Abstract
Precise control over how and where actin filaments are created leads to the construction of unique cytoskeletal arrays within a common cytoplasm. Actin filament nucleators are key players in this activity and include the conserved actin-related protein 2/3 (Arp2/3) complex as well as a large family of formins. In some eukaryotic cells, these nucleators compete for a common pool of actin monomers and loss of one favors the activity of the other. To test whether this mechanism is conserved, we combined the ability to image single filament dynamics in the homeostatic cortical actin array of living Arabidopsis (Arabidopsis thaliana) epidermal cells with genetic and/or small molecule inhibitor approaches to stably or acutely disrupt nucleator activity. We found that Arp2/3 mutants or acute CK-666 treatment markedly reduced the frequency of side-branched nucleation events as well as overall actin filament abundance. We also confirmed that plant formins contribute to side-branched filament nucleation in vivo. Surprisingly, simultaneous inhibition of both classes of nucleator increased overall actin filament abundance and enhanced the frequency of de novo nucleation events by an unknown mechanism. Collectively, our findings suggest that multiple actin nucleation mechanisms cooperate to generate and maintain the homeostatic cortical array of plant epidermal cells.
Advanced live-cell imaging along with genetic mutation and small molecule inhibitor approaches reveal the roles of 2 classes of actin filament nucleator in maintaining the homeostatic cortical cytoskeleton array.
Introduction
The actin cytoskeleton comprises a dynamic network of filaments that participates in a wide variety of cellular activities, including vesicle/organelle trafficking, cell expansion, cellulose synthesis, tissue/organelle development, and resistance to microbial pathogen invasion (Gao et al. 2009; Sampathkumar et al. 2013; Peremyslov et al. 2015; Scheuring et al. 2016; Qi and Greb 2017; Li and Staiger 2018; Zhang et al. 2019). Plant cells rearrange their actin cytoskeleton on timescales of seconds to minutes in response to biotic or abiotic stimuli, which suggests not only extreme dynamicity but also precise control over filament formation and turnover (Staiger et al. 2009; Henty-Ridilla, Shimono, et al. 2013). Regulation of cytoskeletal organization and dynamics involves dozens of actin-binding proteins that participate in filament nucleation, elongation, severing, and depolymerization (Blanchoin and Michelot 2012). Nucleation factors or nucleators initiate the formation of new actin filaments at a much faster rate than spontaneous nucleation from free actin monomers and are therefore considered key regulators of array formation and actin dynamics (Kadzik et al. 2020; Rosenbloom et al. 2021). Previous in vitro studies, primarily from nonplant systems, have identified and characterized 2 classes of conserved actin nucleator, the actin-related protein 2/3 (Arp2/3) complex and formins, with different specific activation mechanisms (Kovar 2006; Pollard 2007; Paul and Pollard 2008; Burke et al. 2014; Suarez et al. 2015).
The Arp2/3 complex is a 7-subunit protein complex responsible for generating branched actin filaments and forming dendritic actin networks in eukaryotic cells (Mullins et al. 1998; Pollard 2007). The ARP2 and ARP3 subunits mimic the structure of actin monomers. Once the Arp2/3 complex is activated, the ARP2 and ARP3 subunits along with a recruited actin monomer serve as the actin nucleus for the polymerization of a new (daughter) filament, which grows from the side of a preexisting (mother) actin filament with a characteristic 70° angle (Mullins et al. 1998; Dayel and Mullins 2004; Pollard 2007; Beltzner and Pollard 2008; Rouiller et al. 2008). This branched actin network structure is critical for many cellular processes in yeast and animal cells, such as cell division, cell migration, exocytosis, endocytosis, and tissue or embryo development (Roh-Johnson and Goldstein 2009; Cabrera et al. 2011; Sun et al. 2013; van der Kammen et al. 2017). In contrast, plants, such as Arabidopsis (Arabidopsis thaliana), with mutations in Arp2/3 subunits have relatively normal morphology and fertility; however, arp2/3 mutants exhibit defects in epidermal cell morphology including distorted trichomes, cell–cell adhesion defects, and fewer lobes on leaf pavement cells (Szymanski et al. 1999; Le et al. 2003; Li et al. 2003; Mathur, Mathur, Kernebeck, and Hulskamp 2003; Harries et al. 2005; Yanagisawa et al. 2015). Recent research shows that the plant Arp2/3 complex is also involved in auxin transport (Peng et al. 2017; Sahi et al. 2018; García-González et al. 2020), guard cell opening and closing (Jiang et al. 2012; Li et al. 2014), resistance to penetration-mediated infection by fungal pathogens (Qin et al. 2021), and possibly autophagy (Wang et al. 2016, 2019). Many of these studies reveal that arp2/3 mutant cells have reduced actin filament abundance or misaligned actin bundles, suggesting that the functions of the Arp2/3 complex may be accomplished through generating specific actin structures. In growing trichome branches, for example, the Arp2/3 complex generates a tip-localized actin filament array constrained by cortical microtubules that coordinate growth (Yanagisawa et al. 2015, 2018). During fungal invasion, the Arp2/3 complex localizes to the site of penetration peg attack in epidermal pavement cells and generates a cortical actin filament patch necessary to suppress invasion (Qin et al. 2021). However, the molecular mechanisms by which the Arp2/3 complex coordinates the organization of cortical actin filament arrays in unstimulated cells or how it generates unique arrays in response to biotic and abiotic stress are poorly understood.
Formins represent another family of evolutionarily conserved actin filament nucleators; they typically generate long linear actin filaments that can bundle together to form actin cables (Kovar 2006; Pollard 2007; Paul and Pollard 2008). These actin cables provide tracks for intracellular organelle/vesicle trafficking. Formins have 2 universally conserved formin homology (FH) domains, FH1 and FH2. The proline-rich FH1 domain allows formins to recruit profilin-bound actin to elongate actin filaments at the barbed ends. The FH2 domain, on the other hand, associates with other regions within itself for autoinhibition or it can interact with the FH2 domain of another formin to form a dimer that interacts with and suitably positions 2 actin monomers, thereby initiating the nucleation of linear actin filaments (Cvrčková et al. 2000; Deeks et al. 2002; Cheung and Wu 2004; Cvrčková et al. 2004; Kovar 2006; Michelot et al. 2006; Pollard 2007; Paul and Pollard 2008). There are 21 FHs in Arabidopsis and these are separated into 2 phylogenetic subclasses (Cvrčková et al. 2000). Based on sequence prediction, Class I formins have a signal peptide and an N-terminal transmembrane domain that target them to the plasma membrane, whereas Class II formins are predicted to have more diverse domain organization (Cvrčková et al. 2000; Deeks et al. 2002; Cheung and Wu 2004; Cvrčková et al. 2004; Michelot et al. 2006). In addition, plant formin orthologs can be categorized according to the ability to associate with filament barbed ends and elongate growing filaments. Processive formins remain attached to the actin filament barbed end and move processively as the filament elongates, whereas nonprocessive formins remain at the filament nucleation site (Cvrčková et al. 2000; Deeks et al. 2002; Cheung and Wu 2004; Cvrčková et al. 2004; Michelot et al. 2005; Michelot et al. 2006; Zhang et al. 2016). Biochemical results reveal that a nonprocessive Class I formin, AtFORMIN1 (AFH1), attaches to the side of a preexisting actin filament to nucleate new filaments, suggesting a far more complicated role for formins in plant cells than just the ability to generate linear filaments (Michelot et al. 2006). In addition, both in vivo and in vitro experiments show that formins are critical for maintaining homeostatic actin array organization as well as regulating polarized cell growth (Vidali et al. 2009; Rosero et al. 2013; Lan et al. 2018). However, the detailed molecular mechanism of how formins regulate the cytoskeletal organization and single actin filament dynamics in plant cells remains unclear. For several excellent reviews on functions and regulation of the Arp2/3 complex and formins, the reader is referred to Breitsprecher and Goode (2013), Rotty et al. (2013), Pollard (2016), and Courtemanche (2018).
The decision of when and where to generate an actin array as well as its specific structure and dynamics is essential for each actin array to specifically choreograph its cellular function and often requires coordination between the Arp2/3 complex and formins. Studies in fission yeast (Schizosaccharomyces pombe) and some animal cells indicate that the Arp2/3 complex and formins compete for a limited supply of actin monomers and this competition prevents excessive activity of one or the other, thereby allowing cells to generate distinct actin structures and dynamics by regulating the balance of activities between these 2 nucleators (Hotulainen and Lappalainen 2006; Lomakin et al. 2015; Suarez et al. 2015; Davidson et al. 2018; Antkowiak et al. 2019; Chan et al. 2019; Kadzik et al. 2020). During actin array assembly, the Arp2/3 complex generates branched filament arrays, whereas formins typically produce linear filament bundles (Carlier and Shekhar 2017). Furthermore, in vitro studies reveal that filaments generated by formins grow faster than Arp2/3-nucleated filaments (Vavylonis et al. 2006; Michelot et al. 2013; Suarez et al. 2015; Funk et al. 2019). Studies with M2 melanoma cells and HeLa cells illustrate that actin arrays generated by different nucleators have distinct properties of filament abundance, filament length, and turnover kinetics and also reveal that both nucleators contribute to the maintenance and turnover of the homeostatic actin cortex (Fritzsche et al. 2016). A study in Arabidopsis shows that actin arrays in cotyledon epidermal pavement cells are slightly more dense and the extent of bundling is unchanged when either a Class I formin, AFH1, or the Arp2/3 subunit, ARPC5, is genetically downregulated; however, single filament dynamics or nucleation events were not evaluated directly (Cifrová et al. 2020). The authors propose that the Arp2/3 complex and formins in plants may have complementary roles to overcome the loss of the other. Therefore, it is important to understand how each nucleator regulates the structure and dynamic properties of actin arrays individually to better understand how they contribute to the cortical actin homeostasis and how they are coordinated to regulate the actin cytoskeleton during different biological processes.
Quantitative analysis of single actin filament dynamics in vivo is relatively hard to conduct in yeast and mammalian cells, because either the cells are too small or the actin arrays are too dense and filament lengths are below the limits of resolution of light microscopy. Epidermal cells from dark-grown Arabidopsis hypocotyls are large in size and have relatively sparse actin arrays in the cortical cytoplasm, making this a powerful model system for quantitative analysis of single actin filament dynamics, especially when combined with high spatial and temporal resolution imaging approaches and genetically encoded fluorescence reporters (Staiger et al. 2009; Henty et al. 2011; Henty-Ridilla, Li, et al. 2013; Cai et al. 2014; Cao et al. 2016; Arieti and Staiger 2020). Previous work demonstrates a remarkably high rate of new filament construction, rapid growth at filament ends, and disassembly by prolific severing activity in a mechanism termed “stochastic dynamics” (Staiger et al. 2009). High rates of polymerization are likely supported by a large pool of monomeric actin that is buffered with an excess of the monomer-binding protein, profilin, to suppress spontaneous nucleation events (Chaudhry et al. 2007; Staiger et al. 2009; Cao et al. 2016). The ability to observe both actin filament architecture and activities of single filaments in living cells allows direct visualization and quantitative analysis of the effects associated with the disruption of actin nucleators on filament activities in vivo (Cao et al. 2016).
In this study, we test the hypothesis that the Arabidopsis Arp2/3 complex mediates nucleation of side-branched filaments in vivo but does not facilitate filament elongation directly. We used both genetic and pharmacological approaches to disrupt Arp2/3 complex activity in plant cells. With quantitative live-cell imaging at single filament resolution, we demonstrated directly that the Arp2/3 complex is responsible for the nucleation of side-branched filaments in vivo. We also characterized the difference between the Arp2/3 complex and formins with respect to the dynamic behaviors of the filaments they generate and evaluated the consequence of losing both classes of filament nucleator on actin organization and dynamics. Surprisingly, living plant cells deficient for 2 classes of nucleator generate comparatively normal and dynamic actin arrays by an apparent de novo filament nucleation mechanism.
Results
Arabidopsis plants have defects in general growth and epidermal cell morphology when deficient for the Arp2/3 complex
Previous studies of Arp2/3 complex mutants reveal defects in Arabidopsis epidermal cell growth and morphology (Szymanski et al. 1999; Le et al. 2003; Li et al. 2003; Mathur, Mathur, Kirik, et al. 2003; Harries et al. 2005; Facette et al. 2015; Yanagisawa et al. 2015; Cifrová et al. 2020). Here, we acquired a T-DNA insertion mutant for ARP2, arp2-1 (or wurm1-2; SALK_003448), and a point mutation of ARPC2, arpc2 (or distorted2-1; El-Din El-Assal et al. 2004), and confirmed the cell and organ growth phenotypes. We observed that both arp2-1 and arpc2 homozygous mutant plants had adhesion defects at end walls of hypocotyl epidermal cells and severely distorted leaf trichomes when compared to wild-type sibling lines (Supplemental Fig. S1A). In addition, etiolated hypocotyls from arp2-1 and arpc2 seedlings were significantly shorter than wild-type seedlings at the same time points (Supplemental Fig. S1, B and C). To test whether these growth defects resulted from defects in cell expansion, we examined epidermal cell length and found that cells in all regions of etiolated arp2/3 hypocotyls were significantly shorter compared to wild-type hypocotyls; however, cell widths were not affected (Supplemental Fig. S1, D and E). Similar results for root growth were observed in light-grown seedlings (Supplemental Fig. S1, F and G). These results confirm that the Arp2/3 complex plays a role in axial cell expansion in both dark-grown hypocotyls and light-grown roots of Arabidopsis.
