Abstract
Apart from serving as a prosthetic group in globins and enzymes, heme is a key regulator controlling a wide range of molecular and cellular processes involved in oxygen sensing and utilization. To gain insights into molecular mechanisms of heme signaling and oxygen sensing in eukaryotes, we investigated the yeast heme-responsive transcriptional activator HAP1. HAP1 activity is regulated precisely and tightly by heme. Here we show that in the absence of heme, HAP1 forms a biochemically distinctive higher-order complex. Our data suggest that this complex contains HAP1 and four other cellular proteins including Hsp82 and Ydj1. The formation of this complex is directly correlated with HAP1 repression in the absence of heme, and mutational or heme disruption of the complex correlates with HAP1 activation, suggesting that this complex is responsible for heme regulation of HAP1 activity. Further, we determined HAP1 domains required for heme regulation: three domains—the dimerization domain, the heme domain, and the HRM7 (heme-responsive motif 7) domain—cooperate to form the higher-order complex and mediate heme regulation. Strikingly, we uncovered a novel function for the HAP1 dimerization domain: it not only allows dimerization but also provides critical functions in heme regulation and transcriptional activation. Our studies provide significant insights into the molecular events leading to heme activation of HAP1 and may shed light on molecular mechanisms of various heme-controlled biological processes in diverse organisms.
Heme plays key roles in oxygen sensing and utilization in all living organisms ranging from bacteria to humans. In mammals, recent experiments suggest that hemoproteins are the oxygen sensors in the induction of the synthesis of erythropoietin and vascular endothelial cell growth factor in response to hypoxia (13, 22, 25). Heme also directly regulates various molecular and cellular processes for systems that use or sense oxygen. For example, heme stimulates the expression of globin chains in erythroid cells and cytochrome P-450 in hepatic cells (6, 9, 36), activates protein synthesis through the heme-regulated inhibitor kinase (7), and regulates the transport of numerous enzymes and the assembly of hemoglobin and cytochrome complexes (24, 31). The roles of heme are largely conserved from yeast to humans, and recent evidence indicates that heme might regulate diverse processes through similar mechanisms (24, 48). For example, recent experiments (32, 37) suggest that heme regulation of gene transcription in mammalian cells is mediated by heme-responsive transcription factors like the yeast transcriptional activator HAP1 (8, 34). However, no mammalian heme-responsive transcriptional regulator has been identified to date. Nonetheless, the yeast HAP1-heme regulatory system provides an excellent model for studying heme signaling in eukaryotic cells.
In the yeast Saccharomyces cerevisiae, heme functions as the internal barometer of oxygen tension; heme synthesis in mitochondria is directly correlated with oxygen levels in the extracellular environment (23, 29). Heme induces the expression of many genes encoding respiratory functions and functions involved in controlling oxidative damage (15, 19, 50). The effect of heme on gene expression is mediated by the transcriptional activator HAP1 (8, 34), which is the only known eukaryotic heme-responsive transcriptional regulator. HAP1 activity is precisely controlled by heme concentrations: HAP1 activity gradually increases as the heme concentration increases and reaches the limit at micromolar heme concentrations (46). Presumably, heme binds to HAP1 and activates HAP1, which in turn promotes transcription from many promoters (8, 34). At low levels of HAP1 expression (as occurs naturally from its own promoter), HAP1 activity can be induced 1,000-fold by heme, while at high levels of expression (from the inducible GAL1-10 promoter), HAP1 activity is induced approximately 50-fold by heme (15, 46, 47).
HAP1 contains five functional domains (33, 44, 48, 49): the C6 zinc cluster, the dimerization domain, the heme domain, the heme-responsive motif 7 (HRM7) domain, and the activation domain. The heme domain and the HRM7 domain contain six HRMs and one HRM, respectively; both domains have been shown to be involved in heme regulation of HAP1 (8, 33, 48). HAP1 DNA binding requires the C6 zinc cluster and the dimerization domain. The C6 zinc cluster is a highly conserved DNA recognition motif found in at least 40 yeast transcriptional activators including GAL4 and PPR1 (12, 27, 28, 38, 39). The HAP1 dimerization domain also contains a coiled-coil dimerization element homologous to that within the dimerization domain of GAL4 or PPR1 (27, 38, 44). HAP1, however, is a unique member of the C6 zinc cluster family: it recognizes asymmetric DNA sites and binds to DNA asymmetrically, whereas GAL4 and PPR1 recognize symmetric DNA sites and bind to DNA symmetrically (27, 28, 39, 49).
More intriguingly, how does heme regulate HAP1 activity? Previous results suggest that heme stimulates HAP1 DNA-binding and transcriptional activity through both the heme domain and the HRM7 domain (33, 48). In addition, it has been shown that in the absence of heme, HAP1 appears as a slowly migrating band when analyzed by DNA mobility shift assays (11, 47). This band was designated a high-molecular-weight complex (HMC) (47). Heme disrupts this putative HMC and permits HAP1 to bind with high affinity to DNA as a dimer (47). This observation raises many questions. For example, is the HMC really a large and biochemically distinctive complex containing multiple proteins? Or is it an artifact of DNA mobility shift assays? Or is it just an aggregate of HAP1 formed in the absence of heme? Further, if the HMC is indeed a biochemically distinctive complex, what is its functional role in mediating heme regulation of HAP1? What HAP1 elements are required for the formation of the HMC and for mediating heme regulation?
To answer these questions and to test the hypothesis that the HMC represses HAP1 activity in the absence of heme, we carried out a series of experiments. Using sucrose gradient centrifugation and affinity and gel filtration chromatography, we show here that the HMC is a biochemically distinctive complex and contains HAP1 and four other cellular proteins including heat shock proteins Hsp82 and Ydj1. We further show that the formation of the HMC is directly correlated with HAP1 heme responsiveness. Our data strongly suggest that the HMC is responsible for heme regulation. We also explored the possibility that the HAP1 dimerization domain is important for heme regulation and transcriptional activation. Strikingly, domain-swapping experiments show that the HAP1 dimerization domain is critical for heme regulation and transcriptional activation. We demonstrate that three domains—the heme domain, the HRM7 domain, and the dimerization domain—are all required for the formation of intact HMC and heme regulation of HAP1 activity.
MATERIALS AND METHODS
Yeast strains and reporters.
Yeast strains used were MHY200 (MATa ura3-52 leu2-3,112 his4-519 ade1-100 hem1-Δ100 hap1::LEU2) (16), L51 (MATa ura3-52 leu2-3,112 his4-519 ade1-100 hap1::LEU2 trp1::hisG) (46), JEL1 (MATα leu2 trp1 ura3-52nprb1-1122 pep4-3 Δhis3::pGAL10-GAL4) (provided by B. Pina), W303 (ade2-1 can1-100 his3-12 leu2-3,112 trp1-1 ura3-1), and GRS4 (derived from W303 with LEU2 marked disruption mutations at the HSP82 and HSC82 genes, and bearing a vector carrying the TRP1 gene and the GAL1 HSP82 fusion gene, which expresses the wild-type level of Hsp82 in galactose medium and 5% of the wild-type level in glucose medium) (35). Yeast cells were grown in YPD or synthetic complete medium. The UAS1/CYC1 (upstream activation sequence 1 [UAS1] of the CYC1 promoter)-lacZ and UAS/CYC7 (UAS of the CYC7 promoter)-lacZ reporter plasmids have been described elsewhere (42). The HAP1-driven UAS1/CYC1-lacZ reporter (15), the HAP2/3/4/5-driven UAS2UP1/CYC1-lacZ reporter (10), and the GCN4 and BAS1/2-driven HIS4-lacZ reporter (18) plasmids are as described elsewhere. The radiolabeled UAS1/CYC1 and UAS/CYC7 were described previously (49).
Construction of expression plasmids.
To construct the expression plasmid for the HAP1-PPR1 hybrid protein, the DNA-binding sequence was first generated by overlapping PCR with appropriate primers (49). The DNA was then cut with SmaI and BstEII and inserted into a HAP1 expression vector (SD5-HAP1) (42) cut with SmaI and BstEII. The sequence of the fused region was confirmed by sequencing.
To construct the expression plasmid for the HAP1 mutant with the heme domain deleted (HAP1Δheme), plasmid pHAP1(Δ247-447) (33) was cut with BstEII and KpnI. The fragment containing HAP1 sequences was then inserted into SD5-HAP1 cut with BstEII and KpnI.
