Abstract
Gabapentinoid drugs for pain and anxiety act on the CaVα2δ-1 and CaVα2δ-2 subunits of high-voltage-activated calcium channels (CaV1s and CaV2s). Here we present the cryo-EM structure of the gabapentin-bound brain and cardiac CaV1.2/CaVβ3/CaVα2δ-1 channel. The data reveal a binding pocket in the CaVα2δ-1 dCache1 domain that completely encapsulates gabapentin and define CaVα2δ isoform sequence variations that explain the gabapentin binding selectivity of CaVα2δ-1 and CaVα2δ-2.
Gabapentin (GBP) (1-(aminomethyl)cyclohexane acetic acid; Neurontin)1 and the related gabapentinoid drugs pregabalin (Lyrica)2,3 and mirogabalin (Tarlige)4,5 are widely used to treat post-herpetic neuralgia, diabetic neuropathy, fibromyalgia, epilepsy, restless leg syndrome and generalized anxiety disorder. These drugs bind to high-voltage-activated calcium channel (CaV) CaVα2δ-1 and CaVα2δ-2 subunits3,4,6–8, but not to the related CaVα2δ-3 and CaVα2δ-4 isoforms2,9,10. Gabapentinoid binding to CaVα2δ-1 and CaVα2δ-2 is thought to affect neuronal excitability by impairing CaV surface membrane expression3,7,8 through a mechanism involving Rab11a endosomal recycling11,12. Although the GBP-binding site has been identified13, structural details of CaVα2δ:GBP interactions have not yet been defined.
High-voltage-gated calcium channels (CaV1 and CaV2)14,15 are multi-subunit voltage-gated ion channels comprising three key components—the pore-forming CaVα1 (refs. 14,15), cytoplasmic CaVβ (ref. 16) and extracellular CaVα2δ (refs. 1,17) subunits. Recent cryo-EM structural studies of the CaV1.2/CaVβ3/Cavα2δ-1 channel complex13 revealed a CaVα2δ-1-bound l-leucine, a known CaVα2δ ligand1,18,19 and GBP competitor1,20, that identified the gabapentinoid binding site1,17. In this Brief Communication we present the 3.1-Å cryo-EM structure of the CaV1.2/CaVβ3/Cavα2δ-1 channel bound to GBP. The data show that GBP occupies the l-Leu binding site13 in the first subdomain of the Cavα2δ-1 dCache1 (Ca2+ channel and chemotaxis receptor) domain21. Structural analysis identifies six binding-site residues that differ between the GBP-sensitive isoforms, CaVα2δ-1 and CaVα2δ-2 (refs. 3,4,6–8), and the GBP-insensitive isoforms, CaVα2δ-3 and CaVα2δ-4 (refs. 2,9,10). These yield steric clashes with GBP and cause binding-site polarity changes that rationalize the GBP-insensitivity of the CaVα2δ-3 and CaVα2δ-4 isoforms.
Structure determination of a recombinant CaV1.2(ΔC)/CaVβ3/Cavα2δ-1 channel complex13 comprising a version of human CaV1.2 truncated after the IQ domain (CaV1.2(ΔC), 186 kDa) having wild-type functional properties13, rabbit CaVβ3 (54 kDa) and rabbit Cavα2δ-1 (125 kDa) in the presence of 11.7 mM GBP revealed a tripartite channel assembly (~370 kDa) (Fig.1a) at an overall resolution of 3.1 Å (Extended Data Figs. 1 and 2a–c, and Table 1) largely similar to the l-Leu bound structure13 (root-mean-square deviation, r.m.s.d.Cα = 0.749 Å). As previously observed13, the sample also contained a chaperone:channel complex of the endoplasmic reticulum (ER) membrane protein complex (EMC)22–24, CaV1.2(ΔC) and CaVβ3 (Extended Data Figs. 1 and 2d,e). The overall structure of the CaV1.2(ΔC)/CaVβ3/Cavα2δ-1:GBP complex is similar to other CaV1 and CaV2 structures13,25–27. Cavα2δ-1 has a multi-domain architecture built from two double Cache domains28, dCache1 and dCache2, and a von Willebrand factor type A (VWA) domain25,28,29 (Fig. 1a and Extended Data Fig. 3a). Importantly, the high Cavα2δ-1 local resolution (2.0–2.5 Å) and map quality (Fig. 1b–d and Extended Data Figs. 2b,c and 3a–f) allowed detailed comparison of the dCache1 domain with the l-Leu bound structure13. A map comparison (Fig. 1b–d and Supplementary Video 1) showed clear differences in the binding pocket density. These included a shape not present in the l-Leu bound maps (Fig. 1b–d) that had features that could be attributed to the GBP cyclohexyl ring and that defined the GBP-binding site.
Fig. 1 |. Structure of the CaV1.2(ΔC)/CaVβ3/Cavα2δ-1:GBP complex.
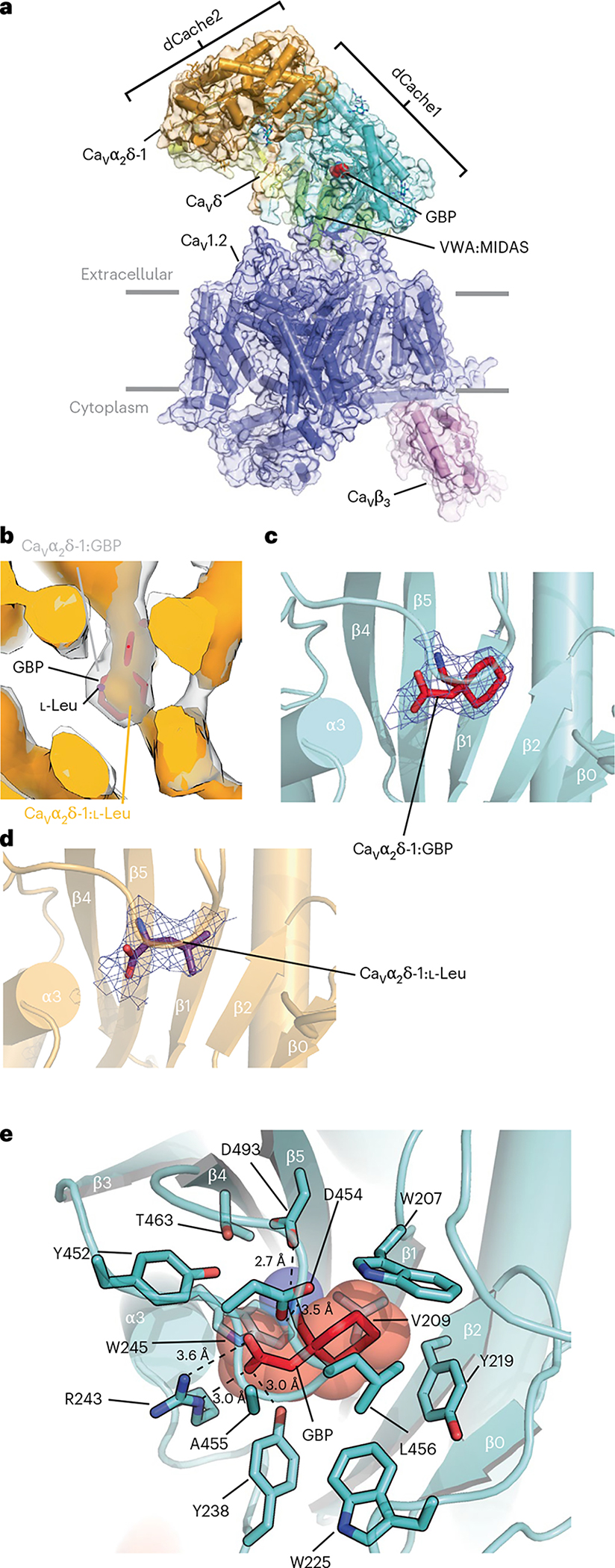
a, Side view of the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP complex. Subunits are colored with CaV1.2(ΔC) in slate and CaVβ3 in violet. CaVα2δ domains are colored with dCache1 in aquamarine, dCache2 in orange, VWA:MIDAS in green and CaVδ in yellow. GBP (in red) is shown as space filling. Gray bars denote the membrane. b, Comparison of CaVα2δ-1 dCache1 binding sites. Map comparison of the dCache1 ligand-binding site for the GBP complex (clear) and the l-Leu complex13 (orange). GBP is shown as red sticks. Map label colors match the respective map colors. c,d, Ligand densities for GBP (10σ, red, c) and l-Leu (7σ, violet purple, d) in the dCache1 domains are shown as cartoons (aquamarine and light orange, respectively). e, CaVα2δ-1 dCache1 ligand-binding site details. GBP (red) and contacting CaVα2δ-1 side chains (green-cyan) are shown as sticks. Distances for the dashed-line interactions are indicated.
