Abstract
The 313-variant polygenic risk score (PRS313) provides a promising tool for breast cancer risk prediction. However, evaluation of the PRS313 across different European populations which could influence risk estimation has not been performed. Here, we explored the distribution of PRS313 across European populations using genotype data from 94,072 females without breast cancer, of European-ancestry from 21 countries participating in the Breast Cancer Association Consortium (BCAC) and 225,105 female participants from the UK Biobank. The mean PRS313 differed markedly across European countries, being highest in south-eastern Europe and lowest in north-western Europe. Using the overall European PRS313 distribution to categorise individuals leads to overestimation and underestimation of risk in some individuals from south-eastern and north-western countries, respectively. Adjustment for principal components explained most of the observed heterogeneity in mean PRS. Country-specific PRS distributions may be used to calibrate risk categories in individuals from different countries.
Introduction
Genetic susceptibility to breast cancer is influenced by multiple genetic variants which contribute different levels of risk to the disease (1–6). Genome-wide Association Studies (GWAS) have identified thus far a large number of common, low-risk variants that each contribute a small risk to the disease but can be combined into Polygenic Risk Scores (PRSs) with larger effect (7, 8). PRSs provide a promising tool for clinical risk prediction of breast cancer by stratifying women into different categories of breast cancer risk (9–11), and may be used to inform targeted screening and prevention strategies (12–20).
Mavaddat et al., (2019) constructed a 313-variant PRS (PRS313) for breast cancer, using data for women of European ancestry from the Breast Cancer Association Consortium (BCAC) (11). In prospective validation studies, this PRS was estimated to be associated with a relative risk for breast cancer of approximately 1.6 per standard deviation increase, and its discriminatory ability, measured in terms of area under the ROC curve (AUC), was 0.63. The lifetime absolute risk of developing breast cancer for individuals in the lowest percentile of the PRS313 risk distribution was estimated to be ~2%, while for those in the highest percentile it was ~33%. PRS313 has been incorporated into the multifactorial BOADICEA (Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm) model which is available via the CanRisk tool (14, 21, 22) ( www.canrisk.org ) and, together with other lifestyle and genetic risk factors, has been shown to improve risk stratification in European and European ancestry populations (14, 23–27). PRS313 has also been shown to be transferrable to women of other ethnic backgrounds, although the strength of the association with breast cancer risk was attenuated compared with that for women of European ancestry (OR per SD (95% CI) 1.52 (1.49–1.56), AUC = 0.61 in women of east Asian ancestry; OR 1.27 (1.23–1.31), AUC = 0.57 in women of African ancestry (28–30)).
Although several studies have investigated the transferability of PRS developed in European ancestry populations to non-European populations (31–34), the PRS distributions across different European countries has not been extensively evaluated. Differences in the PRS distribution, if not appropriately accounted for, could lead to inappropriate risk classification, with implications for clinical management.
In this study, we aimed to examine the distribution of the PRS313 across 17 countries in Europe, together with individuals of European ancestry from Australia, Canada, Israel and the USA. Similar analyses were performed using data from the UK Biobank, stratifying individuals by country of birth. We explored different approaches to account for the differences in the distribution, and investigated the implications of distribution differences across countries in breast cancer risk prediction.
Materials and methods
Study populations and Genotyping
Breast Cancer Association Consortium dataset
The BCAC dataset used here consisted of 110,260 female invasive breast cancer cases and 94,072 female healthy controls of European ancestry, recruited into 84 studies from 21 countries participating in the BCAC (Supplementary Table 1A). For simplicity and with attempt to explore the effect on the general female population, only the control data were used in these analyses as the distribution of the PRS in cases might vary between studies due to differences in study design (in particular oversampling of cases with a family history of disease). Samples from participating individuals were genotyped using the iCOGS (1) or OncoArray (3, 35) genotyping array. For samples genotyped using both arrays, the OncoArray genotype data were used. The iCOGS and the OncoArray datasets were imputed separately in a two-step manner using SHAPEIT (36) for phasing and IMPUTE2 for imputation. The Phase 3 (October 2014) release of the 1000 Genomes data (37) was used as the reference panel. More details on genotyping, quality control and imputation are given elsewhere (2, 3, 35). Ancestry-informative principal components (PCs), derived separately from the iCOGS and the OncoArray genotypes, were also calculated for all the samples, as previously described (3).
UK Biobank dataset
UK Biobank, is a prospective cohort study including more than 500,000 participants from England, Wales and Scotland, with age at recruitment between 40 to 69 years old, more details can be found elsewhere (38, 39). For the analyses in this study, genotype data from females (genetic reported sex) participating in the UK Biobank were used. Individuals were excluded if they had a recorded breast cancer diagnosis (malignant neoplasm of breast or carcinoma in situ of breast) or had a personal history of malignant neoplasm of breast. Genetic ancestry was inferred using the FastPop software (40). Individuals self-reported “white” and with an estimated European ancestry proportion ≥ 80% were retained in the analysis. Then, individuals were stratified by the “country of birth” field in the UK Biobank; only countries with at least 100 participants were used. After filtering, 225,105 females from 17 countries of Europe and from Australia, Canada, New Zealand, and the USA were used in the analyses (more details in Supplementary Table 1B). Samples were genotyped using the Affymetrix UK BiLEVE Axiom array and the Affymetrix UK Biobank Axiom array. Imputation data used were based on the Haplotype Reference Consortium (41), the UK10K +1000 Genomes panel references. More details on genotyping, quality control and imputation are given elsewhere (38). Ancestry informative PCs were also available (38).
