Abstract
Oncogenic Ras mutants such as v-Ha-Ras cause a rapid rearrangement of actin cytoskeleton during malignant transformation of fibroblasts or epithelial cells. Both PI-3 kinase and Rac are required for Ras-induced malignant transformation and membrane ruffling. However, the signal transduction pathway(s) downstream of Rac that leads to membrane ruffling and other cytoskeletal change(s) as well as the exact biochemical nature of the cytoskeletal change remain unknown. Cortactin/EMS1 is the first identified molecule that is dissociated in a Rac–phosphatidylinositol 4,5-biphosphate (PIP2)-dependent manner from the actin-myosin II complex during Ras-induced malignant transformation; either the PIP2 binder HS1 or the Rac blocker SCH51344 restores the ability of EMS1 to bind the complex and suppresses the oncogenicity of Ras. Furthermore, while PIP2 inhibits the actin-EMS1 interaction, HS1 reverses the PIP2 effect. Thus, we propose that PIP2, an end-product of the oncogenic Ras/PI-3 kinase/Rac pathway, serves as a second messenger in the Ras/Rac-induced disruption of the actin cytoskeleton and discuss the anticancer drug potential of PIP2-binding molecules.
The nonmuscle actin cytoskeleton plays the major role in both maintenance of cell shape and variety of cell motility (32). More than 2 decades ago it was demonstrated that the actin cytoskeleton is rearranged rapidly when normal fibroblasts or epithelial cells are transformed with simian virus 40 (SV40) (48). A variety of oncoproteins (e.g., Src and Ras) also cause changes in the actin cytoskeleton during malignant transformation, typically disruption of actin stress fibers followed by induction of membrane ruffling (1). The actin cytoskeleton microfilament is an actomyosin (a complex of actin filament and myosin ATPases)-based contractile apparatus which is associated with a number of other actin- or myosin-binding proteins. These actin- or myosin-binding proteins form a variety of cytoskeleton structures, such as actin stress fibers, membrane ruffles, contractile ring, and microspikes. The transition from the stress fibers to membrane ruffles involves both the dissociation of stress-fiber-specific proteins from and the association of membrane-ruffle-specific proteins with actin filament. For example, single-headed myosin ATPases (myosins I) are associated with membrane ruffles but not with stress fibers or contractile ring, whereas double-headed myosin ATPases (myosins II) are mainly associated with stress fibers and contractile ring (10).
Interestingly, the expression of several genes encoding actin-binding proteins (ABPs), including alpha-actinin, vinculin, gelsolin, and tropomyosin, is suppressed during malignant transformation caused by SV40 or Ras (45). Furthermore, overexpression of alpha-actinin, vinculin, gelsolin, and tropomyosin genes suppresses SV40/Ras-induced malignant transformation of NIH 3T3 normal murine embryonic fibroblasts (6, 12, 13, 19, 30, 33). However, the molecular mechanism underlying the antioncogenic action of these ABPs remains to be determined. In addition to F-actin, at least the first three proteins of these multifunctional ABPs bind phosphatidylinositol 4,5-bisphosphate (PIP2). Production of PIP2 is induced by oncogenic Ras through PI-3 kinase and the G protein Rac, both of which are essential for Ras-induced malignant transformation (14, 34, 35). Ras activates PI-3 kinase, which in turn activates Rac (35) through an as-yet-uncharacterized Rac GDP dissociation stimulator. Rac then activates PI-4 and PI-5 kinases that produce PIP2 (14). This unique acidic phosphoinositide then binds F-actin plus-end capping proteins, such as tensin, thereby causing the uncapping of actin filament and inducing a rapid actin polymerization at this end (14). Since cytochalasins, which cap the plus-end of actin filament and block membrane ruffling (50), and the drug SCH51344, which blocks Ras/Rac-induced membrane ruffling (46), suppress Ras/Rac-induced malignant transformation (21, 27, 46), it is likely that the Rac-induced PIP2 production, uncapping, and membrane ruffling play a critical role in Ras/Rac-induced malignancy.
HS1 is closely related to an F-actin cross-linking protein called cortactin/EMS1, both structurally and functionally, particularly in the N-terminal F-actin binding and the C-terminal SH3 domains (15, 17, 20, 25, 26, 39, 49). However, unlike EMS1, which is expressed ubiquitously, HS1 is normally expressed only in hematopoietic cells (20, 39) and is required for B-cell surface antigen receptor-mediated apoptosis (15, 52). Using two distinct anti-Ras tumor suppressors, i.e., the cytoplasmic protein HS1, which binds PIP2, and the drug SCH51344, which blocks downstream of Rac, we have identified the first biochemically defined pathway in which Ras causes the disruption of an actomyosin complex through Rac and PIP2, without suppressing the expression levels of its protein components, during malignant transformation, suggesting the anticancer potential of various PIP2-binding molecules.
MATERIALS AND METHODS
Detection of the EMS1-actomyosin complex.
Cells (107) cultured on a petri dish (150 mm in diameter) were disrupted with 1 ml of radioimmunoprecipitation assay buffer (47). The cell lysates were cleared by spinning at 13,000 rpm for 10 min. The resultant supernatants were incubated with the 1:500 antiserum RA23 against EMS1 (39), and the immunocomplex was precipitated by protein A beads with or without 5 mM ATP. The pellets were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) followed by staining with 0.1% Coomassie blue. In control experiments with protein A beads alone, nonimmune serum, or antiserum against HS1 (51), no actomyosin complex was detected in the pellets by SDS-PAGE.
SCH51344 treatment of Ras-transformed cells.
To examine the effect of SCH51344, an anti-Ras cancer drug (21), v-Ha-Ras-transformed cells (106) were cultured for 60 h in the presence of 50 μM SCH51344 or 0.08% dimethyl sulfoxide alone, the solvent for dissolving the drug. This drug has no effect on the growth rate of these cells in a liquid culture but flattens the cells and blocks their focus formation as previously described (21). The confluent cells were then harvested and subjected to the analysis of the EMS1-actomyosin complex formation as described above.
Identification of the 200-kDa protein as a nonmuscle myosin IIA heavy chain by microsequencing.
The SDS-PAGE-purified 200-kDa protein (1.5 μg) immunoprecipitated with an EMS1-actin complex from the NIH 3T3 cells (see Fig. 1A) was digested by trypsin (29), and the resulting peptides were separated by high-performance liquid chromatography (28) and identified as the fragments of murine nonmuscle myosin IIA heavy chain by the microsequencing procedures previously described (2, 5, 28, 29) and comparison with the amino acid sequences of the rat and human nonmuscle myosin IIA heavy chains that were determined previously (3, 36, 40, 44) (Table 1).
