Significance
Clonal hematopoiesis (CH) prevalence increases with age. However, not all CH evolves into hematological malignancies or other diseases. Risk assessment of CH is critical in disease prevention and clinical practices. Here, we show that the prevalence of CH is significantly higher in the longevous elderly group, suggesting that CH could be beneficial to prolong life in certain conditions. Interestingly, the size of CH, which has been shown to be associated with higher disease risks, correlates significantly with the number of mutations per individual, but not with age, in the range of 60 to 110 y old. The findings provide a risk assessment biomarker for CH and suggest that the evolution of CH is influenced by factor(s) in addition to age.
Keywords: clonal hematopoiesis, aging, mutations, targeted DNA sequencing
Abstract
Clonal hematopoiesis (CH) represents the clonal expansion of hematopoietic stem cells and their progeny driven by somatic mutations. Accurate risk assessment of CH is critical for disease prevention and clinical decision-making. The size of CH has been showed to associate with higher disease risk, yet, factors influencing the size of CH are unknown. In addition, the characteristics of CH in long-lived individuals are not well documented. Here, we report an in-depth analysis of CH in longevous (≥90 y old) and common (60~89 y old) elderly groups. Utilizing targeted deep sequencing, we found that the development of CH is closely related to age and the expression of aging biomarkers. The longevous elderly group exhibited a significantly higher incidence of CH and significantly higher frequency of TET2 and ASXL1 mutations, suggesting that certain CH could be beneficial to prolong life. Intriguingly, the size of CH neither correlates significantly to age, in the range of 60 to 110 y old, nor to the expression of aging biomarkers. Instead, we identified a strong correlation between large CH size and the number of mutations per individual. These findings provide a risk assessment biomarker for CH and also suggest that the evolution of the CH is influenced by factor(s) in addition to age.
Hematopoietic stem cells (HSCs) gradually lose their proliferation potential in the process of aging (1). Meanwhile, during their self-renewal process, the HSCs continually accumulate DNA mutations (2). Certain mutations in HSCs can cause increased proliferation and abnormal differentiation. The clonal expansion of HSCs and their progeny driven by somatic driver mutations is termed as clonal hematopoiesis (CH) (3, 4). CH becomes increasingly common with age and is referred to as age-related clonal hematopoiesis (ARCH). The presence of somatic mutations in malignancy-related genes in blood, without evidence of hematologic malignancy, dysplasia, or cytopenia, along with variant allele frequencies (VAF) ≥2% is termed as clonal hematopoiesis of indeterminate potential (CHIP) (5).
Previous studies have demonstrated a steady increase in the prevalence of CHIP with age (3, 4, 6, 7). Commonly observed mutations in CHIP affect multiple genes, such as DNMT3A, TET2, PPM1D, TP53, STAT3, IDH1, IDH2, and ASXL1, most of which are known to be associated with hematological malignancies. These mutated genes can be categorized as epigenetic regulators, splicing factors, DNA damage response factors, and hematopoietic transcription factors (2, 8, 9). In addition to age, the development of CHIP is also influenced by factors such as inflammation, cancer-related treatments, smoking, and genetic factors (10–14).
CHIP is associated with an increased risk of hematological malignancies such as acute myeloid leukemia (AML), clonal cytopenia of undetermined significance, myelodysplastic syndromes (15, 16), and non-hematological diseases, such as cardiovascular diseases (17, 18), solid tumors (19), and autoimmune diseases (20), as well as increased overall mortality (21). CHIP can also provoke an inflammatory response in the body and display strong tolerance to this inflammatory environment, enhancing the adaptability of CH (22) and increasing the risk of sepsis, severe coronavirus infections (COVID-19) (23), and other infections (24).
Not all CH evolves into hematological malignancies or other diseases. Risk assessment of CH is therefore critical in disease prevention and clinical practices. It has been shown that the evolution of CH is influenced by both mutation types and non-mutation-related factors (25, 26). Studies have also revealed that the size of CH is linked to disease progression and prognosis (27), with larger clones associated with higher disease risks. Previous studies have found that individuals with high VAF (≥10%) have a significantly increased risk of lung cancer (28), gout (29), major adverse cardiac events (MACE) (30). The size of CH is also included in the CH risk score for prediction of myeloid malignancies (26, 31). However, the factors influencing the size of CH and its underlying molecular mechanisms remain unknown.
Clonal expansion driven by somatic mutations not only occurs in the hematopoietic system but also commonly develops in normal tissues throughout the body (32). Although some clonal expansion can evolve into cancer and/or facilitate the development of other diseases, certain clonal expansion may also play beneficial homeostatic roles in certain states by countering stress and aging conditions (33). The hematopoietic capacity of HSCs declines gradually as people age, and such change becomes more dramatic after 70 y of age (1). Although CH has been detected in various age groups, it has not been systematically studied in the longevous populations (90 y of age and above).
