ABSTRACT
The escalation of antibiotic resistance and the diminishing antimicrobial pipeline have emerged as significant threats to public health. The ESKAPE pathogens – Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. – were initially identified as critical multidrug-resistant bacteria, demanding urgently effective therapies. Despite the introduction of various new antibiotics and antibiotic adjuvants, such as innovative β-lactamase inhibitors, these organisms continue to pose substantial therapeutic challenges. People’s Republic of China, as a country facing a severe bacterial resistance situation, has undergone a series of changes and findings in recent years in terms of the prevalence, transmission characteristics and resistance mechanisms of antibiotic resistant bacteria. The increasing levels of population mobility have not only shaped the unique characteristics of antibiotic resistance prevalence and transmission within People’s Republic of China but have also indirectly reflected global patterns of antibiotic-resistant dissemination. What’s more, as a vast nation, People’s Republic of China exhibits significant variations in the levels of antibiotic resistance and the prevalence characteristics of antibiotic resistant bacteria across different provinces and regions. In this review, we examine the current epidemiology and characteristics of this important group of bacterial pathogens, delving into relevant mechanisms of resistance to recently introduced antibiotics that impact their clinical utility in China.
KEYWORDS: ESKAPE, antimicrobial resistance, epidemiology, China
Introduction
Antimicrobial resistance (AMR) has emerged as a major public health threat in the twenty-first century and caused serious societal and economic burden [1,2]. If AMR is not controlled, 10 million people could die from bacterial infections by 2050 [3]. The irrational use of antimicrobial agents is a significant and critical issue in China, contributing to the development of severe bacterial resistance [4]. In recent years, China has issued two “National Action Plans for Combating Bacterial Resistance (2016–2020, 2022–2025)”, implementing effective antimicrobial stewardship, rational use policies and surveillance [5,6]. China's efforts to combat antimicrobial resistance have shown significant results. The rate of antibiotic use has notably decreased, and the detection rate of some commonly encountered clinically resistant bacteria has either decreased or remained stable [7,8]. However, numerous challenges persist in China's current landscape of AMR, including the escalating antibiotic resistance rates of certain common bacteria [9], continual discovery of new resistance mechanisms [10–12], and dynamic changes in prevalent clones [13,14]. Comprehensive control of nosocomial infections is still a pending task.
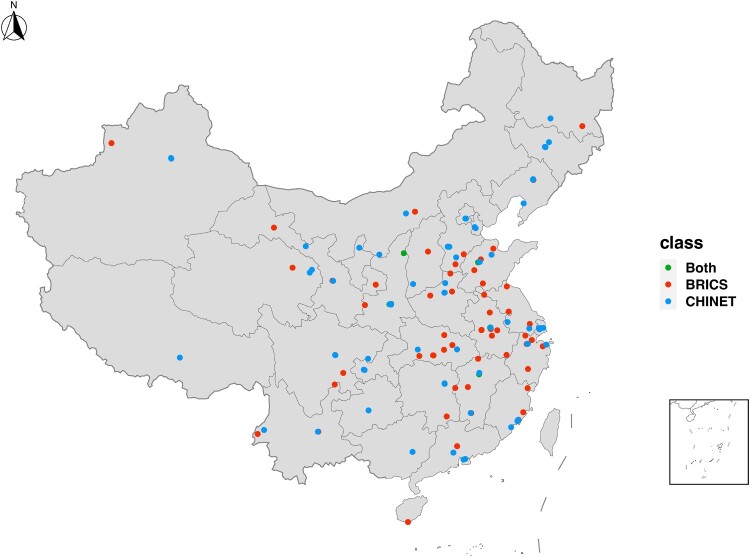
Surveillance and mechanistic studies of bacterial resistance are the most important measures in terms of controlling the spread of resistant bacteria [15–17]. There are several nationwide surveillance networks in China nowadays. CARSS (China Antimicrobial Resistance Surveillance System), currently managed by the National Health Commission, evolved from Mohnarin [18]. BRICS established by Zhejiang University, focused on surveillance of antimicrobial resistance in national bloodstream infection [9]. CHINET established by Fudan University, included more than 70 tertiary and second-class hospitals in China [18,19] (Figure 1). Besides the national surveillance networks, there are several independently established hospitals and local surveillance programmes, and some other special surveys including children hospitals [20]. These initiatives contribute valuable data that can aid in formulating strategies for the rational use of antimicrobial agents. With the establishment and development of these antimicrobial resistance surveillance networks, the quantity and quality of research output on antimicrobial resistance in China has remarkably increased [18], including molecular epidemiology and the underlying and genetic basis and evolution of antimicrobial resistance [21,22]. These studies make a significant and valuable contribution to the scientific community engaged in researching antimicrobial resistance, offering insights that inform strategies for mitigating antimicrobial resistance.
Figure 1.
Surveillance networks. Geographical distribution of sentinel hospitals of BRICS and CHINET. Due to the extensive number of sentinel hospitals in CARSS (exceeding 1,500), it is impractical to display them individually on the map. Therefore, CARSS sentinel hospitals have not been marked on the map.
We published a review paper related to epidemiology and characteristics of antimicrobial resistance in China in the first decade of 21th [23]. From the second decade of 21th to now, dynamic changes have made in epidemiology, underlying mechanisms and evolution of antimicrobial resistance. To comprehensively illustrate the extent and dynamics of antimicrobial resistance in China over the last decade, we examined data mainly on ESKAPE pathogens (including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) that often trigger severe nosocomial infections in critically ill and immunocompromised individuals [24] (Figure 2), and are notable for their propensity to develop antibiotic resistance mechanisms. Most of the data came from CARSS, BRICS and CHINET, which covered all the regions of China.
Figure 2.
Trends in the prevalence of the predominant antimicrobial resistant bacteria in China. (A) data from CHINET. (B) data from BRICS.
Enterococcus faecium
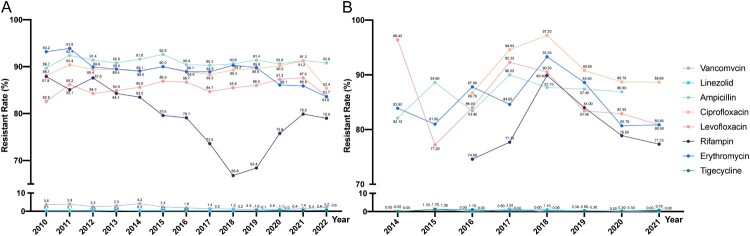
Enterococcus, mainly consisted by Enterococcus faecalis and Enterococcus faecium, is a globally important opportunistic pathogen, and can persist in the gastrointestinal tract for an extended period without causing any symptoms of infection, similarly enduring in the hospital environment [25,26]. The widespread use of antimicrobial agents in clinical treatment has led to the emergence and global spread of antibiotic-resistant enterococci, particularly multi-drug resistant (MDR) isolates such as vancomycin-resistant Enterococci (VRE) and linezolid-resistant Enterococci (LRE) [25]. Although ten years ago the isolation rate of E. faecium was significantly lower than that of E. faecalis in China, now the two are comparable as each around 50% (CARSS data in 2022). The antimicrobial resistance of E. faecium was more severe than that of E. faecalis, such as fosfomycin (30.3% vs. 4.8%), ciprofloxacin (85.4% vs. 30.6%) and nitrofurantoin (46.4% vs. 1.6%), except for chloramphenicol (5.2% vs. 22.6%), linezolid (0.6% vs. 3.4%) and contezolid (0% vs. 1.1%) (CHINET data in 2022) (Figure 3).
Figure 3.
Antimicrobial resistance trends for E. faecium. (A). data from CHINET. (B). data from BRICS.
The prevalence of VRE in China is relatively low, with a stable low resistance around 0.1% in E. faecalis, while the nationwide incidence of vancomycin-resistant E. faecium (VREfm) was fluctuate downtrend from 2010 (3.6%) to 2018 (1.2%), and a slow upward trend from 2019 (1.0%) to 2022 (2.2%) [27–30] (Figure 3). Of note, the prevalence of VREfm in Beijing and Guangdong province were markedly higher, and increased to 11.7% and 8% in 2022, respectively (CARSS data). Yan et al., reported an extremely high prevalence of VRE in patients admitted to ICUs in tertiary hospitals in Beijing, with 31.1% (46/148) patients carried VRE and 91.3% among them being E. faecium [32]. Compared to the globally predominant vanA and vanB genes associated with vancomycin resistance, the most prevalent vancomycin resistance genes in China are vanA and vanM [30–32]. Among these, vanA has the highest prevalence, reported to be in the range of 78.3%−81.7% [30,32]. vanM has been predominant in VRE strains since 2011 and showing a greater prevalence than vanA in Shanghai, and later spread to other regions such as Zhejiang and Beijing, becoming the second most prevalent vancomycin resistance gene in China [30,32]. The isolation rate of vanM has gradually increased, with recent reports indicating around 15.3%−16.7% [30,32]. Generally, both vanA and vanM genotype are characterized by an acquired high-level of resistance to both vancomycin and teicoplanin. The horizontal transfer capability of antibiotic resistance plasmids plays a crucial role in the dissemination of VRE strains. Multi-locus sequence type (ST) 78 VREfm are predominant in China, ranging from 47.6% to 57.1% in studies [30,32]. In contrast to Europe and Australia, where the VRE subtype ST80 has isolation rates ranging from 40% to 67.1%, ST80 has been rarely reported in clinical cases in China. However, recent findings by Li et al. have documented an outbreak of ST80 in a hospital in Shenzhen, suggesting that ST80 may be emerging in China [34]. Clonal expansion and horizontal transfer of resistance genes have contributed to VREfm increased prevalence in hospitals in China, which deserved further study.
Linezolid serves as one of the final lines of defense against Gram-positive bacteria. Before 2010, linezolid-resistant enterococci (LRE) was not found in any national survey. In recent years, with the widespread application of linezolid in clinics, the gradual increasing reports of linezolid resistant gram-positive pathogens highlights the enhanced risk of linezolid resistance transmission [35,36]. From 2010 on, the escalating emergence of LRE has evolved into a significant concern in nosocomial infections. LREfm was 0.1% in 2011 increased to 0.6% in 2022, and LREfs was 0.2% from 2011 increase to 3.4% in 2022 (CHINET data) (Figure 3). Several genes are related to linezolid resistance, but optrA and poxtA, is a growing concern, as it is being increasingly reported in both human and veterinary medicine [37]. OptrA was primary mechanism in LRE that was first detected in a clinical E. faecalis from China in 2015, and was commonly localized within chromosomes or plasmids and can be transmitted through horizontal gene transfer (HGT) via mobile genetic elements such as transposons, integrative and conjugative elements, and insertion sequences [38,39]. The recently reported poxtA gene, mediating resistance to oxazolidinones, tetracyclines, and phenicols, was first described in an MRSA isolate from an Italian patient [40]. Yi et al. reported that 46.7% (7/15) of linezolid-resistant clinical isolated E. faecium were positive for optrA, and 13.3% (2/15) of isolates had poxtA in China [41]. Besides, Tn558 and IS1216Es may play an important role in the dissemination of optrA and poxtA, respectively [41]. poxtA is found more in the environment or food-producing animals than human samples and E. faecium has higher prevalence than E. faecalis, indicating the necessity of applying the “One Health” perspective to analyse the evolutionary dynamics of poxtA-positive E. faecium in clinical and non-clinical settings worldwide [42,43]. Although linezolid is currently effective in the treatment of Enterococcal infections, advanced monitoring of changes in the resistance mechanism of linezolid is needed in the future.
Staphylococcus aureus
S. aureus is a commensal bacterium in humans, but it also serves as a pathogen responsible for various infections, ranging from mild skin and soft tissue infections to more severe conditions like endocarditis, pneumonia, and sepsis. Approximately 30% of healthy individuals harbour S. aureus in their anterior nares. The global prevalence of S. aureus in human populations has been on the rise, potentially attributed to the continual emergence of antibiotic-resistant strains, particularly methicillin-resistant S. aureus (MRSA).