To construct the expression plasmids for His6-HAP1 and His6-HAP1ΔKpn, a DNA fragment encoding Met-Arg-Gly-Ser-His-His-His-His-His-His [MRGS(H)6] was synthesized and inserted into the HAP1 and HAP1ΔKpn expression plasmids, SD5HAP1 and SD5HAP1ΔKpn, cut with SmaI. The corrected clones were identified by colony hybridization and verified by DNA sequencing. The resulting plasmids express HAP1 and HAP1ΔKpn with the MRGS(H)6 tag at their N termini.
Preparation of yeast extracts and DNA mobility shift assays.
Yeast JEL1 cells bearing expression plasmids were grown to an optical density (OD) of 0.5 and induced with 2% galactose for 6 h. Cells were harvested and resuspended in 3 packed cell volumes of buffer (20 mM Tris, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT], 0.3 M NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of pepstatin per ml, 1 μg of leupeptin per ml). Cells were then permeabilized by agitation with 4 packed cell volumes of glass beads, and extracts were collected as described previously (34). This method consistently yielded extracts with protein concentrations at approximately 10 mg/ml.
DNA-binding reactions were carried out in a 20-μl volume with 5% glycerol, 4 mM Tris (pH 8), 40 mM NaCl, 4 mM MgCl2, 10 mM DTT, 3 μg of salmon sperm DNA, 10 μM zinc acetate, and 300 μg of bovine serum albumin per ml as described previously (47). Approximately 0.01 pmol of labeled UAS1/CYC1 or UAS/CYC7 and 20 μg of protein extracts were used in each reaction. The reaction mixtures were incubated at room temperature for 1 h and then loaded onto 4% polyacrylamide gels in 0.5× Tris-borate-EDTA for polyacrylamide gel electrophoresis (PAGE) at 4°C.
Generation of HAP1 antibody and Western blotting.
HAP1 antiserum was generated by injecting purified glutathione S-transferase (GST)–HAP1-171 (containing HAP1 residues 1-171) from bacteria into rabbits according to established protocols (17). To purify HAP1 antibody, purified GST–HAP1-171 was conjugated to CNBr-activated agarose beads. Then, HAP1 antiserum was loaded onto the GST–HAP1-171 column and washed extensively. Purified HAP1 antibody was eluted as described previously (17).
For Western blotting, approximately 150 μg of whole-cell extracts was first separated on sodium dodecyl sulfate (SDS)–7% polyacrylamide gels and then transferred to polyvinylidene difluoride or nitrocellulose membranes. HAP1 was visualized by using a purified antibody against GST-HAP1 1-171 and a chemiluminescence Western blotting kit (Boehringer Mannheim). Antisera specific to Hsp82, Ydj1, and Ssn6 were provided by S. Lindquist (5, 35), A. J. Caplan (3, 4), and R. Trumbly (43), respectively. Antibody specific to MRGS(H)6 was purchased from Qiagen.
Sucrose gradient centrifugation.
Extracts (150 μl) containing HAP1 or its derivatives were loaded directly onto an approximately 11-ml 10 to 45% sucrose gradient in 25 mM Tris-HCl (pH 8)–50 mM NaCl–6 mM MgCl2–1 mM EDTA–10 μM zinc acetate–1 mM PMSF–10 mM DTT. Centrifugation was for 15 h at 32,000 rpm in a Beckman SW50.1 rotor at 5°C; 500-μl fractions were collected, and HAP1 or its derivatives were analyzed in the presence of heme by DNA mobility shift assays. The amounts of HAP1 and its derivatives in the fractions were detected by measuring the amounts of radioactivity in the bands containing HAP1-DNA complexes with a PhosphorImager (Molecular Dynamics).
Purification of the HMC.
Yeast JEL1 cells bearing plasmids expressing MRGS(H)6-HAP1 or MRGS(H)6-HAP1ΔKpn were grown in raffinose complete selective medium to an OD of 0.5 and then induced with 2% galactose for 5 to 6 h. Ten-liter volumes of cells were routinely collected, and extracts were prepared as described above or by using a Mini-Bead Beater (Biospec). We often obtained 50 to 100 ml of extracts with a protein concentration of 10 mg/ml.
To purify the HMC, 2 to 3 ml of Ni-nitrilotriacetic acid (NTA) superflow beads (Qiagen) was packed in a column and equilibrated with buffer containing 25 mM Tris-HCl (pH 8), 100 mM NaCl, 6 mM MgCl2, 0.5 mM EDTA, 1 mM PMSF, and 10 mM DTT. Then, extracts were loaded into the column at the rate of approximately 15 ml/h. Columns were subsequently washed with 150 to 200 column volumes of the equilibration buffer with 20 mM imidazole. The HMCs were eluted with buffer containing 250 mM imidazole, and four 2- to 3-ml fractions were collected. The fractions were further concentrated on Centricon 10 (Amicon) and analyzed by SDS-PAGE and DNA mobility shift assays. The HMC was subjected to sucrose gradient centrifugation, but no significant improvement was observed, presumably because the HMC eluted from Ni-NTA was of high purity already. When analyzed on SDS-PAGE, the components of the HMC are identical whether the extracts contain MRGS(H)6-HAP1 or MRGS(H)6-HAP1ΔKpn.
To further fractionate the eluate from the Ni-NTA column, 500 μl of eluate was loaded onto an FPLC Superose 6 HR10/30 column (Pharmacia). Proteins were eluted with buffer containing 20 mM Tris, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.3 M NaCl, 1 mM PMSF, 1 μg of pepstatin per ml, and 1 μg of leupeptin per ml. Fractions of 400 μl were collected, trichloroacetic acid precipitated, and analyzed by SDS-PAGE.
β-Galactosidase assays.
To determine β-galactosidase levels from reporter genes in cells containing wild-type HAP1, the HAP1-PPR1 hybrid protein, and other mutants in the presence and absence of heme, yeast high-copy-number 2μm replicating plasmids expressing HAP1 and mutants from the GAL1-10 promoter were transformed into the strain MHY200 (16) also bearing the UAS1/CYC1-lacZ reporter. Cells were grown in synthetic complete medium containing 2% raffinose with limiting amounts of the heme precursor δ-aminolevulinate (2 μg/ml) or high amounts of δ-aminolevulinate (250 μg/ml) to an OD of approximately 0.5. Cells were then induced with 2% galactose for 7 h and harvested for determination of β-galactosidase as described previously (47).
To determine β-galactosidase levels from reporter genes in cells containing HAP1, HAP1-18, and the HAP1-PPR1 hybrid protein at UAS1/CYC1 or UAS/CYC7, 2μm plasmids expressing wild-type HAP1, HAP1-18, and HAP1-PPR1 from the GAL1-10 promoter were transformed into the strain L51 also bearing the UAS1/CYC1-lacZ or UAS/CYC7-lacZ reporter. Cells were grown in 2% raffinose to an OD of 0.3 and then induced with 2% galactose prior to β-galactosidase assays as described previously (46). β-Galactosidase levels from various reporter genes in GRS4 and W303 cells were detected as described previously (35).
RESULTS
The HMC is a biochemically distinctive complex.
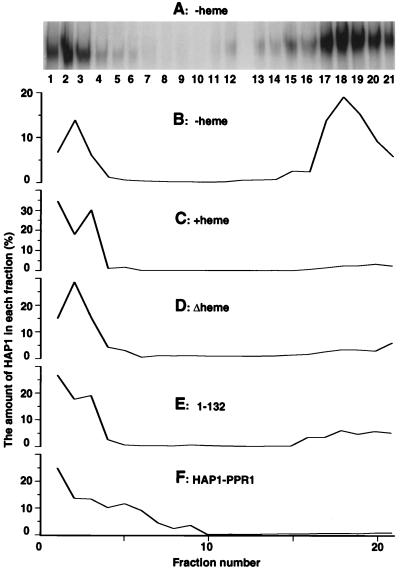
Previous studies have shown that in the absence of heme, HAP1 appears on nondenaturing polyacrylamide gels as a slowly migrating but distinctive band, designated an HMC (11, 47). Based on indirect evidence from titration experiments, it was hypothesized that in the absence of heme, HAP1 is bound by a cellular factor in the HMC (11, 47). Because the evidence for the HMC came only from DNA mobility shift assays, the hypothesis that the HMC is a distinctive biochemical entity and contains multiple cellular factors must be further tested. To demonstrate that the HMC indeed represents a biochemically distinctive complex containing multiple proteins, we carried out gel filtration and gradient sedimentation analyses of HAP1 complexes and the eventual purification of the HMC (described below). Sucrose gradient centrifugation (Fig. 1A to C) showed that HAP1 is indeed in distinctive forms with or without heme (we also attempted to separate the HMC and HAP1 complexes formed with heme on Superose 6 and Sepharose CL-4B columns, but the two complexes had very similar elution volumes).
