Asp (no G1 arrest or apoptosis). Analysis of the series of mutants showed that there were other mutants with alterations at residue 175 which could behave like 175Pro and that the series of mutants could be broadly classified into three groups (Fig. 1B): class I mutants, which retained essentially wild-type cell cycle arrest and apoptotic functions (175Cys); class II mutants, which showed specific loss of apoptotic activity but retained cell cycle arrest function (175Lys, 175Pro, 175Ile, and 175Ser); and class III mutants, which showed loss of both activities (175Tyr, 175Trp, 175Asp, and 175Phe), giving profiles identical to untransfected cells or cells transfected with vector alone. This is an extension of previous studies which identified the class I and II mutants as wild type in cell growth suppression assays (32). In addition, some mutants showed an intermediate phenotype: 175Asn and 175Thr showed wild-type cell cycle arrest and impaired but not defective apoptotic function (class I/II), and 175Leu and 175Gln showed loss of apoptotic function and impaired cell cycle arrest activity (class II/III) (Fig. 1B). This series of mutants, therefore, afforded an excellent system in which to analyze the underlying defects in p53 function leading to these different phenotypes.
FIG. 1.
Induction of cell cycle arrest and apoptosis by p53 proteins. (A) Flow cytometry analyses of Saos-2 cells transiently transfected with wild-type p53 and two p53 mutants, 175Pro and 175Asp. Cell cycle arrest function is indicated as the percentage of cells in the G1 phase of the cell cycle relative to those in the S and G2/M phases. Apoptotic rates are given as the percentage of cells with sub-G1 DNA content above that observed in the untransfected population. (B) Representation of the cell cycle arrest and apoptotic functions of a series of mutants with alterations at codon 175 of p53.
Transcriptional activation by residue 175 mutants.
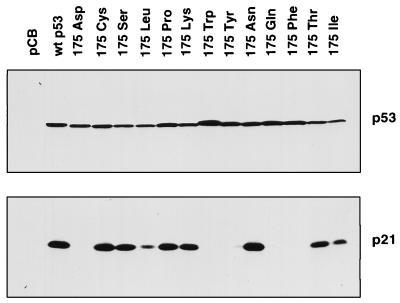
Activation of p21Waf1/Cip1 plays a principal role in mediating p53 cell cycle arrest, and we analyzed the ability of each of the mutants to activate expression of the endogenous p21Waf1/Cip1 protein (Fig. 2). As expected, the ability to activate expression of p21Waf1/Cip1 correlated with cell cycle arrest function, with all class I and II mutants activating expression of p21Waf1/Cip1 to levels comparable to the wild-type p53 protein. Interestingly, one of the intermediate class II/III mutants, 175Leu, also showed low but detectable ability to activate expression of p21Waf1/Cip1, which correlated with the ability to induce modest cell cycle arrest. However, the other class II/III mutant, 175Gln, which showed G1 arrest comparable to 175Leu, failed to activate detectable expression of p21Waf1/Cip1. Essentially similar results were also seen with a p21Waf1/Cip1promoter-driven reporter gene (data not shown).
FIG. 2.
Western blot analyses of lysates from Saos-2 cells which were transfected with p53 constructs as indicated. The nitrocellulose membrane from one polyacrylamide gel was probed with both p53- and p21Waf1/Cip1-specific monoclonal antibodies. The figure shown is representative of three independent experiments.
