Abstract
Mutation of the p53 tumor suppressor gene is the most common genetic alteration in human cancer, and tumors that express mutant p53 may be more aggressive and have a worse prognosis than p53-null cancers. Mutant p53 enhances tumorigenicity in the absence of a transdominant negative mechanism, and this tumor-promoting activity correlates with its ability to transactivate reporter genes in transient transfection assays. However, the mechanism by which mutant p53 functions in transactivation and its endogenous cellular targets that promote tumorigenicity are unknown. Here we report that (i) mutant p53 can regulate the expression of the endogenous c-myc gene and is a potent activator of the c-myc promoter; (ii) the region of mutant p53 responsiveness in the c-myc gene has been mapped to the 3′ end of exon 1; (iii) the mutant p53 response region is position and orientation dependent and therefore does not function as an enhancer; and (iv) transactivation by mutant p53 requires the C terminus, which is not essential for wild-type p53 transactivation. These data suggest that it may be possible to selectively inhibit mutant p53 gain of function and consequently reduce the tumorigenic potential of cancer cells. A possible mechanism for transactivation of the c-myc gene by mutant p53 is proposed.
Wild-type p53 plays a key role in tumor suppression by monitoring DNA damage and executing pathways that negatively control cell growth, either by blocking cells in the G1 phase of the cell cycle or by inducing apoptosis (for a review, see reference 51). Wild-type p53 regulates these processes, at least in part, by functioning as a transactivator of gene expression. Wild-type p53 binds DNA in a sequence-specific fashion and interacts with various components of the transcription complex (22, 43, 48) to activate the expression of target genes associated with either growth arrest (Waf1 and GADD45) or cell death (Bax) (8, 18, 26).
Tumor suppressors are formally defined by a loss of function involved in blocking tumor progression. Consistent with this definition, naturally occurring mutants of p53 are generally defective in sequence-specific DNA binding and consequently do not induce the appropriate target genes, cause cell cycle arrest, or mediate cell death (51). However, in contrast to a classical tumor suppressor, mutation of the p53 gene leads not only to a loss of function but also to a gain of function that promotes the tumorigenicity of various p53-null cell types. Overexpression of mutant p53 in pre-B cells (37, 38, 44), fibroblasts (6), and osteosarcomas (42) dramatically enhances the tumorigenicity of these cells independent of a transdominant negative mechanism. In addition, stable expression of naturally occurring mutant p53 alleles in human T-cell acute lymphoblastic leukemia cells increases tissue invasiveness and enhances tumor formation (15).
The gain of function for mutant p53 is also supported by the findings that mutant p53 selectively transactivates several viral and cellular promoters, including the human MDR1 promoter, that are not regulated or are repressed by wild-type p53 (3, 5, 6, 12, 21). Importantly, enhancement of tumorigenicity by mutant p53 correlates with its ability to localize to the nucleus (37) and to transactivate the MDR1 promoter (21), as point mutation in the acidic domain of mutant p53 abolishes its transactivation and tumor-promoting functions.
The high frequency of p53 mutations in human cancer and the data indicating that mutant p53 promotes tumorigenicity warrant a detailed analysis of the molecular mechanism of the gain of function of mutant p53 and the cellular targets that may be regulated by this activity. The studies presented here demonstrate that mutant p53 regulates endogenous c-myc gene expression and promoter activity and provide insight into the mechanism of mutant p53 transactivation. These results lead to the novel hypotheses that mutant p53 transactivates the c-myc gene through a single-strand DNA or RNA intermediate and that c-myc is an important target in mutant p53-mediated tumorigenicity.
MATERIALS AND METHODS
Cell lines and DNA.
The culture conditions for the various cell lines used have been described previously (49, 50). The (10)1 murine fibroblast cell line (13) and SaOs-2 human osteosarcoma cells (24) contain deletions in the p53 gene and do not express endogenous p53. LAPCx3Ras cells are transformed rat embryo fibroblasts that express high levels of human mutant p53-143A under the control of the lac activator protein (LAP) transactivation system (49). Treatment of these cells with isopropyl-β-d-thiogalactopyranoside (IPTG) inhibits the LAP transactivator and down-regulates mutant p53 gene expression. Cx3Ras transformed rat embryo fibroblast cells constitutively express high levels of p53-143A under the regulation of a cytomegalovirus (CMV) promoter, which is unaffected by IPTG (49). Cx3Ras-LAP cells were derived from the Cx3Ras cell line and express a regulated LAP transactivator. Treatment of Cx3Ras-LAP cells with IPTG efficiently represses LAP activity but does not alter expression of mutant p53.