Actin filament abundance and bundling are reduced in arp2/3 mutants
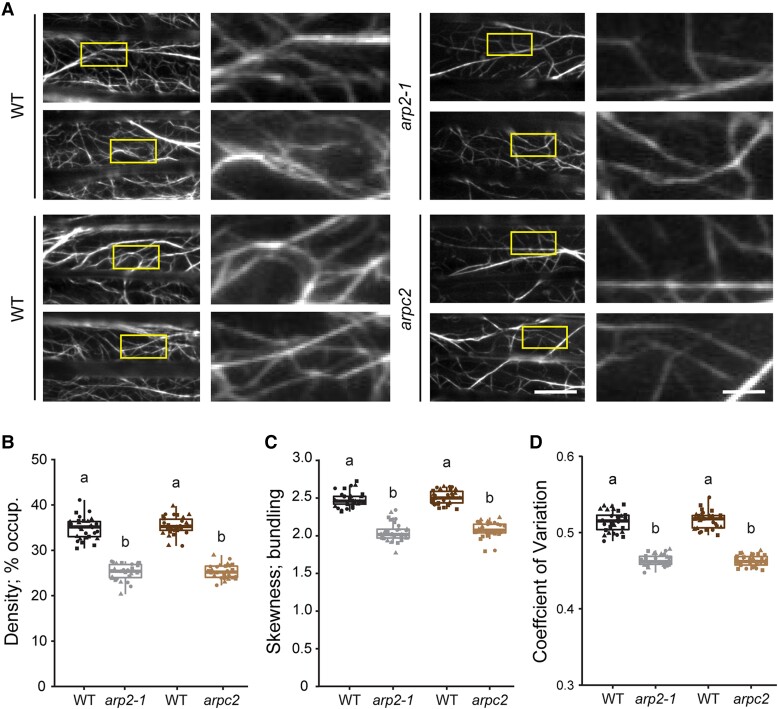
To assess the influence of the Arp2/3 complex on cortical actin cytoskeletal organization, we compared images of epidermal cells from the apical region of 5-d-old etiolated wild-type arp2-1 and arpc2 hypocotyls collected by variable-angle epifluorescence microscopy (VAEM) (Staiger et al. 2009). Homozygous mutant and isogenic wild-type sibling lines that express the actin reporter, GFP fused with FIMBRIN1 actin-binding domain 2 or GFP-fABD2 (Sheahan et al. 2004), were prepared by crossing to facilitate the observation and measurement of actin cytoskeleton organization and filament dynamics (Staiger et al. 2009). The actin array structure in arp2-1 and arpc2 cells had fewer, thinner, and more scattered filaments compared to corresponding wild-type cells (Fig. 1A).
Figure 1.
Genetic disruption of the Arp2/3 complex leads to reduced actin filament density and bundling. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls expressing GFP-fABD2 imaged by VAEM are shown in the left columns. Scale bar: 20 μm. ROIs (boxes) were magnified and shown in the right columns. Scale bar: 5 μm. B to D) Quantitative analysis of the percentage of occupancy or density of actin filament arrays B) and the extent of filament bundling as measured by skewness C) and coefficient of variance D) analyses. Both the density and the bundling of actin arrays in arp2-1 and arpc2 cells were significantly decreased compared to those in the respective wild-type cells. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 3 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 30 seedlings, 10 seedlings per biological repeat). Letters a and b denote groups that show statistically significant differences with other genotypes by 1-way ANOVA with Tukey's post hoc test (P < 0.05). WT, wild type.
To compare these differences quantitatively, we used density, skewness, and the coefficient of variation (CV) as 3 key parameters established previously for standardized descriptions of actin cytoskeleton array organization in living cells (Higaki et al. 2010; Ueda et al. 2010; Henty et al. 2011; Higaki et al. 2020). Density measures the percentage of occupancy of actin filaments in an array, whereas both skewness and CV define the extent of bundling of actin filaments. CV uses a different calculation method compared to skewness and is reportedly a better indicator of bundling, especially for VAEM images (Higaki et al. 2020). Actin density in arp2-1 and arpc2 cells was significantly decreased compared to the respective wild-type cells (Fig. 1B), and the extent of filament bundling was significantly reduced as well when measured by skewness or CV methods (Fig. 1, C and D). These results demonstrate that the Arp2/3 complex contributes to the generation of both individual filaments and filament bundles and is necessary for maintaining the homeostatic organization of the cortical actin cytoskeleton in epidermal cells.
The Arp2/3 complex plays a significant role in filament nucleation and generation of side-branched filaments
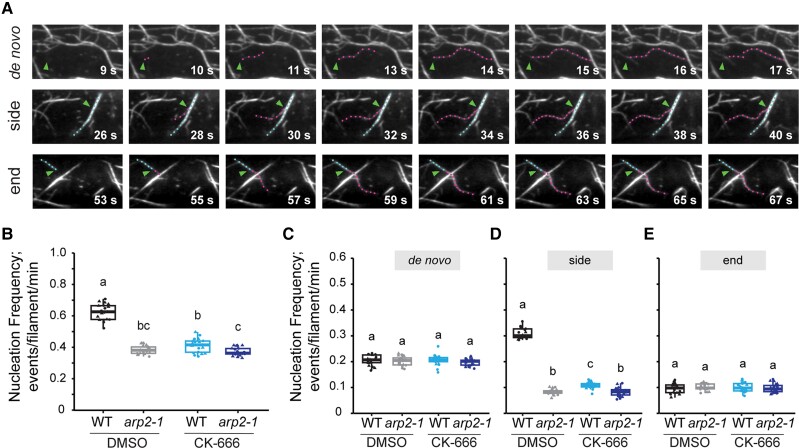
To investigate whether the Arp2/3 complex participates in actin filament nucleation, we measured single actin filament formation and dynamics in wild-type and arp2/3 mutant cells by collecting time-lapse movies from the apical regions of 5-d-old dark-grown hypocotyls with VAEM. The sparse nature of the cortical actin array in hypocotyl epidermal cells and the high spatial and temporal resolution afforded by VAEM allowed us to visualize (Fig. 2A) and quantify individual nucleation events with a previously described assay (Cao et al. 2016). Briefly, we counted all new or regrowing filaments identified in multiple 400-µm2 regions of interest (ROIs) within a cell during a 100-s time-lapse movie. The nucleation frequency was normalized to the average filament number within each ROI to minimize the influence of filament abundance differences between ROIs and genotypes. The overall nucleation frequency in arp2-1 cells (0.38 ± 0.03 events/filament/min) was significantly decreased compared to wild-type cells (0.62 ± 0.05 events/filament/min) (Fig. 2B; Supplemental Movies S1 and S2) and coincided with our observation that loss of function of the Arp2/3 complex resulted in a significant reduction in filament abundance (Fig. 1B). Similar reductions in overall nucleation frequency were also observed for arpc2 compared to wild-type cells (Supplemental Fig. S2A).
Figure 2.
Actin filament nucleation frequency is decreased by chemical or genetic inhibition of the Arp2/3 complex. A) Representative time-lapse series showing 3 subclasses of actin filament origin identified in hypocotyl epidermal cells: actin filaments initiated de novo in the cytoplasm (top), from the side of a preexisting filament (middle), or from the end of a preexisting filament (bottom row). Blue dots: preexisting filament. Magenta dots: new growing filament. Arrowheads: nucleation site. Scale bar: 5 μm. B to E) Quantitative analysis of actin filament nucleation frequency, both overall B) and by subclass of origin C to E). Hypocotyls were treated with DMSO solution (0.05% DMSO) or 10 µM CK-666 for 5 min prior to imaging with VAEM. The overall nucleation frequency B) for each genotype or treatment was defined as the total number of filament origins per filament per minute in a 400-μm2 ROI. Total nucleation frequency in DMSO-treated wild-type cells was significantly higher than DMSO-treated arp2-1, CK-666-treated wild-type, or CK-666-treated arp2-1 cells. When filament origin events were categorized into de novo C), side D), and end populations E), only the side-branching nucleation events showed a significant reduction in arp2-1 and CK-666-treated cells compared to DMSO-treated wild-type cells. It should be noted that experiments reported here were conducted at the same time as those testing SMIFH2; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 2 and 6. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to c denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). WT, wild type.
Next, we classified nucleation events into 3 different subpopulations based on filament origin: de novo from the cytoplasm, from the side of a preexisting filament or bundle, or from the end of a preexisting filament (Fig. 2A; Supplemental Movies S3 to S5) (Staiger et al. 2009; Henty-Ridilla, Li, et al. 2013; Cao et al. 2016). Regrowth from preexisting filament ends is not a conventional nucleation with respect to actin seed trimer formation; however, for simplicity and semantic purposes, we include these measurements as a subclass of nucleation event. Only the side-branched nucleation frequency was significantly decreased in arp2-1 and arpc2 cells compared to the respective wild-type sibling cells (Fig. 2, C to E; Supplemental Movies S1 and S2 and Fig. S2, C to E). Side-branched nucleation events contribute about half of all new filament origins and these were reduced by 60% to 70% in the arp2/3 mutants.
A previous study reports different results regarding the actin organization in arp2/3 mutants; instead of significantly decreased filament abundance as observed here, a minor increase in actin filament density in arpc5 cotyledon pavement cells was reported (Cifrová et al. 2020). These differences could result from the use of a different arp2/3 complex subunit mutation, the investigation of epidermal cells from a different tissue, the choice of actin cytoskeleton reporter, or all of the above. Several previous publications show that LifeAct reduces both the actin reorganization rate and actin polymerization rate (van der Honing et al. 2011; Spracklen et al. 2014; Courtemanche et al. 2016) and displays longer and thicker filaments (Flores et al. 2019). To test which factors led to differing results between the current work and that of Cifrová et al. (2020), we conducted a direct comparison between the ability of GFP-fABD2 or GFP-LifeAct to report actin architecture and single filament dynamics in either etiolated hypocotyls (Supplemental Fig. S3) or light-grown cotyledons (Supplemental Fig. S4). Moreover, we introduced GFP-LifeAct into arp2-1 to compare directly with the GFP-fABD2 arp2-1 reporter line described above. Our results showed that arp2-1 expressing either GFP-fABD2 or GFP-LifeAct had significantly lower actin filament density in the cortical array of both etiolated hypocotyl and light-grown cotyledon epidermal cells (Supplemental Figs. S3B and S4B). Similarly, overall actin filament nucleation frequency and the side-branched nucleation subclass were significantly reduced in arp2-1 epidermal cells from both reporter lines, in either hypocotyls or cotyledons, compared to the corresponding wild-type lines (Supplemental Fig. S3, E to H; Supplemental Fig. S4, E to H). These results suggest that using GFP-fABD2 as the actin reporter, or imaging epidermal cells of the etiolated hypocotyl, is not causal for the actin phenotypes we observed in arp2/3 mutants. Because it is beyond the scope of the current investigation, we did not seek to determine whether the previous findings are specific to the arpc5 mutant allele used, but note that we find similar quantitative differences in actin architecture and nucleation frequency with mutations in 2 different subunits (arp2-1 and arpc2) of the Arp2/3 complex. Considering the nature of the LifeAct marker in stabilizing actin structures and our primary goal of observing and analyzing the dynamic behaviors of single actin filaments, we used the GFP-fABD2 marker lines for the remainder of the experiments in this study.
Actin filaments generated by the Arp2/3 complex have unique dynamic properties
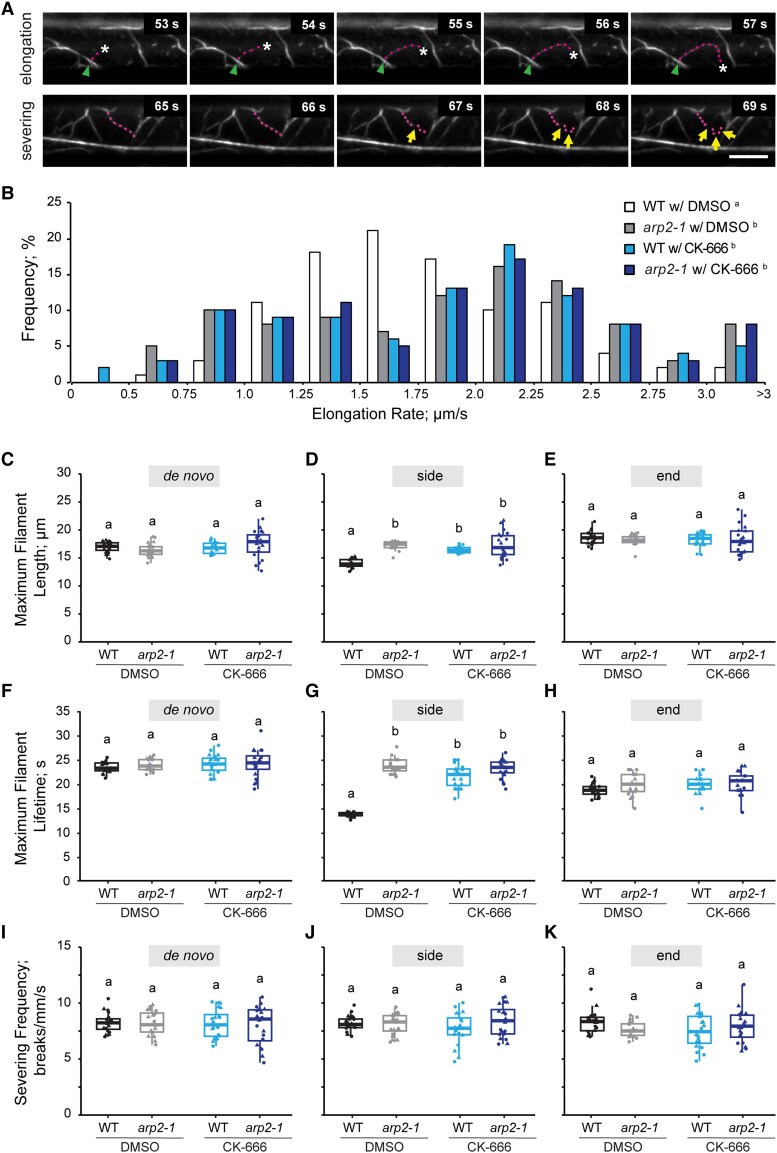
To understand how the Arp2/3 complex influences the dynamics of actin filaments, we tracked many dozens of individual filaments from their first appearance to complete disappearance and measured several parameters that were previously established to describe actin filament turnover (Fig. 3A; Table 1; Supplemental Movies S6 and S7) (Staiger et al. 2009; Henty et al. 2011). Compared to wild type, the population distribution of actin filament elongation rates in arp2-1 cells was more left skewed, and the average filament elongation rate was also higher (Fig. 3B; Table 1), indicating that actin filaments were growing significantly faster in arp2-1 cells. It is also clear that there were 2 or 3 distinct populations of filament elongation rates with peaks at 0.75 to 1.25, 2.0 to 2.25, and >3 µm/s with the latter 2 categories becoming more prevalent in arp2-1 cells (Fig. 3B). Similar differences in overall elongation rate and fast-growing filament populations were observed in arpc2 (Supplemental Table S1 and Fig. S2B). Filaments in both arp2-1 and arpc2 mutant cells also had significantly longer filament lengths and lifetimes than the ones in respective wild-type cells (Table 1; Supplemental Table S1).
