Abstract
Cellular condensates are usually ribonucleoprotein assemblies with liquid- or solid-like properties. Because these subcellular structures lack a delineating membrane, determining their compositions is difficult. Here we describe a proximity-biotinylation approach for capturing the RNAs of the condensates known as processing bodies (PBs) in Arabidopsis (Arabidopsis thaliana). By combining this approach with RNA detection, in silico, and high-resolution imaging approaches, we studied PBs under normal conditions and heat stress. PBs showed a much more dynamic RNA composition than the total transcriptome. RNAs involved in cell wall development and regeneration, plant hormonal signaling, secondary metabolism/defense, and RNA metabolism were enriched in PBs. RNA-binding proteins and the liquidity of PBs modulated RNA recruitment, while RNAs were frequently recruited together with their encoded proteins. In PBs, RNAs follow distinct fates: in small liquid-like PBs, RNAs get degraded while in more solid-like larger ones, they are stored. PB properties can be regulated by the actin-polymerizing SCAR (suppressor of the cyclic AMP)-WAVE (WASP family verprolin homologous) complex. SCAR/WAVE modulates the shuttling of RNAs between PBs and the translational machinery, thereby adjusting ethylene signaling. In summary, we provide an approach to identify RNAs in condensates that allowed us to reveal a mechanism for regulating RNA fate.
A proxitome-RNA-capture approach captures the transcriptome of the archetypal condensates known as processing bodies and reveals a translational hub based on liquid–liquid phase separation.
IN A NUTSHELL.
Background: Interactions between proteins and nucleic acids can promote the generation of droplet-like ribonucleoprotein condensates. One example of condensate is processing bodies (PBs), which are found in the cytoplasm and contain many proteins and RNA molecules. Previously, we defined the proteins residing in PBs using an approach known as proximity biotinylation, which can capture weak and transient interactions, such as those occurring in condensates. We did that in 2 conditions, normal conditions, and heat stress, showing that PBs are highly dynamic. This approach allowed us to identify a link between the core PB component known as DECAPPING 1 (DCP1) and the SCAR/WAVE actin-nucleating complex.
Question: Having defined the PB proteome, we asked whether the proximity biotinylation approach can also be used to capture associated RNA molecules and what kind of RNA molecules reside in PBs.
Findings: Herein, we used Arabidopsis thaliana seedlings and especially roots. We showed that the proximity biotinylation approach can indeed capture RNAs stably to more transiently associated with the PBs. These mainly include RNAs involved in cell wall development and regeneration, plant hormonal signaling, secondary metabolism/defense, and RNA metabolism. Furthermore, we showed that RNAs in PBs follow different paths: in small PBs, RNAs get degraded; in larger PBs, RNAs are stored. The stored RNAs are not translated until the actin-related SCAR/WAVE complex dissolves PBs and releases specific RNAs. One example of such RNA is WOUND INDUCED DEDIFFERENTIATION 1/RELATED TO AP2 4 (RAP2.4), which affects responses to the phytohormone ethylene.
Next steps: We could use the findings from this study to define PB functions in a cell-specific manner during the development of the Arabidopsis root. The approaches developed here can also be used to decipher the composition of numerous other condensates.
Introduction
Intra- or intermolecular interactions between proteins and nucleic acids can promote their demixing from the bulk cytoplasmic pool to droplet-like ribonucleoprotein (RNP) condensates. RNP condensates can form through liquid–liquid phase separation (LLPS), which is usually driven by multivalent (i.e. many low-affinity binding sites) interactions between unstructured protein segments and/or nucleic acids (Beutel et al. 2019). Furthermore, LLPS is sensitive to concentration; only when they are above a certain threshold (saturation concentration) do competent RNAs and proteins form or enter condensates (Alberti et al. 2019; Hatzianestis et al. 2023). Because many condensates form through LLPS, they show properties of liquids, but given enough time, they can solidify whereby they show a high internal viscosity and reduced exchange of molecules (Jawerth et al. 2020). In nonplants, these liquid-to-solid transitions are functional and regulate processes such as cell division, signaling, and immunity (Lasker et al. 2022).
The unstructured protein segments driving LLPS are low-complexity regions (LCRs), such as intrinsically disordered regions (IDRs) and prion-like domains (PrLDs) (Wiedner and Giudice 2021). IDRs and PrLDs can have charged amino acids (such as arginine, lysine, and histidine) often punctuated by aromatic ones (Zaccara and Jaffrey 2020). In addition, folded/structured segments of proteins can contribute to LLPS (Dignon et al. 2020). We know comparably little about what drives LLPS of RNA molecules.
Examples of RNP condensates are the evolutionarily conserved processing bodies (PBs), and stress granules (SGs) (Solis-Miranda et al. 2023). The numbers and sizes of PBs and SGs depend on conditions; furthermore, the size of SGs depends on small metabolic molecules (Chodasiewicz et al. 2022). Under heat stress (HS), PB size can increase (Gutierrez-Beltran et al. 2015). In yeast, plants, and animals, PBs contain RNA decapping, exosome, and deadenylase complexes and RNA-binding proteins (RBPs) (Liu et al. 2023). PBs have been associated with RNA decay, as they are enriched in the RNA decapping complex, comprising among others the plant DECAPPING 1 (DCP1), DCP2/Trident, as well as DCP5, the scaffold VARICOSE, and EXORIBONUCLEASE 4 (XRN4). XRN4 is a processive 5′-3′ exonuclease that executes RNA decay by trimming RNAs decapped by the active pair DCP1–DCP2. Arabidopsis mutants of DCP1, DCP2, and VARICOSE are dwarf with trichome abnormalities and disorganized veins (Xu et al. 2006; Xu and Chua 2009). Yet, XRN4 mutants do not show similar phenotypes, suggesting that PBs may execute additional functions or use additional ribonucleases. Although we know little about plant PBs, animal PBs not only degrade RNAs but can store them for conditional translation (Cairo et al. 2022). Yet, the mechanisms releasing RNAs from PBs for translation, as well as those modulating RNA fate in PBs, are under intense scrutiny (e.g. Blake et al. 2023).
Studies on the functions of condensates would benefit from the ability to isolate these subcellular compartments. As condensates lack a delineating membrane, their isolation is challenging due to their liquid nature (Liu et al. 2023). Nevertheless, in nonplants, the catalog of proteins and RNAs associated with PBs has greatly increased in the past few years. Protein cataloging for PBs was recently done by proximity-dependent ligation of biotin (hereafter PDL) in nonplants (Youn et al. 2018) and plants (Liu et al. 2023). PDL harnesses covalent biotinylation of proteins interacting with or near-neighbors of a certain prey protein in vivo. Through PDL, we recently showed that the PB component DCP1 co-condenses with the actin-nucleating SCAR (suppressor of the cyclic AMP)–WAVE (WASP family verprolin homologous) complex in differentiating cells (Liu et al. 2023). This co-condensation takes place specifically at cell edges or vertices (the angular point where 3 cell edges meet), mainly at the root transition and differentiation zones. This SCAR/WAVE-DCP1-axis is important for proper development, but how it works remains unclear.
Inspired by the successful application of PDL for the identification of the PB proteome, here, we repurposed PDL to isolate PB-associated RNAs from Arabidopsis (Arabidopsis thaliana). We show that PB-associated RNAs function mainly in development and secondary metabolism, immunity, cell wall-wounding/regeneration, and plant hormonal signaling. Large PBs are solidified and store RNAs together with their cognate proteins, keeping them translationally inert. By contrast, smaller PBs show high liquidity and can degrade these RNAs. The SCAR/WAVE-DCP1-axis controls the dissolution of PBs, and by dissolving PBs, allows the release of stored RNAs for translation or the reduction of their decay. This function of the SCAR/WAVE-DCP1-axis modulates ethylene responses.
Results
A TurboID-RIP approach captures RNAs associated with PBs
PDL with the biotin ligase BirA* has been used for the efficient purification of RNPs in animals (Leidal et al. 2020). Inspired by such applications, we assumed that the biotin ligase TurboID, which has been successfully used in plants (reviewed in Xu et al. 2023), can also biotinylate proteins bound to RNAs. In turn, these biotinylated proteins could theoretically be crosslinked with their bound RNAs using formaldehyde. For this approach, referred to hereafter as Turbo-RNA Immunoprecipitation (T-RIP; experimental approach detailed in Fig. 1A; Materials and methods), we used Arabidopsis seedlings carrying transgenes encoding either a chimeric DCP1-TurboID-6xHis-3xFLAG (DCP1-TurboID-HF) or the superfolder green fluorescent protein (GFP); in both cases, the transgenes were overexpressed in Arabidopsis under the control of the 35S promoter (GFP-TurboID-HF, Liu et al. 2023), which showed ca. 2-fold increased levels of DCP1 transcript compared with the wild type (Supplemental Fig. S1A). DCP1-TurboID-HF could rescue the weak dcp1-3 allele mutant phenotype (see below Fig. 10; Martinez de Alba et al. 2015; Liu et al. 2023) and showed sufficient biotinylation in immunoblots under the conditions used here (Supplemental Fig. S1B). However, we did not see sufficient biotinylation using the engineered and codon-optimized biotin ligase evolved from the soybean (Glycine max) ascorbate peroxidase (“APEX2” Supplemental Fig. S1, C to F). This result contrasts with what has been observed in nonplant models, where APEX2 directly biotinylates RNA and proteins (Fazal et al. 2019). We thus proceeded with the T-RIP approach.
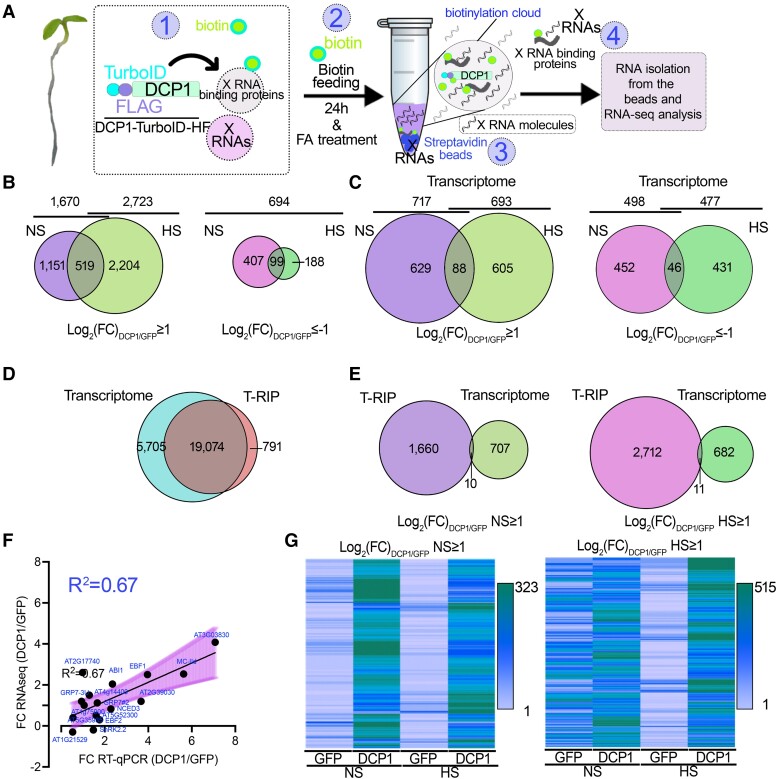
Figure 1.
T-RIP approach for the capture of PBs-associated RNAs. A) Pipeline for T-RIP: (i) Plants were submerged in 50 μM biotin for 1 min under a vacuum. The biotin solution was removed, and plants were placed back in the growth chamber. After 24 h, plants were left at the mock condition denoted as NS or treated with HS (22°C to 37°C for 2 h). (ii) 1% (v/v) formaldehyde was used as an RNA-protein crosslinker (fed to tissues similarly to biotin). Formaldehyde was then quenched by glycine. (iii) Proteins/RNAs were extracted as described for the APEAL approach (Liu et al. 2023); the extraction buffer was supplemented with an RNase inhibitor. The supernatants were loaded on PD-10 columns to deplete excess biotin and incubated with streptavidin-coated beads. (iv) Protease treatments were used to elute RNP complexes from the beads, and RNA was extracted. All the T-RIP samples were subjected to DNase I treatment and ribosomal RNA depletion. B) Venn diagrams showing the number of RNAs identified for T-RIP (left; log2FC≥1) and those with a high probability to be excluded from PBs (right; log2FC≤−−1; 506 RNAs in NS and 287 RNAs in HS). C) Venn diagrams showing the number of genes identified for the transcriptome (total RNA-seq) compared with the DCP1/GFP expressing lines (left, log2FC≥1; right, log2FC≤−1). Note that similar quantitative and small differences were found between samples regarding differential expression. D) Venn diagram showing the overlap between unfiltered RNAs identified for the Transcriptome (from GFP and DCP1 samples) and T-RIP. E) Venn diagram comparing the overlap between the filtered enriched RNAs in NS (log2FC≥1) of the T-RIP dataset and the corresponding transcriptome dataset in NS (left) and HS (right). Note: For esthetic reasons and readability, the Venn diagrams in B to E) are not proportional throughout and the analogies are kept only between the 2 conditions compared each time. F) Correlation between RNA levels determined by RNA-seq or RT-qPCR for the indicated genes. ACTIN7 and PP2A were used for normalization. Data are from 3 independent experiments with 2 technical duplicates (n = 2 assays). The fitted line is shown, along with the confidence intervals as a shaded area (95%, deviation from zero P = 0.0008), with a fitted equation Y = 0.4686*X + 0.2534. FC, fold change between DCP1/GFP samples. G) Heat maps showing sample gene clusters of T-RIP. RNAs were filtered either using a log2FC≥1 for genes enriched in DCP1 NS (left) or HS (right) from GFP and DCP1 samples. The color-coding bars indicate the number of RNAs.
Figure 10.
Modulation of RAP2.4 levels and the ethylene response by the SCAR/WAVE-DCP1 axis. A) Micrographs from smFISH for the detection of RAP2.4. mRNA (gray) in the root epidermal cells of the MZ and TZ zones in the corresponding mutants [wild type (WT), dcp1-3, scar1234, and xrn4-5]. The yellow arrowheads denote RAP2.4 signal. The experiment was replicated 3 times. Scale bars, 12 μm. Right: corresponding quantifications with the number of smFISH spots in WT, dcp1-3, scar1234, and xrn4-5. Data are means of 3 independent experiments with 2 technical replicates each (n = 2 roots each with 10 MZ or TZ randomly selected cells at 5 DAG), and significance was determined by ordinary 1-way ANOVA. ns, not significant. B) Photos showing growth in the absence or presence of the ethylene precursor ACC (10 μM 5 DAG). The experiment was replicated 3 times (n = 10 seedlings in each replicate). Arrowheads denote the elongated hypocotyls in scar1234 and brk1. Also, note the complementation of DCP1-TurboID expressing dcp1-3 (mainly upon ACC treatment). Scale bars, 1 cm. Right: corresponding quantifications of hypocotyl and root length in the presence or absence of ACC. Data are means ± Sd (N = 3 biological replicates with n = 10 roots/hypocotyls each); significance was determined by ordinary 1-way ANOVA with Dunnett's corrections.
Previously, using Arabidopsis lines overexpressing DCP1-TurboID-HF, we successfully captured the proteome of PBs using a tandem approach called APEAL [for Affinity Purification with Proximity-dEpendent LigAtion steps (Liu et al. 2023)]. For T-RIP, we refined APEAL using seedlings grown under normal conditions and transferred to nonstress (NS, 22°C, normal Arabidopsis growth temperature) or HS (37°C) conditions for 2 h. We justify the use of HS considering that it mainly leads to an increase in the size of PBs (20% to 30%) and induces quantitative and qualitative changes in the PB proteome (Liu et al. 2023 and below). Seedlings were submerged in biotin and after 24 h were treated with formaldehyde to fix biotinylated proteins in PBs with adjacent or interacting RNAs (Fig. 1A, details in Materials and methods). Streptavidin-coated beads were used to enrich and purify these PB-associated RNPs, and the corresponding RNA was extracted and subjected to RNA-Seq.
Due to the HS-induced changes in the PB proteome and considering that many proteins in PBs are RNA-binders, we speculated that HS could also induce changes in the RNA composition of PBs. In accordance with the notion, although HS led to a decrease in the number of unique proteins identified in PBs (i.e. reducing their diversity from 16% to 6% of the proteins in APEAL), it induced the accumulation of RBPs (Supplemental Data Set 1); more than >10% of the proteins identified were RNA-binders, which is 4-fold higher than the value observed in NS. Considering these dynamic changes, we decided to do RNA-seq on both NS and HS samples and considered only high-confidence enrichments [fold enrichment (“FC”) log2FCDCP1/GFP ≥ 1 or <−1, i.e. enrichment/depletion when comparing DCP1 to GFP samples; n = 2, false-discovery rate (FDR) = 0.05, Supplemental Data Set 2].
