Abstract
Background
The study aims to explore MRI phenotypes that predict glioblastoma’s (GBM) methylation status of the promoter region of MGMT gene (pMGMT) by qualitatively assessing contrast-enhanced T1-weighted intensity images.
Methods
A total of 193 histologically and molecularly confirmed GBMs at the Kansai Network for Molecular Diagnosis of Central Nervous Tumors (KANSAI) were used as an exploratory cohort. From the Cancer Imaging Archive/Cancer Genome Atlas (TCGA) 93 patients were used as validation cohorts. “Thickened structure” was defined as the solid tumor component presenting circumferential extension or occupying >50% of the tumor volume. “Methylated contrast phenotype” was defined as indistinct enhancing circumferential border, heterogenous enhancement, or nodular enhancement. Inter-rater agreement was assessed, followed by an investigation of the relationship between radiological findings and pMGMT methylation status.
Results
Fleiss’s Kappa coefficient for “Thickened structure” was 0.68 for the exploratory and 0.55 for the validation cohort, and for “Methylated contrast phenotype,” 0.30 and 0.39, respectively. The imaging feature, the presence of “Thickened structure” and absence of “Methylated contrast phenotype,” was significantly predictive of pMGMT unmethylation both for the exploratory (p = .015, odds ratio = 2.44) and for the validation cohort (p = .006, odds ratio = 7.83). The sensitivities and specificities of the imaging feature, the presence of “Thickened structure,” and the absence of “Methylated contrast phenotype” for predicting pMGMT unmethylation were 0.29 and 0.86 for the exploratory and 0.25 and 0.96 for the validation cohort.
Conclusions
The present study showed that qualitative assessment of contrast-enhanced T1-weighted intensity images helps predict GBM’s pMGMT methylation status.
Keywords: glioblastoma, magnetic resonance image, MGMT, MGMT promoter methylation, radiogenomics
Key Points.
MRI features, the “Thickened structure” and the “Methylated contrast phenotype,” help predict GBM’s pMGMT methylation.
The presence of “Thickened structure,” and absence of “Methylated contrast phenotype” has a specificity of 0.86–0.96 favoring pMGMT unmethylation.
Importance of the Study.
Noninvasive prediction of glioblastoma’s (GBM) pMGMT methylation status is still a challenging research topic despite recent technological advancements in image analysis. Thus, this study explored a clinically feasible imaging biomarker that represents GBM’s pMGMT methylation status with external validation. Two qualitative imaging features, namely the “Thickened structure” and the “Methylated contrast phenotype,” were identified as valuable to this means. GBMs presenting the imaging feature, the presence of “Thickened structure” and absence of “Methylated contrast phenotype” exhibited a significantly high specificity, favoring pMGMT unmethylation in the exploratory and validation cohorts with a sensitivity and specificity of approximately 0.3 and 0.9. The easy clinical application of the proposed imaging features is expected to facilitate better preoperative GBM characterization.
Glioblastoma (GBM) is one of the most common malignant brain tumors with an abysmal prognosis despite multimodal treatments consisting of maximal safe resection followed by radiation and chemotherapy with temozolomide.1 Methylation of the gene’s promoter region encoding the O-6-methylguanine-DNA methyltransferase (MGMT) is a predictive and prognostic factor in GBM patients.2–4 Although the benefit of presurgical identification of MGMT promoter methylation (pMGMT-met) of GBMs is debatable, it may help clinicians and patients choose the most appropriate treatment strategy. For instance, aggressive surgical intervention could be proposed more strongly for MGMT promotor unmethylated (pMGMT-unmet) than for pMGMT-met GBM patients due to the expected minimal benefit from temozolomide.5 In contrast, an optimal balance between maximal resection and preservation of the patient’s quality of daily life could be considered for pMGMT-met GBM patients.
Great efforts have been made to meet this research community’s demand to develop methods for noninvasive prediction of the pMGMT methylation status in GBM by magnetic resonance image (MRI). Experimented techniques include qualitative image assessments,6,7 texture features,8–10 and deep learning architectures.11–14 Despite these efforts, the prediction of GBM’s pMGMT methylation status is still troubled by inconsistent research results,7,15 insufficient diagnostic performance, with sensitivity and specificity widely ranging from 55.6% to 93%, and 39.0% to 76.0%, respectively,8,16 and possible overfitting of the deep learning algorithm.12,13 Furthermore, texture feature analyses and deep learning architectures are still far from being incorporated into routine clinics, as they require sophisticated procedures, such as segmentation, manual intervention, in-house analytic pipeline, and lengthy processing time.17
We previously reported that the diagnostic performance of pMGMT methylation status was low, with a sensitivity of 67% and a specificity of 66% by structural MRI-based radiomics.9 However, tumors with irregular shapes pose a challenge in acquiring consistent radiomic data. Thus, qualitative evaluation of radiological images may still be a valuable approach to predict the molecular status of GBM preoperatively. Previous research harnessing deep learning algorithms reported that pMGMT-unmet gliomas tended to demonstrate thick enhancement with central necrosis. In contrast, heterogenous or nodular enhancement were features characteristic of pMGMT-met gliomas.13 These imaging features identified by the deep learning algorithm could potentially be applied to predict GBM’s pMGMT methylation status by qualitative evaluation of MRI. The current study attempted to discover qualitative MRI characteristics corresponding to GBM’s pMGMT methylation status and to test the hypothesis that conventional qualitative evaluation of MRI remains valid for predicting brain tumors’ molecular characteristics.
