Abstract
DNA damage recognition by basal transcription factors follows different mechanisms. Using transcription-competition, nitrocellulose filter binding, and DNase I footprinting assays, we show that, although the general transcription factor TFIIH is able to target any kind of lesion which can be repaired by the nucleotide excision repair pathway, TATA binding protein (TBP)-TFIID is more selective in damage recognition. Only genotoxic agents which are able to induce kinked DNA structures similar to the one for the TATA box in its TBP complex are recognized. Indeed, DNase I footprinting patterns reveal that TBP protects equally 4 nucleotides upstream and 6 nucleotides downstream from the A-T (at position −29 of the noncoding strand) of the adenovirus major late promoter and from the G-G of a cisplatin-induced 1,2-d(GpG) cross-link. Together, our results may partially explain differences in transcription inhibition rates following DNA damage.
Many carcinogens and antitumor agents structurally modify DNA, often at specific DNA sequences, with as a consequence the disturbance of mechanisms which govern cell life. Following DNA damage, one observes cell cycle arrests in G2/M and G1 phases and a decrease in the rate of transcription. Investigations aimed at elucidating how cells respond to DNA damage evidenced a transcriptionally connected subpathway of DNA repair, called nucleotide excision repair (NER), in which lesions in transcribed genes were preferentially repaired (4, 28). This connection between the two mechanisms was further and definitively established when it was demonstrated that the multiprotein complex TFIIH, essential for protein-encoding gene transcription, was also fundamental for NER (for reviews, see references 16 and 36).
One of the first steps of any repair process is recognition of the damage, and thus, significant studies have been devoted to identifying proteins able to bind specifically to cisplatin- or UV-induced lesions (for a review, see reference 5). In an effort to understand how TFIIH shuttles between the transcription template and the DNA lesion, we surprisingly demonstrated that TATA binding protein (TBP)-TFIID, another essential basal transcription factor which normally recognizes the TATA box sequence located 30 bp upstream from the transcription start site, also interacts with damaged DNA (41).
Before trying to understand the putative role of TBP in DNA repair, we considered it worthwhile to expand our investigations examining the connection between these two essential transcription components, TFIIH and TFIID, and several damaged DNAs. Seven different drugs (Fig. 1 and Table 1) which bind covalently to DNA were chosen. The platinum derivatives form mono- or bifunctional adducts with DNA. First, the diethylenetriaminedichloroplatinum(II) derivative (Dien) (24) was chosen for its ability to form a monofunctional adduct recognized by the NER pathway in bacteria (2). Cisplatin (CDDP), transplatin (TDDP), and dachplatin (Dach) were used for their capacity to induce monofunctional adducts which evolve into intrastrand or interstrand cross-links. CDDP reacts mainly with adjacent purines, leading to the formation of 1,2-d(GpG) and 1,2-d(ApG) intrastrand cross-links in the range of 60 to 65 and 25 to 30%, respectively. Minor adducts are interstrand and 1,3-d(GpXpG) intrastrand cross-links between two guanines and represent 5 to 10% (9, 10, 23). The Dach derivative produces similar adducts, despite the fact that nonleaving groups are different and may, once bound to DNA, induce different steric hindrance (17, 22). In contrast, the isomer TDDP induces the formation of 1,3-d(GpXpG) and interstrand cross-links. The methylmethanesulfonate (MMS) provokes the methylation of guanine, mainly to N7, resulting in damage which is recognized by glycosylases belonging to the base excision repair process (BER). The N2-acetylaminofluorene (AAF) is a synthetic carcinogen for liver and breast tissue which, once activated, binds to C8 and N2 of the guanine (29). AAF adduct is recognized by the NER proteins. Finally, ethidium azide (EtAz), an intercalating dye which exhibits a GC preference, covalently binds to DNA upon photoactivation. The resulting lesion is recognized by the NER and BER processes in vitro.
FIG. 1.
Chemical structure of mono- and bifunctional drugs reacting with DNA.
TABLE 1.
Genotoxic agent effects on DNA structure, DNA repair, and transcription
| Genotoxic agent | DNA adduct | Bendinga (major groove)(°) | Repairb | Transcription inhibitionc | Transcription factor bindingc
|
Toxicityd | |
|---|---|---|---|---|---|---|---|
| TFIIH (p62) | TFIID (TBP) | ||||||
| CDDP | 1,2-d(GpG) | 32–58 | + (NER) | + | + | + | +++ |
| 1,3-d(GpXpG) | 20–35 | ++ (NER) | NDe | + | ± | ||
| TDDP | 1,3-d(GpXpG) | 26 | ++ (NER) | ± | + | ± | ± |
| Dach | 1,2-d(GpG) | ND | + (NER) | + | + | + | +++ |
| 1,3-d(GpXpG) | 20–35 | ++ (NER) | ND | + | ± | ||
| Dien | 1N7G | 0 | + (NER) | − | ± | − | − |
| AAF | 1C8G | ND | ++ (NER) | + | + | + | ND |
| MMS | 1N7G | ND | +++ (BER) | − | − | − | +++ |
| EtAz | ND | + (NER)/+ (BER) | ± | + | − | ND | |
| UVC | T-T | 30 | ++ (NER) | + | + | + | +++ |
Bending angle was calculated by nuclear magnetic resonance, X-ray diffraction, and gel shift assay (3, 27, 43).
+ to +++, lowest to highest level, respectively.
+, inhibition or binding; −, no inhibition or binding; ±, mixed result.
+++, highly cytotoxic; ±, weakly cytotoxic; −, no effect. The sensitivity of HeLa cells to the genotoxic agents used in this study was assessed by colony formation assay.
ND, not determined.
Our results indicate that only specific damage leads to significant inhibition of RNA polymerase II (RNA Pol II) transcription, although almost all damaged DNAs tested so far are repaired by the NER machinery. Inhibition of transcription is due to selective sequestration of TBP by damaged DNA according to nitrocellulose filter binding assays, as well as DNase I footprinting analysis. These results may at least partially explain the decrease in transcription and the delay in NER response observed upon treatment of cells with certain drugs, two events which could be the specific cellular response to these genotoxic agents, leading to cell death.