Multiplexed libraries averaged 20 million reads, with >90% mapping to the Arabidopsis genome. T-RIP captured 19,865 RNAs, of which 1,670 (8.4%) and 2,723 (13.7%) were enriched in DCP1 samples in NS and HS conditions, respectively (Fig. 1B). We found that 519 RNAs were common among NS and HS, representing those that are associated with PBs at both temperatures (Supplemental Data Set 3 for gene names). The T-RIP DCP1 datasets had more enriched RNAs compared with GFP; only 694 RNAs were enriched in the T-RIP GFP datasets (Fig. 1B, log2FCDCP1/GFP < −1; i.e. 3.49% from HS + NS). At the transcriptome level, however, HS induced similar quantitative changes in RNA levels for both NS and HS for up- and downregulated genes; ca. 500 RNAs were differentially expressed between DCP1 and GFP-TurboID in NS and HS (Fig. 1C). This differential expression suggests that DCP1 overexpression may affect the transcriptome, at least to some extent. We could not identify 5,705 RNAs in the T-RIP dataset (unfiltered dataset including all RNAs from both NS and HS and DCP1 compared with GFP samples) that could be identified for the transcriptome, while 791 were exclusively identified following T-RIP (Fig. 1D). Many RNAs that were enriched in the T-RIP dataset were not significantly enriched in the total RNA sequencing dataset from the same samples using the same enrichment criteria; only 10 and 11 RNAs were enriched in both transcriptome and T-RIP data for NS and HS, respectively (Fig. 1E). Hence, enriched RNAs in the T-RIP dataset did not necessarily follow the corresponding enrichments of total RNA. In turn, among transcripts with a high enrichment in T-RIP were RNAs with a low read coverage (or absent) in the total RNA data set. Taken together, these results validate the notion that HS not only remodels the PBs’ proteome but also their RNAs.
To further validate the RNA-seq data from T-RIP, we performed reverse transcription quantitative PCR (RT-qPCR) on T-RIP samples to test the relative levels of 16 RNAs showing various enrichment levels (log2FC from −0.3 to 4.1). The RNA levels determined by RT-qPCR correlated relatively well with the corresponding FC (fold change) values obtained in T-RIP (Fig. 1F, R2 = 0.67; Supplemental Data Set 4 for the genes used). We also confirmed the presence in our T-RIP datasets of the few known RNA molecules localizing at PBs: EBF1 (EIN3 BINDING F-BOX PROTEIN1) and EBF2 (Li et al. 2015; Merchante et al. 2015; Fig. 1F; Supplemental Data Set 4). Furthermore, T-RIP-enriched RNAs in the DCP1 samples showed certain enrichment clusters in heat maps that were not observed in the corresponding GFP controls (Fig. 1G).
As a cautionary note here, we used T-RIP lines overexpressing DCP1 (DCP1 mRNA levels increased by ∼30%, Liu et al. 2023). We thus compared whether DCP1 expressed under the control of its native promoter (DCP1pro) could also lead to the enrichment of selected RNAs. We used formaldehyde to fix seedlings expressing DCP1-GFP driven by 35Spro or DCP1pro under NS conditions (similar conditions used for T-RIP). We further purified enriched RNAs following a refined approach for GFP-trapping (with GFP-beads). For this assay, we selected RT-qPCR results from 5 RNAs enriched and 3 depleted from T-RIP [with a T-RIP log2FC ranging from 0 (no enrichment) to 3.9 (high enrichment)] as a readout. We observed similar enrichment patterns between 35Spro and DCP1pro lines when normalized to a line expressing GFP (Supplemental Fig. S2). Yet, these enrichments were moderate when compared with those obtained using T-RIP (e.g. for EBF2 2.0 for T-RIP vs. 1.3 for GFP-trap), which is consistent with the shorter radius of formaldehyde fixation (2.3 to 2.7 Å; Sutherland et al. 2008) than the predicted biotinylation radius of TurboID; we have suggested that TurboID can indeed identify interactions not identified by formaldehyde fixations or simple affinity purifications (Liu et al. 2023). Notably, we identified a high enrichment of the actin-related ACTIN DEPOLYMERIZING FACTOR 9 (ADF9), which is consistent with our previous results on the actin-related SCAR-WAVE-DCP1 axis. Collectively, based on these findings, we propose that T-RIP can capture at least a portion of RNAs localizing in PBs; we refer to these RNAs as “PBs-enriched RNAs” hereafter.
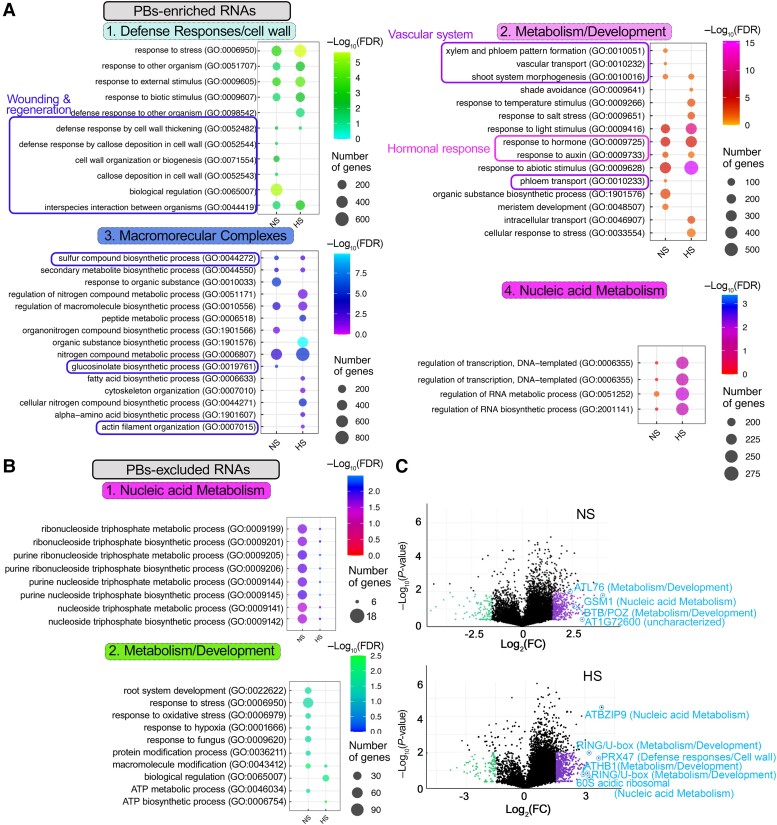
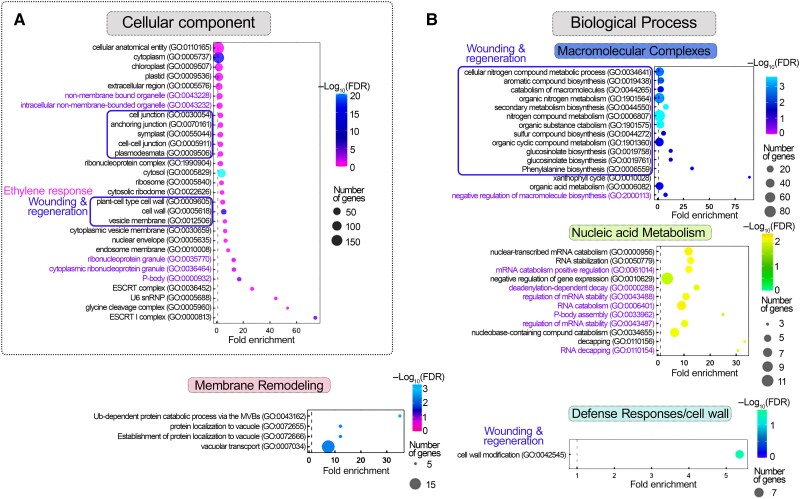
We classified the putative PBs-enriched RNAs according to their gene ontology (GO, biological process) using PANTHER (protein analysis through evolutionary relationships; cutoff P < 0.05 for the −log2FDR; datasets in Supplemental Data Sets 3 and 5, where redundant GO terms have been retained). Most RNAs in NS and HS could fit in 4 GO subnetworks: (i) membrane/cell wall, (ii) nucleic acid metabolism organization/RNA localization or transport, (iii) assembly of macromolecular complexes, and (iv) metabolism/development (e.g. regulation of hormonal responses, auxin, and ethylene; Fig. 2A). Enriched GO terms within these 4 subnetworks that mostly appeared under NS conditions were related to wounding/regeneration [Fig. 2A; indicated with blue boxes, i.e. defense response, secondary metabolism, glucosinolates-cell wall (FDR = 3.97e−3), callose deposition (FDR = 3.46e−3), and actin organization (FDR = 4.58e−2)]. On the contrary, GO terms such as light signaling were observed under HS conditions [Fig. 2A; shade avoidance and response to light stimulus (FDRs = 3.12e−2 and 4.59e−4)]. The involvement of PBs in the regulation of light signaling has been previously described and is consistent with our results here (Jang et al. 2019). We also observed common GO terms between NS and HS, e.g. of hormonal responses and actin (Fig. 2A; indicated with magenta boxes, FDR, NS = 5.15e−5, HS = 6.04e−5). The PBs-depleted RNAs fit into 2 subnetworks related to the housekeeping processes of nucleic acid and metabolism (fitting to a broader metabolism term compared with the respective network of enriched RNAs; e.g. “root system development,” “ATP metabolic process” etc.) (Fig. 2B). These GO terms were not identified in the corresponding total transcriptome dataset (Supplemental Data Set 5). The enrichment distribution of RNAs in PBs is shown in Fig. 2C, where we highlight enriched RNAs from the 4 subnetworks. Altogether, we conclude that PBs-enriched RNAs can fit into a small number of subnetworks.
Figure 2.
Subnetworks of RNAs enriched in PBs. A) Enriched GO terms of PBs-enriched RNAs fitting into 4 subnetworks. Boxes annotate clusters: wounding and regeneration, vascular system, and plant hormonal responses. FDR, false discovery rate. B) Enriched GO terms of PBs-excluded RNAs fitting into 2 subnetworks. C) PB-enriched/depleted RNAs (log2FCDCP1/GFP) in NS (upper) or HS (lower), visualized by volcano plots. RNAs from the 4 subnetworks defined in A are indicated.
RNAs with shorter coding regions tend to localize in PBs
Thus far, we have defined the PBs-enriched RNAs under 2 conditions. Our next question was whether these RNAs have specific features that promote their recruitment to PBs. In animals, RNA size seems to modulate PB localization (Wang et al. 2018). We thus first established a pipeline for comparisons between PBs-enriched/depleted RNAs and the total transcriptome for 3′- or 5′-UTR, coding sequences (CDS) lengths, and GC% content. By examining and analyzing these parameters, we found a negative correlation between CDS length and PBs association (r = ∼−0.25 in NS and HS, while r > 0.2 for total; Supplemental Fig. S3, A to I). Hence, smaller RNAs tend to localize in PBs under NS and HS conditions.
We next queried the free energy (folding energies) of the secondary structures of PBs-enriched RNAs following the pipeline described above (for comparisons). In yeast and human cells, the secondary structure of the 5′-UTR regulates mRNA localization (Ringner and Krogh 2005). Accordingly, considering that the size of UTRs does not affect mRNA localization in PBs, we opted to use a normalized metric, the “minimum free energy (MFE) density,” which discounts size and works well for >40 nt RNA (Trotta 2014). This metric accounts for local free energy, which could be important for RNA functions in PBs. High structural stability (i.e. local free energy) is compatible with specific functions, for example, the necessity of RNAs to maintain a stem-loop structure for recognition by RBPs (Ha and Kim 2014). MFE density is likely more adequate for comparisons in PBs-enriched RNAs with distinct lengths, as it correlated well with GC% but not, as expected, with length (Supplemental Fig. S4, A to D). We observed that increased stability of the 5′-UTR (lower MFE density) was well-associated with increased half-life of RNAs [half-life defined in (Sorenson et al. 2018)]; yet PBs-depleted RNAs had increased 5′-UTR stability (Supplemental Fig. S5). This result suggests that PBs may recruit RNAs with less-structured UTRs. Accordingly, from the same datasets, we observed that HS was associated with higher stability than NS for most PB-enriched RNAs, while the small size of CDSs in NS (<800 nt) was associated with reduced RNA stability. However, we did not find a general correlation between PBs-enrichment levels and MFE density (r < 0.01, in NS and HS). During these analyses, we also observed that PBs-enriched UTRs did not show a composition bias for increased adenine and guanidine, which correlates with decay (Supplemental Fig. S6; Sorenson et al. 2018).
As PBs can compete with polysomes for RNAs (Sheth and Parker 2003), we also tested whether PBs-enriched RNAs have rare codons that could halt translation and shuttle RNAs to PBs for decay. Using the Relative Codon Bias Strength (RCBS) index to calculate the difference between the observed vs. the expected codon frequency (Sahoo et al. 2019; Parvathy et al. 2022), we observed only a marginally increased RCBS in HS (Supplemental Fig. S7, A to C). Overall, these results suggest that the small size of RNA and a looser 5′-UTR structure are associated with localization to PBs.
N 6-methyladenosine modification is not a prerequisite for PBs-RNA enrichment
Thus far, our results predict a moderate link between RNA size and secondary structure with PB localization. We thus asked whether PBs-enriched RNAs show modifications that could modulate their localization with PBs. We focused here on N6-methyladenosine (m6A), owing to its high abundance (25% of all RNAs are modified) and the ability to modulate PB dynamics in nonplants (Zaccara and Jaffrey 2020). To check whether PBs RNAs are m6A-enriched, we compared PBs-enriched/depleted RNAs to the dataset described by Parker et al. (2020). In this study, to extract information about m6A-enriched RNAs, authors compared RNA-seq reads of the Arabidopsis virilizer (vir-1) mutant, which is impaired in a conserved m6A component of the writer complex, to a corresponding complementation line (VIR-GFP, denoted as “virC”). This comparison confirmed that PBs-enriched RNAs can have m6A-modifications, mainly during HS (Supplemental Fig. S8).
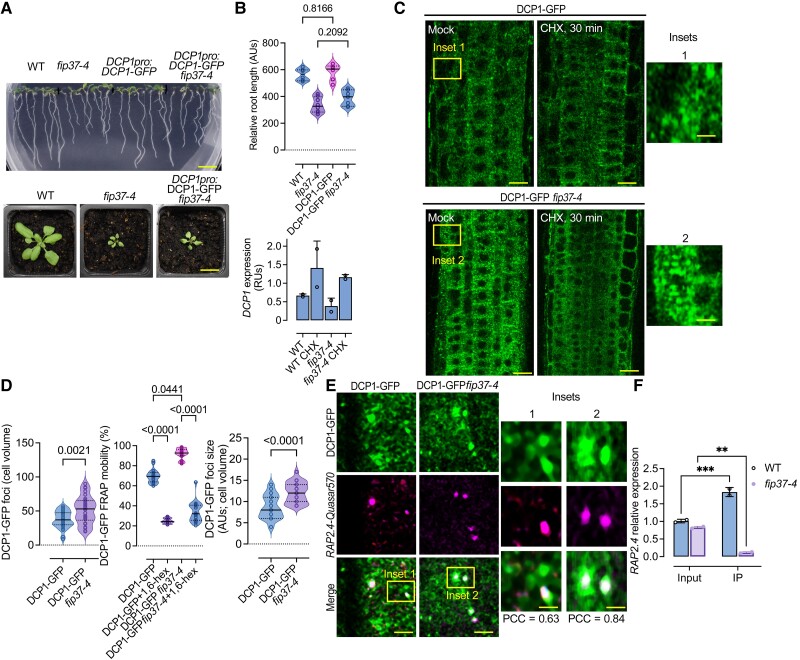
This prediction implied a possible requirement for RNA m6A for localization in PBs. To experimentally test this requirement, we used a mutant allele of the m6A writer FIP37 [FKBP12 Interacting protein 37; fip37-4 (Shen et al. 2016)], a major component of the m6A writer complex, showing an ∼85% reduction in m6A. To follow PB dynamics in this background, we introduced DCP1pro:DCP1-GFP into this mutant by crossing. The DCP1-GFP fip37-4 mutant did not differ phenotypically from fip37-4 and showed wild type-like levels of DCP1 (Fig. 3, A and B). We observed reduced stability of PBs in DCP1-GFP fip37-4, as determined by a short treatment with cycloheximide (CHX) (30 min), which is known to disassemble PBs (Gutierrez-Beltran et al. 2015), and an increase in PBs sizes/numbers and fluorescence recovery after photobleaching (FRAP; Fig. 3C). Rapid rates in FRAP are indicative of high liquidity and likely reduced stability of condensates (Alberti et al. 2019). Furthermore, treatments with 1,6-hexanediol (which dissolves liquid condensates while leaving the more solid ones intact) (Alberti et al. 2019) showed that DCP1-GFP fip37-4 PBs could be dissolved in a manner similar to wild-type ones (Fig. 3D). Hence, our results suggest that m6A-modifications on RNAs may modulate the stability but not formation of PBs.
