Abstract
The Saccharomyces cerevisiae SBA1 gene was cloned by PCR amplification from yeast genomic DNA following its identification as encoding an ortholog of human p23, an Hsp90 cochaperone. The SBA1 gene product is constitutively expressed and nonessential, although a disruption mutant grew more slowly than the wild type at both 18 and 37°C. A double deletion of SBA1 and STI1, encoding an Hsp90 cochaperone, displayed synthetic growth defects. Affinity isolation of histidine-tagged Sba1p (Sba1His6) after expression in yeast led to coisolation of Hsp90 and the cyclophilin homolog Cpr6. Using an in vitro assembly assay, purified Sba1His6 bound to Hsp90 only in the presence of adenosine 5′-O-(3-thiotriphosphate) or adenyl-imidodiphosphate. Furthermore, interaction between purified Sba1His6 and Hsp90 in yeast extracts was inhibited by the benzoquinoid ansamycins geldanamycin and macbecin. The in vitro assay was also used to identify residues in Hsp90 that are important for complex formation with Sba1His6, and residues in both the N-terminal nucleotide binding domain and C-terminal half were characterized. In vivo analysis of known Hsp90 substrate proteins revealed that Sba1 loss of function had only a mild effect on the activity of the tyrosine kinase v-Src and steroid hormone receptors.
Polypeptide folding occurs in vivo with the aid of molecular chaperones. Members of this diverse protein family have been proposed to function by protecting nascent chains from off-pathway interactions that might otherwise lead to aggregation and to stabilize conformations from which productive folding can proceed (25, 29). The two major classes of molecular chaperone that exist in the eukaryotic cytosol under normal growth conditions are Hsp70 and Hsp90. Both of these chaperones function in protein folding reactions with the aid of several factors that act as cochaperones or modulators. In the case of eukaryotic Hsp70, the cochaperones include members of the DnaJ family, p48/Hip, p60/Sti1p, and Bag1 (10, 21, 26, 50). Each of these cochaperones has been reported to modulate the ATPase activity of Hsp70 by either stimulating ATP hydrolysis or regulating nucleotide exchange (10, 21, 24, 26). The Hsp70 chaperone machine also appears to function in association with Hsp90 via the action of the p60 gene, an ortholog of the yeast STI1, which binds to both chaperone proteins (9, 11, 36, 46). Hsp90 is an abundant and highly conserved molecular chaperone. It exists in several discrete subcomplexes that together comprise the Hsp90 chaperone machine. One of these subcomplexes contains Hsp90/Hsp70/Hip and p60/Sti1 (46), while another contains one of several immunophilins and the acidic protein p23 (31).
The p23 protein was first identified in association with the progesterone receptor (PR) (45) and was later shown to be an Hsp90 binding protein (30, 32, 49). In addition to being found in protein complexes with several unliganded steroid hormone receptors, p23 has been found to associate via Hsp90 with several proteins including transcription factors, protein kinases, and a viral reverse transcriptase (27, 37, 56). In all of these interactions, p23 is thought to be indirectly associated via Hsp90. There is also evidence, however, that p23 can bind independently to polypeptides and prevent them from aggregating (5, 20, 27).
Purified p23 binds to Hsp90 in a manner that is stabilized by nonhydrolyzable ATP and by molybdate ions (49). In addition, complex formation between p23 and Hsp90 is inhibited by geldanamycin, a benzoquinoid ansamycin that was originally defined as a tyrosine kinase inhibitor but was recently shown to specifically interact with Hsp90 (54). Geldanamycin negatively affects the activity of steroid hormone receptors and protein kinases and stimulates their degradation (43, 44, 47, 53). By contrast, p23 appears to be important for stabilizing a high-affinity hormone binding conformation of the glucocorticoid receptor (GR) and PR (14, 47).
Recent studies have demonstrated the conserved nature of yeast and animal cell Hsp90 protein complexes (8, 9; see reference 6 for a review), but to date there has been no report of a yeast p23 homolog. In this report, we present a genetic and biochemical characterization of a p23 homolog from the yeast Saccharomyces cerevisiae.
MATERIALS AND METHODS
Materials.
Geldanamycin and macbecin were obtained from the Drug Synthesis & Chemistry Branch, Developmental Theraputics Program, Division of Cancer Treatment, National Cancer Institute. Both compounds were stored at 2 mg/ml in dimethyl sulfoxide at −20°C. Monoclonal antibodies to phosphotyrosine (4G10) and v-Src (GD11) were purchased from Upstate Biotechnology (Lake Placid, N.Y.). Antiserum to Hsp90 was described previously (12), and antiserum to Hsp70 was purchased from Affinity Bioreagents. Antisera to Cpr6 and Sti1 were the kind gifts of D. Picard and D. Toft, respectively. Isogenic yeast strains encoding wild-type and G170D mutant forms of HSP82 were the kind gifts of S. Lindquist. Plasmids encoding the A97I (pts38RV), T101I (pcs2-3RV), and S485Y (pts33BE) hsp82 mutants were the kind gift of Y. Kimura (33), and those encoding the E431K (pTCA/hsp82 E431K) and T525I (pTCA/hsp82 T525I) mutants of hsp82 were the gift of K. Yamamoto (3). Plasmid pGVSRC (CEN/ARS URA3 v-src) was the kind gift of Frank Boschelli.
Yeast methods and strains.
