Abstract
Many physiological osteocalcin-regulated functions are affected in adult offspring of mothers experiencing unhealthy pregnancy. Furthermore, osteocalcin signaling during gestation influences cognition and adrenal steroidogenesis in adult mice. Together these observations suggest that osteocalcin may broadly function during pregnancy to determine organismal homeostasis in adult mammals. To test this hypothesis, we analyzed in unchallenged wildtype and Osteocalcin-deficient, newborn and adult mice of various genotypes and origin maintained on different genetic backgrounds, the functions of osteocalcin in the pancreas, liver and testes and their molecular underpinnings. This analysis revealed that providing mothers are Osteocalcin-deficient, Osteocalcin haploinsufficiency in embryos hampers insulin secretion, liver gluconeogenesis, glucose homeostasis, testes steroidogenesis in adult offspring; inhibits cell proliferation in developing pancreatic islets and testes; and disrupts distinct programs of gene expression in these organs and in the brain. This study indicates that osteocalcin exerts dominant functions in most organs it influences. Furthermore, through their synergistic regulation of multiple physiological functions, osteocalcin of maternal and embryonic origins contributes to the establishment and maintenance of organismal homeostasis in newborn and adult offspring.
Keywords: Osteocalcin, Developmental Effect, Postnatal Physiology
Subject terms: Development, Metabolism
Synopsis

In three distinct Osteocalcin-deficient mouse strains, osteocalcin signaling during pregnancy determines homeostasis in adult offspring.
Osteocalcin functions are dominant.
Maternal and embryonic osteocalcin influences homeostasis postnatally.
In three distinct Osteocalcin-deficient mouse strains, osteocalcin signaling during pregnancy determines homeostasis in adult offspring.

Introduction
From energy metabolism to fertility and from musculoskeletal functions to behaviors, hormones regulate most physiological functions in adult mammals. As a result, they are essential contributors to the establishment and maintenance of organismal homeostasis (Drucker, 2018; Friedman, 2019; Kliewer and Mangelsdorf, 2019; Kreymann et al, 1987; Lo et al, 2014; Wu et al, 1995). The observation that children born from mothers who experienced an unhealthy pregnancy have a high propension to develop metabolic, endocrine, and neuropsychiatric diseases later in life (Burger et al, 1948; Hales and Barker, 2001) has long suggested that hormones made in the mother and crossing the placenta and/or made by the embryo might influence energy metabolism and other physiological processes in adult progenies (Chida et al, 2007; Choi et al, 2016; Karpac et al, 2005). Addressing this critical question of metabolism and endocrinology has been challenging because several hormones that could explain such an influence of gestation on postnatal energy metabolism and organismal homeostasis are either necessary for life (insulin), fertility (leptin), and/or are synthesized by the placenta (leptin, steroid hormones) (Accili et al, 1996; Ashworth et al, 2000; Hummel et al, 1966; Osinski, 1960; Oury et al, 2013b).
Osteocalcin is a peptide hormone that, following its binding to one of its three known receptors, regulates a broad array of physiological functions (Karsenty and Ferron, 2012; Karsenty and Olson, 2016; Pi et al, 2021; Pi et al, 2017). Those include glucose homeostasis, energy expenditure, exercise capacity, electrolyte homeostasis, blood pressure, male fertility, cognition, anxiety, and the acute stress response (Berger et al, 2019; De Toni et al, 2014; Glatigny et al, 2019; Gupte et al, 2014; Lee et al, 2007; Mao et al, 2021; Mera et al, 2016a; Oury et al, 2013b; Pi et al, 2016; Qian et al, 2021; Yadav et al, 2022). Remarkably, many physiological processes that are regulated by osteocalcin such as glucose homeostasis, cognition and even fertility are hampered in adult animals born from mothers experiencing an unhealthy pregnancy (Burger et al, 1948; Lee et al, 2007; Oury et al, 2013b; Oury et al, 2011). Furthermore, by signaling through the same one of the three osteocalcin receptors, Gpr158, in the adrenal glands and developing brain, the embryonic and maternal (i.e., developmental) pools of osteocalcin regulate adrenal steroidogenesis and behaviors in adult offspring (Oury et al, 2013b; Yadav et al, 2022). When considered together, these observations raise the prospect that osteocalcin of maternal and/or embryonic origins may be a more significant and global regulator of physiological functions and organismal homeostasis in adult offspring than anticipated. Moreover, the facts that osteocalcin is not synthesized in the placenta but crosses it and that inactivation of Osteocalcin does not cause perinatal lethality or sterility (Oury et al, 2011) make this hormone a good candidate to assess, in a proof-of-principle study, to what extent the functions of hormones during gestation influence organismal homeostasis in adult animals. Moreover, the fact that osteocalcin of both maternal and embryonic origins contribute to post-natal physiology allows one to further refine the question, and through rigorous crosses to determine which functions of osteocalcin are of maternal origin and which ones are of embryonic origins (Oury et al, 2013b; Yadav et al, 2022).
We addressed this question through physiological, cellular, and molecular means in newborn and adult Osteocalcin (Ocn)-deficient mice of different genotypes or origins and maintained on diverse genetic backgrounds. This analysis revealed that if mothers are Ocn haplo-insufficient, haploinsufficiency for Ocn in embryos hampers cell proliferation in developing pancreas and testes, testes steroidogenesis, insulin secretion, liver gluconeogenesis and altogether glucose homeostasis in newborn and adult heterozygous offspring. Molecular analyses revealed that, providing that mothers are Osteocalcin-deficient, Osteocalcin haploinsufficiency in offspring affects in testes, liver and pancreas, where osteocalcin signals through the same receptor, Gprc6a, programs of gene expression that differ from those affected in the brain, an organ in which osteocalcin signals through different receptors. Furthermore, there is also an influence of embryonic osteocalcin on physiology postnatally. Hence, an interplay between Osteocalcin expression in mothers and embryos contributes to determine multiple physiological functions and homeostasis in offspring.
Results
Definition of the models and genotypes used in this study
To test whether maternal and/or embryonic osteocalcin exerts regulatory functions during pregnancy that affect physiology in adult offspring we analyzed Osteocalcin (Ocn)+/− mice born from either WT, Ocn+/− or Ocn−/− mothers using as negative controls WT mice born from WT mothers and as positive controls Ocn−/− mice born from Ocn+/− or Ocn−/− mothers (Fig. 1A). We also analyzed Ocn+/− mice born from WT mothers. Our reasoning in analyzing offspring of these various crosses was the following. First, if an osteocalcin-regulated function is affected in adult Ocn+/− offspring born from either Ocn+/− or Ocn−/− mothers, it shows that Osteocalcin haploinsufficiency suffices to hamper this function, in other words that this function is dominant. Second, if Ocn+/− mice born from Ocn+/− or Ocn−/− mothers display the phenotype and/or molecular perturbations of interests but Ocn+/− mice born from WT mothers do not, it indicates that maternal but not embryonic osteocalcin determines in part the extent of this function in adult offspring. Third, if this osteocalcin-regulated function or molecular event is affected in Ocn+/− mice born from WT mothers, it reveals an influence of embryonic osteocalcin in setting up this function in adult mice. Fourth, if this osteocalcin-regulated function is equally affected in Ocn+/− mice whether they are born from Ocn+/− or Ocn−/− mothers, it indicates that a single allele of maternal Ocn is insufficient to rescue the deleterious consequences of Ocn haploinsufficiency in embryos on this function. The same is true if Ocn−/− mice born from Ocn+/− or Ocn−/− mothers have a phenotype of equal severity.
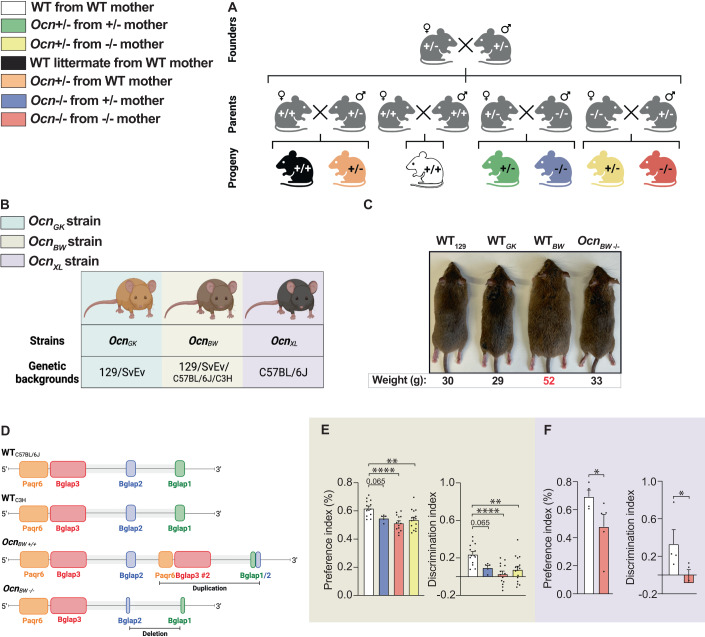
Figure 1. Strategy used to determine the role of gestational osteocalcin in regulating adult physiological functions.
(A) Schematic representation of the breeding strategy used for this study. Ocn+/+, Ocn+/− or Ocn−/− mothers were crossed with Ocn+/+, Ocn+/−, Ocn−/− or Ocn+/− fathers to generate progenies (Ocn+/+, Ocn+/−, or Ocn−/−) for analysis. (B) Ocn-deficient mouse strains (OcnGK, OcnBW and OcnXL) and corresponding genetic backgrounds. (C) Photomicrographs of Ocn-deficient mouse strains and corresponding genetic backgrounds illustrating an obese phenotype in WT mice of OcnBW strain. Body weight of corresponding genotypes is indicated below each photomicrograph. (D) Genome organization at the Ocn locus in WT mice of OcnBW strain. (E) Preference index [left panel] and discrimination index in a novel object recognition test [right panel] in female mice of various genotypes of the OcnBW strain. n = 4 or more mice per genotype analyzed. (F) Preference index [left panel] and discrimination index in a novel object recognition test [right panel] in female mice of various genotypes of the OcnXL strain. n = 3 or more mice per genotype analyzed. Data information: In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05, **p < 0.01, ****p < 0.0001. Source data are available online for this figure.
To reach conclusions of broad physiological significance, this analysis was performed in mice unchallenged by any diet manipulation and in up to three Ocn-deficient mouse strains generated through different means, maintained on distinct genetic backgrounds and studied at two different sites (Vagelos College of Physicians and Surgeons, New York City, USA; Chinese Academy of Science, Shenzen, China) (Fig. 1B). The first strain, OcnGK, was generated through homologous recombination in embryonic stem cells and has been maintained on a 129/SvEv genetic background since 1995 (Ducy et al, 1996). The genomic architecture of the Osteocalcin locus in this mouse strain has been previously reported (Ducy et al, 1996). The second strain termed OcnBW was generated by CRISPR-Cas9 genome editing on a mixed C57BL/6 J/C3H background, backcrossed twice on C57BL/6J background and then four times on a 129/SvEv background (Diegel et al, 2020). Hence, OcnBW mice are on a mixed 129/SvEv with components of C57BL/6J and C3H background. Analysis of energy metabolism in this strain was affected by the existence of an overt obesity restricted to WT mice (Figs. 1C and EV1A,B). A whole genome sequencing analysis initiated to identify molecular bases of this obesity revealed the presence of over 11,000 insertions or deletions that were over 50 base-pairs (bp) long, and of over a million smaller length variants in the genome of WT OcnBW mouse when compared to the one of a WT 129/SvEv mouse (Fig. 1D). Conceivably, any of these changes might contribute to the obesity of these mice. Hence, to insure that the results we would obtain when analyzing glucose homeostasis parameters in this strain will not be confounded by the obesity of WT mice, we used two types of control mice, WT OcnBW mice and WT129/SvEv mice. For all other physiological functions analyzed in this strain we used WT OcnBW mice as controls. The third strain, OcnXL, also generated by CRISPR-Cas9 genome editing, has been maintained on a C57BL/6J background for over 10 generations (Qian et al, 2021). The sequence of the Osteocalcin locus in this strain is available on the National Center for Biotechnology Information web site. As for the OcnGK strain, WT mice in this strain were lean and therefore used as controls (Qian et al, 2021). OcnXL−/− mice were analyzed in the Chinese Academy of Science for static metabolic tests. In each strain, the absence of circulating osteocalcin in Ocn−/− mice was verified (Figure EV1E–G).
