Significance
Disordered Th17/Treg cell balance in vivo is closely related to the pathogenesis and progression of a variety of autoimmune diseases. In this study, we reveal that the epigenetic modulator Setd2 directly targets the phospholipid remodeling enzyme Lpcat4 to promote Lpcat4-mediated phosphatidylcholine production. Phosphatidylcholine metabolite PC(16:0, 18:2) can limit endoplasmic reticulum stress and suppress HIF-1α transcriptional activity, thereby alleviating autoimmunity triggered by Th17/Treg cell imbalance. Consistently, in vivo deficiency of Setd2 aggravates the pathological progression of mouse experimental autoimmune encephalomyelitis. Our study reveals a phosphatidylcholine metabolite PC(16:0, 18:2) in controlling Th17/Treg cell balance and provides mechanistic insight into the epigenetic control of metabolic processes in T cell–mediated autoimmunity.
Keywords: Setd2, lysophosphatidylcholine acyltransferase 4, phosphatidylcholine, autoimmunity
Abstract
Coordinated metabolic reprogramming and epigenetic remodeling are critical for modulating T cell function and differentiation. However, how the epigenetic modification controls Th17/Treg cell balance via metabolic reprogramming remains obscure. Here, we find that Setd2, a histone H3K36 trimethyltransferase, suppresses Th17 development but promotes iTreg cell polarization via phospholipid remodeling. Mechanistically, Setd2 up-regulates transcriptional expression of lysophosphatidylcholine acyltransferase 4 (Lpcat4) via directly catalyzing H3K36me3 of Lpcat4 gene promoter in T cells. Lpcat4-mediated phosphatidylcholine PC(16:0,18:2) generation in turn limits endoplasmic reticulum stress and oxidative stress. These changes decrease HIF-1α transcriptional activity and thus suppress Th17 but enhance Treg development. Consistent with this regulatory paradigm, T cell deficiency of Setd2 aggravates neuroinflammation and demyelination in experimental autoimmune encephalomyelitis due to imbalanced Th17/Treg cell differentiation. Overall, our data reveal that Setd2 acts as an epigenetic brake for T cell–mediated autoimmunity through phospholipid remodeling, suggesting potential targets for treating neuroinflammatory diseases.
Pathological autoimmunity is regulated by complex interactions among epigenetic, metabolic, and immune factors (1, 2). T cell dysfunction is a crucial mediator of autoimmune pathogenesis, promoting neuroinflammatory disorders such as multiple sclerosis (MS) (3, 4). Cross-talk between epigenetic and metabolic programs has emerged as a theme in the regulation of T cell fate and function in different pathophysiological conditions (5–8). However, the specific mechanisms by which epigenetic modifiers impact genes encoding metabolic enzymes to determine T cell differentiation remain unknown. Dissecting the interplay between epigenetics and metabolic programs in T cells will foster a better understanding of the pathogenesis underlying autoimmune disease and facilitate the development of disease intervention strategies.
Metabolic networks play variable roles in determining T cell proliferation, activation, differentiation, and survival (5, 8). Effector T cells mainly depend on glycolysis and fatty acid synthesis for differentiation, function, and survival, while Treg cells depend upon oxidative phosphorylation (OXPHOS) and fatty acid oxidation (FAO) (6). Metabolic reprogramming involving glycolysis, glutaminolysis, and lipid synthesis is closely related to T cell dysfunction during autoimmune diseases and tumors (9–12). Lipid metabolism has emerged as a key regulator of T cell fate and function and is closely associated with the pathogenesis of cancer and autoimmunity (8). In particular, phosphatidylcholine (PC) is the most abundant phospholipid in cell membranes and has been linked to inflammatory processes by shaping membrane composition and fluidity (13–15). The lysophosphatidylcholine acyltransferases (LPCATs) are critical for phospholipid remodeling as they catalyze the conversion of lysophosphatidylcholine (LPC) into PC. However, whether reprogramming of LPCAT-mediated phosphatidylcholine metabolism and phospholipid remodeling is linked to T cell polarization and orchestration of Th17/Treg balance remains unknown.
Epigenetic remodeling plays a critical role in immunity and inflammation (16–18). Modulation of chromatin structure and function has emerged as a critical mechanism for coordinating T cell differentiation programs (6, 19). Genes encoding effector cytokines and lineage-determining transcription factors are dynamically controlled by epigenetic mechanisms involving DNA methylation, histone modifications, and noncoding RNAs (20–22). H3K36 methylation is a histone modification linked to active transcription and implicated in immunity and inflammation (23, 24). However, the link between the dynamic reprogramming of H3K36 in Th17/Treg cell differentiation and autoimmune inflammation remains unclear. Setd2 is the only characterized H3K36 trimethyltransferase (H3K36me3) in mammals and has been linked to immunological, developmental, and pathological processes including antiviral responses, intestinal epithelial integrity, and embryonic cell development (24–26). So far, the role of Setd2 in coordinating T cell balance and its link to metabolic regulation remains unclear. Here, we reported a role for Setd2 in controlling the balance of Th17/Treg cells via phospholipid remodeling, thus adding insights to the interplay between epigenetics and metabolisms in T cell–dependent autoimmunity.
Results
Setd2 Expression Is Down-Regulated in T Cell–Mediated Autoimmune Diseases.
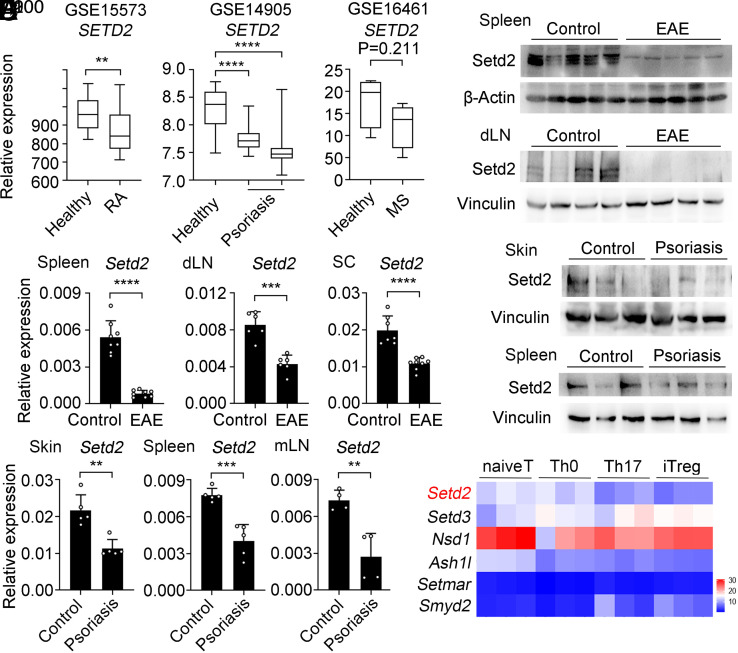
In order to test whether H3K36 methylation might be linked to autoimmune disease, we analyzed SETD2 expression in publicly available sequencing datasets from peripheral blood mononuclear cells isolated from rheumatoid arthritis, skins from psoriasis, and CD4+ T cells from MS patients (NCBI Gene Expression Omnibus: GSE15573, GSE14905, and GSE16461). We found that SETD2 expression was down-regulated compared to healthy controls across these autoimmune disease specimens (Fig. 1A). We further validated these results in well-established autoimmune mouse models with experimental autoimmune encephalitis (EAE) and psoriasis. We found that Setd2 protein and mRNA levels were significantly decreased in CD4+ T cells from EAE (Fig. 1 B and C) and psoriasis mice (Fig. 1 D and E). Therefore, Setd2 gene and protein expression levels are reduced in CD4+ T cells during the development of a range of autoimmune diseases.