Figure 3.
Actin filament dynamic properties are altered when the Arp2/3 complex is inhibited. A) A representative actin filament was tracked during the elongation phase (top) and another filament was fragmented into pieces during the severing phase (bottom). Magenta dots: growing actin filament. Arrowheads: nucleation site. Asterisk: elongating filament end. Yellow arrows: severing events. Scale bar: 10 μm. B) Quantitative analysis of the population distribution and average elongation rate of actin filaments in hypocotyl epidermal cells. Hypocotyls were treated with 0.05% DMSO solution or 10 µM CK-666 for 5 min prior to imaging with VAEM. The elongation rate distribution of DMSO-treated wild type had a single peak at 1.25 to 1.75 μm/s, whereas the Arp2/3-inhibited groups had 3 peaks at 0.75 to 1.25, 2.0 to 2.5, and >3 μm/s. It should be noted that experiments reported here were conducted at the same time as those testing SMIFH2; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 3 and 6. n ≥ 100 single filaments from 2 individual biological repeats (for 1 biological repeat, 5 single filaments were counted in 1 hypocotyl from at least 10 hypocotyls per genotype or treatment). Letters a and b denote genotypes or treatments that show statistically significant differences with other groups by chi-squared test, P < 0.05. C to E) The average maximum length of side-branching filaments in DMSO-treated wild-type cells was significantly shorter than that in DMSO-treated arp2-1, CK-666-treated wild-type, or CK-666-treated arp2-1 cells; however, filaments that originated de novo or from preexisting ends did not show any significant difference. F to H) The average maximum lifetime of side-branching filaments in DMSO-treated wild-type cells was significantly shorter than that in DMSO-treated arp2-1, CK-666-treated wild-type, or CK-666-treated arp2-1 cells; however, filaments that originated de novo or from preexisting ends did not show any significant difference. I to K) The severing frequency did not show any significant difference between different genotypes or treatments. It should be noted that experiments reported here were conducted at the same time as those testing SMIFH2; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 3 and 6. For box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a and b denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05).WT, wild type.
Table 1.
Single actin filament dynamics in wild-type (ARP2) and arp2-1 mutant with or without CK-666
| Genotype and treatment | ||||
|---|---|---|---|---|
| Stochastic dynamic parameters |
ARP2 w/ DMSO |
arp2-1 w/ DMSO |
ARP2 w/ CK-666 |
arp2-1 w/ CK-666 |
| Elongation rate; µm/s | 1.79 ± 0.05a | 1.90 ± 0.07b | 1.89 ± 0.07b | 1.90 ± 0.07b |
| Max. filament length; µm | 16.6 ± 0.3a | 17.4 ± 0.2b | 17.2 ± 0.2b | 17.8 ± 0.3b |
| Max. filament lifetime; s | 18.7 ± 0.4a | 22.6 ± 0.3b | 22.0 ± 0.3b | 22.9 ± 0.3b |
| Severing frequency; breaks/mm/s | 8.20 ± 0.11a | 7.93 ± 0.12a | 7.60 ± 0.19b | 8.18 ± 0.20a |
Measurements were taken from epidermal cells in the elongating apical region of 5-d-old dark-grown hypocotyls. Values represent mean ± Se. n ≥ 100 filaments from 2 biological repeats conducted with independent plant materials (for 1 biological repeat, 5 filaments were counted in 1 hypocotyl from at least 10 hypocotyls per treatment/genotype). By 2-way ANOVA with Tukey's post hoc test, letters a and b denote genotypes or treatments that show statistically significant differences with other genotypes or treatments, P < 0.05.
Previous in vitro data suggest that filament ends generated by the Arp2/3 complex and formins have different elongation rates; many formins are processive polymerases that use profilin-actin to add monomers to growing filaments (Kovar et al. 2003; Romero et al. 2004; Akin and Mullins 2008; Zhang et al. 2016). To test whether the dynamic properties of filaments with different nucleation patterns are distinct from each other, we measured the full suite of dynamic parameters for each population. We found that side-branched filaments in arp2-1 cells were significantly longer (Fig. 3, C to E) and filament lifetime was prolonged (Fig. 3, F to H) compared to the side-branched filaments in wild-type cells; however, the severing frequency did not show a significant difference between wild-type and arp2-1 cells (Fig. 3, I to K). These dynamic differences between side-branched filaments in arpc2 mutant and wild-type sibling cells were conserved (Supplemental Fig. S2, C to K).
Collectively, these data demonstrate that the Arp2/3 complex is responsible for generating new filaments from the side of preexisting filaments and is necessary for cells to maintain the homeostatic actin cytoskeleton architecture. Moreover, Arp2/3-nucleated filaments have distinct dynamic properties compared with those generated de novo or from the end of a preexisting filament; in particular, they grow slower and are shorter and have reduced filament lifetimes.
The small molecule inhibitor CK-666 phenocopies actin-based defects in arp2 and arpc2 mutants
Several studies report that CK-666, a small molecule inhibitor of the Arp2/3 complex that is effective on yeast and animal cells (Nolen et al. 2009; Hetrick et al. 2013), phenocopies the effects of arp2/3 mutants on tomato pollen tube growth (Liu et al. 2020) and Arabidopsis sperm nuclear migration (Ali et al. 2020), both of which depend upon the actin cytoskeleton. Moreover, a recent study demonstrates that CK-666 treatment does not influence actin redistribution between the basal and apical cell region in hypocotyl epidermal cells that occurs during early embryo growth (Cui et al. 2023). Because the earlier studies (Ali et al. 2020; Liu et al. 2020) did not directly demonstrate the effects of CK-666 on actin organization or filament nucleation, it is necessary to validate whether CK-666 influences the function of the Arp2/3 complex in plant cells, as a side-branched actin filament nucleator, before we use it as an effective plant Arp2/3 inhibitor. We hypothesized that applying CK-666 to wild-type Arabidopsis cells would inhibit the Arp2/3 complex activity and mimic the arp2/3 mutant phenotypes described above. To test the effects of CK-666, we applied a dose series (0, 1, 5, 10, 50, and 100 µM) for 5 min and collected snapshots of epidermal cells from the apical region of treated hypocotyls (Supplemental Fig. S5). Compared to the mock treatment (0 µM), CK-666 showed a significant reduction in both filament density and the extent of bundling starting at 5 µM and reached its maximum effect at 50 to 100 µM (Supplemental Fig. S5, B to D). A time course (0, 5, 10, 30, and 60 min) of 10 µM CK-666 treatment showed that 5 min was sufficient to alter actin array organization (Supplemental Fig. S5, E to G). Finally, cells were able to recover normal actin array architecture from a 5-min, 10 µM CK-666 application after a 30-min washout (Supplemental Fig. S5, H to J).
To further examine whether CK-666 phenocopies arp2/3 mutants, we treated 5-d-old etiolated hypocotyls of wild type and arp2-1 with either DMSO (mock) solution or 10 μM CK-666 for 5 min and measured actin organization and single filament dynamics. Compared to DMSO-treated wild-type cells, the CK-666-treated wild-type cells had significantly decreased filament abundance (Fig. 4B), extent of filament bundling (Fig. 4, C and D), and overall nucleation frequency (Fig. 2B), and these changes mirrored actin organization in DMSO-treated arp2-1 cells. Moreover, treatment of arp2-1 with CK-666 showed no differences in terms of filament organization or dynamics compared to arp2-1 alone (Figs. 3 and 4). CK-666 treatment also potently suppressed side-branched filament nucleation in wild-type cells but not end or de novo filament origins (Fig. 2, C to E; Supplemental Movies S1 and S8), thereby fully phenocopying the effects of arp2-1. Finally, treatment of arp2-1 with CK-666 did not further reduce filament side-branch nucleation events, indicating that its likely mode of action is to inhibit daughter filament formation from a mother filament by the Arp2/3 complex (Fig. 2D; Supplemental Movies S8 and S9).
Figure 4.
A small molecule inhibitor of the Arp2/3 complex, CK-666, reduces actin filament density and bundling. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls are shown in the left columns. Scale bar: 20 μm. ROIs (boxes) were magnified and displayed in the right columns. Scale bar: 5 μm. Hypocotyls were treated with 0.05% DMSO solution or 10 µM CK-666 for 5 min prior to imaging with VAEM. Actin filament arrays in DMSO-treated arp2-1, CK-666-treated ARP2, and CK-666-treated arp2-1 cells appeared to be less dense and less bundled compared to DMSO-treated wild-type cells. B to D) Quantitative analysis of the percentage of occupancy or density of actin filament arrays B), and the extent of filament bundling as measured by skewness C) and CV D) analyses. Both the density and bundling of actin arrays in DMSO-treated arp2-1, CK-666-treated wild-type, and CK-666-treated arp2-1 cells were significantly decreased compared to DMSO-treated wild-type cells. It should be noted that experiments reported here were conducted at the same time as those testing SMIFH2; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 4 and 5. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 3 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 30 seedlings, 10 seedlings per biological repeat). Letters a and b denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). WT, wild type.
When we compared the CK-666 treatment of arpc2 with its wild-type siblings, there were similar changes in actin architecture and filament nucleation frequency (Supplemental Figs. S2 and S6). Therefore, we validated CK-666 as a small molecule inhibitor that targets the Arp2/3 complex and demonstrated its utility as an acute but reversible inhibitor for studying the Arp2/3 complex in Arabidopsis.
The Arp2/3 complex and formins are both capable of nucleating side-branched actin filaments
Our previous work shows that profilin-bound actin monomers favor formin-mediated fast actin filament elongation at rates of >2 µm/s (Cao et al. 2016), and the data above suggest that Arp2/3-nucleated filament barbed ends elongated at intermediate rates of 1.25 to 1.5 µm/s (Fig. 3B; Supplemental Fig. S2B). We previously demonstrated the effectiveness of small molecule inhibitor of formin homology 2 (SMIFH2) in Arabidopsis both in vivo and in vitro (Cao et al. 2016). The actin array organization is altered and filament nucleation frequency is significantly reduced when wild-type hypocotyl epidermal cells were treated with 25 μM SMIFH2 for 5 min (Cao et al. 2016), and similar results were obtained here (Figs. 5 and 6, A and C to E; Supplemental Movie S10).
Figure 5.
Actin filament density increases after treatment of arp2-1 with the formin inhibitor SMIFH2. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls are shown in the left column. Scale bar: 20 μm. ROIs (boxes) were magnified and displayed in the right column. Scale bar: 5 μm. Hypocotyls were treated with 0.05% DMSO solution or 25 µM SMIFH2 for 5 min prior to imaging with VAEM. Actin filament arrays in DMSO-treated arp2-1 and SMIFH2-treated wild-type cells appeared to be less dense and less bundled compared to DMSO-treated wild-type cells, but SMIFH2-treated arp2-1 cells have a significantly increased actin abundance. B to D) Quantitative analysis of the percentage of occupancy or density of actin filament arrays B), and the extent of filament bundling as measured by skewness C) and CV D) analyses. The density of actin arrays in DMSO-treated arp2-1 and SMIFH2-treated wild-type cells was significantly decreased compared to DMSO-treated wild type; however, SMIFH2-treated arp2-1 cells had significantly increased actin density compared to all other genotypes and treatments B). Actin arrays in arp2-1 or SMIFH2-treated cells were significantly less bundled compared to DMSO-treated wild-type cells C, D). It should be noted that experiments reported here were conducted at the same time as those testing CK-666; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 4 and 5. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 3 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 30 seedlings, 10 seedlings per biological repeat). Letters a to d denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). WT, wild type.
Figure 6.
