Significance
Fetal development in placental mammals relies on formation of the maternal–fetal exchange interface, which ensures nutrient supply to the developing fetus. Syncytiotrophoblast (SynT), a special cell type of the placenta, is the major constituent of the maternal–fetal exchange interface. However, molecular mechanisms that control SynT development are incompletely understood. Here, using genetic mouse models and human trophoblast stem cells, we showed that GATA2 and GATA3, two transcription factors conserved in placental mammals, are essential for SynT development. In addition, we identified gene regulatory mechanisms through which GATA2 and GATA3 ensure SynT development in humans. Thus, we uncovered an essential mechanism for the development of the maternal–fetal exchange interface and the progression of pregnancy.
Keywords: placenta, trophoblast progenitors, syncytiotrophoblast, GATA2, GATA3
Abstract
The placenta establishes a maternal–fetal exchange interface to transport nutrients and gases between the mother and the fetus. Establishment of this exchange interface relies on the development of multinucleated syncytiotrophoblasts (SynT) from trophoblast progenitors, and defect in SynT development often leads to pregnancy failure and impaired embryonic development. Here, we show that mouse embryos with conditional deletion of transcription factors GATA2 and GATA3 in labyrinth trophoblast progenitors (LaTPs) have underdeveloped placenta and die by ~embryonic day 9.5. Single-cell RNA sequencing analysis revealed excessive accumulation of multipotent LaTPs upon conditional deletion of GATA factors. The GATA factor–deleted multipotent progenitors were unable to differentiate into matured SynTs. We also show that the GATA factor–mediated priming of trophoblast progenitors for SynT differentiation is a conserved event during human placentation. Loss of either GATA2 or GATA3 in cytotrophoblast-derived human trophoblast stem cells (human TSCs) drastically inhibits SynT differentiation potential. Identification of GATA2 and GATA3 target genes along with comparative bioinformatics analyses revealed that GATA factors directly regulate hundreds of common genes in human TSCs, including genes that are essential for SynT development and implicated in preeclampsia and fetal growth retardation. Thus, our study uncovers a conserved molecular mechanism, in which coordinated function of GATA2 and GATA3 promotes trophoblast progenitor-to-SynT commitment, ensuring establishment of the maternal–fetal exchange interface.
Rodents and humans have hemochorial placentation, which is characterized by two vascular systems, maternal and fetal, and the maternal blood comes in direct contact with multinucleated SynTs (1–3). SynTs ensure the barrier function and facilitate nutrient and gas exchange between the maternal blood and the developing fetus (4). Establishment of this exchange interface is essential for the successful progression of pregnancy and mammalian reproduction.
In a postimplantation mouse embryo, multipotent trophoblast progenitor cells (TPCs) reside within the ectoplacental cone (EPC) and the chorion (5). Eventually, the TPCs within the EPC and chorion give rise to lineage-specific trophoblast precursors, which differentiate into trophoblast cells of specialized functions of a matured placenta. The EPC and the chorion give rise to a functional placenta that comprises two structural regions—the junctional zone (JZ) and the labyrinth zone (LZ), which contains SynTs and fetal vasculature. The JZ comprises a compact layer of cells sandwiched between the labyrinth and the parietal trophoblast giant cell (p-TGC) layer. The TPCs within the EPC contribute to differentiated trophoblast cell types: spongiotrophoblast cells (SpT) (6), glycogen cells (GC), and TGCs (7), including invasive TGCs that invade the uterine wall and maternal vessels (8–10). The mouse placental labyrinth lies beneath the JZ, contains multinucleated SynTs, and establishes the exchange interface between the mother and the fetus (11).
The labyrinth development in the mouse placenta is initiated ~E8.0 when the allantoic mesoderm attaches to the chorionic ectoderm (12). At the onset of labyrinth formation, glial cells missing 1 (Gcm1) expression is induced in a few cells within the chorionic ectoderm (13–15). Postattachment, the chorioallantoic layer starts invaginating at the sites of GCM1 expression (14), thereby creating branch point initiation. The labyrinth undergoes extensive branching morphogenesis and expands from E9.5 to E14.5 to establish a functional maternal–fetal interface with enormous surface area for nutrient and gas exchange (12, 13, 15). The maternal blood sinusoids and fetal blood capillaries within a mouse labyrinth are separated by three differentiated trophoblast cell layers, the mononuclear sinusoidal trophoblast giant cells (S-TGCs) and two multinucleated Syncytiotrophoblast (SynT) layers, SynTI and SynTII (3, 11, 16–18). S-TGCs, lining the maternal blood sinusoids of the LZ, express hormones and growth factors and loosely attach to the SynTI layer (11, 12, 19). SynTI layer comes in direct contact with maternal blood due to the perforated nature of the S-TGC layer. The SynTII layer lies between the SynTI layer and the fetal endothelium. Thus, the three trophoblast layers along with the fetal endothelium form the maternal–fetal exchange interface, which is integral for the barrier and exchange function of the placenta to assure embryonic growth and viability.
Studies on mutant mouse embryos have identified key molecular regulators for SynT development. These molecular regulators include peroxisome proliferator-activated receptor gamma (PPARG), GCM1, distal-less homeobox 3 (DLX3), CAAT enhancer binding protein alpha and beta (CEBPA/B) and hepatocyte growth factor (HGF) receptor, the Met tyrosine kinase (c-MET) (14, 20–24). Expression analyses also identified markers that are specifically expressed in distinct trophoblast cell layers of the developing placental labyrinth. For example, messenger RNA (mRNA) expressions of nuclear receptor subfamily 6 group A member 1 (Nr6a1), ESX homeobox 1 (Esx1) along with Gcm1, Dlx3, Cebpa/b, and c-Met can be specifically detected in the SynT cells during early stages of labyrinth development (11). Several trophoblast layer–specific markers for three labyrinth trophoblast cell types have also been identified. Retroviral gene, Syncytin a (Syna), the Monocarboxylic acid transporter 1 (MCT1, encoded by Slc16a1 gene), and transferrin receptor (Tfrc) are selectively expressed in SynTI, whereas Syncytin b (Synb), MCT4 (encoded by the Slc16A3 gene), and iron transporter ferroportin, MTP1 (encoded by the gene Slc40a1), are selectively expressed in the SynTII population (25–27). Similarly, the S-TGCs selectively express Heart and neural crest derivatives expressed 1 (Hand1) and cathepsin Q (Ctsq) (7). Thus, we have a significant insight about the structural architecture, molecular regulators and specific markers of three trophoblast cell layers in the mouse labyrinth. However, we still have a poor understanding about the molecular processes through which these cells develop during placentation.
The presence of a common labyrinth progenitor in mouse was first reported by Ueno et al. (16), showing that the mouse labyrinth contains an Epcam(hi) multipotent labyrinth trophoblast progenitor (LaTP), which can differentiate to all labyrinth trophoblast subtypes when cultured at a clonal level. A recent study by Marsh et al. (18), which utilized unbiased single-nuclei RNA sequencing (snRNA-seq) followed by RNA-velocity mapping, further confirmed that LaTPs of a mouse placenta have trilineage potential to form—SynTI, SynTII, and S-TGCs. The transition of LaTPs to the corresponding mature differentiated trophoblast populations rely on establishment of distinct gene expression patterns (18). The snRNA-Seq analyses showed that LaTPs that start expressing higher levels of c-Met get committed to the SynTII lineage and are identified as SynTII precursors. A similar observation had previously been reported showing that the HGF/C-MET axis and WNT signaling mechanism are essential for the polarization and the terminal differentiation of the SynTII cells (16). LaTPs that express higher levels of Egfr and Epcam are biased toward forming the SynTI precursors, whereas the expression of Lifr and Podxl marks the transition and commitment of LaTPs to the S-TGC lineage. Interestingly, Natale et al. (28) reported the existence of a SCA1 (encoded by Ly6a gene)-expressing multipotent trophoblast progenitor population in the midgestation mouse placenta. These SCA1-expressing progenitors are reported to have the potential to differentiate to trophoblast cell types of both the LZ and the JZ. Collectively, these studies provide evidence that trophoblast development in the mouse labyrinth is associated with establishment of multiple progenitor cell types that could adopt distinct differentiation fates based on gene expression programs. However, transcriptional mechanisms that control differentiation fates in these progenitors remain incompletely understood.
In humans, two types of SynT arise during different stages of placentation. At the site of blastocyst implantation, a primitive syncytium is formed (29–31) that erodes the surface of the uterine epithelium. This primitive syncytium lays the groundwork for the columns of cytotrophoblasts (CTBs) to penetrate the primitive syncytium and form the primary villi. Subsequently, proliferation and differentiation of the CTBs cause the primary villi to branch and mature into a villous placenta that comprises two types of mature villi—the floating villi and the anchoring villi. The floating villi is bathed in maternal blood and consists of an underlying layer of CTBs that eventually fuse to form a multinucleated outer layer of SynT (32, 33). Like in mice, the SynT layer in humans is multinucleated and comes in direct contact with maternal blood. Thus, formation of differentiated, multinucleated SynTs from their committed trophoblast progenitors for the establishment of the placental exchange surface is a conserved adaptation in both mice and humans (32, 34). Furthermore, several transcription factors, such as PPARG, GCM1, and DLX3, which are essential for placental labyrinth development in mice, have also been implicated in human SynT development (35–38). The CTB progenitors in anchoring villi, which anchors to the maternal decidua through CTB cell columns, adopt a different differentiation fate and develop into invasive extravillious trophoblast cells (EVTs) (39). Interestingly, recent single-cell RNA sequencing (scRNA-seq) studies discovered the existence of multiple CTB subpopulations with distinct gene expression patterns, in a developing human placenta (40, 41). Thus, the presence of distinct CTB subpopulations and their altered differentiation fates in floating vs. anchoring villi indicate that the CTB to SynT and CTB to EVT transition during human placentation requires fine-tuning of the gene expression program in CTB progenitors. However, transcriptional mechanisms that regulate gene expression programs in CTB progenitors to promote either SynT differentiation or EVT differentiation are incompletely understood.
Earlier, we and other laboratories showed that GATA family transcription factors GATA2 and GATA3 (henceforth mentioned as GATA factors) are conserved in the trophoblast cells of both mouse and human placentae (42–46). Individual deletion of either Gata2 or Gata3 in mice does not overtly affect placental development. Gata3-null mouse embryos die at ~E11.5 due to defective neuroendocrine system development (47, 48) and Gata2-null mouse embryos die at ~E10.5 due to defective hematopoiesis (49). However, combinatorial deletion of both Gata2 and Gata3 genes in an early postimplantation mouse embryo (GATA-DKO embryo) abrogates placentation, leading to embryonic death at around E9.0 (50). Despite this importance of GATA factors in early placentation, it was unknown whether cell-autonomous GATA factor function in labyrinth progenitors is essential for the SynT/LZ development and progression of pregnancy. Also, both GATA3 and GATA2 expressions are conserved in the human trophectoderm (51) as well as in CTB progenitors and in differentiated SynTs within human placental villi. However, the importance of GATA factors during human SynT development and their contribution in human pregnancy-associated disorders is unknown.