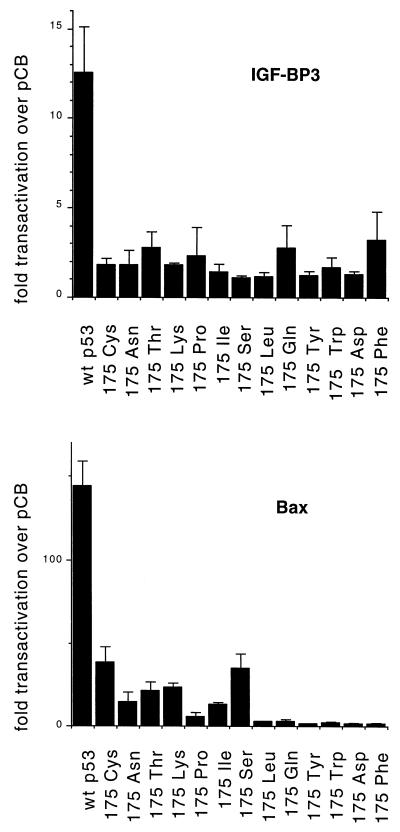
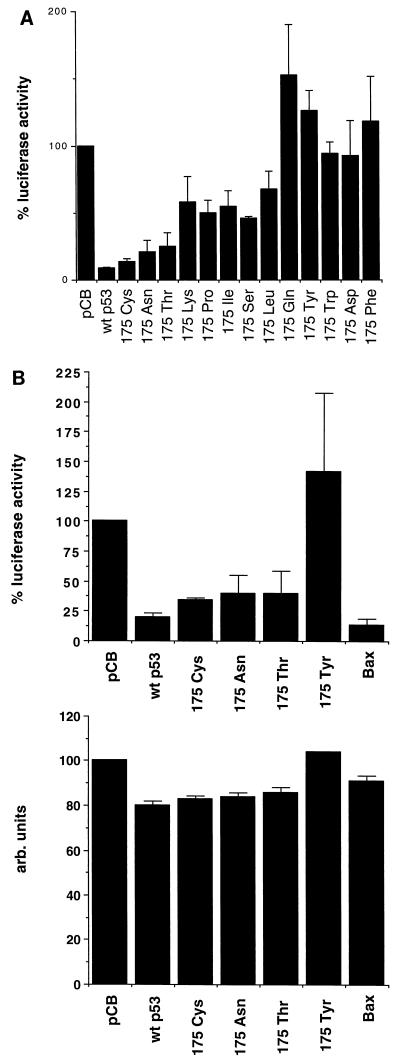
Since our previous analysis of p53 175Pro indicated a correlation between failure to activate expression of a Bax- or IGF-BP3-derived promoter and loss of apoptotic activity (26), we also tested all of the mutants for the ability to activate transcription of a luciferase gene under the regulation of promoters containing p53-responsive elements from Bax and IGF-BP3. As shown in Fig. 3, all of the residue 175 mutants showed substantial defects in the ability to activate expression of either reporter construct. The ability of all of the mutants to activate the IGF-BP3 reporter was essentially indistinguishable; activation of the Bax reporter was slightly higher in the class I and II mutants but clearly significantly reduced compared to the activity of the wild-type protein. It is particularly noteworthy that class I and I/II mutants (175Cys, 175Asn, and 175Thr), which retained significant apoptotic activity, did not retain enhanced ability to activate expression of IGF-BP3 or Bax compared to mutants with severely impaired apoptotic function (such as 175Ser) (Fig. 1B and Fig. 3). Therefore, although loss of the ability to activate expression from bax or IGF-BP3 promoters correlated with loss of apoptotic function in class II mutants, it would appear that the retention of wild-type ability to activate expression of these genes is not necessary for a significant apoptotic response.
FIG. 3.
Transcriptional activation by wild-type p53 and the indicated mutants of the p53-responsive promoter plasmids Bax-luc and boxB-luc (IGF-BP3). Assays were carried out following transfection of 5 μg of reporter plasmid and 50 ng of p53 expression plasmid into Saos-2 cells. The values shown are fold trans-activation of the reporters relative to values observed for cells transfected with vector alone.
Transcriptional repression by residue 175 mutants.
