Abstract
Localization signals are RNA regulatory elements that direct the localization of mRNAs to subcellular sites. Localization signals presumably function by mediating RNA recognition events through which the mRNA becomes associated with the localization machinery. At present little is known about individual RNA recognition events, which in turn has limited progress in identifying the trans-acting binding factors involved in these events. Here we describe a detailed characterization of the RNA elements required for the RNA recognition event, event A, that initiates localization of bicoid mRNA in the Drosophila ovary. One element is a helix in which nucleotide identities are not important, suggesting that it plays a primarily structural role. Immediately adjacent to the helix is a recognition domain in which the identities of some, but not all, nucleotides are important for function. Comparison of two related but different RNAs that both support recognition event A further defines the important features of the recognition domain.
Control of gene expression occurs by a variety of mechanisms. Some of these act to regulate transcription and can restrict expression to a particular tissue or cell type. Greater precision in spatial control of gene activity, at a subcellular level, can be achieved only posttranscriptionally. One mechanism that provides such precision is mRNA localization, the process by which certain mRNAs are selectively targeted to specific regions within the cytoplasm of an individual cell. Once localized, an mRNA can serve as a source for local translation, allowing the encoded protein to be concentrated at or even restricted to a single site within the cell. Many examples of localized mRNAs from animal cells have been described: some were derived from the germ line gametes, where mRNA localization can play a crucial role in organization of the basic body plan, and others were derived from specialized somatic cells, where the localized mRNAs often contribute to cellular asymmetries (reviewed in reference 21). Recent work has established that the phenomenon of mRNA localization is not restricted to animals and has provided examples from both plants and yeast (1, 12, 24). Most, if not all, of these mRNAs contain a localization signal, the regulatory element or elements that direct localization. The signals commonly appear in the mRNA 3′ untranslated regions (UTRs) and must direct association with the localization machinery (13). One approach to identifying the factors that mediate this association has been to first characterize the localization signals; RNA sequences or structures that are required for localization are the likely binding sites for the localization factors.
One mRNA localization signal that has been characterized in some detail is that of the Drosophila bicoid (bcd) mRNA. The program of bcd mRNA localization involves multiple steps and is carried out during oogenesis and early embryogenesis (23). In Drosophila melanogaster, the oocyte develops while connected via cytoplasmic bridges to 15 sister cells, the nurse cells. The nurse cells synthesize mRNAs and proteins for transport to the oocyte, whose nucleus is transcriptionally inactive (19). Transcription of bcd mRNA begins in the nurse cells during stages 4 and 5 of oogenesis, and the mRNA is immediately concentrated in the oocyte. As oogenesis proceeds, transport to the oocyte continues, and starting at stage 8 bcd mRNA becomes localized within the oocyte at the anterior margin, flanking the nurse cells. Anterior localization persists into embryogenesis, until the mRNA disappears shortly after formation of the cellular blastoderm (23). Sequences both necessary and sufficient for this program of localization are found in the bcd mRNA 3′ UTR (17). Progress in understanding how these sequences act has focused attention on specific RNA recognition events and on RNA elements that mediate the recognition events (4, 5, 14–16).
Two redundant RNA recognition events, designated event A and event B, serve to initiate largely overlapping programs of bcd mRNA localization (Fig. 1) (14). Event A occurs first and is solely responsible for the earliest transport to the oocyte during stages 4 to 6 of oogenesis. Subsequently, event B-dependent localization is initiated, and either RNA recognition event is sufficient for continued localization. Identification of these events was achieved by mutating the bcd localization signal, rather than analysis of mutants lacking trans-acting factors; the genes known to act in localization of bcd mRNA all contribute only to the later steps in the process (14, 20, 23), and factors that mediate RNA recognition events A and B remain to be identified. Elimination of event A, by either a point mutation (change from G to U at position 4496 [4496 G→U]; numbering from GenBank accession no. X51741) or a small deletion (Δ14S, which removes nucleotides 4490 to 4507) in the stem-loop V region of the predicted structure of the bcd 3′ UTR, prevents early (stages 4 to 6) localization, but all later steps proceed normally. Elimination of event B (whose sequence requirements remain poorly understood) is achieved through use of a subdomain of the localization signal consisting of stem-loops IV and V (IV-V) and has no detectable effect on localization during oogenesis, although a final step in embryogenesis is defective. When events A and B are both prevented through use of a mutated form of IV-V, all steps of localization are almost completely abolished. Whether events A and B contribute directly to later stages of localization or only indirectly as prerequisites remains uncertain (14).
FIG. 1.