Epidemiology of antimicrobial resistance
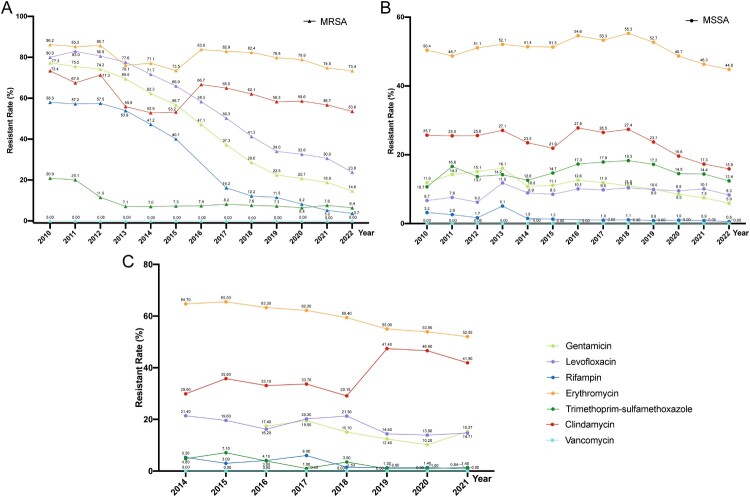
The high prevalence of MRSA was once a critical antimicrobial resistance issue for S. aureus in China, with an isolation rate exceeding 50% before 2010 [23] (Figure 4). This is attributed to the effective implementation of antimicrobial stewardship and rational use policies in China [4]. The isolation rates of MRSA vary geographically, with rates in regions such as Tibet, Shanghai, and Jiangsu remaining above 40% in recent years (CARSS data). The resistance rate of MRSA to antimicrobial agents is higher than that of methicillin-sensitive S. aureus (MSSA), except for trimethoprim-sulfamethoxazole (6.4% vs. 12.4% in 2022, CHINET data). MRSA is higher resistance rates to macrolides, clindamycin, aminoglycosides and quinolones compared with MSSA [44]. The rate of rifampicin resistance in MRSA, which was about 50% in 2010, has sharply declined to about 3% in 2022, though it remains higher than in MSSA (Figure 4). The rate of sulfamethoxazole-trimethoprim resistance in MSSA has remained stable around 15% from 2011 to 2022, whereas in MRSA, it decreased from 20.1% to 6.4% (CHINET data). S. aureus is still highly sensitive to vancomycin, linezolid, teicoplanin, contizolamide and tigecycline. Among the major treatment options for MRSA, no vancomycin, linezolid or teicoplanin resistant strains were reported in the surveillance data in China. Whereas in-host evolution of daptomyxin resistance and heteroresistance in MRSA were reported with mutations in mprF and yycH [45,46]. The role of mprF mutations in seesaw effect of daptomyxin resistance MRSA provides fundamental insights into the combination therapy of beta-lactam antibiotics and daptomycin for treating MRSA infections [46]. Nevertheless, as these antibiotics are widely utilized, there has been a rising number of reports on the emergence of drug non-susceptible strains among these antibiotics called the “MIC creep” phenomenon [47,48].
Figure 4.
Antimicrobial resistance trends for S. aureus. (A, B). data from CHINET. (C). data from BRICS.
Susceptibility tests on four major MRSA STs (ST239, ST5, ST59, and ST398) isolated from 61 hospitals in China from 2014 to 2019 discovered that all strains maintained resistance to penicillin and all MRSA exhibited sensitivity to vancomycin, daptomycin, and linezolid [49]. The susceptibility of other antibiotics changed between STs [49]. Overall, ST239 clone showed a high (>66%) and increasing trend of resistance rate in tetracycline and quinolones. The ST5 clone maintained a consistently high resistance rate (>78%) to quinolones and erythromycin. The ST59 clone exhibited high resistance rates (>80%) only to erythromycin and clindamycin, with an overall decreasing trend for moxifloxacin, and the resistance rates to amikacin and tetracycline began to decline after reaching their peak in 2016. The ST398 clone showed relatively low overall resistance rates to antimicrobial drugs, with a generally increasing trend for clindamycin and high fluctuation for erythromycin and tetracycline [49].
To date, at least 15 major staphylococcal cassette chromosome mec (SCCmec) types (I–XV) have been identified [49–51]. Over the past decade, MRSA-SCCmec IV and SCCmec V has spread widely in China, gradually replacing previously prevalent hospital-acquired MRSA-SCCmec II and III types [53,53]. While MRSA-SCCmec II has shown a declining trend in recent years, it still dominates in the East China region [54,55]. SCCmec elements IV (21–24 kb) and V (27 kb), being relatively smaller, facilitate their transfer among S. aureus strains, enhancing their potential for dissemination. MRSA carrying IV and V SCCmec elements typically show sensitivity to many non-β-lactam antibiotics and are predominantly found in community-acquired MRSA (CA-MRSA) in China [49]. In China, the recently prevalent ST59 clone is primarily associated with SCCmec IV [56,57]. IV and V elements are also present in some widely distributed healthcare-aquired (HA)-MRSA clones, such as ST22-MRSA-IV, ST45-MRSA-IV, and ST5-MRSA-VI [58]. A rising number of variants of SCCmec elements that harbour composite cassettes and pseudo-SCCmec elements but do not harbour ccr genes, have also been detected in recent years. This reflected the ongoing intra/interspecies genetic rearrangements [59]. Among the 1,006 MRSA strains collected between 2014 and 2020 in China, a total of 25 non-duplicate MRSA strains with non-typeable SCCmec elements were detected, provide evidence for the ongoing evolution of SCCmec elements within MRSA [60]. mecA-positive but phenotypically susceptible to oxacillin, suggesting potential frameshift mutations or single base substitutions in nucleotide repeat regions within mecA, with the capability to revert to resistance when exposed to antibiotics [61].
Reshaping the epidemiology of MRSA
Between 2005 and 2010, the ST239 clone dominated MRSA clones in China (50–80.8%), followed by the ST5 clone (15.5%) [54,61]. Since 2010, multiple studies have confirmed a decrease in the prevalence of the dominant MRSA ST239 and ST5 clones in China, while ST59 has been steadily increasing [21,62]. The isolation rate of ST59-MRSA increased from 25.09% in 2014 to 35.58% in 2019 [63,64]. Jin et al. discovered that in bloodstream infections, the ST59 clone becoming the dominant clone in most Chinese hospitals (33%−36.98%), with varying prevalence rates among different provinces ranging from 17.86% to 80.95%; the ST239 clone causing decreased from 10.29% in 2014 to 3.05% in 2019, while ST5 sharply declined from 39.71% to 9.64% during the same period [49].
In the past decade, the “main battlefield” of MRSA has shifted from hospitals to the community, with a reciprocal influence from the community to hospitals [65–67]. The boundary between HA-MRSA and CA-MRSA has become increasingly blurred. With the rapid development of whole-genome sequencing, CA-MRSA is defined based on microbiological and molecular characteristics, utilizing SCCmec typing and phylogenetic methods [21,68]. The prevalence of CA-MRSA in China has become significant, reaching a rate of 24%, with the unique Chinese CA-MRSA clone CC59 (mainly ST59) has been steadily increasing [52,62]. Studies reported that HA-MRSA strains belonged to the CC59-MRSA-IV/V-t437 clone, exhibiting characteristics of traditional Chinese CA-MRSA, carrying SCCmec IVa, and being PVL-positive [52,63]. However, HA-MRSA ST59 displayed higher resistance to compound sulfamethoxazole and gentamicin, while CA-MRSA ST59 was more likely to be resistant to clindamycin and levofloxacin. The sak and chp genes carried by MRSA ST59 and higher toxin gene content may be associated with the increased transmission success of this clone [49,64]. It's worth noting that Chen et al. found that almost all CC59 isolates in China (91%) exhibited sensitivity to the combination of penicillin and clavulanic acid due to the susceptible genotype [52]. The increasing prevalence of ST59 clones in Chinese hospitals highlights the multifaceted adaptive evolution of MRSA as a pathogen, extending beyond the community healthcare environment.
ST5 HA-MRSA clone is considered a representative epidemic clone in East China, with high prevalence reported in Shanghai and Zhejiang [55,69]. Wu et al. reported that ST5-t311-II lineage has become the predominant ST5 clone in Zhejiang Province (65%) [55]. The expansion and structurally unstable nature of the CC5 population were noted, with ST5-t311-II, ST764-t1084-II, ST5-t2460-II and ST764-t002-II existing complex competition [63]. The intra-hospital dissemination of ST5-MRSA has led to nosocomial outbreaks [70]. Simultaneously, the proportion of some sporadic STs, such as ST30, ST4513, ST1821, and ST5529, causing bloodstream infections in China increased from 5.88% in 2014 to 20.30% in 2019, demonstrating a rising diversity of MRSA strains over the years. It is noteworthy that, despite its lower prevalence, the ST398 clone has shown an increasing trend; as reported, an increase in the isolation rate of the ST398 clone from 1.47% in 2014 to 9.14% in 2019 [49,69].
As the isolation rate of HA-MRSA declines, increasing attention is being directed towards MRSA's animal hosts [71]. The livestock-associated MRSA (LA-MRSA) is predominantly associated with CC398 (in Europe) and CC9 (in Asia), causing infections primarily among individuals engaged in livestock farming [71]. However, recent observations indicate that LA-MRSA can also infect some individuals who have not had direct contact with animals. Phylogenetic analysis along with the distribution of resistance genes demonstrated that HA-ST9 MRSA and LA-ST9 MRSA share similar genetic backgrounds, which provides evidence that multidrug-resistant LA-MRSA-ST9, evolved in the livestock industry, spreads to communities and clinical settings through trade chains [60].
Vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA)
hVISA/VISA infections, reported since 1996, have been linked to poor patient outcomes. A meta-analysis reveals that Vancomycin-resistant S. aureus (VRSA), VISA and hVISA isolates have a worldwide prevalence of 1.5%, 1.7% and 4.6% in S. aureus, respectively [72]. No VRSA cases have been reported in China; however, VISA and hVISA are present at rates of 0.5% and 10.0%, respectively, with hVISA showing a significantly higher isolation rate compared to global levels [72]. Moreover, there is a growing concern due to the increasing prevalence of hVISA in recent years. Shen et al. identified 7.6% (36/476) hVISA among S. aureus isolated in 2010–2011 [73], while Liang et al. discovered 10.8% (22/204) hVISA among isolates from inpatients and outpatients in 2019–2021 [74]. With the recent decrease in MRSA isolation rates, the proportion of MRSA among VISA/hVISA has also declined [73,74]. Liang et al. reported 22.7% of hVISA isolates belonged to ST72 and CC5 (ST5/965/7197), followed by CC59 (ST59) and ST25 (18.2%, respectively), which indicated that previous dominant hVISA/VISA clone ST239 in China is changed [74].
Klebsiella pneumoniae
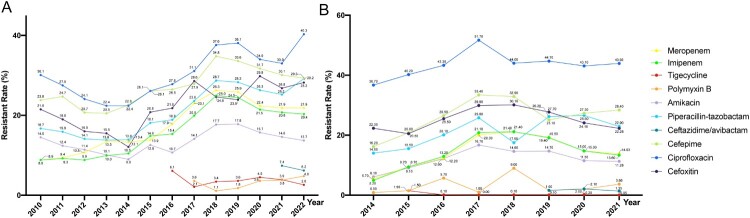
Klebsiella pneumoniae rose from about 15% to around 20% in all the clinical isolated bacteria nationwide (CARSS data). In the nationwide bloodstream infections surveillance (BRICS), K. pneumoniae increased from 9.91% in 2014–18.9% in 2021 (Figure 2). Over the past decade, no matter in all clinical samples or in bloodstream infection samples, K. pneumoniae has exhibited an overall steady rise of resistance to cephalosporins, aminoglycosides, fluoroquinolones, polymyxins and tigecycline [9,19]. In special, the increasing isolation rate of carbapenem-resistant K. pneumoniae (CRKP) is concerning [75]. Furthermore, K. pneumoniae demonstrates relatively high sensitivity to tigecycline, polymyxin, and ceftazidime-avibactam, with nationwide surveillance in 2022 indicating resistance rates of 2.6%, 2.8%, and 6.2% for these antibiotics (CHINET data) (Figure 5), respectively. The issue of drug resistance in K. pneumoniae has become one of the most challenging among Enterobacteriaceae worldwide [76].
Figure 5.
Antimicrobal resistance trends for K. pneumoniae. (A). data from CHINET. (B). data from BRICS.
Resistance to carbapenems
With widespread clinical use, resistance to carbapenems among Enterobacteriaceae, particularly CRKP, has become increasingly serious in China [76–78]. According to CHINET, the resistance rates of K. pneumoniae to imipenem and meropenem have straightly climbed from 8.8% and 8.9% in 2010–25.0% and 26.3% in 2018. However, a stabilizing trend is observed from 2019 onwards, with rates of 22.6% and 24.2% in 2022. This suggests a plateauing of carbapenem resistance development, potentially influenced by optimized antimicrobial drug use in China and a reduction in nosocomial infections due to the COVID-19 pandemic since 2019 [19]. Regional variations in resistance rates are substantial, with resistance predominantly concentrated in eastern China [19,79]. Similarly, the incidence of CRKP in BRICS countries sharply increased from 2014 to 2018 (5.2% to 21.4%) and has been slowly declining from 2018 to 2021 (21.4% to 15.8%) [9] (Figure 5).