Fig. 1.
Setd2 expression is reduced in autoimmune diseases. (A) Boxplot of SETD2 expression levels in healthy control groups and autoimmune disease groups, rheumatoid arthritis (RA), psoriasis, and multiple sclerosis (MS). (B) Immunoblot analysis of Setd2 levels of CD4+ T cells in the spleen and draining lymph node (dLN) from healthy controls and EAE mice. (C) qPCR analysis of Setd2 mRNA in the spleen, dLN, and spinal cord of EAE specimens and controls (n = 6 to 8). (D) Immunoblot analysis of Setd2 levels from healthy control and imiquimod induced-psoriasis mice. (E) qPCR analysis of Setd2 mRNA in the skin, spleen, and mesenteric lymph node (mLN) of healthy controls and psoriasis specimens (n = 4 to 5). (F) The heatmap shows the expression levels of histone H3K36 methyltransferases in naive CD4+ T, Th0 (α-CD3/CD28), Th17, and iTreg cells. Data represent one of three independent experiments. Results are presented as means ± SD (A, C, and E). **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We next used RNA sequencing (RNA-seq) (27) to analyze the expression of 8 reported H3K36 methyltransferases (23) in naive CD4+ T, Th0 (αCD3/CD28), Th17, and induced Treg (iTreg) cells. Setd2, Nsd1, and Ash1l were down-regulated in Th17 and iTreg cells compared with that in naive CD4+ T cells, while Smyd2 and Setd3 expression levels were increased in iTreg cells (Fig. 1F). Compared to that in naive CD4+ T cells, the Setd2 mRNA and protein levels in Th17 and iTreg cells were decreased (SI Appendix, Fig. S1). The downregulation of Setd2 both in CD4+ T cells from autoimmune diseases and in differentiated Th17 cells indicates a potential correlation between Setd2 and autoimmune pathogenesis.
Setd2 Suppresses Th17 and Promotes Treg Differentiation.
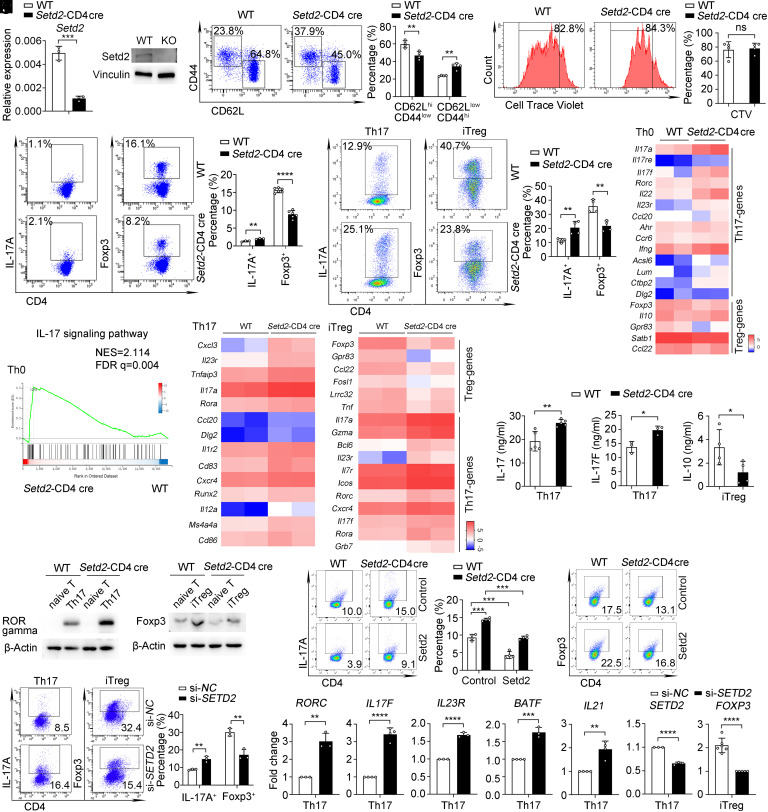
The above results inspired us to further investigate the function of Setd2 in T cell–mediated autoimmunity. To examine the function of Setd2 in regulating T cells and autoimmunity in vivo, we crossed Setd2-flox mice (Setd2f/f) (24) with CD4-Cre mice to obtain CD4-specific Setd2 knockout mice (named as Setd2-CD4 cre mice), and the expression of Setd2 in naive T cells of Setd2-CD4 cre mice was significantly lower as compared to the control group (Fig. 2A). We found that Setd2-CD4 cre mice had normal populations of lymphocytes and naturally regulated T cells (nTregs) in the thymus (SI Appendix, Fig. S2A). The percentages of T cells, B cells, and CD4+ and CD8+ T cells (SI Appendix, Fig. S2 B–E) in the spleen and lymph nodes also resembled controls in Setd2-CD4 cre mice. These results indicate that the peripheral T lymphocytes of Setd2-CD4 cre mice develop normally.
Fig. 2.
Deletion of Setd2 promotes Th17 but suppresses Treg cell polarization. (A) mRNA and protein levels of Setd2 expression in CD4+ T cells of wild-type (WT) and Setd2-CD4 cre mice (n = 3). (B) Flow cytometric analysis of CD62L+ and CD44+ in splenic CD4+ T cells from WT and Setd2-CD4 cre mice (n = 4). (C) CD4+ T cells proliferation after treatment with αCD3 (5 μg/mL) and αCD28 (2 μg/mL) for 72 h (n = 4). (D and E) Flow cytometric analysis and quantification of IL-17A+ and Foxp3+ cells in splenic CD4+ T cells (D) and differentiated Th17 and iTreg cells for 3 d (E) (n = 3 to 6). (F) Heatmap shows the differential genes from Th0 cells (fold change > 1.5, P < 0.05). (G) GSEA analysis of enrichment in IL-17 signaling pathway-related genes from Th0 cells. (H) Heatmap showing genes differentially expressed in Th17 cells and iTreg cells (fold change ≥ 2, P < 0.05). (I) CBA analysis of IL-17, IL-17F, and IL-10 in Th17 or iTreg cell supernatants (n = 3 to 5). (J) Immunoblot analysis of RORγt and Foxp3 levels in Th17 and iTreg cells. (K and L) Flow cytometric analysis of IL-17A+ and Foxp3+ cells transfected with Setd2 expressing lentivirus (K) and SETD2 siRNA (si-SETD2) or control siRNA (si-NC) (L) in Th17 and iTreg cells (n = 3 to 4). (M) qPCR analysis of Th17 and Treg-genes expression in human Th17 or iTreg cells transfected with si-SETD2 or si-NC (n = 3 to 6). Data show one of three independent experiments. Results are presented as means ± SD (A–E, I, and K–M). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns, not significant. FDR, false discovery rate. NES, normalized enrichment scores.
Next, we investigated the changes in T cell proliferation and activation. Notably, the percentages of naive CD4+ (CD62LhiCD44low) cells were decreased, and effector/memory CD4+ (CD62LlowCD44hi) cells were increased in Setd2-CD4 cre mice (Fig. 2B), implying that Setd2-deficient T cells have increased effector function. However, the proliferation of CD4+ T cells after treatment with T cell receptor (TCR) was similar in WT and Setd2-CD4 cre mice (Fig. 2C).