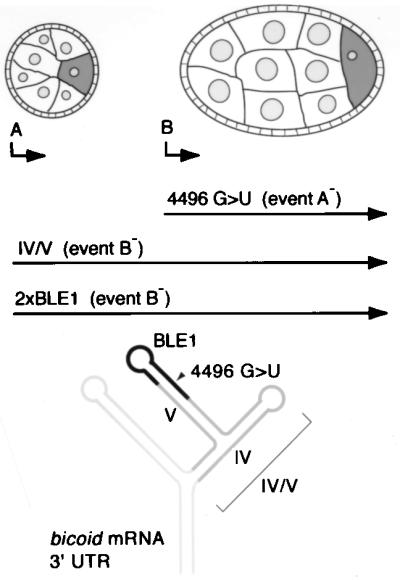
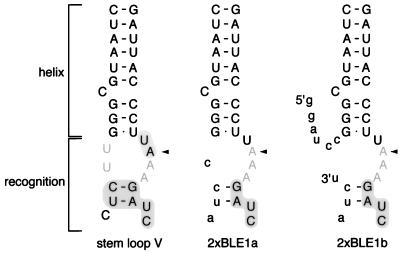
RNA recognition events and localization signals that direct bcd mRNA localization. The upper diagrams show stage 5 (left) and stage 7 (right) egg chambers. The germ line nurse cells and oocyte are surrounded by a layer of somatic follicle cells. At these stages bcd mRNA (dark shading) is localized to a single cell, the oocyte. RNA recognition events, A and B, are indicated at the developmental stages when they first arise. The three horizontal arrows indicate the programs of bcd mRNA localization directed by mutant forms of the bcd localization signal. Mutant 4496 G→U is defective in event A and initiates localization late, when event B occurs. The IV-V localization signal is competent only for event A, but its localization appears like that of the wild type because events A and B are redundant from stage 7 onwards. The 2×BLE1 RNA localization signal acts indistinguishably from that of IV-V during these stages and also does not support event B. Both IV-V and 2×BLE1 have later defects: 2×BLE1 fails to support localization beyond stage 10 of oogenesis, and IV-V displays a defect in embryogenesis. The diagram at the bottom shows the predicted structure of the bcd mRNA 3′ UTR. Stem-loops IV and V together make up the IV-V localization signal. A portion of the IV-V region includes BLE1, which when dimerized makes 2×BLE1. Note that although the existence of certain parts of the structure now has experimental support (this work and reference 5), this diagram is only a model and may not be completely correct.
The sequences required for event A are contained in the IV-V region of the 3′ UTR. Shorter RNA segments from IV-V lack localization activity, suggesting that sequences contributing directly or indirectly to binding sites are dispersed in the primary IV-V sequence (14). One such binding site is likely to be in the terminal portion of stem-loop V, the site of the point mutation (4496 G→U) that eliminates event A. Notably, a short sequence (BLE1; nucleotides 4464 to 4517) from the terminal portion of stem-loop V by itself lacks localization activity but can, when dimerized to form 2×BLE1, support a partial program of mRNA localization (Fig. 1; see Fig. 5 for a diagram of dimerized BLE1) (15). Localization begins normally at stage 4 or 5, coincident with the onset of event A-dependent localization, but ceases prematurely at stage 10. Although the question of why dimerization allows the isolated BLE1 to function remains open, the fact that 2×BLE1 is active clearly implies the presence of an important binding site within the sequence. Here we describe the results of extensive mutational analyses of the terminal portion of stem-loop V and of 2×BLE1 to better characterize the putative binding site. Our results reveal essential structural features as well as nucleotides that appear to form the actual binding site for a factor involved in recognition event A. This detailed information about the binding site should facilitate identification of the protein or proteins with which it interacts.
FIG. 5.
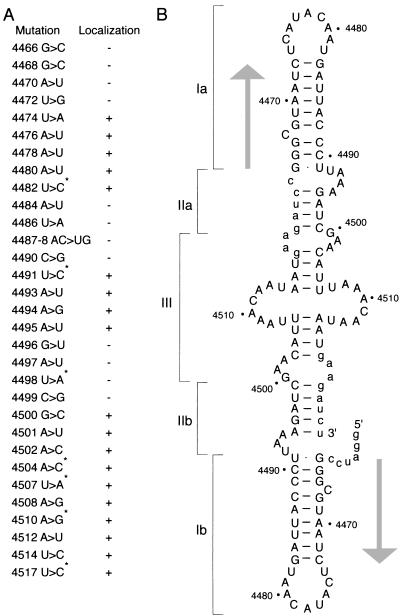
Summary of mutagenesis of 2×BLE1. (A) Summary of 2×BLE1 mutant analysis. Data for seven of the mutations (those noted with an asterisk) are from reference 16. Localization was monitored at stages 5 to 9 of oogenesis. A plus sign indicates mRNA localization to the oocyte that is similar or identical to that of wild-type 2×BLE1. A minus sign indicates little or no oocyte localization. (Some of the mutants labeled with a plus sign, i.e., those that fail to bind Exl protein, do have a localization defect, but it occurs in a later step in the process [16].) (B) Predicted structure of 2×BLE1 RNA. Nucleotides from the bcd mRNA are in uppercase, and linker sequences that flank and join the two copies of BLE are in lowercase. The 5′ and 3′ ends are indicated in the lower right quadrant. The structure is divided into regions, labeled I, II, and III, to facilitate description of the mutations in the text. The regions corresponding to the helix domain of stem-loop V are indicated by arrows. Notably, all mutations that would eliminate a base pair of these helices disrupt localization, while mutation 4491 U→C, which greatly strengthens a base pair, retains normal localization. The regions corresponding to the recognition domain of stem-loop V are shown with several additional base pairings between bcd and linker sequences, as predicted by computer analysis. We argue that this region adopts a slightly different structure, that shown in Fig. 6 (see text and legend to Fig. 6).
MATERIALS AND METHODS
Localization signals.