KPC enzymes account for more than 70% of CRKP, followed by NDM-producing strains, while the prevalence of IMP, VIM, and OXA-48 enzymes is relatively low in China [79,80]. China reported the first KPC-producing CRKP in 2007 [81], and the prevalent genotype is blaKPC-2, most commonly associated with the epidemic clone ST11, accounting for approximately 60–80% of the CRKP isolation rate [82]. Different from the classical Tn4401 background reported in the United States, China identified unique genetic backgrounds for blaKPC-2, such as Tn3 transposon and partial Tn4401 fragments [83]. New structures like Tn1721-blaKPC-2-Tn3 and Tn1721-blaKPC-2-ΔTn3-IS26 have been discovered in China [84,85]. The predominant plasmid carrying blaKPC-2 in ST11 CRKP in China is IncFII type [86,87]. In contrast, in other international regions, diverse plasmids like IncFIA, IncX, IncP, IncN, and IncC are common carriers of blaKPC-2, indicating the diversity and widespread dissemination of KPC-2 resistance plasmids between regions [88,89]. These plasmids, containing blaKPC-2, often carry genes conferring resistance to other antibiotics, resulting in the multidrug resistance of CRKP [89].
Another representative of the epidemic CRKP strains carries NDM enzymes, belonging to class B metallo-β-lactamases dependent on zinc. These enzymes can hydrolyze all β-lactam antibiotics except aztreonam and are not inhibited by β-lactamase inhibitors. In China, the main genotypes are blaNDM-1 and blaNDM-5, and although the isolation proportion is lower than that of KPC-2, it is more common in CRKP isolated from pediatric patients [90–92]. Furthermore, there has been an increase in the reported incidence of blaOXA-48-like genes in China in recent years. Individual regions have experienced small-scale outbreaks, with blaOXA-232 being the most prevalent, mainly associated with ST15 CRKP and carried by ColKP3-type plasmids [93]. In Taiwan, blaOXA-48 has been reported in ST11 K. pneumoniae [94]. Moreover, in China, the combination of blaNDM and blaKPC is prevalent among K. pneumoniae [78,95].
In recent years, it has been observed that CRKP in China is undergoing evolution with the acquisition of high virulence traits, and there is an increasing number of reports on highly virulent CRKP [96–102]. Current studies suggest that the emergence of highly virulent CRKP is primarily the result of bidirectional evolution involving both high virulence and high resistance [14,103,104]. One pathway involves classic high-virulence K. pneumoniae (hvKP) acquiring plasmids or mobile elements containing carbapenem-resistant genes. Another pathway is CRKP acquiring virulence plasmids or mobile elements containing virulence genes [83,105–107]. It is noteworthy that through widespread transposon-mediated transposition or the fusion of resistance plasmids and virulence plasmids, virulence and resistance genes can be located on the same conjugative plasmid [108,109]. In recent years, an alternating trend of sub-clones has been observed among the prevalent ST11-type CRKP strains in China, with KL64 gradually replacing the earlier KL47 type [14,104]. KL64 carries a series of virulence genes associated with high pathogenicity, contributing to a high mortality rate in clinical infections [14]. An analysis of 1,052 CRKP strains from 19 regions and 56 centres in China between 2015 and 2017 revealed that 34.2% of CRKP strains carried virulence genes, primarily iron carrier genes (iucA, iroN), and capsular polysaccharide expression regulatory genes (rmpA and rmpA2). 80% of these strains belonged to the ST11-KL64 clone carrying the blaKPC-2 gene [96]. Zhou et al. revealed the gradual replacement of the once prevalent sub-clone OL101:KL47 by O2v1: KL64, with the latter exhibiting an increased abundance of mobile genetic elements. Additionally, a specific point mutation in the recC of O2v1: KL64 significantly enhances recombination proficiency [104]. The prevalent ST11 clone of CRKP in China is undergoing rapid evolution, forming a pathogenic and highly transmissible high-risk sub-clone, posing significant challenges to clinical diagnosis and treatment as well as infection control in healthcare settings [110,111].
Resistance to ceftazidime/avibactam
Ceftazidime/avibactam (CAZ/AVI) is currently the first novel β-lactam/β-lactamase inhibitor combination available for the treatment of infections caused by KPC-producing Enterobacteriaceae [112]. It was officially launched in China in early September 2019. Avibactam, within this combination, inhibits KPC and OXA-48 enzymes but does not inhibit metallo-enzymes like NDM. With its clinical application, there have been reported cases of non-metallo-enzyme CRKP resistance to CAZ/AVI [113]. The resistance rates to CAZ/AVI in China have been monitored since 2020, with resistance rates decreasing from 11.8% and 14.9% in 2020 to 6.2% and 9.1% in 2022 for K. pneumoniae and CRKP, respectively. Moreover, resistance rates were higher in children compared to adults, possibly due to a higher prevalence of metallo-enzymes like NDM in children.
CAZ/AVI resistance primarily due to mutations in the blaKPC gene and alterations in outer membrane porins [75,113]. To date, over 150 variants of blaKPC have been reported globally, with the majority of new variants identified in the last three years, warranting public concern [75]. The variant KPC protein increases its affinity to ceftazidime while reducing its affinity to avibactam through structural changes, thereby facilitating bacterial resistance to CZA [75]. Zhang et al. reported that resistance against CAZ/AVI in CRKP emerged before clinical use of CAZ/AVI in China, although most of the CRKP isolates maintained the susceptibility [113]. Among the resistant isolates, 53.1% (17/32) were metallo-blactamase-producing K. pneumoniae (MBL-KP), 40.6% (13/32) were KPC-producing K. pneumoniae (KPC-KP) and 6.3% (2/32) produced both MBL and KPC [113]. MBL production, blaKPC-2 point mutation and high KPC expression played an important role in CAZ/AVI resistance [113]. Shi et al. reported CAZ/AVI resistance caused by the change from KPC-2 to KPC-33 carbapenemase via the D179Y variant [12]. Plasmid-mediated novel CMY-type AmpC β-lactamase, leading to high-level resistance to CAZ/AVI, was discovered [114]. Therefore, enhanced monitoring is essential to prevent the spread of novel CAZ/AVI -resistant strains in China.
Resistance to polymyxins
Polymyxins (polymyxin B and polymyxin E) were officially introduced into clinical use in China in 2018. Prior to its clinical use, there was a lack of attention to polymyxins resistance in China's clinical antimicrobial resistance monitoring system. However, it had been closely monitored in the country's animal bacterial resistance monitoring system because polymyxins had been widely used in livestock farming before being reintroduced for clinical use [10,115]. Due to prolonged exposure to low concentrations of polymyxins, there has been a selection pressure leading to the prevalence and spread of low-level polymyxin-resistant strains mediated by the mobile resistance gene mcr [115].
The continuous increase in polymyxins resistance among clinically isolated strains, especially in countries with a high proportion of MDR and carbapenem-resistant strains, is a particularly concerning issue [116]. Research data from the European Antimicrobial Resistance Surveillance Network (EARS-Net) indicates that CRKP exhibits a much higher resistance rate to polymyxins than carbapenem-sensitive strains, especially in countries with a high prevalence of CRKP, such as Greece and Italy. In China, the resistance rates of K. pneumoniae to polymyxin B and polymyxin E have increased from 1.1% and 1.2% in 2018–4.8% and 2.8% in 2022, respectively. The resistance rate of CRKP to polymyxins has increased from 3.6% in 2020–8.2% in 2022 (CHINET data). In recent years, the increase in polymyxins resistance among K. pneumoniae has been significantly greater than that observed in other Gram-negative bacteria (BRICS and CHINET data) (Figure 5). It can be anticipated that without the proper use of polymyxins, the resistance rates of Gram-negative bacteria in China, especially CRKP, to polymyxins will continue to rise, leading to a further loss of effective antibiotic treatment options [116].
The resistance mechanisms of different bacterial species to polymyxin can be broadly classified into chromosome gene mutation-mediated and plasmid mcr gene-mediated [117]. Chromosome gene mutations associated with polymyxin resistance that have been reported include pmrCAB, phoPQ, mgrB, pmrD, etc [118]. Additionally, mutations in the crrAB two-component regulatory system have been reported in K. pneumoniae [119,120]. What’s more, one significant factor contributing to the increasing trend of polymyxins resistant CRKP is the high proportion of heteroresistance characteristics in K. pneumoniae, making them prone to mutations that induce full-resistance [121]. Therefore, the resistance of clinical isolates to polymyxins underscores the importance of addressing chromosomal gene mutations leading to heteroresistance as a consequence of clinical polymyxin use.
Resistance to tigecycline
In recent years, the heavy use of tigecycline (TGC) in clinical settings has resulted in the global emergence of resistance [122]. Tigecycline was introduced to the clinical in China at the end of 2011. The tigecycline resistance rate of clinical isolated K. pneumoniae in China has increased from 2.1% in 2017–3.9% in 2022. Tigecycline resistant rate of CRKP was higher, reaching to 5% in 2022 (Figure 5).
The mechanisms underlying TGC resistance in Enterobacteriaceae are complicated and have not been fully elucidated. Studies revealed that TGC resistance is primarily due to chromosome-encoding mechanisms, including overexpression of efflux pumps and ribosome protection [123–125]. An uncommon mechanism of TGC resistance is provided by enzymatic inactivation mediated by the flavin-dependent monooxygenase Tet(X) [11,126]. Fortunately, the mobile tigecycline-resistance tet(X) genes currently are rarely found in clinical isolates. A nationwide epidemiological analysis discovered no K. pneumoniae isolates were positive to tet(X) genes though 4% isolates were resistant to TGC [127].
Resistance to aminoglycosides
Aminoglycosides are important options for treating life-threatening infections and are generally administered in combination with β-lactam agents [128]. In China, the resistance rate of K. pneumoniae to gentamicin decreased from 34.0% in 2010–25.1% in 2022, possibly attributed to a reduction in the usage of gentamicin. In contrast, the resistance rate to amikacin showed a slow decrease from 14.4% in 2010–13.7% in 2022. Additionally, in 2022, CRKP exhibited resistance rates of 75.8% to gentamicin and 62.2% to amikacin (CHINET data) (Figure 5).
The predominant mechanism of aminoglycoside resistance involves the activity of aminoglycoside-modifying enzymes (AMEs) [129]. However, a significant mechanism mediating resistance to nearly all clinically available aminoglycosides is the production of 16S rRNA methyltransferase (16S-RMTase) [130]. rmtB and armA are the most widespread 16S rRNA methylase genes [131]. Furthermore, there is an emerging trend of high-level aminoglycoside resistance (HLAR) in K. pneumoniae isolates in China, where they carry 16S rRNA methylase genes (armA and rmtB), resulting in aminoglycoside MIC values exceeding 256 μg/mL [132]. Zhang et al. reported an incidence of 13.7% (40/292) HLAR strains, with rmtB being the predominant resistance gene (70%, 28/40), identified in a Chinese teaching hospital, and discovered conjugative armA and AME genes carrying plasmid [132].
Resistance to fosfomycin
Fosfomycin, an old antibiotic, has been re-introduced to fight infections caused by CRKP. However, the trend of fosfomycin resistance among CRKP is increasing dramatically in China, and becomes an emerging problem. CHINET data indicates that the resistance rate of CRKP to fosfomycin has remained consistently around 50% from 2019 to 2022. However, data from BRICS reveals that the resistance rate of all bloodstream K. pneumoniae isolates to fosfomycin has shown a linear increase, rising from 0.4% in 2014–11.57% in 2021 (Figure 5). Huang et al. reported that 80.0% (64/80) carbapenemase-producing K. pneumoniae (KPC-KP) were resistant to fosfomycin from three tertiary hospital in China (2014–2017) [133], and is similar to the report by a systematic review of the literature (73.5%) [134].
Bacterial resistance to fosfomycin can be caused by multiple mechanisms, including transporter defect (i.e. GlpT- or Uhp-deficient strains), target modification and FosA-mediated inactivation [133]. Most fosA3-carrying KPC-KP strains are highly resistant to fosfomycin (MIC > 2048mg/L) [133]. Huang et al. and Xiang et al. reported that 36.3% (29/80) and 45.4% (44/97) of the KPC-KP were positive for the mobile fosfomycin resistance gene fosA3, respectively, in China [133,135]. All fosA3-positive strains belonged to the dominant ST11-pulse type [135]. Findings indicate that the two major mechanisms of resistance identified were plasmid-mediated fosfomycin resistance gene fosA3 and mutation of the target gene glpT [133,135].