Figure 3.
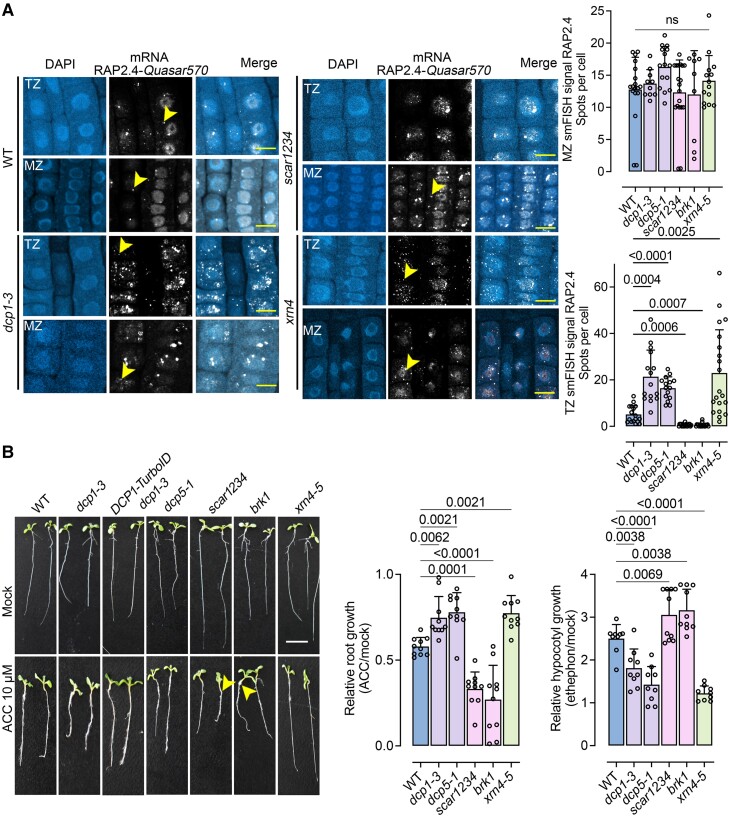
The m6A writer FIP37 controls stability but not the formation of PBs. A) Phenotypes of fip37-4 and fip37-4 DCP1pro:DCP1-GFP seedlings [7 d after germination (DAG)]. Lower: corresponding phenotypes of rosettes. Scale bars, 1 cm. B) Relative root length of wild type (WT), fip37-4 or fip37-4 DCP1pro:DCP1-GFP seedlings. Data are means (±Sd) of 3 independent experiments with 2 technical replicates (n = 2 with 10 roots from 7 DAG seedlings), and significance was determined by ordinary 1-way ANOVA. Lower: RT-qPCR of DCP1 expression in WT or fip37-4 DCP1pro:DCP1-GFP seedlings (7 DAG) in the presence or absence of CHX (30 μM, 30 min). TUA4 was used for normalization. Data are from 2 independent experiments with 2 technical duplicates (n = 2 assays). AUs, arbitrary units; RUs, relative units (normalized to TUA4). C) Micrographs from meristematic epidermis root cells 5 DAG expressing DCP1pro:DCP1-GFP in WT or fip37-4 in the presence or absence of CHX (30 μM, 30 min). The experiment was replicated 3 times. Scale bars, 10 μm. D) Quantification of the number of DCP1-GFP foci per cell (cell volume in μm3), corresponding FRAP mobility (%; corresponding to the initial signal recovery), and diameter (size) of PBs in WT or fip37-4. For FRAP mobility, seedlings were treated with 1,6-hexanediol (“hex”) to dissolve PBs showing liquidity. Data are means (±Sd) of 3 independent experiments with 2 technical replicates each (n = 2 with 6 to 10 roots from 5 DAG); significance was determined by ordinary 1-way ANOVA. AUs, arbitrary units. E) Micrographs from meristematic epidermis root cells 7 DAG showing smFISH detection of RAP2.4-Quasar570 mRNA (in magenta) expressing DCP1pro:DCP1-GFP in WT or fip37-4. Two independent experiments with (n = 6 to 10 cells randomly picked) showed similar results. PCC values to show colocalization of the 2 signals are noted in each panel. Scale bars, 2 μm (insets, 0.2 μm). F) IP-RT-qPCR of m6A-modified RAP2.4 in WT and fip37-4 seedlings. Data are means of 2 independent experiments with 2 technical replicates each (n = 2 IP assays from 5 DAG seedlings). Data were normalized against TUA4 (reference gene) calculated from the input sample (Parker et al. 2020). Significance was determined by ordinary 1-way ANOVA (asterisks indicate significance).
We further queried whether PBs in DCP1-GFP fip37-4 showed reduced localization of m6A-modified PBs-enriched RNAs, for example, the mRNA of WOUND INDUCED DEDIFFERENTIATION 1/RELATED TO AP2 4 [RAP2.4, log2(FC)NS, HS = 1.586/1.943] (Fig. 3E). We justify this selection based on the association of RAP2.4 to 2 out of 4 enriched GO terms, hormonal responses and regeneration, processes followed up later in this article. We thus adopted and refined a single-molecule fluorescence in situ hybridization (smFISH) approach for RNA detection in root meristem cells, enabling the observation of intact tissues (Zhao et al. 2023). For smFISH, we used probes labeled with the fluorescence dye Quasar 570 spanning the whole CDS (see also Materials and methods). We observed what appeared to be a slight increase in the RAP2.4 signal in DCP1-GFP fip37-4 PBs when compared with the corresponding wild type (Fig. 3E). Furthermore, RAP2.4 is m6A modified under our conditions, as shown using m6A-immunoprecipitation (IP) combined with RT-qPCR in wild type and fip37-4 mutant (Fig. 3F; Shen et al. 2016). These results suggest that m6A-modification levels may not necessarily affect RNA localization to PBs.
The sizes of PBs inversely correlate with their RNA degradation capacity
Because RNA decay factors accumulate in PBs, it was proposed that the function of PBs is RNA decay (Luo et al. 2018). On the other hand, our 4 observations so far suggest that PBs may also store RNAs in plants: (i) m6A-RNAs, which are likely enriched in PBs are generally more stable (e.g. Song et al. 2023). Furthermore, PBs-enriched UTRs were not rich in adenine and guanidine, which are associated with decay (Supplemental Fig. S6; Sorenson et al. 2018); (ii) During HS, when PBs increase in size, we found more RNAs in PBs compared with NS conditions (Fig. 1B). Furthermore, smaller RNAs, which are mainly associated with PBs, show reduced decay in HS. This finding is consistent with observations in yeast, where PBs store RNAs upon glucose starvation for later translation (Wang et al. 2018); (iii) The RNA-seq read distributions, calculated using a refined version of “metagene plot” to provide profiles of average coverage of PBs-enriched RNAs, showed a slight reduction toward the 3′-UTR. This observation is consistent with the association of the de-adenylation machinery with PBs that trims the 3′-UTR (Supplemental Data Set 6 and Fig. S9). On the other hand, we did not find the expected reduction of 5′-UTR reads (especially during HS), as decapping is expected to trim 5′-UTRs through XRN4 or other exonucleases (Goeres et al. 2007). In accordance, we did not find a correlation between the levels of our previously obtained decapping dataset (from NS and HS) with the enrichment level of PBs-enriched RNAs; we note that we found a moderate negative correlation with the levels of PBs-excluded RNAs confirming that PBs are involved in decapping [Supplemental Fig. S10A; dataset from (Gutierrez-Beltran et al. 2015)]; (iv) Finally, when comparing the degradation rates for the total transcriptome defined in Sorenson et al. (2018) (for both wild type and varicose mutant) of PBs-enriched RNAs with excluded RNAs, we found that under HS, when PBs increased in size, PBs-enriched RNAs were rather more stable (Supplemental Fig. S10B).
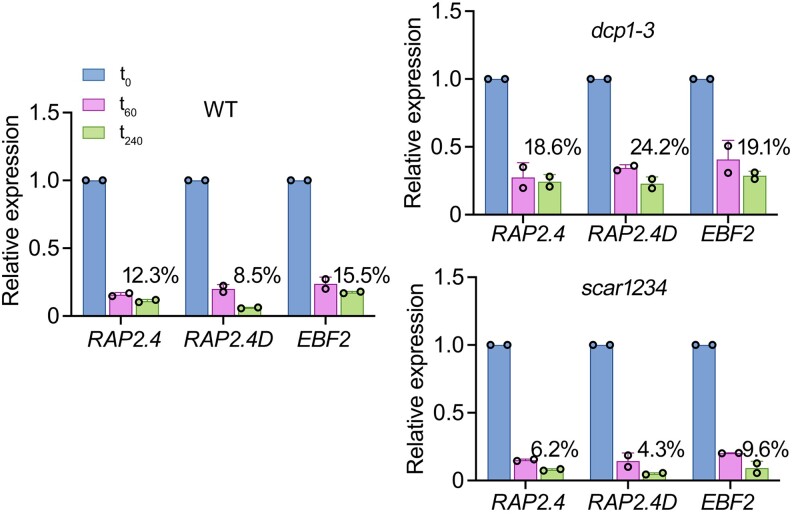
We thus asked when PBs might function in RNA decay by measuring the effect of PB numbers/sizes of RNA half-lives. To address this question, we first used 5-d-old seedlings treated with a transcription inhibitor (cordycepin) to block de novo RNA synthesis and collected samples across a 240 min time course [as done in (Sorenson et al. 2018)]. We compared RNA decay between wild type and the dcp1-3 mutant, which shows fewer PBs, or the SCAR/WAVE complex mutant scar1234, which shows more and larger PBs than the wild type (Supplemental Fig. S11; Liu et al. 2023). We followed the degradation rates of 3 PBs-enriched RNAs using RT-qPCR based on their relevance to plant hormonal responses and regeneration, processes followed up later in this article: RAP2.4, EBF2 [log2(FC)NS, HS = 1.015/1.205], and RELATED TO AP2.4 [RAP2.4D/WIND2; log2(FC)NS, HS = 1.426/1.550; Class 2 of CDS length, 800 to 1,600 close to the expected 1.2]. Compared with the wild type, scar1234 showed increased RNA degradation rates, while dcp1-3 showed reduced RNA degradation rates (mainly for WIND2; Fig. 4).
Figure 4.
Decay rates modulation by the number and size of PBs of 3 PBs-enriched RNAs. RT-qPCR determination of RNA decay levels for RAP2.4, EBF2, and RAP2.4D in wild type (WT), dcp1-3, and scar1234 5 DAG and in 3-time points (0, 60, and 240 min) upon treatment with the transcription inhibitor cordycepin [1 mM, (Sorenson et al. 2018)]. The percentages show the levels of RNAs at 240 min compared with 0 min. Data are means (±Sd) of 2 independent experiments with 2 technical replicates each (n = 2 roots from 5 DAG seedlings).
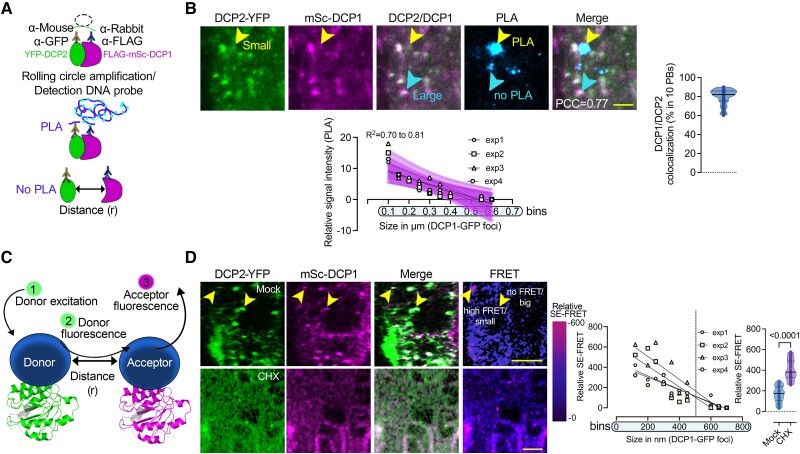
As scar1234 showed both increased numbers and sizes of PBs, we asked which of these 2 parameters could contribute to the increase in RNA decay. As the association of DCP1 and DCP2 correlates well with active RNA-degradation (Tibble et al. 2021; Liu et al. 2023), we aimed to use DCP1–DCP2 interaction as a proxy for decapping/decay. To achieve this aim, we established a quantitative 3D proximity ligation assay (PLA; Teale et al. 2021). PLA uses complementary oligonucleotides fused to antibodies to determine the frequency with which proteins of interest find themselves nearby (Fig. 5A). PLA allows in situ detection of endogenous protein interactions with high sensitivity and single-molecule resolution (at distances < 40 nm). Typically, 2 primary antibodies against epitope tags are used to detect 2 unique protein targets. A pair of oligonucleotide-labeled secondary antibodies (PLA probes) bind to the primary antibodies. Next, hybridizing connector oligos join the PLA probes only if they are near each other, and ligase forms a closed, circle DNA template that is required for rolling-circle amplification (RCA). The PLA probe then acts as a primer for a DNA polymerase, generating concatemeric sequences during RCA. This RCA step allows up to a 1,000-fold signal amplification that is still tethered to the PLA probe, allowing the detection of even very transient interactions. Finally, labeled oligos hybridize to the amplicon, which is then visualized and quantified as discrete spots (PLA signals). Contrary to our expectations, we discovered that DCP1–DCP2 interacted well in small PBs of root epidermal meristematic cells (i.e. on the limit of or below detection in our super-resolution confocal setting, 120 nm). By correlating DCP1–DCP2 interactions (PLA signals) with the sizes of PBs through regression analyses, we found a strong negative correlation (Fig. 5B; R2 = −0.6). As DCP1–DCP2 colocalized in most PBs (Fig. 5B; ∼80%), this result suggests that DCP1–DCP2 usually dissociate in larger PBs (>∼400 nm).
Figure 5.
Correlation between DCP1/DCP2 interaction with the size of PBs in root cells. A) Illustration of the PLA approach principle (see details in the main text). B) PLA signal produced by α-GFP/α-FLAG of a line co-expressing RPS5apro:HF-mScarlet-DCP1 (mSc-DCP1) and 35Spro:DCP2-YFP. The “spots” (violet “PLA”) do not connote physiologically relevant puncta (e.g. condensates). The colocalization of PLA spots with DCP1–DCP2 signals is also shown (noted as PCC). Scale bars, 5 μm. Lower: correlation between the size of PBs (DCP1–GFP foci) and PLA spot number (DCP1/DCP2 interaction), in 100 nm PB size bins. Data are means of 4 independent experiments with 2 technical replicates each (n = 2 roots with 5 meristematic epidermal cells 5 to 7 DAG each). The simple regression analysis includes data from the 4 independent experiments denoted as “exp.” All fitted lines are shown along with the overlaid confidence intervals (purple stripe; 95%, deviation from zero P = 0.0009 to 0.0049), with a fitted equation Y = –20.54*X + 11.06). Nonlinear regression through one-phase decay predicted relevant models here. Right: colocalization of DCP1/DCP2 per 10 PBs. Data are means of 4 independent experiments with 2 technical replicates each (n = 2 roots with 5 randomly selected meristematic epidermal cells 5 to 7 DAG). C) Sensitized emission FRET principle (SE-FRET), where the emission spectrum of the donor (i) overlaps with the excitation spectrum of the acceptor (ii), and if the distance (r) between the 2 molecules is sufficient, energy is transferred (iii). D) Micrographs of root meristematic epidermal cells showing SE-FRET efficiency between mScarlet-DCP1 and DCP2–YFP in the absence or presence of CHX (10 μM 20 min). The arrowheads denote small (at the detection limit) or big PBs (upper micrograph). Scale bars, 10 μm. Right: correlation between PB size and SE-FRET efficiencies in 50 nm PBs size bins and SE-FRET efficiency in the presence or absence of CHX. Data are means of 4 independent experiments with 2 technical replicates each (n = 2 roots in each replicate with 10 randomly picked meristematic epidermal cells 5 to 7 DAG). The simple regression analysis includes data from 4 independent experiments denoted as “exp” (n = 16 to 33 cells each). All fitted lines are shown.