Methods
Patient Cohort
This study was performed per the principles of the Helsinki Declaration, and it was approved by the internal ethical review boards (Approval number 21040) and all collaborative institutes from the Kansai Molecular Diagnosis Network (KANSAI) for Central Nervous System Tumors, the list of which can be found in the acknowledgment section. Written informed consent was obtained from patients or their families for the prospectively recruited cohort. We also used the Cancer Imaging Archive (TCIA)/ Cancer Genome Atlas (TCGA) data set accessed on September 16, 2022,18–20 as an external validation cohort.
The Inclusion criteria for the present study were as follows: newly diagnosed GBM according to WHO Classification of Tumours Fifth Edition (WHO2021),21 available for both tumor’s pMGMT methylation status and preoperative gadolinium-enhanced T1-weighted images (T1WI-Gd). Cases without pMGMT methylation or IDH mutation status information, lack of postoperative images, or insufficient or atypical images were excluded from this study. There were 193 GBM, IDH-wildtype from 12 KANSAI institutions, with 97 being pMGMT-met and 96 being pMGMT-unmet GBM. The TCIA/ TCGA validation cohort comprised 93 GBM, IDH-wildtype with 49 pMGMT-met and 44 pMGMT-unmet GBM patients.
A supplementary cohort was established following the WHO Classification of Tumours, Revised Fourth Edition (WHO2016), specifically including IDH-mutant tumors.22 This cohort aims to cater to those interested in this subgroup. Detailed information is shown in Supplementary Tables 1 and 2. The KANSAI cohort consisted of 202 subjects, comprising 103 pMGMT-methylated and 99 pMGMT-unmethylated GBM cases. The TCIA/TCGA validation cohort consisted of 104 subjects, with 59 pMGMT-methylated and 45 pMGMT-unmethylated GBM.
Genetic Analysis
Frozen or fresh tumor samples were obtained during surgery, and tumor genomic DNA was extracted from those tissues for genetic analysis. All Genetic analyses were performed at the Osaka National Hospital according to previously described procedures. Briefly, the methylation status of pMGMT was analyzed by quantitative methylation-specific PCR after bisulfite modification of genomic DNA, and a threshold of ≥1% was used for pMGMT methylation. The presence of hotspot mutations in IDH1 (R132) and IDH2 (R172) genes was analyzed by Sanger sequencing. A senior board-certified neuropathologist performed a central pathology review. Patient characteristics are described in Supplementary Tables 1 and 2. We obtained genetic information regarding IDH genes and pMGMT for the TCIA/ TCGA data set from the report by Cameron et al.23
