Abstract
Nucleosome organization is important for chromatin compaction and accessibility. Profiling nucleosome positioning genome-wide in single cells provides critical information to understand the cell-to-cell heterogeneity of chromatin states within a cell population. This protocol describes single-cell micrococcal nuclease sequencing (scMNase-seq), a method for detecting genome-wide nucleosome positioning and chromatin accessibility simultaneously from a small number of cells or single cells. To generate scMNase-seq libraries, single cells are isolated by FACS sorting, lysed and digested by MNase. DNA is purified, end-repaired and ligated to Y-shaped adaptors. Following PCR amplification with indexing primers, the subnucleosome-sized (fragments with a length of ≤80 bp) and mononucleosome-sized (fragments with a length between 140 and 180 bp) DNA fragments are recovered and sequenced on Illumina HiSeq platforms. On average, 0.5–1 million unique mapped reads are obtained for each single cell. The mononucleosome-sized DNA fragments precisely define genome-wide nucleosome positions in single cells, while the subnucleosome-sized DNA fragments provide information on chromatin accessibility. Library preparation of scMNase-seq takes only 2 d, requires only standard molecular biology techniques and does not require sophisticated laboratory equipment. Processing of high-throughput sequencing data requires basic bioinformatics skills and uses publicly available bioinformatics software.
Introduction
In eukaryotes, DNA is packaged into chromatin by wrapping 146 bp of DNA around a histone octamer, consisting of two copies each of H2A, H2B, H3 and H4, to form a nucleosome structure, which generally suppresses transcription, replication, repair and recombination1–8. Measuring nucleosome position helps to decipher chromatin states9. Genome-wide nucleosome position and occupancy can be measured by micrococcal nuclease sequencing (MNase-seq)10, which involves MNase digestion to preferentially degrade the linker DNA between nucleosomes and sequencing of the protected nucleosome core particles by next-generation sequencing techniques. The MNase-seq technique detects the nucleosome profiles from a population of cells and reveals an average pattern of nucleosome positions across different cells9,10. Recent studies have indicated an apparent heterogeneity in chromatin states across different cells even in a homogeneous cell population11–13. As nucleosome organization may underlie chromatin accessibility, it will be important to identify the positions of nucleosomes at a single-cell level.
Our recent work highlighted the use of single-cell MNase-seq (scMNase-seq) for mapping genome-wide nucleosome positioning and chromatin accessibility simultaneously at the single-cell level in NIH3T3 cells, embryonic stem cells and naive CD4 T cells9. At the single-cell level, scMNase-seq can reproducibly detect on average ~3, 0.9 and 0.7 million unique fragments, respectively, for each cell type. Fragments with a length between 140 and 180 bp are considered as canonical nucleosomes and fragments with a length of ≤80 bp as subnucleosome-sized particles. The midpoints of the nucleosomal and subnucleosomal DNA are defined as their positions on the genome. Based on nucleosome positions, nucleosome space is calculated and refers to the distance between adjacent nucleosomes. Genome-wide nucleosome space patterns are poorly understood because previous knowledge about nucleosome spacing revealed by bulk cell MNase-seq data is limited to the positioned nucleosomes, that is, nucleosomes that adopt the same genome position in every cell9,14. The bulk-cell MNase-seq data failed to reveal the actual spacing pattern due to the mixture of non-positioned nucleosomes from a population of different cells14. scMNase-seq circumvents this problem by detecting nucleosome positions in genomic regions with both positioned and non-positioned nucleosomes at the single-chromosome level9. Analysis of these scMNase-seq data revealed that (i) subnucleosome fragments are enriched in DNase I hypersensitive sites (DHSs); and (ii) nucleosomes have regular spacing of ~180 bp between neighboring nucleosomes but random positions relative to the underlying DNA across different cells in heterochromatic regions, while they have heterogeneous spacing between neighboring nucleosomes but occupy the same positions across different cells in active chromatin regions9. Furthermore, nucleosome organization patterns at cell-specific enhancers predict cell fate during development9. scMNase-seq offers a method of identifying nucleosome positioning and chromatin accessibility in single-cell or low-input materials.
Development and overview of scMNase-seq
The traditional MNase-seq requires millions of cells as starting material and isolation of mononucleosome-sized DNA for library preparation10. Owing to the low quantity of DNA per single cell, the key for single-cell techniques is to minimize DNA loss and efficiently recover target DNA15. Thus, for developing scMNase-seq, we adopted a strategy similar to our scDNase-seq technique by including a circular plasmid during DNA purification, and constructed libraries without isolating the mononucleosome-sized DNA12. The spike-in of circular plasmid makes the DNA pellet visible during DNA purification and helps to minimize DNA loss during library preparation. The final expected success rate of single-cell libraries by this protocol is >90% according to our filtering criteria that the library should provide >0.5 million unique reads per single cell and present the nucleosome depletion and subnucleosome enrichment at the DHSs and the transcription start sites (TSSs) of active genes9.
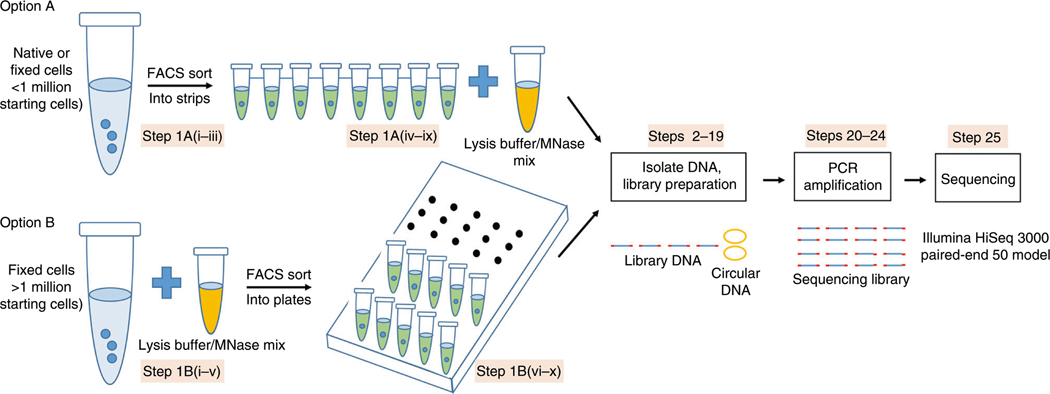
There are two options for when to perform MNase digestion (Fig. 1): (A) MNase digestion in single cells; and (B) MNase digestion on bulk cells; the user can choose the appropriate option depending on the starting cell number, cell fixation, experimental design and how many single-cell libraries need to be prepared. For option A, both native cells and formaldehyde-crosslinked cells can be sorted by FACS to single cells in PCR tube strips, digested and processed for library preparation individually. In this option, since MNase digestion is performed in single cells and careful planning and handling are needed to keep the same extent of MNase digestion for each single cell, the cell throughput is limited (<100) in one batch. For option B, however, cells crosslinked with formaldehyde can be digested by MNase in bulk, sorted into single cells in 96-well plates and processed for library preparation, which provides not only a uniform extent of MNase digestion for each sorted single cell but also increases the cell throughput by sorting hundreds of digested single cells into several 96-well plates. Formaldehyde-crosslinking of the cells helps to (i) stabilize protein–DNA interactions and avoid loss of nucleosomes or transcription factors during cell manipulation and MNase digestion16; (ii) minimize DNA loss from nuclei after MNase digestion during the sorting process; and (iii) store precious samples for a long time. In our experience, we recommend performing FACS sorting of single cells from small starting cell numbers (at least 100,000) by option A, generating limited numbers of single-cell libraries for sequencing. If the starting cell number is >1 million and higher cell throughput is required, option B is recommended. After MNase digestion following either option A or option B, circular plasmid is spiked-in and DNA from single cells is purified and collected for library preparation individually without fractionation of DNA fragments. Sequencing is performed on Illumina HiSeq 3000 by paired-end 50 cycles. Sequencing data are processed, mapped to the relevant genomes and analyzed using established procedures.
Fig. 1 |. Workflow illustration of the scMNase-seq protocol.