Point mutations were incorporated into oligonucleotides and introduced into bcd DNA by PCR; all mutations were confirmed by sequencing. Mutant nomenclature is based on the original numbering of the bcd genomic DNA sequence (GenBank accession no. X51741). Mutations in stem-loop V were tested in the context of the complete 3′ UTR, using two different reporter transgenes. One reporter, bcd+lacZ, is adapted directly from the bcd gene by insertion, near the end of the 3′ UTR, of a portion of the Escherichia coli lacZ gene (15). The second reporter is osk/gfp, which consists of the oskar promoter, the region coding for green fluorescent protein, and a minimal 3′ UTR bearing a unique XbaI site and a polyadenylation signal (14). An EcoRV-StuI bcd genomic DNA fragment containing almost all of the 3′ UTR was first modified by addition of XbaI linkers and then cloned into the XbaI site of the osk/gfp reporter. Mutants tested in the bcd+lacZ reporter were those with affected nucleotides at 4470 and 4471, at 4472 and 4473, at 4474 to 4482, at 4483 and 4484, at 4472 and 4473 as well as at 4483 and 4484, at 4470 and 4471 as well as at 4485 and 4486, between 4488 and 4489, and at 4492 and 4493. All other mutations were tested in the osk/gfp transgene.
Altered versions of 2×BLE1 (with both copies of BLE1 bearing the same mutation) were incorporated into the bcd+lacZ reporter transgene as described previously (16).
Flies.
Transgenic Drosophila stocks were created by P-element-mediated transformation. Multiple independent lines were obtained and analyzed for each transgene, all in the w1118 genetic background.
Analysis of mRNA localization.
Ovaries were dissected from healthy well-fed 3- to 4-day-old females and prepared for in situ hybridization as described previously (8). Digoxigenin-labeled RNA hybridization probes complementary to the lacZ and green fluorescent protein reporter mRNAs were prepared by in vitro transcription (25, 27).
RNA folding.
Computer prediction of RNA folding (28) was performed over the Internet on Michael Zuker’s mfold server (http://www.ibc.wustl.edu/~zuker/rna/form1.cgi). Foldings were made by using parameters at default settings, except that the temperature was set at 30°C. For 2×BLE1, only two possible folds within 10% of the most optimal folding were predicted. The optimal fold was used to draw the structure in Fig. 5B. The other fold differs in that the large central bulge now forms two terminal loops, while the terminal loops now form a large central bulge. The helices of the favored structure still form, but now have the opposite orientation, i.e., directed towards the central bulge rather than towards the terminal loops.
RESULTS
The point and small deletion mutations known to affect RNA recognition event A lie near the end of stem-loop V in the predicted structure of the bcd mRNA 3′ UTR (14). To better define the features of stem-loop V involved in event A, we created additional mutations and determined their effects on mRNA localization. All mutations were tested in the context of reporter mRNAs carrying the complete bcd mRNA 3′ UTR, such that event A was monitored in the presence of event B. Thus, event A activity was revealed by localization of mRNA to the oocyte during the earlier stages of oogenesis (Fig. 1). When event A is defective, localization is dependent on event B and begins later; the event B-dependent localization provides a simple control to ensure that the transgenic mRNA is expressed and is competent for localization.
Each of the mutations tested alters a particular feature of the predicted structure of the RNA, which includes a terminal loop and an extensive helical region that is distorted at several positions by bulged nucleotides (see Fig. 3). Examples of the data are shown in Fig. 2, and all of the results are summarized in Fig. 3.
FIG. 3.
Summary of mutagenesis of stem-loop V. The RNA structure is that predicted for the stem-loop V region. For convenience, the extensive predicted helical region is divided into three parts, outer, middle and inner, as discussed in the text. Replacement nucleotides of primary mutations are indicated by arrowheads at the immediate left and right of the structure. At the far right, arrows point to secondary compensatory mutations that restore predicted base pairing interactions disrupted by certain primary mutations. All but four of the mutations are simple substitutions in which there is no change in the total number of nucleotides. For the four mutations that involve either addition or deletion of nucleotides, the affected regions are indicated by brackets at the immediate right of the structure. Mutations defective in RNA recognition event A-dependent mRNA localization are shaded, while those retaining event A activity are not. The mutation involving positions 4460 and 4461 as well as 4496 and 4497 has partial event A activity and is indicated by an asterisk.
FIG. 2.
