Summary
A generic level of chromatin organization generated by the interplay between cohesin and CTCF suffices to limit promiscuous interactions between regulatory elements, but a lineage-specific chromatin assembly that supersedes these constraints is required to configure the genome to guide gene expression changes that drive faithful lineage progression. Loss-of-function approaches in B cell precursors show that IKAROS assembles interactions across megabase distances in preparation for lymphoid development. Interactions emanating from IKAROS-bound enhancers override CTCF-imposed boundaries to assemble lineage-specific regulatory units built on a backbone of smaller invariant topological domains. Gain-of-function in epithelial cells confirms IKAROS’ ability to reconfigure chromatin architecture at multiple scales. While the compaction of the Igκ locus required for genome editing represents a function of IKAROS unique to lymphocytes, the more general function to preconfigure the genome to support lineage-specific gene expression and suppress activation of extra-lineage genes provides a paradigm for lineage restriction.
In brief
IKAROS mediates long-distance interactions both within and across domains and compartments to assemble a lineage-specific 3D genome organization required for immune cell development and function.
Graphical Abstract
Introduction
Emerging evidence connects higher order chromatin structure to temporal and cell type-specific regulation of gene expression, lineage identity, and cell homeostasis.1,2 Regulatory sites, such as enhancers and promoters, are co-assembled in space in a manner that both facilitates and restricts their regulation of target genes. These form discrete topologically self-associated domains (TADs).3–6 Cohesin complex-mediated loop extrusion contributes to TAD formation both by bringing distant sites in proximity where stable contacts may form, and more directly when loop extrusion is halted at CTCF bound sites.7–14 CTCF structural loops can act as TAD boundaries that are rapidly disassembled when either CTCF or cohesin is depleted11,12,15, albeit with minimal effects on gene expression or regulatory site contacts.11,15–17 Thus although cohesin may facilitate regulatory interactions, these may also form by cohesin-independent mechanisms.18–22 Interactions between distal genomic regions organize chromatin into euchromatic (A) or heterochromatic (B) compartments. This process of nuclear compartmentalization is developmentally regulated and correlates with epigenetic state, transcription potential of associated genes, lamin association, and replication timing.6,23–28 Homotypic interactions between biochemically similar chromatin regions are considered a driving force for segregation into A or B compartments with positive regulators of this process beginning to be identified.29–31
The developing immune cell system provides a unique opportunity to study how regulatory sites scattered over mega-base regions come together to support the gene expression and recombination events that define the divergent immune cell fates.32–34 One of the earliest regulators of lymphoid lineage identity is IKAROS, a zinc finger DNA-binding factor encoded by Ikzf1.35–37 IKAROS sets the stage for lymphocyte differentiation in multipotent hematopoietic progenitors by priming genes required for lymphoid lineage differentiation.38,39 Conditional inactivation of IKAROS after lymphoid lineage specification blocks the transition from a highly proliferative large pre-B to a quiescent small pre-B cell. Similar developmental effects and gene expression changes are seen with either conditional deletion of the IKAROS DNA binding domain or a conditional IKAROS null allele.40,41 IKAROS controls B cell differentiation by regulating the activity of transcriptional enhancers with distinct lineage affiliations in a reciprocal manner, by promoting expression of B cell lineage genes while suppressing extra-lineage genes.42 The gene expression defects and block in B cell differentiation occurs despite expression and chromatin occupancy by other transcription factors (TFs), critical for B cell development.42 Notably, heterozygous mutations that interfere with IKAROS activity are frequent in human B cell precursor lymphoblastic leukemias (B-ALL) and are associated with poor disease prognosis.43,44 The most frequently reported are deletions that impair DNA binding45–48and exert a dominant negative effect on the DNA-binding activity of wild-type IKAROS isoforms and IKAROS family members still expressed in these cells through dimerization.49–52 IKAROS proteins dimerize and oligomerize making the IKAROS gene a prime candidate for regulating chromatin structure and spatial organization.49,53–57
Using complementary chromosome conformation capture (3C) approaches, we interrogated the chromatin interactions that control early B cell differentiation. With gain and loss of function studies, we show that IKAROS directly and positively configures regulatory interactions that presage lymphoid gene expression. IKAROS-enriched enhancer clusters make contacts that span CTCF-mediated TAD boundaries to assemble superTAD structures that harbor lymphoid-specific genes and maintain euchromatic localization of the associated genomic regions.
Results
IKAROS regulation of enhancer-based spatial interactions
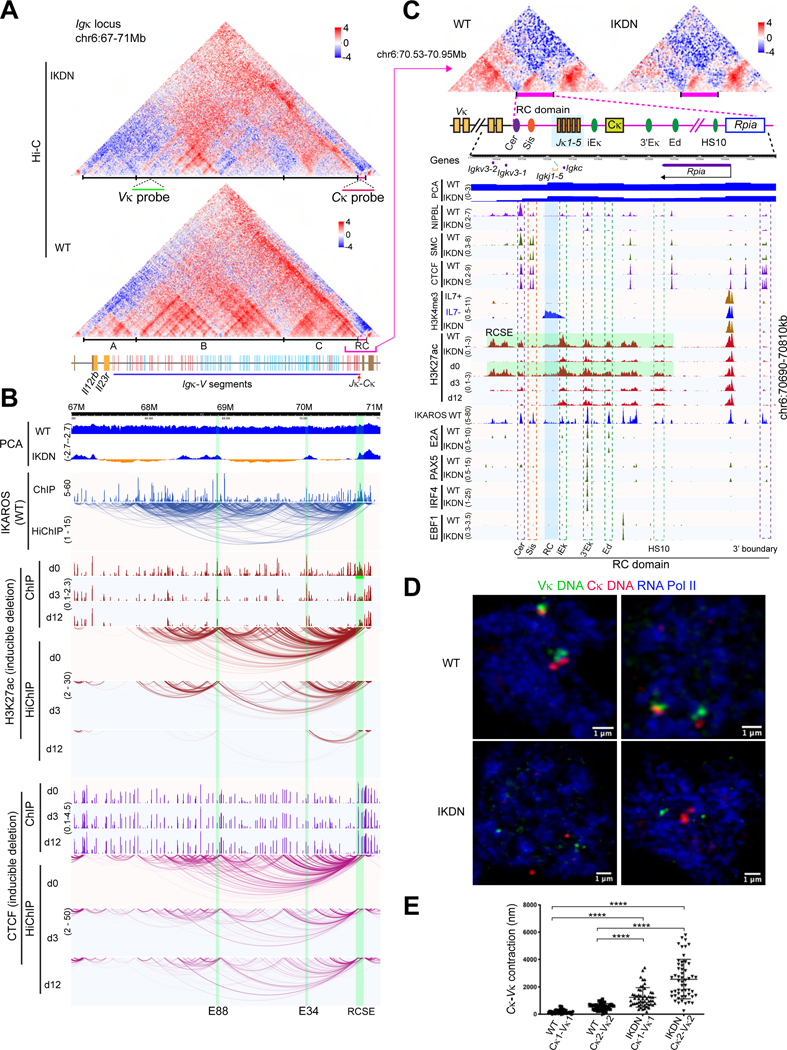
Establishing communication between distant regulatory sites may be a key mechanism by which IKAROS controls transcriptional programs that support lymphoid cell fate decisions. We tested this hypothesis by performing high-resolution 3C assays in combination with gene expression profiling (RNA-seq) and chromatin immunoprecipitations (ChIP-seq) in large pre-B cells. These committed B cell precursors that have rearranged the Ig heavy chain but not the light chain differentiate to small pre-B cells in wild type (WT) but accumulate when IKAROS is deleted (Figures 1A, S1A-B).40,41 IKAROS mutant large pre-B cells were generated in vivo after deletion of Ikzf1 exon 5 encoding the IKAROS DNA binding domain starting at the common lymphoid progenitor58 (IkE5fl/fl hCD2-Cre: hereafter IKDN) (Figures 1A, S1C). This loss-of-function mutation also interferes through dimerization with the activity of AIOLOS/Ikzf3 that compensate for loss of IKAROS during B cell differentiation.41,42
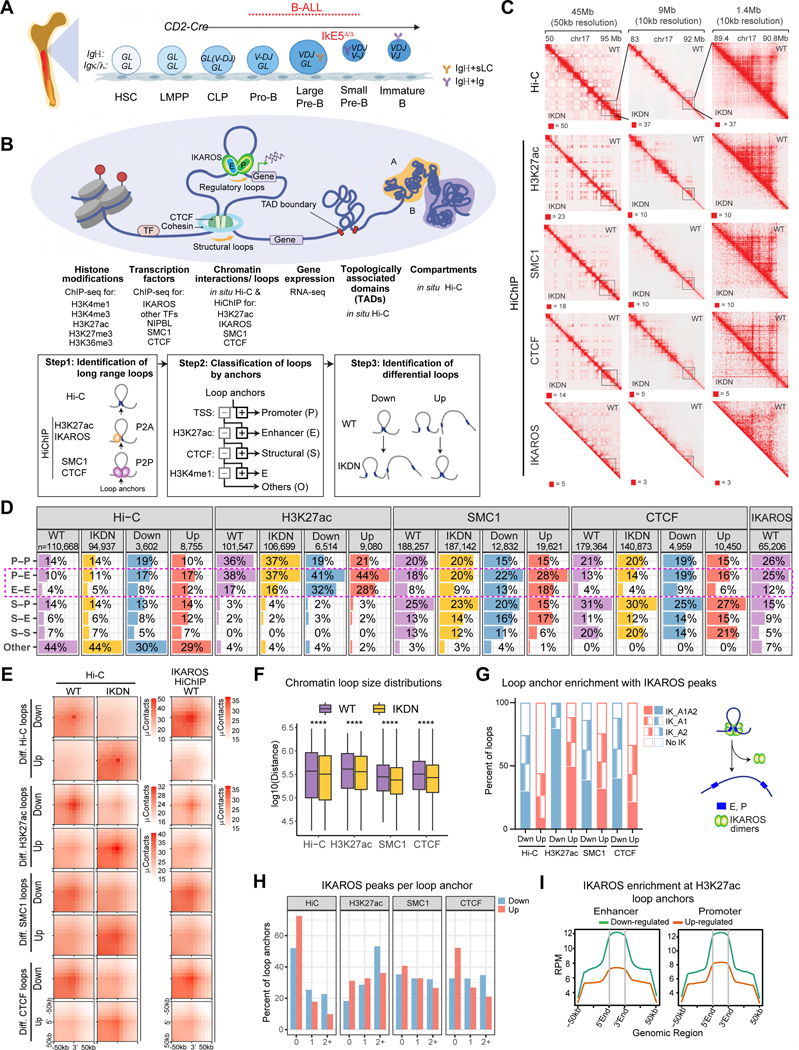
Figure 1. IKAROS controls spatial organization of large pre-B cells.
(A) Schematic of early B-cell differentiation and conditional deletion of the IKAROS DNA binding domain (IKDN). (B) Experimental strategy to profile WT and IKDN large pre-B cells. Schematic (bottom) illustrates the classification of loops, anchors and differential loops (n=2 replicates). (C) Hi-C and SMC1, H3K27ac, CTCF and IKAROS HiChIP experiments shown at 50kb and 10kb resolution around Nrxn1 in WT (upper triangle) and IKDN (lower triangle). Pixel intensity represents the observed number of contacts/valid read pairs. (D) The number and percentage of Hi-C and HiChIP interactions (WT, IKDN) and differential loops (Down, Up) identified at 10kb resolution stratified by anchor classification. (E) Aggregate peak analysis (APA) of differential chromatin loops. Top labels indicate contact maps from which the aggregate pattern is derived. (F) Distance distributions of all significant chromatin loops identified by Hi-C and HiChIP in WT or IKDN. **** p < 0.0001. (G) The percentage of differential HiChIP loops that overlap with IKAROS ChIP-seq peaks at both (IK_A1A2), only one (IK_A1, IK_A2) or none of the anchors (No IK). Model of long-distance interactions between regulatory sites supported by IKAROS-bound dimers. (H) Distribution of IKAROS peaks that overlap anchors of differential loops identified by HiChIP and Hi-C. (I) Average IKAROS ChIP-seq enrichment (RPM) at enhancer and promoter anchors (10kb) from differential H3K27ac loops as in Figure S1H. Source: TableS1-3.
The differences in 3D chromatin organization associated with IKAROS deficiency were assessed using Hi-C and HiChIP (Figures 1B, S1D-E). 1.7B in situ Hi-C valid pair reads provided uniform coverage of chromatin contacts genome-wide, while HiChIP experiments targeting H3K27ac, CTCF, the cohesin core component SMC1 and IKAROS enriched for contacts associated with these factors. Hi-C and HiChIP revealed chromatin organization into compartments, TADs, and specific long-range chromatin interactions (henceforth referred to as loops) and highlighted striking changes for each of these organizational features between WT and IKDN large pre-B cells (Figure 1C). Chromatin loops were stratified by annotation of their anchors as promoters (P), enhancers (E), structural (S), or other (O) using ChIP-seq datasets (Figures 1B,1D, S1E). Loops identified from different 3C techniques showed a high level of overlap and strong peak patterns (Figure S1F). Although different 3C approaches enriched for different categories of loops, e.g., H3K27ac HiChIP selectively identified loops involving enhancers and promoters, the distribution of loop types identified by an approach was similar between WT and IKDN (Figure 1D).
10%−17% of all chromatin loops identified by different 3C approaches were differential between the two genotypes and the same loops identified by different methods showed concordant changes (Figure 1D). APA and heatmap analysis of Hi-C contact counts confirmed that differential HiChIP loops reflect change in contacts and not merely changes in the precipitated antigen (Figure 1E). Notably, loops involving enhancers, especially at both anchors, were preferentially sensitive to disruption in IKDN across all 3C approaches (Figures 1D, S1G). For all 3C experiments, loops were longer-range in WT than IKDN suggesting that IKAROS supports longer-distance interactions (Figure 1F).
Loops downregulated in IKDN overlapped loops detected by IKAROS HiChIP in WT (Figure 1E). Approximately 80% of the downregulated chromatin loops identified by H3K27ac HiChIP had IKAROS at both anchors in WT and the rest had IKAROS at one anchor (Figure 1G). Moreover, >50% of these downregulated loop anchors had multiple IKAROS peaks in one anchor (Figure 1H). Loss of IKAROS binding from downregulated regulatory loop anchors correlated with loss in permissive histone modifications, the cohesin loader NIPBL, and SMC1 at these sites (Figure S1H).
Motif analysis of IKAROS peaks associated with downregulated H3K27ac loop anchors confirmed binding through cognate IKAROS sites and suggested co-occupancy with other B cell lineage TFs (e.g., E2A and EBF1) at a subset of sites (Figure S1I). While EBF1 motifs were enriched at anchors of loops both downregulated and preserved in IKDN, E2A motifs were enriched specifically among the downregulated. ChIP-seq of E2A, EBF1 and LEF1 confirmed their presence at downregulated H3K27ac loop anchors, albeit at a lower level relative to IKAROS (Figure S1J). A major fraction (67%) of downregulated CTCF loop anchors was also enriched for IKAROS binding but not for other B cell lineage TFs (Figures 1H, S1J). While in the T cell lineage CTCF was shown to interact with TCF1,59,60 Lef-1, its paralog expressed in B cells, showed a modest enrichment at 27 % of the differential CTCF loop anchors compared to 20% of unchanged anchors (Figure S1J).
Compared with downregulated chromatin loops, loops induced in IKDN were less likely to involve an anchor bound by IKAROS in WT cells (Figure 1G-H). Those that did showed a lower level of IKAROS enrichment compared to downregulated loop anchors (Figures 1I, S1H). Instead, anchors of loops gained in IKDN were strongly enriched with induced extra-lineage TFs42, such as TEAD1, LMO2 and LHX2 (Figure S1J-K). Chromatin modifications detected at these anchors in WT suggests priming that is permissive for binding of extra-lineage TFs in IKDN with some of these anchors exhibiting erasure of a WT Polycomb mark H3K27me3 (Figure S1H).
IKAROS also conferred selectivity for interaction partners. 9.4% of IKAROS-bound regulatory anchors from WT loops downregulated in IKDN were assembled with a different partner in a de novo loop in IKDN (Figure S1L). Consistent with the role of IKAROS in assembling longer-range interactions, the new loop formed in IKDN was shorter than the lost WT loop (Figure S1L). This redirection of loop anchors was also observed by CTCF HiChIP (21% of anchors) and 12% of this group were structural loops where an anchor not bound by IKAROS changed partner (Figure S1L).
Together these high-resolution chromatin organization studies establish a direct role of IKAROS in the maintenance and specificity of long-range interactions in WT large pre-B cells, particularly those that involve enhancers.
Control of B cell identity by IKAROS-dependent chromatin organization
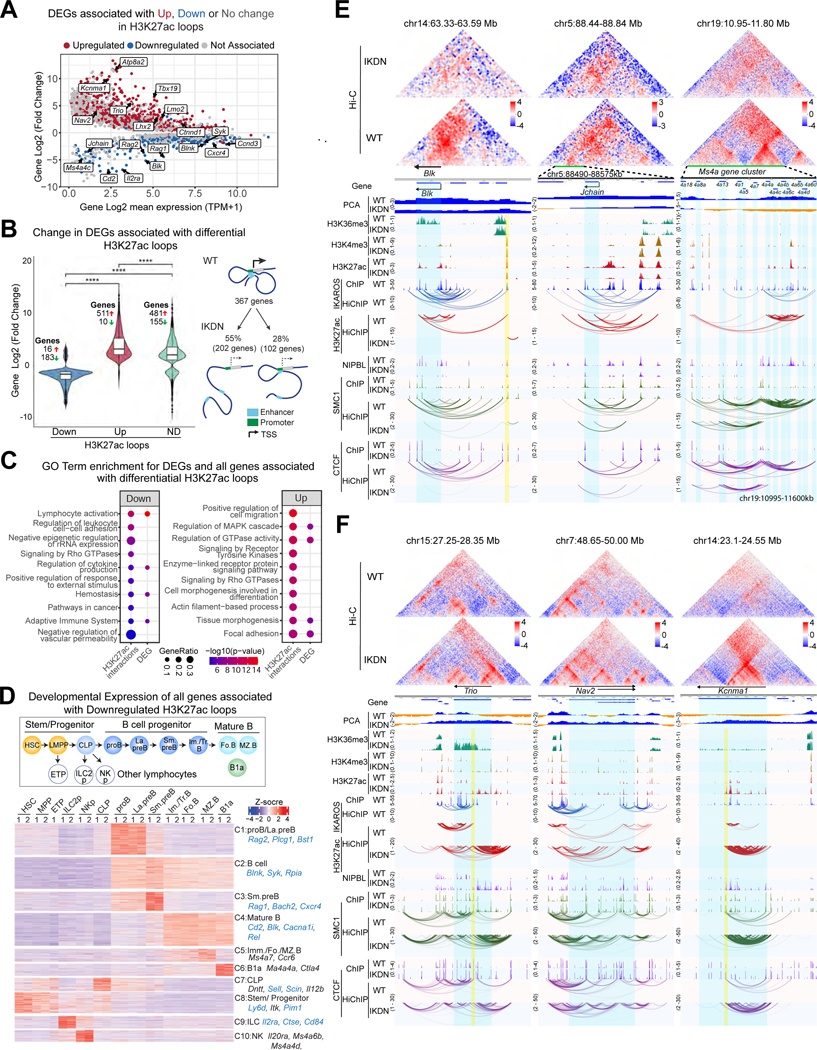
Differential analysis of gene expression identified 1,254 upregulated and 404 downregulated genes in IKDN large pre-B cells (Figure 2A). Among genes exclusively associated with an up or down E-P contact (H3K27ac HiChIP), there was high concordance between loss or gain of loops and gene expression (Figures 2A-B, S2A). Genes downregulated by loss of IKAROS were enriched for functional pathways that support B cell differentiation (Figure 2C). Of the 404 genes downregulated in IKDN, 367 had a loop to the promoter in WT cells. Of these, 53% were associated with loss of a promoter contact in IKDN and an additional 28% had loss of a loop to an anchor that also contacted the promoter in WT (Figure 2B). CTCF HiChIP revealed a similar albeit smaller overlap of downregulated genes with WT-specific CTCF loops (Figure S2A-C).
Figure 2. IKAROS-dependent 3D chromatin organization controls B cell identity.