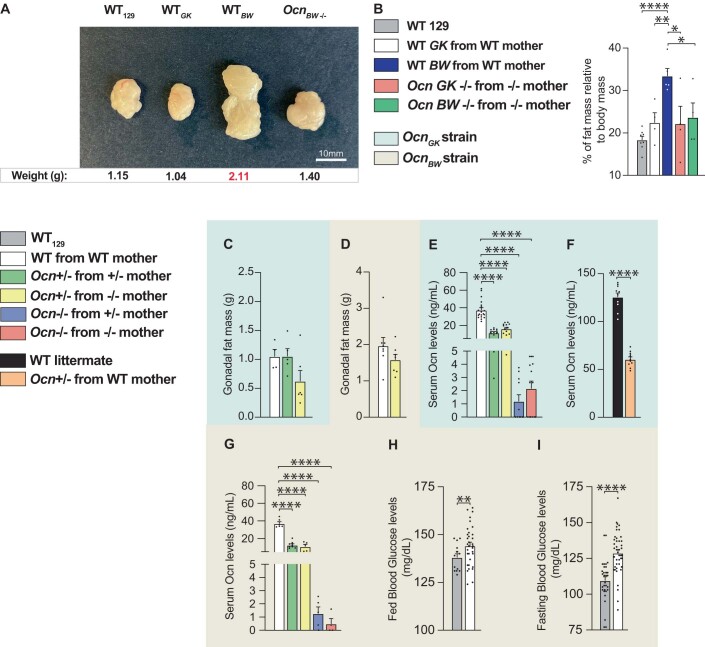
Figure EV1. Characterization of three mouse models of Ocn deletion.
(A) Photomicrographs of gonadal fat pads from WT129, WTBW, and OcnBW−/− mice. Weight for each group is shown below each representative fat pad image. (B) Percent fat mass relative to body weight in WT129, WTBW, and OcnBW−/− mice assessed by EchoMRI. n = 4 or more mice per genotype analyzed. (C, D) Gonadal fat mass in different genotypes of 12 weeks-old OcnGK mice (C). n = 4 or more mice per genotype analyzed. Gonadal fat mass in different genotypes of 12 weeks-old OcnBW mice (D). n = 7 mice per genotype analyzed. (E–G) Serum Osteocalcin levels in different genotypes of 12 weeks-old (E) and 5 weeks-old (F) OcnGK mice. n = 11 or more mice per genotype analyzed. Osteocalcin levels in different genotypes of 12 weeks-old OcnBW mice (G). n = 4 or more mice per genotype analyzed. (H, I) Fed (H) and fasting (I) glucose levels in OcnBW WT and WT129 mice. n = 14 or more mice per genotype analyzed. In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05; **p < 0.01, ****p < 0.0001. Source data are available online for this figure.
Prior to initiate a broad and time-consuming phenotypic analysis we verified that regardless of their mode of generation, genetic background, or body weight, disruption of a function of osteocalcin could be detected in these three mutant mouse strains. Specifically, we found that as seen in OcnGK mice, spatial learning and memory were significantly decreased in adult OcnBW+/− (Fig. 1E) and OcnXL−/− (Fig. 1F) mice born from their respective Ocn−/− mothers when analyzed by a novel object recognition test (Oury et al, 2013b).
The osteocalcin genotype of the mothers determines whether Osteocalcin haploinsufficiency in offspring hampers glucose homeostasis
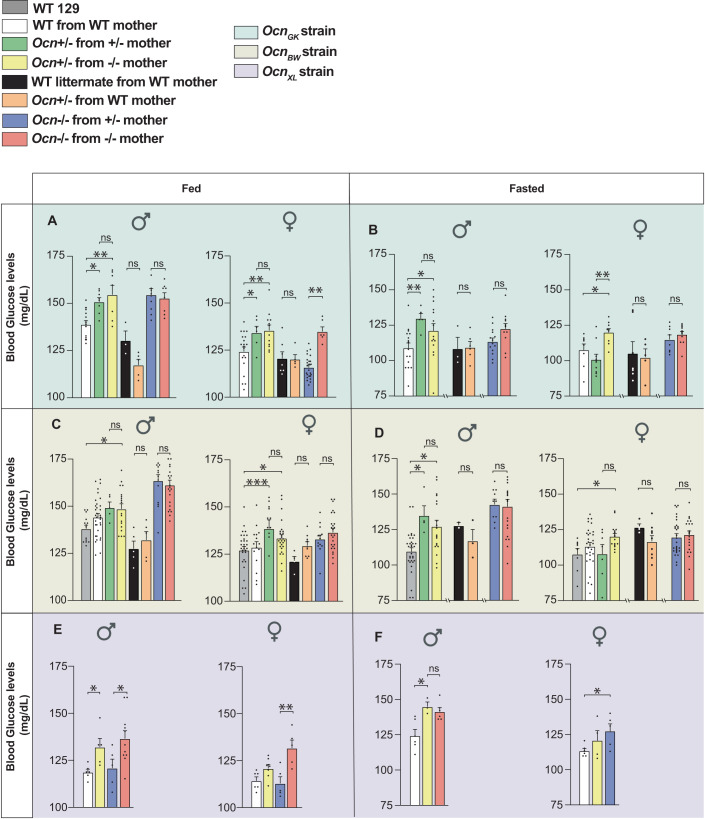
The first physiological function we assayed to look for a possible influence of osteocalcin of maternal and/or embryonic origin on adult physiology was glucose homeostasis as assessed by the measurement of random fed and fasting blood glucose levels in adult (3-month-old) mice (Lee et al, 2007). We observed that random fed blood glucose levels were significantly higher in OcnGK+/− mice born from OcnGK+/− and OcnGK−/− mothers than in control WT mice whereas OcnGK+/− mice born from WT mothers were normoglycemic (Fig. 2A,B). These data revealed that Ocn haploinsufficiency in offspring results in hyperglycemia only if their mothers also lack at least one allele of Ocn. When mothers were WT, the amount of maternal osteocalcin passing through the placenta sufficed to prevent any deleterious and long-lasting effect on glucose homeostasis post-natally that Ocn haploinsufficiency in offspring may have caused. In adult OcnGK−/− mice, the hyperglycemia was also of similar severity whether their mothers were OcnGK+/− or OcnGK−/− (Fig. 2A,B), verifying that a single Ocn allele in mothers cannot overcome the deleterious consequences on glucose homeostasis in offspring resulting from the absence of Ocn expression in embryos.
Figure 2. Osteocalcin of developmental origin regulates blood glucose levels in adult mice.
(A, C, E) fed and (B, D, F) fasting blood glucose levels in male and female mice from various genotypes of the OcnGK (A, B). n = 3 or more mice per genotype analyzed. OcnBW (C, D). n = 3 or more mice per genotype analyzed. OcnXL strains (E, F). n = 5 or more mice per genotype analyzed. Data information: In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are available online for this figure.
This analysis was extended to other Ocn-deficient strains. As anticipated given their obesity, WT OcnBW mice were hyperglycemic compared to WT 129 Sv/Ev mice and could not be used as controls (Figure EV1H,I). When WT129Sv/Ev mice bought from a vendor were instead used as controls, we observed that OcnBW+/− mice born from OcnBW+/− or OcnBW−/− mothers were hyperglycemic whereas OcnBW+/− mice born from WT mothers were not (Fig. 2C,D). Thus, in this mouse strain too, Ocn haploinsufficiency impairs glucose homeostasis in adult offspring only if their mothers are Ocn-deficient. As observed in the OcnGK strain too, OcnBW−/− mice showed equally high fasting and random fed blood glucose levels whether they were born from OcnBW+/− or OcnBW−/− mothers, illustrating that maternal osteocalcin cannot alleviate the deleterious consequences that the total absence of Ocn expression in embryos has on glucose homeostasis in adult offspring (Fig. 2C,D). In the third strain, the OcnXL-deficient mice (Qian et al, 2021), blood glucose levels were higher in OcnXL−/− and OcnXL+/− mice born from OcnXL−/− mothers than in WT mice (Fig. 2E,F). These dominant abnormalities of glucose homeostasis were not observed when we studied fat mass. Indeed, Ocn+/− mice of either strain tested did not harbor an increase in the weight of gonadal fat pads (Figure EV1C,D).
The osteocalcin genotype of the mothers determines whether Osteocalcin haploinsufficiency in offspring hampers insulin secretion and β-cell proliferation
Osteocalcin regulates glucose homeostasis in part by signaling via its receptor Gprc6a in pancreatic β-cells to enhance insulin secretion and proliferation of these cells (Ferron et al, 2008; Lee et al, 2007; Pi et al, 2011; Sabek et al, 2015; Wei et al, 2014). Thus, we asked whether these two parameters are affected in Ocn-deficient offspring depending on the Osteocalcin genotype of the mother.
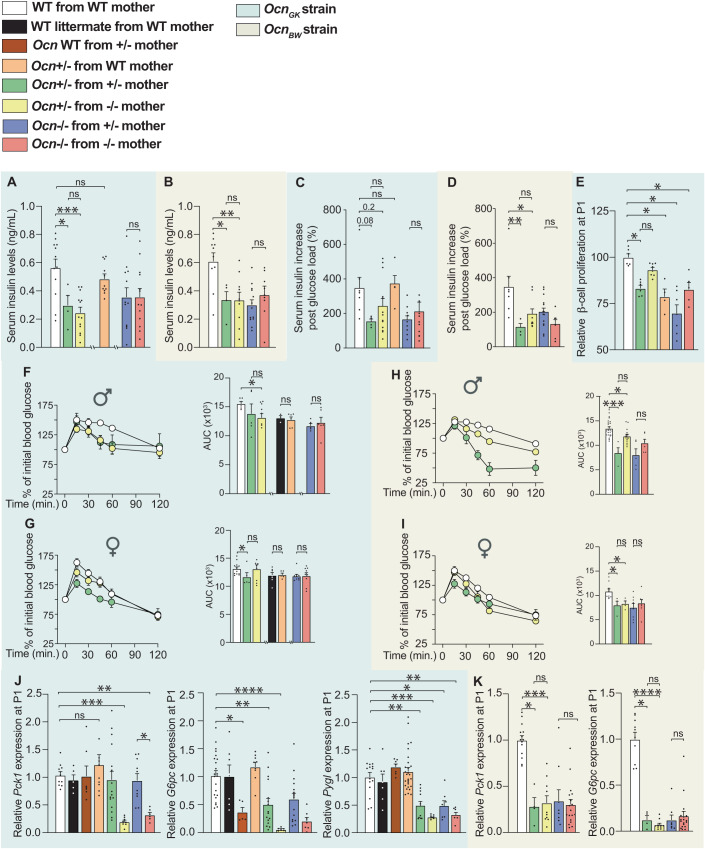
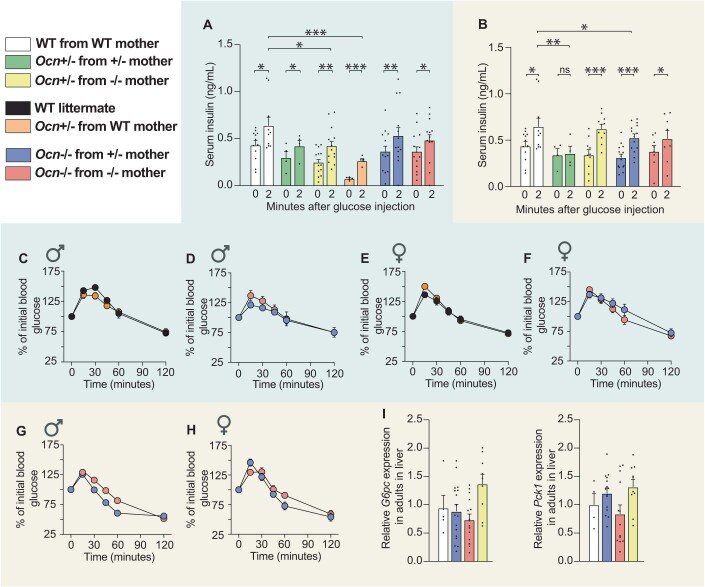
In both the OcnGK and OcnBW strains, fasting circulating insulin levels were significantly lower in adult Ocn+/− mice whether they were born from Ocn+/− or Ocn−/− mothers compared to what it was in WT mice (Fig. 3A,B). In contrast and as observed for blood glucose levels, this decrease in circulating insulin levels was not observed if Ocn+/− mice were born from WT mothers (Fig. 3A). These observations verified that Ocn haploinsufficiency in embryos affects circulating insulin levels in adult offspring only if their mothers are also Ocn-deficient. In both strains, circulating insulin levels were equally decreased in adult Ocn−/− offspring whether they were born from Ocn+/− or Ocn−/− mothers (Fig. 3A,B). The fact that circulating insulin levels are decreased by half in OcnBW−/− mice provides a plausible element of explanation for why the obesity characterizing WT OcnBW mice is not observed in mutant mice of these strain. In agreement with these results, glucose-stimulated insulin release was consistently hampered, albeit these differences did not reach statistical significance because of variability between samples, in OcnGK+/− mice born from OcnGK+/− or OcnGK−/− mothers but not in those born from OcnGK WT mice (Figs. 3A and EV2A). Adult OcnGK−/− mice were equally poor glucose responder whether their mothers were Ocn+/− or Ocn−/− in both the OcnGK and OcnBW strains (Figs. 3C,D and EV2A,B).