To discount the possibility that PLA antibodies could not identify some DCP1/DCP2 molecules due, e.g. to stereochemical hindrance in the crowded environment of PBs, we utilized a refined live cell imaging approach. We used the stoichiometry-sensitive type of Försters resonance energy transfer called “sensitized emission” (Fig. 5C, SE-FRET). FRET is a nonradiative interaction between 2 molecules that happens at distances in the range of a few nm. In SE-FRET, the sensitized emission (i.e. the acceptor fluorescence resulting from energy transfer from excited donor molecules) from separately acquired donor and acceptor images is calculated, following leak-through corrections (van Rheenen et al. 2004). The SE-FRET approach allows FRET measurements to be performed on moving condensates (e.g. PBs) and could in principle provide stoichiometric information about DCP1–DCP2 interactions within a shorter range than PLA (i.e. 10 nm). Using SE-FRET, we confirmed the PLA results (Fig. 5D). Furthermore, using a short treatment with CHX to partially reduce the size and dissolve PBs, we observed a transient increase in DCP1–DCP2 interactions (Fig. 5D, CHX). Together, the PLA and SE-FRET results suggest that RNA decay is mainly executed by small PBs (or even in submicroscopic diffused decapping complexes).
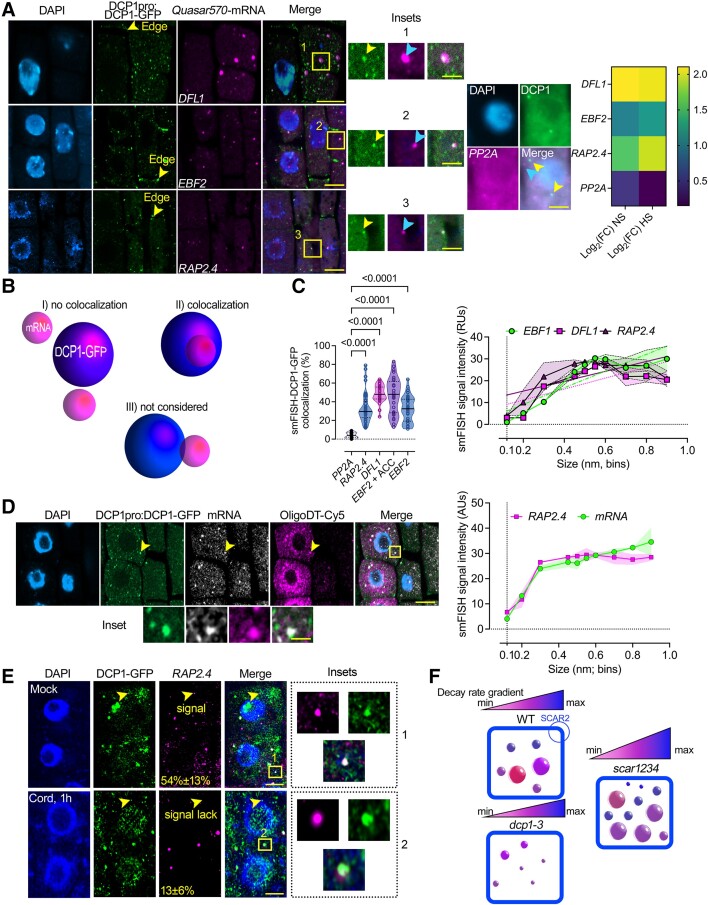
To validate this suggestion, we directly queried RNA fate in PBs using smFISH. We performed quantitative smFISH colocalizations between PBs labeled by DCP1-GFP (driven by the DCP1 promoter) and 3 RNAs enriched in PBs with sets of Quasar 570 labeled mRNA probes [EBF2, DWARF IN LIGHT 1 (DFL1), or RAP2.4; Fig. 6A]. As a negative control, we used the PBs-depleted RNA SERINE/THREONINE PROTEIN PHOSPHATASE 2A [PP2A; log2(FC)NS/HS = 0.46/0.15; Supplemental Data Set 7 for enrichments of RNAs used for probes]. We confirmed the colocalization between PBs and EBF2, DFL1, or RAP2.4 and the lack of colocalization with PP2A (Fig. 6, A and B). Using high-resolution imaging, we found a positive correlation between large PBs (>400 nm) and localization of the smFISH signal with size up to a point (Fig. 6C). We also applied a short treatment with an ethylene precursor (10 μM 1-aminocyclopropane-1-carboxylic acid, ACC) for EBF2, as it can increase the localization of EBF2 mRNA with PBs (Fig. 6C; see also Supplemental Fig. S15; Li et al. 2015; Merchante et al. 2015). mRNAs, in general, showed a similar trend of accumulation in PBs to that observed using the above probes, as revealed using costaining of RAP2.4 mRNA with Cy5-labeled oligoDT probes (Fig. 6D). Furthermore, treatments with cordycepin affected the accumulation of RAP2.4 mRNA only in smaller PBs and had no effect for larger ones (Fig. 6E). Overall, we showed that small PBs execute RNA decay in vivo (the corresponding model is presented in Fig. 6F).
Figure 6.
In vivo correlation between PB size and RNA levels. A) Micrographs from smFISH for detection of DFL1, EFB2, RAP2.4, and PP2A mRNAs (in magenta) in root meristematic epidermal wild type (WT) cells at 5 DAG expressing DCP1pro:DCP1-GFP. Insets: details of DCP1-GFP with each mRNA probe and with the negative control PP2a mRNA probe showing puncta colocalization (black box; arrows). Nuclei were stained with DAPI (violet). Right: DCP1-GFP/PP2A lack of colocalization and heat map showing enrichment levels for the 4 mRNAs. The experiment was replicated 3 times (n = 10 randomly selected cells). Scale bars, 2 μm (insets, 0.5 μm). B) Cartoon displaying the strategy for quantification of the colocalization between DCP1-GFP and mRNAs smFISH signal. Three major classes of colocalization are shown (I–III). Case III was not considered as colocalization since confocal microscopy may underestimate distances. C) Colocalization efficiency between mRNAs/DCP1-GFP (number of mRNA smFISH puncta in PBs/total number of mRNA puncta expressed as a percentage). Data are means of 3 independent experiments with 2 technical replicates each (n = 2 roots with 10 meristematic epidermal cells 5 DAG), and significance was determined by ordinary 1-way ANOVA. ACC, ethylene in the form of 10 μM ACC (see also Supplemental Fig. S15). Right: corresponding quantifications of the correlation between mRNAs/DCP1-GFP and PBs size in 100 nm PBs size bins. Data are means of 3 independent experiments with 2 technical replicates each (n = 2 roots with 10 randomly selected meristematic epidermal cells 5 DAG each); significance for the violin plots was determined by ordinary 1-way ANOVA. For the line plot, data are means ± Sd (SD: denoted as shaded area; N = 3 biological replicates with n = 10 randomly selected meristematic epidermal cells 5 DAG each). Fitted lines are also shown. RUs, relative units. D) Micrographs from RAP2.4 smFISH signal (gray) counterstained with oligoDT-Cy5 in root meristematic epidermal wild-type cells expressing DCP1pro:DCP1-GFP. Nuclei were stained with DAPI (violet). Arrowheads denote an example of the colocalization between RAP2.4, OligoDT, and DCP1-GFP, and the inset below denotes a detail of this colocalization. Scale bars, 10 µm. Right: quantification of the correlation between PBs size (DCP1-GFP foci) and RAP2.4/OligoDT signals in 100 nm PBs size bins. For the line plot, data are means ± Sd denoted as the shaded area (N = 3 biological replicates with n = 10 randomly selected meristematic epidermal cells 5 DAG each). Fitted lines are also shown. AUs, arbitrary units. E) Micrographs from smFISH for detection of RAP2.4 (in magenta) counterstained in the presence or absence of cordycepin (cord, 1 h), in root meristematic epidermal WT cells 5 DAG expressing DCP1pro:DCP1-GFP. Nuclei were stained with DAPI (violet). Arrowheads denote small PBs with or without smFISH signal for mock or cordycepin treatments. Percentages on micrographs indicate small PBs (∼0.2 μm), with smFISH signal ± Sd. The 2 values were statistically different at P < 0.05 (n = 10 randomly selected meristematic epidermal cells; 1-way ANOVA). Insets on the right, indicate large PBs with smFISH RAP2.4 signal for mock and cordycepin treatments. The experiment was replicated 3 times (n = 10 randomly selected cells). Scale bars, 5 μm (insets, 0.2 μm). F) Graphical representation of PBs sizes in WT, dcp1-3, and scar1234 meristematic, epidermal root cells. Above each model, there is a graphical representation of the decay rate. scar1234 contains a larger range of turnover. SCAR2 is the main protein responsible for retracting DCP1 from PBs and thus, their dissolution (Liu et al. 2023).
An increase in protein disorder and RBPs in PBs correlates with RNA storability
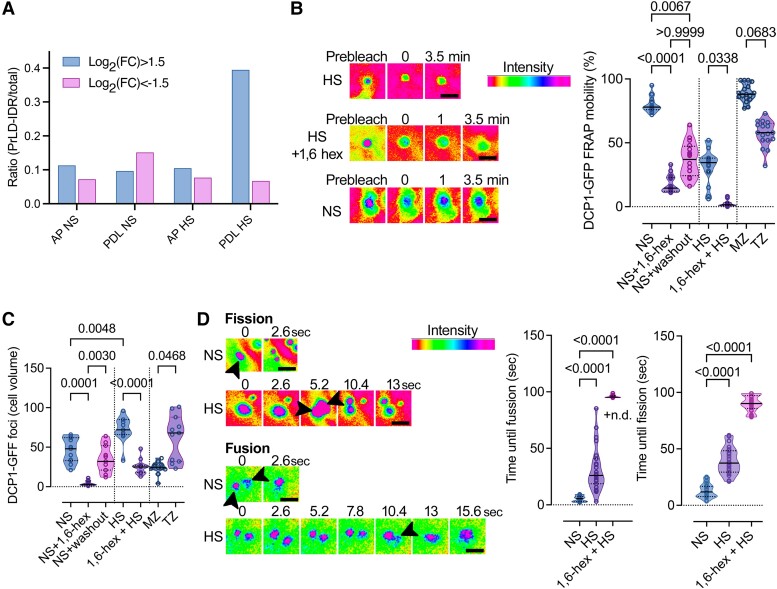
We next aimed to determine which features of PBs could be associated with increased PB size and RNA storage. In silico analyses of the PB proteome using the PLAAC algorithm (Dosztanyi et al. 2005) obtained through APEAL (Liu et al. 2023) revealed that only during HS, when PBs mainly increase in size, were they were enriched in LCRs. For these analyses, we used as an index the LCR length (in amino acids) vs. the corresponding total protein length (Fig. 7A; Supplemental File 1 for proteins with enriched LCRs). This result is consistent with the increase in levels of RBPs in PBs during HS (Supplemental Data Set 1); these proteins are known to contain LCRs in their RNA-interacting interfaces (Zagrovic et al. 2018). Hence, although disorder has been suggested as key for LLPS, we did not find that under NS conditions, PBs are richer in protein disorder than the corresponding controls.
Figure 7.
Content of PBs in proteins with LCRs during NS or HS and regulation of their dynamics. A) PrLDs and IDRs (LCRs) composition of APEAL (proteome)-enriched (log2FC > 1.5)/excluded(log2FC < −1.5) proteins [from (Liu et al. 2023)]. The ratio here represents the LCR length sum (PrLDs + IDRs) divided by the total protein length. The PDL step can identify the disordered part of the proteome in PBs (Liu et al. 2023). AP, affinity purification; PDL, proximity-dependent ligation of biotin. B) FRAP assays in the presence or absence of 1,6-hexanediol (“hex”) in NS or HS from wild type (WT) expressing DCP1pro:DCP1-GFP. The experiment was replicated more than 10 times. Note the lack of FRAP in the 1,6-hexanediol-treated sample. Right: relevant quantifications of mobile DCP1-GFP fraction from FRAP in the presence or absence of 1,6-hexanediol in NS or HS (and in washout experiments), and in MZ (meristematic) and TZ (transition) zones of the root. Data are means ± Sd (N = 6 biological replicates with n = 3 randomly selected meristematic epidermal cells 5 DAG each), and significance was calculated by an unpaired t-test (vs. the NS; 2-tailed P values are indicated). Scale bars, 400 nm. C) Quantifications of PBs in the presence or absence of 1,6-hexanediol (“hex”) in NS or HS from WT expressing DCP1pro:DCP1-GFP (cell volume in μm3). Data are means ± Sd (N = 3 biological replicates with n = 20 randomly selected meristematic epidermal cells 5 DAG each); significance determined by unpaired t-test (vs. the NS; 2-tailed P values are indicated). D) Super-resolution spinning disc microscopy images (combined with image deconvolution, ∼120 nm axial resolution at maximum acquisition speed of 0.1 s per frame) of DCP1 fusion and fission dynamics, in NS or HS conditions from WT expressing DCP1pro:DCP1-GFP. Arrowheads indicate PBs; in fission, the 2 produced PBs are indicated for HS, while in fusion the arrowhead indicates in HS the coalescence. Scale bars, 200 nm. Right: quantification of corresponding fusion and fission events in the presence or absence of 1,6-hexanediol (“hex”) in NS or HS (N = 10 biological replicates with n = 3 randomly selected meristematic epidermal cells 5 DAG each; for 1,6-hexanediol N = 2 biological replicates with n = 1 randomly selected meristematic epidermal cells 5 DAG each; significance determined by Mann–Whitney). n.d., not detected.
As HS increased the LCR contents of PBs, we reasoned that this could lead to a change in their properties. We compared the properties of large vs. small PBs during NS or HS and in different root epidermal zones (i.e. meristematic vs. differentiating) through FRAP and upon 1,6-hexanediol treatments. PBs under HS (or large ones, >300 nm) showed reduced FRAP rates and partial insensitivity to 1,6-hexanediol (Fig. 7, B and C). To further confirm this finding, we followed the fusion/fission dynamics of PBs in both NS and HS, as high rates of these 2 parameters indicate high liquidity (Linsenmeier et al. 2022). Accordingly, cell differentiation (i.e. transition zone) or HS significantly increased the time needed for fusion/fission, suggesting that HS (and thus size) associated with reduced liquidity of PBs (Fig. 7D). Furthermore, as HS induced an increase in the levels of proteins in PBs containing canonical RNA-binding domains (Supplemental Data Set 1), a combination of increased RNA-binding capacity, RNA content, and LCRs could contribute to PB solidification.
RNAs can localize together with their encoded proteins in PBs
During the analysis of LCR content and its link to RNA storability, we serendipitously observed an enrichment of the m6A-readers YTH-homologs EVOLUTIONARY CONSERVED C-TERMINAL REGION (ECTs) mRNAs in PBs and at the same time, the corresponding homologous encoded proteins (Supplemental Data Set 8; Liu et al. 2023). Intriguingly, this observation is reminiscent of observations in animals that many RNAs and their corresponding encoded proteins are stored in PBs (Hubstenberger et al. 2017). Hence, our observation prompted us to examine whether plant PBs also have the ability to store RNAs/cognate proteins and to what extent. As was done previously (Hubstenberger et al. 2017), we integrated the datasets for the PB proteome from APEAL (PBs-enriched proteins) and T-RIP (PBs-enriched RNAs) using the GO categories “biological processes” or “cellular components.” We next ran a GO analysis of the common proteins/cognate RNAs (Fig. 8, A and B). We evaluated hierarchical linkages between these terms using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins; FDR < 0.05). Through GO-STRING, we assigned hubs of proteins/cognate RNAs involved in cell wall remodeling and membrane remodeling and wounding/ethylene responses associated with secondary metabolism for defense. As expected, we identified a high enrichment of PBs-related GO terms (more than the ones observed for subnetworks above) related to RNA/nucleic acid metabolism and decapping [Fig. 8, A and B (green); Supplemental Fig. S12, A and B]. On the other hand, we did not identify a similar enrichment of proteins/cognate RNAs from APEAL and T-RIP datasets using depleted proteins/RNAs or when using a permutated analysis between enriched/depleted datasets or with randomly enriched RNA/protein molecules. Examples of highly enriched cognate RNA/proteins include those related to actin-dynamics (e.g. ADFs, log2FC = 3.95; see also ADF9 above), veins and associated peptides [e.g. CLAVATA3/Embryo Surrounding Region-Related (CLE), log2FC = 2.7], and cell wall [e.g. pectin methyl-esterase inhibitor 9 (PMEI9), log2FC = 2.5 (Sorek et al. 2015)]. We also noticed GO terms related to pectin, diverse (a)biotic stresses (i.e. bacterial, fungi, insects, heat, and salt stress), and a wounding/regeneration cluster with processes related to immunity and ethylene. These GO terms are consistent with the phenotypes reported so far for plant decapping mutants, showing phenotypes relevant to actin dynamics (e.g. trichome defects), veins, and cell wall remodeling (Xu et al. 2006; Xu and Chua 2009).