In this study, we compared the CH in longevous (≥90 y old) and common (60~89 y old) elderly groups and found that the prevalence of CH is significantly higher in the longevous elderly group, with significant increase of TET2 and ASXL1 mutations. Unexpectedly, the size of CH exhibited no significant correlation with age, in the range of 60 to 110 y old, nor with the expression of aging biomarkers. Instead, we observed a significant correlation between the size of CH and the number of mutations per individual. These findings provide a risk assessment biomarker for CH and suggest that the evolution of the CH is influenced by factor(s) in addition to age.
Results
Elevated Incidence of CH in the Longevous Elderly Group.
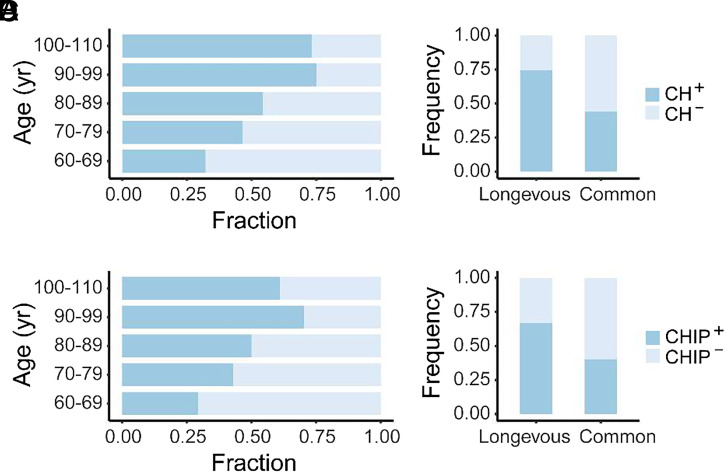
We collected blood samples from 105 longevous elderly individuals aged 90 to 110 y, referred to as the longevous elderly group; and 126 common elderly individuals aged 60 to 89 y, referred to as the common elderly group, as the control. To identify individuals with CH, we analyzed somatic mutations in 46 CH-related genes (SI Appendix, Table S1) by targeted next-generation sequencing (NGS). In this context, CH with a VAF ≥2% is referred to as CHIP, and that with all VAFs is referred to as CH. The incidence of both CH and CHIP increased continuously with age until 100 y old (Fig. 1 A and B). Although the incidence of CH and CHIP was trending lower in individuals aged 100 to 110 y (Fig. 1 A and B), statistical analysis using the chi-square test indicates that the difference in CH and CHIP incidence between individuals aged 100 to 110 y and those aged 90 to 99 y is statistically insignificant (P = 0.834 and P = 0.437, respectively).
Fig. 1.
CH in longevous elderly group and common elderly group. (A) Proportion of CH in different age intervals. (B) Proportion of CHIP in different age intervals. (C) Stacked bar plot depicting individuals with CH (CH+) and individuals without CH (CH−) in the longevous versus common elderly groups. Chi-square test indicates a significant difference between the two age groups (P < 0.001). (D) Stacked bar plot illustrating individuals with (CHIP+) and individuals without CHIP (CHIP−) in the longevous versus common elderly groups. Chi-square test indicates a significant difference between the two age groups (P < 0.001).
Next, we compared the occurrence of CH between the two elderly groups. Within the longevous group, 74.3% (78 out of 105) exhibited CH and 66.7% (70 out of 105) harbored CHIP. In contrast, the common elderly group had 44.4% (56 out of 126) with CH and 39.7% (50 out of 126) with CHIP (Fig. 1 C and D). The fraction of individuals with CH (CH+) and CHIP (CHIP+) in the longevous group was significantly higher than that of the common elderly group (P < 0.001). There was no significant difference in the incidence of CH or CHIP between two genders in the tested individuals from both age populations (SI Appendix, Fig. S1).
Significantly More TET2 and ASXL1 Mutations in the Longevous Elderly Group.