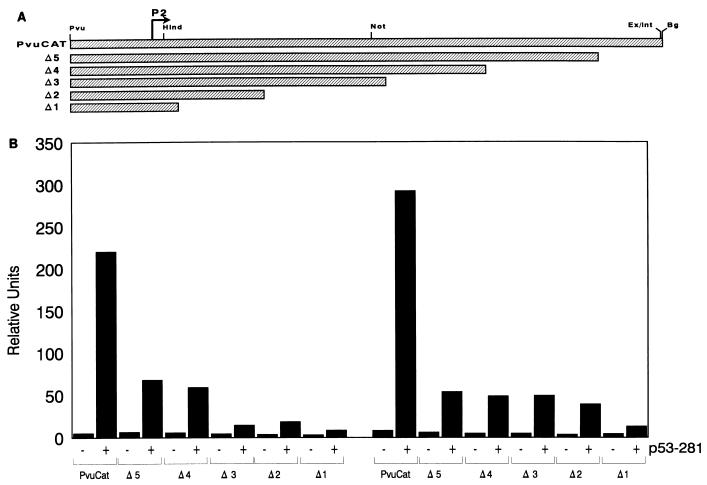
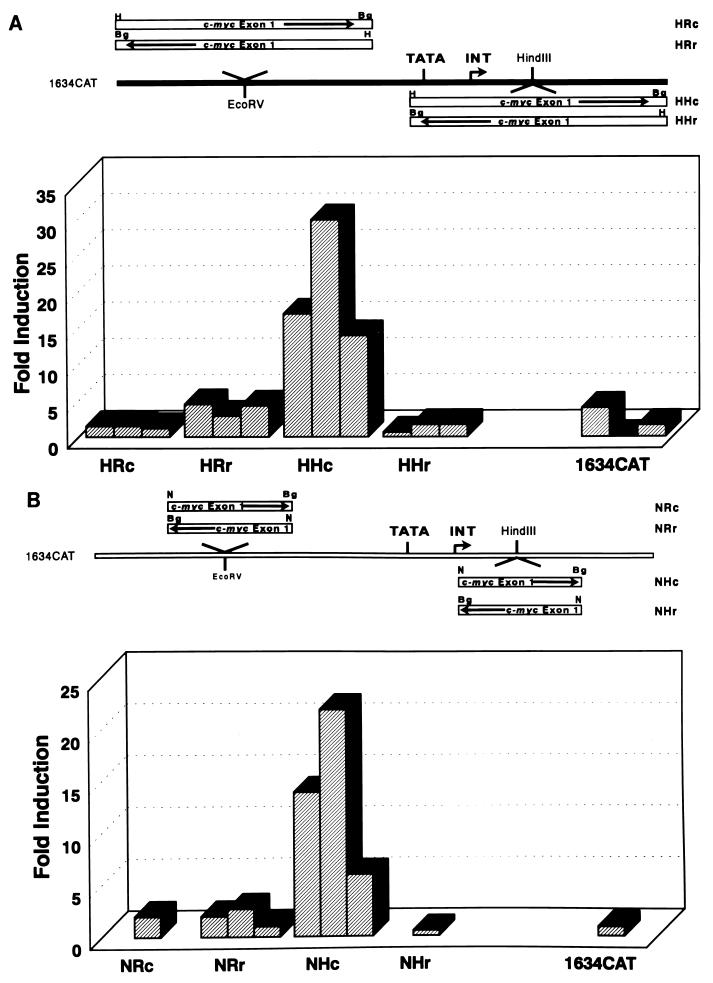
The human p53 expression vectors are regulated by the CMV promoter and have been previously described (14). Wild-type and mutant p53-281G carboxy-terminal truncation mutants were generated by inserting three tandem translation stop codons (TGATAGTAA) at various selected positions by using PCR methodology. The mutations were confirmed by DNA sequence analysis. The murine p53 miniprotein expression plasmids, pCMVDD (DD1) and pCMVDΔSS (deltaSS), have been previously described (35). The murine c-myc promoter–chloramphenicol acetyltransferase (CAT) reporter construct (BgCAT) extends from approximately 1 kb upstream from the P1 promoter to 413 bp downstream from the P2 promoter and was described previously (7). The PvuCAT c-myc promoter-CAT reporter, which deletes the P1 promoter, extends from −141 to +413 (in relation to the P2 promoter) and was generated in this study by subcloning the PvuI-to-BglII fragment from the c-myc promoter into the SmaI site of the p22 vector (7). The 3′ c-myc promoter deletion series was derived from BgCAT and PvuCAT by using PCR methodology, and the resulting reporters terminate at the following positions: +358 (Δ5), +271 (Δ4), +189 (Δ3), +79 (Δ2), and +7 (Δ1) (see Fig. 5). The heterologous minimal promoter-CAT reporter, p1634CAT, contains the adenovirus major late promoter and the terminal deoxynucleotidyltransferase (tdt) initiator element (34, 50). The HindIII-BglII (H) and NcoI-BglII (N) fragments from BgCAT (exon 1 of c-myc) were separately ligated into the HindIII site of p1634CAT, which is downstream (H) of the tdt element, in either the correct (c; as found in exon 1) or reverse (r) orientation, resulting in the constructs HHc, NHc, HHr, and NHr (see Fig. 6). We similarly cloned the H and N fragments into the EcoRV site upstream (R) of the TATA box, yielding HRc, NRc, HRr, and NRr.
FIG. 5.
Mapping of the mutant p53 response region in c-myc to the 3′ end of exon 1. PvuCAT c-myc promoter reporter deletion constructs were transfected into (10)1 cells with and without mutant p53-281, and promoter activity was measured by CAT assay as described in Materials and Methods. (A) Schematic diagram of the c-myc–CAT reporter constructs. The PvuCAT reporter spans −140 to +413 and contains the P2 promoter, nontranslated exon 1, and the exon 1/intron 1 (Ex/Int) junction. The c-myc promoter 3′ deletion constructs span the regions from −140 to +358 (Δ5), +271 (Δ4), +189 (Δ3), +79 (Δ2), and +7 (Δ1). (B) Basal and p53-281-induced promoter activities are presented from two independent experiments that were each conducted in duplicate. Similar results were also obtained when the BgCAT and derivative 3′ deletion promoter-reporter constructs were used (data not shown).
FIG. 6.
The mutant p53 response region in c-myc is position and orientation dependent. The c-myc-heterologous minimal promoter reporters were constructed as described in Materials and Methods and are schematically diagrammed. The heterologous reporters were transfected into (10)1 cells with and without mutant p53-281, and transactivation was measured by CAT assays as described in Materials and Methods. Results are presented as the fold induction of reporter activity with mutant p53-281 compared to basal expression of the reporter only. The basal promoter activities of p1634CAT and each of the heterologous promoters are approximately equal but close to background. c-myc exon 1 sequences spanning −22 to +413 (H fragment) (A) and c-myc exon 1 sequences from +174 to +413 (N fragment) (B) cloned into the p1634CAT minimal heterologous promoter reporter. Results of multiple independent experiments are presented. INT, tdt initiator element.