Figure 8.
GO terms identified for common proteins/cognate RNAs in PBs. A) and B) GO analysis of enriched cellular component (A) and biological process (B) terms of common proteins/cognate of PBs-enriched RNAs and proteins. Cocluster analyses of APEAL and T-RIP enrichments reveal hubs of cell wall remodeling, membrane remodeling, and wounding/ethylene responses associated with secondary metabolism for defense and RNA metabolism (GO terms denoted with green text). FDR, false discovery rate.
We next queried whether cognate RNA/protein interactions in PBs depend on the affinity of certain amino acids to codons enriched in pyrimidines (PYR) or purines (PUR). The direct interactions between codons (in RNA) and the amino acids they code for have been proposed as a potential mechanism that drives cognate RNA/protein interactions (Zagrovic et al. 2018). These interactions are based on a proposed affinity scale of PUR/PYR-amino acids interactions. Moreover, the lack of protein order (an increase of LCRs) during HS motivated further this analysis; residues in LCRs are not occupied in interactions aiming at retaining a rigid 3D structure but could be free for interactions with codons (Zagrovic et al. 2018). Yet, we did not observe differences between PYR/PUR matching with distributions of residues belonging to IDRs or other protein segments when comparing PBs-enriched/non-enriched RNAs/protein pairs (Supplemental Fig. S13, A to C). Furthermore, LCRs in RBPs may promote the association of RNAs with PBs. In accordance, we observed that RNA targets of the RBP GRP7 [Gly Rich Protein 7, AT2G21660, (Meyer et al. 2017)], which is rich in LCRs and localizes to PBs (Liu et al. 2023), could also be found in PBs (Supplemental Data Set 9). Our results thus suggest that the recruitment of RBPs and the increase in LCRs in PBs are associated with a marked increase in RNAs. The exact mechanism by which RNAs and cognate proteins localize in PBs, while others escape from PBs (in HS), merits further investigation.
The SCAR/WAVE-DCP1 axis regulates RNA dynamics
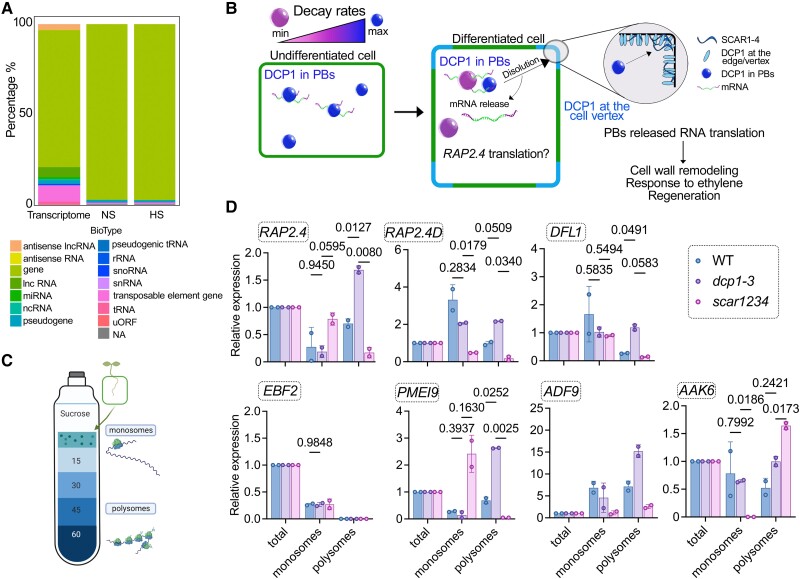
In nonplants, PBs compete with polysomes for RNAs (Cougot et al. 2013). Accordingly, we observed that most RNAs stored in PBs encode proteins, both in NS and HS, and thus could in principle affect translation (Fig. 9A; 95% in PBs vs. 80% in the total transcriptome). We thus aimed to evaluate whether PBs could affect their translation dynamics. As we previously showed that the dissolution of PBs is at least partially controlled by the SCAR/WAVE-DCP1 axis, we assumed that this axis could modulate the PBs-to-translation shuttling for at least a portion of PBs-enriched RNAs (Fig. 9B, model). We thus queried whether PBs-enriched RNAs from seedlings of wild type, the scar1234 mutant (more/larger PBs), or the weak allele dcp1-3 (fewer PBs) are depleted of polysomes where RNAs are translationally active (Jang et al. 2019). To isolate polysomes, we refined a ribosome profiling approach on a sucrose cushion with 12 fractions (Yanguez et al. 2013). Density ribosome profiles of scar1234 and dcp1-3 matched that of the wild type (Supplemental Fig. S14, A and B; A260), suggesting that profiling would be rather specific and not reflect a broad translational modulation shown for other mutants (Cho et al. 2022). We used RT-qPCR of combined fractions of monosomes (first 4 fractions of the 12 collected in total) and polysomes (fractions 5 to 12) to determine the levels of mRNAs in wild type, scar1234, and dcp1-3 compared with the total mRNA levels of the same samples corresponding to the 10% (v/v) region of the initial polysome profiling extract (Fig. 9, C and D). We selected the PBs-enriched RNAs with varying enrichment levels and the relevant networks wounding/regeneration and ethylene: EBF2, DFL1, RAP2.4, RAP2.4D, AAK6 (ARABIDOPSIS ADENYLATE KINASE 6), and PMEI9 (Supplemental Data Set 10). For normalization of RNA levels between fractions, we used TUBULIN ALPHA-4 CHAIN, which was excluded from PBs and showed equivalent levels among fractions (Supplemental Fig. S14B). We observed a relatively good inverse correlation between polysome occupancy and PBs-enrichment for most but not all RNAs [Fig. 9D, e.g. ADF9 had a log2(FC)NS = 3.96 and was enriched in polysomes at the same time in the WT].
Figure 9.
Regulation of translational dynamics by the SCAR/WAVE-DCP1 axis. A) RNA classes of PBs-enriched RNAs (%). Total RNA corresponds to the distribution of RNAs in the whole transcriptome. NA, not available or not defined. Lnc, long noncoding; mi, micro; nc, noncoding; sno, small nucleolar; uORF, upstream open reading frame. B) Model for the dissolution of PBs by the SCAR/WAVE-DCP1 axis. The SCAR/WAVE (mainly through SCAR2) entraps DCP1 at the edge/vertex (through SCAR2), which leads to PBs dissolution and consecutive mRNA release for translation, e.g. of RAP2.4. Processes like cell wall remodeling-related processes, such as response to ethylene and regeneration could be affected. The color-coding for PBs corresponds to their ability for RNA decay. C) Cartoon showing the polysome profiling approach. The denser part [60% (w/v) sucrose] corresponds to RNAs associated with polysomes where they are mostly translated (Jang et al. 2019). D) RT-qPCR of RAP2.4, APUM, EBF2, DFL1, and PMEI9 from total RNA, monosome, or polysome fractions in wild type (WT), dcp1-3, or scar1234. Data are means of 2 independent experiments with 2 technical replicates (n = 2 RT-qPCR assays). The data were normalized against TUBULIN 4 (TUA4, reference gene) and the levels of RNAs in the input, which is denoted as “total”; significance was determined by 2-way ANOVA using a Geisser-Greenhouse correction (due to unequal variance) and Fisher's exact test for multiple comparisons.
In this context, we decided to further study RAP2.4 because (i) it is rapidly decayed through the decapping complex [results herein and (Sorenson et al. 2018)], (ii) showed enrichment in GFP-trap experiment with the DCP1pro:GFP-DCP1 lines (i.e. in “native PBs”), (iii) correlated well with PBs-depletion/translation, and (iv) as mentioned above, fits in 2 central PBs subnetworks but also in an RNA/cognate protein hub, “response to wounding/regeneration” and “ethylene” [Fig. 2 for GO analysis and Fig. 7; (Lin et al. 2008; Iwase et al. 2011)]. According to the polysome and decay results, the RAP2.4 smFISH signal inversely correlated to the activity of the SCAR/WAVE-DCP1-axis; this axis is more active at the root transition zone onwards [i.e. dissolving PBs (Liu et al. 2023); Fig. 10A]. In more detail, dcp1-3 (or dcp5-1) had increased cytoplasmic levels of RAP2.4 mRNA throughout the root, unlike wild type, whose RAP2.4 mRNA level in this region was reduced. On the other hand, scar1234 also showed reduced RAP2.4 mRNA levels at the transition zone but to a greater extent than wild type. This observation is consistent with the finding that this background contains larger PBs with greater storability, as well as an increased number of PBs that execute decay (see also Fig. 4). Hence, the SCAR/WAVE-DCP1-axis could also target and dissolve small PBs to modulate decay. We also observed high nuclear levels of RAP2.4 mRNA in scar1234, suggesting that high decay rates of RAP2.4 mRNA induce feedback regulation of transcription, as suggested for other RNAs in plants (Manavella et al. 2023). Accordingly, RAP2.4 mRNA decay depends on the PB-associated nuclease XRN4, as in the loss-of-function xrn4-5 mutants, RAP2.4 mRNA levels increased (Fig. 10A). Overall, these results suggest that an RNA-specific combination of storage, translation, and decay by PBs modulate RNA turnover.
The SCAR/WAVE-DCP1 axis modulates ethylene signaling
Given the link of RAP2.4 with ethylene and PBs, we asked whether the SCAR/WAVE-DCP1-axis modulates ethylene signaling. PBs sequester and degrade EBF1 and EBF2 through XRN4 [also known as ETHYLENE INSENSITIVE 5; (Li et al. 2015; Merchante et al. 2015)]. Accordingly, EBF2 mRNA levels increased by 50% in dcp1-3 vs. the wild type, as has been observed in xrn4 (Potuschak et al. 2006), as both mutants showed reduced PBs-depended RNA decay. As predicted above using GO-STRING (Fig. 8), we observed that ethylene or HS led to the localization of both the cognate RNA/protein RAP2.4 pair to PBs (Supplemental Fig. S15, A and B). Furthermore, ethylene could induce the removal of SCAR/WAVE from the vertex in the transition zone (and above) of the root, leading to an increase in the size/number of PBs (Supplemental Fig. S16). Accordingly, scar1234 and brk1 (BRICK1), another SCAR/WAVE component mutant, were sensitive to the application of the ethylene precursors ACC or Ethephon [(2-chloroethyl)phosphonic acid, known also as “Ethrel”], as revealed by increased hypocotyl elongation under light conditions [as in the rap2.4 mutant, (Lin et al. 2008)], unlike decapping mutants, which showed reduced sensitivity to ethylene (Fig. 10B). These results provide a long-sought link between ethylene and PBs.
Discussion
Here we propose T-RIP as an approach to capture RNAs in plant condensates using PBs as a test case. T-RIP is a TurboID proximity biotinylation approach that is preferred over the APEX2-RIP, which showed poor specificity under our conditions [as suggested also in (Xu et al. 2023)]. Through T-RIP, we identified the PBs-enriched RNAs and classified them into 5 subnetworks. Except for the expected networks, given the known functions of PBs in RNA turnover (Xu et al. 2006; Xu and Chua 2009), we identified networks related to plant hormones, the plasma membrane/cell wall, and wounding-tissue regeneration. The composition of PBs highly depended on conditions, as under HS, we observed significantly more and different RNAs associated with PBs compared with NS (ca. 2-fold). Furthermore, PBs-enriched RNAs are usually shorter than other RNAs, with less complex 5′-UTRs. This result suggests that PBs may stabilize RNAs by recruiting those with less structured UTRs, which are likely less stable or more accessible to regulators (Hubstenberger et al. 2017).
Beyond the aforementioned primary sequence features, PBs were rich in proteins involved in reading the post-transcriptional modification m6A (i.e. ECTs) and in RNAs carrying this modification during HS. m6A has been linked to the modulation of RNA structure, stability, and translation (Reichel et al. 2019). Although we did not find a correlation between the localization of RNAs to PBs and m6A-RNA modifications using the m6A writer mutant fip37-4, we found that m6A may promote the stability of PBs. We thus confirm recent findings in plants suggesting that m6A could affect the stability of condensates (Lee et al. 2022).
Could the readers of m6A ECTs instruct to some extent the composition of PBs in RNA by selecting the RNAs recruited in PBs? Although ECTs could in principle affect to some extent the PBs-enriched RNAs, we favor the idea that their main function is to “read” m6A and use it as a scaffold to promote the stability of PBs. Our interpretations do not contradict earlier findings about the regulation of turnover of RNAs by m6A (Anderson et al. 2018), as proteins such as ECTs may bind methylated RNAs and further stabilize them (leading also to an increased size of PBs). However, the exact mechanism of this modulation merits further investigation.
PBs have long been considered to be major RNA degradation sites because they contain decay factors (Xu et al. 2006; Xu and Chua 2009). Contrary to this common belief, we showed that PBs comprise 2 distinct populations: small and large ones. Using smFISH, we showed that large PBs store RNA while the smaller ones degrade it. Interestingly, the stored RNAs have shorter poly(A) tails likely due to de-adenylation. In plants and other eukaryotes, de-adenylation is required for the association of RNA with PBs (Chen and Shyu 2013). It would thus be interesting to study the links of PBs with the de-adenylation machinery, considering the association of this machinery with PBs (Liu et al. 2023).
The decay-to-storage transition of PBs (except for a change in size) likely also involves changes in the assembly of an active DCP1–DCP2 complex. In smaller PBs, the DCP1–DCP2 complex forms efficiently, while in larger PBs, DCP1 fails to interact with DCP2. This observation is consistent with the results of in vitro reconstitution of RNA decay complexes for nonplants, suggesting that small PBs carry most of the RNA decay (Tibble et al. 2021). Furthermore, as PBs grow, for example under HS, their LCR content increases, likely contributing to increased stability, further interactions with RNAs, and likely stereochemical hindrances that do not allow for efficient DCP1–DCP2 interactions. Further interactions with RNA in PBs during HS are likely due to an increased content of RBPs and are further promoted by the increased protein disorder found at RNA-binding interfaces (Zagrovic et al. 2018). The structural reconfigurations of DCP1, DCP2, or other components of the degradation complex could be important for active assembly of the decay complex. These reconfigurations could be halted by an increased LCR content, which in time brings increased solidity to PBs and thus reduced molecular movements.
The stored RNAs in PBs are likely dormant and they are not translated. However, certain stimuli (e.g. development) could promote the dissolution of PBs for the release and translation of RNAs. This release involves the SCAR/WAVE complex, which dissolves PBs. Accordingly, the lack of SCAR/WAVE (e.g. in scar1234 mutant) leads to increased PB number and size. Similarly, in nonplants, PBs-stored RNAs can be kept stable and translationally inert (Hubstenberger et al. 2017), but dissolution mechanisms that release these RNAs have remained unclear. Translational regulation from PBs, however, can be more complex. For example, PBs can exchange RNAs with SGs, where in nonplants translation can take place (Pitchiaya et al. 2019; Baumann 2021). Hence, PBs-enriched RNAs could be channeled to translation through SGs.
Subnetworks formed by RNAs and their encoded proteins in PBs could drive their storage and translational repression in mammalian cells (Hubstenberger et al. 2017). We showed that plant PBs are also rich in RNAs-cognate proteins. Most importantly, these networks are related to responses to diverse stresses (i.e. bacterial, fungi, insects, heat, and salt stress) and to a wounding/regeneration network that incorporates processes related to immunity, the cytoskeleton, and ethylene. These networks explain well the associated phenotypes for plant decapping mutants related to actin dynamics (e.g. trichome defects), veins, and cell wall remodeling (Xu et al. 2006; Xu and Chua 2009). Although interactions between codons (in RNA) and the amino acids they code for could drive this RNAs-cognate protein matching (Zagrovic et al. 2018), using bioinformatics, we refuted this hypothesis. However, low-dielectric environments like those present in PBs could make interactions between codons and amino acids important (Zagrovic et al. 2018). However, we still lack information about the dielectric environment of PBs.