Table 1.
Statistics for data collection, refinement and validation
| CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP | ||
|---|---|---|
| Data collection and processing | ||
| Magnification | 105,000 | |
| Voltage (kV) | 300 | |
| Electron dose (e−/Å2) | 46 | |
| Defocus range (μm) | −0.9~−1.7 | |
| Pixel size (Å) | 0.835 | |
| Symmetry | C1 | |
| Initial particle images (no.) | 9,703,873 | |
| CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP (PDB 8FD7; EMD-29004) | EMC:CaV1.2(ΔC)/CaVβ3 (EMD-29006) | |
| Final particle images (no.) | 259,107 | 383,185 |
| Map resolution (Å) | 3.1 | 3.1 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | ~2.4–8.0 | ~2.4–8.0 |
| Refinement | ||
| Initial model used (PDB code) | 8EOG | |
| Model resolution (Å) | 3.3 | |
| FSC threshold | 0.5 | |
| Map sharpening B factor (Å2) | −20 | |
| Model composition | ||
| Nonhydrogen atoms | 19,701 | |
| Protein residues | 2,416 | |
| Ligands | 20 | |
| B factors (Å2) | ||
| Protein | 118.63 | |
| Ligand | 42.23 | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | |
| Bond angles (°) | 0.479 | |
| Validation | ||
| MolProbity score | 2.16 | |
| Clashscore | 7.54 | |
| Rotamer outliers (%) | 2.76 | |
| Ramachandran plot | ||
| Favored (%) | 93.78 | |
| Allowed (%) | 6.10 | |
| Outliers (%) | 0.13 |
Similar to l-Leu, GBP occupies a pocket in the first Cavα2δ-1 dCache1 β-barrel lined by Trp207, Val209, Tyr219, Trp225, Tyr238, Arg243, Trp245, Tyr452, Asp454, Ala455, Leu456, Thr463 and Asp493 (Fig. 1e and Extended Data Fig. 3f) that is closed to solvent access. In line with the similar affinities of the two ligands18, there are no large conformational differences in the dCache1 binding site relative to the Cavα2δ-1:l-Leu complex (r.m.s.d.Cα = 0.155 Å; Extended Data Fig. 4a). The structure shows that GBP buries a larger total surface area than l-Leu (409 Å2 versus 367 Å2 for GBP and l-Leu, respectively) and that the different sizes and shapes of the ligands alter the details of the hydrogen-bond network surrounding the carboxylate and amino groups found in both ligands (Extended Data Fig. 4b,c). As with the l-Leu complex, Arg243 (Arg217 in some numbering schemes), a key residue for GBP binding to Cavα2δ-1 (ref. 30), is central to the coordination of the GBP carboxylate and makes bidentate interactions to the ligand through its guanidinium group (Fig. 1e and Extended Data Fig. 4b). The importance of this interaction is supported by the observation that an R→A change at this site is known to eliminate GBP binding3,30, abolish the analgesic effects of GBP and mirogabalin on pain3,31, and mitigate the effects of pregabalin on seizure and anxiety32,33. The Asp493 side chain makes a salt bridge with the GBP amino group that is shorter than the similar interaction in the l-Leu complex (2.7 Å versus 3.2 Å, GBP and Leu, respectively; Fig. 1e and Extended Data Fig. 4b,c). Disruption of this interaction by mutation to alanine strongly reduces the effect of GBP on CaV function28. Previous studies30 also showed that two large deletions of 24 and 23 residues (called ΔH and ΔI, respectively30) and multiple mutations in these same segments compromise GBP binding. The structure shows that these changes disrupt key Cavα2δ-1 elements that contribute to the integrity of the dCache1 domain (Extended Data Fig. 4d). Together with the previously demonstrated potent effects of R243A3,30–33 and D493A28 mutations on gabapentinoid binding and function, the structure underscores the importance of the dCache1 hydrogen-bond network that coordinates the amino and carboxylate moieties shared by amino acid and gabapentinoid Cavα2δ ligands.
The first subdomain of dCache1 repeats is a common binding site for free amino acids in many archaeal, bacterial and eukaryotic dCache domain containing proteins28,34, as exemplified by structures of the Pseudomonas aeruginosa chemoreceptor PctA34. The overall fold of the first Cavα2δ-1 dCache1 domain is largely similar to the amino acid binding the dCache1 domain of bacterial PctA34 (r.m.s.d.Cα = 4.123 Å). However, a structural comparison reveals key differences, including the long Cavα2δ-1 β2-α3 loop, absent from bacterial dCache domains (Extended Data Fig. 4e), the relative position of the β3-β4 loop that covers the ligand-binding pocket, and positional changes in the residues that encircle the ligand (Extended Data Fig. 4f). Nevertheless, Cavα2δ-1 uses the signature dCache1-domain amino acids that recognize the carboxylate (YxxxxRxW) and amino (Y[x~27–34]D) groups of various amino acid-derived ligands in bacterial dCache domains28 to coordinate the GBP and l-Leu carboxylate (Y238, R243, W245) and amino moieties (Y452, D493) through common hydrogen-bond networks (Extended Data Fig. 4b,c). This network is augmented in Cavα2δ-1 by Ala455, a β3-β4 loop residue (Figs. 1e and 2a) that contributes to carboxylate coordination (Extended Data Fig. 4b,c). In bacterial dCache domains34, the equivalent of the β3-β4 loop that covers the GBP-binding site moves to open access to the amino acid binding pocket, suggesting that similar Cavα2δ-1 dCache1 motions could provide a means for l-Leu, GBP and other gabapentinoids to access the Cavα2δ-1 binding pocket.
Fig. 2 |. Comparison of CaVα2δ-1 GBP-binding sites.