The Definition and Classification of Image Findings
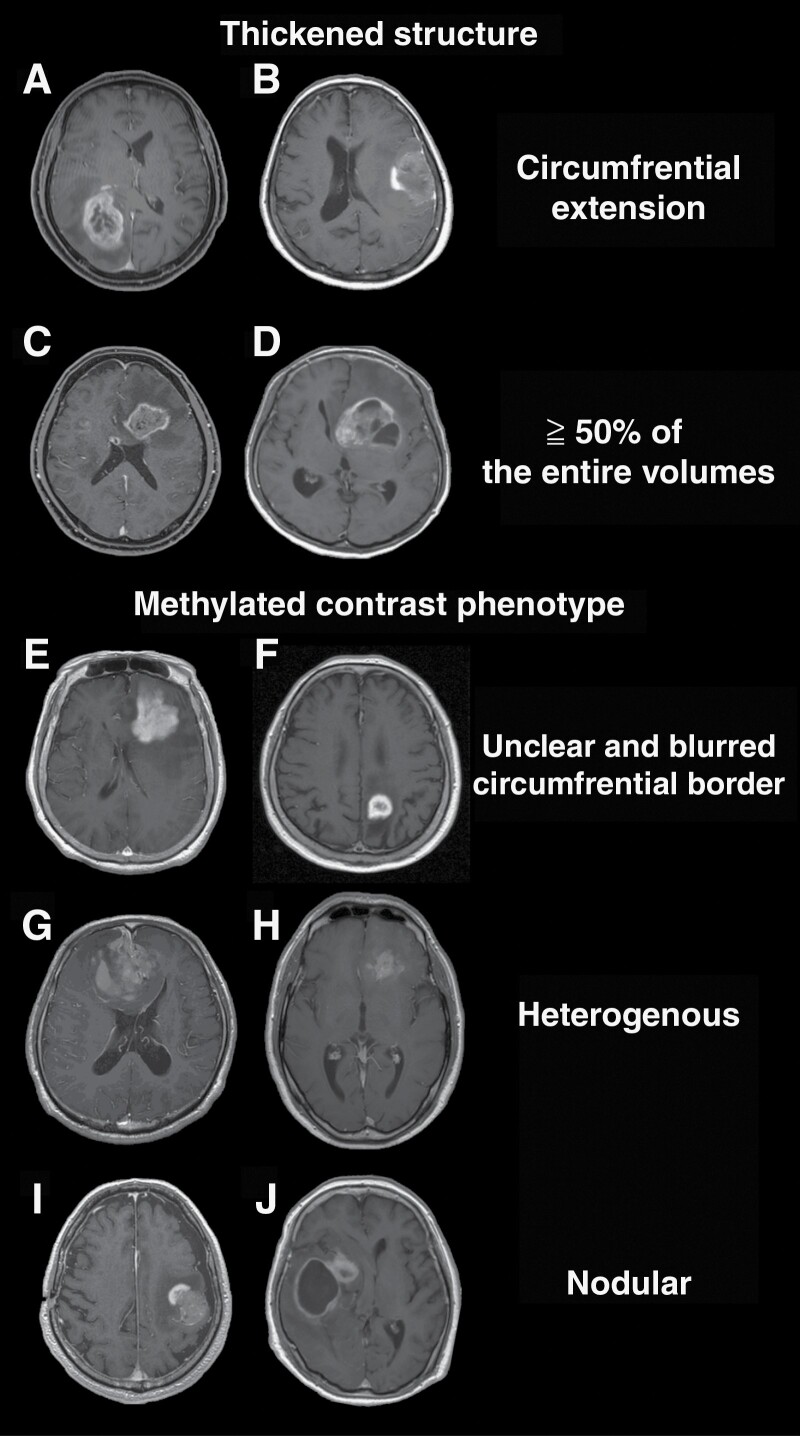
MRIs were independently evaluated by 3 board-certified neurosurgeons with 7, 8, and 12 years of experience blinded to tumors’ genetic information. The readers assessed the presence or absence of the following 2 T1WI-Gd characteristics; “Thickened structure” and “Methylated contrast phenotype” (Figure 1). These features were initially discovered as imaging characteristics useful for predicting glioma’s pMGMT methylation status by a deep learning algorithm in a previous study.13 The current study included the following definitions to clarify the description of imaging characteristics further. The presence of a “Thickened structure” was defined when the contrast-enhancing compartment of the tumor had either a circumferential extension (Figure 1A and B) or occupied more than 50% of the entire volume of the tumor (Figure 1C and D). A thin contrast-enhancing rim was defined as an absence of a “Thickened structure” (Figure 1A and B). The presence of a “Methylated contrast phenotype” was determined when the tumor showed either one of the following 3 features: 1. the entire enhancing circumferential border is unclear and blurred in relation to the surrounding structure (Figure 1E and F), 2. the enhancement is heterogenous (Figure 1G and H), 3. a nodular contrast enhancement is present (Figure 1I and J). Microvasculature-like minor sequential enhancements were not defined as “Methylated contrast phenotype” present (Figure 2C). The inter-rater consistencies among the 3 evaluators were evaluated using Fleiss’s Kappa coefficient. The final image findings were then determined by majority voting, and the cases were classified into 4 image types (Figure 2).
Figure 1.
The “Thickened structure” was defined based on the presence of any of the 2 criteria: a solid circumferential component with central necrosis (A: KANSAI, Image 1D 35, pMGMT-met, B: KANSAI, Image 1D 15, pMGMT-met) or a solid component occupying more than 50% of the entire volume of the tumor (C: KANSAI, Image 1D 233, pMGMT-unmet, D: KANSAI, Image 1D 17, pMGMT-unmet). Methylated contrast phenotype” was defined by imaging findings with 1 of the 3 definitions: the entire enhancing circumferential border is unclear and blurred in relation to the surrounding brain (E: KANSAI, Image 1D 18, pMGMT-met, F: KANSAI, Image 1D 111, pMGMT-met), the enhancement is heterogenous (G: KANSAI, Image 1D 139, MGMT-met, H: KANSAI, Image 1D 218, pMGMT-met), or the enhancement is nodular (I: KANSAI, Image 1D 35, pMGMT-met, J: KANSAI, Image 1D 15, pMGMT-met).
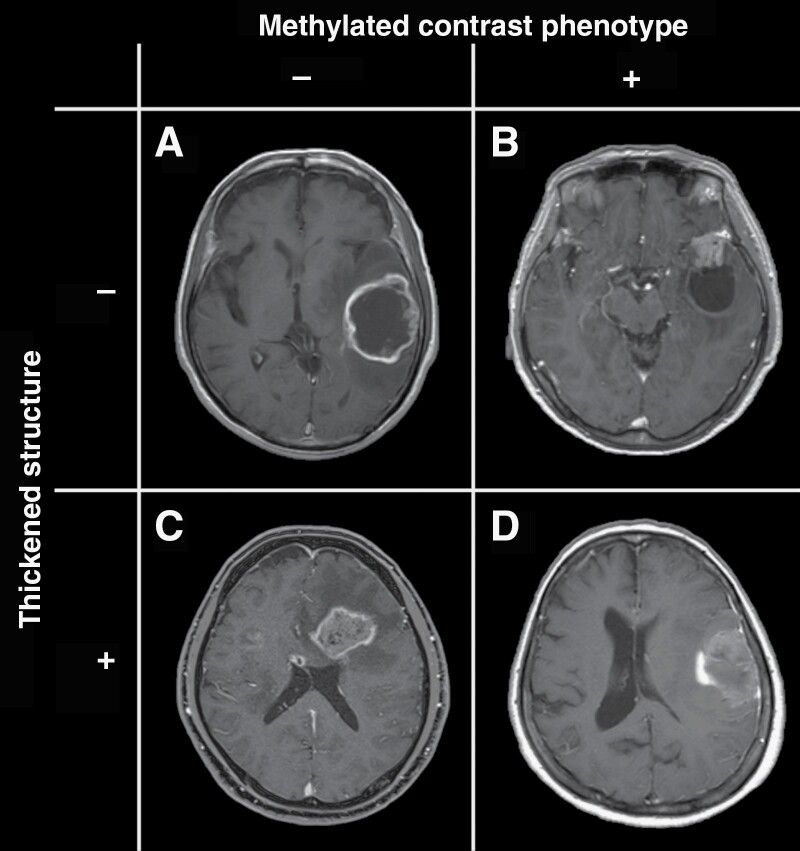
Figure 2.