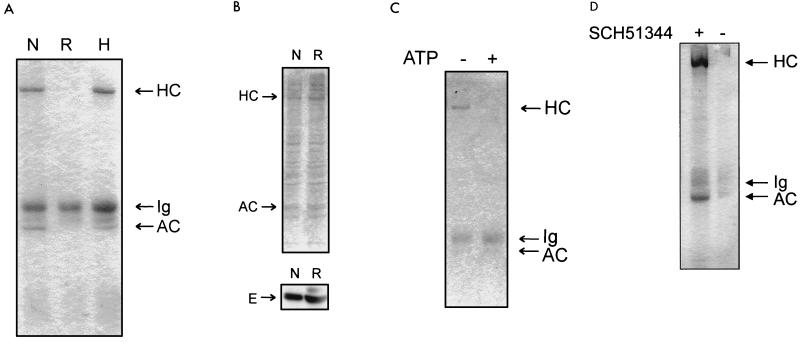
FIG. 1.
Ras induces the disruption of an EMS1-actomyosin complex. (A) Proteins immunoprecipitated with EMS1. Normal (N), Ras-transformed (R), and HS1-transfected Ras-transformed (H) NIH 3T3 cells stained with Coomassie blue. HC, a protein of 200 kDa comigrating with myosin II heavy chain; AC, actin of 42 kDa. (B) No effect of Ras transformation on the expression levels of myosin II heavy chain (HC), actin (AC), and EMS1 (E). (Top panel), Coomassie-blue stained gel of the lysates from normal (N) and Ras-transformed (R) NIH 3T3 cells. (Bottom panel) Immunoblot showing the level of EMS1 in normal (N) and Ras-transformed (R) NIH 3T3 cells. EMS1 had a size of 85 kDa. Fifty micrograms of protein was loaded in each lane. (C) Effect of ATP. Dissociation of HC from the actin-EMS1 complex in the presence of 5 mM ATP (+). (D) Effect of SCH51344. Both myosin IIA heavy chain (HC) and actin (AC) were immunoprecipitated with EMS1 from Ras-transformed cells treated with SCH51344 (+) but not with cells without (−) drug treatments. Ig, immunoglobulin.
TABLE 1.
The 200-kDa protein immunoprecipitated with an EMS1-F-actin complex is murine myosin IIA heavy chaina
| Source and fragment no. |
Sequence |
|---|---|
| 15 | |
| Murine | NFINNPLAQADWAAK |
| Rat | NFINNPLAQADCGAK |
| Human | NFINNPLAQADWAAK |
| 290 | |
| Murine | TDLLLEPYNK |
| Rat | TDLLLEPYNK |
| Human | TDLLLEPYNK |
| 843 | |
| Murine | HEDELLAK |
| Rat | HEDELLAK |
| Human | QEEEMMAK |
| 1325 | |
| Murine | LSLSTK |
| Rat | LSLSTK |
| Human | LSLSTK |
| 1394 | |
| Murine | DLEGLSQR |
| Rat | DLEGLSQR |
| Human | DLEGLSQR |
| 1479 | |
| Murine | ALSLAR |
| Rat | ALSLAR |
| Human | ALSLAR |
| 1756 | |
| Murine | ANLQIDQINTDLNLER |
| Rat | ANLQIDQINTDLNLER |
| Human | ANLQIDQINADLNLER |
| 1817 | |
| Murine | IAQLEEQLDNETK |
| Rat | IAQLEEQLDNETK |
| Human | IAQLEEQLDNETK |
The primary sequences of tryptic fragments derived from the murine 200-kDa protein are compared with those of rat and human myosin IIA heavy chains (3, 36, 40, 44). The number of each fragment indicates the first residue of the corresponding fragment from the human nonmuscle myosin IIA heavy chain (36, 40, 44).
Preparation of muscle actin and HS1 or EMS1 fragments.
Actin was purified from acetone powder of rabbit skeletal muscle (Sigma Chemicals) as described previously (41). Full-length human HS1 and the actin-binding domain (residues 82 to 192) were produced in bacteria as thrombin-cleavable glutathione S-transferase (GST) fusion proteins by subcloning the corresponding PCR DNAs (EcoRI fragments) into the vector pGEX-2TH. Full-length human EMS1-GST fusion protein (17) and other GST fusion proteins were affinity purified by glutathione beads (24).
F-actin cross-linking assay.
F-actin cross-linking activity of EMS1 or HS1 was measured by the superprecipitation procedure (17) with a slight modification in the presence or absence of PIP2. Purified GST-fusion proteins of EMS1 or HS1 were preincubated with PIP2 on ice for 30 min. F-actin was then added into the above-mentioned mixtures and incubated for another 30 min on ice. The mixture was then centrifuged at 15,000 × g for 10 min at 4°C, and the resultant pellets were subjected to SDS-PAGE. The Coomassie blue-stained gels were then subjected to densitometry for quantitation of actin bands. Under these conditions, the control GST alone caused no cross-linking of F-actin.
PIP2-binding assay.
Purified GST fusion proteins of EMS1 or HS1 were fixed onto a nitrocellulose membrane with a dot blot apparatus. The membrane was then blocked with 10% skim milk and washed (47). The membrane was then incubated with 10 mg of PIP2 per ml in TSBT (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% bovine serum albumin, and 0.02% Tween 20) at 25°C for 30 min. After being washed, the membrane was incubated with the 1:1,000 rabbit antiserum against PIP2 (7) at 25°C for 60 min, washed three times, and immunoblotted with 1:10,000 goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Bio-Rad, Richmond, Calif.) at 25°C for 60 min. The membrane was incubated with enhanced chemiluminescence detection solution (Amersham, Little Chalfont, United Kingdom) at 25°C for 1 min and exposed to XAR5 film (Kodak, Rochester, N.Y.) for 2 min. The density of each dot was quantitated by a densitometer. Under these conditions, the control GST alone did not bind PIP2.
Construction of plasmids expressing human HS1 (wild type and mutants) in mammalian cells.
For expression of wild-type HS1 and its mutants in mammalian cells, EcoRI DNA fragments containing the HindIII site followed by a Kozak consensus sequence (AAGCTTGCCGCCACCATG) at the 5′ end and encoding full-length human HS1 (HS1F) or its SH3-negative (HS1-SH3) actin-binding domain-negative mutants (HS1ad) and HS1-EMS1 chimera (HEaH) were prepared by PCR and subcloned into the retroviral vector pMV7 as described previously (24). The orientation of the inserts was determined by HindIII digestion (a second HindIII site is located 100 bp downstream of the EcoRI site in the vector). The resultant plasmids were purified by a Plasmid Maxi Kit Qiagen (Hilden, Germany) according to the manufacturer’s instructions and used for transfection. Each plasmid expresses a neomycin G418 resistance selectable marker in addition to HS1 (wild type or mutants).