FIG. 1.
Analysis of HAP1 complexes by sucrose gradient centrifugation. (A) Analysis of HAP1 complexes in fractions from a 10 to 45% sucrose gradient in the absence of heme. The fraction numbers were counted from top to bottom (i.e., fraction 1 corresponds to 10% sucrose, while fraction 21 corresponds to 45% sucrose). The amounts of HAP1 in fractions were detected in the presence of heme by DNA mobility shift assays. The low level of dimeric complexes in fractions 1 to 3 is due to HAP1 overexpression or disruption of the HMC (47). (B) Distribution profile of HAP1 in the sucrose gradient in the absence of heme. The fractions are the same as those shown in panel A, but HAP1 amounts were calculated and plotted as percentages of the total HAP1 amount in all fractions. (C) Distribution profile of HAP1 in the sucrose gradient in the presence of heme. (D) Distribution profile of HAP1Δheme in the sucrose gradient. (E) Distribution profile of HAP1-132 in the sucrose gradient. (F) Distribution profile of HAP1-PPR1 in the sucrose gradient. No heme was included in the extracts loaded onto gradients shown in panels D to F. Note that different scales, especially in panels B and C, are used in the graphs in order to make them identical in size. The same HAP1 distribution profiles were obtained when Western blotting instead of DNA mobility shift assay was used to quantify HAP1 amounts. All extracts used here were prepared from JEL1 cells under previously established conditions (11, 47).
Whole-cell extracts containing HAP1 were subjected to centrifugation in a 10 to 45% linear sucrose gradient in the presence or absence of heme. We then analyzed the amounts of HAP1 in the fractions by DNA mobility shift assays in the presence of heme. When centrifugation was carried out in the presence of heme, the majority (82% of total HAP1 in all fractions) of HAP1 was found in fractions 1 to 3 (Fig. 1C). However, in the absence of heme (Fig. 1B), we detected a high level of HAP1 in fractions 17 to 21 (60%) and a low level of HAP1 in fractions 1 to 3 (25%; the same results were obtained by Western blotting). When heme was not included in the DNA-binding reactions, as shown by DNA mobility shift assays, HAP1 in fractions 17 to 21 was in the form of the HMC while HAP1 in fractions 1 to 3 was in the form of dimeric complexes (data not shown). The low level of dimeric complexes formed in the absence of heme is due to HAP1 overexpression and/or disruption of the HMC (when HAP1 is expressed at a lower level, all HAP1 forms the HMC [47]). These results show that HAP1 is in distinctive forms with different sedimentation properties, depending on whether heme is present or absent. In the absence of heme, HAP1 is in the form of HMC, and so it sediments faster. In the presence of heme, the majority of HAP1 is monomeric (in the absence of DNA [45]), so it sediments at a lower rate. In other words, heme induces a drastic redistribution of HAP1 in the fractions (the distributions of total proteins in fractions with and without heme are identical [not shown]). Based on the distribution of the HMC and molecular markers on the sucrose gradient, we estimate the apparent molecular mass of the HMC at approximately 1,000 kDa. Collectively, these results show that the HMC is not an artifact but a biochemically distinctive complex that can be detected by DNA mobility shift assays and sucrose gradient centrifugation.
Purification of the HMC shows that the HMC contains four other cellular proteins in addition to HAP1.
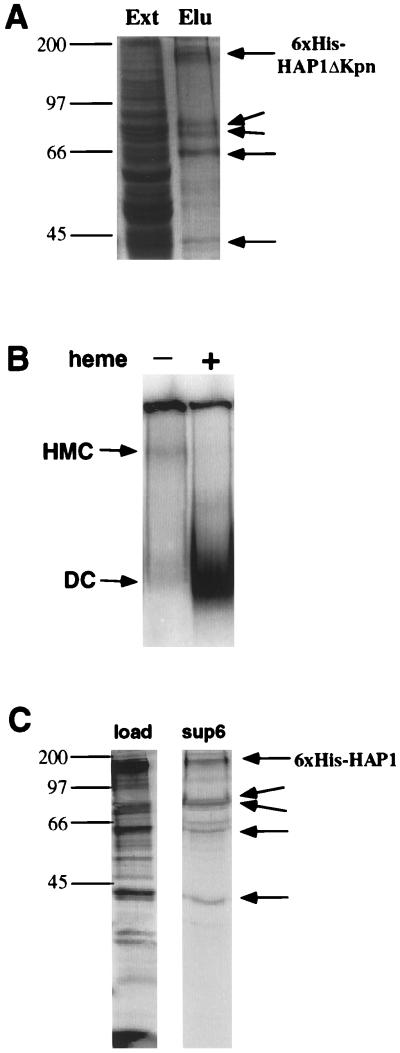
To further test the idea that the HMC is a distinctive biochemical entity and contains proteins other than HAP1, we purified the complex and analyzed its components. We found that the fusion of the His6 tag to the N terminus of HAP1 has no effect on the formation of the HMC and HAP1 activity. We transformed His6-HAP1 and HAP1 with the UAS1/CYC1-lacZ or UAS/CYC7-lacZ reporter into yeast cells, detected the β-galactosidase activity in the absence and presence of heme, and found that His6-HAP1 and HAP1 behaved identically. Therefore, we used His6-HAP1 or His6-HAP1ΔKpn in our experiments. Large quantities of cells expressing His6-HAP1 or His6-HAP1ΔKpn were collected, and extracts were prepared as described in Materials and Methods. His6-HAP1ΔKpn lacks the HAP1 activation domain encompassing residues 1309 to 1483. We often expressed His6-HAP1ΔKpn instead of His6-HAP1 because the former is less toxic than the latter when overexpressed, and because it also forms the HMC. To purify the HMC, we used Ni-NTA columns. The HMC was eluted from the columns by imidazole, and the fractions were further concentrated and analyzed by SDS-PAGE and DNA mobility shift assays. Sucrose gradient centrifugation was used to further purify the complex, but it did not significantly improve the purification (not shown), presumably because the complex was quite pure already (Fig. 2A).
FIG. 2.
Purification of the HMC indicates that HAP1 is bound to multiple cellular proteins in the absence of heme. (A) Purification of the HMC on Ni-NTA columns. JEL1 cells bearing a plasmid expressing His6-HAP1ΔKpn under the control of the GAL1-10 promoter were induced by 2% galactose for 5 h (47), and whole-cell extracts were prepared as described in Materials and Methods. The extracts were purified on Ni-NTA columns, and the eluted fractions were analyzed by SDS-PAGE (10% gels) and visualized by Coomassie blue staining. Shown are the eluted peak fraction (Elu) and unpurified whole-cell extracts (Ext). Four proteins (shown by arrows) are bound to HAP1 in the absence of heme. Based on the positions of molecular weight standards, we estimate that the sizes of these proteins are 79, 73, 60, and 42 kDa. The identity of the His6-HAP1ΔKpn band was verified by Western blotting using antibodies against MRGS(H)6 (Qiagen) and GST-HAP1 (see Materials and Methods). (B) DNA-binding complexes formed by the purified HMC in the presence and absence of heme. Approximately 0.2 μg (total protein amount in the fraction) of purified HMC was incubated with radiolabeled DNA in the absence and presence of heme (5 ng/μl) and then analyzed on a 3.5% polyacrylamide gel. DC, dimeric complex. (C) The four proteins coelute with HAP1 during chromatography on an FPLC Superose 6 column. A 500-μl fraction eluted from a Ni-NTA column bound with extracts containing His6-HAP1 was loaded onto an FPLC Superose 6 HR10/30 column (Pharmacia). The loaded fraction is less pure than the one shown in panel A because no imidazole was included for washing the Ni-NTA column during this preparation. Fractions of 400 μl were collected from the Superose column, trichloroacetic acid precipitated, and analyzed by SDS-PAGE (12% gel). Shown are the loaded fraction from the Ni-NTA column (load) and the peak fraction (sup6) from the Superose 6 column containing the highest amount of His6-HAP1. The elution volume of the peak fraction was between that (void volume) of dextran (2,000 kDa) and that of thyroglobulin (670 kDa; Bio-Rad). The four proteins shown in panel A are also marked here by arrows. The minor band above the 60-kDa protein is due to HAP1 degradation. Note that the 79/73-kDa bands are closer together here than in panel A, very likely because this gel contains a higher percentage of acrylamide.