The genotypes of S. cerevisiae strains used in this study are shown in Table 1. Standard genetic methods were used for growth and manipulation. Yeast strains were grown in either rich medium (YPD) or selective minimal medium (0.67% yeast nitrogen base) with 2% glucose (or raffinose or galactose) and supplemented with adenine, uracil, and amino acids, depending on auxotrophy. Transformation of yeast strains was performed as described previously (22). Strain ACY77 was constructed from W3031b by one-step gene replacement (42) using a DNA fragment containing the sti1-1::HIS3 allele that was amplified by PCR from the genomic DNA of strain CN11 (38), using the primers 5′ GCGCGCAAGCTTAATATTCTATGTCTGGCAACT 3′ and 5′ GCGCGCTCTAGACTTATTATCTTCAAGTTCCGA 3′ in a 30-cycle reaction at an annealing temperature of 50°C. sti1-1/sba1-1 double mutants were prepared from ACY77 by one-step gene replacement using the 2.4-kb sba1-1::URA3 fragment as described below.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W3031b | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 | 51 |
| CN11 | a Δtrp1 lys1 lys2 ura3-52 leu2-3,112 his3-11,15 sti1-1::HIS3 | 39 |
| ACY77 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sti1-1::HIS3 | This study |
| YF233 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sba1-1::URA3 | This study |
| YF240 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sba1-1::ura3 | This study |
| YF256 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sba1-1::ura3 pGAL-SBA1 | This study |
| YF257 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sti1-1::HIS3 sba1-1::URA3 | This study |
| YF265 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 pRS426 | This study |
| YF266 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 pJR10 | This study |
| AFY200 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 pGVSRC | This study |
| AFY202 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sba1-1::ura 3 pGVSRC | This study |
| YF267 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 pARH, pPGKarelacZC | This study |
| YF268 | α ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 sba1-1::ura3 pARH, pPGKarelacZC | This study |
| p82a | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pTGPDHsp82 | 38 |
| G170Da | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pTGPpd/T1-101 | 38 |
| ACY 2W | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pTGPDHsp82, pJR1 | This study |
| ACY 1WU | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pJR1 | This study |
| AFYA97I | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pts38RV | This study |
| AFYT101I | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pcs2-3RV | This study |
| AFYE431K | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pTCA/hsp82 E431K | This study |
| AFYS485Y | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pts33BE | This study |
| AFYT525I | a ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 (hsc82::LEU2) (hsp82::LEU2) pTCA/hsp82 T525I | This study |
The hsp82 mutant strains (except for G170D, which was the gift of S. Lindquist) were constructed from strain p82a (38). This strain was transformed with pJR1 (CEN6/ARS4/HSP82/URA3), and the resulting strain (AFY2W) was grown in rich medium to deselect for pTGPD (HSP82/TRP1). Resulting Trp auxotrophs were designated AFY1WU. This strain was transformed with plasmids containing different hsp82 mutant genes (see above) and subsequently plated on 5-fluoro-orotic acid to select for loss of pJR1. The resulting strains all contained a single mutant hsp82 allele (Table 1).
SBA1 cloning and gene disruption.
SBA1 was cloned by a 30-cycle PCR amplification (using Taq polymerase; annealing temperature of 37°C) from S. cerevisiae W3031b genomic DNA, using the primers 5′ GCGCGCCTCGAGGATAATCATCCAGCA 3′ and 5′ GCGCGCTCTAGAACTGCCTTTTATGAA 3′. The 1,269-bp product was gel purified and cloned into the pCR-Script vector (Stratagene) to generate the clone designated p2323. The SBA1 disruption allele was generated by insertion of a 1.1-kb blunt-ended DNA fragment containing the URA3 gene into HpaI-digested p2323, which linearized the plasmid in the SBA1 open reading frame (ORF). A 2.4-kb DNA fragment containing the sba1-1::URA3 disruption allele (called sba1-1 hereafter) was released from this plasmid (pΔ23URA3) by digestion with restriction endonucleases XhoI and XbaI. This linear 2.4-kb fragment was transformed into haploid wild-type yeast (W3031b) for one-step gene replacement of SBA1.
A gene encoding His6-tagged Sba1 (Sba1His6) under control of the yeast GAL1 promoter was cloned by PCR amplification (using Pfu polymerase at an annealing temperature of 55°C) from p2323 by using the primers 5′ ATAGTCATGCACCACCATCACCACCACTCCGATAAAGTTATTAAC 3′ and 5′ GCGCGCCTCGAGGATAATCATCCAGCA 3′. This amplification resulted in addition of six histidine residues just downstream of the N-terminal methionine. The 1-kb product was first cloned into pCR-Script (Stratagene) and then released from this vector by BamHI and SacI digestion before ligation into pRS316.GAL (CEN/ARS URA3 GAL1-10 promoter) digested with the same restriction endonucleases. The resulting plasmid (pGAL-SBA1) was transformed into an sba1-1 strain that had been plated on medium containing 5-fluoro-orotic acid (2) to select for reversion to uracil auxotrophy (YF240).
Purification of Sba1His6 and preparation of Sba1 antiserum.
A gene encoding recombinant Sba1His6 for expression in Escherichia coli was generated by PCR amplification from p2323 (as above), using the primers 5′ GCGCGCCATATGTCCGATAAAGTTATTAAC 3′ and 5′ GCGCGCGGATCCAAGTTACTCATTCTAGCA 3′. The 1-kb product was cloned into pCR-Script (Stratagene), excised by using NdeI and BamHI, and subcloned into similarly digested pET15b (Novagen). Construction of SBA1 in this vector resulted in an N-terminal sequence extension juxtaposed to wild-type SBA1 sequence as follows: MGSSHHHHHHSSGLVPRGSH (single-letter amino acid code). The resulting plasmid, p23ET, was transformed into E. coli BL21(DE3). Expression of Sba1His6 was induced as follows. BL21(DE3) containing p23ET was grown overnight at 37°C in 100 ml of LB medium plus 50 μg of ampicillin per ml. The culture was diluted 1:10 into fresh LB medium and incubated for 1 h at 37°C before addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. The culture was incubated for a further 2 h before harvesting the cells. The cells were resuspended in 5 ml of BL buffer (10 mM Tris-HCl [pH 7.5], 50 mM Sba1, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail containing equimolar amounts of aprotinin, chymostatin, leupeptin, and pepstatin [each at 2 μg/ml]) and broken by sonication (10 times, 10-s bursts with cooling on ice for 30 s between bursts). The lysate was cleared by centrifugation at 14,000 × g for 30 min at 4°C. The protein concentration was adjusted to 10 mg/ml with lysis buffer.
Sba1His6 was purified by a two-step procedure. First, Sba1His6 present in the crude bacterial lysate was bound to Ni-nitrilotriacetic acid (NTA) resin (Qiagen) in batch. This was performed by adding 2 ml of Ni-NTA resin (equilibrated in lysis buffer) to 20 ml of crude lysate (at 10 mg/ml) supplemented with 5 mM imidazole (Sigma); this mixture was incubated on a nutator for 30 min at 4°C. The resin was collected by centrifugation at 1,000 × g and washed twice in lysis buffer plus 10 mM imidazole and twice with lysis buffer plus 20 mM imidazole. Bound proteins were eluted in lysis buffer plus 150 mM imidazole in a 3-ml volume for 10 min at 4°C. The eluate was dialyzed overnight against 20 mM HEPES (pH 7.2)–0.2 mM EDTA–0.5 mM PMSF. The dialyzed eluate was concentrated in a Centricon-10 centrifugal concentrator before loading onto a 5-ml Mono-Q column. Bound proteins were eluted with a 0 to 500 mM KCl gradient.
