Summary
Idebenone, the only approved treatment for Leber hereditary optic neuropathy (LHON), promotes recovery of visual function in up to 50% of patients, but we can neither predict nor understand the non-responders. Idebenone is reduced by the cytosolic NAD(P)H oxidoreductase I (NQO1) and directly shuttles electrons to respiratory complex III, bypassing complex I affected in LHON. We show here that two polymorphic variants drastically reduce NQO1 protein levels when homozygous or compound heterozygous. This hampers idebenone reduction. In its oxidized form, idebenone inhibits complex I, decreasing respiratory function in cells. By retrospectively analyzing a large cohort of idebenone-treated LHON patients, classified by their response to therapy, we show that patients with homozygous or compound heterozygous NQO1 variants have the poorest therapy response, particularly if carrying the m.3460G>A/MT-ND1 LHON mutation. These results suggest consideration of patient NQO1 genotype and mitochondrial DNA mutation in the context of idebenone therapy.
Keywords: idebenone, NQO1, complex I, LHON, Leber hereditary optic neuropathy, mtDNA, cybrids, fibroblasts, retinal ganglion cells
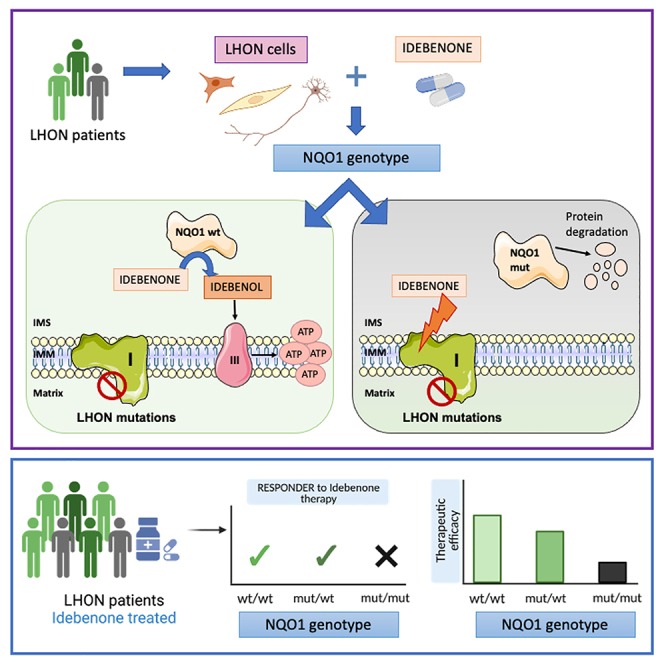
Graphical abstract
Highlights
-
•
NQO1 activity drives idebenone ability to bypass complex I defect in LHON cells
-
•
NQO1 polymorphic variants impact therapeutic capacity of idebenone
-
•
NQO1 genotypes associate with idebenone responder analysis of LHON patients
-
•
NQO1 genotype mostly impact the m.3460G>A/MT-ND1 LHON mutation in vivo
Aleo et al. investigate the impact of NQO1 protein levels on the ability of idebenone to bypass the respiratory complex I deficiency in LHON. NQO1 expression depends on two polymorphic variants and strictly correlates with the therapeutic effectiveness of idebenone both in LHON cellular models and in clinical response.
Introduction
Idebenone (2-(10-hydroxydecyl)-5,6-dimethoxy-3-methyl-cyclohexa-2,5-diene-1,4-dione) is a benzoquinone compound and synthetic analog of CoQ10 developed in the early 1980s for the treatment of neurodegenerative disorders. Due to its shorter and less lipophilic tail, it shows better penetration across mitochondrial membranes and the blood-brain barrier.1,2 It has been demonstrated that, upon reduction by the cytosolic NAD(P)H oxidoreductase I (NQO1), idebenone can be oxidized by complex III (CIII), promoting downstream mitochondrial respiration and ATP synthesis.3,4,5 A fraction of NQO1 is located in the mitochondria, justifying the NQO1/idebenone/CIII electron flow.6,7 Idebenone reduction is key for its effect on bypassing complex I (CI). In its oxidized form, idebenone may exert a deleterious inhibitory effect on both the redox and proton-pumping activity of CI.8,9 Therefore, idebenone in its reduced form exerts an antioxidant function, while in its oxidized form it acts as a pro-oxidant, prompting a debate when idebenone was tested in Friedreich ataxia with conflicting results.6,10,11,12
Leber hereditary optic neuropathy (LHON) is a mitochondrial disease characterized by CI impairment due to maternally inherited mitochondrial DNA (mtDNA) missense mutations in CI subunits. Affected patients present with optic nerve atrophy and low vision.13 Recently, other rare recessive genetic defects were also associated with LHON but affecting CI.14 Peculiar and not completely understood features of LHON are incomplete penetrance and male prevalence.15 Both also apply to the biallelic recessive mutations.14 Convincing evidence showing that reduced idebenone bypasses defective CI,3,4,5 together with the results of an LHON clinical trial16 and retrospective patient case series,17 collectively provided encouraging data regarding its clinical efficacy, leading to its approval for LHON in 2015 by the European Medicines Agency (EMA; https://www.ema.europa.eu/en/medicines/human/EPAR/raxone), becoming the only approved therapy to date for a mitochondrial disease.18 LHON’s natural history is unusual for a neurodegenerative disorder; the optic nerve degeneration develops subacutely within weeks to months, and by about 1 year after onset, the patients enter a chronic phase, usually characterized by severely reduced visual acuity.19 Only a limited subset of patients (e.g., about 11% for the m.11778G>A/MT-ND4 mutation) may spontaneously recover to some extent, depending on the age of onset and the mutation type.20,21 Idebenone treatment significantly increases the proportion of patients with clinically relevant recovery (CRR) to almost 50%.21 However, idebenone seems to be unable to mitigate the visual deterioration of many patients, who reach the so-called “nadir,” the lowest point of visual function, including color vision, contrast sensitivity, visual acuity, and visual field.22,23 From the “nadir,” patients may regain visual acuity after prolonged treatment with idebenone at 900 mg/day, ameliorating their visual fields.16,17,21,24,25
Unfortunately, about half of LHON patients remain unresponsive, and there is no clear explanation for why idebenone may be ineffective in these patients. Also, in vitro contradictory effects of idebenone have been reported by studying LHON-derived fibroblasts.26,27,28
NQO1 is a cytosolic homodimeric flavoprotein that catalyzes the reduction of highly reactive quinones and their derivates by complete two-electron reduction, avoiding the production of reactive semiquinones and favoring the quinol antioxidant activity.29 NQO1 has been reported to reduce idebenone, thus promoting its antioxidant properties and rescuing ATP in cells with pharmacologically inhibited CI.3,4,5 A demonstration showing that this also occurs in cells with a genetic defect of CI is still missing.
We here hypothesize that NQO1 protein levels may affect the efficacy of idebenone treatment in LHON patients. We investigated this hypothesis in LHON cell models, carried out studies on histological retinal specimens, and retrospectively assessed the visual outcome in idebenone-treated LHON patients, providing multiple lines of evidence supporting the notion that NQO1 is a key protein for idebenone efficacy in LHON.
Results
Idebenone effectiveness in cybrids depends on NQO1 expression
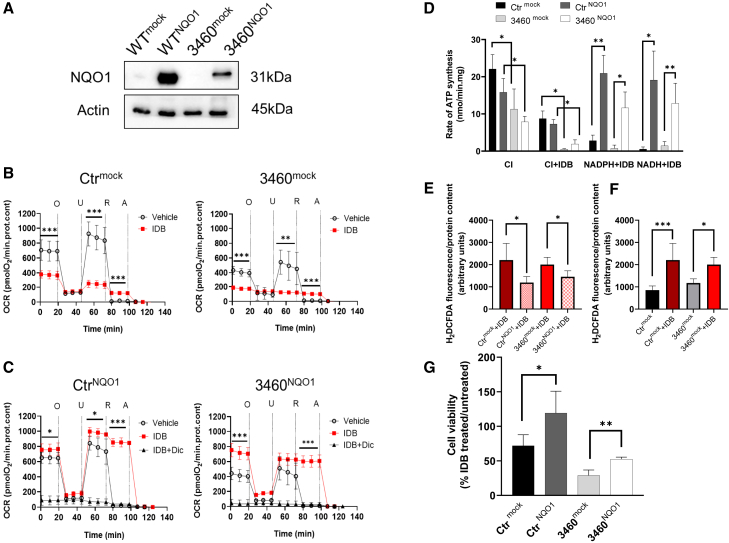
We have previously reported that idebenone treatment was not able to enhance the viability of LHON cybrids when oxidative phosphorylation was forced by galactose medium.30 This could be related to the very low levels of NQO1 expression in osteosarcoma-derived (143B.TK−) cybrids, as shown in Figure 1A (control [Ctr]mock and 3460mock cells). Thus, we decided to overexpress NQO1 in a Ctr and in an m.3460G>A/MT-ND1 LHON-mutated cybrid cell line (CtrNQO1 and 3460NQO1 cells; Figure 1A), testing the effect of idebenone treatment on mitochondrial respiration in the presence or absence of NQO1. First, we measured the oxygen consumption rates (OCR) of cybrids with different concentrations of idebenone (10, 20, 40, and 80 μM) and established that, in NQO1-overexpressing cells, the most effective concentration of idebenone was 10 μM (Supplemental results; Figure S1). Figure 1B shows that, in Ctrmock cells treated with vehicle, the OCR displayed an expected profile of inhibition after the addition of oligomycin (O), rotenone (R), and antimycin (A) and enhanced respiration after addition of the uncoupler FCCP (U) (Figure 1B). In agreement with the literature, 3460mock cybrids showed respiration reduced by about 50% due to the mutation,31 and in mock cells treated with idebenone, the OCR was significantly inhibited in both cell lines, suggesting that, in cybrids lacking NQO1, the oxidized idebenone blocks respiration, likely through CI inhibition, as shown previously (Figure 1B).8,9 Cybrids overexpressing NQO1 showed an OCR profile similar to the mock cells when treated with vehicle only, indicating that the enhanced expression of NQO1 did not alter mitochondrial respiration per se (Figure 1C). However, the addition of idebenone significantly increased the basal OCR in NQO1-overexpressing cells, specifically restoring 3460NQO1 cellular respiration similar to Ctr levels. Furthermore, under these experimental conditions, R addition did not inhibit the OCR, indicating that idebenone caused CI-independent respiration. Finally, the OCR was sensitive to A and dicoumarol, inhibitors of CIII and NQO1, respectively, indicating that the oxygen consumption was dependent on these two enzymes (Figure 1C; Supplemental results; Figure S2). Together, these results demonstrate that idebenone is able to restore respiratory chain defects linked to CI deficiency when NQO1 is overexpressed in cybrids.
Figure 1.
IDB effectiveness in cybrids depends on NQO1 expression
(A) Western blot analysis of NQO1 expression level in cellular lysates from control (Ctr) and m.3460G>A/MT-ND1 (3460) cybrids carrying an empty plasmid (mock) or human NQO1 (NQO1). Actin was used as a loading Ctr. A representative blot of two similar ones is shown.
(B) OCR measurements of Ctr (Ctrmock) and m.3460G>A/MT-ND1 LHON cybrids (3460mock). OCR values were expressed as pmolO2 consumed/min normalized for cellular protein content under resting conditions and after oligomycin (O), FCCP (U), rotenone (R), and antimycin A (A) addition as detailed under STAR Methods. Empty circles correspond to cells treated with the vehicle DMSO (10 and seven independent experiments for Ctrmock and 3460mock, respectively) and red squares to cells treated with 10 μM idebenone (IDB; six and five independent experiments for Ctrmock and 3460mock, respectively). Data are reported as mean ± SD, and statistical analysis was performed using a t test as detailed under STAR Methods. ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) OCR measurements of Ctr (CtrNQO1) and m.3460G>A/MT-ND1 LHON cybrids (3460NQO1) overexpressing NQO1. Open circles correspond to cells treated with the vehicle DMSO (eight and five experiments for CtrNQO1 and 3460NQO1, respectively), red squares to cells treated with 10 μM IDB (six and five experiments for CtrNQO1 and 3460NQO1, respectively), and black triangles to cells treated with 10 μM IDB and 10 μM dicoumarol (IDB+DIC). Statistical analysis was performed for cells treated with vehicle and IDB as described in B).
(D) Rate of ATP synthesis measured in digitonin-permeabilized cells driven by malate and pyruvate (CI substrates) or exogenous NAD(P)H. Where indicated, IDB was added at the final concentration of 10 μM as detailed under STAR Methods. Data are means ± SD of at least three to five independent experiments. Statistical analysis was performed using unpaired t test. ∗p < 0.05, ∗∗p < 0.01.
(E) Rate of H2O2 production in mock or NOQ1-overexpressing cells grown in the presence of 10 μM IDB as described under STAR Methods. Data are reported as mean ± SEM of four to six independent experiments. Statistical analysis was performed using a paired t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(F) Rate of H2O2 production in mock cell lines in the presence or absence of 10 μM IDB. The measurement and data analysis were performed as described in (E).
(G) Cell viability measurements were performed in the presence or absence of 10 μM IDB as detailed under STAR Methods. Values are expressed as percentage of the ratio between IDB treated and non-treated cells. Data are reported as mean ± SD of four independent experiments. Statistical analysis was performed using unpaired t test. ∗p < 0.05, ∗∗p < 0.01.
To confirm the role of NQO1 in modulating the idebenone-dependent restoration of mitochondrial energetics, we also measured in cybrids the rate of ATP synthesis driven by NADH/NADPH, the natural substrates of NQO1. Figure 1D shows the rate of ATP synthesis driven by CI-linked substrates (pyruvate/malate) or by exogenous NAD(P)H in digitonin-permeabilized mock and NQO1-overexpressing cybrids in the presence or absence of idebenone. As expected, NQO1 overexpression did not significantly alter CI-driven ATP synthesis (CI) under both Ctr and m.3460G>A/MT-ND1-mutated cells, which showed a reduced ATP synthesis due to the LHON mutation (Figure 1D). The addition of idebenone strongly inhibited CI-driven ATP synthesis in all cell lines, suggesting that idebenone is present in its oxidized form and cannot be reduced by NADH-linked substrates inside the mitochondria (CI+idebenone [IDB] condition). However, the addition of exogenous NAD(P)H together with IDB fully supported ATP synthesis in cells overexpressing NQO1 (NADPH+IDB and NADH+IDB), reaching levels similar to CI-driven ATP synthesis (CI). These results demonstrate that NQO1 oxidizes the NAD(P)H provided externally to mitochondria to reduce IDB and ultimately sustains CI-independent mitochondrial ATP synthesis.