In this study, we show that conditional deletion of GATA factors in mouse labyrinth progenitors arrests them in a multipotent state leading to defective placental labyrinth development. The loss of GATA factors leads to defective lineage segregation and inability of the labyrinth progenitors to differentiate into mature SynTs. Similarly, loss of function of either GATA2 or GATA3 in human trophoblast stem cells (human TSCs) arrests them in an intermediate stage where they are poised for differentiation but unable to undergo SynT differentiation. Thus, we highlight a developmental stage-specific transcriptional program established by conserved GATA factors that prime differentiation of multipotent trophoblast progenitor to SynT lineage at the maternal–fetal exchange interface.
Results
GATA Factors Function in Mouse LaTPs Is Essential for Placentation and Fetal Development.
As GATA factors are expressed extensively in the mouse LZ and their global loss abrogates LZ development (50), we wanted to investigate the importance of GATA factors in a labyrinth progenitor-specific manner. To restrict GATA factor deletion to the labyrinth progenitors, we used a previously established Tg(Gcm1-cre)1Chrn (Gcm1Cre) mouse model (52), where the Cre recombinase expression is restricted to the Gcm1 expressing cells. During mouse placentation, Gcm1 is first expressed in trophoblast progenitors of the chorionic ectoderm at ~E8.0 (13, 14). After chorionic attachment, the GCM1 expressing progenitors initiate the branching morphogenesis of the labyrinth and contribute to the development of both SynTI and SynTII cells (13, 14, 16, 18).
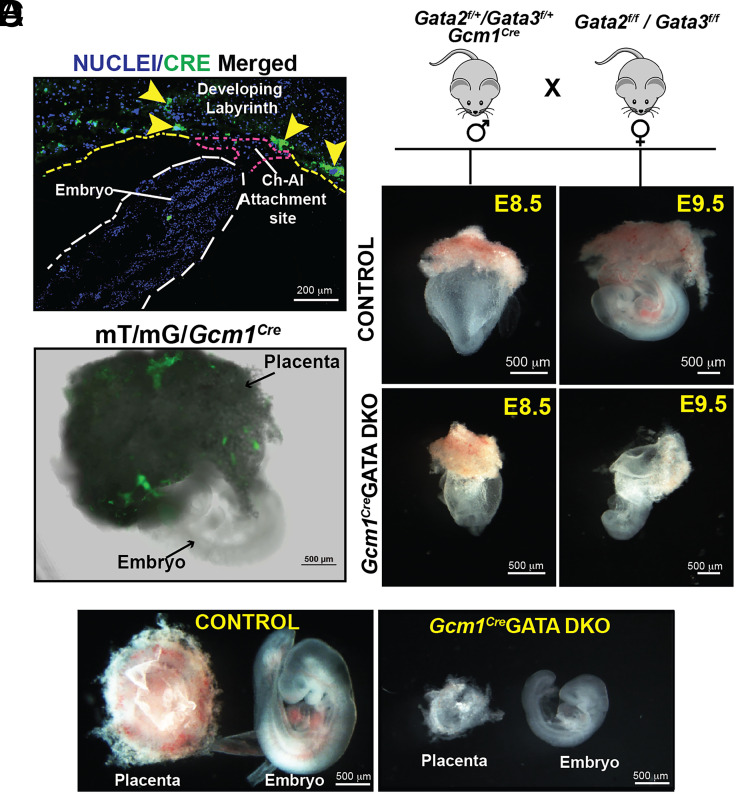
To validate the specificity of the Cre expression, we crossed Gcm1Cre mice with Gt(ROSA)26Sortm4(ACTB-tdTomato, -EGFP)Luo/J mice (also known as mT/mG mice), which possess loxP flanked membrane-targeted tdTomato (mT) cassette and express strong red fluorescence in all tissues (53). Upon breeding with the Gcm1Cre recombinase expressing mice, the resulting progenies have the mT cassette deleted in the Cre expressing tissue(s), allowing expression of the membrane-targeted EGFP (mG) cassette located in-frame immediately downstream. We analyzed E8.5 to E9.5 conceptuses from cross between Gcm1Cre male and mT/mG female to confirm the specificity of the CRE recombinase expression and activity within the developing placenta. Immunostaining analyses of histological sections confirmed that the CRE recombinase expression from the Gcm1-Cre driver mimics endogenous Gcm1 expression pattern at E8.5 (14, 15) and is predominantly expressed in small clusters of cells at the chorioallantoic interface (Fig. 1A). Furthermore, EGFP expression due to CRE recombinase activity was only detected in the developing placenta and not within the embryo proper in E8.5 to E9.5 conceptuses (Fig. 1B and SI Appendix, Fig. S1A). These observations confirmed the specificity of Gcm1Cre recombinase expression and activity only within labyrinth progenitors at the onset of labyrinth development.
Fig. 1.
Conditional deletion of Gata2 and Gata3 in mouse labyrinth progenitors abrogates placental and embryonic development. (A) and (B) Validation of CRE recombinase expression and activity specifically in labyrinth progenitors within E8.5 to E9.0 Gcm1cremT/mG conceptuses. The immunostaining panel in (A) shows that at E8.5, CRE expression from the Gcm1-Cre driver is mainly induced in clusters of labyrinth progenitors at the chorioallantoic interface. Panel (B) shows that the CRE-mediated recombination of the mT/mG locus leads to EGFP expression (green fluorescence) only within the developing placenta of an E9.0 Gcm1cremT/mG conceptus and not within the embryo proper. (C) (Top) Mating strategy used to determine the importance of Gata2 and Gata3 in labyrinth trophoblasts affecting placentation. Homozygous Gata2f/f/Gata3f/f female mice were crossed with heterozygous Gata2f/+/Gata3f/+/Gcm1Cre male mice to generate homozygous Gcm1CreGATA DKO (Gata2f/f/Gata3f/f/Gcm1Cre). Gata2f/f/Gata3f/fconceptuses were used as control. Embryonic and placental developments were analyzed at E8.5 and E9.5, representative images are shown. At E8.5, no obvious defect was observed in Gcm1CreGATADKO embryos. However, defect in placentation and embryonic development was obvious in Gcm1CreGATA DKO conceptuses at E9.5. (Scale bars, 500 μm). (D) Representative images showcase developing placenta and embryos from control and Gcm1CreGATA DKO. The Gcm1CreGATA DKO placentae and embryos show varying severity in the phenotype depending on the penetrance of the Cre activity. (Scale bars, 500 μm.)
To delete GATA factors in labyrinth progenitors we studied mouse models, harboring conditional knockout alleles of either Gata2 (Gata2f/f) or Gata3 (Gata3f/f) or both (Gata2f/f/Gata3f/f) (50). To confine the homozygous deletion of Gata2 and Gata3 gene only within the developing embryos, we crossed floxed female mice with male mice harboring Gcm1Cre allele along with respective floxed alleles (SI Appendix, Fig. S1B). We noticed that individual loss of either Gata2 or Gata3 in labyrinth progenitors did not affect embryonic development as homozygous mutant pups were born with normal Mendelian ratios and without noticeable defects. However, when we crossed Gata2 f/f/Gata3f/f (henceforth mentioned as Control Floxed) female mice with male mice harboring Gcm1Cre allele and heterozygous floxed alleles of both Gata2 and Gata3 genes (Gcm1Cre/Gata2f/+/Gata3f/+), the embryos with the genotype Gata2 f/f/Gata3f/f/Gcm1Cre (henceforth mentioned as Gcm1CreGATA DKO) showed prominent developmental defect at E9.5 and were resorbed by E10.5 (Fig. 1 C and D and SI Appendix, Fig. S2 A–D). At E9.5, the Gcm1CreGATA DKO placentae were significantly smaller in size in comparison to littermate controls (Fig. 1D and SI Appendix, Fig. S2B). The embryo proper of the Gcm1CreGATA DKO embryos also showed arrested development (Fig. 1D). Thus, our gene knockout studies confirmed that GATA factor functions in the labyrinth progenitors of a developing mouse embryo are essential for progression of placentation and embryonic survival beyond E9.5.
Loss of GATA Factors in the Labyrinth Progenitors Impairs Mature Syncytia Formation and Disorganizes the Trilaminar Architecture at the Maternal–Fetal Interface.
The mouse maternal–fetal interface contains three layers of trophoblast cells, SynTI, SynTII, and S-TGCs, that make up the trilaminar architecture (3, 11, 17). These layers facilitate the separation of maternal blood from the endothelial cells of the fetal vasculature. Thus, the two SynT layers along with the fetal endothelium are integral for the barrier and exchange function of the placenta to assure embryonic growth and viability. As we noticed defective placentation in Gcm1CreGATA DKO conceptuses, we further analyzed the labyrinth architecture in Gcm1CreGATA DKO placentae.