MATERIALS AND METHODS
Preparation of plasmid DNA substrates.
Treatments of pHM plasmid DNA (100 μg/ml), a 3,738-bp derivative of pBluescript KS+ plasmid DNA (Stratagene), with platinum compounds were performed in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at 37°C overnight in the dark. Platinum compounds were diluted to obtain different ratios of platinum bound to nucleotide (rbs), assuming that under these conditions, approximately 50 and 70% of the total amount of CDDP and TDDP, respectively, react with DNA (13). Dach was used in the same conditions as those for CDDP, and Dien was assumed to react completely with DNA. Reactions were stopped by addition of NaCl to 0.5 M, and plasmids were recovered by ethanol precipitation, dried, and redissolved in TE buffer. The DNA concentration was quantified by UV spectrometry, and total platinum content was determined by atomic absorption spectroscopy.
pHM plasmids (200 μg/ml in 20 mM Tris-HCl [pH 7.2]–2 mM EDTA) were mixed per volume with EtAz (diluted in water at various concentrations) and incubated for 10 min on ice in the dark to allow intercalation. Photolytic coupling was subsequently performed by irradiation of the reaction mixture with white light (20 cm from a 200-W source) for 10 min on ice, and adducted plasmids were recovered by ethanol precipitation. The overall coupling efficiency was calculated to be 50% (14); final EtAz concentrations (0.8, 1.6, and 2.4 μM) were calculated to yield approximately 10, 20, and 30 adducts per 3,738-bp plasmid, respectively.
Plasmid DNA (100 ng/μl) was incubated with MMS (Sigma) at various concentrations in water for 30 min at 30°C. Methylated plasmids were subsequently separated from the unreacted MMS by centrifugation through G-50 microcolumns (Pharmacia), and DNA concentrations were determined. Based on previous results (38) and taking into account the size of the DNA molecules and the incubation time and temperature, we calculated the methylation rate to be 30, 60, and 90 methyls per plasmid.
Plasmid pBKS (3.0 kbp) was treated with N-acetoxy-2-AAF, inducing mainly N-(guanine-8-yl)-AAF adducts to obtain 15 to 20 AAF-guanine adducts per damaged plasmid (39).
Transcription-competition assay.
Approximately 30 μg of HeLa whole-cell extract (WCE) was incubated with varying amounts of competitor DNA in a 50 mM Tris-HCl (pH 7.9) buffer containing 10% glycerol, 1 mM EDTA, 0.5 mM dithiothreitol, and 5 mM MgCl2. Reaction mixtures were incubated for 15 min at 25°C, at which point 50 ng of linear adenovirus major late promoter (AdMLP) template was added, and preinitiation of transcription was allowed to continue for 15 min. Transcription was then initiated by addition of nucleoside triphosphates including [α-32P]CTP (400 Ci/mmol). The final volume of the reaction was 25 μl, and transcription was carried out for 45 min at 25°C. The RNA transcripts were analyzed by autoradiography and quantified directly by counting them on a PhosphoImage analyzer or indirectly by densitometric scanning of autoradiograms with a Bio-Rad GS700 imaging densitometer.
The reconstituted transcription system (RTS) containing purified transcription factors, TBP, TFIIA, TFIIB, TFIIE, TFIIH, TFIIF, and RNA Pol II (12), was modified to include an initial incubation step to allow the potential binding of transcription factors to either nontreated (NT) or damaged competitors.
Analysis of DNA-associated proteins.
Protein-DNA interactions were analyzed with a derivative of a previously described in vitro repair assay (35) with several DNA substrates adsorbed in microtitration wells as follows. One hundred nanograms of damaged or undamaged (NT) plasmid as a control was adsorbed on polylysine-sensitized 96-well microtiter plates (Microlite II; Dynatech) in 10 mM phosphate buffer, pH 7, for 30 min at 30°C with shaking, before being incubated with 200 μg of HeLa WCE (50 μl) in 40 mM HEPES-KOH (pH 7.6) buffer, containing 60 mM KCl, 7 mM MgCl2, 2 mM ATP, 0.5 mM dithiothreitol, 10 mM phosphocreatine, 2.5 μg of creatine phosphokinase type I (Sigma), 2 mM EGTA, and 18 μg of bovine serum albumin. After a 2-h incubation at 30°C, the wells were washed three times with PBST (phosphate-buffered saline [pH 7.4] plus 0.01% Tween 20), and the protein fractions bound to DNA were analyzed by Western blotting, with anti-TBP (3G3), anti-p62-TFIIH (3C9), or anti-TFIIEα (2A1) antibodies.
Filter binding assay.
Purified recombinant human TBP was combined with various DNA probes, such as linearized damaged pHM plasmid, labelled with [α-32P]dATP (3,000 Ci/mmol) with the Klenow fragment of DNA polymerase. One nanogram of probe (5,000 cpm) was combined with various amounts of TBP, in 20 μl of the transcription buffer containing 60 μg of bovine serum albumin per ml, 500 ng of poly(dG-dC), and 5 mM MgCl2, for 30 min at 30°C. Reaction mixtures were applied to nitrocellulose membranes (0.45-mm pore size; Millipore) with the 96-well Hybri-Dot Manifold (BRL), presoaked in 0.4 mM KOH, washed with distilled water, and preequilibrated in the reaction buffer without bovine serum albumin. Filters were air dried and directly exposed to a PhosphoImage screen for quantification or Biomax film (Kodak). One microliter of input DNA corresponding to the same volume used in each reaction was spotted on Whatman filter paper as a control for determination of the percentage of DNA retained on nitrocellulose filters. The amount of radioactivity retained in the presence of TBP was measured, background counts (radioactivity retained in the absence of protein) were subtracted, and the amount was divided by the level of radioactivity present in the input.
DNase I footprinting of TBP.