Antiserum to Sba1His6 was prepared by resolving crude bacterial lysate as described above in a denaturing 10% polyacrylamide gel. The band containing Sba1His6 was identified after Coomassie blue staining and excised with a razor blade. Antiserum to Sba1His6 was prepared from these gel slices in rabbits by Covance Research Products Inc. (Denver, Pa.).
Affinity isolation of Sba1 from yeast.
One-step affinity chromatography of Sba1His6 expressed in yeast was performed by a modification of the procedure of Fang et al. (18). Briefly, 100-ml yeast cultures were harvested, the pellet was washed once with water and once with YL buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail [2 μg/ml] as described above). The pellet was resuspended in 0.8 ml of lysis buffer plus 20 mM sodium molybdate, and the cells were broken in the presence of 1.5 g of 0.4-mm-diameter glass beads by 5 30-s bursts in a mini-BeadBeater (Biospec Products) at 4°C, with cooling on ice for 30 s between bursts. The crude lysate was cleared at 14,000 × g for 30 min at 4°C. The protein concentration was adjusted to 3 mg/ml, and 0.5 ml of protein extract was incubated with 50 μl of Ni-NTA resin (equilibrated in lysis buffer) for 15 min at 4°C on a nutator. The resin was collected at 500 × g and washed three times in 1 ml of lysis buffer plus 20 mM sodium molybdate and 10 mM imidazole. The resin was incubated with 50 μl of elution buffer (lysis buffer plus 150 mM imidazole) at 4°C for 10 min. The eluted protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Coomassie blue staining or by Western blotting.
In vitro assembly assay.
Sba1His6 purified from E. coli was bound to Ni-NTA resin (30 μl of packed beads, equilibrated in YL buffer as described above) by direct addition and incubation for 15 min at 4°C. Beads containing 1 μg of Sba1His6 were added to 100 μl of desalted yeast whole-cell extract (at a concentration of 1 mg/ml; prepared by using a 5-ml HiTrap desalting column [Pharmacia]). The volume was adjusted to 250 μl with YL buffer, and the mixture was incubated for 30 min at 30°C. The beads were collected by centrifugation at 1,000 × g and washed three times with 1 ml of YL buffer plus 10 mM imidazole. Protein was eluted from the beads YL buffer plus 150 mM imidazole in a 30-μl volume for 10 min at 4°C and analyzed by SDS-PAGE and Western blotting.
v-Src expression, immunoblot analysis, and steroid hormone receptor assays.
Lysates of yeast expressing v-Src were prepared as follows. Yeast cultures were grown at 30°C in minimal medium containing 2% (wt/vol) raffinose to late log phase. v-Src expression was induced by the addition of 2% (wt/vol) galactose to the medium. Induction was continued for 6 h, at which time cells were harvested, resuspended in YLS buffer (20 mM Tris-HCl [pH 7.4], 0.14 M NaCl, 5 mM EDTA, 1% Nonidet P-40, 1 mM sodium vanadate, 5 mM dithiothreitol, 1 mM PMSF, a mixture of aprotinin, chymostatin, leupeptin, and pepstatin [each at 2 μg/ml]), and lysed with glass beads as described above. Lysates were cleared of broken cell debris at 14,000 × g.
The levels of tyrosine phosphorylation and v-Src protein were assayed by immunoblot analysis using either antiphosphotyrosine or anti-v-Src monoclonal antibodies (see Materials and Methods). Lysates (100 μg of total protein) were resolved by SDS-PAGE and transferred to nitrocellulose membranes (0.45 μm; MSI). Filters were washed briefly with TTBS (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 0.05% Tween 20) and blocked overnight with TTBS containing 5% nonfat dry milk. Filters were incubated with antibodies to v-Src or phosphotyrosine (diluted 1:1,000 in antibody dilution buffer [1× phosphate-buffered saline, 3% bovine serum albumin, 0.05% Tween 20, 0.1% thimerosal) for 2 h. Filters were washed three times for 10 min each in TTBS. Filters were incubated with secondary antibody (horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G, diluted 1:10,000 in antibody dilution buffer) for 1 h, and subsequently washed as for the primary antibody. Filters were then treated with the chemiluminescence reagent (Pierce) and exposed to X-ray film for detection.
Immunoblots for detecting Sba1, Cpr6, Sti1, and Hsp90 were processed by standard procedures after transfer of the proteins in denaturing gels to nitrocellulose membranes. The filters were typically blocked overnight in TTBS plus 5% nonfat dried milk and incubated with primary and secondary antibodies for 1 to 2 h. Filters were washed between antibody incubations with TTBS (three times, 5 min each), and proteins were detected as described above. Assays for steroid hormone receptor activation were performed as previously described (7, 18).
RESULTS
YKL117W is orthologous to vertebrate p23 genes.
A BLAST search of the GenBank database with the sequence of human p23 protein revealed a single orthologous ORF in S. cerevisiae displaying 24% identical amino acids (28). This gene (YKL117W) encodes a putative protein of 24.1 kDa, which is 5.4 kDa larger than the human p23 protein (18.7 kDa) (Fig. 1 shows protein sequence alignments). YKL117W resides on the left arm of chromosome XI between coordinates 219970 and 220620, just downstream of a tRNAAla gene and upstream of a hypothetical ORF, YKL116C.
FIG. 1.
Sequence comparison of Sba1 with chicken (Cp23) and human (Hp23) p23 proteins. Identical matches are denoted by asterisks; gaps in the sequence are denoted by periods.