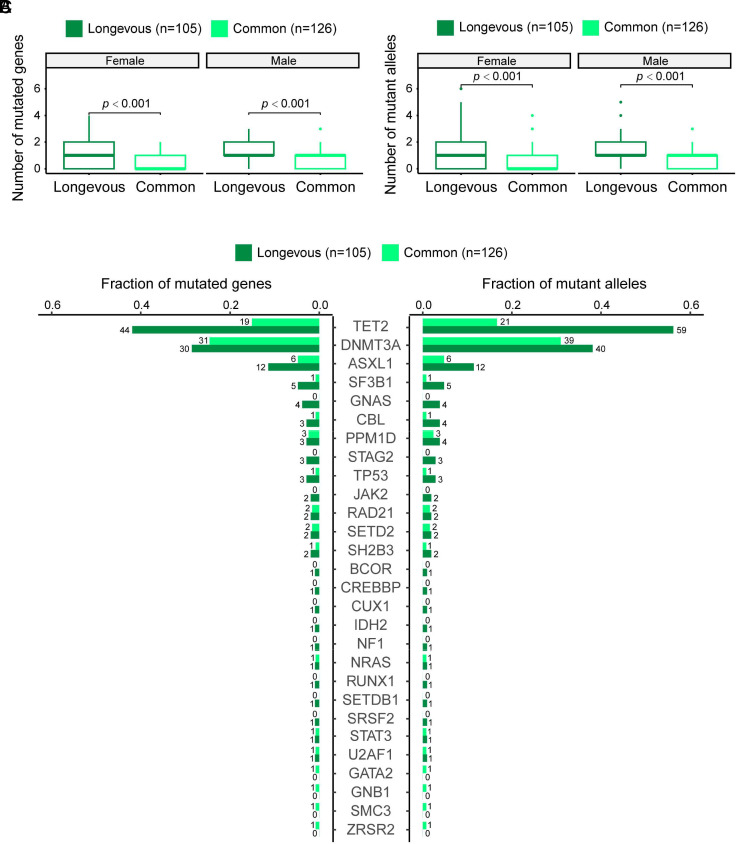
Through the analysis of CH in both longevous and common elderly groups, a total of 200 mutated genes and a total of 237 mutant alleles in 133 individuals were identified (SI Appendix, Table S2). To elucidate the relationship between the number of mutations and CH, we assigned a value to the mutation count, with each mutated gene or mutant allele receiving one point. We then compared both the number of mutated genes and the number of mutant alleles between the longevous elderly group and the common elderly group and found that the longevous elderly group had a significantly higher number of mutated genes per individual (Fig. 2A), as well as a significantly higher number of mutant alleles per individual (Fig. 2B) compared to the common elderly group, both in females and males.
Fig. 2.
CH mutations in the longevous elderly group versus the common elderly group. (A) Comparison of number of mutated genes per individual in the female and male longevous elderlies versus the female and male common elderlies. Statistical analysis was conducted using the Wilcoxon’s rank-sum test. (B) Comparison of the number of mutant alleles per individual in the female and male longevous elderlies versus the female and male common elderlies. Wilcoxon’s rank-sum test was used for statistical analysis. (C) Distribution of mutated genes (Left) and mutant alleles (Right) in the longevous elderly group versus the common elderly groups. The numbers labeled represent the number of individuals harboring the corresponding mutated genes and mutant alleles, respectively.
Like previous reports, TET2, DNMT3A, and ASXL1 are the most frequently mutated genes in the tested individuals with CH. Fig. 2C shows the distribution of mutated genes (Left) and mutant alleles (Right) in the longevous elderly group versus the common elderly group. Univariate logistic regression analysis, accounting for gender, revealed that the frequency of mutated TET2 and ASXL1 was significantly higher in the longevous elderly group than the common elderly group (P < 0.0001 and P = 0.021, respectively). Similarly, significantly more mutated TET2 alleles were found in the longevous elderly group compared to the common elderly group (P < 0.0001).
Occurrence of CH Is Positively Associated with the Level of Aging Biomarkers.
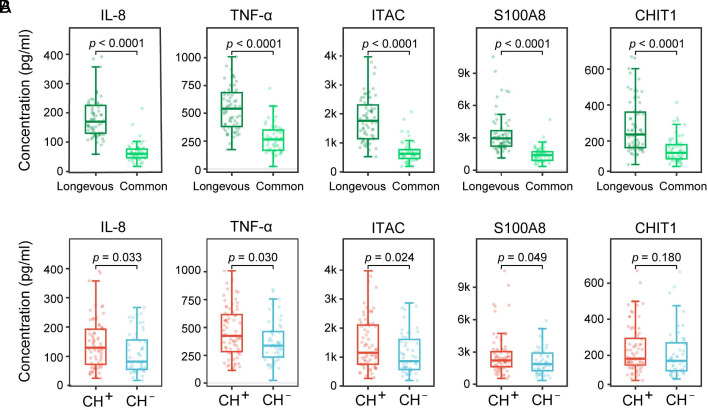
Although aging is age-related, chronological age does not always accurately reflect biological aging. The serum levels of IL-8, TNF-α, ITAC, S100A8, and CHIT1 have been reported to be associated with aging (34–38). To check whether the occurrence of CH is also associated with aging, we examined the serum level of these aging biomarkers in the longevous and common elderly groups. Consistent with their association with aging, the serum level of all these proteins was significantly higher in the longevous elderly group than the common elderly group (Fig. 3A). Subsequently, we analyzed the association between the expression of these aging biomarkers and the occurrence of CH. IL-8, TNF-α, ITAC, and S100A8 were found to be significantly associated with CH (P = 0.033, 0.030, 0.024, 0.049, respectively) (Fig. 3B).