The −194 to +33 fragment of AdMLP and the +5914 to +6410 fragment of the cisplatin-damaged (M13mp18GG) or nondamaged (NTGG) plasmid DNA (30) were used for footprinting experiments. The AdMLP TATA box DNA probe was obtained by PCR amplification with a unique end-labelled primer and purification by G-50 gel filtration. The plasmid DNA containing a single cisplatin-induced 1,2-d(GpG) intrastrand cross-link was digested with AvaII (New England Biolabs), and the 5′ extremity was labelled with the Klenow fragment of DNA polymerase in the presence of [α-32P]dCTP (3,000 Ci/mmol) (Amersham). The plasmid was then digested with PvuI (New England Biolabs), and the AvaII-PvuI fragment as well as the control nondamaged DNA probe (NT) was purified on a 5% polyacrylamide gel. The standard binding reaction mixtures contained 1 to 2 ng of end-labelled probe (20,000 cpm), 500 ng of poly(dG-dC), 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 50 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, 5 mM MgCl2, 2.5 mM CaCl2, 4 mM spermidine, 0.2% Nonidet P-40, and 15% glycerol. After addition of 40 ng of purified recombinant yeast TBP and, when indicated, 200 ng of human TFIIB, the mixture was incubated for 20 to 30 min at room temperature in the absence or presence of competitor DNA. Freshly diluted DNase I was added to the binding reaction mixtures, and digestion was allowed to proceed for 1 min at room temperature. The reactions were stopped by adding stop solution containing 0.5% (wt/vol) sodium dodecyl sulfate, 50 mM sodium acetate, and 50 mg of tRNA per ml. Following phenol-chloroform extraction and ethanol precipitation, samples were electrophoresed on an 8% polyacrylamide sequencing gel.
RESULTS
Inhibition of AdMLP transcription by specific damaged DNA competitors.
The abilities of different damaged DNA competitors to inhibit transcription of an AdMLP reporter template in vitro were examined by a transcription competition assay. Either HeLa WCE or purified fractions containing all the basal transcription factors, in addition to RNA Pol II, necessary for an in vitro RTS were first preincubated with various damaged DNAs. After a 15-min preincubation to favor the recognition of damage by specific proteins, one of the first steps of NER, AdMLP was introduced and the reaction was continued for an additional 15 min to allow the redistribution of the various factors and to promote the formation of the preinitiation transcription complex. RNA synthesis was then initiated by addition of nucleoside triphosphates and quantified by the production of a 309-nucleotide (nt) transcript. As such, WCE or RTS fractions (Fig. 2A and B) were mixed with increasing amounts of various competitor DNAs containing a fixed ratio of lesions per plasmid (about 100 lesions per molecule). CDDP-damaged DNA, as well as Dach-platinated competitor, inhibited transcription from AdMLP (Fig. 2A, compare lane 2 with lanes 3 to 5 and lanes 9 to 11, respectively) whereas TDDP- and Dien-treated competitors (compare lane 2 with lanes 6 to 8 and lanes 12 to 14) as well as the control NT DNA had only a slight effect on the transcription of the reporter template. A quantitative analysis demonstrated that we had about 75% inhibition of AdMLP (0.67 kbp, 50 ng/assay, one promoter per fragment) transcription in the presence of 25 ng of CDDP- and Dach-damaged plasmid (3.7 kbp, 100 lesions per plasmid). This means that, in our in vitro experimental conditions, such competition was obtained in a ratio of eight lesions to one promoter.
FIG. 2.
Inhibition of in vitro transcription from AdMLP by the presence of damaged DNA. (A) Transcription of AdMLP (50 ng) was performed with 30 μg of HeLa WCE or with RTS fractions in the presence of increasing amounts (25, 50, and 100 ng) of plasmid pHM untreated (NT) (lane 2) or treated with CDDP (lanes 3 to 5), TDDP (lanes 6 to 8), Dach (lanes 9 to 11), or Dien (lanes 12 to 14) (100 lesions/plasmid). Lane 1, absence of DNA competitor. (B) (Left panel) Densitometric quantification of the above autoradiogram (309-nt band) as well as additional experiments (at least three, when standard deviation is indicated), presented as the percentage of transcription from AdMLP as a function of DNA competitor. Lanes correspond to those in panel A. (Right panel) Quantification of experiments performed as described for panel A, with DNA competitors previously treated with AAF (50 and 100 ng), MMS (25 and 50 ng), or EtAz (100 and 200 ng) (lanes 17 to 22, respectively). Transcription in the absence of competitor DNA equals 100%. (C) Transcription of AdMLP was performed with WCE in the presence of 50 ng of different damaged DNA competitors containing 100 (lanes 3, 5, 7, and 9) or 200 (lanes 4, 6, 8, and 10) lesions per molecule as indicated at the top of the figure. Lane 1 represents transcription without DNA competitor; lane 2 (+) contains 50 ng of undamaged pHM plasmid. (D) Recovery of transcription inhibited in an RTS by CDDP (lanes 1 to 8)-, Dach (lanes 9 to 12)-, and AAF (lanes 13 to 16)-damaged plasmid (containing 100 lesions/plasmid for CDDP and Dach and 15 to 20 lesions/plasmid for AAF; the amount of each competitor is indicated in the figure in nanograms). TBP and TFIIB added to restore transcription were as described in reference 41.
As summarized in Fig. 2B, when AAF-treated DNA is used as a competitor, it also inhibits transcription of AdMLP whereas MMS- and EtAz-treated DNA competitors have no specific inhibitory effect. Consistent with previous observations (41), the capacities of CDDP-, Dach-, or AAF-treated DNA to inhibit the transcription of AdMLP with a WCE as well as an RTS fraction (free of repair factors) certainly account, at least partially, for the capacity of TBP-TFIID to directly interact with lesions. To further demonstrate that inhibition of the 309-nt transcript synthesis was due to the presence of lesions and not to increasing concentrations of DNA itself, we included a fixed concentration of DNA competitor (50 ng) with either 100 or 200 lesions per molecule of competitor. In these conditions, CDDP and Dach adducts inhibited transcription of the AdMLP template as a function of the number of lesions (Fig. 2C, compare lane 2 with lanes 3 and 4 and 7 and 8, respectively), whereas TDDP and Dien had a slight effect (lanes 5 and 6 and 9 and 10).