As mentioned above, the production of ethylene is highly linked to wounding/regeneration (Marhavy et al. 2019). These processes likely converge on RAP2.4, which seems to modulate both these processes (Lin et al. 2008; Iwase et al. 2011). Ethylene directly modulates PB dissolution by inducing the degradation of the SCAR/WAVE complex. We speculate that this degradation depends on the ethylene-induced activation of the E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), which targets the SCAR/WAVE complex (Dyachok et al. 2011). Hence, ethylene may spatially restrict RAP2.4 by reducing PB dissolution. Furthermore, an additional regulatory layer could be present at the transcriptional level, pointing to crosstalk between PBs and transcription. The scar1234 mutant showed high nuclear levels of RAP2.4 mRNA, suggesting that high decay rates of RAP2.4 mRNA undergo feedback to transcription as a homeostatic mechanism. Similar feedback regulation has been suggested for other RNAs in plants (Manavella et al. 2023). The underlying mechanism, however, as well as how SCAR/WAVE induces PBs dissolution merit further investigation.
Collectively, we presented approaches for the study of RNA composition, properties, and activities of condensates in plants. We further suggest that certain properties of PBs regulate their ability to store RNA. This ability directly affects cellular protein levels and phenotypes by regulating the translation and turnover of selected RNAs. In animals, the fragile X mental retardation protein (FMRP, neuron generation) inhibits translation by binding to accessory proteins of the SCAR/WAVE complex (Kim et al. 2019). Another FMRP member form condensates that activate translation (Kang et al. 2022). We thus offer a different model for translational regulation by the SCAR/WAVE-DCP1 axis through the release of RNAs from processing “storage bodies.”
Our study has certain limitations. The use of an overexpression promoter to drive the expression of DCP1, which serves as bait in T-RIP, may result in a skewed determination of RNA levels in PBs. DCP1 overexpression leads to moderate changes at the transcriptome level, which could affect PBs enrichment. Although we confirmed the localization of selected RNAs in PBs (RAP2.4, DFL1, EBF2) through imaging and RT-qPCR using DCP1 driven by its native promoter, additional confirmatory studies are needed at a wider scale. Furthermore, through T-RIP, we also likely capture RNAs transiently associating with soluble and preformed DCP1–DCP2 complexes outside the PBs. These transient interactions may increase the number of RNAs classified as PBs-enriched. Finally, our approach is an indirect one, in the sense that it biotinylates proteins in PBs and does not directly biotinylate RNAs, which can lead to the loss of some RNAs.
Materials and methods
Plant material
All the plant lines used in this study were in the Arabidopsis (A. thaliana) Columbia-0 (Col-0) ecotype or Nicotiana benthamiana unless stated otherwise. Primers used for genotyping and cloning are described in Supplemental Table S1. The following mutants and transgenic lines used in this study were described previously: dcp1-1 (Xu et al. 2006), dcp1-3 (Martinez de Alba et al. 2015), dcp2-1 (Chantarachot et al. 2020), dcp5-1 (Xu and Chua 2009), xrn4-5 (Souret et al. 2004), brk1 (Dyachok et al. 2008), scar1234 (Dyachok et al. 2008), 35Spro:GFP-DCP1 (Gutierrez-Beltran et al. 2015), 35Spro:DCP2-YFP (Jang et al. 2019), fip37-4 (Bodi et al. 2012) and DCP1pro:DCP1-GFP (Yu et al. 2019), SCAR2pro:mCherry-SCAR2 (Chin et al. 2021). Seedlings carrying the TurboID (35Spro:sGFP-TurboID-HF) construct have been previously described (Liu et al. 2023). In all experiments, plants from the T1/F1 (colocalization experiments), T2/F2, or T3 (for physiological experiments) generations were used.
Plant growth and pharmacological treatments
Arabidopsis seeds were sterilized and germinated on half-strength Murashige and Skoog (MS) agar medium under long-day conditions (16 h light/8 h dark). In all experiments involving the use of mutants or pharmacological treatments, the medium was supplemented with 1% (w/v) sucrose or as otherwise specified. Arabidopsis plants for crosses, phenotyping of the above-ground parts, and seed collection were grown in soil (comprising of a blend of sphagnum moss peat, compost, worm castings, perlite, and a special “natural plant booster”) in an Aralab (FITOCLIMA D1200 PLH configuration) plant chamber at 22°C/19°C, 14-h-light/10-h-dark or 16-h-light/8-h-dark cycles, and a light intensity of 150 µE m−2 s−1 (4 × 11 W bulbs, tubular white LED). N. benthamiana plants were grown in Aralab (FITOCLIMA D1200 PLH configuration) at 22°C, 16-h-light/8-h-dark cycles, and a light intensity of 150 µE m−2 s−1(4 × 11 W bulbs, tubular white LED). Vertically grown 4 to 5-d-old Arabidopsis seedlings were transferred to and incubated in half-strength liquid MS medium containing the corresponding drugs for each specific time course treatment as indicated. Ethephon treatment was performed at a final concentration of 100 μM (from a 100 mM stock) in seedlings or N. benthamiana slices (leaf slices of the infiltrated area in the corresponding transient experiment) for 20 min. Long ethephon or 1-aminocyclopropane-1-carboxylate (ACC) treatment was performed on half-strength MS plates in a 10 μM final concentration (under light or in the dark). For 1,6-hexanediol treatments, 10% (v/v) aqueous solution was used. The stock solutions of 50 mM biotin, 100 mg/mL CHX were dissolved in DMSO. Cordycepin treatments were performed at a 1 mM final concentration in incubation buffer [15 mM sucrose, 1 mM piperazine-N, N′-bis(2-ethane sulfonic acid (PIPES) pH 6.25, 1 mM KCl, 1 mM sodium citrate] (Sorenson et al. 2018).
Bacterial strains, cloning, and constructs
Cloning of RAP2.4pro:RAP2.4-venusYFP was done according to standard In-fusion cloning procedures (Takara In-Fusion Snap Assembly Master Mix #638948), using the linearized pGWB601 vector. Primers used for this cloning can be found in Supplemental Table S1. The Escherichia coli strains NEB10 (New England Biolabs #C3019H) or NEB stable (New England Biolabs #C3040H; CRISPR constructs) were used for cloning. Electrocompetent Agrobacterium tumefaciens C58C1 RifR (pMP90) or GV3101 RifR bacterial cells (i.e. a cured nopaline strain commonly used for N. benthamiana infiltration) were used for electroporation and N. benthamiana infiltration.
Transient transformation of N. benthamiana
N. benthamiana plants were grown under normal light and dark regimes at 25°C and 70% relative humidity. Three- to 4-wk-old N. benthamiana plants were watered from the bottom ∼24 h before infiltration. Transformed Agrobacterium strain C58C1 RifR (pMP90) or GV3101 RifR harboring the constructs of interest were used to infiltrate N. benthamiana leaves and for transient expression of binary constructs by Agrobacterium-mediated transient infiltration of lower epidermal leaf cells. Transformed Agrobacterium colonies were grown for ∼20 h in a shaking incubator (200 rpm) at 28°C in 5 mL of yeast extract peptone medium (10 g/L yeast extract, 10 g/L peptone, 5 g/L NaCl and 15 g/L bacterial agar) supplemented with the appropriate antibiotics (i.e. 100 g/L spectinomycin). After incubation, the bacterial culture was transferred to 15-mL Falcon tubes and centrifuged (10 min, 3,500 × g, RT). The pellets were washed with 5 mL of infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.7), and the final pellet was resuspended in infiltration buffer supplemented with 200 μM acetosyringone. The bacterial suspension was diluted with infiltration buffer to adjust the inoculum cell density to a final OD600 value of 0.4. The inoculum was incubated for 2 h with agitation at 28°C before infiltration into N. benthamiana leaves by gentle pressure infiltration of the lower epidermis of leaves (fourth and older true leaves were used, and about 4/5-1/1 of their full size) with a 1-mL hypodermic syringe without a needle.
T-RIP method
All materials were RNase-free. Seven-day-old seedlings were submerged in 50 mL of 50 μM biotin (in 10 mM MgCl2, 10 mM MES pH 5.7) for 1 min under a vacuum. After a 24 h incubation on a new ½ MS plate, the plates were treated with HS (37°C, 2 h). Around 1 to 2 g of seedlings’ fresh weight was harvested in 1% (v/v) formaldehyde (FA) solution and a 15 min vacuum was applied. FA was quenched by adding glycine to a final concentration of 125 mM for 5 min under a vacuum. The plant material was rinsed 2× with RNase-free H2O and pulverized in liquid N2. Proteins were extracted as described for APEAL (Liu et al. 2023); the extraction buffer was supplemented with 20 U/mL RNase inhibitor (ThermoFisher Scientific, EO0381). The supernatant was loaded on a PD-10 column (Cytiva, GE17-0851 to 0801) for the removal of the excess biotin and incubated with equilibrated 100 μL Dynabeads MyOne Streptavidin (ThermoFisher Scientific, 65601) at 4°C for 2 h with gentle rotation. The beads were precipitated using a magnetic rack and washed 4 times with extraction buffer and 2 times with dilution buffer (extraction buffer without detergent). The RNA-protein complexes were eluted from the beads with 100 μL protease buffer (0.5 mg/mL Proteinase K, 20 U/mL RNase inhibitor in 30 mM Tris-HCl, pH 8.0) at 55°C for 30 min. RNA was extracted from the eluted beads with phenol: chloroform: iso-amyl alcohol (25:24:1) (v/v/v) (VWR, 136112-00-0), followed by precipitation with 1:1 (v/v) isopropanol with 0.1 volume of 3 M NaAc, pH 5.2 and 2 μL Glycoblue (ThermoFisher Scientific, AM9515) for 1 h. After centrifugation at 4°C, 16,000 × g for 20 min, the blue (due to the presence of Glycoblue) pellet was washed 2× with 75% (v/v) ethanol. The air-dried pellet was resuspended in 15 μL of DNase-, RNase-, and protease-free water. The concentration of RNA was determined using Qubit RNA HS Assay Kit (New England BioNordika BioLab, Q32852). All the RIP-RNA samples were treated with DNase I (ThermoFisher Scientific, EN0521) and a Ribosomal RNA depletion RiboMinus Plant Kit (ThermoFisher Scientific, A1083808). The RNA was measured with Qubit RNA HS Assay Kits again, and libraries were prepared with NEBNext Ultra II RNA Library Prep with Sample Purification Beads [Invitrogen Life Technologies (Ambion Applied Biosystem), E7775S] and NEBNext Multiplex Oligos for Illumina (Dual Index Primers Set 1) (New England BioNordika BioLab, E7600S). cDNA library quality was monitored with an Agilent DNA 7500 Kit (Agilent Technologies Sweden AB, 5067-1506). cDNA libraries were sequenced with a paired-end sequencing strategy to produce 2 × 150-bp reads using Novogene sequencers (Novogene, England).
GFP-RIP method
A modified T-RIP protocol was followed. Following fixation and formaldehyde quenching steps, as described for T-RIP, the lysates of 7-d-old seedlings of 35Spro:DCP1-GFP, DCP1pro:DCP1-GFP/dcp1-1, 35Spro:TAP-GFP (NS and HS) was incubated with ChromoTek GFP-Trap Agarose beads (Cat No. gta) for 2 h with gentle rotation at 4°C. The beads were precipitated and washed by centrifugation at 1,000 × g for 2 min. After Proteinase K treatment and RNA extraction, RNA samples were cleaned up with a Monarch RNA Cleanup Kit (50 μg, T2040L). Reverse-transcription was performed using a Minotech RT [Catalogue No. 801-1(10 KU)] kit following the manufacturer's instructions. The starting RNA material for the RT reaction was 300 ng in total per technical replicate, and the qPCR was done using SYBR Fast Green Master mix (Thermo Fisher, 4385612), following the exact protocol from the manufacturer, in a Bio-Rad CFX machine (Model No. CFX connect Optics Module; Serial No. 788BR01848). RT-qPCR experiments were performed according to MIQE guidelines (Bustin et al. 2009).
Immunoblotting
Infiltrated N. benthamiana leaves or Arabidopsis leaves and seedlings were harvested, and proteins were extracted. The tissue samples were flash-frozen in liquid N2 and kept at −80°C until further processing (not more than a month). The samples were crushed using a liquid N2-cooled mortar and pestle, and the pulverized powder was transferred to a 1.5 mL tube. Extraction buffer (EB), 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 2 mM ethylene diamine tetraacetic acid (EDTA), 5 mM dithiothreitol (DTT), 1 mM phenylmethylsulphonyl fluoride (PMSF), Protease Inhibitor Cocktail (Sigma-Aldrich, P9599), and 0.5% (v/v) octyl phenyl-polyethylene glycol (IGEPAL CA-630, Sigma-Aldrich) was added to the sample. The lysates were precleared by centrifugation at 16,000 × g at 4°C, for 15 min, and the supernatant was transferred to a new 1.5 mL tube. This step was repeated 2×, and the protein concentration was determined using an RC DC Protein Assay Kit II (Bio-Rad, 5000122). Laemmli buffer was added at a 1:2 ratio, and equivalent amounts of protein (∼30 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (1.0 mm thick 4% to 12% gradient polyacrylamide Criterion Bio-Rad) in 3-(N-morpholino) propane sulfonic acid (MOPS) buffer (Bio-Rad) at 150 V. Subsequently, the proteins were transferred onto a polyvinylidene fluoride (Bio-Rad) membrane with a 0.22 μm pore size. The membrane was blocked with 3% (w/v) BSA fraction V (ThermoScientific) in Phosphate Buffer Saline-Tween 20 (PBS-T) for 1 h at room temperature, followed by incubation with horseradish peroxidase (HRP)-conjugated primary antibody at room temperature for 2 h (or primary antibody for at room temperature for 2 h followed by the corresponding secondary antibody at RT for 2 h). The following antibodies were used: streptavidin-HRP (Sigma-Aldrich; 1:25,000), and FLAG-HRP (Sigma-Aldrich, A8592, 1:2,000). Chemiluminescence was detected with ECL Prime Western Blotting Detection Reagent (Cytiva, GERPN2232) and SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific, 34094).
Detection of RNA molecules by whole-mount-smFISH
The mRNA probes used were selected following the criteria (i) roughly 18 to 22 bases long, and (ii) GC content close to 45%. Furthermore, individual probes within a set had at least 2 nucleotides of space between their target regions. The Stellaris Probe Designer version 4.2 (Biosearch Technologies) was used for probe design and ordering (https://www.biosearchtech.com/stellaris-designer). Localization was performed following the protocols from Stellaris RNA fish (Protocol for Arabidopsis root meristems) and (Duncan et al. 2016). The roots of 4 to 5-d-old Arabidopsis seedlings were fixed for 2 h in fixation buffer (50 mM piperazine-N,N'-bis(2-ethane sulfonic acid (PIPES), pH 7.2, with 20 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N', N'-tetraacetic acid) and 20 mM MgSO4 containing 2% w/v paraformaldehyde, 0.1% Triton X-100 and 400 μM Maleimidobenzoyl-N-hydroxy succinimide ester (ThermoFisher Scientific, 22311), washed 2 times with 1× Phosphate-buffered saline (PBS) buffer, and incubated for 4 d in ClearSee solution (xylitol powder [10% (w/v)], sodium deoxycholate [15% (w/v)] and urea [final 25% (w/v)] in RNase-free water) (Kurihara et al. 2015). The seedlings were washed 2× with 1× Phosphate-buffered saline (PBS) buffer, followed by cell wall digestion at 37°C for 30 min [in 0.2% (w/v) driselase and 0.15% (w/v) macerozyme]. The roots were washed 2 times with washing buffer (50 mM piperazine-N, N'-bis(2-ethane sulfonic acid (PIPES), pH 7.2, with 20 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid) and 20 mM MgSO4) and incubated in permeabilization buffer (3% v/v octylphenyl-polyethylene glycol IGEPAL CA-630,10% dimethyl sulfoxide, 50 mM piperazine-N,N'-bis(2-ethane sulfonic acid (PIPES), pH 7.2, with 20 mM EGTA and 20 mM MgSO4) at 37°C for 30 min. The roots were washed 2× with wash buffer [10% formamide and 2 × saline-sodium citrate (SSC)], 5 min per wash. The slides were incubated with 100 μL 250 nM of probes (in hybridization solution: 10% dextran sulfate, 2 × SSC, and 10% formamide) at 37°C for 4 h or overnight hybridization at 4°C in the dark. Samples were washed 2× with wash buffer for 5 min each. The slides were incubated with 100 μL nuclear stain 4,6-diamidino-2-phenylindole (DAPI; 100 ng mL−1) for 30 min at 37°C in the dark. Excess DAPI was removed and 100 μL 2 × SSC was added and then washed away. A drop of Vectashield antifade mounting medium was added and sealed with a coverslip and nail polish. A Zeiss 800 confocal microscope was used for imaging, using a 63× NA = 1.4 water-corrected immersion objective, and the wide-field mode was used to obtain all images with standard settings. The following wavelengths were used for fluorescence detection: for probes labeled with Quasar 570, an excitation line of 561 nm was used and the signal was detected at wavelengths 570 to 640 nm; for DCP1-GFP an excitation line of 488 nm was used and the signal was detected at wavelengths of 530 to 550 nm; for DAPI an excitation line of 405 nm was used and the signal was detected at wavelengths of 420 to 460 nm.
