SUMMARY
Cancer cell plasticity enables cell survival in harsh physiological environments and fate transitions such as the epithelial-to-mesenchymal transition (EMT) that underlies invasion and metastasis. Using genome-wide transcriptomic and translatomic studies, an alternate mechanism of cap-dependent mRNA translation by the DAP5/eIF3d complex is shown to be essential for metastasis, EMT, and tumor directed angiogenesis. DAP5/eIF3d carries out selective translation of mRNAs encoding EMT transcription factors and regulators, cell migration integrins, metalloproteinases, and cell survival and angiogenesis factors. DAP5 is overexpressed in metastatic human breast cancers associated with poor metastasis-free survival. In human and murine breast cancer animal models, DAP5 is not required for primary tumor growth but is essential for EMT, cell migration, invasion, metastasis, angiogenesis, and resistance to anoikis. Thus, cancer cell mRNA translation involves two cap-dependent mRNA translation mechanisms, eIF4E/mTORC1 and DAP5/ eIF3d. These findings highlight a surprising level of plasticity in mRNA translation during cancer progression and metastasis.
In brief
Alard et al. show that cancer cell mRNA translation involves two distinct cap-dependent mRNA translation mechanisms, one directed by canonical eIF4E/mTORC1 and the other by specialized DAP5/eIF3d. A surprising level of plasticity exists in mRNA translation during cancer progression and metastasis.
Graphical abstract
INTRODUCTION
Metastasis is responsible for 90% of cancer-related deaths, yet at a molecular level remains poorly understood, particularly with respect to translational regulation. Selective mRNA translation is important in cancer development, progression, and metastasis, although a molecular understanding is incomplete.1–3 While the proliferation of transformed cancer cells necessitates high levels of protein synthesis and an increased number of ribosomes, highly malignant and metastatic tumor cells are also under considerable physiological stresses that strongly downregulate protein synthesis in response to energy depletion resulting from hypoxia and nutrient deprivation.4 Cancer cells therefore need to maintain physiological plasticity to survive and adapt quickly to microenvironmental alterations, including that of mRNA translation.5,6
Cancer cell metastasis presents a particularly severe physiological cellular challenge that also involves major changes to cancer cell identity. Metastasis involves the loss of cell adherence, which requires suppression of apoptosis because of loss of adherence, a process known as anoikis,7,8 inhibition of cell cycling, and acquisition of migratory cell properties collectively known as the epithelial-to-mesenchymal transition (EMT).9,10 Roughly 80% of human malignant cancers from a wide range of tissues and organs are derived from epithelial cells, making EMT a central process in metastasis.9,10
Mesenchymal, highly migratory cancer cells survive under significant physiological stress that downregulates canonical cap-dependent mRNA translation, raising the possibility that they utilize specialized mechanisms for mRNA translation that have not yet been described.1,11,12 The canonical translation initiation machinery is well characterized, consisting of cap binding protein eIF4E that locates the m7GTP 5′ capped end of the mRNA and recruits the pre-initiation complex (PIC), which consists of eIF4E, the large scaffolding protein eIF4G that binds eIF4E, the ATP-dependent RNA helicase eIF4A, the multi-subunit complex eIF3 that recruits the 40S ribosome subunit, the ternary complex consisting of eIF2, GTP, and the initiator methionyl tRNA, among a number of additional factors.13 eIF4E, eIF4G, and eIF4A can all be increased in expression in cancer cells, thereby selectively enabling increased translation initiation of mRNAs that are limited by their strong 5′ secondary structure, including oncogenic mRNAs.14–16
eIF4E-mediated cap-dependent mRNA translation constitutes the majority of mRNA translation initiation activity, which is regulated by eIF4E availability. eIF4E availability is downregulated through sequestration by the eIF4E binding proteins (4E-BPs), which compete with eIF4G for eIF4E binding, and are inhibited by the mammalian target of rapamycin complex 1 (mTORC1) through phosphorylation.13 Importantly, mTORC1 kinase activity itself is downregulated by a number of physiological stresses common to cancer cells, including hypoxia, nutrient deprivation and growth factor restriction, among others, which results in dephosphorylation of 4E-BPs, their sequestration of eIF4E and inhibition of eIF4E-mediated cap-dependent mRNA translation initiation. Thus, canonical cap-dependent mRNA translation directed by eIF4E is dependent on active mTORC1. While some tumors uncouple responses that would downregulate mTORC1 activity or sequestration of eIF4E by 4E-BPs under stress, many do not.17 Moreover, cancer cells undergoing EMT must cease proliferation which involves downregulation of mTORC1 activity and therefore eIF4E-mediated mRNA translation.11 This raises the paradoxical question as to how mRNAs encoding essential functions for cancer cell survival, the EMT program and metastasis are able to translate despite the downregulation of mTORC1 activity and sequestration of eIF4E by the 4E-BPs. As only 3%–5% of cellular mRNAs contain internal ribosome entry sites (IRESs), the majority of mRNA translation during cancer EMT and metastasis must be cap-dependent despite inhibition of mTORC1 activity and eIF4E. Thus, there must be additional mechanisms for cap-dependent mRNA translation that are important in EMT and metastasis but have not yet been identified.
Here we show that an alternate mechanism of cap-dependent mRNA translation mediated by the DAP5/eIF3d complex, that does not utilize eIF4E and mTORC1 activity,18 is required for EMT and metastasis. We previously showed that the eIF4G homolog DAP5, along with translation initiation factor eIF3d, which has cap binding activity,19 comprises an alternate mechanism to facilitate cap-dependent, but eIF4E/mTORC1-independent translation of up to 20% of capped mRNAs.18 eIF4G consists of three family members: eIF4GI (gene: eIFG1), eIF4GII (gene: eIF4G3) and DAP5 (gene: eIF4G2). DAP5 is 65% homologous to the C-terminal domain of eIF4GI, but lacks the N-terminal domain found in eIF4GI and II, which binds eIF4E and polyA binding protein (PABP).13 While DAP5 can promote IRES-dependent and cap-independent mRNA translation,20–22 by binding eIF3d, DAP5 can also carry out cap-dependent mRNA translation.18 Thus, many mRNAs previously thought to be “cap independent” because they do not require mTORC1 activity nor eIF4E for their translation, may instead be cap-dependent and use the DAP5/eIF3d complex.
Here we demonstrate that DAP5/eIF3d-mediated mRNA translation is essential for EMT, cell migration, invasion, metastasis, and suppression of apoptosis in non-adherent cells. DAP5/eIF3d selectively translates key mRNAs involved in breast cancer cell invasion and metastasis, including EMT transcription factors and regulators, cell survival factors, integrins (ITGs) that promote cell invasion, and others. Silencing DAP5 in animal models of breast cancer has little effect on primary tumor growth, but substantially impairs cancer cell invasion, migration, metastasis, and survival of established metastases. Engineered overexpression of DAP5 enhances EMT, cancer cell invasion, and metastasis.
RESULTS
Increased expression of eIF4G2/DAP5 mRNA and protein is associated with increased triple-negative breast cancer distant metastasis and poor survival
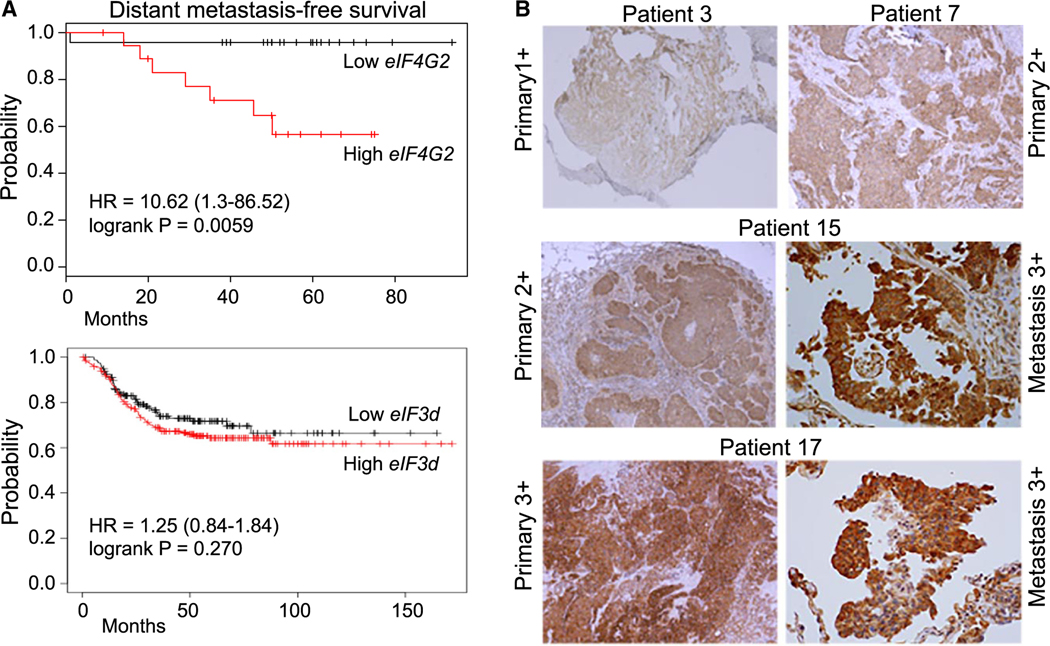
We first asked whether DAP5 expression levels are correlated with human metastatic breast cancer by analyzing the National Cancer Institute (NCI) human Cancer Genome Atlas (TCGA). Increased expression of eIF4G2 mRNA is significantly associated with poor overall survival and metastasis-free survival in estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2/neu receptor-negative breast cancer (Figure 1A). There was no correlation between higher levels of eIF3d expression and poorer overall survival in triple-negative breast cancer (TNBC) nor with poorer metastasis-free survival. We therefore focused on the association of higher expression levels of DAP5 and increased metastatic potential of TNBC.
Figure 1. Increased eIF4G2/DAP5 mRNA and protein expression are strongly correlated with metastasis and reduced survival in triple-negative breast cancer patients.
(A) TCGA cohort for metastasis-free survival for ER−/PR−/Her2− (triple-negative) breast cancer (TNBC) patients. Kaplan-Meier estimates derived from mRNA expression data on the basis of high and low expression levels of eIF4G2 mRNA (top) and eIF3d (bottom). Univariate analysis was performed using the log rank test. Cox proportional hazard regression models were also used.
(B) Representative IHC staining showing score for DAP5 protein levels of TNBC primary tumors that did not metastasize during 8-year follow up, and primary tumor and recurrent matched metastases that recurred within 8 years. See also Tables S1 and S2 for IHC scoring summaries, statistical analysis, patient demographics, and tumor characteristics. Patient demographics are representative of first clinical presentation for the overall U.S. TNBC population.
We also characterized DAP5 protein levels in highly metastatic triple negative invasive ductal breast cancers (IDCs) compared with those that did not metastasize in 8 years from clinical presentation and treatment. In TNBC, most patients who recur do so within 5 years.23–26 We identified a small set of matched pretreatment biopsy specimens from primary tumors, and used a conservative parameter of absence of metastasis within 8 years for non-metastatic tumors. Similarly staged IDC TNBCs were identified with matched post-surgical/post-treatment metastasis biopsies that recurred with distant metastasis within 8 years (mostly to lung). Tumor characteristics and immunohistochemical (IHC) scoring for DAP5 are shown (Table S1). Statistical analysis of IHC DAP5 staining was compared between primary tumors that did not metastasize to those that metastasized within 8 years (Table S2). All non-metastatic primary tumors had much lower expression of DAP5 (100% 1+/2+) compared with metastatic primary tumors (50% 1+/2+, 50% 3+; p = 0.0325). The metastases derived from primary tumors all expressed higher levels of DAP5 than their respective primary tumors (100% 3+/4+; p = 0.0867). Representative IHC images show higher levels of DAP5 expression in metastases compared with respective matched primary tumors, and lower levels of expression in non-metastatic primary tumors (Figure 1B). These data suggest that increased expression of eIF4G homolog DAP5 is strongly correlated with increased TNBC metastatic capacity. We were not able to perform similar analyses for eIF3d protein because all available antibodies tested had a poor dynamic range in IHC. We therefore next investigated the effect of reduction of DAP5 levels on genome-wide transcription (mRNA levels) and translation in triple negative human and murine breast cancer cells and tumor models.