CAT assays.
Cells were transfected overnight by calcium phosphate precipitation and grown at 37°C for an additional 24 to 48 h as previously described (50). The cells were lysed by freeze-thawing, and the protein concentration was quantitated by using a modified Bradford assay (Bio-Rad Laboratories). Normalized amounts of protein were analyzed for CAT activity, and results were quantitated by using Molecular Dynamics PhosphorImager analysis and ImageQuant software.
Northern blot analysis.
To ensure exponential growth, the various cell lines were replated daily 2 days prior to the experiment. The cells were then grown at various densities in Dulbecco modified Eagle medium containing 10% fetal bovine serum, glutamine (2 mM), penicillin (100 IU/ml), and streptomycin (100 μg/ml) with or without 5 mM IPTG. The cells were harvested at various time points, and total RNA was purified as previously described (4). RNA (10 μg) was separated on 1% agarose–6% formaldehyde gels by electrophoresis and transferred to nylon membranes. The filters were probed with 32P-labeled c-myc cDNA and analyzed by autoradiography. The membranes were stripped and reprobed with radiolabeled histone H4, proliferating cell nuclear antigen (PCNA), and β-actin cDNAs.
RNase protection analysis.
Total RNA from transiently transfected cells was evaluated by RNase protection analysis by using an RPA II kit (Ambion) according to the manufacturer’s recommendations. The 32P-labeled c-myc-CAT RNase protection probe is 1,058 nucleotides (nt) and includes sequences from the EcoRI site in the CAT coding sequences to the ScaI site in c-myc, which is upstream of the P1 TATA box. The probe was hybridized with 20 μg of RNA in annealing buffer for 16 to 20 h at 43°C. The samples were digested with 0.25 U of RNase A and 10 U of RNase T1 for 30 min at 37°C, separated on a 5% acrylamide–8 M urea gel, and visualized by autoradiography.
Western blot analysis.
Transfected cells were lysed in radioimmunoprecipitation assay buffer, and equal amounts of protein were analyzed on denaturing 12% polyacrylamide gels as previously described (27). The proteins were transferred to nylon membranes and probed with either sheep polyclonal antibody Ab-7 (panspecific for p53) or with murine monoclonal antibody DO-1 (specific for human p53) (Oncogene Science). The blot was probed with either rabbit anti-sheep or donkey anti-mouse immunoglobulin G-horseradish peroxidase conjugate (Pierce) and developed by using an enhanced chemiluminescence detection kit (Amersham).
RESULTS
Mutant p53 regulates expression of the endogenous c-myc gene.
To study the mechanism by which mutant p53 regulates gene expression and induces tumorigenicity, we used LAPCx3Ras cells, which conditionally express the tumor-derived human mutant p53-143A gene under tight regulation by the LAP transactivator (49). In these cells, mutant p53 gene expression is high during normal growth conditions and is efficiently inhibited after addition of IPTG, which inactivates the LAP transactivator. As controls, we also examined gene expression in Cx3Ras-LAP and Cx3Ras cells, which express high constitutive levels of the human mutant p53-143A from a CMV promoter, with and without the regulated LAP transactivator, respectively.
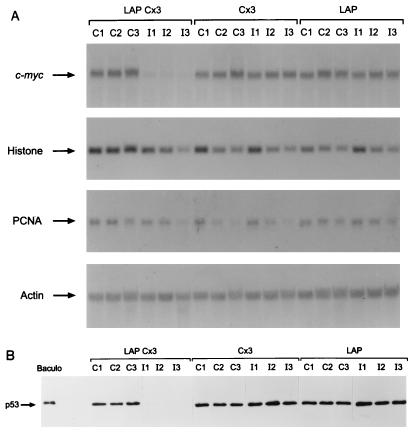
Inhibition of mutant p53 gene expression in the LAPCx3Ras cell line with IPTG leads to a dramatic decrease in steady-state levels of c-myc mRNA (Fig. 1A). Loss of c-myc gene expression in these cells was specific, as the levels of histone H4, PCNA, and β-actin were relatively unaffected compared to those in IPTG-treated Cx3Ras-Lap and Cx3Ras cells (Fig. 1A). These results suggest that the selective decrease in c-myc gene expression is not due to general transrepression by endogenous wild-type p53, as the PCNA and β-actin promoters have been previously reported to be suppressed by wild-type p53 (10, 32, 41). Furthermore, the loss of c-myc gene expression was coincident with the loss of mutant p53 gene expression, as determined by Western blot analysis (Fig. 1B). In contrast, c-myc and mutant p53 gene expression remained elevated in the IPTG-treated Cx3Ras and Cx3Ras-LAP cells (Fig. 1). Therefore, the expression of c-myc in these cells was dependent on sustained expression of mutant p53 and was not due to inhibition of the LAP transactivator per se or to the drug treatment.
FIG. 1.
Mutant p53 regulates c-myc gene expression. (A) Northern blot analysis of total RNA (10 μg/sample) isolated from LAPCx3Ras (LAP Cx3), Cx3Ras (Cx3), and Cx3Ras-LAP (LAP) cells on days 1 to 3 from control (C1 to C3) and IPTG-treated (I1 to I3) samples. Northern filters were sequentially probed for expression of c-myc, histone H4, PCNA, and β-actin. (B) Western blot analysis of mutant p53 expression, using total-cell extracts (5 μg/sample) prepared on days 1 to 3 from control (C1 to C3) and IPTG-treated (I1 to I3) cell lines. Samples were analyzed by using murine monoclonal antibody DO-1 as described in Materials and Methods. The experiments were repeated at least three times, and representative data are presented. Baculo, baculovirus.