The therapeutic action of IDB has also been associated with the antioxidant effect of the reduced form.4 Therefore, we measured hydrogen peroxide production in cybrid cell lines overexpressing NQO1 or otherwise in the presence of IDB. Figure 1E shows that, in both NQO1-overexpressing cell lines, the treatment with IDB significantly decreased H2O2 production compared with the corresponding mock cells, showing that NQO1 promotes the antioxidant effect of IDB (Figure 1E). Contrarily, IDB treatment of both 3460mock and Ctrmock cells increased H2O2 production compared with untreated cells, indicating that oxidized IDB induces oxidative stress in cells (Figure 1F). Cells with the m.3460G>A/MT-ND1 LHON mutation also showed increased levels of H2O2 in the absence of IDB (Figure S3), congruent with what has been reported previously in LHON-mutated cybrids.30,32,33 Measurements in the presence of the well-known antioxidant N-acetylcysteine were performed as a positive Ctr (Supplemental results; Figure S3).
Finally, we evaluated the long-term effect of IDB treatment in cells overexpressing NQO1. To this end, we tested the cell viability of mock and NQO1-overexpressing cybrids grown under the same condition as used for enhancing respiration in OCR assays. Clearly, IDB treatment was toxic in cells with the empty vector, with 70% and 30% residual cell viability for Ctr and LHON-mutant cells, respectively (Figure 1G). Conversely, IDB treatment significantly increased cell viability when NQO1 was overexpressed. These results confirm that the effectiveness of IDB treatment in vitro depends on the NQO1 protein level and its ability to maintain reduced IDB.
IDB effectiveness depends on NQO1 protein levels in fibroblasts from Ctrs as well as in LHON patients
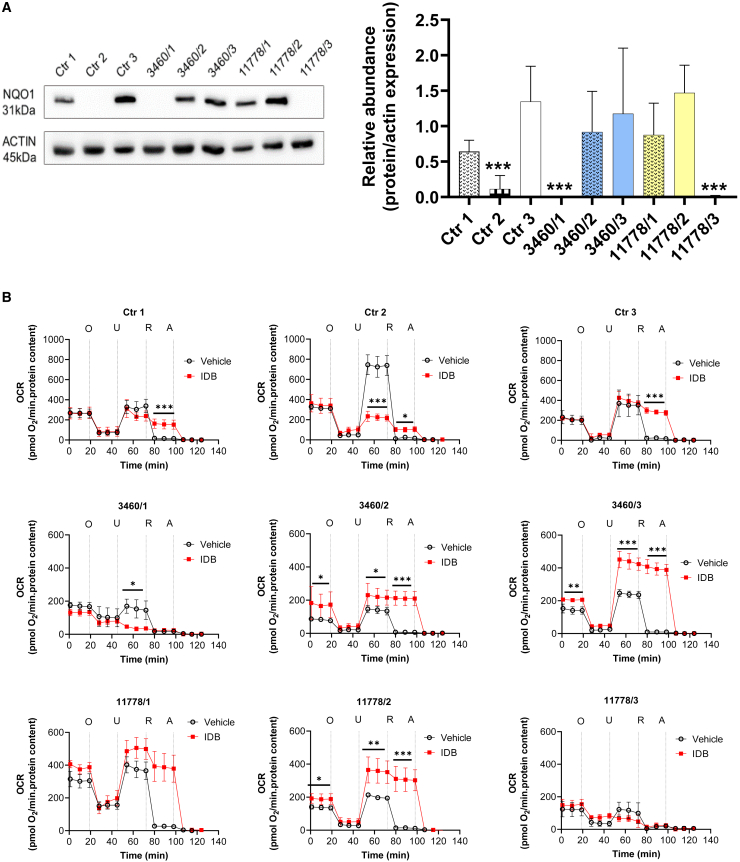
Because cybrids are constructed by transferring only the mtDNA from donor cells,34 the effect of IDB was studied in fibroblasts derived from healthy Ctrs and LHON patients, which present the full genetic complement of original nuclear and mitochondrial genomes. Therefore, we assessed whether the physiological levels of NQO1 expression could maintain mitochondrial respiration in the presence of the LHON mtDNA mutation or pharmacological inhibition of CI. For this purpose, we carried out experiments using three fibroblast cell lines from healthy donors, three from LHON patients with the m.3460G>A/MT-ND1 mutation, and three with the m.11778G>A/MT-ND4 mutation. We first evaluated NQO1 protein levels in the different cell lines, showing that this enzyme is differently expressed in fibroblasts. Considering Ctr3 as a reference, being the Ctr cell line with the highest levels of NQO1, in three samples (Ctr2; 3460/1 and 11778/3), the protein was almost undetectable (Figure 2A). Afterward, we measured the OCR in the presence or absence of IDB in all fibroblast cell lines. Figure 2B shows that, in the presence of IDB, cells expressing NQO1 were able to increase or maintain respiration under uncoupled conditions and in the presence of R. Conversely, IDB inhibited uncoupled respiration, and the residual OCR was R sensitive in fibroblasts lacking NQO1 expression (Ctr2; 3460/1 and 11778/3). Remarkably, IDB treatment increased both basal and uncoupled respiration, enhancing the respiratory-chain capacity of LHON patient cells (3460/2, 3460/3, 11778/1, and 11778/2) expressing NQO1.
Figure 2.
IDB effectiveness correlates with NQO1 protein levels in Ctr and LHON fibroblasts
(A) Western blot analysis and quantification of NQO1 expression in cellular lysates from Ctr (WT1, WT2, and WT3) and LHON (3460/1, 3460/2, 3460/3, 11778/1, 11778/2, and 11778/3) fibroblasts. Actin was used as a loading Ctr. A representative blot of three similar ones is shown. Data are reported as means ± SD of protein abundance of at least three to five independent experiments. Statistical analysis was performed using one-way ANOVA with a Dunnett’s post hoc test comparing the mean of each column with the mean of WT3 as a Ctr column. ∗∗∗p < 0.001.
(B) OCR measurements in Ctr and LHON fibroblasts in the presence or absence of IDB (as in Figure 1B). Open circles correspond to OCR values measured in cells treated with the vehicle DMSO and red squares to OCR values measured in cells treated with 10 μM IDB. Data are reported as mean ± SD of at least three to five independent experiments (Ctr1: n = 5 vehicle, n = 4 IDB; Ctr2: n = 5 vehicle, n = 4 IDB; Ctr3: n = 3 vehicle, n = 3 IDB; 3460/1: n = 3 vehicle, n = 3 IDB; 3460/2, n = 3 vehicle, n = 3 IDB; 3460/3: n = 4 vehicle, n = 3 IDB; 11778/1: n = 3 vehicle, n = 2 IDB; 1177/2: n = 3 vehicle, n = 3 IDB; 11778/3: n = 5 vehicle, n = 4 IDB. Statistical analysis is the same as reported in Figure 1B and STAR Methods. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To match these in vitro results with the in vivo real-word experience of IDB treatment, we summarized, in Table S1, the clinical information of these six LHON patients. All patients were treated with IDB except for one treated with a different compound (EPI-743). This latter compound, however, has also been reported to be cycled by the same NQO1 pathway.4 According to the so-called CRR,21 a definition currently used in LHON clinical trials,17,20 we classified these patients as responders or non-responders. We observed that non-responders fitted the lack of NQO1 expression, except for the patient treated with EPI-743 (3460/3) and the patient with LHON (11778/1), for whom we have reported previously that the disease was triggered by toxic chemicals.35 Taken together, these results suggest that the NQO1-IDB -CIII pathway may recover the energy deficit in LHON cells bypassing the CI defect, mirroring the clinical response to IDB therapy in four of six cases.
Common polymorphic variants in NQO1 influence its levels in fibroblasts from Ctrs and LHON patients
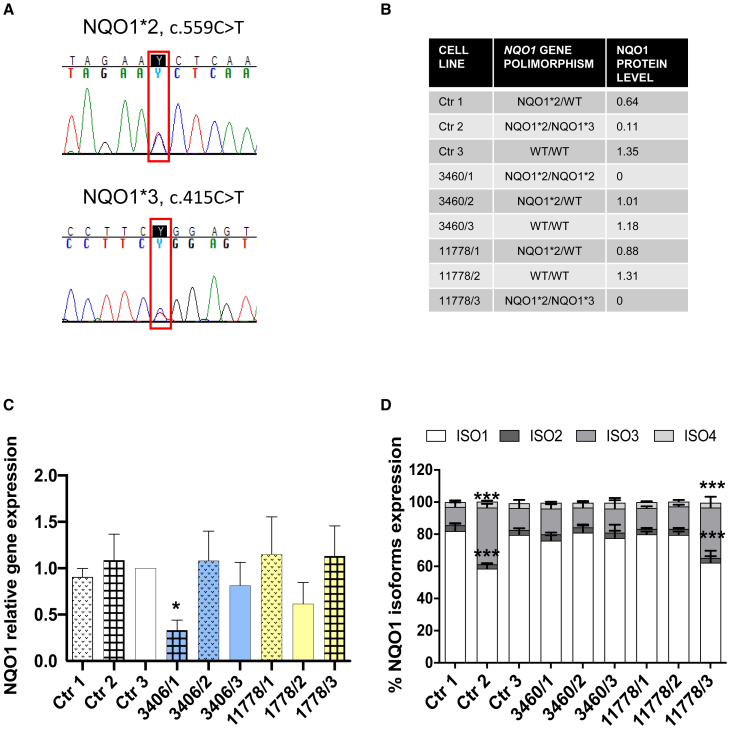
To explain the different levels of NQO1 expression in fibroblasts, affecting either Ctr cells (Ctr2) or LHON mutant cells (3460/1 and 11778/3), we next sequenced the NQO1 gene. We detected two known variants: GenBank: NM_000903.3: c.559C>T, p.P187S (dbSNP: rs1800566, known by old nomenclature as c.609C>T) defining the NQO1∗2 allele and GenBank: NM_000903.3: c.415C>T, p.R139W (rs1131341, known by old nomenclature as c.465C>T) defining the NQO1∗3 allele (Figure 3A). Both variants are known as relatively common population polymorphisms, with a frequency of 18.9% for NQO1∗2 and 3.7% for NQO1∗3 in the European (non-Finnish) population, as reported in the GnomAD v.2.1.1 database.36 Western blot analysis indicated that the presence of one of these two variants decreased at 25%–50% the NQO1 protein level (Ctr1, 3460/2, 11778/1 compared with Ctr3, considered as reference), whereas the 3 cases with compound heterozygosity or homozygosity of the two variants (Ctr2, 3460/1, 11778/3) were essentially equivalent to null alleles (Figure 3B). Previous studies have documented that NQO1∗2 induces instability of the protein, which leads to degradation,37,38 whereas NQO1∗3 causes increased skipping of exon 4, leading to higher isoform 3 expression, which, in turn, is not translated or rapidly degraded.39,40 Indeed, we did not observe major alterations in NQO1 global mRNA expression in our fibroblast cell lines compared with Ctr3 (our internal reference) except for 3460/1 (Figure 3C). The analysis of the four NQO1 isoforms (Figure S4A) revealed that, in the NQO1∗3 cell lines (Ctr2 and 11778/3), the expression of isoform 3 was increased, whereas isoform 1 was reduced (Figure 3D). NRF2, as a major regulator of NQO1 expression,41,42 was also assessed, noting that 3460/1 had the lowest level, which is concordant with the lowest NQO1 mRNA level (Figure S4B).
Figure 3.
Common polymorphic variants in NQO1 influence its levels in Ctr and LHON fibroblasts
(A) Representative electropherograms showing the presence of both WT and variants in fibroblast DNA: the NQO1∗2 allele (top) and the NQO1∗3 allele (bottom).
(B) Table summarizing the NQO1 polymorphism identified and NQO1 protein levels for each of the nine fibroblast cell lines analyzed, normalized on WT3.
(C) NQO1 gene expression evaluated by qPCR. GAPDH was used as a reference gene. Fold changes are normalized to WT3 and are expressed as means ± SD of three independent experiments. ∗p < 0.05 using Dunnett’s post hoc test.
(D) Gene expression of the four isoforms of NQO1 evaluated by qPCR. GAPDH was used as a reference gene. Data are expressed as means ± SD of three independent experiments. ∗∗∗p < 0.001 using Dunnett’s post hoc test.
NQO1 is minimally expressed in blood but highly expressed in retinal ganglion cells (RGCs), neuronal precursor cells (NPCs), and induced pluripotent stem cell (iPSC)-derived neurons, driving IDB efficacy on respiration of neuronal cells
To verify whether NQO1 levels could be detected in blood samples, we assessed NQO1 mRNA expression and protein levels in peripheral blood mononuclear cells (PBMCs) from available patients. NQO1 expression in PBMCs, although not different between the three samples, was more than 100-fold lower compared with fibroblasts (Figures S5A and S5B), whereas the protein was essentially undetectable by western blot analysis (Figure S5C).