There are two options for generating scMNase libraries according to the starting cell number. For <1 million starting cells, single cells are isolated first by FACS sorting, and then each cell is individually digested by MNase digestion. For >1 million starting cells, bulk cells are fixed and digested by MNase first, and then single cells are isolated by FACS sorting. Both options follow the same DNA extraction procedure by phenol–chloroform. Circular DNA is spiked-in to prevent DNA loss during processing of the samples. End-repair, A/T ligation and PCR amplification are performed to prepare scMNase-seq libraries. After isolation of desired DNA fragments, the libraries are sequenced on an Illumina sequencing platform.
Comparison with other methods
A number of single-cell epigenomics techniques have been developed to probe different aspects of chromatin states at single-cell resolution17,18, including scDNase-seq13, single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq)19, scMNase-seq9, single-cell nucleosome occupancy and methylome-sequencing (scNOMe-seq)20, single-cell nucleosome, methylation and transcription sequencing (scNMT-seq)21 and single-cell chromatin overall omic-scale landscape sequencing (scCOOL-seq)22. Different enzymes are employed in these different methods. Specifically, scDNase-seq uses DNase I to cut the open chromatin DNA into single-stranded and double-stranded nicks; scATAC-seq makes use of Tn5 transposase to insert sequencing adaptors into DNA at open chromatin regions; and the scNOMe-seq, scNMT-seq and scCOOL-seq techniques use GpC methyltransferase to probe chromatin accessibility by methylating cytosines in GpC dinucleotides in non-nucleosomal DNA in vitro followed by bisulfite conversion and next-generation sequencing, thus detecting the methylation states of GpC sites (exogenous DNA methylation catalyzed by GpC methyltransferase, reflecting chromatin accessibility) and CpG sites (reflecting endogenous DNA methylation).
ATAC-seq can also be used to detect nucleosome positions within accessible chromatin in bulk cells23,24. This is achieved by hyperactive Tn5 transposase-mediated insertion of adaptors into the linker region between nucleosomes. As Tn5 transposase preferentially integrates into active regulatory elements, the nucleosome profiles revealed by ATAC-seq are mainly enriched in open chromatin. Besides, the characteristic phasing of nucleosome detected by ATAC-seq is achieved by averaging nucleosome signals by defining the central position of nucleosome from bulk cell data, as Tn5 transposase randomly inserts its adaptor at the linker DNA region. Similar to Tn5 transposase, GpC methyltransferase labels the linker DNA only at the open chromatin sites in a GpC dinucleotide-dependent manner. Compared to Tn5 transposase and GpC methyltransferase, only the MNase enzyme can cut linker DNA in all parts of the genome, precisely define the boundary of nucleosomes, and thus provide critical nucleosome-positioning information in single cells. In addition, the sub-nucleosomal fragments detected by scMNase-seq provide information on chromatin accessibility, comparable to scATAC-seq, scDNase-seq, and scNOMe-seq, scNMT-seq or scCOOL-seq.
Among these different single-cell epigenomics techniques, scATAC-seq measures chromatin accessibility, scMNase-seq measures chromatin accessibility and nucleosome position, and scNMT-seq measures chromatin accessibility, DNA methylation and transcriptome at the single-cell level. Users need to choose the desired techniques for their studies based on their specific goal of investigation.
Applications
scMNase-seq enables the detection of genome-wide nucleosome positions and chromatin accessibility simultaneously in a wide range of cell types and tissues with either a small number of pooled cells or at a single-cell resolution. We have applied scMNase-seq to the human NIH3T3 cell line, mouse primary naive CD4 T cells and mouse embryonic stem cells9. This method does not require expensive machines and can be easily adapted by any biomedical labs with adequate molecular biology capabilities. It can be applied to any tissue or cell type that can form a single-cell suspension from dissociated tissues. Because scMNase-seq is a robust single-cell epigenomics technique that measures both chromatin accessibility and nucleosome position, we expect that it will contribute to revealing epigenetic heterogeneity during normal cell development or pathological processes such as cancer and provide important information for the underlining molecular mechanisms at the single-chromosome level14.
Limitations
Capture rate
Missing information is unavoidable for this method, which is a common challenge to all single-cell sequencing methods. Owing to the multiple steps in library preparation and the high levels of PCR duplicates, only a fraction of nucleosome and subnucleosome fragments are detected in a single cell (~3–10% of capture rate, assuming a total of 30 million nucleosomes in one single cell).
Automation and cell throughput
Similar to scDNase-seq developed by our lab, scMNase-seq has not been adapted to an automated or semi-automated format because of the need for phenol–chloroform DNA extraction from single cells. Compared to the thousands of single cells analyzed by scATAC-seq25 or droplet-microfluidic methods for single-cell RNA sequencing (Drop-seq)26 using microfluidics or cell barcode labeling, scMNase-seq has a lower throughput and can handle ~100 single cells in one experiment by a single person in 1 week. However, as our method provides high coverage (0.5–1 million unique reads per cell), we could make inferences about nucleosome positioning and chromatin accessibility with 50–100 single cells in general. In the future, it will be important to develop a high throughput and less time-consuming method to survey more single cells.
Experimental design
Cell preparation
Either primary or cultured cells can be used, and both native and formaldehyde-crosslinked cells are acceptable. Single-cell suspensions should be prepared before FACS sorting by trypsinization of adherent cultured cells or dissociation of primary tissues. The cell suspension should be filtered using a cell strainer to remove cell clumps before FACS sorting. For <1 million starting cells (at least 100,000), we recommend processing the cells by FACS sorting and MNase digestion (option A). If the number of starting cells is >1 million and more libraries of single cells are needed, we suggest fixing the cells by formaldehyde first, which can be stored in a −80 °C freezer for up to 1 year, and then follow option B of the procedure.
Starting cell quality
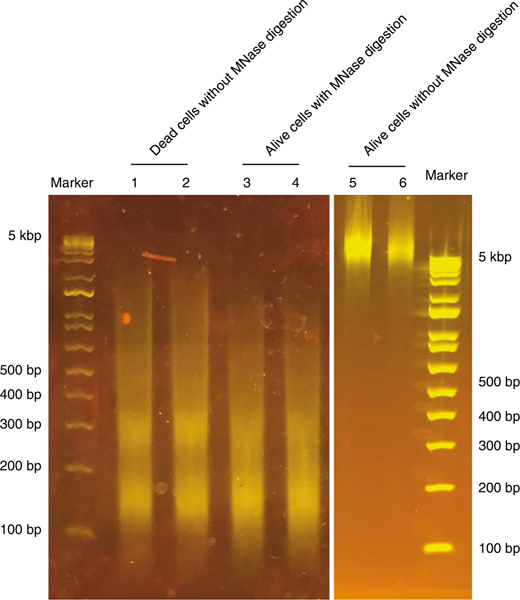
For both option A and option B, it is critical to make sure that >95% of the cells used are viable (before cell fixation) because chromatin in dead or dying cells is fragmented into nucleosome-sized DNA (Fig. 2), which will influence the optimal MNase titration. Use of a control sample without MNase digestion in the MNase titration experiment is essential to confirm that the sample does not contain any fragmented DNA from dead cells (Fig. 2). Cultured cells should be collected at the exponential growth phase, and primary cells should be dissociated as soon as possible following collection. A commercial dead cell removal kit is recommended if there are abundant starting materials. When performing single-cell FACS sorting, Annexin V plus DAPI staining for native cells and amine viability dye staining before formaldehyde fixation are recommended to exclude the dead or dying cells27.
Fig. 2 |. Dead or apoptotic cells exhibit chromatin fragmentation without MNase digestion.
DNA from 10,000 NIH3T3 cells treated with 10% (vol/vol) dimethyl sulfoxide during culture to induce cell death was isolated and resolved on a 2% E-Gel (lanes 1 and 2). As controls, 10,000 live NIH3T3 cells with (lanes 3 and 4) or without (lanes 5 and 6) MNase digestion were resolved on E-Gel. Even without MNase digestion, genomic DNA from apoptotic or dead cells shows chromatin fragmentation with a size range similar to nucleosomal DNA. Lanes 1 and 2, 3 and 4, and 5 and 6 are technical replicates.