Downregulation of DAP5 expression selectively impairs translation of mRNAs involved in EMT, cell migration, and anoikis survival responses
We investigated which mRNAs are highly dependent on DAP5 for their translation. We developed both human and murine TNBC cell lines that express doxycycline (Dox)-inducible DAP5-specific or non-silencing (Nsi) control short hairpin RNAs (shRNAs), using highly transformed and metastatic human MDA-MB-231 LM2 cells (herein MB-231 cells) and murine 4T1 cells. The DAP5 shRNA and its control Nsi shRNA were previously validated.18 At 4 days post-silencing, DAP5 was reduced by ~75% in both 4T1 and MB-231 cells at the protein and mRNA levels (Figures 2A and S1A). DAP5 silencing reduced global protein synthesis rates by 15%–20% (Figure S1B), similar to previous reports,18,27 with no substantial change in cell proliferation rates in culture (Figure S1C).
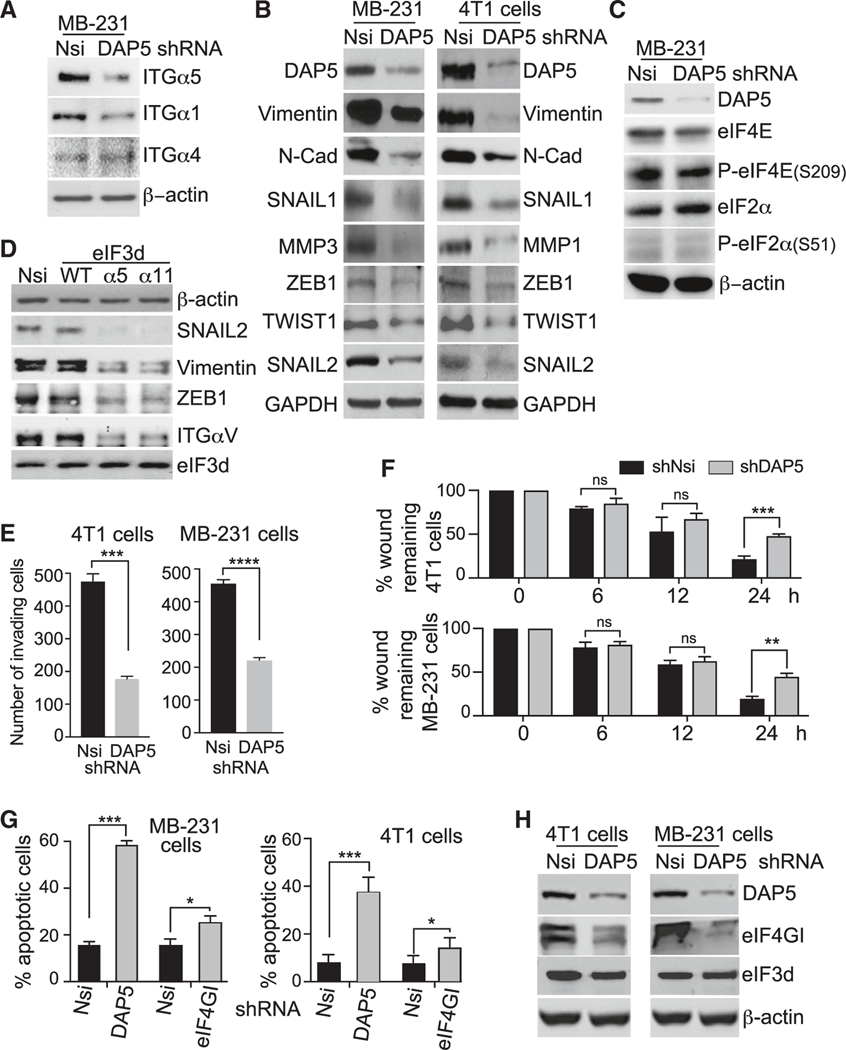
Figure 2. DAP5 expression promotes breast cancer cell migration, EMT, and resistance to anoikis.
(A) Representative immunoblots of 3 independent studies of ITG proteins in Nsi and DAP5-silenced MB-231 cells. Dox induction of shRNAs for 4 days followed by immunoblot analysis of equal protein amounts.
(B) Representative immunoblots of 3 independent studies of EMT biomarker protein levels in Nsi and DAP5-silenced 4T1 and MB-231 cells carried out as in (A).
(C) Representative immunoblots of 3 independent studies of eIF4E and eIF2α proteins in Nsi and DAP5-silenced MB-231 cells carried out as in (A).
(D) Representative immunoblots of 3 independent studies to test requirement for eIF3d cap binding activity for DAP5-dependent mRNAs in MB-231 cells. Cells were small interfering RNA (siRNA) silenced for eIF3d for 1 day and transfected with vectors expressing WT, α5, or α11 cap binding mutants of eIF3d for 2 days, and equal protein amounts analyzed using immunoblot.
(E) Matrigel Transwell invasion assays performed with 4T1 and MB-231 cells silenced for 24 h with Nsi or DAP5 Dox-inducible shRNAs. Mean number of invading cells/field with SEM from 3 replicates for each cell line. See also Figure S4.
(F) Cell migration wound healing assays performed with 4T1 and MB-231 cells silenced for 24 h with Nsi or DAP5 Dox-inducible shRNAs. Time 0 represents 100% wound separation of the cell layer. Mean of percentage of migrated cell surface areas (percentage closure) with SEM from 3 replicates for each cell line. See also Figure S4.
(G) Induction of apoptosis by cell detachment (anoikis) determined in 4T1 and MB-231 cells silenced for 24 and 48 h for Nsi, DAP5, or eIF4GI, comparing cells maintained on adherent or ultra- low-adherence plates. Percentage cell apoptosis determined by annexin V-fluorescein isothiocyanate (FITC) staining and flow cytometry, quantified using FlowJo software. Mean with SEM of 3 independent studies per cell line.
(H) Representative immunoblot of 3 independent studies of DAP5, eIF4G1, eIF3d, and control β-actin protein levels in Nsi or DAP5-silenced 4T1 and MB-231 cells, silenced for 2 days with Dox-inducible shRNAs.
Data are represented as mean ± SEM. n.s., not significant. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by unpaired parametric two-tailed t test.
We next characterized translation activity of individual mRNAs on a genome-wide scale, comparing non-silenced with cells silenced for DAP5 by ~75% (Figure S2A). Total mRNA and well-translated (≥4 ribosome) fractions that represent the majority of mRNA translation activity were subjected to RNA sequencing (RNA-seq). Data were analyzed for translation activity on the basis of translation efficiency (TE), which represents log2 ratios of ≥4 ribosome mRNAs individually normalized to the total level of each mRNA. TE therefore represents authentic changes in individual mRNA translation-specific activity. We also recorded total translation activity of the ≥4 ribosome fraction, which is not normalized to changes in mRNA abundance (represents transcription plus translation), which is important, for instance, in identifying translation changes resulting from altered translation of transcription factor mRNAs. Principal-component analysis (PCA) showed acceptable reproducibility for Nsi control and DAP5-silenced datasets (Figure S2B). Polysome absorbance profiles of DAP5-silenced 4T1 and MB-231 cells demonstrated modest reduction in polysome content, consistent with the modest reduction in protein synthesis activity (Figure S2C). In both murine 4T1 and human MB-231 breast cancer cells, 15%–20% of mRNAs were translationally downregulated with DAP5 silencing (reduced in TE) in the well-translated heavy polysome fraction (≥4 ribosome) (Figure S2D; 4T1 cell Data S1). The MB-231 cell TE and total translation data analyzed here for comparison purposes was previously published (GSE115142).18 Of these mRNAs, 632 mRNAs (21% of 4T1, 26% of MB-231 cells) were in common, supporting a high level of identity for DAP5 translation dependence across species (Figure S2E), particularly given the difference in species and cell types. Scatterplots comparing log2 fold changes at q < 0.05 showed that the majority of translation-only changes comprised large reductions in mRNA content in the well-translated polysome fraction (from 3- to 5-fold on average; Figure S2F), consistent with a strong DAP5 requirement.
Ingenuity Pathway Analysis (IPA) and Gene Ontology (GO) analysis of genome-wide TE data from both 4T1 and MB-231 cells was used to develop categorical classifications for mRNAs highly dependent on DAP5 (Figure S2G). The top functional categories for DAP5-dependent mRNA translation in both mouse and human breast cancer cells were cell migration, cell movement and cell survival functions. Gene set enrichment analysis (GSEA) (Figures S2H and S3) of the common murine and human breast cancer datasets identified top categories for DAP5 translationally dependent mRNAs. Of the common DAP5-dependent mRNAs between human breast cancer cells and murine mammary carcinoma cells, the majority of these DAP5-dependent mRNAs also defined functional categories in common. DAP5 silencing increased expression of cell death, hypoxia, DNA repair cell and cycle checkpoint pathways. This is consistent with the importance of DAP5 in maintenance of cell viability during EMT and cell migration.
We analyzed mRNAs downregulated in TE by DAP5 silencing by ≥0.6 log2 fold changes, in common between 4T1 and MB-231 cells for functions involved in EMT, metastasis, and cell survival functions. Of the >600 mRNAs analyzed, ~65% are involved in EMT, metastasis, and survival functions (Table S3). Among these mRNAs that were solely translationally downregulated with DAP5 silencing (TE), were a group of ITGs that are involved in cancer cell migration and EMT, particularly ITGs α1, α3, α5, α6, and α8. Immunoblot analysis of MB-231 cells demonstrated much lower levels of these ITGs with DAP5 silencing (Figure 2A). ITG proteins α5 and α6, whose mRNAs are translationally DAP5 dependent, were reduced with DAP5 silencing, but not the ITGα4 mRNA which is not a target of DAP5 and was unchanged. Other EMT protein migratory factors and EMT-mediating transcription factors showed only modest reduction in mRNA levels of 20%–50% but strong reduction in protein levels that represents translational downregulation (Figures 2B, S4A, and S4B). These include SNAIL1, SNAIL2 (SLUG), ZEB1, TWIST1, and factors that promote the loss of epithelial characteristics that aid in reprogramming to the mesenchymal phenotype such as vimentin, N-cadherin, and MMPs.9,28 DAP5 silencing, as shown in MB-231 cells, was not accompanied by changes in eIF4E levels or phosphorylation at S209 (Figure 2C), are therefore not involved in the effects of DAP5 reduction. There was also no evidence for cell stress unfolded protein responses with DAP5 silencing, such as eIF2α S51 phosphorylation that could reduce translation rates or alter coding region selection with DAP5 silencing (Figure 2C).
DAP5-dependent mRNAs required the cap-binding activity of eIF3d, shown for several key EMT mRNAs. MB-231 cells were silenced for endogenous eIF3d, then 1 day later, transfected with plasmids expressing wild-type (WT) or either of two cap-binding deficient mutated forms of eIF3d.29 Loss of eIF3d cap binding activity resulted in strong reduction in levels of all EMT proteins tested, but not the actin control and eIF3d itself (Figure 2D), whose mRNAs use canonical eIF4E/eIF4G-mediated mRNA translated, not the DAP5/eIF3d complex.18
DAP5/eIF3d-directed mRNA translation promotes breast cancer cell invasion, migration, and resistance to anoikis-mediated cell death
To test the importance of DAP5/eIF3d-mediated mRNA translation on cancer cell invasion, it was inducibly silenced in 4T1 and MB-231 cells, which were then seeded on Matrigel Transwell plates and quantified for invasion activity. Non-silenced control and DAP5-silenced 4T1 and MB-231 cells displayed similarly high levels of invasion activity that were reduced 2.5–3.0 fold with DAP5 silencing (Figures 2E and S4C), with no effect on adherent cell viability (Figure S4D). DAP5 silencing also impaired cancer cell migration in in vitro wound healing assays (Figures 2F and S4E). Thus, DAP5/eIF3d-dependent mRNA translation is important for both cancer cell migration and invasion.
Epithelial cancer cells that acquire migratory mesenchymal characteristics and become metastatic lose attachment to the extracellular matrix (ECM) and must have survival mechanisms to resist loss of adherence-mediated anoikis while migrating through the lymphatic and circulatory systems.30 Moreover, detachment from the ECM and acquisition of a migratory cell phenotype involves downregulation of mTORC1 and inhibition of eIF4E-mediated mRNA translation.6,17 We therefore determined whether DAP5/eIF3d-mediated mRNA translation is required for resistance to anoikis. 4T1 and MB-231 cells were inducibly silenced with Nsi or DAP5 shRNAs while grown on adherent plates, then seeded onto ultra-low attachment plates. Flow cytometry analysis showed that 4-fold silencing of DAP5 for 48 h induced apoptotic death in ~60% of MB-231 cells compared with ~17% for Nsi controls and ~40% of 4T1 cells compared with ~7% for Nsi controls (Figures 2G and 2H). In contrast, silencing canonical eIF4GI, only slightly increased apoptosis in non-adherent MB-231 and 4T1 cells comparative to Nsi controls. DAP5 therefore maintains cancer cell survival during ECM detachment and protects against anoikis-mediated cell death.