All cases were classified into 4 types: absence of both “Methylated contrast phenotype” and “Thickened structure” (A: KANSAI, Image 1D 210, pMGMT-unmet); presence of “Methylated contrast phenotype” and absence of “Thickened structure” (B: KANSAI, Image 1D 54, pMGMT-met); absence of “Methylated contrast phenotype” and presence of “Thickened structure” (C: KANSAI, Image 1D 233, pMGMT-unmet). Presence of both “Methylated contrast phenotype” and “Thickened structure” (D: KANSAI, Image 1D 35, pMGMT-met).
Fleiss’s Kappa Coefficient and Statistical Analysis
Fleiss’s kappa coefficient was calculated using the “irr” package version 0.84.1 for R with default parameters (https://cran.r-project.org/web/packages/irr/irr.pdf). Fleiss’s kappa coefficient of 0.00 to 0.20 was considered as slight agreement, 0.21 to 0.40 as fair agreement, 0.41 to 0.60 as moderate agreement, and larger than 0.6 as substantial agreement.24 Statistical analysis was performed using Prism 9 for macOS (GraphPad Software, San Diego, CA, USA). The relationship between image characteristics and the pMGMT methylation status was investigated by the Fisher exact test or multiple logistic regression analysis. A p value of less than .05 was considered significant.
Results
Inter-rater Reliability of the “Thickened Structure” and the “Methylated Contrast Phenotype”
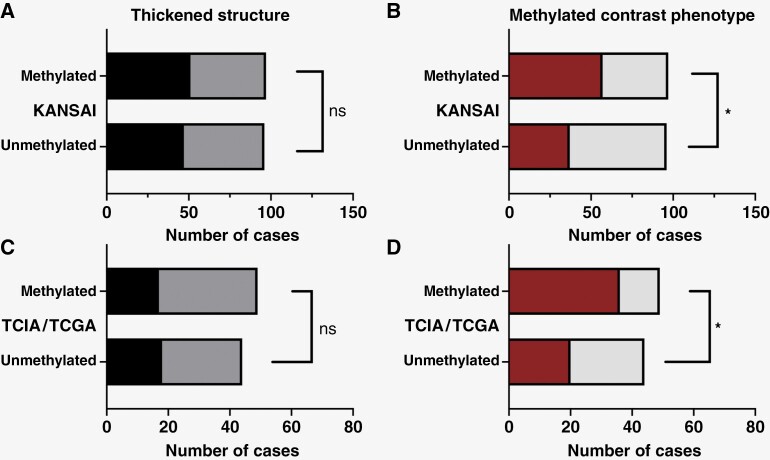
Inter-rater reliability of 3 evaluators assessing the “Thickened structure” was 0.68 for the KANSAI exploratory and 0.55 for the TCIA/ TCGA validation cohorts, suggesting substantial, and moderate agreements by Fleiss’s kappa coefficient, respectively. Regarding “Methylated contrast phenotype,” Fleiss’s kappa coefficients were 0.30 for the KANSAI and 0.39 for the TCIA/ TCGA cohorts, both of which implied fair agreement. The information regarding the agreement or disagreement between each reader can be referred to in Supplementary Tables 1 and 2.
Correlation of Single Qualitative Image Phenotypes and pMGMT Methylation Both for the Exploratory and Validation Cohorts
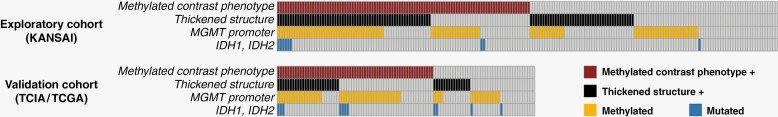
Figure 3 shows the presence or absence of qualitative image phenotypes and molecular status of the 2 cohorts. A multiple logistic analysis for the exploratory cohort (KANSAI) following the WHO2021 criteria revealed that pMGMT methylation was predicted by the following equation:
Figure 3.
Overall study cohort. The study was conducted in 2 stages, an exploratory cohort study followed by a validation cohort study, to investigate the relationship between the imaging characteristics and the pMGMT-methylation status of histologically confirmed GBM. KANSAI, Kansai Molecular Diagnosis Network; TCIA/TCGA, Cancer Imaging Archive/ Cancer Genome Atlas.
MGMT (unmethy:0, methy:1) approximately −0.39 + 0.01 x “Thickened structure (absent:0, present:1)” + 0.82 x “Methylation contrast phenotype (absent:0, present:1)”
Although the “Thickened structure” did not significantly contribute to the model construction (p = .98, Figure 4A), “Methylation contrast phenotype” was considered significant (p = .006, Figure 4B). This trend was consistently observed in the validation cohort as well (TCIA/ TCGA; Figure 4C and D).
Figure 4.