In addition, the ability of TBP to restore transcription activity inhibited by CDDP-, Dach-, and AAF-damaged plasmids was tested in an RTS. Addition of TBP restored transcription activity of the AdMLP reporter template inhibited by damaged DNA (Fig. 2D, lanes 6, 12, and 16), whereas addition of TFIIE, TFIIA, TFIIF, or RNA Pol II had no effect (reference 41 and data not shown). Interestingly, we also observed that, although addition of TFIIB alone to transcription inhibited by CDDP-damaged competitor had no effect (lane 7), addition of TFIIB and TBP led to a significantly greater degree of restoration than that with TBP alone (compare lane 8 with lane 6).
Together, these data demonstrate that one can define two groups of DNA-adducting drugs. The first group, consisting of CDDP, Dach, and AAF, inhibits the in vitro transcription of AdMLP by sequestrating basal transcription factors such as TBP-TFIID, whereas the second group, which includes TDDP and Dien as well as MMS or EtAz, has little or no effect on transcription.
Damaged DNAs selectively sequester transcription factors.
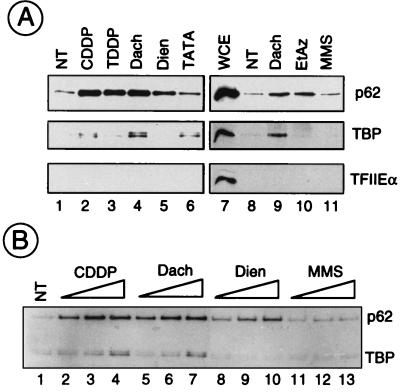
To evaluate the nature of proteins associated with DNA damage, we took advantage of a recently developed technology (25). The damaged plasmid DNAs containing equivalent amounts of lesions per plasmid (except for EtAz) were immobilized on sensitized microplate wells, before being incubated with HeLa WCE. Under these conditions, damaged DNA can be repaired (35), thus demonstrating that active NER proteins have the ability to bind DNA damage. Preliminary studies using radiolabelled damaged DNA plasmids have shown, however, that, independently of the nature of the damage, damaged DNAs were adsorbed equally well to microtiter dishes. Furthermore, in order to compare the relative affinities of the factors for the lesions, WCE was not in excess in the reaction. After a 1-h preincubation period and extensive washing of the microplates, the adsorbed proteins were analyzed by sodium dodecyl sul- fate-polyacrylamide gel electrophoresis followed by Western blotting (Fig. 3). The amount of each factor adsorbed onto damaged DNA varied as a function of the nature of lesion presented. Keeping as a reference the binding of TFIIH and TBP on the TATA box (lane 6), we observed that TFIIH, as visualized by the detection of its p62 subunit, was retained on CDDP-, TDDP-, Dach-, EtAz-, and, to a lesser extent, Dien-treated DNAs (Fig. 3A). In contrast, TBP selectively bound to CDDP- and Dach-treated DNAs (lanes 2 to 5 and 9 to 11). As a control, we noticed that almost nothing was retained on MMS-treated DNA compared to untreated DNA (lanes 11 and 1, respectively). Under these experimental conditions, each of the damaged DNAs did not sequester other transcription factors such as TFIIE (Fig. 3A), TFIIF, and RNA Pol II (data not shown). This latter observation is not surprising, since we know that formation of a stable preinitiation complex on a promoter requires only the presence of TFIID, TFIIA, and TFIIB (7, 11).
FIG. 3.
Analysis of damaged-DNA-associated proteins. Aliquots of HeLa WCE were incubated with damaged pHM plasmids adsorbed in microwells under conditions described in Materials and Methods. After a 2-h incubation, the protein fraction bound to DNA was recovered and analyzed by Western blotting. (A) One hundred nanograms of pHM was either NT or treated with CDDP, TDDP, Dach, or Dien to an rb value of ∼100 Pt atoms/plasmid (lanes 2 to 5 and 9); EtAz (2.4 μM, leading to ∼30 adducts/plasmid [lane 10]); or MMS (15 mM, leading to ∼90 methylations/plasmid [lane 11]). Lane 6, as a positive control, 200 ng of a TATA box-containing plasmid (pUC309, 3.4 kb) was adsorbed in the well. Lane 7, 50 μg of WCE was analyzed in parallel. The presence of the p62 subunit of TFIIH, TBP, and TFIIEα was analyzed with 3C9, 3G3, and 2A1 monoclonal antibodies, respectively. (B) pHM was either NT; was platinated with CDDP, TDDP, Dach, or Dien to rb values of ∼50, ∼100, and ∼200 Pt atoms/plasmid; or was methylated with MMS (5, 10, and 15 mM, leading to ∼30, 60, and 90 methylations/plasmid, respectively).
Both TFIIH and TBP-TFIID factors were retained as a function of the number of CDDP- and Dach-damaged sites per plasmid, whereas alkylated DNA did not attract basal transcription factors (Fig. 3B). Again, we noticed that increasing the number of either CDDP or Dach adducts per DNA molecule concurrently increased the number of TBPs retained (lanes 2 to 4 for CDDP and lanes 5 to 7 for Dach). This was not the case for Dien-damaged DNA (lanes 8 to 10). The saturating level of TFIIH bound to CDDP- and Dach-treated DNA compared to the level of TBP may reflect the excess of free TFIIH compared to TBP in the cell. Even in the presence of WCE, TBP-TFIID was not highly retained, compared to TFIIH, on Dien-platinated DNA, which suggests that binding of these two transcriptional complexes to damaged DNA does not follow the same mechanism.
TBP as a selector of specific three-dimensional DNA structure.
