Summary
Prior observational studies suggest an association between intra-pancreatic fat deposition (IPFD) and pancreatic ductal adenocarcinoma (PDAC); however, the causal relationship is unclear. To elucidate causality, we conduct a prospective observational study using magnetic resonance imaging (MRI)-measured IPFD data and also perform a Mendelian randomization study using genetic instruments for IPFD. In the observational study, we use UK Biobank data (N = 29,463, median follow-up: 4.5 years) and find that high IPFD (>10%) is associated with PDAC risk (adjusted hazard ratio [HR]: 3.35, 95% confidence interval [95% CI]: 1.60–7.00). In the Mendelian randomization study, we leverage eight out of nine IPFD-associated genetic variants (p < 5 × 10−8) from a genome-wide association study in the UK Biobank (N = 25,617) and find that genetically determined IPFD is associated with PDAC (odds ratio [OR] per 1-standard deviation [SD] increase in IPFD: 2.46, 95% CI: 1.38–4.40) in the Pancreatic Cancer Cohort Consortium I, II, III (PanScan I-III)/Pancreatic Cancer Case-Control Consortium (PanC4) dataset (8,275 PDAC cases and 6,723 non-cases). This study provides evidence for a potential causal role of IPFD in the pathogenesis of PDAC. Thus, reducing IPFD may lower PDAC risk.
Keywords: pancreas fat, pancreatic fat, fatty pancreas, pancreatic steatosis, pancreatic adenocarcinoma, pancreas cancer
Graphical abstract

Highlights
-
•
Cohort study of 29,463 individuals assessed for intra-pancreatic fat deposition (IPFD)
-
•
Individuals with >10% IPFD face a three times higher risk of pancreatic cancer
-
•
Mendelian randomization (MR) analysis used 8 genetic variants linked to IPFD
-
•
MR analysis provides evidence of causal link between IPFD and pancreatic cancer
Yamazaki et al. investigate the causal relationship between intra-pancreatic fat deposition (IPFD) and pancreatic cancer. Their observational cohort study (N = 29,463) shows that individuals with >10% IPFD have a 3-fold increased risk of pancreatic cancer. Furthermore, their Mendelian randomization study provides evidence suggesting a causal link between IPFD and pancreatic cancer.
Introduction
Pancreatic ductal adenocarcinoma (PDAC), responsible for 90% of all pancreatic cancers, is highly lethal, with only a 10% 5-year survival rate.1 It is expected to become the second leading cause of cancer-related mortality by 2030.1 Thus, understanding risk factors and etiologic mechanisms is imperative for identifying susceptible populations and reducing the burden of disease. One important modifiable risk factor for many cancers is body fat,2 which can be stored in the classical subcutaneous and visceral depots, as well as accumulate within various organs.3 For instance, excessive fat storage in the liver is a well-established risk factor for liver cancer, presumably through pro-inflammatory and pro-fibrotic mechanisms.4 While fat deposition in the pancreas, also known as intra-pancreatic fat deposition (IPFD), is a long-known phenomenon,5 it has received less attention due to past challenges in accurately quantifying IPFD in this small and irregularly shaped organ.6 Histologically, fat in the pancreas is present in adipocytes that reside between pancreas cells.6 Adipocytes in the pancreas secrete a variety of proteins, including chemokines and cytokines, thereby promoting tissue inflammation.7
Recent developments in imaging methods such as magnetic resonance imaging (MRI) have not only revealed that IPFD is common8,9 but have also enabled a better understanding of the clinical significance of IPFD.10,11,12 Several imaging studies have shown that individuals with higher levels of IPFD have an increased diabetes risk, presumably due to signals from local adipocytes impairing insulin secretion.13,14 Most importantly, higher levels of IPFD are also hypothesized to cause PDAC.6,15
In the current epidemiologic literature, it has been suggested that IPFD is associated with precancerous lesions and PDAC.16,17 However, these results are based on several cross-sectional studies and a retrospective case-control study with a short observational period (i.e., 1–36 months). Because of the long latency of tumorigenesis, the results from these studies could be potentially biased due to reverse causation (i.e., IPFD could be a consequence, rather than a risk factor, of PDAC). Thus, a prospective assessment of IPFD and PDAC in a cohort with substantial follow-up time is needed to overcome this limitation and provide stronger evidence of a causal association.
Causality can further be evaluated using Mendelian randomization, a method that uses genetic variants (e.g., single-nucleotide polymorphisms [SNPs]) to assess the causal effect of an exposure (e.g., IPFD) on a disease (e.g., PDAC).18 Mendelian randomization studies are considered natural randomized trials, in which the random inheritance of genetic variants works as random treatment assignments. Because genetic variants are assigned randomly at conception and are not affected by acquired diseases or environmental factors, Mendelian randomization is less prone to reverse causation and confounding.19,20
We conducted a prospective observational study of MRI-measured IPFD and PDAC incidence and performed a Mendelian randomization study to evaluate the causal association of genetically determined IPFD with PDAC. We thereby aimed to clarify if IPFD is indeed a causal contributor to PDAC, which could ultimately lead to improvement in prevention, early detection, or treatment of this highly fatal cancer.