Based on the above findings, we assessed whether Setd2 functions in CD4+ T cell differentiation and function. Setd2-deficient mice showed increased IL-17+ cells and decreased Foxp3+ cells in their spleens (Fig. 2D). Consistently, Setd2 deficient CD4+ T cells from the spleen (Fig. 2E) and mLN (SI Appendix, Fig. S3A) displayed increased differentiation toward Th17 cells but decreased differentiation toward iTreg cells under in vitro polarization conditions. In addition, Setd2-deficient mice showed increased IFN-γ+ cells but unchanged IL-4+ cells (SI Appendix, Fig. S3 B and C). RNA-seq analysis revealed the upregulation of Th17-related genes, including Il17a, Il17f, etc. In contrast, the Treg-related genes Foxp3, Il10, and Gpr83 were down-regulated in TCR-activated T cells from Setd2-deficient mice (Fig. 2F and SI Appendix, Fig. S3 D and E). Gene set enrichment analysis (GSEA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the up-regulated genes in Setd2-deficient T cells were enriched in IL-17 signaling pathway components (Fig. 2G and SI Appendix, Fig. S3F). These results suggest that Setd2 plays an important role in inhibiting Th17 but promoting Treg cell differentiation in vitro and in vivo.
We next explored the function of Setd2 in shaping the gene signatures of polarized Th17 and iTreg cells. RNA-seq showed that signature Th17-related genes were increased and Treg-related signature genes were reduced in Setd2-deficient T cells (Fig. 2H and SI Appendix, Fig. S3 G and H). qPCR analysis further verified this phenomenon (SI Appendix, Fig. S3 I and J). In addition, the KEGG pathway analysis identified that the IL-17 signaling pathway and glycerolipid metabolism pathways were enriched in Th17 and iTreg cells (SI Appendix, Fig. S3K). In line with transcript levels, Setd2-deficient Th17 cells produced more IL-17 and IL-17F cytokines, while Setd2-deficient iTreg cells produced less IL-10 (Fig. 2I). Furthermore, Setd2 deficiency increased RORγt and decreased Foxp3 expression (Fig. 2J). Moreover, overexpression of Setd2 decreased Th17 and increased iTreg differentiation in both WT and Setd2-CD4 cre naive T cells in vitro (Fig. 2K).
Consistent with mouse T cells, silencing of SETD2 in human peripheral blood naive CD4+ T cells also promoted Th17 differentiation and decreased iTreg differentiation (Fig. 2 L and M and SI Appendix, Fig. S3L). Together, these data demonstrate that Setd2 specifically inhibits Th17 differentiation and promotes iTreg differentiation.
Setd2-Mediated H3K36me3 Promotes Lpcat4 Expression to Mediate Phosphatidylcholine Generation.
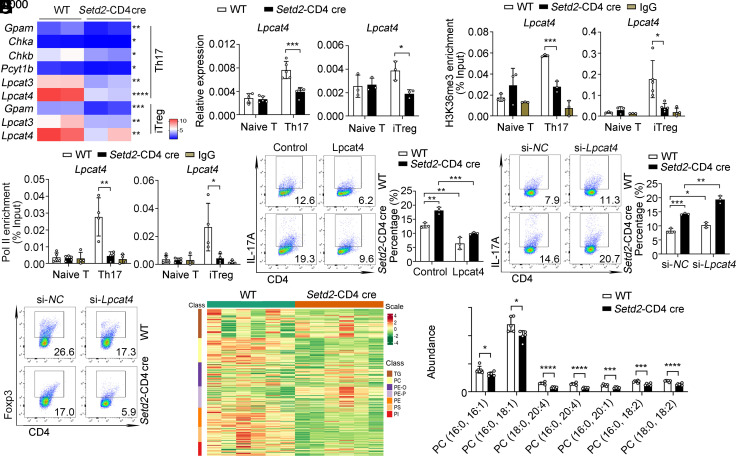
To explore the molecular mechanism underlying the imbalance in Th17/Treg cells associated with Setd2 loss, we further analyzed differentially expressed genes in Th17 cells using transcriptome analysis. RNA-seq analysis revealed that the expression of phosphatidylcholine synthesis-related genes, including Gpam, Lpcat3, and Lpcat4, was decreased in Setd2-deficient T cells (Fig. 3A). qPCR confirmed that the expression of phosphatidylcholine synthesis-related genes was down-regulated in Setd2-deficient T cells (Fig. 3B and SI Appendix, Fig. S4 A and B), indicating that loss of Setd2 impairs the phosphatidylcholine synthesis pathway. Phosphatidylcholine is synthesized by two distinct pathways: the de novo and remodeling pathways. Four lysophosphatidylcholine acyltransferase family members, Lpcat1, Lpcat2, Lpcat3, and Lpcat4 are responsible for catalyzing PC remodeling (13). Intriguingly, the mRNA and protein expression levels of Lpcat4 were markedly down-regulated in Setd2-deficient Th17 and iTreg cells (Fig. 3B and SI Appendix, Fig. S4C). Thus, Setd2 up-regulates Lpcat4 expression in T cells.
Fig. 3.
Setd2-mediated H3K36me3 promotes Lpcat4 transcription and phosphatidylcholine generation. (A) Heatmap shows the down-regulated phosphatidylcholine synthesis-related genes in Th17 and iTreg cells (fold change > 1.5, P < 0.05). (B) mRNA levels of Lpcat4 expression in T cells (n = 3 to 5). (C and D) ChIP-qPCR analysis of the enrichment of H3K36me3 (C) or RNA polymerase II (Pol II) (D) at the Lpcat4 promoter region in T cells (n = 3 to 5). (E) Flow cytometric analysis of IL-17A+ cells transfected with control and Lpcat4 expressing lentivirus in Th17 cells (n = 3). (F and G) Flow cytometric analysis of IL-17A+ and Foxp3+ cells transfected with Lpcat4 siRNA (si-Lpcat4) or control siRNA (si-NC) in Th17 and iTreg cells (n = 3). (H) Cluster analysis of lipidomics in Th17 cells from WT and Setd2-CD4 cre mice. Six independent samples were analyzed for each group. (I) Lipidomics analysis and quantification of differential phosphatidylcholine species in Th17 cells (n = 5 to 6). Data show one of three independent experiments. Results are presented as means ± SD (A–F and I). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Setd2 trimethylates H3K36, a mark associated with transcriptional activation (23). To test whether Setd2 controls Th17/Treg cell balance through changes in the Setd2-mediated H3K36me3 profile, we measured H3K36me3 levels in T cells. As expected, H3K36me3 levels were increased in Th1, Th17, and iTreg cells (SI Appendix, Fig. S4D), suggesting that H3K36me3 is involved in T cell differentiation. Since Setd2 deficiency significantly reduces Lpcat4 expression, we wondered whether Setd2 regulates Lpcat4 gene expression through its histone methyltransferase activity. ChIP-qPCR results showed that the H3K36me3 modification was enriched at the promoter region of Lpcat4 in WT cells and was lost in Setd2-deficient cells (Fig. 3C). RNA polymerase II (Pol II) can initiate transcription and synthesize mRNA (28). ChIP-qPCR revealed that Pol II occupancy was decreased at the Lpcat4 promoter region in Setd2-deficient T cells (Fig. 3D). Therefore, Setd2 mediates H3K36me3 modification at the Lpcat4 gene promoter region, thereby promoting the transcription of the Lpcat4 gene.