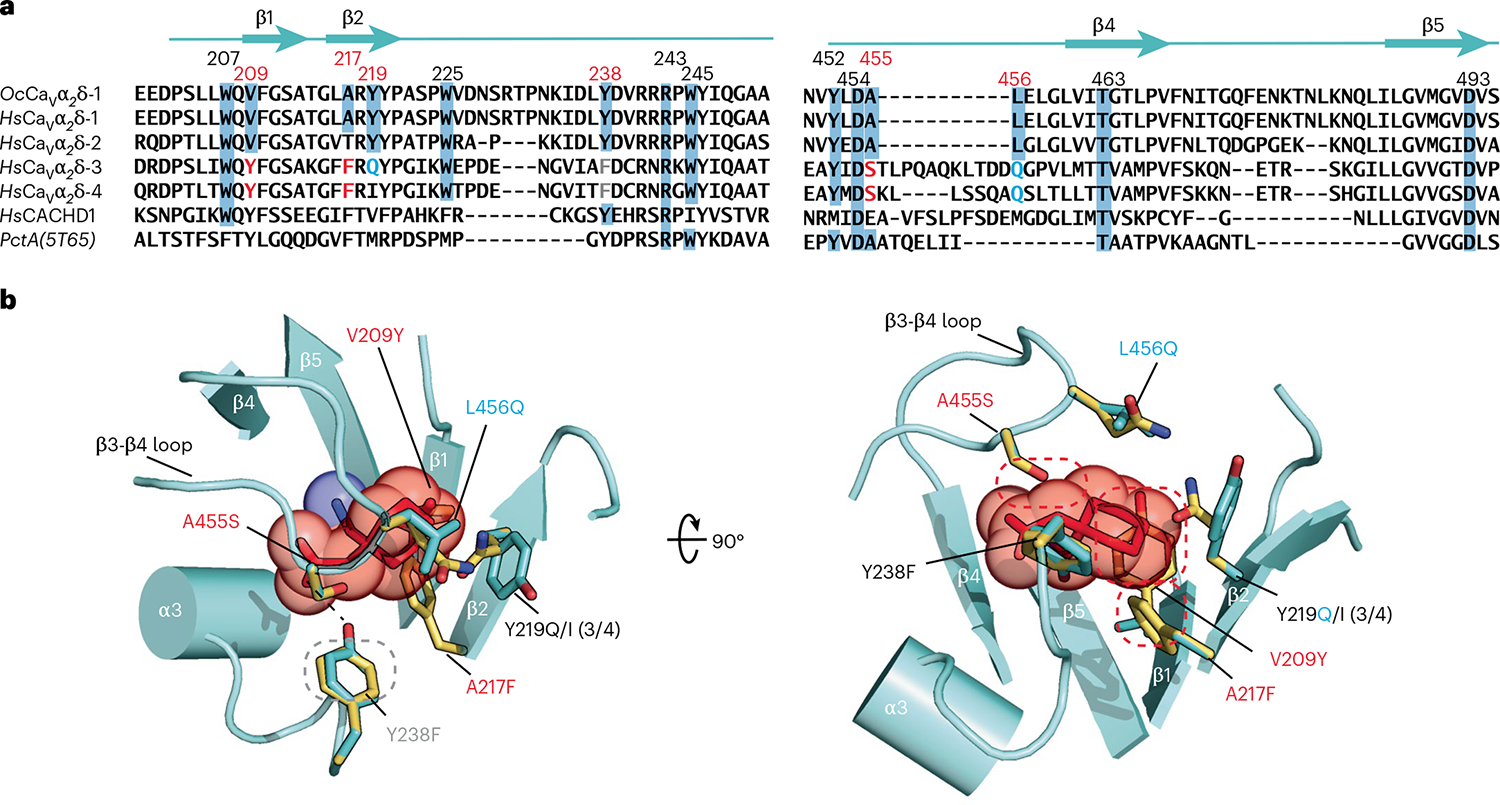
a, Sequence comparison of rabbit OcCaVα2δ-1 (NCBI, NP_001075745.1), human HsCaVα2δ-1 (NCBI, NP_001353796.1), HsCaVα2δ-2 (NCBI, NP_006021.2), HsCaVα2δ-3 (NCBI, NP_060868.2), HsCaVα2δ-4 (NCBI, NP_758952.4) and HsCACHD1 (NCBI, NP_065976.3), and bacterial PctA (PDB 5T65)34 dCache1 domain sequences. Numbers indicate residues interacting with l-Leu and GBP. Red numbers indicate sites that differ between GBP-sensitive and GBP-insensitive isoforms. Residue colors indicate steric clash (red), hydrogen bond loss (gray), and polarity changes (blue) between GBP-sensitive and GBP-insensitive isoforms. b, Structural context for amino acid differences between GBP-sensitive (aquamarine) and GBP-insensitive (yellow) CaVα2δ-1 Cache1 ligand-binding sites. GBP-insensitive residues are modeled on the CaVα2δ-1:GBP structure. Dashed ovals denote hydrogen bond loss (gray) and steric clashes (red). (3/4) indicates amino acid changes for CaVα2δ-3 and CaVα2δ-4.
The four different mammalian Cavα2δ isoforms share a common structure28, yet GBP and the related gabapentinoids bind to CaVα2δ-1 and CaVα2δ-2 (refs. 3,4,6–8), but not to CaVα2δ-3 and CaVα2δ-4 (refs. 2,9,10). Structure-based sequence comparisons identify six sites in the GBP-binding pocket that differ between the gabapentinoid-sensitive (CaVα2δ-1 and CaVα2δ-23,4,6–8) and gabapentinoid-insensitive (CaVα2δ-3 and CaVα2δ-4, refs. 2,9,10) CaVα2δ isoforms (Fig. 2a). Mapping these on the Cavα2δ-1:GBP structure (Fig. 2b) reveals numerous alterations expected to interfere with GBP binding to CaVα2δ-3 and CaVα2δ-4, including (1) loss of a hydrogen-bond donor to the GBP carboxylate (Y238F), (2) introduction of steric clashes (V209Y, A217F35 and A455S), two of which (V209Y and A217F) would occupy the same space as the GBP cyclohexyl ring, and (3) changes in contact residue size (Y219I) and polarity (Y219Q, L456Q and A455S) that reshape binding pocket physiochemical characteristics. The CaVα2δ-related protein CACHD1 also lacks most of the key GBP-binding residues (Fig. 2a), indicating that the effects of this protein on CaV surface expression36 are likely to be GBP-independent. Taken together, the multiple changes between the GBP-sensitive and GBP-insensitive isoforms that remove hydrogen bonds, introduce steric clashes and reduce the hydrophobicity of the binding pocket provide a structural rationalization for the differences in gabapentinoid binding properties among the CaVα2δ isoforms2–4,7–10.
Binding of GBP and gabapentinoids to the CaVα2δ subunit of CaVs is critical for the pharmacological effects of this drug class3,31–33. The structural identity of the l-Leu13 and GBP-bound CaVα2δ complexes (Extended Data Fig. 4a) suggests that the effects of GBP on CaV function may arise from changes in the dynamic behavior of CaVα2δ when it is bound to different ligands. Structural definition of the CaVα2δ gabapentinoid binding site provides a platform to dissect the mechanisms by which these drugs affect CaV function and should guide the development of next-generation CaVα2δ-directed drugs for the treatment of pain and anxiety.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-023-00951-7.