The YKL117W protein sequence displays weak identity and similarity throughout its length with human and chicken p23 proteins. The strongest region of identity is between seven identical consecutive amino acids starting at position 104 of the yeast sequence: Trp Pro Arg Leu Thr Lys Glu (three-letter code). The sequences of vertebrate and yeast p23 proteins diverge toward the C-terminal end, with the yeast protein having additional sequence that is rich in hydrophobic amino acids. This region contains the motif Gly Gly Ala repeated five times from amino acids 150 to 174. Like human and chicken p23 proteins, YKL117W has an acidic C terminus. We have named this yeast gene SBA1 because the binding of Sba1 to Hsp90 is sensitive to benzoquinoid ansamycins, as will be shown below.
SBA1 is nonessential and constitutively expressed.
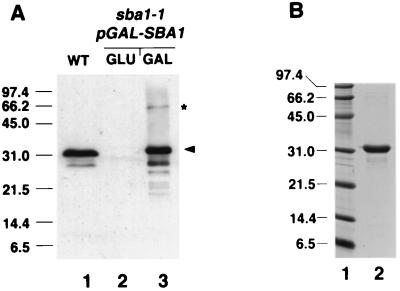
The SBA1 gene was cloned by PCR amplification from wild-type yeast genomic DNA. Northern blot analysis revealed that SBA1 is constitutively expressed as previously found (17) but not heat shock inducible under the same conditions when STI1 mRNA is induced (reference 38 and data not shown). Sba1 protein was detected in wild-type yeast whole-cell extracts by Western blot analysis using antisera prepared from recombinant Sba1His6 expressed in E. coli. Wild-type Sba1 migrates anomalously at 32 kDa, approximately 8 kDa larger than the predicted size (Fig. 2A, lane 1). Sba1His6 also migrated anomalously at 32 kDa after inducible expression in yeast or after purification (to approximately 90%) from E. coli (Fig. 2).
FIG. 2.
Characterization of Sba1 by gel electrophoresis. (A) Western blot analysis of Sba1 in yeast whole-cell extracts using polyclonal anti-Sba1. Lane 1, 50 μg of whole-cell extract from wild-type (WT) cells; lane 2, 50 μg of whole-cell extract from sba1-1 cells containing pGAL-SBA1 (YF256) grown in glucose-containing medium; lane 3, 5 μg of whole-cell extract from sba1-1 cells containing pGAL-SBA1 grown in galactose-containing medium. The arrowhead shows positions of Sba1 (lane 1) and Sba1His6 (lane 3). The asterisk shows the band at 60 kDa that reacts with anti-Sba1 in cells overexpressing Sba1His6. (B) Coomassie blue-stained gel of 2 μg of Sba1His6 (lane 2) after overexpression in E. coli and protein purification. Molecular size markers (in kilodaltons) shown in lane 1.
A disruption allele was constructed by inserting the URA3 gene into the SBA1 ORF. Replacement of wild-type genomic SBA1 with the disruption allele (sba1-1) resulted in complete loss of Sba1 protein expression as assessed by Western blotting (Fig. 2A), although this did not result in a very severe growth phenotype. Using a plate assay to determine the relative growth rate of wild-type and sba1-1 cells, there was very little difference in the growth rate at 30 or 37°C, although the sba1-1 cells grew noticeably more slowly at 18°C (Fig. 3). In liquid media, sba1-1 cells had a doubling time similar to that of the wild type at 30°C but grew 1.6 times more slowly than the wild type at 37°C. Thus, deletion of Sba1 protein resulted in mild growth defects at both low and high temperatures but not at 30°C.
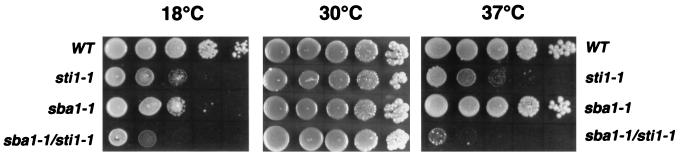
FIG. 3.
Growth phenotype of sba1-1 cells. Each panel shows a serial dilution of yeast cells spotted (3 μl of culture starting at 2 × 108 cells/ml) from left to right of each plate, using wild-type (WT), sti1-1, sba1-1, and double-mutant (sba1-1/sti1-1) strains. Each plate was incubated at the indicated temperature for either 4 days (30 and 37°C) or 7 days (18°C).
To investigate the relationship between Sba1 and other components of the molecular chaperone machinery, we constructed a double mutant that was deleted for SBA1 and also for STI1, a gene encoding an Hsp90 cochaperone that is orthologous to p60 of vertebrate cells (8, 9, 36, 38, 46). As previously shown, STI1 deletion results in reduced growth at both low and high temperatures (38) (Fig. 3). While the double mutant grew normally at 30°C, it exhibited synthetic defects by having more restrictive growth at both 18 and 37°C compared with either mutant alone (Fig. 3). These synthetic defects suggest that Sba1 and Sti1 either interact with each other or function in the same or redundant processes. Expression of wild-type SBA1 on a low-copy-number plasmid in the double mutant resulted in a growth phenotype similar to that of the sti1-1 mutant (data not shown). The temperature-sensitive growth phenotype of sba1-1/sti1-1 double mutants was somewhat strain dependent, being more severe in the W303 background having the ade2 mutation than in strains that were ADE2 wild type.
Sba1 interacts with Hsp90.
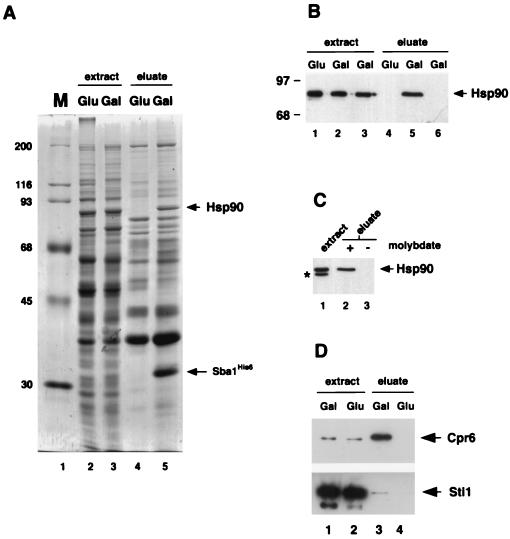
Vertebrate p23 proteins bind to Hsp90 in a nucleotide-dependent manner (49). To determine whether Sba1 also interacts with Hsp90, Sba1His6 was inducibly expressed from the yeast GAL1 promoter at 30°C in the sba1-1 disruption strain (Fig. 2A). Sba1His6 was isolated from whole-cell extracts by affinity chromatography using Ni-NTA resin. Examination of the protein profiles after affinity chromatography revealed that many yeast proteins bound nonspecifically to the Ni-NTA resin. However, comparison of the eluates from induced and noninduced cells revealed the presence of two major bands at 32 and 86 kDa that were observed only after Sba1His6 induction (Fig. 4A; compare lanes 4 and 5). The band at 32 kDa corresponds to Sba1His6 itself and reacted with Sba1 antiserum (data not shown), while the 86-kDa protein (which appears as a very tight doublet in Fig. 4A) reacted with antiserum to Hsp90 (Fig. 4B). The isolation of Hsp90 with Ni-NTA resin was dependent on the presence of Sba1His6, since it was not isolated from extracts of wild-type cells not containing plasmid pGAL-SBA1 (Fig. 4B). Complex formation between Hsp90 and Sba1His6 was stabilized by the presence of sodium molybdate in the binding and wash buffers; omission of sodium molybdate from the wash buffer destabilized the complex and Hsp90 dissociated from Sba1His6 (Fig. 4C).