Silencing DAP5 impairs metastasis but not primary tumor growth
We explored the role of DAP5/eIF3d-dependent mRNA translation on tumorigenesis, using murine 4T1 and human MB-231 cells in syngeneic and immune impaired animals, respectively. Both cell lines were engineered to express Firefly luciferase and turbo red fluorescent protein (tRFP) for bioluminescence and fluorescent imaging of tumor growth and metastasis. For MB-231 cell studies, cells were implanted in the fourth mammary fat pad, and DAP5 silencing induced by Dox addition to animal drinking water 28 days post-tumor cell implantation (Figure 3A). Primary tumor growth and metastasis were quantified by precision caliper measurements of primary tumor volumes, bioluminescence imaging and H&E staining of lungs. Mice were sacrificed at 49 days and tumors and lungs collected.
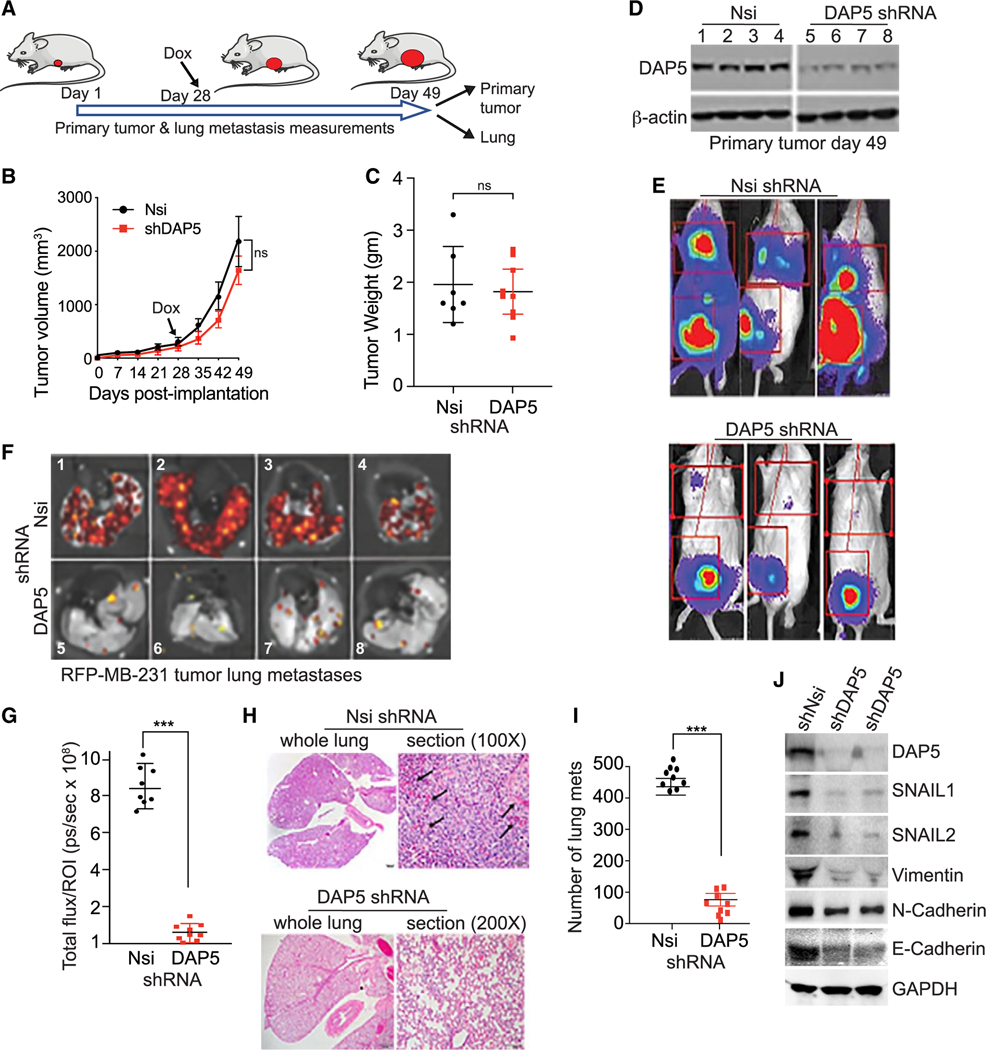
Figure 3. Silencing eIF4G2 mRNA in human MB-231 cell tumors does not impair primary tumor growth but strongly blocks metastasis.
(A) NOD/SCID/γ mice orthotopically injected with 5 × 106 MB-231 cells stably transformed to express Firefly luciferase and red fluorescent protein (RFP), Dox-inducible Nsi or shDAP5 RNAs. Silencing initiated by Dox addition to drinking water 28 days after tumor cell implantation, tumors ~100–150 mm3 in size. At 49 days, mice were sacrificed, tumors and lungs collected.
(B) Growth of tumors was recorded every 7 days by quantitative caliper measurement. n = 8 or 9 mice per group repeated twice. Volumes of non-silenced control compared with DAP5-silenced tumors was not statistically significant.
(C) Primary tumors weighed at excision 49 days. Non-silenced control compared with DAP5-silenced tumors not statistically significant. n = 7–9 mice/group. Representative of 2 trials.
(D) Equal protein amounts of whole tumor lysates at 49 days post-excision, subjected to immunoblot analysis for DAP5 and control actin levels. n = 4 tumors (of 8 or 9 mice) per arm were evaluated.
(E) Representative Firefly bioluminescent images from 8 or 9 mice per group (3 shown), silenced for Nsi or DAP5 as in (A) immediately prior to sacrifice at 49 days. Repeated 3 times.
(F) Lungs excised, subjected to bioluminescent imaging for RFP. Representative images of 8 or 9 mice per group (4 shown) of mice silenced for Nsi or DAP5 as in (A). Repeated twice.
(G) Quantification of metastatic burden in lungs of mice silenced for Nsi or DAP5 as in (A). Total RFP fluorescent flux quantified (photons [ps]/sec), representative of total lung tumor burden, at 49 days. n = 8 or 9 mice per group. Repeated twice.
(H) Representative H&E-stained images of lungs of 8 or 9 mice per group harvested at 49 days. Arrows indicate dark staining tumor masses.
(I) Quantification of number of lung metastases (mets) per field at 1003 magnification of H&E-stained lungs as shown in (H). A minimum of 5 fields per lung were quantified from 9 mice/group, mean plus SEM.
(J) DAP5-silenced tumors have reduced levels of EMT proteins. Immunoblot of equal protein amounts from control Nsi and shDAP5-silenced tumors harvested at 49 days. Representative results of 6 tumors are shown.
Data are mean plus SEM. Statistical significance was assessed by two-way ANOVA with Dunnett post-ANOVA test determination for analysis of repeated measures. n.s., not significant. ***p < 0.001. See also Figures S2 and S3.
A strong reduction (4- to 5-fold) in DAP5 expression by inducible silencing in MB-231 cells starting at 28 days when tumors were 100–150 mm3 in size (Figure 3B) did not significantly reduce primary tumor growth or weight (Figures 3B and 3C). DAP5 silencing was maintained throughout the 49 days of tumor growth (Figure 3D). DAP5 silencing did not alter expression in tumors of eIF4E, eIF2α, or their regulatory phosphorylation (Figure S4F). Whole body bioluminescence imaging of Firefly luciferase carried out immediately prior to termination of the study at 49 days showed that DAP5 silencing strongly reduced metastasis from the primary tumor site (Figure 3E). Representative red fluorescent protein (RFP) images of lungs from 4 animals per group of 8 or 9 mice showed a striking reduction in metastasis with DAP5 silencing (Figure 3F), averaging 7-fold (Figure 3G). H&E analysis of lungs showed a 9-fold reduction in metastatic lesions in lungs with DAP5 silencing (Figures 3H and 3I). DAP5-silenced primary tumors were strongly reduced in translation of key DAP5/eIF3d-dependent EMT mRNAs, with no significant reduction in encoding mRNA levels (Figures 3J and S4G). DAP5 deficiency therefore impairs translation of DAP5/eIF3d-dependent EMT mRNAs in tumors.
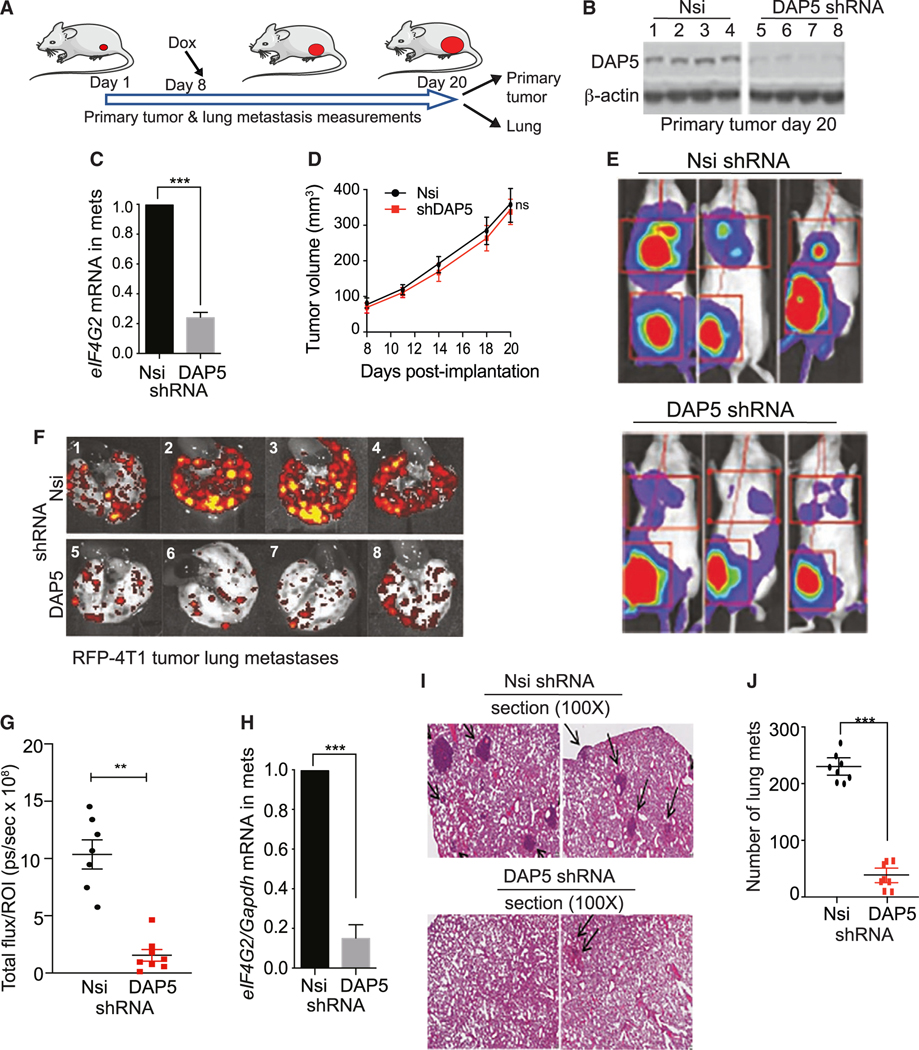
4T1 cells silenced for DAP5 also demonstrated no effect on primary tumor growth but strong inhibition of metastasis. Because 4T1 cells implanted subcutaneously into the flank of immune competent Balb/c mice proliferate and metastasize very rapidly, DAP5 silencing was initiated at 8 days post-implantation, when tumors were ~75–100 mm3. Tumor growth was measured by precision caliper for 20 days, mice sacrificed, and tumors and lungs collected (Figure 4A). As observed for MB-231 cells, strong downregulation of DAP5 protein and mRNA (Figures 4B and 4C) did not significantly affect primary tumor growth (Figure 4D), but strongly reduced metastasis (Figure 4E). Lung metastatic burden quantified by tumor cell RFP fluorescence (Figure 4F), demonstrated that DAP5 silencing reduced 4T1 metastasis to lung by an average of 5- to 6-fold (Figure 4G). DAP5 mRNA remained silenced in metastases, shown by fluorescence-activated cell sorting (FACS) isolation of metastatic 4T1 cells from the lung using RFP followed by qRT-PCR quantification of eIF4G2 mRNA (Figure 4H). Lung metastases quantified by H&E staining (Figures 4I and 4J), showed that DAP5 silencing reduced lung metastatic burden by 5- to 6-fold.