All study participants gave written informed consent, and all the studies were approved by the relevant ethics committees. The use of the UK Biobank has been approved under application ID102655.
Statistical Analyses
PRS313 was developed previously using a hard-thresholding stepwise forward regression approach, and included variants independently associated with breast cancer risk at a p-value cut off < 10−5 (11). PRS313 was calculated in each study participant using the following formula:
Where is the PRS of individual , is the estimated effect allele dosage for carried by individual and can take values between 0 and 2, and is the weight for in the PRS for overall breast cancer, as derived by Mavaddat et al. (11). PRS313 was standardized to have unit SD in controls in the pooled dataset. Mavaddat et al. (11) also derived ER-specific versions of PRS313, with weights optimised for predicting ER-positive or ER-negative breast cancer risk (Supplementary Table 2).
The main analyses focused on calculating the mean standardized PRS313 in BCAC controls, using both the iCOGS and OncoArray datasets. These values were derived using linear regression with array type as a covariate and no intercept (so that estimates were generated for every country). Heterogeneity in the mean PRS313 between countries was assessed using I2 statistics and Q statistic p-values.
We also evaluated the distribution of the mean PRS by country of birth in female participants in the UK Biobank. Seven of the 313 variants were not available from the UK Biobank data and thus we used the remaining 306 variants in the analysis (PRS306) (Supplementary Table 2). We also evaluated a “standard” breast cancer PRS available in the UK Biobank data, previously generated from external GWAS data (42), and was available for 224,776 individuals (Supplementary Table 1B).
Potential sources of the variability in the mean PRS313 across the countries were explored in the BCAC dataset using three approaches. The PRS was first recalculated excluding variants in the CHEK2 region. The protein truncating variant CHEK2 c.1100delC is a relatively common founder variant that exhibits a large variation in frequency across Europe (43). Although it is not included in PRS313, other variants in the PRS313 are correlated with this variant. For this reason, the four variants in the CHEK2 region included in PRS313 (the CHEK2 p. Ile157Thr variant, and variants at positions 29135543, 29203724 and 29551872 on chromosome 22, positions based on build 37) were removed, resulting in a 309-variant PRS (PRS309). Mean and SE by country were recalculated for PRS309, as described above.
Second, we examined the effect of removing variants with the most variable frequency across countries. For this analysis, the mean and SD of the effect allele frequency in controls of the pooled dataset was calculated for each of the 313 variants by country. Variants with a coefficient of variation (SD/mean) greater than 0.3 were removed. Means and SE of the newly constructed PRS were recalculated by country as described above.
Third, we explored the effect of adjusting for up to 10 ancestry-informative PCs, in addition to type of array. As the PCs derived from the iCOGS and OncoArray are not comparable, separate PCs for each were included in the regression. We explored the number of PCs that were required to eliminate the heterogeneity in the adjusted mean PRS313, using the thresholds I2 < 10% and p-value > 0.05. Similarly, for the UK Biobank dataset, PRS306 was adjusted for up to 10 PCs, which were available in the UK Biobank.
As a complementary approach to generating population-specific estimates, we explored an empirical Bayes approach similar to that described by Clayton and Kaldor (44) for mapping disease rates. The motivation of this approach is that, if some of the variation in means among countries is genuine, while some is due to sampling variation, better estimates of the country-specific means can be obtained by “shrinking” the country-specific estimates towards the overall mean, by an amount depending on the sample size. In our implementation, we allowed the PRS means to be correlated between countries, using the autocorrelation matrix proposed in Clayton and Kaldor. A detailed description is given in Supplementary Methods.
To investigate the implications of PRS distribution differences in breast cancer risk prediction, we explored the proportion of women by country by percentile (<1%, 1%−5%, 5%−10%, 10%−20%, 20%−40%, 40%−60%, 60%−80%, 80%−90%, 90%−95%, 95%−99%, ≥99% percentiles), based on the distribution cut-offs of either the full dataset or country-specific estimates. We also examined a specific risk estimation example using the CanRisk tool (14, 21, 22).
All analyses were performed in R (version 4.2.1) (45). Forest plots were generated using the metafor package (46). Maps were generated using the packages world map data from natural earth (rnaturalearth) (47), the world vector map data from natural earth used in ‘rnaturalearth’ (rnaturalearthdata) (48), simple features for R (sf) (49) and interface to geometry engine (rgeos) (50).