Results
Observational study for the association of IPFD with PDAC
We conducted an observational prospective cohort study to investigate the association between IPFD and PDAC in 29,463 UK Biobank participants who underwent pancreas MRI.21 Characteristics of the study participants are shown in Table 1. The high-IPFD group (i.e., participants whose IPFD values were more than the mean IPFD value of 10%) tended to be older, predominantly male, and had a higher body mass index (BMI) compared to the low-IPFD group (IPFD ≤ 10%). Correlation between IPFD and BMI was moderate (Pearson correlation coefficient = 0.40). During the median follow-up period of 4.5 years (interquartile range: 3.8–5.4), the cumulative incidence of PDAC was 0.28% (32 cases out of 11,485 individuals) in the high-IPFD group and 0.07% (12 cases out of 17,978 individuals) in the low-IPFD group. High IPFD was associated with a 3-fold increased risk of PDAC (multivariable adjusted hazard ratio: 3.35, 95% confidence interval [95% CI]: 1.60–7.00, p = 0.001). Similar results were observed when IPFD was analyzed as tertiles, while the analysis using continuous IPFD showed a comparable trend (Table 2).
Table 1.
Characteristics of the 29,463 participants in the prospective observational study of IPFD and PDAC in the UK Biobank, stratified by high vs. low IPFD
| High-IPFD group (N = 11,485) | Low-IPFD group (N = 17,978) | |
|---|---|---|
| Age (years) | 67 (61–71) | 63 (57–69) |
| Male, N (%) | 7,390 (64.3) | 6,882 (38.3) |
| White ethnicity, N (%) | 11,221 (98.0) | 17,316 (96.6) |
| BMI (kg/m2) | 27.9 (25.5–30.8) | 24.7 (22.6–27.2) |
| Intra-pancreatic fat deposition (%) | 15.8 (12.4–22.3) | 5.5 (3.8–7.4) |
| Hepatic fat deposition (%) | 4.3 (2.8–7.9) | 2.5 (1.9–3.9) |
| Current smoker, N (%) | 420 (3.7) | 536 (3.0) |
| Daily drinker, N (%) | 2,027 (17.8) | 2,787 (15.6) |
| Follow-up period (years) | 4.5 (3.8–5.4) | 4.5 (3.8–5.4) |
High and low IPFD were defined based on the mean IPFD among all participants (10%). Continuous data were expressed as median (interquartile range). Missing data: ethnicity (N = 82), BMI (N = 862), hepatic fat deposition (N = 2,255), current smoker (N = 273), and daily drinker (N = 185). IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; BMI, body mass index.
Table 2.
Results from the prospective observational study of the association between IPFD and incidence of PDAC in the UK Biobank
| Cumulative incidence,a % |
Crude |
Age and BMI adjusted |
Multivariable adjustedb |
||||
|---|---|---|---|---|---|---|---|
| (N cases/N total) | HR (95% CI)c | p value | HR (95% CI)c | p value | HR (95% CI)c | p value | |
| Categorization (mean value)d | |||||||
| Low IPFD (≤10%) | 0.07 (12/17,978) | 1.00 (reference) | – | 1.00 (reference) | – | 1.00 (reference) | – |
| High IPFD (>10%) | 0.28 (32/11,485) | 4.20 (2.16–8.15) | <0.001 | 3.53 (1.72–7.26) | 0.001 | 3.35 (1.60–7.00) | 0.001 |
| Categorization (tertiles) | |||||||
| Low IPFD (≤5.8%) | 0.06 (6/9,821) | 1.00 (reference) | – | 1.00 (reference) | – | 1.00 (reference) | – |
| Moderate IPFD (5.8–11%) | 0.09 (9/9,821) | 1.49 (0.53–4.19) | 0.45 | 1.34 (0.47–3.87) | 0.58 | 1.28 (0.44–3.73) | 0.65 |
| High IPFD (>11%) | 0.30 (29/9,821) | 4.83 (2.01–11.6) | <0.001 | 3.85 (1.47–10.1) | 0.006 | 3.57 (1.32–9.62) | 0.012 |
| Continuous IPFDe | |||||||
| IPFD (per 1-SD increase) | 0.15 (44/29,463) | 1.69 (1.25–2.27) | 0.001 | 1.48 (1.05–2.08) | 0.025 | 1.42 (0.99–2.02) | 0.056 |
IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; HR, hazard ratio; CI, confidence interval; BMI, body mass index; SD, standard deviation.
Median follow-up period was 4.5 years (interquartile range: 3.8–5.4).
The multivariable analysis was adjusted for age, gender, BMI, current smoking status, and daily drinking.
HR and 95% CI estimated using Cox regression models.
High and low IPFD were defined based on the mean IPFD among all participants (10%).
Log transformation was applied to continuous IPFD to correct for skewness. Models evaluated 1-SD increase in the log-transformed value.
Mendelian randomization study
We conducted a two-sample Mendelian randomization study using data from two large-scale genome-wide association studies (GWASs) for MRI-measured IPFD and PDAC in individuals of European ancestry.21,22,23 The details of assumptions required in Mendelian randomization are shown in Figure S1. Data sources and selection of genetic instruments are shown in Figure 1. Table 3 shows descriptive information of the GWAS datasets used in this Mendelian randomization study. We leveraged eight out of nine genetic variants associated with IPFD (p < 5 × 10−8) from a GWAS in the UK Biobank (N = 25,617 individuals)21 and assessed their association with PDAC in the Pancreatic Cancer Cohort Consortium I, II, III (PanScan I-III) and Pancreatic Cancer Case-Control Consortium (PanC4) dataset (8,275 PDAC cases and 6,723 non-cases).22,23
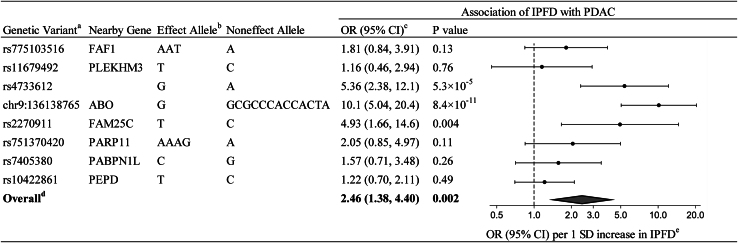
Figure 1.