Figure 2A shows a typical result from the purification: four cellular proteins appear to bind to HAP1 in the absence of heme. We estimated that the apparent molecular masses of these four proteins are 79, 73, 60, and 42 kDa (Fig. 2A). The same results were obtained whether His6-HAP1 or His6-HAP1ΔKpn was expressed, consistent with previous results showing that HAP1ΔKpn also forms the HMC. When analyzed by DNA mobility shift assays, as expected, the purified complex migrated very slowly and bound to DNA with low affinity, while in the presence of heme, HAP1 dimeric complexes were formed and bound to DNA with high affinity (Fig. 2B).
To further ascertain that four proteins are bound to HAP1 and form one complex, we fractionated the eluate from Ni-NTA columns by gel filtration chromatography. We took a peak fraction containing His6-HAP1 from a Ni-NTA column (Fig. 2C, load) and loaded it onto an FPLC Superose 6 column. The loaded fraction is less pure than the one shown in Fig. 2A because less stringent washing conditions (no imidazole included) were used during this purification. Nonetheless, the four proteins shown in Fig. 2A clearly coeluted with HAP1 from the Superose 6 column (Fig. 2C). The elution volume for the complex containing HAP1 and four other proteins was slightly smaller than that of thyroglobulin (670 kDa). This elution volume is consistent with the estimated molecular mass (1,000 kDa) of the HMC based on sucrose gradient centrifugation. Other proteins were eluted out at higher elution volumes. Further, these four proteins were selectively cross-linked to HAP1 by glutaraldehyde (not shown). Collectively, these results strongly support the conclusion that these four proteins are bound to HAP1 and form a distinctive complex, HMC.
Hsp82 associates with HAP1 and affects HAP1 transcriptional activity.
Heme regulation of HAP1 may resemble the regulation of steroid hormone receptors by steroids (33, 47, 48). In the absence of ligands, steroid hormone receptors are bound to heat shock proteins such as Hsp90 (Hsp82 in yeast) and Ydj1, so that the receptors are repressed and are maintained in an activatable state (2, 3, 21, 35, 40). Heat shock proteins are also required in the subsequent ligand-dependent activation of the receptors. In a strain that expresses a low level of Hsp82, steroid hormone receptors are activated much less efficiently by hormonal ligands (35). Likewise, similar proteins might bind to and repress HAP1 in the absence of heme and facilitate HAP1 activation in the presence of heme.
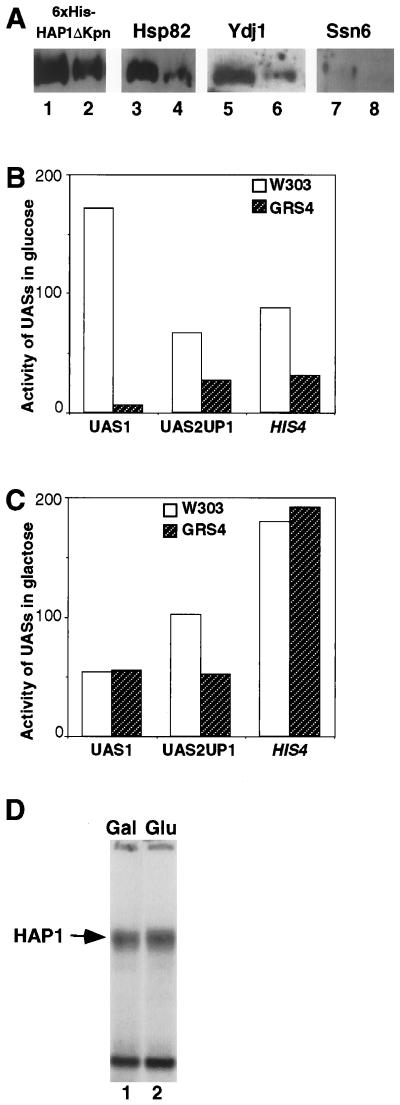
To test the idea that heat shock proteins are components of the HMC, we used antiserum specific to Hsp82 or Ydj1 to probe whether they are present in the purified HMC. We took two purified fractions (the peak fraction and the subsequent fraction) containing His6-HAP1ΔKpn (Fig. 2A) from the Ni-NTA column and probed with antiserum specific to His6-HAP1ΔKpn, Hsp82 (35), Ydj1 (3), or Ssn6 (43) (these antibodies exhibited similar sensitivities in detecting corresponding proteins in crude extracts). As shown in Fig. 3A, Hsp82 (lanes 3 and 4) and Ydj1 (lanes 5 and 6) were indeed present in the fractions at significant levels similar to the level of His6-HAP1ΔKpn (lanes 1 and 2). The levels of Hsp82 and Ydj1 decreased as the level of His6-HAP1ΔKpn decreased, and no signal was detected in fractions containing no HMC or HAP1 (Fig. 3A [compare lanes 2 and 1, 4 and 3, and 6 and 5] and data not shown). When extracts containing wild-type HAP1 (without the His tag) were loaded onto the Ni-NTA column and washed, no Hsp82 or Ydj1 was detected in the eluted fractions, suggesting that Hsp82 and Ydj1 specifically interact with His6-HAP1 and do not bind to the Ni-NTA column nonspecifically. As a further control, lanes 7 and 8 in Fig. 3A show that no significant level of Ssn6 (perhaps a trace amount is present due to contamination [lane 7]), a protein previously shown to be absent in the HMC (46), is detected in the fractions. The specificity of these antibodies has been demonstrated in previous experiments (4, 5, 43). In our experiments, we also detected only a strong single band at the expected position in the fractions and extracts when using antiserum specific to Hsp82, Ydj1, or Ssn6. Together, these lines of evidence strongly suggest that Hsp82 and Ydj1 specifically associate with HAP1. Based on approximate positions and molecular masses of the proteins, we infer that Hsp82 corresponds to the 79-kDa band whereas Ydj1 corresponds to the 42-kDa band in Fig. 2.
FIG. 3.
Hsp82 is present in the HMC and greatly affects HAP1 transcriptional activity. (A) Hsp82 and Ydj1 are found in the purified HMC. Two purified HMC fractions from a Ni-NTA column (Fig. 2A), the peak fraction (lanes 1, 3, 5, and 7) and the subsequent fraction (lanes 2, 4, 6, and 8), were electrophoresed on 6% (lanes 1 and 2), 7% (lanes 3 and 4), 10% (lanes 5 and 6), and 7% (lanes 7 and 8) SDS-polyacrylamide gels. Then, proteins on these gels were transferred to polyvinylidene difluoride membranes and probed with antibodies specific for MRGS(H)6 (lanes 1 and 2), Hsp82 (lanes 3 and 4), Ydj1 (lanes 5 and 6), and Ssn6 (lanes 7 and 8) by Western blotting. These detected bands were all at the expected positions according their molecular weights. No significant signals were detected in lanes 7 and 8 even after extended exposure. These antibodies exhibited similar sensitivity (at the nanogram level) when used to probe proteins in crude extracts. (B) Hsp82 greatly affects HAP1 activity selectively. Yeast wild-type W303 and Hsp82/Hsc82-deficient GRS4 cells (35) bearing the HAP1-driven UAS1/CYC1-lacZ reporter (15), the HAP2/3/4/5-driven UAS2UP1/CYC1-lacZ reporter (10), or the GCN4 and BAS1/2-driven HIS4-lacZ reporter (18) were grown in glucose medium at ambient temperature, and β-galactosidase activities were detected as described previously (35). The β-galactosidase activity detected from cells bearing a reporter plasmid is plotted. (C) HAP1 activity is not affected in the GRS4 strain (35) when the wild-type level of Hsp82 is expressed. β-Galactosidase activities were detected in the same way as for panel B except that cells were grown in galactose medium. (D) The reduced level of Hsp82 does not affect HAP1 DNA-binding activity. Extracts were prepared from GRS4 cells grown in galactose medium (lane 1) and glucose medium (lane 2) as described previously (34). DNA-binding reactions were carried out in the presence of heme as described in Materials and Methods. The position of the HAP1-DNA complex is marked by an arrow. The bottom band is due to DNA binding by an uncharacterized protein (34).