FIG. 4.
Sba1His6 binding to Hsp90. (A) Coomassie blue-stained gel of whole-cell extracts from strain YF256 (sba1-1 pGAL-SBA1) (lanes 2 and 3) and eluates after affinity chromatography with Ni-NTA resin (lanes 4 and 5). Extracts were prepared from cells grown in glucose-containing medium (lanes 2 and 4) or galactose-containing medium (lanes 3 and 5). Molecular size markers (M, in kilodaltons) are shown in lane 1. Bands corresponding to Sba1His6 and Hsp90 are indicated by arrows. (B) Detection of Hsp90 by Western blot analysis in whole-cell extracts (lanes 1 to 3) and eluates after Ni-NTA resin affinity chromatography (lanes 4 to 6). Extracts and eluates were prepared from strain YF256 grown in glucose (lanes 1 and 4)- or galactose (lanes 2 and 5)-containing medium and wild-type strain W3031b grown in galactose-containing medium (lanes 3 and 6). (C). Western blot analysis of Hsp90 after isolation of Sba1His6 protein complexes on Ni-NTA resin and washing in buffers plus (lane 2) or minus (lane 3) sodium molybdate. A Western blot of the whole-cell extract is shown in lane 1; the band labeled with an asterisk is an Hsp90 degradation product. Data are from nonconsecutive lanes of the same gel and Western blot. (D) Western blot analysis of Cpr6 (upper panel) and Sti1 (lower panel) after isolation of Sba1His6 protein complexes on Ni-NTA resin. Lanes 1 and 2, whole-cell extracts of cells grown in galactose or glucose; lanes 3 and 4, eluates after affinity chromatography on Ni-NTA resin.
In vertebrate cells, p23 binds to Hsp90 in a complex with immunophilins such as cyclophilin 40. S. cerevisiae contains cyclophilin homologs encoded by the CPR6 and CPR7 genes that also bind to Hsp90 (16, 52). We therefore analyzed whether one of these proteins (Cpr6) was present in the Sba1His6-Hsp90 complexes isolated by the affinity chromatography procedure described above. Cpr6 was identified in this experiment by using specific antisera (52), and as shown in Fig. 4C, it is highly enriched in the eluates containing Sba1His6 and Hsp90. We also observed a small amount of Sti1p to coisolate with Sba1His6 (Fig. 4D, lower panel), although the amount is very small compared with Cpr6, as judged by the relative band intensity of the eluted sample (Fig. 4D, lane 3) compared with the corresponding extract sample (Fig. 4D, lane 1).
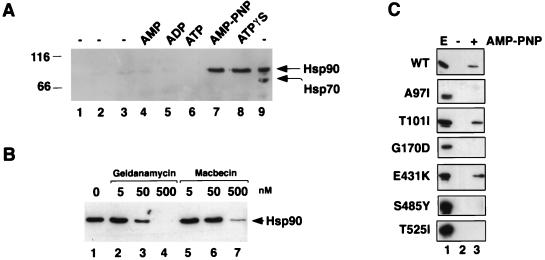
The formation of Sba1His6-Hsp90 complexes was further investigated by an in vitro assembly assay. Purified recombinant Sba1His6 was first bound to Ni-NTA resin and then incubated at 30°C with desalted whole-cell extracts from a wild-type yeast strain. Complex formation with Hsp90 was assayed after reisolation of the resin bound Sba1His6, several rounds of washing, and subsequent protein elution with a high concentration of imidazole. The presence of Hsp90 was determined after SDS-PAGE and Western blot analysis. As shown in Fig. 5, complex formation between Hsp90 and Sba1His6 was extremely weak in the absence of nucleotide or in the presence of AMP, ADP, or ATP. Complex formation between Hsp90 and Sba1His6 was readily observed, however, when the extracts were supplemented with adenyl-imidodiphosphate (AMP-PNP) or adenosine 5′-O-(3-thiotriphosphate) (ATPγS), suggesting that this interaction is strongest when ATP is unable to be hydrolyzed. The binding of Sba1His6 to Hsp90 is apparently specific since there was no interaction with Hsp70 under any of the conditions used, as revealed by reprobing the Western blot with antiserum to Hsp70 as shown in Fig. 5A. The interaction between Sba1His6 and Hsp90 can also occur with either the Hsc82 or the Hsp82 isoform, as judged by the formation of complexes in strains having deletions in either the HSC82 or HSP82 gene (data not shown). This interaction with both isoforms may account for the presence of a doublet at 86 kDa (4) after affinity isolation of Sba1His6 from wild-type yeast cell extracts after in vivo expression (Fig. 5A).
FIG. 5.