Figure 4. Silencing eIF4G2 mRNA in murine 4T1 cell tumors does not impair primary tumor growth but strongly blocks metastasis.
(A) Schema of animal study. Balb/c mice subcutaneously injected in the flank with 5 × 105 4T1 cells stably transformed to express Firefly luciferase and red fluorescent protein (RFP), Dox-inducible Nsi or shDAP5 RNAs. Silencing initiated 8 days after tumor cell implantation, tumors at 75–100 mm3 in size, by Dox addition to drinking water. At 20 days, mice were sacrificed and tumors and lungs collected.
(B) Equal protein amounts of whole tumor lysates at 20 days post-excision subjected to immunoblot analysis for DAP5 protein and control actin levels. n = 4 tumors per condition were evaluated.
(C) mRNA extracted from tumors removed at 20 days and equal RNA amounts used to determine eIF4G2 mRNA levels normalized to Gapdh mRNA. n = 3 tumors per condition; mean and SEM are shown.
(D) Growth of tumors recorded every 2–4 days by quantitative caliper measurement. n = 8 or 9 mice per group repeated twice. Volumes of Nsi control compared with DAP5-silenced tumors not statistically significant.
(E) Representative Firefly bioluminescent images of 8 mice per group (4 shown), silenced for Nsi or DAP5 as in (A), immediately prior to sacrifice at 20 days. Repeated 3 times.
(F) Lungs excised from mice and subjected to fluorescence imaging for RFP. Representative images of 8 or 9 mice per group (4 shown) of mice silenced for Nsi or DAP5 as in (A). Repeated twice.
(G) Quantification of metastatic burden in lungs of mice silenced for Nsi or DAP5 as in (A). Total RFP fluorescence flux quantified (photons [ps]/sec), representative of total lung tumor burden harvested at 20 days. n = 7 or 8 mice per group. Repeated twice.
(H) Quantification of eIF4G2 mRNA levels in Nsi and DAP5-silenced metastases in lungs of mice. n = 3 per group. Lungs excised, tumor cells dispersed, isolated by RFP FACS, subjected to qRT-PCR for eIF4G2 mRNA normalized to Gapdh mRNA.
(I) Representative H&E-stained images of lungs of 8 mice/group harvested from mice bearing Nsi or shDAP5 tumors. Arrows indicate dark staining tumor masses.
(J) Quantification of number of lung metastases (mets) per field at 100× magnification of H&E-stained lungs as shown in (I). A minimum of 5 fields per lung quantified, mean plus SEM of n = 8 mice per condition.
Data are mean plus SEM. Statistical significance was assessed by two-tailed Student’s t test for unpaired experimental values (C and H), or two-way ANOVA with Dunnett post-ANOVA test determination for analysis of repeated measures with (D, G, and J). n.s., not significant. ***p < 0.001. See also Figure S4.
DAP5/eIF3d-directed mRNA translation promotes breast tumor cell stromal invasion and angiogenesis in mice
A hallmark of EMT is the invasion of the peri-tumoral stroma by cancer cells. MB-231 cells expressing Dox-inducible shRNAs to DAP5 or control Nsi and an RFP tag were implanted in the fourth mammary fat pad of 8 week old NOD/SCID/γ female mice as in Figure 3, and silencing initiated at 28 days post-tumor cell implantation when tumors were 100–150 mm3 in size. Tumors along with peri-tumoral stroma were excised at 49 days, fixed/embedded, sectioned, stained with DAPI to identify nuclei and RFP for tumor cells. There was a strong reduction in MB-231 cell stromal invasion with DAP5 silencing (Figure 5A). Quantification of 3 fields chosen at random per section of 4 different tumors indicates a 4.5-fold reduction in MB-231 stromal invasion with DAP5 silencing (Figure 5B), consistent with a requirement for DAP5/eIF3d-mediated translation of EMT, cell migration and survival mRNAs.
Figure 5. DAP5 promotes breast cancer cell peri-tumoral stromal invasion and tumor angiogenesis and reduced metastatic disease survival.
(A) MB-231 cells (1 × 107) expressing RFP and Dox-inducible control Nsi or shRNA to DAP5 implanted in mammary fat pad of NOD/SCID/γ mice, Dox induced at 28 days, when tumors 100–150 mm3 in size, maintained for 20 days, tumors with stroma excised, embedded, sectioned for immunofluorescence microscopy for RFP (red) and DAPI (blue) stained nuclei. Representative images of RFP expressing MB-231 cells beyond the periphery of tumor, two different tumors analyzed, 3 fields each. Scale bar, 200 μm. Arrows indicate direction of tumor cell stromal migration away from tumor capsule.
(B) Quantitation of images in (A). Quantitation determined from 2 independent tumors, 3 fields each (n = 6), using ImageJ software.
(C) Tumors obtained as in (A) from non-silenced Nsi control and DAP5-silenced tumor specimens sectioned, IHC stained for CD31 to highlight tumor vasculature. Representative sections shown from 5 different tumors. Scale bar, 1,000 μm.
(D) Quantitation of CD31 stained MB-231 cells from images in (C). Quantitation determined from 3 different tumors, 3 fields each.
(E) Schema of animal study for survival determination with DAP5 tumor-specific silencing. Animals subcutaneously injected in flank with 1 × 105 4T1-Nsi or 4T1-shDAP5 cells. At 12 days, Dox added to the drinking water to induce eIF4G2 mRNA silencing.
(F) Tumor growth recorded every 2–4 days by quantitative caliper measurement until 22 days as control mice began to die. n = 8–10 mice per group, repeated twice.
(G) Survival of mice with 4T1-Nsi or 4T1-shDAP5 tumors silenced starting at 13 days. n = 15 mice/group. Animals either died or were sacrificed when terminally moribund.
Statistical significance was determined using unpaired non-parametric log rank Mantel-Cox test. Data are mean with SEM. Statistical significance (B, D, and F) assessed using two-tailed Student’s t test.
Silencing DAP5 also reduced the translation of certain mRNAs that encode proteins involved in tumor angiogenesis (Table S3; Data S1). These include A disintegrin and metalloproteinases (ADAMs), 9, 10, 15, 17, TS3, angiogenesis promoting ITGs, and other mRNAs. We therefore determined the effect of DAP5 silencing on tumor angiogenesis using tumors developed in Figure 3, that were sectioned and stained by immunohistochemistry for endothelial cell vascular marker CD31. There was a 2-fold reduction in tumor vascularization in DAP5-silenced MB-231 tumors (Figures 5C and 5D).
DAP5 silencing in 4T1 cells significantly improves overall cancer survival in mice
We asked whether DAP5 silencing improves survival during 4T1 cell tumorigenesis. Mice were implanted in the flank with 4T1-shDAP5 or 4T1-Nsi cells, and DAP5 silencing induced by Dox addition to the drinking water 12 days later when tumors averaged 75–100 mm3 in size (Figure 5E). Mice were maintained until terminally moribund or death. Primary tumor size quantified by precision caliper showed no statistically significant difference between Nsi and DAP5-silenced animals (Figure 5F). However, survival curves demonstrated that DAP5 silencing significantly increased mouse overall survival by as much as one-third compared with control mice (p < 0.0001, Mantel-Cox test; Figure 5G).
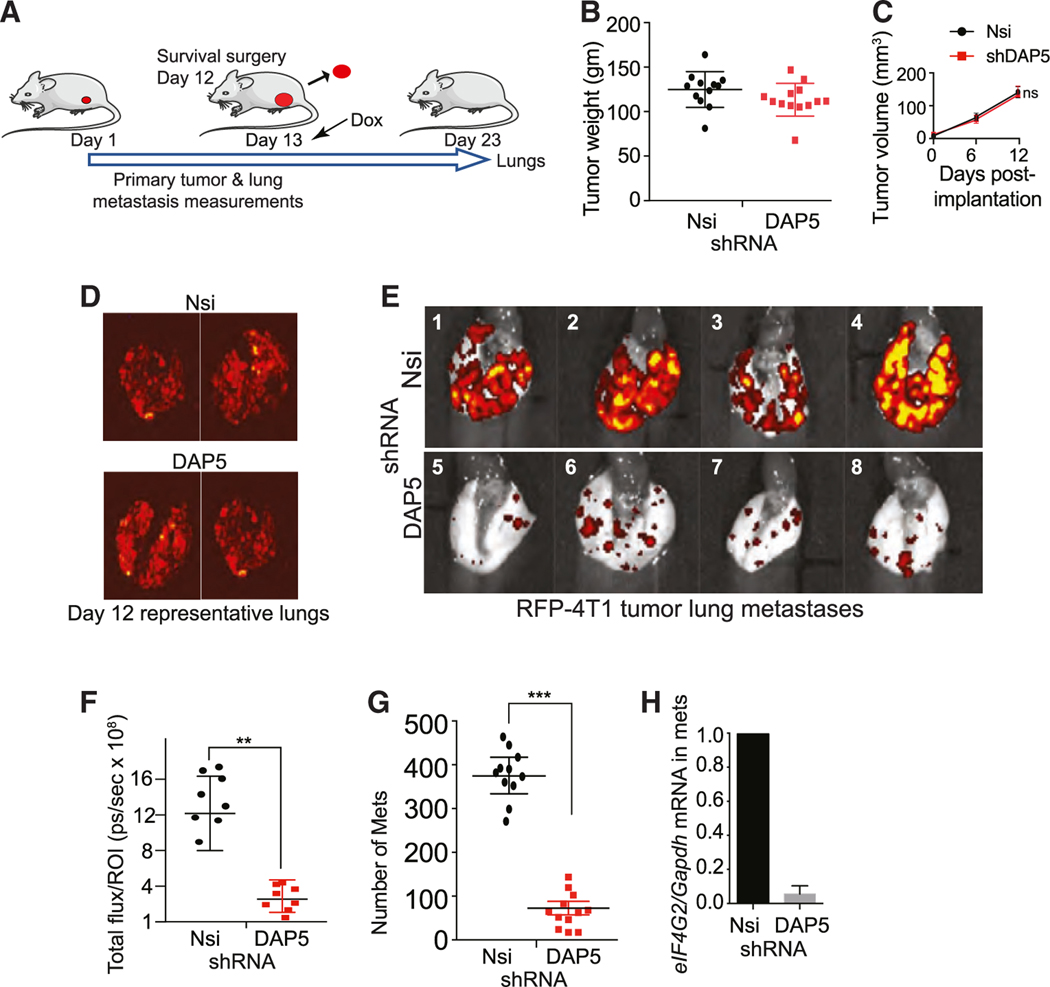
Established metastases require DAP5 for survival
Because the DAP5/eIF3d complex is required for translation of tumor cell EMT, survival, migration, angiogenesis, and metastasis mRNAs, we determined whether DAP5 is required for the survival of established metastases. 4T1 primary tumors were generated in the flank of Balb/c mice until they were ~100 mm3 in size, then aseptically removed, followed by Dox-mediated initiation of silencing of DAP5 in tumor cells that had metastasized (Figure 6A). Nsi and DAP5 shRNA primary tumors prior to silencing were of similar weight and size (Figures 6B and 6C). Representative RFP bioluminescent imaging of excised lungs at the time of primary tumor surgical removal, indicated strong establishment of metastases in lungs in Nsi and DAP5 shRNA mice, prior to shRNA induction (Figure 6D). Ten days after initiation of DAP5 silencing, lung metastases were quantified by RFP fluorescence imaging (Figure 6E). DAP5 silencing reduced established metastases in the lung by ~4- to 5-fold on the basis of total RFP signal (Figure 6F). Quantification of lung metastases by H&E staining also indicated an ~5-fold reduction in the number of metastatic lesions in DAP5-silenced mice (Figure 6G). DAP5 silencing in metastatic cancer cells in lungs was maintained, as shown by qRT-PCR of eIF4G2 mRNA in RFP FACS isolated lung tumor cells (Figure 6H). These data indicate that expression of DAP5/eIF3d-mediated mRNA translation is required for both the process of metastasis and the maintenance of established metastases.
Figure 6. Silencing eIF4G2 mRNA in established metastases strongly reduces metastatic burden.