Fig. 3.
Association analysis of CH and age-related biomarkers. (A) Comparison of the expression of aging biomarkers between the longevous elderly group and the common elderly group. Wilcoxon’s rank-sum test was used for statistical analysis, with the resulting P-value labeled for each comparison. (B) Comparison of the expression of aging biomarkers between individuals with CH (CH+) and individuals without CH (CH−). Wilcoxon’s rank-sum test was used for statistical analysis, with the resulting P-value labeled for each comparison.
The Size of CH Does Not Associate Significantly with Age and the Level of Aging Biomarkers in Elderly Individuals.
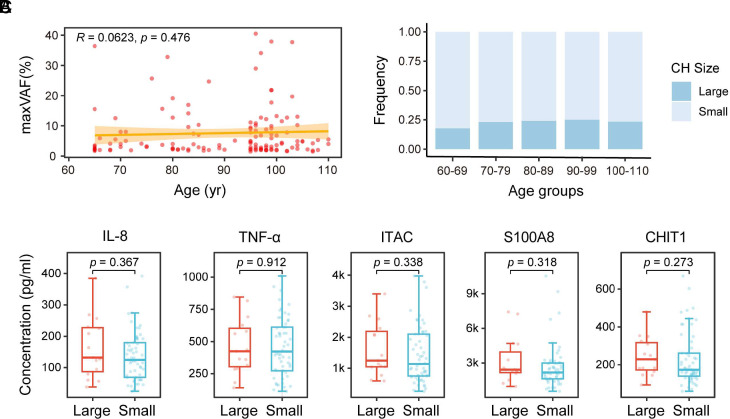
Not all CH develop into hematological malignancies or other related diseases. However, large size of CH (VAF ≥ 10%) has been found to be associated with an increased risk of developing diseases such as lung cancer (28), gout (29), and MACE (30). In this study, we categorized the CH clones with VAF < 10% as small clones and those with VAF ≥ 10% as large clones. In individuals with multiple clones, the VAF of the largest clone (maxVAF) is used in the analysis. Both large and small clones were observed in all ages. The relationship between the size of CH and age assessed by Spearman’s correlation showed no significant difference (Fig. 4A, P = 0.476). The ratio of large to small size of CH also showed no significant difference between all age intervals (Fig. 4B, P = 0.983). We also analyzed the association between the size of CH and aging biomarkers and found that there was no significant correlation between the size of CH and the expression of IL-8, TNF-α, ITAC, S100A8, and CHIT1 (Fig. 4C).
Fig. 4.
Correlation between the size of CH versus age and aging biomarkers. (A) Scatter plot illustrating the size of CH (reflected by maxVAF) in each CH-positive individual across age. The relationship between the size of CH and age was assessed by Spearman’s correlation. Calculated correlation coefficient and P-value are labeled. The orange-yellow band represents the 95% CI for the linear regression. (B) Stacked bar plot depicting CH with VAF ≥ 10% (Large) and CH with VAF < 10% (Small) in 5 age groups. Statistical analysis using the chi-square test indicates no significant difference between the groups (P = 0.983). (C) Comparison of the expression of aging biomarkers between individuals harboring CH with VAF ≥ 10% (Large) and individuals harboring CH with VAF < 10% (Small). Wilcoxon’s rank-sum test was used for statistical analysis, with the resulting P-value labeled for each comparison.
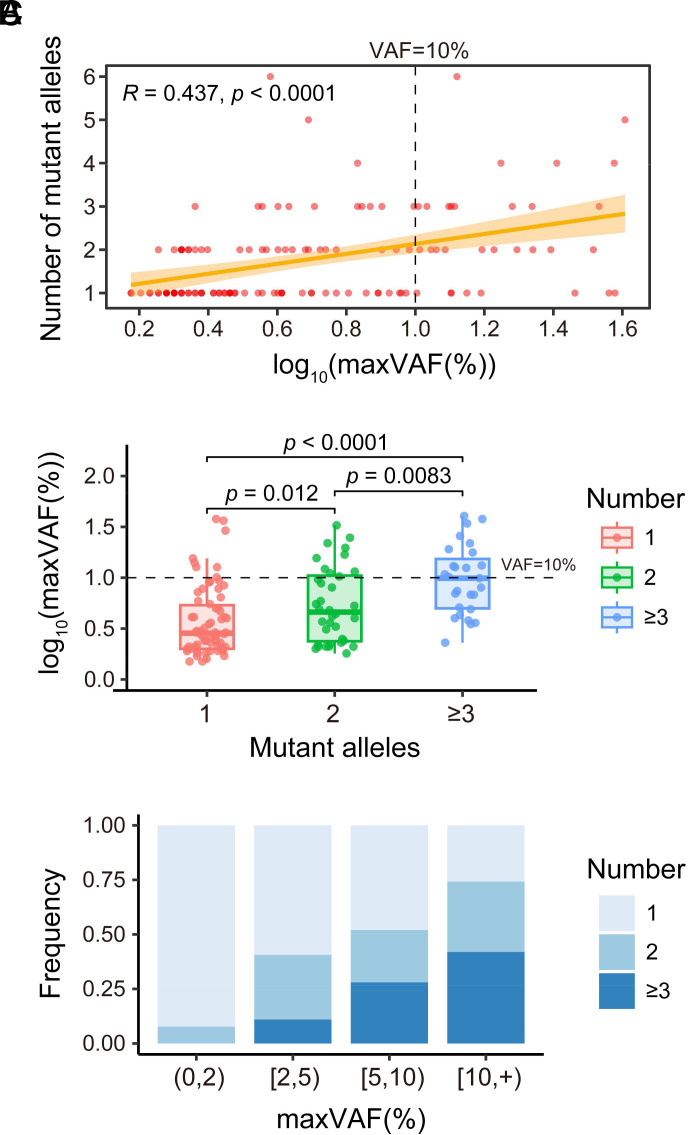
The Number of Mutations Per Individual Correlates Positively with the Size of CH.