Effect of wild-type HS1 or its mutants on the colony-forming ability of v-Ha-Ras transformants in soft agar.
v-Ha-Ras-transformed NIH 3T3 cells were transfected with HS1F or other plasmids as complexes with liposomes (31). A parallel transfection with the vector alone was also carried out as a negative control. The resultant transfectants were cloned in the presence of 400 μg of G418 per ml (24). The whole-cell lysate of each G418-resistant clone was then subjected to immunoblotting with the rabbit antiserum against a human HS1 peptide (residues 306 to 320) (51) for selecting out the clones that overexpress the wild-type HS1 or its mutants. The colony formation in a soft agar (anchorage-independent growth) of these HS1 transfectants in comparison with that for the clones with the control vector alone was examined by incubating 1,000 cells/plate at 37°C for 18 days under standard culture conditions (24). The colonies were stained with 0.005% crystal violet and counted.
Autokinase assay for v-Ha-Ras GTPase.
Unlike normal Ras GTPases, v-Ha-Ras GTPase, which uniquely contains Thr at position 59, is autophosphorylated at this position during the hydrolysis of GTP (11). According to the previously described autokinase assay procedures (4), the cell lysate containing 100 μg of protein prepared from the parental v-Ha-Ras transformants or HS1 transfectants was immunoprecipitated with the anti-Ras antibody Y13-259, and then each immunoprecipitate resuspended in the kinase buffer (4) was subjected to the autokinase assay with 10 nM [γ-32P]GTP at 37°C for 30 min and to SDS–15% PAGE. The autophosphorylated v-Ha-Ras band was visualized by autoradiography.
F-actin binding assay.
The assay was performed according to the F-actin cosedimentation procedure (49). Briefly, F-actin (4 μM) was mixed with the fibroblast lysate containing the native full-length HS1 or HS1-EMS1 chimera. The mixture (100 μl) was kept on ice for 60 min and then centrifuged at 100,000 × g for 30 min at 4°C in a Beckman model TL-100 ultracentrifuge. The resultant supernatants and pellets were subjected to SDS-PAGE, followed by immunoblotting with the antibody against HS1.
RESULTS AND DISCUSSION
Ras-induced disruption of an EMS1-actomyosin complex.
Microinjection of oncogenic Ras mutants into normal fibroblasts causes the disruption of actin stress fibers, followed by the induction of membrane ruffles in half an hour (1). To understand how Ras causes such a rapid change in the actomyosin complexes, presumably without any change in the levels of its protein components, we have examined the interaction of EMS1 with F-actin in normal and v-Ha-Ras-transformed NIH 3T3 cells. EMS1 is an F-actin cross-linking SH3 protein with 550 amino acids (17). With an antibody specific for EMS1, proteins associated with EMS1 were immunoprecipitated from the cytosol of both normal and Ras-transformed cells and separated by SDS-PAGE. In normal cells, EMS1 forms a stable complex with F-actin and an additional protein of 200 kDa, whereas in Ras-transformed cells, such a complex fails to coprecipitate with EMS1 (Fig. 1A). Furthermore, overexpression of HS1 in the Ras transformants restores the complex with EMS1 and suppresses Ras-induced malignancy as described later. Under these conditions, either F-actin or the 200-kDa protein is not coprecipitated with protein A beads alone, nonimmune serum, or an antibody specific for HS1 (data not shown), confirming that these two proteins are coprecipitated only when they form a complex with EMS1. Since EMS1 is an F-actin cross-linker, even a few EMS1 molecules per actin filament consisting of an average of 1,000 actin monomers are sufficient for the anti-EMS1 to coprecipitate the F-actin 200-kDa protein complex. Ras has no effect on the expression level of either EMS1 or actin (Fig. 1B), indicating that Ras causes a disassembly of the EMS1-actin filament complex.
We suspected that the 200-kDa protein is a heavy chain of a nonmuscle myosin II isozyme, because it is one of the major cytoplasmic proteins in the fibroblasts and comigrates with the heavy chain of skeletal muscle myosin (SDS-PAGE). In support of this notion, 5 mM ATP, which dissociates any myosin from actin filament (23), causes a dissociation of the 200-kDa protein from the EMS1-actin complex (Fig. 1C). Finally, the microsequencing of several tryptic peptides derived from the 200-kDa protein confirmed that it is the heavy chain of a nonmuscle myosin II type A (Table 1). The cytoplasmic myosin II level is not affected by Ras transformation (Fig. 1B). This notion was further confirmed by immunoblotting with an antibody specific for myosin IIA heavy chain (data not shown). Thus, it is clear that Ras transformation is associated with the disruption of an EMS1-actomyosin II complex, and the reversion of Ras transformation by HS1 overexpression restores the ability of EMS1 to bind actomyosin II. This is the first biochemical demonstration of the disruption by Ras of the specific interaction of actomyosin with ubiquitous cellular proteins such as EMS1. However, so far, no clear-cut change has been detected in the overall intracellular localization pattern of EMS1 upon Ras transformation by fluorescent EMS1 staining (44a).
SCH51344, which blocks Rac-induced membrane ruffling suppresses Ras-induced disruption of the EMS1-actomyosin II complex.
We have shown previously that the drug SCH51344 blocks both Ras/Rac-induced membrane ruffling and malignant transformation and stimulates stress fiber formation (21, 46). However, this drug has no effect on either Raf-induced activation of MEK/mitogen-activated protein kinase (extracellular signal-regulated protein kinases) or Rac-induced activation of JNK kinase pathways which are not essential for membrane ruffling (21, 46). Interestingly, the treatment of v-Ha-Ras-transformed cells with 50 μM SCH51344 completely restores the EMS1-actomyosin II complex (Fig. 1D), clearly indicating that Ras requires a component(s) acting downstream of Rac to both disrupt the EMS1-actomyosin II complex and induce membrane ruffling.
PIP2 inhibits the F-actin cross-linking activity of either EMS1 or HS1 in vitro.
