Abstract
Inflammasome activation and pyroptotic cell death are known to contribute to the pathogenesis of cardiovascular diseases, such as myocardial ischemia-reperfusion (I/R) injury, although the underlying regulatory mechanisms remain poorly understood. Here we report that expression levels of the E3 ubiquitin ligase membrane-associated RING finger protein 2 (MARCH2) were elevated in ischemic human hearts or mouse hearts upon I/R injury. Genetic ablation of MARCH2 aggravated myocardial infarction and cardiac dysfunction upon myocardial I/R injury. Single-cell RNA-seq analysis suggested that loss of MARCH2 prompted activation of NLRP3 inflammasome in cardiomyocytes. Mechanistically, phosphoglycerate mutase 5 (PGAM5) was found to act as a novel regulator of MAVS-NLRP3 signaling by forming liquid-liquid phase separation condensates with MAVS and fostering the recruitment of NLRP3. MARCH2 directly interacts with PGAM5 to promote its K48-linked polyubiquitination and proteasomal degradation, resulting in reduced PGAM5–MAVS co-condensation, and consequently inhibition of NLRP3 inflammasome activation and cardiomyocyte pyroptosis. AAV-based re-introduction of MARCH2 significantly ameliorated I/R-induced mouse heart dysfunction. Altogether, our findings reveal a novel mechanism where MARCH2-mediated ubiquitination negatively regulates the PGAM5/MAVS/NLRP3 axis to protect against cardiomyocyte pyroptosis and myocardial I/R injury.
Subject terms: Ubiquitylation, Mechanisms of disease, Necroptosis
Introduction
The inflammasome is a macromolecular structure responsible for sensing injury and eliciting a cascade of inflammatory responses. The most well-characterized sensor of inflammasomes is NACHT, LRR, and PYD domains-containing protein 3 (NLRP3), which is activated in response to sterile inflammation1. Activation of inflammasome sensor NLRP3 recruits the adaptor apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) and the effector caspase-1, triggering severe tissue damage directly by promoting pyroptosis and indirectly by IL-18 secretion2,3. NLRP3 inflammasome activation contributes to the pathophysiology of numerous inflammatory diseases4. For example, NLRP3 inflammasome activated by damage-associated molecular patterns (DAMPs) participates in the pathogenesis of nonalcoholic fatty liver disease5. Additionally, expression levels of NLRP3 and inflammatory cytokines IL-18 and IL-1β were found to be elevated in atherosclerosis plaque development6. Besides, knockout (KO) of NLRP3 protects against high-fat diet-elicited obesity and insulin resistance by abrogating reactive oxygen species (ROS)- and mitochondrial injury-evoked inflammasome activation7.
Myocardial infarction (MI) remains one of the leading causes of disability and mortality worldwide8. Timely restoration of blood flow through occluded coronary arteries represents an optimal maneuver for patients with myocardial infarction9. However, such reperfusion may activate cascades of detrimental injuries to the myocardium known as ischemia-reperfusion (I/R) injury9,10. Emerging evidence has indicated the activation of inflammasomes as an integral part of inflammation-driven myocardial I/R injury1,11. Suppressing NLRP3 inflammasome activation is known to restrict infarct size and cardiac dysfunction following myocardial I/R injury12,13. Therefore, the identification of endogenous modulator(s) for NLRP3 inflammasome to suppress its activation might be clinically beneficial.
The ubiquitin (Ub)-proteasome system regulates protein degradation and homeostasis in numerous cellular processes14. Therefore, identifying pivotal ubiquitination-related modulators may alleviate myocardial I/R injury. For this purpose, we performed single-cell RNA sequencing (scRNA-seq) on heart samples from mice with myocardial I/R injury. We found upregulation of the E3 ligase membrane-associated RING finger protein 2 (MARCH2), a member of the MARCH E3 Ub ligase subfamily including 11 members and localizing primarily to the endoplasmic reticulum, endosome, Golgi apparatus, mitochondria, and plasma membranes15. MARCH2 is known to regulate vesicular trafficking between the endosomes and trans-Golgi network as well as recycle endosomes through interacting with syntaxin-616. MARCH2 was also shown to negatively regulate autophagy in tumor cells by promoting ubiquitination and degradation of CFTR and activating the PIK3CA-AKT-mTOR pathway17.
Recent evidence has demonstrated an essential role for MARCH2 in the regulation of inflammatory responses, such as antiviral defense and bacterial infection, as well as T-cell development18. MARCH2 is upregulated upon HIV infection, which restricts the production and infection of HIV-119. In addition, MARCH2 catalyzes K48-linked ubiquitination on NF-κB essential modulator, a critical modulator functioning in NF-κB and interferon-mediated signaling. MARCH2 KO leads to robust production of proinflammatory cytokines (IL-6, CXCL10, TNF-α and IL-1β) upon lipopolysaccharides (LPS) challenge20. Nevertheless, whether MARCH2 plays a role in myocardial I/R injury remains elusive.
Taking advantage of the MARCH2 KO and AAV9-cTnT-MARCH2 transfection murine models, our present study showed that MARCH2 ameliorates myocardial I/R injury and suppresses NLRP3 inflammasome activation. MARCH2 was found to interact with phosphoglycerate mutase family member 5 (PGAM5) to promote its K48-linked ubiquitination and degradation. Ubiquitin-dependent degradation of PGAM5 leads to inhibition of phase-separated condensates of PGAM5-mitochondrial anti-viral-signaling protein (MAVS) and activation of downstream NLRP3 inflammasome. Taken together, these findings demonstrate the vital role of MARCH2 as a novel therapeutic target for myocardial I/R injury.
Results
Characterization of MARCH2 expression during myocardial I/R
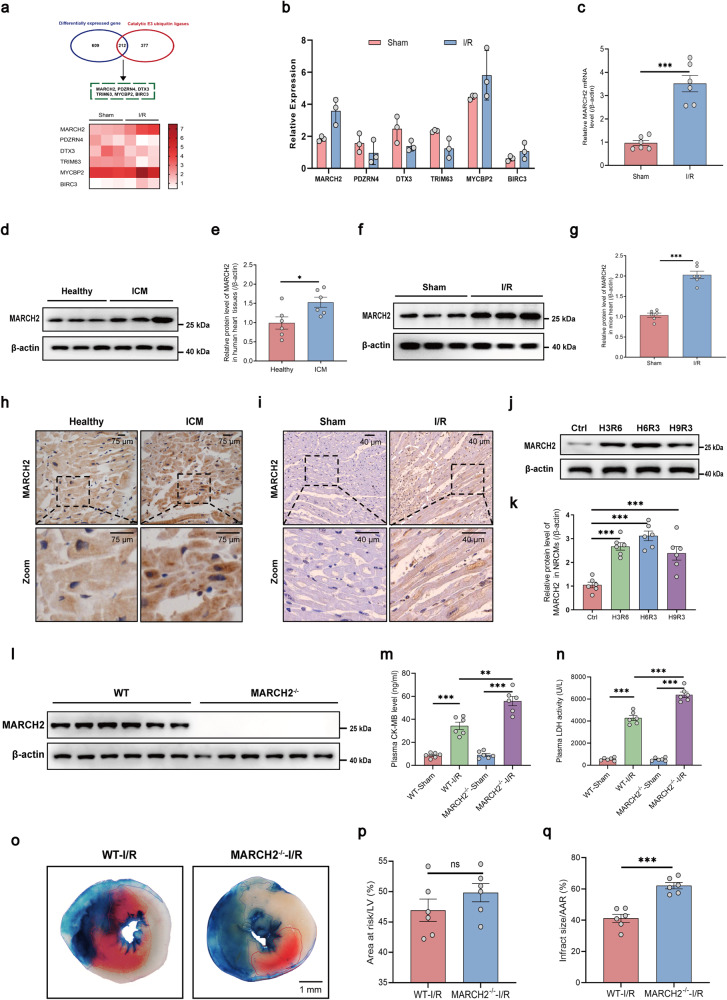
To reveal possible ubiquitination-related candidate molecules that might participate in the pathogenesis of myocardial I/R injury, we performed scRNA-seq for wild-type (WT) mice subjected to sham or I/R surgery. Upregulated or downregulated differentially expressed genes (DEGs) were screened (Supplementary Fig. S1a). We found that six E3 ligases reached the threshold of screening, namely MARCH2, PDZRN4, DTX3, TRIM63, MYCBP2, and BIRC3, among which MARCH2 exhibited the most pronounced transcriptional change (Fig. 1a, b). To confirm the results of scRNA-seq, we next performed qPCR analyses and found that mRNA levels of MARCH2 was increased following I/R injury (Fig. 1c), suggesting a potential role of MARCH2 in myocardial I/R injury. Levels of MARCH2 were evaluated in heart tissues from patients with ischemic cardiomyopathy (ICM) (Supplementary Table S1). Western blotting and immunohistochemistry data revealed elevated MARCH2 levels in heart tissues from ICM patients in comparison with healthy controls (Fig. 1d, e, h). Consistently, western blotting and immunohistochemistry results revealed increased myocardial MARCH2 levels following I/R injury (45 min/9 h) in mice (Fig. 1f, g, i). A similar pattern was observed in primary neonatal mouse cardiomyocytes (NMCMs) and primary adult mouse cardiomyocytes during hypoxia and reoxygenation (H/R) (Fig. 1j, k; Supplementary Fig. S1b). Altogether, these data indicate an integral role for MARCH2 in myocardial I/R injury.
Fig. 1. MARCH2 protein level is increased in hearts of patients with ICM and mice during myocardial I/R.
a Heatmap showing six differentially expressed E3 ligases in mouse hearts following I/R injury. b Histogram showing the relative mRNA expression of the six ubiquitin-related genes in the indicated groups. c mRNA levels of MARCH2 following I/R injury (45 min/9 h) were analyzed by RT-qPCR (normalized to β-actin). d Western blotting analysis of MARCH2 expression in healthy human hearts and patients with ICM. e Quantitated MARCH2 levels in hearts of healthy human and patients with ICM (n = 6 for each group). f Western blotting analysis of MARCH2 protein levels following I/R injury (45 min/9 h). g Quantitated MARCH2 levels following I/R injury (45 min/9 h). h Representative immunohistochemistry images showing MARCH2 protein levels in human hearts. i Representative immunohistochemistry images showing MARCH2 protein levels in sham- or I/R-treated mouse hearts. Dotted white lines indicate the boundary of the infarct area and border area. j, k Representative western blotting image (j) and quantification analysis (k) of MARCH2 expression in NMCMs subjected to hypoxia (6 h)/reoxygenation (3 h) (n = 6 for each group). l Western blotting analysis showed that MARCH2 protein could not be detected following the deletion of MARCH2 gene. m, n CK-MB level (m) and LDH activity (n) of WT and MARCH2 KO mice with or without myocardial I/R (45 min/24 h) (n = 6 for each group). o Representative images of heart sections by TTC/Evans Blue staining depicting infracted area. p Ratios of area at risk (AAR) to left ventricular (LV) area. q Infarct area normalized to AAR. Data are shown as means ± SEM. Statistical significance was examined by two-way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, ***P < 0.001.
MARCH2 deficiency exacerbates cardiac dysfunction during I/R injury
To elucidate potential role of MARCH2 in I/R injury, MARCH2 KO mice were generated and genotype was confirmed (Fig. 1l; Supplementary Fig. S2a). MARCH2 homozygous KO mice showed normal baseline heart rate and systolic blood pressure comparable to those from heterozygous or WT mice (Supplementary Fig. S2b, c). WT and MARCH2 KO mice were then subjected to I/R surgery. Myocardial I/R challenge increased levels of creatine phosphokinase-MB (CK-MB) and lactic dehydrogenase (LDH), two cardiac injury markers, with more pronounced rises in MARCH2 KO mice (Fig. 1m, n). Evans blue/TTC double staining revealed comparable area at risk (AAR) between WT and MARCH2 KO mice following myocardial I/R (45 min/24 h), with a more pronounced infarct size response in MARCH2 KO mice (Fig. 1o–q).
