Abstract
Previous in vitro studies have shown that initiation of transcription of ribosomal DNA (rDNA) in the yeast Saccharomyces cerevisiae involves an interaction of upstream activation factor (UAF) with the upstream element of the promoter, forming a stable UAF-template complex; together with TATA-binding protein (TBP), UAF then recruits an essential factor, core factor (CF), to the promoter, forming a stable preinitiation complex. TBP interacts with both UAF and CF in vitro. In addition, a subunit of UAF, Rrn9p, interacts with TBP in vitro and in the two-hybrid system, suggesting the possible importance of this interaction for UAF function. Using the yeast two-hybrid system, we have identified three mutations in RRN9 that abolish the interaction of Rrn9p with TBP without affecting its interaction with Rrn10p, another subunit of UAF. Yeast cells containing any one of these individual mutations, L110S, L269P, or L274Q, did not show any growth defects. However, cells containing a combination of L110S with one of the other two mutations showed a temperature-sensitive phenotype, and this phenotype was suppressed by fusing the mutant genes to SPT15, which encodes TBP. In addition, another mutation (F186S), which disrupts both Rrn9p-TBP and Rrn9p-Rrn10p interactions in the two-hybrid system, abolished UAF function in vivo, and this mutational defect was suppressed by fusion of the mutant gene to SPT15 combined with overexpression of Rrn10p. These experiments demonstrate that the interaction of UAF with TBP, which is presumably achieved by the interaction of Rrn9p with TBP, is indeed important for high-level transcription of rDNA by RNA polymerase I in vivo.
Transcription of ribosomal DNA (rDNA) by RNA polymerase I (Pol I) in Saccharomyces cerevisiae (referred to as yeast in this paper) utilizes at least four factors other than Pol I: upstream activation factor (UAF) (which includes Rrn5p, Rrn9p, Rrn10p, and at least two more proteins) (11), core factor (CF) (which contains Rrn6p, Rrn7p, and Rrn11p) (12, 15, 17), TATA-binding protein (TBP) (6, 23, 25), and Rrn3p (30). Like other eukaryotic rDNA promoters studied previously, the yeast rDNA promoter consists of two cis elements, the upstream element and the core promoter. The upstream element is required for a high level of transcription but is dispensable for a basal level of transcription, whereas the core promoter is essential for the accurate initiation of transcription (5, 11, 14, 18). Our previous in vitro studies have demonstrated that UAF interacts with the upstream element of the rDNA promoter, forming a stable UAF-template complex and committing the template to transcription. UAF then recruits CF to the promoter, and TBP is required for this recruitment to form a stable preinitiation complex containing UAF, CF, and presumably TBP. Finally, with the aid of Rrn3p, Pol I joins this preinitiation complex, and the system becomes ready for transcription initiation (11, 25, 30).
In agreement with the conclusion that UAF mediates the stimulatory function of the upstream element, UAF is not required for the basal level of transcription from the template missing the upstream element in vitro, and the genes (RRN5, RRN9, and RRN10) for the three characterized subunits of UAF (Rrn5p, Rrn9p, and Rrn10p) are not absolutely required for a very low level of in vivo rRNA transcription and cell viability. In contrast, the genes (RRN6, RRN7, and RRN11) encoding the three subunits of CF are essential for cell viability and appear to be absolutely required for rDNA transcription by Pol I both in vitro and in vivo (11). Thus, UAF appears to be analogous to activator proteins that mediate stimulatory functions of upstream elements in many genes transcribed by RNA polymerase II (Pol II). Extensive studies on the activation of Pol II transcription have shown that activators in general contain a DNA-binding domain (DBD) and one or more activation domains (ADs) and that stimulation of transcription appears to take place through interactions of the AD(s) with one or more components of the general Pol II transcription machinery. Such interactions would stimulate recruitment of the general transcription factors to the promoter or, alternatively, change the conformation of an inactive transcriptional machinery (or its subassembly) on the promoter to its active form (for reviews, see references 9, 22, 26, and 28).
We have previously shown that TBP interacts specifically with both UAF and CF in vitro, with the interaction with UAF being stronger than that with CF (25). We have also shown that a subunit of UAF, Rrn9p, interacts strongly with TBP in vitro and in the yeast two-hybrid system in vivo and have suggested that this interaction might be important for the function of UAF to recruit CF, leading to a high level of rDNA transcription (25). However, as often noted in connection with studies on Pol II transcriptional activators, interactions between activators and general transcription factors such as TBP observed in vitro do not necessarily mean that these interactions operate in vivo (see, e.g., reference 27). Interactions observed in the yeast two-hybrid system also do not guarantee a functional significance of the interactions in vivo. In this paper, we describe the results of mutational analysis of Rrn9p-TBP interactions designed to examine the functional significance of the observed interactions. The results demonstrate that the interaction of UAF with TBP, which is presumably mediated by the interaction of the Rrn9p subunit with TBP, is indeed important for the activated transcription of rDNA by Pol I in vivo.
MATERIALS AND METHODS
Media, strains, and plasmids.
YEP-glucose medium, YEP-galactose medium, synthetic galactose medium, and synthetic glucose medium were used to grow yeast cells as described previously (12, 19). The strains and plasmids used in this study are listed in Table 1. Restriction enzyme sites within RRN9, which were used for plasmid construction, are shown in Fig. 1. Plasmids used to study protein-protein interactions with the yeast two-hybrid system were prepared as derivatives of pAS2, pACT2, or pGAD424. For construction of pNOY410, the 435-bp coding region of RRN10 from pNOY337 (11) was inserted in frame between the SmaI and SalI sites of pGAD424. pNOY411 and pNOY412 were constructed by ligating the 1.1-kb NcoI/BamHI fragments from pNOY3271 and pNOY3272, respectively, into the NcoI and BamHI sites of pAS2. Plasmids pNOY413, pNOY414, and pNOY415 were generated by PCR mutagenesis with pNOY355 as described below. Plasmid pNOY417 was constructed by ligating the 2.2-kb BamHI/SacI fragment from pNOY337 (11) containing RRN10 into the BamHI and SacI sites of YEp351. Plasmid pNOY420 was constructed by inserting a 1.7-kb SnaBI/NaeI fragment encoding triple-HA1-tagged Rrn9p from pNOY332 (11) between a blunt-ended XhoI site and the NaeI site of pRS314. Plasmids pNOY421, pNOY422, and pNOY423 were constructed by replacing the wild-type 435-bp SalI/EcoRI fragment in pNOY420 with the corresponding mutant SalI/EcoRI fragments from pNOY3271, pNOY3272, and pNOY413, respectively. Plasmids pNOY424 and pNOY425 were similarly constructed by replacing the wild-type 508-bp EcoI/BamHI fragment in NOY420 with the corresponding EcoRI/BamHI fragments from pNOY414 and pNOY415, respectively. Plasmids pNOY426 and pNOY427 were constructed by ligating the 660-bp BglII fragments from pNOY414 and pNOY415, respectively, into the place of the wild-type BglII fragment in pNOY423. Plasmids pNOY428, pNOY429, and pNOY430 were constructed from pNOY426, pNOY427, and pNOY422, respectively, by ligating a 720-bp BamHI PCR fragment encoding TBP (which was made with the oligonucleotide primers 5′-GGA TTC ATG GCC GAT GAG GAA CGT TTA AAG-3 and 5′-GGA TCC TCT ACT CCT TCC CCA TCA C-3′ by using DNA carrying SPT15 as the template) into the BamHI site within the region encoding the triple HA1 tag at the carboxy terminus of mutant Rrn9p. Plasmid pNOY3266 was constructed by cutting pNOY3246 with EcoRI, dropping out the 508-bp fragment, and religating. Plasmid pNOY3267 was constructed by cutting pNOY3246 with XbaI, dropping out the 348-bp fragment, and religating. Plasmid pNOY3268 was created by cutting pNOY3246 with BstBI, treating the linearized vector with T4 DNA polymerase, and then recutting the linear vector with StyI and treating this recut vector with mung bean nuclease. The vector was then ligated, dropping out the 90-bp fragment between StyI and BstBI. Plasmids pNOY3269 and pNOY3270 were created by cutting pNOY3246 and pNOY3268, respectively, with HindIII and AccI, treating the linear vector with T4 DNA polymerase, and then ligating the vector, dropping out the 337-bpHindIII/AccI fragment. Plasmids pNOY3271 and pNOY3272 were created by site-directed mutagenesis on template pNOY3246 as described below.