Mutant p53 transactivates the c-myc promoter.
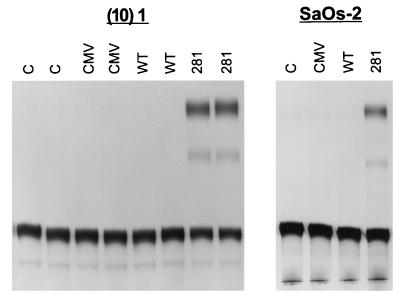
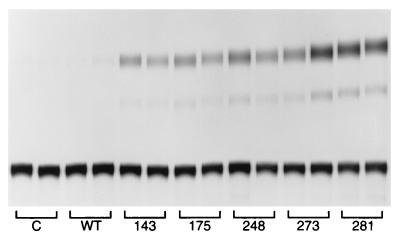
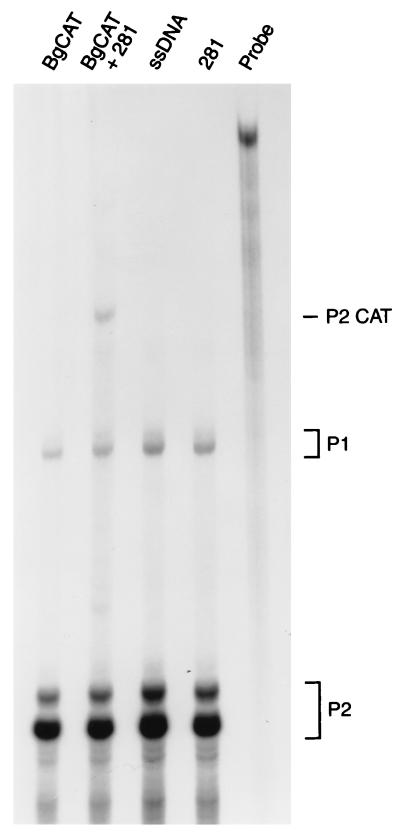
To determine whether mutant p53 regulation of c-myc gene expression was transcriptional, we examined the effects of mutant p53 on the activity of various c-myc promoter–CAT reporter constructs. The BgCAT reporter extends approximately 1 kb upstream from the murine c-myc P1 promoter to 413 bp downstream of the P2 transcription start site (7). The reporter was transiently transfected with and without human mutant p53-281 (isolated from a human colon carcinoma) into murine (10)1 fibroblasts, which are null for endogenous p53 (13). As shown in Fig. 2, mutant p53-281 dramatically induced activity of the BgCAT reporter 40- to 50-fold, whereas wild-type p53 had no effect or repressed activity. No induction was evident when BgCAT was cotransfected with the CMV vector, which does not express p53. Transactivation of the c-myc promoter by mutant p53 in p53-null cells demonstrates that this activity is due to a gain of function and not to a transdominant negative mechanism. Induction of murine c-myc promoter activity by mutant p53-281 was also observed in the human SaOs-2 osteosarcoma cell line, which does not express endogenous p53 protein (Fig. 2). Each of the naturally occurring hot-spot mutant p53 alleles transactivated the c-myc promoter to various degrees (Fig. 3), as previously reported for the MDR1 promoter (6). The human c-myc promoter was also transactivated by mutant p53 in the (10)1 fibroblasts (data not shown). The increase in BgCAT expression due to mutant p53 in (10)1 cells is associated with an increase in steady-state levels of P2-derived c-myc–CAT transcripts, as determined by RNase protection analysis (Fig. 4). Therefore, mutant p53 transactivates the c-myc promoter independently of a transdominant mechanism, and this activity is not species specific for cell type or promoter.
FIG. 2.
Mutant p53 transactivates the c-myc promoter. (10)1 murine fibroblasts and SaOs-2 human osteosarcoma cells were transfected with 0.25 μg of BgCAT alone (C) or cotransfected with BgCAT and 1 μg of the p53 empty-vector plasmid (CMV), wild-type p53 (WT), or mutant p53-281 (281) and assayed for CAT activity as described in Materials and Methods. Transactivation function is demonstrated by conversion of unacetylated [14C]chloramphenicol (lower band) to acetylated forms (upper bands). The experiments were repeated at least three times, and representative data are presented.
FIG. 3.
Panel of tumor-derived human mutant p53 alleles transactivate the c-myc promoter. SaOs-2 cells were transfected with the BgCAT reporter alone (C) and with the indicated human wild-type (WT) and mutant p53 expression vectors and then assayed for CAT activity as described in Materials and Methods. Samples were transfected in duplicate, and the experiment was repeated at least three times; representative data are presented. Similar results were obtained when the human MDR1 promoter reporter construct was used.
FIG. 4.
Mutant p53 increases steady-state levels of c-myc–CAT mRNA. (10)1 cells were transiently transfected with salmon sperm DNA (ssDNA), the c-myc reporter (BgCAT), mutant p53 (281), or BgCAT and p53-281 (BgCAT + 281) as described for Fig. 2. Two days after transfection, total RNA was isolated and analyzed for c-myc expression by RNase protection assays as described in Materials and Methods. Protected fragments (expected sizes) are as follows: undigested full-length probe (1,058 nt), endogenous c-myc P1 (573 nt) and P2 (413 nt), P2CAT (639 nt), and P1CAT (799 nt). As expected (40), endogenous c-myc mRNA derived from the predominant P2 promoter (accounting for ∼90% of c-myc transcripts) and the minor P1 promoter is evident in all samples. Unspliced P1 and P2 c-myc mRNA transcripts run approximately 10 nt slower than the spliced form. Transactivation of the c-myc promoter reporter by mutant p53 is demonstrated by the appearance of c-myc–CAT mRNA in the BgCAT–p53-281 cotransfected samples but not in the ssDNA-, BgCAT-, or p53-281-only samples. The c-myc-CAT mRNA is derived from the P2 promoter, based on multiple RNase protection assays using various radiolabeled probes and RNA size standards.