Quantification of fluorescence intensity, FRAP, and FRET
To create the most comparable lines to measure the fluorescence intensity of reporters in multiple mutant backgrounds, we crossed homozygous mutant bearing the marker with either a WT plant (outcross to yield progeny heterozygous for the recessive mutant alleles and the reporter) or crossed to a mutant only plant (backcross to yield progeny homozygous for the recessive mutant alleles and heterozygous for the reporter). Fluorescence was measured as a mean integrated density in regions of interest (ROIs) with the subtraction of the background (a proximal region that was unbleached and had less signal intensity than the signal of the ROI region). FRAP mode of Zeiss 780 ZEN software was used for the acquisition of 3 prebleach images, one bleach scan, and 96 postbleach scans (or more). Bleaching was performed using a 488 nm laser line at 100% transmittance and 20 to 40 iterations depending on the region and the axial resolution (iterations increased in deeper tissues to compensate for the increased light scattering). In FRAP, the width of the bleached ROI was set at 2 to 10 µm. Pre- and postbleach scans were at minimum possible laser power (0.8% transmittance) for 458 or 514 nm (4.7%) and 5% for 561 nm; 512 × 512 8-bit pixel format; pinhole of 181 μm (>2 Airy units) and zoom factor of 2.0. The background values were subtracted from the fluorescence recovery values, and the resulting values were normalized by the first postbleach time point and divided by the maximum fluorescent time-point set maximum intensity as 1. The objective used was a Plan-Apochromat 20× with NA = 0.8 M27 (Zeiss). The following settings were used for photobleaching of DCP1: 10 to 20 iterations for DCP1-GFP; 10 to 60 s per frame; 100% transmittance with the 458- to 514-nm laser lines of an argon laser. Pre- and postbleach scans were at the minimum possible laser power (1.4% to 20% transmittance) for the 488 nm and 0% for all other laser lines, 512 × 512 pixel format, and zoom factor of 5.1. The fluorescence intensity recovery values were determined, then the background values were subtracted from the fluorescence recovery values, and the resulting values were normalized against the first postbleach time point. FRET analyses were done as described previously (Liu et al. 2023).
PLA protocol
For PLA immunolocalization, the primary antibody combinations were diluted 1:200 for α-FLAG mouse (Sigma-Aldrich, F1804) and α-GFP rabbit (Millipore, AB10145). The antibodies were incubated at 4°C. Roots were then washed with microtubule-stabilizing buffer (MTSB): 50 mM piperazine-N,N'-bis(2-ethane sulfonic acid (PIPES), 5 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N-tetraacetic acid), 2 mM MgSO4, 0.1% [v/v] Triton X-100) and incubated at 37°C for 3 h either with α-mouse plus and α-rabbit minus for the PLA assay (Sigma-Aldrich, 681 Duolink). PLA samples were then washed with MTSB and incubated for 3 h at 37°C with ligase solution as described (Teale et al. 2021). Roots were then washed 2× with buffer A (Sigma-Aldrich, Duolink) and treated at 37°C for 4 h in a polymerase solution containing fluorescent nucleotides as described (Sigma-Aldrich, Duolink). Samples were then washed 2× with buffer B (Sigma-Aldrich, Duolink), with 1% (v/v) buffer B for another 5 min, and then the specimens were mounted in Vectashield (Vector Laboratories) medium. Root tips were imaged under a Zeiss 780 confocal laser scanning microscope using a 63× NA = 1.6 oil-corrected immersion objective.
Ribosome profiling
A previously described protocol was used (Yanguez et al. 2013) with some modifications. All materials were RNase-free, and all buffers were autoclaved for 15 min. Two hundred milligrams of seedlings grown on half-strength MS plates were used as a starting material. Tissue lysis was performed with a modified Polysome Extraction Buffer (0.2 M Tris-Cl pH 9.0, 0.2 M KCl, 25 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid) 35 mM MgCl2, 1% detergent mix from a 20% stock solution; 20% (w/v) polyoxyethylene(23)lauryl ether (Brij-35), 20% (v/v) Triton X-100, 20% (v/v) octyl phenyl-polyethylene glycol (IGEPAL CA-630), 20% (v/v) polyoxyethylene sorbitan monolaurate 20 (Tween 20), 1% (v/v) polyoxyethylene 10 tridecyl ether (PTE), 5 mM DTT, 1 mM PMSF, 50 μg/mL CHX, 50 μg/mL chloramphenicol). The ultracentrifuge step was performed in 3 mL final volume tubes and with a Sorvall TST60.4 swing-out rotor. RNA extraction was done with TriZol from the 12 fractions collected, and the samples were cleaned up using a Monarch RNA Cleanup Kit (50 μg) from New England Biolabs (NEB). Optical densities (OD260) were used to calculate RNA concentration. Reverse-transcription was performed using Minotech RT [Catalogue No. 801-1(10 KU)] kit following manufacturer instructions. The starting RNA material for the RT reaction was at 40 ng in total per technical replicate, and the qPCR was done using SYBR Fast Green Master mix (Thermo Fisher, 4385612), following the exact protocol from the manufacturer, in a Bio-Rad CFX machine (Model No. CFX connect Optics Module; Serial No. 788BR01848). RT-qPCR experiments were performed according to MIQE guidelines (Bustin et al. 2009).
Determination of decay levels
Five-day-old seedlings were treated with cordycepin (Sigma, C3394) according to (Sorenson et al. 2018) and for the following time points: t0, 60 and 240 min. RNA was extracted with a Spectrum Plant Total RNA Kit (Sigma, STRN250). Reverse-transcription was performed using a Minotech RT [Catalogue No. 801-1(10 KU)] kit following manufacturer instructions. The starting RNA material for the RT reaction was at 500 ng in total per technical replicate, and the qPCR was done using SYBR Fast Green Master mix (Thermo Fisher, 4385612), following the exact protocol from the manufacturer, in a Bio-Rad CFX machine (Model No. CFX connect Optics Module; Serial No. 788BR01848). RT-qPCR experiments were performed according to MIQE guidelines (Bustin et al. 2009).
N 6-methyladenosine (m6A) RNA purification
m6A IP was performed with 5-d-old seedlings using an EpiMark N6-methyladenosine enrichment kit (NEB) starting from 4 μg of total RNA. The technical control for the IP was the one suggested by the manufacturer (m6A modified control RNA from Gaussia luciferase). Reverse-transcription was performed using a Minotech RT [Catalogue No. 801-1(10 KU)] kit following the manufacturer's instructions. The starting RNA material for the RT reaction was at 45 ng in total per technical replicate, and the qPCR was done using SYBR Fast Green Master mix (Thermo Fisher, 4385612), following the exact protocol from the manufacturer, in a Bio-Rad CFX machine (Model No. CFX connect Optics Module; Serial No. 788BR01848). RT-qPCR experiments were performed according to MIQE guidelines (Bustin et al. 2009).
Visualization of networks and analyses
For visualization, Cytoscape 3.5.1 was used. Tab-delimited files containing the input data were uploaded. The default layout was an edge-weighted spring-embedded layout, with the NormSpec used as edge weight. Nodes were manually rearranged from this layout to increase visibility and highlight specific proximity interactions. The layout was exported as a PDF and converted to a TIFF file.
In silico data analyses
For RNAs that are enriched in PBs, the log2 (DCP1/GFP) value (from the RNA-seq dataset) was used, as an indicator of the enrichment of each RNA with PBs. The “Fold_enrichment” values from the graphs, which are associated with m6A modifications, are referred to as the average “Fold_enrichment” value of each RNA from the given dataset (Parker et al. 2020). In brief, the m6A modifications in the Arabidopsis VIRILIZER (vir-1) mutant, which is impaired in a conserved m6A component of the writer complex, was compared with a line expressing VIR fused to GFP (VIR complemented; virC), which restores VIR activity in the vir-1 background. To provide the m6A sites, the differential error rate analysis approach was used (using the base-calling) to compare the mutant (defective m6A) and VIR-complemented lines. Through this approach, 17,491 sites with an FC of >2 error rate (compared with the TAIR10 reference base) in the VIR-complemented line were identified. The “Fold_enrichment” values indicate the statistical importance of the detected RNA to be methylated to one or more sites. Information about the total length of the GC% of 5′ and 3′ UTR was retrieved from the TAIR database (https://www.arabidopsis.org/) and plotted against the corresponding log2(DCP1/GFP) values.
Quantification and statistics
Statistical data and the corresponding measurements used for each graph in this study can be found in Supplemental Data Set 11. All statistical data show the mean ± Sd of at least 3 biologically independent experiments or samples unless otherwise stated. N denotes biological replicates, and “n” technical replicates or population size. Statistical analyses were performed in GraphPad (https://graphpad.com/) or R studio (R-project.org). Each data set was tested to determine whether it followed normal distribution when N ≥ 3 using the Shapiro normality test. The significance threshold was set at P < 0.05 (significance claim), and the exact values are shown in the graphs. Graphs were generated by GraphPad Prism or R. Details of the statistical tests applied, including the choice of the statistical method, are shown in the corresponding figure legends. In boxplots or violin plots, upper and lower box boundaries represent the first and third quantiles, respectively, horizontal lines mark the median and whiskers mark the highest and lowest values. Figures relating to sequence properties i.e. Codon Usage Bias, were created using custom Python (https://www.python.org/) and R scripts. Associated boxplots follow the same characteristics as stated previously, and any overlying data points denote the included values. In histograms, vertical lines refer to the median of the presented distributions whereas boxplots, when present, are used as a visual alternative for statistical purposes. In scatterplots, existing lines represent the loess-smoothed representations of the underlying points. Kernel density plots were created using a Gaussian kernel. Finally, the height of related bar plots accounts for the median value in all cases. Displayed P-values in the case of multiple-comparison testing were calculated using a Dunn's post hoc (Bonferroni; FDR < 0.05) test after a statistically significant Kruskal–Wallis ANOVA (P-value < 0.05). In the case of pairwise hypothesis testing, a Mann–Whitney U test was used instead (P-value < 0.05).
Image analyses and preparation
Image analyses and intensity measurements (Integrated Density) were done using Fiji v. 1.52 software (rsb.info.nih.gov/ij). The intensities were normalized for relative values against the background signal intensity in raw images/micrographs. The calculations were done with arbitrary units (denoted as AUs). The dwell time rate of tagged proteins in FRAP experiments was calculated by the single exponential fit (Moschou et al. 2016; Deli et al. 2022). Colocalization was analyzed using Pearson correlation coefficient (PCC) statistics Spearman or Manders analyses produced similar results (French et al. 2008). Images were prepared in Adobe Photoshop v. 2023. Time series movies were compressed, corrected, and exported as.avi extension files. The nonspecific fluorescence decay was corrected using Fiji and default options using the bleaching correction tool. smFISH images were corrected using Dust and Scratches filter with pixel width set to 1.0 and threshold to 0. Levels were corrected in the RGB channel and midtones were adjusted to increase the signal-to-noise ratio.
Accession numbers
Accession numbers and names of the genes mostly referred to this study are available in the Supplemental Data Set 10. The RNA sequencing data of the T-RIP and total RNA datasets have been submitted to the BioStudies database (Sarkans et al. 2018) under the accession number S-BSST1096 and can be accessed through the following link: https://www.ebi.ac.uk/biostudies/studies/S-BSST1096?key=58afa48a-c6c2-4366-b059-d2504392a2b2. Python scripts may be retrieved from https://github.com/Nwntastz/RNA_proxitome_paper.
Supplementary Material
Acknowledgments
We thank Elison B Blancaflor, Rupert Frey, Laurie Smith, Pamela J Green, Nam-Hai Chua, Julia Bailey-Serres, Alexis Maizel, Ping He, Shu-Hsing Wu for kindly providing published materials and Fanourios Mountourakis from Moschou's lab for fruitful discussions.
Contributor Information
Chen Liu, Department of Biology, University of Crete, Heraklion 70013, Greece; Department of Molecular Sciences, Uppsala BioCenter, Swedish University of Agricultural Sciences and Linnean Center for Plant Biology, Uppsala 75007, Sweden.
Andriani Mentzelopoulou, Department of Biology, University of Crete, Heraklion 70013, Greece; Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece.
Ioannis H Hatzianestis, Department of Biology, University of Crete, Heraklion 70013, Greece; Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece.
Epameinondas Tzagkarakis, Department of Biology, University of Crete, Heraklion 70013, Greece.
Vasileios Skaltsogiannis, Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece.
Xuemin Ma, Umeå Plant Science Centre (UPSC), Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), Umeå, Sweden.
Vassiliki A Michalopoulou, Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece.
Francisco J Romero-Campero, Department of Computer Science and Artificial Intelligence, Universidad de Sevilla, Avenida Reina Mercedes s/n, Seville 41012, Spain; Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla and Consejo Superior de Investigaciones Científicas (CSIC), Seville, Spain.
Ana B Romero-Losada, Department of Computer Science and Artificial Intelligence, Universidad de Sevilla, Avenida Reina Mercedes s/n, Seville 41012, Spain; Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla and Consejo Superior de Investigaciones Científicas (CSIC), Seville, Spain.
Panagiotis F Sarris, Department of Biology, University of Crete, Heraklion 70013, Greece; Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece; Biosciences, University of Exeter, Exeter, UK.
Peter Marhavy, Umeå Plant Science Centre (UPSC), Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), Umeå, Sweden.
Bettina Bölter, Ludwig Maximilians University Munich, Plant Biochemistry, Großhadernerstr. 2-4, Planegg-Martinsried 82152, Germany.
Alexandros Kanterakis, Institute of Computer Science, Foundation for Research and Technology-Hellas, Heraklion, Greece.
Emilio Gutierrez-Beltran, Instituto de Bioquímica Vegetal y Fotosíntesis, Universidad de Sevilla and Consejo Superior de Investigaciones Científicas (CSIC), Seville, Spain; Departamento de Bioquímica Vegetal y Biología Molecular, Facultad de Biología, Universidad de Sevilla, Sevilla, Spain.
Panagiotis N Moschou, Department of Biology, University of Crete, Heraklion 70013, Greece; Department of Molecular Sciences, Uppsala BioCenter, Swedish University of Agricultural Sciences and Linnean Center for Plant Biology, Uppsala 75007, Sweden; Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Heraklion 70013, Greece.
Author contributions
Conceptualization: P.N.M.; Methodology: C.L., A.M., I.H.H., E.T., V.S., A.B.R.-L., B.B.; Investigation: C.L., A.M., F.J.R.-C., V.S., A.B.R.-L., E.G.-B., X.M., V.A.M., A.K.; Visualization: C.L., A.M., F.J.R.-C., P.N.M.; Funding acquisition: C.L., F.J.R.-C., E.G.-B., P.N.M.; Project administration: P.N.M., P.M., P.F.S., E.G.-B.; Supervision: P.N.M.; Writing—original draft: P.N.M.; Writing—review & editing: C.L., A.M., E.G.-B., V.AM., P.F.S.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. APEX2 biotinylation efficiency.
Supplemental Figure S2. Quantification of selected RNA levels captured using as bait DCP1-GFP expressed under DCP1pro or 35Spro.
Supplemental Figure S3. Correlation between UTRs and CDS length or GC% with PB enrichment/depletion levels under NS or HS.
Supplemental Figure S4. Correlation between UTRs length and GC% with free energy in NS and HS.
Supplemental Figure S5. Correlation between half-life with free energy, complexity, and lengths of UTRs of PB-enriched/depleted RNAs in NS and HS.
Supplemental Figure S6. Nucleotide composition in UTRs of PBs-enriched RNAs.
Supplemental Figure S7. Codon usage bias of RNAs enriched in PBs.
Supplemental Figure S8. m6A content of PBs-enriched RNAs in NS or HS.
Supplemental Figure S9. RNA-seq read distributions of PBs-enriched RNAs and association of the adenylation-regulating machinery with PBs.