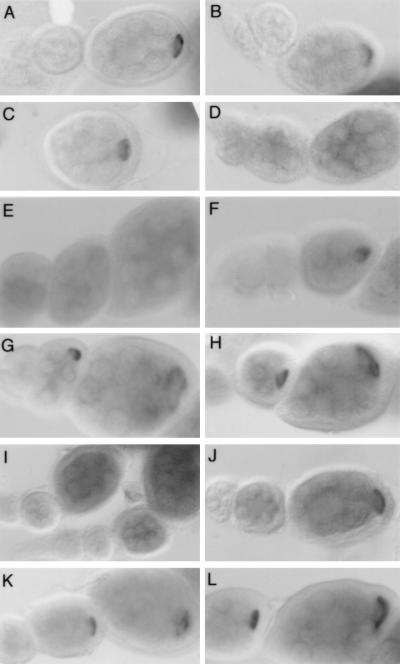
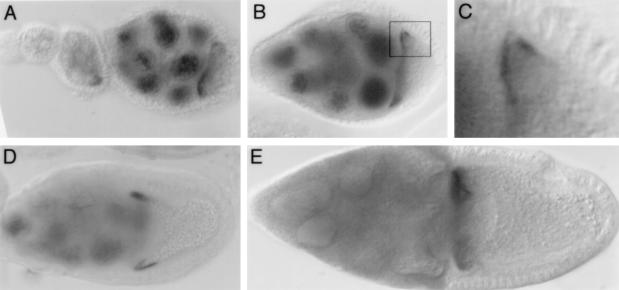
Localization of mRNAs bearing mutations in the stem-loop V region of the bcd 3′ UTR. In all panels transgenic mRNAs were detected by in situ hybridization to whole-mount ovaries. Developmental stages advance from left to right. Structural features mentioned here can be seen in Fig. 3. (A) The wild-type mRNA localization signal directs localization to the oocyte as soon as the mRNA appears, beginning at stage 4 or 5. (B) Localization of the mRNA bearing the tetraloop mutation at positions 4474 to 4482 is normal. (C) Elimination of the distal bulge in mutation of position 4488b has no effect on localization. (D) Elimination of the central bulge in RNA bearing a mutation at positions 4492 and 4493 completely abolishes the event A-dependent localization step. The mRNA accumulates in the nurse cells (grey staining) but is not concentrated in the oocyte. (E and F) Evidence of base pairing in the predicted stem structure. The 4483,4484 GA→CU mutation disrupts two predicted base pairs and is defective in event A-dependent localization (E), but restoration of base pairing through compensatory changes in RNA bearing the 4472,4473 UC→AG and 4483,4484 GA→CU mutations restores localization (F). (G to J) RNAs bearing mutations that define a proposed recognition domain. The upper two predicted base pairs are not essential, as mutation at positions 4494 and 4495 (G) or at positions 4462,4463,4494, and 4495 (H) has no effect on event A. In contrast, alteration of nucleotides at positions 4496 and 4497 eliminates localization (I). Introduction of compensatory changes in RNA bearing mutations at positions 4460 and 4461 as well as at positions 4496 and 4497 partially restores event A-dependent localization (J). Note the accumulation of the mutant mRNA (grey staining) in the nurse cells of the central egg chamber in panel J, with no obvious concentration in the oocyte. Slightly later the mRNA does become localized to the oocyte, as seen in the egg chamber at the right. This delay in event A-dependent localization is observed consistently and reveals that simple base pairing at these positions in the structure is not sufficient for complete activity. Rather, both base pairing and nucleotide identity are important, implicating this region of the structure in recognition (i.e., protein binding). (K and L) Mutations in the proximal portion of stem-loop V have no effect on event A. Mutations at positions 4457 and 4458 (K) and 4500 and 4501 (L) disrupt a base pair of the predicted helix below the recognition domain but produce normal mRNA localization.
The role of the terminal loop was tested by replacement with a GAAA tetranucleotide. In structurally characterized RNAs a tetraloop forms a stable loop when adjacent to a helix (2, 3, 26). The tetraloop mutation had no effect on event A, as the early localization to the oocyte remained normal (Fig. 2B).
Each of the three predicted bulges was eliminated by deletion or by substitutions that would support base pairing. The outer bulge (closest to the loop) consists of an unpaired C in the left (5′-most) strand of the helix. Its importance was tested by introducing a complementary G in the right strand of the helix. There was no effect on event A-dependent localization (Fig. 2C), revealing that the bulge is not required for that process. In contrast, mutations altering the other two bulges eliminated event A-dependent localization, and the mRNA appeared in the nurse cells during the early stages of oogenesis, rather than in the oocyte (Fig. 2D and 3). The central bulge involves a UA dinucleotide on the right strand of the helix and was mutated by deletion of both nucleotides. The inner bulge consists of a C on the left strand of the helix and a UC on the right strand. In the mutated RNA the UC was replaced with a G, to allow base pairing with the bulged C on the opposite strand. Because these mutations disrupt event A, we conclude that features of this region are essential for RNA recognition by a localization factor or factors.
Portions of the predicted helix were mutated in two nucleotide units, using compensatory mutations to test for base pairing. The predicted helical region is considered in three parts: an outer segment between the central bulge and the loop, a middle segment between the central and inner bulges, and an inner segment proximal to the inner bulge. For the outer segment all of the mutational data support the existence of the helix. Each dinucleotide mutation tested (all of which collectively cover six of the nine predicted base pairs) eliminated event A (Fig. 2E and 3). Compensatory mutations restored the predicted base pairing while exchanging nucleotides between the two strands of the helix. In each case event A localization was regained for these doubly mutated RNAs (Fig. 2F and 3). These results lead to three conclusions. First, the region of stem-loop V from the central bulge to the loop is extensively base paired and thus largely helical. Second, this helical region is required for RNA recognition event A. Third, the identities of particular base pairs within the helical region are not crucial for event A, as base pair replacements (i.e., the compensatory mutations) have no marked effect on event A (at least for the six base pairs tested). Notably, the only predicted bulge within the outer segment of the predicted helix can be converted to a base pair with no effect on event A (see above).
The middle segment of the predicted helix lies between the central and inner bulges and includes the site of a point mutation (4496 G→U) previously shown to eliminate event A (14). A dinucleotide mutation (4494,4495 AA→UU) that would disrupt the two distal base pairs has no effect on event A (Fig. 2G). Similarly, a mutation that exchanges bases across the helix at these positions does not impair event A (Fig. 2H). Therefore, event A does not require specific nucleotides or base pairing at these positions. In contrast, either of two dinucleotide mutations (4460,4461 UC→AG and 4496,4497 GA→CU) that would disrupt the two proximal base pairs abolishes event A (Fig. 2I and 3). When these dinucleotide mutations are combined to introduce compensatory changes that would restore base pairing, event A is only partially restored (Fig. 2J and 3). The results indicate (i) that nucleotides at these positions must be base paired and (ii) that the identities of some, if not all, of the four affected nucleotides are important for RNA recognition event A (see Discussion).