Figure 3. Osteocalcin of developmental origin influences insulin secretion, b-cell proliferation and liver gluconeogenesis in newborn and adult mice.
(A, B) Fasting serum insulin levels in mice of different genotypes of the OcnGK mouse strain (A). n = 4 or more mice per genotype analyzed. OcnBW mouse strain (B). n = 4 or more mice per genotype analyzed. (C, D) Relative increases in serum insulin levels between baseline and 2 minutes after glucose administration in a glucose stimulated insulin secretion test in male mice of various genotypes in OcnGK mouse strain (C). n = 4 or more mice per genotype analyzed. OcnBW mouse strain (D). n = 3 or more mice per genotype analyzed. Values were normalized to WT values for comparison. (E) Relative β-cell proliferation in pancreas at postnatal day 1 (P1) pups of different genotypes in OcnGK mouse measured after Insulin/BrdU immunostaining. Values were normalized to WT values for comparison. n = 4 or more mice per genotype analyzed. (F–I) Percent of initial blood glucose levels and corresponding area under the curve (AUC) in male (F, H) and female (G, I) mice of different genotypes in the OcnGK mouse strain (F, G). n = 3 or more mice per genotype analyzed. OcnBW (H, I) mouse strain following a Pyruvate tolerance test. n = 4 or more mice per genotype analyzed. (J) Relative Pck1, G6pc, and Pygl expression in the liver from P1 pups of different genotypes in the OcnGK strain. n = 5 or more mice per genotype analyzed. (K) Pck1 and G6pc relative expression in the liver of P1 pups in the OcnBW strain. n = 4 or more mice per genotype analyzed. Data information: In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, ns: not significant. Source data are available online for this figure.
Figure EV2. Dynamic analysis of glucose metabolism in Ocn-deficient mouse strains.
(A, B) Serum insulin levels at 0 and 2 min after glucose injection in glucose stimulated insulin secretion test in mice of indicated genotypes of OcnGK mouse strain (A). n = 4 or more mice per genotype analyzed. Serum insulin levels at 0 and 2 min after glucose injection in glucose stimulated insulin secretion test in mice of indicated genotypes of OcnBW (B) mouse strain. n = 7 or more mice per genotype analyzed. (C–H) Serum glucose levels at different time points post injection of pyruvate in a pyruvate tolerance test in mice of indicated genotypes in the OcnGK (C–F) mouse strain. Serum glucose levels at different time points post injection of pyruvate in a pyruvate tolerance test in mice of indicated genotypes in OcnBW (G, H) mouse strain. n = 6 or more mice per genotype analyzed. (I) Relative Pck1 and G6pc expression in the liver from adult mice of different genotypes in the OcnBW strain. n = 4 or more mice per genotype analyzed. In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001 ns: not significant. Source data are available online for this figure.
To assess whether maternal osteocalcin influences the ability of adult Ocn-deficient offspring to secrete insulin because it affects β-cell endowment, we performed in vivo BrdU labeling in newborn mice (P1) of various genotypes and quantified β-cell proliferation by double insulin/BrdU immunohisto-fluorescence (IHF). We found that β-cell proliferation was significantly decreased in pancreata of OcnGK+/− newborn mice compared to what was seen in pancreata of WT pups regardless of the genotype of the mother (Fig. 3E). β-cell proliferation was also significantly decreased in pancreata of OcnGK−/− pups born from OcnGK+/− or OcnGK−/− mothers (Fig. 3E). Thus, as it is the case for blood glucose levels and insulin secretion, a single Ocn allele in the mother cannot rescue a defect in perinatal β-cell proliferation caused by Ocn haploinsufficiency (or complete absence) in the embryo.
Osteocalcin genotype of the mother determines whether osteocalcin haploinsufficiency in offspring hampers liver gluconeogenesis
Osteocalcin also regulates glucose homeostasis through insulin-independent mechanisms. In particular, osteocalcin signals in hepatocytes through Gprc6a to promote liver gluconeogenesis, a process that is inhibited by insulin signaling (Pi et al, 2020). Therefore, we asked whether liver gluconeogenesis in Ocn-deficient offspring is affected by the genotype of the mother.
In both the OcnGK and OcnBW strains, adult Ocn+/− born from WT mothers showed a similar increase of blood glucose levels as WT controls following a pyruvate challenge (Figs. 3F–I and EV2C,E). In contrast, when mothers were Ocn+/− or Ocn−/−, the increase in blood glucose levels during a pyruvate challenge was lower in adult Ocn+/− than in WT controls indicating that Ocn haploinsufficiency in the mother determines whether Ocn haploinsufficiency in the progeny will hamper liver gluconeogenesis (Fig. 3F–I). In contrast, as observed for blood glucose levels and insulin secretion, adult Ocn−/− mice experienced a defect in pyruvate stimulated blood glucose levels of similar severity whether their mothers were Ocn+/− or Ocn−/− (Figs. 3F–I and EV2D,F–H).
To ascertain the existence of a developmental defect, we also assessed in newborn WT and Ocn-deficient mice born from mothers of different genotypes the expression in the liver of Glycogen Phosphorylase that encodes an enzyme that breaks down glycogen (Pygl) and of Phosphoenolpyruvate carboxykinase (Pck1) and Glucose-6-phosphatase (G6pc), two genes that are necessary for liver gluconeogenesis and whose expression is regulated by signaling through Gprc6a, the receptor for osteocalcin (Pi et al, 2011). Expression of these 3 genes was decreased in the liver of Ocn+/− and Ocn−/− pups compared to WT only if mutant pups were born from Ocn-deficient mothers (Fig. 3J,K). These results confirmed that Ocn haploinsufficiency in embryos hampers liver gluconeogenesis and glucose availability intracellularly in adult offspring only if their mothers lack at least one allele of Ocn. This decrease in gene expression was more severe in Ocn+/− and Ocn−/− pups when mothers were Ocn−/− than when they were Ocn+/− (Fig. 3J,K). We also observed that G6pc expression and to a lesser extent Pepck expression were decreased in postnatal day 1 (P1) Ocn+/− pups born from WT mothers, an observation indicating that embryonic osteocalcin contributes to setting up the expression of these genes in newborn mice (Fig. 3K). The decrease in expression of these three genes was as or more severe in Ocn+/− P1 pups born from Ocn−/− mothers than in those born from Ocn+/− mothers, but was normal in adult Ocn+/− mice regardless of the genotype of their mothers (Compare Figs. 3J,K and EV2I). These results suggest that the abnormal gluconeogenesis observed in adult Ocn+/− mice is due to a deficit in osteocalcin signaling during pregnancy.
When taken together, the analysis of these various aspects of glucose homeostasis in Ocn-deficient mice of various genotypes, origins, and genetic backgrounds provide convincing evidence that maternal osteocalcin is a determinant of pancreatic β-cell proliferation, insulin secretion, liver gluconeogenesis and overall, glucose homeostasis in adult mice provided it is delivered to embryos at WT levels.
The osteocalcin genotype of the mother determines whether osteocalcin haploinsufficiency in offspring hampers testes steroidogenesis
Osteocalcin signaling through Gprc6a in Leydig cells of the testes promotes testosterone biosynthesis in cell culture and maintains normal circulating testosterone levels in adult male mice and humans (Oury et al, 2013a; Oury et al, 2011). At the time these data were collected, it was not known that Osteocalcin haploinsufficiency in the mother may determine the consequence of Osteocalcin haplo-insufficiency on physiology in the offspring (Oury et al, 2013a; Oury et al, 2011). In view of the results observed when studying various aspects of the regulation of glucose homeostasis by osteocalcin, this became an important question to address.
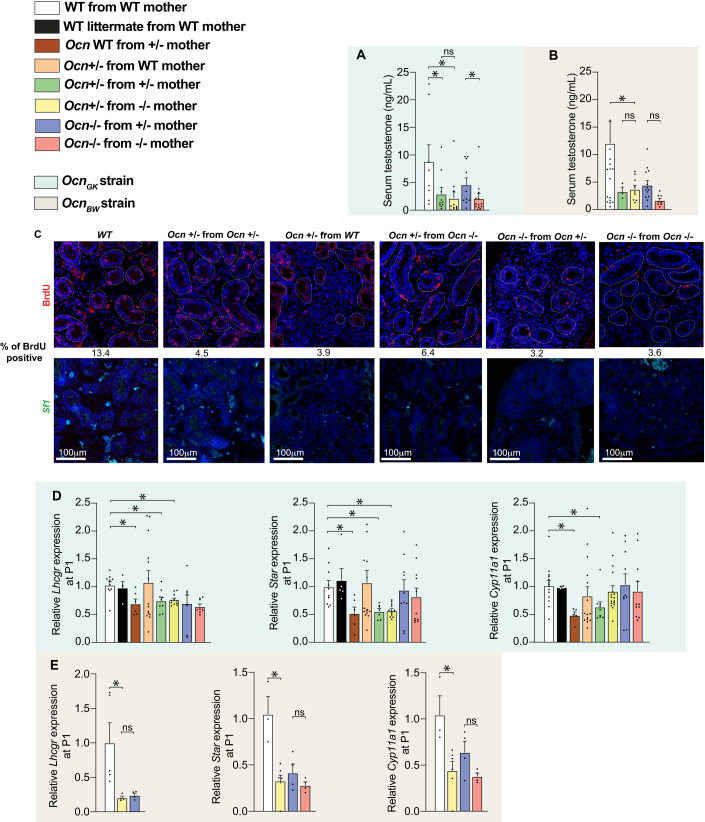
In both the OcnGK and OcnBW strains, circulating testosterone levels were significantly decreased in male adult Ocn+/− mice born from Ocn−/− mothers and to a lower extent in those born from Ocn+/− mothers thus verifying that Osteocalcin deficiency in mothers determines whether Ocn haploinsufficiency in the adult offspring hampers testes endocrine functions (Fig. 4A,B). The same was true for adult Ocn−/− mice whether they were born from Ocn+/− or Ocn−/− mothers (Fig. 4A,B). To assess whether this effect of Ocn haploinsufficiency in mothers and embryos on testes steroidogenesis originates in part from a defect of Leydig cell proliferation, we performed in vivo BrdU labeling in newborn mice and quantified Leydig cell proliferation on testes sections using analysis of Steroidogenic Factor 1 (Sf1) expression as a molecular marker of these cells. This analysis revealed that Leydig cell proliferation was markedly decreased in testes of OcnGK+/− or OcnGK−/− newborn mice regardless of the genotype of the mother (Fig. 4C). These data indicate that Ocn haploinsufficiency in embryos alone hampers proliferation of Leydig cells.