Data sources and selection of genetic instruments for the Mendelian randomization analysis
aPalindromic SNPs are those where the alleles are complementary (G/C or A/T).
bProxy genetic variants were used when selected genetic variants did not exist in PanScan I–III or PanC4.
cAssociation of each genetic variant with BMI was evaluated using summary statistics obtained from meta-analysis results of the UK Biobank and GIANT consortium. Although none of the eight genetic variants were associated with BMI at the genome-wide significance, we further conducted a sensitivity analysis excluding the three genetic variants with a nominal BMI association.
dThe estimates for the association of each genetic variant with PDAC were combined using the inverse-variance weighted method, with summary statistics for PDAC obtained from PanScan I–III and PanC4. IPFD, intra-pancreatic fat deposition; SNP, single-nucleotide polymorphism; MAF, minor allele frequency; GIANT, genetic investigation of anthropometric traits; BMI, body mass index; PanScan, Pancreatic Cancer Cohort Consortium; PanC4, Pancreatic Cancer Case-Control Consortium; PDAC, pancreatic ductal adenocarcinoma.
Table 3.
Descriptive information of the GWAS used in the Mendelian randomization study
| UK Biobank21 | PanScan I–III + PanC423 | |
|---|---|---|
| Aim of GWAS | IPFD | PDAC |
| Participants, N | 25,617 | 14,998 |
| (8,275 cases and 6,723 non-cases) | ||
| Age (years) | ||
| Mean (SD) | 64.2 (7.5) | N/A |
| <50; 50–60; 60–70; 70–80; >80, N | N/A | 1,159; 3,088; 5,275; 4,354; 1,122 |
| Female (%) | 51.2 | 45.8 |
| European ancestry (%) | 100 | 100 |
| BMI (kg/m2), mean (SD) | 26.5 (4.3) | N/A |
| IPFD (%), mean (SD) | 10.4 (7.9) | N/A |
IPFD (i.e., fat fraction percentage within the pancreas) was measured on MRI. IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; PanScan, Pancreatic Cancer Cohort Consortium; PanC4, Pancreatic Cancer Case-Control Consortium; GWAS, genome-wide association study; SD, standard deviation; BMI, body mass index; N/A, not applicable; MRI, magnetic resonance imaging.
Primary Mendelian randomization analysis for the association of IPFD with PDAC
Characteristics of the eight IPFD-associated genetic variants are shown in Table 4. All eight genetic variants were strongly associated with IPFD: mean F-statistics 54 (min 33, max 103). The odds ratios (ORs) of PDAC for each of the eight genetic variants were greater than the reference value of one.
Table 4.
Characteristics of the IPFD-associated genetic variants
| Genetic variant | Nearby gene | Chromosome | Position | Effect allelea | Proxy SNP | Association with IPFD |
Association with PDAC |
||
|---|---|---|---|---|---|---|---|---|---|
| Beta (%)b | p valueb | OR (95% CI)c | p valuec | ||||||
| rs775103516 | FAF1 | 1 | 51397564 | AAT | rs113170275 | 0.52 | 3.4 × 10−13 | 1.04 (0.99, 1.09) | 0.13 |
| rs11679492 | PLEKHM3 | 2 | 208834477 | T | – | 0.39 | 1.3 × 10−8 | 1.01 (0.96, 1.05) | 0.77 |
| rs4733612 | – | 8 | 129569999 | G | – | 0.52 | 8.8 × 10−12 | 1.12 (1.06, 1.18) | 6.1 × 10−5 |
| chr9: 136138765 | ABO | 9 | 136138765 | G | rs495828 | 0.64 | 2.7 × 10−13 | 1.21 (1.14, 1.27) | 8.4 × 10−11 |
| rs2270911 | FAM25C | 10 | 49313245 | T | – | 0.49 | 1.8 × 10−8 | 1.10 (1.03, 1.18) | 0.0041 |
| rs751370420 | PARP11 | 12 | 4122179 | AAAG | rs7307879 | 0.46 | 2.4 × 10−11 | 1.04 (0.99, 1.10) | 0.11 |
| rs7405380 | PABPN1L | 16 | 88975910 | C | rs12444726 | 0.5 | 6.1 × 10−13 | 1.03 (0.98, 1.08) | 0.26 |
| rs10422861 | PEPD | 19 | 33894846 | T | – | 0.69 | 2.1 × 10−22 | 1.02 (0.97, 1.07) | 0.5 |
IPFD results were obtained from a genome-wide association study of 25,617 individuals of European ancestry in the UK Biobank.21 PDAC results were obtained from the PanScan I–III and PanC4, comprising a total of 8,275 PDAC cases and 6,723 non-cases of European ancestry.23 IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; SNP, single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Allele associated with increasing IPFD levels.
Effect size estimates and p values for the association between each effect allele and IPFD levels (i.e., fat fraction percentage within the pancreas).
Effect size estimates and p values for the association between each effect allele and PDAC.
In the primary Mendelian randomization analysis using the inverse-variance weighted (IVW) method, genetically determined IPFD levels were associated with PDAC risk (Figure 2). The OR of PDAC per 1-standard deviation (SD) increase in genetically determined IPFD level (i.e., per 7.9% increase in fat fraction percentage within the pancreas) was 2.46 (95% CI: 1.38, 4.40; p = 0.002), an average 146% increased risk of PDAC per 1-SD (7.9%) increase in IPFD.