Exploration of the relationship between the imaging characteristics and the pMGMT-methylation status of GBMs. “Thickened structure” was not significantly associated with pMGMT-met GBMs (KANSAI cohort (A), p = 0.98 and TCIA/ TCGA cohort (C), p = 0.44). “Methylated contrast phenotype” was significantly associated with pMGMT-met of GBMs (KANSAI cohort (B), p = 0.006 and TCIA/ TCGA cohort (D), p = 0.006)
Correlation of the Combined Qualitative Image Phenotypes and pMGMT Methylation Both for the Exploratory and Validation Cohorts
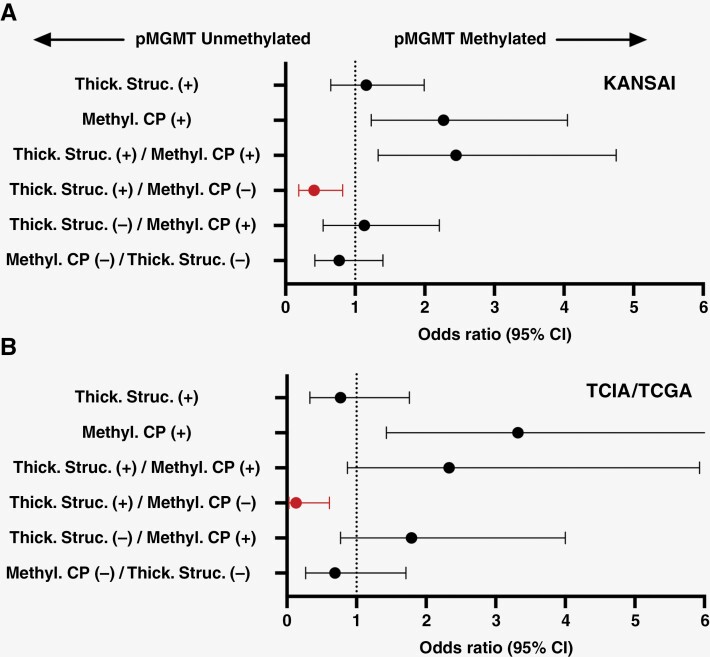
Fisher exact test revealed that the presence of both the “Thickened structure” and “Methylated contrast phenotype” was significantly predictive of pMGMT-met GBM for the KANSAI exploratory (Figure 5A and Supplementary Figure 1A; p = .007, odds ratio = 2.50, 95% confidence interval [CI] = 1.33–4.76). However, there was no significant difference in TCIA/ TCGA validation cohorts (Figure 5B and Supplementary Figure 2A; p = .14, odds ratio = 2.32, 95% CI = 0.87–5.93). The sensitivities and specificities of the “Thickened structure” and “Methylated contrast phenotype” double positive for correctly predicting pMGMT-met GBM were 0.66 and 0.80 for the KANSAI exploratory and 0.31 and 0.84 for the TCIA/ TCGA validation cohort (Table 1).
Figure 5.
The odds ratios (OR) of the imaging characteristics for predicting GBM’s pMGMT methylation status are presented. The OR of the “Methylated contrast phenotype” (Methyl. CP) was significantly higher than 1.0 in both the KANSAI exploratory and TCIA/ TCGA validation cohorts. The OR of the presence of “Thickened structure” (Thick. Struc.) and the absence of “Methylated contrast phenotype” was significantly lower than 1.0 both in the KANSAI exploratory and TCIA/ TCGA validation cohort.
Table 1.
Odds Ratio and Diagnostic Performance of the “Thickened Structure” and “Methylated Contrast Phenotype” for pMGMT Methylation Status in GBM. † Indicates p < 0.05.
| Positive number | p Value | pMGMT-Met GBM prediction | pMGMT-Unmet GBM prediction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Odds ratio | 95% CI | Sensitivity | Specificity | Odds ratio | 95% CI | Sensitivity | Specificity | ||||
| KANSAI cohort (exploratory) Total 193 subjects |
Thick. Struc. (+) | 98 | 0.67 | 1.16 | 0.65–1.99 | 52.0% | 51.0% | 0.87 | 0.50–1.55 | 49.0% | 47.4% |
| Methyl. CP (+) | 94 | 0.01† | 2.27 | 1.23–4.05 | 60.6% | 61.5% | 0.44 | 0.25–0.80 | 38.5% | 41.2% | |
| Thick. Struc.(+)/ Methyl. CP (+) | 56 | 0.01† | 2.45 | 1.33–4.75 | 66.1% | 80.2% | 0.40 | 0.21–0.75 | 19.8% | 61.9% | |
| Thick. Struc.(+)/ Methyl. CP (-) | 42 | 0.01† | 0.41 | 0.19–0.82 | 33.3% | 70.8% | 2.44 | 1.21–5.15 | 29.2% | 85.6% | |
| Thick. Struc.(-)/ Methyl. CP (+) | 38 | 0.86 | 1.13 | 0.54–2.21 | 52.6% | 81.3% | 0.89 | 0.45–1.85 | 18.8% | 79.4% | |
| Thick. Struc.(-)/ Methyl. CP (-) | 57 | 0.43 | 0.77 | 0.42–1.40 | 45.6% | 67.7% | 1.30 | 0.72–2.41 | 32.3% | 73.2% | |
| TCIA/ TCGA cohort (validation) Total 93 subjects |
Thick. Struc. (+) | 35 | 0.67 | 0.77 | 0.33–1.76 | 34.7% | 59.1% | 1.30 | 0.57–3.03 | 40.9% | 65.3% |
| Methyl. CP (+) | 56 | 0.01† | 3.32 | 1.43–7.63 | 73.5% | 54.6% | 0.30 | 0.13–0.70 | 45.5% | 26.5% | |
| Thick. Struc.(+)/ Methyl. CP (+) | 22 | 0.14 | 2.33 | 0.87 – 5.93 | 30.6% | 84.1% | 0.43 | 0.17–1.15 | 15.9% | 69.4% | |
| Thick. Struc.(+)/ Methyl. CP (-) | 13 | 0.01† | 0.13 | 0.03–0.61 | 4.1% | 75.0% | 7.83 | 1.63–36.56 | 25.0% | 95.9% | |
| Thick. Struc.(-)/ Methyl. CP (+) | 34 | 0.20 | 1.79 | 0.77–4.00 | 42.9% | 70.5% | 0.56 | 0.25–1.30 | 29.6% | 57.1% | |
| Thick. Struc.(-)/ Methyl. CP (-) | 24 | 0.48 | 0.69 | 0.27–1.71 | 22.5% | 70.5% | 1.45 | 0.58–3.77 | 29.6% | 77.6% | |
Further investigating the correlation of the 2 qualitative image features to GBM’s pMGMT methylation status, the presence of the “Thickened structure” and absence of Methylated contrast phenotype” significantly correlated with GBM’s pMGMT unmethylation both for the KANSAI exploratory cohort (Figure 5A red colored datapoint and Supplementary Figure 1B, p = .015, odds ratio = 2.44, 95% CI = 1.21–5.15) and the TCIA/ TCGA validation cohort (Figure 5B red colored datapoint and Supplementary Figure 2B, p = .006, odds ratio = 7.83, 95% CI = 1.63–36.56). The sensitivities and specificities for correctly predicting pMGMT-unmet GBM referring to the presence of the “Thickened structure” and absence of Methylated contrast phenotype” were 0.29 and 0.86 for the KANSAI exploratory and 0.25 and 0.96 for the TCIA/ TCGA validation cohort (Table 1) based on the WHO2021 criteria. On the other hand, different combinations of the qualitative image features did not correlate with GBM’s pMGMT methylation status (Supplementary Figures 3 and 4). The reestablished cohort based on the WHO2016 criteria also confirmed these tendencies (Supplementary Figures 3 and 4; Supplementary Table 3).