Figure 4. Osteocalcin of developmental origin influences testicular steroidogenesis in newborn and adult mice.
(A, B) Relative testosterone levels in adult mice of different genotypes in OcnGK mouse strain (A). n = 8 or more mice per genotype analyzed. OcnBW (B) mouse strain. n = 3 or more mice per genotype analyzed. Results were normalized to WT levels for comparison. (C) BrdU immunostaining and quantification of % BrdU positive cells in testes at P1 in pups of different genotypes in OcnGK [top panel] mouse strain. In situ hybridization analysis of Sf1 in testes at P1 pups of different genotypes in OcnGK [bottom panel] mouse strain. (D, E) Relative Lhcgr, Star and Cyp11a1 expression in testes at P1 of different genotypes in OcnGK (D) mouse strain. n = 4 or more mice per genotype analyzed. OcnBW (E) mouse strain. n = 4 or more mice per genotype analyzed. Data information: In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05; ns: not significant. Source data are available online for this figure.
Thus, to determine whether maternal osteocalcin also influences testes development in embryos, we measured expression of Steroidogenic acute regulatory protein (Star) and Cytochrome P450 family 11 subfamily A member 1 (Cyp11a1), two genes that are necessary for testosterone biosynthesis and whose expression is regulated by Osteocalcin (Oury et al, 2011), and of Luteinizing hormone/choriogonadotropin receptor (Lhcgr), which encodes the receptor of the pituitary hormone luteinizing hormone (LH) that is the canonical regulator of testosterone biosynthesis in Leydig cells (Zirkin and Papadopoulos, 2018). We observed that the expression of Star and Cyp11a1 were decreased in testes of newborn male Ocn+/− pups born from Ocn+/− or Ocn−/− mothers but not in those born from WT mothers (Fig. 4D,E). These results support the notion that Ocn expression in mothers and embryos are both necessary and synergize to regulate testes development and thereby of testosterone biosynthesis in offspring.
Genetic pathways regulated by developmental osteocalcin in testes
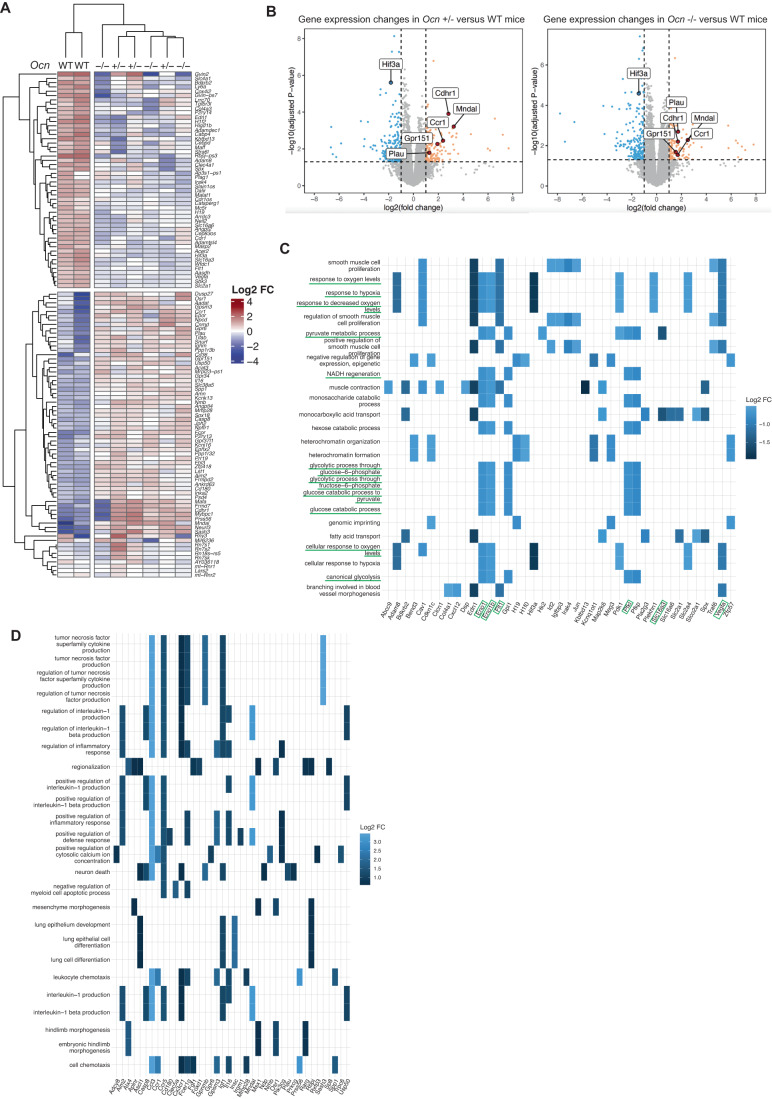
The observation that osteocalcin of embryonic and maternal origins cooperates to determine the quality of physiological processes taking place in the pancreas, liver, and testes of adult offspring, raises the question of the molecular modes of action of osteocalcin during gestation. To begin addressing this question, we performed an RNA sequencing (RNA-seq) analysis in testes of newborn WT mice born from WT mothers and in those of newborn Ocn+/− and Ocn−/− mice born from Ocn−/− mothers. Given that circulating testosterone levels were similarly decreased in Ocn+/− mice in both the OcnGK- and OcnBW-deficient mouse strains, this analysis was performed only in the former strain.
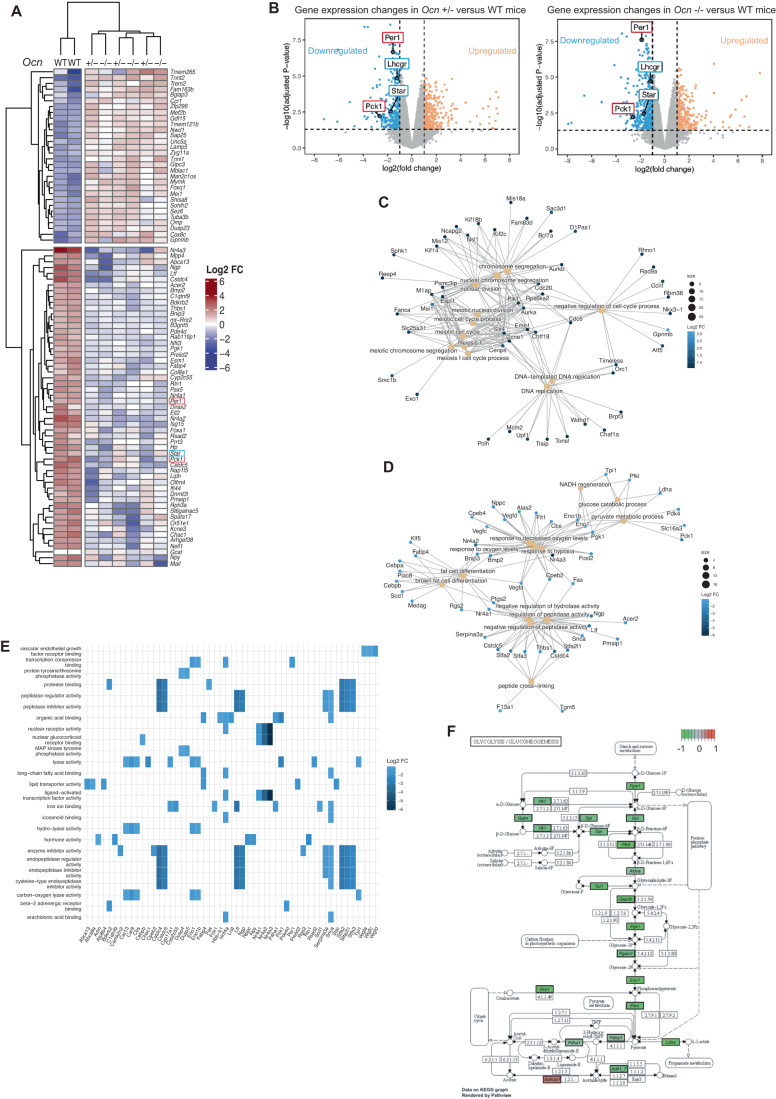
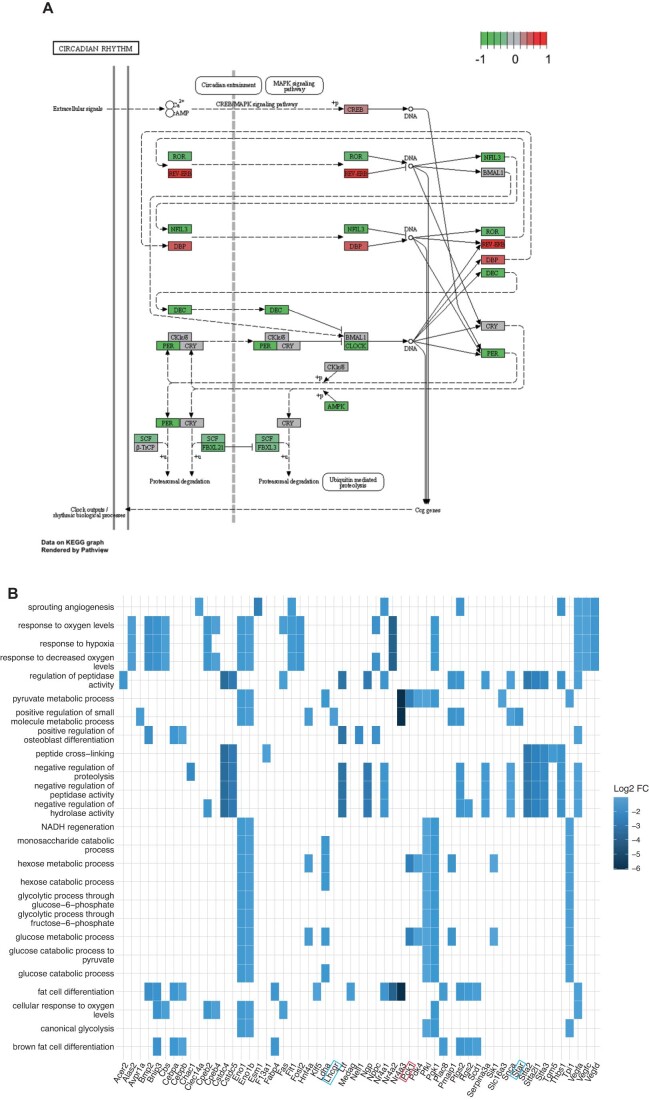
Consistent with results obtained when analyzing testes functions in Ocn+/− and Ocn−/− adult mice, a clustering of the most dysregulated genes in testes of Ocn+/− and Ocn−/− newborn mice compared to those of WT ones, failed to distinguish heterozygotes from homozygotes samples (Fig. 5A). Among the most downregulated genes in testes of both Ocn+/− and Ocn−/− newborn mice we found, as expected, Star and Lhcgr, two genes involved in testes steroidogenesis (Fig. 5B). Moreover, a gene ontology (GO) term analysis of the genes most significantly downregulated (FC < 0.5) identified a decrease in the expression of markers of hormonal activity in testes of both Ocn+/− and Ocn−/− newborn mice (GO:0005179) (Fig. 5C–E). Those include Adrenomedullin (Adm) and Natriuretic peptide C (NppC) that have been implicated in Leydig cell functions and testes development, respectively (Chan et al, 2008; Xia et al, 2007; Fig. 5C–E). We also observed that expression of Period circadian protein homolog 1 (Per1), a core gene involved in circadian rhythm, and of Pck1, a main control point for gluconeogenesis, was decreased in testes of Ocn+/− and Ocn−/− newborn mice (Figs. 5A,B and EV3A). In agreement with this latter observation, the GO term analyses based on the genes significantly downregulated (FC < 0.5) highlighted numerous pathways involved in glucose homeostasis such as glucose (GO:0006006), hexose (GO:0019318) and pyruvate (GO:0006090) metabolism (Figs. 5D and EV3B). Furthermore, a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis identified gluconeogenesis (mmu00010) as the most downregulated pathway in testes of both Ocn+/− (stat.mean = −3.6; p value = 3.9e-04, q-value = 3.4e-02) and Ocn−/− newborn mice (stat.mean = −3.6; p value = 2.2e-04, q-value = 5.0e-02) (Fig. 5F). Taken together these results indicate that, when mothers are Ocn-deficient, Ocn haploinsufficiency in embryos disrupts testes steroidogenesis, gluconeogenesis, and possibly, the molecular clock in newborn mice.