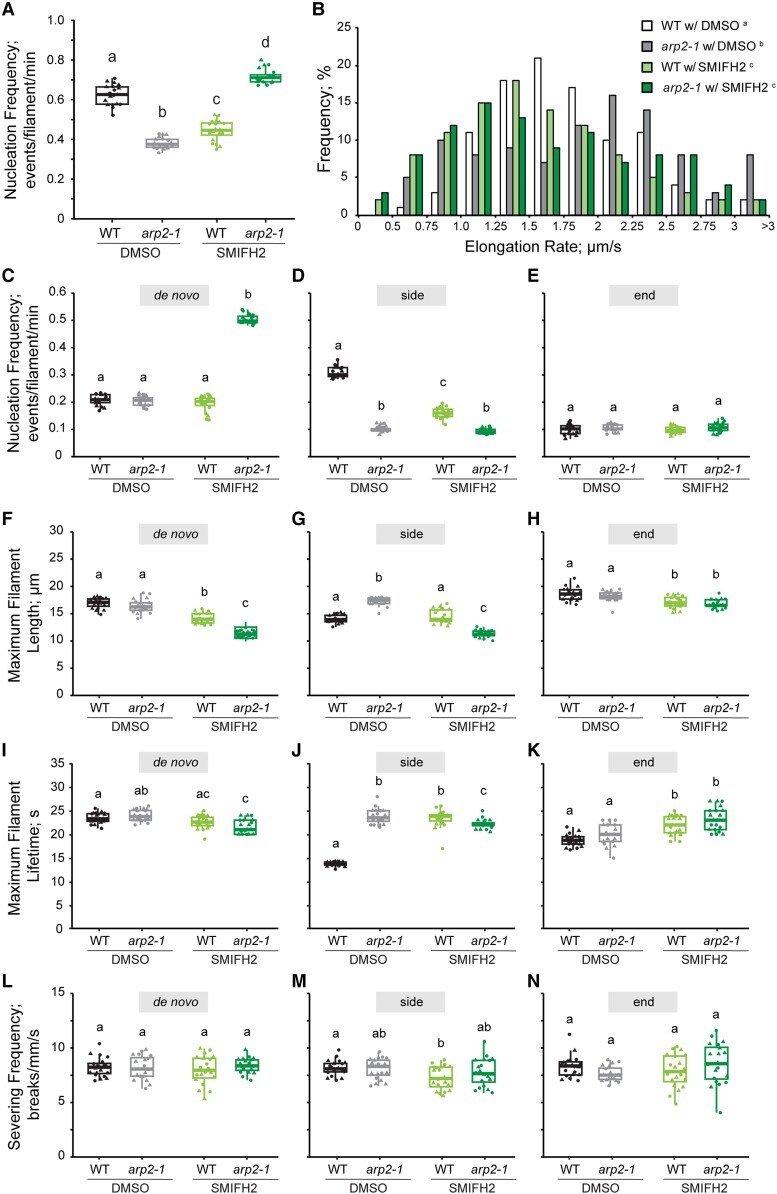
Overall and de novo filament nucleation increases when Arp2/3 and formin activity are simultaneously inhibited. A, C to E) Quantitative analysis of actin filament nucleation frequency, both overall A) and by the subclass of origin C to E). The total nucleation frequency in DMSO-treated arp2-1 and SMIFH2-treated wild-type cells was significantly reduced compared to DMSO-treated wild-type cells. However, the total nucleation frequency of SMIFH2-treated arp2-1 cells was significantly higher than all other genotypes and treatments A) and this correlated with increased de novo nucleation events C). It should be noted that experiments reported here were conducted at the same time as those testing CK-666; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 2 and 6. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to c denote groups that show statistically significant differences with other genotypes or treatments (within the same filament nucleation subclass) by 2-way ANOVA with Tukey's post hoc test (P < 0.05). B) Analysis of the population distribution of actin filament elongation rates. The elongation rate distribution of DMSO-treated wild type had a single peak at 1.25 to 1.75 μm/s, the DMSO-treated arp2-1 had 3 peaks at 0.75 to 1.0, 2.0 to 2.5, and >3 µm/s, the SMIFH2-treated wild type had a peak at 1.0 to 1.75 μm/s, but SMIFH2-treated arp2-1 had peaks at 1.0 to 1.25 and 1.75 to 2.0 μm/s. It should be noted that experiments reported here were conducted at the same time as those testing CK-666; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 3 and 6. n ≥ 100 single filaments from 2 individual biological repeats (for 1 biological repeat, 5 single filaments were counted in 1 hypocotyl from at least 10 hypocotyls per genotype or treatment). Letters a to d denote genotypes or treatments that show statistically significant differences with other groups by chi-squared test, P < 0.05. F to H) The average maximum length of filaments that originated de novo or from side-branching events in SMIFH2-treated arp2-1 was significantly shorter than that in other cells, but filaments that originated from preexisting ends did not show a difference between any genotype and treatment. I to K) The average maximum lifetime of side-branching filaments in DMSO-treated arp2-1, SMIFH2-treated arp2-1, and SMIFH2-treated wild-type cells were all significantly longer than the ones from DMSO-treated wild-type cells; however, filaments that originated de novo did not show any significant difference, and filaments that elongated from preexisting ends in DMSO-treated wild-type cells were slightly shorter than SMIFH2-treated cells. L to N) The severing frequency did not show any significant difference between different genotypes or treatments. It should be noted that experiments reported here were conducted at the same time as those testing CK-666; therefore, control data sets for DMSO-treated wild-type and DMSO-treated arp2-1 cells are the same in Figs. 3 and 6. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to c denote groups that show statistically significant differences with other genotypes or treatments (within the same filament nucleation subclass) by 2-way ANOVA with Tukey's post hoc test (P < 0.05). WT, wild type.
To test whether the homeostatic actin filament array generated by these 2 nucleators depends upon the Arp2/3 complex or formins, or both, we genetically and/or chemically inhibited the Arp2/3 complex and formins. Both Arp2/3-inhibited (DMSO-treated arp2-1 or arpc2) and formin-inhibited (SMIFH2-treated wild-type) cells had a significant decrease in actin filament abundance as well as in the extent of filament bundling compared to DMSO-treated wild type (Fig. 5; Supplemental Fig. S7). Thus, both the Arp2/3 complex and formins are required to generate dense arrays of individual actin filaments in the homeostatic actin cortex. To test whether filament dynamics or the types and extent of filament nucleation are distinctly due to the Arp2/3 complex or formins, we examined single filament dynamics in genetically and/or chemically inhibited material. The nucleation frequency of both total and side-branching filaments was significantly decreased in both Arp2/3-inhibited (DMSO-treated arp2-1) and formin-inhibited (SMIFH2-treated wild-type) cells compared to wild-type (Fig. 6, A and C to E; Supplemental Movies S2 and S10), and the nucleation frequency of side-branched filaments in Arp2/3-inhibited cells (0.09 ± 0.01 events/filament/min) was ∼45% less than in formin-inhibited cells (0.16 ± 0.02 events/filament/min; Fig. 6D). However, the nucleation frequency due to de novo filament origins was not influenced when either the Arp2/3 complex or formin was suppressed (Fig. 6C). The average filament elongation rate in Arp2/3-inhibited cells (1.89 ± 0.07 μm/s) was also significantly higher than the rate in formin-inhibited cells (1.57 ± 0.06 μm/s) or in wild-type cells (1.79 ± 0.05 μm/s). In addition, the population distribution of filament elongation rates in Arp2/3-inhibited cells was significantly left skewed (peak value at 2.0 to 2.5 μm/s) compared to wild type (peak value at 1.25 to 1.75 μm/s), whereas formin-inhibited cells lost a large portion of filaments elongating at 2.0 to 2.5 μm/s but the distribution had a prominent peak at 1.0 to 1.5 μm/s (Fig. 6B). In addition, filaments in Arp2/3-inhibited cells had a longer average length compared to formin-inhibited cells (Fig. 6, F to H), but we did not observe any significant difference in filament lifetime or severing frequency when parameters of side-branched filaments in Arp2/3-inhibited cells and formin-inhibited cells were compared (Fig. 6, I to N). Again, we found similar results in both actin architecture (Supplemental Fig. S7) and single filament dynamics (Supplemental Fig. S8) when arpc2 and its respective wild-type line treated with DMSO solution or SMIFH2 were compared. These data indicated that both the Arp2/3 complex and formin predominantly generate side-branched actin filaments and formin-nucleated filaments elongate faster and longer than Arp2/3-nucleated ones.
To test whether SMIFH2 exhibits off-target inhibition of myosin superfamily members, as reported in Drosophila melanogaster (Nishimura et al. 2021), we evaluated the influence of SMIFH2 (and CK-666) on the activity of myosin XIK, the major myosin isoform mediating the delivery, vesicle tethering, and exocytosis of secretory vesicles in Arabidopsis cells (Zhang et al. 2021). We found that neither SMIFH2 nor CK-666 treatment reduced the speed of myosin XIK-YFP motility compared to pentabromopseudilin (PBP), an effective myosin inhibitor in plant cells (Supplemental Fig. S9, D to F) (Zhang et al. 2019). In addition, we also measured filament convolutedness and the rate of change of convolutedness of actin filaments, which are parameters describing the buckling and straightening of filaments (Staiger et al. 2009; Cai et al. 2014). Both parameters were significantly decreased after treatments with SMIFH2 or PBP but unchanged by CK-666, indicating that SMIFH2 does indeed alter filament buckling and straightening (Supplemental Fig. S9, A to C). These data suggest that SMIFH2 may not have an off-target effect on the activity of myosin XI based on the lack of effect on the motility of YFP-XIK; however, it remains a formal possibility that SMIFH2 targets myosin isoforms other than myosin XIK. Although the inhibition of filament buckling and straightening could indicate an off-target effect of SMIFH2 on myosin XI, it seems equally likely that filament buckling is caused by the processive assembly of actin filaments by Arabidopsis formins (Supplemental Fig. S9, A to C).
Simultaneous genetic and/or chemical inhibition of Arp2/3 and formins increases spontaneous filament nucleation and overall actin filament abundance
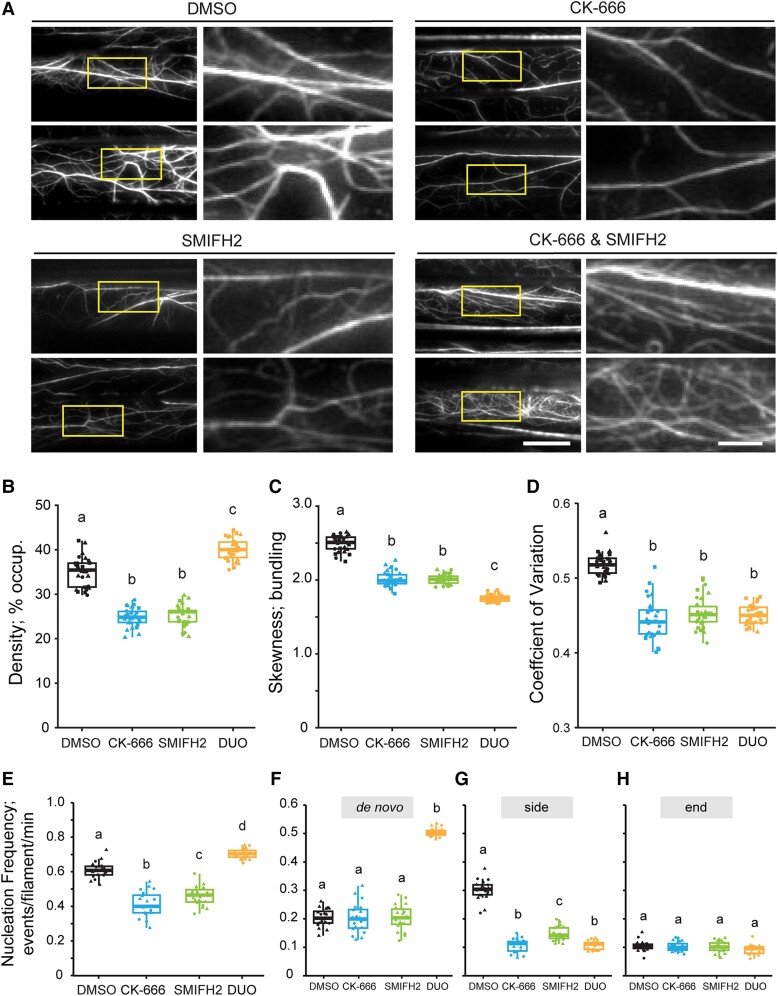
To test whether 2 classes of nucleator cooperate to generate and maintain the homeostatic actin cortical array in plant epidermal cells, we explored means to simultaneously suppress the activity of both the Arp2/3 complex and formins and quantitatively evaluate the effects on actin organization and dynamics. Specifically, using chemical and genetic tools to suppress the activity of both the Arp2/3 complex and formins simultaneously, we applied SMIFH2 to arp2-1 or arpc2 cells, treated wild-type cells with a combination of CK-666 and SMIFH2, and applied CK-666 to the fh1-2 mutant. Since the Arp2/3 complex and formins are, so far, the only 2 actin filament nucleators identified in plant cells, we predicted that simultaneously inhibiting the Arp2/3 complex and formin would markedly suppress actin filament nucleation activities and significantly reduce filament abundance. Surprisingly, in arp2-1 cells treated with SMIFH2, the actin filament abundance was significantly increased compared to cells with normal Arp2/3 complex and formin activities or with only 1 nucleator inhibited (Fig. 5B). Notably, SMIFH2-treated arp2-1 cells exhibited a marked abundance of short, single actin filaments in the cortical array (Fig. 5A). Unlike actin array density, the extent of actin filament bundling was not restored to wild-type levels when both the Arp2/3 complex and formins were simultaneously inhibited (Fig. 5, C and D). In addition to increased filament abundance, total filament nucleation frequency was significantly increased when both the Arp2/3 complex and formin activities were reduced, and a 2.5-fold enhancement of de novo filament origins was responsible for this increase (Fig. 6, A and C to E; Supplemental Movie S11). However, the lengths of filaments generated de novo or from the side-branching nucleation events were both significantly reduced in SMIFH2-treated arp2-1 cells compared to other treatments and genotypes (Fig. 6, F to H). Similar results were obtained when arp2-1 epidermal cells from light-grown cotyledons were treated with SMIFH2 (Supplemental Fig. S4), demonstrating that enhanced filament abundance and a 2.5-fold increase in de novo filament nucleation were not unique to etiolated hypocotyls. Moreover, the findings were identical when GFP-LifeAct was used as the reporter instead of GFP-fABD2 (Supplemental Fig. S3), indicating that the reporter plays little or no role in the results obtained. Finally, we also observed similar phenotypes in both actin architecture and filament dynamics when we applied SMIFH2 to arpc2 (Supplemental Figs. S7 and S8) or simultaneously applied CK-666 and SMIFH2 to wild type (Fig. 7). These results indicate that a marked increase in de novo filament nucleation results when both the Arp2/3 complex and formins are inhibited in plant epidermal cells.
Figure 7.