Escherichia coli
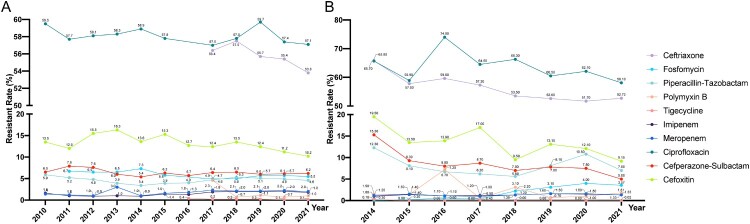
Although E. coli is not included in the ESKAPE pathogens, its clinical isolation rate (accounting for stable approximately 29% of Gram-negative bacteria, CARSS data) has consistently been 8–10 percentage points higher than that of K. pneumoniae in China. It has remained the most prevalent species in terms of isolation rates among Gram-negative bacteria and displayed important MDR phenotypes. Therefore, we included E. coli in this review. The bloodstream infection isolation rate of E. coli has increased in recent years, rising from 29.13% in 2014–37.60% in 2021 (BRICS data) (Figure 2). However, this growth trend, while notable, has not been as rapid as the nearly twofold increase observed in K. pneumoniae during the same period.
As two of the most representative species within the Enterobacteriaceae family, E. coli exhibits similar antibiotic resistance characteristics compared with K. pneumoniae. However, in recent years, the resistance trend in E. coli has shown a more moderate increase compared to K. pneumoniae. From various clinical isolation sources, E. coli has shown a generally stable or decreasing trend in resistance rates to most antibiotics in recent years. In 2022, E. coli exhibited high resistance rates to ciprofloxacin (61.4%) and cefepime (25.1%). However, resistance rates to ceftriaxone, fosfomycin, piperacillin-tazobactam, polymyxin B, and tigecycline were relatively low, at 9.7%, 4.4%, 4.3%, 2.2%, 1.0%, and 0.1% in 2022, respectively (CHINET data). From 2014 to 2021, the resistance rates of bloodstream-isolated E. coli to imipenem and meropenem have consistently remained between 1% and 1.6%. The resistance to polymyxin B (0.5% to 3.1%) and ceftazidime-avibactam (0.56% to 1.2%), and tigecycline (0% to 0.3%) have consistently been low. The resistance to ciprofloxacin varied from 27% to 30.4%. Over the same period, resistance to cefperazone-sulbactam (15.3% to 5%) and cefoxitin (19.5% to 9.15%) decreased, while resistance to fosfomycin increased from 0.1% to 3.57% in bloodstream-isolated E. coli. The bloodstream-isolated E. coli has exhibited a notable upward trend in resistance to fosfomycin, mirroring the trend observed in bloodstream-isolated K. pneumoniae (BRICS data) (Figure 6). This contrasts with the generally stable or decreasing resistance rates to most antibiotics, highlighting a divergent pattern that warrants continued attention and further research into the mechanisms underlying fosfomycin resistance.
Figure 6.
Antimicrobial resistance trends for E. coli. (A). data from CHINET. (B). data from BRICS.
Carbapenem-resistant E. coli (CRECO) in China typically carry the NDM gene. Bi et al. reported that 70.83% (102/144) CRECO isolates that produced various NDMs. In addition, CRECO also produced KPC-2 (n = 15), IMP-4 (n = 3), and OXA-48 (n = 3) [136]. The blaNDM gene is rarely found on the chromosome but is predominantly located on plasmids, with plasmid types covering IncX3, IncFII, IncC, IncFIB, IncHI1B, and others [137]. This indicates that various plasmids can acquire blaNDM and facilitate horizontal transfer through plasmid mediation [137]. In China, IncX3 is the most common plasmid type carrying blaNDM, suggesting that IncX3 plasmids may be the primary vehicles mediating the dissemination of blaNDM in China [138,139]. An analysis of all NDM-1 gene sequences revealed the consistent presence of the ISAba125 sequence in the upstream 100 bp region of the blaNDM-1 gene, indicating that the blaNDM-1 gene likely first appeared in A. baumannii, mediated by ISAba125 [140,141]. Furthermore, downstream of the blaNDM-1 gene, there is a consistent presence of the bleMBL gene, conferring resistance to bleomycin [140]. Other blaNDM variants share a similar genetic background with blaNDM-1, often associated with upstream ISAba125 (complete or truncated) and downstream bleMBL [142]. E. coli exhibits resistance mechanisms to antibiotics such as polymyxins, aminoglycosides, and fosfomycin that are similar to those observed in K. pneumoniae [143]. Reports suggest that the prevalence of the fosA3 gene in fosfomycin-resistant E. coli may be higher than K. pneumoniae. Li et al. reported that 83.3% of fosfomycin-resistant E. coli carry the fosA3 gene, with 70% of the fosA3 genes located on transferable resistant plasmids [144]. This indicates that the dissemination of fosfomycin resistance in E. coli is likely to be a significant concern.
Acinetobacter resistance
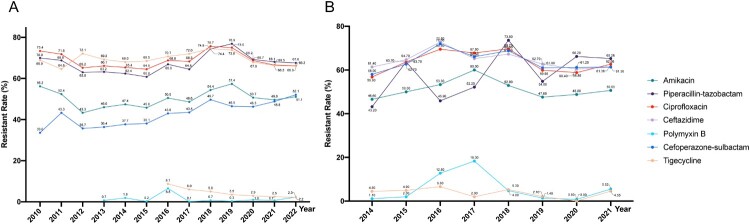
Acinetobacter baumannii is an opportunistic human pathogen that infected particularly in critically ill patients, mainly causing hospital-acquired (HAP) and ventilator-associated pneumonia (VAP) [145,146]. A. baumannii is now considered a global threat in the healthcare setting mainly owing to its propensity to acquire multidrug resistance phenotypes and to thrive in the healthcare environment at previously unforeseen rates [147,148]. What’s more, A. baumannii globally exhibits a high MDR rate, reaching approximately 45% [149]. The overall mortality rate estimate for A. baumannii infection reach up to 45% and is a major concern for nosocomial infection control [149,150]. Studies estimated that in 2019, carbapenem-resistant A. baumannii (CRAB) ranked as the fourth most burdensome pathogen–drug combination in high-income regions and the third in South Asia [2,151]. In China, CRAB consistently demonstrating pan-drug resistance and susceptibility only to polymyxins and tigecycline, has an isolation rate of around 70%, which has been increasing in recent years [152].
According to the CHINET monitoring results from 2015 to 2021, A. baumannii maintains an absolute dominance among all clinically isolated Acinetobacter species, with an average composition ratio of 89.6% over the seven years [19]. Apart from minocycline, tigecycline, and polymyxin B, A. baumannii exhibit higher resistance rates to other antimicrobial agents such as β-lactams, aminoglycosides, and fluoroquinolones [9]. From 2015 to 2021, there was an increasing trend in the resistance of A. baumannii to cefoperazone-sulbactam (from 40.6% to 52.8%) and piperacillin-tazobactam (from 64.5% to 73.2%), while a decreasing trend was observed in resistance to tigecycline (from 10.3% to 2.8%) and minocycline (from 30.4% to 21.0%) (Figure 7).
Figure 7.
Antimicrobial resistance trends for A. baumannii. (A). data from CHINET. (B). data from BRICS.
A study of nationwide and genome-based surveillance on CRAB strains collected from intensive care units (ICUs) in hospitals in different provinces in China found that CRAB strains were prevalent in 71.4% (55/77) of the ICUs surveyed. The study further indicated that CRAB isolates exhibited a notably low resistance rate to colistin (0.4%) and tigecycline (2.5%), while displaying a high resistance rate to ceftazidime–avibactam (70.2%) [153]. 59% among the GenBank available genomes of A. baumannii belongs of global clone (GC)2 according to the Institut Pasteur MLST scheme [154], which is mainly clonal complex (CC)92 according to the Oxford MLST. ST195, ST191 and ST208 are the most prevalent A. baumannii STs belonging to CC92 [155–158]. Clonal dissemination of CRAB was identified in 37.6% (29/77) of ICUs, revealing a total of 22 distinct clones and majority of these clones demonstrated transmissibility within a single ICU [153].
The major mechanism of carbapenem resistance in A. baumannii occurs through the action of acquired OXA-type carbapenem-hydrolyzing class D β-lactamases, which are encoded by blaOXA-23-like, blaOXA-40-like, blaOXA-58-like, blaOXA-143-like, and blaOXA-235-like genes [159,160]. CRAB is less commonly mediated by MBLs and only exceptionally by class A KPC and GES beta-lactamases [160]. The AdeABC efflux pump system contributed to CRAB isolates, as evidenced by the high expression of some of its encoding genes [150]. CRAB from the global T.E.S.T. worldwide surveillance between 2012 and 2016 (data from hospitals in 47 countries representing a total population of around 2.2 billion people spread over five geographical regions) that acquired OXA-type carbapenemase genes were found in 300 (96%) isolates with blaOXA-23-like and blaOXA-40-like predominating, which constitutes a significant increase compared to their findings from 2010 [161].
Polymxyins and tigecycline are commonly regarded as potent first-line agents for treating infections caused by CRAB [162]. While most CRAB strains remain highly sensitive to colistin and tigecycline, there has been a growing number of reports on A. baumannii strains resistant to these antibiotics in recent years. In 2022, CHINET revealed CRAB exhibited 1.2% resistance to polymyxins and 3.1% resistance to tigecycline (Figure 7). However, currently, over 50% of clinical A. baumannii strains demonstrate resistance to CAZ-AVI [112]. Presently, mutations in the pmrA/pmrB and lpxACD genes are the primary mechanisms of A. baumannii resistance to colistin [163,164]. Conversely, mutational changes in the tet(X) gene and the overexpression of resistance-nodulation-cell division (RND) efflux pumps are the major mechanisms observed in tigecycline-resistant A. baumannii isolates [165].
Pseudomonas aeruginosa
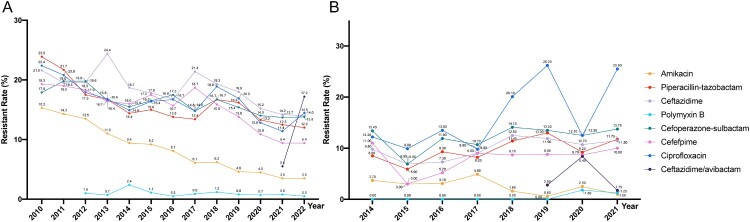
The isolation rate of P. aeruginosa has seen a relatively stable trend in China, from 12.7% in 2014 to 11.9% in 2022, and still ranks third among clinically isolated Gram-negative bacteria in China. MDR and extensively drug-resistant XDR P. aeruginosa poses an increasing global threat of nosocomial infections [166]. According to CHINET, P. aeruginosa shows a generally stable or decreasing trend in resistance to most drugs in China, and decreased for almost all antibiotics, during the five-year period. Resistance to imipenem and meropenem decreased in P. aeruginosa nationwide. From 2010 to 2022, the rates of imipenem and meropenem resistant isolates decreased from 30.8% to 22.1% and from 25.8% to 17.6%, respectively. However, CRPA in Zhejiang Province increased annually from 22% in 2015–38.67% in 2017 and that Zhejiang had the highest rates of CRPA of all provinces in China in 2017 [167]. Resistance to CAZ/AVI in P. aeruginosa significantly decreased from 11.1% in 2018–6.3% in 2022 [19]. Furthermore, in the recent five-years, the resistance rates to cefoperazone-sulbactam, piperacillin-tazobactam, ceftazidime, ciprofloxacin and amikacin decreased from 17.1% to 15%, 16.7% to 13.5%, 19.3% to 14.9%, 24.1% to 15.3%, and 6.2% to 3.8%, respectively. The resistance rates of colistin and polymyxin B fluctuated around 1.5% and 0.5% respectively (CHINET data). Nevertheless, the nationwide bloodstream surveillance system BRICS discovered an increase of P. aeruginosa in resistance to piperacillin-tazobactam (from 8.5% in 2014 to 11.75% in 2021), fosfomycin (from 0% in 2014 to 6.5% in 2021) and meropenem (from 6.1% in 2014 to 13.5% in 2021) (BRICS data) (Figure 8). Therefore, we still cannot relax our vigilance against the antibiotic resistance of P. aeruginosa.
Figure 8.
Antimicrobial resistance trends for P. aeruginosa. (A). data from CHINET. (B). data from BRICS.