Results
Geographic diversity in the mean PRS313 across European ancestry populations
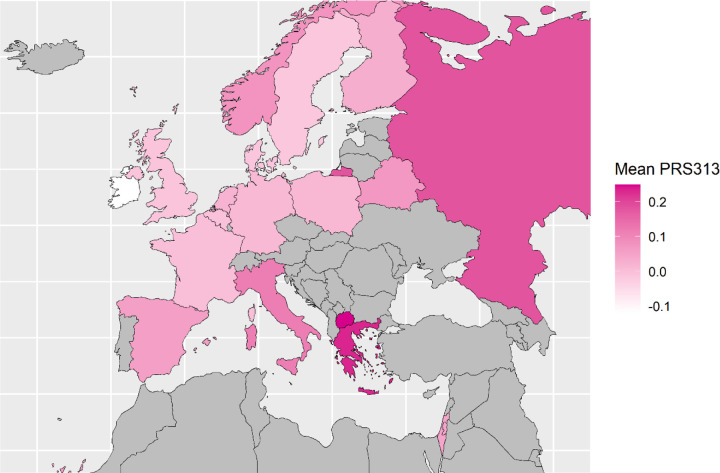
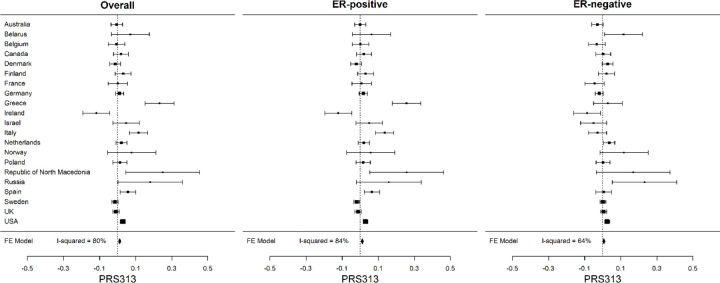
The mean PRS313 in the BCAC controls differed markedly across European countries, with heterogeneity I2 = 80% (p-value = 5.6 × 10−13). The mean was highest in the Republic of North Macedonia (0.25), Greece (0.23), Russia (0.18) and Italy (0.12), and lowest in Ireland (−0.12). The mean estimates for Australia, Canada, Israel and the USA were close to the overall mean (Figure 1; Figure 2; Table 1; Supplementary Table 3A). A similar level of heterogeneity was observed for the ER-positive (I2 = 84%) and ER-negative PRS (I2 = 64%) (Figure 2; Supplementary Table 3B). There was no evidence of a difference in the SD of the PRS between countries. (Supplementary Table 3A).
Figure 1:
Map of the European countries of origin of BCAC study participants included in the analysis. Countries were coloured based on their mean standardized PRS313 in control dataset of BCAC. Countries with higher mean are represented with darker colour while those with lower mean with lighter colour.
Figure 2:
Distribution of the standardized PRS313 across country of origin for overall, ER-positive and ER-negative breast cancer in control dataset of BCAC. The squares represent the mean PRS by country and the error bars represent the corresponding 95% confidence intervals (FE Model: Fixed effect Model).
Table 1:
Mean standardized PRS313 by country in controls in the pooled BCAC dataset, estimated when adjusted for array, 6 PCs country and array, using fitted values adjusted for 6 PCs and array and when using an Empirical Bayes approach adjusted for array.
| Country | Number of Controls | Mean PRS3131 | Mean PRS adjusted for array and 6 PCs | PRS adjusted for 6 PCs, fitted values2 | Empirical Bayes Posterior Mean3 |
|---|---|---|---|---|---|
| Australia | 4049 | −0.005 | 0.01 | −0.005 | −0.003 |
| Belarus | 342 | 0.07 | 0.071 | 0.016 | 0.064 |
| Belgium | 1823 | −0.006 | −0.007 | 0.010 | −0.002 |
| Canada | 2277 | 0.018 | 0.019 | 0.013 | 0.02 |
| Denmark | 5241 | −0.013 | 0.012 | −0.031 | −0.012 |
| Finland | 2083 | 0.031 | 0.008 | 0.010 | 0.032 |
| France | 1372 | 0.0003 | −0.008 | 0.008 | 0.004 |
| Germany | 8563 | 0.011 | 0.004 | 0.013 | 0.011 |
| Greece | 607 | 0.232 | 0.043 | 0.208 | 0.199 |
| Ireland | 719 | −0.118 | −0.015 | −0.112 | −0.092 |
| Israel | 724 | 0.047 | 0.001 | 0.062 | 0.047 |
| Italy | 1554 | 0.115 | −0.007 | 0.131 | 0.11 |
| Netherlands | 4407 | 0.021 | 0.043 | −0.019 | 0.022 |
| Norway | 217 | 0.077 | 0.094 | −0.027 | 0.066 |
| Poland | 2554 | 0.013 | 0.025 | 0.010 | 0.015 |
| Republic of North Macedonia | 92 | 0.25 | 0.134 | 0.140 | 0.129 |
| Russia | 120 | 0.18 | 0.166 | 0.044 | 0.11 |
| Spain | 2098 | 0.057 | −0.006 | 0.057 | 0.056 |
| Sweden | 16680 | −0.015 | 0.005 | −0.017 | −0.014 |
| UK | 16854 | −0.01 | 0.019 | −0.023 | −0.01 |
| USA | 21696 | 0.029 | 0.033 | 0.013 | 0.029 |
Mean PRS313 adjusted for array
Mean PRS313 by country using predicted PRS of each individual; estimated using linear predictor of PRS vs 6 PCs and the command predict () in R.
Country-specific estimates, means β, using the Empirical Bayes approach, adjusted for array
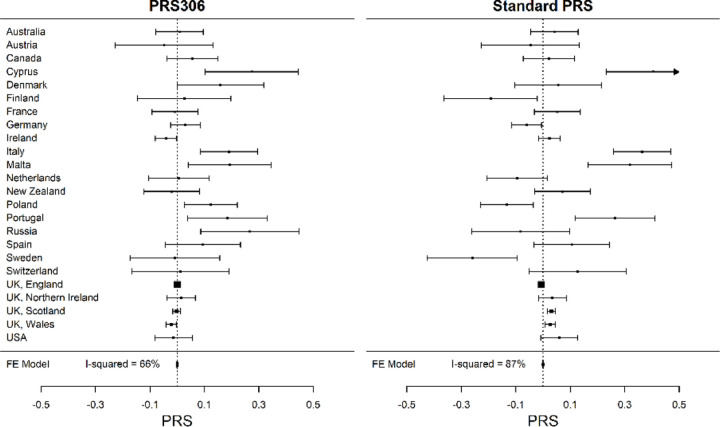
The mean PRS306 in female UK Biobank participants, stratified by country of birth, was also calculated (Figure 3 and Supplementary Table 4). There was strong evidence of heterogeneity in the PRS distribution (I2 = 66%, p-value = 2.3 × 10−06). The pattern was generally similar to that seen in the BCAC dataset, with a higher PRS in individuals born in southern and eastern Europe (e.g. Cyprus, Russia, Italy) and lower in western Europe (e.g. Ireland). Similar results were found for the “standard” UK Biobank PRS (I2 = 87%, p-value = 1.4 × 10−25) (Figure 3 and Supplementary table 4).