(A) Change in gene expression between WT and IKDN (DEGs) and overlap with H3K27ac HiChIP loops that are up (red), down (blue) or unchanged (grey). (B) DEGs associated with differential (Down and Up) and non-differential (ND) loops identified by H3K27Ac HiChIP. **** p < 0.0001. (C) GO pathway enrichments for all genes associated with differential H3K27ac loops or DEGs. Statistical significance (−log10p-value) is shown by color scale and gene ratio by circle size. (D) K-means clustering of expression during WT hemo-lymphopoiesis and B cell differentiation of all genes associated with downregulated loop anchors. Examples downregulated in IKDN pre-B cells are highlighted in blue. (E) Examples of down-regulated and (F) up-regulated genes with changes in long-distance interactions. Hi-C contact maps (Observed/Expected, 5kb resolution). Compartment affiliation (PCA): euchromatin-blue, heterochromatin-orange. Normalized histone modification and transcription factor ChIP-seq tracks are shown. Interactions identified by FitHiChIP for IKAROS, H3K27ac, SMC1 and CTCF are shown. Highlights: blue -genes, yellow-re-directed loop anchors. Source: Tables S2-4.
Many genes whose expression was unchanged by IKAROS deletion were associated with differential loops. Analysis of their expression during hemo-lymphopoiesis61,62 revealed 10 clusters (Figure 2D). In addition to genes upregulated at the pre-B cell stage and sustained or downregulated in subsequent developmental stages (clusters 1–3), three clusters (4–6) expressed predominantly in mature B cells included “poised” and “primed” genes with no and low expression in pre-B cells respectively. A fourth category represented lymphoid lineage genes not expressed in B cells (clusters 8–10). These were likely primed at the chromatin level in multipotent progenitors for later expression in non-B cell lymphoid lineages that maintained long distance interactions in WT pre-B cells after lineage divergence. GO analysis of the clusters supports their inferred affiliation with hematopoietic lineage branches (Figure S2D).
Blk, required for pre-BCR signaling63, illustrates the effects of IKAROS loss (Figure 2E). Interactions between a cluster of enhancers, identified in WT, were lost in IKDN as were enrichment for H3K27ac, NIPBL and SMC1 at regulatory sites, the elongation mark H3K36me3, and Blk mRNA expression (Figure 2E). CTCF loops were lost despite minimal change in CTCF occupancy, although SMC1 binding was reduced at most anchors and NIPBL was reduced at internal regulatory sites. At the Rag1/2, Ahr, Jchain, Ms4a and TCRγ loci, a cluster of IKAROS-enriched sites connected enhancers, promoters and CTCF sites despite lack of transcriptional activity (Figures 2E, S2E). Loss in H3K27ac, reduction in NIPBL/SMC1 and the minimal change in CTCF occupancy correlated with lost interactions between regulatory and/or CTCF sites and a change in compartment affiliation.
GO terms for the 1254 genes upregulated in IKDN included focal adhesion and tissue morphogenesis (Figure 2C). Examples were Trio, ATP82a, Nav2, Kcnma1, Lpl and Abca1 implicated in focal adhesion dynamics64,neurodevelopmental disorders, channelopathies, and lipid metabolism65–69 (Figure 2A). 1021 of these genes had a P-E contact detected in IKDN, and of these 53% were associated with a novel regulatory loop to the promoter and an additional 27% had a novel loop to an anchor that contacted the promoter. IKAROS was not detected in WT cells at many of the IKDN-specific loop anchors associated with upregulated genes, indicating an indirect role of IKAROS loss in their induction (Figures 2F, S2F).
Thus, both priming and expression of lymphoid-specific genes correlated with establishment of a permissive chromatin landscape and 3D organization that involved spatial contacts between enhancers, promoters and structural sites, topological features that were dependent on IKAROS. Induction of de novo chromatin topological features upon IKAROS loss related in part to repurposing of regulatory sites normally bound by IKAROS and in part to indirect mechanisms.
Cross-boundary enhancer interactions organize lineage-specific TADs
IKAROS loss led to substantial changes in TAD organization. 22% of TADs in WT were either split or merged in IKDN and the boundaries of another 18% were shifted (Figures 3A, S3A). WT-specific TADs that split into smaller TADs in IKDN were on average twice the size of invariant TADs, suggesting that they were composed of two underlying TADs connected by IKAROS-dependent contacts that spanned a common border (Figures 3A, 3B:1). Consistent with this idea of a cryptic TAD boundary, 97% of the CTCF loops connecting the boundary anchors in the split TADs in IKDN were also present in the merged wild-type TADs. Filtering differential aggregate TAD analysis (ATA) to identify WT TADs split in IKDN with lost boundary-spanning contacts in IKDN revealed that this was the dominant mechanism (55%) of TAD splitting (Figures 3B:2, S3B:2). The inverse process was different mechanistically. Assembly of new loops spanning the WT TAD borders to join them in a merged TAD in IKDN was largely absent and weakening of boundary region interactions within the WT TADs dominated the pattern (Figure 3B:3, S3B:3). The Igκ locus provided examples of both TAD splitting and merging between the two genotypes (Figure 3C). Changes in TAD configuration were associated with extensive loss in inter- and intra-TAD interactions. Examples of lineage-specific TADs that split or merged in IKDN respectively are also shown for Ms4a and Ptpre (Figure S3C). Other TADs identified in WT cells displayed a change in interactions that redefined one of their boundaries in IKDN. Boundary contractions in IKDN were driven by loss of contacts that spanned an internal structural anchor to connect to the boundary region. In their absence the internal structural anchor became the new boundary (Figures 3B:4, 3D, S3C). Boundary expansions showed a more complex pattern of changes across the TAD (Figure S3B:4).
Figure 3. IKAROS regulates TAD and sub-TAD organization.
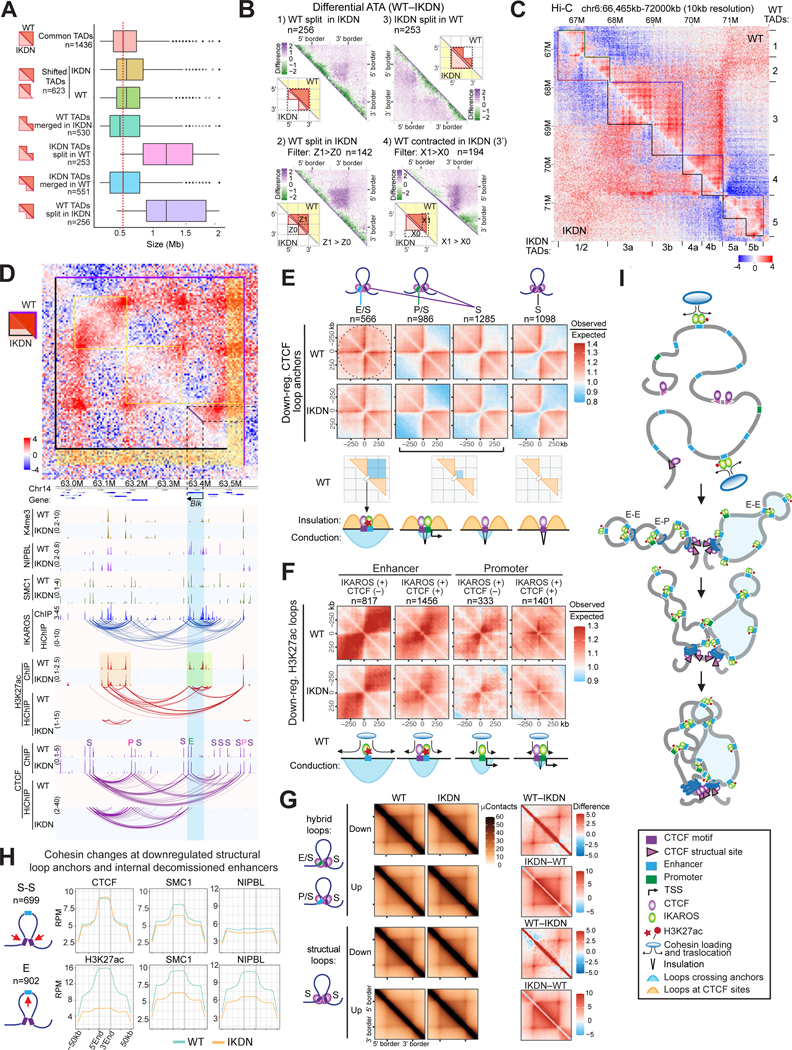
(A) Size distribution of common and differential TADs. Red dashed line shows the median size of common TADs. (B) Differential aggregate TAD analysis (ATA) of TADs classified into split (1,2) and contracted (4) in IKDN or split in WT (3). WT-IKDN score is shown in all panels. A schematic of TADs in WT and IKDN is shown with the perspective employed for analysis shaded yellow. (2) subset of TADs in (1) with (Z1 > Z0). (3) Note absence of signal in the Z0 sector. (4) Filter X1>X0 reveals the subset of contracted TADs where loss of border-spanning contacts drives the shift. (C) Merged and split TADs at the Igκ locus. O/E Hi-C contacts and TAD calls for WT and IKDN. (D) Loss in CTCF hybrid loops leading to TAD boundary shift around the Blk locus (yellow stripe). (E) Aggregate regional analysis (ARA) of WT and IKDN O/E Hi-C signal at anchors of downregulated hybrid and structural CTCF HiChIP loops. Schematics illustrate the insulation and conduction properties of each anchor type. (F) ARA of WT and IKDN O/E Hi-C signal at enhancer and promoter anchors of downregulated H3K27ac HiChIP loops with or without CTCF (+ or −) binding. Schematics illustrate interactions between adjacent domains and potential mechanisms involved. (G) Aggregate WT and IKDN Hi-C signals of downregulated and upregulated hybrid and structural CTCF HiChIP loops. Difference between WT and IKDN aggregate signals is shown on the right. (H) Average ChIP-seq enrichment (RPM) at downregulated structural loop anchors and at internal decommissioned enhancers. (I) Model by which IKAROS organizes TADs. Source: Tables S2-3, S5.
CTCF loop anchors can constrain interactions across them and play a significant role in TAD organization and boundary insulation. Anchors bound only by CTCF, whether assembled in differential structural (S-S) or hybrid (S-E, S-P) loops, exhibited strong insulating activity in both WT and IKDN (Figure 3E). In contrast, numerous interactions across enhancer-containing CTCF anchors were observed, indicating a role of enhancers in countering CTCF-mediated insulation (i.e., conduction) to allow long-range interactions between adjacent regions. This property was not specific to IKAROS-bound enhancer anchors as the newly formed hybrid loops in IKDN exhibited conduction across enhancer/CTCF anchors as well (Figure S3D). However, both the number of long-range interactions and the distance between contact points were greater across the IKAROS-bound enhancer/CTCF anchors. Enhancer anchors of downregulated H3K27ac loops without CTCF showed a dominant enrichment of long-range interactions spanning them (Figure 3F). This pattern was reduced at anchors that also bound CTCF, but a positive observed/expected ratio indicated a lack of effective insulation at these sites (Figure 3E-F). In contrast to enhancers, promoter anchors centered only short-range interactions, which were diminished by CTCF and eliminated by assembly with a second CTCF anchor (Figures 3E-F, S3E). Thus, enhancers can counteract insulating effects of CTCF-bound sites, and enhancers bound by IKAROS are particularly potent at doing so.
IKAROS loss also resulted in disruption and reconfiguration of some CTCF structural loops (Figures 1D, 3G). CTCF levels did not change at the anchors of downregulated structural loops and IKAROS was not present at these sites in WT (Figures 3H:S-S, S3G). However, all of these structural loops (n=699) spanned an enhancer that was inactivated by IKAROS removal. The internal enhancers that showed loss of H3K27ac also showed high levels of SMC1 and NIPBL in WT that were lost in IKDN (Figure 3H:E). This and the lack of NIPBL at structural anchors suggests that reduced cohesin loading at internal enhancers leads to the observed reduction of cohesin at the CTCF anchors and weakening of these structural loops. The Blk locus (Figures 2D, 3D) illustrates that lost CTCF structural loops show no change in CTCF at their anchors but loss of H3K27ac, NIPBL and SMC1 at internal enhancers. CTCF was also unaffected at disrupted hybrid and regulatory loop anchors, but IKAROS was enriched at the enhancer and promoter anchors and its loss correlated with reduced H3K27ac, NIPBL and cohesin levels consistent with a direct role in loop assembly (Figure S3F). In contrast to disrupted loops, both the structural and regulatory anchors of new CTCF loops in IKDN showed increase in both CTCF and cohesin occupancy (Figure S3F).
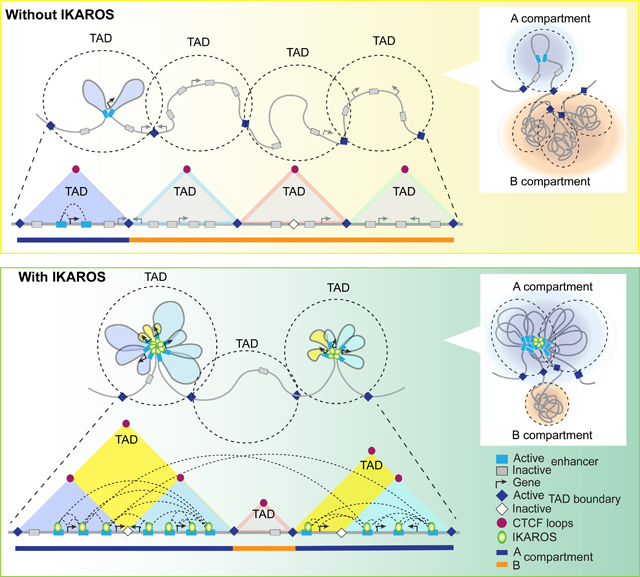
Taken together our studies demonstrate two ways in which IKAROS functionally interacts with cohesin-CTCF to organize chromatin. IKAROS binding at internal enhancers is necessary for cohesin loading required for maintenance of CTCF structural loops whose insulating properties are one component of TAD formation. IKAROS is also effective at overcoming CTCF insulating effects to assemble interactions that cross over CTCF-defined boundaries. These two activities based on IKAROS’ presence at the same enhancers combine to superimpose a lineage-specific TAD organization on a backbone of invariant structural loops (Figure 3I).
Long-range regulatory loops control lineage-specific nuclear compartmentalization
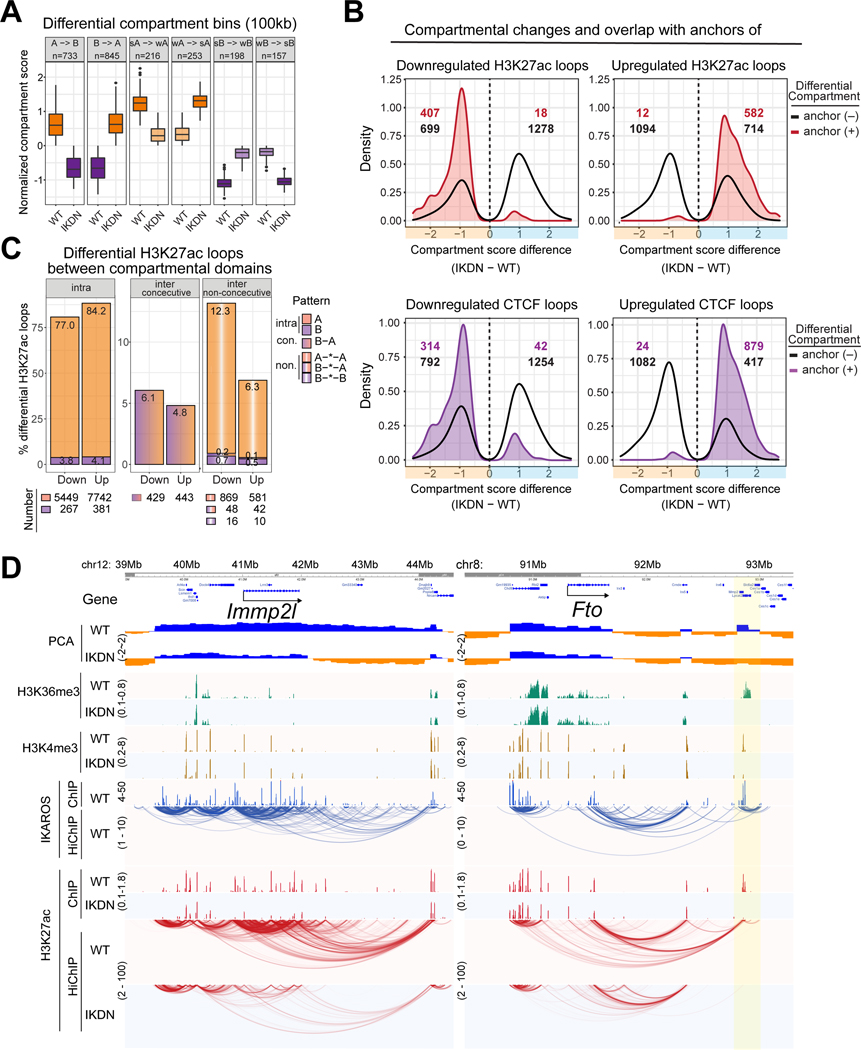
IKAROS loss influenced segregation of domains into nuclear compartments. ~10% (2402 100kb bins) of the genome showed a significant change in compartmental affiliation score with 3% and 3.5% showing a switch from A-to-B or B-to-A, respectively (Figure 4A). Overall, nearly half of the significant heterochromatic shifts (515/1106) overlapped a downregulated H3K27ac or CTCF loop (Figure 4B). IKAROS appeared to play a direct role in maintaining these regions in euchromatin since virtually all the H3K27ac and 90% of the CTCF downregulated loops had IKAROS bound at one or both anchors (Figure 1G). IKAROS also indirectly regulates the heterochromatic compartment and novel loops formed in its absence were associated with a shift towards euchromatin.
Figure 4. Higher-order chromatin compartmentalization is controlled by IKAROS.
(A) Bins with different compartment affiliation scores between IKDN and WT classified by affiliation shift. (B) Distributions of differential compartmental bins with (colored) or without (black outline) overlap with differential loop anchors identified by indicated HiChIP and numbers are shown. (C) Differential H3K27ac regulatory loops classified in WT as intra-compartmental, inter-compartmental and consecutive. (D) Examples of compartmental changes associated with intra-(left) and inter-(right) compartmental regulatory loops downregulated in IKDN. Source: Tables S2-3, S6.
Although most (~80%) differential regulatory loops were interactions within one A compartment (intra), 12% represented interactions between non-adjacent A compartments (Figure 4C). These inter-compartmental regulatory loops lost in IKDN were higher in frequency by 2-fold compared to the same type of regulatory loops induced in IKDN. This and the fact that a greater number of long-range loops (>600kb) are lost than formed in IKDN suggest that IKAROS’ contribution to assembling loops may include a mechanism that overcomes impediments to loop formation between discrete compartmental domains (Figure 4C, S4C).
The Immp2l region illustrates the flip to heterochromatin that occurs when IKAROS-dependent enhancers are decommissioned and associated long-range (>500kb) regulatory loops are lost in IKDN (Figure 4D). At the Fto locus, specific loss of an H3K27ac IKAROS-enriched peak and its inter-compartmental contacts flips an isolated euchromatic island (92.9Mb) to heterochromatin while another such island (92.3Mb) with no loss of loops stays euchromatic. These examples and genome-wide prevalence of such cases suggest that long-distance interactions between lymphoid-specific enhancers bound by IKAROS keep the associated regions affiliated with euchromatin (Figure S4D).