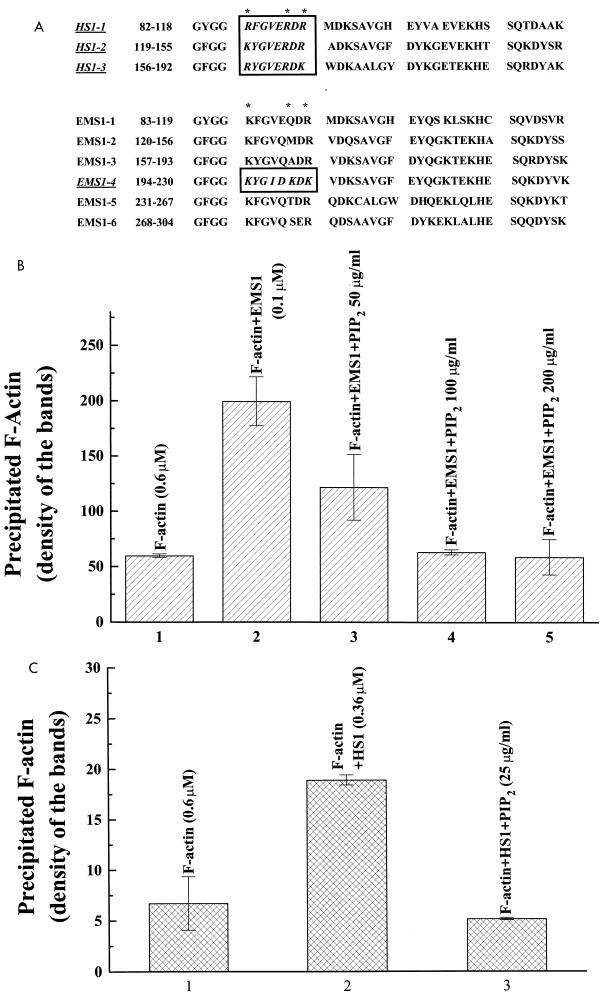
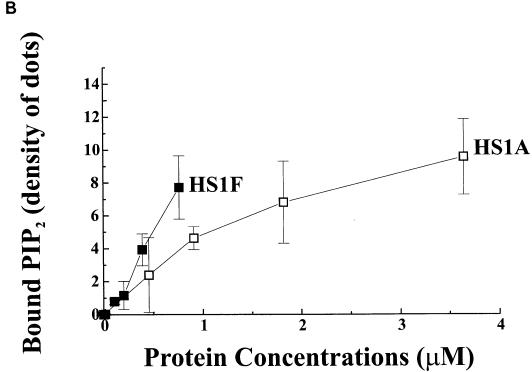
The G protein Rac that is activated by Ras through PI-3 kinase (35) induces overproduction of PIP2 by activating PI-4 and PI-5 kinases (14). Interestingly, as shown in Fig. 2A, we found that both EMS1 and HS1 contain a putative PIP2-binding motif (R/K YG V/I E/D R/K D R/K) in their N-terminal 37-amino-acid repeats (20, 39, 54) as do other F-actin cross-linking proteins, such as alpha-actinin and vinculin (8, 9), and F-actin-severing proteins, such as gelsolin and cofilin (18, 53). In fact, PIP2 clearly inhibits the F-actin cross-linking activity of EMS1 in a dose-dependent manner in vitro (Fig. 2B). While the F-actin cross-linking activity of HS1 requires higher concentrations, the cross-linking is inhibited more effectively by PIP2 (Fig. 2C). As expected, HS1’s apparent affinity for PIP2 appears to be at least several fold higher than that of EMS1 (Fig. 3A). EMS1 contains only one putative PIP2-binding motif, whereas HS1 contains three (Fig. 2A).
FIG. 2.
PIP2 inhibits F-actin cross-linking activity of both EMS1 and HS1. (A) The putative PIP2-binding motifs (in boxes) within the actin-binding repeats (underlined) of EMS1 and HS1. Asterisks indicate the critical basic residues for PIP2 binding. (B) Inhibition by PIP2 of EMS1-induced F-actin cross-linking. F-actin alone or a mixture of F-actin and EMS1 with or without PIP2 was centrifuged under conditions in which only the cross-linked F-actin was significantly precipitated. PIP2, at 100 μg/ml, completely abolished the F-actin cross-linking activity of EMS1. The data presented are the averages of three duplicated samples. (C) Inhibition by PIP2 of HS1-induced F-actin cross-linking. Much less PIP2 (25 μg/ml) was sufficient to inhibit the F-actin cross-linking activity of HS1. Under these conditions, GST alone did not cross-link F-actin. The data are the averages of two duplicated samples.
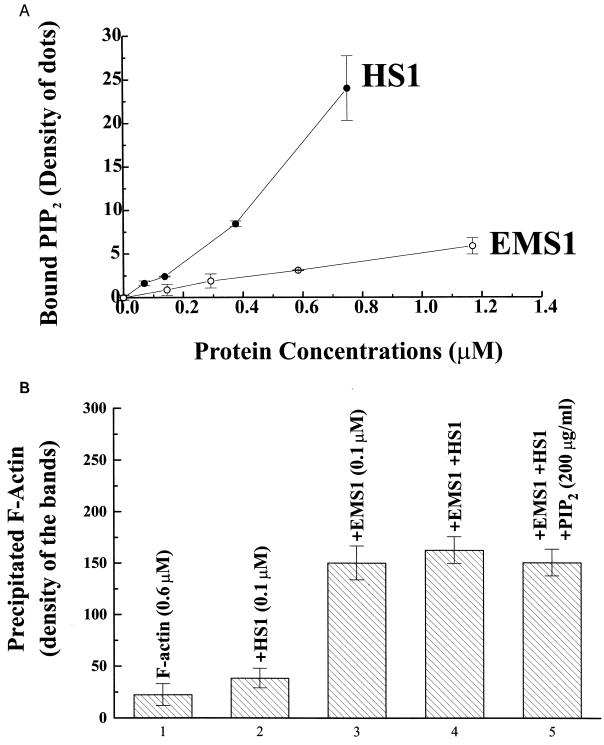
FIG. 3.
(A) HS1 has a much higher affinity for PIP2 than EMS1. PIP2-binding assay was performed with GST fusion proteins of full-length HS1 or EMS1 fixed on a nitrocellulose membrane. Under these conditions, GST alone did not bind PIP2. The data are the averages of three duplicated samples. (B) HS1 reverses the effect of PIP2 that inhibits the F-actin cross-linking activity of EMS1. HS1 (0.1 μM) that does not cause F-actin cross-linking abolished the inhibition by PIP2 (200 μg/ml) of EMS1-induced F-actin cross-linking, as the assay was performed under the essentially same conditions as those described in the legend for Fig. 2B. The data presented are the averages of two duplicated samples.