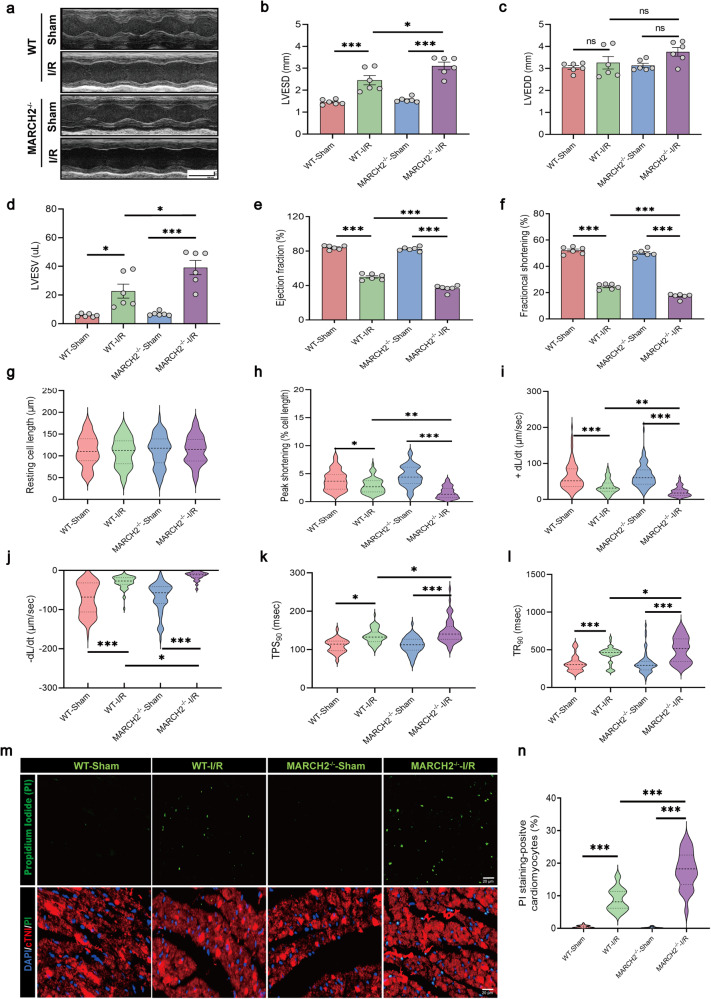
Echocardiographic analysis showed comparable cardiac function and morphology between MARCH2 KO and WT mice under physiological conditions. I/R procedure evoked markedly decreases in left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), and increases in left ventricular end-systolic diameter (LVESD), left ventricular end-systolic volume (LVESV) in WT mice, and these dysfunctions were accentuated by MARCH2 KO (Fig. 2a–f). Chronological echocardiographic assessment of cardiac structure and function was performed. The aggravation caused by MARCH2 KO sustained from 24 h after reperfusion to 28 days (Supplementary Fig. S2d–g).
Fig. 2. MARCH2 KO exacerbates myocardial dysfunction after myocardial I/R injury.
a Representative M-mode recordings of echocardiography of WT and MARCH2 KO mice subjected to I/R. b–f Quantitative analyses of echocardiographic measurements performed after I/R injury. n = 6 mice per group. Left ventricular end-systolic diameter (LVESD, b), left ventricular end-diastolic diameter (LVEDD, c), left ventricular end-systolic volume (LVESV, d), ejection fraction (EF, e), and fractional shortening (FS, f). g–l Evaluation of mechanical properties of single, acutely isolated cardiomyocytes from I/R-treated mice. Resting cell length (g), peak shortening (h), +dL/dt: maximal velocity of shortening (i), -dL/dt: maximal velocity of re-lengthening (j), TPS90: time-to-90% peak shortening (k); TR90: time-to-90% re-lengthening (l). n = 60 cells from 3 mice per group. m Representative PI-stained images of myocardial sections from WT and MARCH2–/– mice subjected to I/R. Green: PI-positive nuclei; Red: cTnI-stained cardiomyocytes; blue, DAPI-stained nuclei; Scale bar = 20 μm. n Quantitative analysis of PI-positive cells. Experiments were repeated three times with similar results. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Contractile properties of adult cardiomyocytes isolated from sham or I/R-treated mice were evaluated. Neither I/R procedure nor MARCH2 ablation exerted any effect on resting cardiomyocyte length (Fig. 2g). However, I/R injury-evoked suppression in peak shortening and maximal velocity of shortening/re-lengthening (±dL/dt) as well as prolonged shortening duration (TPS90) and prolonged re-lengthening duration (TR90). MARCH2 ablation markedly aggravated I/R-induced cardiomyocyte contractile anomalies without eliciting any notable effect itself (Fig. 2h–l). Besides, cardiomyocyte necrosis was monitored using propidium iodid (PI) staining. The PI-positive cardiomyocytes were notably increased following I/R procedure, which was significantly exacerbated in MARCH2 KO mice (Fig. 2m, n). These data suggest that MARCH2 deficiency accentuates I/R-elicited myocardial dysfunction and cardiomyocyte death.
MARCH2 deletion induces NLRP3 inflammasome activation following myocardial I/R
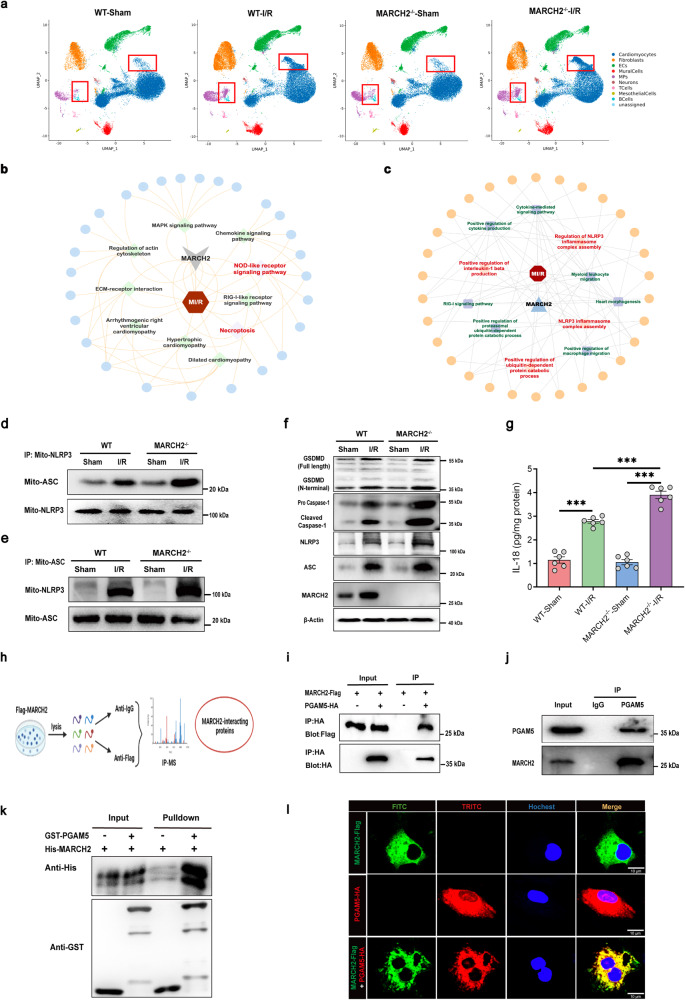
To explore the mechanisms through which MARCH2 KO accentuates cardiac I/R injury, scRNA-seq was performed in WT and MARCH2 KO mice that underwent sham or I/R surgery. Uniform Manifold Approximation and Projection (UMAP) analysis of scRNA-seq data (n = 178,274 cells) revealed cell types including cardiomyocytes, fibroblasts, endothelial cells (ECs), mural cells, macrophages (MPs), neurons, T cells, mesothelial cells, and B cells (Supplementary Fig. S3a–c). Cell type distribution comparison among four groups revealed significant changes in MP and cardiomyocyte clusters after the I/R procedure (Fig. 3a). WT-I/R group exhibited significant upregulation of NLRP3 inflammasome assembly-related genes in B cells, cardiomyocytes, ECs, fibroblasts, and MPs compared with WT-sham group, indicating a role of NLRP3 inflammasome assembly in myocardial I/R injury response (Supplementary Fig. S3d).
Fig. 3. PGAM5 is identified to interact with MARCH2.
a Comparison of cell type distributions among different groups showing changes in MP and cardiomyocyte clusters after I/R procedure. b, c KEGG enrichment analysis (b) and GO (c) analysis of scRNA-seq data from WT-I/R and MARCH2–/–-I/R hearts. d, e Interaction between NLRP3 and ASC in mitochondria (mito-NLRP3 and mito-ASC) of cardiac tissues of MARCH2 KO and WT mice subjected to I/R was examined by IP-western blotting assay. IP with NLRP3 antibody (d) and IP with ASC antibody (e). f Representative Western blotting analyses of MARCH2, NLRP3, ASC, caspase-1 (procaspase1; cleaved caspase-1), GSDMD (full-length; N-terminal) in cardiac tissues of MARCH2 KO and WT mice subjected to I/R (45 min/9 h). g IL-18 release was measured by enzyme-linked immunosorbent assay (ELISA) in cardiac tissues of MARCH2 KO and WT mice subjected to I/R (45 min/9 h). h Schematic diagram showing MS analysis workflow for identifying targets of MARCH2. i IP analysis with anti-HA antibody and immunoblotting with antibodies of anti-Flag and anti-HA, respectively, in NMCMs transfected with PGAM5-HA or control vector along with MARCH2-Flag. j Endogenous IP of MARCH2 and PGAM5 in NMCMs. k Direct interaction between GST-PGAM5 and His-MARCH2 demonstrated by GST pull-down assays. Both input and pull-down samples were immunoblotted with anti-GST and anti-His antibodies. GST-PGAM5 and His-MARCH2 proteins were expressed in vitro. l Representative immunofluorescence images of MARCH2 and PGAM5 in NMCMs. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis found that genes involved in the NOD-like receptor signaling pathway, necroptosis, hypertrophic cardiomyopathy, and dilated cardiomyopathy were upregulated in hearts of MARCH2 KO mice subjected to I/R injury (Fig. 3b). Gene ontology (GO) analysis revealed NLRP3 inflammasome complex assembly, positive regulation of cytokine production, and positive regulation of ubiquitin-dependent protein catabolic process as the top enriched pathways in MARCH2 KO-I/R mice compared with WT-I/R group (Fig. 3c), suggesting an essential role of MARCH2 in the regulation of inflammatory response. To further discern the cell types involved in MARCH2 deficiency-elicited responses, expression levels of essential genes in this signaling cascade including NLRP3, caspase-1, IL-18 and GSDMD were examined in MARCH2 KO-I/R and WT-I/R groups. Our data showed that MARCH2 KO exhibited higher expression levels of these genes primarily in cardiomyocytes (Supplementary Fig. S4). Furthermore, comparative gene set variation analysis (GSVA) also suggested significant upregulation of NOD-like receptor (Supplementary Fig. S5a) and NLRP3 inflammasome complex assembly (Supplementary Fig. S5b) in MARCH2 KO-I/R cardiomyocytes compared with WT-I/R cardiomyocytes. Next, we verified the effect of MARCH2 on NLRP3 inflammasome complex assembly signaling. As shown in Fig. 3d, interaction between NLRP3 and the inflammasome adaptor protein ASC in mitochondria was increased in MARCH2 KO mice compared to WT mice following I/R injury. Consistently, ASC interaction with NLRP3 was enhanced in MARCH2 KO mice upon I/R stress (Fig. 3e). Expression levels of NLRP3 and ASC were upregulated in the hearts of WT mice subjected to I/R surgery, with a more pronounced response in MARCH2 KO mice (Fig. 3f; Supplementary Fig. S6a, b). Moreover, increased caspase-1 and GSDMD cleavage (Fig. 3f; Supplementary Fig. S6c, d) and downstream proinflammatory IL-18 secretion after I/R (Fig. 3g) were intensified by MARCH2 KO.
MARCH2 directly interacts with PGAM5
In an effort to elucidate the molecular mechanism of MARCH2 in the regulation of myocardial I/R injury, we screened for MARCH2-binding proteins using immunoprecipitation-mass spectrometry (IP-MS) (Fig. 3h; Supplementary Fig. S7a), and focused on genes involved in both necroptosis and NOD-like receptor signaling pathway. Among the precipitated proteins, PGAM5 is implicated in NOD-like receptor signaling pathway and inflammasome activation21,22. Deletion of PGAM5 attenuated I/R-induced necroptosis-like cell death and inflammation response23. To investigate whether MARCH2 protects against I/R injury and negatively regulates NLRP3 inflammasome complex assembly via a direct interaction with PGAM5, a co-IP assay was performed. NMCMs were transfected with HA-tagged PGAM5 or control vector along with Flag-tagged MARCH2. Flag-tagged MARCH2 was found in the HA-PGAM5 IP complexes (Fig. 3i). More importantly, co-IP in NMCMs revealed that endogenous MARCH2 interacts with PGAM5 but not control IgG (Fig. 3j). Furthermore, purified glutathione S-transferase (GST)-fused PGAM5 was capable of pulling down MARCH2 (Fig. 3k), suggesting a direct interaction between PGAM5 and MARCH2. Consistently, immunofluorescence showed colocalization between Flag-MARCH2 and HA-PGAM5 in NMCMs (Fig. 3l). These results suggest a direct interaction between MARCH2 and PGAM5 in cardiomyocytes under physiological conditions.