In vitro association of Sba1His6 with Hsp90. (A) Purified Sba1His6 was prebound to Ni-NTA resin and added to desalted whole-cell extracts from wild-type yeast cells and incubated at 30°C for 30 min. Hsp90 binding to Sba1His6 (lanes 1 to 8; lane 9 is whole-cell extract loaded directly onto the gel) was determined after reisolation of Sba1His6 and Western blotting using anti-Hsp90. Lane 1, Ni-NTA-Sba1His6 incubated in buffer; lane 2, Ni-NTA resin without prebound Sba1His6 incubated with extract; lane 3; Ni-NTA-Sba1His6 incubated with extract but without further addition; lane 4, Ni-NTA-Sba1His6 incubated with extract plus 5 mM AMP; lane 5, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ADP; lane 6, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ATP; lane 7, Ni-NTA-Sba1His6 incubated with extract plus 5 mM AMP-PNP; lane 8, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ATPγS; lane 9, 2.5 μg of desalted whole-cell extract. The blot was first probed with anti-Hsp90 and subsequently reprobed with anti-Hsp70. Molecular size markers are shown in kilodaltons at left. (B) Binding of Hsp90 to Sba1His6 is competed by benzoquinoid ansamycins. Ni-NTA-Sba1His6 resin was incubated with desalted extracts as described above plus 5 mM AMP-PNP in the presence of solvent (dimethyl sulfoxide; lane 1), geldanamycin (lanes 2 to 4), or macbecin (lanes 5 to 7) at the concentrations indicated. After 30 min at 30°C, the resin was reisolated and washed, and bound proteins were resolved by SDS-PAGE. Hsp90 was detected by Western blotting. (C) Binding of wild-type (wt) and different mutant forms of Hsp90 to Ni-NTA-Sba1His6 resin. Lane 1, whole-cell extract (E); lane 2, incubation of Ni-NTA-Sba1His6 resin with extracts in the absence of AMP-PNP; lane 3, incubation of Ni-NTA-Sba1His6 resin with extracts in the presence of AMP-PNP. The Western blots were probed with anti-Hsp90.
We also tested whether binding of Sba1His6 to Hsp90 was sensitive to benzoquinoid ansamycins, which have been shown to inhibit complex formation between p23 and Hsp90 (23, 32, 47). Similar inhibition occurs between yeast Hsp90 and Sba1. As shown in Fig. 5B, both geldanamycin and the related compound macbecin inhibit the association of purified Sba1 with Hsp90 in yeast extracts supplemented with AMP-PNP. The inhibition by macbecin is 5- to 10-fold less sensitive than was found for geldanamycin.
The in vitro binding assay was used to identify residues in Hsp90 that are critical to Sba1 binding. This experiment was performed by incubating purified Sba1His6 with extracts from yeast cells expressing different mutant forms of Hsp90 (specifically the Hsp82 isoform expressed from a constitutive promoter). Six different point mutants were used in this experiment (Fig. 5C), and in the presence of AMP-PNP, only two of these mutant forms of Hsp90 (T101I and E431K) were able to bind to Sba1His6. Of those that failed to bind Sba1His6, two have their mutations in the N-terminal nucleotide binding domain and two (S485Y and T525I) reside in the C-terminal half of Hsp90, suggesting that this region is also important for Sba1His6 binding.
These combined results demonstrate the similarity in structure and function between Sba1 and vertebrate p23 proteins. We next performed a functional analysis in yeast to determine whether Sba1 loss of function affected processes known to require the Hsp90 chaperone machine: the activity of the tyrosine kinase v-Src and steroid hormone receptors.
Role of Sba1 in v-Src maturation and steroid hormone receptor activation.
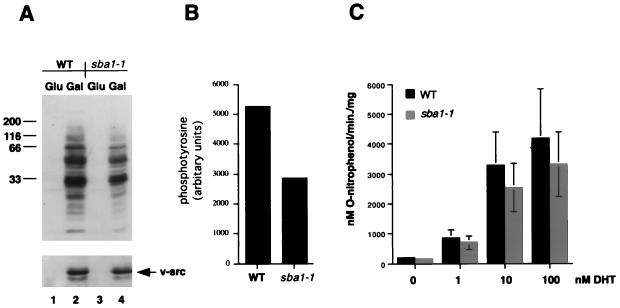
After heterologous expression in yeast, the tyrosine kinase v-Src is active in a manner that is dependent on Hsp90 and its cochaperones. In wild-type cells, expression of v-Src from a galactose-inducible promoter leads to greatly increased levels of phosphotyrosine (Fig. 6A) which result in a lethal phenotype (reference 19 and data not shown). Previous studies have shown that mutation or deletion of yeast genes encoding Hsp90 or other components of the Hsp90 chaperone machine resulted in decreased levels of v-Src activity, often suppressing the lethal phenotype accompanying it (9, 12, 13, 16, 34, 35, 55). Disruption of the SBA1 gene, however, did not suppress this lethal phenotype upon inducible expression of v-Src. Furthermore, v-Src gene induction from a GAL1 promoter led to similar accumulation of v-Src protein (Fig. 6A, lower panel), and the level of protein tyrosine phosphorylation, as assessed by Western blotting with antiphosphotyrosine, revealed significant v-Src activity in sba1-1 cells (Fig. 6A), although the levels were approximately twofold lower in this strain than in the wild type (Fig. 6B).
FIG. 6.
v-Src and AR activities in wild-type and sba1-1 mutant yeast cells. (A) Western blot analysis of phosphotyrosine activity (upper panel) and the level of v-Src protein (lower panel) in wild-type (WT; strain AFY200; lanes 1 and 2) or sba1-1 (strain AFY202; lanes 3 and 4) cells after growth in glucose (lanes 1 and 3) or galactose (lanes 2 and 4) for 6 h. Molecular size markers are shown in kilodaltons at the left. (B) Quantitation of the level of phosphotyrosine in wild-type and sba1-1 cells from the Western blot shown in panel A. (C) lacZ gene expression in wild-type (strain YF267) and sba1-1 cells (strain YF268) after addition of dihydrotestosterone (DHT) as indicated. Cells were assayed for β-galactosidase 2 h after addition of hormone. Data are means of three independent assays.
We also tested whether Sba1 was important for signaling by steroid hormone receptors. For these experiments, androgen receptors (AR) were assayed for hormone-dependent transactivation of a lacZ reporter gene in wild-type and sba1-1 strains. However, there was only a slight decrease in signaling in the mutant compared to the wild type (Fig. 6C), corresponding to a mean of 82% of wild-type activity in the sba1-1 mutant. Similar results were obtained by assaying GR activity in the same strains (data not shown). Thus, Sba1 loss of function did not significantly affect the activity of proteins known to require other components of the Hsp90 chaperone machine.