HS1 rescues EMS1 from the PIP2 effect that blocks its F-actin cross-linking.
Since the F-actin cross-linking activity of HS1 is much weaker than that of EMS1, HS1 (0.1 μM) does not significantly cross-link F-actin (0.6 μM), whereas EMS1 at the same concentration is a strong actin cross-linker (Fig. 3B). In the presence of both HS1 and EMS1 (0.1 μM), PIP2, even at 200 μg/ml, fails to inhibit F-actin cross-linking by EMS1 (Fig. 3B). These observations clearly indicate that the high-affinity PIP2 binder HS1 is able to abolish completely the action of PIP2 that inhibits the F-actin cross-linking activity of EMS1 at least in vitro. These in vivo and in vitro effects of the PIP2-binding protein HS1 and the Rac blocker SCH51344 on the EMS1-actin interaction support the notion that PIP2 mediates Ras-induced disruption of the EMS1-actomyosin II complex, at least in part, as a factor acting downstream of Rac.
HS1 reverses Ras-induced malignant phenotype.
Two distinct PIP2-binding or F-actin cross-linking proteins, alpha-actinin and vinculin, suppress SV40-induced malignant transformation of NIH 3T3 cells when they are overexpressed (6, 13). Furthermore, a mutant of gelsolin, the PIP2-binding or F-actin-severing or -capping protein, also suppresses v-Ha-Ras-induced malignancy of the same fibroblasts (30). In addition, we have shown that NF2/Merlin, which is closely related to the Ezrin-Radixin-Moesin (ERM) family of PIP2-binding and F-actin-capping proteins (16, 38), reverses v-Ha-Ras-induced malignancy (43). These previous observations prompted us to examine the anticancer potential of the newly identified PIP2-binding and F-actin cross-linking protein HS1.
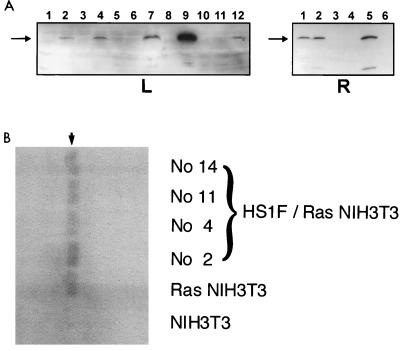
v-Ha-Ras-transformed fibroblasts were transfected with a DNA encoding full-length human HS1 with a retroviral vector (24). Several neomycin-resistant HS1-overexpressing clones were screened by immunoblot analysis with an antibody specific for the HS1 epitope which is not present in EMS1 (Fig. 4A). Unlike the parental Ras transformants, none of HS1 overexpressors forms any focus at their confluence (data not shown), suggesting that HS1 restores contact inhibition of growth. The colony-forming ability in soft agar (anchorage-independent growth) of these HS1-overexpressing clones is greatly reduced (by 70 to 80%) compared with that of the parental or vector-alone-transfected v-Ha-Ras transformants (Table 2). This reduction was not due to any loss of v-Ha-Ras gene expression, because all HS1 overexpressors show the basically same autophosphorylating activity of v-Ha-Ras as that of the parental cells (Fig. 4B). These observations clearly indicate that overexpression of HS1 suppresses Ras transformation.
FIG. 4.
Characterization of HS1 overexpressors derived from v-Ha-Ras-transformed NIH 3T3 cells. (A) Screening of HS1 overexpressors by immunoblot analysis. The supernatants (50 μl each) of cell lysates prepared from G418-resistant transfectants were subjected to SDS-PAGE and blotted by the antiserum against the human HS1 fragment. L, blotted with 1:1,000 antiserum; lanes 2, 4, 7, 9, and 12 correspond to the full-length HS1 overexpressors no. 2, 4, 7, 11, and 14, respectively. R, blotted with 1:2,000 antiserum; lanes 1, 2, and 5 correspond to the full-length HS1 overexpressors no. 15, 16, and 11. Arrow, HS1 of 75 kDa. (B) Autophosphorylation of v-Ha-Ras GTPase in HS1 revertants. Arrowhead, v-Ha-Ras GTPase of 21 kDa. There was no significant reduction in the v-Ha-Ras GTPase level of Ras-transformed cells transfected with HS1F (full-length HS1).
TABLE 2.
Full-length HS1 suppresses anchorage-independent growth of v-Ha-RAS transformation
| Clone | No. of coloniesa
|
Suppression (%) | |||
|---|---|---|---|---|---|
| Large | Medium | Small | Total | ||
| Vector alone | 31 | 334 | 357 | 721 | 0 |
| HS1 transfectants | |||||
| Clone 14 | 2 | 25 | 94 | 119 | 83 |
| Clone 04 | 2 | 47 | 89 | 134 | 81 |
| Clone 02 | 1 | 25 | 126 | 152 | 79 |
| Clone 11 | 3 | 47 | 138 | 188 | 74 |
A total of 103 cells from each clone were plated in soft agar. After 18 days, the number of colonies formed in each dish was counted. Colony size: large, more than 100 cells; medium, 30 cells; small, fewer than 10 cells. Each value is the average of data from triplicate assay, and the standard deviation of each was less than 5%.
Both the SH3 domain and the F-actin- and PIP2-binding motifs of HS1 are required for the anti-Ras action.
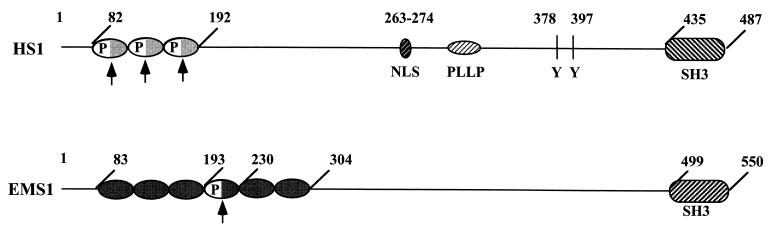
Full-length HS1 contains several distinct functional domains (Fig. 5): (i) the N-terminal three repeats of 37 amino acids, each of which binds F-actin (25, 26) and contains a putative PIP2-binding motif, (ii) the Pro-rich domain (PLLP) that binds the SH3 domain of the Tyr-kinase Lck (42), (iii) two critical Tyr residues at positions 378 and 397, whose phosphorylation by the kinase Syk is required for the apoptosis of B lymphocytes (52), (iv) a nuclear localization sequence (residues 263 to 274), and (v) the C-terminal SH3 domain, deletion of which causes the nuclear localization of HS1 (15, 26). As summarized in Table 3, we found that deletion of either the SH3 domain or the entire F-actin- and PIP2-binding motifs abolishes the ability of HS1 to suppress Ras-induced malignancy, although both deletion mutants are expressed as much as the full-length HS1 (data not shown). Here we describe the biological properties of only the following two distinct mutants of HS1 lacking all three actin-binding 37-amino-acid repeats of HS1 (ACT−) that no longer bind either F-actin or PIP2.