Methods
Expression and purification of human Cav1.2
Expression and purification of CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 was carried out, as previously described13 using an HEK293S GnTI− (ATCC #CRL-3022) ‘CaVβ3-stable’ cell line expressing rabbit CaVβ3 (477 residues, UniProt P54286) bearing a Strep-tag II sequence38. The codon-optimized construct of human Cav1.2 bore a C-terminal truncation at residue 1648 (denoted CaV1.2(ΔC)), a site located 13 residues after the end of the IQ domain (Δ1649–2138, UniProt Q13936–20, 1,648 residues), followed by a 3C protease cleavage site, monomeric enhanced green fluorescent protein, and a His8 tag. The second construct carried unmodified rabbit Cavα2δ-1 (1,105 residues, UniProt P13806–1). Both constructs were cloned into modified pFastBac expression vectors having the polyhedrin promoter replaced by a mammalian cell active CMV promoter39. All constructs were sequenced completely.
Chemically competent DH10EmBacY (Geneva Biotech) were used to generate the recombinant bacmid DNA, which was then used to transfect Spodoptera frugiperda (Sf9; Expression Systems, #94–001F) cells to make baculoviruses for each subunit40. Cav1.2 was expressed in CaVβ3-stable cells together with Cavα2δ-1 using a baculovirus transduction-based system40. CaVβ3-stable cells were grown in suspension at 37 °C while supplied with 8% CO2 in FreeStyle 293 Expression Medium (Gibco) supplemented with 2% fetal bovine serum (Peak Serum), and were transduced with 5% (vol/vol) baculovirus for each target subunit when the cell density reached ~2.5 × 106 cells per ml. Sodium butyrate (10 mM) was added to the cell culture 16–24 h post-transduction, and the cells were subsequently grown at 30 °C. Cells were collected 48 h post-transduction by centrifugation at 5,000g for 30 min. The pellet was washed with Dulbecco’s phosphate buffered saline (Gibco) and stored at −80 °C.
A cell pellet (from ~3.6 l of culture) was resuspended in 200 ml of resuspension buffer containing 0.3 M sucrose, 1 mM ethylenediaminetetraacetic acid, 10 mM Tris-HCl, pH 8.0 supplemented with 1 mM phenylmethylsulfonyl fluoride, and four Pierce protease inhibitor tablets (Thermo Scientific), then stirred gently on a Variomag magnetic stirrer (Mono Direct, Thermo Scientific) at 4 °C for 30 min. The membrane fraction was collected by centrifugation at 185,500g for 1 h and subsequently solubilized in 200 ml of solubilization buffer (buffer S) containing 500 mM NaCl, 5% glycerol (vol/vol), 0.5 mM CaCl2, 20 mM Tris-HCl, pH 8.0, and supplemented with 1% (wt/vol) glycol-diosgenin (GDN) and rotated on an Orbitron rotator II (speed mode S; Boekel Scientific) at 4 °C for 2 h. The supernatant, collected by centrifugation at 185,500g for 1 h, was diluted with an equal volume of buffer S to a final concentration of 0.5% GDN and incubated with anti-GFP nanobody Sepharose resin41 at 4 °C overnight. The resin was loaded on an Econo-Column chromatography column (BioRad) and was then washed stepwise with 20 column volumes (CV) of buffer S supplemented with 0.1% (wt/vol) GDN, 20 CV of buffer S supplemented with 0.02% (wt/vol) GDN, and 20 CV of elution buffer (buffer E) containing 150 mM NaCl, 0.5 mM CaCl2, 0.02% (wt/vol) GDN, 20 mM Tris-HCl pH 8.0. The protein was eluted with 3C protease42 and subsequently incubated at 4 °C for 2 h with 4 ml of Strep-tactin Superflow Plus beads (Qiagen) pre-equilibrated with buffer E. The beads were washed with 20 CV of buffer E, and the protein was eluted with buffer E supplemented with 2.5 mM desthiobiotin. The eluent was concentrated using an Amicon Ultra-15 100-kDa-cutoff centrifugal filter unit (Merck Millipore) before purification on a Superose 6 Increase 10/300 GL gel filtration column (GE Healthcare) pre-equilibrated in buffer E. Concentrated Cav1.2(ΔC)/Cavβ3/Cavα2δ-1 sample was immediately incubated with GBP (final concentration 2 mg ml−1, 11.7 mM; Sigma-Aldrich) on ice for 4 h before cryo-EM sample preparation, denoted Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP.
Sample preparation and cryo-EM data acquisition
For cryo-EM, 3.5 μl of 2.0 mg ml−1 Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP was applied to Quantifoil R1.2/1.3 300 mesh Au holey-carbon grids, blotted for 4–6 s at 4 °C and 100% humidity using a FEI Vitrobot Mark IV (Thermo Fisher Scientific), and plunge-frozen in liquid ethane. The cryo-EM grids were screened on an FEI Talos Arctica cryo-TEM system (Thermo Fisher Scientific; at University of California, San Francisco (UCSF) EM Facility) operated at 200 kV and equipped with a K3 direct detector camera (Gatan), and then imaged on a 300-kV FEI Titan Krios microscope (Thermo Fisher Scientific) with a K3 direct detector camera (Gatan; UCSF). Cryo-EM datasets were collected in super-resolution counting mode at a nominal magnification of ×105,000 with a super-resolution pixel size of 0.4175 Å (physical pixel size of 0.835 Å) using a SerialEM v4.1 (ref. 43). Images were recorded with a 2.024-s exposure over 81 frames with a dose rate of 0.57 e− Å−2 per frame. The defocus range was set from −0.9 μm to −1.7 μm.
Image processing and 3D reconstruction
A total of 26,928 movies were collected for Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP. Initial image processing was carried out in cryoSPARC-3.344. Raw movies were corrected for motion and Fourier binned by a factor of two (final pixel size of 0.834 Å) with the patch motion correction program. Contrast transfer function parameters of the resulting micrographs were estimated with the Patch contrast transfer function estimation program in cryoSPARC-3.3. Particles were picked by blob picking, extracted using a box size of 440 pixel (2× binned to 220 pixels), and 2D-classified using a circular mask diameter of 260 Å. Selected particles represented by good 2D classes were subjected to one round of ab initio reconstruction and heterogeneous refinement with C1 symmetry. Particles having reasonable 3D reconstructions (as judged by the Fourier shell correlation (FSC) curve) were re-extracted to physical pixel size and subjected to iterative rounds of ab initio reconstruction and heterogeneous refinement. Further, non-uniform refinements were performed to achieve high-resolution reconstruction.
To improve the Cavα2δ-1 3D reconstruction, multibody refinement was carried out in RELION-3.145. In total, 259,107 refined particles for the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP complex were exported from cryoSPARC-3.3 to RELION-3.1 using the csparc2star.py (UCSF pyem v0.5. Zenodo) suite of conversion scripts (https://doi.org/10.5281/zenodo.3576630). Following a 3D refinement in RELION-3.1 using the refined map from cryoSPARC-3.3 and the exported particles, an overall 3.1-Å EM density map (consensus map) was obtained (Extended Data Fig. 1 presents processing flow charts). Multibody refinement was performed in RELION-3.1 to improve the features of the lumenal domain and the transmembrane region for the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP complex. We used the phenix.combine_focused_maps program to combine the improved features of the segments from multibody refinement and those of the consensus map to obtain the final map with best features for the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP complex46.