TABLE 1.
Yeast strains and plasmids
| Strain or plasmid | Description |
|---|---|
| Strains | |
| SFY526 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canrgal4-542, gal80-538 URA3::GAL1-lacZ (Clontech) |
| NOY703 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103 |
| NOY817 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY421 |
| NOY818 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY422 |
| NOY819 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY430 |
| NOY821 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY422, pNOY417 |
| NOY823 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY422, YEp351 |
| NOY825 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY430, pNOY417 |
| NOY827 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY430, YEp351 |
| NOY828 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pRS314, YEp351 |
| NOY829 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY420, YEp351 |
| NOY830 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY423 |
| NOY831 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY424 |
| NOY832 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY425 |
| NOY833 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY426 |
| NOY834 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY427 |
| NOY835 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY428 |
| NOY836 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY429 |
| NOY838 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, YEp351, pNOY426 |
| NOY840 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, YEp351, pNOY427 |
| NOY841 | MATa ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11 can1-100 rrn9Δ::HIS3 pNOY103, pNOY420 |
| Plasmids | |
| pBS(−) | A 3,204-bp phagemid derived from pUC19 Ampr (Stratagene Cloning Systems, La Jolla, Calif.) |
| YEp351 | E. coli-yeast shuttle vector carrying 2μm, LEU2, Ampr |
| pRS314 | E. coli-yeast shuttle vector carrying TRP1, CEN6, ARSH4 (24) |
| PACT2 | GAL4 AD vector; LEU2, 2μm, Ampr and carries the GAL4 transcriptional AD [GAL4(768-881), encoding Gal4p(768-881)], which is under the control of the ADH promoter and is followed by the region encoding an HA1-flu epitope tag and multicloning sites (1) |
| pAS2 | GAL4 DBD vector; TRP1, CYH2, 2μm, Ampr and carries the GAL4 DBD [GAL4(1-147), encoding GAL4p(1-147)], which is under control of the ADH promoter and is followed by the region encoding an HA1-flu epitope tag and multicloning sites (1) |
| pGAD424 | GAL4 AD vector [GAL4(768-881)]; LEU2, 2μm, Ampr (Clontech) |
| pNOY103 | Multicopy plasmid carrying GAL7-35S rDNA, ADE3, URA3, 2μm, and Ampr (19) |
| pNOY355 | Derivative of pAS2 carrying a GAL4(1-147)-RRN9 fusion gene (25) |
| pNOY359 | Derivative of pACT2 carrying a GAL4(768-881)-SPT15 fusion gene (SPT15 encodes TBP) (25) |
| pNOY410 | Derivative of pGAD424 carrying a GAL4(768-881)-RRN10 fusion gene |
| pNOY411 | Derivative of pAS2 carrying a GAL4(1-147)-rrn9(L185S) fusion gene |
| pNOY412 | Derivative of pAS2 carrying a GAL4(1-147)-rrn9(F186S) fusion gene |
| pNOY413 | Derivative of pAS2 carrying a GAL4(1-147)-rrn9(L110S) fusion gene |
| pNOY414 | Derivative of pAS2 carrying a GAL4(1-147)-rrn9(L269P) fusion gene |
| pNOY415 | Derivative of pAS2 carrying a GAL4(1-147)-rrn9(L274Q) fusion gene |
| pNOY417 | Derivative of YEp351 carrying RRN10 |
| pNOY420 | Derivative of pRS314 carrying triple-HA1-tagged RRN9 |
| pNOY421 | Derivative of pNOY420 carrying rrn9(L185S) |
| pNOY422 | Derivative of pNOY420 carrying rrn9(F186S) |
| pNOY423 | Derivative of pNOY420 carrying rrn9(L110S) |
| pNOY424 | Derivative of pNOY420 carrying rrn9(L269P) |
| pNOY425 | Derivative of pNOY420 carrying rrn9(L274Q) |
| pNOY426 | Derivative of pNOY420 carrying rrn9(L110S/L269P) |
| pNOY427 | Derivative of pNOY420 carrying rrn9(L110S/L274Q) |
| pNOY428 | Derivative of pNOY420 carrying an rrn9(L110S/L269P)-SPT15 fusion gene |
| pNOY429 | Derivative of pNOY420 carrying an rrn9(L110S/L274Q)-SPT15 fusion gene |
| pNOY430 | Derivative of pNOY420 carrying an rrn9(F186S)-SPT15 fusion gene |
| pNOY3246 | Derivative of pBS(−) carrying RRN9 (25) |
| pNOY3247 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ48-197) (25) |
| pNOY3248 | Derivative of pGEX-1 carrying SPT15 fused to the GST-coding region (a gift from J. C. Reese [20]) |
| pNOY3266 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ199-365) |
| pNOY3267 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ48-163) |
| pNOY3268 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ157-186) |
| pNOY3269 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ1-133) |
| pNOY3270 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(Δ1-133/Δ157-186) |
| pNOY3271 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(L185S) |
| pNOY3272 | Derivative of pBS(−) carrying the mutant rrn9 gene encoding Rrn9p(F186S) |
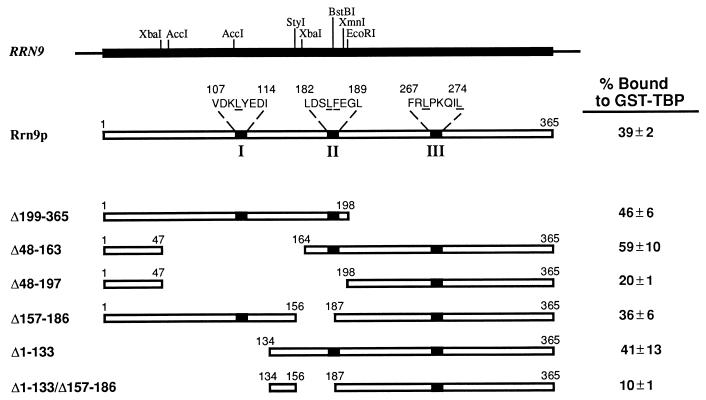
FIG. 1.