The ability of p53 to repress several cell and virus promoters has been linked to apoptotic activity (36, 37). One promoter which has been shown to be markedly repressed by p53 is c-fos (14), and we tested the series of mutants for the ability to repress expression from this promoter. As shown in Fig. 4A, a perfect correlation between the ability of each mutant to repress transcription and the ability to induce apoptosis (Fig. 1B) was seen. Class I and I/II mutants, which retained significant apoptotic activity, showed strong repressor functions, comparable to the wild-type protein. Class II mutants, which showed severely reduced apoptotic activity, also showed significant defects in the ability to repress transcription from the c-fos promoter; class III mutants, which were completely inactive for apoptosis, not only failed to repress expression from the c-fos promoter but in some cases showed an apparent activation of expression.
FIG. 4.
Transcriptional repression of the human c-fos promoter. (A) Wild-type p53 and the indicated mutants were cotransfected into Saos-2 cells together with the reporter plasmid fos-luc. The percent luciferase activity caused by each p53 protein was calculated relative to that observed in cells transfected with the empty CMV expression plasmid alone. (B) Comparison of the effects of expression of wild-type p53, the p53 mutants 175Cys, 175Asn, 175Thr, and 175Tyr and Bax on the reporter constructs fos-luc and pJ4Ωβgal. Saos-2 cells were transfected with 5 μg of each reporter construct together with either 5 μg of CMV-driven wild-type or mutant p53 or 1 μg of CMV-driven Bax.
Although the correlation between transcriptional repression and apoptosis was striking, we were concerned that this may not represent a causal step in p53-induced apoptosis. One possibility was that the repression was simply the consequence of overexpression of transcriptionally active p53 in transient transfection assays. Although all of these p53 mutants retain the trans-activation domain at the N terminus of the protein, it is possible that the transcriptionally inactive mutants are locked in a conformation which masks this domain. However, high-level expression of the VP16 trans-activation domain, which clearly showed transcriptional activation activity, failed to repress expression from the c-fos promoter (data not shown), indicating that simple overexpression of a transcription factor and titration of the basal transcriptional machinery (squelching) is not sufficient for repression.
Our principal concern, however, was that the transcriptional repression was a consequence, rather than a cause, of apoptotic cell death. To address this point, we examined the effect of induction of apoptosis in the same cells following expression of Bax, a cytoplasmic protein with no recorded transcriptional function (1). Expression of Bax resulted in apoptotic rates similar to those seen following p53 expression (data not shown), and Bax expression also efficiently repressed expression from the c-fos promoter (Fig. 4B). As a control, expression from the Moloney murine leukemia virus long terminal repeat was unaffected by Bax, indicating that, like p53, transcriptional repression in response to Bax expression showed some degree of promoter specificity.
Temperature sensitivity of residue 175 mutants.
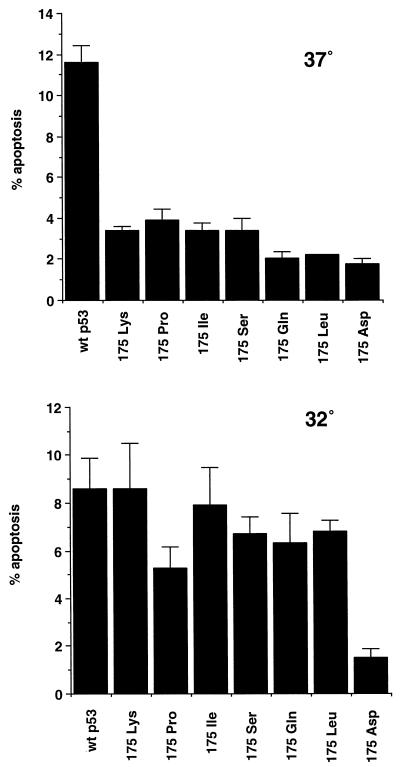
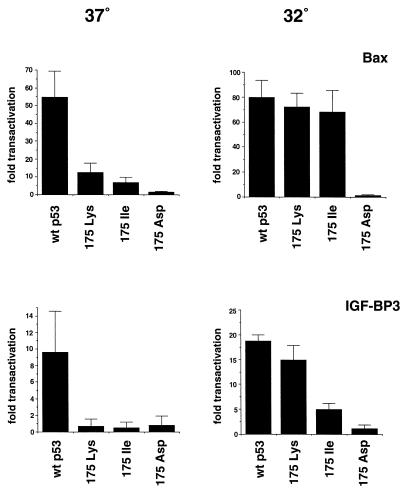