Titration of MNase concentration
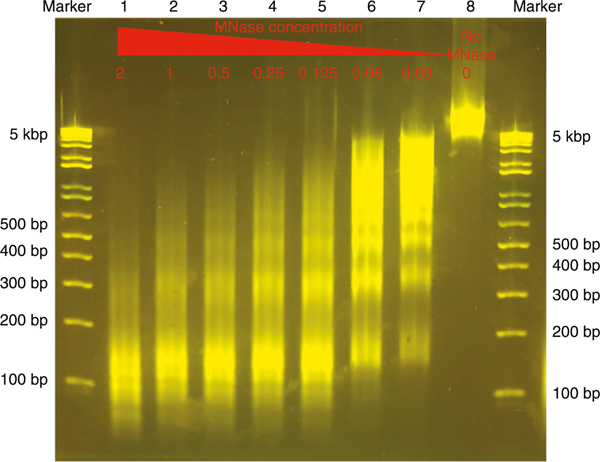
Different cell types may have different chromatin states and thus exhibit different sensitivity toward MNase digestion. We recommend performing the MNase titration before working with a new cell type or a new lot of MNase enzyme. The goal of the titration is to obtain as many mononucleosomal DNA fragments as possible to determine the optimal MNase concentration for the MNase-seq experiment28. We avoid selecting under-digested or over-digested DNA for library construction. Under-digestion is characterized by a large amount of over-sized DNA fragments, which represent oligonucleosomes. Over-digestion is characterized by a lot of oligonucleotide-sized DNA fragments (usually <100 bp). Since over-digestion by MNase destroys nucleosomal DNA while under-digestion reduces the number of detectable nucleosomal particles, it is necessary to titrate the amount of MNase using bulk cells before starting the single-cell experiment. For titrating MNase, we usually start with 10,000 cells per reaction and test different concentrations of MNase (2, 1, 0.5, 0.25, 0.125, 0.06, 0.03 and 0 U/ml) in a 50-μl reaction volume. After incubation of the reaction mixture at 37 °C for 5 min, the DNA is purified and loaded on E-Gel directly to visualize the nucleosome ladders (Fig. 3). The optimal MNase digestion should generate a DNA ladder with the majority as mononucleosomal DNA (with bright band of ~147 bp). Thus, chromatin was under-digested in lanes 6 and 7 as the majority of DNA are above triple-nucleosomes, while it was over-digested in lanes 1 and 2 because there were too many DNA bands below the mononucleosome-sized DNA, indicating degradation of nucleosomal DNA. It appears that MNase digestion may be appropriate in lanes 3 and 4. MNase concentration selected by this strategy works well even with a single cell in the same 50-μl reaction volume.
Fig. 3 |. MNase titration using bulk cells.
MNase titration is necessary before preparing scMNase-seq libraries. Formaldehyde-fixed human peripheral blood mononuclear cells (10,000) per lane were digested with decreasing MNase concentration as indicated on the gel image from 2, 1, 0.5, 0.25, 0.125, 0.06, 0.03 to 0 U/ml; DNA was purified and resolved on E-Gel. Without MNase digestion, lane 8 shows only genomic DNA fragments above 5 kbp. With an increase in MNase concentration (lanes 1–7), fragmented genomic DNA is detected as subnucleosomes (<150 bp), mononucleosomes (~150 bp), dinucleosomes (~300 bp), triple-nucleosomes (~550 bp) and larger nucleosomal particles. In this experiment, lanes 3 and 4 represent an optimal range of MNase concentration generating the most mononucleosome-sized fragments.
Starting cell number
For the single-cell experiments, if flow cytometry is used to sort single cells, cell loss at the FACS-sorting step should be considered, and we recommend using at least 100,000 cells for FACS sorting. Considering the cells required for titrating MNase concentration (100,000) and the cell loss at FACS sorting, the minimal starting cell number should be 200,000 (option A procedure). While other methods may be used to isolate single cells (see the ‘Single-cell isolation’ section), such as laser microdissection or manual cell picking, the starting cell number needs to be experimentally tested for each method.
Single-cell isolation
There are several ways to isolate single cells, such as sorting by flow cytometry, laser microdissection, manual cell picking, random seeding/dilution, and microfluidics/lab-on-a-chip devices29. We used FACS sorting by forward versus side scatter (FSC versus SSC) gating to collect single cells of interest based on size and granularity (complexity) without antibody staining (Supplementary Fig. 1). A preferred method may be chosen based on the starting materials and the availability of equipment.
Library preparation and amplification
Following MNase digestion of single cells, DNA is purified by phenol–chloroform extraction and ethanol precipitated in the presence of circular plasmid DNA that is added before phenol–chloroform extraction. Next, the DNA ends are repaired, and an A base is added to the 3′ end, followed by subsequent ligation to Y-shaped adaptors. After ligation, DNA is amplified using multi-indexing PCR primers. The ‘A-tailing’, adaptor ligation and PCR amplification steps are performed in the same tube without purifying the DNA to minimize DNA loss. Indexed single-cell libraries can be pooled for sequencing on the Illumina platform. Our protocol offers the choice of including ‘i7’ index by indexed Illumina primer 2.0; however, using ‘i5’ index by including another indexed Illumina primer 1.0 is recommended to pool more library samples and decrease the false-assignment rate of the Illumina platform30.
The number of PCR cycles depends on the number of cells used. For single cells, we recommend 25 cycles for visualization of the DNA smear on gel. As the number of cells increases, the number of PCR cycles should be reduced to minimize over-amplification, which reduces library complexity. Starting with 100 cells or 1,000 cells, we perform 20 or 18 cycles of PCR, respectively. Further amplification with 3–4 additional cycles can be performed if there is insufficient DNA after the initial PCR steps.
Fractionation of amplified DNA and sequencing
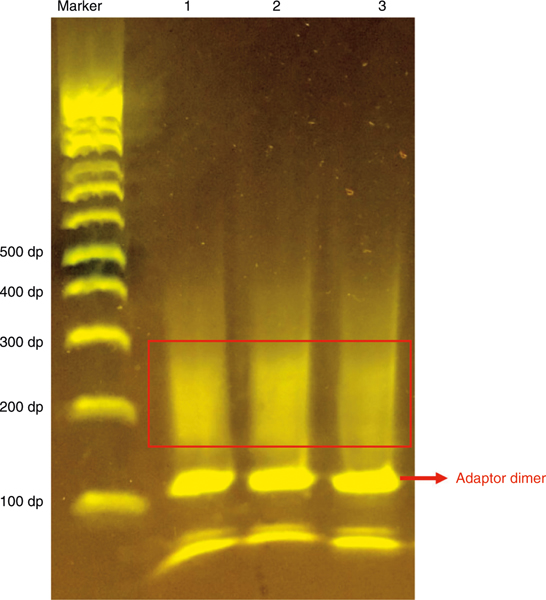
Subnucleosomal and nucleosomal DNA can be isolated from a 2% E-Gel for sequencing. Figure 4 shows three samples of scMNase-seq libraries generated from single mouse naive T cells with formaldehyde fixation following option B of the procedure. The majority of meaningful DNA fragments should be between 150 and 320 bp, which contain the adaptor sequence and a genomic DNA fragment of 30–200 bp. The DNA band at ~120 bp is the adaptor dimer, which should be excluded carefully when cutting E-Gel as the dimer will decrease the mappability of data. scMNase-seq libraries are then sequenced on Illumina HiSeq platforms. In our protocol, we prefer to use Invitrogen 2% agarose E-Gel for visualization and isolation of PCR products to minimize cross-contamination, although homemade 2% agarose gels are also acceptable for this purpose. However, precaution must be taken to avoid cross-contamination of PCR products when using homemade agarose gels. The sequencing efficiency of any single-cell experiment is low, and we usually start an initial round of sequencing to generate a few million reads, estimating library complexity (mappability and redundancy) and quality (nucleosome/subnucleosome-sized particle density relative to the TSS/DHS region). Libraries predicted to have adequate complexity (>500,000 estimated unique nonredundant reads per single cell) and good quality (nucleosome depletion and subnucleosome enrichment at the TSS/DHS) can be selected for deeper sequencing. Typically, we sequence each single-cell library to 5–10 million paired-end reads of 50 bp with a mappability of ~40% and a redundancy of ~70–80%, which would provide >0.5 million unique reads per single cell.