Figure 2.
Primary Mendelian randomization estimates of the association between IPFD and PDAC
aProxy SNPs were used for rs775103516 (rs113170275), chr9:136138765 (rs495828), rs751370420 (rs7307879), and rs7405380 (rs12444726).
bAllele associated with increasing IPFD levels.
cOR (95% CI) of PDAC per-1 SD increase in genetically determined IPFD levels (i.e., per 7.9% increase in fat fraction percentage within the pancreas).
dRandom-effects inverse-variance weighted method was used to obtain the overall estimate for the association of genetically determined IPFD with PDAC.
eData markers indicate the OR for the association of genetically determined IPFD with PDAC, which was estimated using each genetic variant. Error bars indicate 95% CIs. IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; OR, odds ratio; CI, confidence interval; SD, standard deviation.
Sensitivity Mendelian randomization analyses for the association of IPFD with PDAC
To account for potential pleiotropy (i.e., a violation of assumptions for Mendelian randomization), we conducted several sensitivity analyses: the IVW method with leave-one-out analysis and pleiotropy-robust statistical methods. These sensitivity analyses also showed consistent associations between genetically determined IPFD levels and PDAC (Figure S2; Table 5). All of the leave-one-out ORs and 95% CIs indicated a statistically significant association with PDAC. The weighted median method and the Mendelian randomization-pleiotropy residual sum and outlier (MR-PRESSO) method showed similar associations (ORs [95% CI]: 1.79 [1.13, 2.83] and 2.29 [1.61, 3.26], respectively), while the MR-Egger method showed a wide CI (OR 4.56 [95% CI: 0.14, 144.9]). Using MR-PRESSO, we found evidence of outliers (pglobal test < 0.001), but the Mendelian randomization estimates for PDAC did not alter the inference of the results after removal of the outliers (pdistortion = 0.57). Although Cochrane’s Q value was high (31.6), there was no evidence of pleiotropy in the MR-Egger method (MR-Egger intercept: −0.042; p = 0.72).
Table 5.
Comprehensive Mendelian randomization estimates of the association between IPFD and PDAC
| No. of genetic variants | Analysis | Association of IPFD with PDAC |
|||
|---|---|---|---|---|---|
| OR (95% CI)a | p value | p for pleiotropyb | p for distortionc | ||
| 8 | IVW | 2.46 (1.38, 4.40) | 0.002 | – | – |
| 8 | weighted median | 1.79 (1.13, 2.83) | 0.013 | – | – |
| 8 | MR-Egger | 4.56 (0.14, 144.9) | 0.39 | 0.72 | – |
| 6 (exclusion of outlier variantsd) | MR-PRESSO | 2.29 (1.61, 3.26) | <0.001 | – | 0.57 |
| 5 (exclusion of variants nominally linked to BMIe) | IVW | 3.79 (1.66, 8.65) | 0.002 | – | – |
IPFD, intra-pancreatic fat deposition; PDAC, pancreatic ductal adenocarcinoma; OR, odds ratio; CI, confidence interval; BMI, body mass index; IVW, inverse-variance weighted method: SD, standard deviation. See also Figure S2.
OR (95% CI) of PDAC per 1-SD increase in genetically determined IPFD levels (i.e., per 7.9% increase in fat fraction percentage within the pancreas).
p for pleiotropy was obtained from p value of MR-Egger intercept. Less than 0.05 indicates a possible pleiotropic effect.
p for distortion was obtained from MR-PRESSO. Less than 0.05 indicates a difference between estimates before and after exclusion of outlier genetic variants.
Two outlier genetic variants (chr9: 136138765 and rs10422861) were detected in MR-PRESSO.
Although none of the eight genetic variants were associated with BMI at genome-wide significance, we further conducted a sensitivity analysis excluding three genetic variants with a nominal BMI association.
Although none of the eight genetic variants were associated with BMI at genome-wide significance in a past GWAS in the UK Biobank and the Genetic Investigation of Anthropometric Traits (GIANT) consortium (N = 806,834 individuals),24 we further conducted a sensitivity analysis excluding genetic variants with a nominal BMI association. After removal of the three genetic variants nominally linked to BMI, the association between genetically determined IPFD levels and PDAC remained significant (OR [95% CI]: 3.79 [1.66, 8.65], p = 0.002) (Table 5).
Additional Mendelian randomization analyses for the association of IPFD with PDAC
We conducted an additional Mendelian randomization analysis using only the significant genetic variants for PDAC risk (rs4733612, chromosome 9 [chr9]: 136138765, rs2270911 as listed in Table 4). The results showed that genetically determined higher levels of IPFD were significantly associated with an increased risk of PDAC (OR [95% CI] per 1-SD increase in IPFD: 7.09 [4.41–11.4], p < 0.001). Furthermore, we performed another Mendelian randomization analysis using only the non-significant genetic variants for PDAC risk (rs775103516, rs11679492, rs751370420, rs7405380, and rs10422861, also in Table 4). The result still indicated a significant association; higher genetically determined levels of IPFD corresponded with an elevated risk of PDAC (OR [95% CI] per 1-SD increase in IPFD: 1.47 [1.05–2.05], p = 0.024).
Discussion
Triangulating evidence from our prospective observational study and Mendelian randomization study suggests that IPFD plays a causal role in increasing the risk of PDAC. Firstly, our observational study (with median follow-up of 4.5 years) found that participants with IPFD >10% had a 3-fold increased risk of PDAC. Secondly, our Mendelian randomization study provided evidence supporting a causal association between IPFD and PDAC. Consequently, IPFD represents a pathogenic contributor of PDAC. This is likely independent of general adiposity, as suggested by the BMI-adjusted result from our observational study and the sensitivity analysis result from our Mendelian randomization study that excluded genetic variants nominally linked to BMI.