Locations of rrn9 deletions analyzed in TBP binding experiments and of rrn9 point mutations affecting the interaction with TBP in the two-hybrid system in vivo. Restriction enzyme sites used to construct various deletions are shown on the RRN9 gene at the top. Three regions containing rrn9 point mutations are indicated as I, II, and III, respectively, on the Rrn9p protein, and their amino acid sequences are shown, with amino acid residues altered by the mutations underlined. Rrn9p derivatives carrying the deletions indicated were labeled with [35S]methionine, and their binding to GST-TBP was studied. Average values of binding, as percentages of input Rrn9p, obtained from three such experiments are shown for each construct. The results of one experiment are shown in Fig. 2.
In vitro interaction analysis of Rrn9p and deletion derivatives of Rrn9p with GST-TBP.
A glutathione-S-transferase (GST)–TBP fusion protein was prepared by induction of Escherichia coli strains carrying pNOY3248 with IPTG (isopropyl-β-d-thiogalactopyranoside) followed by affinity purification of fusion proteins from extracts by using glutathione-agarose beads (Sigma, St. Louis, Mo.). Protein concentrations on the beads were determined by Bradford analysis (2) and then adjusted to 5 mg/ml in a 50% slurry.
[35S]methionine-labeled Rrn9p and deletion mutants of Rrn9p were synthesized in vitro by using rabbit reticulocyte systems (Promega, Madison, Wis.) with pNOY3246, pNOY3247, pNOY3266, pNOY3267, pNOY3268, pNOY3269, pNOY3270, pNOY3271, and pNOY3272 as templates. The labeled proteins were added in approximately equal molar amounts to GST-TBP bound to glutathione-agarose beads (10 μl) preequilibrated with buffer A (20 mM Tris-acetate [pH 8.0], 10 mM Mg acetate, 200 mM KCl, 30% glycerol, 0.2% bovine serum albumin, 0.5% Tween 20, and 1 mM phenylmethylsulfonyl fluoride) in a final volume of 200 μl. After 30 min at room temperature with gentle agitation, the beads were recovered and washed three times in 1 ml of buffer A. The bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography and were quantified by cutting out pertinent protein bands and determining radioactivity with a liquid scintillation counter.
Site-directed mutagenesis of RRN9.
Site-directed mutagenesis to create mutations L185S and L186S in RRN9 was done by using a Transformer Mutagenesis kit (Clontech, Palo Alto, Calif.). The template used for the mutagenesis was pNOY3246, which is a pBS(−) derivative containing the region of DNA encoding the full-length Rrn9p. To make both mutations, the HindIII site in the polylinker of pBS(−) was changed to an MluI site with the oligonucleotide 5′-AAA GGG AAC AAA CGC GTG CAT GCC TG-3′. To create pNOY3271, containing the DNA encoding Rrn9p(L185S), the oligonucleotide 5′-CTG GAC AGC TCA TTC GAA GGC-3′ was used. To create pNOY3272, containing DNA encoding Rrn9p(F186S), the oligonucleotide 5′-G GAC AGC TTA TCC GAA GGG TTG-3′ was used. The presence of these mutations in the final constructs was confirmed by DNA sequencing with the Tag DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif.) and by the sensitivity of the constructs to MluI but not to HindIII. The mutant rrn9 genes were then cloned into both pAS2 and pRS314 as described above, to create (i) pNOY411 and pNOY412 and (ii) pNOY421 and pNOY422, respectively.
Analysis of the interaction of wild-type and mutant Rrn9p with TBP and Rrn10p by use of the two-hybrid system.
Plasmid pAS2, carrying the gene for the Gal4p DBD DNA-binding domain (residues 1 to 147), and pACT2, carrying the gene for the Gal4p AD activation domain (residues 768 to 881), were obtained from S. J. Elledge (1) and were used as control vectors. The wild-type RRN9 gene and rrn9 mutant genes carrying point mutations, i.e., L185S, F186S, L110S, L269P, and L274Q, were cloned into pAS2 to create the following plasmids carrying DBD-RRN9 (or DBD-rrn9) fusion genes, respectively: pNOY355, pNOY411, pNOY412, pNOY413, pNOY414, and pNOY415. DBD-Rrn9p fusion proteins encoded by these fusion genes were tested for interaction with AD-Rrn10p and AD-TBP fusion proteins encoded by pNOY410 and pNOY359, respectively. The plasmids carrying DBD-RRN9 or DBD-rrn9 fusion genes mentioned above were transformed together with either pNOY410, pNOY359, or control vector (pACT2) into the two-hybrid reporter strain SFY526. In addition, pNOY410 and pNOY359 were cotransformed with pAS2 into SFY526 as negative controls. Eight independent transformants for each plasmid combination were restreaked and grown for 2 days on synthetic medium containing glucose and lacking tryptophan and leucine. A toothpick-full of colonies from each restreak was placed in a grid pattern on Whatman no. 50 filters on agar plates. Each filter was lifted off the plate, dipped in liquid nitrogen for 5 s, and then placed, after thawing, onto a second Whatman filter wet with buffer Z (100 mM Na phosphate [pH 7.0], 10 mM KCl, 1 mM MgSO4) containing 39 mM β-mercaptoethanol and 1 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. The filters were placed at 37°C, and the rate of blue color development was noted on a scale of 1 (+) to 4 (++++), with 1 being development of good blue color at 1 h and 4 being the development of good blue color in less than 15 min. When liquid β-galactosidase assays were done as previously described (25), the activity obtained for transformants showing a score of 1 corresponded to approximately 2 U, while the activity obtained for those showing a score of 4 corresponded to approximately 100 U (units were calculated as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein).
PCR mutagenesis and screening for rrn9 mutations which affect the interaction of Rrn9p with TBP in the yeast two-hybrid system.
In the two-hybrid system, DBD-Rrn9p interacts strongly with TBP. When pNOY355 and pNOY359 encoding each of these fusion proteins were cotransformed into the reporter strain SFY526, a plate assay showed a color development score of 4. The background for transformants obtained by cotransformation with pNOY355 and pACT2 showed a color development score of 1. We took advantage of the large difference in color development between the positive interaction and the background and carried out PCR mutagenesis to isolate mutants of Rrn9p which no longer interacted with TBP. The scheme for this PCR mutagenesis with the two-hybrid system is based on a method described previously (16) and is shown graphically in Fig. 5.
FIG. 5.
Strategy used to screen for rrn9 mutations that abolish the interaction of Rrn9p with TBP in the yeast two-hybrid system. Structures of the DNA fragment generated after PCR mutagenesis (RRN9 PCR fragment) and the linearized form of pAS2 are shown as DBD-Rrn9p, indicating that a homologous recombination in vivo will form the structure identical to that of pNOY355, encoding Rrn9p fused to the DBD of Gal4p (DBD-Rrn9p). The structure of pNOY359 present in the reporter strain SFY526 is also shown (AD-TBP); pNOY359 carries a fusion gene encoding TBP fused to the AD of Gal4p. This strategy is based on that described in reference 16. For details of screening of mutants, see Materials and Methods.