The inner segment of the predicted helix extends proximally from the inner bulge. Dinucleotide mutations that disrupt the first four base pairs in either strand of the predicted helix have no effect on event A-dependent mRNA localization (Fig. 2K and L and 3). Similarly, exchanging bases across the predicted helix does not interfere with event A (Fig. 3). Thus, neither nucleotide identity nor base pairing is required in this region for event A.
We conclude that the RNA element involved in recognition event A consists of two parts. One is a recognition domain, made up of the proximal two predicted bulges and the sequences between them (the middle part of the predicted helix shown in Fig. 3). The identities of some, but not all, of the nucleotides in this region are important for recognition. Nucleotides whose identities are not important could still play a structural role, as in maintaining correct spacing. The second part is a flanking helix (the outer part of the predicted helix shown in Fig. 3). The role of the helix could be structural, e.g., facilitating correct folding of the recognition domain. The helix could also be a substrate for binding of a protein that recognizes double-stranded RNA, although this protein could not be solely responsible for event A. These roles are not mutually exclusive, and the helix could contribute to both structure and protein binding.
Mutational analysis of BLE1.
2×BLE1 is a dimerized portion of stem-loop V from the bcd mRNA 3′ UTR that directs a large part of the bcd localization program. Although 2×BLE1 is an artificial localization signal, it nevertheless retains key features of the intact bcd localization signal, including the ability to direct mRNA localization to the anterior of the oocyte and a dependence on exu for a specific phase in its activity (15). 2×BLE1 has served as an attractive model for the analysis of localization signals, as its activity is sensitive to point mutations, allowing for the study of correlations between in vivo activity and in vitro binding to proteins (16). There are two phases in the action of 2×BLE1: the initial recognition by the localization machinery beginning at stage 5 of oogenesis that directs localization to and within the oocyte and the Exl-binding-protein-dependent persistence of localization during stage 9. Only the latter step has been studied in any detail (16). The early step appears to be identical to the recently defined RNA recognition event A-dependent step of bcd mRNA localization (14), as both direct mRNA localization from the nurse cells to the oocyte, they occur with the same developmental timing, and both depend on sequences from stem-loop V.
To better appreciate how BLE1 is initially recognized by the mRNA localization machinery, and as an aid in understanding RNA recognition event A, we performed a systematic mutagenesis of the element. Single nucleotide mutations were introduced into both copies of BLE1 in a 2×BLE1 reporter transgene, and mRNA localization was monitored during stages 5 to 9 of oogenesis, when 2×BLE1 is active but not yet dependent on Exl protein. Examples of the data are shown in Fig. 4, and all of the results are summarized in Fig. 5A. As for stem-loop V, interpretation of the mutants of 2×BLE1 is facilitated by comparison with an RNA structure. Computer prediction suggests a structure containing RNA elements very similar to those of the stem-loop V region but present in two copies. We describe the effects of the mutations in three groupings according to their positions within the predicted 2×BLE1 structure.
FIG. 4.
Mutational analysis of the 2×BLE1 localization element. (A to D) Localization patterns directed by wild-type and mutant forms of 2×BLE1 during stages 5 to 9 of oogenesis. (A) Wild type. The mRNA is concentrated in the oocyte beginning at stages 4 and 5. Localization is less efficient than that directed by the complete bcd 3′ UTR, and nurse cell staining is evident. (B) Mutant bearing 4496 G→U. No oocyte localization is detected, and the mRNA is dispersed throughout the nurse cells. (C) Mutant bearing 4514 U→C. Localization is indistinguishable from that of the wild type. (D) Mutant bearing 4484 A→U. Localization to the oocyte (arrows) is extremely weak, with most of the mRNA dispersed throughout the nurse cells.
Mutations of one group (region III in Fig. 5B) are from the central portion of the 2×BLE1 structure, including the internal loop and immediately flanking helical regions. All of these sequences lie outside of the region of stem-loop V implicated in RNA recognition event A. Not surprisingly, none of these mutations has any effect on the early, Exl-independent localization of the reporter mRNA (Fig. 4C and 5). There is no evidence to suggest that the predicted base pairs exist, and this portion of the RNA may be entirely unstructured. This region includes the binding site for Exl protein (16).
Mutations of another group (regions Ia and Ib in Fig. 5B) lie within a domain identical to the terminal loop and the adjacent helical region of stem-loop V. These mutations mimic similar mutations in stem-loop V: no changes in the loop have any effect on localization (Fig. 5), while all changes that disrupt predicted base pairings greatly reduce or eliminate localization (Fig. 4D and 5). Thus, it is very likely that this region adopts the structure demonstrated for stem-loop V.