(A) Schema of animal study. Balb/c mice subcutaneously injected in the flank with 1 × 107 4T1 cells stably transformed to express Firefly luciferase and RFP, Dox-inducible Nsi or shDAP5 RNAs. Tumors aseptically excised at 12 days, ~100–125 mm3 in size, Dox added to drinking water at 13 days to express shRNAs. At 23 days, mice were sacrificed, and lungs were collected.
(B) Primary tumors weighed at excision at 12 days, weight of non-silenced control compared with DAP5-silenced tumors. n = 10–13 mice/group.
(C) Growth of tumors recorded 6 and 12 days prior to surgical excision of tumors, by quantitative caliper measurement. n = 8 mice per group repeated twice.
(D) Lungs excised from 2 mice per group at 12 days prior to shRNA induction and subjected to fluorescent imaging for RFP. Images show qualitatively similar metastatic tumor burden in lungs of mice prior to Nsi or DAP5 silencing.
(E) Lungs excised from mice at 23 days, subjected to fluorescent imaging for RFP. Representative images of 8 mice per group (4 shown) for mice silenced for Nsi or DAP5 as in (A). n = 8 mice/group. Repeated twice.
(F) Quantification of metastatic burden in lungs at 23 days of mice silenced for Nsi or DAP5 as in (A). Total RFP fluorescence flux quantified (photons [ps]/sec), representative of total lung tumor burden. n = 8–10 mice per group. Repeated twice.
(G) Quantification of number of lung metastases (mets) per field at 100× magnification of H&E-stained lungs at 31 days. A minimum of 5 fields per lung quantified, mean plus SEM. n = 11 or 12 mice per condition.
(H) Quantification of eIF4G2 mRNA levels in Nsi and DAP5-silenced metastases in lungs of mice at 31 days. n = 3 per group. Lungs excised, tumor cells dispersed, isolated by FACS gating on RFP and subjected to qRT-PCR for eIF4G2 mRNA normalized to Gapdh mRNA.
Data are mean plus SEM. Statistical significance by two-way ANOVA with Dunnett post-ANOVA test determination for analysis of repeated measures. n.s., not significant. **p < 0.01 and ***p < 0.001.
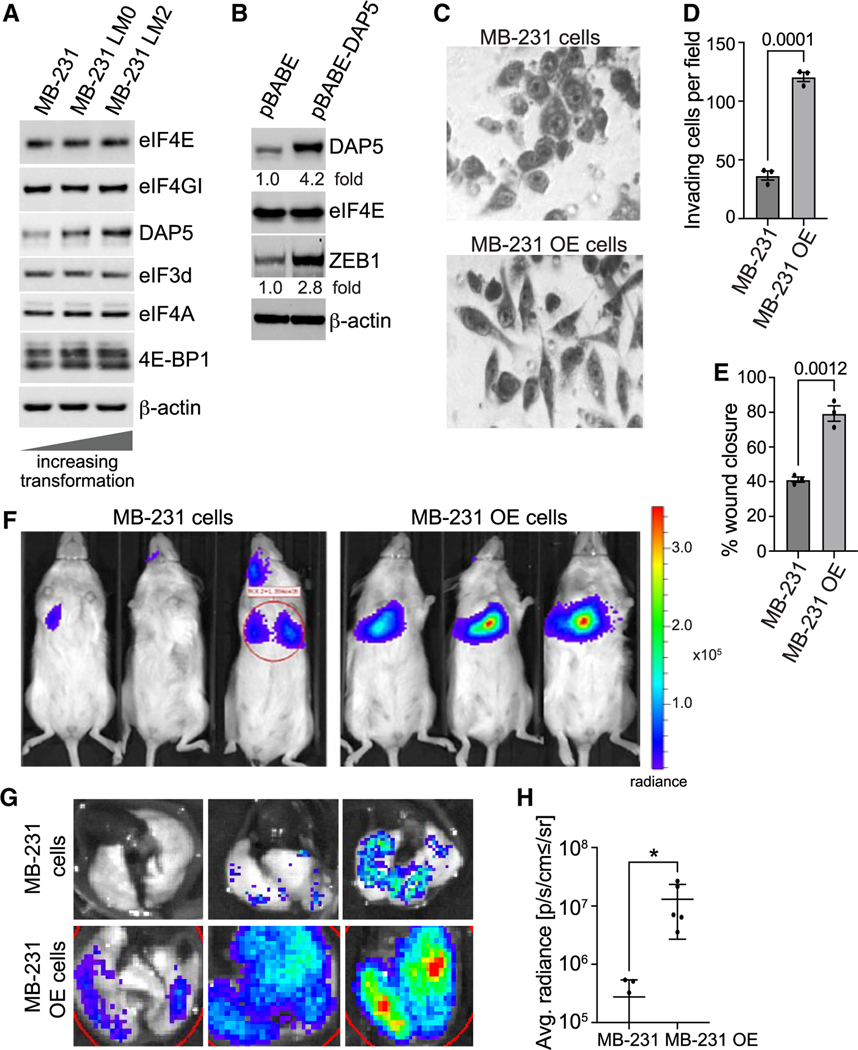
Increased expression of DAP5 promotes increased invasion and metastatic colonization
We next carried out studies to ask whether increased expression of DAP5 increases cancer cell invasion and metastatic activity. MB-231 cells that are increasingly transformed31 demonstrate increased expression of DAP5 protein, but not other initiation factors, including eIF3d (Figure 7A). We therefore stably expressed a cDNA of human eIF4G2 mRNA32 in the least transformed parental MB-231 cell line by ~4-fold to create MB-231 overexpressing (OE) cells (Figure 7B). There was no increase in eIF4E, whereas ZEB1 expression increased almost 3-fold with higher expression of DAP5, indicative of increased mesenchymal characteristics. This was observed in MB-231 OE cells which acquired an enhanced mesenchymal cell phenotype demonstrated by elongated fibroblast and ameboid morphology, and loss of the epithelial rounded aggregated non-motile cell morphology that comprised the major morphology of parental MB-231 cells (Figure 7C). Compared with parental MB-231 cells, MB-231 OE cells showed 4-fold increase in Matrigel cell invasion activity and 2-fold increase cell migration activity (Figures 7D, 7E, S5A, and S5B), without any change in cell proliferation (Figure S5C). We compared the metastatic colonization potential of parental and DAP5 OE cell lines. An equal number of parental and OE MB-231 cells expressing Firefly luciferase were injected into the retro-orbital sinus of NOD/SCIDγ mice and imaged 10 days later for lung colonization. Whereas bioluminescence imaging showed only a low level of lung metastasis for parental MB-231 cells, MB-231 OE cells strongly colonized the lung (Figures 7F and 7G), quantified in excised lungs as a 20-fold increase (Figure 7H). Taken collectively, these results confirm that DAP5/eIF3d-mediated mRNA translation is important for EMT, cell invasion, and metastatic activity.
Figure 7. Overexpression of DAP5 promotes increased metastatic colonization.
(A) Representative immunoblot of select translation factor proteins in equal amounts of lysates from less transformed parental MB-231 cells, and two increasingly more transformed and more metastatic variants, LM0 and ML2 cells.
(B) Representative immunoblot of 3 independent studies of parental MB-231 cells and parental MB-231 cells stably transfected with DAP5 cDNA (MB-231 OE cells).
(C) Representative light field microscopic images of trypan blue stained parental MB-231 cells and MB-231 OE cells. n = 3 independent plating of cells, images representative from 12 different fields chosen at random.
(D) Matrigel Transwell invasion assays performed with parental MB-231 cells or MB-231 OE cells, carried out as in Figure 2E. Results represent the mean of invading cells/field with SEM from 3 independent studies. Statistical analysis by unpaired two-tailed t test.
(E) Cell migration wound healing assay. performed with parental MB-231 cells and MB-231 OE cells carried out as in Figure 2F. Statistical analysis by unpaired two-tailed t test.
(F) Mice were injected in the retro-orbital (RO) sinus with 103 parental MB-231 cells or MB-231 OE cells, both expressing Firefly luciferase. Ten days later representative Firefly bioluminescent images were obtained from 3–5 mice per group (3 shown). Repeated 2 times.
(G) Lungs excised at 10 days post-RO injection of mice described in (E), subjected to bioluminescent imaging for Firefly luciferase, repeated twice.
(H) Quantification of metastatic burden in lungs of mice from (E). Total luciferase fluorescent flux quantified (photons [ps]/sec). *p < 0.05 by unpaired two-tailed t-test.
DISCUSSION
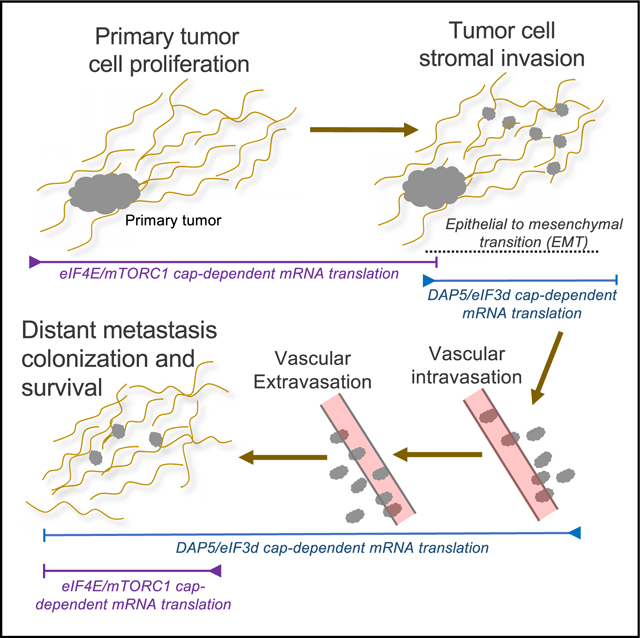
There is increasing evidence that different mechanisms of mRNA translation are essential for cancer development and malignant progression, as well as cancer cell survival under changing and often harsh microenvironmental settings.5,6 Different mRNA translation mechanisms can provide cancer cells with the plasticity necessary to commit to major dynamic phenotypic changes such as the EMT and metastasis.5,6 What remains poorly understood are the specialized mechanisms that selectively translate the reprogramming mRNAs. The phenotypic and physiologic changes that cells undergo during EMT and migration, whether during embryonic development, wound healing, or cancer metastasis, involves multiple levels of cellular reprogramming, including ribosome biogenesis and translational control, the latter only incompletely characterized to date.5,6,33–43 In fact, the downregulation of protein synthesis during EMT, cell migration, and metastasis suggests that there must be specialized mechanisms for selective translation that drives these events, including that of survival factor and other mRNAs that are used at this time.
It had long been thought that almost all eukaryotic cellular mRNA translation involves cap recognition by a single factor, eIF4E, that promotes cap-dependent assembly of the PIC which directs ribosome loading onto mRNA. Perhaps 3% of cellular mRNAs can carry out internal ribosome entry directed by IRES elements, which is independent of eIF4E and mTORC1 activity.44,45 It has therefore been puzzling that despite quantitative silencing of eIF4E, and/or its sequestration by the 4E-BPs because of pharmacologic inhibition of mTORC1 activity, that 20% or more of capped mRNA translation remains active.18,27,46 Moreover, studies show that despite inhibition of eIF4E expression and/or mTORC1 activity, cancer cell metastasis persists in both animal models of cancer and in human clinical trials conducted with eIF4E or mTORC1 inhibitors.2,47–49
Our study sought to understand the mechanism by which mRNAs that drive the EMT, invasion, metastasis, and cancer cell survival are translated despite strong downregulation of mTORC1 activity and eIF4E sequestration in these settings. There are three conclusions that can be drawn from our findings. First, DAP5 expression in human TNBCs is elevated in metastatic tumors and their metastases compared with non-metastatic tumors, and associated with poor metastasis-free survival, although the small sample size warrants future validation studies. Second, the DAP5/eIF3d complex is required for translation of capped mRNAs involved in the EMT, cell migration, invasion, suppression of anoikis and survival in breast cancer cells. Third, DAP5/eIF3d-mediated mRNA translation is essential for metastasis and survival of established metastases.