To further explore the idea that Hsp82 is important for heme regulation of HAP1, we took advantage of the previously generated GRS4 strain (35), which produces 5% of the wild-type level of Hsp82/Hsc82 in glucose medium and the wild-type level in galactose medium but is sufficient in heme synthesis. We detected β-galactosidase activities of three reporters—the HAP1-driven UAS1/CYC1-lacZ reporter (15), the HAP2/3/4/5-driven UAS2UP1/CYC1-lacZ reporter (10), and the GCN4 and BAS1/2-driven HIS4-lacZ reporter (18)—in glucose medium in the GRS4 strain and the wild-type W303 strain. We found that the activity of HAP1-driven UAS1/CYC1-lacZ reporter was reduced approximately 30-fold in the GRS4 strain compared to the wild-type W303 strain, while the activities of the UAS2UP1/CYC1-lacZ reporter and the HIS4-lacZ reporter were generally reduced only 2- to 3-fold (Fig. 3B). As a control, Fig. 3C shows that all three reporters exhibited similar levels of activity in both strains in galactose medium, in which GRS4 produces the wild-type level of Hsp82 (35). The result suggests that a reduced level of Hsp82 has a strong and selective effect on HAP1 activity in heme-sufficient cells.
To rule out the possibility that the effect of Hsp82 on HAP1 activity is due to its effect on HAP1 expression level or stability, we detected HAP1 levels in extracts prepared from cells producing the wild-type level of Hsp82 and 5% of the wild-type level by DNA mobility shift assays. We found that extracts prepared from cells expressing the wild-type level of Hsp82 (GRS4 in galactose [Fig. 3D, lane 1]) and 5% of the wild-type level (GRS4 in glucose [Fig. 3D, lane 2]) exhibited the same level of DNA-binding activity in vitro in the presence of heme, suggesting that the reduced level of Hsp82 did not affect HAP1 expression levels or stability. Because of the difficulty in expressing sufficient levels of HAP1 transiently in GRS4 in glucose medium and in growing the GRS4 cells in glucose medium under heme-deficient conditions, we could not examine the effect of the reduced level of Hsp82 on the HMC and HAP1 activity in the absence of heme. However, we suspect that the HMC can form at the reduced level of Hsp82 because the HAP1 level is also low when expressed from its own promoter on the chromosome (47). Very likely, HAP1 is inactive in the absence of heme at the reduced level of Hsp82, because HAP1 is largely inactive even in heme-sufficient cells—the activity of the UAS1/CYC1-lacZ reporter in GRS4 in glucose medium (Fig. 3B) is at the same level as the basal transcriptional activity of the reporter detected in cells lacking HAP1 (46). In any case, the data in Fig. 3 suggest that Hsp82 associates with HAP1 and that the level of Hsp82 strongly affects HAP1 activity. The result that Hsp82 has a positive effect on HAP1 activity might seem to be puzzling and contradictory to the role of the HMC in HAP1 repression. However, this is not entirely inconceivable because HAP1, like steroid hormone receptors (2, 3, 21, 35, 40), might require Hsp82 and other heat shock proteins for repression in the absence of the ligand, and for subsequent activation by the ligand (see also Discussion).
The HMC is functionally linked to heme regulation.
Biochemical data described above have established the fact that the HMC is a biochemically distinctive complex containing HAP1 and four other proteins including Hsp82 and Ydj1, not an artifact or an aggregate of HAP1. The next important task is to demonstrate its functional significance in heme regulation of HAP1. We asked whether the HMC is critical for heme regulation of HAP1 activity and how the heme domain and the HRM7 domain may affect the HMC and heme regulation. To answer these questions, we took advantage of HAP1 mutants: we examined the effects of mutations in the heme and HRM7 domains on heme regulation and on the formation of the HMC and determined whether there is a direct correlation between the formation of the HMC and HAP1 heme responsiveness.
We characterized a HAP1 derivative with the heme domain deleted (HAP1Δheme; residues 245 to 444 are deleted) and a HAP1 mutant (HAP1-132) with a mutation at amino acid position 1048 (Cys to Tyr) (16) in the HRM7 domain (Fig. 4). Although the HAP1Δheme mutant has previously been shown to bind to DNA constitutively as a dimer in the absence of heme (11, 47), it has not been shown that it causes constitutive transcriptional activity due to its toxicity under heme-deficient conditions. We overcame this problem by expressing the protein from the inducible GAL1-10 promoter. We placed HAP1, HAP1Δheme, and HAP1-132 under the control of the inducible GAL1-10 promoter, so that we could analyze their transcriptional and DNA-binding activities in parallel. We detected the activities of wild-type HAP1 and mutants in heme-deficient (2 μg of the heme precursor δ-aminolevulinate per ml) and heme-sufficient (250 μg of δ-aminolevulinate per ml) cells. We analyzed the HMC formed in the absence of heme and HAP1 dimeric complexes formed in the presence of heme by wild-type HAP1 and mutants.
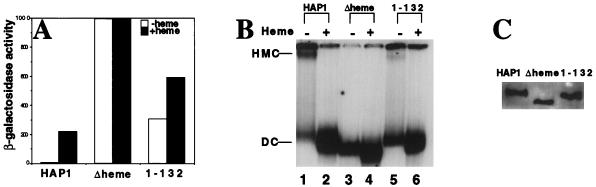
FIG. 4.
Formation of the HMC is critical for HAP1 repression in the absence of heme. (A) β-Galactosidase activities of HAP1, HAP1Δheme (Δheme, the derivative lacking the heme domain), and HAP1-132 in heme-deficient cells (open bars) and heme-sufficient cells (shaded bars). 2μm plasmids expressing wild-type HAP1 and mutants from the GAL1-10 promoter were transformed into strain MHY200 bearing the UAS1/CYC1-lacZ reporter. β-Galactosidase assays were carried out as described in Materials and Methods. (B) DNA-binding complexes formed by HAP1 and mutants in the presence or absence of heme. Extracts containing HAP1 (lanes 1 and 2), HAP1Δheme (lanes 3 and 4), and HAP1-132 (lanes 5 and 6) were incubated with radiolabeled DNA in the absence (lanes 1, 3, and 5) and presence (lanes 2, 4, and 6) of heme (5 ng/μl). The reaction mixtures were analyzed on a 4% polyacrylamide gel. A low level of wild-type HAP1 forms dimeric complexes (DC) in the absence of heme due to the titration of other cellular proteins in the HMC. All extracts used here were prepared from JEL1 cells under previously established conditions (11, 47). (C) Western blot showing that virtually identical amounts of HAP1 and mutant proteins are expressed in vivo.
As shown in Fig. 4A and B, we found that HAP1 forms a high level of HMC (Fig. 4B, lane 1), and its transcription-activating activity is induced at least 30-fold by heme (Fig. 4A); HAP1Δheme does not form the HMC (Fig. 4B, lane 3) and is constitutively active regardless of heme concentrations (Fig. 4A); and the HAP1-132 mutant forms a very low level of HMC (Fig. 4B, lane 5) and causes constitutivity in the absence of heme and 2-fold hyperinduction by heme (Fig. 4A). Western blotting shows that all three proteins were expressed at the same level (Fig. 4C; the same conclusion can be made based on the dimeric complexes formed in the presence of heme [Fig. 4B]). The low level of the HMC formed by the mutants is therefore not due to differences in protein levels. These results show that the disappearance of the HMC coincides with the disappearance of HAP1 heme responsiveness. The dimeric complex formed by wild-type HAP1 in the absence of heme is due to HAP1 overexpression (46). The overexpression of wild-type HAP1 and formation of dimeric complexes allow HAP1 to gain higher than basal but low levels of activity in the absence of heme (46), indicating that wild-type HAP1 dimeric complexes are largely inactive, very likely because dimeric complexes in vivo can rapidly reassociate with other cellular proteins to form the HMC, and remain inactive (see Discussion). Note that the shape and the mobility of the dimeric complex band (Fig. 4B) appear to be different in the absence and presence of heme, very likely because of conformational changes induced by heme binding (the mutants can still bind to heme through HRMs, although the binding is not important for activity).
To confirm the effects of HAP1Δheme and the HAP1-132 mutant on the HMC, we examined the distribution of mutant complexes in the sucrose gradient. As shown in Fig. 1D, HAP1Δheme, like wild-type HAP1 in the presence of heme, is present predominantly in fractions 1 to 3. Figure 1E shows that the majority of HAP1-132 is in fractions 1 to 3, while a small portion is also found in fractions 17 to 21. These results provide another line of strong evidence supporting the conclusion that HAP1Δheme cannot form the HMC whereas HAP1-132 forms a low level of the HMC. Together, the data strongly suggest that the HMC plays a critical role in heme regulation and that the heme and HRM7 domains effect heme regulation by allowing the formation of the HMC.
The HAP1 dimerization domain is critical for heme regulation and the formation of intact HMC.