Although mutations of amino acid 175 have been shown to perturb sequence-specific DNA binding of p53, crystallographic studies have shown that this residue does not directly contact DNA (7). It appears that this residue is important in maintaining the conformation of the DNA binding domain, and disruption of the wild-type conformation can result in a loss of DNA binding activity. Our results so far suggest that the conformational alterations induced by the class II mutants might be rather subtle, allowing retention of at least some wild-type function. We therefore tested whether these mutants might regain a wild-type conformation and apoptotic function at reduced temperatures by analyzing them at 32°C. Analysis of apoptosis showed a slight reduction in the apoptotic rate induced by wild-type p53 at 32°C, which may be a consequence of a reduced growth rate, and the class III 175Asp mutant remained unable to induce apoptosis at either temperature (Fig. 5). All of the class II mutants, however, showed some evidence for temperature-sensitive recovery of apoptotic activity. In particular, 175Lys and 175Ile induced apoptotic rates at 32°C which were indistinguishable from the wild-type protein. Analysis of the transcriptional activity of these mutants at the two temperatures showed, as expected, that 175Lys and 175Ile activated expression from the p21Waf1/Cip1 and Mdm2 promoters as efficiently as the wild type at both temperatures (data not shown). Examination of the two potential apoptotic targets showed that both temperature-sensitive mutants regained the ability to activate expression. 175Lys showed substantially enhanced ability to activate the IGF-BP3-derived promoter, although 175Ile showed only a modest enhancement in the ability to activate this promoter. Both 175Lys and 175Ile, however, regained full wild-type ability to activate expression from the bax-derived promoter at 32°C (Fig. 6).
FIG. 5.
Induction of apoptosis by wild-type p53 and the indicated mutants at 37°C and 32°C. Saos-2 cells were transfected with 5 μg of p53 expression plasmid, left for 16 h, washed, and then placed at the indicated temperatures for 24 h before being harvested.
FIG. 6.
Transcriptional activation of the p53-responsive reporter plasmids Bax-luc and boxB-luc (IGF-BP3) by wild-type p53 and the indicated mutants at 37°C and 32°C. Saos-2 cells were transfected with 5 μg of reporter plasmid and 50 ng of the indicated p53 expression plasmid. Following transfection, the cells were incubated at 37°C for 16 h, washed, and moved to the appropriate temperature for 24 h before being harvested for luciferase and β-galactosidase assays. The values shown represent fold trans-activation relative to cells transfected with the empty CMV expression plasmid alone.
DISCUSSION
Transcriptional activation of target genes is one of the key mechanisms by which p53 induces cell cycle arrest and apoptosis. Many lines of evidence support a role for the activation of the cyclin-dependent kinase inhibitor p21Waf1/Cip1 in p53-induced cell cycle arrest (35, 42), and we show further evidence here of the excellent correlation between activation of p21Waf1/Cip1 expression and the ability of several p53 mutants to induce G1 arrest. Nevertheless, cells from p21Waf1/Cip1-defective mice do not show complete loss of p53-induced cell cycle arrest, and other p53 target genes probably also contribute to this response (3, 11). The phenotype of the 175Gln mutant supports the concept that mechanisms other than activation of p21Waf1/Cip1 can contribute to p53-induced cell cycle arrest, since this mutant retains some ability to activate G1 arrest without a detectable increase in p21Waf1/Cip1 expression. This is in agreement with the observation that cells from p21Waf1/Cip1-deficient mice retain some G1 arrest function in response to p53 activation (3, 11). Of particular interest is our observation that the rare tumor-derived codon 175 mutants (175Ser, 175Leu, and 175Pro) all show a clear defect in apoptotic function, supporting the importance of loss of this activity to tumor progression.