IKAROS maintains lineage-specific chromatin architecture during B cell differentiation
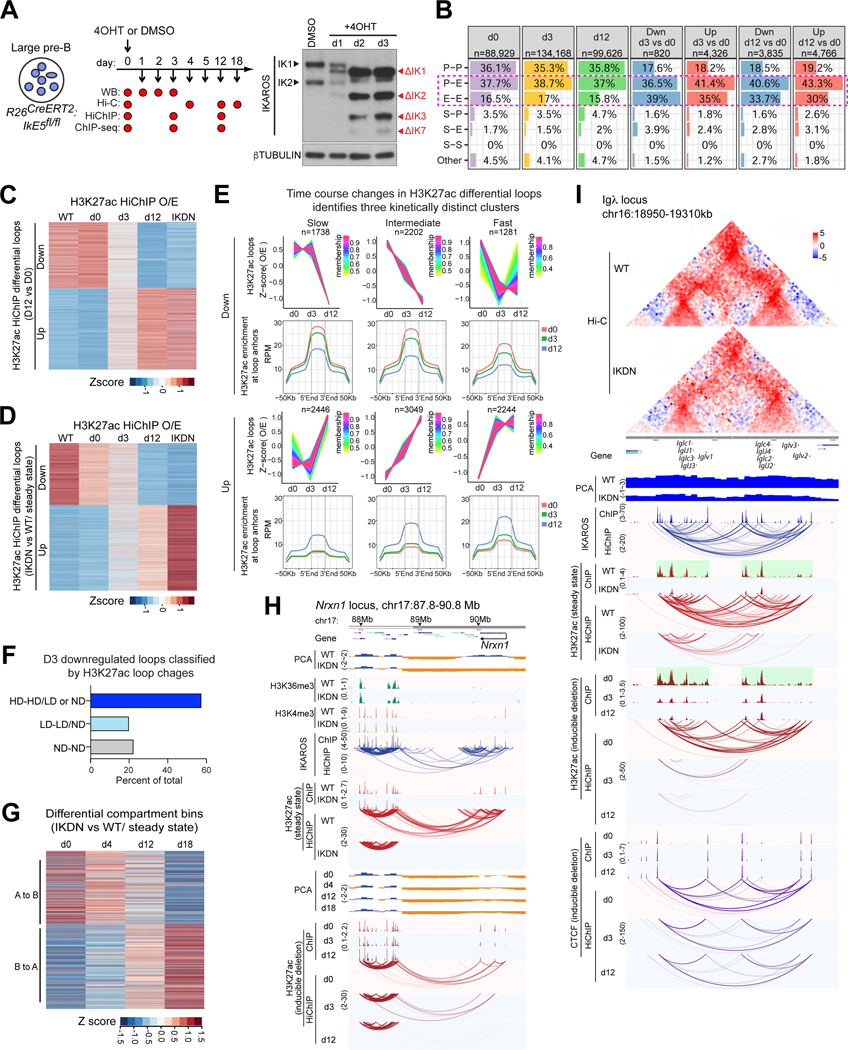
The effects of IKAROS deletion in pre-B cells could be secondary to developmental effects as Cre recombinase activity initiates at the CLP stage (Figure 1A). However, deletion of the IKAROS DNA binding domain in WT large pre-B cell cultures (Figure 5A) had similar effects on chromatin structure and organization to those observed in vivo (hereafter “steady state”). WT IKAROS protein was undetectable 2 days after Rosa26-Cre-ERT2 induction. HiChIP at d3 identified early responses after several rounds of cell division in the absence of IKAROS and at d12 and d18 approximate final configurations for different aspects of chromatin structure and organization (Figure S5A). Virtually all of the differential loops identified in vitro showed concordant changes in contact strength in steady state (WT vs IKDN; Figure 5C) and most of those identified in steady state (Figure 5D) showed the expected change in vitro, confirming that these are mainly expeditious responses to IKAROS removal rather than a developmental defect. Similar to steady state, enhancers were substantially overrepresented in anchors of differential regulatory loops identified in vitro (Figure 5B). The changes in regulatory loops after either in vivo or in vitro IKAROS deletion were confirmed by APA plots of Hi-C data from both systems (Figure S5B-E).
Figure 5. IKAROS maintains lineage-specific chromatin organization.
(A) Cultured large pre-B cells harvested at indicated time points after Cre-ERT2 induction and used for Hi-C, HiChIP and ChIP-seq. Detection of IKAROS WT (IK1, IK2) and mutant isoforms (ΔIK1, 2, 3, 7) is shown. (B) The number and percentage of loops (d0, d3, d12) and differential loops identified by H3K27ac HiChIP (d3 vs. d0 and d12 vs. d0), stratified according to anchor classification. (C) Contact counts for the differential H3K27ac HiChIP loops identified after in vitro deletion (d12 vs d0) are displayed for d0, d3, d12 and for WT and IKDN (in vivo). (D) Contact counts for the differential H3K27ac HiChIP loops identified after in vivo deletion (WT vs. IKDN) displayed as in (C). (E) Sites of differential H3K27ac interactions identified in vivo analyzed after in vitro deletion and segregated into clusters of slow, intermediate, and fast changes in contact strength. Average H3K27ac enrichment at the loop anchors and flanking regions is shown across the time course for each group. (F) Downregulated H3K27ac loops at d3 classified according to change of H3K27ac ChIP-seq signal at their anchors (HD high difference, LD low difference, ND no difference). (G) Z-score transformed compartment scores derived from Hi-C data of d0-d18 after in vitro IKAROS deletion were plotted for the differential compartment bins identified in vivo. Positive z-scores = stronger euchromatic affiliation in WT. (H, I) Steady-state (WT, IKDN upper tracks) and in vitro deletion data (d0, d3, d12, lower tracks) are compared for the Nrxn1 (H) and Igλ (I) locus. Source: Tables S2-3, S6-7.
In vitro deletion allowed timing of changes after IKAROS depletion. For almost all the differential loops identified at d12 change was detectable at d3 although it did not reach the threshold for loop call (Figure 5C). In the steady state, the correlated change of regulatory loops with local loss or gain of H3K27ac and other histone modifications links loop formation to enhancer commissioning (Figure S1H). The differential loop anchors identified in steady state were clustered according to their temporal behavior after in vitro IKAROS deletion and classified as slow, intermediate, or fast, based on interaction strength at d3 relative to d12 (Figure 5E). Loops fully established at d3 showed on average only 14% of the final H3K27ac gain at their anchors, and loops broken by d3 showed only 43% of the final reduction of H3K27ac. Inspection of individual loops suggested that these aggregate figures reflected a range of behaviors. To confirm this, we analyzed the H3K27ac peak heights at both anchors of all downregulated loops at d3 (Figure 5F). While 57% showed a significant change in acetylation peak in at least one anchor, 22% showed no detected change at either anchor. Thus, although changes in H3K27ac are often concomitant with loop changes, they are not obligatory for them (e.g., Figure S5G).
Genomic regions that underwent a compartmental change after IKAROS deletion in vivo exhibited a consistent change in compartmental score after deletion in vitro (Figure 5G). A to B transitions observed in steady state were detected at day 4 and further decreased to a negative value by day 18. B to A transitions occurred more slowly with smaller changes seen at day 4 but increasing scores at d12–18 (Figure S5F). This is consistent with the slow increase to histone acetylation levels that may promote a switch in compartment affiliation through homotypic interactions. Thus, the compartmental changes observed in steady state experiments initiate rapidly in response to IKAROS removal and continue to consolidate as 3D contacts and changes in the 1D chromatin landscape evolve over the ensuing period.
Genes exhibiting different orders of change are shown (Figure 5H-I, S5G-H). Inter-compartmental interactions associated with the Tgfbr2 locus are lost when IKAROS is deleted despite maintenance of euchromatin and H3K27ac at the anchors (Figure S5G). The Nrxn1 gene resides in an A compartment with both intra-and inter-compartmental interactions in WT cells (Figure 5H). For this locus, H3K27ac levels persisted to an extent at d3 after deletion but some loops were lost with accompanying change in compartmental affiliation. Loss of strong interactions between the constituent enhancers70 of two IKAROS-enriched super-enhancers of the Igλ locus was matched by H3K27ac loss at d3, while other loops were lost despite persistence of strong H3K27ac peaks (Figure 5I). The Ahr locus exhibits a decrease in regulatory loops and loss in A compartment affiliation, but in this case H3K27ac loss, loop breakage and compartment changes are closely correlated across the time course (Figure S5H).
Thus, IKAROS is required to actively maintain lineage-specific fine- and higher-order genome organization during early B cell differentiation by controlling enhancer interactions that mediate chromatin folding into domains and compartments (Figure 3I, S4D). Both formation and disruption of physical interactions between regulatory sites caused by IKAROS removal can precede changes in permissive chromatin marks.
IKAROS controls chromatin structure, organization, and locus contraction at the Igκ locus
Lymphocytes generate the capacity to respond to diverse antigens by genomic rearrangement that combines one element from an array of genetically related but functionally distinct variable segments with one of a small number of junctional segments and the constant region. In WT large pre-B cells, the 3.2MB Igκ locus is poised for recombination by assembly into a highly contracted configuration (Figures 3C, 6A-B, S6A). Three large topological domains, 700–2200 kB in size, harbor 100 Vκ regions (Figure 6A). The five Jκ regions that constitute the recombination center (RC) and the Cκ region reside within a 95kb boundary domain (the RC domain) within TAD-C (Figure 6A-C). In addition to extensive intra-domain contacts, the three topological domains show extensive inter-domain interactions that configure a 3.6MB superTAD (Figures 6A, S6A). FISH imaging with probes derived from the distal Vκ and Cκ confirmed the spatial proximity of the two ends of the locus in WT and demonstrated that removal of IKAROS and the loops sustained by it resulted in de-contraction that eliminated that close spatial association (Figure 6D-E).
Figure 6. Regulation of chromatin organization and locus contraction at the Igκ locus.
(A) Hi-C at the Igκ locus for WT and IKDN large pre-B cells (10kb). TADs (A-C) and FISH probes are marked. Orientation of Igκ-V segments relative to the J segments is indicated (red-forward and blue-reverse). (B) Genomic tracks as in Figure 5H. (C) The 3’ boundary domain is shown with ChIPseq peaks for indicated chromatin marks, B lineage TFs and other factors. Regulatory (iEκ, 3’Eκ, Ed, HS10) and structural (Cer, Sis) elements and the RCSE (green) that spans them are highlighted. (D) Igκ locus DNA-Immuno FISH using the Vκ (green) and Cκ (red) probes in WT and IKDN pre-B cells (pSer2 RNA Pol II (blue). (E) Distance between Vκ-Cκ regions in WT and IKDN large pre-B cells, **** p < 0.0001). Source: Tables S2-3, S6-7.
IKAROS binding was seen at numerous active enhancers that anchored both intra- and inter-TAD interactions (Figure 6B). These included the well-characterized Igκ iEκ, 3’Eκ, Ed and HS10 elements (Figure 6C).71–77 These conserved enhancers form an 83kb IKAROS-enriched super-enhancer (RCSE) in WT and engage in long-range interactions with other upstream IKAROS-enriched enhancers located in the three Vκ-containing TADs (Figure 6B-C). As H3K4me3 enrichment at the RC is required for RAG1/2 recruitment78, its absence is consistent with lack of Igκ recombination despite Rag1/2 expression in WT large pre-B cells. Induction of differentiation by IL-7 withdrawal induced both H3K4me3 over the RC and V-J rearrangements (Figures 6C, S1B). Thus, despite lack of active promoters and transcriptional activity in the WT large pre-B cell, the Igκ locus is pre-assembled by numerous long-distance interactions anchored at IKAROS-enriched active enhancers and poised for recombination.
Loss of IKAROS caused loss of interactions observed by all 3C approaches, including loops within and between TADs (Figure 6A-B, S6A-B, 3F). Lost interactions correlated with loss in H3K27ac and NIPBL/SMC1 and occurred despite little change in CTCF occupancy (Figure 6B-C, S6A-B). Notably, the RC domain anchored ~1/4 of all IKAROS-dependent enhancer loops and ~2/3 of the longer-range (> 0.9MB) enhancer loops lost at the Igκ locus in IKDN (Figure S6B-C). Loops connecting the RCSE cross over the CTCF-based structural Cer-Sis elements,79,80and upstream TAD boundaries, to connect to Vκ enhancers up to 2.5Mb upstream (Figure 6B-C, S6C). These include loops that assemble the RCSE with the E88 and E34 enhancers.81,82 Inducible deletion of IKAROS confirmed IKAROS’ active role in maintaining Igκ locus organization (Figures 6B, S6B-C, S6D). Loop changes identified by CTCF HiChIP were mostly complete by d3 and were not associated with loss of CTCF binding (Figure 6B). Many regulatory loops identified by H3K27ac HiChIP were also lost by d3, but loss continued through d12 and was confirmed by Hi-C contacts obtained after in vitro deletion (Figure 6A-B, S6A-B, D).
Most variable regions switched from euchromatin to heterochromatin in IKDN (Figure 6A-B, S6A). The switch correlated with loss of regulatory loops both within and across the Vκ−containing TADs. A small domain near the boundary of each WT variable region TAD remained euchromatic in IKDN, bracketing the large heterochromatic domains of the variable region in the de-contracted Igκ locus. These euchromatic regions displayed continued enrichment for the B cell lineage-specific TFs E2A, PAX5, IRF4 and EBF1 in IKDN at enhancers although H3K27ac was diminished at these sites (Figure 6C, S6B). These factors may contribute to residual long-range interactions and euchromatic localization of these regions in IKDN, but they are not sufficient for contraction of the Igκ locus or euchromatic retention.
IKAROS, by establishing long range interactions between active enhancers, organizes the locus into a configuration that supports deployment of the full range of Vκ rearrangements required for the generation of an effective humoral immune system.
Chromatin re-organization in skin epithelial cells.
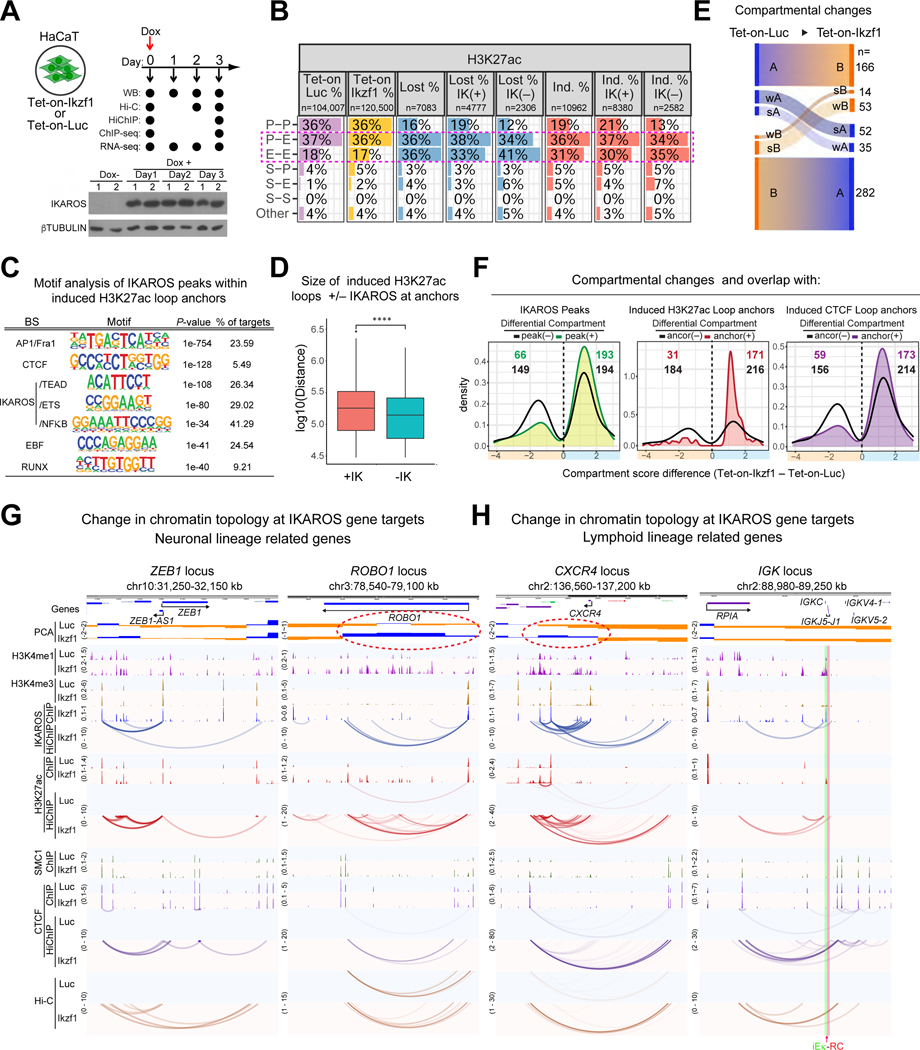
From loss of function, we infer that IKAROS binds to and assembles regulatory sites in 3D space to generate and maintain a chromatin conformation that is permissive for lineage-appropriate gene expression. These inferences and the sufficiency of IKAROS to exert these functions outside the hemo-lymphoid system were tested by gain-of-function. IKAROS expression was induced in HaCaT, a human skin epithelial cell line, and these were profiled 1–3 days later using 3C, ChIP-seq and RNA-seq (Figure 7A).
Figure 7. Extra-lineage chromatin organization induced by IKAROS in skin epithelial cells.
(A) Experimental strategy and Western blot of IKAROS induction. (B) Number and percentage of loops and differential loops (Lost, Induced (Ind.)) from H3K27ac HiChIP at d3 stratified according to anchor classification and presence/absence of IKAROS ChIP-seq peaks in at least one anchor (IK(+), IK(−)). (C) De novo TF-binding motif enrichment of IKAROS peaks within upregulated H3K27ac loop anchors. (D) Size distribution of induced H3K27ac loops at d3 with (+IK) or without (-IK) IKAROS at their anchors. **** p <= 0.0001. (E) Compartmental changes between Luciferase- and IKAROS-expressing cells at d2. (F) Distributions of differential compartmental regions with and without overlap with IKAROS peaks or induced H3K27ac or CTCF loop anchors. (G-H) Examples of IKAROS-mediated induction of chromatin topology. The IGK RC (red) and iEκ (green) are highlighted. Source: Tables S3-4, S6-7.
Interactions between enhancers (E-E) or enhancers and promoters (P-E) dominated both up and downregulated H3K27ac HiChIP loops as seen in lymphocytes (Figure 7B, 1D). Most of induced H3K27ac HiChIP loops (8380/10962) at d3 were bound by IKAROS in at least one anchor and were supported by changes in Hi-C contacts as early as d2 (Figures 7B, S7A). Motif analysis of IKAROS peaks at upregulated H3K27ac loop anchors confirmed binding through cognate sites and suggested co-occupancy with AP1 and RUNX (Figure 7C). Consistent with IKAROS’ role in longer-range loops in lymphocytes, induced loops with IKAROS at their anchors were on average larger than induced loops without (Figures 7D, 1F). Changes in compartment affiliation were detected two days after IKAROS induction (Figure 7E). Most were B to A transitions, and these euchromatic shifts were selectively enriched for IKAROS peaks and for induced H3K27ac and CTCF HiChIP loop anchors (Figures 7E-F, S7B).
Induction of IKAROS in HaCaT cells caused rapid downregulation of genes active in keratinocytes and associated pathways as early as d1 (Figure S7C-D). Genes associated with neuronal functions were prominent among upregulated genes and ~58% of upregulated genes were associated with induced regulatory loops. ZEB1 is not expressed in keratinocytes but is expressed in the CNS and in T cells.83,84 It resides in heterochromatin in HaCaT and is not involved in loops. IKAROS bound to H3K4me3-enriched ZEB1 promoters and generated loops among them, observed by HiChIP and Hi-C (Figure 7G). Little or no change in H3K27ac and CTCF binding at the novel anchor sites was observed despite robust loop formation but ZEB1 shifted to weaker heterochromatin. The axon guidance gene ROBO1 resides in a B compartment in HaCaT prior to IKAROS expression. IKAROS bound to the ROBO1 promoter and an array of intragenic enhancers that exhibited increased H3K27ac and SMC1. These elements and flanking structural sites not bound by IKAROS were assembled into novel loops as the locus switched to an A compartment and gene expression was induced (Figure 7G).
Although IKAROS ectopic expression does not confer lymphoid lineage identity to these ectodermal cells, it does organize a subset of lymphoid-specific genes in a lymphoid chromatin conformation (Figures 7H, S7E). Genes that support lymphoid cell functions showed priming of a permissive chromatin environment, newly assembled chromatin loops and in some cases, induced expression. CXCR4 and the linked CD28 and CTLA4 genes are expressed in pre-B cells and T cells respectively even though they exhibit extensive IKAROS-dependent contacts in pre-B cells (Figure S7E). In HaCaT, IKAROS bound to and promoted contacts between enhancers spanning the CD28 gene that resides in weak heterochromatin with concomitant strengthening of structural loops spanning the region, and a flip to euchromatin in the absence of H3K4me3 at promoters or induced gene expression. However, it did not access the IKAROS loop anchor sites observed in pre-B cells that surround the adjacent CTLA4 gene, which resides in stronger heterochromatin in HaCaT, or assemble loops involving this region. The IGK locus is buried in heterochromatin in HaCaT. While most of the contacts across the locus detected in pre-B cells remained absent, IKAROS was able to assemble the short-range contacts in the RC region that resides at the edge of the heterochromatic domain (Figure 7H). IKAROS bound to conserved enhancers including iEκ, H3K4me1 and SMC1 peaks were increased, and de novo regulatory and structural loops were assembled.