LPCAT family members can catalyze the conversion of LPC to PC and have been linked to both pathological and physiological processes (13). To explore the role of Lpcat4 in T cell differentiation, we used a pan-lysophospholipid acyltransferase inhibitor, CI-976, that can disrupt the phospholipid remodeling pathway (29). We found that the expression of Th17-related genes was up-regulated and Treg-related genes were down-regulated after CI-976 treatment (SI Appendix, Fig. S4E). Furthermore, overexpression of Lpcat4 significantly down-regulated Th17 and up-regulated iTreg differentiation in both WT and Setd2-deficient T cells (Fig. 3E and SI Appendix, Fig. S4F). Consistently, silencing of Lpcat4 also promoted Th17 differentiation and decreased iTreg differentiation (Fig. 3 F and G). These data suggest that Setd2 targets Lpcat4 to regulate Th17/Treg cell balance and PC remodeling is involved in regulating Th17/Treg cell differentiation.
Next, we performed lipidomics to analyze the lipid composition in Th17 cells. Consistent with transcriptome results, lipidomics analysis revealed that the up-regulated lipid components were significantly enriched in phospholipid biosynthesis and glycerolipid metabolism as analyzed using the Small Molecule Pathway Database (SI Appendix, Fig. S4G). Many classes of lipids such as PC, phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylinositol (PI), were lower in Setd2-deficient Th17 cells (Fig. 3H). Lpcat4 uses acyl CoAs (18:2 or 20:4-acyl-CoA) and oleoyl-CoA (18:1-acyl-CoA) as substrates to form the corresponding PC (13). Our lipidomics data revealed that Lpcat4-mediated production of the PC species PC(16:0, 18:1), PC(16:0, 18:2), PC(16:0, 20:4), PC(18:0, 18:2), and PC(18:0, 20:4) was significantly decreased in Setd2-deficient Th17 cells (Fig. 3I). Overall, our data indicate that Setd2 promotes H3K36 trimethylation at the Lpcat4 promoter, thus increasing Lpcat4 expression and PC generation.
Lpcat4-Mediated PC(16:0, 18:2) Generation Controls Th17/Treg Cell Balance.
Next, we tested the proposal that Setd2-mediated PC generation is involved in Th17/Treg balance. We measured the effect of stimulating CD4+ T cells with two commercially available PCs: PC(16:0, 18:1) or PC(16:0, 18:2). PC(16:0, 18:1) treatment did not affect Th17 and iTreg cell polarization or cytokine expression (SI Appendix, Fig. S5). These data suggest that PC(16:0, 18:1) does not affect Th17 or iTreg differentiation.
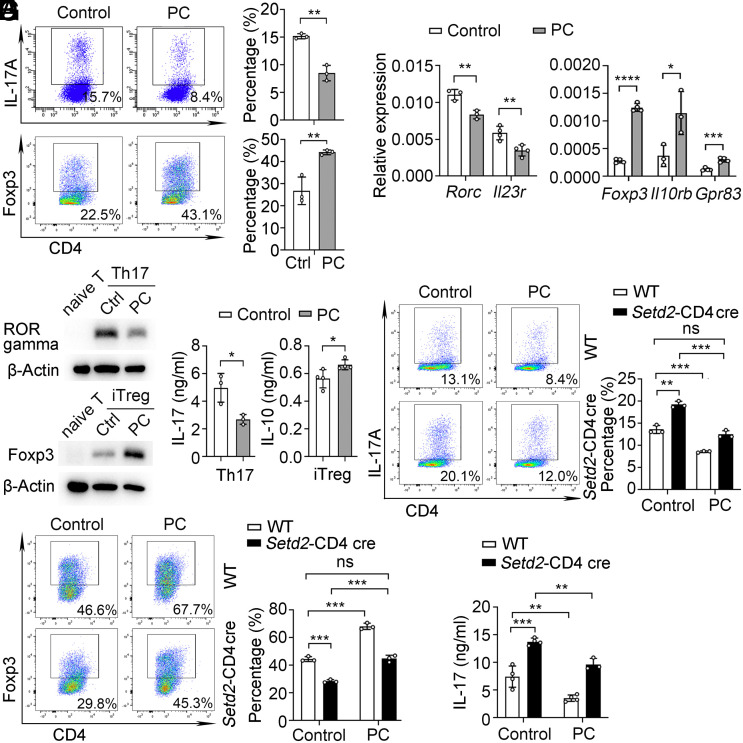
In contrast, the proportion of Th17 cells was decreased and iTreg cells were increased compared to the control group after PC(16:0, 18:2) treatment (Fig. 4A). Consistently, Th17 differentiation markers were down-regulated and iTreg markers were increased in the presence of PC(16:0, 18:2) (Fig. 4 B and C). In addition, the PC(16:0, 18:2)-treated Th17 cells produced less IL-17 and more IL-10 in PC(16:0, 18:2)-treated iTreg cells (Fig. 4D). Thus, PC(16:0, 18:2) can contribute to regulating Th17/Treg cell balance.
Fig. 4.
Lpcat4-mediated PC(16:0, 18:2) controls Th17/Treg cell balance. (A) Flow cytometric analysis of IL-17A+ in Th17 (Top) and Foxp3+ in iTreg (Below) cells after treatment with solvent control (Ctrl) or PC(16:0, 18:2) (PC, 50 μM) (n = 3). (B) qPCR analysis of Rorc, Il23r, Foxp3, Il10rb, and Gpr83 mRNA expression in Th17 and iTreg cells after stimulation with control or PC (n = 3 to 4). (C) Immunoblot analysis of RORγt and Foxp3 levels in Th17 and iTreg cells after PC and Ctrl treatments. (D) CBA analysis of IL-17 in Th17 and IL-10 in iTreg cells after PC and Ctrl treatment (n = 3 to 4). (E and F) Flow cytometric analysis of IL-17A+ and Foxp3+ in Th17 and iTreg cells from WT and Setd2-CD4 cre mice (n = 3) after PC or Ctrl treatment. (G) CBA analysis of IL-17 in Th17 cell after PC or Ctrl treatment (n = 3 to 4). Data show one of three independent experiments. Results are presented as means ± SD (A, B, and D–G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns, not significant. PC referred to PC(16:0, 18:2).
We further examined whether PC(16:0, 18:2) is responsible for Setd2-mediated T cell differentiation. We found that PC(16:0, 18:2) can reduce the differentiation of Setd2-deficient Th17 cells and enhance the differentiation of Setd2-deficient iTreg cells (Fig. 4 E–G). Collectively, these data demonstrate that Setd2 promotes Lpcat4-mediated PC(16:0, 18:2) generation to control Th17/Treg cell balance.
Setd2 Promotes PC(16:0, 18:2) Generation to Limit ER Stress and Oxidative Stress.