Identification of the apoptotic targets of p53 has been less straightforward, although there is convincing evidence that Bax plays at least some role in this process. Cells from Bax-deficient mice show some defect in the p53-dependent apoptotic response (28, 43), although the retention of significant levels of apoptosis in these cells (19) strongly implies a role for other p53 activities. These are likely to encompass activation of expression of other p53 target genes, repression of transcriptional activation by p53, and other, transcriptionally independent activities of the p53 protein. In this study, we show that loss of apoptotic function correlates with loss of the ability to activate expression of promoters derived from the Bax and IGF-BP3 genes. The correlation between apoptosis and activation of the Bax promoter was particularly close, with mutants that retain very low apoptotic activity (class II mutants) also retaining a low-level ability to activate the Bax promoter (Fig. 3). Similarly, mutants that were temperature sensitive for apoptosis regained wild-type ability to activate the Bax-derived promoter but showed only partial restoration of transcriptional activation of the IGF-BP3 promoter at 32°C, again underscoring the closer association between activation of Bax and apoptosis. However, analysis of the series of mutants showed that apoptotic function could be retained by mutants which failed to efficiently activate transcription from either IGF-BP3- or Bax-derived promoters. Taken together, these results support the suggestion that the activation of apoptosis by p53 may depend in part on activation of Bax and IGF-BP3 but may also be mediated by activation or repression of other target genes or through transcriptionally independent mechanisms. This collection of mutants may be of use in the further identification of such apoptotic activities of p53.
We have shown previously, and confirm here, that the differential loss of apoptotic function but the the retention of cell cycle arrest activity shown by some of these residue 175 mutants is accompanied by selective loss of the ability to activate some, but not all, p53-responsive promoters. Residue 175 participates in maintaining the conformation of the p53 DNA binding domain, and it seems likely that the promoter selectivity displayed by some of the residue 175 mutants reflects differences in the magnitude of the conformational shift induced by each amino acid substitution. It is possible that slight changes, induced by the class II mutants, may lead to failure to bind the Bax- and IGF-BP3-derived binding sites but retaining of binding to the p21Waf1/Cip1 promoter, maybe reflecting higher-affinity binding of p53 to the sites in the p21Waf1/Cip1 promoter than in those found in Bax or IGF-BP3. Alternatively, the different abilities of p53 and the mutants to activate transcription may depend on the DNA conformation of the consensus p53 binding site within the promoter (18). Recognition of different DNA conformations has been suggested to depend on the ability of the p53 protein itself to adopt different conformations, a property that may be differentially compromised in the various mutants. The concept that cell cycle arrest targets of p53, such as p21Waf1/Cip1, are more efficiently activated than apoptotic targets, such as Bax, might explain the observation that low levels of p53 activate cell cycle arrest while higher levels of p53 expression are necessary for the induction of apoptosis (6). This might imply that in normal cells the initial response to stress such as DNA damage is modest elevation of p53 levels, resulting in reversible cell cycle arrest, with apoptosis being activated only as p53 continues to accumulate, perhaps due to a prolonged or more severe exposure to the p53-activating event.
Despite the importance of transcriptional activation by p53 to the apoptotic response, it is also evident that other activities of p53 play a role in this response. One activity which has been suggested to be important in the induction of apoptosis is the ability of p53 to repress transcription of certain genes. Recent identification of a cell gene, MAP4, which is clearly transcriptionally repressed in cells following activation of p53, supports the importance of this function (31), although the exact contribution of transcriptional repression to each of the p53 activities remains unclear. Many viral and cellular promoters have been shown to be repressed in transient assays, and this activity has frequently been linked to the apoptotic response to p53 (36, 37). Although it remains likely that repression of specific cellular genes plays a role in mediating the p53 apoptotic response, our results showing a similar repression of the c-fos promoter following Bax expression indicate that detection of transcriptional repression by p53 should be interpreted with caution, since this may be a consequence as well as a cause of the apoptotic response.