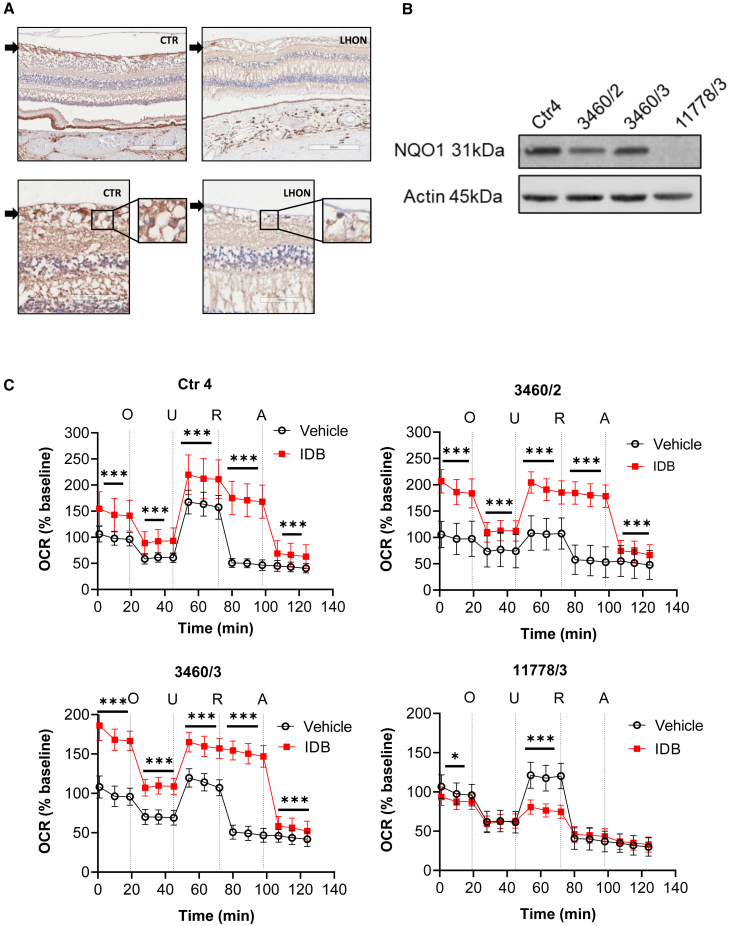
Furthermore, NQO1 is variably expressed in human tissues (www.proteinatlas.org), and it is expressed in human retina and RGCs, as also reported recently (https://data.iob.ch/?ds=Adult_human_foveal_retina&gene=NQO1).43,44 We took advantage of available human postmortem formalin-fixed specimens from a Ctr (NQO1 genotype not available) and an LHON patient carrying the m.11778G>A/MT-ND4 mutation (NQO1∗2 heterozygous variant) to verify NQO1 expression by immunohistochemistry. We found that RGCs and RNFL robustly expressed NQO1 in the Ctr retina, whereas the NQO1 signal was partially reduced in the LHON patient retina, in consideration of the profound loss of RGCs and NQO1∗2 heterozygous variant (Figure 4A). NQO1 was also robustly expressed in photoreceptors, rods, and cones in both Ctr and LHON specimens, indicating that NQO1 levels match the dependence from mitochondrial metabolism in different retinal subtypes. Overall, these results confirm that NQO1 is highly expressed in RGCs, which are the disease and therapeutic target in LHON.
Figure 4.
NQO1 expression in blood, RGCs, and iPSC-derived NPCs, driving IDB efficacy on respiration of NPCs
(A) Immunoperoxidase stain with anti-NQO1 (A180) on horizontal retinal sections from a normal individual (Ctr: male, 74 years old) and an LHON patient carrying the m.11778G>A/MT-ND4 mutation (LHON: male, 52 years old). In the normal individual, a positive stain is observed in the unmyelinated portion of the axons in the retinal nerve fiber layer (arrow) and in the somata of RGCs. In the patient, a faint positivity is present both in the atrophic retinal nerve fiber layer and in residual RGCs (insets: higher magnification).
(B) Western blot analysis of NQO1 levels in cellular lysates from a Ctr (Ctr4), 3460/2, 3460/3, and 11778/3 LHON NPCs. Actin was used as a loading Ctr. Due to the very low number of NPCs, the western blot was performed only once.
(C) OCR measurements in Ctr (Ctr4) and LHON (3460/2, 3460/3, and 11778/3) NPCs in the presence or absence of IDB. OCR data were normalized on cell counts and expressed as a percentage of the baseline measurement of untreated lines. Open circles correspond to OCR values measured in cells treated with the vehicle DMSO and red squares to OCR values measured in cells treated with 10 μM IDB. Data are reported as mean ± SD of two independent experiments (12 technical replicates per line for each experiment). Statistical analysis is the same as reported in Figure 1B and STAR Methods. ∗p < 0.05, ∗∗∗p < 0.001.
To further confirm that the influence of NQO1 on IDB efficacy documented in cybrids and fibroblasts also occurs in the setting of neuronal cells, we took advantage of already generated iPSCs from Ctr (Ctr4; NQO1 genotype wild-type [WT]/WT) and some of the LHON fibroblasts studied previously (3460/2, 3460/3, and 11778/3).45 From iPSCs, we generated NPCs, and we eventually obtained terminally differentiated neurons, which we demonstrated to be the prevalent cell type compared with astrocytes in our neuronal culture (Figure S6A). We analyzed the NQO1 levels in NPCs and differentiated neurons and found that NQO1 protein levels were dependent on the NQO1 polymorphic variants (Figures 4B and S6B). We thus evaluated the functional effect of these NQO1 variants on NPCs and differentiated neurons by assessing mitochondrial respiration in the absence/presence of IDB (Figures 4C and S6C, respectively). The results obtained in NPCs (Figure 4C) mirrored the previous results in fibroblasts (Figure 2B). This also applied to the iPSC-derived neurons from 3460/3 and 11778/3 LHON cases (Figure S6C). The only iPSCs-derived Ctr neurons used in this experiment (Ctr4) had mixed behavior at 10 μM IDB (Figure S6D). This concentration of IDB was used for all mitochondrial respiration experiments independent from the cell types analyzed. However, when tested with 1 μM IDB, the effect of the drug was reproduced in Ctr4 iPSCs-derived neurons, similar to what was observed in cells expressing NQO1, including both fibroblasts (Ctr1 and Ctr3) and NPCs (Ctr4) (Figure S6D).
Considering the whole set of results, the NQO1 polymorphic variants clearly impinge on the NQO1 protein amount, determining the fate of IDB efficacy on cell respiration. This applies not only to peripheral tissues such as fibroblast cell lines but also to neuronal cells and possibly to RGCs because, physiologically, they clearly show robust NQO1 expression.
Homozygous NQO1 polymorphisms result in reduced NQO1 protein levels
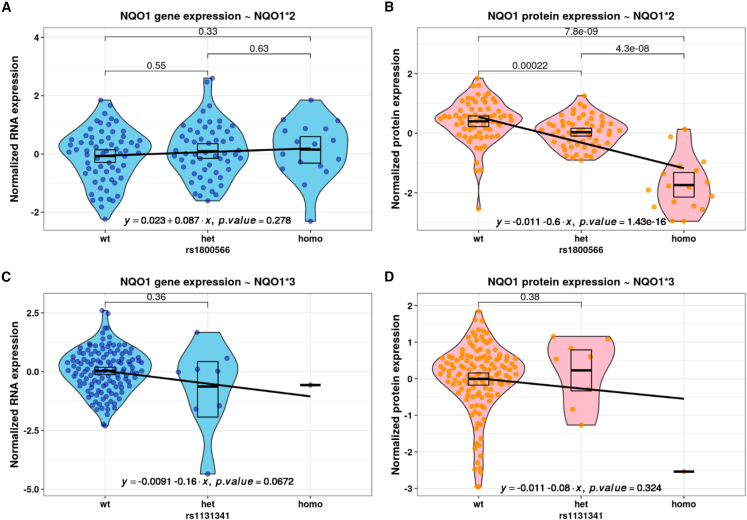
To further investigate the effects of NQO1 polymorphisms on its RNA and protein expression, we performed candidate association analysis in the combined fibroblast RNA sequencing (RNA-seq) and quantitative tandem mass tag (TMT) proteomics dataset.46,47 The polymorphic variants NQO1∗2 and NQO1∗3 were detected in 52% (80 of 147) of individuals and did not show any significant effects on NQO1 transcript levels (Figures 5A and 5C), in accordance with previously published results.48 On the other hand, the NQO1∗2 polymorphism had a significant effect on protein levels in both heterozygous and homozygous states (Figure 5B), in line with western blot analysis. Although we observed underexpression of NQO1 protein for the homozygous NQO1∗3 carrier, due to the availability of only a single observation, we cannot generalize from this result. The heterozygous NQO1∗3 (dbSNP: rs1131341) polymorphism did not show any significant effect on protein level (Figure 5D). However, a splicing quantitative trait locus (sQTL) analysis performed earlier48 demonstrated significant multitissue effects on NQO1 splicing of this polymorphism even in the heterozygous state. In our homozygous case, this resulted in exon 4 skipping in one-third of transcripts (Figure S7).
Figure 5.
Homozygous NQO1 polymorphisms result in reduced NQO1 protein levels
(A and B) Violin plots of normalized RNA expression (A) and protein expression (B), stratified by genotypes of NQO1∗2.
(C and D) Violin plots of normalized RNA expression (C) and protein expression (D), stratified by genotypes of NQO1∗3.
Each dot represents an individual observation. The black box indicates mean and 95% confidence intervals. Blue color corresponds to RNA-seq analysis and red-spectrum colors to proteomics. The black line was fitted using linear regression, with the equation and significance specified in the bottom.
NQO1 variants leading to very low protein levels match the IDB responder/non-responder analysis, impinging the final outcome in LHON patients
Having established the effects of polymorphic variants (NQO1∗2 and NQO1∗3) on NQO1 protein levels and their functional relevance across cell types, we checked their impact on treatment response by retrospectively investigating a large IDB -treated LHON cohort (n = 118) combining Italian and German patients. In both countries, IDB is currently the standard of care for LHON patients. We considered as the primary visual outcome (responder/non-responder to IDB therapy) the most used criterion adopted in LHON clinical trials, the so-called CRR, with minor modification as detailed in the STAR Methods.21 The demographic and clinical features of the investigated LHON cohort are reported in Table S2.
Considering this visual outcome (responder/non-responder to IDB therapy), CRR to IDB therapy occurred in 27% of eyes (7 of 26) in the NQO1 mut/mut genotype subgroup (homozygous or compound heterozygous for the polymorphic variants), and in 52% and 57% of eyes in NQO1 mut/WT and NQO1 WT/WT, respectively (heterozygous for one polymorphic variant or homozygous WT). These results are highlighted in Table S2.
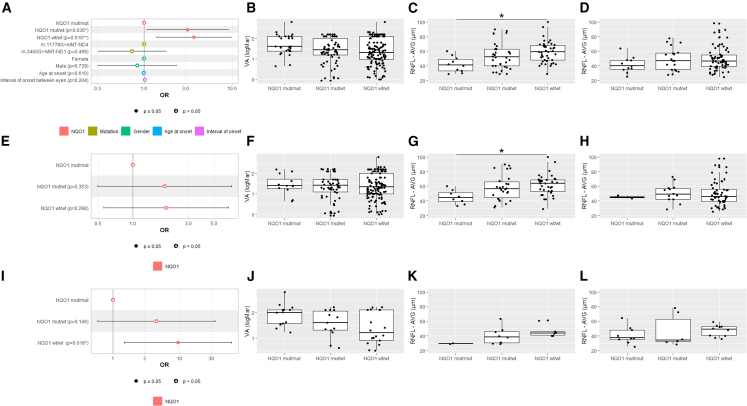
Generalized estimating equation (GEE) model-based analysis investigating the association between the binary visual outcome (responder/non-responder to IDB therapy) and the NQO1 genotype showed that both NQO1 mut/WT (odds ratio [OR]Crude = 3.2, 95% confidence interval = 1.1–9.4, p = 0.03; ORAdjusted = 3.1, 95% confidence interval = 1.1–8.9, p = 0.036) and NQO1 WT/WT (ORCrude = 3.8, 95% confidence interval = 1.4–10.2, p = 0.008; ORAdjusted = 3.7, 95% confidence interval = 1.4–9.9, p = 0.01) were significantly associated with the responder status (Figure 6A).
Figure 6.
NQO1 variants leading to very low protein levels match the IDB responder/non-responder analysis
(A, E, and I) Forest plots of odds ratios (ORs) for the binary visual outcome (responder/non-responder to IDB therapy).
(A) ORs calculated by multivariable GEE modeling on the 118 LHON patients treated with IDB therapy.
(E) ORs calculated by univariable GEE modeling on the LHON patients treated with IDB therapy carrying the m.11778G>A/MT-ND4 mutation.
(I) ORs calculated by univariable GEE modeling on the LHON patients treated with IDB therapy carrying the m.3460G>A/MT-ND1 mutation.
(B, F, and J) Boxplots of VA at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes of all 118 LHON patients (B), LHON patients carrying the m.11778G>A/MT-ND4 (F), and LHON patients carrying m.3460G>A/MT-ND1 (J).
(C, G, and K) All Italian LHON patients (C), Italian LHON patients with m.11778G>A/MT-ND4 (G), and Italian LHON patients with m.3460G>A/MT-ND1 (K). Shown are boxplots of average RNFL thickness at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes. All of these patients were evaluated with swept-source OCT (DRI Triton OCT, Topcon, Tokyo, Japan). ∗p < 0.05.
(D, H, and L) All German LHON patients (D), German LHON patients with m.11778G>A/MT-ND4 (H), and German LHON patients with m.3460G>A/MT-ND1 (L). Shown are boxplots of average RNFL thickness at last visit, with solid lines representing median values for NQO1 mut/mut, NQO1 mut/WT, and NQO1 WT/WT genotypes. All of these patients were evaluated with spectral-domain OCT (Heidelberg Spectralis OCT, Heidelberg Engineering, Heidelberg, Germany).
As secondary visual outcomes, we compared visual acuity (VA) and optical coherence tomography (OCT) average RNFL measures at last visit by stratifying patients according to the three NQO1 genotype subgroups as for the aforementioned responder/non-responder analysis. We did not observe any differences at last visit in terms of VA (Figure 6B). The average RNFL thickness (Figures 6C and 6D), however, assessed separately for the Italian and German patients considering that different OCT machines were used, was significantly thinner for the NQO1 mut/mut genotype compared with NQO1 WT/WT in the Italian cohort (Figure 6C; p = 0.02). Although the NQO1 mut/mut genotype in German patients also had the lowest average RNFL thickness (Figure 6D), it did not reach statistical significance. A further exploratory analysis, focusing on sectorial RNFL thickness data limited to the LHON cases from the IRCCS Institute of Neurological Sciences of Bologna (Figure S8), highlighted that, for most quadrants (nasal/inferior/superior), there was an overall trend toward the lowest RNFL thickness in the mut/mut and highest in the WT/WT NQO1 genotypes, with heterozygote mut/WT being intermediate. Specifically, by comparing the mut/mut with the WT/WT NQO1 genotype, mut/mut showed significantly decreased RNFL thickness in the nasal (p = 0.03) and inferior (p = 0.009) sectors (Figures S8A and S8B). Then, comparing the mut/WT with the WT/WT NQO1 genotype, the heterozygotes also had significantly decreased inferior (p = 0.019) and temporal (p = 0.016) RNFL thicknesses (Figures S8B and S8C) and thinner temporal fibers (p = 0.03) compared with the mut/mut NQO1 genotype (Figure S8C). Next, we wanted to determine whether the association between the NQO1 genotype and the main visual outcome (responder/non-responder to IDB therapy) interacts with the LHON mutation (Tables S3 and S4). For the m.11778G>A/MT-ND4 mutation the previously noted association between the NQO1 genotypes and the responder status was lost (Figure 6E), and VA did not display differences among NQO1 genotypes (Figure 6F). However, the average RNFL thickness was the lowest in the NQO1 mut/mut genotype (Figures 6G and 6H), again significant only for the Italian patients (Figure 6G; p = 0.03). For the m.3460G>A/MT-ND1 mutation, NQO1 WT/WT was significantly associated with the responder outcome (ORCrude = 9.4, 95% confidence interval = 1.5–54.6, p = 0.018) (Figure 6I). None of the secondary visual outcomes (VA and average RNFL thickness) reached statistical significance (Figures 6J–6L).