MARCH2 mediates ubiquitination and proteasomal degradation of PGAM5 by catalyzing K48-linked polyubiquitination
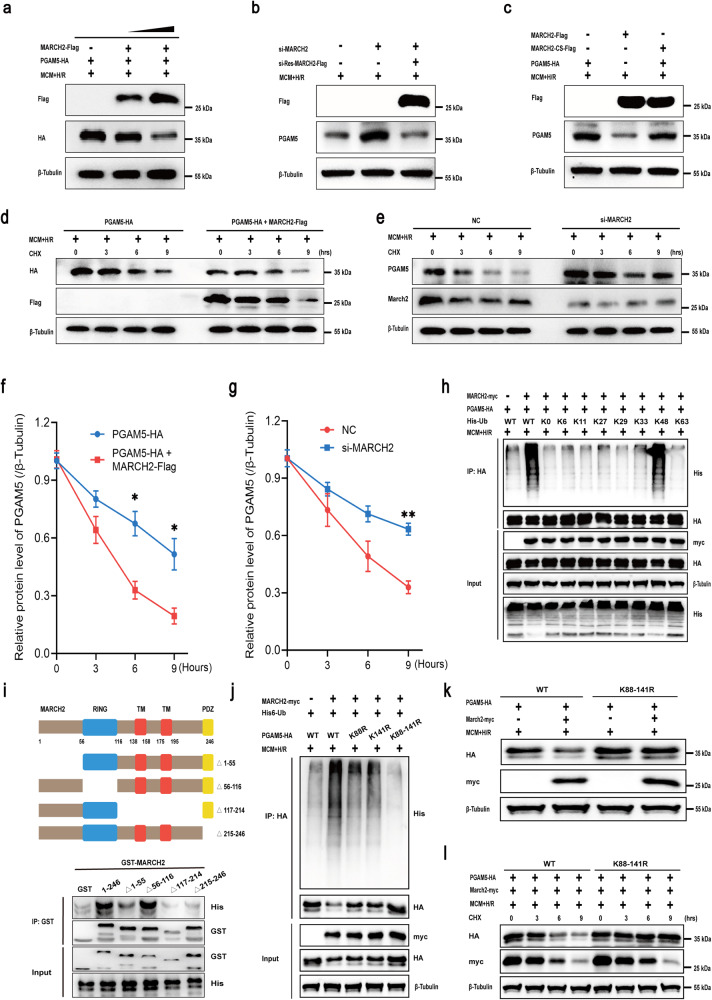
Cell–cell interactions in hearts of WT and MARCH2 KO mice subjected to I/R were predicted using Cellphone DB. As shown in Supplementary Fig. S7b, interaction scores between MPs and cardiomyocytes were increased in MARCH2 KO-I/R hearts compared with WT-I/R group. To mimic H/R injury and proinflammatory environment in vitro, cardiomyocytes were treated with macrophage-conditioned medium (MCM) and H/R (6 h/3 h). Given the biochemical property of MARCH2 as an E3 Ub ligase, we tested whether PGAM5 may serve as a previously unidentified substrate of MARCH2. HL-1 cardiomyocytes were transfected with plasmids expressing MARCH2-Flag and PGAM5-HA or transfected with empty plasmid vectors (as control) prior to MCM + H/R challenge. Levels of PGAM5 were downregulated in a dose-dependent manner following rises in MARCH2 protein levels in MCM + H/R-treated cardiomyocytes (Fig. 4a), whereas knockdown of MARCH2 upregulated PGAM5 levels and the siRNA-resistant MARCH2 exhibited an opposite effect (Fig. 4b). Nonetheless, knockdown or overexpression of MARCH2 did not affect the mRNA level of PGAM5 in MCM + H/R-exposed NMCMs (Supplementary Fig. S8a, b). Notably, transfection of a catalytically inactive mutant (C64/67 S, abbreviated as CS) of MARCH2 abolished MARCH2-accelerated degradation of PGAM5 (Fig. 4c), suggesting an important effect of catalytic activity of MARCH2 in decreasing PGAM5 protein level in the setting of MCM + H/R insult. The degradation dynamics of PGAM5 was examined following cycloheximide (CHX) treatment for various durations. Overexpression of MARCH2 significantly promoted degradation of PGAM5 in comparison to that in control (Fig. 4d, f), while knockdown of MARCH2 exhibited the opposite effect in MCM + H/R-treated HL-1 cardiomyocytes (Fig. 4e–g) and NMCMs under MCM + H/R treatment (Supplementary Fig. S8c). Consistently, the proteasome inhibitor MG132 partially reversed PGAM5 level under MARCH2 overexpression in MCM + H/R-treated NMCMs (Supplementary Fig. S8d). Together, these data suggest that MARCH2 promotes proteasome-dependent degradation of PGAM5.
Fig. 4. MARCH2 E3 ligase promotes degradation of PGAM5 through K48-linked polyUb.
a Western blotting analysis of PGAM5-HA and MARCH2-Flag. HL-1 cardiomyocytes were transfected with vector expressing PGAM5-HA or MARCH2-Flag and treated with MCM + H/R. b Western blotting analysis of MARCH2 and PGAM5 proteins in HL-1 cardiomyocytes transfected with MARCH2 siRNA, siRNA-resistant MARCH2 or negative control and treated with MCM + H/R. c Analysis of PGAM5-HA protein levels in MCM + H/R-treated HL-1 cardiomyocytes transfected with either MARCH2 WT or its catalytically inactive mutant (C64/67 S, abbreviated CS). d, e Western blotting analysis of MARCH2 and PGAM5 protein levels and quantification analysis of PGAM5 level. HL-1 cardiomyocytes were transfected with MARCH2 (d) or siRNA-MARCH2 (e) and then treated with MCM + H/R and cycloheximide (CHX; 30 μm). f, g Corresponding quantitation graphs of relative PGAM5 degradation in MCM + H/R-treated HL-1 cardiomyocytes transfected with MARCH2 (f) or siRNA-MARCH2 (g) and CHX. The experiments were repeated three times. Error bars represent standard deviation. h Effects of the indicated polyUb on MARCH2-mediated PGAM5 ubiquitination. HL-1 cardiomyocytes were transfected with the indicated ubiquitin under MCM + H/R treatment. IP analysis with anti-HA antibody and immunoblotting with antibodies of anti-His and anti-HA. i Western blotting of the mapping analysis showing the binding domains of MARCH2 to PGAM5. Various truncated forms of MARCH2 (△1–55, △56–116, △117–214 and △215–246) were expressed and purified. j–l Effects of the indicated PGAM5 KR mutants (K88R, K141R, or combined K88-141R) on MARCH2-mediated PGAM5 ubiquitination (j), protein level (k), and degradation (l) under MCM + H/R treatment. PGAM5–/– HL-1 cardiomyocytes were transfected with the indicated constructs, and PGAM5 protein levels were analyzed. Statistical differences were analyzed by unpaired Student’s t-test for comparison between two groups. *P < 0.05, **P < 0.01.
Furthermore, WT MARCH2 but not the E3 ligase-inactive mutant mediated PGAM5 polyubiquitination (polyUb) upon MCM + H/R treatment (Supplementary Fig. S9a). PolyUb chains of different Lys (K) linkages are known to confer distinct signaling outcomes24. To determine the linkage type of the polyUb chains that MARCH2 conjugates onto PGAM5, a series of Ub mutants were generated to preserve Lys on specific residues (Kx = K6, K11, K27, K29, K33, K48, and K63, with the other Lys residues mutated into Arginine (Arg), while K0 refers to the Ub mutant with all Lys substituted with Arg thus being Lys-free) or that have Lys mutated into Arg on these individual sites (KyR, y = 6, 11, 27, 29, 33, 48, 63). As shown in Fig. 4h, K48-linked polyUb chains were most efficiently conjugated by MARCH2-mediated chains onto PGAM5, while MARCH2 appeared to conjugate K48R (Lys48-Arg)-linked polyUb chain poorly onto PGAM5 under MCM + H/R challenge (Supplementary Fig. S9b). Moreover, truncation mutants of MARCH2 were used to explore the nature of interacting domains. GST pull-down assay showed that deletion of amino acids 117–214 or 215–246 disengaged the binding of MARCH2 to PGAM5, suggesting crucial roles for TM and PDZ domains of MARCH2 in direct interaction with PGAM5 (Fig. 4i). Next, MS analysis was performed to map ubiquitination sites on PGAM5 and two lysine residues of PGAM5 capable of being ubiquitinated (K88 and K141) were yielded. Then, we generated PGAM5 KO HL-1 cardiomyocyte cell line by CRISPR/Cas9-mediated genome editing technique to discern the ubiquitination sites of PGAM5. Remarkably, the combined mutants of K88R and K141R of PGAM5, PGAM5K88,141R, were slightly ubiquitinated by MARCH2 in MCM + H/R-challenged PGAM5 KO cardiomyocytes (Fig. 4j). Furthermore, compared to WT PGAM5, the homeostatic level of PGAM5K88,141R accumulated to a much higher level (Fig. 4k), most likely due to its longer half-life, as revealed by the pulse/chase experiment in the presence of CHX (Fig. 4l). Collectively, our findings suggest that MARCH2 promotes ubiquitination and degradation of PGAM5 by conjugating K48-linked polyUb onto K88 and K141 sites.
MARCH2 suppresses PGAM5-dependent NLRP3 inflammasome activation following myocardial I/R
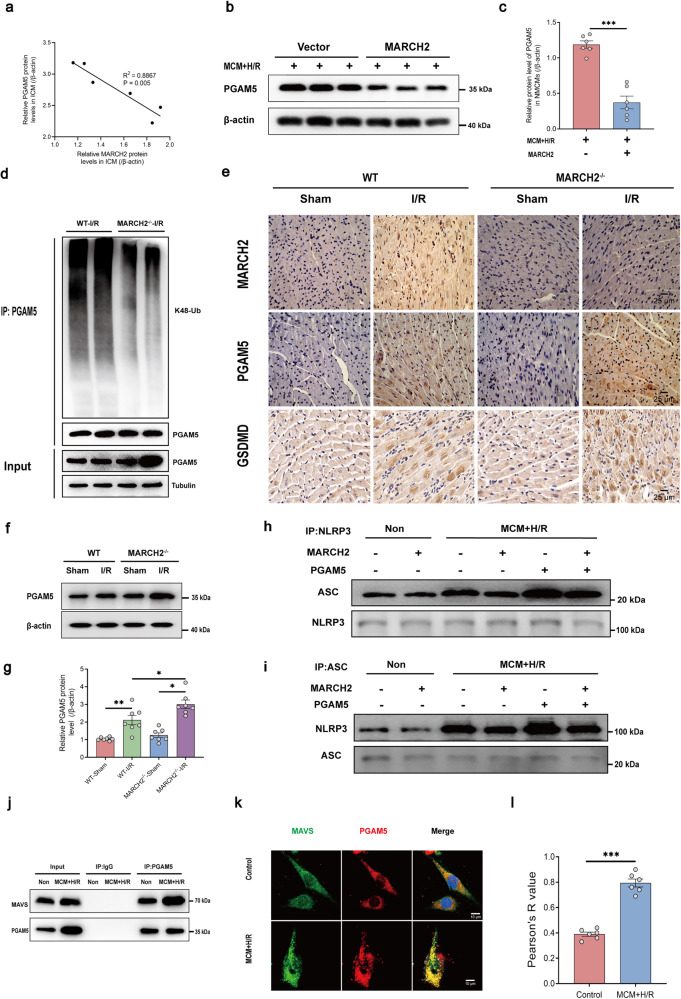
We then explored the potential roles of MARCH2-mediated PGAM5 degradation in cardiac I/R injury. Protein level of PGAM5 was negatively correlated with that of MARCH2 in patients with ICM (Fig. 5a). Protein level of PGAM5 was decreased in MARCH2-overexpressing NMCMs treated with MCM + H/R (Fig. 5b, c). Moreover, PGAM5 K48-linked ubiquitination was decreased in MARCH2 KO hearts compared with WT mice after I/R injury (Fig. 5d). PGAM5 was also elevated in mouse hearts following I/R procedure, and such effect was further augmented by genetic ablation of MARCH2 (Fig. 5e–g).