Two observations of note are shared by these examples. First, many of the loops assembled by IKAROS on lymphoid-specific genes in HaCaT are very similar to those observed in large pre-B cells. Figure S7E compares the loop structure in mouse pre-B cells and human HaCaT cells for two examples. The IKAROS peaks associated with anchor elements in contact in mouse pre-B cells and assembled in the induced loops in HaCaT are bound at homologous stretches of DNA in both cell types. Some of these loops were not attenuated in IKDN pre-B cells even though IKAROS is bound to them in pre-B cells and capable of assembling them in HaCaT. Second, in many examples, IKAROS formed new loops in weaker heterochromatin in HaCaT but did not assemble nearby sites that it binds to in pre-B cells when these had a stronger heterochromatin affiliation. In aggregate, newly assembled IKAROS anchors were largely restricted to euchromatin or weak heterochromatin.
In sum, IKAROS gain-of-function in cells where it is not normally expressed confirmed the ability of IKAROS to promote interactions between distant enhancers and promoters and CTCF structural sites and confer euchromatic localization to regions associated with induced loop anchors. When those activities were exerted at genes normally expressed in lymphocytes but not keratinocytes, IKAROS often bound to homologous regulatory sites and generated similar chromatin contacts.
Discussion
Assembly of a lymphoid lineage-defining chromatin architecture.
These observations document the capacity of IKAROS to assemble loops required for pre-B cell gene expression, its physical presence at loop anchors associated with much of the B cell lineage gene signature, and its ongoing requirement to maintain the pre-B cell configuration at multiple scales of chromatin structure. For actively expressed genes, these IKAROS-dependent contacts promote expression, for inactive genes, IKAROS loops maintain a poised state in accessible chromatin. While derived from an intermediate point in the lineage, several observations imply that IKAROS configures the genome earlier in the lineage, possibly in the HSC, and actively supports differentiation during progression through it. The genes associated with IKAROS-dependent loop anchors include those expressed in earlier progenitors but no longer required by the pre-B cell stage, and genes required in other branches of the lymphoid lineage that are not expressed in the B cell branch.
In ectopic expression studies, IKAROS could assemble lymphoid-specific loops between sites in euchromatin and shallow heterochromatin but not deep heterochromatin. It also assembled lymphoid lineage contacts that did not require IKAROS’s continued presence to maintain them in pre-B cells. Both observations, and the widespread presence of IKAROS at loop anchors that remain in contact after IKAROS depletion suggest that IKAROS may act early in the lineage when lymphoid genes remain in accessible chromatin. While many critical B cell genes require sustained IKAROS activity to maintain their accessibility and function, the initial configuration constructed by IKAROS in early progenitors, particularly at the order of TADs and compartments, may permit other lineage TFs, induced subsequently, to maintain many contacts and restore them after replication in the absence of IKAROS. The architecture assembled by IKAROS imparts the ability to contribute to alternative lineage choices in a multi-potent hemo-lymphoid progenitor. In contrast, IKAROS’ normal role in the ectodermal lineage is late and restricted to terminal branches of the lineage arbor. The selective upregulation of neuronal genes when IKAROS is expressed in keratinocytes may reflect lineage-appropriate access to these sites and the presence of cooperating TFs.
The suppression of chromatin configurations that support alternative lineages may be as important to lineage fidelity as the construction of lymphoid-appropriate contacts. Some of that suppression is direct, as many sites bound by IKAROS in WT assemble with new partners in IKDN. Some reflects the fact that an IKAROS assembled configuration constrains other interactions, as demonstrated by the many loops broken by forced IKAROS expression that do bind IKAROS at either anchor. This conformational restraint may contribute to a third mechanism whereby IKAROS represses the expression of DNA binding factors not normally expressed in pre-B cells that support assembly of novel chromatin contacts in the deleted cells when expressed.42,85–87 While the relative contributions of these mechanisms remain unclear, the striking fact is that the configuration of a lineage-appropriate genomic architecture includes the widespread suppression of contacts that would promote lineage-inappropriate gene expression. This activity to ensure lineage fidelity is important for normal development and its absence contributes to the pathological consequences of IKAROS loss. The frequent presence of IKAROS mutations in B-ALL and their association with poor prognosis can be attributed in part to the derepression of extra-lineage survival pathways that allow escape from the feedback mechanisms that regulate normal pre-B cells.
Mechanisms by which IKAROS configures the genome for lymphoid gene expression.
These results suggest the direct effects of IKAROS on genome configuration are mediated by binding to its cognate sites, particularly at enhancers and that most IKAROS-dependent loops positively influence either expression of actively transcribed genes or a poised configuration for future expression. Motif analysis and ChIP-seq studies revealed co-occupancy of other B lineage factors but these were not obligatory partners for IKAROS in either loop formation or stimulation of transcriptional activity.
IKAROS homotypic interactions are required for DNA binding, and IKAROS proteins are assembled in chromatin remodeling complexes with multiple IKAROS molecules.49,53,54 These self-associative properties and the frequent presence of IKAROS peaks at both anchors of regulatory loops lead us to speculate that multivalent IKAROS complexes directly mediate contacts between apposed cognate DNA sites. Higher-order local concentrations of IKAROS proteins are observed in the nucleus by immunohistochemistry88,89, suggesting additional mechanisms such as phase separation by which IKAROS proteins may be co-segregated to promote assembly of interacting regulatory sites into 3D superclusters.
The activities of IKAROS to configure the genome must be considered in the context of the cohesin loop extrusion and CTCF blockade mechanisms. IKAROS cooperates with this system in two ways. IKAROS bound at internal enhancers is associated with increased occupancy of the cohesin loader NIPBL and higher levels of cohesin at these sites. In its absence, cohesin loading internal to the structural anchors is diminished. This in turn can lead to decreased accumulation of cohesin at the structural anchors and disruption of the structural loop, despite no change in CTCF occupancy and no presence of IKAROS peaks at structural sites. The abundant regulatory contacts associated with IKAROS within a TAD may also depend on loop extrusion to bring regulatory anchors into apposition where interactions between IKAROS molecules may directly stabilize contact.
While the dominant role of IKAROS in maintaining the polarized interactions within TAD boundaries may be shared with other TFs that regulate 3D chromatin structure, a noteworthy aspect of IKAROS activity was evasion of the mechanism by which CTCF structural anchors constrain chromatin interactions within structurally defined TADs. The dramatic changes in TAD structure that occurred upon loss of IKAROS were not caused by the appearance of novel loops between CTCF anchors to define a new boundary. Instead, these CTCF loops were present in wild type, but the TAD boundary was rendered cryptic by enhancer contacts spanning the boundary. Loss of IKAROS removed these loops to reveal the underlying TAD structure.
This capacity of IKAROS-bound enhancers to assemble contacts over long-intervening stretches of DNA that can span strong CTCF structural loop anchors and also connect anchors in different euchromatic domains separated by intervening stretches of heterochromatin suggests that cohesin is not directly responsible for the generation of this class of loops. Instead, although cohesin assembled structural loops may bring DNA anchors in rough proximity, the physical association of these sites relies on IKAROS. As suggested by the model in Figures 3I and S4D, successive formation of stabilized regulatory contacts anchored in different structural loops both brings adjacent sites into proximity (zippering) and drives the formation of superclusters of IKAROS-bound sites which may then contribute to the strength of self-associative forces to attract additional elements including those separated by flanking stretches of heterochromatin.
While we postulate a direct role for IKAROS in mediating stable DNA contacts, we cannot exclude an indirect mechanism by which changes in histone marks after IKAROS loss cause changes in 3D contacts. However, based on the small average H3K27ac change at time points when loops are gained or lost and on inspection of examples spanning a range of behaviors, we conclude that although the accrual or loss of permissive marks associated with enhancer commissioning is correlated with changes in chromatin loops, the lack of an obligate order suggests that their interdependence does not reflect a strictly hierarchical mechanistic link. Assembly of loops in HaCaT cells in the absence of changes in histone marks further support that conclusion. Similarly, changes in compartment affiliation could occur rapidly and before histone changes were detected. Thus, although IKAROS can support changes in permissive histone modifications at chromatin loop anchors it may also operate independently to promote their spatial contact. Similarly, heterochromatin segregation and consolidation may be driven by spreading epigenetic marks, but this mechanism can be counteracted by IKAROS-mediated loop assembly to anchor otherwise susceptible regions into euchromatin.
The Igκ locus illustrates the effect of IKAROS-based interactions at different levels of chromatin organization. The remarkable contraction of this locus may be attributed to the reiterated Vκ regions which result in an extremely dense cluster of IKAROS-bound enhancers whose extensive contacts suggest a hierarchical assembly into superclusters. Abundant IKAROS-mediated interactions between these superclusters and the super-enhancer at the RC ignore TAD boundaries to assemble the locus into a highly compact euchromatic structure that allows deployment of the full repertoire of Vκ regions.90,91 The sustained binding of B cell lineage TFs and CTCF could not maintain this compact architecture when IKAROS was removed.
Conclusions
IKAROS is not unique in its capacity to overcome the insulating effects of CTCF and assemble domains that harbor lineage-specific genes59. It is notable in its capacity to do so from enhancers, across great distances and intervening regions of heterochromatin, and for the scope of its activity to pre-configure so much of the genome at the base of the lineage to enable the different branches of lymphoid development. IKAROS’ possible cooperation with factors such as TCF1, E2A or EBF1 to refine this configuration in its ongoing role as lineal branches diverge merits investigation. These observations provide a paradigm for how lineage-defining DNA binding proteins can direct the machineries that configure the genome in nuclear space to specify lineage potential and guide appropriate development along its divergent branches.
Limitations of the study
The aggregate analyses may obscure heterogeneous mechanisms and behaviors that are locus- or subset-specific. We mitigate this by stratification of organizational elements, temporal analysis, and direct evaluation of individual loci. These studies, while an accurate characterization of “acute” responses, are limited by the temporal resolution of induced genetic deletion.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Katia Georgopoulos (katia.georgopoulos@cbrc2.mgh.harvard.edu)
Materials availability
All plasmids and cell lines generated in this study are available from the lead contact without restriction.
Data and code availability
All sequencing data were deposited at NCBI Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Zenodo. The DOI is listed in the key resources table. Microscopy data reported in this paper is available from the lead contact upon request.
All original code is publicly available in GitHub. Links to Github and all software used as described in the STAR Methods are listed in the key resources table. A static snapshot of all original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| H3K4me1 | Active Motif | Cat#AM39297 |
| H3K4me3 | Active Motif | Cat#AM39155 |
| H3K27ac | Abcam | Cat#ab4729 |
| H3K27me3 | EMD Millipore | Cat#07–449 |
| H3K36me3 | Abcam | Cat#ab9050 |
| IKAROS | EMD Millipore | Cat#MABE912 |
| IKAROS | EMD Millipore | Cat#MABE913 |
| IKAROS | Abcam | Cat#ab191394 |
| CTCF | Abcam | Cat#ab188488 |
| NIPBL | Proteintech | Cat#18792–1-AP |
| SMC1a | Bethyl | Cat#A300–055A |
| GAPDH | Millipore | Cat#MAB374 |
| βTubulin | Santa Cruz Biotech | Cat#sc-9104 |
| FITC anti-mouse CD19 | Biolegend | Cat#152403 |
| Alexa Fluor 647 anti-mouse CD19 | Biolegend | Cat#115525 |
| APC anti-mouse CD43 | BD Biosciences | Cat#560663 |
| PE anti-mouse BP1 | Southern Biotech | Cat#1770–09L |
| APC anti-mouse B220 | Thermo Fisher Scientific | Cat#17–0452-81 |
| BV510 anti-mouse CD21 | BD Biosciences | Cat#563175 |
| PE anti-mouse CD23 | Thermo Fisher Scientific | Cat#12–0232-81 |
| BV421 anti-mouse CD5 | Biolegend | Cat#100629 |
| FITC-Ig-μ | BD Biosciences | Cat#553408 |
| FITC-Ig-κ | BD Biosciences | Cat#550003 |
| TER119 | BD Biosciences | Cat#553669 |
| Mac-1 (M1/70) | BD Biosciences | Cat#553307 |
| Gr-1 (RB6–8C5) | BD Biosciences | Cat#553123 |
| IgM (11/41) | BD Biosciences | Cat#562032 |
| CD3 (17A2) | BD Biosciences | Cat#557869 |
| CD8α (53–6.7) | BD Biosciences | Cat#553027 |
| TCRβ (H57–597) | BD Biosciences | Cat#553174 |
| CD49b (DX5) | BD Biosciences | Cat#553855 |
| Bacterial and virus strains | ||
| NEB® Stable Competent E. coli | New England Biolabs | C3040H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Hydroxytamoxifen (4OHT) | Sigma | Cat#H7904 |
| Biotin-14-dATP | ThermoFisher Scientific | Cat#19524016 |
| Doxycycline (Dox) | ThermoFisher Scientific | Cat#BP26531 |
| Equisul-SDT | Aurora Pharmaceutical | Cat#28000 |
| Recombinant Human IL-7 | Biolegend | Cat#715302 |
| KAPA HiFi HotStart ReadyMix | Roche | Cat#KK2601 |
| Protease inhibitor cocktail | Roche | Cat#11873580001 |
| Critical commercial assays | ||
| Pan B cell isolation kit II (mouse) | Miltenyi Biotec | Cat#130-104-443 |
| CFSE Cell Division Tracker Kit | Biolegend | Cat#423801 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23227 |
| ChIP DNA Clean and Concentrator | Zymo Research | Cat#D5205 |
| Quick-DNA Miniprep Plus kit | Zymo Research | Cat#D4068 |
| Direct-zol RNA Miniprep kit | Zymo Research | Cat#R2050 |
| Lenti-X Tet-On Advanced Inducible Expression System | Clontech | Cat# 630930 |
| TruSeq® Stranded mRNA Library Prep kit | Illumina | Cat#20020594 |
| Deposited Data | ||
| Raw and analyzed data | This study | GSE232490 |
| Western blot images | This study | https://doi.org/10.5281/zenodo.8339089 |
| Analyzed data | This study | https://doi.org/10.5281/zenodo.8339291 |
| RNAseq datasets for HSC, MPP, CLP,ILC2p and NKp cells | Shih et al, 201661 | GSE77695 |
| RNAseq datasets for pro-B, large pre-B and small pre-B cells | Kashiwagi et al, 202262 | GSE211079 |
| ChIP-seq datasets for B cell and extra-lineage transcription factors | Hu et al, 201642 | GSE86897 |
| Experimental Models: Cell lines | ||
| Human: HEK 293T/17 cells | ATCC | Cat#CRL-11268 |
| Human: HaCaT cells | J.M. Park | N/A |
| Mouse: OP9 bone marrow stromal cells | ATCC | Cat#CRL-2749 |
| Mouse: Passage 4 IkE5fl/fl hCD2-Cre (+/−) large pre-B cells |
This study |
N/A |
| Mouse: Passage 2 IkE5fl/fl Rosa26-Cre-ERT2 large pre-B cells | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| IkE5fl/fl hCD2-Cre (+/−) | Joshi et al, 201441 | N/A |
| IkE5fl/fl Rosa26-Cre-ERT2 | Hu et al, 201642 | N/A |
| Oligonnucleotides | ||
| Igk forward primer (Vκ) | Intergrated DNA Technologies (IDT) | GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC |
| Igk reverse primer (Jκ5) | IDT | ATGCGACGTCAACTGATAATGAGCCCTCTCC |
| Igk probe (upstream of Jκ5) | IDT | ACCAAGCTGGAGCTGAAACGTAAGTACAC |
| Dntt forward primer | IDT | CCCAAGTTCACTGTTCTTTCTGTC |
| Dntt reverse primer | IDT | CTCACCAGCAAACCCATAGGTA |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clontech | Cat# 630930 |
| pTRE-Tight-Luc control vector | Clontech | Cat# 630930 |
| pLVX-Tet-On Advanced | Clontech | Cat# 630930 |
| pLVX-Tight-FH-IKZF1 | This study | N/A |
| Software and Algorithms | ||
| Code for data analysis | This study | https://github.com/ay-lab/3D-IKAROS |
| FlowJo v10.9 | BD Biosciences | https://www.flowjo.com |
| bedGraphToBigWig v4 | Kent et al., 201096 | https://github.com/ENCODE-DCC/kentUtils |
| CALDER v1.0 | Liu et al., 2021107 | https://github.com/CSOgroup/CALDER |
| dcHiC v2.1 | Chakraborty et al., 2022106 | https://github.com/ay-lab/dcHiC |
| DEseq2 | Love et al., 201499 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FitHiC v.2.0.7 | Kaul et al., 2020104 | https://github.com/ay-lab/fithic |
| FitHiChIP v9.1 | Bhattacharyya et al., 2019105 | https://ay-lab.github.io/FitHiChIP/html/index.html |
| GENOVA v1.0.0.9 | van der Weide et al., 2021109 | https://github.com/robinweide/GENOVA |
| GenomicRanges v1.42.0 | Lawrence et al., 201398 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| Gene Ontology (GO) analysis | Zhou et al., 2019101 | https://metascape.org |
| HiCExplorer v3.5.1 | Wolff et al., 2020108 | https://hicexplorer.readthedocs.io/en/latest/index.html |
| HiC-Pro 2.8.0 | Servant et al., 2015102 | https://github.com/nservant/HiC-Pro |
| hicreppy v0.0.6 | Yang et al., 2017112 | https://github.com/cmdoret/hicreppy |
| Homer v4.11 | Heinz et al., 2010100 | http://homer.ucsd.edu/homer/ |
| Juicebox v1.11.08 | Durand et al., 2016103 | https://github.com/aidenlab/Juicebox |
| Liftover v2 | Hinrichs et al., 2006111 | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| MACS2 v2.11 | Zhang et al., 200895 | https://github.com/macs3-project/MACS |
| ngs.plot v2.63 | Shen et al., 201497 | https://github.com/shenlab-sinai/ngsplot |
| STAR v5.02 | Dobin et al., 201394 | https://github.com/alexdobin/STAR |
| TCseq v1.14 | Wu & Gu, 2022110 | https://doi.org/10.18129/B9.bioc.TCseq |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
The IkE5fl/fl mouse line was generated by insertion of loxP sites flanking Ikzf1 exon 5 by a standard gene-targeting method 41. The hCd2-Cre transgenic line (Strain #:027406), and the Rosa26-Cre-ERT2 (Strain #:008463) mice were purchased form the Jackson Laboratory. All mice were bred and maintained under pathogen-free conditions with antibiotics (Equisul-SDT 1:100) in water under standard husbandry conditions in the animal facility at Massachusetts General Hospital (building 149–8). At the time of analysis, mice were 6–10 weeks of age, healthy, tested naïve, and not involved in any previous procedure.
WT, IkE5fl/fl CD2-Cre and IkE5fl/fl Rosa26-ERT2-Cre mouse strains were used for isolation of bone marrow large pre-B cells. WT and IkE5fl/fl large pre-B cells deleted during B cell development using CD2-Cre were prepared from two female (WT_R1& IKDN_R1) and two male mice (WT_R2 & IKDN_R2). Large pre-B cells used for in vitro IKAROS deletion were prepared from two male IkE5fl/fl Rosa26-ERT2-Cre mice. No difference in phenotypes described in this study were observed between male and female mice. All animal experiments were done according to protocols approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital (Charlestown, MA) and in accordance with the guidelines set forth by the National Institutes of Health.
Large pre-B cell culture
Experiments were performed with large pre-B cells (sorted from two male and two female mice as described above) that were WT or conditionally deleted for the IKAROS DNA binding domain during B cell development (IkE5fl/fl CD2-Cre, n = 2 biological replicates). WT and IkE5Δ/Δ large pre-B cells (also referred to as IKDN) were sorted and cultured with OP9 stroma in DMEM (D-5671; Sigma) supplemented with 10% FBS (2442; Sigma), 10 mM HEPES (156–30-80; Gibco) 1× Glutamax (35050–062; Gibco), 1mM sodium pyruvate (11360–070; Gibco), 50 Units/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol and 5ng/ml IL-7 at 37°C. Stroma adherent large pre-B cells from WT or IKDN cultures were detached with 0.2% trypsin (EDTA free) for 2 minutes at room temperature. Large pre-B cells were ~ 99% of the cells in suspension as determined by flow cytometry. To investigate the effects of IL7 withdrawal-induced pre-B cell differentiation on Igκ recombination and histone modifications, WT large pre-B cells were first cultured in the presence of 5ng/ml IL7, then the culture medium was replaced with ‘IL7-’ medium and cultured for 1–2 days.