Fig. 4 |. Gel image showing the size range of scMNase-seq library DNA after PCR amplification.
Three scMNase-seq libraries from formaldehyde-fixed single mouse naive CD4 T cells were prepared following the scMNase-seq protocol with 25 cycles of PCR amplification. The single-cell libraries were resolved on 2% E-Gel. The subnucleosomal and mononucleosomal DNA are recovered from the size range of 150–350 bp (indicated by a red rectangle). Be careful to exclude the adaptor dimer band at 120 bp (indicated by a red arrow) when excising the gel slice.
Data analysis
Paired-end sequencing reads for scMNase-seq are mapped against a reference genome using Bowtie 2 (ref. 31) with default parameters. Redundant reads with identical coordinates should be removed from further analysis. The fragments with a range of 140–180 bp are selected as canonical nucleosomes, and the short fragments (≤80 bp) are selected as subnucleosome-sized particles for further analysis. The midpoints of nucleosomes and subnucleosome-sized particles are defined as their positions. Please refer to the paper of Lai et al.9 for further downstream analysis, such as quantifying nucleosome spacing uniformness, nucleosome variance, nucleosome spacing flanking DHSs, bimodal nucleosome spacing across DHS and its links to cell–cell variation of nucleosome positioning and accessibility, and cell clustering based on the cell-to-cell nucleosome positioning dissimilarity score matrix.
Expertise needed to implement the protocol
scMNase-seq does not require advanced technical skills and is easily accessible to almost any molecular biology laboratory. Processing of high-throughput sequencing data uses existing software packages for sequence alignment, marking of duplicate reads, generation of genome browser visualizations and peak calling. Complex analysis of genome-wide nucleosome position requires additional bioinformatics expertise.
Materials
Biological materials
NIH3T3 cells (American Type Culture Collection, cat. no. CRL-1658; see Reagent setup for details) ! CAUTION The cell lines used in your research should be regularly checked to ensure that they are authentic and are not infected with mycoplasma ▲CRITICAL Other cultured or primary eukaryotic cells can be used.
Reagents
▲CRITICAL All reagents must be kept DNase-free.
Culture medium DMEM (Invitrogen, cat. no. 10566–016)
FBS (Sigma-Aldrich, cat. no. F4135–500ML)
Penicillin–streptomycin (Invitrogen, cat. no.15140–122)
Pierce 16% (wt/vol) formaldehyde (methanol free; Thermo Fisher Scientific, cat. no. 28906)
Glycine (Sigma-Aldrich, cat. no. 50046–250G)
1× PBS (155 mM NaCl, 2.7 mM Na2HPO4-7H2O, 1.54 mM KH2PO4, without calcium and magnesium, pH 7.4; Thermo Fisher Scientific, cat. no. 10010–023)
Dead Cell Removal Kit (Miltenyi Biotec, cat. no. 130–090-101)
LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Thermo Fisher Scientific, cat. no. L34963)
Nuclease-free water (Life Technologies, cat. no. AM9930)
1 M Tris-HCl (pH 7.4; K-D Medical, cat. no. RGF-3340)
Triton X-100 (Quality Biological, cat. no. A611-M143–13)
2 M calcium chloride (Quality Biological, cat. no. 351–130-721EA)
EGTA (molecular biology grade; Quality Biological, cat. no. A611–0732-88)
5 M sodium chloride (molecular biology grade; Promega, cat. no. V4221)
3 M sodium acetate (pH 5.2, molecular biology grade; Quality Biological, cat. no. 351–035-721)
10% (vol/vol) SDS (Quality Biological, cat. no. 351–032-101)
Proteinase K (recombinant, PCR grade; Sigma-Aldrich, cat. no. 3115836001)
pUC19 plasmid (New England BioLabs, cat. no. N3041S)
Phenol–chloroform (pH 6.7/8.0; Amresco, cat. no. 0883) ! CAUTION Phenol–chloroform can burn eyes and skin. Use in a chemical hood and dispose of waste in accordance with institutional regulations.
Ethanol (99.5% (vol/vol); Warner-Graham, cat. no. 64–17-5) ! CAUTION Ethanol is highly flammable and volatile. All manipulation should be carried out under a fume hood for flammables.
Glycogen (molecular biology grade; Sigma-Aldrich, cat. no. 10901393001)
MNase (Sigma-Aldrich, cat. no. N3755–500UN) ▲CRITICAL We divide MNase (1 U/μl) into a 5-μl aliquot for single use and store at −80 °C for up to 1 year.
End-It DNA End-Repair Kit (Lucigen, cat. no. ER0720)
dATP (10 mM; Thermo Fisher Scientific, cat. no. 18252015)
Klenow Fragment (3′→5′exo-; 5,000 U/ml; New England BioLabs, cat. no. M0212S)
NEB buffer 2 (New England BioLabs, cat. no. B7002S)
T4 DNA ligase (400,000 U/ml; New England BioLabs, cat. no. M0202S)
Phusion High-Fidelity PCR Master Mix with HF Buffer (New England BioLabs, cat. no. M0531S)
MinElute Reaction Cleanup Kit (Qiagen, cat. no. 28206)
Elution buffer (EB buffer) (Qiagen, cat. no. 19086)
Illumina adaptor oligo mix (see Reagent setup for details)
Illumina common PCR primer 1.0 (see Reagent setup for details)
Illumina indexing PCR primer 2.0 (see Reagent setup for details)
E-Gel EX Agarose Gels (2%; Thermo Fisher Scientific, cat. no. G401002)
E-Gel 1 kb Plus DNA Ladder (Thermo Fisher Scientific, cat. no. 10488090)
MinElute Gel Extraction Kit (Qiagen, cat. no. 28604)
Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, cat. no.Q32854)
Equipment
0.2-ml PCR tubes (Molecular BioProducts, cat. no. 3418)
Eppendorf tubes, 1.5 mL (Eppendorf, cat. no. 0030125150)
Falcon 50-ml conical centrifuge tubes (Corning, cat. no. 14–432-22)
Costar TC-treated multiple-well plates (96 wells, round bottom; Corning, cat. no. 3799)
LightCycler 480 Sealing Foil (Roche, cat. no. 04729757001)
Microtube (1.5 ml, DNA LowBind; Sarstedt, cat. no. 72.706.700)
Tabletop microcentrifuge (model no. MicroStar 17R; VWR, cat. no. 521–1647)
Precision general-purpose water baths (Thermo Fisher Scientific, cat. no. 2824)
Thermal cycler (model no. PTC-200; MJ Research, cat. no. 8252–30-0001)
Freezers, −20 and −80 °C (Liebherr, cat. no. GPESF1476)
E-Gel Power Snap Electrophoresis System (Thermo Fisher Scientific, cat. no. G8300)
Qubit fluorometer (Life Technologies, cat. no. Q32866)
Cell sorter (BD Biosciences, model no. FACSAria II)
Sequencing system (Illumina, model no. HiSeq 3000)
Cell strainer
Microscope
Software
Linux/UNIX environment with GCC compiler, R and Python installed. A memory of at least 8 GB is required.
Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)31
samtools (https://github.com/samtools/samtools)32
bedtools (https://bedtools.readthedocs.io/en/latest/)33
Custom codes (https://github.com/binbinlai2012/scMNase)
Reagent setup
Cell culture
Seed NIH3T3 cells in a 10-cm Petri dish containing complete DMEM medium supplemented with 10% (vol/vol) FBS and 100 U/ml penicillin–streptomycin, grow the cells to ~80% confluency (~1 × 107 cells per dish) at 37 °C in a humidified and aseptic atmosphere containing 5% CO2 for ~2 d.