Overall, these results indicate that the homozygous status or the compound heterozygous combination of NQO1 polymorphic variants, the genotypes that lead to drastic decrease in NQO1 protein levels, are associated with the poorest clinical response in IDB-treated LHON patients. This was most evident with the m.3460G>A/MT-ND1 mutation (Figure 6I).
Molecular docking predicts differential binding for IDB to the WT and m.3460G>A/MT-ND1 mutation at the Q-binding site of CI
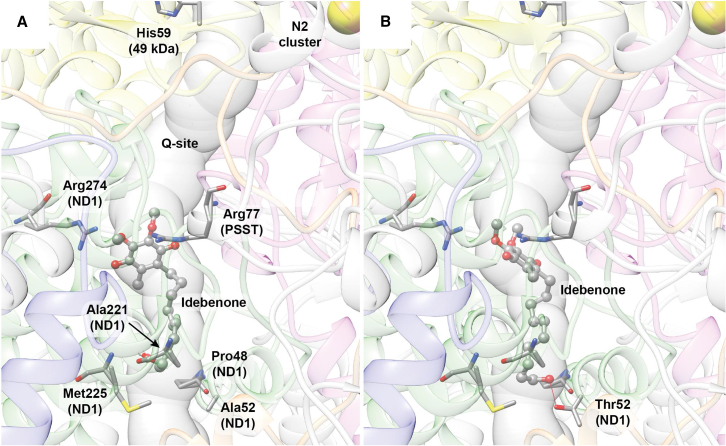
The previous results suggest that the m.3460G>A/MT-ND1 mutation seems to be the most affected by the NQO1 polymorphisms when patients are treated with IDB. Therefore, we analyzed the structure/function relationship of oxidized IDB interaction on WT CI and in the presence of the m.3460G>A/MT-ND1 (p.A52T) mutation. The investigation was performed using docking calculations and taking advantage of the recently released high-resolution structures of ovine CI.49 The use of the latter structure was justified by the high conservation between ovine and human ND subunits, in particular for the ND1 (78% identity), in which, at position 52, there is an alanine in both species. To analyze the effect of the LHON mutation, we generated the p.A52T variant in silico. Considering that IDB is suggested to act at the Q-binding site as a quinone analog,8 we focused on this internal tunnel formed by ovine subunits PSST (NDUFS7 in human CI), 49 kDa (NDUFS2 in human CI), and ND1. An analysis of the Q site performed with MOLEonline showed that the Q site assumes different shapes depending on the conformation of the CI itself and on the bound molecule(s).50 The results of the docking calculations are reported in Figure 7, while an analysis of the IDB interactions in the Q site is reported in Figure S9. In the WT and mutant protein, the best IDB binding pose is located in proximity of the so-called shallow site, which is at the entrance of the Q-binding site, with the aromatic head of the molecule oriented toward the inside of the protein and in the proximity of Arg77 (PSST/NDUFS7). However, the ligand assumes a slightly different pose in the region of the hydroxylic tail. Indeed, in the WT CI, the IDB molecule forms two H bonds with the backbone of Ala221 and Met225 of the ND1 subunit. The latter residues are conserved in humans. On the other hand, in the case of the p.A52T ND1 mutant, IDB is able to form two further H bonds: one with the backbone of Pro48, another ND1conserved residue, and the second with the side chain of the mutated Thr52 (Figures 7B and S9B). The formation of new H bonds in the case of the mutated CI does not preclude the possibility of forming the intermolecular interactions observed in the case of the WT protein. Thus, there are more possibilities for IDB to establish contacts with the Q site in the shallow region, with a putative increase in the stability of the overall interaction.
Figure 7.
Molecular docking of IDB in the Q-binding site of CI carrying the m.3460G>A/MT-ND1 mutation
Details of the best docking poses for IDB in the WT (A) and mutated (B) CI. The Q-site channel is reported in transparent light blue to show the position of the ligand in the cavity with the entrance of the channel at the bottom. IDB is shown in ball-and-stick representation, colored according to the atom type, while CI residues forming specific interaction with the ligands are shown as sticks colored accordingly to atom type. Residues are labeled in (A) only, except for A52T. H bonds are shown using red lines. Protein backbones are reported in transparent cartoons colored in gray, except for the 49 kDa/NDUFS2, PSST/NDUFS7, ND1, ND3 and ND6 subunits, which are shown in yellow, purple, green, orange, and blue, respectively. The N2 [4Fe4S] cluster is shown as spheres colored according to the atom type.
Discussion
This study documents that common homozygous or compound heterozygous NQO1 polymorphic variants (NQO1∗2 and NQO1∗3) lower the NQO1 protein levels in human cells. In vitro, using patient-derived cells, this impacts the therapeutic capacity of IDB to bypass defective CI in LHON. In vivo, reduced NQO1 protein levels reflected as poor CRR of LHON-treated patients. Thus, we here provide compelling evidence that incomplete IDB efficacy in LHON can be partially explained by NQO1 pharmacogenetics.
Our in vitro studies, using multiple cell models, convincingly reconciled the current in vivo therapeutic use of IDB with the old observations showing that oxidized IDB acts as a CI inhibitor8,9 and that it is ineffective in LHON cybrids.30 We demonstrated that, in NQO1-overexpressing cybrids, the NAD(P)H-reduced IDB positively impacts ATP synthesis and contributes to re-establishing R -insensitive cell respiration, proving that electrons are shuttled directly to CIII. Furthermore, NQO1 overexpression also contributed to lower reactive oxygen species (ROS) production and improved LHON cybrid viability when treated with IDB.
This was further corroborated by the results in patient-derived fibroblast cell lines that mostly correlated with the in vivo clinical response to IDB treatment. Previous fibroblast studies concluded that IDB acts in a patient-specific fashion, in some cases improving oxidative phosphorylation (OXPHOS) efficiency and decreasing cell respiration in others.26,27 The explanation resides in the occurrence of individuals displaying low expression of NQO1 protein, which is caused by specific combinations of the two common polymorphic variants (NQO1∗2 and NQO1∗3), both in Ctr and LHON fibroblasts. Both variants did not impact NQO1 mRNA expression levels, as shown previously.38,39,40 NQO1∗2 causes destabilization of the interactions between the core and the C-terminal domains of NQO1 protein, leading to reduced affinity for the FAD cofactor, decreased enzymatic activity, and rapid ubiquitin-dependent degradation.37,38,51 Indeed, cells homozygous for this variant lack NQO1 activity and display only traces of NQO1 protein (e.g., the 3460/1 line analyzed here). The NQO1∗3 variant produces an enzyme harboring the p.R139W missense change, which has been shown to maintain the biochemical and structural characteristics of the WT protein.37 However, the main molecular defect of NQO1∗3 comes from the alteration of exon 4 splicing, reducing the amount of active, full-length protein encoded by isoform 1 to favor the expression of the inactive exon 4-deleted isoform 3.39,40,51 Accordingly, we observed an altered ratio of isoform 1 and 3 expression in fibroblast cell lines harboring NQO1∗3 (the WT2 and 11778/3 lines analyzed here). Moreover, the interaction between the unstable NQO1∗2 product and the NQO1∗3 full-length protein may perturb the NQO1 homodimer, inducing its complete degradation, thus enhancing the severe defect shown by NQO1∗2/NQO1∗3 compound heterozygotes.
We also showed that NQO1 is highly expressed in RGCs and RNFL from postmortem human retinas, as shown previously for the mouse retina.5 To test that NQO1 expression is needed for IDB efficacy, we retrospectively analyzed a cohort of 118 LHON patients treated with IDB from centers in Italy and Germany. The responder/non-responder definition followed the same criteria as in the clinical trials.16,21 We showed that the NQO1 mut/WT and WT/WT genotypes were significantly associated with IDB responders. In contrast, the NQO1 mut/mut genotype not only had the highest rate of non-responders but was also associated with thinner RNFL thickness, confirming the poorest outcome. These results, stratified by the individual LHON pathogenic mutations, indicated that m.3460G>A/MT-ND1 was more profoundly affected by the NQO1 genotype, suggesting a possible direct negative interaction on the outcome of IDB administration for this mutation. We are conscious of the study limitation imposed by the low number of m.3460G>A/MT-ND1 patients included, which is expected due to the lower frequency of this mutation.52 Therefore, the modifying effect of the m.3460G>A/MT-ND1 mutation on the association between IDB response and NQO1 genotype needs to be confirmed by a rigorously designed, larger clinical study. This might reveal that even NQO1 mut/WT heterozygous patients, partially affected by a variable reduction in NQO1 protein levels as seen in fibroblast experiments, may be associated with an increased rate of non-responders to IDB administration. A rationale for this may be envisaged by considering the biochemical phenotype of the m.3460G>A/MT-ND1 mutation. In fact, this mutation affects the quinone site of CI, reducing CI enzymatic activity and potentially directly interfering with IDB binding, as already discussed for CoQ.53,54 This is further strengthened by our structural analysis of mutant ND1 and its interaction with IDB, suggesting that the p.A52T amino acid change introduces further possible H bonds with IDB, ultimately favoring its inhibitory action. This is consistent with a recently published study modeling the interaction of CoQ10 with CI in the setting of the m.3460G>A/MT-ND1 mutation.55 Most remarkably, our current results, pointing to m.3460G>A/MT-ND1 mutation as the most sensible target of the NQO1 pharmacogenetics, are corroborated by the recently released results of the LEROS clinical trial (Study to Assess the Efficacy and Safety of Raxone in LHON Patients, ClinicalTrials.gov ID NCT02774005). In fact, the IDB-treated m.3460G>A/MT-ND1 patients performed worse than natural history untreated LHON patients, whereas significant improvement of VA characterized the IDB-treated patients with the m.11778G>A/MT-ND4 mutation (presented at the ARVO - Association for Research in Vision and Ophthalmology 2023 meeting). In addition, we also encountered difficulty in obtaining in vitro terminally differentiated neurons derived from patient 3460/3, further reinforcing the observations reported above regarding the detrimental effect of this mutation. Overall, the results obtained on the iPSC-derived neurons were based on a limited set of experiments and suffered, in some cases, a variability in the sensitivity to IDB concentration, as we highlighted for Ctr4 iPSCs-derived neurons, which had different behavior at 1 or 10 μM IDB supplementation.
Intriguingly, along the same line, it must be noted that even some patients carrying the NQO1 genotypes associated with strongly reduced NQO1 protein levels were classified as responders. This suggests that NQO1 absence or minimal expression may be compensated for by other mechanisms, such as the activation of survival pathways or reduction of cellular inflammation.12 Moreover, because IDB is rapidly converted into downstream active metabolites, some therapeutic effects have been suggested for its metabolites, such as QS10.56 Further investigation is required to define its precise mechanism of action, considering that this quinone is too soluble to act only at the mitochondrial membrane level.3,4,12 Conversely, even some NQO1 WT patients had poor IDB response, again pointing to a multifactorial mechanism for being a non-responder to IDB treatment.
NQO1 is also a well-known enzyme involved in cancer therapy response.29 Several studies have demonstrated IDB antioxidant activity and decreased lipid peroxidation both in vitro and in vivo.12 Here, we show that overexpression of NQO1 in LHON cells reduced ROS production, but its effect was enhanced in the presence of IDB, confirming an antioxidant role of this quinone in its reduced form. The impact of NQO1 protein levels in smokers may be another aspect that deserves further investigation, considering the known detrimental effect of tobacco smoke on LHON penetrance and severity, probably related to increased oxidative stress.57 Furthermore, there is emerging evidence showing that IDB impacts cell metabolism with long-range effects, implicating signaling upstream or collateral to OXPHOS, such as Shc- and Lin28-related pathways.12,58,59 Our recent work supports the capacity of IDB on rebalancing autophagy and mitophagy and consequently reducing the cell propensity to undergo apoptosis, which goes well beyond the effect on bypassing CI dysfunction.28
Considering the importance of NQO1 on the IDB therapeutic effect, co-administering drugs able to increase NQO1 expression/stability may be considered to improve IDB efficacy. For example, it has been shown that carnosic acid or other compounds may upregulate NQO1 expression.6 On the other hand, screening for the NQO1 genotype may lead to personalized guidelines for IDB administration. In fact, despite the generally well-established good safety profile of in vivo IDB administration, the absence of or very low NQO1 protein levels is harmful in vitro because IDB turns into a toxic form that inhibits CI.8,9
Further considerations include evidence showing that NQO1 expression is greater in glial cells than in neurons,6,60 which raises the question of how IDB may affect different cell types. For example, concerning myelin maintenance by oligodendrocytes in the retrobulbar optic nerve of LHON patients, there is histological evidence of demyelination and myelin remodeling in LHON,61 and we do not yet know how IDB may affect these aspects of the disease mechanism.
Remarkably, NQO1 expression may also be relevant for other drugs currently investigated in clinical trials for several mitochondrial diseases (including LHON), such as EPI-74362,63 and KL1333.64
In conclusion, we provide important insights into the pharmacogenetics of IDB that have explanatory power for the poor response to IDB treatment in a genetically defined subset of LHON patients. This understanding has clinical implications for more effective use of IDB in LHON therapy.
Limitations of the study
One experimental limitation in differentiated neurons was the mixed nature of the cell culture, including neuronal and non-neuronal cells. However, we carefully checked for neurons vs. astrocytes markers and showed that neurons are four to five times more abundant compared with astrocytes, reducing the potential bias.