The HAP1 dimerization domain is adjacent to the heme domain, and it has been suggested that dimerization is a determining step in heme activation of HAP1 (44). Therefore, we speculated that the HAP1 dimerization domain may play regulatory roles in heme regulation in addition to allowing dimerization. Perhaps the HAP1 dimerization domain is also involved in the formation of the HMC and participates in interactions critical for heme regulation. To explore this possibility, we substituted the HAP1 dimerization domain with that of PPR1. Both HAP1 and PPR1 contain a C6 zinc cluster and a coiled-coil dimerization element in their DNA-binding sequences (28, 44, 49). Previously, we showed that replacing the HAP1 dimerization domain with that of PPR1 has no effect on HAP1 dimeric binding to its cognate DNA sites including UAS1/CYC1 and UAS/CYC7 (45). Therefore, we infer that this substitution should have no effect on HAP1 DNA binding in the presence of heme.
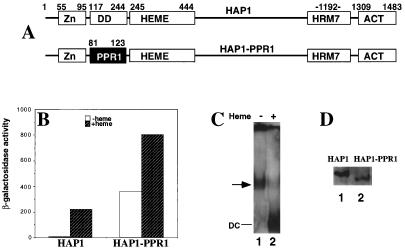
We made a HAP1-PPR1 hybrid protein with the HAP1 dimerization domain (residues 117 to 244) (44, 49) replaced by the PPR1 dimerization domain (PPR1 residues 81 to 123) (Fig. 5A) (28). Strikingly, as shown in Fig. 5B, this hybrid protein causes constitutivity in the absence of heme. Its activity is further induced by heme by only 2- to 3-fold, while HAP1 activity is induced more than 30-fold by heme. This result shows that the HAP1 dimerization domain is indeed critical for heme regulation. Further, we found that the chimeric protein, unlike HAP1, can no longer form the HMC (Fig. 5C, lane 1). Rather, in the absence of heme, it forms a complex intermediate in mobility between the HMC and the dimeric complex (compare lanes 1 and 2 in Fig. 4B with those in Fig. 5C). Although the level of the chimeric protein in extracts is less than half of that of wild-type HAP1 (Fig. 5D), we reason that the low protein level per se should not affect the formation of the HMC, because our previous experiments show that HAP1 forms the HMC well even when HAP1 is expressed at a low level from its own promoter (47). In other words, any change in the HMC formed by the chimeric protein is not attributable to the lower amount of the chimeric protein in the extracts. We suspect that this complex with an intermediate mobility is part of a disassembled HMC, since evidence presented above suggests that multiple proteins are associated with HAP1 in the absence of heme. Note that in the presence of heme, the hybrid protein forms a dimeric complex (Fig. 5C, lane 2) like that formed by HAP1 (Fig. 4B, lane 2).
FIG. 5.
The HAP1 dimerization domain is critical for heme regulation. (A) Domain structures of HAP1 and the HAP1-PPR1 chimeric protein. Shown are the C6 zinc cluster (Zn, residues 55 to 95), the dimerization domain (DD, residues 117 to 244), the heme domain (HEME, residues 245 to 444), the HRM7 domain (HRM7, encompassing the sequence surrounding residue 1192), and the activation domain (ACT, residues 1309 to 1483). The HAP1-PPR1 chimera contains all domains of HAP1 except that the HAP1 dimerization domain (residues 117 to 244) is replaced by that of PPR1 (residues 81 to 123). (B) β-Galactosidase activities of HAP1 and the HAP1-PPR1 hybrid protein in heme-sufficient (shaded bars) and heme-deficient (open bars) cells. 2μm plasmids expressing HAP1 and the HAP1-PPR1 hybrid protein from the GAL1-10 promoter were transformed into strain MHY200 bearing the UAS1/CYC1-lacZ reporter. (C) Complexes formed by the HAP1-PPR1 hybrid protein. The hybrid protein forms dimeric complexes (DC) (lane 2) in the presence of heme, while in the absence of heme it forms a complex (shown by an arrow in lane 1) that migrates much faster than the HMC (Fig. 4B) but slower than the dimeric complex. (D) Expression levels of the HAP1-PPR1 hybrid protein and wild-type HAP1 detected by Western blotting. All extracts used were prepared from JEL1 cells under previously established conditions (11, 47).
Data from sucrose gradient centrifugation further support the conclusion that HAP1-PPR1 forms a complex smaller than the HMC. As shown in Fig. 1F, approximately half of the HAP1-PPR1 protein is present in fractions 4 to 9, suggesting the presence of a complex that is smaller than the HMC (in fractions 17 to 21 [Fig. 1A and B]) but larger than the HAP1 complex formed in the presence of heme (in fractions 1 to 3 [Fig. 1C]). A significant portion of HAP1-PPR1 is present in fractions 1 to 3, very likely because this complex is labile and easily disrupted. These results again show a strong correlation between the disappearance of the HMC and the disappearance of heme regulation, indicating the importance of the HMC in heme regulation of HAP1. Taken together, the data show that the HAP1 dimerization domain is critical for heme regulation of HAP1 and for formation of the HMC.
The HAP1 dimerization domain is also critical for differential transcriptional activation.
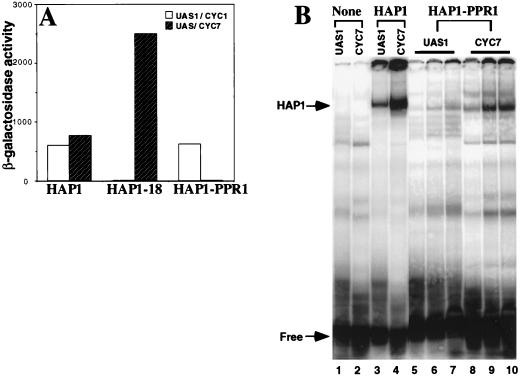
HAP1 activates transcription at two distinctive UASs, UAS/CYC7 and UAS1/CYC1 (34). Previous experiments have suggested that the HAP1 DNA-binding domain has an impact on transcriptional activation (20, 42). For example, a previously identified mutant, HAP1-18, with a mutation in the zinc cluster (Ser-63 to Arg) binds to UAS/CYC7 with the same affinity as HAP1 but activates transcription at a much higher level than HAP1 (Fig. 6A) (20). It was hypothesized that the HAP1 DNA-binding domain contacts a coactivator or a general transcription factor at UAS/CYC7 (20, 42).
FIG. 6.
The HAP1 dimerization domain plays an important role in transcriptional activation. (A) β-Galactosidase activities of HAP1, HAP1-18, and HAP1-PPR1 at UAS1/CYC1 (open bars) and UAS/CYC7 (shaded bars). HAP1-18 is shown as a control; it has a much higher activity at UAS/CYC7 than HAP1 but is inactive at UAS1/CYC1 (42). In contrast, the HAP1-PPR1 hybrid protein is active at UAS1/CYC1 but not at UAS/CYC7. (B) Binding of the HAP1-PPR1 hybrid protein to UAS1/CYC1 and UAS/CYC7. Approximately 300,000 cpm of radiolabeled UAS1/CYC1 (UAS1; lanes 1, 3, 5, 6, and 7) and UAS/CYC7 (CYC7; lanes 2, 4, 8, 9, and 10) was incubated with 20 μg of protein extracts prepared from cells expressing no HAP1 or HAP1-PPR1 (lanes 1 and 2), with 10 μg of extracts containing HAP1 (lanes 3 and 4) or the HAP1-PPR1 hybrid protein (lanes 5 and 8), with 20 μg of extracts containing the HAP1-PPR1 hybrid protein (lanes 6 and 9), or with 40 μg of extracts containing the HAP1-PPR1 hybrid protein (lanes 7 and 10). Shown by arrows are the positions of HAP1/HAP1-PPR1-DNA complexes (HAP1) and unbound DNA (Free). In the presence of heme, the HAP1-PPR1 hybrid protein binds to the UAS/CYC7 with the same affinity or higher affinity than to UAS/CYC1, although the hybrid protein is inactive at UAS/CYC7. Note that the HAP1-PPR1 fusion protein is very unstable in extracts, and significant degradation often occurs during DNA-binding reactions and electrophoresis.