Filament abundance, total nucleation events, and de novo filament formation increase when the Arp2/3 complex and formin activity are simultaneously reduced with chemical inhibitors. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls are shown in the left columns. Scale bar: 20 μm. ROIs (boxes) were magnified and displayed in the right columns. Scale bar: 5 μm. Hypocotyls were treated with 0.05% DMSO solution, 10 µM CK-666, 25 µM SMIFH2, or both inhibitors for 5 min prior to imaging with VAEM. Actin filament arrays in CK-666-treated cells and SMIFH2-treated cells appeared to be less dense and less bundled compared to mock-treated cells. However, dual treatment markedly increased actin filament abundance. B to D) Quantitative analysis of actin filament density B) and extent of bundling by skewness C) and CV D) analyses. The density of actin arrays in CK-666-treated and SMIFH2-treated cells was decreased compared to mock-treated cells; however, dual-treated cells had significantly increased actin density compared to all other treatments. Actin arrays in CK-666- or dual-treated cells were significantly less bundled than in mock-treated cells. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 3 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 30 seedlings, 10 seedlings per biological repeat). Letters a to c denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). E to H) Quantitative analysis of actin filament nucleation frequency, both overall E) and by subclass of origin F to H). The total nucleation frequency in mock-treated cells was higher than CK-666-treated and SMIFH2-treated cells. However, the total nucleation frequency of dual-treated cells was significantly higher than all other treatments and this correlated with increased de novo nucleation events. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to c denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). DUO, dual-treated; WT, wild type.
Besides the possible inhibition of myosin XI, another potential issue with using SMIFH2 as a formin inhibitor on plant cells is that Arabidopsis has 21 FORMIN homologs, and it remains unclear whether SMIFH2 can inhibit all of them. To investigate the efficacy of SMIFH2 for suppressing formin activity, as well as its ability to phenocopy the effects of plant formin mutants, we prepared a homozygous AtFORMIN1 mutant line, fh1-2, expressing GFP-fABD2. AtFORMIN1 is a major housekeeping FH in Arabidopsis vegetative tissues (Cvrčková et al. 2000; Blanchoin and Staiger 2010; Rosero et al. 2013, 2016). Previously, we demonstrated that SMIFH2 inhibits the nucleation and assembly activity of recombinant FH2 domain from AtFORMIN1 in vitro (Cao et al. 2016). Here, we found that the density of actin filament arrays was significantly reduced but the extent of filament bundling was higher in DMSO-treated fh1-2 cells compared to DMSO-treated wild-type cells (Fig. 8, A to D). The nucleation frequency, specifically the side-branched filament subclass, was also significantly decreased in DMSO-treated fh1-2 cells compared to DMSO-treated wild-type cells (Fig. 8, E to H). SMIFH2 treatment caused an additional reduction in both actin filament abundance and nucleation activity in fh1-2 cells (Fig. 8). These results suggest that SMIFH2 targets more FHs than just AtFORMIN1; however, we cannot rule out the possibility that SMIFH2 does not inhibit all formins. Consistent with previous biochemical results (Michelot et al. 2007), these findings provide genetic evidence that AtFORMIN1 is a filament nucleator that generates new daughter filaments from the side of a mother filament or bundle.
Figure 8.
SMIFH2 treatment results in further reduction of both actin density and actin filament nucleation frequency in the fh1-2 mutant. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls are shown in the left columns. Scale bar: 20 μm. ROIs (boxes) were magnified and displayed in the right columns. Scale bar: 5 μm. Hypocotyls were treated with 0.05% DMSO solution, 10 µM CK-666, 25 µM SMIFH2, or both inhibitors for 5 min prior to imaging. Actin filament arrays in either DMSO-treated or single-inhibitor-treated fh1-2 cells all appeared to be less dense compared to DMSO-treated wild-type cells, but both dual-inhibitor-treated wild-type and fh1-2 cells appeared to have more dense actin arrays compared to DMSO-treated wild-type cells. B to D) Quantitative analysis of the percentage of occupancy or density of actin filament arrays B) and the extent of filament bundling as measured by skewness C) and coefficient of variance D) analyses. The density of actin arrays in DMSO-treated fh1-2 cells was significantly decreased but the extent of filament bundling was increased compared to DMSO-treated wild-type cells. Treatment with either CK-666 or SMIFH2 caused an additional decrease in actin density in fh1-2 cells and filaments were less bundled compared to DMSO-treated wild type. However, simultaneous treatment with both inhibitors significantly increased actin array density in both wild-type and fh1-2 cells. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to d denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). E to H) Quantitative analysis of actin filament nucleation frequency, both overall E) and by subclass of origin F to H). The total nucleation frequency in DMSO-treated fh1-2 cells was significantly reduced compared to DMSO-treated wild-type cells, and either CK-666 or SMIFH2 treatment caused an additional decrease in overall filament nucleation in fh1-2 cells E). However, the total nucleation frequency of dual-inhibitor-treated fh1-2 or wild-type cells was significantly higher than all other genotypes and treatments E) and this correlated with increased de novo nucleation events F). In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to d denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). DUO, dual-treated; WT, wild type.
To further test whether loss of a single FH could recapitulate the effects of SMIFH2, we treated fh1-2 with CK-666 or a combination of CK-666 and SMIFH2, as shown in Fig. 8. Treatment of fh1-2 with CK-666 led to a further significant reduction in filament density (Fig. 8B) as well as side-branched nucleation frequency (Fig. 8G), but no change in de novo nucleation events (Fig. 8F), compared to DMSO-treated fh1-2. This does not resemble the dual inhibition of wild type with SMIFH2 and CK-666 (Fig. 7), suggesting that the loss of a single major formin is not responsible for the phenotype. Further, we found that identical to wild-type cells simultaneously treated with SMIFH2 and CK-666, fh1-2 responds to dual inhibition with significantly increased filament abundance (Fig. 8B) as well as enhanced total (Fig. 8E) and de novo nucleation frequency (Fig. 8F). These results indicate that loss of AtFH1 does not play a role in the rapid response to inhibition of 2 classes of nucleator in epidermal cells resulting in enhanced filament formation through de novo nucleation.
PRF1 does not play a role in the enhanced de novo nucleation in response to simultaneous inhibition of 2 classes of nucleator
Profilins are actin monomer-binding proteins that suppress spontaneous nucleation, prevent subunit addition onto filament pointed ends (Cao et al. 2016; Sun et al. 2018), and are reportedly present at up to a 3-fold molar excess to total actin protein in several plant tissues (Chaudhry et al. 2007). To test whether the enhanced de novo actin filament nucleation, when the Arp2/3 complex and formins are both inhibited, was due to spontaneous nucleation, we introduced a PROFILIN1 (PRF1) mutant, prf1-2, to suppress one of the major profilin homologs in Arabidopsis vegetative tissues (Sun et al. 2018; Qiao et al. 2019). Consistent with our previous findings (Cao et al. 2016), prf1-2 had significantly reduced actin filament abundance (Fig. 9, A and B), decreased total and side-branched nucleation (Fig. 9, E and G), but modestly elevated de novo nucleation frequency (Fig. 9F) compared to mock-treated wild type. If the enhanced de novo nucleation frequency following simultaneous inhibition of both nucleators is due to the spontaneous nucleation resulting from increased free actin monomers, then we expect that prf1-2 would be less responsive to these conditions as it presumably already has an elevated free monomer concentration. We found that the triple inhibition of Arp2/3, formins, and PRF1 caused a significantly increased actin density (Fig. 9, A and B), as well as enhanced total and de novo nucleation frequency (Fig. 9, E to H), all of which were slightly elevated compared with dual-inhibitor-treated wild type. However, either CK-666 or SMIFH2 single-inhibitor treatment on prf1-2 did not cause a 2.5-fold higher de novo nucleation frequency (Fig. 9E) as observed in dual-inhibitor-treated prf1-2 cells. These results suggest that a change in free actin monomer concentration or the function of PRF1 are is the cause of the elevated de novo nucleation when both the Arp2/3 complex and formins are inhibited.
Figure 9.
Increased filament abundance, total nucleation events, and de novo filament formation following simultaneous CK-666 and SMIFH2 treatment are further enhanced in the prf1-2 mutant. A) Representative images of epidermal cells from the apical region of 5-d-old etiolated hypocotyls are shown in the left columns. Scale bar: 20 μm. ROIs (boxes) were magnified and displayed in the right columns. Scale bar: 5 μm. Hypocotyls were treated with 0.05% DMSO solution, 10 µM CK-666, 25 µM SMIFH2, or both inhibitors for 5 min prior to imaging with VAEM. Actin filament arrays in either DMSO-treated or single-inhibitor-treated prf1-2 cells all appeared to be less dense and less bundled compared to DMSO-treated wild-type cells, but both dual-inhibitor-treated wild-type and prf1-2 cells appeared to have more dense actin arrays compared to DMSO-treated wild-type cells. B to D) Quantitative analysis of actin filament density B) and extent of bundling by skewness C) and CV D) analyses. The density of actin arrays in either DMSO-treated or single-inhibitor-treated prf1-2 cells was significantly decreased compared to DMSO-treated wild-type cells; however, the treatment with both inhibitors significantly increased the actin density in both wild-type and prf1-2 cells. Actin arrays were less bundled when either Arp2/3, formins, or PRF1 was inhibited compared to DMSO-wild-type cells. In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 3 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 30 seedlings, 10 seedlings per biological repeat). Letters a to f denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). E to H) Quantitative analysis of actin filament nucleation frequency, both overall E) and by subclass of origin F to H). The total nucleation frequency in DMSO-treated, CK-666-treated, and SMIFH2-treated prf1-2 cells was significantly reduced compared to DMSO-treated wild-type cells. However, the total nucleation frequency of dual-inhibitor-treated prf1-2E) was significantly higher than all other genotypes and treatments and this corresponded to a significant increase in de novo nucleation events F). In box-and-whisker plots, boxes show the interquartile range and the median, and whiskers show the maximum–minimum interval of 2 biological repeats with independent populations of plants. Individual biological repeats are represented with different shapes (n = 20 seedlings, 10 seedlings per biological repeat). Letters a to e denote groups that show statistically significant differences with other genotypes or treatments by 2-way ANOVA with Tukey's post hoc test (P < 0.05). DUO, dual-treated; WT, wild type.
Discussion
In this study, we utilized a combination of genetic mutations or small molecule inhibitors along with high spatiotemporal resolution fluorescence microscopy to demonstrate, for the first time, that the Arp2/3 complex nucleates side-branched actin filaments in living Arabidopsis epidermal cells. We found that the Arp2/3 complex mutants, arp2-1 and arpc2, had reduced overall abundance of actin filaments in the cortical array, a reduction in the extent of filament bundles, and a significantly decreased frequency of side-branched nucleation events. In addition, acute treatment of wild-type plants with CK-666 phenocopied the actin-based defects observed in arp2/3 mutants, whereas applying CK-666 to arp2/3 mutants did not have any additional effect on overall actin structure or filament dynamics. Thus, we confirmed that these actin-based defects correlated with the loss of functional Arp2/3 complex and established that CK-666 is an effective tool for inhibition of the Arp2/3 complex in plant cells. Through experiments comparing genetic and chemical inhibition of the Arp2/3 complex or formins, we observed that both proteins were capable of nucleating side-branched actin filaments but that Arp2/3-nucleated filaments grew slower and were shorter than formin-nucleated ones. Surprisingly, simultaneous inhibition of both the Arp2/3 complex and formins led to an increase in actin filament abundance and dramatically promoted de novo nucleation events. By examining the consequences of the loss of a major housekeeping profilin isoform, PRF1, we rule out the role of this monomer-binding protein in the enhanced de novo nucleation following simultaneous inhibition of 2 classes of nucleator. These observations indicate that the regulatory mechanisms for actin filament nucleation in the homeostatic cortical array of plant cells may be unique and reveal a failsafe mechanism that maintains filament abundance and dynamics when both nucleators are inactivated.
The Arp2/3 complex is a major nucleator of side-branched actin filaments in Arabidopsis epidermal cells
Biochemical studies demonstrate that the Arp2/3 complex and formins use different molecular mechanisms to overcome the rate-limiting step for filament formation, and they typically generate different types of actin arrays. After creating an actin nucleus, the Arp2/3 complex remains attached to the pointed end of the new filament so that the barbed end is free for the addition of actin monomers, profilin-actin, or capping protein (Mullins et al. 1998; Amann and Pollard 2001; Fujiwara et al. 2002; Ichetovkin et al. 2002; Rouiller et al. 2008). In contrast, many formins remain associated at the filament barbed end after the nucleation and processively facilitate filament elongation through the addition of profilin-actin. This allows formin-associated filaments to grow much faster and longer compared to filaments with free barbed ends that are typically bound by heterodimeric capping protein soon after initiation (Kovar and Pollard 2004; Romero et al. 2004; Kovar 2006). Based on these observations, the conventional model of actin nucleation posits that formins generate long, unbranched filaments with a fast filament elongation rate, whereas the Arp2/3 complex nucleates short, branched actin networks with a slower growth rate. The Arp2/3 complex is accepted as a major nucleator of side-branched actin filaments in yeast and animal cells (Mullins et al. 1998; Amann and Pollard 2001; Fujiwara et al. 2002; Ichetovkin et al. 2002; Rouiller et al. 2008). Even though the Arp2/3 complex is also shown to be critical for plant cells to maintain their homeostatic actin organization and is required for a wide variety of cellular activities, little is known about the exact function of plant Arp2/3 complex in terms of its contribution to actin filament dynamics.
In this study, we characterized the function of the Arp2/3 complex in coordinating the organization of the cortical actin array as well as single actin filament dynamics in living Arabidopsis cells. Epidermal cells of etiolated hypocotyls, as well as light-grown cotyledons, with genetically or chemically inhibited Arp2/3 complex showed a quantifiable decrease in both actin filament abundance and extent of filament bundling, and, more importantly, there was a 50% to 70% reduction in the nucleation frequency of side-branched actin filaments. These results provide a compelling live-cell view of the formation of branched actin filaments by active Arp2/3 complex at single filament resolution. Furthermore, our results reveal that there are at least 2 populations of filaments with different dynamic properties, likely due to different filament growth mechanisms and/or the availability of filament barbed ends for monomer addition, that contribute to the organization of the homeostatic actin cortical array in epidermal cells. By examining filament elongation rate frequencies when formin alone or both the Arp2/3 complex and formins were inhibited, filaments with elongation rates of 1 to 1.25 µm/s predominated. We expect this represents the population of filaments with free barbed ends extending by monomer addition and is consistent with the in vitro mechanism of branched filament nucleation by the Arp2/3 complex.