The retrospective analysis of the effect of IDB in patients suffers from all of the limitations intrinsic to retrieving clinical data. However, we made a special effort to harmonize data from three centers, accounting for the different OCT machines. Finally, as stated under Discussion, we are conscious that the subset of m.3460G>A/MT-ND1 patients was of limited size.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-NQO1 | Thermo fisher Scientific | Cat#39–3700, clone A180; RRID: AB_2533410 |
| Anti-ACTIN | Sigma-Aldrich | Cat#SAB5500001, clone SP124 |
| Anti-GAPDH | Sigma-Aldrich | Cat#G8795; RRID:AB_1078991 |
| Rabbit anti-MAP2 | Cell Signaling Technology | Cat# 4542, RRID:AB_10693782 |
| Rabbit anti-βIIITUBULIN | Cell Signaling Technology | Cat# 5568, RRID:AB_10694505 |
| Mouse anti-GFAP | Millipore | Cat# MAB360, RRID:AB_11212597 |
| Goat anti-Rabbit IgG Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11034, RRID:AB_2576217 |
| Goat anti-Mouse IgG Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11030, RRID:AB_2534089 |
| Hoechst 33342 | Invitrogen | Cat#H1399 |
| Chemicals, peptides, and recombinant proteins | ||
| Idebenone | Santhera Pharmaceuticals | Gift |
| H2DCFDA | Thermo fisher Scientific | Cat#D399 |
| Sulforhodamine B (SRB) | Merck | Cat#S1402 |
| Dicoumarol | Sigma-Aldrich | 287897 |
| N-acetyl-cysteine (NAC) | Sigma-Aldrich | A7250 |
| Critical commercial assays | ||
| Seahorse XFe Cell Mito Stress Test Kit | Seahorse Bioscience | Cat#102340-100 |
| Seahorse XFe96 FluxPak mini | Seahorse Bioscience | Cat#102601-100 |
| XF medium pH 7.4 | Seahorse Bioscience | Cat#103575-100 |
| CyQuant Direct Cell Proliferation Assay Kit | Life Technologies | Cat#C35011 |
| Lentiviral Packaging Kit | Origene | Cat#TR30037 |
| Nextera XT DNA Library Preparation Kit (96 samples) | Illumina | Cat#FC-131-1096 |
| Nextera XT Index Kit v2 Set A (96 indexes, 384 samples) | Illumina | Cat#FC-131-2001 |
| MiSeq Reagent Kit v2 (300-cycles) | Illumina | Cat#MS-102-2002 |
| SNaPshot Multiplex Kit | Applied Biosystems | Cat#4323159 |
| NucleoSpin Tissue, Mini kit for DNA | MACHEREY-NAGEL | Cat#740952 |
| Qubit 1X dsDNA HS Assay Kit | Invitrogen | Cat#Q33231 |
| SureSelect Human All Exon V5/V6 kits | Agilent Technologies | Cat#5190-8863 |
| Pure Link RNA mini kit | Thermo Fisher Scientific | Cat#12183018A |
| SuperScript™ VILO™ cDNA Synthesis Kit | Thermo Fisher Scientific | Cat#11754050 |
| LightCycler® 480 SYBR Green I | Roche | Cat# 04887352001 |
| Deposited data | ||
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Genome Aggregation Database (gnomAD) v2.1.1 | Broad Institute | http://gnomad.broadinstitute.org/; RRID:SCR_014964 |
| Experimental models: Cell lines | ||
| Skin fibroblasts (control, carrying the 3460/ND1 or the 11778/ND4 mutation) | This paper | N/A |
| Wild-type and 3460/ND1 cybrids | This paper, Strobe et al.,65 and Ghelli et al.66 | N/A |
| Neural precursors cells (control, carrying the 3460/ND1 or the 11778/ND4 mutation) | This paper and Peron et al.45 | N/A |
| Oligonucleotides | ||
| Primers for qPCR | This paper | See supplemental information |
| Primers for NQO1 sequencing | This paper | See supplemental information |
| Primers for NQO1 SNaPshot assay | This paper | See supplemental information |
| Recombinant DNA | ||
| pLenti-C-Myc-DDK-P2A-Puro | Origene | Plasmid #PS100092 |
| human NQO1 isoform 1 in pLenti-C-Myc-DDK-P2A-Puro | Origene | Plasmid #RC200620L3 |
| Software and algorithms | ||
| ANNOVAR | Wang et al.67 | https://annovar.openbioinformatics.org/en/latest/; RRID:SCR_012821 |
| bcl2fastq v2.20 | Illumina | https://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html; RRID:SCR_015058 |
| Burrows-Wheeler Aligner v0.7.17 | Li and Durbin68 | http://bio-bwa.sourceforge.net/; RRID:SCR_010910 |
| GeneMapper Software v4.1 | Applied Biosystems | RRID:SCR_014290 |
| Genome Analysis Toolkit v3.8 | Van der Auwera GA, O’Connor BD. Genomics in the Cloud. In: Genomics in the Cloud. O’Reilly Media, Inc.; 2020. | https://gatk.broadinstitute.org/hc/en-us; RRID:SCR_001876 |
| Graphpad Prism v9.3.1 | GraphPad | RRID: SCR_002798 |
| R version 4.1.0 | R Project for Statistical Computing | RRID:SCR_001905 |
| Picard v2.26.0 | Broad Institute | http://broadinstitute.github.io/picard/; RRID:SCR_006525 |
| SAMtools v1.13 | Li et al.69 | http://www.htslib.org/; RRID:SCR_002105 |
| UCSF Chimera 1.16 | Pettersen et al.70 | https://www.cgl.ucsf.edu/chimera/ |
| MOLEonline update 2018 (MOLE 2.5 web interface) | Sehnal et al. and Pravda et al.71,72 | https://mole.upol.cz/online/ |
| AutoDock Vina 1.0 | Trott et al.73 | https://vina.scripps.edu/ |
| LigPlot 4.5.3 | Wallace et al.74 | https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/ |
| GATK v3.8 | Auwera and O’Connor75 | https://gatk.broadinstitute.org/hc/en-us |
| Other | ||
| MagSi-NGSPREP Plus | Magtivio | Cat#MDKT00010075 |
| dNTP Mix, 10mM each | Thermo Scientific | Cat#10610851 |
| Phusion™ High-Fidelity DNA Polymerase (2 U/μL) | Thermo Scientific | Cat#F-530XL |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anna Maria Ghelli (annamaria.ghelli@unibo.it).
Materials availability
All mock and NQO1 overexpressing cybrids cell lines generated in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
This paper does not report original code. All software utilized is freely or commercially available and is listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request. Fastq files with complete sequencing of NQO1 gene after PCR amplification for cell lines harboring m.11778G>A/MT-ND4 or m.3460A>G/MT-ND1 and controls ara availabe on Zenodo.org (https://doi.org/10.5281/zenodo.10422520). Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
LHON cohorts
A cohort of 118 LHON patients treated with idebenone in accordance with routine clinical practice were retrospectively evaluated. Four clinical sites contributed to this aggregated cohort: IRCCS Istituto delle Scienze Neurologiche di Bologna, Bellaria Hospital (Bologna, Italy), IRCCS Ospedale San Raffaele (Milan, Italy), Fondazione IRCCS Istituto Neurologico “Carlo Besta” (Milan, Italy) and Department of Neurology, Friedrich-Baur-Institute, University Hospital of the Ludwig-Maximilians-University (Munich, Germany). Table S2 summarizes all descriptive data of this aggregated LHON cohort.
This study has a historical cohort design and follows the STROBE guidelines. The study was conducted in agreement with the declaration of Helsinki and approved by the local ethics committee (EC#121/2019/OSS/AUSLBO – 19012).
Inclusion criteria were: i) molecularly (m.3460G>A/MT-ND1 or m.11778G>A/MT-ND4 mutations) and clinically defined LHON diagnosis, ii) idebenone therapy within 5 years of disease onset, iii) idebenone treatment for at least 6 months, iv) disease duration of at least 1 year. Exclusion criteria were: i) childhood onset of LHON (LHON onset earlier than 12 years of age), ii) LHON with the m.14484C>T/MT-ND6 mutation or other rare mutations, due to the confounder of high rate of spontaneous visual recovery, iii) reasons other than LHON for having suffered optic atrophy (in particular glaucoma and optic neuritis), iv) participation in gene therapy clinical trials.
A total of 118 patients had been enrolled and 233 eyes had been analyzed (3 patients had monocular disease, 2 wild-type and 1 heterozygous for a NQO1 variant).
All patients underwent extensive neuro-ophthalmological evaluation including visual acuity (VA) testing, tonometry, fundus examination, Ishihara color vision test and optical coherence tomography [OCT, DRI Triton (Topcon, Tokyo, Japan) or Heidelberg Spectralis (Heidelberg Engineering, Heidelberg, Germany) instruments] as detailed elsewhere.76 VA was assessed using ETDRS (Early Treatment Diabetic Retinopathy Study) charts with logMar values as unit or, in cases where VA was assessed using Snellen or decimal scale, logMar values where converted for analysis purposes. For each patient, data on VA were gathered at baseline (the available visit immediately before starting idebenone therapy), nadir (the visit with worst VA) and at last visit (the last visit before withdrawing idebenone, or the last visit available if therapy was still ongoing). Moreover, for each patient we collected the OCT assessment peripapillary retinal nerve fiber layer, RNFL, average thickness at last visit, where available.
We adopted as primary outcome the responder/non-responder analysis as previously defined for clinical trials with idebenone21 with minor modifications. Specifically, we defined the responder/non-responder status as assessed between nadir and last visit: responders were those eyes that experienced a VA improvement from off-chart to on-chart of at least 1 full line (5 letters), or an on-chart VA improvement of at least 2 lines (10 letters = 0.2 logMar), as for the definition of CRR.21
Cell lines and culture conditions
Skin fibroblasts were derived, following informed consent, from 3 healthy donors and 6 LHON patients carrying 3460/ND1 or 11778/ND4 mutations, as assessed by targetSNaPshot Multiplex System (Thermo Fisher Scientific)77 and complete mtDNA sequence analysis.78 The two transmitochondrial cytoplasmic hybrids (cybrids) lines control and 3460/ND1 were generated as previously reported79 and the mitogenome sequences and GeneBank accession numbers were already published,65,66 respectively. Cybrids and fibroblasts were grown at 37°C in Dulbecco’s modified Eagle’s medium (DMEM-high glucose) supplemented with 10% fetal bovine serum (FBS) (South America source from Gibco, Invitrogen, Italy), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/mL streptomycin, in an incubator with a humidified atmosphere of 5% CO2.
Induced pluripotent stem cells (iPSC) were generated, from control and patient fibroblasts, as previously described.45 Neural precursors cells (NPCs) were obtained through embryoid body formation and fully characterized by PCR and immunofluorescence as described.28
To obtain iPSC-derived neurons, we followed previously published protocol.28
Peripheral blood cells isolation
Ten mL of venous blood was collected in EDTA from 1 healthy control and two LHON-affected patients, corresponding to Crt3, 11778/1 and 11778/3 fibroblast’s. PBMCs were isolated using a density gradient cell separation medium (Sigma Aldrich: Histopaque-1077), following manufacturer’s instructions.
Method details
Generation of cybrids overexpressing NQO1
Lentivirus particles containing a pLenti-C-Myc-DDK-P2A-Puro plasmid empty (OriGene, PS100092) or with human NQO1 isoform 1 (OriGene, RC200620L3) were prepared using the Lentiviral Packaging Kit (OriGene, TR30037), following manufacturer’s instructions and used to infect the cybrid cell lines (H42A and RJ206). Infected cells were selected with 0.25μg/mL and 0.75μg/mL puromycin for H42A and RJ206, respectively. Once the overexpression was assessed by Western blot, clonal selection was performed by diluting cells suspensions. For each cell line, three clones with higher NQO1 overexpression evaluated by Western blot analysis were pooled together. Three clones were also pooled for the mock cell lines.
SDS-PAGE and Western blot
Whole lysates of cultured cells and PBMC pellet were prepared in lysis buffer (Triton X-100 1%, EDTA 0.5 mM, PMSF 0.6 mM in Phosphate Buffered saline solution 1X) supplemented with protease inhibitors (Roche #04693116001), incubated on ice for 15 min followed by two freeze/thaw cycles. Then samples were sonicated in a bath at high frequency for 10 min and centrifuged at 10000g for 10 min at 4°C. The supernatant was collected, quantified by Bradford protein assay80 and stored at −80°C. Samples (50 μg of protein) were separated by SDS-PAGE and transferred onto nitrocellulose membrane. Membranes were blocked 60 min at 37°C in 5% milk dissolved in TBS (25mM Tris-HCl pH 7.5, 137mM NaCl)-Tween 0.05% and incubated with primary antibodies (NQO1 Thermo fisher Scientific #39-370-0 1:1000; ACTIN Sigma-Aldrich #SAB5500001 1:5000; GAPDH Sigma-Aldrich #G8795 1:10000) 2 h at RT or 4°C overnight. After washes in TBS-Tween 0.05%, the secondary antibodies (Horseradish peroxidase-conjugated secondary antibodies from Jackson Immuno Research Mouse 115-035-146, Rabbit 111-035-144, 1:5000; or Licor fluorescent secondary antibodies anti-rabbit 926–32210, anti-mouse 926–68071 1:7000) were incubated for 1 h at RT. The chemiluminescence signals were revealed by an ECL western blotting kit and measured with Gel Logic 1500 Imaging System (Kodak, Rochester, NY, USA). The fluorescence signals were detected with Licor Odyssey instrument.
Sulphorhodamine B assay
Cell content for the different assays was determined by using the colorimetric Sulforhodamine B (SRB) assay.81 After treatments, the medium was removed, fresh medium was added and cells were fixed adding 10% trichloro-acetic acid for 1h at 4°C. Then cells were washed five times with H2O and dried for 1h at room temperature. Cells were stained with 0.4% SRB in 1% acetic acid for 30 min and subsequently unbound dye was removed with four washes in 1% acetic acid. SRB was solubilized by addition of 10 mM Tris pH 10 and absorbance was measured at 564 nm by using a micro-plate reader [VICTOR3 Multilabel Plate Counter (PerkinElmer Life and Analytical Sciences, Zaventem Belgium)].
Oxygen consumption rate in fibroblasts and cybrids
Mitochondrial respiration was evaluated using the Seahorse XFe Cell Mito Stress Test Kit (Seahorse Bioscience #101706-100) following the manufacturer instructions. Cells were seeded (3 × 104 cells/well as for fibroblasts, 2.5 × 104, 3 × 104 cells/well as for H42A and RJ206 cybrids respectively) into XFe24 cell culture plate and allowed to attach for 24 h. Cell culture media was replaced with XF medium (Seahorse Bioscience #102353-100). OCR was measured over a 3 min period, followed by 3 min of mixing and 2 min of re-oxygenation of the media. For Mito Stress Test, the complete growth medium was replaced with unbuffered XF medium supplemented with 10 mM glucose, 5 mM pyruvate, and 1 mM glutamine pH 7.4 pre-warmed at 37°C. Cells were incubated with 10 μM idebenone or the same volume of vehicle at 37°C for 60 min to allow temperature and pH equilibration. After an OCR baseline measurement, oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), rotenone and antimycin A were sequentially added to each well to reach final concentrations of 1 μM oligomycin, 0.5 μM and 1 μM FCCP for cybrids and fibroblasts respectively, and 1 μM rotenone and antimycin A. At the end of each experiment, the medium was removed and SRB assay was performed to determine the amount of total cell proteins as previously described.81 OCR data were normalized to total protein levels in each well. Each cell line was represented in five wells per experiment (at least n = 3 biological replicate). Data are normalized on SRB absorbance and expressed as pmoles of O2/minute x protein content.