It is well established that mTORC1/eIF4E-mediated cap-dependent mRNA translation is required for primary tumor cell growth and proliferation, protects against mitotic catastrophe associated with oncogene activation and promotes metastasis.2,47,50–52 But other mRNA translation mechanisms are also essential for translation of mRNAs in non-proliferating cells, for phenotypic and physiologic reprogramming, and to respond to cell stresses, which often inhibit mTORC1 activity and eIF4E availability. In this regard, it is notable that there is little evidence to support translation of the large group of hypoxia or EMT induced mRNAs by cap-independent ribosome internal initiation.5 Notably, reference to cap-independent translation has more correctly been shown to be eIF4E-independent translation, and might in fact be cap mediated by the DAP5/eIF3d cap-binding complex in many of these settings. Cellular IRES containing mRNAs can be DAP5 dependent,22,53,54 though they are uncommon and cannot account for the majority of DAP5-dependent mRNAs in our data. While DAP5 has also been shown to promote leaky scanning translation initiation at alternate start codons, in these cases translation initiation is still cap dependent,55,56 which would require eIF3d. Thus, these and other studies taken collectively with our findings, indicate that there are multiple distinct mechanisms of mRNA translation, all of which are important in cancer development, progression, metastasis, and cancer cell survival, including canonical eIF4E/TORC1 mediated cap-dependent mRNA translation, DAP5/eIF3d cap-dependent mRNA translation, IRES-mediated mRNA translation, and DAP5 or eIF4GI mediated mechanisms of re-initiation and leaky scanning.
Limitations of the study
There are several limitations that should involve additional follow-up studies. While we demonstrated that both eIF4G2 mRNA and its encoded DAP5 protein are increased in expression in pro-metastatic primary tumors and matched metastases of TNBC, it was a small patient cohort that needs to be expanded in future studies. In addition, our data only explored TNBC, not more prevalent hormone receptor positive breast cancers, which also needs to be examined. However, this is more challenging given typical 10 year adjuvant anti-endocrine treatment and lifetime recurrence rates. With respect to animal studies, it is likely that the vast majority of mRNAs that demonstrate DAP5-dependent translation also require eIF3d cap-binding activity, although this has been shown only for selected mRNAs. Finally, our findings relate to female breast cancer and have not involved much more rare male breast cancers.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Robert J. Schneider (Robert.schneider@nyulangone.org).
Materials availability
All materials and reagents used in this study, apart from patient tissue samples, are available upon request from the authors.
Data and code availability
All sequencing data have been deposited in the NCBI GEO database. Previously published and deposited data for MB-231 cells can be found under accession number GSE115142 which was involved in this study. New data for 4T1 cells generated in this study can be found under GSE188733. No software was generated for this project. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND STUDY PARTICIPANT DETAILS
Patient cohorts and human tissues
Archival breast tumor tissue biopsy specimens were obtained with prior Institutional Review Board approval for patients ≥18 years of age with hormone receptor negative (ER/PR) and Her2 receptor negative breast cancer (triple negative breast cancer, TNBC) at stages I-IV generally with invasive ductal carcinoma (IDC), either untreated or treated with neoadjuvant therapy (Table S1). Cases were obtained from the voluntary tissue donation registry at New York University (NYU) Langone Medical Center. A pathology database of all available breast cancer specimens was queried to identify cases with 8 years follow up that had a clinical description of TNBC. For all cases, clinical and survival data were obtained in a de-identified manner including age, date of clinical and pathological diagnosis, estrogen receptor (ER), progesterone receptor (PR), HER2 status, treatment history including details of systemic therapy, date of recurrence, date of last follow up and survival status. TNBCs were determined to be non-recurrent if there was no recurrence within 8 years of clinical diagnosis and treatment, in patients that had not been lost to follow up. Metastasis biopsy specimens (mostly lung, one brain) were obtained from the archive for patients who recurred within 8 years, for whom primary breast cancer specimens were available (matched metastasis).
Ethical compliance
All experiments involving live animals were carried out at NYU School of Medicine in full compliance with ethical regulations approved by the NYU Langone IACUC Committee. The protocol for collection of human breast cancer tissue specimens was approved by the NYU Langone Health/NYU School of Medicine Institutional Review Board and obtained from de-identified and consented individuals in compliance with all ethical regulations.
Cells and cell culture
The human breast cancer cell lines parental MDA-MB-231, MDA-MB-231 LM0 and MDA-MB-231 LM2, and the murine mammary carcinoma cell line 4T1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS, Laboratory Disposable Products, Towaco, NJ, USA) at 37°C in 5% CO2. Cell lines were authenticated by short tandem repeat profiling and routinely checked for mycoplasma contamination. Generation and characterization of MB-231 and 4T1 cells with stable inducible silencing of DAP5 (shDAP5) and non-silencing (shNsi) control are described below and in detail.18
Mouse tumor studies
All studies were approved by the NYU Grossman School of Medicine Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with IACUC guidelines. Female 6–8 week old NOD/SCID immunodeficient mice were used for human MB-231 tumors studies, and Balb/cJ mice for murine 4T1 tumor studies (Jackson Laboratories, Bar Harbor, ME). Mice were injected subcutaneously (s.c.) in the flank with 5×105 4T1 cells unless otherwise indicated in total volume of 200 μL with CORNING Matrigel matrix (Corning) or orthotopically in the fourth mammary fat pad with 5×106 MB-231 cells unless otherwise indicated. Mice were randomized to treatment groups to receive placebo or Dox in their drinking water. To obtain tumor growth curves, perpendicular length measurements and tumor volumes were calculated using the formula (π/6 x L x W2). Mouse tumor volumes were scored throughout the trial by precision caliper as indicated in figure legends. Mice were sacrificed and tumors excised for analysis at the times indicated in figure legends.
METHOD DETAILS
Immunohistochemistry and scoring of human breast cancer specimens
All specimens were analyzed using immunohistochemistry (IHC) and scored by two independent pathologists. Paraffin-embedded sections were warmed and serially de-paraffinized in Xylene and ethanol, introduced into an antigen unmasking solution, blocked and sections incubated overnight at 4°C with antibodies. Antigen retrieval requirements were first determined and then primary antibody was serially diluted to determine the optimum dilution/concentration. Briefly, sections were deparaffinized in xylene (3 changes), rehydrated through graded alcohols (3 changes 100% ethanol, 3 changes 95% ethanol) and rinsed in distilled water. Heat induced epitope retrieval was performed in 10 mM citrate buffer pH 6.0 in a 1200-Watt microwave oven at 90% power. Sections were allowed to cool for 30 min and then rinsed in distilled water. Antibody incubations and detection were carried out on a NEXes platform (Ventana Medical Systems Tucson, AZ USA) using Ventana’s buffer and detection system unless otherwise noted. Levels of DAP5 protein were detected using DAP5 antibody. Control studies included staining in the absence of primary antibody, titration of primary antibody to determine minimum amount required for strongest signal to noise ratio, and normalization to MB-231 cells that were pelleted, embedded in paraffin, sectioned and stained for DAP5, and included in tumor specimen staining. Specimens were scored using traditional categorical standards that reflect the range of expression and allowed the middle quantile to represent the median of the data spread, assuring a full dynamic range.57 Quartiles were therefore used for scoring DAP5 which had a large dynamic range. Quartile scoring used: 1+: <5%; 2+ ≥6% but <33%; 3+ >33% but <65%; 4+ >66%. Scoring was carried out in a blinded manner by a board-certified breast cancer pathologist.
Aseptic survival surgery
Animals were anesthetized by isoflurane inhalation, first be placed into an induction box, with 2.0–3.0% isoflurane delivered in 100% oxygen. Once the animal was anesthetized, it was removed from the induction box and attached to a nose-cone mask and anesthesia maintained with 1.0–3.0% isoflurane in 100% oxygen. Depth of anesthesia was continuously monitored using animal toepinch, palpebral reflex, and respiration rate. Animals were lain ventral side up and kept warm by indirect heating with a heat lamp, provided during and after the surgery. The surgical site was denuded of fur, the skin scrubbed with Betadine solution, then 70% ethanol. Beginning from the center to the incision site and working out toward the perimeter, a surgical excision was made with the surgical site covered with a sterile drape and using sterile autoclaved equipment. A Germinator (hot beads sterilization) was used to sterilize instruments between surgical procedures on multiple animals. The incision did not exceed 0.5 to 1 cm and was cleaned with Betadine before closure. Stainless steel wound clips or Vetbond glue was used to close the incision. After surgery, animals were placed in a clean cage with indirect heat from a heat lamp to avoid hypothermia and to avoid over-heating, and monitored closely until recovery from anesthesia.
Bioluminescent imaging of tumors
Tumor growth was assessed using bioluminescent imaging (IVIS Lumina III) until flux over the defined region of interest (ROI) of the entire mouse abdomen was in the approximate order of magnitude of 108 photons/second. For imaging luciferase, animals were injected by IP with 200 μL of luciferin (diluted in PBS per manufacturer instructions). For imaging RFP no additional preparation was necessary. Animals were imaged in groups of 3. Images were analyzed using Living Image software. The ROI was defined as an ellipse surrounding the mouse abdomen. Flux was calculated within the ROI per mouse and averaged across all mice within each treatment group. Average flux was compared across groups over time and normalized to baseline flux levels prior to initiation of treatment (Dox). This allowed for assessment of relative tumor growth, independent of starting volume. Mice were sacrificed if they appeared ill as per IACUC protocol. To image lungs ex vivo, immediately after surgical excision of lungs, lungs were inflated with 1x cold PBS prior to imaging.
DAP5 short hairpin RNA (shRNA) silencing
Lentiviruses were produced by transient transfection of human embryonic kidney (HEK) 293FT cells with individual lentiviral vectors containing the pTRIPZ Doxycycline (Dox)-inducible transgene with packaging plasmids pMD2G and psPAX2 (Addgene, Cambridge, MA, USA) using Lipofectamine 2000 (Promega, Madison, WI, USA). Supernatants containing viral particles were collected 48 h post-transfection, filtered through 0.45 μm filters and viral particles concentrated using the PEG-itTM Virus Precipitation Solution Kit (System Biosciences, Mountain View, CA, USA). Cloning of the shRNA cassette sequences for eIF4G2/DAP5 (5′-TACCTCTAGTAATGGGCTTTA-3′) and non-silencing sequence control Nsi (5′-AATTCTCCGAACGTGTCACGT-3′) were previously described.18 Cells were infected with viral particles, selected with puromycin (1 μg/mL), and transformed with reporter plasmid expressing tRFP (Turbo Red Fluorescent Protein). The lentiviral vectors used for shNsi and shDAP5 contains a constitutively expressed transcript encoding the puromycin resistance gene, and a doxycycline-inducible cassette that expresses target shRNA sequence as well as the reporter protein tRFP (Turbo Red Fluorescent Protein). Cells were treated with 0.1–2 μg/mL of Dox and FACs isolated gating on RFP+. Silencing of DAP5 expression was confirmed by qRT-PCR and immunoblot analysis.
Cell viability/proliferation assays
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed using the CellTiter 96 Non-Radioactive Cell Proliferation Assay according to manufacturer instructions (Promega, Madison, WI, USA). Cells were seeded onto 96-well plates at an optimal density of 5×103 cells per well and incubated overnight at 37°C. During a period of 1–6 d cells were treated with Dox (1 μg/mL for MB-231, 0.5 μg/mL for 4T1) to measure the effect of silencing DAP5 on cell proliferation. Cells were treated with 15 μL MTT dye and cultures were re-incubated for an additional 4 h. Following removal of the supernatant, 100 μL of the Solubilization Solution/Stop Mix was added to each well to completely dissolve the crystals and absorbance was measured at 570 nm using a 96-well plate reader.
Matrigel invasion assay
Invasion assays were performed using Corning BioCoat Matrigel Invasion Chamber with Corning Matrigel Matrix (Thermo Fisher Scientific) per manufacturer instructions. After Dox treatment as described above, a suspension of 1×105 cells in 500 μL of serum-free media were added to the upper chambers, and 10% FBS added to the lower chambers. After 22 h 37°C, 5% CO2, cells that had migrated through the membrane were fixed with methanol and stained with 1% of crystal violet. Invading cells per field were scored and performed in triplicate (n = 12) using a ZEISS Axio light microscope (100× magnification) and ZEISS software (Carl Zeiss Meditec, Inc., San Diego, CA, USA).
In vitro wound healing assay
Cell migration was measured using CytoSelect Wound Healing Inserts (Cell Biolabs, Inc., San Diego, CA, USA). After treatment with Dox as above, cells were collected in a suspension containing 5×105 cells/mL, dispensed into inserts, incubated at 37°C in 5% CO2 overnight, inserts removed, media from wells slowly aspirated, wells washed with media, new media added and wound healing closure was quantified at 0, 6, 12, and 24 h with a digital inverted microscope (40× magnification) and imaging software (EVOS FL Imaging System, Thermo Fisher Scientific). The total surface area was measured by 0.9 mm x length (mm). The migrated cell surface area was measured by the length of cell migration (mm) x 2 x length (mm). The percent of closure measured by Percent Closure (%) = Migrated Cell Surface Area/Total Surface Area x 100.