The crystal structures of DNA complexes formed by C6 zinc cluster proteins including GAL4 and PPR1 show that the residues in the dimerization domains of these proteins are largely exposed (27, 28). Therefore, we speculated that the residues in the HAP1 dimerization domain might be exposed and could participate in protein-protein interactions that are important for transcriptional activation. To test this possibility, we determined the ability of the HAP1-PPR1 hybrid protein to activate transcription at UAS/CYC7 and UAS1/CYC1. We found that the HAP1-PPR1 chimera cannot activate transcription at UAS/CYC7 but retains a high level of activity at UAS1/CYC1 (Fig. 6A). Figure 6B further shows that the hybrid protein, like HAP1, binds to UAS/CYC7 with the same or even higher affinity than to UAS1/CYC1 (Fig. 6B; compare lanes 8 to 10 with lanes 5 to 7). The position of the HAP1- and HAP1-PPR1-DNA complexes is marked by an arrow in Fig. 6B. Note that the HAP1- and HAP1-PPR1-DNA complexes were not present when extracts containing no HAP1 or HAP1-PPR1 were used (Fig. 6B, lanes 1 and 2). The same result is obtained when the corresponding purified proteins containing only the DNA-binding sequences are used (45). Therefore, the differential transcriptional activity of the hybrid protein at UAS/CYC7 and UAS1/CYC1 is not due to the lack of HAP1 binding at UAS/CYC7. These results show that the HAP1 dimerization domain is indeed critical for transcriptional activation at UAS/CYC7 and may participate in interactions critical for transcriptional activation at the site.
DISCUSSION
Multiple domains cooperate to form the HMC and mediate heme regulation.
Here we show, by combining biochemical and genetic analyses, that heme regulation of HAP1 is mediated by a higher-order complex, termed HMC. Our biochemical data from DNA mobility shift assays, sucrose gradient centrifugation, and affinity and gel filtration chromatography convincingly show that the HMC is indeed a biochemically distinctive complex containing multiple cellular proteins including Hsp82 and Ydj1, not just an aggregate of HAP1 or an artifact of DNA mobility shift assays. Further biochemical and functional analyses of HAP1 mutants provide compelling evidence for the conclusion that the HMC is critical for heme regulation of HAP1. We have shown that the formation of the HMC is closely linked to HAP1 heme responsiveness; the amount of intact HMC formed by HAP1 and mutants is proportional to the degree of HAP1 heme responsiveness (Fig. 1 and 4). Our data provide a direct functional link between the HMC and heme regulation of HAP1 activity.
Further, our data show that three distinct domains—the heme domain, the HRM7 domain, and the dimerization domain—are all required for the formation of an intact HMC and for heme regulation. Mutation or deletion of any one of these domains abrogates the formation of the HMC and abolishes heme regulation of HAP1 activity (Fig. 1, 4, and 5). The effect of the heme domain on the HMC and HAP1 heme responsiveness is the most drastic (Fig. 4), suggesting that it plays a major role in heme regulation and perhaps provides a strong interaction between HAP1 and other components of the HMC.
How is the precise regulation of HAP1 activity by heme accomplished? Our data provide significant insights into the mechanism by which HAP1 is repressed in the absence of heme and subsequently activated by heme. The formation of the HMC involving multiple domains very likely plays a key role in regulating HAP1 activity. Three domains—the dimerization domain, the heme domain, and the HRM7 domain—are all required for heme regulation. A high magnitude of heme regulation is observed only if all three domains are intact and a high level of HMC is formed (Fig. 1, 4, and 5). Neither the heme domain nor the HRM7 domain alone confers significant activity increases in response to heme (48). We postulate that a dynamic equilibrium exists between the transcriptionally inactive HMC and the active dimeric HAP1 complex. At low heme concentrations, the majority of HAP1 is in the form of the HMC and is transcriptionally inactive, whereas at high heme concentrations, the majority is in the form of dimeric complexes and is transcriptionally active. When the heme concentration changes, a redistribution of the HMC and the dimeric complex ensues, causing a change in HAP1 activity. All three heme regulatory domains—the dimerization domain, the heme domain, and the HRM7 domain—may help bind or bind directly to heme, sense heme concentrations, and mediate HAP1 conformational changes. Therefore, the formation of the HMC and the equilibrium between the HMC and the dimeric complex can be highly sensitive to changes of heme concentration. Consequently, cooperation of multiple domains and formation of a dynamic complex highly sensitive to heme allow the precise and tight regulation of HAP1 activity.
This model explains why overexpression of wild-type HAP1 in vivo in the absence of heme causes only a low level of HAP1 activation (Fig. 4 and reference 47), even though considerable amounts of dimeric complexes are observed. Because in vivo, in the absence of heme, the equilibrium is predominantly toward the formation of the HMC, any transiently formed dimeric complexes would be rapidly rebound by other cellular proteins and thus inactivated (considerable amounts of dimeric complexes were observed by in vitro assays because dimeric complexes cannot reassociate with other cellular proteins once separated by force). The mutants, HAP1Δheme, HAP1-132, and HAP1-PPR1, however, have little or no potential to form the HMC even in the absence of heme (Fig. 1, 4, and 5). These mutants are therefore poorly repressed and gain high levels of activity in the absence of heme. The wild-type HAP1 appears to be partially repressed in vivo even at high heme concentrations (Fig. 4A; the activity of wild-type HAP1 at the high heme concentration is still lower than that of HAP1Δheme), very likely because wild-type HAP1 still possesses the potential to form the HMC even at high levels of heme. Our proposed model is consistent with all the existing data and should provide valuable guidance in future studies, even though many questions about heme regulation of HAP1 remain to be answered. Interestingly, although the HMC allows HAP1 to be repressed in the absence of heme, our data also indicate that it might play a positive role in transcriptional activation by HAP1, as discussed below.
Roles of Hsp82 in heme regulation of HAP1.
Our data suggest that Hsp82 is a component of the HMC that allows HAP1 to be repressed in the absence of heme (Fig. 3A), and we have further shown that a reduced level of Hsp82 causes a drastic and selective decrease in HAP1 transcriptional activity (Fig. 3B). These results might seem to be contradictory to each other. However, this effect of Hsp82 is analogous to the effect of Hsp90 on steroid hormone receptors. Hsp90 is bound to and reversibly inactivates steroid hormone receptors in the absence of hormone, and it maintains them in an activatable conformation (2, 35). Hsp90 is essential for ligand binding and response; the activity of steroid hormone receptors under the reduced level of Hsp90 is greatly diminished (2, 35). We suspect that Hsp82 might affect HAP1 in a similar manner. As a component of the HMC, Hsp82 should contribute to HAP1 repression in the absence of heme, but it might also be required for HAP1 activation by heme. Thus, a reduced level of Hsp82 diminishes HAP1 activity in heme-sufficient cells. HAP1 constitutive mutants also behave in a manner similar to constitutive steroid hormone receptors. Constitutive steroid hormone receptors do not bind to and do not require Hsp82 for activity (35). Likewise, HAP1 constitutive mutants (Fig. 4 and 5) do not form intact HMC and thus very likely do not require Hsp82 for transcriptional activity. The analogy between the HAP1-heme signaling pathway and the steroid hormone signaling pathway has long been suspected (33, 47, 48). The data in Fig. 3 provide the preliminary biochemical and genetic evidence supporting the analogy of these pathways. However, whether Hsp82 affects HAP1 through the same mechanism as that by which Hsp90 affects steroid hormone receptors is not yet clear. Further investigations must be carried out in order to understand exactly how Hsp82 affects HAP1 repression in the absence of heme and subsequent activation by heme.
The involvement of heat shock proteins in heme regulation of HAP1 may explain why the steroid hormone receptor regulatory system is so highly conserved in mammals and yeast (2, 35), although steroid hormone receptors do not exist naturally in yeast. The heat shock regulatory system in yeast may serve to control HAP1 activity, and steroid hormone receptors may be analogs of HAP1 in yeast. Therefore, this system can serve to regulate steroid hormone receptors when expressed artificially in yeast. This regulatory system may also act to control the activity of other yeast transcriptional activators, such as LEU3 (41) and PPR1 (26).
A versatile dimerization domain.
Dimerization is critical for the functions of a wide array of regulatory proteins, particularly transcription factors. The dimerization domains of numerous transcription factors promote the formation of homo- or heterodimers that are able to bind to DNA with high affinity. Likewise, the HAP1 dimerization domain is required for HAP1 to bind to DNA with high affinity (44). Remarkably, we uncovered a novel function for the HAP1 dimerization domain: it is not only required for HAP1 dimerization and high-affinity DNA binding but also essential for heme regulation and transcriptional activation at certain DNA sites.