The IKAROS DNA binding domain was also deleted in vitro in pre-established WT large Pre-B cell cultures. WT large pre-B cells were sorted from IkE5fl/fl Rosa26-ERT2-Cre mice and expanded for 6 days on bone marrow derived stromal cells (OP9) in the presence of 5ng/ml of IL7 and 50μM 2-Mercaptoethanol (n = 2 biological replicates). They were then re-plated onto stromal cells with 0.2 μM of 4-OHT (Sigma H7904) or with an equal volume of DMSO as control. Cells were harvested at different time points (1–18 days) of culture in the presence of 4-OHT or DMSO. Pooled DMSO samples were used as undeleted (d0) control.
Human cell cultures
HaCaT (male) human skin keratinocyte cells and HEK 293/17 (female) cells were cultured in DMEM (D-5671; Sigma) supplemented with 10% FBS (2442; Sigma), 1× Glutamax (35050–062; Gibco), 1mM sodium pyruvate (11360–070; Gibco), 50 units/ml penicillin and 50 μg/ml streptomycin at 37°C.
METHOD DETAILS
Large pre-B cell isolation
For isolation of large pre-B cells, bone marrow (BM) cell preparations were depleted of cells binding to anti-Ter119, anti-Mac-1, anti-Gr-1, anti-IgM, anti-CD3, anti-CD8α, anti-TCRβ and anti-DX5 by removal with magnetic beads conjugated to BioMag goat anti-rat IgG (310107; Qiagen). The cells remaining after depletion were labeled with fluorochrome-conjugated monoclonal antibodies against B cell markers (anti-CD19, anti-CD43 and anti-BP1). Large pre-B cells from WT, IkE5fl/fl:CD2-Cre, or IkE5 fl/fl: Rosa26-ERT2-Cre were sorted as CD19+CD43+BP1+ using a three-laser FACS Fortessa (BD Biosciences) or a three-laser SONY SH800 Cell sorter (SONY Biotechnology).
Splenic B cell isolation
B cell subsets were obtained from the spleen of 10-week-old female mice. B cells were enriched with the pan B cell Isolation kit II (Miltenyi Biotech), stained for B cell surface markers (FITC anti-CD19, APC anti-B220, BV510 anti-CD21, PE anti-CD23, and BV421 anti-CD5), and sorted. Follicular B (CD19+, CD23+, CD21mid), marginal zone B (CD19+, CD23−, CD21hi), splenic B1a B (CD19+, CD5+, B220lo) and transitional/immature B (CD19+, B220+, CD5−, CD23−, CD21low/−) cell subsets were sorted with a Five-laser FACSAria Fusion Cell Sorter (BD Biosciences).
Flow cytometry for intracellular Igμ and Igκ
Large pre-B cells or splenic cells were fixed for 20 min at room temperature with 2% paraformaldehyde (Electron Microscopy Sciences) in PBS. After two washes with PBS, the cells were subsequently permeabilized for 20 min at 4 °C with 0.5% saponin and 2% FCS in PBS. The cells were stained for an additional 30 min at 4 °C with fluorescein isothiocyanate–conjugated anti-CD19 (115525, Biolegend) and anti-Igμ (553408; BD) or anti-CD19 (115525, Biolegend) and anti-Igκ (550003; BD), then were washed twice in 2% FCS in PBS before analysis on a FACSCanto (BD). The resulting files were analyzed by Flowjo v10 (BD Biosciences).
Ig kappa gene-rearrangement analysis.
Genomic DNA was isolated from sorted large pre-B cells or splenic B cells using Quick-DNA miniprep plus kit (Zymo Research). Rearrangements of genes encoding the immunoglobulin light chains were assessed by PCR with primers specific for Vκ -Jκ rearrangements as described 92. Southern blot hybridization of PCR products for V-J rearrangement was done with probes that bind upstream of the Jκ 5 region.
CFSE tracking of cell proliferation
Purified IkE5fl/fl, Rosa26-ERT2-Cre large pre-B cells were washed and resuspended at 5×106 cells/ml in pre-warmed labeling solution (0.1% FBS in PBS) in a 15ml conical tube. For CFSE labeling, equal volume of freshly prepared pre-warmed 10 μM CFSE solution was added to the tube, mixed well, and incubated in the dark for 10 min at 37°C with interval mixing. To stop the reaction, the tube was filled with ice-cold complete medium, and spined down at 350g for 5 min at 4°C. After another wash with ice-cold complete medium, labelled cells were resuspended with culture medium and seeded into a 6-well plate at 5×105 cells/well with OP9 stroma and 5ng/ml IL7. Fourteen hours later (d0), 4OHT was added to the medium to a final concentration of 0.2 μM, then cells were harvested every 24 hours and CFSE fluorescence was measured on a BD Canto flow cytometer. The resulting files were analyzed by Flowjo v10 (BD Biosciences).
Ectopic IKAROS expression in human keratinocytes
The “Lenti-X Tet-On Advanced Inducible Expression System” (Clontech) was used for tetracycline-controlled expression of IKAROS. A Flag-HA-Ikzf1 coding sequence was inserted between the XhoI and NotI cloning sites of the vector ‘pLVX-Tight-Puro’, and sequence verified. The resulting IKAROS response vector, ‘pLVX-Tight-FH-Ikzf1’, and a separate regulator vector ‘pLVX-Tet-On Advanced’ were packaged into lentiviruses using HEK 293T/17 cells. HEK 293T/17 cells were purchased from ATCC (ATCC CRL-11268) and cultured in DMEM with 10% FBS, 4.5 g/L Glucose, 4 mM L-Glutamine, 1.5 g/L NaHCO3, 1.0 mM Sodium pyruvate and 1% P/S at 37°C.
The HaCaT human skin keratinocyte cell line (male) was used for IKAROS ectopic expression (gift from Dr. Jin Mo Park/CBRC/MGH). HaCaT cells cultured in DMEM (D-5671; Sigma) and supplemented with 10% FBS (2442; Sigma), 1× Glutamax (35050–062; Gibco), 1mM sodium pyruvate (11360–070; Gibco), 50 units/ml penicillin and 50 μg/ml streptomycin at 37°C were simultaneously co-transduced with the two lentiviruses. Cells stably expressing both vectors were selected with puromycin (1 μg/ml) and G418 (200 μg/ml) and the inducibility of IKAROS with Doxycycline (Dox) was verified (as shown in Figure 7A). Luciferase inducible HaCaT cells were also generated as a control. The cells were cultured in DMEM medium with 10% ‘Tet System Approved FBS’ (Clontech), and then separated into ‘Dox+’ (0.1μg/ml) or ‘Dox-’ (DMSO) conditions for induction and non-induction pairs. Cells were harvested at day 1, day 2 and day 3 post-induction for downstream analyses.
Western blot analysis
Cells were washed with ice-cold PBS and then lysed in SDS sample buffer (50mM Tris-HCl, pH8, 1%SDS, 10%(v/v) Glycerol, protease inhibitors (Roche)). Protein concentration of the cell lysate was determined by BCA protein assay kit (Thermo Fisher). Equal amounts of lysate proteins were fractionated by SDS-PAGE and transferred to PVDF membranes (Millipore) and probed with anti-IKAROS, β−tubulin and GAPDH antibodies.
DNA-Immuno FISH
For DNA-Immuno FISH, Igkv (RP23–182E6) and Igkc (RP24–387E13) BACs were purchased from Children’s Hospital Oakland Research Institute (CHORI) and were labeled using nick translation. Cells were fixed with 2% paraformaldehyde (PFA) for 10 minutes on ice, treated with 0.2 μg/ml RNaseA for 30 minutes 37° C followed by 0.7% Triton/0.1 M HCl for 10 min on ice. During this time, the probes were denatured for 5 min at 95° C and allowed to pre-anneal for 30 min at 37°C. The cells were then denatured at 80° C for 10 min in 50% Formamide/2X SSC (adjusted to pH 7), then hybridized with the probes for 16 hr. The next day, the samples were washed, blocked and stained for pSer2 RNA Pol II (clone 3E10, EMD Millipore). Images were collected on a Leica TCS SP5 II STED laser scanning confocal microscope and image processing was performed using ImageJ. Distances between Vκ and Cκ were calculated using Euclidean Distance Transformation on Imaris.
ChIP-seq
For ChIPs cells were crosslinked in 1% fresh formaldehyde for 10 min at room temperature. For ChIP of NIPBL cells were first crosslinked with 2mM DSG for 45 min and then fixed with 1% formaldehyde for 10 min at RT. Crosslinked cells were then quenched with glycine and washed twice with ice-cold PBS. Cells were resuspended in lysis buffer 1 (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% IGEPAL CA-630, 0.25% Triton X-100 1x protease inhibitor cocktail [Roche]) for 10 min at 4°C, pelleted, and then resuspended in lysis buffer 2 (200mM NaCl, 1mM EDTA, 0.5mM EGTA, 10 mM Tris at pH 8, 1x protease inhibitor cocktail) for 5 min at 4°C. Cells were then resuspended in sonication buffer (50 mM HEPES at pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.2% SDS, 0.5mMPMSF, 1x protease inhibitor cocktail) and sonicated to an average size of 350 bp. Chromatin was cleared by centrifugation at 18,000g for 15 min, incubated with 5–10 μg of antibodies prebound to Dynabeads protein G (Life Technologies), and rotated overnight at 4°C. The antibodies used for ChIP are listed in the key resource table. ChIPs for each histone modifications and transcription factor were performed in parallel with wild-type and IKDN pre-B-cell chromatins, or with chromatins from different time points of inducible Ikzf1 deletion. After extensive washing on beads, bound chromatin was eluted and de-crosslinked in 300μl of elution buffer (50mMTris at pH 8, 10mMEDTA, 1% SDS, 0.3 M NaCl) overnight at 65°C. Proteinases K was added to a final concentration of 200 μg/mL and incubated for 2 h at 45°C. DNA was ethanol-precipitated, resuspended in TE, and purified using the DNA Clean and Concentrator-5 kit (Zymo Research).
Purified ChIP DNA was end repaired, end adenylated, and then ligated with Illumina Truseq indexed adaptors. The ligated DNA was purified with AMPure XP beads (Beckman Coulter) and then amplified with KAPA HiFi DNA Polymerase (KAPA Biosystems) for 8 to 13 cycles. After amplification, the library DNA was size selected with AMPurex XP beads to 200–600 bp range, and the purified libraries were multiplexed for sequencing in house with an Illumina NextSeq 550 system.
RNA-seq
Total RNAs from WT and IKDN primary large pre-B cells cultures or from HaCaT cells after IKAROS or Luciferase (control) induction were prepared using the Direct-zol RNA Miniprep (Zymo Research) and cDNA libraries were constructed for sequencing. The TruSeq Stranded mRNA Library prep kit was used for construction of cDNA libraries for RNA-sequencing (Illumina). The cDNA libraries were ligated with indexed primers and amplified by PCR for 10 cycles, multiplexed and sequenced in house with an Illumina NextSeq550 system.
In situ Hi-C
Large pre-B cells or HaCaT cells were crosslinked with 1% formaldehyde at RT for 10 minutes, and then quenched by the addition of 0.125 M glycine for 10 minutes. In situ Hi-C was performed following the established protocol6 with minor modifications. Nuclei were permeabilized, and DNA was digested overnight with 100 U MboI (for large pre-B cells) or DpnII (for HaCaT cells). The ends of the restriction fragments were labeled using biotin-14-dATP and then ligated in a 1.2 mL final volume. After reversal of crosslinks, ligated DNA was purified and sheared to a length of ~400 bp with a Covaris E220 evolution instrument (Covaris, Woburn, MA). Successful digestion, ligation and chromatin sheering were confirmed with an Agilent TapeStation 4200 system before library construction. Biotin labelled ligation junctions were pulled down with streptavidin beads, and DNA fragments were end-repaired, dA-tailed and Illumina adapters ligated. Libraries were produced by 8 cycles of PCR amplification with KAPA Hifi DNA polymerase (Roche). The Hi-C libraries were size selected (300–600bp) with AMPure XP beads and sequenced in house with an Illumina NEXTseq550 system or with a Novaseq 6000 system at the Knapp Center for Biomedical Discovery, University of Chicago.
HiChIP
HiChIP assays were performed according to the previously published protocol93 with minor modifications. ~10 million cells were crosslinked with 1% Formaldehyde for 10 mins at room temperature and lysed with Hi-C lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.2% NP40) supplemented with protease inhibitors for 30 mins at 4C. Chromatin was digested by the MboI (for WT and CD2-Cre IkE5Δ/Δ large pre-B cells) or DpnII (for ERT2-cre Ikzf1fl/fl large pre-B cells (4OHT or DMSO induction) and HaCaT cells) restriction enzyme for two hours at 37C. The 5’ overhang of the digested DNA were filled with dCTP, dGTP, dTTP and biotin-l14-dATP by DNA Polymerase I, Large Klenow fragment, and then ligated by T4 DNA ligase for 4 hours at room temperature. Chromatin was then sonicated in Nuclear Lysis buffer (50 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.6% SDS) with a Covaris E220evolution to an average size of 600bp. Sonicated chromatin was cleared by centrifugation at 18,000g for 15 min, diluted 1:2 in ChIP Dilution buffer (2% Triton X-100, 300 mM NaCl, 0.2% NaDoc), and then incubated with 5–10 μg of antibodies prebound to Dynabeads protein G (Life Technologies), and rotated overnight at 4°C. The antibodies used for ChIP are listed in the key resource table. After extensive washing on beads, bound chromatin was eluted and de-crosslinked in 200μl of elution buffer (50mMTris at pH 8, 10mMEDTA, 1% SDS, 0.3 M NaCl) for 3 hours at 67°C. Proteinases K was added to a final concentration of 200 μg/mL and incubated for 45 minutes at 45°C, and DNA was purified using the ChIP DNA Clean and Concentrator-5 kit (Zymo Research). The ChIP DNA containing Biotin-14-dATP labelled junctions was then purified by Streptavidin C1 magnetic, Tn5 tagmented and amplified with KAPA Hifi DNA polymerase (Roche) for 8–12 PCR cycles. The DNA libraries were size selected (300–600bp) with AMPure XP beads, quantified by both Qubit and qPCR analysis, and then sequenced in house with an Illumina NEXTseq550 system or with a Novaseq 6000 system at the Knapp Center for Biomedical Discovery, University of Chicago.
QUANTIFICATION AND STATISTICAL ANALYSIS
ChIP-seq analysis
Read alignment was performed on the mm10 assembly of the mouse mm10 or human hg19 assembly with the STAR 5.02 genome alignment algorithm94 (--alignIntronMax 1 --alignEndsType EndToEnd)on the DNAnexus platform (https://www.dnanexus.com/). Picard-tools 4.0.3 (Broad Institute) were used to remove PCR duplicates. Sequencing depth was tested by analyzing high quality biological replicates and by determining if independent data sets when combined yielded a significant higher number of new peaks. Peak calling was performed using MACS2 2.1.1 95 with input chromatin as control and with a q-value (minimum FDR) cutoff of 0.05. Model building with --mfold of 5, 50 was used for transcription factors, H3K4me3and H3K27ac. The –broad flag was used to identify enriched regions for spreading histone marks such as H3K4me1, H3K27ac, H3K9me3. Normalized “pileup.bedgraphs” were used after indexing for browser visualization. Bigwigs were obtained from the “pileup” files for visualization using bedGraphToBigWig v4.96 Peak overlap of at least 1 bp was performed to check the reproducibility of replicates (see Supplementary Fig. 1E). Heatmaps and enrichment profiles of average Reads Per Million mapped reads (RPM) were performed on indexed bam files using ngs.plot v2.6397. Significant peaks in at least one replicate were used for downstream analysis. The presence of ChIP-seq peak was assessed at loop anchors and A/B compartments by extracting the peaks that are located within these regions. Overlap of genomic regions and peaks contained within them was determined with the R package GenomicRanges v1.42.0.98
Motif analysis
To do the enrichment analysis for TF motif overlapping ChIP-seq peaks within loop anchors, we first extracted the peaks that were fully contained within H3K27ac loop anchors and then used the Homer script findMotifsGenome with default parameters.
RNA-seq and GO analysis
Alignment to the mouse mm10 or human hg19 assembly was conducted by STAR 5.0294 on the DNAnexus platform. Normalization and differential expression by DESeq299 were performed through a Homer v4.10.4100 implementation of the package in R. An adjusted p-value of < 0.05 and an absolute log2 fold change > 1 was used to identify differentially expressed genes. Metascape, a free gene annotation and analysis resource, was used for Gene Ontology (GO) analysis (https://metascape.org).101 To define genes associated with Hi-C and HiChIP loops, we searched for genes that had its transcription starts sites (TSS) within any of the loop anchors.
Hi-C analysis
Sequencing reads were aligned to the mouse mm10 (for mouse large pre-B cells) or human hg19 (for HaCaT cells) and processed into normalized contact maps with the HiC-Pro 2.8.0 pipeline102 on the DNAnexus platform. Default settings were used to remove duplicate reads, assign reads to restriction fragments, filter for valid pairs, and generate raw and ICE normalized interaction matrices at a range of resolutions. For visualization by Juicebox (v1.11.08)103 valid pairs were converted to .hic files using the script “hicpro2juicebox.sh”. For FitHiC analysis104, three contact matrix files (raw.matrix, abs.bed and iced.matrix.bias) generated by the HiC-Pro pipeline at 10kb resolution were converted using HiCPro2FitHiC.py to the three files (i.e., fragment.mapability, interaction.Count and .biases) required for running the algorithm. Statistically significant interactions were called using FitHiC v.2.0.7 (parameters: -L 20000 -U 3000000 -p 2 -b 200) and a false discovery rate (FDR) threshold of 0.0001 for each sample. The reproducibility of replicates at 10 kb resolution was verified using hicreppy v0.0.6 (parameter: --max-dist 5000000).
HiChIP analysis
HiChIP paired-end reads were aligned to the mouse mm10 (for mouse pre-B cells) or human hg19 (for HaCaT cells) using the HiC-Pro 2.8.0 pipeline as described under the Hi-C Data processing section. Valid pairs were combined across replicates and FitHiChIP v9.1 105 was used to identify statistically significant HiChIP interactions, using a bin size of 10 kb and reference peaks (i.e., H3K27ac, SMC1, CTCF, IKAROS) obtained from ChIP-seq data generated independently from the same cells/conditions as the HiChIP libraries. Statistically significant HiChIP interactions (q < 0.001) were called separately in each experiment with a minimum distance of 20 kb. For H3K27ac HiChIP, the UseP2PBackgrnd parameter was set to 0 and interactions were assessed for statistical significance if a ChIP-seq peak was present in at least one anchor (Peak-to-all interactions with maximum distance threshold of 4Mb). For SMC1 and CTCF HiChIP, the interactions were assessed for significance only when both anchors overlapped a ChIP-seq peak (Peak-to-peak interactions with maximum distance of 3Mb). For IKAROS HiChIP, the UseP2PBackgrnd parameter was set to 0 and interactions were assessed for significance if a peak was found in at least one anchor (Peak-to-all interactions with maximum distance of 3Mb and q-value less than 0.01). The reproducibility of replicates at 10 kb resolution was verified using hicreppy v0.0.6 (https://github.com/cmdoret/hicreppy) (parameter: --max-dist 5000000). Loop overlap between pairs of HiChIP datasets was performed by using a slack/extension of 5 kb on both sets of loops (+/− 1 bin around each loop anchor). To address loop anchor redirection, anchor overlap of at least 1 bp was used between the upregulated and downregulated loop anchors. Upregulated and downregulated loops in which at least one of its anchors correspond to a redirected anchor were selected for downstream analysis. The overlap of regions was performed using the R package GenomicRanges v1.42.0.