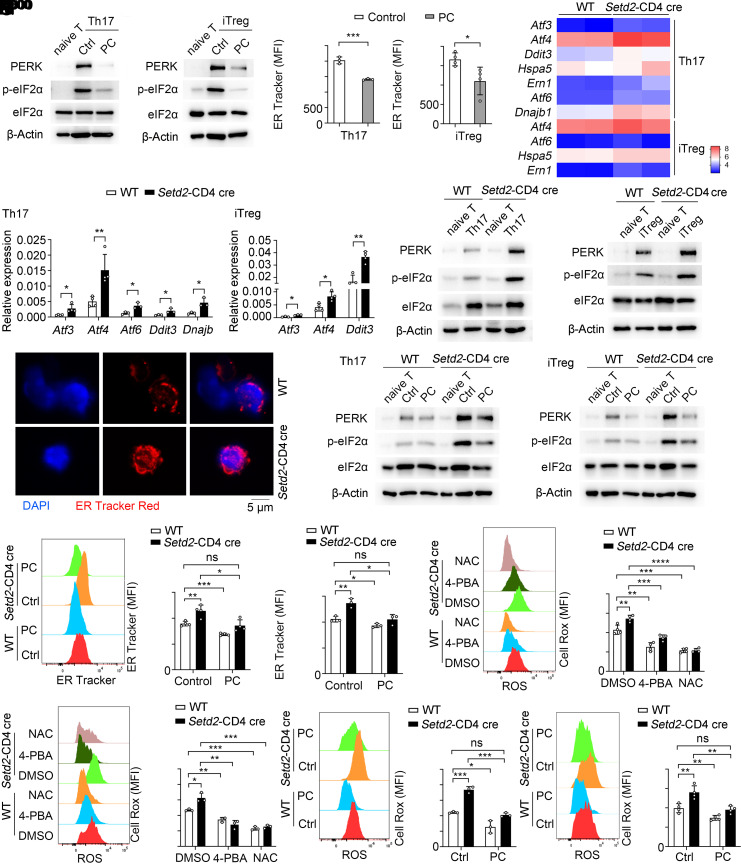
We next sought to further define the molecular mechanisms underlying PC-mediated control of T cell balance. Distinct PCs have unique acyl-chain composition and membrane remodeling properties that play variable roles in regulating inflammation (30, 31). LPCAT family members were previously shown to regulate tissue PC acyl-chain composition: for example, Lpcat3 can change the composition of membrane phospholipids and ameliorate endoplasmic reticulum (ER) stress and tissue inflammation (31). We therefore hypothesized that the Setd2/PC(16:0, 18:2) axis may regulate ER stress during T cell differentiation. Interestingly, PC(16:0, 18:2) significantly reduced the mRNA expression of ER stress makers and ER stress-related proteins in Th17 and iTreg cells (Fig. 5A and SI Appendix, Fig. S6 A and B). Cells undergoing ER stress can be identified by increased ER-tracker probe uptake (32). In addition, treatment with PC(16:0, 18:2) was associated with a reduction in ER-tracker mean fluorescence intensity (MFI) in Th17 and iTreg cells (Fig. 5B). These data indicate that Lpcat4-mediated synthetic product PC(16:0, 18:2) can restrain ER stress signaling pathway.
Fig. 5.
Setd2 promotes PC(16:0, 18:2) generation to limit ER stress and the oxidative stress response. (A) PERK and phospho-eIF2α levels in T cells treated with PC(16:0, 18:2) and controls. (B) Flow cytometric analysis of ER tracker mean fluorescence intensity (MFI) in T cells treated with control or PC (n = 3 to 4). (C) Heatmap showing differentially expressed ER stress markers in T cells (fold change > 1.5, P < 0.05). (D and E) ER stress markers mRNA and protein expression in T cells (n = 3 to 4). (F) Immunofluorescence analysis of ER Tracker Red (red) and DAPI (blue) in Th17 cells. (Scale bar, 5 μm.) (G) The protein levels of PERK and phospho-eIF2α in WT and Setd2-deficient Th17 and iTreg cells stimulated with control or PC. (H and I) Analysis of ER tracker MFI in Th17 (H) and iTreg (I) cells treated with control or PC (n = 3 to 4). (J and K) Flow cytometric analysis of Cell Rox MFI in Th17 (J) and iTreg (K) cells stimulated with DMSO, 4-PBA (1 mM), and NAC (1 mM) (n = 3 to 4). (L and M) Flow cytometric analysis of Cell Rox MFI in Th17 (L) and iTreg (M) cells treated with control or PC(16:0, 18:2) (n = 3 to 4). Data show one of three independent experiments. Results are presented as means ± SD (B, D, and H–M). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns, not significant.
The observation that PC(16:0, 18:2) can restrain ER stress and regulate Th17/Treg cell balance, prompted us to test whether Setd2 feeds into the same pathway and is linked to ER stress-associated T cell differentiation. In our RNA-seq data, several ER stress markers were up-regulated in Setd2-deficient T cells (Fig. 5C). These ER stress markers and signaling proteins were also increased in Setd2-deficient T cells (Fig. 5 D and E). ER stress is generally associated with enlarged ER lumen size in cells that can be detected with an ER tracker probe using confocal microscopy (32). Confocal microscopy imaging showed that the ER lumen was larger in Setd2-deficient Th17 cells (Fig. 5F). The data indicate that similar to PC(16:0, 18:2), Setd2 can inhibit the ER stress response in Th17 and iTreg cells.
Given the comparable role of Setd2 and PC(16:0, 18:2) in inhibiting ER stress and regulating Th17/Treg differentiation, we speculated that Setd2 may control Th17/Treg cell balance by promoting PC(16:0, 18:2)-mediated inhibition of ER stress. As expected, PC(16:0, 18:2) could largely reverse the up-regulated ER stress signaling (Fig. 5G) and ER abundance (Fig. 5 H and I and SI Appendix, Fig. S6C) in Setd2-deficient Th17 or iTreg cells. By using 4-phenylbutyric acid (4-PBA), an ER stress inhibitor, we showed that 4-PBA significantly reduced Th17 differentiation in both WT and Setd2-CD4 cre T cells (SI Appendix, Fig. S6D). Together, these results demonstrate that Setd2 deficiency suppresses PC(16:0, 18:2) to restrict the ER stress response, thereby controlling Th17/Treg cell balance.
ER stress is accompanied by oxidative stress and reactive oxygen species (ROS) production, which plays an important role in Th17/Treg imbalance-mediated autoimmune diseases (33, 34). Therefore, we investigated whether Setd2/PC(16:0, 18:2)-mediated inhibition of ER stress may alter the oxidative stress response of Th17/Treg cells. We found that the deletion of Setd2 promotes the production of ROS in Th17 and iTreg cells, while treatment with 4-PBA significantly reduces the production of ROS in Setd2-deficient T cells (Fig. 5 J and K). N-acetyl cysteine (NAC), is a scavenger of ROS (35). ROS is known to function in autophagy and phagocytosis (36–38). We also observed a significant enrichment in the expression of genes linked to phospholipid phagocytosis in Setd2-deficient T cells (SI Appendix, Fig. S6E), further supporting a model where Setd2 inhibits ROS production by regulating phospholipid production. Next, we tested the role of PC in regulating ROS production and found that PC(16:0, 18:2) treatment significantly reduced ROS levels in WT and Setd2-deficient T cells (Fig. 5 L and M). Endoplasmic reticulum oxidoreductin 1-alpha (Ero1α) is an enzyme in ROS production. Consistently, PC(16:0, 18:2) also down-regulated the expression of Ero1α in WT and Setd2-deficient Th17 cells (SI Appendix, Fig. S6F). These results demonstrate that Setd2 inhibits ER stress and the subsequent oxidative stress response by promoting PC(16:0, 18:2) generation.