We used two oligonucleotides as primers, oligonucleotides I (5′-GCC TCT AAC ATT GAG ACA GC-3′) and II (5′-CCT ACA GGA AAG AGT TAC TC-3′). Oligonucleotide I is a sequence in pAS2 approximately 100 bp upstream of the polylinker. Oligonucleotide II is the opposite strand of the sequence approximately 150 bp downstream of the polylinker of pAS2. PCRs were carried out with these primers in buffer B (15 mM MgCl2, 10 mM Tris-HCl [pH 8.3], 50 mM KCl, 400 nM oligonucleotide I, 400 nM oligonucleotide II, 1 μg of pNOY355 DNA per ml, and 100 U of Fisher Taq DNA polymerase per ml) under the following conditions: 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and 10 min at 72°C. Under the conditions used, Taq DNA polymerase is expected to produce about one base pair change per 1,000 base pairs (16). The PCR product containing a library of mutations in RRN9 was cotransformed with pAS2 linearized with NcoI and BamHI into the reporter strain SFY526, which already contained pNOY359 (the AD-TBP plasmid), and incubated for 1 week at 30°C. The PCR fragment was ligated into the linearized vector by homologous recombination in vivo (16) (see Fig. 5). Approximately 4,300 colonies on 20 plates were screened by the plate β-galactosidase assay. Sterile Whatman no. 50 filters were laid onto the colonies; the filters were removed from the plates, and assays were conducted as described above. Most colonies on the filters showed a score of 4 in plate assays. Colonies which gave a score of 1 in plate assays, like the control background, were picked off the original plates and were restreaked and retested. Western immunoblot analysis with anti-HA1 antibodies was carried out to confirm expression of full-length DBD-Rrn9p and AD-TBP, both of which carried epitopes as programmed in the parent plasmids pAS2 and pACT2 (1). Plasmids containing potential DBD-Rrn9p mutants were rescued from strains expressing both proteins. These rescued plasmids were retransformed into SFY526 expressing AD-TBP. Plate assays were repeated, and if the colonies still gave a color test score of 1, DNA sequencing was carried out to identify mutational alterations. Plasmids with mutations interfering with the interaction with TBP were then each cotransformed with pNOY410 (AD-Rrn10p) and with pACT2 (vector). Plasmids from strains expressing mutant Rrn9p which still interacted with Rrn10p like the wild-type Rrn9p as judged by plate assay were kept for further experiments.
RESULTS
Interaction of Rrn9p and its deletion derivatives with GST-TBP in vitro.
In order to obtain information on the regions of Rrn9p involved in the interaction with TBP and to help design mutagenesis for isolation of rrn9 mutants defective in this interaction, we first studied the interaction of Rrn9p and its deletion derivatives with TBP in vitro. 35S-labeled Rrn9p and its deletion derivatives (Δ199-365, Δ48-163, Δ48-197, Δ157-186, Δ1-133, and Δ1-133/Δ157-186 [the numbers indicate the amino acid residues which have been deleted] (Fig. 1) were prepared by using a reticulocyte lysate in vitro transcription-translation system. These radioactive proteins were incubated with GST-TBP fusion protein bound to glutathione-agarose beads. The beads were washed with a buffer containing 200 mM KCl, and the fractions of 35S-labeled proteins that were bound to GST-TBP were determined by SDS-PAGE followed by autoradiography. Measurements of radioactivity in pertinent protein bands were made with a liquid scintillation counter. Average values obtained from three experiments are given in Fig. 1, and an example of the autoradiograms is shown in Fig. 2.
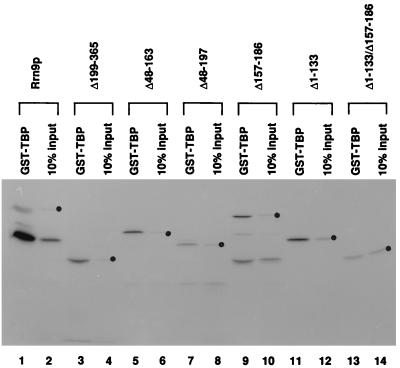
FIG. 2.
Binding of [35S]methionine-labeled Rrn9p and its deletion derivatives to GST-TBP. Equivalent amounts of [35S]methionine-labeled Rrn9p and its deletion derivatives were mixed with GST-TBP attached to glutathione-agarose beads for 30 min at room temperature. Beads were washed three times, and the labeled proteins bound to the beads were analyzed by SDS-PAGE; 10% of each of the input of the labeled proteins was also analyzed. An autoradiogram of the gel is shown. Positions of each intact labeled protein in the radioactive protein preparations used as input are shown by dots. Smaller radioactive proteins found in some of the input protein preparations, especially in Rrn9p and the Δ157-186 protein, may have been produced by incorrect translation initiation, premature termination of translation, or degradation of full-sized proteins during their preparation in reticulocyte lysate. In other similar experiments, binding of 35S-labeled Rrn9p and its deletion derivatives to GST was analyzed in parallel to binding to GST-TBP. None of the labeled proteins were bound to GST control beads to any significant extent.
As can be seen in Fig. 1, deletion of approximately half (45%) of the Rrn9p protein from the C terminus (Δ199-365) or deletion of approximately one-third (36%) of the protein from the N-terminal half (Δ1-133) did not decrease the binding to GST-TBP. In addition, two other internal deletions, Δ48-163 and Δ157-186, also did not decrease the binding. These four deletions cover almost the entire region of Rrn9p (except the region from residue 187 to 198), suggesting that the binding of Rrn9p to GST-TBP in vitro involves multiple regions and that disruption of an interaction of TBP with one region is not sufficient to affect the binding in vitro. In fact, a combination of two of the above-mentioned four deletions, Δ1-133 (41% binding) and Δ157-186 (36% binding) (the control without deletion had 39% binding), led to a significant decrease in binding (Δ1-133/Δ157-186; 10% binding). This result suggests that each of these deleted segments may contain at least one specific region involved in the binding reaction. The importance of the second segment, Δ157-186, was also supported by comparison of two deletion constructs, Δ48-163 (59% binding) and Δ48-197 (20% binding); that is, an extension of deletion from residue 164 to 197 caused a threefold decrease in binding, indicating the importance of the segment from residue 164 to 197 for binding. (Since we have identified at least one small segment of Rrn9p, residues 157 to 186 or 164 to 197, we did not carry out further studies to answer the question of whether the apparent weak binding shown by Δ48-197 [20% binding] or by Δ1-133/Δ157-186 [10% binding] represents a significant contribution of the regions which have remained in these deletion constructs, e.g., the C-terminal segment from residue 198 to 365, to binding to TBP. Mutational analysis described below suggests that a region in this segment may indeed be important for interaction with TBP. We also note that the small deletion Δ157-186, which covers the region that appeared to play a role in the in vitro interaction of Rrn9p with TBP, abolished the function of RRN9 in vivo as judged by complementation assay [data not shown]. The effects of other, larger deletions on the in vivo function were not examined.)
Site-directed mutagenesis of a middle region of Rrn9p resembling ADs of Pol II transcriptional activators.
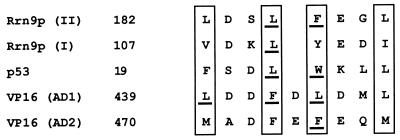
A visual inspection of the middle small segment (residues 157 to 197 [see above]) of Rrn9p involved in the in vitro interaction with GST-TBP revealed a similarity of amino acid residues 182 to 189 to those in ADs in some Pol II transcriptional activators studied previously (Fig. 3). It has been previously pointed out that functionally important bulky hydrophobic amino acids are arranged in a similar way in several ADs of Pol II transcriptional activators (7). The conserved arrangement for ADs of p53 and VP16 shown in Fig. 3 is taken from previous publications (3, 7) (see the legend to Fig. 3), and the sequence found in the middle segment of Rrn9p (residues 182 to 189; Rrn9p(II) in Fig. 3] is aligned to follow the pattern published previously.