Figure 3:
Distribution of the mean PRS306, and “standard” PRS for breast cancer, as defined in the UK Biobank, across countries of origin of participating white females. The squares represent the mean PRS by country and the error bars represent the corresponding 95% confidence intervals (FE Model: Fixed effect Model).
Exploring potential reasons for differences in mean standardized PRS between countries
Potential sources of the variability in the mean PRS313 across the countries were explored in the BCAC dataset, using three approaches. We first evaluated the effect of removing variants in the CHEK2 region on the distribution of the mean PRS313 for the countries. After removing these four variants, the variation in the mean PRS309 across countries in the controls remained similar that for PRS313 (I2 = 83%, p-value = 9.4 × 10−16). We next identified the variants with the most variable frequency from countries in the control dataset. Seventeen of the 313 variants had a coefficient of variation greater than 0.3 (Supplementary Table 5). Excluding these 17 variants did not reduced the variation in the mean PRS (I2 = 80%, p-value = 2.4 × 10−12).
We next explored the effect of adjusting for PCs. When individuals in the BCAC dataset genotyped with OncoArray were plotted by the first two PCs, those from the same country separated clearly, in a pattern consistent with their geographical relationship (Supplementary Figure 1). This suggests that adjusting for PCs maybe an effective approach to reducing the variation in PRS distribution. When we adjusted the PRS for the leading PCs in the BCAC dataset, the I2 reduced as each PC was added in the model and reached < 10% when adjusted for the first six PCs (I2 = 69%, 54%, 47%, 39%, 22%, 0%, and 0% when including 1, 2, 3, 4, 5, 6, and 10 PCs respectively) (Table 1; Supplementary Table 3A; Supplementary Figure 2). A similar result was obtained for the ER-positive PRS (Supplementary Table 3B), when adjusted for the first 6 PCs (Heterogeneity: I2 = 0%, p-value = 0.69). For the ER-negative PRS, however, the heterogeneity was not eliminated even when the PRS was adjusted for 10 PCs (Heterogeneity: I2 = 56%, p-value = 0.001) (Supplementary Table 3B). The predicted PRS of each individual, as derived from the fitted values of the linear regression model of PRS adjusted for the first 6 PCs and array, were then used to calculate a predicted mean PRS313 by country (Table 1 and Supplementary Table 3A).
We repeated these analyses for PRS306 using the UK Biobank dataset. I2 reduced as each PC was added in the model and reached < 10% when adjusted for the first eight PCs (Supplementary Table 4 and Supplementary Figure 3).
Mean PRS estimates by country calculated using an Empirical Bayes approach
The empirical Bayes estimates by country for the mean PRS in controls of the BCAC dataset are given in Table 1 and Supplementary Table 6. Compared with the unadjusted estimates, the estimates shrunk towards the overall mean, with the shrinkage being greatest for countries that had small available sample sizes, such as Republic of North Macedonia and Russia (Table 1). The adjusted mean PRS by country were generally similar to those predicted by the model adjusting for six PCs (Supplementary Table 6). When PRSs were adjusted for the first 6 PCs, applying the empirical Bayes approach makes little difference to the estimates (Supplementary Table 6).
Implications for Breast Cancer Risk Prediction
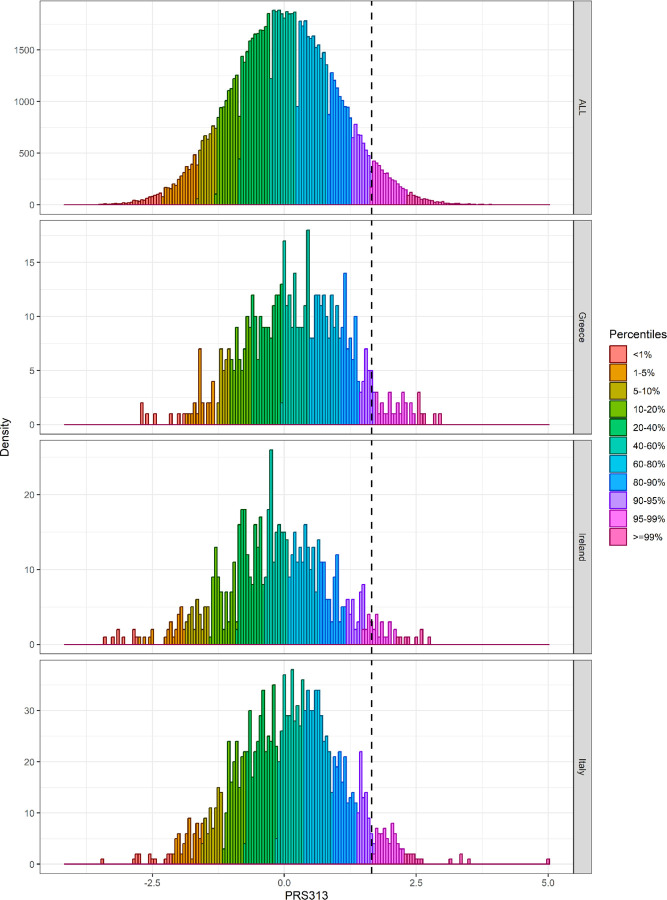
To explore the effect of these differences in PRS distribution between different European populations on risk stratification, we first defined risks thresholds based on the distribution of the controls in the full BCAC dataset (Supplementary Table 7A). We then calculated the percentage of controls by country that would be categorized in the 90–95th, 95–99th, and >99th percentile categories, based on the distribution in the full dataset, and compared these to the percentages based on the country-specific distributions (Supplementary Tables 7B-D). Based on the overall distribution, approximately 4.1%, 3.7%, 1.3% and 0.5% additional women from Belarus, Republic of North Macedonia, Greece, and Italy, respectively, would be incorrectly classified in the 95–99th percentile instead at the 90–95th percentile; while 1.1% and 1.4% additional women from France and Ireland, respectively, would be incorrectly classified in the 90–95th instead of the 95–99th percentile (Supplementary Table 7C). Figure 4 and Supplementary Table 8 illustrate the PRS313 percentile distribution in the full dataset, Greece, Italy (countries with the highest PRS313 and including more than 100 controls) and Ireland (lowest PRS313).