As with an earlier report13, purification of CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 yielded a sample that also had a substantial portions of particles (383,185 refined particles) comprising the EMC:CaV1.2(ΔC)/CaVβ3 complex. These refined particles were exported from cryoSPARC-3.3 to RELION-3.1, and subsequent 3D refinement resulted in a 3.1-Å consensus map. Comparison of this 3.1-Å consensus map for the EMC:CaV1.2(ΔC)/CaVβ3 complex with the one reported from the previous study13 revealed no apparent conformational difference (cross correlation = 0.9554), so the model was not docked for this complex.
Model building and refinement
We used phenix.dock_in_map46 to dock the Cav1.2(ΔC)/Cavβ3/Cavα2δ-1 model (PDB 8EOG) into the Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP map. As the density for Cavβ3 was weaker than for other parts of the channel, we rendered the CaVβ region of the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 complex at a threshold of 2.25 (>5 was used for the rest of map) and placed CaVβ3 from the de novo modeled EMC:CaV1.2(ΔC)/CaVβ3 structure (EMD-28376; PDB 8EOI) in the density, followed by rigid-body refinement of the Cav1.2(ΔC)/Cavβ3/Cavα2δ-1 complex. The docked model and maps were manually checked and fitted in Coot47. Iterative structure refinement and model building were performed using the phenix.real_space_refine program46. Restraint files necessary for refinement were generated using phenix.elbow46,48. The final statistics of 3D reconstruction and model refinement are provided in Table 1. The per-residue B factors, after final refinement against the overall map, were rendered on the refined model and are presented in Extended Data Fig. 2c. The final models were evaluated using MolProbity49. All figures and movies were generated using ChimeraX50 and the Pymol package (v2.4.0; http://www.pymol.org/pymol). Close-contact interaction analyses were performed using LIGPLOT and DIMPLOT37,51.
Extended Data
Extended Data Fig. 1 |. CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP Cryo-EM analysis.
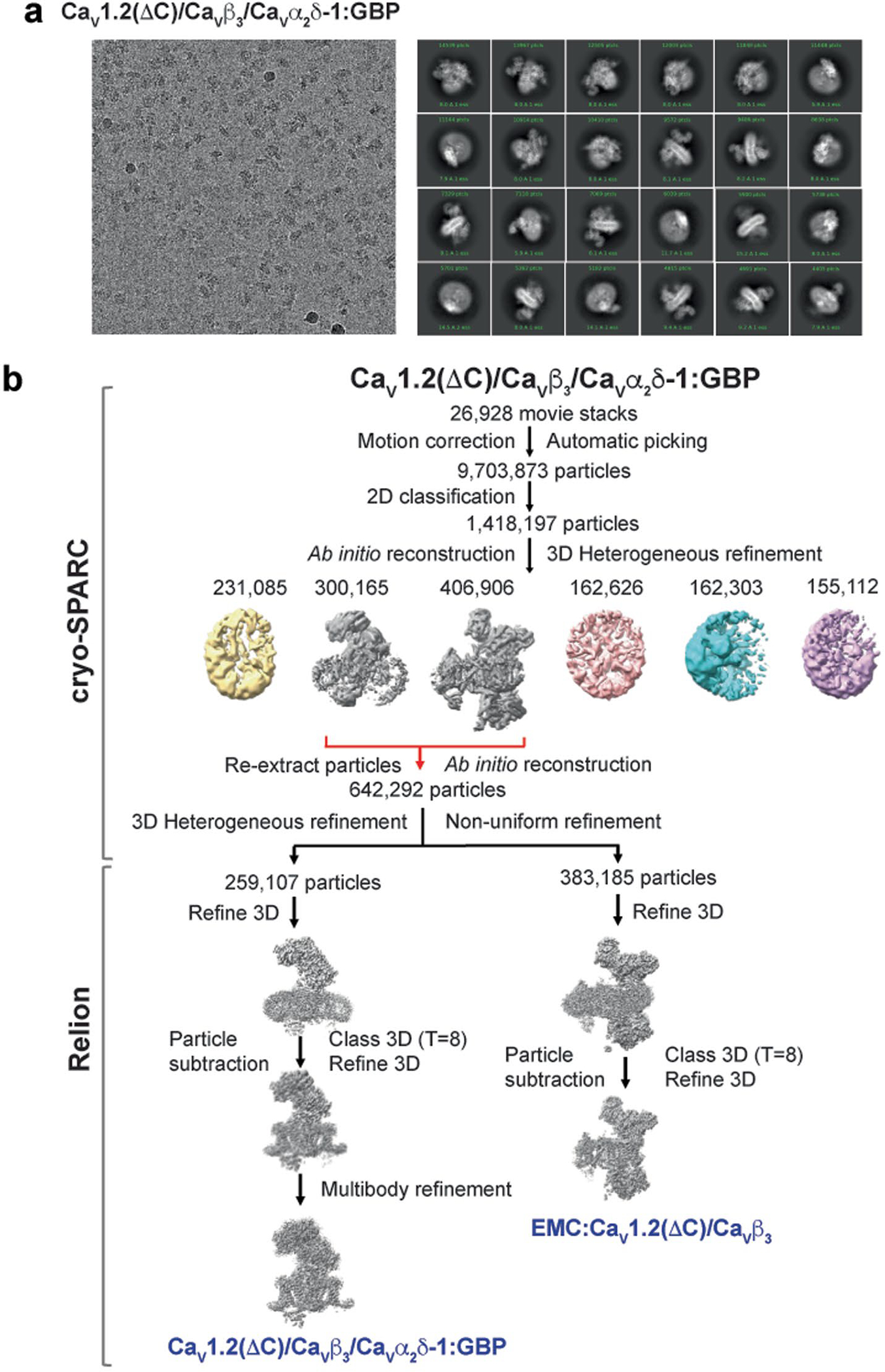
a, Exemplar CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP electron micrograph (~105,000x magnification) and 2D class averages. N = 3. b, Workflow for electron microscopy data processing for CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP sample. Initial cryoSPARC-3.2 Ab initio reconstruction identified a population of particles containing the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 and EMC:CaV1.2(ΔC)/CaVβ3 complexes, similar to prior studies13. Red arrows indicate the two classes that were re-extracted, subjected to multiple rounds of 3D heterogeneous classification, and exported from cryoSPARC-3.2 for further 3D refinement in RELION-3.1. Particle subtraction was performed for both the refined maps in Relion-3.1 followed by 3D classification with single class and 3D refinement to get the final consensus maps. Multibody refinement was performed to enhance features of CaVα2δ-1, which was used for the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP composite map. The composite map was used for model building and refinement.
Extended Data Fig. 2 |. CaV1.2(ΔC)/CaV β3/CaVα2δ-1:GBP map and model quality.