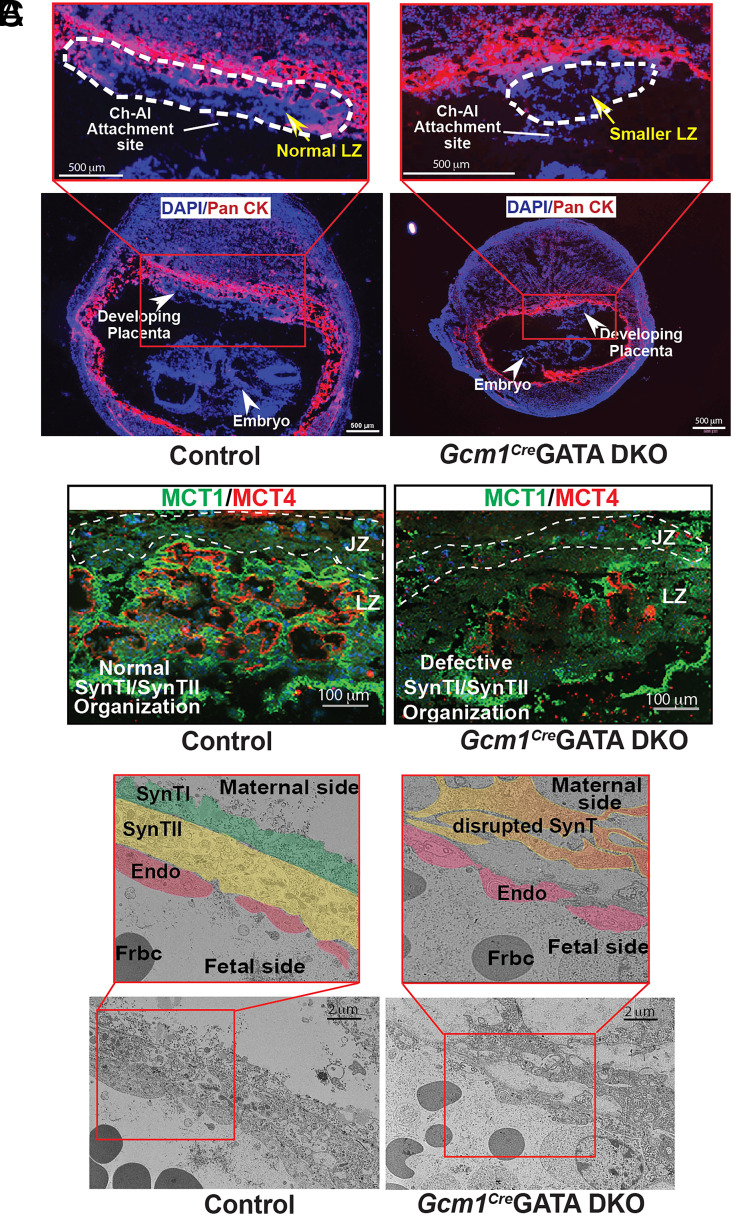
Immunofluorescence analyses showed that Gcm1CreGATA DKO conceptuses are growth retarded prior to the onset of labyrinth formation (Fig. 2A). Hence, we tested developmental status of the two mature SynT layers. We analyzed expressions of MCT1 and MCT4, which are specifically expressed in SynTI and SynTII cell, respectively (54). Unlike the control placentae, where tight juxtaposition of the two SynT layers was evident from MCT1 and MCT4 expressions, the Gcm1CreGATA DKO placentae showed gross disruptions of both SynTI and SynTII layers (Fig. 2B and SI Appendix, Fig. S3A). The MCT4-expressing SynTII population was drastically reduced (Fig. 2B and SI Appendix, Fig. S3A). In addition, strong reduction of MCT1-expressing SynTI population was also evident in Gcm1CreGATA DKO placentae (Fig. 2B and SI Appendix, Fig. S3A). In contrast to SynT layers, development of p-TGCs, which border the placental-uterine interface, was not affected in Gcm1CreGATA DKO placentae (SI Appendix, Fig. S3B)
Fig. 2.
GATA factor function in LaTPs is required for SynT maturation. (A) Placentation at control and Gcm1CreGATA DKO implantation sites were analyzed at approximately E9.5 via immunostaining with anti–pan-cytokeratin antibody (red, trophoblast marker). The developing Gcm1CreGATA DKO placenta lacks a well-developed LZ (white dashed line). Also, unlike control embryos, the Gcm1CreGATA DKO embryos are underdeveloped and have a much smaller surface area. (Scale bars, 500 μm.) (B) Immunofluorescent analysis of SynTI and SynTII layers was done in control and Gcm1CreGATA DKO placental labyrinth with MCT1 (green, marks SynTI layer) and MCT4 (red, marks SynTII layer) antibodies. The SynTI and SynTII formed nicely juxtaposed layered organization in the LZ of the control placenta. In contrast, both SynTI and SynTII development were impaired in the Gcm1CreGATA DKO placenta (JZ, Scale bars, 100 μm). (C) The trilaminar architecture of a developing labyrinth comprising SynTI, SynTII, and the fetal endothelium (endo), was captured using an electron microscope. In the control placenta, we observe the organized trilayered architecture—SynTI (pseudocolor green), SynTII (pseudocolor yellow), and fetal endothelium (pseudocolor red), but in the Gcm1CreGATA DKO, both the SynT layers (pseudocolor orange) are completely disrupted. The endothelial layer in Gcm1Cre GATA DKO was not as severely affected, indicating that the mature SynT layers do not form in Gcm1Cre GATA DKO placenta. (Scale bars, 2 μm.) Frbc-fetal red blood cells, MCT1-Monocarboxylate transporter 1, MCT4-Monocarboxylate transporter 4, SynTI &SynTII-SynT layer I & II.
To further confirm the disruption of the labyrinth architecture we analyzed the knockout and control labyrinth under the electron microscope, which allowed us to visualize the trilaminar architecture at high resolution. Electron microscope analyses also revealed disrupted trilaminar structure in Gcm1CreGATA DKO placentae (Fig. 2C). Instead of two closely positioned, mature SynT layers, the knockout labyrinth contained only one SynT layer that was discontinuous. Thus, from our immunofluorescence and EM studies, we concluded that the labyrinth progenitor-specific loss of GATA factors leads to defective development of the mature SynT lineages, resulting in abrogation of placental labyrinth formation.
scRNA-seq Revealed the Importance of GATA Factors in Committing the Labyrinth Progenitor to the SynT Lineage
As loss of GATA factors impaired formation of mature SynT cells, we then inquired whether the distribution of trophoblast progenitors is altered in Gcm1CreGATA DKO placentae. As labyrinth progenitors are single-cell populations, we performed scRNA-seq to capture distribution of distinct progenitor populations with E9.5 control and Gcm1CreGATA DKO placentae. Mouse placenta from E9.5 embryos were carefully isolated and the embryo proper and decidual tissue were dissected out (SI Appendix, Fig. S4A). The embryo was used for genotyping to confirm Gcm1CreGATA DKO placentae. The pooled control and Gcm1CreGATA DKO placental tissues were used to generate single-cell suspensions and subjected for scRNA-seq analyses.
Dimensionality reduction and clustering of all single cells was done using Seurat package and projected using t-SNE plots showcasing visible proximity between clusters. The Seurat package identified 21 clusters (SI Appendix, Fig. S4B). Based on the gene expression profile and cell type–specific markers (mentioned in SI Appendix, Table S1), these clusters were classified into 5 broad groups: endodermal, endothelial, stromal, trophoblast and blood cells. Clusters 2, 4, 13, and 14 were identified as trophoblast cells as they highly expressed trophoblast markers Epcam, Ly6a, Prl3d1, Prl2c2, and Tpbpa (SI Appendix, Fig. S4B).
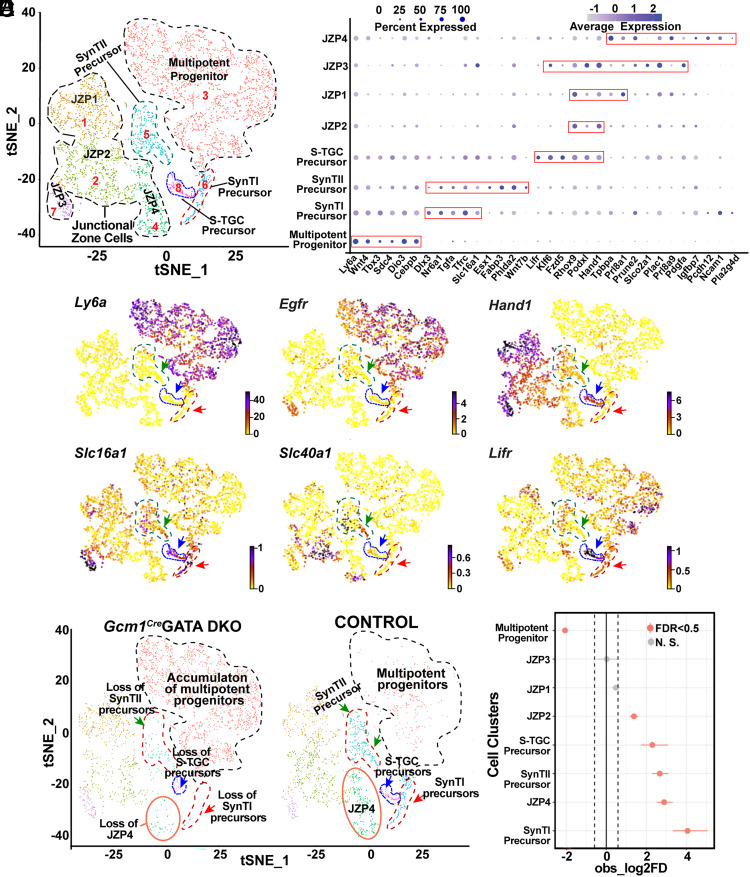
For a better understanding of the status of the trophoblast progenitors, we reannotated only trophoblast cells using the Seurat package and identified eight distinct cell clusters based on marker gene expressions (Fig. 3A and SI Appendix, Table S1). We observed a cell population (cluster 3, Fig. 3A) with high-level expression of Ly6a (Fig. 3 B and C). These cells also highly express several other labyrinth progenitor-specific genes, such as Wnt4, T-Box transcription factor 3 (Tbx3), Cebpb, imprinted gene Iodothyronine Deiodinase 3 (Dio3), and Tfrc (Fig. 3B and SI Appendix, Fig. S5A). Interestingly, the Ly6a expressing cell population also showed high-level expressions of Epidermal Growth Factor Receptor (Egfr) (Fig. 3C), which were earlier identified to be expressed in multipotent LaTPs by Marsh et al. (18). Thus, we concluded that the Ly6a, Cebpb, and Egfr expressing cells make up the multipotent LaTP compartment within the E9.5 labyrinth. We also identified more committed SynTI (cluster 6, Fig. 3A), SynTII (cluster 5, Fig. 3A), and S-TGC (cluster 8, Fig. 3A) precursors, through which these multipotent progenitors transition to the corresponding mature differentiated populations. The committed SynTI precursors showed high expressions of characteristic SynTI markers, Tfrc and Slc16a1 (MCT1) (Fig. 3 B and C and SI Appendix, Fig. S5A). Nr6a1 and Dlx3, which are expressed in SynT cells during early labyrinth development (11), were detected in both SynTI and SynTII precursors (SI Appendix, Fig. S5B). The SynTII precursors highly express Slc40A1 (MTP1), Esx1, Fabp3, Phlda2, Wnt7b, and Slc16a3 (MCT4) (Fig. 3 B and C and SI Appendix, Fig. S5C). In addition, the S-TGC precursors express Lifr and Podxl (Fig. 3 B and C and SI Appendix, Fig. S5D), which were earlier reported to be induced during S-TGC commitment (18). S-TGC precursors also highly express Hand1, which was mostly suppressed in multipotent labyrinth progenitors and in SynT precursors (Fig. 3 B and C). Similar to the snRNA-seq analyses by Marsh et al. (18), our scRNA-seq analyses detected high level of Met expression only in SynTII precursors (SI Appendix, Fig. S5C). The single-cell trophoblast populations also contained four JZ progenitor trophoblasts (JZP1-4, Fig. 3A). JZP1-3 showed high-level Hand1 expression along with placenta-specific gene Placenta Enriched 1 (Plac1) (Fig. 3 B and C), whereas JZP4 cells showed high-level expression of SpTs marker, Tpbpa, along with other JZ trophoblast-specific genes Prl8a9, Plac1, Pcdh12, and Ncam1 (Fig. 3 A and B and SI Appendix, Fig. S4E).