Figure 4:
PRS313 distribution in controls by percentiles in the pooled BCAC dataset, Greece, Ireland and Italy. The dashed line corresponds to the 95th percentile of the PRS313 distribution in controls of the pooled BCAC dataset.
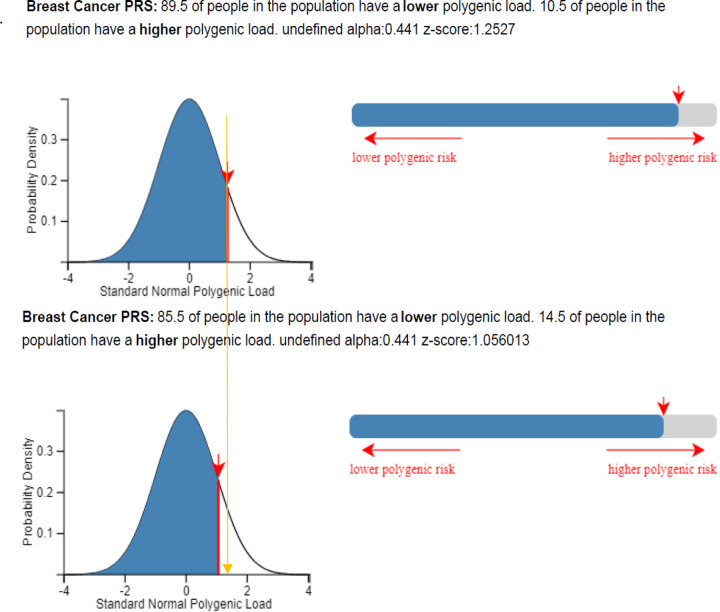
We next considered as an example a 50-year-old female from Greece with a raw PRS313 of 0.3414 (falling into the 90th – 95th percentile category in the full BCAC dataset) and no data on family history or other known risk factors. As Greek incidence rates are not available and not currently implemented in CanRisk, we used the UK incidence rates for the calculations. If this PRS was standardized based on the mean and SD used in the CanRisk tool (when a variant call format (vcf) file is uploaded to the CanRisk tool, a raw PRS313 can be calculated and standardized using the mean: −0.424; SD: 0.611), the individual would (assuming UK incidence rates) be given an estimate of 14.1% risk of developing breast cancer by the age of 80 and classified in the moderate risk category (Table 2). On the other hand, if the PRS were standardized based on the mean and SD of the controls of Greece (mean: −0.305; SD: 0.612-raw values), she would fall in the 80–90% percentile category with an estimated 13.3% risk of developing breast cancer by the age of 80, and be classified into the population risk category (Figure 5 and Table 2). Similarly, if the PRS were standardized based on the mean and SD of PRS for Greece predicted by adjustment for the first 6 PCs (mean: −0.42, SD: 0.696), she would also be classified in the population risk category (Table 2). Finally, if the PRS were standardized based on the mean and SD of the empirical Bayes approach (mean: −0.325 SD: 0.554), will have an estimated 13.9% risk of developing breast cancer by the age of 80, and be classified into the moderate risk category (Table 2).
Table 2:
Mean and SD used to standardize PRS313 of a 50-year-old woman with raw PRS313 equal to 0.341 from Greece and another 50-year-old woman with raw PRS313 equal 0.273 from Ireland, and the risk estimation and categorization when using the CanRisk tool, Greek and Ireland values.