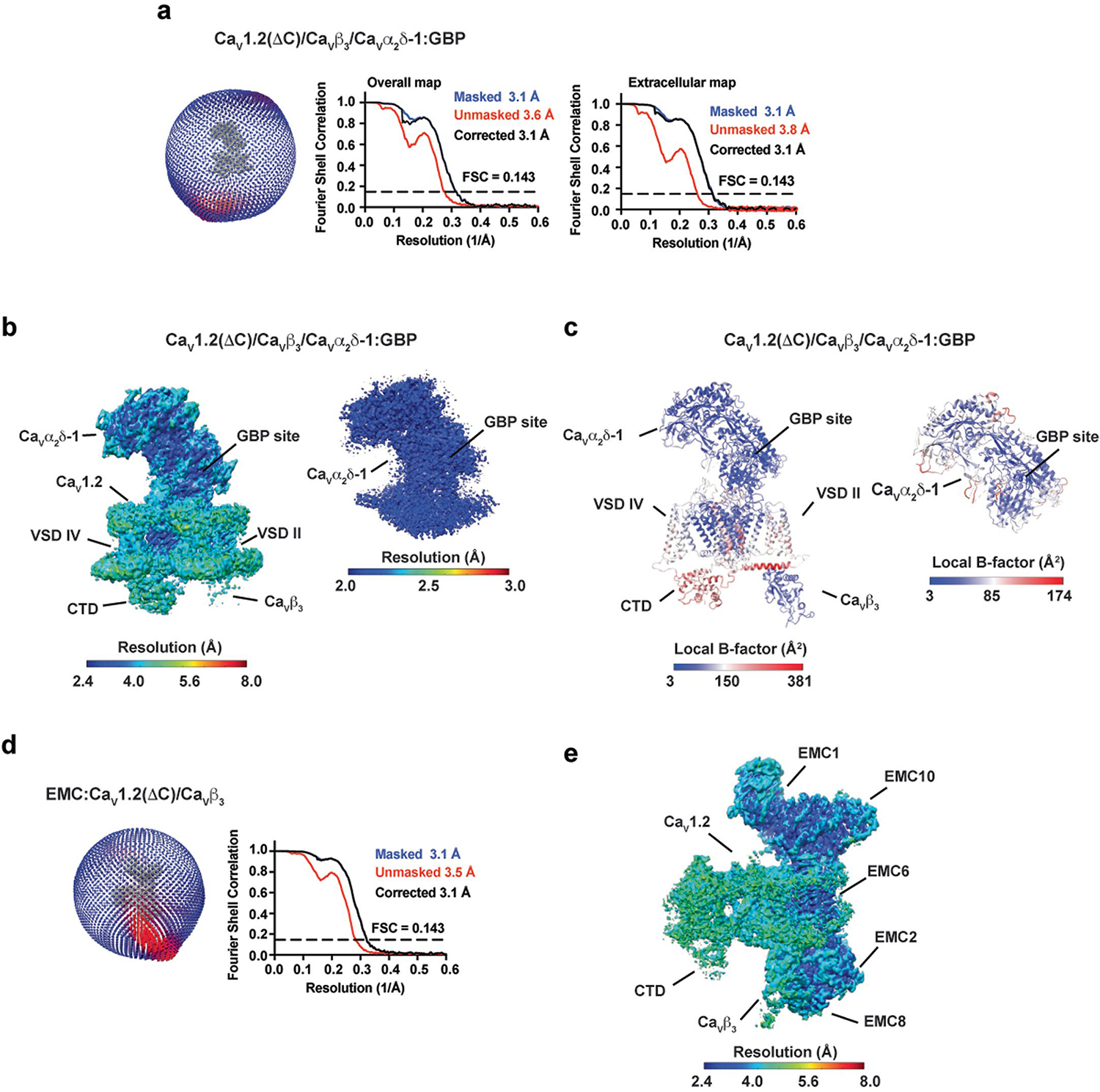
a, Particle distribution plot and gold-standard Fourier shell correlation (FSC) curve for the overall CaV1.2(ΔC)/CaV β3/CaVα2δ-1:GBP complex map and the extracellular map containing CaVα2δ-1:GBP. b, local resolution for the overall CaV1.2(ΔC)/CaV β3/CaVα2δ-1:GBP map and the extracellular map containing CaVα2δ-1:GBP. c, local B-factor for the overall CaV1.2(ΔC)/CaV β3/CaVα2δ-1:GBP model and the CaVα2δ-1:GBP subunit. d, Particle distribution plot and gold-standard Fourier shell correlation (FSC) curve for the EMC:CaV1.2(ΔC)/CaVβ3 complex from the CaV1.2(ΔC)/CaV β3/CaVα2δ-1:GBP sample. e, EMC:CaV1.2(ΔC)/CaVβ3 complex local resolution. Select elements of each complex are labeled.
Extended Data Fig. 3 |. CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP Cryo-EM maps.
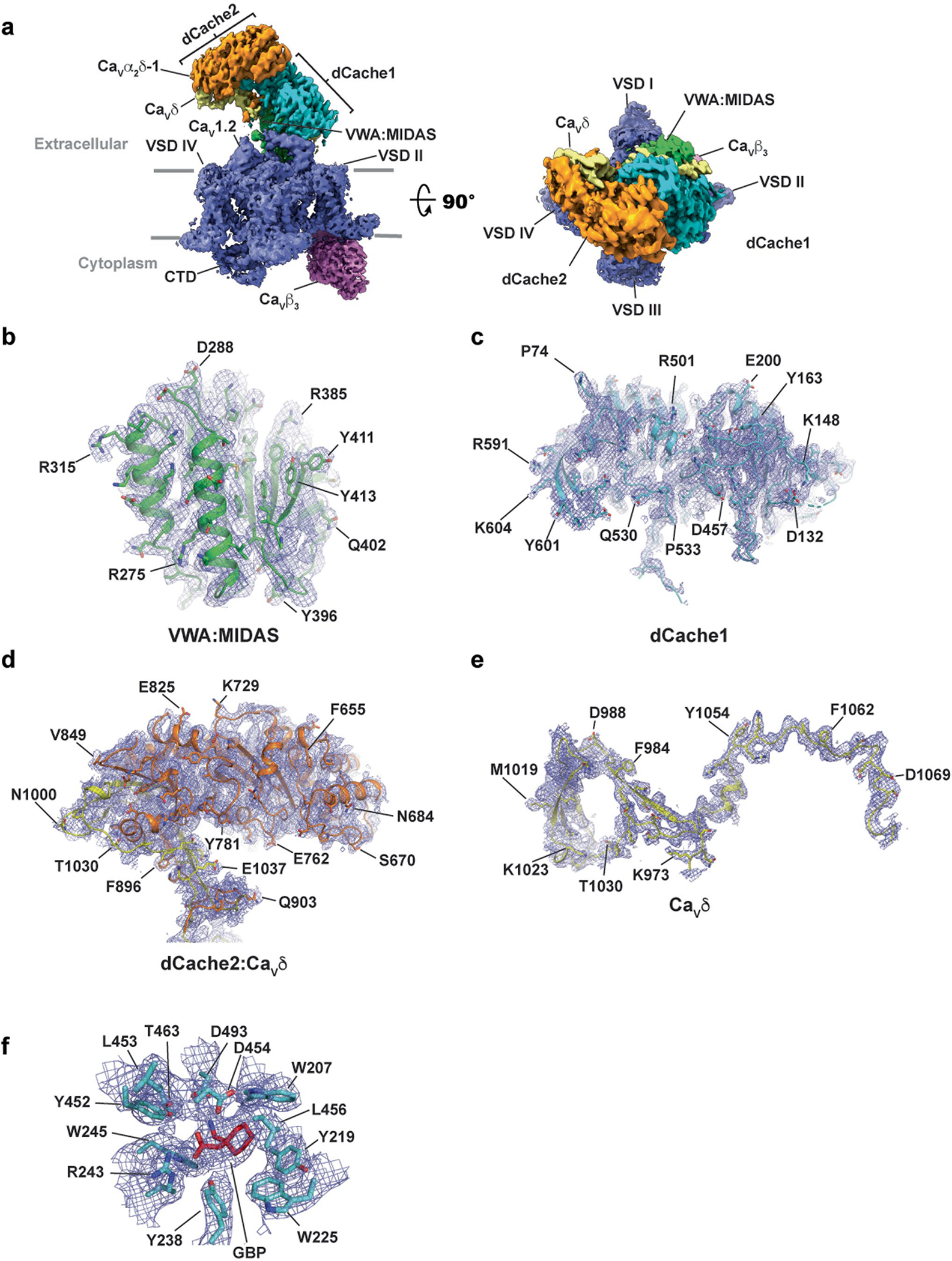
a, CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 side view (left) and extracellular (right) view. Subunits are colored: CaV1.2 (slate) and CaVβ3 (violet). CaVα2δ domains are colored as: dCache1 (aquamarine), dCache2 (orange), VWA:MIDAS (green), and CaVδ (yellow). Grey bars denote the membrane. b-e, CaVα2δ-1 subdomain representative maps for b, VWA:MIDAS domain, c, dCache1, d, dCache2:CaVδ. Part of CaVδ completes the second β-barrel subdomain of dCache2, e, CaVδ. f, GBP-binding site. Maps are rendered at 9–10σ. Domain colors are as in a.