FIG. 3.
Similarity of two regions of Rrn9p to ADs of some Pol II transcriptional activators. Regions I and II of Rrn9p (Fig. 1) are aligned to follow the conserved pattern published for ADs of p53 and VP16 (3, 7). Bulky hydrophobic amino acids used to align the sequences are boxed. Amino acid residues which have been shown to be important for activation by mutational analysis (3, 7, 10, 21; this study) are underlined. The position of the first amino acid of each sequence is shown. It should be noted that in the previous papers (3, 7), the amino acid sequences of the ADs of quite a few other transcription factors were also aligned with the sequences of VP16 (7) and p53 (3), and the boxed hydrophobic amino acids were suggested as a consensus sequence. Here, only the sequences of VP16 and p53, for which there is strong experimental support for the significance of these amino acids, are given.
The residue phenylalanine at position 442 (F442) in AD1 of VP16 (7, 10) and the comparable residue L22 as well as W23 in p53 (3) were demonstrated by previous mutagenesis experiments to be important for both the in vitro interaction with TBP and the in vivo activation function of these proteins. In addition, other mutagenesis work showed that F475 in AD2 of VP16 as well as L439 and L444 in AD1 are also important for the in vivo activation function of VP16 (21). These residues in VP16 and p53 are underlined in Fig. 3. From the sequence alignment shown in Fig. 3, we selected L185 and F186 of Rrn9p for mutagenesis as possibly important residues involved in the interaction of Rrn9p with TBP. We changed these residues to serine by site-directed mutagenesis and examined the interaction of mutant Rrn9p with TBP in vitro and in the yeast two-hybrid system in vivo.
Although the full-length 35S-labeled Rrn9p containing one of these mutations, L185S or F186S, did not show a decrease in the binding to GST-TBP compared to that of the control Rrn9p in the in vitro binding experiments (data not shown), these mutations did abolish the interaction with TBP in the yeast two-hybrid system, as shown in Table 2. The results confirm the inference of the importance of these residues in the interaction with TBP. However, these mutations also abolished the interaction of Rrn9p with Rrn10p, another subunit of UAF, in the two-hybrid system (Table 2). Thus, both L185 and F186 appear to be important in the interactions of Rrn9p with Rrn10p as well as with TBP. However, since a single amino acid alteration abolishes two different protein-protein interactions, it is difficult to determine whether L185 or F186 is involved directly in the Rrn9p-TBP interaction, is involved directly in the Rrn9p-Rrn10p interaction, or is simply important for the maintenance of a protein conformation required for the interactions with both TBP and Rrn10p. Nevertheless, we proceeded to study the effects of the two mutations constructed on cell growth in connection with UAF function in vivo.
TABLE 2.
Interactions of wild-type and mutant Rrn9 proteins with TBP and Rrn10p analyzed by the two-hybrid systemsa
| DBD fused to: | β-Galactosidase activityb with AD fused to:
|
||
|---|---|---|---|
| — | TBP | Rrn10p | |
| — | − | − | − |
| WT | + | ++++ | +++ |
| L185S | + | + | + |
| F186S | + | + | + |
| L110S | + | + | +++ |
| L269P | + | + | +++ |
| L274Q | + | + | +++ |
Plasmids carrying genes encoding Gal4 DBDs without fusion (—) or fused to the wild-type Rrn9p (WT), Rrn9p(F185S), Rrn9p(L186S), Rrn9p(L110S), Rrn9p(L269P), or Rrn9p(L274Q) are pAS2, pNOY355, pNOY411, pNOY412, pNOY413, pNOY414, and pNOY415, respectively. Plasmids carrying genes encoding Gal4 ADs without fusion (—) or fused to TBP or Rrn10p are pACT2, pNOY359, and pNOY410, respectively. Reporter strain SFY526 was cotransformed by these plasmids in various combinations as indicated, β-galactosidase activities of transformants were analyzed by the plate colony assay, and the activities were scored on a scale of 1 to 4, as described in Materials and Methods.
−, +, +++, and ++++, negative, weakly positive (score of 1), strongly positive (score of 3), and very strongly positive (score of 4), respectively.
Effects of the rrn9(F186S) mutation in vivo and their suppression by overexpression of RRN10 and by fusion of the mutant gene to the gene for TBP.
Effects of the two rrn9 mutations L185S and F186S, described in the previous section, were studied in vivo in the following way. NOY703 carries a chromosomal rrn9 deletion and can grow well only on a galactose media because it carries the GAL7-35S rDNA fusion gene on plasmid pNOY103 (11, 19). On glucose media, where the transcription of the GAL7-35S rDNA gene by Pol II is repressed, the strain grows extremely poorly. The presence of a derivative of a CEN plasmid (pRS314) carrying RRN9 (pNOY420) complements mutational defects, allowing good growth on glucose media. We constructed a derivative of pRS314 carrying rrn9(L185S) (pNOY421) or rrn9(F186S) (pNOY422) and examined the ability of these mutant genes to complement growth defects on glucose by introducing these plasmids into NOY703. The strain carrying pNOY421 (NOY817) showed a slow-growth phenotype on glucose at 30°C, indicating weak complementation by the mutant (L185S) gene. The strain carrying pNOY422 (NOY818) showed the same phenotype as NOY703, i.e., growth on galactose and extremely poor growth on glucose, indicating that the F186S mutation abolishes the function of RRN9 to maintain a high level of rDNA transcription in vivo.
Because the Rrn9p(F186S) protein failed to interact with TBP and also with Rrn10p in the yeast two-hybrid system, we investigated the possibility that the defects in these interactions are responsible for the inability of the mutant protein to function in vivo in place of the wild-type Rrn9p. We found that overexpression of Rrn10p from a multicopy plasmid, pNOY417 (in NOY821), suppresses the growth defects of NOY818 carrying rrn9(F186S) partially at 30°C but barely at 36°C (Fig. 4, compare NOY821 to NOY823). It appears that a weakened interaction between the mutant Rrn9p(F186S) and Rrn10p may cause a decrease in the amounts of UAF complex containing all the subunits and that increasing the cellular concentration of Rrn10p may increase the amounts of (mutant) UAF, leading to a partial suppression of the mutational defect. In contrast to the suppression by RRN10 overexpression, overexpression of TBP did not show any suppression of this mutation (data not shown).
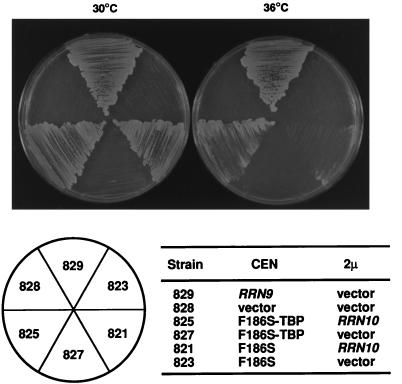
FIG. 4.