Figure 5. Transcriptomic analysis of osteocalcin signaling in testes of newborn mice of different genotypes.
(A) Heatmap illustrating results of unsupervised hierarchical clustering of testes obtained from WT (n = 2), Ocn+/− (n = 3) and Ocn−/− (n = 3) mice. (B) Volcano plots of the genes differentially expressed in testes from Ocn+/− [left panel] and Ocn−/− [right panel] mutant compared to WT mice. Each dot represents one gene or pseudogene (n = 3). (C, D) Category netplots depicting the linkages between the top 12 biological processes (GO term) and genes that are significantly (C) upregulated or (D) downregulated in the testes of both Ocn+/− and Ocn−/− compared to WT mice. (E) Heatplot of the top 25 GO molecular functions (GO term) that are significantly downregulated in both Ocn+/− and Ocn−/− compared to WT mice. (F) KEGG pathview graph of glycolysis/gluconeogenesis pathways comparing the transcriptomic data obtained from Ocn+/− mice to those from WT mice. Data information: Values close to −1 (green) indicate a downregulation of the gene in Ocn+/− compared to WT, values close to 0 (gray) indicate no difference and values close to 1 (red) indicate an upregulation in Ocn+/− compared to WT.
Figure EV3. KEGG analysis of transcriptomic studies of testes gene expression.
(A) KEGG pathview graph of the molecular clock comparing the transcriptomic data obtained from Ocn−/− mice to those from WT mice. Values close to −1 (green) indicate a downregulation of the gene in Ocn−/− compared to WT, values close to 0 (gray) indicate no difference and values close to 1 (red) indicate an upregulation in Ocn−/− compared to WT. (B) Heatplots of the top 25 GO biological processes (GO term) that are significantly downregulated in both Ocn+/− and Ocn−/− compared to WT mice. n = 2-3 mice per genotype analyzed.
Developmental osteocalcin regulates similar genetic pathways in the liver, pancreas, and testes
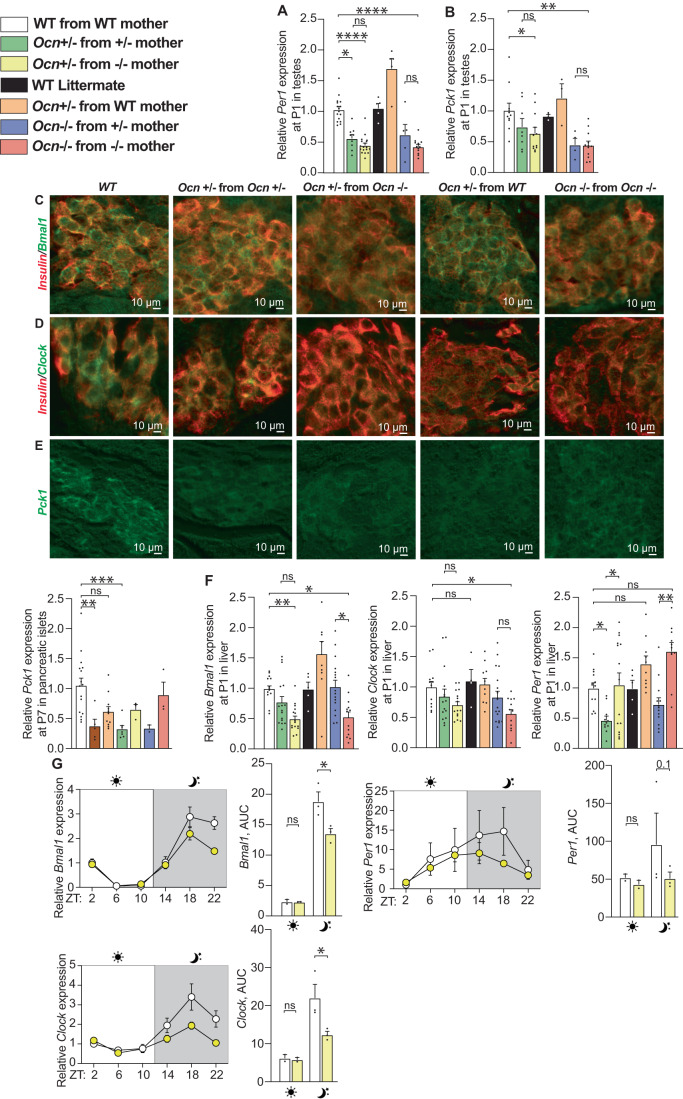
The results of the transcriptomic analysis performed in testes provided us with the opportunity to ask whether the same genes are regulated by the interplay between maternal and embryonic osteocalcin in the pancreas and the liver, two organs where osteocalcin also signals through Gprc6a. Prior to performing this analysis, we verified by qPCR that the expression of a few genes identified as dysregulated by the transcriptomic analysis were indeed affected in testes of Ocn-deficient newborn mice. This survey showed that expression of Per1 and Pck1 was significantly decreased in testes of Ocn−/− and Ocn+/− pups whether they were born from Ocn+/− or Ocn−/− mothers (Fig. 6A,B). In contrast, the expression of these two genes was not decreased in testes of Ocn+/− pups born from WT mothers (Fig. 6A,B). These results further establish that Ocn haploinsufficiency in embryo affects programs of gene expression in testes only if mothers are Ocn-deficient too.
Figure 6. Expression of components of molecular clock and a gluconeogenic enzyme in the pancreas, liver and testes depends on the Ocn genotype of the mother.
(A) Relative expression of circadian rhythm-related gene (Per1) in testes of P1 pups in different genotypes from the OcnGK mouse strain. n = 5 or more mice per genotype analyzed. (B) Relative expression of glucose homeostasis-related gene (Pck1) in testes at P1 pups of different genotypes from the OcnGK mouse strain. n = 4 or more mice per genotype analyzed. (C–E) Immunohistofluorescence analysis of Bmal1 (C), Clock (D), and Pck1 (E) levels in pancreata of P1 pups of different genotypes in the OcnGK strain. In (C, D), co-immunolocalization of insulin levels was used to identify β-cells. (F) Relative expression of circadian rhythm-related gene (Bmal1, Clock, and Per1) in liver of P1 pups of different genotypes in the OcnGK mouse strain. n = 4 or more mice per genotype analyzed. (G) Relative expressions and quantifications of area under the curve (AUC) of circadian rhythm-related genes Bmal1, Clock, Per1 during the course of 24 h in the liver of adult mice of different genotypes in the OcnGK strain. n = 3 or more mice per genotype analyzed. Data information: In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001, ns: not significant. Source data are available online for this figure.
Having verified the validity of the RNA-Seq study in testes, we turned our attention to the pancreas and liver. We first asked through immunofluorescence whether the accumulation of components of the molecular clock or implicated in gluconeogenesis in pancreatic islets of newborn mutant mice was modified (Fig. 6C–E). The absence of an anti-Per1 antibody that could be used on paraffin sections prevented the analysis of its expression, but the levels of Bmal1 and Clock, two other key components of the molecular clock, were lower in pancreatic islets of Ocn+/− pups born from Ocn+/− or Ocn−/− mothers than in those of WT pups (Fig. 6C,D). For Bmal1, the decrease in its accumulation was not seen in Ocn+/− pups born from WT mothers (Fig. 6C). The accumulation of Pck1 was also decreased in islets of Ocn+/− newborn mice regardless of the genotype of their mothers (Fig. 6E). These results are consistent with the notion that embryonic and maternal osteocalcin synergize to influence the expression of key components of the molecular clock and of the gluconeogenic pathway in pancreatic islets in newborn mice.
In the liver, we relied on gene expression analysis and observed that the expression of Per1, Bmal1 and Clock, was significantly decreased in newborn Ocn−/− and Ocn+/− mice born from Ocn−/− mothers (Fig. 6F). In contrast, only Per1 expression was significantly decreased in Ocn+/− mice born from Ocn−/− mothers suggesting a greater sensitivity of this gene’s regulation to osteocalcin levels (Fig. 6F). Expression of these three genes was normal in the liver of Ocn+/− newborn pups born from WT mothers, verifying that Ocn haploinsufficiency in embryos affects liver gene expression in offspring only if their mothers were Ocn-deficient too (Fig. 6F). We also observed that the rhythmic expression of Per1, Bmal1, and Clock was perturbed in the liver of adult Ocn+/− mice born from Ocn−/− mothers (Fig. 6G). These results are consistent with the notion that osteocalcin of maternal and embryonic origins synergizes to determine expression of the same key components of the molecular clock and gluconeogenic pathways in the liver, pancreas, and testes of newborn and adult offspring.
Developmental osteocalcin regulates distinct genetic pathways in the brain
The results presented above raised the question as to whether osteocalcin regulates the same genes and genetic pathways in all organs independently of the receptor through which it signals. To address this question, we performed an RNA-seq analysis in the forebrain (including the hippocampus) where osteocalcin signals through Gpr158 and Gpr37 (Khrimian et al, 2017; Qian et al, 2021). Here again, this analysis was performed in newborn WT, Ocn+/− and Ocn−/− mice born from Ocn−/− mothers.
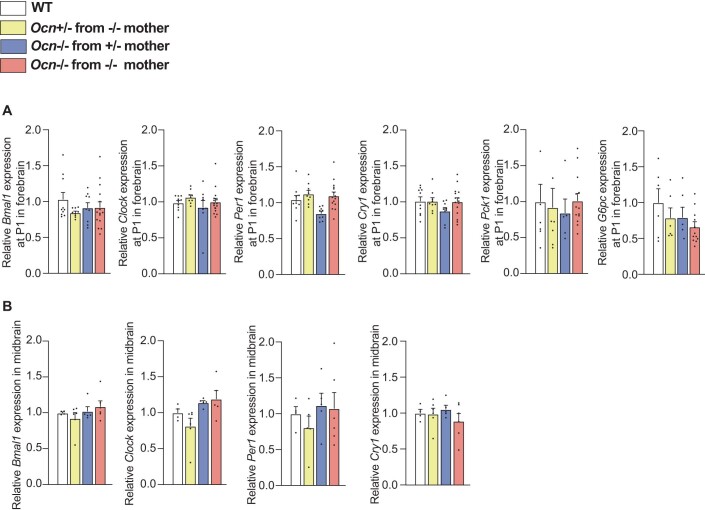
As it is the case in the testes, a clustering analysis based on the identification of the most dysregulated genes, failed to distinguish any differences between forebrains of Ocn+/− and Ocn−/− pups (Fig. 7A,B). The expression of Pck1 that was downregulated in testes, pancreas and liver of newborn Ocn-deficient pups was normal in their forebrain (Figure EV4A). Expression of key components of the molecular clock was also similar in the forebrain and adult midbrain of Ocn-deficient and WT newborn mice (Figure EV4A,B). These results suggested that osteocalcin regulates different genes and genetic pathways depending on the organ where it signals.
Figure 7. Transcriptomic analysis of osteocalcin signaling in the forebrain of newborn mice of different genotypes.
(A) Heatmap illustrating results of unsupervised hierarchical clustering of forebrains obtained from WT (n = 2), Ocn+/− (n = 3) and Ocn−/− (n = 3) mice. (B) Volcano plots of the genes differentially expressed in forebrains from Ocn+/− [left panel] and Ocn−/− [right panel] mutant compared to WT mice. Each dot represents one gene or pseudogene (n = 3). (C, D) Heatplots of the top 25 GO molecular functions (GO term) that are significantly (C) downregulated or (D) upregulated in both Ocn+/− and Ocn−/− compared to WT mice. Pathways and genes that are underlined in green have also been identified in the molecular signature of osteocalcin in testes.