Extended Data Fig. 4 |. CaVα2δ-1 GBP-binding site analysis and comparisons.
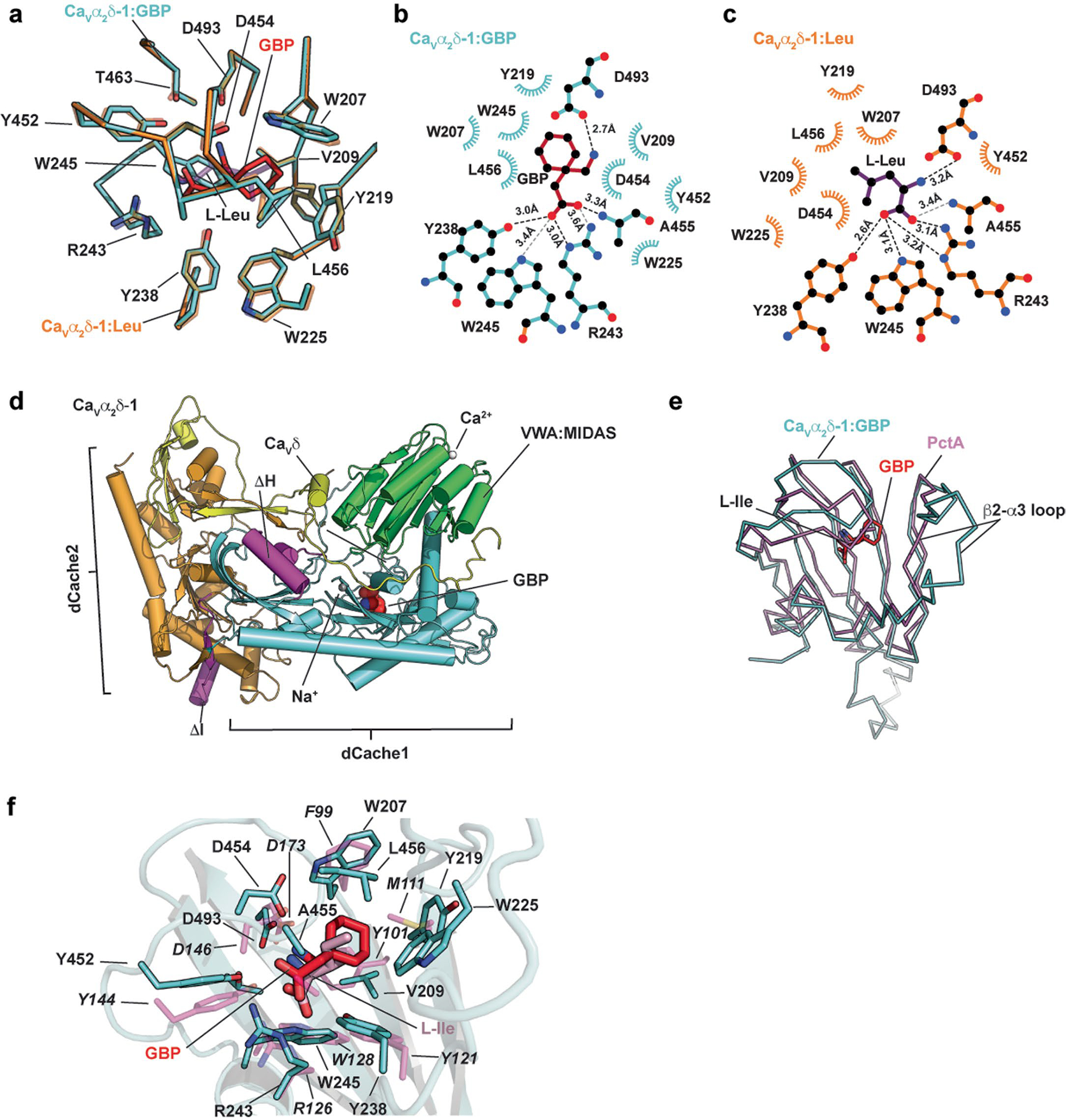
a, Superposition of the CaVα2δ-1:GBP (aquamarine) and CaVα2δ-1:l-Leu (orange) (PDB:8EOG)13 binding sites. GBP is red. l-Leu is purple. b and c, LigPLOT37 diagrams of the b, CaVα2δ-1:GBP (aquamarine) and c, CaVα2δ-1:l-Leu (orange) (PDB:8EOG)13 binding sites showing hydrogen bonds and ionic interactions (dashed lines) and van der Waals contacts ≤ 5 Å. GBP is red. l-Leu is purple. d, Superposition of the first dCache1 repeats from CaVα2δ-1:GBP (aquamarine) and the PctA:l-Ile complex (magenta) (PDB: 5T65)34. GBP is red. e,f, Closeup view of superposition from ‘d’ showing ligand contact residues. CaVα2δ-1 is shown as a cartoon. GBP is red. Corresponding sidechains of PctA are magenta. l-Ile form the PctA complex is pink. PctA residues are labeled in italics.
Supplementary Material
Acknowledgements
We thank D. Bulkley for technical help and K. Brejc for comments on the manuscript. This work was supported by grant no. NIH R01 HL080050 to D.L.M.
Footnotes
Competing interests
The authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this Article.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41594-023-00951-7.
Peer review information Nature Structural & Molecular Biology thanks Rachelle Gaudet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41594-023-00951-7.
Data availability
Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP coordinates and maps (PDB 8FD7, EMD-29004, EMD-29007 and EMD-29015) and the map of the EMC:Cav1.2(ΔC)/Cavβ3 complex (EMD-29006) are deposited with the RCSB and EMDB. Requests for materials should be addressed to D.L.M.