Lysis buffer
Prepare the lysis buffer consisting of 20 mM Tris-HCl (pH 7.4), 0.1% (vol/vol) Triton X-100, 10 mM NaCl, 2 mM CaCl2 and nuclease-free water. Store the buffer at 4 °C for 1 year. ▲CRITICAL Ca2+ is essential to activate the MNase enzyme.
Lysis buffer/MNase enzyme mix
Immediately before MNase digestion, add ice-cold lysis buffer and dilute the MNase stock solution to optimal concentration on ice. ▲CRITICAL Do not refreeze or reuse diluted MNase enzyme.
Stop buffer
Prepare the stop buffer consisting of 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10 mM EGTA, 0.1% (vol/vol) Triton X-100 and nuclease-free water and store at room temperature (20–25 °C) for 1 year. ▲CRITICAL 0.25 mg/ml proteinase K and 0.15% (vol/vol) SDS are freshly added into the stop buffer before MNase digestion. EGTA is essential to inactivate the MNase enzyme.
Illumina adaptor and primers
Oligonucleotides (see Supplementary Table 1 for oligo sequence) are synthesized by Integrated DNA Technology and suspended by EB buffer to a final concentration of 100 μM. For primers, dilute in EB buffer to 10 μM concentration, prepare aliquots of 50 μl per tube and store at −20 °C for 1 year. The adaptor is generated by annealing single-stranded oligonucleotides to obtain a final concentration of 50 μM in EB buffer as follows. Mix 10 μl of the top and 10 μl of the bottom adaptor oligonucleotides (100 μM) in equal molar amounts, heat the mixture in a 95 °C water bath for 5 min, and then let the mixture and hot water cool down to room temperature slowly. Dilute the double-stranded adaptor to 15 μM in EB buffer. The mixture can be prepared into aliquots of 50 μl per tube for use and stored at −80 °C for 1 year.
Procedure
Cell collection, lysis and MNase digestion (day 1) ● Timing 3–24 h
-
1If the amount of starting material is limiting (100,000–1 million cells), follow option A. If initial cells are enough (>1 million cells) for formaldehyde fixation, follow option B.
- (A) MNase digestion on single cells ● Timing 3–24 h
-
(i) Prepare single-cell suspension by trypsin digestion of adherent cell lines or dissociation of tissues according to the status of the desired samples (see Cell preparation in Experimental design). Resuspend the cells in 1× PBS containing 1% (vol/vol) FBS, filter the cells using a cell strainer to remove cell clumps before FACS sorting and place the tubes on ice for subsequent FACS sorting.▲CRITICAL STEP The cell pellet may not be visible. Leave 2–5 μl of liquid at the bottom of the tube to avoid aspiration of the cell pellet.
-
(ii) Prepare 10–50 0.2-ml PCR 8-tube strips with each tube containing 32 μl of ice-cold lysis buffer (see Reagent setup) on ice.▲CRITICAL STEP For single-cell sorting, we use the smallest tubes possible to minimize the chance of the cell adhering to the wall of the tube. After adding the lysis buffer to the tubes, vortex to wet the tube wall in case of single cells hitting the wall instead of the bottom of the tube during sorting.
-
(iii) Single-cell sorting. Use a cell sorter to distribute one viable single cell per tube. Briefly centrifuge the tubes in a tabletop microcentrifuge and return the tubes on ice for 10–30 min.▲CRITICAL STEP After sorting, centrifugation is needed to ensure that the single cells stay in the lysis buffer. Make sure that apoptotic or dead cells and cell doublets are removed by FACS sorting (see Starting cell quality in Experimental design).
- (iv) Prepare stop buffer containing 0.25 mg/ml proteinase K and 0.15% (vol/vol) SDS, which will be used in Step 1A(vi).
-
(v) MNase digestion. Dilute the 1-U/μl MNase stock in ice-cold lysis buffer to desired MNase concentration and put it on ice. Add an 8-μl aliquot of diluted MNase solution into each tube on ice. Gently pipette the mixture up and down several times. Incubate the samples at 37 °C in a water bath for exactly 5 min.▲CRITICAL STEP The MNase concentration for single cells depends on the pilot experiments of MNase titration. In our experience, the final MNase concentration (0.33 U/ml) worked well for both single cells of mouse native NIH3T3 and human peripheral blood mononuclear cells with or without formaldehyde fixation. If processing more than eight samples, consider handling them in batches to ensure that digestion occurs for exactly 5 min. Keep the tubes on ice before MNase digestion.
- (vi) Add 80 μl of stop buffer containing proteinase K and SDS prepared in Step 1A(iv) to each tube containing a single cell (from Step 1A(v)). Mix well and spin the PCR strip at room temperature in a tabletop microcentrifuge for 1 min.
-
(vii) Add 1 ng of circular carrier DNA (pUC19 vector) into each tube.▲CRITICAL STEP Circular DNA should be added after inactivation of MNase by the stop buffer to prevent its degradation by MNase, and 1 ng of circular DNA is sufficient to reduce DNA loss in subsequent steps.
-
(viii) Proteinase K digestion. Incubate the strip tubes in a PCR machine at 65 °C for 1 h with lid heating for cells without formaldehyde fixation. For formaldehyde-fixed single-cell samples, extend the incubation time to overnight for reverse crosslinking of chromatin.■PAUSE POINT For native cells, overnight incubation is also acceptable. For formaldehyde-fixed cells, incubation at 65 °C overnight is necessary for reverse crosslinking.
-
(ix) Transfer the samples from 0.2-ml PCR strip tubes to 1.5-ml DNA LowBind tubes for the following DNA extraction by phenol–chloroform.■PAUSE POINT The samples can be stored at −80 °C for up to several weeks.
-
- (B) MNase digestion on bulk cells ● Timing 8–24 h
-
(i) Prepare a single-cell suspension (see Cell preparation) and exclude dead cells using a commercial kit such as the Dead Cell Removal Kit (see Starting cell quality).▲CRITICAL STEP Staining with amine viability dyes using the LIVE/DEAD Fixable Violet Dead Cell Stain Kit before formaldehyde fixation is recommended to remove apoptotic or dead cells during FACS sorting.
- (ii) Cell fixation by formaldehyde. Start with 1–50 million cells suspended in culture medium supplemented with 10% (vol/vol) FBS at a concentration of 1 million cells per ml medium. Add 16% (vol/vol) formaldehyde to a final concentration of 1% and rotate at room temperature for 10 min in Falcon 50-ml conical centrifuge tubes.
-
(iii) Add 1/10 volume of 1.25 M glycine to stop the reaction of formaldehyde fixation. Rotate for 5 min at room temperature. Centrifuge for 7 min at 400g at 4 °C. Remove supernatant and wash cells by resuspending the cell pellets with 1 ml of PBS per million cells and centrifuge for 7 min at 400g at 4 °C; repeat once. Divide 1 million cells per 1.5-ml Eppendorf tube, freeze on dry-ice and store at −80°C.■PAUSE POINT Formaldehyde-fixed cells can be stored in a −80 °C freezer for at least 1 year.
-
(iv) MNase digestion. Resuspend 1 million fixed cells in 0.9 ml of lysis buffer and incubate the cells on ice for 10 min. Add diluted MNase to a final concentration of 0.33 U/ml on ice. Gently pipette the mixture up and down several times on ice. Incubate the samples at 37 °C in a water bath for exactly 5 min.▲CRITICAL STEP The optimal MNase concentration needs to be titrated in advance. Manipulate the tube on ice before MNase digestion.
- (v) Add 20 μl of 100 mM EGTA to stop MNase digestion, mix well and centrifuge for 7 min at 400g at 4 °C. Aspirate the supernatant completely and wash the cell pellets twice with 500 μl of PBS (final 1 mM EGTA).
-
(vi) Resuspend the cell pellet in 1 ml of PBS containing 1 mM EGTA and filter the cell suspension by cell strainer before FACS sorting. Put cells on ice.▲CRITICAL STEP Make sure cells used for FACS sorting do not include cell aggregates; this can be checked by a microscope.