Supplemental Figure S10. Correlations between localization to PBs and RNA degradation.
Supplemental Figure S11. PB sizes and numbers in WT or scar1234.
Supplemental Figure S12. Cocluster analyses of APEAL and T-RIP enrichments.
Supplemental Figure S13. Complementarity hypothesis testing between amino acid residues and RNA codons for PB-enriched/excluded RNAs.
Supplemental Figure S14. Monosomes and Polysomes profiling in WT, dcp1-3, and scar1234.
Supplemental Figure S15. RAP2.4 localization upon HS or ACC (precursor of ethylene) treatment.
Supplemental Figure S16. Effect of ACC on SCAR/WAVE-DCP1 axis and PBs.
Supplemental Table S1. Primer lists.
Supplemental Data Set 1. RBPs enriched in T-RIP under NS or HS.
Supplemental Data Set 2. RNAs enriched/depleted from T-RIP.
Supplemental Data Set 3. RNA and GO terms enriched in T-RIP in both NS and HS (519 genes).
Supplemental Data Set 4. RT-qPCR RNAs used for correlation with T-RIP.
Supplemental Data Set 5. GO terms enriched/depleted (from T-RIP and transcriptome) with FDR values.
Supplemental Data Set 6. Polyadenylation enrichment in PBs.
Supplemental Data Set 7. smFISH probes used (EBF2, DFL1, RAP2.4, and PP2A).
Supplemental Data Set 8. Enrichment of ECTs and corresponding proteins in both APEAL and T-RIP datasets.
Supplemental Data Set 9. Enriched RNA targets of GRP7.
Supplemental Data Set 10. Enrichments for polysome profiling RNAs.
Supplemental Data Set 11. Statistical data and the corresponding measurements are used for each graph.
Supplemental File 1. IDR levels in the APEAL dataset, both in NS and HS.
Funding
This work was supported, by the Carl Tryggers Stiftelse för Vetenskaplig Forskning (15:336 and 17:336; P.N.M.), Svenska Forskningsrådet Formas (2017-00938; PNM), the HORIZON EUROPE EXCELLENT SCIENCE—Marie Skłodowska-Curie Actions (“PANTHEON,” 872969; P.N.M.), Helge Ax:son Johnsons stiftelse (F19-0419; C.L.), Hellenic Foundation of Research and Innovation Ph.D. scholarships (06526; A.M. and 05947; I.H.H.), Foundation for Research and Technology-Hellas (start-up funding from the Institute of Molecular Biology and Biotechnology; P.N.M.), Ministerio de Ciencia e Innovación, Grant MINOTAUR BIO2017-84066-R (F.J.R.-C. and A.B.R.-L.), Grant PID2020-119737GA-I00 (MCIN/AEI/10.13039/501100011033; E.G.-B.), Junta de Andalucia (ProyExcel_00587; E.G.-B.), Vetenskapsrådet (2019-05634; P.M.), and Kempestiftelserna (JCK-2011; P.M.).
Data availability
The raw RNAseq data underlying this article are available in BioStudies database, at https://www.ebi.ac.uk/biostudies/studies/S—BSST1096?key=58afa48a—c6c2—4366—b059—d2504392a2b2. Other data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019:176(3):419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson ADL, Anderson ZD, Beilstein MA, Fray RG, Lyons E, et al. N(6)-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 2018:25(5):1146–1157.e1143. 10.1016/j.celrep.2018.10.020 [DOI] [PubMed] [Google Scholar]
- Baumann K. mRNA translation in stress granules is not uncommon. Nat Rev Mol Cell Biol. 2021:22(3):164. 10.1038/s41580-021-00331-3 [DOI] [PubMed] [Google Scholar]
- Beutel O, Maraspini R, Pombo-Garcia K, Martin-Lemaitre C, Honigmann A. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell. 2019:179(4):923–936.e911. 10.1016/j.cell.2019.10.011 [DOI] [PubMed] [Google Scholar]
- Blake LA, Liu Y, Inoue T, Wu B. A rapid inducible RNA decay system reveals fast mRNA decay in P-bodies. bioRxiv 538452. 10.1101/2023.04.26.538452, 26 April 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG. Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front Plant Sci. 2012:3:48. 10.3389/fpls.2012.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009:55(4):611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Cairo A, Vargova A, Shukla N, Capitao C, Mikulkova P, Valuchova S, Pecinkova J, Bulankova P, Riha K. Meiotic exit in Arabidopsis is driven by P-body-mediated inhibition of translation. Science. 2022:377(6606):629–634. 10.1126/science.abo0904 [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Sorenson RS, Hummel M, Ke H, Kettenburg AT, Chen D, Aiyetiwa K, Dehesh K, Eulgem T, Sieburth LE, et al. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat Plants. 2020:6(6):675–685. 10.1038/s41477-020-0681-8 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. Deadenylation and P-bodies. Adv Exp Med Biol. 2013:768:183–195. 10.1007/978-1-4614-5107-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S, Kwon T, Khan BR, Sparks JA, Mallery EL, Szymanski DB, Blancaflor EB. The BEACH domain-containing protein SPIRRIG modulates actin-dependent root hair development in coordination with the WAVE/SCAR and ARP2/3 complexes. bioRxiv 381848. 10.1101/2020.11.13.381848, 14 November 2020, preprint: not peer reviewed. [DOI]
- Cho HY, Chou MY, Ho HY, Chen WC, Shih MC. Ethylene modulates translation dynamics in Arabidopsis under submergence via GCN2 and EIN2. Sci Adv. 2022:8(22):eabm7863. 10.1126/sciadv.abm7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodasiewicz M, Kerber O, Gorka M, Moreno JC, Maruri-Lopez I, Minen RI, Sampathkumar A, Nelson ADL, Skirycz A. 2′,3′-cAMP treatment mimics the stress molecular response in Arabidopsis thaliana. Plant Physiol. 2022:188(4):1966–1978. 10.1093/plphys/kiac013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Molza AE, Giudice E, Cavalier A, Thomas D, Gillet R. Structural organization of the polysomes adjacent to mammalian processing bodies (P-bodies). RNA Biol. 2013:10(2):314–320. 10.4161/rna.23342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deli A, Tympa LE, Moschou PN. Analyses of protein turnover at the cell plate by fluorescence recovery after photobleaching during cytokinesis. Methods Mol Biol. 2022:2382:233–243. 10.1007/978-1-0716-1744-1_14 [DOI] [PubMed] [Google Scholar]
- Dignon GL, Best RB, Mittal J. Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu Rev Phys Chem. 2020:71(1):53–75. 10.1146/annurev-physchem-071819-113553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005:21(16):3433–3434. 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- Duncan S, Olsson TSG, Hartley M, Dean C, Rosa S. A method for detecting single mRNA molecules in Arabidopsis thaliana. Plant Methods. 2016:12(1):13. 10.1186/s13007-016-0114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok J, Shao MR, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L. Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant. 2008:1(6):990–1006. 10.1093/mp/ssn059 [DOI] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB. SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell. 2011:23(10):3610–3626. 10.1105/tpc.111.088823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY. Atlas of subcellular RNA localization revealed by APEX-seq. Cell. 2019:178(2):473–490.e426. 10.1016/j.cell.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008:3(4):619–628. 10.1038/nprot.2008.31 [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007:19(5):1549–1564. 10.1105/tpc.106.047621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell. 2015:27(3):926–943. 10.1105/tpc.114.134494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014:15(8):509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hatzianestis IH, Mountourakis F, Stavridou S, Moschou PN. Plant condensates: no longer membrane-less? Trends Plant Sci. 2023:28(10):1101–1112. 10.1016/j.tplants.2023.04.006 [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017:68(1):144–157.e145. 10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011:21(6):508–514. 10.1016/j.cub.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Jang GJ, Yang JY, Hsieh HL, Wu SH. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc Natl Acad Sci U S A. 2019:116(13):6451–6456. 10.1073/pnas.1900084116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawerth L, Fischer-Friedrich E, Saha S, Wang J, Franzmann T, Zhang X, Sachweh J, Ruer M, Ijavi M, Saha S, et al. Protein condensates as aging Maxwell fluids. Science. 2020:370(6522):1317–1323. 10.1126/science.aaw4951 [DOI] [PubMed] [Google Scholar]
- Kang JY, Wen Z, Pan D, Zhang Y, Li Q, Zhong A, Yu X, Wu YC, Chen Y, Zhang X, et al. LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs. Science. 2022:377(6607):eabj6647. 10.1126/science.abj6647 [DOI] [PubMed] [Google Scholar]
- Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019:365(6455):825–829. 10.1126/science.aax4240 [DOI] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015:142(23):4168–4179. 10.1242/dev.127613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Boeynaems S, Lam V, Scholl D, Stainton E, Briner A, Jacquemyn M, Daelemans D, Deniz A, Villa E, et al. The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems. Nat Commun. 2022:13(1):5643. 10.1038/s41467-022-33221-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Kim J, Seo PJ. N(6)-methyladenosine-modified RNA acts as a molecular glue that drives liquid-liquid phase separation in plants. Plant Signal Behav. 2022:17(1):2079308. 10.1080/15592324.2022.2079308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol. 2020:22(2):187–199. 10.1038/s41556-019-0450-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma M, Feng Y, Li H, Wang Y, Ma Y, Li M, An F, Guo H. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell. 2015:163(3):670–683. 10.1016/j.cell.2015.09.037 [DOI] [PubMed] [Google Scholar]
- Lin RC, Park HJ, Wang HY. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant. 2008:1(1):42–57. 10.1093/mp/ssm004 [DOI] [PubMed] [Google Scholar]
- Linsenmeier M, Hondele M, Grigolato F, Secchi E, Weis K, Arosio P. Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat Commun. 2022:13(1):3030. 10.1038/s41467-022-30521-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mentzelopoulou A, Muhammad A, Volkov A, Weijers D, Gutierrez-Beltran E, Moschou PN. An actin remodeling role for Arabidopsis processing bodies revealed by their proximity interactome. EMBO J. 2023:42(9):e111885. 10.15252/embj.2022111885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Na Z, Slavoff SA. P-bodies: composition, properties, and functions. Biochemistry. 2018:57(17):2424–2431. 10.1021/acs.biochem.7b01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Godoy Herz MA, Kornblihtt AR, Sorenson R, Sieburth LE, Nakaminami K, Seki M, Ding Y, Sun Q, Kang H, et al. Beyond transcription: compelling open questions in plant RNA biology. Plant Cell. 2023:35(6):1626–1653. 10.1093/plcell/koac346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Kurenda A, Siddique S, Denervaud Tendon V, Zhou F, Holbein J, Hasan MS, Grundler FM, Farmer EE, Geldner N. Single-cell damage elicits regional, nematode-restricting ethylene responses in roots. EMBO J. 2019:38(10):e100972. 10.15252/embj.2018100972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Alba AE, Moreno AB, Gabriel M, Mallory AC, Christ A, Bounon R, Balzergue S, Aubourg S, Gautheret D, Crespi MD, et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015:43(5):2902–2913. 10.1093/nar/gkv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell. 2015:163(3):684–697. 10.1016/j.cell.2015.09.036 [DOI] [PubMed] [Google Scholar]
- Meyer K, Koster T, Nolte C, Weinholdt C, Lewinski M, Grosse I, Staiger D. Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol. 2017:18(1):204. 10.1186/s13059-017-1332-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschou PN, Gutierrez-Beltran E, Bozhkov PV, Smertenko A. Separase promotes microtubule polymerization by activating CENP-E-related kinesin Kin7. Dev Cell. 2016:37(4):350–361. 10.1016/j.devcel.2016.04.015 [DOI] [PubMed] [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. Elife. 2020:9:e49658. 10.7554/eLife.49658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathy ST, Udayasuriyan V, Bhadana V. Codon usage bias. Mol Biol Rep. 2022:49(1):539–565. 10.1007/s11033-021-06749-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S, Mourao MDA, Jalihal AP, Xiao L, Jiang X, Chinnaiyan AM, Schnell S, Walter NG. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol Cell. 2019:74(3):521–533.e526. 10.1016/j.molcel.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006:18(11):3047–3057. 10.1105/tpc.106.046508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M, Koster T, Staiger D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J Mol Cell Biol. 2019:11(10):899–910. 10.1093/jmcb/mjz085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringner M, Krogh M. Folding free energies of 5′-UTRs impact post-transcriptional regulation on a genomic scale in yeast. PLoS Comput Biol. 2005:1(7):e72. 10.1371/journal.pcbi.0010072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S, Das SS, Rakshit R. Codon usage pattern and predicted gene expression in Arabidopsis thaliana. Gene X. 2019:2:100012. 10.1016/j.gene.2019.100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkans U, Gostev M, Athar A, Behrangi E, Melnichuk O, Ali A, Minguet J, Rada JC, Snow C, Tikhonov A, et al. The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 2018:46(D1):D1266–D1270. 10.1093/nar/gkx965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H. N(6)-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell. 2016:38(2):186–200. 10.1016/j.devcel.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003:300(5620):805–808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Miranda J, Chodasiewicz M, Skirycz A, Fernie AR, Moschou PN, Bozhkov PV, Gutierrez-Beltran E. Stress-related biomolecular condensates in plants. Plant Cell. 2023:35(9):3187–3204. 10.1093/plcell/koad127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Wei L, Chen Z, Cai Z, Lu Q, Wang C, Tian E, Jia G. M(6)A readers ECT2/ECT3/ECT4 enhance mRNA stability through direct recruitment of the poly(A) binding proteins in Arabidopsis. Genome Biol. 2023:24(1):103. 10.1186/s13059-023-02947-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Szemenyei H, Sorek H, Landers A, Knight H, Bauer S, Wemmer DE, Somerville CR. Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1. Proc Natl Acad Sci U S A. 2015:112(52):16048–16053. 10.1073/pnas.1521675112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson RS, Deshotel MJ, Johnson K, Adler FR, Sieburth LE. Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc Natl Acad Sci U S A. 2018:115(7):E1485–E1494. 10.1073/pnas.1712312115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004:15(2):173–183. 10.1016/j.molcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom. 2008:43(6):699–715. 10.1002/jms.1415 [DOI] [PubMed] [Google Scholar]
- Teale WD, Pasternak T, Dal Bosco C, Dovzhenko A, Kratzat K, Bildl W, Schworer M, Falk T, Ruperti B, Schaefer JV, et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 2021:40(1):e104416. 10.15252/embj.2020104416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibble RW, Depaix A, Kowalska J, Jemielity J, Gross JD. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat Chem Biol. 2021:17(5):615–623. 10.1038/s41589-021-00774-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta E. On the normalization of the minimum free energy of RNAs by sequence length. PLoS One. 2014:9(11):e113380. 10.1371/journal.pone.0113380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J. 2004:86(4):2517–2529. 10.1016/S0006-3495(04)74307-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, Spang A. Context-dependent deposition and regulation of mRNAs in P-bodies. Elife. 2018:7:e29815. 10.7554/eLife.29815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedner HJ, Giudice J. It's not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat Struct Mol Biol. 2021:28(6):465–473. 10.1038/s41594-021-00601-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009:21(10):3270–3279. 10.1105/tpc.109.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006:18(12):3386–3398. 10.1105/tpc.106.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SL, Shrestha R, Karunadasa SS, Xie PQ. Proximity labeling in plants. Annu Rev Plant Biol. 2023:74(1):285–312. 10.1146/annurev-arplant-070522-052132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanguez E, Castro-Sanz AB, Fernandez-Bautista N, Oliveros JC, Castellano MM. Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One. 2013:8(8):e71425. 10.1371/journal.pone.0071425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018:69(3):517–532.e511. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Yu X, Li B, Jang GJ, Jiang S, Jiang D, Jang JC, Wu SH, Shan L, He P. Orchestration of processing body dynamics and mRNA decay in Arabidopsis immunity. Cell Rep. 2019:28(8):2194–2205.e2196. 10.1016/j.celrep.2019.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020:181(7):1582–1595.e1518. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrovic B, Bartonek L, Polyansky AA. RNA-protein interactions in an unstructured context. FEBS Lett. 2018:592(17):2901–2916. 10.1002/1873-3468.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Fonseca A, Meschichi A, Sicard A, Rosa S. Whole-mount smFISH allows combining RNA and protein quantification at cellular and subcellular resolution. Nat Plants. 2023:9(7):1094–1102. 10.1038/s41477-023-01442-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNAseq data underlying this article are available in BioStudies database, at https://www.ebi.ac.uk/biostudies/studies/S—BSST1096?key=58afa48a—c6c2—4366—b059—d2504392a2b2. Other data underlying this article are available in the article and in its online supplementary material.