Fig. 5. PGAM5 mediates the regulation of MARCH2 on NLRP3 inflammasome assembly following myocardial I/R.
a Pearson correlation coefficients between MARCH2 and PGAM5 protein levels in ICM. b, c Representative western blotting (b) and quantitated analysis (c) of PGAM5 expression in NMCMs infected with vector or MARCH2 under MCM + H/R treatment. d Ubiquitination assays determining the ubiquitination of endogenous PGAM5 in the hearts of MARCH2 KO and WT mice subjected to I/R injury. e Immunohistochemistry for MARCH2, PGAM5, and GSDMD in hearts of MARCH2 KO and WT mice following I/R injury. f, g Representative western blotting image (f) and quantitated analysis (g) of PGAM5 in cardiac tissues of MARCH2 KO and WT mice subjected to I/R (45 min/9 h). h, i Interaction between NLRP3 and ASC was examined by IP-western blotting assay in cardiomyocytes with MARCH2 or PGAM5 overexpression following MCM + H/R challenge. IP with NLRP3 antibody (h), IP with ASC antibody (i). j Interaction between PGAM5 and MAVS was examined by IP-western blotting assay in cardiomyocytes with or without MCM + H/R treatment. k Representative immunofluorescence images of MAVS and PGAM5 in HL-1 cells. l Colocalization analysis of PGAM5–MAVS. Pearson’s R value (no threshold) was calculated by ImageJ Fiji software. n = 6 images from 3 biological replicates. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We next asked how MARCH2-mediated ubiquitin signaling affects PGAM5-mediated NLRP3 inflammasome. Co-IP assay with an anti-NLRP3 antibody in HL-1 cardiomyocytes indicated that MCM + H/R-elicited elevation of ASC was ablated by MARCH2 overexpression, which was counteracted by co-expression of PGAM5 (Fig. 5h, i). Moreover, the inhibition of MARCH2 on MCM + H/R-induced ASC oligomerization was mediated by PGAM5 (Supplementary Fig. S10).
The formation of PGAM5–MAVS co-condensates under MCM + H/R treatment and PGAM5 promotes MAVS-dependent NLRP3 activation
Given the pivotal role of PGAM5 in regulating activation of NLRP3 inflammasome21, we went on to screen for potential PGAM5-interacting proteins starting from clues found in a public protein–protein interaction database (https://www.ebi.ac.uk/intact/). Indeed, PGAM5 was capable of interacting with MAVS, an adapter of NLRP3 inflammasome that is required for NLRP3 inflammasome activity25. We found that endogenous MAVS interacts with PGAM5 protein, as anti-PGAM5 enriched MAVS along with PGAM5 but the scramble IgG did not (Supplementary Fig. S11a). Co-IP assay was then performed in HL-1 cardiomyocytes expressing Flag-tagged MAVS and HA-PGAM5. Flag-MAVS was found in the HA-PGAM5 immune complexes (Supplementary Fig. S11b).
Next, we sought to evaluate potential role of PGAM5–MAVS interaction in response to MCM + H/R treatment. Ectopically expressed PGAM5 did not elicit notable change in the total cellular level of MAVS protein in H/R-challenged NMCMs (Supplementary Fig. S11c, d). A recent study reported that PGAM5 may serve as a scaffolding protein to allow the formation of PGAM5–MAVS complex for immunity activation26. Furthermore, PGAM5 and MAVS had both been shown to form puncta structures26–28. Therefore, we speculated that PGAM5 may serve as a scaffold protein co-condensed with the client protein MAVS to evoke MAVS-mediated activation of NLRP3 inflammasome29. We observed that MCM + H/R induced a more robust interaction between endogenous PGAM5 and MAVS in HL-1 cardiomyocytes (Fig. 5j) and NMCMs (Supplementary Fig. S12a). Likewise, MCM + H/R also promoted PGAM5–MAVS colocalization in the phase-separated condensates in HL-1 cardiomyocytes (Fig. 5k, l) and NMCMs (Supplementary Fig. S12b, c).
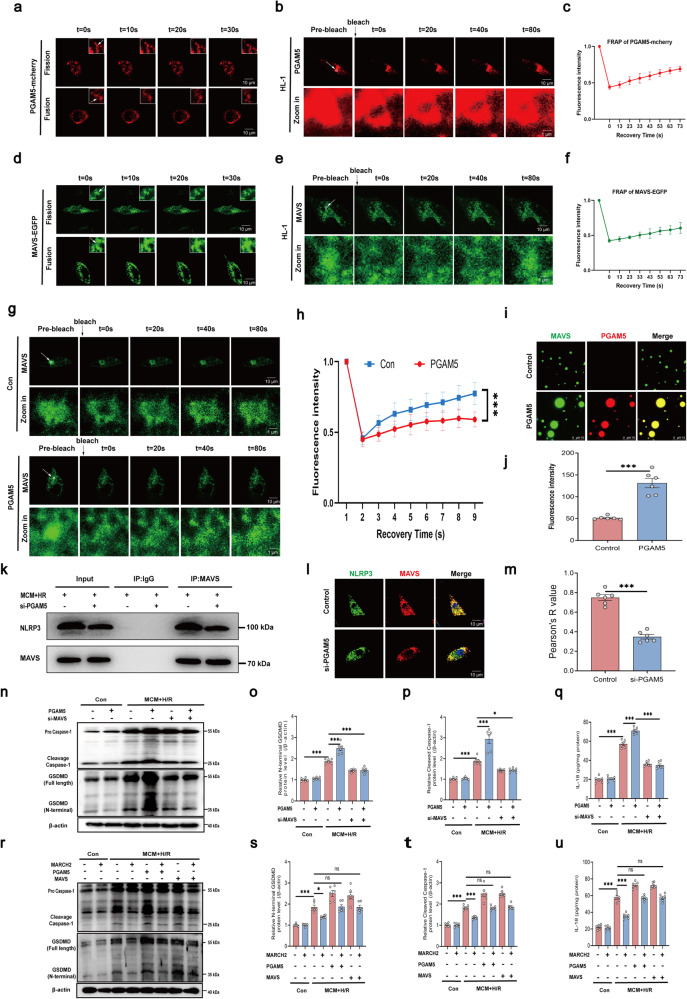
Our results indicated that PGAM5 condensates undergo fusion and fission in HL-1 cardiomyocytes (Fig. 6a) and HEK293T cells (Supplementary Fig. S13a) following MCM + H/R challenge, indicating liquid-like property of the PGAM5 condensates. Furthermore, fluorescence of PGAM5-mcherry also displayed a faster recovery following photobleaching under MCM + H/R challenge in HL-1 cardiomyocytes (Fig. 6b, c) and HEK293T cells (Supplementary Fig. S13b, c). Next, we reconstituted PGAM5 LLPS in vitro with purified full-length PGAM5-mcherry protein. As expected, PGAM5 droplets merged to grow with rising PGAM5 concentrations (Supplementary Fig. S13d), while the droplets disappeared with treatment of NaCl (Supplementary Fig. S13e). Moreover, MAVS-EGFP also formed condensates shown by droplet fission and fusion dynamically following MCM + H/R treatment in HL-1 cardiomyocytes (Fig. 6d) and HEK293T cells (Supplementary Fig. S13f). Upon photobleaching, MAVS-EGFP fluorescence rapidly recovered in HL-1 cardiomyocytes (Fig. 6e, f) and HEK293T cells (Supplementary Fig. S13g, h). Similarly, we reconstituted MAVS LLPS in vitro using purified full-length MAVS-EGFP protein. The formation of the MAVS-EGFP protein droplets was concentration-dependent (Supplementary Fig. S13i), and NaCl treatment abolished the formation of MAVS-EGFP droplets (Supplementary Fig. S13j). These results suggest that MAVS exhibits a dynamic liquid-like state under MCM + H/R treatment.
Fig. 6. PGAM5–MAVS co-condensates form under MCM + H/R challenge and PGAM5 promotes MAVS-dependent NLRP3 activation.
a Time-lapse images of HL-1 cells expressing PGAM5-mcherry. PGAM5 condensate fission and fusion is presented in the boxes. b FRAP analysis of PGAM5-mcherry condensates in HL-1 cells. c Quantification of FRAP in the bleached region of PGAM5-mcherry condensates, show as means ± SD (n = 6). d Time-lapse images of HL-1 cells expressing MAVS-EGFP. MAVS condensate fission and fusion is presented in the boxes. e FRAP analysis of MAVS-EGFP condensates in HL-1 cells. f Quantification of FRAP in the bleached region of PGAM5-mcherry condensates, show as means ± SD (n = 6). g FRAP analysis of MAVS-EGFP condensates in HL-1 cardiomyocytes without (upper) or with (lower) the presence of PGAM5. h Quantification of FRAP in the bleached region of MAVS-EGFP condensates with or without PGAM5, show as means ± SD (n = 6). i Representative fluorescent images of MAVS-EGFP droplets in the presence or absence of PGAM5. j Quantification of fluorescence intensity of MAVS-EGFP liquid droplets in i. k Interaction between NLRP3 and MAVS was examined by IP-western blotting assay in cardiomyocytes in the presence or absence of si-PGAM5 under MCM + H/R treatment. l Immunofluorescence of NLRP3 and MAVS in cardiomyocytes with or without PGAM5 knockdown under MCM + H/R treatment. m Colocalization analysis of NLRP3–MAVS. Pearson’s R value (no threshold) was calculated by ImageJ Fiji software. n = 6 images from 3 biological replicates. n–p Representative western blotting (n) and quantitated analyses of GSDMD (o) and caspase-1 (p) in cardiomyocytes infected with PGAM5 or si-MAVS under MCM + H/R treatment. q IL-18 release measured by ELISA in cardiomyocytes of the indicated groups underwent MCM + H/R treatment. r–t Representative western blotting (r) and quantitated analyses of GSDMD (s) and caspase-1 (t) in cardiomyocytes infected with MARCH2, PGAM5, or MAVS under MCM + H/R treatment. u IL-18 release measured by ELISA in cardiomyocytes of the indicated groups underwent MCM + H/R treatment. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
To test whether PGAM5 influences formation of MAVS condensate, fluorescence recovery after photobleaching (FRAP) assays were conducted in live HL-1 cardiomyocytes. The presence of PGAM5 prolonged MAVS-EGFP recovery process following photobleaching, suggesting that PGAM5–MAVS interaction might prompt the MAVS gel-like transition (Fig. 6g, h). Next, we examined whether PGAM5 induces MAVS phase separation in a cell-free condition using purified PGAM5-mcherry and MAVS-EGFP proteins. As expected, MAVS (30 μmol/L) formed more abundant and larger liquid droplets in the presence of PGAM5 (30 μmol/L) (Fig. 6i, j). To investigate whether PGAM5 can promote MAVS-mediated NLRP3 inflammasome activation, co-IP assay was performed in HL-1 cardiomyocytes in the presence or absence of PGAM5 following MCM + H/R treatment. Indeed, PGAM5 knockdown decreased binding between MAVS and NLRP3 in HL-1 cardiomyocytes (Fig. 6k) and NMCMs (Supplementary Fig. S14a). Furthermore, colocalization of NLRP3 and MAVS was significantly disrupted under PGAM5 knockdown in HL-1 cardiomyocytes (Fig. 6l, m) and NMCMs (Supplementary Fig. S14b, c). Of note, PGAM5 overexpression caused activation of NLRP3 inflammasome as shown by caspase-1 and GSDMD cleavage as well as IL-18 release in HL-1 cardiomyocytes following MCM + H/R treatment, the response to which was negated by MAVS knockdown (Fig. 6n–q; Supplementary Fig. S15a, b). To examine the formation of PGAM5–MAVS condensates in ischemic hearts in vivo, PGAM5 and MAVS were stained in sham and I/R mouse hearts and were found to exhibit an obvious condensation in I/R but not the sham hearts (Supplementary Fig. S16a, b). The number of PGMA5–MAVS condensates positively correlated with the PI-positive cardiomyocytes in hearts subjected to I/R injury (Supplementary Fig. S16c).
Moreover, MCM + H/R-evoked mitochondria-localization of inflammasome (colocalization of ASC and TOM20, Supplementary Fig. S17a), elevation of caspase-1 and GSDMD cleavage (Fig. 6r–t) as well as IL-18 release (Fig. 6u; Supplementary Fig. S17b, c) in HL-1 cardiomyocytes were blocked by MARCH2 overexpression, while PGAM5 or MAVS overexpression abrogated cardioprotective effects of MARCH2. Collectively, these results suggest a role of MARCH2 in inhibiting NLRP3 inflammasome activation in a PGAM5/MAVS-dependent manner.