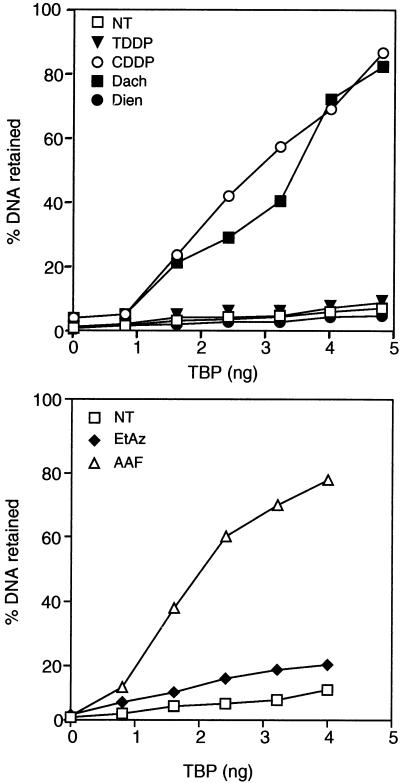
TFIIH alone does not bind damaged DNA without the help of additional repair factors (18, 31). We then wondered whether TBP alone may recognize and differentiate between the various types of damage on DNA. Damaged plasmids were tested for their capacity to retain highly purified recombinant TBP by the standard nitrocellulose filter binding assay (Fig. 4). We observed that TBP recognized CDDP- and Dach- as well as AAF-treated DNA, whereas almost no interaction was observed with either TDDP-, Dien-, EtAz-, or MMS-treated DNA.
FIG. 4.
Preferential binding of TBP to damaged DNA. CDDP-, TDDP-, Dach-, or Dien-treated (100 lesions/plasmid); EtAz-treated (30 lesions/plasmid); or AAF-treated (15 to 20 lesions/plasmid) DNA or untreated (NT) DNA was labelled with [α-32P]dATP. NT or damaged DNA was then incubated with increasing amounts of TBP and tested for retention on nitrocellulose filters. Graphs represent the percentages of DNA retained on the filters as a function of the amount of TBP. One hundred percent represents the total counts obtained when 1 μl of each DNA probe (input) was spotted onto Whatman paper and simultaneously exposed with the nitrocellulose filter.
The apparent discrepancy between the fewfold effect of CDDP-damaged DNA shown in Fig. 3A and its more than 20-fold effect shown by nitrocellulose filter binding probably reflects the differences in the conduct of the assays. Indeed, in the first case we used crude extract and we did not use saturating conditions for the damaged DNA.
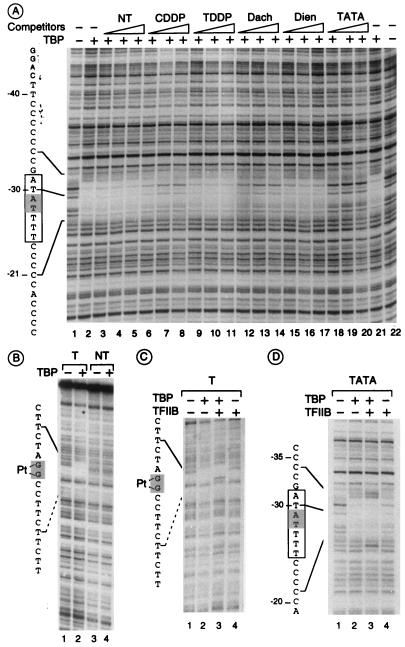
Having demonstrated that TBP is able to recognize different lesions, we thus wondered if these various damaged plasmids compete for TBP binding on a class II TATA box-containing promoter (Fig. 5A). In these studies, yeast TBP was preferentially chosen because it was easier for us to produce highly pure full-length yeast TBP than to produce human TBP. The addition of TBP showed clear protection between nt −34 and −21 on the noncoding strand (compare lanes 1 and 22 with lanes 2 and 21) and between nt −38 and −22 on the coding strand (data not shown) of the AdMLP. The footprint pattern on the noncoding strand was conserved upon addition of increasing amounts of either NT (lanes 3 to 5), TDDP-treated (lanes 9 to 11), or Dien-treated (lanes 15 to 17) DNA. In contrast, the TBP footprint was reduced upon addition of increasing amounts of either CDDP-treated (lanes 7 to 9) or Dach-treated (lanes 12 to 14) DNA (10 ng of competitor represents a 60-fold excess of lesions compared to promoters) and of course upon addition of the AdMLP consensus TATA box (lanes 18 to 20). Similar results were obtained with human TBP (data not shown). The above data strongly suggest that TBP is hijacked from its natural DNA promoter site by specific damaged DNA structures induced by bifunctional agents such as CDDP or Dach or a monofunctional and intercalating agent such as AAF, all of which induce a peculiar kinked DNA structure (see Discussion).
FIG. 5.
Footprinting of TBP onto DNA containing either a TATA box or CDDP damage. (A) TBP footprint on the TATA box could be competed by adding increasing amounts (10, 20, and 30 ng) of either undamaged (NT) or CDDP-, TDDP-, Dach-, or Dien-damaged DNA (containing 100 lesions per molecule) or TATA box-containing fragment (AdMLP). Lanes 1 and 22, naked DNA; lanes 2 and 21, no DNA competitor. (B) Footprint of TBP onto a 1,2-d(GpG) cisplatin lesion (T) compared with NT DNA. (C and D) Footprint of TBP onto a 1,2-d(GpG) cisplatin lesion (T) compared with a TATA box-containing fragment in the presence or absence of TFIIB as indicated. The TATA box is boxed and the 1,2-d(GpG) cisplatin lesion position is shaded on the left of the figure, and TBP footprint protection is determined upon sequencing.