FIG. 5.
Schematic structures of HS1 and EMS1. Numbers indicate amino acid residues. Arrowheads indicate only the 37-amino-acid motifs that bind F-actin. P, putative PIP2-binding motif; NLS, nuclear localization sequence; PLLP, Pro-rich domain; Y, critical Tyr residue.
TABLE 3.
SH3 and actin-binding domains are required for HS1 to suppress Ras-induced malignancy
| v-Ha-Ras-transformed NIH 3T3 cellsa | Colonies formed in soft agar/103 cellsb |
|---|---|
| Vector alone | 648 ± 122 |
| HS1 (full length) | 148 ± 30 |
| Vector alone | 600 ± 43 |
| HS1 (SH3 negative) | 565 ± 103 |
| Vector alone | 865 ± 143 |
| HS1 (ACT negative) | 645 ± 97 |
| Vector alone | 728 ± 19 |
| HS1-EMS1 chimera (ACT negative) | 678 ± 25 |
HS1 (SH3 negative), lacking SH3 domain; HS1 (ACT negative), lacking the actin-binding domain; HS1-EMS1, non-actin-binding chimera.
Each value is the average of data from at least four individual clones in duplicate assays ± standard deviation.
The N-terminal F-actin-binding domain of HS1 (residues 82 to 192) contains three unique tandem repeats, and each repeat comprises 37 amino acids and binds F-actin (25, 26). Although the corresponding domain of EMS1 (residues 83 to 193) also contains three similar tandem repeats of 37 amino acids possessing 60 to 70% sequence identity with the HS1 repeats, this EMS1 domain does not bind F-actin. Instead, only the fourth 37-amino-acid repeat of EMS1 (residue 194 to 230) binds the F-actin (25, 26). EMS1 appears to form a homodimer for cross-linking F-actin (17). Only these three F-actin-binding 37-amino-acid motifs of HS1 and the single F-actin-binding motif of EMS1 contain a putative PIP2-binding motif (Fig. 2A).
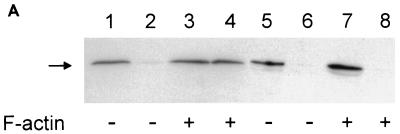
To examine whether the three F-actin binding motifs of HS1 are involved in the anti-Ras action of HS1, we have generated an HS1-EMS1 chimera in which the whole F-actin-binding domain (residues 82 to 192) of HS1 is replaced by the corresponding domain of EMS1 (residues 83 to 193), which contains the highly homologous three 37-amino-acid motifs but does not bind F-actin. Unlike the wild-type HS1, the HS1-EMS1 chimera overexpressed in v-Ha-Ras-transformants fails to bind F-actin (Fig. 6A). The HS1-EMS1 chimera does not suppress the colony-forming ability of Ras transformants in soft agar (Table 3). Similarly, an internal deletion mutant of HS1 that lacks all three actin-binding motifs (ACT−) no longer suppresses Ras transformation (Table 3). These observations clearly indicate that the actin-binding domain of HS1 is also essential for the anti-Ras action. A reverse HS1 mutant alone, consisting of only three actin-binding 37-amino-acid motifs, binds PIP2 (Fig. 6B), strongly supporting the notion that the putative PIP2-binding motifs within these actin-binding repeats are responsible for the PIP2 binding. Thus, it remains to be clarified whether F-actin-binding or PIP2-binding activity (or both) is required for the anti-Ras action of HS1.
FIG. 6.
Characterization of HS1 mutant (HS1ad) lacking the actin-binding domain and HS1-EMS1 chimera (HEaH). (A) F-actin binds the full-length HS1 but not the HS1-EMS1 chimera. v-Ha-Ras-transformed NIH 3T3 transfectants (106 cells) that overexpress the wild-type HS1 (lane 1 to 4) or chimera (lane 5 to 8) were disrupted, and the cleared cell lysate was incubated with (+) or without (−) F-actin and then centrifuged at 100,000 × g. The immunoblot of resultant supernatants (lanes 1, 3, 5, and 7) and pellets (lanes 2, 4, 6, and 8) indicates that the wild-type HS1 coprecipitates with F-actin (lane 4), whereas the chimera does not coprecipitate with F-actin (lane 8). (B) Actin-binding domain of HS1 binds PIP2. A GST fusion protein of actin-binding domain (residues 82 to 192) of HS1 fixed on a nitrocellulose membrane was subjected to a PIP2-binding assay as described in the legend for Fig. 3A. The actin-binding domain alone (HS1A) is sufficient to bind PIP2, as is the full-length HS1 (HS1F). The data presented are the averages of two duplicated samples.
Interestingly, like HS1, all other actin-binding tumor suppressors, such as alpha-actinin, vinculin, NF2, tensin, and gelsolin or cofilin mutants, bind PIP2 (26, 27). However, a mutant of cofilin (Gln112 Gln114) that no longer binds F-actin but still binds PIP2 is able to suppress Ras-induced malignancy (26, 27). In addition, we have previously demonstrated that microinjection of the antibody specific for PIP2 into Ras-transformed NIH 3T3 cells inhibits their proliferation and reverts their morphology to the normal flat phenotype, although these effects are both partial and transient (7), suggesting an essential role for PIP2 in Ras transformation. It would be of great interest to examine whether other PIP2-binding proteins, such as alpha-actinin and vinculin, also restore the EMS1-actomyosin II complex in Ras-transformed cells and suppress Ras transformation.
To test more directly the notion that the PIP2 binding alone is sufficient for both restoring the EMS1-actomyosin II complex and suppressing the malignancy, we are currently examining the effects of PIP2-binding antibiotics such as neomycin, which inhibits thrombin-induced actin polymerization in vivo (22), presumably by blocking the PIP2-induced uncapping of actin filament plus-ends (14). If some PIP2-binding drugs or peptides prove to have potent anticancer activities, the screening for various other PIP2-binding compounds or PI-4 and PI-5 kinase inhibitors such as ribofuranosyl derivatives of echiguanine analogs (37) could lead to the development of novel anticancer therapeutics useful for the treatment of Ras-associated tumors, which represent more than 30% of all human cancers.