| Samples used for the standardization: | Raw PRS; Mean (SD) | Standardized PRS1 | Percentage based on CanRisk tool | Lifetime risk based on CanRisk tool2 | NICE Risk category |
|---|---|---|---|---|---|
| Individual from Greece with raw PRS313 = 0.341 (falling into the 90–95% percentile category in the full BCAC dataset) | |||||
| CanRisk tool3 | −0.424 (0.611) | 1.253 | 89.5% | 14.1% | Moderate |
| Controls Greece (raw)4 | −0.305 (0.612) | 1.056 | 85.5% | 13.3% | Population |
| Controls Greece adjusted for 6 PCs (raw) | −0.420 (0.696) | 1.094 | 86.3% | 13.5% | Population |
| Controls Greece, using Empirical Bayes method | −0.325 (0.554) | 1.204 | 88.6% | 13.9% | Moderate |
| Individual from Ireland with raw PRS313 = 0.273 (falling into the 85–90% percentile category in the full BCAC dataset) | |||||
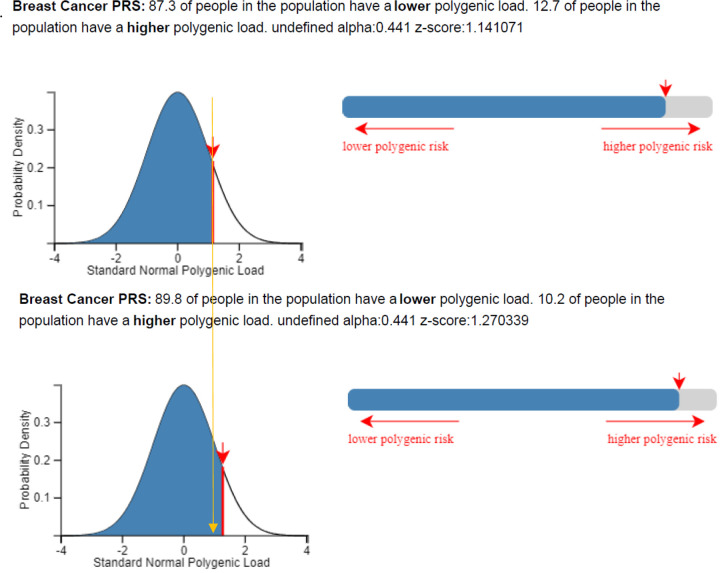
| CanRisk tool3 | −0.424 (0.611) | 1.14 | 87.3% | 13.7% | Population |
| Controls Ireland (raw)1 | −0.519 (0.624) | 1.27 | 89.8% | 14.2% | Moderate |
| Controls Ireland adjusted for 6 PCs (raw) | −0.456 (0.74) | 0.985 | 83.8% | 13% | Population |
| Controls Ireland, using EB | −0.503 (0.562) | 1.38 | 91.7% | 14.7% | Moderate |
Standardised based on the mean and SD specified in the second column
Absolute risk of developing breast cancer by the age of 80
When a variant call format (vcf) file is uploaded to the CanRisk tool, a raw PRS313 can be calculated and standardized using the mean (SD) −0.424 (0.611)
Adjusted for array type
Figure 5:
Classification of a 50-year-old woman from Greece when her raw PRS313, which is equal to 0.34 is standardized based on the mean and SD of the controls of BOADICEA model (upper panel) and Greece (lower panel), using the CanRisk tool. Plots were generated using the CanRisk tool (www.canrisk.org).
A second example is illustrated in Table 2 and Figure 6, based on a 50-year-old female from Ireland with raw PRS313 equal to 0.273 (equivalent to the 85th – 90th percentile-in the full BCAC dataset) and no other risk factors known. Using the CanRisk tool and assuming UK incidence rates, she would be classified in the 87.3% percentile with an estimated 13.7% absolute risk of developing breast cancer by the age of 80, which according to the NICE guidelines would be classified in the population risk category. If the PRS was standardized based on the mean and SD of PRS313 as derived from the controls in Ireland (mean for Ireland: −0.519, and SD: 0.624 -raw values), then she would be classified in the 89.8% percentile with estimated 14.2% risk of developing breast cancer by the age of 80, classified in the moderate risk category (Figure 6). If the PRS were standardized based on the mean and SD of PRS for Ireland predicted by adjustment for the first 6 PCs (mean: −0.456, SD: 0.74), she would also be classified in the population risk category (Table 2). Finally, if the PRS were standardized based on the mean and SD of the empirical Bayes approach (mean: −0.503, SD: 0.562), will have an estimated 14.7% risk of developing breast cancer by the age of 80, and be classified into the moderate risk category (Table 2).
Figure 6:
Classification of a 50-year-old woman from Ireland when her raw PRS313, which is equal to 0.27 is standardized based on the mean and SD of the controls BOADICEA (upper panel) model and Ireland (lower panel), using the CanRisk tool. Plots were generated using the CanRisk tool (www.canrisk.org).
Discussion
Transferability of PRSs across different populations remains a major challenge in the field of personalized cancer risk prediction (31, 51). In this study, we explored the distribution of PRS313 for breast cancer in European ancestry women from 21 countries, using data from studies participating in the BCAC, and further investigated how the observed variability might be accounted for in breast cancer risk prediction.
The results indicated that the PRS313 distribution varies markedly even within Europe, with a higher mean in south-east Europe (e.g. Republic of North Macedonia, Greece, Italy) and a lower mean in western Europe (e.g. Ireland). We observed a very similar pattern in females participating in the UK Biobank, based on country of birth. If not accounted for, these differences would lead to an over- or under-estimation of risk, thus affecting the risk categorization and possibly the clinical management of some women. This may be important not only at the individual country level but also for individuals living in a different country to that of their origin.
The variability in the mean PRS313 could not be explained by removing variants with the most variable frequency, indicating that a large number of variants may contribute to this difference. Removing such variants to reduce the heterogeneity would not in any case be desirable as it would reduce the risk discrimination provided by the PRS. The results do, however, indicate that most if not all of the variability in the mean PRS313 across countries in controls can be explained by adjusting for the leading ancestry informative PCs (6 PCs in the BCAC datasets, based on the OncoArray or iCOGS arrays, 8 PCs in UK Biobank). An advantage of using PCs is that they do not require any prior data from the population in question. A disadvantage, however, is that PCs require array genotyping data to generate, making them less attractive when implemented using sequencing panels. Moreover, the PCs generated using different genotyping arrays are not necessarily comparable. One interesting observation is that the heterogeneity of the ER-negative specific PRS was not eliminated even with the adjustment for 10 PCs.