P. aeruginosa demonstrates diverse characteristics in the prevalence of its clones [168]. Several STs of P. aeruginosa, including ST235, ST111, ST463 etc, are of special concern [168,169]. These infamous STs are strongly linked to adverse clinical outcomes and harbour a multitude of horizontally acquired resistance determinants. In recent years, prevalent clones have emerged in various regions across China. In Zhejiang, ST463 poses a potential health threat due to its unique combination of pyocyanin production and virulence genes [170]. In Shenzhen, the detection of blaNDM-1 and mcr-1 in MDR P. aeruginosa underscores the need for continuous surveillance in pediatric patients [171]. In Shanghai, a low prevalence of polymyxin-resistant P. aeruginosa, including the globally disseminated ST277, was observed following polymyxin administration in a tertiary teaching hospital [172]. Furthermore, CRPA strains were reported in Shandong (ST244), Sichuan (ST277), and Hainan (ST357), respectively [173].
Enterobacter species
Enterobacter species rank fifth in the isolation rate of Gram-negative bacteria, comprising 3–5% of all bacterial isolates in China (CARSS data). Enterobacter species extracted from clinical samples are usually reported as E. cloacae, and sometimes E. asburiae, E. hormaechei, or E. kobei. They are all members of E. cloacae complex (ECC) [174,175]. The resistance rates of Enterobater species to imipenem and meropenem increased from 5.2% and 4.8% in 2010 to 9.7% each in 2022. Enterobacter species are highly sensitive to amikacin (resistant rate 1.6% in 2022), and tigecycline (resistant rate 1.9% in 2022) (CHINET data). Additionally, Enterobacter species exhibit a very high resistant rate (93.6%) to cefotaxime. Regarding the resistance of Enterobacter species to polymyxins, there is a discrepancy between the CHINET and BRICS data. CHINET data indicates that the resistance rate of Enterobacter species to polymyxin ranged from 2% to 5% from 2019 to 2022. However, BRICS data shows a different trend, with the resistance rate increasing from 1.5% in 2014 to 29.58% in 2021 (Figure 9). Although the exact prevalence of polymyxins resistance is uncertain, Enterobacter appears to exhibit a higher resistance rate than Escherichia and Klebsiella (usually <2% for both) [176,177]. What’s more, Enterobacter species often exhibit heteroresistance to polymyxins [178], may cause the “skipped wells” phenomenon in broth microdilution, potentially influencing the observed resistance rate.
Figure 9.
Antimicrobial resistance trends for Enterobacter species. (A). data from CHINET. (B). data from BRICS.
Strains of carbapenem-resistant Enterobacter (CR-Ent) have emerged in the last two decades that contributing majorly by carbapenemases, with 99.9% of Enterobacter clinical strains reported to be carbapenem-susceptible prior to 2001 [177]. A variety of carbapenemases have been found in Enterobacter with geographic specificity. KPC is prevalent in the Americas, VIM is most common in Europe, NDM is mostly encountered in India and mainland China [177]. Enterobacter has demonstrated faster carbapenem penetration via porins compared with Klebsiella, suggesting that porins may play an important role in carbapenem resistance in Enterobacter [179].
Clinical laboratories commonly categorize Enterobacter species as the ECC, leading to challenges in correlating antimicrobial resistance with specific Enterobacter species. The widespread adoption of genomic sequencing analysis technology has made the classification ECC strains more convenient and accurate. Zong et al. analysed 4,899 Enterobacter genomes from GenBank, and revealed that E. xiangfangensis is the most prevalent species worldwide, with the blaNDM gene predominantly found in China [177]. Study revealed that the most common human-source CR-Ent species in China was E. xiangfangensis (66/92, 71.93%), and the proportion of carbapenemase-producing Enterobacter (CP-Ent) in CR-Ent was high (72/92, 78.26%) in comparison to other global regions [177]. The high risk of carbapenemase-producing ST171 and ST116 E. xiangfangensis, and the blaNDM-harbouring IncX3-type plasmid were detected and emphasized in China [180]. Furthermore, globally distributed strains, such as ST90, ST93, and ST114 E. xiangfangensis, and ST78 E. hoffmannii, are associated with multiple carbapenemases (VIM, NDM, KPC, and OXA-48) [181]. ECC is conventionally regarded as a low-virulence pathogen. However, an epidemic hypervirulent clone of Enterobacter hormaechei, ST133, showing high invasiveness and mortality in clinical settings was reported [182,183].
Concluding remarks and future directions
From the second decade of 21th, although the resistance rates of clinically isolated bacteria in China to common antibiotics continue to exhibit an increasing trend, there has been a slight stabilization or even a modest decline in resistance to certain antibiotics among some bacteria in recent years [19]. Moreover, the detection rates of critically important carbapenem-resistant bacteria, such as CRKP and CRPA, which have shown a continuous increase over many years, have recently demonstrated a consecutive decline [9,19]. This suggests the importance of enhancing bacterial resistance monitoring and implementing multidisciplinary collaboration, along with hospital infection prevention and control measures, as effective strategies to curb the epidemic spread of antibiotic resistant bacteria [4,5]. What’s more, the variation in antibiotic resistant rates in China during the three-year period of COVID-19 pandemic control (from late 2019 to the end of 2022) may be influenced by differences in nosocomial infection control strategies [184]. Therefore, ongoing monitoring and attention are needed to assess the trend in antibiotic resistant rates in Chinese clinical settings starting from 2023.
In recent years, there has been a continuous evolution of prevalent antibiotic resistant bacterial clones. For instance, the prevalence of CA-MRSA strain ST59 invading clinical settings has gradually surpassed that of HA-MRSA ST239 and ST5 [49,53,62]. The proportion of A. baumannii ST208 in bloodstream infections has been steadily increasing [185,186], while the prevalence of high-virulence CRKP strains, such as ST11-KL64, is on the rise, with a simultaneous decrease in the prevalence of ST11-KL47 strains [104]. The emergence of next-generation sequencing has facilitated the development of advanced tools for monitoring and addressing the dissemination of antibiotic resistant bacteria [187,188]. Leveraging the advancements in high-throughput sequencing technology to study their systematic evolutionary dynamics and resistance evolution trends will aid in predicting the future direction of antibiotic resistant bacteria, providing essential insights for proactive monitoring and effective resistance control.
Data from the Centre for Antibacterial Surveillance in China indicates that in recent years, the utilization of antibiotics among outpatients and inpatients has significantly decreased. However, there is a continuous increase in the use or intensity of cephalosporin/beta-lactamase inhibitor combinations, carbapenems, and tigecycline [189]. The persistent high concentration of antibiotic usage is likely to have a direct impact on bacterial resistance [189]. Ongoing efforts in the antibiotic stewardship will play a crucial role in preventing and controlling antibiotic resistance.
The prevention and control of antibiotic resistance are not limited to clinical concerns; they also involve issues related to animal farming, environmental pollution, food safety, and more [189,190]. Resistant bacteria in areas such as animals, the environment, and food can be transmitted to humans, as seen in the case of MRSA [191,192]. The use of antibiotics in animal farming and other sectors can lead to resistant bacteria, impacting the selection of antibiotics in clinical settings, such as the resistance issue with polymyxins [10,115]. Therefore, it is essential to adopt a One Health strategy to address antibiotic resistance in clinical settings, fostering collaboration across various sectors and optimizing the use of antibiotics in both human and animal contexts.
Funding Statement
This work was supported by The National Key Research and Development Program of China (2021YFC2300300). This work was partially supported by grants from the Key Research and Development program of Zhejiang province (No. 2021C03068), the Natural Science Foundation of Zhejiang Province, China (No. LY24H190002), the National Natural Science Foundation of China (No. 81971984).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Jee Y, Carlson J, Rafai E, et al. Antimicrobial resistance: a threat to global health. Lancet Infect Dis. 2018;18(9):939–940. doi: 10.1016/S1473-3099(18)30471-7 [DOI] [PubMed] [Google Scholar]
- 2.Antimicrobial Resistance C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.<apo-nid63983.pdf>.
- 4.Xiao Y. Antimicrobial stewardship in China: systems, actions and future strategies. Clin Infect Dis. 2018;67(suppl_2):S135–S141. doi: 10.1093/cid/ciy641 [DOI] [PubMed] [Google Scholar]
- 5.Xiao Y, Li L.. A national action plan to contain antimicrobial resistance in China: contents, actions and expectations. AMR Control. 2017;15(3):17–20. [Google Scholar]
- 6.Ding L, Hu F.. China's new national action plan to combat antimicrobial resistance (2022–25). J Antimicrob Chemother. 2023;78(2):558–560. doi: 10.1093/jac/dkac435 [DOI] [PubMed] [Google Scholar]
- 7.Zong Z, Wu A, Hu B.. Infection control in the era of antimicrobial resistance in China: progress, challenges, and opportunities. Clin Infect Dis. 2020;71(Suppl 4):S372–S378. doi: 10.1093/cid/ciaa1514 [DOI] [PubMed] [Google Scholar]
- 8.Hu F, Zhu D, Wang F, et al. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67(suppl_2):S128–S134. doi: 10.1093/cid/ciy657 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Ji J, Ying C, et al. Blood bacterial resistant investigation collaborative system (BRICS) report: a national surveillance in China from 2014 to 2019. Antimicrob Resist Infect Control. 2022;11(1):17. doi: 10.1186/s13756-022-01055-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 11.Fang L-X, Chen C, Cui C-Y, et al. Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile Tet(X). Bioessays. 2020;42(8):e2000014. doi: 10.1002/bies.202000014 [DOI] [PubMed] [Google Scholar]
- 12.Shi Q, Yin D, Han R, et al. Emergence and recovery of ceftazidime-avibactam resistance in blaKPC-33-Harboring Klebsiella pneumoniae Sequence Type 11 Isolates in China. Clin Infect Dis. 2020;71(Suppl 4):S436–S439. doi: 10.1093/cid/ciaa1521 [DOI] [PubMed] [Google Scholar]
- 13.Chen H, et al. Drivers of methicillin-resistant staphylococcus aureus (MRSA) lineage replacement in China. Genome Med. 2021;13:1–14. doi: 10.1186/s13073-020-00808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi: 10.3201/eid2602.190594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tacconelli E, Sifakis F, Harbarth S, et al. Surveillance for control of antimicrobial resistance. Lancet Infect Dis. 2018;18(3):e99–e106. doi: 10.1016/S1473-3099(17)30485-1 [DOI] [PubMed] [Google Scholar]
- 16.Cornaglia G, Hryniewicz W, Jarlier V, et al. European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect. 2004;10(4):349–383. doi: 10.1111/j.1198-743X.2004.00887.x [DOI] [PubMed] [Google Scholar]
- 17.Organization, W.H. GLASS whole-genome sequencing for surveillance of antimicrobial resistance; 2020.