These observations prompted us to investigate TBP footprinting on the CDDP-damaged DNA, this bifunctional chemical adduct being chosen because its chemical synthesis was relatively easy to implement. As shown in Fig. 5B, highly purified TBP was able to protect a DNA sequence encompassing a unique cisplatin-induced 1,2-d(GpG) intrastrand cross-link. Using a shorter probe (60 nt) containing the same surrounding sequences, we determined more precisely that TBP protects the radiolabelled DNA fragment 4 nt upstream and at least 6 nt downstream (5′→3′) from the 1,2-d(GpG) lesion (data not shown). As a control, the same untreated DNA probe did not show protection of any DNA sequence upon addition of TBP. It is also worth noting the similarities between DNase I footprinting patterns of TBP when bound either on AdMLP TATA box or on cisplatin-damaged DNA: TBP protects equally 4 nt upstream and 6 nt downstream from A-T (at position −29) of the AdMLP and from the G-G of the 1,2-d(GpG) adduct. Moreover, we were not able to observe any significant footprint of TBP on a DNA containing a unique 1,3-d(GpXpG) lesion (data not shown), indicating a stronger affinity of TBP for 1,2-d(GpG) than for the 1,3-d(GpXpG) cross-link.
Since TBP is binding to the lesion as well as to the TATA box (Fig. 2 and 3), in addition to the partial requirement for TFIIB in restoring transcription inhibition (Fig. 2D), we thus wondered what could be the behavior of other factors such as TFIIB which form a ternary complex with TBP and the TATA box (6). Interestingly, the DNase I footprint pattern of TBP on damaged DNA was modified upon addition of TFIIB. Indeed, we observed a partial extension of TBP protection upstream of the cisplatin lesion, with the appearance of two hypersensitive sites on both sides of the lesion (Fig. 5C, lane 3), a situation similar to what is found on the TATA box (Fig. 5D, lane 3). It should be noted that TFIIB per se does not interact with DNA (Fig. 5D, lane 4, and Fig. 2E).
DISCUSSION
In this study, we demonstrated that DNA damage recognition by basal transcription factors follows different mechanisms: although TFIIH is able to target any kind of lesion which can be repaired by NER machinery, TBP-TFIID is more selective in damage recognition. Only kinked structures similar to the one in the TATA box in its TBP complex are recognized. This may partially explain differences in transcription inhibition rates following DNA damage.
Binding of TFIIH and that of TBP-TFIID on damaged DNA are disconnected.
The in vitro transcription challenge competition assay and the sensitized microplate assay allowed discrimination between the effects of the various drugs which damaged the DNA. Firstly, we observed that DNAs damaged by the platinum derivatives CDDP, TDDP, and Dach and the two aromatic drugs AAF and EtAz (the present study) as well as by UV irradiation (41) sequestered TFIIH, whereas MMS-methylated DNA did not interact with TFIIH at all. This observation is not surprising when one considers that those types of DNA damage, with the exception of methylated bases, are repaired by the NER machinery and as such require the presence of TFIIH. Secondly, according to the nitrocellulose filter binding assay, only some of the damaged DNAs bound TBP. This was the case for DNAs which were damaged either by the two bifunctional platinum derivatives CDDP and Dach or by the monofunctional intercalating agent AAF.
Although it was previously shown first that arrival of TFIIH on the promoter is directed by the presence of TBP-TFIID and second that both factors interact with each other upon the formation of the transcription initiation complex (12, 45), the recognition mechanism of DNA damage of TBP-TFIID and that of TFIIH do not seem to be directly related. Indeed, binding of TFIIH to damaged DNA occurs through interactions with other repair factors such as xeroderma pigmentosum group A and/or replication protein A (18, 31, 32) whereas TBP does not require additional proteins to bind damaged DNA. This is further confirmed by the fact that the presence and the requirement for TFIIH around the DNA lesion do not compulsorily imply the arrival of TBP-TFIID.
TBP recognizes selectively kinked DNA structure.
Only some of the damaged DNAs which are repaired by the NER machinery are recognized by TBP-TFIID. Moreover, crystallographic studies have established that DNA containing a 1,2-d(GpG) cisplatin cross-link (resulting from CDDP treatment) or a cyclobutane thymine dimer (resulting from UV irradiation) mimics the previously described TATA box configuration upon TBP binding (19, 20, 37, 40). These studies show that cisplatin adduct and thymine dimer induce a kink on the DNA, similar to the one of the TATA box in its TBP complex. It is also worth noting the surprising similarities between DNase I footprints of TBP either on AdMLP TATA box or on cisplatin-damaged DNA: TBP protects equally 4 nt upstream and 6 nt downstream from A-T (at position −29) and 1,2-d(GpG) adduct, respectively (Fig. 5). Instead of using a special mechanism to increase the bending of DNA by intercalating the two phenylalanine rings of TBP into the DNA, at either end of the TATA box, which already has a propensity to bend, TBP easily interacts with damaged DNA which already possesses an optimal fit (shape complementarity). The bendability of DNA, as well as the nature of the adduct, is the determinant for TBP binding. In addition to CDDP, Dach, a cisplatin derivative which also binds to two consecutive guanines, and AAF, which binds DNA as a monofunctional adduct and is inserted within the helix (29), confer a kinked structure on the DNA (Table 1). In contrast, the drugs we tested, which bind DNA only through one functional group (EtAz, MMS, or Dien), as well as TDDP, which favors interstrand linkage and consequently does not induce similar kinked structure (24) (Table 1), do not bind TBP. It can easily be understood that the nonleaving groups of the platinum adduct and/or the overall structure of any drug, which can induce a kinked structure, determine the affinity of TBP and the associated proteins for the damaged DNA.
More interestingly, TBP is also able to strongly discriminate between the 1,2-d(GpG) adduct on which it binds and the 1,3-d(GpXpG) cross-link lesions (Fig. 4) (our unpublished results). In the latter case, the affinity of TBP for the lesion is insufficient to observe a DNase I footprint pattern. The conformational change in DNA induced by the 1,3-d(GpXpG) cross-link has been reported to be closer in structure to that induced by TDDP lesions (21, 24). This indicates that TBP is able to bind selectively to specific three-dimensional structure, the model of which is the TATA box in its TBP-associated complex.
Possible biological consequences of TBP binding to lesions.