Suppression of the rrn9(F186S) mutant phenotype by fusion of the mutant gene to the SPT15 gene combined with overexpression of Rrn10p. Yeast strains were streaked on a synthetic glucose medium lacking tryptophan and leucine, and the plates were incubated at 30 and 36°C for 5 days. Positions of the strains on plates are indicated below the photograph. They are designated by numbers that follow NOY in the complete names. All strains carry the chromosomal rrn9Δ::HIS3 deletion and two plasmids in addition to pNOY103. One (CEN) is pRS314 (vector) or its derivatives carrying RRN9, rrn9(F186S), or the rrn9(F186S)-SPT15 fusion as indicated. The second plasmid (2 μm) is YEp351 (vector) or its derivative carrying RRN10 as indicated.
We then asked whether the suspected defect in the interaction of UAF (containing the mutant Rrn9p) with TBP can be improved by fusing the mutant Rrn9p to TBP. It was previously shown that in the yeast S. cerevisiae, activation of specific transcription by certain Pol II activators takes place through their interaction with TBP and that recruitment of TBP to the promoter through its attachment to a heterologous DBD is sufficient for transcriptional activation (4, 13, 29). Therefore, we considered the possibility that the suspected defect in the ability of mutant Rrn9p might be suppressed by fusion of the mutant protein to TBP. We constructed a plasmid (pNOY430) carrying an rrn9(F186S)-SPT15 fusion gene in which the coding region of the gene for TBP (SPT15) is fused to the C terminus of the protein-coding region of rrn9(F186S) in frame. Introduction of this plasmid into the rrn9 deletion strain (yielding NOY827) did not complement the growth defect; that is, there was no difference in the growth phenotype between the resultant strain (NOY827) and the strain carrying the mutant rrn9(F186S) gene (NOY823) or the rrn9 deletion mutant (NOY828) (Fig. 4, compare NOY827 with NOY823 and NOY828). However, the introduction of the plasmid carrying rrn9(F186S)-SPT15 together with pNOY417 (which overexpresses Rrn10p) yielded a strain (NOY825) whose growth at 36°C is significantly better than that of the control strain, NOY821, which carries the mutant rrn9(F186S) gene and the plasmid overexpressing Rrn10p (pNOY417) (Fig. 4, compare NOY825 with NOY821 at 36°C). These results support the original inference from the two-hybrid system, namely, that the F186S mutation weakens the interaction of Rrn9p with TBP as well as Rrn10p; physical fusion of the mutant Rrn9p with TBP improves the cell growth (i.e., in vivo rDNA transcription by Pol I) under the condition of excess production of Rrn10p.
Isolation of rrn9 mutants that specifically affect the interaction of Rrn9p with TBP in the two-hybrid system.
The F186S and L185S mutations discussed above affected the interaction of Rrn9p with Rrn10p as well as with TBP in the two-hybrid system, and the suppressor analysis was not as simple as we wished. We therefore looked for rrn9 mutants with a defect in the Rrn9p-TBP interaction and without a defect in the Rrn9p-Rrn10p interaction. As described above and in a previous paper (25), Rrn9p interacts with both TBP and Rrn10p in the two-hybrid system, with the interaction with TBP being especially strong. We used PCR mutagenesis to create mutations in RRN9 (in the form of the fusion gene encoding Rrn9p fused to the DBD of Gal4p [DBD-Rrn9p]). PCR fragments containing rrn9 mutations were cotransformed with linearized DBD vector (pAS2) into a reporter yeast strain, a derivative of SFY526 carrying pNOY359, which encodes TBP fused to the Gal4 AD (AD-TBP) (Fig. 5). The PCR fragment was ligated into the linearized vector by homologous recombination in vivo (Fig. 5) (16). Plate β-galactosidase assays were carried out, and colonies that failed to turn blue as quickly as control colonies expressing the wild-type DBD-Rrn9p and AD-TBP were picked. Western immunoblot analysis of cell extracts was used to confirm expression of full-length DBD-Rrn9p and AD-TBP in the potential mutant strains.
Plasmids containing suspected rrn9 mutations were recovered and reintroduced into the SFY526 derivative expressing AD-TBP to confirm the mutational defect and into another SFY526 derivative expressing AD-Rrn10p to examine the interaction of Rrn9p with Rrn10p. Three mutations that abolish the Rrn9p-TBP interaction without affecting the Rrn9p-Rrn10p interaction were identified in this way, and DNA sequencing has shown that they are L110S, L269P, and L274Q, respectively (Table 2).
Suppression of mutational defects of rrn9 by gene fusion to SPT15.
Among the three mutations described above, L110S was found to be in a sequence of Rrn9p (amino acids 107 to 114) which is similar to another region (amino acids 182 to 189) of Rrn9p and to the sequences in some Pol II activators discussed above [Rrn9p(I) (Fig. 3)]. The other two mutations, L269P and L274Q, are localized close together in a region within the C-terminal segment, residues 198 to 365, discussed in connection with the in vitro interaction experiments described above (region III [Fig. 1]).
The three rrn9 mutations, L110S, L269P, and L274Q, were introduced into the RRN9 gene in the native context on a CEN-containing plasmid (creating plasmids pNOY423, pNOY424, and pNOY425, respectively) and transformed into the rrn9 deletion mutant NOY703 to create strains NOY830, NOY831, and NOY832, respectively. These yeast strains did not show any defects in growth. We then constructed strains containing double mutations. One strain carrying L110S and L269P double mutations (NOY833) and another strain carrying L110S and L274Q double mutations (NOY834) were found to be temperature sensitive. Growth of NOY833 was clearly defective at 35°C, and growth of NOY834 was clearly defective at 36°C, as examined on glucose medium (Fig. 6). We were then able to demonstrate that fusion of each mutant gene to the SPT15 gene, encoding TBP (in strains NOY835 and NOY836), suppressed the mutational defects, although not completely (Fig. 6). It should be noted that the introduction of both the rrn9(L110S/L269P) [or rrn9(L110/L274Q)] mutant gene and the SPT15 gene each on a CEN plasmid without gene fusion did not suppress growth defects caused by the rrn9(L110S/L269P) mutation to any significant extent (data not shown). In addition, we also considered the possibility that the mutant proteins might be unstable and suppression by fusion to the SPT15 gene might be due to increased stability of the fusion proteins. By Western immunoblot analysis with anti-HA1 epitope antibodies, we examined cellular amounts of the mutant proteins in these temperature-sensitive mutants at 3 h after a temperature shift from 30 to 37°C, when a decline in growth rate relative to that of the control strain had already started. No significant difference in the amounts of Rrn9p (HA1-tagged Rrn9p) was observed between the control strain and the two mutants (data not shown), indicating that the above-mentioned possibility is unlikely. These results demonstrate that physical proximity of TBP to Rrn9p, and therefore the interaction of TBP with UAF as observed in vitro (25), is indeed required for activated Pol I transcription in vivo. It appears that this TBP-UAF interaction in vivo is probably mediated through interaction of Rrn9p with TBP, as inferred from the results of the two-hybrid experiments and the demonstration of suppression by fusion of the TBP gene to the mutant rrn9 genes.