Figure EV4. Gene expression analysis in forebrain (newborn) and midbrain (adults).
(A) Relative expressions of circadian rhythm-related genes, Bmal1, Clock, Per1, and Cry1, and glucose homeostasis genes, Pck1 and G6pc in forebrain at P1 in different genotypes from OcnGK mouse strain. n = 9 or more mice per genotype analyzed. (B) Relative expressions of circadian rhythm-related genes Bmal1, Clock, Per1 and Cry1 in midbrain of adult mice of different genotypes from OcnGK mouse strain. n = 4 or more mice per genotype analyzed. In bar plots, each dot represents an individual mouse. All data are shown as mean ± SEM. Statistical significance was determined by one-way Kruskal–Wallis test followed by post hoc multiple comparisons test. Source data are available online for this figure.
Consistent with this notion, we noticed that the most upregulated GO pathways identified through this analysis included those involved in neuronal death, and in inflammation such as the production of interleukin-1 and of the tumor necrosis factor superfamily (Fig. 7C,D). The same GO analysis also identified several genetic pathways involved in intracellular energy metabolism such as pyruvate metabolic process (GO:0006090), and most notably, glucose catabolism (GO:0006007) and canonical glycolysis (GO:0061621, all adjusted p values <0.006, Fig. 7C). Importantly, virtually all genes downregulated in these specific pathways in the forebrain (35 out of 41 genes) were distinct from those involved in glucose homeostasis and downregulated in the testes (compare Figs. 5E and 7C). Taken together these results suggest that osteocalcin regulates different genes and genetic pathways in organs where it signals through Gprc6a and in those like the brain where it signals through Gpr158 or Gpr37.
Discussion
The goals of this study were several. One of them was to determine whether functions of osteocalcin are dominant or recessive. A second one was to determine the respective importance of embryonic and maternal osteocalcin in defining homeostasis postnatally. To address these goals, we analyzed its functions and their molecular underpinnings in the liver, pancreas, testes and for the transcriptomic analysis, the brain of WT and Ocn-deficient unchallenged mice of various genotypes and origins. We made two observations. The first is that osteocalcin exerts dominant functions in the pancreas, liver, and testes as it does in the brain (Oury et al, 2013a) and that maternal and embryonic osteocalcin synergize to regulate multiple physiological processes and thereby contribute to establishing organismal homeostasis in adult offspring. This was observed regardless of the genetic background of the animals analyzed. A third observation obtained through a molecular analysis conducted in peripheral organs and in the brain is that osteocalcin affects distinct genetic pathways in peripheral organs where it signals through Gprc6a and in the brain where it signals through Grp158 and Gpr37.
Our analysis of the OcnBW mouse strain generated results that differ from those reported or mentioned by Diegel et al (2020). Because of its exhaustive genomic, phenotypic, and physiological analyses, the present study, performed on a large number of mice at birth and in adulthood, fasting or fed, provides explanations for the apparent discrepancies between results published by Diegel et al (2020) and those published by others (Lee et al, 2007; Oury et al, 2013a; Pi et al, 2011; Pi et al, 2020). One explanation certainly resides in the small number of mice analyzed by Diegel et al (2020) and the presence of an overt hyperglycemia at baseline in their WT controls that had been overlooked. In addition to technical considerations, the massive genomic rearrangements observed in what should have been WT controls weakens the credibility of the original analysis of this mouse strain (Diegel et al, 2020).
Analyses of newborn and adult Ocn+/− mice of various maternal origins and distinct genetic backgrounds show that developmental osteocalcin, i.e., maternal and embryonic, contributes to the regulation of pancreatic β-cell proliferation, insulin secretion, liver gluconeogenesis and altogether, glucose homeostasis in newborn or adult mice. Developmental osteocalcin also promotes testicular cell proliferation during development and testosterone biosynthesis in adult mice. That the regulation of several physiological processes in adult offspring significantly relies on osteocalcin signaling during gestation does not exclude that osteocalcin of postnatal origin contributes to the regulation of the same physiological functions as well as of others that do not seem to be affected by developmental osteocalcin. Indeed, we have previously shown that it is osteocalcin made postnatally that regulates muscle function during exercise (Berger et al, 2019; Mera et al, 2016a). Moreover, we verified here that the postnatal pool of osteocalcin regulates glucose homeostasis and testes steroidogenesis too. Thus, this work identifies two groups of osteocalcin functions, a first one is that most osteocalcin regulatory functions are dominant and fulfilled in part by osteocalcin signaling during gestation. A smaller group of physiological functions such as muscle function during exercise and the acute stress response that are regulated by osteocalcin of postnatal origin. In view of the observation that energy metabolism and organismal homeostasis in adult animals are regulated in part by osteocalcin acting during gestation, we believe it is likely that additional hormones signaling during pregnancy may contribute to the establishment of homeostasis in adult offspring.
Our results show that maternal and embryonic osteocalcin signaling during gestation synergize to influence physiological functions analyzed postnatally. Indeed, Ocn haploinsufficiency in the embryos affects physiology in peripheral organs and in the brain only if the mother is also Ocn haplo-insufficient or null (Oury et al, 2013b). These observations may have direct relevance to our understanding of the pathogenesis of some human pathological situations. Specifically, the deleterious influence that an unhealthy pregnancy has on energy metabolism, cognitive functions and other physiologies in adult offspring is reminiscent of phenotypes observed in animals lacking Ocn or its receptors. This suggests that a dysregulation in Ocn expression or circulating levels during a difficult or stressful pregnancy may affect physiology postnatally (Burger et al, 1948; Hales and Barker, 2001; Karsenty and Ferron, 2012; Karsenty and Olson, 2016; Pi et al, 2021; Pi et al, 2017). This hypothesis does not exclude by any means that other hormonal and molecular events contribute to the consequences of unhealthy pregnancy on homeostasis in the offspring.
When put in an evolutionary context, the fact that haploinsufficiency for Osteocalcin affects several physiological processes in newborn and adult offspring that are unchallenged by a diet or any other manipulations, together with the observation that osteocalcin often acts as a regulator of regulatory molecules, whether they are hormones (insulin), cytokines (interleukin-6) or neurotransmitters (monoamines, dopamine, acetylcholine), is consistent with the notion that the appearance of bone during evolution has coincided with the acquisition of another necessary mechanism of regulation of physiology that is housed in bone (Berger et al, 2019; Lee et al, 2007; Mera et al, 2016b; Oury et al, 2011).
Another purpose of this study was to begin unraveling cellular and molecular mechanisms of osteocalcin action during gestation in multiple organs. At the cellular level, our analysis reveals that developmental osteocalcin promotes cell proliferation in pancreas and testes and thereby the size of the pools of hormones that can be released by these organs. In contrast, in neurons of the brain, it prevents cell death. In addition, and besides its own direct regulation of testicular steroidogenesis, developmental osteocalcin promotes in testes the expression of the receptor for luteinizing hormone (LH) and in that way modulates the signaling of LH in Leydig cells. This function of developmental osteocalcin in the testes is reminiscent of the function it exerts in the developing adrenal glands, where it promotes expression of the receptor of the adrenocorticotrophic hormone (Yadav et al, 2022).
To draw a roadmap of molecular signaling by developmental osteocalcin in various target organs, we performed a transcriptomic analysis in testes and forebrains of Ocn+/− and Ocn−/− newborn mice. We chose these two organs because osteocalcin signals in them through distinct receptors and they were never subjected to this type of survey. Consistent with the notion that osteocalcin exerts dominant functions, this analysis revealed that the same genes and genetic pathways were dysregulated in Ocn−/− and, albeit to a lesser extent, in Ocn+/− newborn mice in testes and other peripheral organs where osteocalcin signals through Gprc6a. Specifically, a group of genes that is consistently downregulated in testes of newborn mice lacking one or two copies of Ocn is the one encoding key component of the molecular clock including Bmal1, Clock, and Per1. Given that through their expression in the liver, clock genes contribute to the regulation of glucose homeostasis and energy metabolism (Perelis et al, 2015), these results suggest that osteocalcin may regulate energy metabolism in part by regulating the molecular clock in the liver. In support of this contention, we note that osteocalcin controls the circadian expression of Pck1 in the liver (Pi et al, 2020). A second class of genes that saw their expression downregulated in testes and the liver of Ocn+/− and Ocn−/− newborn and adult mice are those implicated in gluconeogenesis such as Pck1. These results highlight how central is the ability of osteocalcin to increase the intracellular availability of glucose to its functions in peripheral organs where it signals. They also distinguish the influence of osteocalcin on glucose metabolism in peripheral organs, where it is anabolic, from the one it exerts in the brain where it is favors glycolysis. We also note that in the brain and unlike what is the case in peripheral organs, in addition to genes controlling cell death, osteocalcin signaling inhibits the expression of genes involved in inflammation. Hence, osteocalcin appears to regulate different sets of genes depending on the receptor it signals through.
Methods
Mouse models
OcnGK−/− (129/SvEv) (Ducy et al, 1996), OcnBW−/− (Diegel et al, 2020), and OcnXL−/− (Qian et al, 2021) have been previously described. Wild-type (WT) mice of indicated ages and genetic backgrounds were obtained as described in Fig. 1A or purchased from Jackson laboratories or from Taconic laboratory for the analysis of OcnBW mice. All animal studies were approved by Columbia University Animals Ethics Committee (Protocol AABH4550). To limit the information bias, the experimentalists were blinded to the genotype of the mice in all experiments.
Blood glucose measurements
Random fed blood glucose levels were measured between 9:00 AM and 10:00 AM by tail blood collection. Fasting blood glucose levels were measured between 4:00 and 5:00 PM after 6 h of fasting. In both cases, glucose levels were monitored using blood glucose strips and the Accu-Check glucometer (Roche).
Hormone measurements and dynamic metabolic tests
Blood samples were obtained at indicated ages and at indicated times mentioned below for each hormone measurement. For measurements of circulating osteocalcin, testosterone, or insulin levels, samples were collected between 9:00 AM and 9:30 AM from facial vein. Blood was collected in serum separating tubes (Microvet 500 Z Gel, Starstedt) allowed to clot for 30 min and centrifuged at 12,000 rpm for 10 min at 4 °C to obtain serum. Mouse circulating osteocalcin was measured by ELISA (Quidel kit Cat #60-1305). Blood testosterone levels were determined by RIA (Testo-US Cisbio Bioassays). Blood insulin levels were measured by ELISA (Crystal Chem Cat #90080). All measurements were performed in duplicate according to the manufacturer’s instructions.
Pyruvate tolerance tests were performed as described previously (Mauvais-Jarvis et al, 2002). Briefly, pyruvate (Sigma #P2256-25G) was injected intraperitoneally (IP) (2.5 g/kg body weight (BW)) after 6 h of fast. Blood glucose was monitored using blood glucose strips, and the Accu-Check glucometer (Roche) at indicated times (Lee et al, 2007). Glucose stimulated insulin tolerance tests (GSIS) were performed as described previously (Lee et al, 2007). Briefly, glucose (3 g/kg BW) was injected IP after an overnight fast. Blood was collected at indicated times from the tail in serum separating tubes (Microvet 100 Z, Starstedt), allowed to clot for 30 min, and centrifuged at 12,000 rpm for 10 min at 4 °C to obtain serum (Lee et al, 2007). After that, insulin levels were measured by ELISA (Crystal Chem Cat #90080).
Behavioral analyses
For all behavioral assessments, mice were handled for at least 20 min over 3 consecutive days. Before the behavioral procedure, mice were maintained in the testing room, for at least 1 h in their home cage prior to testing. Behavior was scored by two observers blind to the groups.