Setd2 Promotes PC(16:0, 18:2) Generation to Restrict HIF-1α Transcription in T Cells.
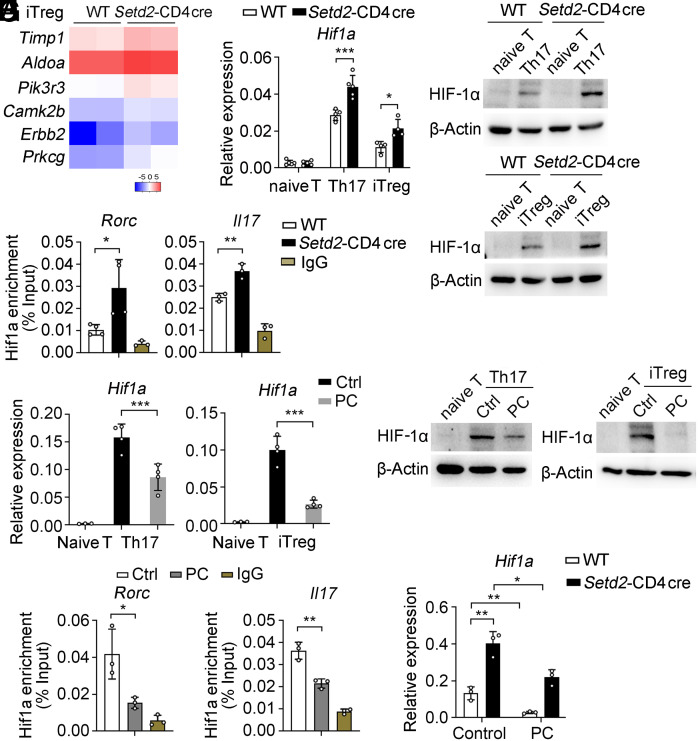
HIF-1α is a key transcription factor that controls the balance between Th17 and Treg differentiation (39). ROS is known to be essential for HIF-1α activation by inhibiting the activity of the prolyl hydroxylase domain and enhancing HIF-1α stability (7, 40). Therefore, we explored whether Setd2 regulates Th17/Treg cell balance by reducing the oxidative stress response and thus altering the activity of HIF-1α. In RNA-seq data, we found that genes linked to the HIF-1 signaling pathway were up-regulated in Setd2-deficient iTreg cells (Fig. 6A). We also detected increased HIF-1α mRNA and protein expression in the Setd2-CD4 cre mice (Fig. 6 B and C). ChIP-qPCR revealed that HIF-1α directly binds to the Rorc and Il17 promoter, and Setd2 deficiency can increase the enrichment of HIF-1α binding to the Rorc and Il17 promoter region (Fig. 6D). These results indicate that Setd2 can inhibit HIF-1α transcriptional activity to suppress Rorc and Il17 expression.
Fig. 6.
Setd2 increases PC(16:0, 18:2) levels to inhibit HIF-1α transcriptional activity. (A) Heatmap showing HIF-1 signaling-linked genes in iTreg cells (fold change ≥ 2, P < 0.05). (B and C) mRNA and protein levels of HIF-1α in T cells (n = 4 to 5). (D) ChIP-qPCR analysis of HIF-1α enrichment at the Rorc and Il17 promoter region in Th17 cells (n = 3 to 4). (E and F) mRNA and protein levels of HIF-1α in Th17 and iTreg cells after stimulation with control or PC(16:0, 18:2) (n = 3 to 4). (G) ChIP-qPCR analysis of the enrichment of Hif1α at the Rorc and Il17 promoter regions in Th17 cells after stimulation with control or PC (n = 3). (H) qPCR analysis of Hif1a mRNA in Th17 cells after stimulation with control or PC (n = 3). Data show one of three independent experiments. Results are presented as means ± SD (B, D, E, G, and H). *P < 0.05, **P < 0.01, and ***P < 0.001.
Moreover, PC(16:0, 18:2) directly inhibits HIF-1α expression in WT Th17 and iTreg cells (Fig. 6 E and F), and ChIP-qPCR results show that PC(16:0, 18:2) can significantly reduce the binding ability of HIF-1α to the Rorc and Il17 promoter region (Fig. 6G). PC(16:0, 18:2) also diminishes Hif1α transcript levels in Setd2-deficient Th17 cells (Fig. 6H), suggesting that Setd2 inhibits the transcriptional activity of HIF-1α by promoting Lpcat4-mediated PC(16:0, 18:2) generation.
Setd2 Deficiency Exacerbates T Cell–Mediated Autoimmunity In Vivo.
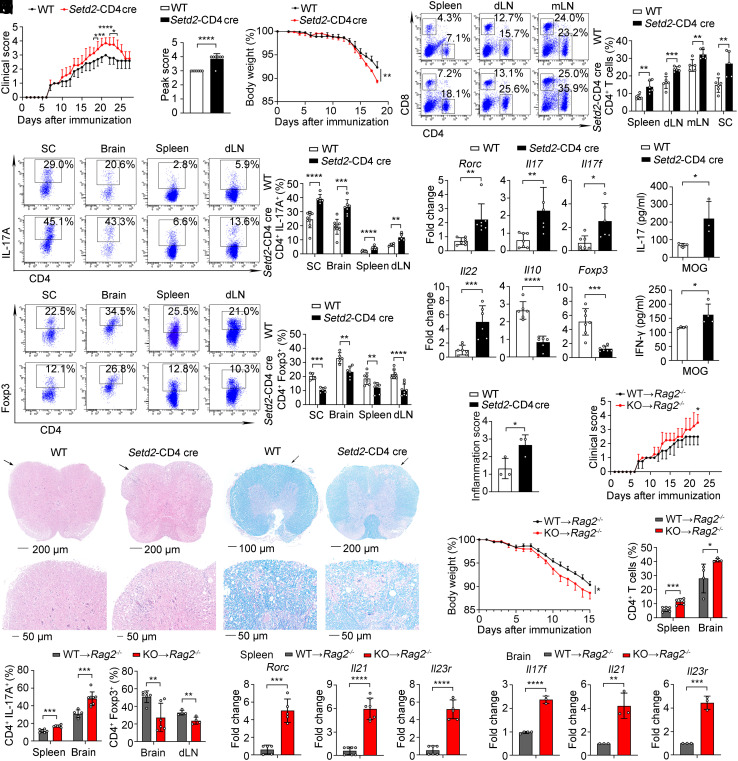
We next explored the role of Setd2 in the development of T cell–mediated autoimmune diseases in a multiple sclerosis mouse model, experimental autoimmune encephalomyelitis. We found that Setd2-deficient mice showed more severe disease symptoms and had a higher clinical score, peak score, and increased loss of body weight compared to WT mice (Fig. 7 A–C). In addition, Setd2-CD4 cre mice showed a significant increase in CD4+ T and Th17 cell infiltration, while the infiltration of Treg cells was reduced in Setd2-CD4 cre mice (Fig. 7 D–F and SI Appendix, Fig. S7A). Consistently, the expression of Rorc, Il17, Il17f, Batf, and Il22 was markedly increased, whereas Foxp3, Il10, and Il10ra were reduced in the spleen and brain (Fig. 7G and SI Appendix, Fig. S7 B and C). Moreover, in vitro MOG35-55 peptide recall analysis substantially elevated IL-17 and IFN-γ production in Setd2-deficient splenocytes (Fig. 7H). Histological analysis showed increased lymphocyte infiltration into the spinal cords of Setd2-CD4 cre mice and more severe demyelination (Fig. 7 I–K). These results demonstrate that CD4+ T cell–specific deletion of Setd2 aggravates EAE immunopathology by regulating the balance of Th17/Treg cell differentiation.