References
- 1.Dooley DJ, Taylor CP, Donevan S & Feltner D Ca2+ channel α2δ ligands: novel modulators of neurotransmission. Trends Pharmacol. Sci. 28, 75–82 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Taylor CP, Angelotti T & Fauman E Pharmacology and mechanism of action of pregabalin: the calcium channel α2-δ (α2-δ) subunit as a target for antiepileptic drug discovery. Epilepsy Res. 73, 137–150 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Field MJ et al. Identification of the α2-δ-1 subunit of voltage-dependent calcium channels as a molecular target for pain mediating the analgesic actions of pregabalin. Proc. Natl Acad. Sci. USA 103, 17537–17542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domon Y et al. Binding characteristics and analgesic effects of Mirogabalin, a novel ligand for the α2δ subunit of voltage-gated calcium channels. J. Pharmacol. Exp. Ther. 365, 573–582 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Kato J, Inoue T, Yokoyama M & Kuroha M A review of a new voltage-gated Ca2+ channel α2δ ligand, mirogabalin, for the treatment of peripheral neuropathic pain. Expert Opin. Pharmacother. 22, 2311–2322 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Gee NS et al. The novel anticonvulsant drug, gabapentin (Neurontin), binds to the α2δ subunit of a calcium channel. J. Biol. Chem. 271, 5768–5776 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Bauer CS et al. The increased trafficking of the calcium channel subunit α2δ-1 to presynaptic terminals in neuropathic pain is inhibited by the α2δ ligand pregabalin. J. Neurosci. 29, 4076–4088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassidy JS, Ferron L, Kadurin I, Pratt WS & Dolphin AC Functional exofacially tagged N-type calcium channels elucidate the interaction with auxiliary α2δ-1 subunits. Proc. Natl Acad. Sci. USA 111, 8979–8984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marais E, Klugbauer N & Hofmann F Calcium channel α2δ subunits-structure and Gabapentin binding. Mol. Pharmacol. 59, 1243–1248 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Qin N, Yagel S, Momplaisir ML, Codd EE & D’Andrea MR Molecular cloning and characterization of the human voltage-gated calcium channel α2δ-4 subunit. Mol. Pharmacol. 62, 485–496 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Meyer JO & Dolphin AC Rab11-dependent recycling of calcium channels is mediated by auxiliary subunit α2δ-1 but not α2δ-3. Sci. Rep. 11, 10256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran-Van-Minh A & Dolphin AC The α2δ ligand gabapentin inhibits the Rab11-dependent recycling of the calcium channel subunit α2δ-2. J. Neurosci. 30, 12856–12867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z et al. EMC holdase:CaV1.2/CaVβ3 complex and CaV1.2 channel structures reveal CaV assembly and drug binding mechanisms. Preprint at bioRxiv 10.1101/2022.10.03.510667 (2022). [DOI] [Google Scholar]
- 14.Zamponi GW, Striessnig J, Koschak A & Dolphin AC The physiology, pathology and pharmacology of voltage-gated calcium channels and their future therapeutic potential. Pharm. Rev. 67, 821–870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanou E & Catterall WA Calcium channels, synaptic plasticity and neuropsychiatric disease. Neuron 98, 466–481 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Buraei Z & Yang J The β subunit of voltage-gated Ca2+ channels. Physiol. Rev. 90, 1461–1506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolphin AC Voltage-gated calcium channels and their auxiliary subunits: physiology and pathophysiology and pharmacology. J. Physiol. 594, 5369–5390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JP, Dissanayake VU, Briggs AR, Milic MR & Gee NS Isolation of the [3H]gabapentin-binding protein/α2δ Ca2+ channel subunit from porcine brain: development of a radioligand binding assay for α2δ subunits using [3H]leucine. Anal. Biochem. 255, 236–243 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Dolphin AC Calcium channel auxiliary α2δ and β subunits: trafficking and one step beyond. Nat. Rev. Neurosci. 13, 542–555 (2012). [DOI] [PubMed] [Google Scholar]
- 20.David DJ, Donovan CM, Meder WP & Whetzel SZ Preferential action of gabapentin and pregabalin at P/Q-type voltage-sensitive calcium channels: inhibition of K+-evoked [3H]-norepinephrine release from rat neocortical slices. Synapse 45, 171–190 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Anantharaman V & Aravind L Cache—a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25, 535–537 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Jonikas MC et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christianson JC et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegde RS The function, structure, and origins of the ER membrane protein complex. Annu. Rev. Biochem. 91, 651–678 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Wu J et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature 537, 191–196 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Gao S & Yan N Structural basis for pore blockade of human voltage-gated calcium channel Cav1.3 by motion sickness drug cinnarizine. Cell Res. 32, 946–948 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S, Yao X & Yan N Structure of human Cav2.2 channel blocked by the painkiller ziconotide. Nature 596, 143–147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gumerov VM et al. Amino acid sensor conserved from bacteria to humans. Proc. Natl Acad. Sci. USA 119, e2110415119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canti C et al. The metal-ion-dependent adhesion site in the von Willebrand factor-A domain of α2δ subunits is key to trafficking voltage-gated Ca2+ channels. Proc. Natl Acad. Sci. USA 102, 11230–11235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Offord J, Oxender DL & Su TZ Structural requirement of the calcium-channel subunit α2δ for gabapentin binding. Biochem. J. 342, 313–320 (1999). [PMC free article] [PubMed] [Google Scholar]
- 31.Oyama M, Watanabe S, Iwai T & Tanabe M Mirogabalin activates the descending noradrenergic system by binding to the α2δ-1 subunit of voltage-gated Ca2+ channels to generate analgesic effects. J. Pharm. Sci. 146, 33–39 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Lotarski S et al. Anticonvulsant activity of pregabalin in the maximal electroshock-induced seizure assay in α2δ1 (R217A) and α2δ2 (R279A) mouse mutants. Epilepsy Res. 108, 833–842 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Lotarski SM et al. Anxiolytic-like activity of pregabalin in the Vogel conflict test in α2δ-1 (R217A) and α2δ-2 (R279A) mouse mutants. J. Pharmacol. Exp. Ther. 338, 615–621 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Gavira JA et al. How bacterial chemoreceptors evolve novel ligand specificities. mBio 10.1128/mBio.03066-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page KM, Gumerov VM, Dahimene S, Zhulin IB & Dolphin AC The importance of cache domains in α2δ proteins and the basis for their gabapentinoid selectivity. Channels (Austin) 17, 2167563 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahimene S et al. The α2δ-like protein Cachd1 increases N-type calcium currents and cell surface expression and competes with α2δ-1. Cell Rep. 25, 1610–1621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 37.Wallace AC, Laskowski RA & Thornton JM LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Schmidt TG & Skerra A The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Liao M, Cao E, Julius D & Cheng Y Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goehring A et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, Lolicato M, Arrigoni C & Minor DL Jr Production of K2P2.1 (TREK-1) for structural studies. Methods Enzymol. 653, 151–188 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Shaya D et al. Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc. Natl Acad. Sci. USA 108, 12313–12318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebschner D et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriarty NW, Grosse-Kunstleve RW & Adams PD Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams CJ et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF et al. UCSF ChimeraX: structure visualization for researchers, educators and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laskowski RA & Swindells MB LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cav1.2(ΔC)/Cavβ3/Cavα2δ-1:GBP coordinates and maps (PDB 8FD7, EMD-29004, EMD-29007 and EMD-29015) and the map of the EMC:Cav1.2(ΔC)/Cavβ3 complex (EMD-29006) are deposited with the RCSB and EMDB. Requests for materials should be addressed to D.L.M.