A/B compartment analysis
dcHiC v2.1106 was applied to contact maps at 100 kb resolution to identify differential A/B compartments bins (adjusted p-value < 0.05) in IKDN and WT. Genomic regions with positive and negative first principal component (PC1) values were classified as compartment A and B, respectively. Compartments bins with a significant increase in PC values in IKDN were further classified as either B→A, when there was a sign change in PC value, or weakA→strongA (wA→sA) and strongB→weakB (sB→wB) when the PC value was higher or lower in IKDN, respectively. Compartment bins with a significant decrease in PC values in IKDN were classified as either A→B, weakB→strongB (wB → sB) or strongA→weakA (sA→wA). Differential loop anchors were assessed for their association with compartment changes by classifying them with respect to their overlap with differential compartmental bins. The difference in compartmentalization for differential anchors was assessed by subtracting normalized PC values of the compartment bins they overlap in IKDN and WT. To define stretches of chromatin compartments, we first called sub-compartments by applying CALDER v1.0107 to contact maps at 100 kb resolution. Then, contiguous A.X.X and B.X.X bins were stitched together to extract stretches of A and B compartments, respectively.
Differential HiChIP loops were classified as intra-compartmental if the central position of each anchor was contained in the same compartment stretch and as inter-compartmental otherwise. Inter-compartmental loops were further subclassified into consecutive (between an A and a B compartment stretch by definition) and non-consecutive depending on whether the loop spans an adjacent compartment or compartments. To address the compartment changes across different time points (0, 4, 12 and 18 days) of in vitro Ikzf1 deletion (inducible system), dcHiC v2.1 106 was first applied to WT, day0, day4, day12, day18 and IKDN PC values derived from 100kb Hi-C contact maps for each. Then, we used the previously defined differential compartments between IKDN and WT and visualized the quantile normalized compartment scores across time points. For each differential compartment identified by comparing day0 to day18 of the inducible system, the relative change of compartment scores was quantified between day0 and day4, day4 and day12, and day12 and day18 relative to the total change between day18 and day0. Then, the median of these relative fractions of change among different bins were calculated for differential B→A and A→B compartments in each timepoint.
Topologically associating domain (TAD) analysis
Contact maps at 50 kb resolution in HDF5 format were generated with the tool hicConvertFormat from HiCExplorer v3.5.1.108 Topologically associating domain (TAD) calling was performed in IKDN and WT contact maps at 50 kb resolution with hicFindTADs from HiCExplorer v3.5.1 (parameters: --minDepth 150000 --maxDepth 500000 --step 50000 --thresholdComparisons 0.05 --delta 0.01 --correctForMultipleTesting fdr). We further classified TADs into merged, split and shifted (either expanded or contracted in IKDN). Regions defined by two or more neighboring TADs in IKDN that were contained in a bigger TAD in WT were classified as IKDN TADs merged in WT while allowing one 50 kb bin of slack at each boundary. WT TADs involved in the previously described pattern were classified as WT TADs split in IKDN, the rest of the differential TADs were classified as shifted. The IKDN TADs contracted at the 3’ were defined as a subgroup of shifted TADs that share one boundary with a WT TAD and a non-shared boundary contained within the larger WT TAD. Other combinations of shifted TADs were defined similarly. We further separated differential TADs into two groups with respect to their difference in interaction density in WT and IKDN. To classify split TAD, we calculated the median of the observed/expected contacts counts of regions Z1 and Z0 in WT and IKDN (Figure 3B, S3B). When the value of the region from the larger TAD (Z1) exceeded the value for the corresponding region in the condition with two smaller TADs (Z0), the TAD was assigned to one group. The same calculation was applied to filter merged TADs. Shifted TAD boundaries were often associated with a general weakening of contacts within the TAD that made boundary calling less reliable. TADs with changed boundary coordinates were filtered for those that showed a change in border defining contacts defined as a positive difference between the median of observed/expected contacts counts between the region incorporated in the larger TAD, (+/− 50 kb) to account for the surrounding region, and the corresponding region in the condition with the smaller TAD.
To show the average contact count distribution of differential TADs and their surroundings, we performed an aggregated TAD analysis (ATA) using the ATA-tool from the R package GENOVA v1.0.0.9.109 We retrieved the normalized contacts counts of differential TADs, including the up- and downstream 500kb regions, from Hi-C contact maps at 10 kb resolution. To match the dimensions of the count matrices across TADs, these were interpolated using bilinear interpolation. Then the aggregated matrix was calculated by taking the average of the bilinearly interpolated values for each pixel.
Loop anchor annotation
The anchors of HiChIP and Hi-C loops were classified as promoters (P), enhancer (E) and structural (S) (Schematic shown in Figure 1B). Anchors that contained a transcription start site (TSS) of GRCm38 (release 97) genes (for mouse large pre-B cells) and GRCh37 (release 19) genes (for HaCaT cells) were classified as promoters. The remaining anchors were first overlapped with H3K27ac ChIP-seq peaks and if the anchor had an overlap of at least 1 bp with one or more peaks, it was classified as an active enhancer. Then, among the remaining anchors, CTCF ChIP-seq peaks were used to define structural anchors following the same procedure. The anchors with absence of a TSS, H3K27ac or CTCF peaks were overlapped with H3K4me1 ChIP-seq peaks to annotate them as poised enhancers. Active and poised enhancer annotated anchors were combined as the latter group had very few elements. Interactions between regulatory anchors (P and E) and structural (S) were classified as hybrid loops. The overlap of regions was performed using the R package GenomicRanges v1.42.0.
Differential Hi-C/HiChIP interactions analysis
Differential loops were identified using 2 methods. The first method is a modified EdgeR script, using FDR with corrected by the Independent Hypothesis Weighting (IHW) and the logCPM as covariates. All the loops called by FitHiChIP in the IKDN, and WT condition were used to obtain differential interactions with an IHW corrected FDR < 0.05 and an absolute log2 fold change > 1. These differential interactions were further filtered and the interactions with raw contact counts > 3 and observed/expected counts > 1 in their respective condition were kept for downstream analysis. The second method referred to as Exclusive better captures longer-range differential interactions using the same data sets. This method identifies as differential interactions those that are called significant by FiTHiChIP in in at least one replicate in one condition but not in any replicate of the other condition. Differential loops called by the second method with raw contact counts > 10 and an absolute log2 fold change > 2 were kept for downstream analyses. The differential loop calls from the second method (Exclusive) were only used for intra- and inter-compartmental comparisons of steady state pre-B cells (WT vs IKDN; Figure 4C) and for analysis of HaCaT cells (Figure 7B, F). The first method (EdgeR), which utilizes replicate information, was used for the rest of the differential loop analyses throughout the paper. Changes in intensity of contacts for differential interactions were visualized by heatmaps of z-score transformed observed/expected HiChIP counts. To define Hi-C domains from differential HiChIP interactions, we used the end position of the 5’ anchor and the start position of the 3’ anchor as domain coordinates.
Decommissioned enhancer analysis
Decommissioned enhancers internal to structural loops were identified by selective H3K27ac enrichment in WT compared to IKDN after filtering out regions that also had H3K4me3. Enrichment for H3K27ac, SMC1 and NIPBL was plotted for the decommissioned enhancers in WT and IKDN. The Homer script findPeaks (parameters: -style histone) was used to obtain differentially enriched H3K27ac peaks between WT and IKDN using one condition as test and the other as control. Overlap of genomic regions was determined by the R package GenomicRanges v1.42.0.
Time course of loop changes upon inducible IKAROS deletion
To study the dynamics of differential loops, differential H3K27ac HiChIP interactions identified between IKDN and WT (steady state) were used to extract the observed/expected HiChIP counts from the d0, d3 and d12 (inducible) H3K27ac HiChIP data sets. Time-course clustering was generated using the R package TCseq v1.14.0110. Differential analysis of d0 vs d3 H3K27ac HiChIP interactions (described above and implemented by FitHiChIP v9.1) was used to classify differential loops according to the change in H3K27ac ChIP-seq coverage at their anchors. Each loop anchor was classified as ND: < 25% difference of H3K27ac signal at an anchor, and not statistically significant, LD: > 25% difference of H3K27ac signal at an anchor, and not statistically significant. HD: Significant difference of H3K27ac signal at an anchor (FDR < 0.05, |logFC|>1).
Aggregated Peak/Region Analysis
To show the average contact count distribution of loops and their surroundings, we performed an aggregate peak analysis (APA) of loop calls using the R package GENOVA v1.0.0.9.109 We used HiChIP and Hi-C contact maps at 10 kb resolution that were normalized by Knight-Ruiz (KR). We retrieved the normalized contact counts of a 100kb x 100kb region centered on each loop coordinate corresponding to a pair of loci: i and j. Without loss of generality, we can assume coordinate i < j and represent their genomic distance by d=d(i,j) for a given loop. The APA then plots the average contact count across all 100kb x 100kb with center pixel depicting (i,j), bottom left pixel corresponding to (i+50kb, j-50kb) and top right pixel denoting (i-50kb, j+50kb) with genomic distances of d, d-100kb and d+100kb, respectively. Due to effect of such difference in genomic distance, APA plots have higher average counts on bottom left and lower average count for the top right corner leading to asymmetry. Only loops having a distance > 150 kb were aggregated to avoid displaying pixels corresponding to very short-range interactions (less than 50kb genomic distance). For the aggregate region analysis (ARA), we retrieved the normalized contact counts of a 1Mb x 1Mb region on the diagonal of contact matrix that is centered on each target region coordinate (the square matrix defined by (i-500kb, i+500kb) for region i). To compute expected values, we first generated the aggregate matrix for the same target region but shifted 1Mb downstream. We then used the shifted matrix and computed the sum per genomic distance d divided by maximal possible contacts: sum(diagonal(d)) / number of elements in diagonal(d). To compute the observed/expected matrix, the normalized contact counts were divided by the expected matrix.
Mouse and human homology analysis
To lift loop anchors between mouse and human genome builds, we first extracted the IKAROS peak with the highest signal that was within the anchor. We then converted the peak coordinates from mm10 to hg19 using liftOver v2.111
Statistics and reproducibility
R v4.0.1 was used for statistical analysis and extraction and plotting of data from NGS-generated data files. The statistical significance between two groups was determined by the Wilcoxon or the t-test.
Supplementary Material
(A) Intracellular staining by flow cytometry for Immunoglobulin μ-chain (Igμ) and κ-chain (Igκ) was performed in IkE5Δ/Δ also referred to as IKDN (blue filled histogram; top rows), WT (red filled histogram; middle rows) large pre-B cells, and WT splenic B cells (black filled histogram, bottom row). White unfilled histograms show staining with isotype-matched control antibody. This analysis confirmed intracellular expression of Igμ and lack of expression of Igκ in WT and IKDN asserting their identity as large pre-B cells. (B) Lack of appreciable Vκ-Jκ recombination events were seen in both WT and IKDN large pre-B cells. Splenic B cells were used as controls. IL7 withdrawal (IL7-) from WT large pre-B cell cultures, normally grown on bone marrow stroma supplemented with IL7, caused cell cycle withdrawal and induced Vκ-Jκ recombination and H3K4me3 deposition at the recombination center (as shown in Figure 6C for the Jκ region). PCR for a Dntt genomic fragment was used as loading control. Autoradiograms for Vκ-Jκ recombination in large pre-B and splenic B cells are shown at the same exposure. (C) Deletion of the IKAROS DNA binding domain (IkE5Δ/Δ/IKDN) in large pre-B cells generated in vivo was confirmed by Western blotting (WB). The IKAROS DNA binding isoforms (IK1, IK2) expressed in WT cells and mutant isoforms lacking the DNA binding domain (ΔIK1,2) detected in IKDN are marked. (D) Distribution of reproducibility scores for WT and IKDN large pre-B cells as well as mouse embryonic stem cells (ESCs – outside control group) for Hi-C and HiChIP replicates. Stratum adjusted correlation coefficient (SCC) that measures reproducibility was computed at 10kb resolution using HiCrep.112 (E) Reproducibility analysis of H3K27ac, SMC1, CTCF and IKAROS ChIP-seq peaks between the two replicates (R1 and R2) of the same condition (WT or IKDN). Peaks were deemed overlapped between replicates if they intersected by at least 1 bp. (F) Aggregate peak analysis (APA) of significant chromatin loops (WT or IKDN) identified from Hi-C or HiChIP data as described in Figure 1D. For each case, the contact counts were aggregated from either WT or IKDN Hi-C data. Hi-C contact counts at 10kb resolution were aggregated +/− 50kb of each loop anchor and loops >150kb distance were used to avoid genomic distance patterns dominating the signal. (G) Heatmap of Observed/Expected (O/E) Hi-C, H3K27ac, SMC1 and CTCF HiChIP contact counts for loops identified as differential from comparison of WT vs IKDN H3K27ac HiChIP data. Each row was z-score transformed and hierarchical clustering was applied on these values to obtain the displayed ordering. (H) Heatmaps of histone modification and transcription factor binding enrichment at enhancer- and promoter-classified anchors (10kb) from differential H3K27ac loops is shown using ChIP-seq data from WT and IKDN large pre-B cells (±50 kb genomic window around the anchors). Scale bars denote Read count Per Million (RPM) as measured from ChIP-seq signals. (I) De novo transcription factor-binding motif enrichment analysis (HOMER) of IKAROS peaks within downregulated H3K27ac loop anchors. Motif enrichment at IKAROS peaks for loop anchors that are associated with differentially expressed genes (DEGs) and those that are not associated with DEGs is shown separately. The top motif hits alongside their significance (p-values) and frequency (percentage) are shown. Note that the core IKAROS binding motif (t/c/a/g G/A G G A A/t A/T) also presents a subset of binding motifs for the ETS, STAT, TEAD and NFkB family of transcription factors.42,113,114 (J) The distribution of the number of ChIP-seq peaks for B cell lineage transcription factors (i.e., E2A, EBF1, LEF1) and extra-lineage transcription factors (i.e., TEAD, LHX2, LMO2) that overlap anchors of differential loops identified from HiChIP and Hi-C experiments. (K) Motif analysis (HOMER) of TEAD, LHX2 and LMO2 peaks enriched at the anchors of induced loops in IKDN. (L) The size of original (WT-downregulated) and de novo (IKDN-induced) loops associated with redirected anchors of downregulated H3K27ac and CTCF loops. *** indicates p-value <= 0.001 using Wilcoxon test. The change in anchor interactions associated with loop redirection is shown in the schematic. Loop anchor depicted in red interacts with blue anchor in WT and with green anchor in IKDN. Source: Tables S1-3.
(A) Volcano plot of the log2 fold change (x-axis) of normalized contact counts vs. statistical significance (-log10FDR) of the difference (y-axis) for differential (Down/blue and Up/red) and non-differential (ND/gray) H3K27ac and CTCF HiChIP loops genome-wide. (B) MA plot illustrating the association of DEGs with different categories of CTCF HiChIP loops (Down, Up or Not Differential) similar to Figure 2A. (C) Log2 fold change of DEGs associated with differential (Down and Up) and non-differential (ND) CTCF loops similar to Figure 2B. Upregulated and downregulated genes associated with both an up and a down regulated anchor were excluded from this plot. (D) GO pathway enrichments for genes associated with the ten developmental clusters identified in Figure 2D and associated with genes whose 3D genome organization is lost upon IKAROS loss in large pre-B cells. Pr: progenitor, Ma: Mature. (E) Examples of downregulated and (F) upregulated genes with changes in long-distance interactions are shown as in Figure 2E and 2F. The Erag enhancer at the Rag1/2 locus that serves as an anchor for chromatin loops identified by different 3C approaches is highlighted. Source: Tables S2-4.
(A) Size distribution of domains defined by the coordinates of the anchors of downregulated and upregulated hybrid (P-S or E-S) and structural (S-S) CTCF HiChIP loops. Red dashed line shows the median size of common TADs. (B) Differential ATA of TADs classified into merged (1) in WT, split in IKDN (2), merged in IKDN (3) and contracted (4) in WT is shown for the two conditions. A schematic of the method used to identify differential TADs and their number are shown on the left side of each panel. This analysis performed from the second condition point of view (Z1<=Z0) complements the one provided in Figure 3B. (C) Examples of split merged and shifted WT TADs that contain the Ms4a, Ptpre, Mapk8 and TCRγ loci are shown, respectively. (D) ARA of WT and IKDN (O/E) Hi-C signal from anchors classified as enhancers (E/S), promoters (P/S) and structural (S) in upregulated hybrid CTCF HiChIP loops. Aggregate signals from anchors (S) of upregulated structural CTCF loops (S-S) are shown as a comparison. The dotted circle in the enhancer anchor ARA plot in IKDN indicates the range of across anchor interactions. This radius is smaller compared to the one shown for active enhancer anchors in WT as shown in Figure 3E. Bottom schematic illustrates the insulation and conduction properties of each anchor type as detailed in the key panel. (E) ARA of WT and IKDN O/E Hi-C signal from enhancer- and promoter-classified anchors of upregulated H3K27ac HiChIP loops. These were further classified for presence or absence of CTCF (+ or −) binding. Schematics below the ARA plots illustrate the range of interactions between adjacent domains and potential mechanisms involved in the process as detailed in the shared key panel with D. (F) Distribution of the average ChIP-seq Read count Per Million (RPM) of factors and histone modifications at anchors (10kb) classified as enhancers, promoters, or structural sites from downregulated and upregulated CTCF HiChIP loops, using a ±50 kb genomic window around the anchors. Source: Tables S2-3, S5.
(A-B) Compartmental score difference between IKDN and WT (IKDN-WT) calculated for regions contained within differential CTCF hybrid loops and H3K27ac differential regulatory loops. (C) Size comparison of down and upregulated regulatory loops is shown by both density vs. size distribution and box plot. **** indicates p-value <= 0.0001 using Wilcoxon test. (D) Schematic illustration of a model mechanism by which IKAROS-mediated spatial interactions between enhancers located at distal compartmental domains supports euchromatic localization. Source: Table S3, S6.
(A) Proliferation profile of cultured WT large pre-B cells isolated from IkE5fl/fl Rosa26-ERT2-Cre mice that were treated (4OHT) or not treated (DMSO) with tamoxifen for in vitro IKAROS deletion. (B) APA using Hi-C contact maps from in vitro IKAROS deletion (d0, d4, d12 and d18) as well as from the in vivo system (WT, IKDN) confirms bona fide interaction changes for differential H3K27ac HiChIP loops identified from comparisons of d3 vs d0 (top) and d12 vs d0 (bottom). (C) Similar APA plots confirm in vitro interaction changes for differential H3K27ac HiChIP loops identified from in vivo comparison (WT vs IKDN). (D) Heatmap of O/E Hi-C contact counts for pooled d0, d3, d12 replicates and WT and IKDN replicates for differential H3K27ac loops at d12 vs d0 is shown as in Figure 5C. (E) Heatmap of O/E Hi-C contact counts for pooled d0, d3, d12 replicates and WT and IKDN replicates at the anchors of differential H3K27ac chromatin loops in IKDN vs WT is shown as in Figure 5D. (F) Relative change in the median values of compartmental scores between d0-d18 is calculated for the compartmental changes established in steady state WT and IKDN as shown in Figure 5G and Figure 4A. (G) Example of loss in long-distance interactions at the Tgfbr2 and (H) Ahr loci detected in vivo and in vitro after IKAROS deletion as described for examples in Figures 5H-I. Source: Table S2-3, S6-7.