Fig. 7.
Deficiency of Setd2 aggravates the severity of the autoimmune disease. (A–C) Clinical scores (A), peak scores (B), and body weight (C) of WT and KO mice after induced EAE (n = 4 to 7). (D) Frequency of CD4+ and CD8+ T cells in the spleen, dLN, mLN, and SC (n = 5 to 6). (E and F) Frequency of IL-17A+ and Foxp3+ cells in the SC, brain, spleen, and dLN (n = 4 to 9). (G) mRNA expression of inflammatory genes in the spleen (n = 5 to 7). (H) Splenocytes were rechallenged with MOG35–55 peptide (20 μg/mL) for 3 d; IL-17 and IFN-γ were assessed by CBA (n = 3 to 4). (I and J) Representative histology of the spinal cord stained with hematoxylin and eosin (I) and Luxol fast blue (J) after EAE induction (day 21). (K) Inflammation scores of spinal cords are shown (n = 3). (L–Q) Adoptive transfer of WT and Setd2-deficient CD4+ T cells into Rag2–/– host mice. (L and M) Clinical scores and body weight of Rag2–/– host mice after EAE inducion (n = 4). (N and O) Quantification of CD4+, IL-17A+, and Foxp3+ cells in the spleen, brain, and dLN (n = 4 to 6). (P and Q) mRNA expression of inflammatory genes in the spleen and brain (n = 3 to 7). Data show one of two or three independent experiments. Results are presented as means ± SD (A–H and K–Q). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. KO referred to Setd2-CD4 cre mice.
To directly examine the cell-intrinsic function of Setd2 in CD4+ T cells, we transferred naive CD4+ T cells from WT and Setd2-CD4 cre mice into Rag2−/− hosts to induce the EAE model. Rag2−/− hosts that received Setd2-deficient CD4+ T cells showed higher clinical scores and increased loss of body weight (Fig. 7 L and M), and significantly increased CD4+ T cells and IL-17A+ cells infiltration and also reduced Foxp3+ cells (Fig. 7 N and O and SI Appendix, Fig. S7D). In addition, expression of the inflammatory cytokines was significantly increased in recipients of Setd2-deficient T cells (Fig. 7 P and Q). Histological analysis showed the infiltration of lymphocytes into the spinal cord was increased along with more severe demyelination in recipients of Setd2-deficient T cells (SI Appendix, Fig. S7E). To investigate whether PC(16:0,18:2) has a therapeutic effect on autoimmune diseases in Setd2-CD4 cre mice, we used an EAE model in vivo to show that PC(16:0,18:2) treatment significantly reduced Th17 cell infiltration and enhanced the numbers of Treg cell both in WT and Setd2-deficient mice, as compared to the control treatment (SI Appendix, Fig. S7 F–I). The data suggest that PC(16:0,18:2) could rescue Setd2-CD4 cre mice from the T cell imbalance during autoimmunity, potentially exerting a therapeutic effect on autoimmune diseases. Together, these data demonstrate that Setd2 deficiency exacerbates CD4+ T cell–mediated autoimmunity that correlates with Th17/Treg cell imbalance.
Discussion
Here, we have uncovered a cross-regulatory mechanism that links epigenetic modification and phospholipid metabolism in autoimmunity. Our study reveals a function for Setd2-mediated H3K36me3 modification in regulating Th17/Treg cell balance via Lpcat4-based phospholipid remodeling. The metabolite PC(16:0, 18:2), produced by Lpcat4, can inhibit endoplasmic reticulum stress and the oxidative stress response, thus reducing HIF-1α transcriptional activity and suppressing Th17 differentiation and promoting Treg differentiation. Our study, therefore, reveals a significant role for the phospholipid PC(16:0, 18:2) in controlling Th17/Treg cell balance and also provides mechanistic insight into the epigenetic control of metabolic processes in T cell–mediated autoimmunity.
H3K36 modifications have been shown to regulate T cell differentiation and function. Setd2, as a H3K36 trimethyltransferase, has been studied primarily in tumors and nonimmune cells (24, 26, 41). Although evidence has shown that Setd2 facilitates RORγt+ Treg homeostasis in group 3 innate lymphoid (ILC3) cells and suppresses intestinal inflammation (41), the role of Setd2 in T cell–mediated autoimmune diseases has remained untested. In our study, we found reduced Setd2 expression in multiple autoimmune diseases, including psoriasis, rheumatoid arthritis, and experimental autoimmune encephalomyelitis. Functionally, Setd2 prevents autoimmune pathogenesis by suppressing Th17 cell differentiation and promoting Treg cell differentiation through H3K36 trimethylation. Therefore, we suggest Setd2 acts as an indispensable brake that usually prevents harmful autoreactive T cell responses. Potential strategies for enforcing Setd2 expression and function may provide opportunities for the treatment of autoimmune and inflammatory diseases.
Dysregulation of lipid metabolism is related to many diseases, including chronic inflammation, autoimmunity, cancer, and metabolic and degenerative diseases (7, 42). The involvement of lipid metabolites as well as their catalyzing enzymes in T cell differentiation is attracting increasing attention. One recent study showed that lysophosphatidylethanolamine synthase Pla2g12a produces 1-Oleoyl-lysophosphatidylethanolamine [LPE (1-18:1)], which can directly bind to RORγt and promote RORγt transcriptional activity (43). Our study revealed a phospholipid remodeling enzyme Lpcat4 is regulated by an epigenetic mechanism, and Lpcat4-mediated metabolite PC(16:0, 18:2) can inhibit the transcriptional activity of HIF-1α to regulate Th17/Treg cell balance. Therefore, phospholipids play widespread roles in determining Th17 cell generation. As LPE (1-18:1) contained sn-1 saturated fatty acids but PC(16:0, 18:2) contained sn-2 unsaturated fatty acid linoleic acid, we suspected that the opposite role of LPE (1-18:1) and PC(16:0, 18:2) was likely due to their differential fatty acyl chain component and property. Whether PC(16:0, 18:2) directly binds to key protein regulators such as HIF1α and RORγt in Th17 cells requires further exploration.
LPCAT family members participate in phospholipid remodeling (13). LPCAT1, LPCAT2, and LPCAT3 are mainly involved in the pathogenesis of cancer and hepatocellular inflammation (44, 45). However, little is known about the molecular mechanisms controlling LPCAT expression or the potential immunological functions of this family. Here, we have revealed a role for Lpcat4 in controlling T cell polarization via PC remodeling. We found that Setd2 directly targets Lpcat4 and thereby promotes LPCAT4-dependent unsaturated PC generation. Since PCs are the main component of the cell membrane, it will be interesting to further explore whether Setd2-regulated unsaturated PC affects membrane fluidity or membrane microdomains in the regulation of T cell immunity.