MARCH2 overexpression protects against myocardial I/R injury
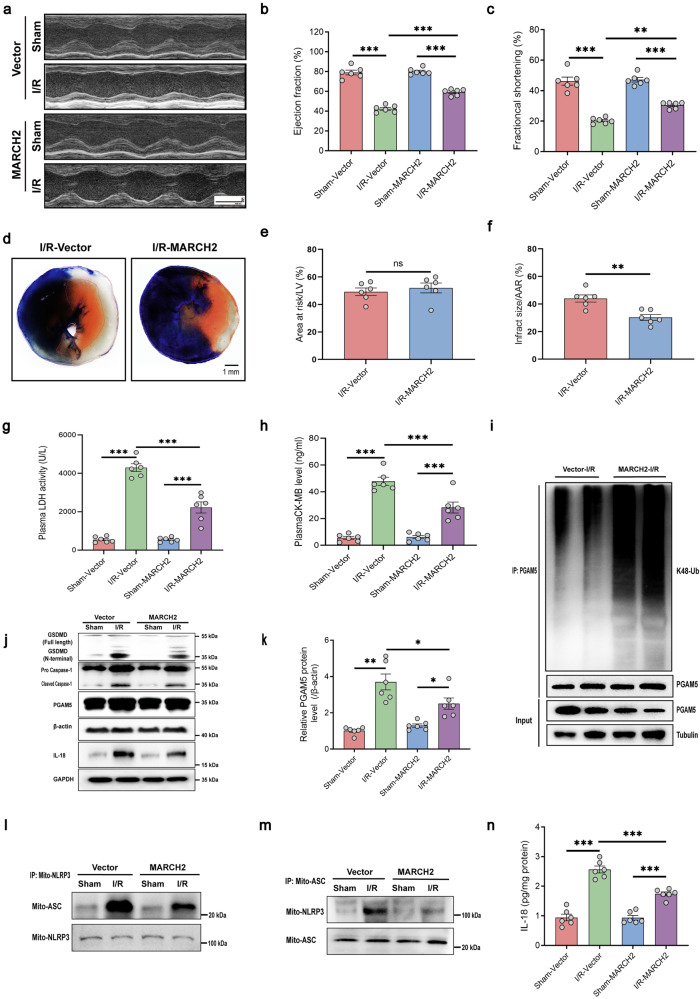
Given the observed role of MARCH2 in suppressing PGAM5/MAVS-mediated NLRP3 inflammasome activation, we explored therapeutic potential of MARCH2 for myocardial I/R injury. To this end, we constructed a viral vector to afford high MARCH2 expression in cardiomyocytes. Mice were delivered with AAV9-cTnT-MARCH2 3 weeks prior to I/R procedure. Notably, AAV9-cTnT-MARCH2 injection significantly elevated MARCH2 protein level in cardiomyocytes compared with AAV9-cTnT-Vector (Supplementary Fig. S18a, b). Remarkably, compared with AAV9-cTnT-Vector-injected mice, AAV9-cTnT-MARCH2-injected mice exhibited an overtly improved response to I/R injury, including improved cardiac function (Fig. 7a–c; Supplementary Fig. S18c–e) and decreased infarct size (Fig. 7d–f), as revealed by echocardiography and Evans blue/TTC double staining. As shown in Fig. 7g, h, levels of LDH and CK-MB, two markers for cardiac injury, were both less evident in the AAV9-cTnT-MARCH2 hearts compared to those received AAV9-cTnT-Vector. The PGAM5 K48-linked ubiquitination was increased (Fig. 7i) and PGAM5 protein level was decreased (Fig. 7j, k) in AAV9-cTnT-MARCH2 hearts compared to AAV9-cTnT-Vector hearts after I/R injury. Moreover, the intensified interaction between mitochondria-localized NLRP3 and ASC (Fig. 7l, m), caspase-1 and GSDMD cleavage (Fig. 7j), downstream proinflammatory IL-18 secretion after I/R (Fig. 7n) were reversed by AAV9-cTnT-MARCH2 injection. Altogether, MARCH2 manifests a previously unidentified protective property in cardiomyocytes against myocardial I/R injury, most likely through suppression of NLRP3 inflammation activation and cardiomyocyte pyroptosis.
Fig. 7. MARCH2 overexpression reduces I/R injury and preserves cardiac function via inhibition of PGAM5–MAVS-NLRP3 inflammasome pathway.
a Representative M-mode recordings of echocardiography of WT mice injected with AAV9-cTnT-NC or AAV9-cTnT-MARCH2 (2 × 1011 V.g/mouse) for 3 weeks and then subjected to I/R (45 min/24 h). b, c Quantitative analysis of echocardiographic measurements performed in AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R (45 min/24 h) injury (n = 6 mice per group). Left ventricular (LV) ejection fraction (EF, b) and fractional shortening (FS, c). d Representative images of heart sections by TTC/Evans Blue staining depicting infracted area. e Ratios of area at risk (AAR) to left ventricular (LV) area. f Infarct area normalized to AAR (n = 6 mice per group). g, h LDH activity and CK-MB level in AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice with or without myocardial I/R (45 min/24 h) (n = 6 mice per group). i Ubiquitination assays determining the ubiquitination of endogenous PGAM5 in the hearts of AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R injury. j Representative western blotting analysis of PGAM5, caspase-1 (procaspase1; cleaved caspase-1), GSDMD (full-length; N-terminal), and IL-18 in hearts of AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R (45 min/9 h). k Quantitated analysis of PGAM5 in hearts of AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R (45 min/9 h). l, m Interaction between NLRP3 and ASC in mitochondria of cardiac tissues of AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R was examined by IP-western blotting assay. n IL-18 release was measured by ELISA in cardiac tissues of AAV9-cTnT-NC or AAV9-cTnT-MARCH2 mice subjected to I/R. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Cardiac PGAM5–MAVS signaling is essential for the protective effect of MARCH2 against myocardial I/R injury
Next, endogenous PGAM5 in cardiomyocytes was knocked down through RNA interference based on AAV9 viral vectors to test whether MARCH2-evoked cardioprotective effect is dependent upon PGAM5. WT and MARCH2 KO mice were delivered with AAV9-cTnT-shPGAM5 3 weeks prior to I/R injury (Supplementary Fig. S19a). Evans blue/TTC double staining and echocardiography showed that cardiomyocyte-specific PGAM5 knockdown alleviated MARCH2 KO-exacerbated effects as shown by infarct size (Supplementary Fig. S19a–c) and cardiac function (Supplementary Fig. S19d, e). Increases in LDH and CK-MB in MARCH2 KO hearts were also restrained after cardiomyocyte-specific PGAM5 knockdown (Supplementary Fig. S19f, g). Consistently, increases in caspase-1 or GSDMD cleavage (Supplementary Fig. S19h–j) and IL-18 secretion (Supplementary Fig. S19k) in MARCH2 KO hearts were decreased when PGAM5 was knocked down. Therefore, PGAM5/MAVS signaling is indispensable for the protective function of MARCH2 against cardiac I/R injury.
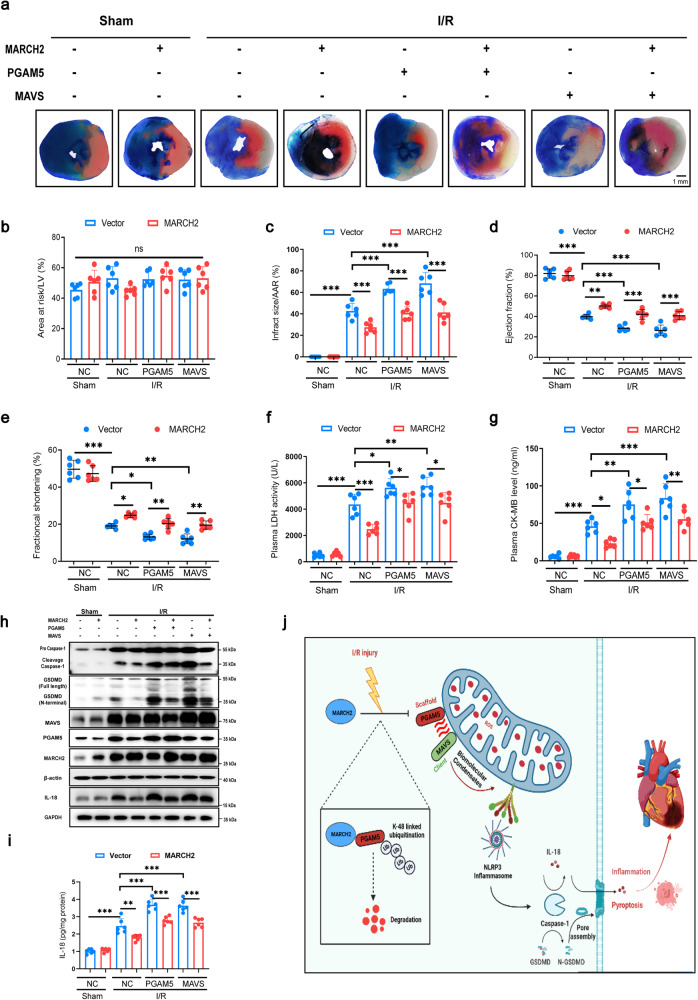
To further pinpoint the essential role of PGAM5/MAVS signaling in regulating myocardial I/R injury by MARCH2, AAV9 viral vectors were employed to afford stable expression of MARCH2, PGAM5 and MAVS, respectively. Mice were delivered with AAV9-cTnT-MARCH2 with or without AAV9-cTnT-PGAM5 and AAV9-cTnT-MAVS for 3 weeks before I/R (Fig. 8a). Evans blue/TTC double staining showed that cardiomyocyte-specific MARCH2 overexpression in mice significantly reduced I/R-induced infarct size, which was blocked by cardiomyocyte-specific PGAM5 or MAVS overexpression (Fig. 8a–c). I/R-elicited dramatic myocardial injury as evidenced by overt decreases in EF and FS (Fig. 8d, e) and simultaneous elevation in cardiac injury markers, including serum LDH and CK-MB (Fig. 8f, g), all of which were off-set by MARCH2 overexpression. In contrast, the PGAM5 or MAVS overexpression caused the opposite effect. Furthermore, MARCH2 overexpression effectively suppressed I/R-evoked caspase-1 and GSDMD cleavage (Fig. 8h), reduced IL-18 secretion (Fig. 8i) and PI-positive cardiomyocytes (Supplementary Fig. S20), which were again abolished by co-expression of PGAM5 and MAVS. Taken together, these results demonstrated that MARCH2 protects against myocardial I/R injury, through suppression of NLRP3 inflammation assembly and pyroptosis, in a PGAM5/MAVS-dependent manner.
Fig. 8. PGAM5/MAVS signaling pathway is essential for the regulation of MARCH2 in I/R injury.
a Prior to I/R injury, WT mice were administrated with AAV9-cTnT-vector, AAV9-cTnT-MARCH2, AAV9-cTnT-PGAM5 or AAV9-cTnT-MAVS (2.0 × 1011 V.g/mouse) for 3 weeks by tail vein injection. TTC/Evans Blue staining is used to depict infracted area. b Ratios of area at risk (AAR) to left ventricular (LV) area. c Infarct area normalized to AAR. No statistical significance for Vector-I/R vs MARCH2-PGAM5-I/R, no statistical significance for Vector-I/R vs MARCH2-MAVS-I/R. d, e Echocardiographic assessment of ejection fraction (EF, d) and fractional shortening (FS, e) in the indicated groups. No statistical significance for Vector-I/R vs MARCH2-PGAM5-I/R, no statistical significance for Vector-I/R vs MARCH2-MAVS-I/R. f, g LDH activity (f) and CK-MB level (g) in mouse with and without myocardial I/R. No statistical significance for Vector-I/R vs MARCH2-PGAM5-I/R; no statistical significance for Vector-I/R vs MARCH2-MAVS-I/R. h Representative western blotting results of MARCH2, PGAM5, mito-MAVS, caspase-1 (procaspase1; cleaved caspase-1), GSDMD (full-length; N-terminal), and IL-18 in hearts of the indicated group mice subjected to I/R (45 min/9 h). i IL-18 release was measured by ELISA in cardiac tissues of the indicated group mice subjected to I/R (45 min/9 h). No statistical significance for Vector-I/R vs MARCH2-PGAM5-I/R, no statistical significance for Vector-I/R vs MARCH2-MAVS-I/R. j A working model for MARCH2 function in I/R injury. MARCH2 interacts with and promotes the degradation of PGAM5 by facilitating its K48-linked polyUb, thus inhibiting the activity of MAVS/NLRP3 inflammasome pathway and cardiomyocyte pyroptosis. Data are shown as the means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Our findings indicated a prominent cardioprotective role for MARCH2 against myocardial I/R injury, inflammasome activation, and cardiomyocyte pyroptosis. MARCH2 is upregulated in ICM patients and I/R-challenged mouse hearts. MARCH2 ablation exacerbated I/R-evoked myocardial damage, as evidenced by infarct size, echocardiographic parameters, and cardiomyocyte mechanics. Our further analyses revealed that MARCH2 promotes degradation of PGAM5 by mediating its K48-linked polyUb, as well as mitigation of inflammatory response and pyroptotic cell death after I/R injury through a PGAM5/MAVS/NLRP3-dependent mechanism (Fig. 8j). In particular, PGAM5–MAVS condensates mediate activation of NLRP3 inflammasome, presumably acting as a PGAM5/MAVS/NLRP3 sponge and promoting caspase-1-dependent cleavage of GSDMD and IL-18. Moreover, ectopic expression of MARCH2 using AAV9-cTnT-MARCH2 effectively ameliorated I/R injury-evoked myocardial damages. In this context, MARCH2 is beneficial for I/R-elicited myocardial injury.