Discussion
The pMGMT methylation status is known to be one of the most important prognostic and predictive factors of GBM,25 and preoperative prediction of the pMGMT methylation status could benefit patient care, providing the possibility of identifying poor prognostic patients and offering them more aggressive or experimental treatments. Although radiomics and deep learning are now under extensive investigation, the diagnostic performance of predicting pMGMT methylation ranges widely in sensitivity and specificity from 55.6% to 93% and from 39.0% to 76.0%, respectively.8–10,12,13,16 There could also be issues related to the data used for training, such as an unbalanced training data set and overestimation of diagnostic performance.13,17 Moreover, applying these relatively complicated procedures to daily clinical practice is also a significant challenge.
A qualitative visual assessment of radiological images does not require any complicated analytical pipeline, which can be readily incorporated into clinical practice. Previous studies reported that ill-defined tumor margin was seen more frequently in high-grade gliomas with pMGMT methylation7 and ring enhancement in pMGMT-unmet GBM.6,8 However, incongruent studies showed no correlation between these findings and the pMGMT methylation status of the tumors.7,15 The present study demonstrated that the newly defined “Methylated contrast phenotype” highly correlated with pMGMT-met GBM in many patients in 2 independent cohorts. Moreover, this is the first report to predict GBM’s pMGMT methylation status by qualitative visual assessment based on the WHO Classification of Tumours Fifth Edition (WHO2021).21
Furthermore, we revealed high specificity for predicting pMGMT-unmet GBM by combining the 2 imaging features: the presence of “Thickened structure” and the absence of “Methylated contrast phenotype.” The specificities were 85.6% in the exploratory cohort and 95.9% in the validation cohort, better than those using radiomics and deep learning.8–10,16 Similar to the T2-FLAIR mismatch sign for detecting IDH-mutant astrocytoma, which is a clinically relevant imaging phenotype with low sensitivity but high specificity,26–29 the current study suggested that a particular GBM population that exhibits specific imaging characteristics can be predicted to be pMGMT unmethylated with high specificity. These findings indicate the potential for predicting the effectiveness of chemotherapeutic agents in GBM patients before surgery,30,31 providing valuable information in the decision-making process regarding intraoperative implantation of BCNU wafers.
It is important to note that the inter-rater agreements for both qualitative assessments were deemed more than “fair,” underscoring their validity as functional imaging characteristics. However, the proposed imaging characteristics in the present study, aimed at predicting GBM’s pMGMT methylation status, fall short of being satisfactory for preoperative assessment in bedside clinics. While additional research incorporating other imaging characteristics retrievable from T2-weighted images, fluid-attenuated inversion recovery (FLAIR), and apparent diffusion coefficient may enhance diagnostic accuracy,7,13 we anticipate challenges in this research domain. A recently published article, which focuses on MRI-based prediction of GBM’s MGMT methylation status powered by deep learning models, highlights the profound difficulty of achieving this task.32 In contrast to previous studies, the current research takes a different approach by attempting to identify a subgroup of GBM where the MGMT methylation status can be readily discerned rather than solely focusing on the overall prediction accuracy of the diagnostic model. Furthermore, it delves into qualitative visual assessment, aiming for an approach easily applicable in daily clinical practice without the need for complicated analytical pipelines.
Several limitations of the present study must be addressed. First, while the present study is the first to evaluate qualitative imaging features to predict GBM’s pMGMT methylation status in a 2-staged fashion with a larger sample size than previous similar studies,6–8,15 the retrospective design requires a prospective study with a larger sample size to validate our findings further. Second, the inter-rater agreement was not excellent, especially in the “Methylated contrast phenotype,” with Fleiss’s kappa coefficient being 0.30 for the KANSAI cohort and 0.39 for the TCIA/ TCGA cohort, which might limit the generalizability of the proposed imaging feature. Furthermore, although this study was based on 2 independent cohorts, the qualitative representation of images may differ among different cohorts. Confounding factors that influence image characteristics, such as MRI vendors and inconsistent MRI acquisition parameters, are another issue that may negatively affect inter-rater agreement. Third, the methods used for detecting pMGMT methylation must also be addressed, as different methods and various cutoffs are available to identify GBM’s pMGMT methylation status.33 This issue could be problematic in generalizing the current finding and could affect the “ground truth” for establishing any diagnostic model. Fourth, the current research used sensitivity and specificity as the endpoint of the analysis due to the binary assignment of each image finding. Many deep learning-based research studies report the area under the curve as its primary outcome,11,14,32,34 which hampers direct diagnostic accuracy comparison with the current research. Last, the present study did not assess the intra-rater consistency over time, which could significantly affect diagnostic reproducibility.