Fig. 3.
Loss of GATA factors impairs differentiation of multipotent LaTPs to more committed precursors. (A) Visualization of the scRNA-seq data with the analysis plotted in two dimensions by transcriptome similarity using t-distributed stochastic neighbor embedding (t-SNE). t-SNE plot represents the cells included in the trophoblast subset, clustered, and plotted according to transcriptome similarity. Clusters were annotated according to canonical marker genes. Each dot represents one cell colored according to assignment by clustering analysis (Materials and Methods). Dotted lines encircle clusters with common properties segregated into five trophoblast subclusters–multipotent progenitors, SynTI precursors, SynTII precursors, S-TGC precursors, and JZ cells. (B) Dot plot showing average expression and percent of cells in each cluster expressing canonical and novel marker genes identified for each cluster. Genes listed on the x-axis and clusters on the y-axis. (C) RNA velocity plots showing the expression of marker genes in specific clusters, the scale represents the level of expression–yellow (low), red (intermediate), and blue (high). The colored arrows mark specific populations–green=SynTII precursors, blue = S-TGC precursors, and red = SynTI precursors. (D) Split by condition, t-SNE plot representing the difference in the trophoblast subclusters between control and Gcm1Cre GATA DKO samples. The t-SNE plot shows accumulation of the multipotent progenitors (cluster marked with black dotted lines) but loss of SynTI precursors, SynTII precursors, and S-TGC precursor (clusters marked by red, green, and blue arrows, respectively) in the Gcm1Cre GATA DKO compared to the control. JZP3 cluster (marked in orange solid line) also diminishes in the Gcm1Cre GATA DKO placenta. (E) Single-cell proportion test R-function was performed to generate a plot that highlights the difference between proportions of cells in each cluster between the control and Gcm1Cre GATA DKO placental samples. The plot shows that cluster multipotent progenitor significantly increases in proportion in the Gcm1Cre GATA DKO sample, whereas SynTI and SynTII precursors, S-TGC precursors, JZP2, and JZP3 show a significant increase in proportion in the control sample. ZP1 and JZP4 do not show significant change in either of the samples.
The comparison of trophoblast progenitor clusters between control and Gcm1Cre GATA DKO placentae revealed two obvious differences. Compared to the control placentae, the Gcm1Cre GATA DKO placentae contained significantly higher number of multipotent progenitor populations (Fig. 3 D and E). In contrast, the committed SynTI, SynTII, and S-TGC precursors, which express high levels of Epcam as reported earlier by Ueno et al. (16) (SI Appendix, Fig. S6A), were strongly diminished in the Gcm1Cre GATA DKO placentae (Fig. 3 D and E and SI Appendix, Fig. S6A). Among the JZPs, JZP1 and JZP3 populations were not significantly altered in Gcm1Cre GATA DKO placentae, whereas cluster comprising JZP2 cells was slightly reduced. However, the JZP4 progenitors, which express Tpbpa, were also reduced in Gcm1Cre GATA DKO placentae (Fig. 3 D and E and SI Appendix, Fig. S6B).
To validate the scRNA-seq observation, we performed additional experiments. First, we confirmed enrichment of the Ly6a expressing multipotent progenitor population in the absence of GATA factors via fluorescence activated cell sorting (FACS) analysis. We generated single-cell preparation from control and Gcm1Cre GATA DKO placentae, removed all the hematopoietic and endothelial cells, and tested the percentage of cells that were SCA1 (encoded by Ly6a) positive. We observed a significantly higher number of trophoblast cells that were SCA1 positive in the knockout placentae (SI Appendix, Fig. S7 A and B). Immunostaining analyses also confirmed the abundance of SCA1-positive cells within the labyrinth of Gcm1Cre GATA DKO placentae (SI Appendix, Fig. S7C).
Next, we performed quantitative RT-PCR analyses to test expression of marker genes for SynTI, SynTII, S-TGC precursors, and JZ trophoblasts in control and Gcm1CreGATA DKO placentae. The RT-PCR analyses confirmed strong loss of Nr6a1, Slc16a1, Fabp3, c-Met, and Dlx3 in E9.5 Gcm1Cre GATA DKO placentae (SI Appendix, Fig. S8). In contrast, mRNA expressions of Podxl (S-TGC precursor marker), Hand1, Prl3d1, and Prl2c2 (TGC markers) were not significantly altered in Gcm1Cre GATA DKO (SI Appendix, Fig. S8). However, RT-PCR analyses also confirmed loss of Tpbpa expression in E9.5 Gcm1Cre GATA DKO placentae. Thus, our scRNA-seq, FACS, and RT-PCR analyses indicated that the loss of the GATA factors in the LaTPs arrests their development at a Ly6a/Egfr expressing uncommitted progenitor state, thereby affecting differentiation to committed SynTI, SynTII, and S-TGC precursors. The strong loss of Tpbpa-expressing JZPs in Gcm1Cre GATA DKO placentae is an interesting observation and indicates that SpT development during mouse placentation may rely on cross talk with LZ trophoblast precursors.
Loss of GATA2 or GATA3 in Human TSCs Impairs SynT Differentiation.
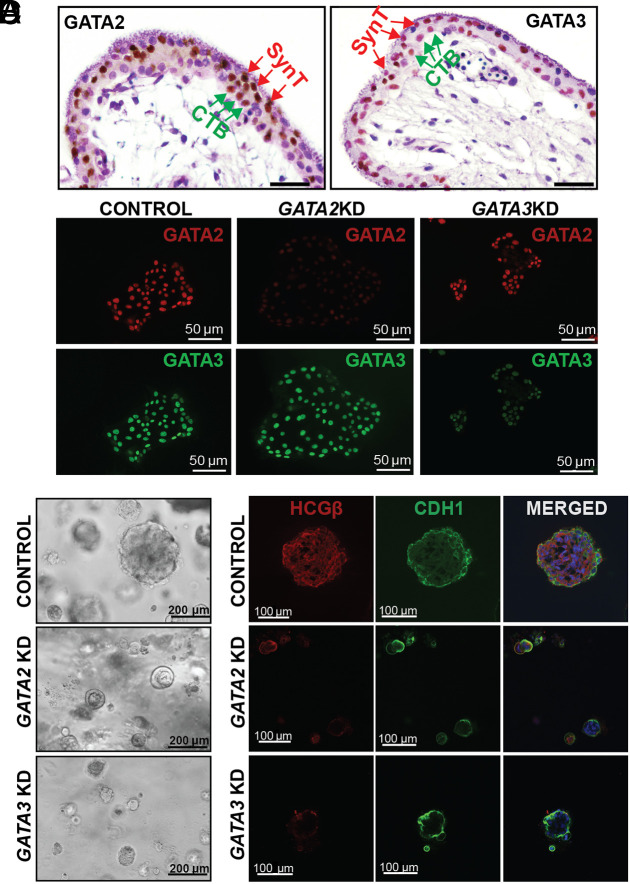
The human placenta is composed of chorionic villi that stem from the chorionic plate. These villous-like structures may anchor onto the uterine decidua (anchoring villi) or remain floating, bathed in maternal blood (floating villi). Floating villi have a characteristic cellular bilayer consisting of two types of trophoblast cells—an inner layer of mononuclear proliferative CTBs and an outer layer of multinucleated, differentiated SynT formed by the fusion of underlying CTBs. In a matured term placenta, the CTB population is diminished and placental villi mainly contain the differentiated SynT population. Our protein expression analyses revealed that both GATA2 and GATA3 are expressed in the CTBs and SynTs of a first-trimester human placenta, and their expression is maintained in SynTs within matured term placentae (Fig. 4A and SI Appendix, Fig. S9A).
Fig. 4.
GATA2 and GATA3 optimize self-renewal ability of human TSCs. (A) Immunohistochemistry showing expression of GATA2 and GATA3 in first-trimester (9 wk) human placenta. GATA2 (Left) and GATA3 (Right) are expressed abundantly in the SynT cells (SynT, red arrows, outer layer) as well as within the villous CTB progenitors (vCTBs, green arrows, inner layer) within the floating villi. (Scale bars, 50 μm.). (B) Immunofluorescent analysis of GATA2 and GATA3 expression in stem state colonies of scrambled, GATA2KD and GATA3KD human TSCs. The expression of GATA2 (red) was depleted in GATA2KD human TSCs but remained unaffected in GATA3KD human TSCs, while GATA3 (green) expression was depleted in GATA3KD human TSCs but remained unaltered in GATA2 KD colonies. Thus, the shRNA molecules targeting the GATA2 had no effect on GATA3 expression and vice-versa. (C) 3D organoid culture was done with control, GATA2 KD, and GATA3 KD human TSCs. The loss of function of GATA2 or GATA3 affected large organoid formations as observed in the control at day 8. The organoids formed with GATA2 KD and GATA3 KD human TSCs were much smaller in size and number. (D) Immunofluorescent analysis of the organoid integrity was done by highlighting its “inside-out” model, with CTBs marked by E-Cadherin antibody (green) on the outside and SynT marked by hCGβ antibody (red) on the inside. The control organoids showcased a distinct inside-out model. The GATA2 KD and GATA3 KD organoids were smaller and underdeveloped; they lacked hCGβ (red) on the inside, indicating no SynT formation, but had E-Cadherin (green) expression suggesting that the CTBs were not affected.
Recent studies showed that bonafide human TSCs, derived from first-trimester CTBs (55), are an excellent model to study molecular mechanisms that regulate CTB to SynT differentiation. Thus, we performed loss-of-function studies with human TSCs to test the importance of GATA2 and GATA3 in SynT development during human placentation. We used lentivirus-mediated delivery of short hairpin RNA (shRNAs) to specifically deplete GATA2 and GATA3 in human TSCs (Fig. 4B and SI Appendix, Fig. S9B). When cultured in 2-dimensional stem state culture conditions [2D stem-state culture, (55) that maintain proliferating cultures, human TSCs with knockdown of either GATA2 (GATA2KD human TSC) or GATA3 (GATA3KD human TSCs) maintained the stem state colony morphology (SI Appendix, Fig. S9C) and did not show significant reduction in cell proliferation ability (SI Appendix, Fig. S9 D and E). However, combinatorial loss of both GATA factors in human TSCs (Dual GATAKD human TSC) impaired stem-state morphology and cell proliferation in a 2D stem-state culture condition (SI Appendix, Fig. S9C).