Novel object recognition paradigm (NOR)
The testing arena consisted of a plastic box (60 × 40 × 40 cm). Mice could not see each other during the experiment. Two different objects were used: (A) a black ceramic pot (diameter 6.5 cm, maximal height 7.5 cm) and (B) a clear/plastic funnel (diameter 8.5 cm, maximal height 8.5 cm). The objects elicited equal levels of exploration as determined in control experiments and training phases. Mice were placed in the center of the arena at the start of each exposure. Sessions were recorded with a video camera. The NOR paradigm consisted of three phases over 2 days. On day 1 (habituation phase), mice were given 5 min to explore the arena devoid of objects and were then taken back to their home cage. On day 2, mice were first allowed to explore for 5 min two identical objects arranged in a symmetric opposite position from the center of the arena and were then transported to their home cage. One hour later (testing phase), mice were given 5 min to explore two objects: a familiar object and a novel one, in the same arena, keeping the same object location. Between exposures, arenas were cleaned with a disinfectant (Phagospore), and the bedding was replaced. The following behaviors were considered as exploration of the objects: sniffing or touching the object with the nose or with the front legs or directing the nose to the object at ≤1 cm distance. Exploration was not scored if the mouse was on top of the object or completely immobile. Total exploration time was quantified during the training and testing phases. The preference index (time spent exploring the new object/the total time spent exploring both objects) and the discrimination index (time spent exploring the new object − time spent exploring the familiar object)/(total time spent exploring both objects) were calculated. As a control, the preference index for the (right/left) object location or for the object A versus B during the training phase of the NOR was measured.
RNA sequencing
Total RNA was isolated from fresh forebrains and testes obtained from one-day-old male mice of the following genotypes: OcnGK WT born from WT mothers, OcnGK−/− born from OcnGK−/− mothers, and OcnGK+/− born from OcnGK−/− mothers. All Testes were collected at same time of the day (between 6 and 7 pm) RNA extraction was performed using the QIAGEN RNeasy isolation kit (Ref#74104) following the manufacturer’s instructions, followed by NanoDrop RNA quantification (Thermo Fisher Scientific). The quality of purified RNA samples was determined by Bioanalyzer 2100 (Agilent) using the RNA Nano kit (Berger et al, 2019). RNA Integrity Number (RIN) was verified for each RNA sample and only samples with a RIN score of 8–10 were only utilized for cDNA synthesis and library preparation. RNA sequencing was performed by the Columbia University Genome Centre. Briefly, following the manufacturer’s manual, sequencing libraries were generated using NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, USA). In all, 2–3 μg RNA per sample was used to purify mRNA using poly-T oligo attached magnetic beads and then the purified mRNA will be fragmented into short fragments (about 200 bp) by the fragmentation buffer. First-strand cDNA was synthesized using random hexamer primer and Reverse Transcriptase. NEBNext Adapter were ligated after the adenylation of 3’ ends of DNA fragments. Complimentary DNA fragments 150–200 bp in length were selected for PCR amplification to create cDNA libraries. Libraries were sequenced on an Illumina Hiseq 2500 platform to generate the sequences.
Bulk RNA-seq data analysis
Quality checks were performed with FastQC version 0.11.9 and processed with Trimgalore version 0.6.6 to exclude low-quality bases. Sequence reads were aligned with STAR version 2.7.8a and calculated read counts with RSEM version 1.3.3 against the mouse reference sequence (GRCm39). Calculating counts per million (CPM) and differentially expressed genes were performed using edgeR package version 3.38.4 in R version 4.2.2. Heatmaps were drawn by using the pheatmap function from the pheatmap package version 1.0.12 in RStudio version 2023.06.0+421. Volcano plots were drawn using the ggplot function from ggplot2 package version 3.4.2 in RStudio. Category netplots and heatplots were respectively drawn using the cnetplot and heatplot functions from enrichplot package version 1.20.0 in RStudio.
Gene expression analysis
All tissues were collected between 6 and 7 pm for all mice of all genotypes and all strains. Total RNA was isolated from the testes, liver, or forebrain using a Qiagen RNA isolation kit (Ref#74104). RNAs were quantified with nanodrop and reverse transcription performed with 1 μg RNA in 20 μl volume. One μl of cDNA was used for qRT-PCR analysis with SYBR green method (Applied Biosystems) of respective genes and 18s ribosomal RNA was used as an internal control. cDNA for the internal control was diluted 50–500× to reach CT value within 5–6 cycles of the gene whose expression was being tested. qRT-PCR end products were run on a 2% agarose gels to confirm specificity of the primers. Gene expression was reanalyzed with standard qRT-PCR in the linear range of amplification and run on a 2% agarose gel to confirm the change observed through the RT-qPCR.
qPCR primer sequences used for SYBR Green-based qPCR assays were as follows: Pck1F: AGAACAAGGAGTGGAGACCG; Pck1R: TCCTACAAACACCCCATGCT; G6pcF: CTGTCTGTCCCGGATCTACC; G6pcR: GCGCGAAACCAAACAAGAAG; StarF: AAGAGCTCAACTGGAGAGCAC; StarR: TACTTAGCACTTCGTCCCCGT; Cyp11a1F: AGGTCCTTCAATGAGATCCCTT; Cyp11a1R: TCCCTGTAAATGGGGCCATAC; 18sF: CGCGGTTCTATTTTGTTGGT; 18sR: AGTCGGCATCGTTTATGGTC; Bmal1F: TCCTCAACCATCAGCGACTT; Bmal1R: TTCAATCTGACTGTGGGCCT; ClockF: ATGCCACAGAACAGTACCCA; ClockR: TTGTGTGGCGAAGGTAGGAT; Cry1F: CAGAGGGCTAGGTCTTCTCG; Cry1R: GTCCCCGTGAGCATAGTGTA; Per1F: AATGGCAAGGACTCAGCTCT; Per1R: CGAAGTTTGAGCTCCCGAAG; PyglF: TACATTCAGGCTGTGCTGGA; PyglR: AAGGCATCAAACACGGTTCC; LhcgrF: ACCCGGTGCTTTTACAAACC; LhcgrR: CGTCGTCCCATTGAATGCAT.
Histological and histomorphometric analysis
P1 pups were intraperitoneally injected with BrdU (100 mg/kg; Sigma-Aldrich). Their pancreata were fixed in 4% PFA in PBS for 12 h at 4 °C followed by dehydration, paraffin embedding, and histologically analyzed as described previously (Lee et al, 2007; Wei et al, 2014). For β-cell proliferation analysis in P1 pups every other section was labeled with guinea pig anti-insulin (1:200; DAKO A0564) and mouse anti-BrdU (1:500 Sigma B2531) antibodies, followed by fluorochrome-coupled secondary antibodies (Millipore) and DAPI counterstaining (Abcam). β-cell proliferation was quantified by counting the number of BrdU/insulin-positive cells over the total number of insulin-positive cells using ImageJ software. An average of 2000 insulin-positive cells per specimen was counted. Bmal1 and Clock levels were assessed on pancreas sections of P1 pups following co-labeling with rabbit anti-Bmal1 (1:500 Abcam ab3350) or rabbit anti-Clock (1:1000 Abcam ab3517) antibodies and a guinea Pig anti-insulin (1:800 DAKO A0564) antibody while Pck1 levels were assessed using a rabbit anti-PCK1 (1:400 Abcam ab70358) antibody, followed by fluorochrome-coupled secondary antibodies (Invitrogen A21206 and A21450).
Whole genome sequencing analysis of CRISPR targeted OcnBW−/− and OcnBW+/+ mice
Library preparation and sequencing
Long read whole-genome sequencing libraries targeting N50:20 kb reads were prepared using the Ligation Sequencing Kit (Oxford Nanopore Technologies, LSK-110) with 10 μg of DNA input to achieve N50:10 kb median read length. Library preparation followed Oxford Nanopore’s recommended procedures, with minor modifications. Samples were aliquoted from the Matrix rack tubes and fragmented using a g-TUBE (Covaris, 520104). After shearing samples were reviewed on the Agilent TapeStation to confirm the desired range, a bead-based cleanup was performed to remove small fragments, followed by another quality control. This was followed by end-prep and nick repair, and then by ligation of sequencing adapters. Final libraries were quantified by Qubit (ThermoFisher, Q32854) prior to being loaded on a 9.4.1 Flow Cell on the Oxford Nanopore Technologies PromethION, one sample per Flow Cell. Nuclease flushes were performed on the PromethION at 24-48 h.
Base calling, assembly and variant calling
The raw sequencing signal data (in FAST5 format) from the Promethion P24 sequencer were converted into DNA base calls (in FASTQ format) using Oxford Nanopore’s Guppy basecaller v6.1.2 in super high accuracy (SUP) mode. The base called sequencing reads were de novo assembled using Flye v2.9-b1768 with default (Lin et al, 2016).
For comparing WT OcnBW and OcnBW−/− against WT 129/SvEv the flye assemblies for WT OcnBW and OcnBW−/− were aligned to the WT 129/SvEv assembly using minimap2 v2.24-r1122 (Li, 2018, 2021). The alignments were then used to call 1. Small variants i.e., single nucleotide polymorphisms (SNPs), insertions and deletions (InDels) less than 50 bp and 2. Large variants, i.e., structural variants (SVs) greater than 50 bp using the minimap2’s “paftools.js call” tool.
In situ hybridization and BrdU labeling analyses in testes
Testes of adult mice were fixed in 4% PFA in PBS for 12 h at 4 °C followed by dehydration and paraffin embedding. Five-7μm sections were used for in situ hybridization analysis using ACD RNAscope kits (RNAscope® Multiplex Fluorescent Reagent Kit v2). Sf1 probe was obtained from ACD RNAscope (catalog # 445731). Serial sections were used for detection of BrdU labeled interstitial cells. All images were taken with identical laser settings and pseudo-color coding in the images was done for better comparative visualization.
Statistics
All analyses described were performed with RStudio v1.2.5033 (RStudio Team (2019), RStudio: Integrated Development for R, RStudio, Inc., Boston, MA, URL http://www.rstudio.com/). For all measurements, samples size was <30 and contained outliers, thus, we could not assume neither the Gaussian distribution of the variables, nor the homoscedasticity of the variances. Consequently, non-parametric tests have been chosen for hypothesis testing. Comparisons of means between two groups were performed by one-sided Mann–Whitney testing with the R function Wilcoxon test from stats package version 4.1.2. For GSIS experiment, one-sided paired Wilcoxon signed-rank test has been used to compare insulin secretion at the baseline and two minutes after glucose injection. To do so, the R function Wilcoxon test from stats package version 4.1.2. has been used. Comparisons of means between more than two groups were done by Kruskal–Wallis testing followed by post hoc Dunn’s multiple comparisons test with the function dunnTest from FSA package version 0.9.3. p value lower than 0.05 was considered significant. Bar plots were drawn with GraphPad Prism 9. Data are presented as means ± standard error of the mean (SEM). In all figures, statistical significance is indicated by stars *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. An absence of star or the presence of “ns” indicates the absence of significant difference.
Supplementary information
Acknowledgements
We thank Dr. Hong Liu for expert technical assistance. Long-read whole-genome sequencing (Oxford Nanopore) and bioinformatics analysis (variant calling and whole-genome assembly) were performed at the New York Genome Center. Histomorphometric analyses were performed by the Histology and Histomorphometry Core. This work was supported by NIH grants P01 AG 032959-13, R01 DE 027887-05, R01 HD 107574-02, and R01 AR 073180-04. JMB was supported by a Druckenmiller fellowship from the NY Stem Cell Foundation. Dr. Xiang Li was supported by a Shenzhen Science and Technology Program (Project number: KQTD20210811090117032).
Expanded view
Author contributions
Danilo Correa Pinto Junior: Investigation; Writing—original draft; Writing—review and editing. Isabella Canal Delgado: Investigation; Writing—original draft; Writing—review and editing. Haiyang Yang: Investigation; Writing—original draft; Writing—review and editing. Alisson Clemenceau: Investigation; Writing—original draft; Writing—review and editing. André Corvelo: Data curation; Formal analysis; Methodology; Writing—original draft. Giuseppe Narzisi: Data curation; Formal analysis; Methodology; Writing—original draft. Rajeeva Musunuri: Formal analysis; Methodology. Julian Meyer Berger: Investigation; Writing—original draft; Writing—review and editing. Lauren E Hendricks: Investigation. Kazuya Tokumura: Formal analysis; Investigation. Na Luo: Investigation. Hongchao Li: Investigation. Franck Oury: Formal analysis; Supervision; Methodology; Writing—original draft; Project administration; Writing—review and editing. Patricia Ducy: Supervision; Funding acquisition; Writing—original draft; Project administration; Writing—review and editing. Vijay K Yadav: Funding acquisition; Writing—original draft; Writing—review and editing. Xiang Li: Data curation; Writing—original draft; Project administration; Writing—review and editing. Gerard Karsenty: Conceptualization; Supervision; Funding acquisition; Writing—original draft; Project administration; Writing—review and editing.