DISCUSSION
Molecular chaperones are thought to play a major role in the folding and assembly of newly synthesized polypeptide chains in both prokaryotes and eukaryotes. Hsp70 and Hsp90 are the most abundant molecular chaperones in the eukaryotic cytosol and are probably responsible for the majority of chaperoning events taking place in this compartment. These activities occur in association with various cochaperones that modulate Hsp70 or Hsp90 action and may also function as molecular chaperones themselves.
The Sba1 protein has properties similar to those of the vertebrate Hsp90 cochaperone, p23. These proteins display several regions of conserved sequence similarity, although the overall amino acid sequence identity between Sba1 and p23 is only 24% (Fig. 1). In addition, both p23 and Sba1 migrate more slowly in denaturing gels than would be predicted from their size, suggesting some shared structural feature (30).
Sba1 and p23 also have functional similarities. Both bind to Hsp90 in a manner that is stabilized by nonhydrolyzable ATP or by molybdate ions (32, 49). This nucleotide requirement is likely to represent the binding of ATP to Hsp90 itself, which has recently been shown to have a nucleotide binding pocket in its N-terminal domain (41). Indeed, two mutations in the HSP82 gene (A97I and G170D) that alter amino acids adjacent to those directly interacting with the nucleotide prevent binding of Sba1p to Hsp90 in vitro (Fig. 5C). Similar inhibition has also been observed between human Hsp90 and p23 when the G170 equivalent (G182 in human Hsp90) is mutated to aspartate (23). Residues at the C-terminal domain of Hsp90 also appear to be important for its interaction with Sba1, since no complex formation was observed with the S485Y or T525I mutant (Fig. 5C). A further similarity between vertebrate and yeast p23 proteins derives from the finding that Sba1 association with Hsp90 is inhibited by geldanamycin and by the related compound macbecin, which has been shown to affect the activity of p53 expressed in yeast (1). Geldanamycin binds in the nucleotide binding pocket of human Hsp90 (48) and also inhibits complex formation with p23 (23, 47, 49). The effect of N-terminal mutations in Hsp90 and the effect of geldanamycin or macbecin may therefore be related to nucleotide-dependent conformations that may allosterically affect the binding of Sba1 elsewhere in Hsp90. The most likely candidate region for Sba1 binding is therefore the C-terminal half of Hsp90, since two of three mutations in this region (S485Y and T525I) blocked complex formation.
The complex between p23 and Hsp90 in animal cells also contains one of several immunophilins, such as cyclophilin 40 (31). Similar interactions also occur in yeast, since the cyclophilin Cpr6 is highly enriched in eluates after affinity chromatography of Sba1His6. By contrast, the Sti1 cochaperone was present in these complexes in much smaller amounts. Previous biochemical studies have concluded that p60 and p23 reside independently of each other on different Hsp90 subcomplexes (30, 46) and that immunophilins and p60 compete with each other for the same binding site on Hsp90 (40). The relationship between p23 and p60/Sti1 may not be so discrete, however, since purified p23 can interact with the Hsp90-Hsp70-p60 complex, suggesting that these proteins may indeed function together, if only in a temporary manner (14, 32). Our results are consistent with there being a functional interaction between Sti1 and Sba1, based on the biochemical evidence that they coexist in the same complexes (albeit to a very small degree) in addition to the synthetic growth defects of the sba1/sti1 double mutant.
Together, these combined data indicate that Sba1 is a yeast homolog of vertebrate p23 proteins. SBA1 disruption did not result in any major growth phenotype, however, although the sba1-1 cells grew more slowly at both low and high temperatures compared to the wild type. These data contrast with the relative importance of Hsp90 for cell growth (4) and suggest that Sba1 is dispensable for general chaperone-mediated protein folding under normal conditions, although it may have a more specific role under nonoptimal growth conditions.
The vertebrate p23 protein has been characterized as a component of Hsp90 containing heterocomplexes with steroid hormone receptors and with v-Src. However, loss of Sba1 function resulted in only mild defects in steroid receptor signaling or v-Src activity. These data contrast with the relative importance of other yeast Hsp90 or Hsp70 cochaperones to both steroid receptor signaling and v-Src activity, such as Ydj1, Cdc37, Cpr7, and Sti1 (9, 12, 13, 16, 34, 35, 55). The results from the sba1-1 strain are especially surprising given the relative importance attributed to p23 for stable complex formation between Hsp90 and steroid hormone receptors (14, 31). However, the near-wild-type activity of AR and GR in the sba1-1 strain is consistent with the recent finding that while p23 is important to stabilize Hsp90-GR complexes in a high-affinity hormone binding state (14), it is not responsible for generating this conformation (15), through which receptor activation is likely to occur in yeast. In the absence of p23, the GR can attain the high-affinity hormone binding state via the action of Hsp90/Hsp70 and p60, although its ability to maintain it is severely compromised. But as observed by Dittmar and Pratt (15), heterocomplex instability does not preclude hormone binding if the ligand is incubated during heterocomplex assembly. Thus, Sba1 may be dispensable for AR and GR signaling because it stabilizes the activatable state rather than generates it. In this manner, Sba1 may contribute to the efficiency with which chaperone-mediated folding of steroid receptors and v-Src take place and hence the relatively mild reductions in activity resulting from its loss of function.
ACKNOWLEDGMENTS
We thank Jie Jin for help with protein purification, Alex Shorstein for help with SBA1 gene cloning, and Robert J. Donnelly (Molecular Resource Facility, New Jersey Medical School, UMDNJ) for DNA sequencing. We also thank F. Boschelli and E. Craig, Y. Kimura, S. Lindquist, D. Picard, D. Toft, and K. Yamamoto for the gifts of reagents, Sean Bohen for discussions and communication of results prior to publication, and Jeanne Hirsch for comments on the manuscript.
This work was supported by grants from the NIH (R01-DK49065) and the Sinsheimer Foundation to A.J.C.