Interestingly, RT-PCR analyses showed that mRNA transcription factor TEAD4, cofactor vestigial like family member 1 (VGLL1), and tumor protein P63 (TP63), which are critical to maintain human TSC stem state (34, 40, 56) as well as cell proliferation regulators polo-like kinase 1 and 2 (PLK1 and PLK2) and Wnt signaling component, WNT5A, were down-regulated in GATA2KD and GATA3KD human TSCs (SI Appendix, Fig. S9F). Although we got strong depletion of GATA2 and GATA3 using shRNA-mediated RNAi strategy, our approach did not lead to a complete absence GATA factors in hTSCs (Fig. 4B). Thus, we reasoned that the residual expression of GATA2 or GATA3 upon individual depletion might be sufficient to maintain hTSC proliferation in 2D culture condition. Therefore, we also tested self-renewal ability of GATA2KD and GATA3KD human TSCs by assessing their ability to form self-renewing 3-dimensional (3D) trophoblast stem cell organoids [3D TSC organoid culture (40, 57)]. In contrast to 2D-stem state culture, the 3D organoid cultures of both GATA2KD and GATA3KD human TSCs showed strong impairment in organoid formation ability (Fig. 4C). Control human TSCs formed large organoids with prolonged culture (7 to 10 d) and could be dissociated and reorganized to form secondary organoids, indicating the self-renewing ability. In contrast, GATA2KD and GATA3KD human TSCs formed much smaller organoids (Fig. 4C), which were not maintained upon passaging. Thus, we concluded that individual functions of GATA2 and GATA3 are important to maintain optimum self-renewal ability in human TSCs, especially when they are cultured in the 3D culture condition. However, our findings in 2D-stem state culture also imply that GATA2 and GATA3 might have complementary functions in human TSCs that allow sustained proliferation and stem-state maintenance when any one of them is depleted in 2D-stem state culture. Whereas, irrespective of culture conditions, combinatorial loss of GATA2 and GATA3 abrogates stem-state maintenance in human TSCs. As human TSCs largely recapitulate the properties of CTB progenitors of a first-trimester human placenta, our findings strongly indicate that GATA factors are important to maintain the self-renewal ability in CTBs of a developing human placenta.
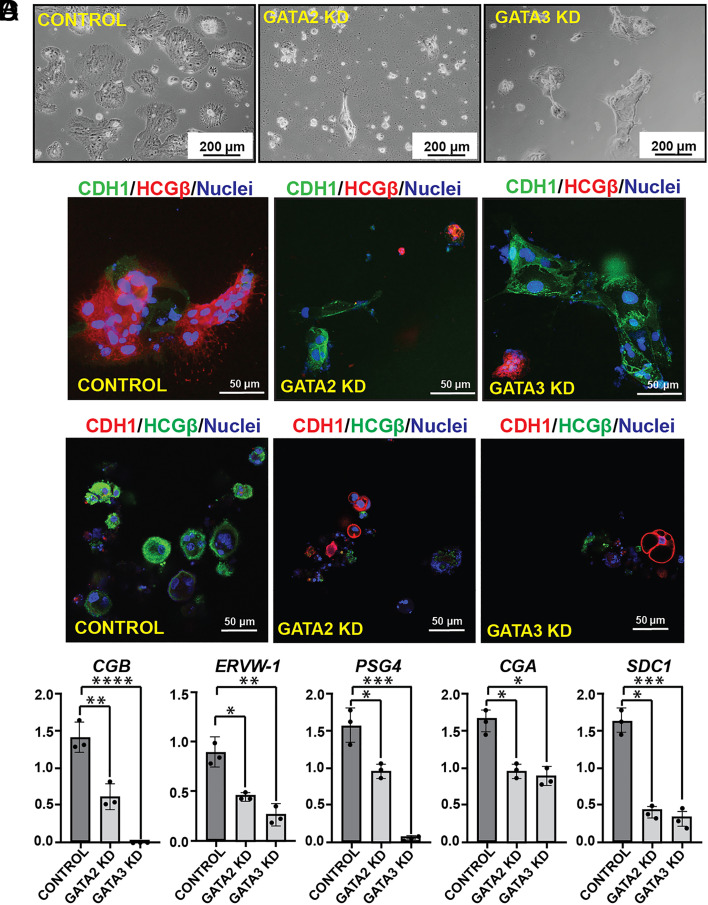
The 3D TSC organoids grow in an inside-out pattern. The undifferentiated TSCs in the outer layer undergo proliferation and express E-Cadherin (CDH1), whereas the inside cells undergo SynT differentiation and highly express HCGβ. We noticed that the defective organoid formations of GATA2KD and GATA3KD human TSCs are associated with near complete loss of HCGβ-expressing SynTs inside those organoids (Fig. 4D). Thus, our 3D TSC organoid culture strongly suggested that loss of either GATA2 or GATA3 in human TSCs might lead to defective SynT development. To confirm this aspect, we tested multinucleated SynT formation ability of GATA2KD and GATA3KD human TSCs in culture conditions that specifically promote SynT differentiation.
Human TSCs, when cultured in the presence of cAMP agonist forskolin, synchronously differentiate and fuse to form 2D syncytia on a high attachment culture plate or 3D cyst-like structures on a low adhesion culture plate. In both 2D and 3D culture conditions, differentiated human TSCs highly express SynT markers such as HCGβ. We noticed that the loss of GATA2 in human TSCs abrogated SynT maturation when cultured in 2D-SynT culture conditions (Fig. 5A). GATA2KD cells failed to fuse and gradually became unhealthy. The loss of GATA3 in human TSCs also impaired SynT maturation in 2D-SynT culture conditions. GATA3KD cells retained their stem state like colony morphology and did not fuse to form multinucleated mature SynTs (Fig. 5A). Impaired SynT differentiation was also confirmed by monitoring HCGβ and CDH1 expression. Unlike in control human TSCs, induction of HCGβ protein expression was strongly inhibited in GATA2KD and GATA3KD human TSCs (Fig. 5B) while expression of CDH1 was maintained along with a near-complete inhibition of cell-to-cell fusion (Fig. 5B).
Fig. 5.
GATA2 and GATA3 are essential regulators for SynT development in humans. (A) Control, GATA2 KD, and GATA3 KD human TSCs were subjected to 2D SynT differentiation on collagen-coated adherent cell culture dishes. Image panels show impaired ST(2D) colony formation in GATA2 KD and GATA3 KD cells. (Scale bars, 200 μm.). (B) Immunofluorescence analysis of the ST(2D) colonies (Left) marked by the loss of E-Cadherin (CDH1, green) expression but induction of HCGβ expression (red). GATA3 KD ST(2D) show altered cellular morphology, maintenance of E-Cadherin (CDH1, green) expression, and impaired induction of HCGβ expression (Right). GATA2 KD ST(2D) colonies (Middle) were much smaller; there was impaired cell fusion as E-Cadherin was maintained compared to the control but lacked HCGβ expression. (C) Immunofluorescence analysis of the ST(3D) colonies revealed loss of E-Cadherin (CDH1, red) expression and induction of HCGβ expression (green). Both GATA2KD and GATA3 KD ST(3D) show altered cellular morphology, maintenance of CDH1 expression, and impaired induction of HCGβ expression. (D) Quantitative RT-PCR analyses (mean ± SE; n = 3, P ≤ 0.001) reveal impaired induction of SynT markers like CGB, ERVW-1, PSG4, CGA, and SDC-1 in GATA2 KD and GATA3 KD human TSCs, undergoing 3D SynT differentiation.
Loss of SynT differentiation potential of GATA2KD and GATA3KD human TCSs was also evident under the 3D-SynT culture condition. Control human TSCs spontaneously formed larger sphere-like structures while the GATA2KD and GATA3KD human TSCs were unable to form larger spheres. Instead, they mainly formed smaller aggregates (Fig. 5C). Furthermore, like in 2D-SynT culture, we confirmed loss of HCGβ protein expression and retainment of CDH1 expression in 3D-SynT cultures of GATA2KD and GATA3KD human TSCs (Fig. 5C). mRNA expression analyses indicated significant inhibition of SynT-specific markers syndecan 1 (SDC1), CGB, endogenous retrovirus group W member 1 (ERVW1), and pregnancy-specific beta-1-Glycoprotein 4 (PSG4) mRNA inductions in the small cell aggregates formed by GATA2KD or GATA3KD human TSCs (Fig. 5D). Thus, both 2D and 3D-SynT differentiation systems revealed impaired SynT differentiation potential of the GATA2KD and GATA3KD human TSCs.
GATA Factors Directly Regulate Expression of Common Genes that Are Important for SynT Differentiation and Are Implicated in Pathological Pregnancies.
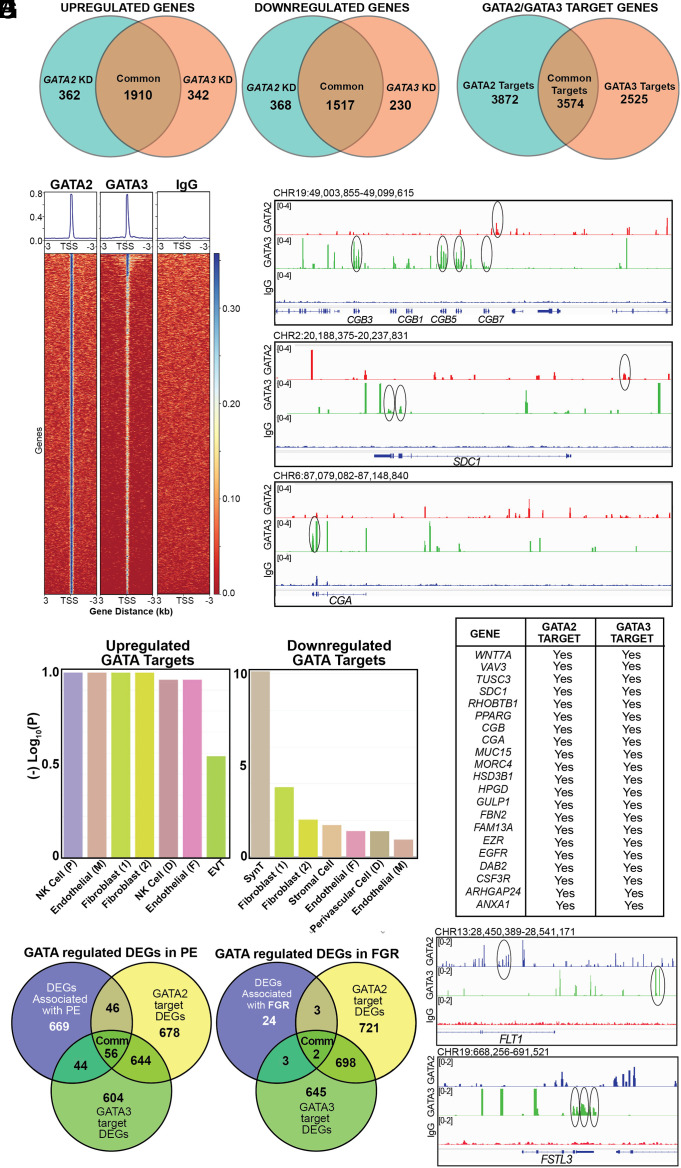
To further understand how GATA2 and GATA3 functions promote SynT development in humans, we performed unbiased genomics studies. We performed RNA-Seq analysis with control, GATA2KD, and GATA3KD human TSCs when they were subjected to SynT differentiation to identify differentially expressed genes (DEGs) upon GATA2 and GATA3 depletion. RNA-seq analyses identified 4,157 DEGs and 3,999 DEGS (with a fold change of ±1.5) in GATA2KD and GATA3KD human TSCs, respectively (Fig. 6A and Dataset S1). A comparison of DEGs revealed a striking overlap between genes that were up- or down-regulated when either GATA2 or GATA3 were depleted in human TSCs (Fig. 6A). We noticed that 1,910 genes were up-regulated and 1,517 genes were down-regulated in both GATA2KD and GATA3KD human TSCs. Whereas only 362 genes were up-regulated and 368 genes were down-regulated specifically in GATA2KD human TSCs. Additionally, 342 genes were up-regulated and 230 genes were down-regulated specifically in GATA3KD TSCs. Thus, the unbiased RNA-seq analyses indicated that GATA2 and GATA3 establish a largely similar gene expression program when promoting SynT development.