ACKNOWLEDGMENTS
We are extremely grateful to Thierry Soussi for the gift of the codon 175 p53 mutants, Moshe Oren and Leonard Buckbinder for the Bax and IGF-BP3 reporter constructs, and Richard Treisman for the c-fos reporter. We also thank members of the Vousden Lab for constructive criticism and advice on the manuscript.
This work was supported by the National Cancer Institute under contract with ABL.
REFERENCES
- 1.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J-J, Maxxei G, Maundrell K, Gambale F, Sadoul R, Martinou J-C. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K H. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:1–7. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 3.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–556. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 4.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 5.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Ko L J, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 7.Cho Y, Gorina S, Jeffery P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 8.Crook T, Fisher C, Masterson P J, Vousden K H. Modulation of transcriptional regulatory properties of p53 by HPV E6. Oncogene. 1994;9:1225–1230. [PubMed] [Google Scholar]
- 9.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 10.Delia D, Goi K, Mizutani S, Yamada T, Aiello A, Fontanella E, Lamorte G, Iwata S, Ishioka C, Krajewski S, Reed J C, Pierotti M A. Dissociation between cell cycle arrest and apoptosis can occur in Li-Fraumeni cells heterozygous for p53 gene mutations. Oncogene. 1997;14:2137–2147. doi: 10.1038/sj.onc.1201050. [DOI] [PubMed] [Google Scholar]
- 11.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 12.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 16.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Induction of apoptosis in HeLa cells by trans-activation deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 17.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sørlie T, Hovig E, Smith-Sørensen B, Montesano R, Harris C C. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 18.Kim E, Albrechtsen N, Deppert W. DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene. 1997;15:857–869. doi: 10.1038/sj.onc.1201412. [DOI] [PubMed] [Google Scholar]
- 19.Knudson M C, Tung K S K, Tourtellotte W G, Brown G A J, Korsmeyer S J. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–98. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 20.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 21.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 22.Lam E W F, Waston R J. An E2F-binding site mediates cell-cycle regulation repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 24.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 25.Lowe S W, Ruley H E, Jacks T, Housman D E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig R L, Bates S, Vousden K H. Differential transcriptional activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack D H, Vartikar J, Pipas J M, Laimins L A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 28.McCurrach M E, Connor T M F, Knudson M C, Korsmeyer S J, Lowe S W. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 30.Momand J, Zambetti G P, George D L, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 31.Murphy M, Hinman A, Levine A J. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 32.Ory K, Legros Y, Auguin C, Soussi T. Analysis of the most representative tumour-derived p53 mutants reveals that changes in protein conformation are not correlated with loss of transactivation or inhibition of cell proliferation. EMBO J. 1994;13:3496–3504. doi: 10.1002/j.1460-2075.1994.tb06656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietenpol J A, Tokino T, Thiagaligam S, El-Deiry W, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polyak K, Waldman T, He T-C, Kinzler K W, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 35.Rowan S, Ludwig R L, Haupt Y, Bates S, Lu X, Oren M, Vousden K H. Specific loss of apoptotic but not cell cycle arrest function in a human tumour derived p53 mutant. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 36.Sabbatini P, Chiou S-K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Y Q, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subler M A, Martin D W, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Symonds H, Krall L, Remington L, Saenzrobles M, Lowe S, Jacks T, Van Dyke T. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell. 1994;78:703–711. doi: 10.1016/0092-8674(94)90534-7. [DOI] [PubMed] [Google Scholar]
- 40.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 41.Wagner A J, Kokontis J M, Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 42.Waldman T, Kinzler K W, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 43.Yin C, Knudson C M, Korsmeyer S J, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385:637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]