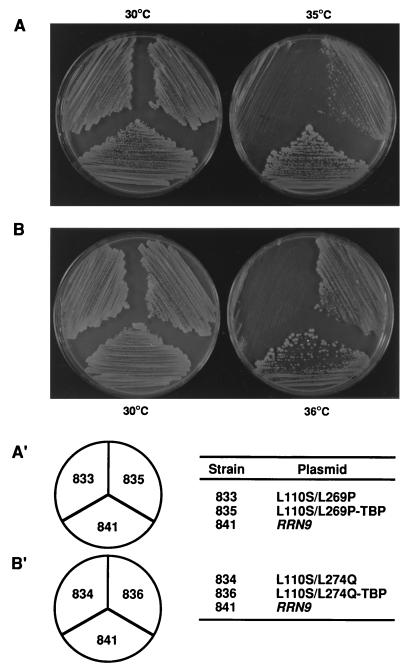
FIG. 6.
Suppression of rrn9(L110S/L269P) and rrn9(L110S/L274Q) mutant phenotypes by fusion of the mutant genes to the SPT15 gene, which encodes TBP. Strains were streaked on a synthetic glucose medium lacking tryptophan, and the plates were incubated at indicated temperatures for 3 days. All the strains carry the chromosomal rrn9Δ::HIS3 mutation and, in addition to pNOY103, a derivative of pRS314 carrying RRN9, rrn9 mutant genes, or the rrn9 mutant genes fused to SPT15, as indicated. The strains are indicated by numbers that follow NOY in the complete names.
In order to establish the specificity of suppression of the rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] mutation by fusion to the TBP gene (SPT15), we carried out additional experiments. First, we examined whether fusion to RRN7 instead of SPT15 suppresses these rrn9 mutations. In addition to the interaction with TBP, it was previously found that Rrn9p interacts with Rrn7p both in vitro and in the two-hybrid system in vivo (25). We used a plasmid construct carrying the RRN9 gene fused to the RRN7 gene (encoding a subunit of CF, Rrn7p) which complements the rrn9 deletion as well as an rrn7 deletion mutation (14a); we introduced the rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] mutation into this fusion gene and found that although these fusion constructs complemented an rrn7 deletion mutation, they did not complement the rrn9 deletion; that is, in contrast to the fusion to the TBP gene, the fusion to RRN7 did not suppress the mutational defect (data not shown).
Suppression of the mutational defect by rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] by overexpression of several genes involved in Pol I transcription was also studied. Overexpression of TBP suppressed the mutational defects (data not shown). This result is consistent with the conclusion obtained from the two-hybrid experiments that the mutations affect the interaction of Rrn9p with TBP. Presumably, increasing the cellular concentration of TBP improves a mutationally weakened interaction between the mutant Rrn9p and TBP, thus improving the UAF function containing the mutant Rrn9p and the cellular growth of the mutant in vivo. Unexpectedly, however, overexpression of RRN10 suppressed the rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] mutation, even though overexpression of another UAF subunit gene (RRN5) as well as other Pol I transcription factor genes (RRN3 and RRN6) analyzed did not show any suppression effects. The observed suppression by RRN10 overexpression is discussed below.
DISCUSSION
Five amino acid substitution mutations in Rrn9p were identified, each of which abolishes the interaction of Rrn9p with TBP in the yeast two-hybrid system. Their locations in Rrn9p are shown in Fig. 1 (see regions I, II, and III; the amino acid residues altered by the mutations are underlined). L185 and F186 were identified by site-directed mutagenesis. As mentioned in Results, these residues are in a sequence context similar to those surrounding hydrophobic amino acids in ADs of the Pol II activators VP16 and p53 that were previously shown to be important for interaction with TBP as well as for transcriptional activation (Fig. 3; see the legend to Fig. 3 and references 3, 7, 10, and 21). In analogy with the importance of F442 of VP16 in the binding to TBP in vitro, which is well correlated to the in vivo activation function of VP16 (10), L185 and F186 might also be directly involved in the interaction with TBP. However, alterations of these amino acids, L185S and F186S, were both found to abolish not only the Rrn9p-TBP interaction but also the Rrn9p-Rrn10p interaction in the two-hybrid system. Therefore, it is difficult to decide whether L185 or F186 is involved directly in the interaction with TBP, is involved directly in the interaction with Rrn10p, or is simply important for the maintenance of a (local or global) protein conformation required for the interactions with both TBP and Rrn10p. Nevertheless, the results of suppressor analysis of the F186S mutation suggest that the interactions of Rrn9p with TBP and Rrn10p detected in the two-hybrid system are both important in vivo for a high level of rDNA transcription by Pol I. (In contrast to the F186S mutation, the L185S mutation showed only a weak phenotype, and suppressor analysis was not carried out. We also note that the mutant strain carrying F186S and growing on galactose with the help of plasmid pNOY103 showed a reduced level of the mutant Rrn9p relative to that of Rrn9p in the control strain. It is possible that the reduced interaction with Rrn10p increased the free form of Rrn9p, which may be more sensitive to proteolytic degradation. Overproduction of Rrn10p may have improved the association of Rrn9p with Rrn10p [and possibly other UAF subunits], leading to partial suppression of the mutational defect as observed in the present work.)
The remaining three mutations are more specific, and each disrupts the Rrn9p-TBP interaction without affecting the Rrn9p-Rrn10p interaction in the two-hybrid system. The mutational alterations do not apparently cause a reduction in the stability of protein in vivo or a significant alteration of protein conformation, at least that involved in the interaction with Rrn10p in the two-hybrid system. Therefore, it is conceivable that the three amino acids identified, L110, L269, and L274, may be directly involved in the interaction with TBP. The sequence context surrounding L110 is similar to that surrounding the well-studied F442 in the AD of VP16 as well as that surrounding L185 and F186 of Rrn9p mentioned above (Fig. 3). The other two amino acids, L269 and L274, are clustered (region III in Fig. 1). Although the sequence context does not follow the pattern given in Fig. 3, the region including these two crucial leucine residues appears to play an important role in the Rrn9p-TBP interaction. As already mentioned, the importance of bulky hydrophobic amino acids in a proper sequence context has been repeatedly emphasized in connection with protein-protein interactions used for activation of Pol II transcription (3, 7, 8, 10, 21). It is interesting that the five amino acid residues identified in Rrn9p to be important in the interaction with TBP, either directly or possibly indirectly through another protein(s) (see below), are all bulky hydrophobic amino acids. Such observations suggest that protein-protein interactions involving bulky hydrophobic amino acids may also be important for activation of Pol I transcription.
Each of the three specific mutations, L110S, L269P, and L274Q, was individually sufficient to disrupt the Rrn9p-TBP interaction in the two-hybrid system, but yeast cells carrying these mutant rrn9 genes in place of the wild-type gene did not show any growth defects. However, combinations of L110S with one of the two mutations in region III led to a temperature-sensitive phenotype. It is probable that the interaction of Rrn9p with TBP in the context of UAF function in vivo is strengthened by other protein-protein interactions involved in the formation of a preinitiation complex, e.g., the interaction of UAF with CF and that of CF with TBP (25), as well as protein-DNA interactions involving these factors; because of these other stabilizing interactions, disruption of both of the two genetically identified Rrn9p-TBP interactions, and not either one alone, is required to show growth defects (temperature-sensitive phenotypes).