Mutations of the final group (regions lla and llb in Fig. 5B) lie within an adjacent region that corresponds to the recognition domain of stem-loop V and has a similar, although not identical, structure. Mutations in this region of 2×BLE1 have effects (Fig. 5) consistent with the notion that it is functionally equivalent to the stem-loop V recognition domain. The interpretation is simplest for mutations at positions 4494 to 4497, where substitution mutations were tested in both 2×BLE1 and stem-loop V. The same effects were observed in both RNAs: mutations at 4494 and 4495 had no effect on localization, while changes at 4496 and 4497 eliminated event A (Fig. 4B and 5). Mutations affecting the proximal bulge were of different types in stem-loop V and 2×BLE1, but they led to the same result: conversion of 4498 and 4499 from UC to G in stem-loop V abolished localization, as did 4498 U→A and 4499 C→G in 2×BLE1. For the final mutation in this region of 2×BLE1 the interpretation is less straightforward, since dinucleotide mutations that alter the same nucleotide in stem-loop V had different effects: in 2×BLE1, 4493 A→U had no effect on localization, while in stem-loop V, deletion of 4492 and 4493 eliminated event A-dependent localization. However, the different results may well reflect differences in the mutations tested: deletion versus substitution and a change of two nucleotides versus one nucleotide. The striking similarities in function and overall structure, together with the mutational data, strongly support the argument that both stem-loop V and 2×BLE1 mediate interaction with the same recognition factor or factors.
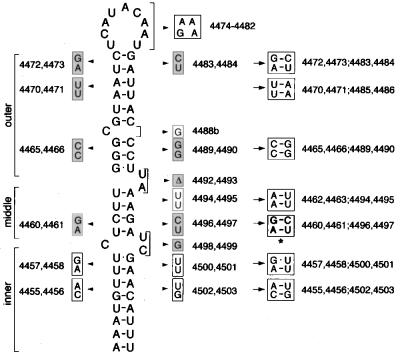
Although the proposed recognition domains of stem-loop V and 2×BLE1 are very similar, there are some differences that may help define the structure of this region (Fig. 6). The differences appear because in 2×BLE1 one strand of the domain is contributed by the synthetic oligonucleotide linkers placed at the ends of BLE1 and used to join the two copies of BLE1. The two nucleotides of this strand shown to be essential in stem-loop V are provided fortuitously by the linker, but all other nucleotides differ. The UU of the left strand of stem-loop V, which can be mutated to AA in stem-loop V with no effect on event A, is replaced by a single C in 2×BLE1. Thus, it appears that neither the identities nor the exact number of nucleotides at this position is important. The bulged C at the base of the recognition region in stem-loop V was not tested by mutation, but the conversion to an A in 2×BLE1 suggests that the identity of this nucleotide is also not important for function. These results allow us to tentatively define the essential features of the RNA element from stem-loop V that acts in RNA recognition event A (Fig. 6).
FIG. 6.
Proposed organization of the stem-loop V recognition domain. Sequences from the helix and recognition domains of stem-loop V, and from the corresponding regions of 2×BLE1, are shown. All of the predicted base pairings in the helix that have been tested by mutation are required for event A. Mutational data for the recognition domain are less straightforward and are summarized by a shading scheme to allow comparison. Nucleotides that when mutated eliminate event A-dependent mRNA localization are indicated by background shading. Nucleotides that can be mutated with no effect on event A are indicated by grey letters. Nucleotides from the recognition domain not tested by mutation are indicated by black letters with no background shading. The results are in complete agreement except for the A indicated by arrowheads. This A was removed in a two-nucleotide deletion from stem-loop V and was replaced with a U in 2×BLE1. Thus, the discrepancy could be due to the different types of mutations tested. The primary differences among the different recognition domains all occur at positions where mutations have no effect or were not tested.
Nuclear accumulation of mutant bcd mRNAs.
A subset of the mutations introduced into the stem-loop V region of the bcd 3′ UTR were associated with an unusual phenotype, nuclear accumulation of the mRNA (Fig. 7). This effect can be attributed to the mutations in the bcd mRNA (rather than to the site of transgene insertion in the genome, for example), as it appears consistently in independent transgenic lines (Table 1). There is no correlation between RNA recognition event A activity and nuclear accumulation: some, but not all, of the mutations that eliminate event A lead to nuclear accumulation; similarly, some mutations with no effect on event A result in the nuclear accumulation phenotype, and some do not (Table 1). Furthermore, although all mutations giving rise to this effect are at the lower portion of the sequences of stem-loop V considered here, there is no obvious explanation of why certain mutations lead to nuclear accumulation while others do not (Table 1).
FIG. 7.
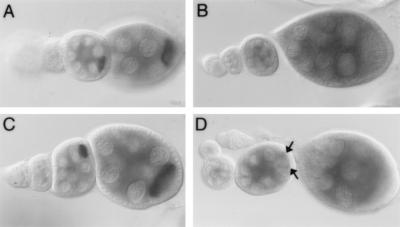
Nuclear accumulation of mutant mRNAs. The transgene, mRNA bearing the 4457,4458 UU→AG mutation is localized to the oocyte in stage 5, displaying no nuclear accumulation (A [center egg chamber]). Subsequently, high levels of the mRNA are detected in the nurse cell nuclei (A [right egg chamber]). Nuclear accumulation remains intense as oogenesis proceeds (B). A portion of the egg chamber shown in panel B, containing the oocyte nucleus, is outlined and is shown at greater magnification in panel C. The oocyte nucleus lies beneath the vertex of mRNA localized to the anterior margin and anterolateral cortex of the oocyte. There is no nuclear accumulation of the mRNA in the oocyte nucleus, which does not transcribe bcd mRNA. Therefore, concentration of the mutant transcripts in the nuclei is more likely the result of nuclear retention or stabilization rather than of nuclear import. The conclusion is only tentative, as the nurse cells and oocyte have quite different functions and nucleocytoplasmic trafficking could differ substantially between them. Later in oogenesis nuclear accumulation is gradually lost (D), disappearing by stage 10b (E).