-
(vii) Prepare 1 ml of stop buffer containing 0.25 mg/ml proteinase K, 0.15% (vol/vol) SDS and 1 ng of circular carrier DNA (pUC19 vector). Divide 120 μl of buffer per well by multi-tip pipette into a 96-well plate.▲CRITICAL STEP For single-cell sorting, we use plates with a round bottom and vertical-wall wells to minimize the chance of the cell adhering to the wall of the well. Circular DNA should be added to reduce DNA loss.
-
(viii) Single-cell sorting. Use the FACS sorter to distribute a single cell into each well.▲CRITICAL STEP Make sure that apoptotic or dead cells and cell doublets are removed by FACS sorting (see Starting cell quality).
-
(ix) Seal the plate completely and incubate overnight at 65 °C.▲CRITICAL STEP Buffer will evaporate, and the DNA may be destroyed by the resulting high concentration of salt if the sealing is not complete.▲CRITICAL STEP Incubation at 65 °C overnight is necessary for reverse crosslinking of formaldehyde-fixed cells.
-
(x) Transfer the reaction mixture from the plate to 1.5-ml DNA LowBind tubes.■PAUSE POINT The samples can be stored at −80 °C for several weeks.
-
DNA extraction by phenol–chloroform (day 2) ● Timing 1 h
▲CRITICAL Consider handling no more than 12 samples in one batch.
-
2
Add 125 μl of phenol–chloroform into each tube from Step 1A(ix) or 1B(x) and vortex the tubes vigorously for 15 s. Spin the tubes at 16,000g at 4 °C in a microcentrifuge for 3 min.
! CAUTION Phenol–chloroform can burn eyes and skin. Use in a chemical hood.
▲CRITICAL STEP Thorough vortexing is important for subsequent phase separation. When vortexing, make sure that the caps are tightly closed.
-
3
Transfer the top aqueous layer to a new 1.5-ml Eppendorf tube. Add 1 μl of 20 mg/ml glycogen, 12.5 μl of 3 M sodium acetate pH 5.2, and 345 μl of 100% (vol/vol) ethanol. Mix them thoroughly by briefly vortexing.
▲CRITICAL STEP Pipette slowly to ensure that the entire aqueous layer is transferred without transferring the phenol layer.
-
4
Incubate the tubes on dry-ice for at least 15 min or in a −80 °C freezer for at least 1 h to precipitate the DNA.
■PAUSE POINT The samples can be stored at −80 °C for several months.
-
5
Spin the tubes in a microcentrifuge at maximum speed at 4 °C for 15 min. Remove the supernatant carefully and discard it.
▲CRITICAL STEP Pipette slowly to avoid disturbing the DNA pellet.
-
6
Wash the pellet with 400 μl of ice-cold 70% (vol/vol) ethanol.
-
7
Spin the tube in a microcentrifuge at maximum speed at 4 °C for 8 min.
-
8
Remove and discard the supernatant without disturbing the pellet.
▲CRITICAL STEP The pellet can be easily disturbed after the 70% (vol/vol) ethanol wash and may become partially transparent.
-
9
Air dry the pellet briefly until the ethanol has evaporated and the DNA pellet becomes colorless (3–5 min).
▲CRITICAL STEP Avoid over-drying the pellet, as it may make the DNA more difficult to dissolve.
-
10
Resuspend the pellet with 13.7 μl of EB buffer.
■PAUSE POINT The samples can be stored at −80 °C overnight.
Library preparation (day 2) ● Timing 7–9 h
-
11Prepare DNA end-repair master mix on ice, and use the Lucigen DNA END-Repair Kit as shown in the following table.
Component Amount (μl) Final concentration Library DNA (Step 10) 13.7 — 10× end repair buffer 2 1× 2.5 mM each dNTPs 2 0.25 mM 10 mM ATP 2 1 mM End-Repair Enzyme Mix (T4 DNA Pol + T4 PNK) 2 — Total volume 21.7 — -
12
Pipette up and down several times to mix well and incubate the sample at room temperature for 45 min. This step generates blunt-ended DNA.
-
13
DNA purification by MinElute column. Follow Qiagen instructions for the MinElute Reaction Cleanup Kit.
▲CRITICAL STEP Avoid DNA loss when transferring the sample from the 1.5-ml tube to the MinElute column.
-
14
Elute each DNA sample by 10 μl of pre-heated EB buffer.
▲CRITICAL STEP Before elution, pre-heat the EB buffer at 65 °C because heated EB buffer facilitates DNA elution. Ensure that pre-heated EB buffer is dispensed directly onto the column membrane for complete elution of bound DNA.
-
15A-tailing. Add adenines to the 3′ ends of DNA templates. Mix ingredients on ice as shown in the following table.
Component Amount (μl) Final concentration Library DNA (Step 14) 10 — 10× NEB buffer 2 1.3 1× 1 mM dATP 1.3 0.1 mM Klenow (3′→5′Exo−) (5,000 U/ml) 0.3 0.12 U/μl Total volume 12.9 — -
16
Transfer to PCR tubes and run the following program on a thermal cycler: 37 °C for 20 min, 75 °C for 20 min.
▲CRITICAL STEP Do not pause after this step as A-tailing may diminish even if DNA is stored at −80 °C.
-
17Adaptor ligation. Mix ingredients on ice as shown in the following table.
Component Amount (μl) Final concentration Library DNA (Step 16) 12.9 — 10× T4 DNA ligase buffer 1.6 1× 15 μM Illumina adaptor oligo mix (Reagent setup) 0.3 0.3 μM 400 U/μl T4 DNA ligase 1 25 U/μl Total volume 15.8 — ▲CRITICAL STEP Add T4 DNA ligase last to minimize the formation of adaptor dimers.
-
18
Incubate at room temperature for 1–3 h.
▲CRITICAL STEP Do not pause after this step as incubation for longer than 3 h favors the formation of adaptor dimer.
-
19Prepare the PCR master mix on ice as shown in the following table.
Component Amount (μl) Final concentration Library DNA (Step 18) 15.8 — 2× Phusion Master Mix 16.6 1× 10 μM common PCR primer 1.0 0.4 0.12 μM 10 μM indexing PCR primer 2.0 0.4 0.12 μM Total volume 33.2 — ▲CRITICAL STEP Each tube from Step 19 requires a unique index to identify each library after sequencing.
-
2020 PCR-amplify the library using the program below. The number of cycles to use depends on the starting cell numbers as shown in the following tables.
Cycle no. Denature Anneal Extend Hold 1 98 °C, 30 s — — — Varies with the starting cell number (see below) 98 °C, 10 s 68 °C, 30 s 72 °C, 30 s Final — — 72 °C, 5 min 4 °C Starting cell number Total PCR cycles 1 25 10 23 100 20 1,000 18 10,000 15 -
21
Load 16 μl of PCR reaction mix and of E-Gel 1 kb Plus DNA Ladder directly and run on a 2% E-Gel for 20 min using the E-Gel Power Snap Electrophoresis System.
▲CRITICAL STEP If the DNA smear is not clearly visible in the range of 150–300 bp, perform three more PCR cycles using the remaining PCR reaction mix and run the product on an E-Gel again. PCR amplification introduces duplications and bias, and the issue will certainly get even worse during over-amplification or when increasing the PCR cycle numbers. To minimize the PCR cycles on a single-cell library, several libraries’ DNA can be pooled and concentrated before running on E-Gel.
? TROUBLESHOOTING
-
22
Excise the smear from the gel, isolating the size range of 150–350 bp.
▲CRITICAL STEP The average size of mononucleosomal DNA plus adaptors should be ~280 bp. Fragments with a size <200 bp are derived from subnucleosomal particles. However, be careful to exclude the adaptor dimer at 120 bp. Contamination by the adaptor dimer will decrease the mappability of reads.
-
23
Excise the gel smear between 150 and 350 bp from each lane following the Qiagen instructions for the MinElute Gel Extraction Kit. Elute the DNA using 15 μl of pre-heated EB buffer.
▲CRITICAL STEP Dissolve gel slices at room temperature with frequent mixing instead of at elevated temperature. This helps to preserve AT-rich DNA that can be denatured easily above 50 °C and could then be lost at the column-binding step.