We also explored generating country-specific mean PRS using an empirical Bayes approach. This approach considers both the uncertainty due to the small available sample size and the true variation in the means across the countries; these country-specific mean PRS were similar to those generated by adjusting for PCs. These values can then be used to standardise the PRS before, for example, implementing in the CanRisk tool. The risk categorization of the females from Greece and Ireland, the two examples in the Result section, was changed depending on the mean and SD of the sample used for the standardization of PRS. According to the NICE, women classified in the moderate risk category (lifetime risk of at least 17% and less than 30%), have different managing guidelines compared to women classified in the population risk category (52).
While adjustment of the PRS distribution at the population level is clearly necessary, the results raise the question as to whether it is appropriate in general to adjust PRS for PCs at the individual level, which gives different scores and potentially different risk classifications. This is a difficult question to address and hinges on whether the PCs should be regarded as nuisance parameters correcting for confounding factors, such as screening or lifestyle factors. Reanalysis of prospective studies with BCAC OncoArray dataset shows that the first two PCs are associated with the PRS (PC1 negatively, PC2 positively) and are also associated with risk (in the same direction). The PRS effect size (OR per 1 SD) is essentially unchanged whether or not adjustment for PCs is made (Supplementary Table 9 and Supplementary Table 10). This implies that risk discrimination would be slightly improved by including the effect of PCs in the PRS, and that adjusting the PRS for PCs further reduces the discrimination. Fortunately, the association between the PC1 and risk is weak and, within a country, the variation in the PC1 is not large enough to materially change risk categories.
The differences in the PRS distribution across Europe are a manifestation, on a continental scale, of the larger intercontinental differences – the mean PRS is higher in both east Asian and African populations than in the European dataset examined here (28, 29, 53). It is interesting to note that the pattern appears unrelated to the population-specific incidence, which is fact lower in south-east than north-west Europe (54), presumably because the effect on disease incidence is counterbalanced by larger effects of lifestyle (or other genetic) factors. It remains unclear whether the differences in the PRS can be attributed purely to random genetic drift or whether selection pressures relevant to breast cancer aetiology are involved.
We would like to acknowledge some potential limitations of our study. The dataset we used was genetically-homogeneous and maybe not completely representative of the population of each country. It remains an important issue how to interpret the PRS in individuals classified as mixed ancestry. In the future, the exploration of the distribution of the mean PRS across the individuals classified as mixed ancestry could be performed. Furthermore, evaluation of the country-specific calibrated PRS in combination with classical breast cancer risk factors should be performed in order to explore the extend to these findings have on final risk prediction.
In summary, these results demonstrate that the implementation of the PRS313 in risk prediction models such as CanRisk/BOADICEA could potentially require country-specific calibration. This can be achieved by genotyping a large control group to obtain population-specific means, by using a principal components adjustment, or the empirical Bayes approach described here.
Supplementary Material
References
- 1.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics. 2013;45(4):353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nature genetics. 2015;47(4):373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551(7678):92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Ahearn TU, Lecarpentier J, Barnes D, Beesley J, Qi G, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nature genetics. 2020;52(6):572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C, et al. Breast Cancer Risk Genes - Association Analysis in More than 113,000 Women. The New England journal of medicine. 2021;384(5):428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA - Journal of the American Medical Association. 2017;317(23):2402–16. [DOI] [PubMed] [Google Scholar]
- 7.Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nature protocols. 2020;15(9):2759–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wand H, Lambert SA, Tamburro C, Iacocca MA, O’Sullivan JW, Sillari C, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature: Nature Research; 2021. p. 211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of breast cancer risk based on profiling with common genetic variants. Journal of the National Cancer Institute. 2015;107(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature genetics. 2018;50(9):1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. American journal of human genetics. 2019;104(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shieh Y, Eklund M, Madlensky L, Sawyer SD, Thompson CK, Stover Fiscalini A, et al. Breast Cancer Screening in the Precision Medicine Era: Risk-Based Screening in a Population-Based Trial. Journal of the National Cancer Institute. 2017;109(5). [DOI] [PubMed] [Google Scholar]
- 13.Pashayan N, Morris S, Gilbert FJ, Pharoah PDP. Cost-effectiveness and Benefit-to-Harm Ratio of Risk-Stratified Screening for Breast Cancer: A Life-Table Model. JAMA Oncol. 2018;4(11):1504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee A, Mavaddat N, Wilcox AN, Cunningham AP, Carver T, Hartley S, et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genetics in medicine : official journal of the American College of Medical Genetics. 2019;21(8):1708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome medicine. 2020;12(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pashayan N, Antoniou AC, Ivanus U, Esserman LJ, Easton DF, French D, et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement. Nature reviews Clinical oncology. 2020;17(11):687–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks JD, Nabi HH, Andrulis IL, Antoniou AC, Chiquette J, Després P, et al. Personalized Risk Assessment for Prevention and Early Detection of Breast Cancer: Integration and Implementation (PERSPECTIVE I&I). Journal of personalized medicine. 2021;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Broek JJ, Schechter CB, van Ravesteyn NT, Janssens A, Wolfson MC, Trentham-Dietz A, et al. Personalizing Breast Cancer Screening Based on Polygenic Risk and Family History. Journal of the National Cancer Institute. 2021;113(4):434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pashayan N, Easton DF, Michailidou K. Polygenic risk scores in cancer screening: a glass half full or half empty? The Lancet Oncology. 2023;24(6):579–81. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Kar S, Antoniou AC, Pharoah PDP. Polygenic scores in cancer. Nature reviews Cancer. 2023;23(9):619–30. [DOI] [PubMed] [Google Scholar]