The aforementioned results indicate that the CH clone size is not correlated with age or aging. We then searched factors that are associated with the expansion of CH. The relationship between the number of mutant alleles per individual and the size of CH was assessed by Spearman’s correlation. This analysis revealed a significant correlation between the number of mutant alleles and the size of CH (Fig. 5A, P < 0.0001). We also divided the tested individuals with CH into three subgroups according to their number of mutant alleles and found that the CH clones are significantly larger in individuals with 2 mutant alleles than those with 1 mutant allele, and the CH clones is significantly larger in individuals with 3 or more mutant alleles than those with 2 or 1 mutant allele(s), respectively (Fig. 5B).
Fig. 5.
Correlation between the size of CH versus the number of mutant alleles per individual. (A) Scatter plot illustrating the number of mutant alleles per individual and the size of CH [expressed as log10(maxVAF(%))]. The relationship between the number of mutant alleles per individual and the size of CH was assessed by Spearman's correlation. Calculated correlation coefficient and P-value are labeled. The orange-yellow band represents the 95% CI for the linear regression. (B) Comparison of the number of mutant alleles per individual, as indicated, and the size of CH [expressed as log10(maxVAF(%))]. (C) Stacked bar plot depicting CH-positive individuals with different numbers of mutant alleles, as indicated, in 4 different CH size (represented as maxVAF) groups. (0,2), [2,5), [5,10), and [10, +) represent the VAF of CH size groups <2%, ≥2% to <5%, ≥5% to <10%, and ≥10%, respectively.
We further categorized the tested individuals with CH into four subgroups based on maxVAF: <2%, ≥2% to <5%, ≥5% to <10%, and ≥10% and found that individuals with larger CH clones have higher proportion of multiple mutant alleles (Fig. 5C).
Discussion
In this study, we found that the development of CH and CHIP increased continuously with age until 100 y old (Fig. 1 A and B). Although the decrease did not reach statistical significance, the incidence of CH and CHIP was trending lower in individuals aged 100 to 110 y old (Fig. 1 A and B). The decline of incidence of CH and CHIP in individuals aged 100 to 110 y may reflect the influence of longevity factors, such as lower mutation rate, on all aspects of health.
Previous studies have revealed that large size of CH (VAF ≥ 10%) correlates more with the development of diseases such as lung cancer (28), gout (29), and MACE (30), and the prognosis of many other diseases (6). Intriguingly, we found that the size of CH does not exhibit significant correlations with age within the range of 60 to 110 y nor to the expression of aging biomarkers (Fig. 4A). This suggests that factors other than age may play pivotal roles in driving the expansion of CH. Our findings align with a large-scale longitudinal study with 3,359 participants, demonstrating that age does not affect clonal expansion in their studied cohort (25).
Our study shows that the size of CH correlates significantly with the number of mutations per individual (Fig. 5). These findings provide a risk assessment biomarker for CH and suggest that the evolution of the CH is influenced by factor(s) in addition to age. The higher number of mutations may reflect a greater mutation rate of the individuals. Defects in DNA repair and the maintenance of genome stability could expedite CH development.
The prevalence of CH in the elderly population in China seems higher than CH in western populations initially reported (3, 4). However, the incidence of CH varies dramatically in recent publications. For example, CH was detected in 305 out of 385 individuals (79.2%) aged 55 to 93 y (7). Although we cannot rule out the possibility that CH incidence is higher in Chinese population, the difference could be due to the depth of sequence, technique, and variant calling.