Oxygen consumption rate in NPCs
Mitochondrial respiration was evaluated using the Seahorse XFe96. NPCs were seeded at 3.5 × 104 cells/well on Poly-L-Ornithine (0.1 mg/ml) - Laminin (2 μg/ml) coated XFe96 cell culture plate (Seahorse XFe96 FluxPak mini #102601-100) and allowed to attach for 24 h. For iPSC-derived neurons, NPCs were seeded 2-3 x 104 cells/well on Poly-L-Ornithine (0.1 mg/ml) - Laminin (2 μg/ml) coated XFe96 cell culture plate and differentiated directly in the plate according to the protocol previously described for 30 days.28 Cell culture media was replaced with XF medium pH 7.4 (Seahorse Bioscience #103575-100) supplemented with 10 mM glucose, 1 mM pyruvate, and 2 mM glutamine pre-warmed at 37°C. Cells were incubated with 10 μM idebenone or with vehicle (DMSO) at 37°C for 60 min to allow temperature and pH equilibration. After an OCR baseline measurement, oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), rotenone and antimycin A were sequentially added to each well to reach final concentrations of 1 μM oligomycin, 2.1 μM FCCP, 1 μM rotenone and 1 μM antimycin A. OCR data were normalized on cell counts, calculated with CyQuant Direct Cell Proliferation Assay Kit (Life Technologies) and expressed as a percentage of the baseline measurement of untreated line. Each NPC cell line was plated in twelve wells per experiment for each condition (treated vs. untreated) and two independent experiments were performed.
Measurement of ATP synthesis rate
The rate of mitochondrial ATP synthesis driven by CI, NADH or NADPH was measured in digitonin-permeabilized cells essentially as described in.9 After trypsinization, cells (10 × 106 mL−1) were suspended in a buffer containing 150 mM KCl, 25 mM Tris–HCl, 2 mM EDTA (ethylenediaminetetraacetic acid), 0.1% bovine serum albumin, 10 mM potassium phosphate, 0.1 mM MgCl2, pH 7.4, kept at room temperature for 15 min, then incubated with 50 μg mL−1 digitonin until 90–100% of cells were positive to erythrosine b staining. Aliquots of 3 × 105 permeabilized cells were incubated in the same buffer and ATP synthesis was assessed. Substrate concentrations were as follows: 1 mM malate plus 1 mM pyruvate for ATP synthesis driven by CI in the presence of 5 mM malonate (complex II inhibitor); 200 μM NADH or NADPH for ATP synthesis driven by NQO1 in the presence of 1 μM rotenone (CI inhibitor) and 5 mM malonate. Idebenone (where indicated) was added at the concentration of 10 μM and incubated for 5 min at RT. After addition of 0.1 mM ADP, chemiluminescence was determined as a function of time with a Sirius L Tube luminometer (Titertek-Berthold, Pforzheim, Germany). The chemiluminescence signal was calibrated using an internal ATP standard after addition of 10 μM oligomycin. The rates of ATP synthesis were normalized to protein content considering that citrate synthase activity was similar in all cell lines.
Reactive oxygen species measurement
To determine the H2O2 production, cells were incubated with 2,7-dichlorodihydrofluorescein diacetate (H2DCFDA) (Life Technologies) as detailed elsewhere.82 Briefly, Cells were seeded 4 × 104 and 6 x104 cells/well for H42A and RJ206 respectively in a 24-wells plate and 48 h after were incubated in DMEM supplemented with H2DCFDA (2μM) 10mM glucose, 5mM pyruvate and 1mM glutamine DMEM in presence or absence of 10μM idebenone. Fluorescence was detected at 535 nm using a multilabel counter Victor3 (PerkinElmer, Turku, Finland) immediately (T0) and after 6h (T6). Fluorescence values were subtracted of blank (cells with medium only) and normalized by protein content (SRB assay) for each well. The difference between the two time points (T6-T0) were calculated and plotted. Each experiment was performed in three wells for each cell line and at least 4 to 6 biological replicates were carried out. As a positive control to indicate what portion of fluorescent signal is due to probe oxidation the increase of H2DCFDA fluorescence (T6-T0), in presence of 10mM N-acetyl-cysteine (NAC) in the assay, was showed in Figure S3.
Cell viability
Viability was determined using the colorimetric sulforhodamine B (SRB) assay.81 Cells were seeded 30.000 cells/well in DMEM-high glucose and after 24 h incubated with 10μM idebenone or the same volume of vehicle (DMSO) in DMEM supplemented with 10mM glucose, 5mM pyruvate and 1mM glutamine. Cellular viability was assessed after 48 h incubation by SRB assay. The SRB absorbance value in the absence of idebenone corresponds to 100% viable cells.
Association analysis in the transcriptomic and proteomic data
For association analysis, combined dataset of whole-exome sequencing (WES), RNA-seq and quantitative tandem mass tag (TMT) proteomics described previously46,47 was used. In brief, DNA for WES was isolated from leukocytes or skin-derived fibroblasts using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and analyzed using the Qubit dsDNA BR Assay Kit. Library preparation was done on 3 μg of DNA with enrichment of exonic regions using the SureSelect Human All Exon V5/V6 kits from Agilent (Agilent Technologies, Santa Clara, CA, USA). Paired-end sequencing was done on Illumina HiSeq2500 or HiSeq4000 platforms (Illumina, San Diego, CA, USA). Reads were aligned to the human reference genome (UCSC build hg19) using Burrows-Wheeler Aligner v0.7.5a68 and variant calling was performed using GATK v3.8.75 Protein coding NQO1∗2 and NQO1∗3 variants were considered called under the following conditions: filter PASS, at least 20 read coverage, and genotype quality greater than 20. Strand-specific, polyA-enriched RNA-seq was performed according to published protocols.83 Following RNA-integrity assessment (BioAnalyzer), libraries were prepared as described in the TruSeq Stranded mRNA Sample Prep Guide (Illumina) and sequenced as 100-bp paired-end runs on a NovaSeq6000. Reads were mapped to GRCh37/hg19 using STAR-aligner. Patient fibroblasts were analyzed in a TMT-batch of 10 cell lines sing previously described sample preparation and data-analysis methods.46 Association analysis was performed using build-in function for linear regression (lm) in R, controlling for sex, age and technical covariates.
NQO1 gene sequencing and analysis of NQO1 polymorphisms
On the fibroblast cell line DNA, the NQO1 coding region was PCR amplified using Phusion High–Fidelity DNA Polymerase (Thermo Fisher Scientific). The purified PCR products were used as input for the Nextera XT DNA Library Preparation Kit (Illumina), and sequenced on a MiSeq System (Illumina) with 2 × 150 bp reads. The obtained FASTQ files were analyzed following GATK Best Practices for SNPs and Indels detection using GATK v3.8, then VCF files were annotated on the GRCh37 build through ANNOVAR.67
We then genotyped for NQO1 a cohort of 118 LHON for NQO1∗2 and ∗3 polymorphisms through a specific assay using the SNaPshot Multiplex System (Thermo Fisher Scientific). The target regions around the SNPs were amplified in multiplex PCR with a GoTaq Hot Start Polymerase (Promega). Purified products were used for the primer extension reaction and then analyzed on a 3500 Dx series Genetic Analyzer (Applied Biosystems) through GeneMapper software (Applied Biosystems). For downstream analysis, patients were categorized in wild type cases (NQO1 w/w), heterozygous (NQO1 mut/wt) and homozygous or compound heterozygous (NQO1 mut/mut) for the two NQO1 polymorphic variants. All primer sequences are available in supplemental information.
Immunohistochemistry
Immunohistochemistry with monoclonal antibody anti-NQO1 (1:100, clone A180, Thermo Fisher Scientific, Waltham, MA, USA) was performed on formalin-fixed, paraffin-embedded human retinal sections obtained from one patient with LHON/11778 (male, aged 52 years) and one control (male, aged 74 years; obtained from the Lions Eye Bank of Oregon, Portland, OR, USA). Primary antibody was visualized using horseradish peroxidase-conjugated secondary antibodies (Dako, Glostrup, Denmark).
Gene expression evaluation
Total RNA was extracted from fibroblasts and PBMC using the Pure Link RNA mini kit (Thermo Fisher Scientific) and then treated with DNAseI enzyme (Sigma Aldrich). 1 μg of RNA was reverse transcribed by Super-Script VILO cDNA synthesis kit (Thermo Fisher Scientific) and analyzed by real-time PCR (LightCycler480, Roche Diagnostics) using SYBR Green I-assay. Primer sequences are listed in supplemental information.
Molecular docking
Among the recently released high resolution CI ovine structures,49 we selected for molecular docking the structure in the close conformation corresponding to the quinone reduction step (PDB ID 6ZKC, in which a molecule of decyl-ubiquinone was solved in both the shallow and the deep Q-sites). The selected CI structure was treated as follows: hydrogen atoms were added by using the addh tool available in UCSF Chimera 1.16,70 the atomic charges were assigned by using the AMBER ff14SB force field84 and the proteins were oriented so that they could contain the Q site within a parallelepiped box of the smallest possible size. The charges for the [Fe4S4] clusters were assigned according to Na et al., 2018.85 The A52T (ND1) mutant was prepared starting from the experimental structure by using the swapaa tool included in the UCSF Chimera suite.70 Swapaa replaces amino acid side chains in order to achieve a different side chain conformation of the same type of amino acid or mutate it into a different type. The conformation of the new side chain is selected from a rotamer library and optimized to reduce the clashes with the neighboring atoms and to optimize the largest possible number of H-bonds. In our case, the side chain conformation has been chosen from the Dunbrack library.86 The idebenone initial conformation was generated by using UCSF Chimera. Idebenone charges were assigned by using the AM1-BCC method87 and the resulting molecule was energy minimized through 1000 step of steepest descent and 100 steps of conjugate gradient using the same software. The obtained idebenone molecule was then docked to the Q-site. The analysis of the Q site channel was performed by using the MOLEonline update 2018 web server.71,72 Re-docking calculations were conducted as described before.50 The docking calculations were performed by using AutoDock Vina 1.073 with default parameters and generating 200 binding poses. The analysis of the results was made by using UCSF Chimera and LigPlot 4.5.3.74
Idebenone titration
Idebenone titration on cybrids showed a dose-dependent inhibitory effect on mock cybrids using 10, 20, 40 80 μM idebenone. Conversely, in NQO1 overexpressing cells the addition of 10 μM idebenone increased both basal and uncoupled respiration, whereas 20, 40, 80 μM progressively lower respiration in a stepwise fashion (Figure S1). Most probably under these conditions of increasing of idebenone concentrations, NQO1 activity is saturated and a corresponding amount of remaining oxidized idebenone progressively inhibits CI.
Mitochondrial respiration in presence or absence of dicoumarol
Dicoumarol is known to act as uncoupler and to be a weak inhibitor of respiratory complex II, III and IV. Therefore, we tried different concentration of dicoumarol 10 and 20μM as reported in literature to avoid the uncoupling effect and inhibition on respiratory chain activity. At the lower concentration 10μM, dicoumarol still partially uncoupled but not inhibited mitochondrial respiration in untreated cells Ctrmock and CtrNQO1. Figure S2, green triangle corresponds to OCR measured in presence of dicoumarol alone in both cell lines, showing a partial uncoupling effect without respiration inhibition. Mitochondrial respiration is dependent on CI activity, being fully inhibited by rotenone. Conversely, when cells were treated with idebenone (red squares), the mitochondrial respiration was partially inhibited in Ctrmock and enhanced in CtrNQO1 cells. In these conditions, particularly in CtrNQO1 cells respiration is not dependent on CI activity, being insensitive to rotenone. This behavior indicates that in presence of idebenone in CtrNQO1 cells is sustained by NQO1 activity and in fact it is completely sensitive to dicoumarol (black triangles).
H2O2 production measurements
In the main text it was shown the H2O2 production in mock and NQO1 overexpressing cells in presence or absence of idebenone. We also measured the effect of an antioxidant (N-acetylcysteine) in mock or overexpressing NQO1 cells as positive control. In Figure S3 it is clearly showed the strong effect of this compound on cells.
Characterization of iPSCs derived neuronal population
To evaluate the fraction of iPSCs derived neurons as compared with iPSCs derived non-neuronal cells, we performed immunofluorescent analysis using antibodies against specific neuronal markers (MAP2 and βIIITUBULIN) and against a specific astrocyte marker (GFAP) on neuronal culture at 30 days of differentiation. Analysis was performed with FIJI software using Analyze Particles plugin after the use of Huang2 auto-threshold adjustment,88 by assessing the contribution of the different fluorochrome (green for MAP2 and βIIITUBULIN; red for GFAP) as the positive fluorescence signal area of the image in micron.28 We evaluated 20 images in which we analyzed in parallel the signals derived from MAP2 and GFAP, and additional 20 images in which we analyzed in parallel the signals derived from βIIITUBULIN and GFAP. The histogram on the right panel show the results of the analysis and demonstrate that MAP2 positive and βIIITUBULIN positive neuronal cells are highly enriched in our neuronal culture.
Immunofluorescence characterization of neuronal culture
Neuronal markers such as MAP2 (Cell Signaling Technology Cat# 4542, RRID:AB_10693782) and βIIITUBULIN (Cell Signaling Technology Cat# 5568, RRID:AB_10694505) and astrocyte specific marker GFAP (Millipore Cat# MAB360, RRID:AB_11212597) were used. The following dilutions were used: anti-MAP2 1:200; anti-βIIITUBULIN 1:500; anti-GFAP 1:1000. Nuclei were colored with 1:1000 Hoechst 33342 (Invitrogen Cat#H1399). Immunofluorescence analysis was performed as already described28 and data evaluated using FIJI software.88
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism v.8 (GraphPad Software Inc., San Diego, CA, USA). Data were expressed as mean ± standard deviation (SD). Statistical analysis of OCR measurements was performed applying a t-test between the measurement in presence (IDB) or absence (vehicle) of idebenone for each point of the time course. Statistical analysis for ATP rate synthesis assay and cell viability assay in presence or absence of idebenone or between mock and NQO1 overexpressing cells was performed as a two-tailed unpaired t-test. ROS production measurements of indicated couple of samples were analyzed as paired t-test. Statistical analysis for NQO1 protein level and relative gene expression quantification of different fibroblast cell lines was carried out using an ordinary one-way ANOVA test with a Dunnett’s post-hoc test, comparing the mean of each column with the mean of WT3 as control column. At least three biological replicates were conducted for each experiment. Statistical significance was defined by ∗ p value <0.05, ∗∗ p value <0.01 and ∗∗∗ p value <0.001. More details on number of experiments are added in the figure legends.