-
24
Measure the library DNA concentration using a Qubit fluorometer according to instructions for the Qubit dsDNA HS Assay Kit. The libraries are then sequenced on Illumina platforms.
■PAUSE POINT The samples can be stored at −80 °C for up to 1 year. However, the library DNA concentration should be remeasured after long-term storage.
Sequencing (day 3) ● Timing 2–14 d
-
25
Sequence the libraries on an Illumina next-generation sequencing platform.
▲CRITICAL STEP An initial sequencing to generate a few million reads by MiSeq is helpful in estimating library complexity and quality. The final sequencing depth is dependent on the library complexity and read redundancy (see Fractionation of amplified DNA and sequencing in Experimental design).
Data analysis ● Timing 1–5 d
▲CRITICAL All comments below are executed in a Terminal Window, and the bullet arrow represents a new line of code.
-
26
Align the reads to the reference genome, remove low-mapping-quality reads (MAPQ < 10) and obtain the resulting bed file.
Software: Bowtie 2 (version 2.3.4), samtools (version 1.9), bedtools (version 2.27.1) Sample data can be downloaded from GEO with accession no. GSE96688.
Example command:
Export BOWTIE2_INDEXES=/path_to_reference_genome
bowtie2 -x genome −1 sample.R1.fastq.gz −2 sample.R2.fastq.gz \
–no-unal | samtools view -bS - | samtools view -bf 0×2 -q 10 |\ bamToBed -i stdin -bedpe | awk ‘BEGIN {OFS=“\t”}’;\
{print $1,$2,$6,$6-$2,$8, “+”} > sample.bed
Parameter: -q cut-off of mappability score for filtering low-mapping-qualilty reads Input: sample.R1.fastq.gz, sample.R2.fastq.gz
Output: sample.bed
-
27
Remove redundant reads and get unique reads.
Custom codes: filter_NR_reads (GCC compiled program; source codes and a Make file can be found at https://github.com/binbinlai2012/scMNase/tree/master/Other_tools/src)
Example command:
filter_NR_reads sample.bed sample_noDup.bed
Input: sample.bed
Output: sample_noDup.bed
-
28
Draw fragment length density.
Custom codes: cal_density_3 (GCC compiled program; source codes and a Make file can be found at https://github.com/binbinlai2012/scMNase/tree/master/fragment_length_density/src), drawprofile.r (R script can be obtained from https://github.com/binbinlai2012/scMNase/tree/master/fragment_length_density/bin).
Example command:
awk ‘{print $4}’ sample_noDup.bed > sample_noDup.len.txt cal_density_3 sample_noDup.len.txt 500 sample_noDup.len_den.txt Rscript drawprofile.r sample_noDup.len_den.txt \sample_noDup.len_den.png
Input: sample_noDup.bed (output from Step 27)
Output: sample_noDup.len_den.png
-
29
Separate nucleosome reads and subnucleosomal reads. This step is independent of Step 28 but needs the output from Step 27.
Custom codes: separate_140_180_reads.py, separate_less80_reads.py (Python scripts; https://github.com/binbinlai2012/scMNase/tree/master/Other_tools/bin)
Example command:
python separate_140_180_reads.py sample_noDup.bed \ sample_noDup. 140_180.bed
python separate_less80.py sample_noDup.bed \ sample_noDup.less80.bed
Input: sample_noDup.bed
Output: sample_noDup.140_180.bed, sample_noDup.less80.bed
-
30
Generate average nucleosome/subnucleosomal particles profile around the TSS.
Custom codes: extprog_readcount_on_tss (GCC compiled program; source codes and a Make file can be found at https://github.com/binbinlai2012/scMNase/tree/master/average_profiling/src), draw_profile.r (R script; can be obtained from https://github.com/binbinlai2012/scMNase/tree/master/average_profiling/bin)
Annotation file: refFlat_wf_mm9.txt (UCSC file for mm9 genome build; it can be obtained from https://github.com/binbinlai2012/scMNase/tree/master/average_profiling/src; for another genome build, please download the corresponding UCSC annotation file from the UCSC genome download website)
Gene list file: gene_rpkm10.txt (pre-selected gene list file; it can be downloaded from https://github.com/binbinlai2012/scMNase/tree/master/average_profiling/src. Genes were selected based on the public RNA-seq data and the gene expression reads per kilobase of transcript per million mapped reads (RPKM) calculation. Genes with RPKM >10 were selected)
Example command:
extprog_readcount_on_tss -U refFlat_wf_mm9.txt \
-g gene_rpkm10.txt -b sample_noDup.140_180.bed -u 1000 -d 1000 \
-w 10 -f 0 -t 1 -m 1 -p sample_noDup.140_180.at_tss_rpkm10 −3 y
Rscript draw_profile.r \ sample_noDup.140_180.at_tss_rpkm10.countaveprofile.txt \ sample_noDup.140_180.at_tss_rpkm10.countaveprofile.png
extprog_readcount_on_tss -U refFlat_wf_mm9.txt \
-g gene_rpkm10.txt -b sample_noDup.less80.bed -u 1000 -d 1000 \
-w 10 -f 0 -t 1 -m 1 -p sample_noDup.less80.at_tss_rpkm10 −3 y
Rscript draw_profile.r \ sample_noDup.less80.at_tss_rpkm10.countaveprofile.txt \ sample_noDup.less80.at_tss_rpkm10.countaveprofile.png
Input: refFlat_wf_mm9.txt, gene_rpkm10.txt, sample_noDup.140_180.bed, sample_noDup. less80.bed
Output: sample_noDup.140_180.at_tss_rpkm10.countaveprofile.png, sample_noDup.less80.at_tss_rpkm10.countaveprofile.png
? TROUBLESHOOTING
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
|
| |||
| 21 | No DNA smear is present after PCR | Inadequate PCR amplification Failure of single-cell isolation DNA loss during processing the whole procedure |
Perform three additional PCR cycles Prepare more single-cell libraries or repeat Sorting Repeat library preparation |
| DNA smear with majority of molecular weight >300 bp | Under-digestion by MNase Over-amplification by PCR |
Perform MNase titration Repeat library preparation and perform <25 PCR cycles |
|
| Excessive adaptor dimer | Ratio of adaptor to insert DNA is too high | Reduce adaptor concentration | |
| DNA is present in no-MNase control | Apoptotic or dead cells | Exclude apoptotic or dead cells | |
| 30 | <40% mappability of reads <0.5 million unique reads with redundancy >80% |
Excessive adaptor dimer DNA loss during processing the whole procedure |
Exclude dimer when excising bands from gel Repeat library preparation |
| No enrichment of subnucleosome fragments at the TSS | Over-digestion by MNase Use of inactive TSS profile |
Perform MNase titration Use expressed genes’ TSS profile |
|
| No depletion of nucleosome fragments at the TSS | Under-digestion by MNase Use of inactive TSS profile |
Perform MNase titration Use expressed genes’ TSS profile |
|
Timing
Step 1A(i–iii), collection of single cells: 1–3 h
Step 1A(iv–vii), MNase digestion on single cells: 1 h
Step 1A(viii, ix), proteinase K digestion of proteins: 1–6 h or overnight
Step 1B(i–iii), cell collection and fixation: 1–3 h
Step 1B(iv–vi), MNase digestion on bulk cells: 0.5 h
Step 1B(vii, viii), single-cell sorting: 0.5 h
Step 1B(ix, x), proteinase K digestion of proteins: 6 h or overnight
Steps 2–10, extraction of DNA: 1 h
Steps 11–14, library preparation (end-repair): 1.5 h
Steps 15 and 16, library preparation (A-tailing): 1.5 h