Although this study mainly compared CH in longevous versus common elderly groups, we also analyzed the CH in 50 to 59 y old (28 individuals) and 40 to 49 y old (33 individuals) groups. CH in age 50s group is about 14%, whereas CH in age 40s group is approximately 6%. These data show again that CH is age-related and develops under the aging condition. To this end, it is of note that in our recent study on the comprehensive genomic profiling of a large cohort of AML, CH-related gene mutations constitute the major genomic abnormalities in elderlies whereas gene fusion events tend to be main disease drivers in younger patients (see accompanying paper by Li JF, et al.)
While some clonal expansion can evolve into cancer and/or facilitate the development for other diseases, certain clonal expansion may also play beneficial homeostatic roles in certain states by countering stress and aging conditions (33). For instance, clonal expansion due to BCOR/BCORL1 and PIGA mutations is beneficial for the prognosis of aplastic anemia treatment (39). Notch1 gene mutations have been found to drive clonal expansion in normal esophageal epithelium but impair esophageal tumor growth (33, 40). In case of CH, it may offer protection against the risk of Alzheimer’s Disease (41). It is well known that with aging, the body exhibits stem cell exhaustion, reduced regenerative abilities (42), and decreased blood cell counts (43). In individuals over 70 y old, due to aging of HSCs, there is a sharp decline in hematopoietic function, leading to an increased risk of anemia, blood cancer, weakened infection resistance, and other age-related conditions (1). Here, we found that CH is common in the longevous group (Fig. 1), suggesting that certain CH could be beneficial to prolong life.
Our study found that individuals aged 90 and above have significantly higher frequency of TET2 and ASXL1 mutations compared to the common elderly population (Fig. 2C). TET2 mutations were also previously found to increase consistently with age in individuals with CH (7). However, there were only a few individuals over 90 y old in the study (7). As cancer incidence peaks approximately age 85 to 89 y (44), one possible explanation of their results is that TET2 mutations may be associated with increased cancer incidence. Our results, showing that TET2 mutations are significantly higher in the longevous group (Fig. 2C), indicate that TET2 mutations may play a protective role in prolonging life. Consistent with the finding that TET2 mutations may play a protective role, more TET2 mutations were found in CH associated with lower risk of Alzheimer’s disease (41). A possible mechanism is that TET2 mutations may promote myeloid cell differentiation and produce more microglial cells (41). The benefitting of clonal expansion might be the result of selection made by the aging organism to maintain stability and counteract aging and stress. These possibilities warrant further exploration in future studies.
Material and Methods
Subject Recruitment.
We enrolled a total of 231 Han Chinese individuals from Lingao County, Hainan Province, China. This cohort is comprised of 105 longevous elderly individuals (≥90 y old, average age: 99.4 ± 4.37 y; gender: 76 females and 29 males) and 126 common elderly individuals (60 to 89 y old, average age: 73.8 ± 8.22 y; gender: 73 females and 53 males). Blood samples of longevous elderlies were collected at participants’ home with the support of Lingao Health Commission. Participants in longevous elderly group all live in normal life without serious illness. Blood samples of common elderlies were collected during their annual physical exam in Lingao People’s Hospital. Participants in common elderly group are healthy individuals confirmed by their blood examination tests (Dataset). To ensure accurate age verification, we recorded the ages of all participants from their identification cards and further corroborated this information by recalling their life events, especially for the longevous elderly participants. The research protocol was approved by the Ethics Committee at Hainan Medical University (HYLL-2023-407). Written informed consent was obtained from each of the participants prior to the study.
Targeted Sequencing, Variant Calling, and Filtering.
Genomic DNA was randomly sheared through ultra-sonication to generate paired-end libraries with an average insert size of ~300 bp and subjected to library construction using KAPA HyperPrep Kits (KAPA Biosystems), followed by hybridization capture using custom IDT xGen Lockdown Probes and Reagents (Integrated DNA Technologies). As the main component, the custom hybridization capture panel targets 120 kb of the human genome containing all coding exons of 46 myeloid and lymphoid driver genes (SI Appendix, Table S1). Targeted NGS was performed using hybridization oligonucleotide probes on the Illumina Nextseq 550 platform (Illumina, San Diego, CA, USA) in Ruijin Hospital, producing 2 × 150 bp paired-end reads. The mean number of aligned consensus reads was 11329801, with a coverage of >1,000× for 84% of all targeted regions. The paired-end reads from whole-exome sequencing were mapped to human genome (version hg19) by BWA aligner (v0.7.17) (45). Mapping results were then sorted and marked for duplications via Picard (v2.23.0, https://broadinstitute.github.io/picard) (46).