How does the HAP1 dimerization domain affect heme regulation and transcriptional activation? Based on the crystal structures of the related PPR1-DNA and GAL4-DNA complexes (27, 28), we suspect that the residues in the HAP1 dimerization domain are exposed and could readily participate in protein-protein interactions that are important for heme regulation and/or transcriptional activation. As shown in Fig. 1F and 5C, the HAP1-PPR1 hybrid protein cannot form an intact HMC. This suggests that the HAP1 dimerization domain may participate in interactions critical for the formation of the HMC, thereby affecting heme regulation. Similarly, the HAP1 dimerization domain may affect transcriptional activation at UAS/CYC7 by contacting a coactivator required for activation at this site. Perhaps even HAP1-18 and other HAP1 activation mutants (42) with mutations located in the DNA-binding sequence affect transcriptional activation through the dimerization domain. Further, the effects of positive control mutants in the DNA-binding domains of other transcriptional activators might also be attributable to the role of dimerization domains on transcriptional activation (1, 14, 30).
Our studies provide an example of how heme controls the activity of a transcriptional activator, how multiple domains act together to confer precise and tight regulation by a signaling molecule, and how a regulatory system may be conserved from mammals to yeast to control the activity of transcriptional activators responsive to signaling molecules. Regulation through the formation of a higher-order complex involving multiple domains may be another general mechanism by which the activity of numerous regulatory proteins can be precisely regulated by small signaling molecules such as heme or oxygen.
ACKNOWLEDGMENTS
We thank W. Jelinek, T. Hon, and A. Murashov for critical reading of the manuscript, S. Lindquist, A. J. Caplan, and R. J. Trumbly for providing antiserum and yeast strains, and M. Haldi, B. Turcotte, and B. Pina for providing strains and plasmids. We also thank D. Tamalis for assistance in preparation of extracts and protein analysis. We are grateful to L. Guarente for supporting the generation of antiserum against GST-HAP1 and stimulating discussions in the course of this study.
This work was supported by grants from NSF (MCB-96174720) and NIH (GM53453) to L.Z.
REFERENCES
- 1.Bengal E, Flores O, Rangarajan P N, Chen A, Weintraub H, Verma I M. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc Natl Acad Sci USA. 1994;91:6221–6225. doi: 10.1073/pnas.91.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohen S P, Kralli A, Yamamoto K R. Hold ’em and fold ’em: chaperones and signal transduction. Science. 1995;268:1303–1304. doi: 10.1126/science.7761850. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 3.Caplan A J, Langley E, Wilson E M, Vidal J. Hormone-dependent transactivation by the human androgen receptor is regulated by a dnaJ protein. J Biol Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- 4.Caplan A J, Tsai J, Casey P J, Douglas M G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 5.Chang H C, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charnay P, Maniatis T. Transcriptional regulation of globin gene expression in the human erythroid cell line K562. Science. 1983;220:1281–1283. doi: 10.1126/science.6574602. [DOI] [PubMed] [Google Scholar]
- 7.Chen J J, Throop M S, Gehrke L, Kuo I, Pal J K, Brodsky M, London I M. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc Natl Acad Sci USA. 1991;88:7729–7733. doi: 10.1073/pnas.88.17.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creusot F, Verdiere J, Gaisne M, Slonimski P P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988;204:263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- 9.Dean A, Ley T J, Humphries R K, Fordis M, Schechter A N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci USA. 1983;80:5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 11.Fytlovich S, Gervais M, Agrimonti C, Guiard B. Evidence for an interaction between the CYP1 (HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 1993;12:1209–1218. doi: 10.1002/j.1460-2075.1993.tb05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner K H, Anderson S F, Coleman J E. Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain. Nat Struct Biol. 1995;2:898–905. doi: 10.1038/nsb1095-898. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg M A, Dunning S P, Bunn H F. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 14.Gosink K K, Gaal T, Bokal A J T, Gourse R L. A positive control mutant of the transcription activator protein FIS. J Bacteriol. 1996;178:5182–5187. doi: 10.1128/jb.178.17.5182-5187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 16.Haldi M L, Guarente L. Multiple domains mediate heme control of the yeast activator HAP1. Mol Gen Genet. 1995;248:229–235. doi: 10.1007/BF02190805. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 18.Hinnebusch A G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hortner H, Ammerer G, Hartter E, Hamilton B, Rytka J, Bilinski T, Ruis H. Regulation of synthesis of catalases and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur J Biochem. 1982;128:179–184. doi: 10.1111/j.1432-1033.1982.tb06949.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim K S, Guarente L. Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI. Nature. 1989;342:200–203. doi: 10.1038/342200a0. [DOI] [PubMed] [Google Scholar]
- 21.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 22.Kuwabara K, Ogawa S, Matsumoto M, Koga S, Clauss M, Pinsky D J, Lyn P, Leavy J, Witte L, Joseph-Silverstein J, et al. Hypoxia-mediated induction of acidic/basic fibroblast growth factor and platelet-derived growth factor in mononuclear phagocytes stimulates growth of hypoxic endothelial cells. Proc Natl Acad Sci USA. 1995;92:4606–4610. doi: 10.1073/pnas.92.10.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbe-Rois R, Labbe P. Tetrapyrrole and heme biosynthesis in the yeast Saccharomyces cerevisiae. In: Dailey H A, editor. Biosynthesis of heme and chlorophylls. New York, N.Y: Green Publishing Associates and Wiley-Interscience; 1990. pp. 235–285. [Google Scholar]
- 24.Lathrop J T, Timko M P. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 25.Levy A P, Levy N S, Loscalzo J, Calderone A, Takahashi N, Yeo K T, Koren G, Colucci W S, Goldberg M A. Regulation of vascular endothelial growth factor in cardiac myocytes. Circ Res. 1995;76:758–766. doi: 10.1161/01.res.76.5.758. [DOI] [PubMed] [Google Scholar]
- 26.Marczak J E, Brandriss M C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991;11:2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 28.Marmorstein R, Harrison S C. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994;8:2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- 29.Mattoon J, Lancashire W, Sanders H, Carvajal E, Malamud D, Braz G, Panek A. Oxygen and catabolite regulation of hemoprotein biosynthesis in the yeast Saccharomyces cerevisiae. In: Caughey W J, editor. Biosynthesis of heme and cholorophylls. New York, N.Y: Academic Press; 1979. pp. 421–435. [Google Scholar]
- 30.Molkentin J D, Black B L, Martin J F, Olson E N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padmanaban G, Venkateswar V, Rangarajan P N. Haem as a multifunctional regulator. Trends Biochem Sci. 1989;14:492–496. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- 32.Palma J F, Gao X, Lin C H, Wu S, Solomon W B. Iron protoporphyrin IX (hemin) but not tin or zinc protoporphyrin IX can stimulate gene expression in K562 cells from enhancer elements containing binding sites for NF-E2. Blood. 1994;84:1288–1297. [PubMed] [Google Scholar]
- 33.Pfeifer K, Kim K S, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer K, Prezant T, Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987;49:19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- 35.Picard D, Khursheed B, Garabedian M J, Fortin M G, Lindquist S, Yamamoto K R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 36.Rangarajan P N, Padmanaban G. Regulation of cytochrome P-450b/e gene expression by a heme- and phenobarbitone-modulated transcription factor. Proc Natl Acad Sci USA. 1989;86:3963–3967. doi: 10.1073/pnas.86.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy S V, Alcantara O, Roodman G D, Boldt D H. Inhibition of tartrate-resistant acid phosphatase gene expression by hemin and protoporphyrin IX. Identification of a hemin-responsive inhibitor of transcription. Blood. 1996;88:2288–2297. [PubMed] [Google Scholar]
- 38.Reece R J, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 39.Siddiqui A H, Brandriss M C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989;9:4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith D F, Faber L E, Toft D O. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J Biol Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 41.Sze J Y, Remboutsika E, Kohlhaw G B. Transcriptional regulator Leu3 of Saccharomyces cerevisiae: separation of activator and repressor functions. Mol Cell Biol. 1993;13:5702–5709. doi: 10.1128/mcb.13.9.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turcotte B, Guarente L. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 1992;6:2001–2009. doi: 10.1101/gad.6.10.2001. [DOI] [PubMed] [Google Scholar]
- 43.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Bermingham M O, Turcotte B, Guarente L. Antibody-promoted dimerization bypasses the regulation of DNA binding by the heme domain of the yeast transcriptional activator HAP1. Proc Natl Acad Sci USA. 1993;90:2851–2855. doi: 10.1073/pnas.90.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Guarente L. The C6 zinc cluster dictates asymmetric binding by HAP1. EMBO J. 1996;15:4676–4681. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Guarente L. Evidence that TUP1/SSN6 has a positive effect on the activity of the yeast activator HAP1. Genetics. 1994;136:813–817. doi: 10.1093/genetics/136.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Guarente L. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J Biol Chem. 1994;269:14643–14647. [PubMed] [Google Scholar]
- 48.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Guarente L. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 1994;8:2110–2119. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]
- 50.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]