- 21.Carver T, Hartley S, Lee A, Cunningham AP, Archer S, Babb de Villiers C, et al. CanRisk Tool-A Web Interface for the Prediction of Breast and Ovarian Cancer Risk and the Likelihood of Carrying Genetic Pathogenic Variants. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2021;30(3):469–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archer S, Babb de Villiers C, Scheibl F, Carver T, Hartley S, Lee A, et al. Evaluating clinician acceptability of the prototype CanRisk tool for predicting risk of breast and ovarian cancer: A multi-methods study. PLoS One. 2020;15(3):e0229999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakeman IMM, Rodríguez-Girondo M, Lee A, Ruiter R, Stricker BH, Wijnant SRA, et al. Validation of the BOADICEA model and a 313-variant polygenic risk score for breast cancer risk prediction in a Dutch prospective cohort. Genetics in Medicine. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal Choudhury P, Brook MN, Hurson AN, Lee A, Mulder CV, Coulson P, et al. Comparative validation of the BOADICEA and Tyrer-Cuzick breast cancer risk models incorporating classical risk factors and polygenic risk in a population-based prospective cohort of women of European ancestry. Breast cancer research : BCR. 2021;23(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li SX, Milne RL, Nguyen-Dumont T, Wang X, English DR, Giles GG, et al. Prospective Evaluation of the Addition of Polygenic Risk Scores to Breast Cancer Risk Models. JNCI cancer spectrum. 2021;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Eriksson M, Czene K, Lee A, Leslie G, Lush M, et al. Prospective validation of the BOADICEA multifactorial breast cancer risk prediction model in a large prospective cohort study. Journal of medical genetics. 2022;59(12):1196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A, Mavaddat N, Cunningham A, Carver T, Ficorella L, Archer S, et al. Enhancing the BOADICEA cancer risk prediction model to incorporate new data on RAD51C, RAD51D, BARD1 updates to tumour pathology and cancer incidence. Journal of medical genetics. 2022;59(12):1206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho WK, Tan MM, Mavaddat N, Tai MC, Mariapun S, Li J, et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nature communications. 2020;11(1):3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du Z, Gao G, Adedokun B, Ahearn T, Lunetta KL, Zirpoli G, et al. Evaluating Polygenic Risk Scores for Breast Cancer in Women of African Ancestry. Journal of the National Cancer Institute. 2021;113(9):1168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Zeinomar N, Chung WK, Kiryluk K, Gharavi AG, Hripcsak G, et al. Generalizability of Polygenic Risk Scores for Breast Cancer Among Women With European, African, and Latinx Ancestry. JAMA network open. 2021;4(8):e2119084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nature genetics. 2019;51(4):584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. American journal of human genetics. 2017;100(4):635–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y, Hou K, Xu Z, Pimplaskar A, Petter E, Boulier K, et al. Polygenic scoring accuracy varies across the genetic ancestry continuum. Nature. 2023;618(7966):774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kachuri L, Chatterjee N, Hirbo J, Schaid DJ, Martin I, Kullo IJ, et al. Principles and methods for transferring polygenic risk scores across global populations. Nature reviews Genetics. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2017;26(1):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS genetics. 2014;10(4):e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Byun J, Cai G, Xiao X, Han Y, Cornelis O, et al. FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics. 2016;48(10):1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson DJ, Wells D, Selzam S, Peneva I, Moore R, Sharp K, et al. UK Biobank release and systematic evaluation of optimised polygenic risk scores for 53 diseases and quantitative traits. medRxiv. 2022:2022.06.16.22276246. [Google Scholar]
- 43.Schmidt MK, Hogervorst F, van Hien R, Cornelissen S, Broeks A, Adank MA, et al. Age- and Tumor Subtype-Specific Breast Cancer Risk Estimates for CHEK2*1100delC Carriers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(23):2750–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clayton D, Kaldor J. Empirical Bayes estimates of age-standardized relative risks for use in disease mapping. Biometrics. 1987;43(3):671–81. [PubMed] [Google Scholar]
- 45.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2023. [Google Scholar]
- 46.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36(3):1 – 48. [Google Scholar]
- 47.South A. Rnaturalearth: world map data from natural earth. R package version 01 0. 2017. [Google Scholar]
- 48.South A. rnaturalearthdata: world vector map data from Natural Earth used in’rnaturalearth’. R package version 0.1. 0. 2017. [Google Scholar]
- 49.Pebesma EJ. Simple features for R: standardized support for spatial vector data. R J. 2018;10(1):439. [Google Scholar]
- 50.Bivand R, Rundel C, Pebesma E, Stuetz R, Hufthammer KO, Bivand MR. Package ‘rgeos’. The Comprehensive R Archive Network (CRAN). 2017. [Google Scholar]
- 51.Wang Y, Tsuo K, Kanai M, Neale BM, Martin AR. Challenges and Opportunities for Developing More Generalizable Polygenic Risk Scores. Annual review of biomedical data science. 2022;5:293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.National Institute for Health and Care Excellence: Guidelines. Familial breast cancer: classification, care and managing breast cancer and related risks in people with a family history of breast cancer. London: National Institute for Health and Care Excellence (NICE) Copyright © NICE 2020.; 2019. [PubMed] [Google Scholar]
- 53.Ho WK, Tai MC, Dennis J, Shu X, Li J, Ho PJ, et al. Polygenic risk scores for prediction of breast cancer risk in Asian populations. Genetics in medicine : official journal of the American College of Medical Genetics. 2022;24(3):586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.