TABLE 1.
Nuclear accumulation of mRNA
| Mutation(s) in bcd mRNAa | % Nuclear accumulationb | Event A |
|---|---|---|
| Tetraloop | 0, 0 | + |
| 4483,4484 GA→CU | 0, 0 | − |
| 4472,4473 UC→AG and 4483,4484 GA→CU | 0, 0, 0 | + |
| 4470,4471 AA→UU | 0, 0, 0 | − |
| 4470,4471 AA→UU and 4485,4486 UU→AA | 0, 0, 0 | + |
| 4488b + G | 0, 0, 0 | + |
| 4465,4466 GG→CC | 0, 0 | − |
| 4489,4490 CC→GG | 0, 0 | − |
| 4465,4466 GG→CC and 4489,4490 CC→GG | 0, 0, 0 | + |
| Δ4492,4493 | 0, 0 | − |
| 4494,4495 AA→UU | 0, 0, 0 | + |
| 4462,4463 UU→AA and 4494,4495 AA→UU | 6, 10, 13 | + |
| 4460,4461 UC→AG | 0, 0, 0 | − |
| 4496,4497 GA→CU | 0, 0 | − |
| 4460,4461 UC→AG and 4496,4497 GA→CU | 74, 66, 57 | +/−c |
| 4498,4499 UC→G | 71, 96, 90 | − |
| 4457,4458 UU→AG | 74, 58 | + |
| 5000,5001 GA→UU | 0, 0, 0 | + |
| 4457,4458 UU→AG and 5000,5001 GA→UU | 0, 0 | + |
| 4455,4456 GU→CA | 6, 2 | + |
| 5002,5003 AC→UG | 0, 4 | + |
| 4455,4456 GU→CA and 5002,5003 AC→UG | 89, 85 | + |
All mutations in each group (indicated by spaces) affect the same part of the structure. Position in the table corresponds to position in stem-loop V.
Multiple independent transgenic lines were examined for each mutation. Values are the percentages of appropriately staged egg chambers displaying nuclear accumulation of the transgene mRNA. At least 100 egg chambers were scored for each transgenic line.
+/−, partial event A activity.
The developmental progression of nuclear accumulation is quite consistent, beginning at stage 6 or 7, becoming most prominent during stages 8 and 9, and disappearing by stage 10b (Fig. 7). RNA levels appear to be transiently increased during the phase of nuclear accumulation, raising the possibility that the mutations stabilize a labile nuclear form of the mRNA. Because there is as yet no indication that the phenotype is related to mRNA localization, we have not attempted to confirm that the mRNA levels are indeed elevated.
DISCUSSION
RNA signals that direct localization of mRNAs during oogenesis and in early-stage embryos tend to be functionally redundant (6, 7, 9, 11, 14, 15), a feature that has hindered progress in defining the essential features of the signals. In previous work we identified a nonredundant form of the bcd localization signal, 2×BLE1, whose entire program of localization could be eliminated by a single point mutation (16). We also showed that the earliest step of localization directed by the complete bcd 3′ UTR did not involve redundant RNA recognition events and that a point mutation in the bcd 3′ UTR could eliminate this early localization, although later steps proceeded normally (14). Given mutations with such dramatic effects on localization, we can begin to define in detail the RNA elements responsible for association of bcd mRNA with the localization machinery. We have taken that approach here, focusing on stem-loop V of the bcd 3′ UTR and on 2×BLE1. These RNAs appear to mediate the same RNA recognition event, event A, and the analysis of both RNAs provided a more elaborate description of the underlying structure than would have been possible with either RNA alone.
From our studies we have defined two RNA structural elements that are required for event A-dependent mRNA localization: a recognition domain and a flanking helix (Fig. 6). Our data place constraints on the structure of the recognition domain, but the mutational approach taken here is not sufficient to provide a complete description of its structure. Although the use of additional mutations would add to our understanding of how the recognition domain is organized, the information available now is sufficient to direct attention to this region in searches for RNA binding proteins expected to act in event A. Further definition of the structure by an approach less dependent on the generation of many transgenic fly stocks might be more easily undertaken once these proteins have been isolated. The second RNA element required for event A is a flanking helix. It may play a purely structural role in promoting correct folding of the recognition domain, thereby facilitating binding of a localization factor. Although the helix could also mediate a protein contact, only a limited degree of binding specificity is attainable, as numerous bases can be exchanged across the helix with no obvious loss of localization activity. However, specificity in binding could be achieved by protein-protein contacts, rather than protein-RNA contacts. Notably, the same helical region has also been implicated in a late, staufen-dependent step of localization. Staufen protein contains multiple double-stranded RNA binding motifs (22), and an indirect assay suggests that Staufen binds to this helix (5). Nevertheless, Staufen is not required for RNA recognition event A (14). Thus, if Staufen actually binds to the helical region of stem-loop V, a factor involved in event A may need to be displaced.
The bcd localization signal is the second for which there is clear evidence of a requirement for highly structured RNA motifs. Characterization of the TLS element of the Drosophila K10 mRNA revealed that a stem-loop structure mediates localization of that mRNA (18). Although the TLS directs nurse cell-to-oocyte mRNA transport, the step requiring bcd stem-loop V, the two elements have substantial differences. The only similarity is the presence of an essential helical region, and the TLS does not have an obvious counterpart to the stem-loop V recognition domain (18). Thus, these two elements are unlikely to interact with the same recognition factors.