Concerning collected clinical data, descriptive statistics were calculated for the entire study population and stratified based on NQO1 genotype subgroups (NQO1 mut/mut; NQO1 mut/wt; NQO1 wt/wt). The association between NQO1 genotype and the primary visual outcome (responder/non-responder to idebenone therapy) was examined by implementing both univariable and multivariable models by means of generalized estimating equations (GEE) (R geepack package, version 1.3.3) under an exchangeable covariance structure. This GEE approach was chosen to account for the existing correlation between outcomes from a person’s two eyes.89 In particular.
-
(1)
for univariable GEE model: the dependent variable was the primary (binary) visual outcome (responder/non-responder to idebenone therapy) and the independent variable was NQO1 genotype (a three-level categorical variable: NQO1 mut/mut; NQO1 mut/wt; NQO1 wt/wt);
-
(2)
for the multivariable GEE model: the dependent variable was the same binary visual outcome, the independent variable was NQO1 genotype, and adjusting confounders were LHON mutation, gender, age at LHON onset and time (months) from LHON onset to start of idebenone therapy. Then, secondary VA/OCT outcome comparisons were also carried out by applying the Clustered Wilcoxon rank-sum test using Rosner-Glynn-Lee method.90 Furthermore, univariable GEE models were also implemented stratifying by LHON mutation to determine whether the association between NQO1 genotype and the investigated visual (responder/non-responder to idebenone therapy) outcome was modified by this third factor (effect modification). In this case, because of the overall reduction of the statistical power, we did not implement multivariable GEE models.
All statistical tests were two-sided and p values of less than 0.05 were considered to indicate statistical significance. All these statistical analyses were carried out by using R software (version 4.1.0).
Acknowledgments
This work was completely or partially supported by MITOCON (to V.C. and A.M.G. [specific grant]) and Italy grant 2018-01 (to V.C. and V.T.), the Italian Ministry of Health as part of the REORION project (RF-2018-12366703 to V.C. and V.T.) and the Italian Ministry of Health (RRC), and a Montevecchi Rita donation and Italian Ministry of University and Research PRIN2020- 2020RRJP5L_004 (to A.M.G. and A.M.P.). V.T. is part of the Center for the Study of Mitochondrial Pediatric Diseases funded by the Mariani Foundation. C. Fiorini is supported by Associazione Luigi Comini, Onlus. A. Cavaliere is a recipient of SG-2021-12374454 from the Italian Ministry of Health. C.L. and A. Catania are supported by RF-2016-02361495. This work was also supported by the German Federal Ministry of Education and Research (BMBF; Bonn, Germany) through a grant to the German Network for Mitochondrial Disorders (mitoNET,01GM1906A to T.K. and 01GM1906D to H.P.) and a grant for the E-Rare project GENOMIT (01GM1920A to H.P. and 01GM1920B to T.K.) and the ERA PerMed project PerMiM (01KU2016A to H.P.). Some authors are members of the European Reference Network for Rare Neurological Diseases (ERN-RND; T.K. and V.C.), the European Reference Network for Neuromuscular Diseases (ERN-EURO-NMD; to C.L., T.K., and V.T.), and the Mitochondrial Inter-ERN Group (C.L., T.K., and V.C.), co-funded by the European Commission. The publication of this article was supported by “Ricerca Corrente” funding from the Italian Ministry of Health.
Author contributions
A.M.G. and V.C., study design; S.J.A. and V.D.D., cellular experiment development and supervision; M.R. and B.B., clinical data analysis and statistical study on idebenone responders/non-responders; C. Fiorini, sequencing of the NQO1 gene; G.C., M. Capristo, C. Fasano, A.M., and C.V.T., cellular experiments on cybrids, fibroblasts, and PBMC samples; C. Peron, A. Catania, and V.T., iPSCs generation and NPC/neuron experiments; L.C., mtDNA sequences; M.G.P. and C.G., immunohistochemistry on retinal sections; F.N.R.-C. and A.A.S. provision and characterization of retinal sections; G.T. and F.M., molecular docking analysis; C.Z., fibroblast cell line generation; A. Catania, S.B.M., C.L., A.D.N., M.L.C., M.B., P.B., M. Carbonelli, G.A., and C.L.M., clinical investigation of Italian LHON patients; D.S., C.V., A.F., B.B., C.B.C., C. Priglinger, B.L., and T.K., clinical investigation of German LHON patients; D.S., C.V., A.F., and H.P., genotyping of the German LHON cohort and transcriptome and proteome analysis; S.J.A., V.D.D., M.R., and C. Fiorini, data analysis and writing of the STAR Methods and Results; A.M.G., V.C., V.T., A.M., and A.M.P., global interpretation; A.M.G., V.C., V.D.D., T.K., H.P., and A.A.S., drafting of the original manuscript. All authors revised and approved the manuscript.
Declaration of interests
M.R., P.B., M. Carbonelli, G.A., C.L.M., C.B.C., T.K., and V.C. are involved in clinical trials with idebenone (Santhera Pharmaceuticals and Chiesi Farmaceutici) in LHON patients. M.R., A.A.S., P.B., M. Carbonelli, G.A., C.L.M., C.B.C., T.K., and V.C. are involved in gene therapy trials with Lumevoq (GenSight Biologics) in LHON patients. A.A.S., C.L.M., T.K., and V.C. have received research support, speaker honoraria, consulting fees, and travel reimbursement from Santhera Pharmaceuticals, Chiesi GmbH, and GenSight Biologics. None of these activities are related to conduct of this study or writing of the manuscript.
Published: January 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101383.
Contributor Information
Valerio Carelli, Email: valerio.carelli@unibo.it.
Anna Maria Ghelli, Email: annamaria.ghelli@unibo.it.
Supplemental information
References
- 1.Zs -Nagy I. Chemistry, toxicology, pharmacology and pharmacokinetics of idebenone: a review. Arch. Gerontol. Geriatr. 1990;11:177–186. doi: 10.1016/0167-4943(90)90063-c. [DOI] [PubMed] [Google Scholar]
- 2.Gillis J.C., Benefield P., McTavish D. Idebenone: A Review of its Pharmacodynamic and Pharmacokinetic Properties, and Therapeutic Use in Age-Related Cognitive Disorders. Drugs Aging. 1994;5:133–152. doi: 10.2165/00002512-199405020-00007. [DOI] [PubMed] [Google Scholar]
- 3.Haefeli R.H., Erb M., Gemperli A.C., Robay D., Courdier Fruh I., Anklin C., Dallmann R., Gueven N. NQO1-dependent redox cycling of idebenone: effects on cellular redox potential and energy levels. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erb M., Hoffmann-Enger B., Deppe H., Soeberdt M., Haefeli R.H., Rummey C., Feurer A., Gueven N. Features of Idebenone and Related Short-Chain Quinones that Rescue ATP Levels under Conditions of Impaired Mitochondrial Complex I. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varricchio C., Beirne K., Heard C., Newland B., Rozanowska M., Brancale A., Votruba M. The ying and yang of idebenone: Not too little, not too much – cell death in NQO1 deficient cells and the mouse retina. Free Radic. Biol. Med. 2020;152:551–560. doi: 10.1016/j.freeradbiomed.2019.11.030. [DOI] [PubMed] [Google Scholar]
- 6.Jaber S., Polster B.M. Idebenone and neuroprotection: antioxidant, pro-oxidant, or electron carrier? J. Bioenerg. Biomembr. 2015;47:111–118. doi: 10.1007/s10863-014-9571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong H., Shertzer H.G., Genter M.B., Gonzalez F.J., Vasiliou V., Jefcoate C., Nebert D.W. Mitochondrial targeting of mouse NQO1 and CYP1B1 proteins. Biochem. Biophys. Res. Commun. 2013;435:727–732. doi: 10.1016/j.bbrc.2013.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esposti M.D., Ngo A., Ghelli A., Benelli B., Carelli V., McLennan H., Linnane A.W. The interaction of Q analogs, particularly hydroxydecyl benzoquinone (idebenone), with the respiratory complexes of heart mitochondria. Arch. Biochem. Biophys. 1996;330:395–400. doi: 10.1006/abbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 9.Giorgio V., Petronilli V., Ghelli A., Carelli V., Rugolo M., Lenaz G., Bernardi P. The effects of idebenone on mitochondrial bioenergetics. Biochim. Biophys. Acta. 2012;1817:363–369. doi: 10.1016/j.bbabio.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geromel V., Darin N., Chrétien D., Bénit P., DeLonlay P., Rötig A., Munnich A., Rustin P. Coenzyme Q(10) and idebenone in the therapy of respiratory chain diseases: rationale and comparative benefits. Mol. Genet. Metabol. 2002;77:21–30. doi: 10.1016/s1096-7192(02)00145-2. [DOI] [PubMed] [Google Scholar]
- 11.Gueven N., Nadikudi M., Daniel A., Chhetri J. Targeting mitochondrial function to treat optic neuropathy. Mitochondrion. 2017;36:7–14. doi: 10.1016/j.mito.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Gueven N., Ravishankar P., Eri R., Rybalka E. Idebenone: When an antioxidant is not an antioxidant. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu-Wai-Man P., Votruba M., Burté F., La Morgia C., Barboni P., Carelli V. A neurodegenerative perspective on mitochondrial optic neuropathies. Acta Neuropathol. 2016;132:789–806. doi: 10.1007/s00401-016-1625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenton S.L., Sheremet N.L., Catarino C.B., Andreeva N.A., Assouline Z., Barboni P., Barel O., Berutti R., Bychkov I., Caporali L., et al. Impaired complex I repair causes recessive Leber’s hereditary optic neuropathy. J. Clin. Invest. 2021;131 doi: 10.1172/JCI138267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporali L., Maresca A., Capristo M., Del Dotto V., Tagliavini F., Valentino M.L., La Morgia C., Carelli V. Incomplete penetrance in mitochondrial optic neuropathies. Mitochondrion. 2017;36:130–137. doi: 10.1016/j.mito.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Klopstock T., Yu-Wai-Man P., Dimitriadis K., Rouleau J., Heck S., Bailie M., Atawan A., Chattopadhyay S., Schubert M., Garip A., et al. A randomized placebo-controlled trial of idebenone in Leber’s hereditary optic neuropathy. Brain. 2011;134:2677–2686. doi: 10.1093/brain/awr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carelli V., La Morgia C., Valentino M.L., Rizzo G., Carbonelli M., De Negri A.M., Sadun F., Carta A., Guerriero S., Simonelli F., et al. Idebenone Treatment In Leber’s Hereditary Optic Neuropathy. Brain. 2011;134:e188. doi: 10.1093/brain/awr180. [DOI] [PubMed] [Google Scholar]
- 18.Amore G., Romagnoli M., Carbonelli M., Barboni P., Carelli V., La Morgia C. Therapeutic Options in Hereditary Optic Neuropathies. Drugs. 2021;81:57–86. doi: 10.1007/s40265-020-01428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carelli V., Carbonelli M., de Coo I.F., Kawasaki A., Klopstock T., Lagrèze W.A., La Morgia C., Newman N.J., Orssaud C., Pott J.W.R., et al. International Consensus Statement on the Clinical and Therapeutic Management of Leber Hereditary Optic Neuropathy. J. Neuro Ophthalmol. 2017;37:371–381. doi: 10.1097/WNO.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 20.Newman N.J., Carelli V., Taiel M., Yu-Wai-Man P. Visual Outcomes in Leber Hereditary Optic Neuropathy Patients With the m.11778G>A (MTND4) Mitochondrial DNA Mutation. J. Neuro Ophthalmol. 2020;40:547–557. doi: 10.1097/WNO.0000000000001045. [DOI] [PubMed] [Google Scholar]
- 21.Catarino C.B., von Livonius B., Priglinger C., Banik R., Matloob S., Tamhankar M.A., Castillo L., Friedburg C., Halfpenny C.A., Lincoln J.A., et al. Real-World Clinical Experience With Idebenone in the Treatment of Leber Hereditary Optic Neuropathy. J. Neuro Ophthalmol. 2020;40:558–565. doi: 10.1097/WNO.0000000000001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carelli V., Ross-Cisneros F.N., Sadun A.A. Mitochondrial dysfunction as a cause of optic neuropathies. Prog. Retin. Eye Res. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Yu-Wai-Man P., Griffiths P.G., Chinnery P.F. Mitochondrial optic neuropathies – Disease mechanisms and therapeutic strategies. Prog. Retin. Eye Res. 2011;30:81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X., Zhang Y., Lu L., Yang H. Therapeutic Effects of Idebenone on Leber Hereditary Optic Neuropathy. Curr. Eye Res. 2020;45:1315–1323. doi: 10.1080/02713683.2020.1736307. [DOI] [PubMed] [Google Scholar]
- 25.Pemp B., Mitsch C., Kircher K., Reitner A. Changes in Visual Function and Correlations with Inner Retinal Structure in Acute and Chronic Leber’s Hereditary Optic Neuropathy Patients after Treatment with Idebenone. J. Clin. Med. 2021;10:151. doi: 10.3390/jcm10010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu-Wai-Man P., Soiferman D., Moore D.G., Burté F., Saada A. Evaluating the therapeutic potential of idebenone and related quinone analogues in Leber hereditary optic neuropathy. Mitochondrion. 2017;36:36–42. doi: 10.1016/j.mito.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angebault C., Gueguen N., Desquiret-Dumas V., Chevrollier A., Guillet V., Verny C., Cassereau J., Ferre M., Milea D., Amati-Bonneau P., et al. Idebenone increases mitochondrial complex I activity in fibroblasts from LHON patients while producing contradictory effects on respiration. BMC Res. Notes. 2011;4:557. doi: 10.1186/1756-0500-4-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danese A., Patergnani S., Maresca A., Peron C., Raimondi A., Caporali L., Marchi S., La Morgia C., Del Dotto V., Zanna C., et al. Pathological mitophagy disrupts mitochondrial homeostasis in Leber’s hereditary optic neuropathy. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pey A.L., Megarity C.F., Timson D.J. NAD(P)H quinone oxidoreductase (NQO1): an enzyme which needs just enough mobility, in just the right places. Biosci. Rep. 2019;39 doi: 10.1042/BSR20180459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghelli A., Porcelli A.M., Zanna C., Martinuzzi A., Carelli V., Rugolo M. Protection against oxidant-induced apoptosis by exogenous glutathione in Leber hereditary optic neuropathy cybrids. Invest. Ophthalmol. Vis. Sci. 2008;49:671–676. doi: 10.1167/iovs.07-0880. [DOI] [PubMed] [Google Scholar]