The mutant p53 response sequences in c-myc are position and orientation dependent.
To define the mutant p53 response element in the murine c-myc promoter, we tested a comprehensive series of 5′ and 3′ deletions of the BgCAT reporter. Large deletions of the 5′ region of c-myc, including the P1 promoter, had little effect on the ability of mutant p53 to activate expression (data not shown). By contrast, deletion of as little as 55 bp from the 3′ end of BgCAT (data not shown) or PvuCAT (Fig. 5) decreased responsiveness to mutant p53 by approximately fourfold, and further deletion of the exon sequences resulted in additional loss of responsiveness. These results suggest that the mutant p53 response element is localized near the exon 1/intron 1 junction and may cooperate with other sequences that reside closer to the transcription start site.
To determine whether mutant p53 responsiveness could be transferred to a heterologous promoter, exon 1 sequences downstream from the c-myc P2 promoter were cloned into a minimal promoter-CAT reporter (p1634CAT). Exon 1 sequences (−22 to +413; H fragment) were cloned upstream and downstream from the minimal promoter and in the correct and reverse orientations (HRc, HRr, HHc, and HHr, respectively). Exon 1 sequences spanning +174 to +413 (N fragment) were also cloned into the minimal promoter at each site and in both orientations. Cotransfection of these heterologous reporters with mutant p53-281 into murine fibroblasts demonstrated that c-myc exon 1 sequences conferred mutant p53 responsiveness in an orientation- and position-dependent manner, as only the HHc and NHc reporters were transactivated by mutant p53 (Fig. 6). Therefore, the mutant p53 response element does not act as a classic enhancer, suggesting that these sequences may need to be transcribed into RNA for mutant p53 to activate c-myc gene expression.
Transactivation by mutant p53 requires the amino and carboxy termini.
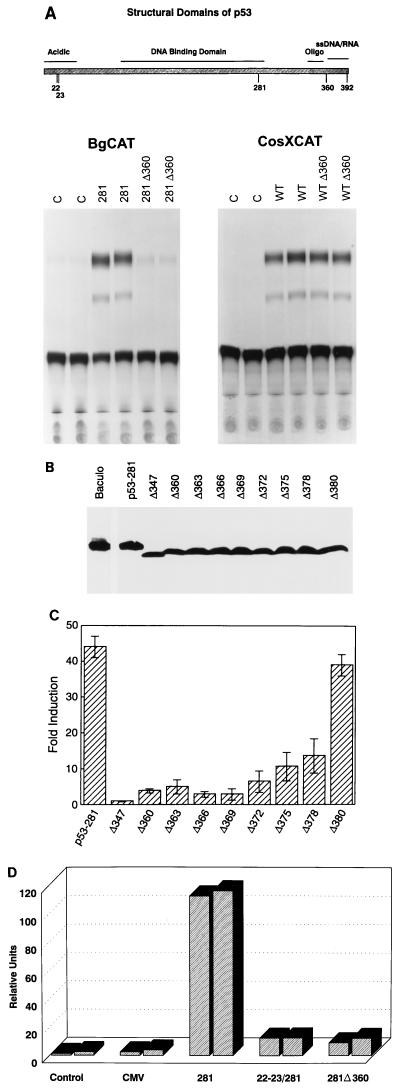
Data from the heterologous reporters suggested that mutant p53 may transactivate the c-myc promoter through an RNA-dependent mechanism, and previous studies have demonstrated that the C terminus of p53 expresses an RNA binding activity (28, 30, 33, 47). To test the role of the C terminus in p53 transactivation, we inserted three tandem in-frame translation stop codons into mutant p53-281 and wild-type p53 at the position corresponding to amino acid 360 (281Δ360 and WTΔ360, respectively). The p53Δ360 proteins are competent for dimerization, according to the crystal structure (16) and our biochemical assays (data not shown). As shown in Fig. 7A, p53-281Δ360 was markedly impaired for transactivation of the c-myc promoter compared to full-length p53-281. In contrast, WTp53Δ360 was functionally comparable to full-length wild-type p53 in transactivation of the wild-type p53-responsive CosXCAT reporter (46) (Fig. 7A). These data demonstrate that the C terminus of p53 is dispensable for wild-type p53 transactivation function but is essential for mutant p53 activity.
FIG. 7.
The N and C termini are required for mutant p53 transactivation. (A) Schematic diagram of p53 highlights the domains for transactivation (acidic), sequence-specific DNA binding, oligomerization (oligo), single-strand DNA/RNA binding, and amino acid 392, which is covalently associated with 5.8S RNA. Point mutations in the acidic domain at amino acids 22 (L→Q), 23 (W→S), and 392 (S→A), as well as the C-terminal truncations, were constructed in wild-type and mutant p53-281 as described in Materials and Methods. Lower left, mutant p53-281 transactivation function requires the C terminus. (10)1 cells were cotransfected with BgCAT and either CMV empty vector (C), mutant p53-281, or mutant p53-281Δ360. Lower right, the C terminus is dispensable for wild-type p53 transactivation function. (10)1 cells were cotransfected with CosXCAT, which is a wild-type p53 response reporter, and either CMV empty vector (C), wild-type p53 (WT), or p53Δ360. (B and C) Amino acids 372 to 380 are essential for mutant p53-281 transactivation function. (10)1 cells were transfected with BgCAT alone and cotransfected with BgCAT and either the p53-281 or p53-281 truncation mutant. (B) Western blot analysis of full-length mutant p53-281 and truncated mutant p53 in cell extracts analyzed for CAT activity in panel C. Equal amounts of protein were analyzed by using polyclonal antibody Ab-7 as described in Materials and Methods. Baculo, purified wild-type p53 protein. Full-length mutant p53-281 and truncated proteins are as indicated. (C) BgCAT reporter activity in response to mutant p53. Data are presented as fold induction of reporter activity with mutant p53 compared to basal promoter expression. (D) Mutant p53 transactivation function requires the N terminus. (10)1 cells were cotransfected with BgCAT and either CMV empty vector, p53-281, triple mutant 22-23/281, or 281Δ360. Samples were transfected in duplicate, and data are presented as fold induction of reporter activity with mutant p53 compared to basal expression. These experiments were repeated at least three times, and representative data are presented.