Fig. 6.
GATA2 and GATA3 directly activate SynT-specific genes. (A) Venn diagrams showing DEGs in GATA2KD and GATA3KD human TSCs with respect to control human TSCs when human TSCs were subjected to SynT differentiation. (B) Venn diagram showing the number of target genes of GATA2 and GATA3 in human TSCs undergoing SynT differentiation. (C) Heat map representing GATA2 or GATA3 binding enrichment at the TSS and 3,000 bp upstream and downstream of the TSS. (D) Integrative Genome Viewer (IGV) tracks showcasing GATA2 and GATA3 CUT&RUN Seq peaks. Statistically significant GATA2 peaks (red) and GATA3 peaks (green) are indicated at the CGB, CGA, and SDC1 loci. Circles indicate statistically significant binding sites. (E) Placenta cell-specific gene enrichment analysis done with up-regulated and down-regulated genes using the PlacentaCellEnrich. Analysis highlights that the down-regulated genes most significantly represent the SynT population. (F) The table shows a list of GATA2/GATA3 target genes, which are important for SynT differentiation and are differentially regulated in both GATA2KD and GATA3KD human TSCs. (G) Venn diagram showing the number of GATA-regulated genes that are also significantly altered in PE. The gene set was curated from the placentome database (58). (H) IGV tracks showcasing GATA2 and GATA3 CUT&RUN Seq peaks at the FLT1 and FSTL3 loci. Circles indicate statistically significant binding sites.
We performed Cleavage Under Targets & Release Using Nuclease (CUT&RUN) with GATA2- and GATA3-specific antibodies to identify genes that are direct targets of GATA factors in human TSCs. Globally, we identified 8,108 GATA2 binding and 5,457 GATA3 binding regions (Dataset S2). HOMER motif analyses identified GATA motifs among most enriched motifs within both GATA2 and GATA3 binding regions (SI Appendix, Fig. S10A). Further analyses of binding regions using Genomic Regions Enrichment of Annotations Tool identified 7,446 putative GATA2 target genes and 6,099 putative GATA3 target genes in human TSCs (Fig. 6B and Dataset S2). We also noticed that 3,574 genes are common targets of GATA2 and GATA3 in human TSCs (Fig. 6B). Further analyses of GATA2 and GATA3 chromatin binding regions of target genes revealed that both GATA2 and GATA3 binding regions were more concentrated near the transcription start site (TSS) (Fig. 6C). Thus, our CUT&RUN analyses revealed that a large number of genes in developing SynTs are targets of GATA2 and GATA3 and the GATA factors might control expression of those genes during SynT development.
A comparative analysis between the CUT&RUN and RNA-Seq data revealed that several genes, such as SDC1, CGA, and CGBs, which are specifically induced in SynTs and are differentially expressed in both GATA2KD and GATA3KD human TSCs, are GATA2/GATA3 targets (Fig. 6D). We identified 355 common up-regulated DEGs and 345 common down-regulated DEGs, that are direct targets of both GATA2 and GATA3 (Dataset S2). We further characterized common GATA target DEGs using placenta cell-specific gene enrichment [PlacentaCellEnrich, (59)] analysis. Interestingly, the enrichment analyses revealed that the common GATA2/GATA3 target genes, which are down-regulated in both GATA2KD and GATA3KD human TSCs, most significantly represent SynTs. In contrast, up-regulated GATA2/GATA3 target genes most significantly represent nontrophoblast cells of a human placenta (Fig. 6 E and F). Thus, our unbiased genomics analyses revealed that GATA2/GATA3-mediated gene activation, rather than gene repression, is critical for human SynT development.
Pathological pregnancies, such as preeclampsia (PE) and fetal growth restriction (FGR), are often associated with altered gene expression in trophoblast cells. Therefore, we next tested the association of GATA factor–regulated genes with respect to pregnancies complicated by PE and FGR. We compared GATA-regulated DEGs with significantly dysregulated (P ≤ 0.05) genes in PE and FGR pregnancies identified by a recent study by Gong et al. [(58) and Dataset S3]. We found that 56 PE-associated DEGs, including GATA2 itself, are directly regulated by both GATA2 and GATA3 in human TSCs during SynT differentiation (Fig. 6G and Dataset S3). In addition, another 46 PE-associated DEGs are specifically regulated by GATA2 and 44 PE-associated DEGs are directly regulated by GATA3 (Fig. 6G and Dataset S3). Among the 32 significantly (P ≤ 0.05) dysregulated FGR-associated genes [(58) and Dataset S3], two genes, Apolipoprotein B (APOB) and Iodothyronine Deiodinase 2 (DIO2), are directly regulated by both GATA2 and GATA3 during SynT differentiation (Fig. 6G and Dataset S3). In addition, three FGR-associated genes, Proteinase 3 (PRTN3), Prostaglandin Reductase 3 (PTGR3), and Triggering Receptor Expressed on Myeloid Cells 1 (TREM1), are directly regulated by GATA2, whereas GATA3 directly regulates three other FGR-associated genes, FSTL3, GPR32 and NOG (Fig. 6G and Dataset S3). Intriguingly, many of the most significantly up-regulated genes in PE [such as Fms Related Receptor Tyrosine Kinase 1 (FLT1), Leptin (LEP), and HtrA Serine Peptidase 4 (HTRA4), (60)] or in both PE and FGR [such as Follistatin Like 3 (FSTL3)] are directly regulated in human TSCs GATA3 or by both GATA2 and GATA3 (e.g., FLT1) and are also up-regulated in both GATA2KD and GATA3KD hTSCs (Fig. 6H and SI Appendix, Fig. S10 B and C). However, we have also noticed that several other significantly up-regulated genes in PE and FGR, such as Endoglin (ENG), DIO2, and TREM1 (60) are directly regulated by GATA factors but down-regulated in both GATA2KD and GATA3KD hTSCs (SI Appendix, Fig. S10 B, C, and D). Nevertheless, our unbiased genomics analyses in human indicated that during SynT development GATA factors directly regulate genes that are not only critical for SynT development but are also implicated in pathological pregnancies, such as PE and FGR.
Discussion
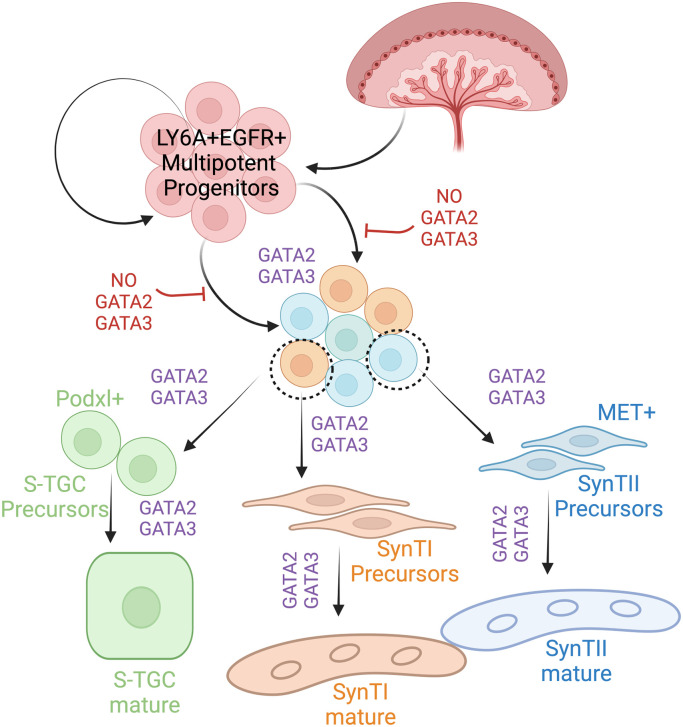
During early mouse development, GATA2 and GATA3 are specifically expressed in the trophoblast lineage and our global knockout studies revealed essential roles of GATA factors in trophoblast cells during preimplantation and early postimplantation embryos. However, specific roles of GATA factors during SynT and LZ development have not been addressed earlier. Our in vivo studies with knockout mice along with our scRNA-seq analysis provided a better understanding of the developmental stages of SynT development from their progenitor populations and the specific role that GATA factors play in that context. We showed that there is a specific need of GATA factors to fine-tune the transcriptional program in SCA1-positive multipotent LaTP-like progenitors for their transition to committed progenitors, which will eventually develop into the terminally differentiated SynT populations to form a functional placental labyrinth (Fig. 7). Interestingly, the lack of GATA function in SCA1-positive multipotent progenitors also affected S-TGC development highlighting the important role of GATA factors during development of all of the three trophoblast subtypes of a mouse placental labyrinth. Our data also indicate that during mouse SynT development, GATA2 and GATA3 mediate redundant roles and the function of one factor can compensate loss of the other factor so that pregnancy and embryonic development can progress.
Fig. 7.
A GATA factor–dependent SynT developmental program during mouse placentation. Schematic representation of midgestation mouse placenta shows GATA factors regulate SynT precursor commitment and differentiation. In the presence of GATA2 and GATA3, the multipotent progenitors of the labyrinth (SCA1+ EGFR+) get committed to either the SynTI, SynTII, or S-TGC lineage forming SynTI, SynTII, and S-TGC precursors, respectively. These precursors under the transcriptional regulation of the GATA factors differentiate into multinucleated mature SynTI and SynTII cells or S-TGCs. In the Gcm1Cre GATA DKO placentae loss of GATA factors prevents multipotent progenitors to commit to the SynT lineage.