- 31.Brown M.D., Trounce I.A., Jun A.S., Allen J.C., Wallace D.C. Functional Analysis of Lymphoblast and Cybrid Mitochondria Containing the 3460, 11778, or 14484 Leber’s Hereditary Optic Neuropathy Mitochondrial DNA Mutation. J. Biol. Chem. 2000;275:39831–39836. doi: 10.1074/jbc.M006476200. [DOI] [PubMed] [Google Scholar]
- 32.Beretta S., Mattavelli L., Sala G., Tremolizzo L., Schapira A.H.V., Martinuzzi A., Carelli V., Ferrarese C. Leber hereditary optic neuropathy mtDNA mutations disrupt glutamate transport in cybrid cell lines. Brain. 2004;127:2183–2192. doi: 10.1093/brain/awh258. [DOI] [PubMed] [Google Scholar]
- 33.Floreani M., Napoli E., Martinuzzi A., Pantano G., De Riva V., Trevisan R., Bisetto E., Valente L., Carelli V., Dabbeni-Sala F. Antioxidant defences in cybrids harboring mtDNA mutations associated with Leber’s hereditary optic neuropathy. FEBS J. 2005;272:1124–1135. doi: 10.1111/j.1742-4658.2004.04542.x. [DOI] [PubMed] [Google Scholar]
- 34.King M.P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 35.Carelli V., Franceschini F., Venturi S., Barboni P., Savini G., Barbieri G., Pirro E., La Morgia C., Valentino M.L., Zanardi F., et al. Grand rounds: could occupational exposure to n-hexane and other solvents precipitate visual failure in leber hereditary optic neuropathy? Environ. Health Perspect. 2007;115:113–115. doi: 10.1289/ehp.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lienhart W.-D., Gudipati V., Uhl M.K., Binter A., Pulido S.A., Saf R., Zangger K., Gruber K., Macheroux P. Collapse of the native structure caused by a single amino acid exchange in human NAD(P)H:quinone oxidoreductase(1.) FEBS J. 2014;281:4691–4704. doi: 10.1111/febs.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel D., Anwar A., Winski S.L., Kepa J.K., Zolman K.L., Ross D. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:quinone oxidoreductase 1. Mol. Pharmacol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 39.Pan S.-S., Han Y., Farabaugh P., Xia H. Implication of alternative splicing for expression of a variant NAD(P)H:quinone oxidoreductase-1 with a single nucleotide polymorphism at 465C>T. Pharmacogenetics. 2002;12:479–488. doi: 10.1097/00008571-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 40.Hu L.T., Stamberg J., Pan S. The NAD(P)H:quinone oxidoreductase locus in human colon carcinoma HCT 116 cells resistant to mitomycin C. Cancer Res. 1996;56:5253–5259. [PubMed] [Google Scholar]
- 41.Dinkova-Kostova A.T., Kostov R.V., Kazantsev A.G. The role of Nrf2 signaling in counteracting neurodegenerative diseases. FEBS J. 2018;285:3576–3590. doi: 10.1111/febs.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaspar J.W., Jaiswal A.K. Antioxidant-induced Phosphorylation of Tyrosine 486 Leads to Rapid Nuclear Export of Bach1 That Allows Nrf2 to Bind to the Antioxidant Response Element and Activate Defensive Gene Expression. J. Biol. Chem. 2010;285:153–162. doi: 10.1074/jbc.M109.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T., et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell. 2020;182:1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schelonka L.P., Siegel D., Wilson M.W., Meininger A., Ross D. Immunohistochemical localization of NQO1 in epithelial dysplasia and neoplasia and in donor eyes. Invest. Ophthalmol. Vis. Sci. 2000;41:1617–1622. [PubMed] [Google Scholar]
- 45.Peron C., Mauceri R., Cabassi T., Segnali A., Maresca A., Iannielli A., Rizzo A., Sciacca F.L., Broccoli V., Carelli V., Tiranti V. Generation of a human iPSC line, FINCBi001-A, carrying a homoplasmic m.G3460A mutation in MT-ND1 associated with Leber’s Hereditary optic Neuropathy (LHON) Stem Cell Res. 2020;48 doi: 10.1016/j.scr.2020.101939. [DOI] [PubMed] [Google Scholar]
- 46.Kopajtich R., Smirnov D., Stenton S.L., Loipfinger S., Meng C., Scheller I.F., Freisinger P., Baski R., Berutti R., Behr J., et al. Integration of proteomics with genomics and transcriptomics increases the diagnostic rate of Mendelian disorders. medRxiv. 2021 doi: 10.1101/2021.03.09.21253187. Preprint at. [DOI] [Google Scholar]
- 47.Dmitrii S., Kopajtich R., Stenton S., Prokisch H. RNA-seq count and proteomics intensity data used in the Kopajtich, Smirnov, Stenton et al study. zenodo. 2021 doi: 10.5281/ZENODO.4501904. Preprint at. [DOI] [Google Scholar]
- 48.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kampjut D., Sazanov L.A. The coupling mechanism of mammalian respiratory complex I. Science. 2020;370 doi: 10.1126/science.abc4209. [DOI] [PubMed] [Google Scholar]
- 50.Kurelac I., Cavina B., Sollazzo M., Miglietta S., Fornasa A., De Luise M., Iorio M., Lama E., Traversa D., Nasiri H.R., et al. NDUFS3 knockout cancer cells and molecular docking reveal specificity and mode of action of anti-cancer respiratory complex I inhibitors. Open Biol. 2022;12 doi: 10.1098/rsob.220198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pey A.L., Megarity C.F., Timson D.J. FAD binding overcomes defects in activity and stability displayed by cancer-associated variants of human NQO1. Biochim. Biophys. Acta. 2014;1842:2163–2173. doi: 10.1016/j.bbadis.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Yu-Wai-Man P., Griffiths P.G., Hudson G., Chinnery P.F. Inherited mitochondrial optic neuropathies. J. Med. Genet. 2009;46:145–158. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baradaran R., Berrisford J.M., Minhas G.S., Sazanov L.A. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung I., Wright J.J., Bridges H.R., Ivanov B.S., Biner O., Pereira C.S., Arantes G.M., Hirst J. Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy. Nat. Commun. 2022;13:2758. doi: 10.1038/s41467-022-30506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuller J.T., Barnes S., Sadun L.A., Ajmera P., Alexandrova A.N., Sadun A.A. Coenzyme Q10 trapping in mitochondrial complex I underlies Leber’s hereditary optic neuropathy. Proc. Natl. Acad. Sci. USAUSA. 2023;120 doi: 10.1073/pnas.2304884120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giorgio V., Schiavone M., Galber C., Carini M., Da Ros T., Petronilli V., Argenton F., Carelli V., Acosta Lopez M.J., Salviati L., et al. The idebenone metabolite QS10 restores electron transfer in complex I and coenzyme Q defects. Biochim. Biophys. Acta Bioenerg. 2018;1859:901–908. doi: 10.1016/j.bbabio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 57.Giordano L., Deceglie S., d’Adamo P., Valentino M.L., La Morgia C., Fracasso F., Roberti M., Cappellari M., Petrosillo G., Ciaravolo S., et al. Cigarette toxicity triggers Leber’s hereditary optic neuropathy by affecting mtDNA copy number, oxidative phosphorylation and ROS detoxification pathways. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomilov A., Allen S., Hui C.K., Bettaieb A., Cortopassi G. Idebenone is a cytoprotective insulin sensitizer whose mechanism is Shc inhibition. Pharmacol. Res. 2018;137:89–103. doi: 10.1016/j.phrs.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Lei D., Shao Z., Zhou X., Yuan H. Synergistic neuroprotective effect of rasagiline and idebenone against retinal ischemia-reperfusion injury via the Lin28-let-7-Dicer pathway. Oncotarget. 2018;9:12137–12153. doi: 10.18632/oncotarget.24343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaber S.M., Ge S.X., Milstein J.L., VanRyzin J.W., Waddell J., Polster B.M. Idebenone Has Distinct Effects on Mitochondrial Respiration in Cortical Astrocytes Compared to Cortical Neurons Due to Differential NQO1 Activity. J. Neurosci. 2020;40:4609–4619. doi: 10.1523/JNEUROSCI.1632-17.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carelli V., La Morgia C., Ross-Cisneros F.N., Sadun A.A. Optic neuropathies: the tip of the neurodegeneration iceberg. Hum. Mol. Genet. 2017;26:R139–R150. doi: 10.1093/hmg/ddx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinelli D., Catteruccia M., Piemonte F., Pastore A., Tozzi G., Dionisi-Vici C., Pontrelli G., Corsetti T., Livadiotti S., Kheifets V., et al. EPI-743 reverses the progression of the pediatric mitochondrial disease--genetically defined Leigh Syndrome. Mol. Genet. Metabol. 2012;107:383–388. doi: 10.1016/j.ymgme.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 63.Sadun A.A., Chicani C.F., Ross-Cisneros F.N., Barboni P., Thoolen M., Shrader W.D., Kubis K., Carelli V., Miller G. Effect of EPI-743 on the Clinical Course of the Mitochondrial Disease Leber Hereditary Optic Neuropathy. Arch. Neurol. 2012;69:331–338. doi: 10.1001/archneurol.2011.2972. [DOI] [PubMed] [Google Scholar]
- 64.Seo K.-S., Kim J.-H., Min K.-N., Moon J.-A., Roh T.-C., Lee M.-J., Lee K.-W., Min J.-E., Lee Y.-M. KL1333, a Novel NAD+ Modulator, Improves Energy Metabolism and Mitochondrial Dysfunction in MELAS Fibroblasts. Front. Neurol. 2018;9:552. doi: 10.3389/fneur.2018.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strobbe D., Caporali L., Iommarini L., Maresca A., Montopoli M., Martinuzzi A., Achilli A., Olivieri A., Torroni A., Carelli V., Ghelli A. Haplogroup J mitogenomes are the most sensitive to the pesticide rotenone: Relevance for human diseases. Neurobiol. Dis. 2018;114:129–139. doi: 10.1016/j.nbd.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 66.Ghelli A., Porcelli A.M., Zanna C., Vidoni S., Mattioli S., Barbieri A., Iommarini L., Pala M., Achilli A., Torroni A., et al. The background of mitochondrial DNA haplogroup J increases the sensitivity of Leber’s hereditary optic neuropathy cells to 2,5-hexanedione toxicity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 71.Sehnal D., Svobodová Vařeková R., Berka K., Pravda L., Navrátilová V., Banáš P., Ionescu C.-M., Otyepka M., Koča J. MOLE 2.0: advanced approach for analysis of biomacromolecular channels. J. Cheminf. 2013;5:39. doi: 10.1186/1758-2946-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pravda L., Sehnal D., Toušek D., Navrátilová V., Bazgier V., Berka K., Svobodová Vareková R., Koča J., Otyepka M. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update) Nucleic Acids Res. 2018;46:W368–W373. doi: 10.1093/nar/gky309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 75.Auwera G. van der, O’Connor B.D. First edition. O’Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 76.Romagnoli M., Stanzani Maserati M., De Matteis M., Capellari S., Carbonelli M., Amore G., Cantalupo G., Zenesini C., Liguori R., Sadun A.A., et al. Chromatic Pupillometry Findings in Alzheimer’s Disease. Front. Neurosci. 2020;14:780. doi: 10.3389/fnins.2020.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez-Iglesias V., Barros F., Carracedo A., Salas A. Minisequencing mitochondrial DNA pathogenic mutations. BMC Med. Genet. 2008;9:26. doi: 10.1186/1471-2350-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caporali L., Iommarini L., La Morgia C., Olivieri A., Achilli A., Maresca A., Valentino M.L., Capristo M., Tagliavini F., Del Dotto V., et al. Peculiar combinations of individually non-pathogenic missense mitochondrial DNA variants cause low penetrance Leber’s hereditary optic neuropathy. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.King M.P., Attadi G. Mitochondria-mediated transformation of human rho(0) cells. Methods Enzymol. 1996;264:313–334. doi: 10.1016/s0076-6879(96)64030-0. [DOI] [PubMed] [Google Scholar]
- 80.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 81.Scarlatti F., Sala G., Somenzi G., Signorelli P., Sacchi N., Ghidoni R. Resveratrol induces growth inhibition and apoptosis in metastatic breast cancer cells via de novo ceramide signaling. Faseb. J. 2003;17:2339–2341. doi: 10.1096/fj.03-0292fje. [DOI] [PubMed] [Google Scholar]
- 82.Iommarini L., Kurelac I., Capristo M., Calvaruso M.A., Giorgio V., Bergamini C., Ghelli A., Nanni P., De Giovanni C., Carelli V., et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum. Mol. Genet. 2014;23:1453–1466. doi: 10.1093/hmg/ddt533. [DOI] [PubMed] [Google Scholar]
- 83.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C., et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8 doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Na S., Jurkovic S., Friedrich T., Koslowski T. Charge transfer through a fragment of the respiratory complex I and its regulation: an atomistic simulation approach. Phys. Chem. Chem. Phys. 2018;20:20023–20032. doi: 10.1039/c8cp02420k. [DOI] [PubMed] [Google Scholar]
- 86.Shapovalov M.V., Dunbrack R.L. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 88.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ying G.-S., Maguire M.G., Glynn R., Rosner B. Tutorial on Biostatistics: Statistical Analysis for Correlated Binary Eye Data. Ophthalmic Epidemiol. 2018;25:1–12. doi: 10.1080/09286586.2017.1320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rosner B., Glynn R.J., Lee M.-L.T. Extension of the rank sum test for clustered data: two-group comparisons with group membership defined at the subunit level. Biometrics. 2006;62:1251–1259. doi: 10.1111/j.1541-0420.2006.00582.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. All software utilized is freely or commercially available and is listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request. Fastq files with complete sequencing of NQO1 gene after PCR amplification for cell lines harboring m.11778G>A/MT-ND4 or m.3460A>G/MT-ND1 and controls ara availabe on Zenodo.org (https://doi.org/10.5281/zenodo.10422520). Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.