In conclusion, the present study showed that qualitative assessment of contrast-enhanced T1-weighted intensity images is useful to predict GBM’s pMGMT methylation status, and the proposed “Thickened structure” and “Methylated contrast phenotype” are valuable image biomarkers to better understand the GBM’s pMGMT methylation status in a preoperative setting.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (https://academic.oup.com/neuro-oncology).
Acknowledgments
This research was approved by the local ethics committees/institutional review boards of the following institutions: Wakayama Rosai Hospital, Wakayama Medical University, Osaka International Cancer Institute, Osaka University Graduate School of Medicine, Osaka National Hospital, Kansai Medical University, Osaka City University Graduate School of Medicine, Kindai University Faculty of Medicine, Kansai Rosai Hospital, Osaka City General Hospital, Toyonaka Municipal Hospital, Tazuke Kofukai Foundation, Medical Research Institute, Kitano Hospital. No natural language processing tool was used to prepare this manuscript.
Contributor Information
Takahiro Sanada, Department of Neurosurgery, Asahikawa Medical University, Asahikawa, Japan.
Manabu Kinoshita, Department of Neurosurgery, Asahikawa Medical University, Asahikawa, Japan; Department of Neurosurgery, Osaka International Cancer Institute, Osaka, Japan.
Takahiro Sasaki, Department of Neurological Surgery, Wakayama Medical University School of Medicine, Wakayama, Japan; Department of Neurosurgery, Wakayama Rosai Hospital, Wakayama, Japan.
Shota Yamamoto, Department of Neurosurgery, Asahikawa Medical University, Asahikawa, Japan; Department of Neurosurgery, Osaka General Medical Center, Osaka, Japan.
Seiya Fujikawa, Department of Neurosurgery, Asahikawa Medical University, Asahikawa, Japan; Department of Neurosurgery, Japanese Red Cross Kitami Hospital, Kitami, Japan.
Shusei Fukuyama, Department of Neurosurgery, Asahikawa Medical University, Asahikawa, Japan.
Nobuhide Hayashi, Department of Neurosurgery, Wakayama Rosai Hospital, Wakayama, Japan.
Junya Fukai, Department of Neurological Surgery, Wakayama Medical University School of Medicine, Wakayama, Japan.
Yoshiko Okita, Department of Neurosurgery, Osaka University Graduate School of Medicine, Osaka, Japan; Department of Neurosurgery, NHO Osaka National Hospital, Osaka, Japan.
Masahiro Nonaka, Department of Neurosurgery, NHO Osaka National Hospital, Osaka, Japan; Department of Neurosurgery, Kansai Medical University, Hirakata, Japan.
Takehiro Uda, Department of Neurosurgery, Osaka Metropolitan University Graduate School of Medicine, Osaka, Japan.
Hideyuki Arita, Department of Neurosurgery, Osaka International Cancer Institute, Osaka, Japan; Department of Neurosurgery, Osaka University Graduate School of Medicine, Osaka, Japan.
Kanji Mori, Department of Neurosurgery, Yao Municipal Hospital, Yao, Japan.
Kenichi Ishibashi, Department of Neurosurgery, Osaka City General Hospital, Osaka, Japan.
Koji Takano, Department of Neurosurgery, Osaka International Cancer Institute, Osaka, Japan; Department of Neurosurgery, Toyonaka Municipal Hospital, Toyonaka, Japan.
Namiko Nishida, Department of Neurosurgery, Tazuke Kofukai Foundation, Medical Research Institute, Kitano Hospital, Osaka, Japan.
Tomoko Shofuda, Department of Biomedical Research and Innovation, Institute for Clinical Research, NHO Osaka National Hospital, Osaka, Japan.
Ema Yoshioka, Department of Biomedical Research and Innovation, Institute for Clinical Research, NHO Osaka National Hospital, Osaka, Japan.
Daisuke Kanematsu, Department of Biomedical Research and Innovation, Institute for Clinical Research, NHO Osaka National Hospital, Osaka, Japan.
Mishie Tanino, Department of Diagnostic Pathology, Asahikawa Medical University Hospital, Asahikawa, Japan.
Yoshinori Kodama, Department of Neurosurgery, NHO Osaka National Hospital, Osaka, Japan; Department of Biomedical Research and Innovation, Institute for Clinical Research, NHO Osaka National Hospital, Osaka, Japan; Department of Diagnostic Pathology and Cytology, Osaka International Cancer Institute, Osaka, Japan.
Masayuki Mano, Department of Central Laboratory and Surgical Pathology, NHO Osaka National Hospital, Osaka, Japan.
Yonehiro Kanemura, Department of Diagnostic Pathology and Cytology, Osaka International Cancer Institute, Osaka, Japan.
Funding
This work was supported by the Japan Society for the Promotion of Science (Nos. 19K09526, 22K09200), the Japan Agency for Medical Research and Development (Japan Cancer Research Project, Nos. 23808466 and 22580474), the Takeda Science Foundation, the Okawa Foundation for Information and Telecommunications and the Akiyama Life Science Foundation. All scientific grants were for M.K.
Conflict of Interest Statement
None declared.
Authorship Statement
Conceptualization: M.K.; Data collection: T.S., M.K., T.S., N.H., J.F., N.O., M.N., T.U., H.A., K.M., K.I., K.T., N.N.; Genetic analysis: T.S., E.Y., D.K., M.T., Y.K., M.M., Y.K.; Image analysis: T.S., S.Y., S.F., S.F.; Manuscript preparation: T.S. and M.K.; All authors approved the manuscript.