MARCH2 protects against I/R-induced injury by reducing inflammatory response
MARCH2, a crucial regulator of innate immune system, curbs the activation of proinflammatory responses. MARCH2 KO evokes excessive release of proinflammatory cytokines and correlates with a poor survival rate in response to bacterial infection and LPS-induced sepsis20. However, specific effect of MARCH2 in cardiovascular diseases remains unknown. Here, we reported that MARCH2 deficiency does not affect normal cardiac function and structure but augments response to myocardial I/R injury. Cardiomyocyte-specific MARCH2 overexpression ameliorates I/R injury by curbing infarct size, improving cardiac function, and minimizing LDH release (Fig. 7a–h). Our data have thus revealed a previously unidentified protective role of MARCH2 in myocardial I/R injury. Mechanically, scRNA-seq results suggested that NOD-like receptor and cytokine-mediated signaling pathway genes were upregulated in cardiomyocytes of MARCH2 KO mice compared to WT mice subjected to I/R, indicating a beneficial role for endogenous MARCH2.
Ample evidence has suggested a critical role for the activation of NLRP3 inflammasomes in myocardial damages and cardiomyocyte pyroptosis during myocardial I/R injury1,11–13. Upon I/R injury, mice with NLRP3 deletion or knockdown with siRNA exhibited reduced infarct size and improved cardiac function compared with WT mice, suggesting a protective role of inhibiting NLRP3 in injury. Noticeably, MARCH2 was reported for its role in negatively regulating NF-κB and type I interferon signalings20. Our present study demonstrated that MARCH2 restrains NLRP3 inflammasome complex assembly and subsequent myocardial pyroptosis. MARCH2 ablation augments I/R-induced elevation in levels of NLRP3, ASC, cleaved caspase-1, N-terminal GSDMD, and IL-18. Consistently, overexpression of MARCH2 blocks activation of NLRP3 inflammasome and cardiomyocyte pyroptosis, suggesting an essential role for MARCH2 in the regulation of inflammasome activation and myocardial damage (Fig. 7j–n).
MARCH2 promotes ubiquitination and degradation of PGAM5 by catalyzing K48-linked polyUb
Protein ubiquitination, a versatile post-translational modification, plays critical roles in cardiovascular system development, whose aberrances either feature or underlie a variety of cardiovascular diseases, including myocardial I/R injury30,31. The functional diversity of ubiquitination stems from structurally intricate polyUb chains, in which a single ubiquitin chain can be further ubiquitinated to form branched chains32–35. K48-linked chains are the prominent linkage form in cells that may target substrate proteins for the 26 S proteasomal degradation36. Previously, the ubiquitin modification of PGAM5 was reportedly catalyzed by E3 ligase RNF5, likely through K48-linked polyUb chains, which is known to inhibit the inflammatory responses in hepatic I/R injury37. In this study, we presented an array of data showing that the E3 Ub ligase MARCH2, which is abundantly expressed in cardiomyocytes and becomes upregulated during myocardial I/R injury, interacts explicitly with PGAM5, where the TM and PDZ domains of MARCH2 were attributed as the binding sites. Furthermore, MARCH2 mediates K48-linked polyUb modifications of PGAM5 under H/R challenge, targeting it to the proteasome for degradation. Interestingly, mutation of Lys residues 88 and 144 of PGAM5 led to decreased degradation of PGAM5 protein. Therefore, these Lys residues may serve as sites on PGAM5 to receive MARCH2-mediated ubiquitin signaling.
MARCH2 inhibits NLRP3 inflammasome activation through the PGAM5/MAVS/NLRP3 pathway
In this study, PGAM5 was found to interact with MARCH2, and MARCH2 conjugates K48-linked polyUb chains onto PGAM5, thus accelerating PGAM5 degradation (Fig. 4). Moreover, PGAM5 negatively correlated with MARCH2 in ICM and mediated regulation of MARCH2 on the MAVS/NLRP3 axis in myocardial I/R injury (Fig. 5). Our results indicate a crucial role for MARCH2-mediated K48-linked PGAM5 polyUb in myocardial I/R injury.
PGAM5 is a serine/threonine-protein phosphatase belonging to the PGAM family. It localizes in outer mitochondrial membrane and participates in a wide range of biological processes, including Wnt/β-catenin signaling38,39, redox regulation40, immune response41, and mitochondrial quality control42,43. Besides, PGAM5 functions at the converging point of several cell death pathways, including necrotic cell death22,44,45 and ROS-induced cell death46. Recently, it was reported that PGAM5 is dispensable for necroptosis although it offers a pivotal role in inflammasome activation and IL-1β release21. Our data suggested that protein level of PGAM5 was elevated in hearts of mice subjected to I/R injury. Cardiac-specific overexpression of PGAM5 exacerbated I/R-elicited infarct size, cardiac dysfunction, NLRP3 inflammasome assembly, and myocardial pyroptosis, indicating PGAM5-aggravated cardiac dysfunction and inflammation responses following I/R injury (Fig. 8). Moreover, inhibition of PGAM5 could also reduce necroptosis in rat hearts following I/R injury through suppression of dynamin-related protein 147, suggesting a key role for PGAM5-regulated other forms of cell death.
The optimal function of NLRP3 inflammasome is achieved through its recruitment to mitochondria by MAVS, a mitochondrial outer membrane-localized, antiviral signaling protein, and plays a pivotal role in innate immune system25. Here, we found that cardiac-specific overexpression of MAVS aggravates the deleterious effects of NLRP3 inflammasome activation and cardiac damages upon myocardial I/R injury, suggesting a deleterious effect of MAVS upregulation during cardiac injury48.
Biomolecular condensate formation regulated by LLPS in biology is now considered an essential mechanism to orchestrate complex cellular processes by concentrating certain molecules or excluding unwanted others49. Aberrant LLPS contributes to inflammasome activation,50 innate immunity51, and pathogenesis of arterial stiffening52. However, the potential roles of biomolecular condensation in cardiac diseases remain poorly understood. Here, we demonstrated that PGAM5 and MAVS are capable of undergoing LLPS in cardiomyocytes following MCM + H/R treatment. The compositions of biocondensates include scaffolds and clients. Scaffold proteins are elementary and essential for condensate formation, and the partition into condensates of client proteins is regulated by the scaffolds29. There is little information concerning how biocondensates are involved in cardiac physiology and pathophysiology. We found that PGAM5 functions as a scaffold protein that co-condenses with the client protein MAVS. It presumably acts like a PGAM5/MAVS/NLRP3 sponge to promote NLRP3 inflammasome activation. Therefore, targeting PGAM5 for proteasomal degradation by MARCH2-mediated proteolytic ubiquitin signaling serves as an endogenous brake on MAVS-NLRP3 inflammasome activation to alleviate myocardial injury during I/R stress. Therapeutic upregulation or activation of MARCH2 thus provides a novel avenue to combat these related diseases.
In summary, findings from our study not only established the E3 ligase MARCH2 as a novel cardioprotective molecule against myocardial I/R injury, but also identified a previously unknown mechanism that negatively regulates PGAM5/MAVS/NLRP3 inflammasome signaling and pyroptosis to maintain homeostasis and normal function of mammalian hearts. Targeting MARCH2 may thus represent a novel opportunity to battle against myocardial injuries.
Materials and methods
Human donors
This study was performed according to the ethical guidelines of the Declaration of Helsinki. Experimental procedures were approved by the institutional review board of the Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University (SYSEC-KY-KS-2021-070). Patients who have undergone heart transplant surgery due to ischemic heart disease were recruited in the study. Control donor hearts were obtained from participants who died in car accidents. Written informed consent was obtained from patients.
Animals
All animal procedures were approved by the institutional animal care and use committee of Zhongshan Hospital, Fudan University. Experiments were performed according to the institutional guidelines.
WT C57BL/6 J mice were purchased from Cyagen Biosciences Inc. (Guangzhou, China). MARCH2 KO mice were generated using CRISPR/Cas9 system. SgRNA targeting the sequence ACGGGATGGCCGGCTGCTC in exon 2 of mouse MARCH2 and Cas9 mRNA were injected into one-cell embryos for producing the offspring. The primers employed for MARCH2 genotyping were 5’-CTGCTGTTGTGACTCACTAATTTTG-3’ and 5’-CCAATGAGCCATTCCTCCAACATC-3’. Genotype was further verified by Sanger sequencing. The founder mouse with a 19 bp deletion in exon 2 of MARCH2 was utilized for our experiments (Supplementary Fig. 2a). Only F3 and beyond were used in this study.
Myocardial I/R procedure
Male mice aged 7–8 weeks were anesthetized with 2% isoflurane with 100% O2 ventilation at 2 L/min. Myocardial ischemia was induced using left anterior descending coronary artery (LAD) ligation by knotting a silk suture (6–0) around LAD. After 45 min of occlusion, the slipknot was released to allow a reperfusion period ranging 3–24 h. Sham surgery was performed by making a left thoracic incision to expose the pericardium except leaving the LAD intact. After reperfusion, blood samples were collected for LDH and CK-MB measurements.
Measurements of infarct area and area at risk
24 h after I/R injury, LAD artery was reoccluded and retied at the same ligation site. Evans blue dye (0.2 mL 1%) was injected into aorta to demarcate the ischemic AAR. Then, the heart was excised and subsequently cross-sectioned into 1-mm slices, which were incubated in 1% 2,3,5-triphenyltetrazolium chloride (TTC) solution for 10 min at 37 °C to delineate infarct size. The percentages of infarcted area (pale), AAR (brick red), and total left ventricular (LV) area of each myocardial sections were measured using Image J software, and the values were averaged.
Echocardiography
Transthoracic echocardiography was performed in mice using a high-resolution micro-imaging system (Visual Sonics Vevo 2100, Toronto, ON, Canada) to evaluate cardiac function and structure. Standard two-dimensional M-mode measurements was performed at 24 h after I/R surgery on mice anesthetized with 2% isoflurane. Measurements of end systole and end diastole LV internal diameters, LV anterior/posterior wall thickness, and interventricular septum were performed using M-mode recordings.
Cardiomyocyte isolation and cell culture
NMCMs were isolated according to the previous report53 with modifications. In brief, hearts from 1-day-old mice were minced and digested with collagenase I (Worthington) for 3 min at 37 °C. The collected cells were incubated in fresh culture medium (DMEM containing 20% fetal calf serum (FBS)) at 37 °C and 5% CO2 for 90 min to separate cardiac fibroblasts from cardiomyocytes.
Adult cardiomyocytes were enzymatically isolated from adult mouse hearts according to a previous report3. Adult cardiomyocytes were plated onto laminin (5 μg/mL, Sigma, St. Louis, USA)-precoated culture plates.
HEK293T and HL-1 murine cardiomyocyte, Raw264.7 murine macrophages were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS, and penicillin/streptomycin under a humidified atmosphere containing 5% CO2 at 37 °C.
MCM was obtained from the medium of Raw264.7 macrophages treated with LPS (200 ng/mL, Sigma) for 6 h and NLRP3 activator ATP (2 nM, Sigma) for 30 min. To mimic the H/R injury and inflammatory environment, HL-1 cardiomyocytes and NMCM cardiomyocytes were treated with MCM in H/R (3 h/6 h) atmosphere.
Cell shortening/re-lengthening
Mechanical properties of cardiomyocytes were assessed using a Soft-edge MyoCam system (IonOptix Corporation, Milton, MA, USA) equipped with an IX-70 Olympus inverted microscope. Single cardiomyocytes were electrically stimulated at 0.5 Hz in a contractile buffer to detect these properties.
Plasmid, small RNA interference, and adenovirus
MARCH2, PGAM5, MAVS, and Flag-Ub were amplified from HEK293T cDNA through standard PCR techniques, and then inserted into pcDNA3.1 (+) vectors. Point mutations of the indicated plasmids of ubiquitin chain were introduced through site-directed mutagenesis.
PGAM5-mcherry, MAVS-EGFP, and the selective siRNAs and negative controls were designed and synthesized by Jiangsu Gencefe Biotechnology Co., Ltd. (Jiangsu, China). Control siRNA was employed as a negative control under similar conditions. siRNAs used in our experiment were: mouse MARCH2 siRNA sequence: 5′-GCCACCUCAAUAUGUAGCACATT-3′; mouse MAVS siRNA sequence: 5′-CCACCUUGAUGCCUGUGAATT-3′. Plasmid or siRNA transfection was conducted using Lipofectamine 3000 reagent (Thermo Fisher Scientific, USA).