REFERENCES
- 1.Blagosklonny M V, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 3.Bohen S P, Yamamoto K R. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 6.Caplan A J. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrine Metab. 1997;8:271–276. doi: 10.1016/s1043-2760(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Caplan A J, Langley E, Wilson E M, Vidal J. Hormone dependent transactivation by the human androgen receptor is regulated by a dnaJ protein. J Biol Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- 8.Chang H-C J, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 9.Chang H-C J, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham M E, Caplan A J. Structure, function and evolution of dnaJ: conservation and adaptation of chaperone function. Cell Stress Chap. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Prapapanich V, Rimerman R A, Honoré B, Smith D F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins Hsp90 and Hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 12.Dey B, Caplan A J, Boschelli F. The YDJ1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol Biol Cell. 1996;7:91–100. doi: 10.1091/mbc.7.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey B, Lightbody J J, Boschelli F. CDC37 is required for p60v-src activity in yeast. Mol Biol Cell. 1996;7:1405–1417. doi: 10.1091/mbc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 15.Dittmar K D, Pratt W B. Folding of the glucocorticoid receptor by the reconstituted hsp90-based chaperone machinery. J Biol Chem. 1997;272:13047–13054. doi: 10.1074/jbc.272.20.13047. [DOI] [PubMed] [Google Scholar]
- 16.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 17.Fairhead C, Dujon B. Transcript map of two regions from chromosome XI of Saccharomyces cerevisiae for interpretation of systematic sequencing results. Yeast. 1994;10:1403–1413. doi: 10.1002/yea.320101103. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y, Fliss A E, Robins D M, Caplan A J. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271:28697–28702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- 19.Florio M, Wilson L K, Trager J B, Thorner J, Martin G S. Aberrant protein phosphorylation at tyrosine is responsible for the growth-inhibitory action of pp60v-src expressed in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:283–296. doi: 10.1091/mbc.5.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman B C, Toft D O, Morimoto R I. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 21.Frydman J, Höhfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- 22.Geitz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 23.Grenert J P, Sullivan W P, Fadden P, Haystead T A J, Clark J, Mimnaugh E, Krutzsch H, Ochel H-J, Schulte T W, Sausville E, Neckers L M, Toft D O. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 24.Gross M, Hessefort S. Purification and characterization of a 66-kDa protein from rabbit reticulocyte lysate which promotes the recycling of Hsp70. J Biol Chem. 1996;271:16833–16841. doi: 10.1074/jbc.271.28.16833. [DOI] [PubMed] [Google Scholar]
- 25.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 26.Höhfeld J, Jentsch S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacquier A, Legrain P, Dujon B. Sequence of a 10.7 kb segment of yeast chromosome XI identifies the APN1 and the BAF1 loci and reveals one tRNA gene and several new open reading frames including homologs to RAD2 and kinases. Yeast. 1992;8:121–132. doi: 10.1002/yea.320080207. [DOI] [PubMed] [Google Scholar]
- 29.Johnson J L, Craig E A. Protein folding in vivo: unraveling complex pathways. Cell. 1997;90:201–204. doi: 10.1016/s0092-8674(00)80327-x. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J L, Beito T G, Krco C J, Toft D O. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson J L, Toft D O. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269:24989–24993. [PubMed] [Google Scholar]
- 32.Johnson J L, Toft D O. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- 33.Kimura Y, Matsumoto S, Yahara I. Temperature-sensitive mutants of hsp82 of the budding yeast Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:517–527. doi: 10.1007/BF00285275. [DOI] [PubMed] [Google Scholar]
- 34.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90 mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 35.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 36.Lässle M, Blatch G L, Kundra V, Takatori T, Zetter B R. Stress inducible, murine protein mSTI1. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 37.Nair S C, Toran E J, Rimerman R A, Hjermstad S, Smithgall T E, Smith D F. A pathway of multi-chaperone interactions common to diverse regulatory proteins; estrogen receptor, fes tyrosine kinase, heat shock transcription factor HSF1 and the arylhydrocarbon receptor. Cell Stress Chap. 1996;1:237–249. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nathan D F, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid hormone receptor and protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owens-Grillo J K, Czar M J, Hutchison K A, Hoffmann K, Perdew G H, Pratt W B. A model of protein targeting mediated by immunophilins and other proteins that bind to hsp90 via tetratricopeptide repeat domains. J Biol Chem. 1996;271:13468–13475. doi: 10.1074/jbc.271.23.13468. [DOI] [PubMed] [Google Scholar]
- 41.Prodromou C, Roe S M, O’Brien R, Ladbury J E, Piper P W, Pearl L H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 42.Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 43.Schulte T W, Blagosklonny M V, Ingui C, Neckers L. Disruption of the raf-1-hsp90 molecular complex results in destabilization of raf-1 and loss of raf-1-ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 44.Segnitz B, Gehring U. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J Biol Chem. 1997;272:18694–18701. doi: 10.1074/jbc.272.30.18694. [DOI] [PubMed] [Google Scholar]
- 45.Smith D F, Faber L E, Toft D O. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J Biol Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 46.Smith D F, Sullivan W P, Marion T N, Zaitsu K, Madden B, McCormick D J, Toft D O. Identification of a 60-kilodalton stress-related protein, p60, which interacts with Hsp90 and Hsp70. Mol Cell Biol. 1993;13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith D F, Whitesell L, Nair S C, Chen S, Prapapanich V, Rimmerman R A. Progesterone receptor structure and function altered by geldanamycin, an Hsp90-binding agent. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stebbins C E, Russo A A, Schneider C, Rosen N, Hartl F U, Pavletich N P. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri E S, Litwack G, Toft D O. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 50.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R I, Reed J C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 52.Warth R, Briand P-A, Picard D. Functional analysis of the yeast 40 kDa cyclophilin Cyp40 homologue and its role for viability and steroid receptor regulation. Biol Chem. 1997;387:381–391. doi: 10.1515/bchm.1997.378.5.381. [DOI] [PubMed] [Google Scholar]
- 53.Whitesell L, Cook P. Stable and specific binding or heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 54.Whitesell L, Mimnaugh E G, De Costa B, Myers C E, Neckers L M. Inhibition of heat shock protein Hsp90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Lindquist S L. Heat-shock protein hsp90 governs the activity of pp60v-src. Proc Natl Acad Sci USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Z, Pal J K, Thulasiraman V, Hahn H P, Chen J J, Matts R L. The role of the 90-kDa heat-shock protein and its associated cohorts in stabilizing the heme-regulated eIF-2alpha kinase in reticulocyte lysates during heat stress. Eur J Biochem. 1997;246:461–470. doi: 10.1111/j.1432-1033.1997.t01-1-00461.x. [DOI] [PubMed] [Google Scholar]