While our study provides genetic evidence for causality using Mendelian randomization, it is well in line with the present prospective observational study and earlier epidemiologic work on the topic.17,25,26,27 These studies reported that IPFD is more frequently found in patients with precancerous lesions (i.e., intra-epithelial neoplasia) or PDAC and that it can even be a predictor for PDAC.17,25,26,27 Similar to our current findings, these prior studies also reported that the relationship between IPFD and PDAC is independent of overall body fat (i.e., BMI).17,25,27 The correlation between IPFD and BMI was only 0.40 in the present cohort study, which indicates that there are individual differences in fat deposition patterns (e.g., high IPFD without obesity and high IPFD without fatty liver), a finding also highlighted in our earlier research.3,14 Furthermore, no colocalization was observed between IPFD traits and hepatic fat deposition traits in a previous GWAS, suggesting that the mechanisms of tumorigenesis related to fat deposition vary between these two organs.21 Thus, our data, along with previous work, support the idea that IPFD may have features that differ from fat deposition in other locations (e.g., the liver).7,21,28
One possible mechanism linking IPFD and PDAC is through the enhanced production of cytokines and adipokines from adipocytes residing within the pancreas.7 By stimulating inflammation, suppressing apoptosis, and promoting cell proliferation and migration, these cytokines and adipokines can contribute to cancer development or progression.9,29 In fact, findings from an observational study showed that ultrasound-measured IPFD is a risk factor for future subclinical chronic pancreatitis,30 supporting the hypothesis that IPFD can contribute to chronic low-grade inflammation of the pancreas, a well-established driver of PDAC.9,15 Another important consideration is the heterogeneous cellular localization of IPFD.15,31 Fat in the pancreas is present in adipocytes that reside between pancreas cells; however, fat also accumulates within pancreas cells such as acinar cells, endocrine cells, and stellate cells.6,15 While imaging modalities such as MRI, computed tomography (CT), and ultrasonography cannot distinguish the cellular location of fat accumulation, it is possible that different types of fat in the pancreas may affect the risk of PDAC differently.15 Our Mendelian randomization results, which show heterogeneity in the effect estimates of each IPFD-related genetic variant on PDAC risk, may reflect this heterogeneous localization of IPFD. Further mechanistic research is warranted to elucidate the effects of IPFD across different cellular compartments on PDAC.
Our current findings can have major clinical implications for reducing PDAC risk, as IPFD is a reversible condition.32 Several randomized controlled trials have demonstrated that IPFD can be reduced through dietary interventions.15,33,34 However, it is possible that a targeted approach to reducing IPFD through a healthy diet may require greater specificity than a general weight reduction diet.15,33,34 For example, a reduction in IPFD was observed following a diet change to a Mediterranean diet with low carbohydrates or an isocaloric multifactorial diet, but not a diet change to a low-fat diet or a diet rich in monounsaturated fatty acids.33,34 Promisingly, emerging pharmacological therapies for weight loss have been shown to dramatically reduce fat mass35,36; however, their potential to reduce IPFD has yet to be proven. Further studies are needed to clarify if and to what extent a reduction of IPFD ultimately translates into decreased PDAC incidence.
One major strength of this study is the use of both a prospective observation investigation and Mendelian randomization. Mendelian randomization, in particular, is a robust approach less susceptible to reverse causation and confounding compared to conventional observational studies. Our observational study and Mendelian randomization study yielded similar findings, supporting the evidence of a causal link between IPFD and PDAC. Secondly, we utilized data from the largest GWAS on IPFD and PDAC to date, providing robust genetic associations for our Mendelian randomization analyses. Third, we also incorporated a third data source (i.e., GIANT) to address potential pleiotropic associations with BMI, a potential confounder, which could have biased our Mendelian randomization findings. Lastly, we used IPFD data measured with MRI, which has been well validated against histologic IPFD measurements and the most sensitive non-invasive modality for detection of IPFD.12
In conclusion, IPFD is a risk factor that could causally contribute to PDAC development, likely independent of overall body fat. Mechanisms linking IPFD to PDAC may include inflammatory and cancer-promoting signals from the local adipocytes. IPFD may serve as a non-invasive biomarker of PDAC risk and may be of even greater utility for individuals who already have elevated risk due to other reasons, such as chronic pancreatitis, adult-onset diabetes, inheritance of predisposing mutations, or family history.1 More importantly, our work further raises the possibility that reduction of IPFD could lower the incidence of PDAC.