Adenovirus expressing MARCH2, shMARCH2, PGAM5, MAVS were conducted by the OBiO Technology Corp., Ltd. (Shanghai, China). For MARCH2, PGAM5, or MAVS overexpression in mice, the entire coding region of the mouse MARCH2, PGAM5, or MAVS cDNA was subcloned into the pHBAAV9-cTNT-MCS vector downstream of the cardiac troponin-T promoter. Mice were given adenovirus (2.0 × 1011 V.g /mouse) using tail intravenous injection. Cardiac-restricted MARCH2, PGAM5, or MAVS overexpression was successfully induced 3 weeks after AAV9 transfection in adult C57BL/6 J mice. The corresponding control mice received pHBAAV9-cTNT-Vector injections at the same dose respectively. For PGAM5 knockdown, AAV9 harboring PGAM5 shRNA (pAAV-cTNT-miR30 shRNA (PGAM5)) was constructed.
Generation of PGAM5 knockout cell lines
PGAM5–/– HL-1 cardiomyocyte cell line was generated by CRISPR/Cas9-mediated genome editing technique as previously described. The sgRNAs targeting PGAM5 were designed using online tool (http://crispr.mit.edu/). The sgRNA-expressing plasmids were transfected into HL-1 cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 24 h, and selected with puromycin (Sigma) for single colonies. Single-cell colonies were picked, amplified, and confirmed by immunoblotting analysis. We then extracted genomic DNAs, amplified the specific target sequences, and performed Sanger sequencing to verify the aimed edits in the genomes of the HL-1 cell line.
Western blotting assay
Total protein was extracted from heart tissues or cells. Equal amounts of protein (generally 25 or 50 μg) were separated by SDS-PAGE gels and subsequently transferred to a nitrocellulose membrane. After blocking with 5% bovine serum albumin (BSA), the membranes were incubated with the indicated primary antibodies at 4 °C overnight.
Antibodies used were anti-MARCH2 (1:1000, Abcam, Ab123136), anti-PGAM5 (1:1000, Abcam, ab126534), anti-MAVS (1:1000, Cell Signaling Technology, #83000), anti-GSDMD (1:1000, Abcam, ab209845), anti-NLRP3 (1:1000, Cell Signaling Technology, #15101), anti-Caspase-1 (1:1000, Proteintech, 22915-1-AP), anti-ASC (1:1000, Cell Signaling Technology, #67824), anti-VDAC1 (1:1000, Abcam, ab154856), anti-VDAC2 (1:1000, Proteintech, 11663-1-AP), anti-HSP90 (1:1000, Proteintech, 11405-1-AP), anti-MFN2 (1:1000, Proteintech, 12186-1-AP), anti-MCU (1:1000, Abcam, ab219827), anti-Ubiquitin (K48) (1:1000, Abcam, ab140601), Anti-Flag (1:1000, Cell Signaling Technology, #14793), Anti-HA (1:1000, Cell Signaling Technology, #2367), Anti-Myc (1:1000, Cell Signaling Technology, #2276). Horseradish-peroxidase (HRP)-conjugated β-actin (1:1000, Proteintech, HRP-60008), HRP-conjugated α-Tubulin (1:1000, Proteintech, HRP-66031).
After washed with TBST for three times, membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 1 h. Bands were scanned by enhanced chemiluminescence luminal reagents (Bio-Rad Laboratories). Gray value was qualified by Image Lab (Bio-Rad, USA).
For chemical cross-linkage of ASC oligomerization, the Triton X-100 insoluble pellet was re-suspended in PBS buffer and chemically crosslinked with 4 mM disuccinimidyl suberate (DSS) at room temperature for 30 min.
RT-qPCR
Total RNA was extracted from heart tissues or cells using Trizol Reagent (Invitrogen). First-strand cDNA was generated from total RNA using the Prime Script™ RT Master Mix Kit (Takara, Tokyo, Japan). RT-qPCR was performed in 25 μL reactions including 0.4 μmol/L primers, 50 ng cDNA, and 12.5 μL TB Green Premix Ex Taq II (Takara) using the CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories). The expression level of targeted genes was normalized to that of belta-actin, which was regarded as an endogenous internal control. Primer sequences were as follows: mouse MARCH2: forward primer 5′-AGGGCTCAGAGGTAGTAGACA-3′, reverse primer 5′-AAATATGTTCACACGGGATCACA-3′; mouse belta-actin: forward primer 5′-GGCTGTATTCCCCTCCATCG-3′, reverse primer 5′-CCAGTTGGTAACAATGCCATGT-3′; mouse PGAM5: forward primer 5′-ATCTGGAGAAGACGAGTTGACA-3′, reverse primer 5′-CCTGTTCCCGACCTAATGGT-3′.
Cell model of H/R injury
Cardiomyocytes were placed in an “ischemic buffer” (118 mM NaCl, 24 mM NaHCO3, 1.0 mM NaH2PO4, 2.5 mM CaCl2-2H2O, 1.2 mM MgCl2, 20 mM sodium lactate, 16 mM KCl, and 10 mM 2-deoxyglucose, pH adjusted to 6.2) and transferred to a controlled hypoxic chamber (1% O2, 94% N2, 5% CO2) for 30 min. Then cardiomyocytes were placed back in a normal humidified atmosphere containing 5% CO2 at 37 °C and replaced with a culture medium for reoxygenation.
Immunofluorescence staining and live cell imaging
Cells grown on confocal dish were washed with PBS, then fixed with 4% paraformaldehyde for 15 min, and permeated with 0.1% Triton X-100 plus 1% BSA for 1 h at room temperature. Subsequently, primary antibodies against Flag (Cell Signaling Technology, Cat. #8146), HA (Cell Signaling Technology, #3724), ASC (Santa Cruz, #SC-514414), TOM20 (Abcam, #ab186735) were used to label cellular proteins at 4 °C overnight. After washed with PBS for three times, cells were then stained with a fluorescent second antibody at 37 °C for 1 h. DNA was stained with DAPI. The fluorescence signals of fixed cells and live cells were acquired using a confocal microscopy (Leica TCS SP8).
Quantification of fluorescence intensity was performed with ImageJ software. Statistic data were acquired from ~40–100 cells from 6 random fields from at least three independent experiments. ImageJ Fiji software (Analyze-colocalization-coloc2) was used to calculate the Pearson colocalization coefficient. Statistic data were obtained from 6 random fields from at least 3 independent experiments.
ELISA
Levels of CK-MB and LDH were determined as markers for cardiac injury. Levels of CK-MB and LDH were measured using CK-MB assay and LDH assay kits (Nanjing Jiancheng, Nanjing, China) according to the manufacturer’s instructions.
Mitochondrial purification
Mitochondria were isolated with a mitochondrial isolation Kit (Abcam, Cambridge, USA). Briefly, cells were harvested and homogenized in isolation buffer. Following centrifugation at 1000 × g for 10 min, the supernatant was collected and was then centrifuged at 12,000 × g for 15 min at 4 °C, yielding enriched mitochondria in pellets. Mitochondrial pellets were re-suspended using a radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail. Cardiac tissues (30–50 mg) were washed in wash buffer and homogenized in isolation buffer. Then homogenized tissue was centrifuged at 1000× g for 10 min, supernatants were collected. After centrifuged at 12,000× g for 15 min, pellets were resuspend using RIPA buffer and were quantified using the Bradford method.
FRAP assay
Cells were grown on glass-bottom dishes until they reached the appropriate density. Cells were then subjected to MCM + H/R treatment. GFP or mcherry signals in regions of interest (ROI) were fully photobleached by using a 488-nm laser on a Leica SP8 confocal microscope. Fluorescence intensity of ROI between pre-bleached and at the start of recovery after bleaching was recorded by microscope. FRAP data were analyzed using GraphPad Prism 8.4.
In vitro droplet assay
Purified MAVS-EGFP and PGAM5-mcherry proteins were diluted to the indicated concentrations with a phase separation buffer containing 20 mM Tris-HCl (pH = 7.5), 75 mmol/L KCl, 10% BSA, 5% PEG, and 1 mmol/L DTT, and the mixtures were incubated at 37 °C for 10 min to induce phase separation.
Protein purification
Plasmid of pGEX-4T-1-GST or pGEX-4T-1-GST-PGAM5, pGEX-4T-1-GST-MARCH2-truncated proteins, pGEX-4T-1-MAVS-EGFP was expressed in Escherichia coli strain BL21 competent cells and induced with IPTG at 16 °C for 12 h. Bacteria were collected by centrifugation and lysed by sonication. Lysate was immobilized on glutathione-agarose beads (GE Healthcare) following the manufacturer’s protocols; pET28a-His-MARCH2, pET28a-His-PGAM5, or pET28a-His-PGAM5-mcherry proteins were purified with Ni-NTA agarose beads (Qiagen) per the manufacturer’s instruction. The pull-down assays of other proteins followed the same procedures.
IP assay
Cells were lysed with 500 μL NP-40 lysis buffer (20 mM Tris-HCl pH 7.4, 1% NP-40, 135 mM NaCl, 10% glycerol, 0.2 mM PMSF, phosphatase inhibitors, and protease inhibitor). Lysates were collected and subjected to centrifugation. After centrifugation, 50 μL of supernatant was aliquoted for input. The remained supernatant was immunoprecipitated with the indicated primary antibodies and protein A/G Agarose beads (Santa Cruz Biotechnology) at 4 °C. Anti-mouse IgG (Santa Cruz Biotechnology, #sc2025) was used as a control. The IP complexes were washed four times with lysis buffer to remove nonspecifically bound protein. Both input and immunoprecipitates were examined by immunoblotting.
IP/MS
Following transfection with Flag-tagged MARCH2 in HEK293T cells, IP was performed using an anti-Flag primary antibody. Precipitates were separated using SDS-PAGE gels and were subsequently stained with Coomassie blue. Bands that differed between the Flag-MARCH2 and control groups were analyzed using liquid chromatograph-electrospray ionization-mass spectrometry (Shanghai Biotechnology, Shanghai, China).
Mass spectrometry analysis to map ubiquitylation modification sites on substrates
Procedures for MS analysis were as previously described34. In brief, the protein pellet was concentrated in 8 M urea, 100 mM Tris-Cl (pH 8.5), followed by TCEP reduction, NEM alkylation, and trypsin digestion. Peptides were separated and analyzed by the EASY-nLC system (Thermo Fisher) and the Q Exactive mass spectrometer (Thermo Fisher), respectively. Ubiquitylation modification sites were determined with Thermo Proteome Discoverer 2.1 (Thermo Fisher) and searched in Uniprot Human database (http://www.uniprot.org/).
PI staining
Mice were injected with PI (Sigma, 10 mg/kg, i.p.) to label necrotic cells following I/R surgery. Briefly, hearts were quickly removed and washed with PBS, embedded in OCT, frozen in liquid nitrogen, and cut into 5-μm sections. Frozen sections were then stained with DAPI. The immunofluorescence staining was conducted as described above.
Ubiquitylation assays
For in vivo PGAM5 ubiquitylation assay, HEK293T and HL-1 cells were transfected with Myc-MARCH2, HA-PGAM5, and Flag-ubiquitin using Lipofectamine 3000. 24 h later, whole cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, with phosphatase and protease inhibitors). The lysates were centrifuged to obtain supernatant proteins and incubated with anti-Flag antibody (Cell Signaling Technology) for 3 h and protein A/G agarose beads (Santa Cruz Biotechnology) for 3 h at 4 °C. Then the immune complex was washed extensively for four times with lysis buffer and boiled with SDS-PAGE sample buffer for 10 min. Ubiquitination was analyzed by immunoblotting with anti-Flag monoclonal antibody (Cell Signaling Technology).