To delineate the sequences of p53-281 that are required for transactivation of c-myc, we prepared additional truncation mutants that terminate at amino acids 347 (Δ347), 380 (Δ380), and 390 (Δ390). In transient transfection studies, p53-281Δ347 and 281Δ360 failed to induce BgCAT reporter activity whereas 281Δ380 and 281Δ390 induced c-myc promoter activity to the same degree as full-length mutant p53-281 (data not shown). These results identify a region between amino acids 360 and 380 that is essential for mutant p53 transactivation. We then constructed a comprehensive series of p53-281 C-terminal truncations that progressively delete sequences spanning amino acids Δ380 to Δ360. Analysis of these mutants in transient transfection assays demonstrated that deletion of as little as two amino acids (379 and 380) decreases mutant p53 transactivation function three- to sixfold (Fig. 7B and C; compare Δ378 to Δ380). Further deletion of sequences from amino acids 380 to 369 decreased activity nearly to background levels. Western blot analysis demonstrated that the marked difference in transactivation by these mutants was a consequence of function and not due to differences in protein expression (Fig. 7B and C). These data clearly identify a C-terminal region between amino acids 370 and 380 that is essential for mutant p53 transactivation of the c-myc promoter.
Previous studies demonstrated that mutation of p53-281 at amino acids 22 and 23, which disrupts its interaction with transcription-associated factors (TAFs) (20, 22, 43), blocked its ability to transactivate the MDR1 promoter (21). Similarly, the p53-22-23/281 triple mutant is defective in transactivation of the murine c-myc promoter and is functionally impaired to the same extent as p53-281Δ360 (Fig. 7D). Therefore, the N- and C-terminal domains of mutant p53 are necessary for transactivation of the c-myc promoter.
Inhibition of mutant p53 gain of function through a transdominant negative mechanism.
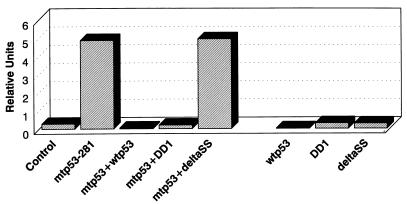
In human cancers, mutation of the p53 gene is almost always reduced to homozygosity. One interpretation of this phenomenon is that the wild-type allele must be inactivated to eliminate residual tumor suppressor activity, despite the ability of the mutant p53 protein to inhibit wild-type p53 functions through a transdominant negative mechanism (14). Alternatively, we propose that wild-type p53 may serve as a dominant negative regulator of mutant p53 gain of function, and that the wild-type allele must be deleted to exert the full tumor-promoting function of mutant p53. To address this issue, we examined the effects of wild-type p53 and derivative proteins on mutant p53-mediated transactivation of the c-myc (BgCAT) promoter (Fig. 8). Consistent with our hypothesis, inclusion of wild-type p53 completely blocked mutant p53 transactivation function (>99% inhibition) in p53-null cells. However, wild-type p53 can indirectly transrepress promoters lacking p53 binding sites (such as c-myc) in transient transfection assays possibly by titrating TATA binding protein (TBP), TAFs, or CBP/p300 (11, 19, 20, 22, 34, 43). To circumvent this limitation, we also tested the effects of murine p53 miniproteins (35), which do not repress c-myc reporter activity under the conditions used in these studies, on mutant p53 transactivation function (Fig. 8). The DD1 miniprotein essentially contains only the C terminus of p53 (amino acids 1 to 14 fused to 302 to 390) and is competent for dimerization with full-length p53 protein (35). By contrast, the deltaSS miniprotein is identical to DD1 but lacks amino acids 330 to 344, which span the oligomerization domain, and is therefore defective in dimerization (35). As shown in Fig. 8, DD1 blocked mutant p53-281 transactivation of the c-myc promoter (>95% inhibition), whereas deltaSS had little or no effect. Similarly, interference with mutant p53-281 transactivation function has also been demonstrated in assays using wild-type p53, the p53 miniproteins, and the MDR1 promoter reporter (data not shown). These results therefore indicate that mutant p53 gain of function can be repressed in a transdominant negative manner through dimerization with inactive forms of p53 and are consistent with our p53 truncation analyses demonstrating that the C terminus is required for transactivation.
FIG. 8.
Inhibition of mutant p53 gain of function through a transdominant negative mechanism. (10)1 cells were transiently cotransfected with the BgCAT reporter (0.5 μg) in combination with CMV vector only (Control; 2 μg), mutant p53-281 (mtp53-281; 2 μg), wild-type p53 (wtp53; 5 μg), DD1 (5 μg), and deltaSS (5 μg) and analyzed for CAT activity as described in Materials and Methods. The experiment was repeated at least three independent times, and representative data are presented. Similar results were obtained when the MDR1-CAT reporter was used.