Data availability
Mouse strains are all publicly available. RNAseq data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248211 and the GSE number is GSE248211. The whole-genome sequencing data are available at http://www.ncbi.nlm.nih.gov/bioproject/1044076 and the Bioproject ID is PRJNA1044076. The data generated in this study are available in this article and in Source data files.
Disclosure and competing interests statement
The authors declare no competing interests.
Footnotes
These authors contributed equally: Danilo Correa Pinto Junior, Isabella Canal Delgado, Haiyang Yang, Alisson Clemenceau.
Contributor Information
Franck Oury, Email: franck.oury@inserm.fr.
Patricia Ducy, Email: pd2193@cumc.columbia.edu.
Vijay K Yadav, Email: vky2101@cumc.columbia.edu.
Xiang Li, Email: xiang.li@siat.ac.cn.
Gerard Karsenty, Email: gk2172@cumc.columbia.edu.
Supplementary information
Expanded view data, supplementary information, appendices are available for this paper at 10.1038/s44319-023-00031-3.
References
- Accili D, Drago J, Lee EJ, Johnson MD, Cool MH, Salvatore P, Asico LD, Jose PA, Taylor SI, Westphal H. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- Ashworth CJ, Hoggard N, Thomas L, Mercer JG, Wallace JM, Lea RG. Placental leptin. Rev Reprod. 2000;5:18–24. doi: 10.1530/ror.0.0050018. [DOI] [PubMed] [Google Scholar]
- Berger JM, Singh P, Khrimian L, Morgan DA, Chowdhury S, Arteaga-Solis E, Horvath TL, Domingos AI, Marsland AL, Yadav VK, et al. Mediation of the acute stress response by the skeleton. Cell Metab. 2019;30:890–902.e8. doi: 10.1016/j.cmet.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Drummond J, Sandstead H (1948) Malnutrition and starvation in Western Netherlands. Part I. General State Printing Office, The Hague
- Chan YF, Wai-Sum O, Tang F. Adrenomedullin in the rat testis. I: Its production, actions on testosterone secretion, regulation by human chorionic gonadotropin, and its interaction with endothelin 1 in the leydig cell. Biol Reprod. 2008;78:773–779. doi: 10.1095/biolreprod.107.060871. [DOI] [PubMed] [Google Scholar]
- Chida D, Nakagawa S, Nagai S, Sagara H, Katsumata H, Imaki T, Suzuki H, Mitani F, Ogishima T, Shimizu C, et al. Melanocortin 2 receptor is required for adrenal gland development, steroidogenesis, and neonatal gluconeogenesis. Proc Natl Acad Sci USA. 2007;104:18205–18210. doi: 10.1073/pnas.0706953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, Huh JR. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toni L, De Filippis V, Tescari S, Ferigo M, Ferlin A, Scattolini V, Avogaro A, Vettor R, Foresta C. Uncarboxylated osteocalcin stimulates 25-hydroxy vitamin D production in Leydig cell line through a GPRC6a-dependent pathway. Endocrinology. 2014;155:4266–4274. doi: 10.1210/en.2014-1283. [DOI] [PubMed] [Google Scholar]
- Diegel CR, Hann S, Ayturk UM, Hu JCW, Lim KE, Droscha CJ, Madaj ZB, Foxa GE, Izaguirre I, Transgenics Core VVA, et al. An osteocalcin-deficient mouse strain without endocrine abnormalities. PLoS Genet. 2020;16:e1008361. doi: 10.1371/journal.pgen.1008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27:740–756. doi: 10.1016/j.cmet.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- Ferron M, Hinoi E, Karsenty G, Ducy P. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc Natl Acad Sci USA. 2008;105:5266–5270. doi: 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM. Leptin and the endocrine control of energy balance. Nat Metab. 2019;1:754–764. doi: 10.1038/s42255-019-0095-y. [DOI] [PubMed] [Google Scholar]
- Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, Shanley MR, Boudarene N, Rousseaud A, Friedman AK, et al. Autophagy is required for memory formation and reverses age-related memory decline. Curr Biol. 2019;29:435–448.e8. doi: 10.1016/j.cub.2018.12.021. [DOI] [PubMed] [Google Scholar]
- Gupte AA, Sabek OM, Fraga D, Minze LJ, Nishimoto SK, Liu JZ, Afshar S, Gaber L, Lyon CJ, Gaber AO, et al. Osteocalcin protects against nonalcoholic steatohepatitis in a mouse model of metabolic syndrome. Endocrinology. 2014;155:4697–4705. doi: 10.1210/en.2014-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- Hummel KP, Dickie MM, Coleman DL. Diabetes, a new mutation in the mouse. Science. 1966;153:1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- Karpac J, Ostwald D, Bui S, Hunnewell P, Shankar M, Hochgeschwender U. Development, maintenance, and function of the adrenal gland in early postnatal proopiomelanocortin-null mutant mice. Endocrinology. 2005;146:2555–2562. doi: 10.1210/en.2004-1290. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Ferron M. The contribution of bone to whole-organism physiology. Nature. 2012;481:314–320. doi: 10.1038/nature10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Olson EN. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell. 2016;164:1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrimian L, Obri A, Ramos-Brossier M, Rousseaud A, Moriceau S, Nicot AS, Mera P, Kosmidis S, Karnavas T, Saudou F, et al. Gpr158 mediates osteocalcin’s regulation of cognition. J Exp Med. 2017;214:2859–2873. doi: 10.1084/jem.20171320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Mangelsdorf DJ. A dozen years of discovery: insights into the physiology and pharmacology of FGF21. Cell Metab. 2019;29:246–253. doi: 10.1016/j.cmet.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreymann B, Williams G, Ghatei MA, Bloom SR. Glucagon-like peptide-1 7-36: a physiological incretin in man. Lancet. 1987;2:1300–1304. doi: 10.1016/s0140-6736(87)91194-9. [DOI] [PubMed] [Google Scholar]
- Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37:4572–4574. doi: 10.1093/bioinformatics/btab705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yuan J, Kolmogorov M, Shen MW, Chaisson M, Pevzner PA. Assembly of long error-prone reads using de Bruijn graphs. Proc Natl Acad Sci USA. 2016;113:E8396–E8405. doi: 10.1073/pnas.1604560113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JC, Ljubicic S, Leibiger B, Kern M, Leibiger IB, Moede T, Kelly ME, Chatterjee Bhowmick D, Murano I, Cohen P, et al. Adipsin is an adipokine that improves beta cell function in diabetes. Cell. 2014;158:41–53. doi: 10.1016/j.cell.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Li L, Fan Q, Angelini A, Saha PK, Coarfa C, Rajapakshe K, Perera D, Cheng J, Wu H. Endothelium-specific depletion of LRP1 improves glucose homeostasis through inducing osteocalcin. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-25673-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais-Jarvis F, Ueki K, Fruman DA, Hirshman MF, Sakamoto K, Goodyear LJ, Iannacone M, Accili D, Cantley LC, Kahn CR. Reduced expression of the murine p85alpha subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J Clin Invest. 2002;109:141–149. doi: 10.1172/JCI13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mera P, Laue K, Ferron M, Confavreux C, Wei J, Galan-Diez M, Lacampagne A, Mitchell SJ, Mattison JA, Chen Y, et al. Osteocalcin signaling in myofibers is necessary and sufficient for optimum adaptation to exercise. Cell Metab. 2016;23:1078–1092. doi: 10.1016/j.cmet.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mera P, Laue K, Wei J, Berger JM, Karsenty G. Osteocalcin is necessary and sufficient to maintain muscle mass in older mice. Mol Metab. 2016;5:1042–1047. doi: 10.1016/j.molmet.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osinski PA. Steroid 11beta-ol dehydrogenase in human placenta. Nature. 1960;187:777. doi: 10.1038/187777a0. [DOI] [PubMed] [Google Scholar]
- Oury F, Ferron M, Huizhen W, Confavreux C, Xu L, Lacombe J, Srinivas P, Chamouni A, Lugani F, Lejeune H, et al. Osteocalcin regulates murine and human fertility through a pancreas-bone-testis axis. J Clin Invest. 2013;123:2421–2433. doi: 10.1172/JCI65952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F, Khrimian L, Denny CA, Gardin A, Chamouni A, Goeden N, Huang YY, Lee H, Srinivas P, Gao XB, et al. Maternal and offspring pools of osteocalcin influence brain development and functions. Cell. 2013;155:228–241. doi: 10.1016/j.cell.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oury F, Sumara G, Sumara O, Ferron M, Chang H, Smith CE, Hermo L, Suarez S, Roth BL, Ducy P, et al. Endocrine regulation of male fertility by the skeleton. Cell. 2011;144:796–809. doi: 10.1016/j.cell.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350:aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi M, Kapoor K, Ye R, Nishimoto SK, Smith JC, Baudry J, Quarles LD. Evidence for osteocalcin binding and activation of GPRC6A in beta-cells. Endocrinology. 2016;157:1866–1880. doi: 10.1210/en.2015-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi M, Nishimoto SK, Darryl Quarles L. Explaining divergent observations regarding osteocalcin/GPRC6A endocrine signaling. Endocrinology. 2021;162:bqab011. doi: 10.1210/endocr/bqab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi M, Nishimoto SK, Quarles LD. GPRC6A: Jack of all metabolism (or master of none) Mol Metab. 2017;6:185–193. doi: 10.1016/j.molmet.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi M, Wu Y, Quarles LD. GPRC6A mediates responses to osteocalcin in beta-cells in vitro and pancreas in vivo. J Bone Miner Res. 2011;26:1680–1683. doi: 10.1002/jbmr.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi M, Xu F, Ye R, Nishimoto SK, Williams RW, Lu L, Darryl Quarles L. Role of GPRC6A in regulating hepatic energy metabolism in mice. Sci Rep. 2020;10:7216. doi: 10.1038/s41598-020-64384-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Li H, Yang H, Yang Q, Lu Z, Wang L, Chen Y, Li X. Osteocalcin attenuates oligodendrocyte differentiation and myelination via GPR37 signaling in the mouse brain. Sci Adv. 2021;7:eabi5811. doi: 10.1126/sciadv.abi5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabek OM, Nishimoto SK, Fraga D, Tejpal N, Ricordi C, Gaber AO. Osteocalcin effect on human beta-cells mass and function. Endocrinology. 2015;156:3137–3146. doi: 10.1210/EN.2015-1143. [DOI] [PubMed] [Google Scholar]
- Wei J, Hanna T, Suda N, Karsenty G, Ducy P. Osteocalcin promotes beta-cell proliferation during development and adulthood through Gprc6a. Diabetes. 2014;63:1021–1031. doi: 10.2337/db13-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- Xia W, Mruk DD, Cheng CY. C-type natriuretic peptide regulates blood-testis barrier dynamics in adult rat testes. Proc Natl Acad Sci USA. 2007;104:3841–3846. doi: 10.1073/pnas.0610100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Berger JM, Singh P, Nagarajan P, Karsenty G. Embryonic osteocalcin signaling determines lifelong adrenal steroidogenesis and homeostasis in the mouse. J Clin Invest. 2022;132:e153752. doi: 10.1172/JCI153752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkin BR, Papadopoulos V. Leydig cells: formation, function, and regulation. Biol Reprod. 2018;99:101–111. doi: 10.1093/biolre/ioy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mouse strains are all publicly available. RNAseq data are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248211 and the GSE number is GSE248211. The whole-genome sequencing data are available at http://www.ncbi.nlm.nih.gov/bioproject/1044076 and the Bioproject ID is PRJNA1044076. The data generated in this study are available in this article and in Source data files.