Steps 17 and 18, library preparation (adaptor ligation): 1–3 h
Steps 19 and 20, library preparation (PCR amplification): 1.5 h
Steps 21–24, gel-size selection of PCR products: 1.5 h
Step 25, sequencing: variable, based on sequencing platform: 2–14 d
Steps 26–30, data analysis: variable
Anticipated results
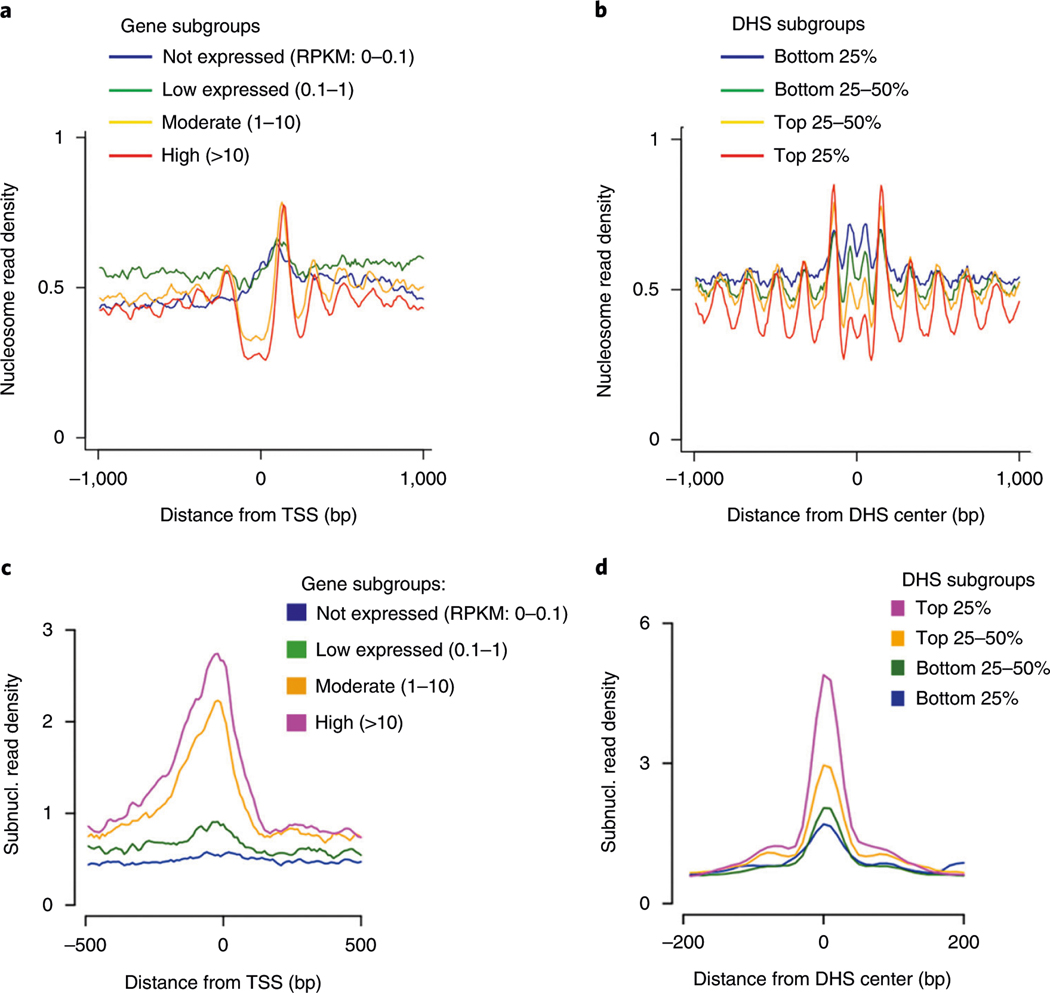
In the MNase titration experiment, the optimal MNase digestion should generate as much mononucleosomal DNA as possible, and there should not be any nucleosomal DNA ladders for the no-MNase-digestion control (Fig. 3). For the single-cell library preparation, a smear of PCR DNA products with the highest intensity between 150 and 300 bp should be observed on the gel (Fig. 4). An initial sequencing of the scMNase-seq libraries to generate a few million reads and plotting nucleosomal read density at active TSS and DHS regions are needed to check the complexity and quality of library data. Successful single-cell libraries should be predicted to generate >0.5 million unique reads per single cell and exhibit the nucleosome depletion and subnucleosome enrichment at the TSS of active genes and surrounding DHSs (Fig. 5).
Fig. 5 |. Average profiling of nucleosomes and subnucleosomes from pooled single cells.
a,b, Nucleosome profiles at TSSs of genes with different expression levels (a) and surrounding DHS centers for four subgroups of DHSs with different strengths (b). c,d, Subnucleosome (Subnucl.) profiles at TSSs of genes (c) and DHS centers (d). Data from 48 libraries of native single NIH3T3 cells following option A procedure were pooled for analysis to evaluate the average profiling of nucleosomes and subnucleosomes at the TSS/DHS sites. Data were downloaded from Lai et al. published previously9. c,d, Duplicated from Extended Data Fig. 2i,j of the previous publication9. Data accession no. GSE96688.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data used in this protocol have been deposited in the Gene Expression Omnibus database with accession number GSE96688. Figure 5 is associated with the raw data. There are no restrictions on data availability.
Code availability
Codes used in this protocol have been deposited in GitHub (https://github.com/binbinlai2012/scMNase). There are no restrictions on code availability.
Supplementary Material
Acknowledgements
We thank the National Heart, Lung, and Blood Institute DNA Sequencing Core Facility for sequencing the libraries and the National Heart, Lung, and Blood Institute Flow Cytometry Core facility for sorting the cells. The work was supported by the Division of Intramural Research, National Heart, Lung and Blood Institute (K.Z.), the National Key Research and Development Project (no. 2016YFA0502203) (B.N.), the General Program of National Natural Science Foundation of China (no. 81670534) (B.N.), and the Medical Scientific and Technological Innovation Funds of Southwest Hospital (no. SWH2016LHYS-04) (B.N.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0243-6.
Peer review information Nature Protocols thanks Gonçalo Castelo-Branco, Emily Hodges and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related link
Key references using this protocol
Lai, B. et al. Nature 562, 281–285 (2018): https://doi.org/10.1038/s41586-018-0567-3
References
- 1.Kornberg RD & Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M. & Workman JL The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Lai WKM & Pugh BF Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol 18, 548–562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodges C. et al. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z. et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332, 977–980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang C. & Pugh BF Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet 10, 161–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev 13, 127–135 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Lai BB et al. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562, 281–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schones DE et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pott S. & Lieb JD Single-cell ATAC-seq: strength in numbers. Genome Biol. 16, 172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper J. et al. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat. Protoc 12, 2342–2354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin WF et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldi S. Nucleosome positioning and spacing: from genome-wide maps to single arrays. Essays Biochem 63, 5–14 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Liang J, Cai W. & Sun ZS Single-cell sequencing technologies: current and future. J. Genet. Genomics 41, 513–528 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hoeijmakers WAM & Bartfai R. Characterization of the nucleosome landscape by micrococcal nuclease-sequencing (MNase-seq). Methods Mol. Biol 1689, 83–101 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Kelsey G, Stegle O. & Reik W. Single-cell epigenomics: recording the past and predicting the future. Science 358, 69–75 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Shema E, Bernstein BE & Buenrostro JD Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet 51, 19–25 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pott S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 6, 061739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark SJ et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L. et al. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol 20, 847–858 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Buenrostro JD et al. Transposition of native chromatin for fast and sensitive mulitmodal analysis of chromatin architecture. Biophys. J 106, 77a (2014). [Google Scholar]
- 24.Buenrostro JD et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusanovich DA et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein AM & Macosko E. InDrops and Drop-seq technologies for single-cell sequencing. Lab Chip 17, 2540–2541 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Perfetto SP et al. Amine-reactive dyes for dead cell discrimination in fixed samples. Curr. Protoc. Cytom 53, 9.34.1–9.34.14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui K. & Zhao K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol 833, 413–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross A. et al. Technologies for single-cell isolation. Int. J. Mol. Sci 16, 16897–16919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kircher M, Sawyer S. & Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B. & Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this protocol have been deposited in the Gene Expression Omnibus database with accession number GSE96688. Figure 5 is associated with the raw data. There are no restrictions on data availability.