ACKNOWLEDGMENTS
We are grateful to Rob Michalides for his gift of human EMS1 cDNA, Henk van Damme and Vera van Buuren for their studies on intracellular localization of EMS1, Anjali Tikoo for her unpublished information on the anti-Ras action of F-actin cappers and cofilin mutants, and Tony Burgess for his consistent support and encouragement throughout this study.
The work has been supported in part by a grant from the Anti-Cancer Council of Victoria.
REFERENCES
- 1.Bar-Sagi D, Feramisco J. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by Ras proteins. Science. 1986;233:1061–1065. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 2.Bleasby A J, Wootton J C. Construction of validated, non-redundant composite protein sequences in a protein database. Protein Eng. 1990;3:153–159. doi: 10.1093/protein/3.3.153. [DOI] [PubMed] [Google Scholar]
- 3.Choi O H, Park C S, Itoh K, Adelstein R S, Beaven M. Cloning of cDNA encoding rat myosin II-A heavy chain and evidence for the absence of myosin II-B heavy chain in cultured rat mast (RBL-2H3) cells. J Muscle Res Cell Motil. 1996;17:69–77. doi: 10.1007/BF00140325. [DOI] [PubMed] [Google Scholar]
- 4.D’Abaco G, Whitehead R, Burgess A W. Synergy between APC (Min) and an activated Ras mutation is sufficient to induce colon carcinomas. Mol Cell Biol. 1996;16:884–891. doi: 10.1128/mcb.16.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eng J K, McCormack A, Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez J L R, Geiger B, Salomon D, Sabanay I, Zoeller M, Ben-Ze’ev A. Suppression of tumorigenicity in SV40-transformed cells after transfection with vinculin cDNA. J Cell Biol. 1992;119:427–438. doi: 10.1083/jcb.119.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukami K, Matsuoka K, Nakanishi O, Yamakawa A, Kawai S, Takenawa T. Antibody to PIP2 inhibits oncogene induced mitogenesis. Proc Natl Acad Sci USA. 1988;85:9057–9061. doi: 10.1073/pnas.85.23.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukami K, Furuhashi K, Inagaki M, Endo T, Hatano S, Takenawa T. Requirement of PIP2 for alpha-actinin function. Nature. 1992;359:150–152. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- 9.Fukami K, Endo T, Imamura M, Tekenawa T. Alpha-actinin and vinculin are PIP2-binding proteins involved in signaling by Tyr kinase. J Biol Chem. 1994;269:1518–1522. [PubMed] [Google Scholar]
- 10.Fukui Y, Lynch T, Brzeska H, Korn E D. Myosin I is located at the leading edges of locomoting Dictyostelium amoeba. Nature. 1989;341:328–331. doi: 10.1038/341328a0. [DOI] [PubMed] [Google Scholar]
- 11.Gibbs J B, Elis R, Scolnick E M. Autophosphorylation of v-Ha-Ras is modulated by amino acid 12. Proc Natl Acad Sci USA. 1984;81:2674–2678. doi: 10.1073/pnas.81.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimona M, Kazzaz J, Helfman D. Forced expression of tropo -myosin2 and 3 in v-Ki-Ras-transformed fibroblasts results in distinct phenotypic effects. Proc Natl Acad Sci USA. 1996;93:9618–9623. doi: 10.1073/pnas.93.18.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glueck U, Kwiatkowski D, Ben-Ze’ev A. Suppression of tumorigenicity in SV40-transformed 3T3 cells transfected with alpha-actinin cDNA. Proc Natl Acad Sci USA. 1993;90:383–387. doi: 10.1073/pnas.90.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwig J, Bokoch G M, Carpenter C L, Janmey P, Taylor L, Toker A, Stossel T. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 15.He H. HS1 and EMS1. Jpn J Cancer Chemother. 1997;24:1448–1453. [PubMed] [Google Scholar]
- 16.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sakaki T, Takai Y, Tsukita S, Tsukita S. Regulation mechanism of ERM (Ezrin/Radixin/Moesin) protein/plasma membrane association: possible involvement of PIP2 turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Ni Y, Wang T, Gao Y, Haudenschild C C, Zhan X. Down-regulation of F-actin cross-linking activity of cortactin/EMS1 by Src-mediated Tyr phosphorylation. J Biol Chem. 1997;272:13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- 18.Janmey P, Stossel T. Modulation of gelsolin function by PIP2. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 19.Janssen R, Mier J. Tropomyosin-2 cDNA lacking the 3′ untranslated region riboregulator induces growth inhibition of v-Ki-Ras-transformed fibroblasts. Mol Biol Cell. 1997;8:897–908. doi: 10.1091/mbc.8.5.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura D, Kaneko H, Miyazgoe Y, Ariyasu T, Watanabe T. Isolation and characterization of a novel human gene expressed specially in the cells of hematopoietic lineage. Nucleic Acids Res. 1989;17:9367–9379. [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar C C, Prorock-Rogers C, Kelley J, Dong Z, Lin J J, Armstrong L, Kung H F, Weber M J, Afonso A. SCH51344 inhibits Ras transformation by a novel mechanism. Cancer Res. 1995;55:5106–5117. [PubMed] [Google Scholar]
- 22.Lassing I, Lindberg U. Evidence that the phosphatidylinositol cycle is linked to cell motility. Exp Cell Res. 1988;174:1–15. doi: 10.1016/0014-4827(88)90136-x. [DOI] [PubMed] [Google Scholar]
- 23.Maruta H, Korn E D. Acanthamoeba myosin II. J Biol Chem. 1977;252:6501–6509. [PubMed] [Google Scholar]
- 24.Maruta H, Holden J, Sizeland A, D’Abaco G. The residues of Ras and Rap proteins that determine their GAP specificities. J Biol Chem. 1991;266:11661–11668. [PubMed] [Google Scholar]
- 25.Maruta H, He H. Cytoskeletal SH3 proteins. Biochemistry (Tokyo) 1995;67:1210–1217. [PubMed] [Google Scholar]
- 26.Maruta H. Downstream of RAS and actin-cytoskeleton. In: Maruta H, Burgess A W, editors. Regulation of the RAS signaling network. Heidelberg, Germany: Springer-Verlag; 1996. pp. 139–180. [Google Scholar]
- 27.Maruta H. F-actin cappers. Jpn J Cancer Chemother. 1997;24:1442–1447. [PubMed] [Google Scholar]
- 28.Moritz R L, Simpson R J. Application of capillary reversed-phase high-performance liquid chromatography to high-sensitive protein sequence analysis. J Chromatogr. 1992;599:119–130. doi: 10.1016/0021-9673(92)85464-5. [DOI] [PubMed] [Google Scholar]