We found that both SynTI and SynTII maturation is impaired in ~E9.5 Gcm1CreGATA DKO placentae. Interestingly, maturation of both SynTI and SynTII is impaired in ~E9.5 Gcm1-KO placentae (14) and also upon conditional deletion of ERK/MAP kinase genes, Map2k1 and Map2k2, in labyrinth progenitors using the Gcm1-Cre driver (54). Thus, it was proposed that the maturation of SynTI depends on the presence of SynTII (54). However, other studies involving knock-in of a LacZ reporter within the Gcm1 loci (15) and our analyses of recently published single-cell RNA Seq data during mouse placentation (61) indicate that Gcm1 expression is induced, albeit at the low level, in many SynT progenitors between E8.5 and E9.5 (SI Appendix, Fig. S11). Thus, it is also possible that between E8.5 and E9.5, CRE recombinase expression is induced from the Gcm1 promoter in many SynT progenitors within Gcm1CreGATA DKO placentae leading to widespread deletion of Gata2 and Gata3 genes and a strong developmental defect at E9.5.
Mouse placentae lacking three other transcription factors, Pparg, Gcm1, and Dlx3 as well as HGF receptor, c-Met, also showed defective SynT maturation. At the onset of labyrinth development, Gcm1 and Dlx3 transcriptions are induced in trophoblast progenitors. Dlx3 is initially induced in basal EPC progenitors, which constitutes a layer in chorion and eventually differentiate to SynTI lineage (11). Later, Dlx3 is broadly expressed in both SynTI and SynTII population. Defect in SynT development and placental labyrinth formation is also evident in the Dlx3 knockout mice by E9.5 (21). Similarly, matured and fused SynT is absent in Pparg and knockout placentae (20, 62). We showed that Dlx3 was repressed in Gcm1Cre GATA DKO placentae. Furthermore, we previously showed that Pparg and Gcm1 are also regulated by GATA factors in developing mouse placentae and Dlx3, Pparg, and Gcm1 are direct target genes of GATA2 and GATA3 in mouse TSCs (50). Thus, GATA factors seem to be the master regulator to induce Pparg, Gcm1, Dlx3, and c-Met during SynT maturation and labyrinth development and loss of expression of these key factors could be the molecular cause for impaired SynT commitment of multipotent labyrinth progenitors in Gcm1CreGATA DKO placentae.
Consistent with our observation in mouse placental development, we observe a severe impairment in SynT differentiation (2D and 3D) of human TSCs due to the loss of function of either GATA2 or GATA3. This observation also indicates a difference in GATA-factor requirements for SynT differentiation in the human and mouse. Individual loss of either GATA2 or GATA3 did not affect SynT development in mice, whereas individual function of either GATA2 or GATA3 seems essential for SynT development in humans. However, unlike our studies in mouse embryos, our experiments in human TSCs do not truly represent the complexities of in vivo systems. Thus, the in vitro human TSC experimental system could be more sensitive to individual loss of GATA factors.
Our RNA-seq and CUT&RUN-seq analyses revealed that GATA2/GATA3-mediated gene activation is critical for human SynT development. Recent genome-wide screening studies have identified key essential genes and growth-restricting genes (GRGs) within hTSCs (63) and transcription factor modules that are associated with SynT differentiation (64). Another CRISPR screening study by Shimizu et al. (65) identified factors that promote SynT differentiation in hTSCs. Our comparative analyses showed that 140 among 619 DRGs, identified by Dong et al. (63), are directly regulated by both GATA2 and GATA3 (Dataset S4). Furthermore, we also found that 38 of the 127 SynT-specific transcription factors, identified by Chen et al. (64), and 14 of the 54 factors, identified by Shimizu et al. (65), are also GATA targets (Dataset S4). Thus, transcription of many of the SynT-specific genes is directly regulated by both GATA2 and GATA3. However, we noticed that GATA factors are also important to maintain self-renewal in human TSCs. Thus, it will be interesting to find out the regulatory axis that causes the switch from the self-renewal mode in the CTB progenitors toward SynT differentiation by modulating the global transcriptional program via GATA factors.
In conclusion, our findings in this study establish that GATA factors are essential for SynT development and establishment of maternal–fetal exchange interface during mammalian placentation. However, our findings in human TSCs do not indicate whether GATA factors are important for the maintenance of various functions that are mediated by differentiated SynTs in a matured placenta. Also, the importance of GATA factors in the context of pathological pregnancies is yet to be defined. Intriguingly, in this study, we found that many of the GATA-regulated genes during SynT development are dysregulated in PE and FGR. In fact, GATA2 itself is significantly down-regulated in PE (58). Thus, it seems that there might be a direct correlation of GATA2/GATA3 transcriptional activity with dysregulated trophoblast gene expression in pathological pregnancies, such as PE and IUGR. The recent success in establishing stem cells from term placenta (66–68) and advancement of single-cell genomics analyses have provided an excellent opportunity to further study the association of GATA factor transcriptional activity in the context of pathological pregnancies. We predict that the establishment of the inducible genome editing system in human TSCs and studies on patient-specific stem cells from pathological pregnancies will further elucidate the importance of GATA factors in the context of human pathological pregnancies.
Materials and Methods
Generation of Conditional Knockout Mice Strains.
All procedures were performed after obtaining IACUC approvals at the Univ. of Kansas Medical Center. Male Gata2flox/flox (Gata2f/f) (69) mice were mated with Tg(Gcm1-cre)1Chrn (Gcm1Cre) female in order to generate Gata2f/+; Gcm1Cre +/wt. In the next step, Gata2f/+; Gcm1Cre +/wt female mice were bred with Gata2f/f males to generate Gata2f/f; Gcm1Cre +/wt. Similarly, Gata3flox/flox (Gata3f/f) (70) mice were used to generate Gata3f/f; Gcm1Cre +/wt. In the next step, Gata2f/f; Gcm1Cre +/wt and Gata3f/f; Gcm1Cre +/wt mice were crossed to generate Gata2f/+; Gata3f/+; Gcm1Cre +/wt. Gata2f/+; Gata3f/+; Gcm1Cre +/wt male mice were used as the Cre driver to restrict the expression of Cre recombinase only in the extraembryonic tissues postmating. Gata2f/f; Gata3f/f generated by our lab in a previous publication (50) was used as the female for the functional analysis and all future experiments.
scRNA-seq Sample Preparation.
Placenta samples were carefully isolated at d9.5, ensuring the decidual layer was peeled off. Individual samples were digested in the presence of collagenase (100 mg/mL) at 37 °C for 30 min, followed by syringe passaging of the cells with 18G, 22G, and 25G needles. One milliliter of PBS supplemented with 10% FBS solution was added to each tube of cell suspension to neutralize the effect of collagenase. Finally, each placental sample was made into single-cell suspensions by passing them through a 40- μm filter. Corresponding embryonic tissues were used to confirm genotypes. Based on the genotyping results, the control samples were pooled together, and the knockout samples were pooled together. These samples were further processed using Debris Removal Solution and Dead Cell Removal Kit (Miltenyi Biotec., Gaithersburg, MD). Red blood cell depletion from cell suspensions was performed using anti-Mouse Ter-119 antibody (BD Biosciences, Franklin Lake, NJ). Cellular viability was checked by automated cell counter (Countess 3, Thermo Fisher Scientific) after staining cells with trypan blue. After library preparation (done by University of Kansas Medical Center (KUMC) genomics core), samples were loaded into the 10× Chromium V3 platform. The raw data for scRNA-seq analyses have been submitted to the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds), with accession No. GSE214499. Additional details of ScRNA-seq data analyses are mentioned in SI Appendix, Supplementary Materials and Methods.
Human TSC Culture.
Human TSC lines were derived from first-trimester CTBs, as described earlier (55). To maintain stem state culture, human TSCs were cultured on collagen IV-coated (5 μg/mL) plate in DMEM/F12 medium, supplemented with 0.1 mM 2-mercaptoethanol, 0.2% FBS, 0.5% penicillin–streptomycin, 0.3% BSA, 1% ITS-X supplement, 1.5 μg/mL L-ascorbic acid, 50 ng/mL EGF, 2 μM CHIR99021, 0.5 μM A83-01, 1 μM SB431542, 0.8 mM valproic acid, and 5 μM Y27632. For SynT(2D) differentiation, plates were coated with 2.5 μg/mL of collagen IV, and TSCs were then cultured in DMEM/F12 medium, supplemented with 0.1 mM 2-mercaptoethanol, 0.5% penicillin–streptomycin, 0.3% BSA, 1% ITS-X supplement, 2.5 mM Y27632, 2 mM forskolin, and 4% KSR. For, SynT(3D) differentiation, no plate coating was required. Cells were cultured in the same media used for SynT(2D) with 50 ng/mL of EGF.
shRNA-Mediated RNAi.
To knockdown GATA factors in human TSCs, shRNA molecules against GATA2 and GATA2 were delivered using lentiviral vectors. We tested multiple shRNA molecules to eliminate the possibility of off-target effects. For data that are presented in this study, shRNA molecule against GATA2 mRNA sequence (GTGCAAATTGTCAGACGACAA) and GATA3 mRNA sequence (TCTGGAGGAGGAATGCCAATG) was used. A scramble shRNA with sequence (CCTAAGGTTAAGTCGCCCTCGC) was used as control. Transduced cells were selected in the presence of puromycin (1.5 to 2 μg/mL). Selected cells were tested for knockdown efficiency and used for further experimental analyses.
CUT&RUN Analyses.
Proliferating semiconfluent 200,000 live hTSC were used per sample for CUT&RUN following the published protocol (71). Cells were captured on Concanavalin A–coated beads (EpiCypher, Durham, NC); cell permeabilization was done using buffers containing 0.5% wt/vol Digitonin before incubation with anti-GATA2 and anti-GATA3 antibodies. Protein A and G fused Micrococcal Nuclease (EpiCypher) was used for DNA digestion. The detailed protocol is mentioned in SI Appendix, Supplementary Materials and Methods.
Statistical Analyses.
Statistical significance was determined for quantitative RT-PCR analyses for mRNA expression and for FACS analyses. We performed at least n = 3 experimental replicates for all these experiments. For statistical significance of generated data, statistical comparisons between two means were determined with Student’s t test, and significantly altered values (P ≤ 0.01) are highlighted in the figures by an asterisk. RNA-seq data were generated with n = 3 experimental replicates per group. The statistical significance of altered gene expression (absolute fold change ≥ 2.0 and false discovery rate q-value ≤ 0.05) was initially confirmed with right-tailed Fisher’s exact test. Independent datasets were analyzed using GraphPad Prism software.