- 18.Ding L, Guo Y, Hu F.. Antimicrobial resistance surveillance: China's nearly 40-year effort. Int J Antimicrob Agents. 2023;62(2):106869. doi: 10.1016/j.ijantimicag.2023.106869 [DOI] [PubMed] [Google Scholar]
- 19.Yang W, Ding L, Han R, et al. Current status and trends of antimicrobial resistance among clinical isolates in China: a retrospective study of CHINET from 2018 to 2022. One Health Advances. 2023;1(1):8. doi: 10.1186/s44280-023-00009-9 [DOI] [Google Scholar]
- 20.Jing C, Wang C.. Surveillance of bacterial resistance at Children's Hospital of Chongqing Medical University in 2015. Chinese J Infect Chemother. 2017;6:413–420. [Google Scholar]
- 21.Lakhundi S, Zhang K.. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020–18. doi: 10.1128/CMR.00020-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christaki E, Marcou M, Tofarides A.. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88:26–40. doi: 10.1007/s00239-019-09914-3 [DOI] [PubMed] [Google Scholar]
- 23.Xiao YH, Giske CG, Wei Z-Q, et al. Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist Updat. 2011;14(4-5):236–250. doi: 10.1016/j.drup.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 24.Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. doi: 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao W, Howden BP, Stinear TP.. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr Opin Microbiol. 2018;41:76–82. doi: 10.1016/j.mib.2017.11.030 [DOI] [PubMed] [Google Scholar]
- 26.Vu J, Carvalho J.. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol (Beijing). 2011;6:357–366. doi: 10.1007/s11515-011-1167-x [DOI] [Google Scholar]
- 27.Shrestha S, Kharel S, Homagain S, et al. Prevalence of vancomycin-resistant enterococci in Asia-A systematic review and meta-analysis. J Clin Pharm Ther. 2021;46(5):1226–1237. doi: 10.1111/jcpt.13383 [DOI] [PubMed] [Google Scholar]
- 28.Zheng B, Tomita H, Xiao YH, et al. Molecular characterization of vancomycin-resistant enterococcus faecium isolates from mainland China. J Clin Microbiol. 2007;45(9):2813–2818. doi: 10.1128/JCM.00457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen J, Long X, Jiang Q, et al. Genomic characterization of a vancomycin-resistant strain of Enterococcus faecium harboring a rep2 Plasmid. Infect Drug Resist. 2023;16:1153–1158. doi: 10.2147/IDR.S398913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Zhou H, Sun Y, et al. Characterization of clinical enterococci isolates, focusing on the vancomycin-resistant enterococci in a tertiary hospital in China: based on the data from 2013 to 2018. BMC Infect Dis. 2020;20(1):356. doi: 10.1186/s12879-020-05078-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan MY, He Y-H, Ruan G-J, et al. The prevalence and molecular epidemiology of vancomycin-resistant Enterococcus (VRE) carriage in patients admitted to intensive care units in Beijing, China. J Microbiol Immunol Infect. 2023;56(2):351–357. doi: 10.1016/j.jmii.2022.07.001 [DOI] [PubMed] [Google Scholar]
- 32.Sun H-L, Liu C, Zhang J-J, et al. Molecular characterization of vancomycin-resistant enterococci isolated from a hospital in Beijing, China. J Microbiol Immunol Infect. 2019;52(3):433–442. doi: 10.1016/j.jmii.2018.12.008 [DOI] [PubMed] [Google Scholar]
- 33.Li L, et al. The first investigation of a nosocomial outbreak caused by ST80 vancomycin-resistant Enterococci faecium in China. J Hosp Infect. 2023. doi: 10.1016/j.jhin.2023.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Dadashi M, Sharifian P, Bostanshirin N, et al. The global prevalence of daptomycin, tigecycline, and linezolid-resistant Enterococcus faecalis and Enterococcus faecium strains from human clinical samples: a systematic review and meta-analysis. Front Med (Lausanne). 2021;8:720647. doi: 10.3389/fmed.2021.720647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma X, Zhang F, Bai B, et al. Linezolid resistance in Enterococcus faecalis associated with urinary tract infections of patients in a tertiary hospitals in China: resistance mechanisms, virulence, and risk factors. Front Public Health. 2021;9:570650. doi: 10.3389/fpubh.2021.570650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun W, Liu H, Liu J, et al. Detection of optrA and poxtA genes in linezolid-resistant Enterococcus isolates from fur animals in China. Lett Appl Microbiol. 2022;75(6):1590–1595. doi: 10.1111/lam.13826 [DOI] [PubMed] [Google Scholar]
- 37.Krause AL, Stinear TP, Monk IR.. Barriers to genetic manipulation of Enterococci: current approaches and future directions. FEMS Microbiol Rev. 2022;46(6):fuac036. doi: 10.1093/femsre/fuac036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadowy E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid. 2018;99:89–98. doi: 10.1016/j.plasmid.2018.09.011 [DOI] [PubMed] [Google Scholar]
- 39.Antonelli A, D’Andrea MM, Brenciani A, et al. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother. 2018;73(7):1763–1769. doi: 10.1093/jac/dky088 [DOI] [PubMed] [Google Scholar]
- 40.Yi M, Zou J, Zhao J, et al. Emergence of optrA-mediated linezolid resistance in Enterococcus faecium: a molecular investigation in a Tertiary Hospital of Southwest China from 2014–2018. Infect Drug Resist. 2022;15:13–20. doi: 10.2147/IDR.S339761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdullahi IN, Lozano C, Juárez-Fernández G, et al. Nasotracheal enterococcal carriage and resistomes: detection of optrA-, poxtA-and cfrD-carrying strains in migratory birds, livestock, pets, and in-contact humans in Spain. Eur J Clin Microbiol Infect Dis. 2023;42(5):569–581. doi: 10.1007/s10096-023-04579-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monteiro Marques J, et al. Dissemination of Enterococcal genetic lineages: a one health perspective. Antibiotics. 2023;12(7):1140. doi: 10.3390/antibiotics12071140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu M, et al. Prevalence of methicillin-resistant Staphylococcus aureus in healthy Chinese population: a system review and meta-analysis. PLoS One. 2019;14(10):e0223599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ji S, Jiang S, Wei X, et al. In-host evolution of daptomycin resistance and heteroresistance in methicillin-resistant staphylococcus aureus strains from three endocarditis patients. J Infect Dis. 2020;221(Suppl 2):S243–S252. doi: 10.1093/infdis/jiz571 [DOI] [PubMed] [Google Scholar]
- 45.Jiang S, Zhuang H, Zhu F, et al. The Role of mprF mutations in Seesaw effect of daptomycin-resistant methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2022;66(1):e01295–21. doi: 10.1128/AAC.01295-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Wang B, Zhao H, et al. In vitro activity of vancomycin, teicoplanin, linezolid and daptomycin against methicillin-resistant staphylococcus aureus Isolates collected from Chinese hospitals in 2018–2020. Infect Drug Resist. 2021;14:5449–5456. doi: 10.2147/IDR.S340623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jian Y, Lv H, Liu J, et al. Dynamic changes of Staphylococcus aureus susceptibility to Vancomycin, Teicoplanin, and Linezolid in a Central Teaching Hospital in Shanghai, China, 2008–2018. Front Microbiol. 2020;11:908. doi: 10.3389/fmicb.2020.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin Y, et al. Genomic epidemiology and characterization of methicillin-resistant Staphylococcus aureus from bloodstream infections in China. MSystems. 2021;6(6):e00837–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uehara Y. Current status of staphylococcal cassette chromosome mec (SCC mec). Antibiotics. 2022;11(1):86. doi: 10.3390/antibiotics11010086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Hu Y, Baker M, et al. Novel SCCmec type XV (7A) and two pseudo-SCCmec variants in foodborne MRSA in China. J Antimicrob Chemother. 2022;77(4):903–909. doi: 10.1093/jac/dkab500 [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Sun L, Ba X, et al. Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: a whole-genome-based survey. Clin Microbiol Infect. 2022;28(1):85–92. doi: 10.1016/j.cmi.2021.05.024 [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Sun L, Wu D, et al. Using core-genome multilocus sequence typing to monitor the changing epidemiology of methicillin-resistant Staphylococcus aureus in a teaching hospital. Clin Infect Dis. 2018;67(suppl_2):S241–S248. doi: 10.1093/cid/ciy644 [DOI] [PubMed] [Google Scholar]
- 53.Li S, Sun S, Yang C, et al. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Front Microbiol. 2018;9:332. doi: 10.3389/fmicb.2018.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao M, Wang H, Zhao Y, et al. National surveillance of methicillin-resistant Staphylococcus aureus in China highlights a still-evolving epidemiology with 15 novel emerging multilocus sequence types. J Clin Microbiol. 2013;51(11):3638–3644. doi: 10.1128/JCM.01375-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D, Wang Z, Wang H, et al. Predominance of ST5-II-t311 clone among healthcare-associated methicillin-resistant Staphylococcus aureus isolates recovered from Zhejiang, China. Int J Infect Dis. 2018;71:107–112. doi: 10.1016/j.ijid.2018.04.798 [DOI] [PubMed] [Google Scholar]
- 56.Hung W-C, et al. Molecular evolutionary pathways toward two successful community-associated but multidrug-resistant ST59 methicillin-resistant Staphylococcus aureus lineages in Taiwan: dynamic modes of mobile genetic element salvages. PLoS One. 2016;11(9):e0162526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X, et al. Multiresistant ST59-SCC mec IV-t437 clone with strong biofilm-forming capacity was identified predominantly in MRSA isolated from Chinese children. BMC Infect Dis. 2017;17:1–12. doi: 10.1186/s12879-016-2122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee AS, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4(1):1–23. [DOI] [PubMed] [Google Scholar]
- 59.Jiang N, Li J, Feßler AT, et al. Novel pseudo-staphylococcal cassette chromosome mec element (φSCCmecT55) in MRSA ST9. J Antimicrob Chemother. 2019;74(3):819–820. doi: 10.1093/jac/dky457 [DOI] [PubMed] [Google Scholar]
- 60.Zhou W, Jin Y, Shen P, et al. Novel SCCmec variants in clonal complex 398 and lineage-specific pseudo-SCC mec identified in ST88 MRSA from invasive bloodstream infections in China. J Antimicrob Chemother. 2023;78(9):2366–2375. doi: 10.1093/jac/dkad250 [DOI] [PubMed] [Google Scholar]
- 61.Proulx MK, Palace SG, Gandra S, et al. Reversion from methicillin susceptibility to methicillin resistance in Staphylococcus aureus during treatment of bacteremia. J Infect Dis. 2016;213(6):1041–1048. doi: 10.1093/infdis/jiv512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li M, Wang Y, Zhu Y, et al. Increased community-associated infections caused by Panton-Valentine Leukocidin-negative MRSA, Shanghai, 2005–2014. Emerg Infect Dis. 2016;22(11):1988–1991. doi: 10.3201/eid2211.160587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang B, Xu Y, Zhao H, et al. Methicillin-resistant Staphylococcus aureus in China: a multicentre longitudinal study and whole-genome sequencing. Emerg Microbes Infect. 2022;11(1):532–542. doi: 10.1080/22221751.2022.2032373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H, Yin Y, van Dorp L, et al. Drivers of methicillin-resistant Staphylococcus aureus (MRSA) lineage replacement in China. Genome Med. 2021;13(1):171. doi: 10.1186/s13073-021-00992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mork RL, Hogan PG, Muenks CE, et al. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect Dis. 2020;20(2):188–198. doi: 10.1016/S1473-3099(19)30570-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hogan PG, Mork RL, Thompson RM, et al. Environmental methicillin-resistant Staphylococcus aureus contamination, persistent colonization, and subsequent skin and soft tissue infection. JAMA Pediatr. 2020;174(6):552–562. doi: 10.1001/jamapediatrics.2020.0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun L, Zhuang H, Di L, et al. Transmission and microevolution of methicillin-resistant Staphylococcus aureus ST88 strain among patients, healthcare workers, and household contacts at a trauma and orthopedic ward. Front Public Health. 2023;10:1053785. doi: 10.3389/fpubh.2022.1053785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richardson EJ, Bacigalupe R, Harrison EM, et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nature Ecology & Evolution. 2018;2(9):1468–1478. doi: 10.1038/s41559-018-0617-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai Y, Liu J, Guo W, et al. Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008–2017. Emerg Microbes Infect. 2019;8(1):471–478. doi: 10.1080/22221751.2019.1595161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jian Y, Zhao L, Zhao N, et al. Increasing prevalence of hypervirulent ST5 methicillin susceptible Staphylococcus aureus subtype poses a serious clinical threat. Emerg Microbes Infect. 2021;10(1):109–122. doi: 10.1080/22221751.2020.1868950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park S, Ronholm J.. Staphylococcus aureus in agriculture: lessons in evolution from a multispecies pathogen. Clin Microbiol Rev. 2021;34(2):e00182–20. doi: 10.1128/CMR.00182-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shariati A, Dadashi M, Moghadam MT, et al. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10(1):12689. doi: 10.1038/s41598-020-69058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen P, Zhou K, Wang Y, et al. High prevalence of a globally disseminated hypervirulent clone, Staphylococcus aureus CC121, with reduced vancomycin susceptibility in community settings in China. J Antimicrob Chemother. 2019;74(9):2537–2543. doi: 10.1093/jac/dkz232 [DOI] [PubMed] [Google Scholar]