Transcription inhibition following DNA damage is not only a consequence of the requirement of the NER machinery for TFIIH to allow the formation of the incision-excision complex but also the consequence of some distortion in the unfolding of transcription. Indeed, following DNA damage due to UV irradiation, it has been observed that the rate of phosphorylation of the large subunit of RNA Pol II (which is necessary for elongation [1, 26]) decreases (15a). Moreover, an analogy was made between UV irradiation and ubiquitination of the large subunit of RNA Pol II, leading to proteolytic degradation and a decrease in the pool of RNA Pol II (5a). It has also to be kept in mind that any drug has side effects: for example, platinum derivatives provoke the formation of very low levels of protein-DNA cross-links in the cell (34).
The present study also strongly suggests that the transcription inhibition could be due to a deficiency of TBP-TFIID, which is diverted from its natural promoter binding site to target some DNA lesions: sequestration of TBP reduces the pool of TFIID available for class II promoter transcription, leading to internal cellular disarray. This sequestration is a function of the nature of the damage (nature of the drug) and the surrounding sequences. At this stage of our investigations, it would be interesting to emphasize the physiological consequences of TBP binding to kinked damaged DNA. For example, cisplatin treatment (80 μg/ml) results in one lesion per 10 to 100 kb. Based on that, of the 5% of sequences which are transcribed in the 3,000 Mb of the human genome, around one-third possess a TATA box. It is thus possible that there would be 0.1 to 1 DNA lesion for every functional TATA box. In this case, TBP could bind either the TATA box or the damaged DNA, its preference being directed by the nature of the TATA box and also by that of the DNA damage. This hypothesis has also to take into account the fact that the nontranscribed regions of the genome are less rapidly repaired than are the transcribed ones and thus may immobilize much longer TBP molecules. The consequence is the formation of a gradient in the requirement of TBP based upon its affinity for any kinked structure, which results in a defect in the transcription of weak promoters, e.g., the one which have no consensus TATA box. Consistent with such a hypothesis, we previously found (41) that microinjection of additional TBP into living fibroblasts alleviates the reduction in RNA synthesis after UV irradiation.
Another possible consequence of the binding of TBP to lesions is the prevention of recognition of these lesions by DNA repair enzymes following binding with TBP. Previous studies have demonstrated the capacities of proteins containing the high-mobility group (HMG) DNA binding domain, known to interact with cisplatin lesions, to specifically inhibit excision repair of the intrastrand 1,2-d(GpG) cross-links (44). Similar protection could be possible for TBP, leading to blocks of replication forks and cell death, since, up to now, there has been no evidence for a possible displacement of TBP from the lesions by proteins involved in NER.
Our present study reveals also a provocative similarity in the types of lesions that are recognized by both TBP-TFIID and the HMG proteins. Both types of proteins selectively and strongly bind to 1,2-d(GpG) lesions rather than 1,3-d(GpXpG) lesions or TDDP-damaged DNA (33). Unfortunately, very little is known about the natural binding sequences of HMG proteins (if any exist); SRY, a transcriptional activator with HMG domains (8), has been previously described as a protein able to interact with a DNA target containing, like the natural binding site of TBP, several A/T base pairs (15). Finally, the three-dimensional structures of SRY (42) and TBP (19, 20), bound to their natural sequences, reveal a common motif of side-chain interchelation driving the deformation of the DNA helix, which may reflect a common way for both proteins to interact with their natural sites.
Our results reveal that TBP-TFIID and some HMG proteins are probably diverted from their natural targets by common effects induced by some drug treatments, which might explain the consequences of particular types of DNA damage for transcription efficiency and maybe cell death. For example, preliminary experiments in the study of the sensitivity of HeLa cells towards the four platinum compounds (CDDP, Dach, TDDP, and Dien) suggest that discrimination between cytotoxic (CDDP and Dach) and ineffective (TDDP and Dien) compounds could at least be partially explained by TBP binding to lesions, with, as a consequence, an inhibition of transcription.
ACKNOWLEDGMENTS
We thank F. Jeffrey Dilworth, D. Moras, and P. Hanawalt for very fruitful discussions. We thank I. Kuraoka and R. Wood for supplying the DNA with a platinum adduct at a specific site and critical reading of the manuscript and A. Fery for her excellent technical assistance.
This work was supported by grants from the Human Frontier Program, the INSERM, the CNRS, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de l’Enseignement Supérieur, the Association pour la Recherche sur le Cancer, and La Ligue Nationale contre le Cancer.
F.C. and P.F. contributed equally to this work.
REFERENCES
- 1.Adamczewski J A, Rossignol M, Tassan J P, Nigg E A, Moncollin V, Egly J M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Alazard R J, Germanier M. Post replication repair in an excision defective strain of Escherichia coli following treatment with cis dichlorodiammineplatinum(II) Biochem Biophys Res Commun. 1982;104:693–700. doi: 10.1016/0006-291x(82)90692-1. [DOI] [PubMed] [Google Scholar]
- 3.Bellon S F, Lippard S J. Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-[Pt(NH3)2(N3-cytosine)Cl]+ Biophys Chem. 1990;35:179–188. doi: 10.1016/0301-4622(90)80007-t. [DOI] [PubMed] [Google Scholar]
- 4.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas T. DNA lesion-recognizing proteins and the p53 connection. Anticancer Res. 1996;16:225–242. [PubMed] [Google Scholar]
- 5a.Bregman, D., et al. Personal communication.