Immunoblot analysis and antibodies
Total protein was extracted by RIPA buffer extraction (150 mM NaCl, 50 mM Tris pH 8.0, 1% NP-40) containing Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), Pierce Protease Inhibitor Tablet (Thermo Fisher Scientific), clarified by microfuge centrifugation at 4°C, and protein concentration determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of denatured protein lysates were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane, blocked with 5% Bovine Serum Albumin (BSA) for 1 h and incubated at 4°C overnight with primary antibodies at 1:1000 dilution against DAP5 (610742; BD Biosciences, Franklin Lakes, NJ, USA), eIF3D (A301–758A; Bethyl Laboratories Inc., Montgomery, TX, USA), E-Cadherin (3195; Cell Signaling Technology), Claudin-1 (ab15098; Abcam, Cambridge, UK), Snail1 (3895; Cell Signaling Technology), Snail2/Slug (9585; Cell Signaling Technology, Danvers, MA, USA), Twist1 (ab50581; Abcam, Cambridge, UK), Zeb1 (A00548–1; BosterBio, Pleasanton, CA, USA), N-Cadherin (610920; BD Biosciences), Vimentin (ab20346; Abcam), eIF4GI (2858, Cell Signaling Technology), MMP1 (ab137332, Abcam), MMP3 (ab52915, Abcam), GAPDH (2118S, Cell Signaling Technology), β-actin (4967; Cell Signaling Technology). All antibodies were diluted in 5% BSA. Proteins were visualized using ECL and anti-Rabbit IgG or anti-Mouse IgG, Horseradish Peroxidase (HRP) linked whole antibody (Catalog no., NA934V and NA931V, respectively; GE Healthcare Bio-sciences Corp., Piscataway, NJ, USA) diluted to 1:10,000 in 1x TBS-T for 1 h at room temperature. Results were quantified using ImageJ software.
Cloning and expression of DAP5/eIF4G2 cDNA
An eIF4G2 cDNA32 was subcloned from a pCDNA3 expression vector into a pBABE retroviral vector to produce pBABE-DAP5 using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB #E5520) and the following primers, according to manufacturer instructions:
eIF4G2-F: 5′-CCAGTGTGGTGGTACGTAGGGTGGAGAGTGCGATTGCAG-3′
eIF4G2-R: 5′-CTGACACACATTCCACAGGGTTAGTCAGCTTCTTCCTCTGATTC-3′.
Plasmids were verified by DNA sequence analysis. 293FT cells were transfected using calcium phosphate precipitation with psPAX2, pMD2.G and pBABE-DAP5 to produce DNA packaged lentiviral particles. Parental MDA-MB-231 cells were infected in the presence of Polybrene. Stable cells lines were generated using puromycin selection 48 h post-infection.
qRT-PCR
Total RNA was extracted from cells using Invitrogen TRIzol Reagent according to manufacturer instructions. RNA concentration and purity were quantified using a NanoDrop Spectrophotometer (Thermo Fisher Scientific). First-strand cDNA was synthesized from 500 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qRT-PCR (qPCR) analysis was performed using 500 ng cDNA, 2 μM primers, and Maxima SYBR Green qPCR Master Mix (2X) (Bio-Rad Laboratories). Samples were analyzed with an Applied Biosystems 7500 Real-Time PCR Systems (Thermo Fisher Scientific).
Anoikis resistance assay
4T1 cells were cultured in DMEM-F12 medium (Corning), supplemented with 5% horse serum (Thermo Fisher Scientific), 0.4% BSA, 5 μg/mL insulin, 40 ng/mL mouse basic fibroblast growth factor (bFGF), 20 ng/mL mouse epidermal growth factor (EGF), 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Thermo Fisher Scientific). MB-231 cells were cultured in DMEM/F12 medium supplemented with B27 (1:50, Thermo Fisher Scientific), 20 ng/mL human bFGF, 20 ng/mL human EGF, 4 μg/mL heparin solution (StemCell Technologies, Vancouver, Canada), 1% antibiotic-antimycotic agent (Thermo Fisher Scientific), and 15 μg/mL gentamicin. Induction of apoptosis by cell detachment (anoikis) from the extracellular matrix (ECM) was determined with and without silencing. Cells were seeded onto ultra-low attachment plates (Corning) at 37°C in 5% CO2, harvested 24 h and 48 h post-transfection, washed twice with 1X Phosphate Buffer Saline (PBS) and 1×105 cells were resuspended in 100 μL binding buffer (0.2 μm sterile filtered 0.1 M HEPES (pH 7.4), 1.4 M NaCl, and 25 mM CaCl2 solution), and cell suspensions incubated with 5 μL Annexin V-FITC and 1x DAPI Fluorescent Stain (#112002; Cell Biolabs, Inc.) for 15 min at room temperature in the dark. The cells were evaluated immediately with a Becton Dickinson BDLSR II UV flow cytometry cell analyzer (Becton, Dickinson and Company, USA). The results were quantified using FlowJo software (Tree Star, Inc., Ashland, OR, USA).
RNA sequencing, data analysis
Sucrose gradient polysome fractions containing 4 or more ribosomes (considered well-translated) were pooled and extracted RNA quality measured by a Bioanalyzer (Agilent Technologies). RNA-seq was carried out by the NYU School of Medicine Genome Technology Core using an Illumina HiSeq 4000 instrument at sequencing depth of 50–60 million reads. Paired end reads were removed from linker and poyA sequences using cutadapt and removed from rRNA reads by Bowtie (ver. 1.2.3) alignment to a rRNA library. Low-quality reads (less than 20) were trimmed with Trimmomatic58 (version 0.36), with the reads lower than 35 nt excluded. The resulting sequences were aligned with STAR59 (version 2.6.0a) to the murine GRCm38 or human hg38 reference genomes in single-end mode. The alignment results were sorted with SAMtools60 (version 1.9), then supplied to HTSeq61 (version 0.10.0) to obtain feature counts. The feature counts tables from different samples were concatenated with a custom R script. To examine differences in transcription and translation, total mRNA and (≥4 ribosome) polysome mRNA were quantile-normalized separately. Regulation by transcription and translation, and accompanying statistical analysis, was performed using the online tool RIVET,62 where significant genes/mRNAs were identified as adjusted p-values q < 0.05. Briefly, we used the Benjamin-Hochberg adjustment63 to reduce the FDR by calculating for each gene an adjusted p value assuming significant all genes with an adjusted p value less than or equal to the gene’s adjusted p value. We analyzed translational efficiency (TE) using the interaction method, analyzing each dataset separately. The statistical model we used is defined as: ~0+ type+ treatment + type:treatment. To perform analysis in DAVID, we also used a more stringent approach setting a cut-off of 0.65 in log fold change on top of q < 0.05. We also used this output to perform comparison analysis in IPA.
RNA sequencing raw data, read counts, read alignments, RNAseq for total mRNA, ≥4 ribosome mRNA, p values, q-values and fold changes developed in this study for 4T1 cells are available at NCBI Genome Expression Omnibus (GEO) accession numbers GSE188733, and previously developed comparative study for MB-231 cells GSE115142. Analyzed transcriptomic/translatomic data analysis are available in Data set S1. TE was quantified from ≥4 ribosome polysome mRNA/total mRNA for each individual mRNA, adjusted for multiple testing using Benjamin and Hochberg adjustment parameters. TE represents authentic translation activity not driven by mRNA abundance and secondary effects.
Reactome pathway analysis was performed on genes/mRNAs that were up- and down-regulated by transcription and translation using Metascape.64 Pathway analysis and enrichment plots of the top genes/mRNAs that were the most regulated by transcription and/or translation were generated using DAVID65 and Metascape. Prediction of transcription factors of the same list of top genes/ mRNAs was performed using Enrichr66 (TRANSFAC and JASPER PWM program) and PASTAA67 online tools.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Distant metastasis-free (recurrence-free) survival (RFS) and overall survival (OS) analysis of human data were determined from the clinical diagnosis data. Human RFS and OS were analyzed using the Kaplan–Meier method with point estimates and 95% confidence intervals (CIs) calculated from Kaplan–Meier curves. Differences in DAP5 protein expression levels in breast cancer specimens used Fisher’s Exact Test. Animal survival data used the unpaired non-parametric log rank Mantel-Cox test for survival determination.
Statistical significance in other studies was assessed by two-tailed Student’s t test for unpaired experimental values or one-way or two-way analysis of variance (ANOVA) tests with Dunnett’s post-ANOVA test determination for analysis of repeated measures as indicated in figure legends. Data are expressed as indicated in figure legends as means with standard error of means (SEM), and when appropriate corrected for sample sizes using Bonferroni corrections to adjust alpha values. Significance difference was defined as p < 0.05. Statistical analyses were performed using GraphPad 7 and 8 software (GraphPad Software, Inc., San Diego, CA, USA).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER/RRID |
|---|---|---|
| Antibodies | ||
|
| ||
| Mouse monoclonal anti-human/mouse DAP5 antibody | BD Biosciences | Cat# 610291/RRID:AB_397685 |
| Mouse monoclonal anti-human/mouse DAP5 antibody | BD Biosciences | Cat# 610742/RRID:AB_398065 |
| Rabbit polyclonal anti-human/mouse eIF3d antibody | Bethyl Laboratories Inc | Cat# A301-758A/RRID:AB_1210970 |
| E-Cadherin 24E10 rabbit monoclonal human/mouse antibody | Cell Signaling Technology | Cat# 3195/RRID:AB_2291471 |
| Mouse monoclonal anti-human/mouse Snail1 antibody | Cell Signaling Technology | Cat# 3895/RRID:AB_2191759 |
| Rabbit monoclonal anti-human/mouse Snail2/Slug antibody | Cell Signaling Technology | Cat# 9585/RRID:AB_2239535 |
| Rabbit polyclonal anti-human/mouse β-actin antibody | Cell Signaling Technology | Cat# 4967/RRID:AB_330288 |
| Rabbit polyclonal anti-human/mouse Claudin-1 antibody | Abcam | Cat# ab15098/RRID:AB_301644 |
| Rabbit polyclonal anti-human/mouse Twist1 antibody | Abcam | Cat# ab50581/RRID:AB_883292 |
| Rabbit polyclonal anti-human/mouse Zeb1 antibody | BosterBio | Cat# A00548-1/RRID N/A |
| Mouse monoclonal anti-human/mouse N-Cadherin antibody | BD Biosciences | Cat# 610920/RRID:AB_2077527 |
| Mouse monoclonal anti-human/mouse vimentin antibody | Abcam | Cat# ab20346/RRID:AB_445527 |
| Donkey anti-Rabbit IgG HRP polyclonal antibodies | GE Healthcare Bio- sciences | Cat# NA934V/RRID N/A |
| Sheep anti-mouse IgG HRP polyclonal antibodies | GE Healthcare Bio- sciences | Cat# NA931V/RRID:AB_772210 |
| Rabbit polyclonal anti-human/mouse eIF4GI antibody | Cell Signaling Technology | Cat# 2858/RRID:AB_2095745 |
| Rabbit monoclonal anti-human/mouse GAPDH antibody | Cell Signaling Technology | Cat# 2118S/RRID:AB_561053 |
| Rabbit polyclonal anti-human/mouse MMP1 antibody | Abcam | Cat# ab137332/RRID:AB_2889296 |
| Rabbit polyclonal anti-human/mouse MMP3 antibody | Abcam | Cat# ab52915/RRID:AB_881243 |
| Mouse monoclonal anti-human CD31 antibody | Abcam | Cat# ab9498/RRID:AB_307284 |
|
| ||
| Biological samples | ||
|
| ||
| Human breast cancer tumor tissues | NYU Langone tissue | N/A |
| Repository | ||
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| human bFGF | StemCell Technologies | Cat# 78003 |
| human EGF | StemCell Technologies | Cat#78006 |
| Heparin solution | StemCell Technologies | Cat# 07980 |
| DAPI fluorescent stain | Cell Biolabs, Inc. | Cat# 112002 |
| D-luciferin | Perkin-Elmer | Cat# 122799 |
| Halt™ Phosphatase inhibitor Cocktail | ThermoFisher | |
| Pierce Protease inhibitor Tablet | ThermoFisher | |
| Lipfectamine™ 2000 | Promega | Cat# 11668019 |
| PEG-itTM Virus precipitation solution Kit | System Biosciences | Cat# LV825A-1 |
|
| ||
| Critical commercial assays | ||
|
| ||
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher | Cat# 4368814 |
| Maxima SYBR Green qPCR Master Mix | Bio-Rad Laboratories | K1070 |
| FITC Annexin V apoptosis kit | BD Biosciences | Cat# 556547 |
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat# 23228 |
| CytoSelect™ Wound Healing Inserts | Cell Biolabs, Inc. | Cat# CBA-120 |
| Corning BioCoat Matrigel Invasion Chamber | ThermoFIsher | Cat# 08–774-122 |
| CellTiter 96® Non-Radioactive Cell Proliferation MTT Assay | Promega | Cat# G9241 |
|
| ||
| Deposited data | ||
|
| ||
| GSE188733 | N/A | |
| GSE115142 | N/A | |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: MDA-MB-231 cells | ATCC | Cat# CRM-HTB-26/RRID:CVCL_006 |
| Murine: 4T1 | ATCC | Cat# CRL-3407/RRID:CVCL_GR31 |
| Human: HEK 293FT | Cellosaurus | RRID:CVCL_6911 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Balb/cJ mice | Jackson Labs | Cat# 000651 |
| NOD SCID mice | Jackson Labs | Cat# 001303 |
|
| ||
| Oligonucleotides (5′ to 3′ shown) | ||
|
| ||
| See Table S4: Oligonucleotides | N/A | |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMD2G | Addgene | Cat# 12259 |
| pTRIPZ | Addgene | Cat# 127696 |
| pMD2G | Addgene | Cat# 12259 |
| psPAX2 | Addgene | Cat# 12260 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 8 & 9 software | GraphPad | https://www.graphpad.com |
| FlowJo | FLowJo | https://www.flowjo.com |
| EVOS™ FL Imaging System | ThermoFisher | https://www.thermofisher.com/us/en/home/technical-resources/software-downloads/evos-fl-cell-imaging-system.html |
Highlights.