DISCUSSION
The results presented here demonstrate that tumor-derived missense mutants of p53 can regulate expression of the c-myc gene. The well-established role of c-myc (40) as a proto-oncogene capable of promoting cell cycle progression and tumorigenesis makes this gene an attractive target for p53 gain-of-function mutants. Furthermore, our analyses demonstrate that the efficient activation of the c-myc promoter by mutant p53 occurs by a mechanism that is distinct from wild-type p53 transactivation.
Induction of murine c-myc promoter activity by mutant p53 in cell lines that are null for endogenous p53 protein is routinely 30- to 60-fold and, to our knowledge, represents the strongest activation of a murine c-myc reporter. Deletion analysis of the murine c-myc promoter demonstrated that a mutant p53 response element resides near the exon 1/intron 1 boundary and that these sequences confer mutant p53 responsiveness to a heterologous minimal promoter only when cloned downstream and in the correct orientation. Preliminary fine mapping data indicate that mutant p53 transactivation of the c-myc promoter is independent of splicing since there are no known splice acceptor sites in the reporter and point mutations in the splice donor site reduces but does not eliminate mutant p53 responsiveness (data not shown). Interestingly, the mutant p53 response region of the c-myc promoter maps near a site implicated in transcription attenuation (1, 29), and a corresponding region of c-myc exon 1 inhibits expression of an α-globin transgene when cloned downstream from the globin transcription start site and in the correct orientation (45). It remains a formal possibility that mutant p53 can transactivate the c-myc promoter by allowing transcription to read through this region (see below).
The orientation- and position-dependent nature of the mutant p53 response element indicates that it does not function as an enhancer and that it may need to be transcribed into RNA. These findings predict that the C terminus of mutant p53, which expresses a single-strand DNA and RNA binding activity (28, 30, 33, 47), may be required for transactivation of the c-myc promoter. Consistent with this concept, truncation of the last 33 amino acids of mutant p53-281 (Δ360) inhibited transactivation by more than an order of magnitude. These results are in agreement with the previous findings that the last 66 amino acids are required for mutant p53-281 activation of the human epidermal growth factor receptor promoter (23). However, the authors of the latter study concluded that the oligomerization domain is required for mutant p53 transactivation function. In our study, we demonstrate that the oligomerization domain is not sufficient for activation of c-myc gene expression and that additional C-terminal sequences are required for transactivation. Detailed mapping of the C terminus demonstrated that the last 13 amino acids, including the 5.8S RNA modification site (9, 25), are dispensable and that amino acids 372 to 380 are critical for mutant p53 transactivation function (Fig. 7). Interestingly, Levine and coworkers have identified the last 27 amino acids of p53 as a necessary domain for promoting the reassociation of single-stranded RNAs and that monoclonal antibody PAb421, which recognizes an epitope encompassing amino acids 372 to 378, completely blocks this annealing activity (47). Taken together, these results raise the possibility that mutant p53 regulates c-myc gene expression through an RNA-dependent mechanism. Additional studies will be necessary to test this hypothesis.
The requirement for the response element to be located downstream from the transcriptional start site and in the correct orientation is not without precedent. In particular, transcriptional regulation of the human immunodeficiency virus long terminal repeat by the Tat transactivator is mediated through the TAR element, which must be downstream from the promoter and in the correct orientation (for a review, see reference 17). Transcription of the long terminal repeat is attenuated near the TAR site, and binding of the Tat transactivator to an RNA stem-loop structure in TAR releases the transcriptional block and promotes elongation. In our studies, Northern blot and RNase protection analysis demonstrated that the steady-state levels of endogenous c-myc and P2-derived c-myc–CAT mRNAs are elevated in response to mutant p53. It is not yet clear whether mutant p53 activates the c-myc promoter by enhancing transcription initiation and/or elongation rates or by increasing mRNA stability. However, since the acidic domain of mutant p53 is strictly required for transactivation of c-myc and presumably for its interaction with various components of the RNA polymerase II complex (TBP, TFIIH, and TAFs) (22, 34, 43, 48), we favor the concept that mutant p53 activates c-myc gene expression at a transcriptional level.
Mutant p53-281, as well as other naturally occurring mutant p53 proteins, activates the c-myc promoter reporter, whereas wild-type p53 represses expression. How does a single missense mutation turn a transrepressor into a potent activator? One possibility is that specific cellular factors cooperate with mutant p53 in transactivation. In support of this hypothesis, a recent study has demonstrated that mutant p53 proteins that are competent for transactivation of the MDR promoter, but not transactivation-defective mutants or wild-type p53, selectively associate with two cell cycle-regulated proteins (p38 and p42) (2). It remains to be determined what functional consequence these proteins or other associated factors may have on mutant p53 gain of function.
A large effort is under way to devise strategies that convert mutant p53 into the wild-type conformation. This approach is ideal in the sense that mutant p53 would lose its tumor-promoting properties while regaining its tumor suppressor activity. However, this goal may be difficult to achieve based on thermodynamic constraints. In light of the previous gain-of-function studies and the results presented here, it may be more reasonable to develop therapies that are designed to inactivate mutant p53. The findings that the C terminus is dispensable for wild-type p53 in transactivation and growth suppression (31, 36, 39) but strictly required for mutant p53 gain of function suggests that it may be possible to selectively target tumors with mutant p53 but not normal tissue expressing wild-type p53.
ACKNOWLEDGMENTS
We thank James Ihle and Amy Frazier for critically reading the manuscript and for their helpful suggestions. We thank Moshe Oren (Rehovot, Israel) for kindly providing the p53 miniprotein expression vectors.