The Arp2/3 complex and formins both mediate side-branched filament nucleation but generate filaments with unique dynamic properties
Nucleation is the rate-limiting step for actin filament formation in vitro and depends on the availability of actin monomers and is suppressed by profilin (Pollard et al. 2000; Sept and McCammon 2001; Michelot et al. 2005). Previous results with fission and budding yeasts, S. pombe and Saccharomyces cerevisiae, as well as mammalian cells, reveal a competition between the Arp2/3 complex and formin for a limited pool of actin monomers; consequently, formin-mediated long filament bundles dominate when the Arp2/3 complex is inhibited, whereas an increased abundance of actin patches created by the Arp2/3 complex is prominent when formins are downregulated (Hotulainen and Lappalainen 2006; Burke et al. 2014; Lomakin et al. 2015; Suarez et al. 2015; Fritzsche et al. 2016; Davidson et al. 2018; Antkowiak et al. 2019; Chan et al. 2019; Kadzik et al. 2020). Plant cells, by contrast, appear to show cooperation between these 2 nucleators rather than a competition to build distinct actin arrays. The cortical cytoplasm of epidermal cells contains a dynamic actin array that is rather disordered and comprises comingled actin filament bundles and individual actin filaments (Staiger et al. 2009; Smertenko et al. 2010). Evidence for actin patches or dense dendritic actin networks is limited to certain cell types, such as guard mother cells (Facette et al. 2015), the apex of trichomes (Yanagisawa et al. 2015), or at focal sites elicited by pathogen attack (Hardham et al. 2007; Qin et al. 2021). Our results revealed that the density of actin filaments in the cortical array, the extent of filament bundling, and filament nucleation frequency all decreased when either the Arp2/3 complex or formins were inhibited, suggesting that the Arp2/3 complex or formins alone are not able to maintain the homeostasis of the cortical actin arrays when the other nucleator is not functional.
Our results are consistent with a previous report showing partial cooperation between plant filament nucleation mechanisms in Arabidopsis cotyledon epidermal cells; however, the consequences of losing either class of nucleator were markedly different (Cifrová et al. 2020). That study showed a minor increase in actin filament density in either the formin fh1 mutant or the arpc5 mutant (Cifrová et al. 2020), rather than the prominent decreases observed here. As demonstrated here, these differences are not due to the use of different actin filament reporters (LifeAct versus fABD2), different developmental states and organs for examining epidermal cells (cotyledons versus dark-grown hypocotyls), or the result of compensation from genetic loss of 1 formin isoform versus acute treatment with a chemical inhibitor.
Our results showed that both the Arp2/3 complex and formins, including AtFORMIN1, are responsible for generating side-branched filaments because the inhibition of either one led to a significant reduction in side-branched filament nucleation and did not promote the other 2 subclasses of filament nucleation. These results also suggest that the Arp2/3 complex and formins are not competing for actin monomers for filament nucleation. Based on the total actin concentration, the ratio of F-actin to total actin, and the ratio of profilin to total actin in other plant tissues, the estimated actin monomer pool in Arabidopsis hypocotyl cells is relatively large, and most monomers are likely to be in a profilin-actin complex (Chaudhry et al. 2007; Staiger et al. 2010); therefore, the Arp2/3 complex and formins may not have to compete for a limited supply of actin monomers in plant cells. Here, if we assume that 1.25 µm/s is the rate of addition of monomers to free barbed ends, that the association rate constant for plant actin is similar to ATP-loaded rabbit skeletal muscle α-actin (k+ = 11.6 µM−1 s−1), and that a micron of actin filament comprises 370 subunits (Pollard et al. 2000), then we estimate the available monomer pool to be 40 µM in hypocotyl epidermal cells using the equation: [G-actin] = rate/k+. In addition, even though both the Arp2/3 complex and formins generate side-branched actin filaments, these filaments have markedly different properties. We found that formin-nucleated filaments grew significantly faster at rates of 2 to 2.25 µm/s, with another small subpopulation growing at rates of >3 µm/s. The different properties of filament barbed ends as well as a large pool of monomers, perhaps buffered with profilin, may facilitate the fast-growing filaments nucleated by side-branching as previously found in Arabidopsis cells (Cao et al. 2016) and are consistent with previous in vitro results (Vavylonis et al. 2006; Michelot et al. 2013; Suarez et al. 2015; Zhang et al. 2016; Funk et al. 2019).
If using the conventional model for how formins generate new actin filaments, we would predict that plant processive formins not only generate growing filaments from the side of a mother filament but also remain attached to the barbed end of the new filament to facilitate the addition of profilin-actin to the growing ends, resulting in faster and longer side-branched filaments compared to Arp2/3-nucleated ones. However, previous in vitro data showed that AtFORMIN1 nucleates actin filaments but remains associated at the pointed end and does not compete with capping protein for filament barbed ends, indicating it is nonprocessive and associates with the side of filaments after nucleation (Michelot et al. 2005, 2006). Further, SMIFH2 application in our studies reduced nucleation frequency but did not reduce the filament length of side-branched filaments compared to the mock treatment. Both in vitro and in vivo results suggest that nonprocessive formins in plant cells may have a weak interaction with filament barbed ends so that these formins move away from the end of the filament to its side to generate a new side-branched filament instead of occupying the barbed end and facilitating filament elongation. However, another analysis showed that the processive formin, AtFORMIN14, promotes de novo filament growth via the association with profilin in vitro (Zhang et al. 2016). All these results suggest that plant formins that nucleate side-branched filaments may have distinct molecular properties compared to formins that nucleate de novo filament formation or formins found in yeast or animal cells. With the exception of enhanced bundling, the loss of a single formin isoform, fh1-2, recapitulates but is less dramatic than the effects of SMIFH2 treatment of epidermal cells, supporting the conclusion that multiple formin isoforms are likely inhibited by this small molecule.
Since SMIFH2 shows off-target inhibition of members of the myosin superfamily (Nishimura et al. 2021), we were compelled to critically evaluate the effects of SMIFH2 treatment on plant cells. Although our data showed that SMIFH2 treatment did not influence the motility of myosin XIK, it did significantly reduce filament convolutedness and the rate of change of convolutedness of actin filaments, suggesting that SMIFH2 alters filament buckling and straightening. However, the change in filament buckling could be the consequence of different filament growth rates and processive elongation by formins (Staiger et al. 2009; Cai et al. 2014; Cao et al. 2016). Even though a recent in vitro study reveals that SMIFH2 is a pan-inhibitor for all human formins (Orman et al. 2022), we should not exclude the possibility that SMIFH2 can only inhibit a subset of plant FHs. Further genetic and biochemical analyses should be designed to dissect the interaction between SMIFH2 and different FHs to determine the specificity of this chemical inhibitor on plant formins.
Simultaneous inhibition of the Arp2/3 complex and formins promotes de novo nucleation
Previous studies demonstrate that the simultaneous inhibition of the Arp2/3 complex and formins results in significantly reduced cortical actin dynamics and turnover in both fission yeast and animal cells (Burke et al. 2014; Fritzsche et al. 2016); however, the application of both CK-666 and SMIFH2 does not lead to any significant difference in the formation of filopodial protrusions in the amoeba Naegleria or in sea urchin (Strongylocentrotus droebachiensis) compared to CK-666 treatment alone (Henson et al. 2015; Velle and Fritz-Laylin 2020). When we combined genetic and chemical methods to downregulate the Arp2/3 complex and formins simultaneously in Arabidopsis epidermal cells, surprisingly, the frequency of actin filament nucleation was significantly enhanced by up to 2.5-fold, with de novo nucleation events appearing to be the exclusive contributor to this enhancement. This difference in actin dynamics upon the loss of multiple major actin filament nucleators suggests a unique stress response mechanism in plant cells. We acknowledge that the use of VAEM to visualize a single optical plane in the cortical cytoplasm could potentially lead to the misidentification of some new filaments growing into the field of view as de novo nucleation events; however, we view this as an unlikely explanation for the dramatic increase in overall nucleation events observed following simultaneous inhibition of 2 major nucleators. The Arp2/3 complex and formins are, to date, the only 2 bona fide actin filament nucleators in plant cells (Blanchoin and Staiger 2010; Vaškovičová et al. 2013; Yanagisawa et al. 2013). Theoretically, when both the Arp2/3 complex and formins are inhibited, the concentration of free actin monomers is predicted to increase, but the ratio of F- to G-actin and the proportion of profilin-actin in the monomer pool should both decrease. It is possible that these changes in the actin monomer pool could increase the nucleation rate of spontaneous filament formation thereby enhancing the total frequency of de novo nucleation events. In support of this model, disruption of AtPRF1 led to a modest increase in de novo nucleation events and a decrease in side-branched events (Cao et al. 2016). However, the prf1-2 mutant exhibits a 2.5-fold increase in de novo nucleation frequency, just like wild-type epidermal cells, when both nucleators are inhibited with chemicals. Perhaps, as suggested by Sun et al. (2018), another profilin isoform, PRF3, is the major regulator of spontaneous filament nucleation and sequesters actin monomers that are poorly utilized by formins. The acute chemical inhibition of nucleation machinery could lead to rapid posttranslational modification and inactivation of PRF3, thereby facilitating spontaneous filament formation from free monomers. This hypothesis needs to be tested through future experiments.
In conclusion, our results provide direct evidence to demonstrate that the Arp2/3 complex nucleates side-branched actin filaments in living plant cells. These studies also confirm that some formin isoforms, including AtFORMIN1, generate side-branched actin filaments in plant cells, which is a unique actin filament nucleation mechanism compared to yeast or animal cells. Moreover, we find that Arp2/3- and formin-nucleated filaments have distinct dynamic properties, which can be applied to future studies about how cells coordinate these 2 nucleators to regulate the actin structure and dynamics for different cellular activities. The simultaneous inhibition of both the Arp2/3 complex and formin surprisingly caused an enhanced de novo filament nucleation, which raises the possibility for a unique actin nucleation mechanism to generate dynamic cortical actin arrays in plant cells.
Materials and methods
Plant materials and growth conditions
All Arabidopsis (A. thaliana) plants in this study were in the Col-0 background. The ARP2 T-DNA insertion line, arp2-1 (SALK_003448), the PRF1 T-DNA insertion line, prf1-2 (SALK_057718; Cao et al. 2016), and the AtFORMIN1 T-DNA insertion line, fh1-2 (SALK_009693; Rosero et al. 2013), were obtained from the Arabidopsis Biological Resource Center (Ohio State University). The ARPC2 point mutation line, arpc2 (G1217A), was kindly provided by Dan Szymanski (Purdue University). Mutant lines, arp2-1, arpc2, and prf1-2, were crossed to wild type (Col-0) expressing the GFP-fABD2 reporter (Sheahan et al. 2004; Staiger et al. 2009), and homozygous mutants and corresponding wild-type siblings were recovered from F2 populations. For the fh1-2 line, homozygous plants were transformed with a 35Spro:GFP-fABD2 construct using the floral dip method, and seeds were screened with kanamycin (Clough and Bent 1998; Sheahan et al. 2004). For lines carry GFP-LifeAct, both Col-0 and arp2-1 mutant were transformed with a UBQ:GFP-LifeAct construct (kindly provided by Weibing Yang from The Sainsbury Laboratory, University of Cambridge, Cambridge, UK) using the floral dip method and transformants selected with BASTA (Clough and Bent 1998).
Seeds were surface sterilized and stratified at 4 °C for 3 d on plates containing half-strength MS medium supplemented with 1% (w/v) sucrose and 1% (w/v) agar. For dark-grown hypocotyl growth, seedlings were grown in continuous darkness after exposing the plates to light for 4 h. For light-grown root and cotyledon growth, seeds were grown vertically on plates comprising half-strength MS medium supplemented with 0% (w/v) sucrose and 0.6% (w/v) agar under long-day conditions (16 h light/8 h dark) at 21 °C with a light intensity of 120 to 140 mmol m−2 s−1 provided by Philips F32T8/L941 Deluxe Cool White bulbs.
Drug treatments
For short-term live-cell treatments, individual seedlings were treated for 5 min in inhibitor solution in 6-well plates in the dark prior to each imaging session. Hypocotyls were mounted in the inhibitor solution and imaged immediately. CK-666 (Sigma-Aldrich; Cat # 182515; St. Louis, MO, USA), SMIFH2 (Sigma-Aldrich; Cat # 344092), and PBP (Adipogen; Cat # BVT-0441) were dissolved in DMSO to prepare a 5 mM stock solution.
Live-cell imaging
For both snapshot and time-lapse image acquisitions in dark-grown hypocotyls, epidermal cells from the apical region of 5-d-old dark-grown hypocotyls were used. In some experiments, the lipophilic dye, FM4-64 (Invitrogen; Cat # T3166; Carlsbad, CA, USA), was dissolved in DMSO and used at 20 mM to label the plasma membrane. All images and movies were collected using VAEM with an Olympus TIRF objective (60×, 1.45 numerical aperture) using SlideBook software (version 5.5; Intelligent Imaging Innovations) as described previously (Staiger et al. 2009; Henty et al. 2011; Li et al. 2012; Cai et al. 2014; Cao et al. 2016).
For experiments with cotyledons, adaxial epidermal cells from 5-d-old light-grown cotyledons were used. An Olympus IX-83 microscope mounted with a spinning-disc confocal unit (Yokogawa CSUX1-A1) and an Andor iXon Ultra 897BV EMCCD camera were used to acquire snapshot and time-lapse images with an Olympus UPlanSApo oil objective (100×, 1.45 numerical aperture). GFP fluorescence was excited with a 488-nm laser line, and emission was collected through the 525/30-nm filter to visualize actin filaments. All images were collected with MetaMorph software (version 7.8.8.0) as described previously (Cao et al. 2022).