Two cap-dependent mRNA translation mechanisms promote tumor progression
eIF4E/mTORC1 is required for cancer cell proliferation and survival
DAP5/eIF3d is required for the cancer cell EMT, cell migration, and metastasis
Inhibition of DAP5 strongly reduces metastasis and survival of metastases
ACKNOWLEDGMENTS
New York University Medical Center (NYUMC) core services were supported by NCI grant P30CA016087 and National Center for Advancing Translational Sciences (NCATS) grant UL1TR00038. This work was supported by BCRF-21-146, NCI grants R01CA178509 and R01CA248397 (R.J.S.), American Cancer Society (ACS) grant PF-16-095-01 (C.D.P.), and ACS grant PF20-031-01 (T.R.-F.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112646.
REFERENCES
- 1.Silvera D, Formenti SC, and Schneider RJ (2010). Translational control in cancer. Nat. Rev. Cancer 10, 254–266. 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 2.Robichaud N, Sonenberg N, Ruggero D, and Schneider RJ (2019). Translational control in cancer. Cold Spring Harbor Perspect. Biol. 11, a032896. 10.1101/cshperspect.a032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truitt ML, and Ruggero D. (2016). New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304. 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robichaud N, and Sonenberg N. (2017). Translational control and the cancer cell response to stress. Curr. Opin. Cell Biol. 45, 102–109. 10.1016/j.ceb.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Lee LJ, Papadopoli D, Jewer M, Del Rincon S, Topisirovic I, Lawrence MG, and Postovit LM (2021). Cancer plasticity: the role of mRNA translation. Trends Cancer 7, 134–145. 10.1016/j.trecan.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bera A, and Lewis SM (2020). Regulation of epithelial-to-mesenchymal transition by alternative translation initiation mechanisms and its implications for cancer metastasis. Int. J. Mol. Sci. 21, 4075. 10.3390/ijms21114075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YN, Koo KH, Sung JY, Yun UJ, and Kim H. (2012). Anoikis resistance: an essential prerequisite for tumor metastasis. Int. J. Cell Biol. 2012, 306879. 10.1155/2012/306879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson CD, Anyiwe K, and Schimmer AD (2008). Anoikis resistance and tumor metastasis. Cancer Lett. 272, 177–185. 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 9.Shibue T, and Weinberg RA (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629. 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto MA, Huang RYJ, Jackson RA, and Thiery JP (2016). Emt: 2016. Cell 166, 21–45. 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Jin L, Chun J, Pan C, Kumar A, Zhang G, Ha Y, Li D, Alesi GN, Kang Y, Zhou L, et al. (2018). The PLAG1-GDH1 Axis promotes anoikis resistance and tumor metastasis through CamKK2-AMPK signaling in LKB1-deficient lung cancer. Mol. Cell. 69, 87–99.e7. 10.1016/j.molcel.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topisirovic I, and Sonenberg N. (2011). mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harbor Symp. Quant. Biol. 76, 355–367. 10.1101/sqb.2011.76.010785. [DOI] [PubMed] [Google Scholar]
- 13.Sonenberg N, and Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. (2016). nanoCAGE reveals 5’ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 26, 636–648. 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu J, and Pelletier J. (2015). Targeting the eIF4A RNA helicase as an anti-neoplastic approach. Biochim. Biophys. Acta 1849, 781–791. 10.1016/j.bbagrm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Feoktistova K, Tuvshintogs E, Do A, and Fraser CS (2013). Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. USA 110, 13339–13344. 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvera D, Formenti SC, and Schneider RJ (2010). Translational control in cancer. Nat. Rev. Cancer 10, 254–266. 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 18.de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, and Schneider RJ (2018). A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 9, 3068. 10.1038/s41467-018-05539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee AS, Kranzusch PJ, Doudna JA, and Cate JHD (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99. 10.1038/nature18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hundsdoerfer P, Thoma C, and Hentze MW (2005). Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 102, 13421–13426. 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, and Kimchi A. (2008). DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell. 30, 447–459. 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, and Kimchi A. (2014). The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 33, 611–618. 10.1038/onc.2012.626. [DOI] [PubMed] [Google Scholar]
- 23.Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, and Gelmon K. (2010). Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 28, 3271–3277. 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 24.Strasser-Weippl K, Badovinac-Crnjevic T, Fan L, and Goss PE (2013). Extended adjuvant endocrine therapy in hormone-receptor positive breast cancer. Breast 22 (Suppl 2), S171–S175. 10.1016/j.breast.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 25.Rosen PR, Groshen S, Saigo PE, Kinne DW, and Hellman S. (1989). A long-term follow-up study of survival in stage I (T1N0M0) and stage II (T1N1M0) breast carcinoma. J. Clin. Oncol. 7, 355–366. 10.1200/JCO.1989.7.3.355. [DOI] [PubMed] [Google Scholar]
- 26.Voduc KD, Cheang MCU, Tyldesley S, Gelmon K, Nielsen TO, and Kennecke H. (2010). Breast cancer subtypes and the risk of local and regional relapse. J. Clin. Oncol. 28, 1684–1691. 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, and Schneider RJ (2008). eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 181, 293–307. 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibue T, Brooks MW, and Weinberg RA (2013). An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell 24, 481–498. 10.1016/j.ccr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamper AM, Fleming RH, Ladd KM, and Lee ASY (2020). A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science 370, 853–856. 10.1126/science.abb0993. [DOI] [PubMed] [Google Scholar]
- 30.Gillis LD, and Lewis SM (2013). Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene 32, 3598–3605. 10.1038/onc.2012.371. [DOI] [PubMed] [Google Scholar]
- 31.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, and Massagué J. (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524. 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SH, and McCormick F. (2006). p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 25, 4008–4019. 10.1038/sj.emboj.7601268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evdokimova V, Tognon CE, and Sorensen PHB (2012). On translational regulation and EMT. Semin. Cancer Biol. 22, 437–445. 10.1016/j.semcancer.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ, and Sorensen PHB (2009). Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15, 402–415. 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Ren M, Zhou C, Liang H, Wang X, and Xu L. (2015). RNAi-mediated silencing of EIF3D alleviates proliferation and migration of glioma U251 and U87MG cells. Chem. Biol. Drug Des. 86, 715–722. 10.1111/cbdd.12542. [DOI] [PubMed] [Google Scholar]
- 36.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, and Beug H. (2006). ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–239. 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 37.Petz M, Kozina D, Huber H, Siwiec T, Seipelt J, Sommergruber W, and Mikulits W. (2007). The leader region of Laminin B1 mRNA confers cap-independent translation. Nucleic Acids Res. 35, 2473–2482. 10.1093/nar/gkm096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhury A, Hussey GS, and Howe PH (2011). 3’-UTR-mediated post-transcriptional regulation of cancer metastasis: beginning at the end. RNA Biol. 8, 595–599. 10.4161/rna.8.4.16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, and Howe PH (2010). TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12, 286–293. 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo B, Wu S, Zhu X, Zhang L, Deng J, Li F, Wang Y, Zhang S, Wu R, Lu J, and Zhou Y. (2020). Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J. 39, e102190. 10.15252/embj.2019102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebright RY, Lee S, Wittner BS, Niederhoffer KL, Nicholson BT, Bardia A, Truesdell S, Wiley DF, Wesley B, Li S, et al. (2020). Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473. 10.1126/science.aay0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, et al. (2019). RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10, 2065. 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Jewer M, Lee L, Leibovitch M, Zhang G, Liu J, Findlay SD, Vincent KM, Tandoc K, Dieters-Castator D, Quail DF, et al. (2020). Translational control of breast cancer plasticity. Nat. Commun. 11, 2498. 10.1038/s41467-020-16352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellen CU, and Sarnow P. (2001). Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 45.Holcik M, Sonenberg N, and Korneluk RG (2000). Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16, 469–473. [DOI] [PubMed] [Google Scholar]
- 46.Badura M, Braunstein S, Zavadil J, and Schneider RJ (2012). DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc. Natl. Acad. Sci. USA 109, 18767–18772. 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS, et al. (2007). Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J. Clin. Invest. 117, 2638–2648. 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh AC, and Ruggero D. (2010). Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin. Cancer Res. 16, 4914–4920. 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Parra C, Walters BA, Geter P, and Schneider RJ (2018). Translation initiation factors and their relevance in cancer. Curr. Opin. Genet. Dev. 48, 82–88. 10.1016/j.gde.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, and Polunovsky VA (2004). Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 5, 553–563. 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 51.Roux PP, and Topisirovic I. (2018). Signaling pathways involved in the regulation of mRNA translation. Mol. Cell Biol. 38, 000700–e118. 10.1128/MCB.00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SMH, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., and Pelletier J. (2008). Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118, 2651–2660. 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoffe Y, David M, Kalaora R, Povodovski L, Friedlander G, Feldmesser E, Ainbinder E, Saada A, Bialik S, and Kimchi A. (2016). Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 30, 1991–2004. 10.1101/gad.285239.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, and Sonenberg N. (2015). DAP5 associates with eIF2beta and eIF4AI to promote Internal Ribosome Entry Site driven translation. Nucleic Acids Res. 43, 3764–3775. 10.1093/nar/gkv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smirnova VV, Shestakova ED, Nogina DS, Mishchenko PA, Prikazchikova TA, Zatsepin TS, Kulakovskiy IV, Shatsky IN, and Terenin IM (2022). Ribosomal leaky scanning through a translated uORF requires eIF4G2. Nucleic Acids Res. 50, 1111–1127. 10.1093/nar/gkab1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber R, Kleemann L, Hirschberg I, Chung M-Y, Valkov E, and Igreja C. (2021). DAP5 enables translation re-initiation on structured messenger RNAs. Preprint at bioRxiv. 10.1101/2021.01.21.427569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyndman RJ, and Fan Y. (1996). Sample quantiles in statistical packages. Am. Statistician 50, 361–365. [Google Scholar]
- 58.Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Pyl PT, and Huber W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ernlund AW, Schneider RJ, and Ruggles KV (2018). RIVET: comprehensive graphic user interface for analysis and exploration of genome-wide translatomics data. BMC Genom. 19, 809. 10.1186/s12864-018-5166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, and Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B 57, 289–300. [Google Scholar]
- 64.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 66.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roider HG, Kanhere A, Manke T, and Vingron M. (2007). Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics 23, 134–141. 10.1093/bioinformatics/btl565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited in the NCBI GEO database. Previously published and deposited data for MB-231 cells can be found under accession number GSE115142 which was involved in this study. New data for 4T1 cells generated in this study can be found under GSE188733. No software was generated for this project. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.