Limitations of the study
There are some limitations to our study. The unknown exact mechanism linking the genetic variants, IPFD, and PDAC could theoretically involve pleiotropic effects that may violate assumptions of Mendelian randomization. To minimize the influence of potential pleiotropic effects, we confirmed the robustness of our results through several sensitivity analyses including leave-one-out analyses, pleiotropy-robust statistical methods, and the analysis excluding genetic variants nominally linked to BMI. Another limitation is the restriction to individuals of European ancestry for our Mendelian randomization study. Future studies should expand this analysis to non-European populations, as well as incorporate genetic variants for IPFD identified in other racial/ethnic groups.37 Lastly, our Mendelian randomization study was unable to evaluate potential non-linear relationships between IPFD and PDAC or the combined effect of IPFD with other risk factors on PDAC. Further Mendelian randomization studies using large-scale individual-level data are warranted to address these issues.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Observational data | UK Biobank | https://www.ukbiobank.ac.uk/ |
| IPFD GWAS summary statics | UK Biobank Liu et al.21 |
https://cdn.elifesciences.org/articles/65554/elife-65554-supp1-v1.xlsx |
| PDAC GWAS summary statics | Pancreatic Cancer Cohort Consortium I-III and the Pancreatic Cancer Case-Control Consortium Liu et al.23 |
https://www.ncbi.nlm.nih.gov/gap/ dbGaP accession phs000206.v5.p3 and phs000648.v1.p1 |
| BMI GWAS summary statics | UK Biobank and Genetic Investigation of Anthropometric Traits consortium Pulit et al.24 |
https://zenodo.org/record/1251813#.Y9n8YnbP1D8 |
| BMI-adjusted waist-to-hip ratio GWAS summary statistics | UK Biobank and Genetic Investigation of Anthropometric Traits consortium Pulit et al.24 |
https://zenodo.org/record/1251813#.Y9n8YnbP1D8 |
| Visceral fat volume GWAS summary statistics | UK Biobank van der Meer et al.38 |
https://www.ebi.ac.uk/gwas/studies/GCST90267357 |
| Software and algorithms | ||
| STATA version 18 | StataCorp, College Station, TX | https://www.stata.com/ |
| R version 4.0.5 | R Foundation for Statistical Computing, Vienna, Austria | https://www.r-project.org/ |
| MendelianRandomization package | R project Broadbent et al.39 |
https://cran.r-project.org/web/packages/MendelianRandomization/index.html |
| MR-PRESSO package | Github Verbanck et al.40 |
https://github.com/rondolab/MR-PRESSO |
| PhenoScanner V2 | Kamat et al.41 | http://www.phenoscanner.medschl.cam.ac.uk/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Hajime Yamazaki (yamazaki.hajime.7n@kyoto-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study uses data from the UK Biobank and dbGaP (accession phs000206.v5.p3 and phs000648.v1.p1). Data can be accessed following approval of a research access application to the UK Biobank and/or dbGaP. This paper also uses publicly available summary statistics from prior GWAS in the UK Biobank and the Genetic Investigation of Anthropometric Traits consortium. The source and identifier of these datasets can be found in the Deposited Data section of the key resources table. This paper does not report the original code. Any additional information required to reanalyze the data reported in this report is available from the lead contact upon request.
Experimental model and study participant details
Source of data
For the prospective observational study using individual-level UK Biobank data, we included 29,463 participants without prevalent PDAC who underwent IPFD measurement using MRI between December 2015 and March 2020. Participants were followed from the date of baseline IPFD measurement to the date of PDAC diagnosis, date of death, or November 2022, when the most recent comprehensive death data was available. Incident cases of PDAC were identified using International Classification of Diseases, 10th Revision (ICD-10) codes C25.0-C25.9 from the national cancer registry, hospital admissions, and cause of death records. We excluded potential neuroendocrine tumors using International Classification of Disease for Oncology, 3rd Edition (ICD-O-3) histology codes (8150; islet cell carcinoma, 8240: carcinoid tumor, 8246: neuroendocrine carcinoma).
For the Mendelian randomization analyses, we obtained IPFD-related genetic variants from a prior GWAS study in the UK Biobank (N = 25,617 individuals).21,42 In the GWAS study, IPFD levels were measured on MRI and shown as fat fraction percentage within the pancreas.21 IPFD fraction measured on MRI represents histological IPFD fraction, defined as the percentage of pancreatic intraparenchymal fat in the total pancreatic parenchyma.12 For PDAC genetic associations, we used information from the PanScan I, II, III and the PanC4 GWAS dataset (N = 14,998 individuals).23
As sensitivity Mendelian randomization analyses to address potential pleiotropic associations with obesity-related measures, we used summary statistics from previous GWAS of BMI, BMI-adjusted waist-to-hip ratio, and visceral fat volume. For BMI and BMI-adjusted waist-to-hip ratio, we used data from a meta-analysis of the UK Biobank and the GIANT consortium (N = 806,834 individuals).24 For visceral fat volume, we used data from a prior GWAS in the UK Biobank (N = 33542).38
The UK Biobank has approval from the North West Multi-centre Research Ethics Committee, and our UK Biobank data usage for this study was also approved by the ethics committee of Kyoto University (R3854). We used publicly available data for GIANT consortium, for which no ethical approval was required. PanScan/PanC4 data usage for this study has been approved by the University of Hawaii Institutional Review Board (2019-00402).