Single nucleotide variants (SNVs) and small insertions and deletions (INDELs) were obtained by taking the union of three callers GATK4 Mutect2 (http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit) (47), VarDict (v1.5.8, https://github.com/AstraZeneca-NGS/VarDict) (48), and MuTect (v1.1.7, https://github.com/broadinstitute/mutect) (49) using default parameters or according to those recommended by the authors. All mutations were annotated by SnpEff (v4.2, https://pcingola.github.io/SnpEff/) and ANNOVAR (v2019Dec03, http://www.openbioinformatics.org/annovar/). All functional mutations, including missense, nonsense, splicing, nonstop SNVs, and INDELs, were obtained. The raw variant sites were screened by the following standards: homemade pipeline was used to filter SNVs and INDELs detected by the above software, excluding 1) mutations called only one software; 2) germline mutations detected from 250 custom control samples; 3) population-related variants reported in 1,000 Genomes (dbSNP 151) as common SNPs and not included in the Catalogue of Somatic Mutations in Cancer version v85; 4) VAFs were <1% and <20 individual mutant reads.
For pathogenic annotation, each filtered mutation annotated by “Pathogenic” and “Likely Pathogenic” with supporting evidence were retrieved from ClinVar database (50). In addition, we use SIFT and PolyPhen-2 to predict the effect of the change on protein function (51, 52).
Measurement of Serum Level of IL-8, TNF-α, ITAC, S100A8, and CHIT1.
The serum concentrations of IL-8, TNF-α, ITAC, S100A8, and CHIT1, in participants were measured with commercial ELISA kits (Shanghai Zhenke biological Technology Co. Ltd.) following the manufacturer’s instructions. The signals were detected at 450 nm using the Spark multimode reader (Tecan, Switzerland). The measurements were normalized to a negative serum control.
Statistics and Reproducibility.
Statistical analyses were performed using the R statistical software (version 4.3.1), with visual plots drawn with the help of R packages “ggplot2” (https://github.com/tidyverse/ggplot2) and “PICH” (https://github.com/hfang-bristol/PICH). All statistical tests were two-sided to compute statistical significance (P-value) and odds ratio where applicable. Categorical variables were compared using the chi-squared tests, while the Wilcoxon’s rank-sum test was employed to compare continuous variables. Univariate logistic regression analysis was utilized to compare differences in the number of mutations between individuals in the longevous elderly group and individuals in the common elderly group. The relationship between CH clone size versus age and the relationship between CH clone size versus the number of mutant alleles were analyzed by Spearman’s correlation. The reproducibility is enabled at http://www.genetictargets.com/CH/index.html, including data, codes, and instructions.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We extend our sincere gratitude to all the elderly volunteers participated in this project. This work was supported by the Key Research and Development Project of Hainan Province (2023ICAC-YANFA), the Science and Technology Platform Construction Project of Hainan Province (2023ICAC-YUNXING), Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01 to R.R.), Shanghai Science and Technology Development Funds (20Z11900200 to R.R.), the Samuel Waxman Cancer Research Foundation (to R.R.), the Innovative Research Team of High-level Local Universities in Shanghai (to R.R.) and National Natural Science Foundation of China (32170663 to H.F.).
Author contributions
Z.C., W.-W.C., and R.-B.R. designed research; K.W., W.Z., P.-Y.L., M.-H.F., and Y.-M.Z. performed research; W.-P.Y. contributed new reagents/analytic tools; K.W., L.Y., J.-F.L., H.F., and R.-B.R. analyzed data; M.Z. designed the targeted NGS panel for CH detection; W.Z., R.L., and W.-W.C. collected blood samples; and K.W., W.Z., L.Y., M.-H.F., and R.-B.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: J.L., University of Florida Cancer and Genetics Research Center; and P.L., NIH.
Contributor Information
Zhu Chen, Email: zchen@stn.sh.cn.
Wang-Wei Cai, Email: caiww591020@163.com.
Rui-Bao Ren, Email: rbren@sjtu.edu.cn.
Data, Materials, and Software Availability
Anonymized targeted DNA sequencing data have been deposited in http://www.genetictargets.com/CH/index.html (53) (open access).