This work was supported by NIH/NCI grant CA63230 (G.P.Z.), NIH/NIDDK grant DK44158 (J.L.C.), NIH/NCI Cancer Center Support CORE grant 5 P30 CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
REFERENCES
- 1.Bentley D, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Chen P-L, Lee W-H. Hot spot mutants interact specifically with two cellular proteins during progression of the cell cycle. Mol Cell Biol. 1994;14:6764–6772. doi: 10.1128/mcb.14.10.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin K-V, Ueda K, Pastan I, Gottesman M M. Modulation of activity of the promoter of the human MDR1 gene by ras and p53. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland J L, Dean M, Rosenberg N, Wang J W-Y, Rapp U R. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol Cell Biol. 1989;9:5685–5695. doi: 10.1128/mcb.9.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deb S, Jackson C T, Subler M A, Martin D W. Modulation of cellular and viral promoters by mutant human p53 proteins found in tumor cells. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittmer D, Pati S, Zambetti G, Chu S, Teresky A K, Moore M, Finlay C, Levine A J. Gain-of-function mutations in p53. Nat Genet. 1994;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 7.Duyao M P, Kessler D J, Spicer D B, Bartholomew C, Cleveland J L, Siekevitz M, Sonenshein G E. Transactivation of the c-myc promoter by human T cell leukemia virus type 1 tax is mediated by NFκB. J Biol Chem. 1992;267:16288–16291. [PubMed] [Google Scholar]
- 8.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 9.Fontoura B M, Sorokina E A, David E, Carrol R B. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, Shi X-L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 12.Gualberto A, Hixon M L, Finco T S, Perkins N D, Nabel G J, Baldwin A S., Jr A proliferative p53-responsive element mediates tumor necrosis factor alpha induction of the human immunodeficiency virus type 1 long terminal repeat. Mol Cell Biol. 1995;15:3450–3459. doi: 10.1128/mcb.15.6.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey D, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/C murine fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 14.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Mutant cDNAs from human colorectal carcinomas can cooperate with ras in transformation of primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 15.Hsiao M, Low J, Dorn E, Ku D, Pattengale P, Yeargin J, Haas M. Gain-of-function mutations of the p53 gene induce lymphohematopoietic metastatic potential and tissue invasiveness. Am J Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffrey P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 17.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 18.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 19.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for the transcriptional activation, binding to mdm-2 and the adenovirus 5E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Teresky A K, Levine A J. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain-of-function phenotypes of human p53 mutants. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 22.Lu H, Levine A J. TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludes-Meyer J, Subler M, Shivakumar C, Munoz R, Jiang P, Bigger J, Brown D, Deb S P, Deb S. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masuda H, Miller C, Koeffler H P, Battifora H, Cline M J. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci USA. 1987;84:7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meek D W, Simon S, Kikkawa U, Eckhart W. The p53 tumor suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 27.Moll U M, Ostermeyer A G, Haladay R, Winkfield B, Frazier M, Zambetti G P. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. doi: 10.1128/mcb.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nepveu A, Marcu K B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986;5:2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellagata N S, Cajot J-F, Standbridge E J. The basic carboxy-terminal domain of human p53 is dispensable for both transcriptional regulation and inhibition of tumor cell growth. Oncogene. 1995;11:337–349. [PubMed] [Google Scholar]
- 32.Santhanam U, Rat A, Sehgal P B. Repression of the interleukin 6 gene promoter by p53 and the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1991;88:7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selivanova G, Iotsova V, Kiseleva E, Strom M, Bakalkin G, Grafstrom R C, Wiman K G. The single-stranded DNA end binding site of p53 coincides with the C-terminal regulatory region. Nucleic Acids Res. 1996;24:3560–3567. doi: 10.1093/nar/24.18.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaulian E, Zauberman A, Milner J, Davies E A, Oren M. Tight DNA binding and oligomerization are dispensable for the ability of p53 to transactivate target genes and suppress transformation. EMBO J. 1993;12:2789–2797. doi: 10.1002/j.1460-2075.1993.tb05940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaulsky G, Goldfinger N, Ben-Ze’ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaulsky G, Goldfinger N, Rotter V. Alterations in tumor development in vivo mediated by expression of wild-type or mutant p53 proteins. Cancer Res. 1991;51:5232–5237. [PubMed] [Google Scholar]
- 39.Slingerland J, Jenkins J, Benchimol S. The transforming and suppressor functions of p53 alleles: effects of mutations that disrupt phosphorylation, oligomerization and nuclear translocation. EMBO J. 1993;12:1029–1037. doi: 10.1002/j.1460-2075.1993.tb05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spencer C A, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 41.Subler M A, Martin D W, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Y, Nakamura K, Wendel E, Colburn N. Progression toward tumor cell phenotype is enhanced by overexpression of a mutant p53 tumor suppressor gene isolated from nasopharyngeal carcinoma. Proc Natl Acad Sci USA. 1993;90:2827–2831. doi: 10.1073/pnas.90.7.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thut C J, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–103. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 44.Wolf D, Harris N, Rotter V. Reconstitution of p53 expression in a nonproducer Ab-MuLV transformed cell line by transfection of a functional p53 gene. Cell. 1984;38:119–126. doi: 10.1016/0092-8674(84)90532-4. [DOI] [PubMed] [Google Scholar]
- 45.Wright S, Bishop J M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci USA. 1989;86:505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 47.Wu L, Bayle J H, Elenbaas B, Pavletich N P, Levine A J. Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acids. Mol Cell Biol. 1995;15:497–504. doi: 10.1128/mcb.15.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zambetti G P, Olson D, Labow M A, Levine A J. A mutant p53 protein is required for maintenance of the transformed phenotype in cells transformed with p53 plus ras cDNAs. Proc Natl Acad Sci USA. 1992;89:3952–3956. doi: 10.1073/pnas.89.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 51.Zambetti G P, Levine A J. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 1993;8:855–865. doi: 10.1096/fasebj.7.10.8344485. [DOI] [PubMed] [Google Scholar]