Data Availability
Anonymized clinical and demographic data of the KANSAI cohort, radiological data, and other relevant data are provided in Supplementary Table 1. The senior authors and the Kansai Network for Molecular Diagnosis of Central Nervous Tumors will review requestable requests for additional data to determine whether they can be fulfilled following the privacy restrictions of each participating institution. Requests for additional materials related to this work should be directed to M.K.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Chinot OL, Barrié M, Fuentes S, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25(12):1470–1475. [DOI] [PubMed] [Google Scholar]
- 3. Arita H, Yamasaki K, Matsushita Y, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binabaj MM, Bahrami A, ShahidSales S, et al. The prognostic value of MGMT promoter methylation in glioblastoma: a meta-analysis of clinical trials. J Cell Physiol. 2018;233(1):378–386. [DOI] [PubMed] [Google Scholar]
- 5. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 6. Eoli M, Menghi F, Bruzzone MG, et al. Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res. 2007;13(9):2606–2613. [DOI] [PubMed] [Google Scholar]
- 7. Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC.. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology. 2012;54(6):555–563. [DOI] [PubMed] [Google Scholar]
- 8. Drabycz S, Roldán G, Robles P de, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010;49(2):1398–1405. [DOI] [PubMed] [Google Scholar]
- 9. Sasaki T, Kinoshita M, Fujita K, et al. Radiomics and MGMT promoter methylation for prognostication of newly diagnosed glioblastoma. Sci Rep-UK. 2019;9(1):14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Y, Patel M, Natarajan K, et al. Machine learning-based radiomic, clinical and semantic feature analysis for predicting overall survival and MGMT promoter methylation status in patients with glioblastoma. Magn Reson Imaging. 2020;74:161–170. [DOI] [PubMed] [Google Scholar]
- 11. Han L, Kamdar MR.. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput Pac Symp Biocomput. 2017;23:331–342. [PMC free article] [PubMed] [Google Scholar]
- 12. Korfiatis P, Kline TL, Lachance DH, et al. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. 2017;30(5):622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang P, Grinband J, Weinberg BD, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39(7):1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saxena S, Jena B, Mohapatra B, et al. Fused deep learning paradigm for the prediction of o6-methylguanine-DNA methyltransferase genotype in glioblastoma patients: a neuro-oncological investigation. Comput Biol Med. 2023;153:106492. [DOI] [PubMed] [Google Scholar]
- 15. Gupta A, Omuro AMP, Shah AD, et al. Continuing the search for MR imaging biomarkers for MGMT promoter methylation status: conventional and perfusion MRI revisited. Neuroradiology. 2012;54(6):641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanas VG, Zacharaki EI, Thomas GA, et al. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed. 2017;140:249–257. [DOI] [PubMed] [Google Scholar]
- 17. Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S.. Emerging applications of artificial intelligence in neuro-oncology. Radiology. 2019;290(3):607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The_Cancer_Imaging_Archive. 2020. https://www.cancerimagingarchive.net. Date accessed February 1, 2020.
- 20. The_Cancer_Genome_Atlas. 2020. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Date accessed February 1, 2020.
- 21. WHO-Classification-of-Tumours-Editorial-Board. Central Nervous System Tumours: WHO Classification of Tumours. (WHO-Classification-of-Tumours-Editorial-Board). Lyon: IARC Press; 2022. [Google Scholar]
- 22. Louis D, Ohgaki H, Wiestler O, Cavenee W.. WHO Classification of Tumours of the Central Nervous System WHO Classification of Tumours. Lyon: IARC Press; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brennan CW, Verhaak RGW, McKenna A, et al.; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landis JR, Koch GG.. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 25. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. [DOI] [PubMed] [Google Scholar]
- 26. Patel SH, Poisson LM, Brat DJ, et al. T2–FLAIR mismatch, an imaging biomarker for IDH and 1p/19q status in lower-grade gliomas: a TCGA/TCIA project. Clin Cancer Res. 2017;23(20):6078–6085. [DOI] [PubMed] [Google Scholar]
- 27. Kinoshita M, Uchikoshi M, Sakai M, et al. T2-FLAIR mismatch sign is caused by long T1 and T2 of IDH-mutant, 1p19q Non-codeleted Astrocytoma. Magn Reson Med Sci. 2021;20(1):119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jain R, Johnson DR, Patel SH, et al. “Real world” use of a highly reliable imaging sign: “T2-FLAIR mismatch” for identification of IDH mutant astrocytomas. Neuro-Oncol. 2020;22(7):936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Broen MPG, Smits M, Wijnenga MMJ, et al. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro-Oncol. 2018;20(10):1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350–1354. [DOI] [PubMed] [Google Scholar]
- 31. Fukai J, Nishibayashi H, Uematsu Y, et al. Rapid regression of glioblastoma following carmustine wafer implantation: a case report. Mol Clinc Oncol. 2016;5(1):153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saeed N, Ridzuan M, Alasmawi H, Sobirov I, Yaqub M.. MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models. Med Image Anal. 2023;90:102989. [DOI] [PubMed] [Google Scholar]
- 33. Butler M, Pongor L, Su YT, et al. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer. 2020;6(5):380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43(6Part1):2835–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized clinical and demographic data of the KANSAI cohort, radiological data, and other relevant data are provided in Supplementary Table 1. The senior authors and the Kansai Network for Molecular Diagnosis of Central Nervous Tumors will review requestable requests for additional data to determine whether they can be fulfilled following the privacy restrictions of each participating institution. Requests for additional materials related to this work should be directed to M.K.