- 74.Liang J, Hu Y, Fu M, et al. Resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus and heterogeneous vancomycin-intermediate Staphylococcus aureus. Infect Drug Resist. 2023;16:379–388. doi: 10.2147/IDR.S392908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding L, et al. Klebsiella pneumoniae carbapenemase variants: the new threat to global public health. Clin Microbiol Rev. 2023;36(4):e0000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang M, Earley M, Chen L, et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22(3):401–412. doi: 10.1016/S1473-3099(21)00399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Z-Q, Huang Y-L, Zhou H-W, et al. Persistent carbapenem-resistant Klebsiella pneumoniae: a Trojan horse. Lancet Infect Dis. 2018;18(1):22–23. doi: 10.1016/S1473-3099(17)30627-8 [DOI] [PubMed] [Google Scholar]
- 78.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi: 10.1080/22221751.2020.1799721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zha L, Li S, Ren Z, et al. Clinical management of infections caused by carbapenem-resistant Klebsiella pneumoniae in critically ill patients: a nationwide survey of tertiary hospitals in mainland China. J Infect. 2022;84(6):e108–e110. doi: 10.1016/j.jinf.2022.03.023 [DOI] [PubMed] [Google Scholar]
- 81.Wei ZQ, et al. Plasmid-Mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–765. doi: 10.1128/AAC.01053-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu J, Yu J, Chen F, et al. Emergence and establishment of KPC-2-producing ST11 Klebsiella pneumoniae in a general hospital in Shanghai, China. Eur J Clin Microbiol Infect Dis. 2018;37(2):293–299. doi: 10.1007/s10096-017-3131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X, Dong N, Chan EW-C, et al. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi: 10.1016/j.tim.2020.04.012 [DOI] [PubMed] [Google Scholar]
- 84.Tang Y, Li G, Liang W, et al. Translocation of Carbapenemase Gene blaKPC-2 both internal and external to transposons occurs via novel structures of Tn 1721 and exhibits distinct movement patterns. Antimicrob Agents Chemother. 2017;61(10):e01151–17. doi: 10.1128/AAC.01151-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu P, Tang Y, Li G, et al. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents. 2019;54(2):117–124. doi: 10.1016/j.ijantimicag.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 86.Yang X, Sun Q, Li J, et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg Microbes Infect. 2022;11(1):841–849. doi: 10.1080/22221751.2022.2049458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y, Yang Y, Chen G, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):700–709. doi: 10.1080/22221751.2021.1906163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raro OHF, da Silva RMC, Filho EMR, et al. Carbapenemase-producing Klebsiella pneumoniae from transplanted patients in Brazil: phylogeny, resistome, virulome and mobile genetic elements harboring bla (KPC-) (2) or bla (NDM-) (1). Front Microbiol. 2020;11:1563. doi: 10.3389/fmicb.2020.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kopotsa K, Osei Sekyere J, Mbelle NM.. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann N Y Acad Sci. 2019;1457(1):61–91. doi: 10.1111/nyas.14223 [DOI] [PubMed] [Google Scholar]
- 90.Yin L, Lu L, He L, et al. Molecular characteristics of carbapenem-resistant gram-negative bacilli in pediatric patients in China. BMC Microbiol. 2023;23(1):136. doi: 10.1186/s12866-023-02875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang X, Xue J, Shen M-J, et al. Molecular typing and drug resistance analysis of carbapenem-resistant Klebsiella pneumoniae from paediatric patients in China. J Infect Dev Ctries. 2022;16(11):1726–1731. doi: 10.3855/jidc.17003 [DOI] [PubMed] [Google Scholar]
- 92.Xiong Z, Zhang C, Sarbandi K, et al. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae in pediatric inpatients in South China. Microbiol Spectr. 2023;11(6):e0283923. doi: 10.1128/spectrum.02839-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu Y, et al. Global phylogeography and genomic epidemiology of carbapenem-resistant bla(OXA-232)-carrying Klebsiella pneumoniae sequence type 15 lineage. Emerg Infect Dis. 2023;29(11):2246–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang C-H, Ma L, Huang L-Y, et al. Molecular epidemiology and resistance patterns of bla(OXA-48)Klebsiella pneumoniae and Escherichia coli: a nationwide multicenter study in Taiwan. J Microbiol Immunol Infect. 2021;54(4):665–672. doi: 10.1016/j.jmii.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 95.Li Z, Ding Z, Yang J, et al. Carbapenem-resistant Klebsiella pneumoniae in Southwest China: molecular characteristics and risk factors caused by KPC and NDM producers. Infect Drug Resist. 2021;14:3145–3158. doi: 10.2147/IDR.S324244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi: 10.1093/jac/dkz446 [DOI] [PubMed] [Google Scholar]
- 97.Tang B, Yang A, Liu P, et al. Outer membrane vesicles transmittingblaNDM-1 Mediate the emergence of Carbapenem-resistant Hypervirulent Klebsiella pneumoniae. Antimicrob Agents Chemother. 2023;67(5):e0144422. doi: 10.1128/aac.01444-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Wang X, Wang S, et al. Emergence of Colistin resistance in Carbapenem-resistant Hypervirulent Klebsiella pneumoniae under the pressure of Tigecycline. Front Microbiol. 2021;12:756580. doi: 10.3389/fmicb.2021.756580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yao H, Qin S, Chen S, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):25. doi: 10.1016/S1473-3099(17)30628-X [DOI] [PubMed] [Google Scholar]
- 100.Wong MHY, Shum H-P, Chen JHK, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):24. doi: 10.1016/S1473-3099(17)30629-1 [DOI] [PubMed] [Google Scholar]
- 101.Zhang R, Lin D, Chan EW-c, et al. Emergence of Carbapenem-resistant Serotype K1 Hypervirulent Klebsiella pneumoniae Strains in China. Antimicrob Agents Chemother. 2016;60(1):709–711. doi: 10.1128/AAC.02173-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi: 10.1016/j.jinf.2015.07.010 [DOI] [PubMed] [Google Scholar]
- 103.Lai Y-C, Lu M-C, Hsueh P-R.. Hypervirulence and carbapenem resistance: two distinct evolutionary directions that led high-risk Klebsiella pneumoniae clones to epidemic success. Expert Rev Mol Diagn. 2019;19(9):825–837. doi: 10.1080/14737159.2019.1649145 [DOI] [PubMed] [Google Scholar]
- 104.Zhou K, Xue C-X, Xu T, et al. A point mutation in recC associated with subclonal replacement of carbapenem-resistant Klebsiella pneumoniae ST11 in China. Nat Commun. 2023;14(1):2464. doi: 10.1038/s41467-023-38061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pu D, Zhao J, Lu B, et al. Within-host resistance evolution of a fatal ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents. 2023;61(4):106747. doi: 10.1016/j.ijantimicag.2023.106747 [DOI] [PubMed] [Google Scholar]
- 106.Xu Y, Zhang J, Wang M, et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021;13(1):119. doi: 10.1186/s13073-021-00936-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang X, Wai-Chi Chan E, Zhang R, et al. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4(12):2039–2043. doi: 10.1038/s41564-019-0566-7 [DOI] [PubMed] [Google Scholar]
- 108.Li R, Cheng J, Dong H, et al. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int J Antimicrob Agents. 2020;55(6):105952. doi: 10.1016/j.ijantimicag.2020.105952 [DOI] [PubMed] [Google Scholar]
- 109.Xia P, et al. Coexistence of multidrug resistance and virulence in a single conjugative plasmid from a Hypervirulent Klebsiella pneumoniae isolate of sequence Type 25. mSphere. 2022;7(6):e0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie M, Yang X, Xu Q, et al. Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun Biol. 2021;4(1):650. doi: 10.1038/s42003-021-02148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou C, Zhang H, Xu M, et al. Within-host resistance and virulence evolution of a hypervirulent Carbapenem-resistant Klebsiella pneumoniae ST11 under antibiotic pressure. Infect Drug Resist. 2023;16:7255–7270. doi: 10.2147/IDR.S436128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yahav D, Giske CG, Grāmatniece A, et al. New beta-Lactam-beta-Lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34(1):e00115–20. doi: 10.1128/CMR.00115-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124.e1–124.e4. doi: 10.1016/j.cmi.2019.08.020 [DOI] [PubMed] [Google Scholar]
- 114.Xu M, Zhao J, Xu L, et al. Emergence of transferable ceftazidime-avibactam resistance in KPC-producing Klebsiella pneumoniae due to a novel CMY AmpC beta-lactamase in China. Clin Microbiol Infect. 2022;28(1):136.e1–136.e6. doi: 10.1016/j.cmi.2021.05.026 [DOI] [PubMed] [Google Scholar]
- 115.Wang Y, Xu C, Zhang R, et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis. 2020;20(10):1161–1171. doi: 10.1016/S1473-3099(20)30149-3 [DOI] [PubMed] [Google Scholar]
- 116.Wang Y, Luo Q, Chen T, et al. Clinical, biological and genome-wide comparison of carbapenem-resistant Klebsiella pneumoniae with susceptibility transformation to polymyxin B during therapy. Clin Microbiol Infect. 2023;29(10):1336.e1–1336.e8. doi: 10.1016/j.cmi.2023.06.029 [DOI] [PubMed] [Google Scholar]
- 117.Luo Q, Yu W, Zhou K, et al. Molecular epidemiology and Colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front Microbiol. 2017;8:2262. doi: 10.3389/fmicb.2017.02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berglund B. Acquired resistance to colistin via chromosomal and plasmid-mediated mechanisms in Klebsiella pneumoniae. Infect Microbes Diseases. 2019;1(1):10–19. doi: 10.1097/IM9.0000000000000002 [DOI] [Google Scholar]
- 119.Kim SJ, Shin JH, Kim H, et al. Roles of crrAB two-component regulatory system in Klebsiella pneumoniae: growth yield, survival in initial colistin treatment stage, and virulence. Int J Antimicrob Agents. 2024;63(1):107011. doi: 10.1016/j.ijantimicag.2023.107011 [DOI] [PubMed] [Google Scholar]
- 120.Wright MS, Suzuki Y, Jones MB, et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother. 2015;59(1):536–543. doi: 10.1128/AAC.04037-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Andersson DI, Nicoloff H, Hjort K.. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17(8):479–496. doi: 10.1038/s41579-019-0218-1 [DOI] [PubMed] [Google Scholar]
- 122.Sun Y, Cai Y, Liu X, et al. The emergence of clinical resistance to tigecycline. Int J Antimicrob Agents. 2013;41(2):110–116. doi: 10.1016/j.ijantimicag.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 123.He F, Shi Q, Fu Y, et al. Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect Genet Evol. 2018;66:188–191. doi: 10.1016/j.meegid.2018.09.025 [DOI] [PubMed] [Google Scholar]
- 124.Bialek-Davenet S, Lavigne J-P, Guyot K, et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):81–88. doi: 10.1093/jac/dku340 [DOI] [PubMed] [Google Scholar]
- 125.Chiu SK, et al. Roles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2017;61(8):e00391–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016;6(4):a025387. doi: 10.1101/cshperspect.a025387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang R, Dong N, Shen Z, et al. Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet(X). Nat Commun. 2020;11(1):4648. doi: 10.1038/s41467-020-18475-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Becker B, Cooper MA.. Aminoglycoside antibiotics in the 21st century. ACS Chem Biol. 2013;8(1):105–115. doi: 10.1021/cb3005116 [DOI] [PubMed] [Google Scholar]
- 129.Garneau-Tsodikova S, Labby KJ.. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm. 2016;7(1):11–27. doi: 10.1039/C5MD00344J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wachino J, Arakawa Y.. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic gram-negative bacteria: an update. Drug Resist Updat. 2012;15(3):133–148. doi: 10.1016/j.drup.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 131.Sacco F, Raponi G, Oliva A, et al. An outbreak sustained by ST15 Klebsiella pneumoniae carrying 16S rRNA methyltransferases and bla(NDM): evaluation of the global dissemination of these resistance determinants. Int J Antimicrob Agents. 2022;60(2):106615. doi: 10.1016/j.ijantimicag.2022.106615 [DOI] [PubMed] [Google Scholar]
- 132.Zhang X, Li Q, Lin H, et al. High-level aminoglycoside resistance in human clinical Klebsiella pneumoniae complex isolates and characteristics of armA-carrying IncHI5 plasmids. Front Microbiol. 2021;12:636396. doi: 10.3389/fmicb.2021.636396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang L, Cao M, Hu Y, et al. Prevalence and mechanisms of fosfomycin resistance among KPC-producing Klebsiella pneumoniae clinical isolates in China. Int J Antimicrob Agents. 2021;57(1):106226. doi: 10.1016/j.ijantimicag.2020.106226 [DOI] [PubMed] [Google Scholar]