- 6.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 7.Davison B L, Egly J M, Mulvihill E R, Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983;301:680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- 8.Dubin R A, Ostrer H. Sry is a transcriptional activator. Mol Endocrinol. 1994;8:1182–1192. doi: 10.1210/mend.8.9.7838151. [DOI] [PubMed] [Google Scholar]
- 9.Eastman A. Reevaluation of interaction of cis-dichloro(ethylenediamine) platinum(II) with DNA. Biochemistry. 1986;25:3912–3915. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 10.Fichtinger-Schepman A M, van Oosterom A T, Lohman P H, Berends F. cis-Diamminedichloroplatinum(II)-induced DNA adducts in peripheral leukocytes from seven cancer patients: quantitative immunochemical detection of the adduct induction and removal after a single dose of cis-diamminedichloroplatinum(II) Cancer Res. 1987;47:3000–3004. [PubMed] [Google Scholar]
- 11.Fire A, Samuels M, Sharp P A. Interactions between RNA polymerase II, factors, and template leading to accurate transcription. J Biol Chem. 1984;259:2509–2516. [PubMed] [Google Scholar]
- 12.Gérard M, Fischer L, Moncollin V, Chipoulet J-M, Chambon P, Egly J-M. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 13.Hansson J, Wood R D. Repair synthesis by human cell extracts in DNA damaged by cis- and trans-diamminedichloroplatinum(II) Nucleic Acids Res. 1989;17:8073–8091. doi: 10.1093/nar/17.20.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardwick J M, von Sprecken R S, Yielding K L, Yielding L W. Ethidium binding sites on plasmid DNA determined by photoaffinity labeling. J Biol Chem. 1984;259:11090–11097. [PubMed] [Google Scholar]
- 15.Harley V R, Jackson D I, Hextall P J, Hawkins J R. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 15a.Herlich, P. Personal communication.
- 16.Hoeijmakers J H J, Egly J M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 17.Jennerwein M M, Eastman A, Khokhar A R. The role of DNA repair in resistance of L1210 cells to isomeric 1,2-diaminocyclohexaneplatinum complexes and ultraviolet irradiation. Mutat Res. 1991;254:89–96. doi: 10.1016/0921-8777(91)90044-p. [DOI] [PubMed] [Google Scholar]
- 18.Jones C J, Wood R D. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 19.Kim J L, Nikolov D B, Burley S W. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 21.Kozelka J, Archer S, Petsko G A, Lippard S J, Quigley G J. Molecular mechanics modeling of oligonucleotide adducts of the antitumor drug cis-diamminedichloroplatinum(II) Biopolymers. 1987;26:1245–1271. doi: 10.1002/bip.360260804. [DOI] [PubMed] [Google Scholar]
- 22.Kraker A J, Moore C W. Accumulation of cis-diamminedichloroplatinum(II) and platinum analogues by platinum-resistant murine leukemia cells in vitro. Cancer Res. 1988;48:9–13. [PubMed] [Google Scholar]
- 23.Lemaire M A, Schwartz A, Rahmouni A R, Leng M. Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum (II) and DNA. Proc Natl Acad Sci USA. 1991;88:1982–1985. doi: 10.1073/pnas.88.5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepre C A, Lippard S J. Interactions of platinum antitumor compounds with DNA. Nucleic Acids Mol Biol. 1990;4:9–38. [Google Scholar]
- 25.Li, R. Y., P. Calsou, C. J. Jones, and B. Salles. Interaction of the transcription/DNA repair factor TFIIH and XPA repair protein with DNA lesions in a cell-free repair assay. Submitted for publication. [DOI] [PubMed]
- 26.Lu H, Zawel L, Fischer L, Egly J M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 27.Malinge J M, Perez C, Leng M. Base sequence-independent distortions induced by interstrand cross-links in cis-diamminedichloroplatinum (II)-modified DNA. Nucleic Acids Res. 1994;22:3834–3839. doi: 10.1093/nar/22.19.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 29.Milhé C, Fuchs R P P, Lefèvre J F. NMR data show that the carcinogen N-2-acetylaminofluorene stabilises an intermediate of −2 frameshift mutagenesis in a region of high mutation frequency. Eur J Biochem. 1996;235:120–127. doi: 10.1111/j.1432-1033.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 30.Moggs J C, Yarema K J, Essigmann J M, Wood R D. Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J Biol Chem. 1996;271:7177–7186. doi: 10.1074/jbc.271.12.7177. [DOI] [PubMed] [Google Scholar]
- 31.Nocentini S, Coin F, Saijo M, Tanaka K, Egly J M. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor IIH. J Biol Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 32.Park C H, Mu D, Reardon J T, Sancar A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 33.Pil P M, Lippard S J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 34.Plooy A C, Fichtinger-Schepman A M, Schutte H H, van Dijk M, Lohman P H. The quantitative detection of various Pt-DNA-adducts in Chinese hamster ovary cells treated with cisplatin: application of immunochemical techniques. Carcinogenesis. 1985;6:561–566. doi: 10.1093/carcin/6.4.561. [DOI] [PubMed] [Google Scholar]
- 35.Salles B, Provot C, Calsou P, Hennebelle I, Gosset I, Fournie G J. A chemiluminescent microplate assay to detect DNA damage induced by genotoxic treatments. Anal Biochem. 1995;232:37–42. doi: 10.1006/abio.1995.9964. [DOI] [PubMed] [Google Scholar]
- 36.Svejstrup J C, Vichi P, Egly J M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 37.Takahara P M, Rosenzweig A C, Frederick C A, Lippard S J. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 38.Tudek B, Boiteux S, Laval J. Biological properties of imidazole ring-opened N7-methylguanine in M13mp18 phage DNA. Nucleic Acids Res. 1992;20:3079–3084. doi: 10.1093/nar/20.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Vuuren A J, Vermeulen W, Ma L, Weeda G, Appeldorn E, Jaspers N G J, van der Eb A J, Hoeijmakers J H J, Humbert S, Shaeffer L, Egly J M. Correction of Xeroderma repair defect by basal transcription factor BTF2/TFIIH. EMBO J. 1994;13:1645–1653. doi: 10.1002/j.1460-2075.1994.tb06428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vassylyev D G, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 41.Vichi P, Coin F, Renaud J P, Vermeulen W, Hoeijmakers J H J, Moras D, Egly J M. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werner M H, Huth J R, Gronenborn A M, Clore G M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 43.Yang D, Van Boom S G E, Reedijk J, Van Boom J H, Wang A H J. Structure and isomerization of an intrastrand cisplatin-cross linked octamer DNA duplex by NMR analysis. Biochemistry. 1995;34:12912–12920. doi: 10.1021/bi00039a054. [DOI] [PubMed] [Google Scholar]
- 44.Zamble D B, Mu D, Reardon J T, Sancar A, Lippard S J. Repair of cisplatin-DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 45.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]