Method details
IPFD measurement
IPFD was quantified using the proton density fat fraction of the pancreas with multi-echo MRI, as described in a prior publication.21 A modified convolutional neural network (CNN) was trained using manually measured IPFD and then applied to the data from all participants.21 In a prior validation study, the CNN was trained on 68 subjects and tested on 30 subjects.43 There were only minor differences between the IPFD measurements obtained using the CNN and those obtained through manual measurements in the test subjects.43
Selection of genetic instruments
Genetic variants used in the Mendelian randomization analyses were selected as follows (Figure 1). All nine independent genetic variants associated with IPFD levels at genome-wide significance (p < 5.0 × 10−8) were selected from the UK Biobank GWAS, in which age, age squared, gender, imaging center, scan date, scan time, genotyping batch, and genetic relatedness were controlled for in the analysis.21 We further excluded one genetic variant (rs13040225) with a palindromic SNP (i.e., those where the alleles are complementary, G/C or A/T) and minor allele frequency (MAF) above 0.4 to avoid ambiguity of effect direction. The remaining eight genetic variants were used for the primary Mendelian randomization analysis. Among the eight genetic variants, four were not found in the GWAS summary statistics for PDAC in the PanScan I-III/PanC4 GWAS. We used the same proxy SNPs in linkage disequilibrium (r2 > 0.7) for these genetic variants (Table 4), as done in a previous Mendelian randomization study for IPFD and diabetes mellitus.42 About 1.6% of the variation in IPFD levels was explained by the eight genetic variants.42 This modest proportion of genetically explained phenotype variation is within the range typically observed in Mendelian randomization studies.44,45 The value of Mendelian randomization lies in elucidating causal associations, not in individual risk prediction, and this modest explained phenotype variation is not a limitation in Mendelian randomization studies.46
Considering possible pleiotropic effects of the genetic variants on potential confounders, we also evaluated the association of the eight genetic variants with obesity-related measures (e.g., BMI, BMI-adjusted waist-to-hip ratio, and visceral fat volume) using GWAS summary statistics from the UK Biobank and the GIANT consortium.24,38 None of the eight genetic variants were associated with BMI, BMI-adjusted waist-to-hip ratio, or visceral fat volume at a genome-wide significance threshold (p < 5.0 × 10−8). However, using the Bonferroni corrected threshold of p < 0.00625 ([p < 0.05]/8 genetic variants) as done in a previous Mendelian randomization study by Larsson, Burgess, and Michaelsson (2017),47 three of the genetic variants (rs775103516, rs751370420, and rs10422861) had a nominal association with BMI. Rs10422861 was also nominally associated with BMI-adjusted waist-to-hip ratio. We conducted a sensitivity analysis excluding these three obesity-related genetic variants to minimize potential residual pleiotropy. In addition to obesity, we also checked pleiotropic association of the eight genetic variants with other potential confounders (current smoking, alcohol intake, and high risk diet1) using PhenoScanner,41,48 but none of the eight genetic variants had a nominal association with these potential confounders.
GWAS data for PDAC
GWAS data for PDAC in PanScan I, PanScan II, PanScan III, and PanC4 were downloaded from dbGaP (study accession nos.: phs000206.v5.p3 and phs000648.v1.p1). The detailed information for these data has been described in previous publications.22,49,50,51,52 In brief, genotyping was performed on the Illumina HumanHap550, 610-Quad, OmniExpress, and OmiExpressExome arrays, respectively. Standard QC was conducted according to the guidelines recommended by the consortia.22,23 Study subjects who were related to each other, had missing information on age or gender, had gender discordance, had non-European ancestry based on genetic estimation, or had a low call rate (less than 94% and 98% in PanScan and PanC4, respectively) were excluded. Duplicated SNPs and those with a high missing call rate (of at least 6% and 2% in PanScan and PanC4, respectively), or violations of Hardy-Weinberg equilibrium (of p < 1 × 10−7 and p < 1 × 10−4 in PanScan and PanC4, respectively) were also excluded. In the PanC4 dataset, we also excluded SNPs that had a MAF <0.005, more than one Mendelian error in HapMap control trios, or more than two discordant calls in study duplicates. SNPs with gender differences in allele frequency >0.2 or in heterozygosity >0.3 for autosomes/XY were further excluded. We conducted the genotype imputation with the Haplotype Reference Consortium reference panel (r1.1 2016), using Minimac4 after phasing with Eagle v2.4.53,54 Imputed SNPs with an imputation quality of >0.3 were retained. All of the genetic variants used in the Mendelian randomization analysis had an imputation quality of >0.8 except for rs2270911, which had an imputation quality of 0.6. The associations between individual SNPs and PDAC risk were further assessed with logistic regression adjusting for age, gender, and the top 10 principal components. In the final analyses, we included 8,275 PDAC cases and 6,723 non-cases of European ancestry.23
Quantification and statistical analysis
Statistical analysis
For the prospective observational study evaluating the association between IPFD and PDAC incidence, we used Cox regression models adjusting for age, gender, BMI, current smoking status, and daily drinking. To account for missing covariates data, we also conducted multiple imputation using multivariate imputation by chained equations. IPFD was dichotomized as high (>10%) and low (≤10%) based on the mean value among all participants. IPFD was also assessed as tertiles and as a continuous measure (per 1 SD increase in log-transformed IPFD).
To evaluate the strength of the association between each genetic variant and IPFD (assumption 1 in Figure S1), we calculated F-statistics. F-statistics should be more than 10 to be valid genetic variants for Mendelian randomization.18 Cochran’s Q value was calculated to evaluate the heterogeneity among estimates obtained using different genetic variants.
For the primary Mendelian randomization analysis, the association of each genetic variant with PDAC was weighted by its association with IPFD, and estimates were combined using the random-effects IVW method. This method is the most efficient and provides valid causal estimates when the average pleiotropic effect is zero. If the genetic variants used in this study are additionally associated with another risk factor for PDAC (i.e., presence of pleiotropic effect), then either assumption 2 or 3 for Mendelian randomization in Figure S1 is violated.
To account for potential pleiotropy, we conducted five sensitivity Mendelian randomization analyses: IVW method with leave-one-out analysis, MR-Egger regression method, weighted median method, MR-PRESSO, and IVW method after exclusion of the three genetic variants nominally linked to BMI. For leave-one-out analysis, each genetic variant was excluded one at a time, and the IVW method was used on the remaining genetic variants to evaluate the causal association of genetically determined IPFD with PDAC. Under the Instrument Strength Independent of Direct Effect (InSIDE) assumption,55 MR-Egger can provide valid causal estimates even if the average pleiotropic effect is not zero, and the intercept in MR-Egger can be tested to judge whether pleiotropic effects exist. However, the drawback of MR-Egger is the wide confidence intervals. Weighted median method can provide valid causal estimates even if up to 50% of genetic variants have pleiotropic effects. MR-PRESSO can detect outlier genetic variants and calculate IVW estimates after exclusion of the outliers.19 Lastly, to address potential pleiotropy with BMI, a potential confounder, we excluded the three genetic variants nominally linked to BMI and conducted IVW analysis.