Dysregulated ER stress can initiate inflammatory responses and metabolic abnormality, leading to a series of human diseases, including autoimmune and neurodegenerative disorders. However, the endogenous pathways that restrain ER stress to maintain immune homeostasis remained unknown. Here, we report that Lpcat4-mediated PC(16:0, 18:2) generation can inhibit ER stress in Th17 and iTreg cells, thus shedding insight into the metabolic control of T cell immunity. Many lipid biosynthesis enzymes are located in the ER membrane, and LPCAT family members are also ER membrane proteins (13, 46). Future studies will clarify whether a specific subcellular organelle is associated with Lpcat4-mediated PC(16:0, 18:2) generation. In addition, the detailed mechanism for PC-mediated modulation of ER stress, as well as the in vivo role of PC(16:0, 18:2) in controlling autoimmunity await future analyses.
In summary, we show that Setd2-mediated H3K36me3 is necessary for maintaining the balance of Th17/Treg cell differentiation to restrain autoimmunity. This process is dependent on Lpcat4-mediated phosphatidylcholine generation. Our research adds to our knowledge of the cross-talk between epigenetics and the phospholipid remodeling pathway and identifies Setd2 as a potential biomarker and therapeutic target for autoimmune disorders.
Materials and Methods
Mice.
C57BL/6J (6 to 8 wk of age) mice were obtained from Joint Ventures Sipper BK Experimental Animal Company (Shanghai, China). Rag2−/− (B6(Cg)-Rag2tm1.1Cgn/J, 008449) and CD4-cre (STOCK Tg(Cd4-cre)1Cwi/BfluJ) mice were obtained from the Jackson Laboratory. Setd2fl/fl mice were prepared as described previously (24). All animal experiments conformed to the NIH Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Naval Medical University, Shanghai.
Detailed materials and methods are presented fully in SI Appendix, Materials and Methods.
Statistical Analysis.
Statistical analysis was performed using software GraphPad Prism 8.3.0. The data were assessed using the unpaired two-tailed Student’s t test; P values ≤ 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Ms. Nayi Weng and Ms. Yiran Wang (Naval Medical University, Shanghai, China) for their technical assistance. This work was supported by Grants from the National Key R&D Program (2018YFA0507400), National Natural Science Foundation of China (82388201, 32370963, 31870909, and 32070903) and CAMS Innovation Fund for Medical Sciences (2021-I2M-1-017) and China Postdoctoral Science Foundation (2023M740318).
Author contributions
X.C. designed research; Y.C., K.C., and H.Z. performed research; H.Q. contributed new reagents/analytic tools; Y.C. and J.L. analyzed data; and Y.C., J.L., and X.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Juan Liu, Email: juanliu@immunol.org.
Xuetao Cao, Email: caoxt@immunol.org.
Data, Materials, and Software Availability
The RNA sequencing data are deposited in the NCBI GEO under accession code GSE235827 (27). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Theofilopoulos A. N., Kono D. H., Baccala R., The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., Cao X., RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 33, 97–115 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charabati M., Wheeler M. A., Weiner H. L., Quintana F. J., Multiple sclerosis: Neuroimmune crosstalk and therapeutic targeting. Cell 186, 1309–1327 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heckmann B. L., et al. , LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178, 536–551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman N. M., Boothby M. R., Chi H., Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Chisolm D. A., Weinmann A. S., Connections between metabolism and epigenetics in programming cellular differentiation. Annu. Rev. Immunol. 36, 221–246 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Green D. R., Galluzzi L., Kroemer G., Cell biology. Metabolic control of cell death. Science 345, 1250256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim S. A., Su W.A.-O., Chapman N. M., Chi H., Lipid metabolism in T cell signaling and function. Nat. Chem. Biol. 18, 470–481 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel C. H., Leone R. D., Horton M. R., Powell J. D., Targeting metabolism to regulate immune responses in autoimmunity and cancer. Nat. Rev. Drug. Discov. 18, 669–688 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Wagner A., et al. , Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo C., et al. , SLC38A2 and glutamine signalling in cDC1s dictate anti-tumour immunity. Nature 620, 200–208 (2023), 10.1038/s41586-023-06299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou W., Green D. R., Beggars banquet: Metabolism in the tumor immune microenvironment and cancer therapy. Cell Metab. 35, 1101–1113 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Tontonoz P., Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 81, 165–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B., et al. , Intestinal phospholipid remodeling is required for dietary-lipid uptake and survival on a high-fat diet. Cell Metab. 23, 492–504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong X., et al. , ER phospholipid composition modulates lipogenesis during feeding and in obesity. J. Clin. Invest. 127, 3640–3651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q., Cao X., Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 19, 417–432 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q., Cao X., Epigenetic remodeling in innate immunity and inflammation. Annu. Rev. Immunol. 39, 279–311 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Cao X., Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Henning A. N., Roychoudhuri R., Restifo N. P., Epigenetic control of CD8(+) T cell differentiation. Nat. Rev. Immunol. 18, 340–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giles J. R., et al. , Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavalli G., Heard E., Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Phan A. T., Goldrath A. W., Glass C. K., Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46, 714–729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner E. J., Carpenter P. B., Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K., et al. , Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506.e414 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Huang H., et al. , Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M., et al. , The histone methyltransferase SETD2 modulates oxidative stress to attenuate experimental colitis. Redox Biol. 43, 102004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Cao X., Data from “Methyltransferase Setd2 prevents T cell-mediated autoimmune diseases via phospholipid remodeling”. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse235827. Deposited 26 June 2023. [DOI] [PMC free article] [PubMed]
- 28.Osman S., Cramer P., Structural biology of RNA polymerase II transcription: 20 years on. Annu. Rev. Cell Dev. Biol. 36, 1–34 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Chambers K., Judson W. J., A unique lysophospholipid acyltransferase (LPAT) antagonist, CI-976, affects secretory and endocytic membrane trafficking pathways. J. Cell Sci. 118, 3061–3071 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Harayama T., et al. , Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 20, 295–305 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Rong X., et al. , LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 18, 685–697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oslowski C. M., Urano F., Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490, 71–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hotamisligil G., Davis R., Cell signaling and stress responses. Cold Spring Harb. Perspect. Biol. 8, a006072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Yang X., Zou H., Li M., Oxidative stress and Treg and Th17 dysfunction in systemic lupus erythematosus. Oxid Med. Cell Longev. 2016, 2526174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atarashi K., et al. , Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernon P. J., Tang D., Eat-me: Autophagy, phagocytosis, and reactive oxygen species signaling. Antioxid Redox Signal. 18, 677–691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galluzzi L., Green D. R., Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moretti J., et al. , STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell 171, 809–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang E. V., et al. , Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaelin W. G. Jr., Ratcliffe P. J., Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell. 30, 393–402 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Chang J., et al. , Setd2 determines distinct properties of intestinal ILC3 subsets to regulate intestinal immunity. Cell Rep. 38, 110530 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Wymann M. P., Schneiter R., Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 9, 162–176 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Endo Y.A.-O., et al. , 1-Oleoyl-lysophosphatidylethanolamine stimulates RORγt activity in T(H)17 cells. Sci. Immunol. 8, eadd4346 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Cotte A. K., et al. , Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat. Commun. 9, 322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfgang M. J., Remodeling glycerophospholipids affects obesity-related insulin signaling in skeletal muscle. J. Clin. Invest. 131, e148176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura T., Stefan C. J., Specialized ER membrane domains for lipid metabolism and transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158492 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The RNA sequencing data are deposited in the NCBI GEO under accession code GSE235827 (27). All other data are included in the manuscript and/or SI Appendix.