The temperature-sensitive phenotypes of rrn9(L110S/L269P) and rrn9(L110S/L274Q) were clearly suppressed by fusing the mutant genes to SPT15, that is, by fusing the mutant proteins to TBP. As mentioned in Results, the suppression must be due to the presence of the fusion protein(s) itself; degradation of the fusion protein into the mutant protein(s) and TBP cannot explain the suppression, because the introduction of both the rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] mutant gene and the SPT15 gene, each on a CEN plasmid without gene fusion, did not suppress growth defects caused by the rrn9(L110S/L269P) mutation. As also mentioned in Results, the mutant proteins are stable in vivo, and therefore, the suppression is not related to the question of protein stability. It appears that the fusion protein(s) carrying an otherwise defective rrn9 mutation [rrn9(L110S/L269P) or rrn9(L110S/L274Q)] can take a conformation resembling the structure of Rrn9p, having presumably a direct contact with TBP within a functional preinitiation complex.
Suppression of the mutation rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] by overproduction of TBP is consistent with the model discussed above, namely, that Rrn9p interacts with TBP directly in vivo and these rrn9 mutations disrupt this interaction. However, suppression of these mutations by overproduction of RRN10 was unexpected. There are two possible alternative explanations. First, even though the rrn9(L110S/L269P) [or rrn9(L110S/L274Q)] mutation did not disrupt the interaction of Rrn9p with Rrn10p as judged by the results in the two-hybrid system, the mutation may have caused some subtle effects on the Rrn9p-Rrn10p interaction in vivo unrecognized by the two-hybrid system assay, and strengthening this interaction by overproduction of Rrn10p might improve the interaction of Rrn9p with TBP, leading to suppression of the mutational defect similar to the suppression by the fusion of the mutant gene to the SPT15 (TBP) gene or by the overproduction of TBP. The second explanation is to invoke an alternative model that the interaction of TBP with UAF in vivo is not through a direct interaction with Rrn9p but through a direct interaction with some other UAF component. According to this model, the mutations in question may cause a local or global change in UAF structures, including a decreased interaction of Rrn9p with Rrn10p, leading to defects in the interaction of UAF with TBP in vivo and consequent defects in cell growth. Overexpression of Rrn10p may increase the amounts of UAF with a functional structure, leading to suppression of growth of the mutants. According to this model, suppression by fusion of Rrn9p to TBP (or overexpression of TBP) simply helps to recruit TBP close to another UAF subunit that normally interacts with TBP directly. Although we favor the first explanation, we cannot rigorously exclude the second explanation based on the alternative model. Nevertheless, it should be emphasized that, regardless of the question of the presence of a direct interaction of Rrn9p with TBP in the normal wild-type cells, a physical fusion of TBP with mutant Rrn9p suppresses defects in UAF function in vivo. Thus, even though we cannot rigorously prove that Rrn9p directly interacts with TBP in vivo, the results described in this paper demonstrate that the improved interaction of TBP with UAF, either through a direct interaction with Rrn9p or through interactions with some other UAF components, leads to suppression caused by mutations in RRN9.
It should be noted that pertinent growth defects observed with various mutations studied here are all rescued by growth on galactose, which allows the cells to synthesize rRNA by transcription of the GAL7-35S rDNA fusion gene on pNOY103 by Pol II (19). Thus, the present results demonstrate that the interaction of UAF with TBP observed in vitro and in the two-hybrid system (25) is indeed important for high-level transcription of rDNA by Pol I in vivo.
As noted in the introduction, protein-protein interactions observed in vitro do not necessarily mean that these interactions operate in vivo. The results of genetic analyses of several point mutations of RRN9 described in this work were more consistent with the results of the two-hybrid system than with those of direct in vitro physical interaction experiments. Several rrn9 mutations, which disrupted the interactions in the two-hybrid system and caused growth defects, failed to show effects on the in vitro interaction assayed by using GST-TBP. One plausible explanation in this case is that the in vitro interaction assayed is between uncomplexed Rrn9p and TBP, whereas Rrn9p is within UAF in vivo. Interactions between TBP and surfaces that are exposed only in uncomplexed Rrn9p would be detected in the in vitro binding experiments. In the present in vitro assay system, this type of interaction may be significant, decreasing the sensitivity of the assay to defects in the specific interaction between Rrn9p and TBP relevant to in vivo UAF function.
Studies on the activation of Pol II transcription have shown that interactions of Pol II activators with general transcription factors other than TBP, e.g., TFIIA, TFIIB, and TAFs, also contribute to activation in various systems (for reviews, see references 22, 26, and 28). Similarly, an interaction of UAF with other factors in the Pol I system, e.g., CF (possibly through the interaction of Rrn9p with Rrn7p, a subunit of CF, as observed in vitro and in the two-hybrid system in vivo [25]), might take place in vivo, contributing to the stimulation of rDNA transcription by UAF. Such possibilities should be examined by appropriate genetic experiments, as was done for the UAF-TBP interaction in the present work. Finally, as Pol II activators bind to specific DNA segments through a DBD, UAF binds to the upstream element (11). We have not studied the question of whether any of the three characterized UAF subunits, Rrn5p, Rrn9p, and Rrn10p, directly participates in the binding to the upstream element. In this respect, we note that one of the previously reported UAF components, p18, has been recently identified as histone H3, and the presence of histone H4 in purified UAF preparations has also been demonstrated (10a). Thus, these histones must surely participate in binding of UAF to DNA. The questions of which other UAF components participate in DNA binding and how the specificity of binding is achieved are also the subject of future studies.
ACKNOWLEDGMENTS
We thank Karen Sutton for technical assistance, Dominique Lalo for providing some plasmid constructs, Marian Waterman and John Keener for critical reading of the manuscript, and D. Semanko for help in preparation of the manuscript.
This work was supported by Public Health Service grant R37GM35949 from the National Institutes of Health.
REFERENCES
- 1.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Chang J, Kim D-H, Lee S W, Choi K Y, Sung Y C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 5.Choe S Y, Schultz M C, Reeder R H. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1992;20:279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 7.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 8.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann A, Oelgescklager T, Roeder R G. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 10a.Keener J, Dodd D A, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor (UAF) required for the high level transcription of yeast rDNA by polymerase I. Proc Natl Acad Sci USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keys D A, Lee B-S, Dodd J A, Nguyen T T, Vu L, Fantino E, Burson L M, Nogi Y, Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 12.Keys D A, Vu L, Steffan J S, Dodd J A, Yamamoto R T, Nogi Y, Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- 13.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 14.Kulkens T, Riggs D L, Heck J D, Planta R J, Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991;19:5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Lalo, D., and M. Nomura. Unpublished data.
- 15.Lalo D, Steffan J S, Dodd J A, Nomura M. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 16.Lehming N, McGuire S, Brickman J M, Ptashne M. Interactions of a Rel protein with its inhibitor. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C H, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder R H. A novel 66-kilodalton protein complexes with Rrn6p, Rrn7p, and the TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musters W, Knol J, Maas P, Dekker A F, van Heerikhuizen H, Planta R J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogi Y, Vu L, Nomura M. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 21.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 23.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 24.Sikorski R A, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffan J S, Keys D A, Dodd J A, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 26.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 27.Tansey W P, Herr W. The ability to associate with activation domains in vitro is not required for the TATA box-binding protein to support activated transcription in vivo. Proc Natl Acad Sci USA. 1995;92:10550–10554. doi: 10.1073/pnas.92.23.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 29.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto R T, Nogi Y, Dodd J A, Nomura M. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]