Supporting Information
References
- 1.Mitchell E., et al. , Clonal dynamics of haematopoiesis across the human lifespan. Nature 606, 343–350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Challen G. A., Goodell M. A., Clonal hematopoiesis: Mechanisms driving dominance of stem cell clones. Blood 136, 1590–1598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G., et al. , Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaiswal S., et al. , Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoury J. D., et al. , The 5th edition of the World Health Organization classification of haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia 36, 1703–1719 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kar S. P., et al. , Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat. Genet. 54, 1155–1166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabre M. A., et al. , The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 606, 335–342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florez M. A., et al. , Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem. Cell 29, 882–904 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscarlet M., et al. , DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130, 753–762 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Coombs C. C., et al. , Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem. Cell 21, 374–382.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton K. L., et al. , Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 52, 1219–1226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bick A. G., et al. , Increased CHIP prevalence amongst people living with HIV. HIV. Sci. Rep. 12, 577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu J. I., et al. , PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem. Cell 23, 700–713.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai Z., et al. , Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem. Cell 23, 833–849.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abelson S., et al. , Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai P., et al. , Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaiswal S., Libby P., Clonal haematopoiesis: Connecting ageing and inflammation in cardiovascular disease. Nat. Rev. Cardiol. 17, 137–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuster J. J., et al. , Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.C. S. Reed, S. Croessmann, B. H. Park, CHIP happens: Clonal hematopoiesis of indeterminate potential and its relationship to solid tumors. Clin Cancer Res. 29, 1403–1411 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., et al. , Clonal hematopoiesis in primary immune thrombocytopenia. Blood Cancer J. 12, 40 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh K., et al. , Clonal hematopoiesis analyses in clinical, epidemiologic, and genetic aging studies to unravel underlying mechanisms of age-related dysfunction in humans. Front. Aging 3, 841796 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Avagyan et al. , Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science 374, 768–772 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Bolton K. L., et al. , Clonal hematopoiesis is associated with risk of severe Covid-19. Nat. Commun. 12, 5975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zekavat S. M., et al. , Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat. Med. 27, 1012–1024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Zeventer I. A., et al. , Evolutionary landscape of clonal hematopoiesis in 3,359 individuals from the general population. Cancer Cell 41, 1017–1031.e4 (2023). [DOI] [PubMed] [Google Scholar]
- 26.L. D. Weeks et al. , Prediction of risk for myeloid malignancy in clonal hematopoiesis. NEJM Evid. 2, 10.1056/evidoa2200310 (2023), 10.1056/evidoa2200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. J. Watson et al. , The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 367, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Tian R., et al. , Clonal hematopoiesis and risk of incident lung cancer. J. Clin. Oncol. 41, 1423–1433 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal M., et al. , TET2-mutant clonal hematopoiesis and risk of gout. Blood 140, 1094–1103 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., et al. , Prevalence and prognostic significance of DNMT3A- and TET2- clonal haematopoiesis-driver mutations in patients presenting with ST-segment elevation myocardial infarction. EBioMedicine 78, 103964 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler M. D., et al. , Common and rare variant associations with clonal haematopoiesis phenotypes. Nature 612, 301–309 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R., et al. , A body map of somatic mutagenesis in morphologically normal human tissues. Nature 597, 398–403 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Venkatachalam A., Pikarsky E., Ben-Neriah Y., Putative homeostatic role of cancer driver mutations. Trends Cell Biol. 32, 8–17 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Ma S., et al. , Caloric restriction reprograms the single-cell transcriptional landscape of rattus norvegicus aging. Cell 180, 984–1001.e22 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Li J., et al. , Determining a multimodal aging clock in a cohort of Chinese women. Med. 4, 825–848.13 (2023). [DOI] [PubMed] [Google Scholar]
- 36.H. Bruunsgaard, P Skinhøj, A. N. Pedersen, M. Schroll, B. K. Pedersen, Ageing, tumour necrosis factor-alpha (TNF-alpha) and atherosclerosis. Clin. Exp. Immunol. 121, 255–260 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markofski M. M., et al. , Evaluation of pro-inflammatory cytokines in frail Tunisian older adults. Plos One 15, e0242152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samson L. D., et al. , Inflammatory marker trajectories associated with frailty and ageing in a 20-year longitudinal study. Clin. Transl. Immunol. 11, e1374 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshizato T., et al. , Somatic mutations and clonal hematopoiesis in aplastic anemia. N. Engl. J. Med. 373, 35–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abby E., et al. , Notch1 mutations drive clonal expansion in normal esophageal epithelium but impair tumor growth. Nat. Genet. 55, 232–245 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouzid H., et al. , Clonal hematopoiesis is associated with protection from Alzheimer’s disease. Nat. Med. 29, 1662–1670 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Guralnik J. M., Eisenstaedt R. S., Ferrucci L., Klein H. G., Woodman R. C., Prevalence of anemia in persons 65 years and older in the United States: Evidence for a high rate of unexplained anemia. Blood 104, 2263–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 44.DeSantis C. E., et al. , Cancer statistics for adults aged 85 years and older, 2019. Cancer J. Clin. 69, 452–467 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.P. toolki, GitHub repository (Broad Institute, 2019). https://broadinstitute.github.io/picard/. Accessed 23 February 2023. [Google Scholar]
- 47.McKenna A., et al. , The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai Z., et al. , VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 44, e108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cibulskis K., et al. , Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landrum M. J., et al. , ClinVar: Improvements to accessing data. Nucleic Acids Res. 48, D835–D844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng P. C., SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adzhubei I. A., et al. , A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren R., et al. , Data and source code in the CH web page. Figshare. 10.6084/m9.figshare.25108277.v1. Deposited 30 January 2024. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
Anonymized targeted DNA sequencing data have been deposited in http://www.genetictargets.com/CH/index.html (53) (open access).