- 29.Moritz R L, Eddes J, Reid G, Simpson R J. S-pyridylethylation of intact polyacrylamide gels and in situ digestion of electrophoretically separated proteins: a rapid mass spectrometric method for identifying cysteine-containing peptides. Electrophoresis. 1996;17:907–917. doi: 10.1002/elps.1150170512. [DOI] [PubMed] [Google Scholar]
- 30.Muellauer L, Fujita H, Shizaki A, Kuzumaki N. Tumor-suppressive function of mutated gelsolin in Ras-transformed cells. Oncogene. 1993;8:2531–2536. [PubMed] [Google Scholar]
- 31.Nur-E-Kamal M S A, Sizeland A, D’Abaco G, Maruta H. Asn26, Glu31, Val45, and Tyr64 of Ras proteins are required for their oncogenicity. J Biol Chem. 1992;267:1415–1418. [PubMed] [Google Scholar]
- 32.Pollard T D, Cooper J A. Actin and actin-binding proteins. A critical evaluation of mechanisms and function. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 33.Prasad G, Fuldner R, Cooper H. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the Ras oncogene. Proc Natl Acad Sci USA. 1993;90:7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu R G, Che J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Viciana P, Warne P, Khwaja A, Marte B, Pappin D, Das P, Waterfield M, Ridley A, Downward J. Role of PI-3 kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 36.Saez C G, Myers J, Shows T, Leinward L. Human non-muscle II heavy chain mRNA: generation of diversity through alternative polyadenylylation. Proc Natl Acad Sci USA. 1990;87:1164–1168. doi: 10.1073/pnas.87.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito Y, Umezawa K. Synthesis of echiguanine analogues and their ribofuranosyl glycosides that inhibit PI-4 kinase. Bioorg Med Chem Lett. 1997;7:861–864. [PubMed] [Google Scholar]
- 38.Sato N, Yonemura S, Obinata T, Tsukita S, Tsukita S. Radixin, an F-actin barbed end-capping protein, is concentrated at the cleavage furrow during cytokinesis. J Cell Biol. 1991;113:321–330. doi: 10.1083/jcb.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuuring E, Verhoeven E, Litvinov S, Michalides R. The product of the EMS1 gene, amplified in human carcinomas, is homologous to a Src substrate and is localized in cell-substratum contact sites. Mol Cell Biol. 1993;13:2891–2898. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons M, Wang M, McBride O, Kawamoto S, Yamakawa K, Gdula D, Adelstein R S, Weir L. Human non-muscle myosin II heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 41.Spudich J A, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of tropomyosin-troponin complex with actin and proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 42.Takemoto Y, Furuta M, Li X K, Strong-Sparks J, Hashimoto Y. LchBP/HS1, expressed in hematopoietic lineage cells, directly associates with the SH3 domain of Lck. EMBO J. 1995;14:3403–3414. doi: 10.1002/j.1460-2075.1995.tb07346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tikoo A, Ramesh V, Gusella J, Maruta H. Anti-Ras function of neurofibromatosis type 2 gene product (NF2/Merlin) J Biol Chem. 1994;269:23387–23390. [PubMed] [Google Scholar]
- 44.Toothaker L E, Gonalez D, Tung N, Lemons R, Lebeau M, Arnaout M, Clayton L, Tenen T. Cellular myosin II heavy chain in human leukocytes: isolation of 5′ cDNA clones, characterization of protein, chromosomal localization, and up-regulation during myeloid differentiation. Blood. 1991;78:1826–1833. [PubMed] [Google Scholar]
- 44a.van Damme, H., V. van Buuren, and E. Schuuring. Unpublished observation.
- 45.Vandekerckhove J, Bauw G, Vancompernolle A, Honore B, Celis J. Gelsolin as one of the most prominent down-regulated markers of transformed human fibroblast and epithelial cells. J Cell Biol. 1990;111:95–102. doi: 10.1083/jcb.111.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh A B, Dhanasekaran M, Bar-Sagi D, Kumar C C. SCH51344-induced reversal of RAS-transformation is accompanied by the specific inhibition of the RAS and RAC-dependent cell morphology pathway. Oncogene. 1997;15:2553–2560. doi: 10.1038/sj.onc.1201424. [DOI] [PubMed] [Google Scholar]
- 47.Wang D Z M, Nur-E-Kamal M S A, Tikoo A, Montague W, Maruta H. The GTPase and Rho GAP domains of p190-A, a tumor suppressor protein that binds the Ras GAP of 120 kDa, independently function as anti-Ras tumor suppressors. Cancer Res. 1997;57:2478–2484. [PubMed] [Google Scholar]
- 48.Weber K, Lazarides E, Goldman R, Vogel A, Pollack R. Localization and distribution of actin fibers in normal, transformed and revertant cells. Cold Spring Harbor Symp Quant Biol. 1974;39:363–369. doi: 10.1101/sqb.1974.039.01.047. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, Parsons T. Cortactin, a Src substrate of 80/85 kDa, is an F-actin binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yahara I, Harada F, Sekita S, Yoshihira K, Natori S. Correlation between effects of 24 different cytochalasins on cellular structures and events and those on actin in vitro. J Cell Biol. 1982;92:69–78. doi: 10.1083/jcb.92.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamanashi Y, Okada M, Semba T, Yamori T, Umemori H, Tsunasawa S, Toyoshima K, Kitamura D, Watanabe T, Yamamoto T. Identification of HS1 as a major substrate of Tyr kinase(s) upon B-cell antigen receptor-mediated signaling. Proc Natl Acad Sci USA. 1993;90:3631–3635. doi: 10.1073/pnas.90.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanashi Y, Fukuda T, Nishizumi H, Ianazu T, Higashi K, Kitamura D, Ishida T, Yamamura H, Watanabe T, Yamamoto T. Role of Tyr phosphorylation of HS1 in B-cell antigen receptor-mediated apoptosis. J Exp Med. 1997;185:1387–1392. doi: 10.1084/jem.185.7.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yonezawa N, Homma Y, Yahara I, Sakai H, Nishida E. A short sequence responsible for both PIP2 binding and actin binding activities of cofilin. J Biol Chem. 1991;266:17218–17221. [PubMed] [Google Scholar]
- 54.Yu F X, Sun H Q, Janmey P, Yin H L. Identification of a PIP2-binding sequence in a G-actin binding domain of gelsolin. J Biol Chem. 1992;267:14616–14621. [PubMed] [Google Scholar]