The RNA element characterized here is required for RNA recognition event A, but in the context of the bcd 3′ UTR it is not sufficient. The IV-V subdomain of the bcd 3′ UTR (consisting of stem-loops IV and V of the predicted structure) supports event A, but stem-loop V alone does not (14). Thus, other regions of IV-V must also contribute to event A. These sequences have been roughly mapped by deletion analysis (reference 15 and unpublished data) and include the distal portion of stem-loop IV. How do the different parts of IV-V contribute to RNA recognition event A? There are two simple options. First, the different regions could provide separate binding sites, either reiterated sites for a single factor (or set of factors) or qualitatively different sites that bind distinct factors. All sites would have to be occupied for localization activity, much as certain transcriptional enhancer elements require binding of homo- or heterodimers for function (10). In this scenario the element we have defined could correspond to a single binding site. Second, the different regions could be folded into a single binding site for one or more factors. For this option the element defined here would be only a part of a larger structure. Comparison with 2×BLE1 suggests that the former option is the more likely. In 2×BLE1 the bcd sequences are from stem-loop V; all stem-loop IV sequences are absent. Nevertheless, 2×BLE1 supports a program of localization whose early steps are indistinguishable from those directed by RNA recognition event A. The unique feature of 2×BLE1 is the presence of two copies of the RNA element—helix and flanking recognition domain—defined here. Thus, the simplest explanation of 2×BLE1 activity is that it provides reiterated binding sites for a localization factor. This factor might normally bind to multiple sites in IV-V. Alternatively, duplication of a site normally present in a single copy might compensate for the lack of a second binding site that appears in IV-V but not 2×BLE1. Examination of the IV-V sequence and predicted structure does not reveal an obvious second copy of the RNA element defined here, but this could reflect our still-incomplete understanding of its structure.
The finding that certain mutations in the bcd 3′ UTR lead to nuclear accumulation of the mRNA is unexpected, as none of the previously characterized 3′ UTR deletion mutants or 2×BLE1 mutants displayed this property (15, 16). Because nuclear accumulation is not correlated with a recognized mRNA localization event, the significance of this phenotype is uncertain. Nuclear accumulation may arise because certain mutations fortuitously increase the affinity of the mRNA for a nuclear protein. However, it seems more likely that a normally labile nuclear population of bcd mRNA is stabilized, as nuclear accumulation appears to involve a transient increase in mRNA levels. An explanation of this phenomenon will clearly require substantial additional analysis.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM42612.
We thank Eric Arn and Ricardo Mancebo for comments on the manuscript and members of the Macdonald lab for discussions.
REFERENCES
- 1.Bouget F-Y, Gerttula S, Shaw S L, Quatrano R S. Localization of actin mRNA during the establishment of cell polarity and early cell divisions in Fucus embryos. Plant Cell. 1996;8:189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 3.Cheong C, Varani G, Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 4.Ferrandon D, Elphick L, Nüsslein-Volhard C, St. Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 5.Ferrandon D, Koch I, Westhof E, Nüsslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautreau D, Cote C A, Mowry K L. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 1997;124:5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- 7.Gavis E R, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 8.Kim-Ha J, Webster P J, Smith J L, Macdonald P M. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 9.Kislauskis E H, Zhu X, Singer R H. Sequences responsible for intracellular localization of b-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb P, McKnight S L. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- 11.Lantz V, Schedl P. Multiple cis-acting targeting sequences are required for orb mRNA localization during Drosophila oogenesis. Mol Cell Biol. 1994;14:2235–2242. doi: 10.1128/mcb.14.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 13.Macdonald P M. The means to the ends: localization of maternal messenger RNAs. Dev Biol. 1992;3:413–424. [Google Scholar]
- 14.Macdonald P M, Kerr K. Redundant RNA recognition events in bicoid mRNA localization. RNA. 1997;3:1413–1420. [PMC free article] [PubMed] [Google Scholar]
- 15.Macdonald P M, Kerr K, Smith J L, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald P M, Leask A, Kerr K. exl protein specifically binds BLE1, a bicoid mRNA localization element, and is required for one phase of its activity. Proc Natl Acad Sci USA. 1995;92:10787–10791. doi: 10.1073/pnas.92.23.10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macdonald P M, Struhl G. cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 18.Serano T L, Cohen R S. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 19.Spradling A C. Developmental genetics of oogenesis. In: Bate M, Arias A M, editors. The development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- 20.Stephenson E C, Chao Y, Fackenthal J D. Molecular analysis of the swallow gene of Drosophila melanogaster. Genes Dev. 1988;2:1655–1665. doi: 10.1101/gad.2.12a.1655. [DOI] [PubMed] [Google Scholar]
- 21.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 22.St Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St Johnston, D., W. Driever, T. Berleth, S. Richstein, and C. Nüsslein-Volhard. 1989. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107(Suppl.):13–19. [DOI] [PubMed]
- 24.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 25.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals a translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 26.Tuerk C, Gauss P, Thermes C, Groebe D, Gayles M, Guild N, Stormo G, D’Aubenton-Carafa Y, Uhlenbeck O C, Tinoco I, Jr, Brody E N, Gold L. CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci USA. 1988;85:1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster P J, Suen J, Macdonald P M. Drosophila virilis oskar transgenes direct body patterning but not pole cell formation or maintenance of mRNA localization in D. melanogaster. Development. 1994;120:2027–2037. doi: 10.1242/dev.120.7.2027. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]