A double-blinded experimental design was used for all image collection, processing, and quantitative analyses.
Image processing and quantitative analysis of actin cytoskeleton architecture and single actin filament dynamics
Images and movies were processed with Fiji Is Just ImageJ (Schindelin et al. 2012). All images used for actin architecture analysis were collected with a fixed exposure time, laser power, and camera gain setting.
To quantify actin architecture, 3 parameters, density, skewness, and CV, were employed in this study; density measures the percentage of occupancy of GFP signal in an image, and skewness and CV measure the extent of actin filament bundling (Higaki et al. 2010; Ueda et al. 2010; Henty et al. 2011; Li et al. 2012; Cai et al. 2014; Cao et al. 2016; Arieti and Staiger 2020). Micrographs were analyzed in Fiji Is Just ImageJ using the methods described previously (Higaki et al. 2010; Henty et al. 2011; Li et al. 2012; Higaki et al. 2020).
To compare single actin filament parameters between genotype or drug treatment, double-blind experiments (data collection and analysis) were performed. Maximum filament length and lifetime, filament severing frequency, elongation rates, filament convolutedness, and the rate of change of convolutedness were measured as described previously (Staiger et al. 2009; Henty et al. 2011; Li et al. 2012; Cai et al. 2014; Cao et al. 2016). For nucleation frequency analysis, a 400-µm2 ROI was randomly selected from movies of epidermal cells at the apical region of the hypocotyl, and all observable nucleation events were counted during a time period of 100 s, according to Cao et al. (2016). To account for differences in filament density in genotypes or drug treatments, the nucleation frequency was normalized against filament numbers in each ROI.
For actin architecture analysis in both hypocotyl and cotyledon epidermal cells, more than 150 ROIs were selected from 50 images of cells collected from at least 10 individual seedlings per genotype or treatment. For nucleation frequency analysis, more than 300 events from at least 10 individual seedlings were observed per genotype or treatment. For single actin filament dynamics analysis, at least 5 newly appeared filaments were tracked from their first appearance to their complete disappearance in each hypocotyl, and more than 50 filaments from at least 10 hypocotyls were measured per genotype or treatment. For all statistical comparisons and plotting, the average value from a single hypocotyl was used as 1 data point. All experiments were conducted with at least 2 independent biological repeats to make conclusions in each figure.
Statistical analyses
Two-way ANOVA with Tukey's post hoc tests was performed in SPSS (version 25) to determine the significance among different genotypes/treatments. Chi-square tests were performed for statistically comparing parametric distributions, and P values were calculated in Excel 15.32. Any difference with a P value less than 0.05 was considered significantly different. Detailed statistical analysis data are shown in Supplemental Data Set S1.
Accession numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (TAIR) under the following accession numbers: ARP2, AT3G27000; ARPC2, AT1G30825; AtFORMIN1, AT3G25500; and PROFILIN1, AT2G19760.
Supplementary Material
Acknowledgments
We thank Hongbing Luo (Purdue University) for excellent care and maintenance of plant materials, Dan Szymanski (Purdue University) for providing the arpc2 line, and David Kovar (University of Chicago) and Laurent Blanchoin (Interdisciplinary Research Institute of Grenoble) for helpful comments on the manuscript. We are also grateful to the Arabidopsis Biological Resource Center (Ohio State University) for supplying the arp2-1, prf1-2, and fh1-2 T-DNA insertion lines.
Contributor Information
Liyuan Xu, Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
Lingyan Cao, Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
Jiejie Li, Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
Christopher J Staiger, Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA; Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN 47907, USA; EMBRIO Institute, Purdue University, West Lafayette, IN 47907, USA.
Author contributions
L.X. acquired and analyzed the data, prepared the figures, and wrote and revised the manuscript; L.C. acquired and analyzed the data; J.L. acquired and analyzed the data; and C.J.S. conceptualized the experiments, acquired the funding, analyzed the data, and wrote and revised the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Disruption of the Arp2/3 complex perturbs epidermal cell morphology and inhibits normal plant growth.
Supplemental Figure S2. Actin filament nucleation frequency is decreased in arpc2 and after treatment of wild type with CK-666.
Supplemental Figure S3. Two different actin cytoskeleton markers, GFP-fABD2 and GFP-LifeAct, report similar changes in actin array organization and nucleation events regardless of the genotype or treatment.
Supplemental Figure S4. Cotyledon epidermal cells show similar phenotypes of reduced actin filament abundance and decreased nucleation frequency in arp2-1 mutants expressing either GFP-fABD2 or GFP-LifeAct cytoskeletal reporters.
Supplemental Figure S5. CK-666 is an acute and reversible chemical inhibitor of the Arp2/3 complex in plant cells.
Supplemental Figure S6. Actin array architecture is perturbed in arpc2 and the reduced filament density and bundling phenotypes are recapitulated by treatment with CK-666.
Supplemental Figure S7. Actin filament density increases after treatment of arpc2 with the formin inhibitor SMIFH2.
Supplemental Figure S8. Overall and de novo filament nucleation increases when ARPC2 and formin activity are simultaneously inhibited.
Supplemental Figure S9. Filament convolutedness decreases when formin or myosin activities are inhibited.
Supplemental Table S1. Single actin filament dynamics in wild type (ARPC2) and arpc2 mutant with or without CK-666.
Supplemental Movie S1. Full time-lapse sequence of actin filament nucleation activities in mock-treated wild-type (ARP2) cells.
Supplemental Movie S2. Full time-lapse sequence of actin filament nucleation activities in mock-treated arp2-1 cells.
Supplemental Movie S3. Representative actin filament nucleated de novo from the cytoplasm (as in Fig. 3A, top row).
Supplemental Movie S4. Representative actin filament nucleated from the side of a preexisting filament (as in Fig. 3A, middle row).
Supplemental Movie S5 . Representative actin filament nucleated from the end of a preexisting filament (as in Fig. 3A, bottom row).
Supplemental Movie S6. Representative example of filament elongation (as in Fig. 4A).
Supplemental Movie S7. Representative example of filament severing (as in Fig. 4A).
Supplemental Movie S8. Full time-lapse sequence of actin filament nucleation activities in CK-666-treated wild-type (ARP2) cells.
Supplemental Movie S9. Full time-lapse sequence of actin filament nucleation activities in CK-666-treated arp2-1 cells.
Supplemental Movie S10. Full time-lapse sequence of actin filament nucleation activities in SMIFH2-treated wild-type (ARP2) cells.
Supplemental Movie S11. Full time-lapse sequence of actin filament nucleation activities in SMIFH2-treated arp2-1 cells.
Supplemental Data Set S1. ANOVA and chi-square test results and parameters.
Funding
The TIRF microscopy facility was supported, in part, by the Bindley Bioscience Center at Purdue University. This work was funded by a grant from the Physical Biosciences Program of the US Department of Energy, Office of Basic Energy Sciences (DE-FG02-09ER15526) to C.J.S. Conceptualization of experiments, data analysis, and manuscript preparation by C.J.S. were also supported by the EMBRIO Institute, a National Science Foundation-Biological Integration Institute under contract no. 2120200.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008:133(5):841–851. 10.1016/j.cell.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MF, Fatema U, Peng X, Hacker SW, Maruyama D, Sun M-X, Kawashima T. ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants. Proc Natl Acad Sci U S A. 2020:117(51):32757–32763. 10.1073/pnas.2015550117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001:98(26):15009–15013. 10.1073/pnas.211556398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antkowiak A, Guillotin A, Boiero Sanders M, Colombo J, Vincentelli R, Michelot A. Sizes of actin networks sharing a common environment are determined by the relative rates of assembly. PLoS Biol. 2019:17(6):e3000317. 10.1371/journal.pbio.3000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti RS, Staiger CJ. Auxin-induced actin cytoskeleton rearrangements require AUX1. New Phytol. 2020:226(2):441–459. 10.1111/nph.16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008:283(11):7135–7144. 10.1074/jbc.M705894200 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Michelot A. Actin cytoskeleton: a team effort during actin assembly. Curr Biol. 2012:22(16):R643–R645. 10.1016/j.cub.2012.07.026 [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Staiger CJ. Plant formins: diverse isoforms and unique molecular mechanism. Biochim Biophys Acta. 2010:1803(2):201–206. 10.1016/j.bbamcr.2008.09.015 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013:126(1):1–7. 10.1242/jcs.107250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014:24(5):579–585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera R, Suo J, Young E, Chang EC. Schizosaccharomyces pombe Arc3 is a conserved subunit of the Arp2/3 complex required for polarity, actin organization, and endocytosis. Yeast. 2011:28(6):495–503. 10.1002/yea.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, Henty-Ridilla JL, Szymanski DB, Staiger CJ. Arabidopsis myosin XI: a motor rules the tracks. Plant Physiol. 2014:166(3):1359–1370. 10.1104/pp.114.244335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Henty-Ridilla JL, Blanchoin L, Staiger CJ. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol. 2016:170(1):220–233. 10.1104/pp.15.01321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Wang W, Zhang W, Staiger CJ. Lipid signaling requires ROS production to elicit actin cytoskeleton remodeling during plant innate immunity. Int J Mol Sci. 2022:23:2447. 10.3390/ijms23052447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M-F, Shekhar S. Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat Rev Mol Cell Biol. 2017:18(6):389–401. 10.1038/nrm.2016.172 [DOI] [PubMed] [Google Scholar]
- Chan FY, Silva AM, Saramago J, Pereira-Sousa J, Brighton HE, Pereira M, Oegema K, Gassmann R, Carvalho AX. The ARP2/3 complex prevents excessive formin activity during cytokinesis. Mol Biol Cell. 2019:30(1):96–107. 10.1091/mbc.E18-07-0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F, Guérin C, Von Witsch M, Blanchoin L, Staiger CJ. Identification of Arabidopsis Cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol Biol Cell. 2007:18(8):3002–3014. 10.1091/mbc.e06-11-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu H-M. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell. 2004:16(1):257–269. 10.1105/tpc.016550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifrová P, Oulehlová D, Kollárová E, Martinek J, Rosero A, Žárský V, Schwarzerová K, Cvrčková F. Division of labor between two actin nucleators-the formin FH1 and the ARP2/3 complex-in Arabidopsis epidermal cell morphogenesis. Front Plant Sci. 2020:11:148. 10.3389/fpls.2020.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacteriummediated transformation of Arabidopsis thaliana. Plant J. 1998:16:735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Courtemanche N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys Rev. 2018:10(6):1553–1569. 10.1007/s12551-018-0468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD, Chen Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat Cell Biol. 2016:18(6):676–683. 10.1038/ncb3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Zou M, Li J. Basally distributed actin array drives embryonic hypocotyl elongation during the seed-to-seedling transition in Arabidopsis. New Phytol. 2023:240(1):191–206. 10.1111/nph.19149 [DOI] [PubMed] [Google Scholar]
- Cvrčková F, Bavlnka B, Žárský V. Identification and cloning of AtFORMIN1, an Arabidopsis thaliana formin homologue. Biochem Soc Trans. 2000:28(5):A208–A208. 10.1042/bst028a208c [DOI] [Google Scholar]
- Cvrčková F, Novotný M, Pícková D, Žárský V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genom. 2004:5(1):44. 10.1186/1471-2164-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Amato C, Thomason PA, Insall RH. WASP family proteins and formins compete in pseudopod- and bleb-based migration. J Cell Biol. 2018:217(2):701–714. 10.1083/jcb.201705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Mullins RD. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004:2(4):476–485. 10.1371/journal.pbio.0020091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Hussey PJ, Davies B. Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 2002:7(11):492–498. 10.1016/s1360-1385(02)02341-5 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Le J, Basu D, Mallery EL, Szymanski DB. DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 2004:38:526–538. 10.1111/j.1365-313X.2004.02065.x [DOI] [PubMed] [Google Scholar]
- Facette MR, Park Y, Sutimantanapi D, Luo A, Cartwright HN, Yang B, Bennett EJ, Sylvester AW, Smith LG. The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat Plants. 2015:1:14024. 10.1038/nplants.2014.24 [DOI] [PubMed] [Google Scholar]
- Flores LR, Keeling MC, Zhang X, Sliogeryte K, Gavara N. Lifeact-TagGFP2 alters F-actin organization, cellular morphology and biophysical behaviour. Sci Rep. 2019:9(1):3241. 10.1038/s41598-019-40092-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M, Erlenkamper C, Moeendarbary E, Charras G, Kruse K. Actin kinetics shapes cortical network structure and mechanics. Sci Adv. 2016:2(4):e1501337. 10.1126/sciadv.1501337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Suetsugu S, Uemura S, Takenawa T, Ishiwata S. Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem Biophys Res Commun. 2002:293(5):1550–1555. 10.1016/S0006-291X(02)00421-7 [DOI] [PubMed] [Google Scholar]
- Funk J, Merino F, Venkova L, Heydenreich L, Kierfeld J, Vargas P, Raunser S, Piel M, Bieling P. Profilin and formin constitute a pacemaker system for robust actin filament growth. eLife. 2019:8:e50963. 10.7554/eLife.50963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X-Q, Wang X-L, Ren F, Chen J, Wang X-C. Dynamics of vacuoles and actin filaments in guard cells and their roles in stomatal movement. Plant Cell Environ. 2009:32(8):1108–1116. 10.1111/j.1365-3040.2009.01993.x [DOI] [PubMed] [Google Scholar]