(A) Chromatin organization at the Igκ locus is shown as in Figure 6A-C that includes significant long-distance interactions identified in ex vivo (steady state) WT and IKDN large pre-B cells by FitHiChIP for IKAROS, H3K27ac, SMC1 and CTCF HiChIP data sets. Chromatin compartmentalization (PCA) and normalized histone modification and transcription factor signals obtained by ChIP-seq analysis under steady state conditions, are also shown. (B) Enlarged view of chromatin 1D and 3D maps at the 3’ end of the Igκ locus highlighting some of the long-range enhancer interactions that connect the elements of a super-enhancer located at the RC boundary domain with enhancers within TAD-C, including the recently characterized E34 enhancer. These E-E interactions cross over the CTCF Cer-Sis elements that define the 3’ boundary of TAD-C and are highly dependent on IKAROS DNA binding for their formation. Significant H3K27ac and CTCF interactions, histone modification and transcription factor enrichment are shown as in Figure 6B and S6A. (C) Size distribution of differential H3K27ac and CTCF interactions at the Igκ locus. The H3K27ac enhancer-enhancer interactions are on average double the size of CTCF chromatin loops reaching over 2.5Mb distances to make contacts between the enhancers at the recombination center (RC) with enhancers proximal to Vκ regions in the three Igκ TADs (A, B, C). A majority (60/84) of long-range interactions > 900kB originate from the RC. These data support a model that the 3.2MB Igκ locus contraction and recombination relies on long-range enhancer-based interactions rather than CTCF-cohesin loop extrusion. (D) Hi-C contact maps (O/E) generated by Juicebox are shown at the Igκ locus at 10 kb resolution prior to and after inducible IKAROS deletion (Rosa26-Cre-ERT2, IKE5 fl/fl) at d0, d4, d12, and d18. Source: Table S2-3, S6-7
(A) Long-distance significant H3K27ac interactions (>150kb) induced at d3 were confirmed by APA of Hi-C contacts at d2 after IKAROS induction stratified by IKAROS presence at anchors. (B) Distributions of compartment score differences calculated for 100kB differential compartment bins that overlap with lost H3K27ac and CTCF loop anchors as described in Figure 7E. (C) GO analysis for DEGs (up and downregulated) at the earliest time point (d1 vs. d0) after IKAROS induction. Downregulated genes enriched for pathways that support keratinocyte differentiation, whereas upregulated genes enriched for pathways that support neuronal functions (highlighted in red). (D) Log2 fold change of DEGs (d2 vs. d0) associated with differential (Upregulated or Downregulated) and Non-Differential H3K27ac loops is shown as detailed in Figure 2B. Up and downregulated genes associated with both an up and a down regulated loop were excluded from the figure. (E) Examples of lymphoid gene targets (BCL6, CD28-CTLA4) that upon IKAROS expression share features in chromatin topology between the two different cell types and species are highlighted. Conserved loop anchors associated with IKAROS peaks were identified with LiftOver111 and highlighted with the same color across mouse and human tracks. Source: Table S3-4, S6-7.
Table S1. Protocol details and mapping statistics for HiC and HiChIP experiments, related to Figures 1–7.
Table S2. Loop calls from HiC and HiChIP in wildtype (WT) and IkE5Δ/Δ (IKDN) large pre-B cells, related to Figures 1–6.
Table S3. Differential loop calls from steady state and inducible HiC and HiChIP in mouse large pre-B cells and human skin epithelial cells, related to Figures 1–7.
Table S4. Differentially expressed genes in large pre-B cells and human skin epithelial cells, related to Figures 2, 7
Table S5. Topologically associating domains in WT and IKDN large pre-B cells, related to Figure 3.
Table S6. Differential compartments in mouse large pre-B cells and human skin epithelial cells, related to Figures 4, 5, 7.
Table S7. Loop calls from inducible IKAROS deletion in mouse large pre-B cells and IKAROS ectopic expression in human skin epithelial cells, related to Figures 5, 7.
Bullets.
Cohesin loading at lymphoid enhancers by IKAROS supports formation of structural loops
IKAROS bound enhancers override CTCF boundaries to assemble lineage-specific domains
IKAROS-based 3D contacts reach over heterochromatin to place domains into euchromatin
Igκ locus contraction in euchromatin relies on IKAROS-dependent enhancer contacts.
Acknowledgements
We thank Drs David Gilbert and Kristin White for manuscript review, Eleanor Wu and Robert Czyzewski for mouse husbandry. Research was supported by NIH R01HL140622 and R01AR069132, R56AR082402 to KG, NIH T32 AI007512 to YH, NIH R35GM128938 to FA, NIH R21AR074748 to BM, NIH R01AI150860 to MC.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Zheng H, and Xie W. (2019). The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20, 535–550. 10.1038/s41580-019-0132-4. [DOI] [PubMed] [Google Scholar]
- 2.Stadhouders R, Filion GJ, and Graf T. (2019). Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569, 345–354. 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- 3.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, and Cavalli G. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, and Aiden EL (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, and Mirny LA (2016). Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049. 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gassler J, Brandao HB, Imakaev M, Flyamer IM, Ladstatter S, Bickmore WA, Peters JM, Mirny LA, and Tachibana K. (2017). A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36, 3600–3618. 10.15252/embj.201798083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. (2015). Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences of the United States of America 112, E6456–6465. 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, and Spitz F. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324. 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wutz G, Varnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, Schoenfelder S, Jessberger G, Muhar M, Hossain MJ, et al. (2017). Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J 36, 3573–3599. 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. (2017). The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707 e614. 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vian L, Pekowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, Huang SC, El Khattabi L, Dose M, Pruett N, et al. (2018). The Energetics and Physiological Impact of Cohesin Extrusion. Cell 175, 292–294. 10.1016/j.cell.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, and Bruneau BG (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169, 930–944 e922. 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo N, Ishii H, Xiong X, Bianco S, Meitinger F, Hu R, Hocker JD, Conte M, Gorkin D, Yu M, et al. (2021). Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat Struct Mol Biol 28, 152–161. 10.1038/s41594-020-00539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh TS, Cattoglio C, Slobodyanyuk E, Hansen AS, Darzacq X, and Tjian R. (2022). Enhancer-promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat Genet 54, 1919–1932. 10.1038/s41588-022-01223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojic A, Cuadrado A, De Koninck M, Gimenez-Llorente D, Rodriguez-Corsino M, Gomez-Lopez G, Le Dily F, Marti-Renom MA, and Losada A. (2018). Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol 25, 496–504. 10.1038/s41594-018-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiecke MJ, Wutz G, Muhar M, Tang W, Bevan S, Malysheva V, Stocsits R, Neumann T, Zuber J, Fraser P, et al. (2020). Cohesin-Dependent and -Independent Mechanisms Mediate Chromosomal Contacts between Promoters and Enhancers. Cell Rep 32, 107929. 10.1016/j.celrep.2020.107929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu NQ, Maresca M, van den Brand T, Braccioli L, Schijns M, Teunissen H, Bruneau BG, Nora EP, and de Wit E. (2021). WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat Genet 53, 100–109. 10.1038/s41588-020-00744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinzema NJ, Sofiadis K, Tjalsma SJD, Verstegen M, Oz Y, Valdes-Quezada C, Felder AK, Filipovska T, van der Elst S, de Andrade Dos Ramos, Z., et al. (2022). Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat Struct Mol Biol 29, 563–574. 10.1038/s41594-022-00787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krivega I, and Dean A. (2017). LDB1-mediated enhancer looping can be established independent of mediator and cohesin. Nucleic Acids Res 45, 8255–8268. 10.1093/nar/gkx433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, and Gilbert DM (2010). Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20, 761–770. 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De S, and Michor F. (2011). DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol 29, 1103–1108. 10.1038/nbt.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, and Corces VG (2017). Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol Cell 67, 837–852 e837. 10.1016/j.molcel.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley MJ, and Corces VG (2018). Organizational principles of 3D genome architecture. Nat Rev Genet 19, 789–800. 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchal C, Sima J, and Gilbert DM (2019). Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol 20, 721–737. 10.1038/s41580-019-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, et al. (2019). Identifying cis Elements for Spatiotemporal Control of Mammalian DNA Replication. Cell 176, 816–830 e818. 10.1016/j.cell.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie L, Dong P, Qi Y, Hsieh TS, English BP, Jung S, Chen X, De Marzio M, Casellas R, Chang HY, et al. (2022). BRD2 compartmentalizes the accessible genome. Nat Genet 54, 481–491. 10.1038/s41588-022-01044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240. 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, and Murre C. (2012). Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nature immunology 13, 1196–1204. 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. (2017). Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 171, 103–119 e118. 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, Cui K, Fang D, Hirose S, Wang X, Wangsa D, Jin W, Ried T, Liu P, Zhu J, et al. (2018). Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 48, 227–242 e228. 10.1016/j.immuni.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgopoulos K, Moore DD, and Derfler B. (1992). Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258, 808–812. [DOI] [PubMed] [Google Scholar]
- 36.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, and Sharpe A. (1994). The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156. 0092–8674(94)90407–3 [pii]. [DOI] [PubMed] [Google Scholar]
- 37.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, and Georgopoulos K. (1996). Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5, 537–549. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida T, Ng SY, Zuniga-Pflucker JC, and Georgopoulos K. (2006). Early hematopoietic lineage restrictions directed by Ikaros. Nature immunology 7, 382–391. 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng SY, Yoshida T, Zhang J, and Georgopoulos K. (2009). Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507. S1074–7613(09)00138–1 [pii] 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwickert TA, Tagoh H, Gultekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. (2014). Stage-specific control of early B cell development by the transcription factor Ikaros. Nature immunology 15, 283–293. 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, and Georgopoulos K. (2014). Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nature immunology. 10.1038/ni.2821. [DOI] [PMC free article] [PubMed]
- 42.Hu Y, Zhang Z, Kashiwagi M, Yoshida T, Joshi I, Jena N, Somasundaram R, Emmanuel AO, Sigvardsson M, Fitamant J, et al. (2016). Superenhancer reprogramming drives a B-cell-epithelial transition and high-risk leukemia. Genes Dev 30, 1971–1990. 10.1101/gad.283762.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. (2009). Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360, 470–480. 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgopoulos K. (2009). Acute Lymphoblastic Leukemia -- On the Wings of IKAROS. N Engl J Med. [DOI] [PubMed]
- 45.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, and Downing JR (2008). Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380. 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, Astolfi A, Chiaretti S, Vitale A, Messa F, et al. (2009). Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 114, 2159–2167. [DOI] [PubMed] [Google Scholar]
- 47.Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, et al. (2009). IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol 27, 5202–5207. [DOI] [PubMed] [Google Scholar]
- 48.Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, and Hoogerbrugge PM (2007). High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 21, 1258–1266. [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Liu A, and Georgopoulos K. (1996). Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. Embo J 15, 5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 50.Molnar A, Wu P, Largespada DA, Vortkamp A, Scherer S, Copeland NG, Jenkins NA, Bruns G, and Georgopoulos K. (1996). The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. Journal of immunology 156, 585–592. [PubMed] [Google Scholar]
- 51.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, and Georgopoulos K. (1997). Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. Embo J 16, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, and Morgan BA (1998). Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol 8, 508–515. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. (1999). Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10, 345–355. [DOI] [PubMed] [Google Scholar]
- 54.McCarty AS, Kleiger G, Eisenberg D, and Smale ST (2003). Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell 11, 459–470. S1097276503000431 [pii]. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. (2012). Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nature immunology 13, 86–94. 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koipally J, Renold A, Kim J, and Georgopoulos K. (1999). Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. Embo J 18, 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida T, Hu Y, Zhang Z, Emmanuel AO, Galani K, Muhire B, Snippert HJ, Williams CJ, Tolstorukov MY, Gounari F, and Georgopoulos K. (2019). Chromatin restriction by the nucleosome remodeler Mi-2beta and functional interplay with lineage-specific transcription regulators control B-cell differentiation. Genes Dev 33, 763–781. 10.1101/gad.321901.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, and Kioussis D. (2003). Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 33, 314–325. [DOI] [PubMed] [Google Scholar]
- 59.Wang W, Chandra A, Goldman N, Yoon S, Ferrari EK, Nguyen SC, Joyce EF, and Vahedi G. (2022). TCF-1 promotes chromatin interactions across topologically associating domains in T cell progenitors. Nature immunology 23, 1052–1062. 10.1038/s41590-022-01232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan Q, Zhu S, Chen X, Liu J, Yuan S, Li X, Peng W, and Xue HH (2022). Tcf1-CTCF cooperativity shapes genomic architecture to promote CD8(+) T cell homeostasis. Nature immunology 23, 1222–1235. 10.1038/s41590-022-01263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, Urban JF Jr., Davis FP, Kanno Y, and O’Shea JJ (2016). Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell 165, 1120–1133. 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kashiwagi M, Figueroa DS, Ay F, Morgan BA, and Georgopoulos K. (2022). A double-negative thymocyte-specific enhancer augments Notch1 signaling to direct early T cell progenitor expansion, lineage restriction and beta-selection. Nature immunology 23, 1628–1643. 10.1038/s41590-022-01322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tretter T, Ross AE, Dordai DI, and Desiderio S. (2003). Mimicry of pre-B cell receptor signaling by activation of the tyrosine kinase Blk. J Exp Med 198, 1863–1873. 10.1084/jem.20030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Medley QG, Buchbinder EG, Tachibana K, Ngo H, Serra-Pages C, and Streuli M. (2003). Signaling between focal adhesion kinase and trio. The Journal of biological chemistry 278, 13265–13270. 10.1074/jbc.M300277200. [DOI] [PubMed] [Google Scholar]
- 65.Bircher JE, and Koleske AJ (2021). Trio family proteins as regulators of cell migration and morphogenesis in development and disease - mechanisms and cellular contexts. J Cell Sci 134. 10.1242/jcs.248393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu X, Libby RT, de Vries WN, Smith RS, Wright DL, Bronson RT, Seburn KL, and John SW (2012). Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet 8, e1002853. 10.1371/journal.pgen.1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McNeill EM, Roos KP, Moechars D, and Clagett-Dame M. (2010). Nav2 is necessary for cranial nerve development and blood pressure regulation. Neural Dev 5, 6. 10.1186/1749-8104-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pardo LA, and Stuhmer W. (2014). The roles of K(+) channels in cancer. Nat Rev Cancer 14, 39–48. 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 69.Schaefer EJ, Geller AS, and Endress G. (2019). The biochemical and genetic diagnosis of lipid disorders. Curr Opin Lipidol 30, 56–62. 10.1097/MOL.0000000000000590. [DOI] [PubMed] [Google Scholar]
- 70.Hagman J, Rudin CM, Haasch D, Chaplin D, and Storb U. (1990). A novel enhancer in the immunoglobulin lambda locus is duplicated and functionally independent of NF kappa B. Genes Dev 4, 978–992. 10.1101/gad.4.6.978. [DOI] [PubMed] [Google Scholar]
- 71.Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, and Alt FW (1996). The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity 5, 241–252. 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 72.Xiang Y, and Garrard WT (2008). The Downstream Transcriptional Enhancer, Ed, positively regulates mouse Ig kappa gene expression and somatic hypermutation. Journal of immunology 180, 6725–6732. 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou X, Xiang Y, Ding X, and Garrard WT (2012). A new hypersensitive site, HS10, and the enhancers, E3’ and Ed, differentially regulate Igkappa gene expression. Journal of immunology 188, 2722–2732. 10.4049/jimmunol.1102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Picard D, and Schaffner W. (1984). A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature 307, 80–82. 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- 75.Meyer KB, and Neuberger MS (1989). The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J 8, 1959–1964. 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, and Garrard WT (2002). Chromatin structural analyses of the mouse Igkappa gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. The Journal of biological chemistry 277, 32640–32649. 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- 77.Takeda S, Zou YR, Bluethmann H, Kitamura D, Muller U, and Rajewsky K. (1993). Deletion of the immunoglobulin kappa chain intron enhancer abolishes kappa chain gene rearrangement in cis but not lambda chain gene rearrangement in trans. EMBO J 12, 2329–2336. 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, and Schatz DG (2010). The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141, 419–431. 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, and Garrard WT (2006). A recombination silencer that specifies heterochromatin positioning and ikaros association in the immunoglobulin kappa locus. Immunity 24, 405–415. [DOI] [PubMed] [Google Scholar]
- 80.Xiang Y, Park SK, and Garrard WT (2013). Vkappa gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vkappa-Jkappa intervening region. Journal of immunology 190, 1819–1826. 10.4049/jimmunol.1203127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barajas-Mora EM, Kleiman E, Xu J, Carrico NC, Lu H, Oltz EM, Murre C, and Feeney AJ (2019). A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol Cell 73, 48–60 e45. 10.1016/j.molcel.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barajas-Mora EM, Lee L, Lu H, Valderrama JA, Bjanes E, Nizet V, Feeney AJ, Hu M, and Murre C. (2023). Enhancer-instructed epigenetic landscape and chromatin compartmentalization dictate a primary antibody repertoire protective against specific bacterial pathogens. Nature immunology 24, 320–336. 10.1038/s41590-022-01402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ 3rd, and Kant JA (1991). Identification of a zinc finger protein that inhibits IL-2 gene expression. Science 254, 1791–1794. 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 84.Poonaki E, Kahlert UD, Meuth SG, and Gorji A. (2022). The role of the ZEB1-neuroinflammation axis in CNS disorders. J Neuroinflammation 19, 275. 10.1186/s12974-022-02636-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schubeler D, and Bauer A. (2015). YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet 11, e1005465. 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Monahan K, Horta A, and Lomvardas S. (2019). LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565, 448–453. 10.1038/s41586-018-0845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Battilana G, Zanconato F, and Piccolo S. (2021). Mechanisms of YAP/TAZ transcriptional control. Cell Stress 5, 167–172. 10.15698/cst2021.11.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, and Fisher AG (1997). Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91, 845–854. [DOI] [PubMed] [Google Scholar]
- 89.Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, and Georgopoulos K. (1999). Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10, 333–343. S1074–7613(00)80033–3 [pii]. [DOI] [PubMed] [Google Scholar]
- 90.Ribeiro de Almeida C, Stadhouders R, de Bruijn MJ, Bergen IM, Thongjuea S, Lenhard B, van Ijcken W, Grosveld F, Galjart N, Soler E, and Hendriks RW (2011). The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity 35, 501–513. 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 91.Ribeiro de Almeida C, Hendriks RW, and Stadhouders R. (2015). Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin kappa Light-Chain Locus. Adv Immunol 128, 183–271. 10.1016/bs.ai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 92.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, and Busslinger M. (2004). Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13, 919–922. 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. gb-2008-9-9-r137 [pii] 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D. (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen L, Shao N, Liu X, and Nestler E. (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, and Barillot E. (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259. 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, and Aiden EL (2016). Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst 3, 99–101. 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaul A, Bhattacharyya S, and Ay F. (2020). Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat Protoc 15, 991–1012. 10.1038/s41596-019-0273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhattacharyya S, Chandra V, Vijayanand P, and Ay F. (2019). Identification of significant chromatin contacts from HiChIP data by FitHiChIP. Nat Commun 10, 4221. 10.1038/s41467-019-11950-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chakraborty A, Wang JG, and Ay F. (2022). dcHiC detects differential compartments across multiple Hi-C datasets. Nat Commun 13, 6827. 10.1038/s41467-022-34626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y, Nanni L, Sungalee S, Zufferey M, Tavernari D, Mina M, Ceri S, Oricchio E, and Ciriello G. (2021). Systematic inference and comparison of multi-scale chromatin sub-compartments connects spatial organization to cell phenotypes. Nat Commun 12, 2439. 10.1038/s41467-021-22666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wolff J, Rabbani L, Gilsbach R, Richard G, Manke T, Backofen R, and Gruning BA (2020). Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res 48, W177–W184. 10.1093/nar/gkaa220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van der Weide RH, van den Brand T, Haarhuis JHI, Teunissen H, Rowland BD, and de Wit E. (2021). Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom Bioinform 3, lqab040. 10.1093/nargab/lqab040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu M, and Gu L. (2022). TCseq: Time course sequencing data analysis. R package version 1.23.0. 10.18129/B9.bioc.TCseq. [DOI]
- 111.Hinrichs D, Wetten M, and Meuwissen TH (2006). An algorithm to compute optimal genetic contributions in selection programs with large numbers of candidates. J Anim Sci 84, 3212–3218. 10.2527/jas.2006-145. [DOI] [PubMed] [Google Scholar]
- 112.Yang T, Zhang F, Yardimci GG, Song F, Hardison RC, Noble WS, Yue F, and Li Q. (2017). HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res 27, 1939–1949. 10.1101/gr.220640.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Molnar A, and Georgopoulos K. (1994). The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol 14, 8292–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. (2011). Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nature immunology 13, 86–94. ni.2150 [pii] 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Intracellular staining by flow cytometry for Immunoglobulin μ-chain (Igμ) and κ-chain (Igκ) was performed in IkE5Δ/Δ also referred to as IKDN (blue filled histogram; top rows), WT (red filled histogram; middle rows) large pre-B cells, and WT splenic B cells (black filled histogram, bottom row). White unfilled histograms show staining with isotype-matched control antibody. This analysis confirmed intracellular expression of Igμ and lack of expression of Igκ in WT and IKDN asserting their identity as large pre-B cells. (B) Lack of appreciable Vκ-Jκ recombination events were seen in both WT and IKDN large pre-B cells. Splenic B cells were used as controls. IL7 withdrawal (IL7-) from WT large pre-B cell cultures, normally grown on bone marrow stroma supplemented with IL7, caused cell cycle withdrawal and induced Vκ-Jκ recombination and H3K4me3 deposition at the recombination center (as shown in Figure 6C for the Jκ region). PCR for a Dntt genomic fragment was used as loading control. Autoradiograms for Vκ-Jκ recombination in large pre-B and splenic B cells are shown at the same exposure. (C) Deletion of the IKAROS DNA binding domain (IkE5Δ/Δ/IKDN) in large pre-B cells generated in vivo was confirmed by Western blotting (WB). The IKAROS DNA binding isoforms (IK1, IK2) expressed in WT cells and mutant isoforms lacking the DNA binding domain (ΔIK1,2) detected in IKDN are marked. (D) Distribution of reproducibility scores for WT and IKDN large pre-B cells as well as mouse embryonic stem cells (ESCs – outside control group) for Hi-C and HiChIP replicates. Stratum adjusted correlation coefficient (SCC) that measures reproducibility was computed at 10kb resolution using HiCrep.112 (E) Reproducibility analysis of H3K27ac, SMC1, CTCF and IKAROS ChIP-seq peaks between the two replicates (R1 and R2) of the same condition (WT or IKDN). Peaks were deemed overlapped between replicates if they intersected by at least 1 bp. (F) Aggregate peak analysis (APA) of significant chromatin loops (WT or IKDN) identified from Hi-C or HiChIP data as described in Figure 1D. For each case, the contact counts were aggregated from either WT or IKDN Hi-C data. Hi-C contact counts at 10kb resolution were aggregated +/− 50kb of each loop anchor and loops >150kb distance were used to avoid genomic distance patterns dominating the signal. (G) Heatmap of Observed/Expected (O/E) Hi-C, H3K27ac, SMC1 and CTCF HiChIP contact counts for loops identified as differential from comparison of WT vs IKDN H3K27ac HiChIP data. Each row was z-score transformed and hierarchical clustering was applied on these values to obtain the displayed ordering. (H) Heatmaps of histone modification and transcription factor binding enrichment at enhancer- and promoter-classified anchors (10kb) from differential H3K27ac loops is shown using ChIP-seq data from WT and IKDN large pre-B cells (±50 kb genomic window around the anchors). Scale bars denote Read count Per Million (RPM) as measured from ChIP-seq signals. (I) De novo transcription factor-binding motif enrichment analysis (HOMER) of IKAROS peaks within downregulated H3K27ac loop anchors. Motif enrichment at IKAROS peaks for loop anchors that are associated with differentially expressed genes (DEGs) and those that are not associated with DEGs is shown separately. The top motif hits alongside their significance (p-values) and frequency (percentage) are shown. Note that the core IKAROS binding motif (t/c/a/g G/A G G A A/t A/T) also presents a subset of binding motifs for the ETS, STAT, TEAD and NFkB family of transcription factors.42,113,114 (J) The distribution of the number of ChIP-seq peaks for B cell lineage transcription factors (i.e., E2A, EBF1, LEF1) and extra-lineage transcription factors (i.e., TEAD, LHX2, LMO2) that overlap anchors of differential loops identified from HiChIP and Hi-C experiments. (K) Motif analysis (HOMER) of TEAD, LHX2 and LMO2 peaks enriched at the anchors of induced loops in IKDN. (L) The size of original (WT-downregulated) and de novo (IKDN-induced) loops associated with redirected anchors of downregulated H3K27ac and CTCF loops. *** indicates p-value <= 0.001 using Wilcoxon test. The change in anchor interactions associated with loop redirection is shown in the schematic. Loop anchor depicted in red interacts with blue anchor in WT and with green anchor in IKDN. Source: Tables S1-3.