ScRNA-seq and data analyses
WT male mice and MARCH2 KO male mice aged 7–8 weeks suffering sham or I/R injury were used for scRNA-seq (3 mice/group). ScRNA-seq was conducted by Singleron Biotechnologies (Shanghai, China). In brief, cells were isolated from heart tissues using GEXSCOPE Nuclear Extraction Buffer (adding RNase inhibitor and DTT before use, the final concentrations of the two are 0.2 U/µL, 1 mM, respectively). Single-cell suspensions at a concentration of 1 × 105 cells/mL in PBS were prepared. Single-cell suspensions were then loaded onto microfluidic devices and scRNA-seq libraries were constructed according to Singleron GEXSCOPE protocol using GEXSCOPE Single-Cell RNA Library Kit (Singleron Biotechnologies), which included cell lysis, mRNA trapping, labeling cells (barcode) and mRNA (UMI), mRNA reverse transcription, cDNA amplification, and finally cDNA fragmentation. Individual libraries were diluted to 4 nM and pooled for sequencing. Pools were sequenced on an Illumina HiSeq X with 150 bp paired-end reads. Raw reads were processed with fastQC and fastp to remove low quality reads. Poly-A tails and adaptor sequences were removed by cutadapt. Reads were mapped to the reference genome GRCh38 (ensembl version 92 annotation) using STAR. Gene counts and UMI counts were acquired by featureCounts software. Cells were filtered by gene counts below 200 and the top 2% gene counts and the top 2% UMI counts. Cells with over 20% mitochondrial content were removed. After filtering, 178,274 cells were retained for the downstream analyses, with on average 1794 genes and 3808 UMIs per cell. Expression matrix files for subsequent analyses were generated based on gene counts and UMI counts. FindMarker (Seurat) was used for DEG analysis. To identify the genes that may participate in the pathogenesis of myocardial I/R injury, we screened candidate genes from the scRNA-seq of WT-sham group and WT-I/R group adopting the following strategy: (1) upregulated DEGs (lnx-FoldChange > 0.25, adjusted P values < 0.05) of the comparison were partitioned; (2) downregulated DEGs (lnx-FoldChange2 < –0.25, adjusted P values < 0.05) of the comparison were partitioned; (3) the filtered upregulated DEGs and downregulated DEGs were then intersected with the E3 Ub ligase database.
Statistical analysis
For statistical analysis, all quantitative data are presented as means ± SEM of at least three independent experiments. Data distribution was first examined using the Shapiro–Wilk normality test. Statistical differences were performed using a two-tailed unpaired Student’s t-test for comparison between two groups. Statistical differences among three or more groups were determined by one-way ANOVA or two-way ANOVA (for two-factor levels), followed by Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001 were considered significant. Statistical analyses were carried out using GraphPad prism 8.0 and SPSS Version 19.0.
Supplementary information
Acknowledgements
We thank the support of Junmeng Zheng from the Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University. This work was supported by the National Key R&D Program of China (2017YFA0506000), the National Natural Science Foundation of China (82270267 and 81900233), the Program of Shanghai Academic/Technology Research Leader (20XD1420900), the Postdoctoral Science Foundation of China (2019M661375), the National Science and Technology Innovation 2030 Major Project of China (2021ZD0203900), Ministry of Science and Technology of China (2021ZD0203900, 2019YFA0802103), Department of Science and Technology of Zhejiang Province (2021C03104), Guangzhou Science Innovation and Development Program (201803010092), Shenzhen-Hong Kong Institute of Brain Science (NYKFKT2019006).
Author contributions
Y.Z., R.H., and J.G. designed the study. S.L., Y.B., T.H., Y.E.L., and C.X. performed the animal experiments and the in vitro experiments. Q.W. and N.N.W. analyzed the data. S.L. wrote the manuscript.
Data availability
Raw scRNA-seq data have been deposited in the National Omics Data Encyclopedia (https://www.biosino.org/node/search) under the accession number: OEP004577.
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shuolin Liu, Yaguang Bi, Tianting Han
Change history
3/18/2024
The incorrect display order for the following two authors Yaguang Bi and Yingmei Zhang was corrected.
Contributor Information
Junbo Ge, Email: ge.junbo@zs-hospital.sh.cn.
Ronggui Hu, Email: coryhu@sibcb.ac.cn.
Yingmei Zhang, Email: zhang.yingmei@zs-hospital.sh.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41421-023-00622-3.
References
- 1.Toldo S, Abbate A. The NLRP3 inflammasome in acute myocardial infarction. Nat. Rev. Cardiol. 2018;15:203–214. doi: 10.1038/nrcardio.2017.161. [DOI] [PubMed] [Google Scholar]
- 2.Deng Y, et al. Targeting mitochondria-inflammation circuit by β-hydroxybutyrate mitigates HFpEF. Circ. Res. 2021;128:232–245. doi: 10.1161/CIRCRESAHA.120.317933. [DOI] [PubMed] [Google Scholar]
- 3.Shi H, et al. GSDMD-mediated cardiomyocyte pyroptosis promotes myocardial I/R injury. Circ. Res. 2021;129:383–396. doi: 10.1161/CIRCRESAHA.120.318629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma BR, Kanneganti TD. NLRP3 inflammasome in cancer and metabolic diseases. Nat. Immunol. 2021;22:550–559. doi: 10.1038/s41590-021-00886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Carvalho Ribeiro M, Szabo G. Role of the inflammasome in liver disease. Annu. Rev. Pathol. 2022;17:345–365. doi: 10.1146/annurev-pathmechdis-032521-102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Heijden T, et al. NLRP3 inflammasome inhibition by MCC950 reduces atherosclerotic lesion development in apolipoprotein E-deficient mice-brief report. Arterioscler. Thromb. Vasc. Biol. 2017;37:1457–1461. doi: 10.1161/ATVBAHA.117.309575. [DOI] [PubMed] [Google Scholar]
- 7.Masters SL, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed GW, Rossi JE, Cannon CP. Acute myocardial infarction. Lancet. 2017;389:197–210. doi: 10.1016/S0140-6736(16)30677-8. [DOI] [PubMed] [Google Scholar]
- 9.Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N. Engl. J. Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 10.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandanger Ø, et al. The NLRP3 inflammasome is up-regulated in cardiac fibroblasts and mediates myocardial ischaemia-reperfusion injury. Cardiovasc. Res. 2013;99:164–174. doi: 10.1093/cvr/cvt091. [DOI] [PubMed] [Google Scholar]
- 12.Toldo S, et al. Inhibition of the NLRP3 inflammasome limits the inflammatory injury following myocardial ischemia-reperfusion in the mouse. Int. J. Cardiol. 2016;209:215–220. doi: 10.1016/j.ijcard.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 13.Sandanger Ø, et al. NLRP3 inflammasome activation during myocardial ischemia reperfusion is cardioprotective. Biochem. Biophys. Res. Commun. 2016;469:1012–1020. doi: 10.1016/j.bbrc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, et al. Targeted protein degradation as a powerful research tool in basic biology and drug target discovery. Nat. Struct. Mol. Biol. 2020;27:605–614. doi: 10.1038/s41594-020-0438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H, Li S, Shu HB. The membrane-associated MARCH E3 ligase family: emerging roles in immune regulation. Front. Immunol. 2019;10:1751. doi: 10.3389/fimmu.2019.01751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura N, Fukuda H, Kato A, Hirose S. MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol. Biol. Cell. 2005;16:1696–1710. doi: 10.1091/mbc.e04-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia D, et al. MARCH2 regulates autophagy by promoting CFTR ubiquitination and degradation and PIK3CA-AKT-MTOR signaling. Autophagy. 2016;12:1614–1630. doi: 10.1080/15548627.2016.1192752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Mintern JD, Villadangos JA. MARCH ligases in immunity. Curr. Opin. Immunol. 2019;58:38–43. doi: 10.1016/j.coi.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Lu J, Liu X. MARCH2 is upregulated in HIV-1 infection and inhibits HIV-1 production through envelope protein translocation or degradation. Virology. 2018;518:293–300. doi: 10.1016/j.virol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Chathuranga K, et al. Negative regulation of NEMO signaling by the ubiquitin E3 ligase MARCH2. EMBO J. 2020;39:e105139. doi: 10.15252/embj.2020105139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriwaki K, et al. The mitochondrial phosphatase PGAM5 is dispensable for necroptosis but promotes inflammasome activation in macrophages. J. Immunol. 2016;196:407–415. doi: 10.4049/jimmunol.1501662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, et al. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, et al. Phosphoglycerate mutase 5 exacerbates cardiac ischemia-reperfusion injury through disrupting mitochondrial quality control. Redox Biol. 2021;38:101777. doi: 10.1016/j.redox.2020.101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng, X., Xia, Z., Yang, H. & Hu, R. The ubiquitin codes in cellular stress responses. Protein Cell10.1093/procel/pwad045 (2023). [DOI] [PMC free article] [PubMed]
- 25.Subramanian N, et al. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu YQ, et al. PGAM5-MAVS interaction regulates TBK1/IRF3 dependent antiviral responses. Sci. Rep. 2020;10:8323. doi: 10.1038/s41598-020-65155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021;23:718–732. doi: 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 28.Hou J, et al. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat. Commun. 2021;12:2970. doi: 10.1038/s41467-021-23219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitrea DM, et al. Modulating biomolecular condensates: a novel approach to drug discovery. Nat. Rev. Drug Discov. 2022;21:841–862. doi: 10.1038/s41573-022-00505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C, et al. Inadequate ubiquitination-proteasome coupling contributes to myocardial ischemia-reperfusion injury. J. Clin. Invest. 2018;128:5294–5306. doi: 10.1172/JCI98287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis MS, et al. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ. Res. 2010;106:463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swatek KN, et al. Insights into ubiquitin chain architecture using Ub-clipping. Nature. 2019;572:533–537. doi: 10.1038/s41586-019-1482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu R, Hochstrasser M. Recent progress in ubiquitin and ubiquitin-like protein (Ubl) signaling. Cell Res. 2016;26:389–390. doi: 10.1038/cr.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, et al. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 2018;28:48–68. doi: 10.1038/cr.2017.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng H, et al. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27:657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding MJ, et al. E3 ubiquitin ligase ring finger protein 5 protects against hepatic ischemia reperfusion injury by mediating phosphoglycerate mutase family member 5 ubiquitination. Hepatology. 2022;76:94–111. doi: 10.1002/hep.32226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernkopf DB, et al. Pgam5 released from damaged mitochondria induces mitochondrial biogenesis via Wnt signaling. J. Cell Biol. 2018;217:1383–1394. doi: 10.1083/jcb.201708191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauschenberger V, et al. The phosphatase Pgam5 antagonizes Wnt/β-Catenin signaling in embryonic anterior-posterior axis patterning. Development. 2017;144:2234–2247. doi: 10.1242/dev.144477. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, et al. KEAP1 is a redox sensitive target that arbitrates the opposing radiosensitive effects of parthenolide in normal and cancer cells. Cancer Res. 2013;73:4406–4417. doi: 10.1158/0008-5472.CAN-12-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang YJ, et al. Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 2015;6:8371. doi: 10.1038/ncomms9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 43.Yu B, et al. Mitochondrial phosphatase PGAM5 modulates cellular senescence by regulating mitochondrial dynamics. Nat. Commun. 2020;11:2549. doi: 10.1038/s41467-020-16312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He GW, et al. PGAM5-mediated programmed necrosis of hepatocytes drives acute liver injury. Gut. 2017;66:716–723. doi: 10.1136/gutjnl-2015-311247. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, et al. Inhibitory effect of melatonin on necroptosis via repressing the Ripk3-PGAM5-CypD-mPTP pathway attenuates cardiac microvascular ischemia-reperfusion injury. J. Pineal Res. 2018;65:e12503. doi: 10.1111/jpi.12503. [DOI] [PubMed] [Google Scholar]
- 46.Holze C, et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 2018;19:130–140. doi: 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.She L, et al. Inhibition of phosphoglycerate mutase 5 reduces necroptosis in rat hearts following ischemia/reperfusion through suppression of dynamin-related protein 1. Cardiovasc. Drugs Ther. 2019;33:13–23. doi: 10.1007/s10557-018-06848-8. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Zhang S, Li F, Qin L. NLRX1 attenuates apoptosis and inflammatory responses in myocardial ischemia by inhibiting MAVS-dependent NLRP3 inflammasome activation. Mol. Immunol. 2016;76:90–97. doi: 10.1016/j.molimm.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Su Q, Mehta S, Zhang J. Liquid-liquid phase separation: Orchestrating cell signaling through time and space. Mol. Cell. 2021;81:4137–4146. doi: 10.1016/j.molcel.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen C, et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell. 2021;184:5759–5774.e5720. doi: 10.1016/j.cell.2021.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, et al. The STING phase-separator suppresses innate immune signalling. Nat. Cell Biol. 2021;23:330–340. doi: 10.1038/s41556-021-00659-0. [DOI] [PubMed] [Google Scholar]
- 52.Liu J, et al. Liquid-liquid phase separation of DDR1 counteracts the Hippo pathway to orchestrate arterial stiffening. Circ. Res. 2023;132:87–105. doi: 10.1161/CIRCRESAHA.122.322113. [DOI] [PubMed] [Google Scholar]
- 53.Yu W, et al. TBC1D15/RAB7-regulated mitochondria-lysosome interaction confers cardioprotection against acute myocardial infarction-induced cardiac injury. Theranostics. 2020;10:11244–11263. doi: 10.7150/thno.46883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw scRNA-seq data have been deposited in the National Omics Data Encyclopedia (https://www.biosino.org/node/search) under the accession number: OEP004577.