- García-González J, Kebrlová S, Semerák M, Lacek J, Kotannal Baby I, Petrášek J, Schwarzerová K. Arp2/3 complex is required for auxin-driven cell expansion through regulation of auxin transporter homeostasis. Front Plant Sci. 2020:11:486. 10.3389/fpls.2020.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham AR, Jones DA, Takemoto D. Cytoskeleton and cell wall function in penetration resistance. Curr Opin Plant Biol. 2007:10(4):342–348. 10.1016/j.pbi.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Harries PA, Pan A, Quatrano RS. Actin-related protein2/3 complex component ARPC1 is required for proper cell morphogenesis and polarized cell growth in Physcomitrella patens. Plant Cell. 2005:17(8):2327–2339. 10.1105/tpc.105.033266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Yeterian M, Weeks RM, Medrano AE, Brown BL, Geist HL, Pais MD, Oldenbourg R, Shuster CB. Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol Biol Cell. 2015:26(5):887–900. 10.1091/mbc.E14-07-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Li J, Blanchoin L, Staiger CJ. Actin dynamics in the cortical array of plant cells. Curr Opin Plant Biol. 2013:16(6):678–687. 10.1016/j.pbi.2013.10.012 [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Shimono M, Li J, Chang JH, Day B, Staiger CJ. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog. 2013:9(4):e1003290. 10.1371/journal.ppat.1003290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty JL, Bledsoe SW, Khurana P, Meagher RB, Day B, Blanchoin L, Staiger CJ. Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell. 2011:23(10):3711–3726. 10.1105/tpc.111.090670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol. 2013:20(5):701–712. 10.1016/j.chembiol.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Akita K, Katoh K. Coefficient of variation as an image-intensity metric for cytoskeleton bundling. Sci Rep. 2020:10(1):22187. 10.1038/s41598-020-79136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S. Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 2010:61(1):156–165. 10.1111/j.1365-313x.2009.04032.x [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Lappalainen P. . Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006:173(3):383–394. 10.1083/jcb.200511093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002:12(1):79–84. 10.1016/s0960-9822(01)00629-7 [DOI] [PubMed] [Google Scholar]
- Jiang K, Sorefan K, Deeks MJ, Bevan MW, Hussey PJ, Hetherington AM. The ARP2/3 complex mediates guard cell actin reorganization and stomatal movement in Arabidopsis. Plant Cell. 2012:24(5):2031–2040. 10.1105/tpc.112.096263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzik RS, Homa KE, Kovar DR. F-actin cytoskeleton network self-organization through competition and cooperation. Annu Rev Cell Dev Biol. 2020:36(1):35–60. 10.1146/annurev-cellbio-032320-094706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006:18(1):11–17. 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003:161(5):875–887. 10.1083/jcb.200211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Progressing actin: formin as a processive elongation machine. Nat Cell Biol. 2004:6(12):1158–1159. 10.1038/ncb1204-1158 [DOI] [PubMed] [Google Scholar]
- Lan Y, Liu X, Fu Y, Huang S. Arabidopsis class I formins control membrane-originated actin polymerization at pollen tube tips. PLoS Genet. 2018:14(11):e1007789. 10.1371/journal.pgen.1007789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, El-Assal SE-D, Basu D, Saad ME, Szymanski DB. Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr Biol. 2003:13(15):1341–1347. 10.1016/s0960-9822(03)00493-7 [DOI] [PubMed] [Google Scholar]
- Li S, Blanchoin L, Yang Z, Lord EM. The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 2003:132(4):2034–2044. 10.1104/pp.103.028563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Huang S, Wang X, Blanchoin L, Staiger CJ. Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell. 2012:24(9):3742–3754. 10.1105/tpc.112.103945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li J-H, Wang W, Chen N-Z, Ma T-S, Xi Y-N, Zhang X-L, Lin H-F, Bai Y, Huang S-J, et al. ARP2/3 complex-mediated actin dynamics is required for hydrogen peroxide-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2014:37(7):1548–1560. 10.1111/pce.12259 [DOI] [PubMed] [Google Scholar]
- Li J, Staiger CJ. Understanding cytoskeletal dynamics during the plant immune response. Annu Rev Phytopathol. 2018:56:513–533. 10.1146/annurev-phyto-080516-035632 [DOI] [PubMed] [Google Scholar]
- Liu H-K, Li Y-J, Wang S-J, Yuan T-L, Huang W-J, Dong X, Pei J-Q, Zhang D, McCormick S, Tang W-H. Kinase partner protein plays a key role in controlling the speed and shape of pollen tube growth in tomato. Plant Physiol. 2020:184(4):1853–1869. 10.1104/pp.20.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin AJ, Lee K-C, Han SJ, Bui DA, Davidson M, Mogilner A, Danuser G. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat Cell Biol. 2015:17(11):1435–1445. 10.1038/ncb3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, Hulskamp M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell. 2003:15(7):1632–1645. 10.1105/tpc.011676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M. Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development. 2003:130(14):3137–3146. 10.1242/dev.00549 [DOI] [PubMed] [Google Scholar]
- Michelot A, Berro J, Guérin C, Boujemaa-Paterski R, Staiger CJ, Martiel J-L, Blanchoin L. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007:17(10):825–833. 10.1016/j.cub.2007.04.037 [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol. 2006:16(19):1924–1930. 10.1016/j.cub.2006.07.054 [DOI] [PubMed] [Google Scholar]
- Michelot A, Grassart A, Okreglak V, Costanzo M, Boone C, Drubin DG. Actin filament elongation in Arp2/3-derived networks is controlled by three distinct mechanisms. Dev Cell. 2013:24(2):182–195. 10.1016/j.devcel.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot AE, Guérin C, Huang S, Ingouff M, Richard SP, Rodiuc N, Staiger CJ, Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell. 2005:17(8):2296–2313. 10.1105/tpc.105.030908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998:95(11):6181–6186. 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shi S, Li Q, Bershadsky AD, Viasnoff V. Crosstalk between myosin II and formin functions in the regulation of force generation and actomyosin dynamics in stress fibers. Cells Dev. 2021:168:203736. 10.1016/j.cdev.2021.203736 [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009:460(7258):1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M, Landis M, Oza A, Nambiar D, Gjeci J, Song K, Huang V, Klestzick A, Hachicho C, Liu SQ, et al. Alterations to the broad-spectrum formin inhibitor SMIFH2 modulate potency but not specificity. Sci. Rep. 2022; 12:13520. 10.1038/s41598-022-17685-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008:18(1):9–19. 10.1016/j.cub.2007.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Yan T, Sun M. The WASP-Arp2/3 complex signal cascade is involved in actin-dependent sperm nuclei migration during double fertilization in tobacco and maize. Sci Rep. 2017:7:43161. 10.1038/srep43161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Cole RA, Fowler JE, Dolja VV. Myosin-powered membrane compartment drives cytoplasmic streaming, cell expansion and plant development. PLoS One. 2015:10(10):e0139331. 10.1371/journal.pone.0139331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007:36:451–477. 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- Pollard TD. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. 2016:8(8):a018226. 10.1101/cshperspect.a018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000:29:545–576. 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- Qi J, Greb T. Cell polarity in plants: the yin and yang of cellular functions. Curr Opin Plant Biol. 2017:35:105–110. 10.1016/j.pbi.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Sun H, Ng JTY, Ma Q, Koh SH, Mu Y, Miao Y, Gao Y-G. Structural and computational examination of the Arabidopsis profilin–Poly-P complex reveals mechanistic details in profilin-regulated actin assembly. J Biol Chem. 2019:294(49):18650–18661. 10.1074/jbc.ra119.011307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Liu L, Tu J, Yang G, Wang S, Quilichini TD, Gao P, Wang H, Peng G, Blancaflor EB, et al. The ARP2/3 complex, acting cooperatively with Class I formins, modulates penetration resistance in Arabidopsis against powdery mildew invasion. Plant Cell. 2021:33(9):3151–3175. 10.1093/plcell/koab170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M, Goldstein B. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell Sci. 2009:122(21):3983–3993. 10.1242/jcs.057562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier M-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004:119(3):419–429. 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- Rosenbloom AD, Kovar EW, Kovar DR, Loew LM, Pollard TD. Mechanism of actin filament nucleation. Biophys J. 2021:120(20):4399–4417. 10.1016/j.bpj.2021.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosero A, Oulehlová D, Stillerová L, Schiebertová P, Grunt M, Žárský V, Cvrčková F. Arabidopsis FH1 formin affects cotyledon pavement cell shape by modulating cytoskeleton dynamics. Plant Cell Physiol. 2016:57(3):488–504. 10.1093/pcp/pcv209 [DOI] [PubMed] [Google Scholar]
- Rosero A, Žárský V, Cvrčková F. AtFH1 formin mutation affects actin filament and microtubule dynamics in Arabidopsis thaliana. J Exp Bot. 2013:64(2):585–597. 10.1093/jxb/ers351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013:14(1):7–12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu X-P, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008:180(5):887–895. 10.1083/jcb.200709092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahi VP, Cifrová P, García-González J, Kotannal Baby I, Mouillé G, Gineau E, Müller K, Baluška FE, Soukup A, Petrášek J, et al. Arabidopsis thaliana plants lacking the ARP2/3 complex show defects in cell wall assembly and auxin distribution. Ann Bot. 2018:122(5):777–789. 10.1093/aob/mcx178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar A, Gutierrez R, McFarlane HE, Bringmann M, Lindeboom J, Emons AM, Samuels L, Ketelaar T, Ehrhardt DW, Persson S. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013:162(2):675–688. 10.1104/pp.113.215277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Lofke C, Kruger F, Kittelmann M, Eisa A, Hughes L, Smith RS, Hawes C, Schumacher K, Kleine-Vehn J. Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc Natl Acad Sci U S A. 2016:113(2):452–457. 10.1073/pnas.1517445113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A . Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012:9:676–682. 10.1038/nmeth [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001:81(2):667–674. 10.1016/s0006-3495(01)75731-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 2004:136(4):3968–3978. 10.1104/pp.104.049411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Deeks MJ, Hussey PJ. Strategies of actin reorganisation in plant cells. J Cell Sci. 2010:123(17):3019–3028. 10.1242/jcs.071126 [DOI] [PubMed] [Google Scholar]
- Spracklen AJ, Fagan TN, Lovander KE, Tootle TL. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol. 2014:393(2):209–226. 10.1016/j.ydbio.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Poulter NS, Henty JL, Franklin-Tong VE, Blanchoin L. Regulation of actin dynamics by actin-binding proteins in pollen. J Exp Bot. 2010:61(7):1969–1986. 10.1093/jxb/erq012 [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L. Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol. 2009:184(2):269–280. 10.1083/jcb.200806185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015:32(1):43–53. 10.1016/j.devcel.2014. doi:10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Qiao Z, Chua KP, Tursic A, Liu X, Gao YG, Mu Y, Hou X, Miao Y. Profilin negatively regulates formin-mediated actin assembly to modulate PAMP-triggered plant immunity. Curr Biol. 2018:28(12):1882–1895.e1887. 10.1016/j.cub.2018.04.045 [DOI] [PubMed] [Google Scholar]
- Sun S-C, Wang Q-L, Gao W-W, Xu Y-N, Liu H-L, Cui X-S, Kim N-H. Actin nucleator Arp2/3 complex is essential for mouse preimplantation embryo development. Reprod Fertil Dev. 2013:25(4):617–623. 10.1071/RD12011 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD, Wick SM. Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell. 1999:11(12):2331–2347. 10.1105/tpc.11.12.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, Hara-Nishimura I. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci U S A. 2010:107(15):6894–6899. 10.1073/pnas.0911482107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing HS, van Bezouwen LS, Emons AM, Ketelaar T. High expression of LifeAct in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskeleton (Hoboken). 2011:68(10):578–587. 10.1002/cm.20534 [DOI] [PubMed] [Google Scholar]
- van der Kammen R, Song J-Y, de Rink I, Janssen H, Madonna S, Scarponi C, Albanesi C, Brugman W, Innocenti M. Knockout of the Arp2/3 complex in epidermis causes a psoriasis-like disease hallmarked by hyperactivation of transcription factor Nrf2. Development. 2017:144(24):4588–4603. 10.1242/dev.156323 [DOI] [PubMed] [Google Scholar]
- Vaškovičová K, Žárský V, Rösel D, Nikolič M, Buccione R, Cvrčková F, Brábek J. Invasive cells in animals and plants: searching for LECA machineries in later eukaryotic life. Biol Direct. 2013:8(1):8. 10.1186/1745-6150-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006:21(4):455–466. 10.1016/j.molcel.2006.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velle KB, Fritz-Laylin LK. Conserved actin machinery drives microtubule-independent motility and phagocytosis in Naegleria. J Cell Biol. 2020:219(11):e202007158. 10.1083/jcb.202007158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PAC, Guerin C, Franco P, Li M, Burkart GM, Augustine RC, Blanchoin L, Bezanilla M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci U S A. 2009:106(32):13341–13346. 10.1073/pnas.0901170106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun. 2019:10(1):5132. 10.1038/s41467-019-12782-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawes C, Hussey PJ. Arabidopsis NAP1 regulates the formation of autophagosomes. Curr Biol. 2016:26(15):2060–2069. 10.1016/j.cub.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Alonso JM, Szymanski DB. Microtubule-dependent confinement of a cell signaling and actin polymerization control module regulates polarized cell growth. Curr Biol. 2018:28(15):2459–2466.e2454. 10.1016/j.cub.2018.05.076 [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Desyatova AS, Belteton SA, Mallery EL, Turner JA, Szymanski DB. Patterning mechanisms of cytoskeletal and cell wall systems during leaf trichome morphogenesis. Nat Plants. 2015:1:15014. 10.1038/nplants.2015.14 [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Zhang C, Szymanski DB. ARP2/3-dependent growth in the plant kingdom: SCARs for life. Front. Plant Sci. 2013:4:166. 10.3389/fpls.2013.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cai C, Staiger CJ. Myosins XI are involved in exocytosis of cellulose synthase complexes. Plant Physiol. 2019:179(4):1537–1555. 10.1104/pp.19.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Huang L, Zhang C, Staiger CJ. Arabidopsis myosin XIK interacts with the exocyst complex to facilitate vesicle tethering during exocytosis. Plant Cell. 2021:33(7):2454–2478. 10.1093/plcell/koab116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu C, Wang J, Ren Z, Staiger CJ, Ren H. A processive Arabidopsis formin modulates actin filament dynamics in association with profilin. Mol Plant. 2016:9(6):900–910. 10.1016/j.molp.2016.03.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.