- 134.Vardakas KZ, Legakis NJ, Triarides N, et al. Susceptibility of contemporary isolates to fosfomycin: a systematic review of the literature. Int J Antimicrob Agents. 2016;47(4):269–285. doi: 10.1016/j.ijantimicag.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 135.Xiang DR, Li J-J, Sheng Z-K, et al. Complete sequence of a Novel IncR-F33:A-:B- plasmid, pKP1034, Harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an Epidemic Klebsiella pneumoniae Sequence Type 11 Strain in China. Antimicrob Agents Chemother. 2016;60(3):1343–1348. doi: 10.1128/AAC.01488-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bi W, Li B, Song J, et al. Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum beta-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou, China. Int J Antimicrob Agents. 2017;50(1):29–34. doi: 10.1016/j.ijantimicag.2017.02.010 [DOI] [PubMed] [Google Scholar]
- 137.Acman M, Wang R, van Dorp L, et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat Commun. 2022;13(1):1131. doi: 10.1038/s41467-022-28819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Qu H, Wang X, Ni Y, et al. NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3-type plasmids may contribute to the dissemination of blaNDM-1. Int J Infect Dis. 2015;34:8–13. doi: 10.1016/j.ijid.2015.02.020 [DOI] [PubMed] [Google Scholar]
- 139.Zhang F, et al. Further spread of bla NDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol. 2016;7:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kikuchi Y, Matsui H, Asami Y, et al. Landscape of bla NDM genes in Enterobacteriaceae. J Antibiot. 2022;75(10):559–566. doi: 10.1038/s41429-022-00553-3 [DOI] [PubMed] [Google Scholar]
- 141.Partridge SR, Iredell JR.. Genetic contexts of bla NDM-1. Antimicrob Agents Chemother. 2012;56(11):6065–6067. doi: 10.1128/AAC.00117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dong H, Li Y, Cheng J, et al. Genomic epidemiology Insights on NDM-producing pathogens revealed the pivotal role of plasmids onblaNDM transmission. Microbiol Spectr. 2022;10(2):e02156–21. doi: 10.1128/spectrum.02156-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Poirel L, et al. Antimicrobial resistance in Escherichia coli. Microbiol Spectr. 2018;6(4):6.4.14. doi: 10.1128/microbiolspec.ARBA-0026-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bi W, Li B, Song J, et al. Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum β-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou, China. Int J Antimicrob Agents. 2017;50(1):29–34. doi: 10.1016/j.ijantimicag.2017.02.010 [DOI] [PubMed] [Google Scholar]
- 145.Peleg AY, Seifert H, Paterson DL.. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Antunes LCS, Visca P, Towner KJ.. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71(3):292–301. doi: 10.1111/2049-632X.12125 [DOI] [PubMed] [Google Scholar]
- 147.Piperaki ET, Tzouvelekis LS, Miriagou V, et al. Carbapenem-resistant Acinetobacter baumannii: in pursuit of an effective treatment. Clin Microbiol Infect. 2019;25(8):951–957. doi: 10.1016/j.cmi.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 148.Peleg AY, Seifert H, Paterson DL.. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, et al. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J Infect. 2019;79(6):593–600. doi: 10.1016/j.jinf.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 150.Gu Y, Zhang W, Lei J, et al. Molecular epidemiology and carbapenem resistance characteristics of Acinetobacter baumannii causing bloodstream infection from 2009 to 2018 in northwest China. Front Microbiol. 2022;13:983963. doi: 10.3389/fmicb.2022.983963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ikuta KS, Swetschinski LR, Robles Aguilar G, et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2022;400(10369):2221–2248. doi: 10.1016/S0140-6736(22)02185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen C-H, Wu P-H, Lu M-C, et al. Geographic patterns of carbapenem-resistant, multi-drug-resistant and difficult-to-treat Acinetobacter baumannii in the Asia-Pacific region: results from the antimicrobial testing leadership and surveillance (ATLAS) program, 2020. Int J Antimicrob Agents. 2023;61(2):106707. doi: 10.1016/j.ijantimicag.2022.106707 [DOI] [PubMed] [Google Scholar]
- 153.Liu C, Chen K, Wu Y, et al. Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg Microbes Infect. 2022;11(1):1730–1741. doi: 10.1080/22221751.2022.2093134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hamidian M, Nigro SJ.. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom. 2019;5(10):e000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen T, Fu Y, Hua X, et al. Acinetobacter baumannii strains isolated from cerebrospinal fluid (CSF) and bloodstream analysed by cgMLST: the dominance of clonal complex CC92 in CSF infections. Int J Antimicrob Agents. 2021;58(4):106404. doi: 10.1016/j.ijantimicag.2021.106404 [DOI] [PubMed] [Google Scholar]
- 156.Jiang Y, Ding Y, Wei Y, et al. Carbapenem-resistant Acinetobacter baumannii: a challenge in the intensive care unit. Front Microbiol. 2022;13:1045206. doi: 10.3389/fmicb.2022.1045206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kang HM, Yun KW, Choi EH.. Molecular epidemiology of Acinetobacter baumannii complex causing invasive infections in Korean children during 2001–2020. Ann Clin Microbiol Antimicrob. 2023;22(1):32. doi: 10.1186/s12941-023-00581-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kong X, Chen T, Guo L, et al. Phenotypic and genomic comparison of dominant and nondominant sequence-type of Acinetobacter baumannii isolated in China. Front Cell Infect Microbiol. 2023;13:1118285. doi: 10.3389/fcimb.2023.1118285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim YJ, Kim SI, Kim YR, et al. Carbapenem-resistant Acinetobacter baumannii: diversity of resistant mechanisms and risk factors for infection. Epidemiol Infect. 2012;140(1):137–145. doi: 10.1017/S0950268811000744 [DOI] [PubMed] [Google Scholar]
- 160.Liu C, Chen K, Wu Y, et al. Epidemiological and genetic characteristics of clinical carbapenem-resistant Acinetobacter baumannii strains collected countrywide from hospital intensive care units (ICUs) in China. Emerg Microbes Infect. 2022;11(1):1730–1741. doi: 10.1080/22221751.2022.2093134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Muller C, Reuter S, Wille J, et al. A global view on carbapenem-resistant Acinetobacter baumannii. mBio. 2023;14(6):e0226023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Isler B, Doi Y, Bonomo RA, et al. New treatment options against Carbapenem-resistant Acinetobacter baumannii Infections. Antimicrob Agents Chemother. 2019;63(1):e01110–18. doi: 10.1128/AAC.01110-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Qureshi ZA, Hittle LE, O'Hara JA, et al. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin Infect Dis. 2015;60(9):1295–1303. doi: 10.1093/cid/civ048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun B, et al. New mutations involved in Colistin resistance in Acinetobacter baumannii. mSphere. 2020;5(2):e00895–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gerson S, Nowak J, Zander E, et al. Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J Antimicrob Chemother. 2018;73(6):1501–1508. doi: 10.1093/jac/dky083 [DOI] [PubMed] [Google Scholar]
- 166.Oliver A, Mulet X, López-Causapé C, et al. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21-22:41–59. doi: 10.1016/j.drup.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 167.Hu Y-Y, Cao J-M, Yang Q, et al. Risk factors for Carbapenem-resistant Pseudomonas aeruginosa, Zhejiang Province, China. Emerg Infect Dis. 2019;25(10):1861–1867. doi: 10.3201/eid2510.181699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Parkins MD, Somayaji R, Waters VJ.. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev. 2018;31(4):e00019–18. doi: 10.1128/CMR.00019-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Treepong P, Kos VN, Guyeux C, et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24(3):258–266. doi: 10.1016/j.cmi.2017.06.018 [DOI] [PubMed] [Google Scholar]
- 170.Zhang B, Xu X, Song X, et al. Emerging and re-emerging KPC-producing hyper virulent Pseudomonas aeruginosaST697 and ST463 between 2010 and 2021. Emerg Microbes Infect. 2022;11(1):2735–2745. doi: 10.1080/22221751.2022.2140609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Patil S, Chen X, Dong S, et al. Resistance genomics and molecular epidemiology of high-risk clones of ESBL-producing Pseudomonas aeruginosa in young children. Front Cell Infect Microbiol. 2023;13:1168096. doi: 10.3389/fcimb.2023.1168096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xiao C, Zhu Y, Yang Z, et al. Prevalence and molecular characteristics of polymyxin-resistant Pseudomonas aeruginosa in a Chinese tertiary teaching hospital. Antibiotics. 2022;11(6):799. doi: 10.3390/antibiotics11060799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhao Y, Xie L, Wang C, et al. Comparative whole-genome analysis of China and global epidemic Pseudomonas aeruginosa high-risk clones. J Glob Antimicrob Resist. 2023;35:149–158. doi: 10.1016/j.jgar.2023.08.020 [DOI] [PubMed] [Google Scholar]
- 174.Wu W, Wei L, Feng Y, et al. Precise species identification by whole-genome sequencing of Enterobacter bloodstream infection, China. Emerg Infect Dis. 2021;27(1):161–169. doi: 10.3201/eid2701.190154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu W, Feng Y, Zong Z.. Precise species identification for Enterobacter: a genome sequence-based study with reporting of two novel species, Enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. mSystems. 2020;5(4):e00527–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Giamarellou H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 2016;48(6):614–621. doi: 10.1016/j.ijantimicag.2016.09.025 [DOI] [PubMed] [Google Scholar]
- 177.Zong Z, Feng Y, McNally A.. Carbapenem and Colistin resistance in Enterobacter: determinants and clones. Trends Microbiol. 2021;29(6):473–476. doi: 10.1016/j.tim.2020.12.009 [DOI] [PubMed] [Google Scholar]
- 178.Guérin F, Isnard C, Sinel C, et al. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J Antimicrob Chemother. 2016;71(11):3058–3061. doi: 10.1093/jac/dkw260 [DOI] [PubMed] [Google Scholar]
- 179.Kim TH, et al. Novel cassette assay to quantify the outer membrane permeability of five β-lactams simultaneously in carbapenem-resistant Klebsiella pneumoniae and Enterobacter cloacae. MBio. 2020;11(1):e03189–19. doi: 10.1128/mbio.03189-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhu Z, Xie X, Yu H, et al. Epidemiological characteristics and molecular features of carbapenem-resistant Enterobacter strains in China: a multicenter genomic study. Emerg Microbes Infect. 2023;12(1):2148562. doi: 10.1080/22221751.2022.2148562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Izdebski R, Biedrzycka M, Urbanowicz P, et al. Genome-based epidemiologic analysis of VIM/IMP Carbapenemase-producing Enterobacter spp., Poland. Emerg Infect Dis. 2023;29(8):1618. doi: 10.3201/eid2908.230199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xu T, Xue C-X, Huang J, et al. Emergence of an epidemic hypervirulent clone of Enterobacter hormaechei coproducing mcr-9 and carbapenemases. Lancet Microbe. 2022;3(7):e474–e475. doi: 10.1016/S2666-5247(22)00122-7 [DOI] [PubMed] [Google Scholar]
- 183.Zhou K, Zhou Y, Xue C-x, et al. Bloodstream infections caused by Enterobacter hormaechei ST133 in China, 2010–22. Lancet Microbe. 2023;4(1):e13. doi: 10.1016/S2666-5247(22)00226-9 [DOI] [PubMed] [Google Scholar]
- 184.Rizvi SG, Ahammad SZ.. COVID-19 and antimicrobial resistance: a cross-study. Sci Total Environ. 2022;807(Pt 2):150873. doi: 10.1016/j.scitotenv.2021.150873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gao Y, et al. Origin, Phylogeny, and Transmission of the Epidemic Clone ST208 of Carbapenem-Resistant Acinetobacter baumannii on a Global Scale. Microbiol Spectr. 2022;10(3):e02604–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X, Du Z, Huang W, et al. Outbreak of multidrug-resistant Acinetobacter baumannii ST208 Producing OXA-23-Like Carbapenemase in a Children's Hospital in Shanghai, China. Microb Drug Resist. 2021;27(6):816–822. doi: 10.1089/mdr.2019.0232 [DOI] [PubMed] [Google Scholar]
- 187.Maljkovic Berry I, et al. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis. 2020;221(Supplement_3):S292–S307. [DOI] [PubMed] [Google Scholar]
- 188.Otto M. Next-generation sequencing to monitor the spread of antimicrobial resistance. Genome Med. 2017;9(1):1–3. doi: 10.1186/s13073-017-0461-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li H, Du Z, Huang W, et al. Trends and patterns of outpatient and inpatient antibiotic use in China’s hospitals: data from the Center for Antibacterial Surveillance, 2012–16. J Antimicrob Chemother. 2019;74(6):1731–1740. doi: 10.1093/jac/dkz062 [DOI] [PubMed] [Google Scholar]
- 190.Qiao M, Ying G-G, Singer AC, et al. Review of antibiotic resistance in China and its environment. Environ Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 191.Algammal AM, Hetta HF, Elkelish A, et al. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255–3265. doi: 10.2147/IDR.S272733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grøntvedt CA, Elstrøm P, Stegger M, et al. Methicillin-resistant Staphylococcus aureus CC398 in humans and pigs in Norway: a “one health” perspective on introduction and transmission. Clin Infect Dis. 2016;63(11):1431–1438. doi: 10.1093/cid/ciw552 [DOI] [PMC free article] [PubMed] [Google Scholar]