GEO Database Accession Codes for Sequencing Data.
The raw genomics data are submitted to the GEO database. Data for scRNA-seq of E9.5 control and Gcm1CreGATA DKO mouse placentae are submitted with GEO accession number GSE214389. Raw data for bulk RNA-seq and CUT & RUN seq in human TSCs are submitted with GEO accession numbers GSE214634 and GSE214486, respectively.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This research was supported by NIH grants HD062546, HD103161, HD102188, and HD101319 to Soumen Paul. This study was also supported by administrative support from the Institute for Reproduction and Developmental Sciences (IRDS), University of Kansas Medical Center (KUMC), and various core facilities, including the Genomics Core, Transgenic and Gene Targeting Institutional Facility, Imaging and Histology Core and the Flow Cytometry Core. We thank Drs. Hiroaki Okae and Takahiro Arima of Tohoku University Graduate School of Medicine, Japan, for sharing human TSC lines. We thank Dr. Pratik Home for his intellectual support about the research project and Ms. Brandi Miller for critical comments on the manuscript.
Author contributions
A.G. and S.P. designed research; A.G., R.P.K., S.R., A.S., N.R., and P.D. performed research; C.M. contributed new reagents/analytic tools; R.K., A.G., and S.P. analyzed data; and A.G. and S.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-seq, CU&RUN Seq, Single Cell RNA seq data have been deposited in GEO database (GSE214634 (72), GSE214486 (73), and GSE214389 (74)).
Supporting Information
References
- 1.Rossant J., Cross J. C., Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Soares M. J., Varberg K. M., Iqbal K., Hemochorial placentation: Development, function, and adaptations. Biol. Reprod. 99, 196–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maltepe E., Fisher S. J., Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523–552 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Hirashima M., Lu Y., Byers L., Rossant J., Trophoblast expression of fms-like tyrosine kinase 1 is not required for the establishment of the maternal-fetal interface in the mouse placenta. Proc. Natl. Acad. Sci. U.S.A. 100, 15637–15642 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossant J., Stem cells in the mammalian blastocyst. Harvey Lect. 97, 17–40 (2001). [PubMed] [Google Scholar]
- 6.Simmons D. G., Cross J. C., Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284, 12–24 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Simmons D. G., Fortier A. L., Cross J. C., Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev. Biol. 304, 567–578 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann P., Black S., Huppertz B., Endovascular trophoblast invasion: Implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol. Reprod. 69, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Rosario G. X., Konno T., Soares M. J., Maternal hypoxia activates endovascular trophoblast cell invasion. Dev. Biol. 314, 362–375 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soares M. J., et al. , Regulatory pathways controlling the endovascular invasive trophoblast cell lineage. J. Reprod. Dev. 58, 283–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons D. G., et al. , Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135, 2083–2091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross J. C., et al. , Genes, development and evolution of the placenta. Placenta 24, 123–130 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Basyuk E., et al. , Murine Gcm1 gene is expressed in a subset of placental trophoblast cells. Dev. Dyn. 214, 303–311 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Anson-Cartwright L., et al. , The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25, 311–314 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Stecca B., et al. , Gcm1 expression defines three stages of chorio-allantoic interaction during placental development. Mech. Dev. 115, 27–34 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Ueno M., et al. , c-Met-dependent multipotent labyrinth trophoblast progenitors establish placental exchange interface. Dev. Cell 27, 373–386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coan P. M., Ferguson-Smith A. C., Burton G. J., Ultrastructural changes in the interhaemal membrane and junctional zone of the murine chorioallantoic placenta across gestation. J. Anat. 207, 783–796 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh B., Blelloch R., Single nuclei RNA-seq of mouse placental labyrinth development. Elife 9, e60266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu D., Cross J. C., Development and function of trophoblast giant cells in the rodent placenta. Int. J. Dev. Biol. 54, 341–354 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Barak Y., et al. , PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4, 585–595 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Morasso M. I., Grinberg A., Robinson G., Sargent T. D., Mahon K. A., Placental failure in mice lacking the homeobox gene Dlx3. Proc. Natl. Acad. Sci. U.S.A. 96, 162–167 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begay V., Smink J., Leutz A., Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol. Cell. Biol. 24, 9744–9751 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uehara Y., et al. , Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature 373, 702–705 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Maina F., et al. , Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell 87, 531–542 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Dupressoir A., et al. , Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U.S.A. 102, 725–730 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai A., Takebe K., Nio-Kobayashi J., Takahashi-Iwanaga H., Iwanaga T., Cellular expression of the monocarboxylate transporter (MCT) family in the placenta of mice. Placenta 31, 126–133 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Walentin K., Hinze C., Schmidt-Ott K. M., The basal chorionic trophoblast cell layer: An emerging coordinator of placenta development. Bioessays 38, 254–265 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Natale B. V., et al. , Sca-1 identifies a trophoblast population with multipotent potential in the mid-gestation mouse placenta. Sci. Rep. 7, 5575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knofler M., et al. , Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell Mol. Life Sci. 76, 3479–3496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James J. L., Carter A. M., Chamley L. W., Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 33, 327–334 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Boss A. L., Chamley L. W., James J. L., Placental formation in early pregnancy: How is the centre of the placenta made? Hum. Reprod. Update 24, 750–760 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Knott J. G., Paul S., Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction 148, R121–R136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M., Lei Z. M., Rao C. V., The central role of human chorionic gonadotropin in the formation of human placental syncytium. Endocrinology 144, 1108–1120 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Soncin F., et al. , Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 145, dev156273 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaiff W. T., et al. , Peroxisome proliferator-activated receptor-gamma modulates differentiation of human trophoblast in a ligand-specific manner. J. Clin. Endocrinol. Metab. 85, 3874–3881 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Tarrade A., et al. , PPAR gamma/RXR alpha heterodimers are involved in human CG beta synthesis and human trophoblast differentiation. Endocrinology 142, 4504–4514 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Jeyarajah M. J., et al. , The multifaceted role of GCM1 during trophoblast differentiation in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 119, e2203071119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberson M. S., Meermann S., Morasso M. I., Mulvaney-Musa J. M., Zhang T., A role for the homeobox protein Distal-less 3 in the activation of the glycoprotein hormone alpha subunit gene in choriocarcinoma cells. J. Biol. Chem. 276, 10016–10024 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Haider S., et al. , Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 113, E7710–E7719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha B., et al. , TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc. Natl. Acad. Sci. U.S.A. 117, 17864–17875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh B., Zhou Y., Kapidzic M., Fisher S., Blelloch R., Regionally distinct trophoblast regulate barrier function and invasion in the human placenta. Elife 11, e78829 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai H., Sakurai T., Godkin J. D., Imakawa K., Expression and potential role of GATA factors in trophoblast development. J. Reprod. Dev. 59, 1–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul S., Home P., Bhattacharya B., Ray S., GATA factors: Master regulators of gene expression in trophoblast progenitors. Placenta 60 (suppl. 1), S61–S66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Home P., et al. , GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729–28737 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray S., et al. , Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 284, 4978–4988 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma G. T., et al. , GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development 124, 907–914 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Pandolfi P. P., et al. , Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 11, 40–44 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Lim K. C., et al. , Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 25, 209–212 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Tsai F. Y., Orkin S. H., Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89, 3636–3643 (1997). [PubMed] [Google Scholar]
- 50.Home P., et al. , Genetic redundancy of GATA factors in the extraembryonic trophoblast lineage ensures the progression of preimplantation and postimplantation mammalian development. Development 144, 876–888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerri C., et al. , Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 587, 443–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadeau V., et al. , Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation. Development 136, 1363–1374 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Nadeau V., Charron J., Essential role of the ERK/MAPK pathway in blood-placental barrier formation. Development 141, 2825–2837 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Li Y., Moretto-Zita M., Leon-Garcia S., Parast M. M., p63 inhibits extravillous trophoblast migration and maintains cells in a cytotrophoblast stem cell-like state. Am. J. Pathol. 184, 3332–3343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karvas R. M., et al. , Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens. Cell Stem Cell 29, 810–825.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong S., et al. , The RNA landscape of the human placenta in health and disease. Nat. Commun. 12, 2639 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain A., Tuteja G., PlacentaCellEnrich: A tool to characterize gene sets using placenta cell-specific gene enrichment analysis. Placenta 103, 164–171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leavey K., Bainbridge S. A., Cox B. J., Large scale aggregate microarray analysis reveals three distinct molecular subclasses of human preeclampsia. PLoS One 10, e0116508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang X., et al. , A differentiation roadmap of murine placentation at single-cell resolution. Cell Discov. 9, 30 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schreiber J., et al. , Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 20, 2466–2474 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong C., et al. , A genome-wide CRISPR-Cas9 knockout screen identifies essential and growth-restricting genes in human trophoblast stem cells. Nat. Commun. 13, 2548 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y., Siriwardena D., Penfold C., Pavlinek A., Boroviak T. E., An integrated atlas of human placental development delineates essential regulators of trophoblast stem cells. Development 149, dev200171 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimizu T., et al. , CRISPR screening in human trophoblast stem cells reveals both shared and distinct aspects of human and mouse placental development. Proc. Natl. Acad. Sci. U.S.A. 120, e2311372120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soncin F., et al. , Derivation of functional trophoblast stem cells from primed human pluripotent stem cells. Stem Cell Rep. 17, 1303–1317 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L. J., et al. , Functional antagonism between DeltaNp63alpha and GCM1 regulates human trophoblast stemness and differentiation. Nat. Commun. 13, 1626 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horii M., et al. , Modeling preeclampsia using human induced pluripotent stem cells. Sci. Rep. 11, 5877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charles M. A., et al. , Pituitary-specific Gata2 knockout: Effects on gonadotrope and thyrotrope function. Mol. Endocrinol. 20, 1366–1377 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Zhu J., et al. , Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5, 1157–1165 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Skene P. J., Henikoff J. G., Henikoff S., Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Paul S., Kumar R., Ghosh A., “RNA-Seq analysis in human trophoblast stem cells (CT27) upon depletion of transcription factor GATA2 or GATA3”. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214634. Deposited 28 September 2022.
- 73.Paul S., Kumar R. P., Ghosh A., Kumar R., “Cut and run analysis in human trophoblast stem cells (CT27) upon pulling down with GATA2 and GATA3 antibody”. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214486. Deposited 29 September 2022.
- 74.Paul S., Kumar R., Ghosh A., “Single cell RNA sequencing analysis using embryonic day 9.5 placenta from control and GATA DKO mice”. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214389. Deposited 26 September 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
RNA-seq, CU&RUN Seq, Single Cell RNA seq data have been deposited in GEO database (GSE214634 (72), GSE214486 (73), and GSE214389 (74)).