(A) Volcano plot of the log2 fold change (x-axis) of normalized contact counts vs. statistical significance (-log10FDR) of the difference (y-axis) for differential (Down/blue and Up/red) and non-differential (ND/gray) H3K27ac and CTCF HiChIP loops genome-wide. (B) MA plot illustrating the association of DEGs with different categories of CTCF HiChIP loops (Down, Up or Not Differential) similar to Figure 2A. (C) Log2 fold change of DEGs associated with differential (Down and Up) and non-differential (ND) CTCF loops similar to Figure 2B. Upregulated and downregulated genes associated with both an up and a down regulated anchor were excluded from this plot. (D) GO pathway enrichments for genes associated with the ten developmental clusters identified in Figure 2D and associated with genes whose 3D genome organization is lost upon IKAROS loss in large pre-B cells. Pr: progenitor, Ma: Mature. (E) Examples of downregulated and (F) upregulated genes with changes in long-distance interactions are shown as in Figure 2E and 2F. The Erag enhancer at the Rag1/2 locus that serves as an anchor for chromatin loops identified by different 3C approaches is highlighted. Source: Tables S2-4.
(A) Size distribution of domains defined by the coordinates of the anchors of downregulated and upregulated hybrid (P-S or E-S) and structural (S-S) CTCF HiChIP loops. Red dashed line shows the median size of common TADs. (B) Differential ATA of TADs classified into merged (1) in WT, split in IKDN (2), merged in IKDN (3) and contracted (4) in WT is shown for the two conditions. A schematic of the method used to identify differential TADs and their number are shown on the left side of each panel. This analysis performed from the second condition point of view (Z1<=Z0) complements the one provided in Figure 3B. (C) Examples of split merged and shifted WT TADs that contain the Ms4a, Ptpre, Mapk8 and TCRγ loci are shown, respectively. (D) ARA of WT and IKDN (O/E) Hi-C signal from anchors classified as enhancers (E/S), promoters (P/S) and structural (S) in upregulated hybrid CTCF HiChIP loops. Aggregate signals from anchors (S) of upregulated structural CTCF loops (S-S) are shown as a comparison. The dotted circle in the enhancer anchor ARA plot in IKDN indicates the range of across anchor interactions. This radius is smaller compared to the one shown for active enhancer anchors in WT as shown in Figure 3E. Bottom schematic illustrates the insulation and conduction properties of each anchor type as detailed in the key panel. (E) ARA of WT and IKDN O/E Hi-C signal from enhancer- and promoter-classified anchors of upregulated H3K27ac HiChIP loops. These were further classified for presence or absence of CTCF (+ or −) binding. Schematics below the ARA plots illustrate the range of interactions between adjacent domains and potential mechanisms involved in the process as detailed in the shared key panel with D. (F) Distribution of the average ChIP-seq Read count Per Million (RPM) of factors and histone modifications at anchors (10kb) classified as enhancers, promoters, or structural sites from downregulated and upregulated CTCF HiChIP loops, using a ±50 kb genomic window around the anchors. Source: Tables S2-3, S5.
(A-B) Compartmental score difference between IKDN and WT (IKDN-WT) calculated for regions contained within differential CTCF hybrid loops and H3K27ac differential regulatory loops. (C) Size comparison of down and upregulated regulatory loops is shown by both density vs. size distribution and box plot. **** indicates p-value <= 0.0001 using Wilcoxon test. (D) Schematic illustration of a model mechanism by which IKAROS-mediated spatial interactions between enhancers located at distal compartmental domains supports euchromatic localization. Source: Table S3, S6.
(A) Proliferation profile of cultured WT large pre-B cells isolated from IkE5fl/fl Rosa26-ERT2-Cre mice that were treated (4OHT) or not treated (DMSO) with tamoxifen for in vitro IKAROS deletion. (B) APA using Hi-C contact maps from in vitro IKAROS deletion (d0, d4, d12 and d18) as well as from the in vivo system (WT, IKDN) confirms bona fide interaction changes for differential H3K27ac HiChIP loops identified from comparisons of d3 vs d0 (top) and d12 vs d0 (bottom). (C) Similar APA plots confirm in vitro interaction changes for differential H3K27ac HiChIP loops identified from in vivo comparison (WT vs IKDN). (D) Heatmap of O/E Hi-C contact counts for pooled d0, d3, d12 replicates and WT and IKDN replicates for differential H3K27ac loops at d12 vs d0 is shown as in Figure 5C. (E) Heatmap of O/E Hi-C contact counts for pooled d0, d3, d12 replicates and WT and IKDN replicates at the anchors of differential H3K27ac chromatin loops in IKDN vs WT is shown as in Figure 5D. (F) Relative change in the median values of compartmental scores between d0-d18 is calculated for the compartmental changes established in steady state WT and IKDN as shown in Figure 5G and Figure 4A. (G) Example of loss in long-distance interactions at the Tgfbr2 and (H) Ahr loci detected in vivo and in vitro after IKAROS deletion as described for examples in Figures 5H-I. Source: Table S2-3, S6-7.
(A) Chromatin organization at the Igκ locus is shown as in Figure 6A-C that includes significant long-distance interactions identified in ex vivo (steady state) WT and IKDN large pre-B cells by FitHiChIP for IKAROS, H3K27ac, SMC1 and CTCF HiChIP data sets. Chromatin compartmentalization (PCA) and normalized histone modification and transcription factor signals obtained by ChIP-seq analysis under steady state conditions, are also shown. (B) Enlarged view of chromatin 1D and 3D maps at the 3’ end of the Igκ locus highlighting some of the long-range enhancer interactions that connect the elements of a super-enhancer located at the RC boundary domain with enhancers within TAD-C, including the recently characterized E34 enhancer. These E-E interactions cross over the CTCF Cer-Sis elements that define the 3’ boundary of TAD-C and are highly dependent on IKAROS DNA binding for their formation. Significant H3K27ac and CTCF interactions, histone modification and transcription factor enrichment are shown as in Figure 6B and S6A. (C) Size distribution of differential H3K27ac and CTCF interactions at the Igκ locus. The H3K27ac enhancer-enhancer interactions are on average double the size of CTCF chromatin loops reaching over 2.5Mb distances to make contacts between the enhancers at the recombination center (RC) with enhancers proximal to Vκ regions in the three Igκ TADs (A, B, C). A majority (60/84) of long-range interactions > 900kB originate from the RC. These data support a model that the 3.2MB Igκ locus contraction and recombination relies on long-range enhancer-based interactions rather than CTCF-cohesin loop extrusion. (D) Hi-C contact maps (O/E) generated by Juicebox are shown at the Igκ locus at 10 kb resolution prior to and after inducible IKAROS deletion (Rosa26-Cre-ERT2, IKE5 fl/fl) at d0, d4, d12, and d18. Source: Table S2-3, S6-7
(A) Long-distance significant H3K27ac interactions (>150kb) induced at d3 were confirmed by APA of Hi-C contacts at d2 after IKAROS induction stratified by IKAROS presence at anchors. (B) Distributions of compartment score differences calculated for 100kB differential compartment bins that overlap with lost H3K27ac and CTCF loop anchors as described in Figure 7E. (C) GO analysis for DEGs (up and downregulated) at the earliest time point (d1 vs. d0) after IKAROS induction. Downregulated genes enriched for pathways that support keratinocyte differentiation, whereas upregulated genes enriched for pathways that support neuronal functions (highlighted in red). (D) Log2 fold change of DEGs (d2 vs. d0) associated with differential (Upregulated or Downregulated) and Non-Differential H3K27ac loops is shown as detailed in Figure 2B. Up and downregulated genes associated with both an up and a down regulated loop were excluded from the figure. (E) Examples of lymphoid gene targets (BCL6, CD28-CTLA4) that upon IKAROS expression share features in chromatin topology between the two different cell types and species are highlighted. Conserved loop anchors associated with IKAROS peaks were identified with LiftOver111 and highlighted with the same color across mouse and human tracks. Source: Table S3-4, S6-7.
Table S1. Protocol details and mapping statistics for HiC and HiChIP experiments, related to Figures 1–7.
Table S2. Loop calls from HiC and HiChIP in wildtype (WT) and IkE5Δ/Δ (IKDN) large pre-B cells, related to Figures 1–6.
Table S3. Differential loop calls from steady state and inducible HiC and HiChIP in mouse large pre-B cells and human skin epithelial cells, related to Figures 1–7.
Table S4. Differentially expressed genes in large pre-B cells and human skin epithelial cells, related to Figures 2, 7
Table S5. Topologically associating domains in WT and IKDN large pre-B cells, related to Figure 3.
Table S6. Differential compartments in mouse large pre-B cells and human skin epithelial cells, related to Figures 4, 5, 7.
Table S7. Loop calls from inducible IKAROS deletion in mouse large pre-B cells and IKAROS ectopic expression in human skin epithelial cells, related to Figures 5, 7.
Data Availability Statement
All sequencing data were deposited at NCBI Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Zenodo. The DOI is listed in the key resources table. Microscopy data reported in this paper is available from the lead contact upon request.
All original code is publicly available in GitHub. Links to Github and all software used as described in the STAR Methods are listed in the key resources table. A static snapshot of all original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
Additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resources Table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| H3K4me1 | Active Motif | Cat#AM39297 |
| H3K4me3 | Active Motif | Cat#AM39155 |
| H3K27ac | Abcam | Cat#ab4729 |
| H3K27me3 | EMD Millipore | Cat#07–449 |
| H3K36me3 | Abcam | Cat#ab9050 |
| IKAROS | EMD Millipore | Cat#MABE912 |
| IKAROS | EMD Millipore | Cat#MABE913 |
| IKAROS | Abcam | Cat#ab191394 |
| CTCF | Abcam | Cat#ab188488 |
| NIPBL | Proteintech | Cat#18792–1-AP |
| SMC1a | Bethyl | Cat#A300–055A |
| GAPDH | Millipore | Cat#MAB374 |
| βTubulin | Santa Cruz Biotech | Cat#sc-9104 |
| FITC anti-mouse CD19 | Biolegend | Cat#152403 |
| Alexa Fluor 647 anti-mouse CD19 | Biolegend | Cat#115525 |
| APC anti-mouse CD43 | BD Biosciences | Cat#560663 |
| PE anti-mouse BP1 | Southern Biotech | Cat#1770–09L |
| APC anti-mouse B220 | Thermo Fisher Scientific | Cat#17–0452-81 |
| BV510 anti-mouse CD21 | BD Biosciences | Cat#563175 |
| PE anti-mouse CD23 | Thermo Fisher Scientific | Cat#12–0232-81 |
| BV421 anti-mouse CD5 | Biolegend | Cat#100629 |
| FITC-Ig-μ | BD Biosciences | Cat#553408 |
| FITC-Ig-κ | BD Biosciences | Cat#550003 |
| TER119 | BD Biosciences | Cat#553669 |
| Mac-1 (M1/70) | BD Biosciences | Cat#553307 |
| Gr-1 (RB6–8C5) | BD Biosciences | Cat#553123 |
| IgM (11/41) | BD Biosciences | Cat#562032 |
| CD3 (17A2) | BD Biosciences | Cat#557869 |
| CD8α (53–6.7) | BD Biosciences | Cat#553027 |
| TCRβ (H57–597) | BD Biosciences | Cat#553174 |
| CD49b (DX5) | BD Biosciences | Cat#553855 |
| Bacterial and virus strains | ||
| NEB® Stable Competent E. coli | New England Biolabs | C3040H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4-Hydroxytamoxifen (4OHT) | Sigma | Cat#H7904 |
| Biotin-14-dATP | ThermoFisher Scientific | Cat#19524016 |
| Doxycycline (Dox) | ThermoFisher Scientific | Cat#BP26531 |
| Equisul-SDT | Aurora Pharmaceutical | Cat#28000 |
| Recombinant Human IL-7 | Biolegend | Cat#715302 |
| KAPA HiFi HotStart ReadyMix | Roche | Cat#KK2601 |
| Protease inhibitor cocktail | Roche | Cat#11873580001 |
| Critical commercial assays | ||
| Pan B cell isolation kit II (mouse) | Miltenyi Biotec | Cat#130-104-443 |
| CFSE Cell Division Tracker Kit | Biolegend | Cat#423801 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23227 |
| ChIP DNA Clean and Concentrator | Zymo Research | Cat#D5205 |
| Quick-DNA Miniprep Plus kit | Zymo Research | Cat#D4068 |
| Direct-zol RNA Miniprep kit | Zymo Research | Cat#R2050 |
| Lenti-X Tet-On Advanced Inducible Expression System | Clontech | Cat# 630930 |
| TruSeq® Stranded mRNA Library Prep kit | Illumina | Cat#20020594 |
| Deposited Data | ||
| Raw and analyzed data | This study | GSE232490 |
| Western blot images | This study | https://doi.org/10.5281/zenodo.8339089 |
| Analyzed data | This study | https://doi.org/10.5281/zenodo.8339291 |
| RNAseq datasets for HSC, MPP, CLP,ILC2p and NKp cells | Shih et al, 201661 | GSE77695 |
| RNAseq datasets for pro-B, large pre-B and small pre-B cells | Kashiwagi et al, 202262 | GSE211079 |
| ChIP-seq datasets for B cell and extra-lineage transcription factors | Hu et al, 201642 | GSE86897 |
| Experimental Models: Cell lines | ||
| Human: HEK 293T/17 cells | ATCC | Cat#CRL-11268 |
| Human: HaCaT cells | J.M. Park | N/A |
| Mouse: OP9 bone marrow stromal cells | ATCC | Cat#CRL-2749 |
| Mouse: Passage 4 IkE5fl/fl hCD2-Cre (+/−) large pre-B cells |
This study |
N/A |
| Mouse: Passage 2 IkE5fl/fl Rosa26-Cre-ERT2 large pre-B cells | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| IkE5fl/fl hCD2-Cre (+/−) | Joshi et al, 201441 | N/A |
| IkE5fl/fl Rosa26-Cre-ERT2 | Hu et al, 201642 | N/A |
| Oligonnucleotides | ||
| Igk forward primer (Vκ) | Intergrated DNA Technologies (IDT) | GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC |
| Igk reverse primer (Jκ5) | IDT | ATGCGACGTCAACTGATAATGAGCCCTCTCC |
| Igk probe (upstream of Jκ5) | IDT | ACCAAGCTGGAGCTGAAACGTAAGTACAC |
| Dntt forward primer | IDT | CCCAAGTTCACTGTTCTTTCTGTC |
| Dntt reverse primer | IDT | CTCACCAGCAAACCCATAGGTA |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clontech | Cat# 630930 |
| pTRE-Tight-Luc control vector | Clontech | Cat# 630930 |
| pLVX-Tet-On Advanced | Clontech | Cat# 630930 |
| pLVX-Tight-FH-IKZF1 | This study | N/A |
| Software and Algorithms | ||
| Code for data analysis | This study | https://github.com/ay-lab/3D-IKAROS |
| FlowJo v10.9 | BD Biosciences | https://www.flowjo.com |
| bedGraphToBigWig v4 | Kent et al., 201096 | https://github.com/ENCODE-DCC/kentUtils |
| CALDER v1.0 | Liu et al., 2021107 | https://github.com/CSOgroup/CALDER |
| dcHiC v2.1 | Chakraborty et al., 2022106 | https://github.com/ay-lab/dcHiC |
| DEseq2 | Love et al., 201499 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FitHiC v.2.0.7 | Kaul et al., 2020104 | https://github.com/ay-lab/fithic |
| FitHiChIP v9.1 | Bhattacharyya et al., 2019105 | https://ay-lab.github.io/FitHiChIP/html/index.html |
| GENOVA v1.0.0.9 | van der Weide et al., 2021109 | https://github.com/robinweide/GENOVA |
| GenomicRanges v1.42.0 | Lawrence et al., 201398 | https://bioconductor.org/packages/release/bioc/html/GenomicRanges.html |
| Gene Ontology (GO) analysis | Zhou et al., 2019101 | https://metascape.org |
| HiCExplorer v3.5.1 | Wolff et al., 2020108 | https://hicexplorer.readthedocs.io/en/latest/index.html |
| HiC-Pro 2.8.0 | Servant et al., 2015102 | https://github.com/nservant/HiC-Pro |
| hicreppy v0.0.6 | Yang et al., 2017112 | https://github.com/cmdoret/hicreppy |
| Homer v4.11 | Heinz et al., 2010100 | http://homer.ucsd.edu/homer/ |
| Juicebox v1.11.08 | Durand et al., 2016103 | https://github.com/aidenlab/Juicebox |
| Liftover v2 | Hinrichs et al., 2006111 | https://genome.ucsc.edu/cgi-bin/hgLiftOver |
| MACS2 v2.11 | Zhang et al., 200895 | https://github.com/macs3-project/MACS |
| ngs.plot v2.63 | Shen et al., 201497 | https://github.com/shenlab-sinai/ngsplot |
| STAR v5.02 | Dobin et al., 201394 | https://github.com/alexdobin/STAR |
| TCseq v1.14 | Wu & Gu, 2022110 | https://doi.org/10.18129/B9.bioc.TCseq |