As additional analyses, we conducted Mendelian randomization analyses using only the significant genetic variants associated with PDAC risk (rs4733612, chr9:136138765, rs2270911 as listed in Table 4), and separately using only the non-significant genetic variants for PDAC risk (rs775103516, rs11679492, rs751370420, rs7405380, rs10422861 also in Table 4).
We considered p < 0.05 as statistically significant for our Mendelian randomization analyses. Statistical analyses were performed using Stata 18 (StataCorp, College Station, TX) and R version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria). We used the MendelianRandomization package39,56 and MR-PRESSO package40 in R to conduct the Mendelian randomization analysis.
Acknowledgments
This work used data provided by patients and collected by the NHS as part of their care and support (Copyright 2024, NHS England. Re-used with the permission of the NHS England and UK Biobank. All rights reserved). This research also used data assets made available by National Safe Haven as part of the Data and Connectivity National Core Study, led by Health Data Research UK in partnership with the Office for National Statistics and funded by UK Research and Innovation (research which commenced between 1st October 2020–31st March 2021 grant ref MC_PC_20029; 1st April 2021–30th September 2022 grant ref MC_PC_20058). This research has been conducted using the UK Biobank Resource under Application Number 93426. We thank the investigators of the UK Biobank and the GIANT consortium for making the GWAS summary statistics publicly available. The authors also would like to thank all the participants in the parent studies and all the researchers, clinicians, technicians, and administrative staff for their contribution to the studies. The PanScan study was funded in whole or in part with federal funds from the National Cancer Institute (NCI), US National Institutes of Health (NIH) under contract number HHSN261200800001E. Additional support was received from NIH/NCI K07 CA140790, the American Society of Clinical Oncology Conquer Cancer Foundation, the Howard Hughes Medical Institute, the Lustgarten Foundation, the Robert T. and Judith B. Hale Fund for Pancreatic Cancer Research, and Promises for Purple. A full list of acknowledgments for each participating study is provided in the supplementary note of the manuscript with PubMed ID 25086665. For the PanC4 GWAS study, the cases and controls were derived from the following PanC4 studies: Johns Hopkins National Familial Pancreas Tumor Registry, Mayo Clinic Biospecimen Resource for Pancreas Research, Ontario Pancreas Cancer Study (OPCS), Yale University, MD Anderson Case Control Study, Queensland Pancreatic Cancer Study, University of California San Francisco Molecular Epidemiology of Pancreatic Cancer Study, International Agency of Cancer Research, and Memorial Sloan Kettering Cancer Center. This work is supported by NCI R01CA154823. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the NIH to Johns Hopkins University, contract number HHSN2682011000111. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors thank Dr. Hua Zhong at University of Hawaii Cancer Center for her help for this study. We also acknowledge the following grant support: NIH/NCI R00CA218892(L.W.), T32CA229110 (S.A.S.), U01CA164973 (L.L.M.), and K99CA256525 and R00CA256525 (B.Z.H.), Japan Society for the Promotion of Science KAKENHI grant JP22K15685, the University of Hawaii Cancer Center, and the V Foundation V Scholar Award (V2021-023).
Author contributions
H.Y., S.A.S., and B.Z.H. designed the study. H.Y., S.A.S., L.W., and B.Z.H. collected the data. H.Y. wrote the draft. H.Y., S.A.S., L.W., and B.Z.H. analyzed the data for Mendelian randomization analyses. H.Y. analyzed individual data on UK Biobank. H.Y., S.A.S., L.W., S.F., R.W., M.H., S.R.G., H.-J.L., V.W.S., L.L.M., and B.Z.H. reviewed, made critical revisions, and approved the article before submission. H.Y. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Declaration of interests
L.W. provided consulting service to Pupil Bio, Inc., and received honorarium.
Published: January 26, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101391.
Supplemental information
and S2
References
- 1.Park W., Chawla A., O'Reilly E.M. Pancreatic Cancer: A Review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown K.F., Rumgay H., Dunlop C., Ryan M., Quartly F., Cox A., Deas A., Elliss-Brookes L., Gavin A., Hounsome L., et al. The fraction of cancer attributable to modifiable risk factors in England, Wales, Scotland, Northern Ireland, and the United Kingdom in 2015. Br. J. Cancer. 2018;118:1130–1141. doi: 10.1038/s41416-018-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki H., Tauchi S., Machann J., Haueise T., Yamamoto Y., Dohke M., Hanawa N., Kodama Y., Katanuma A., Stefan N., et al. Fat Distribution Patterns and Future Type 2 Diabetes. Diabetes. 2022;71:1937–1945. doi: 10.2337/db22-0315. [DOI] [PubMed] [Google Scholar]
- 4.Powell E.E., Wong V.W.S., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. doi: 10.1016/S0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 5.Ogilvie R.F. The islands of langerhans in 19 cases of obesity. J. Pathol. Bacteriol. 1933;37:473–481. [Google Scholar]
- 6.Wagner R., Eckstein S.S., Yamazaki H., Gerst F., Machann J., Jaghutriz B.A., Schürmann A., Solimena M., Singer S., Königsrainer A., et al. Metabolic implications of pancreatic fat accumulation. Nat. Rev. Endocrinol. 2022;18:43–54. doi: 10.1038/s41574-021-00573-3. [DOI] [PubMed] [Google Scholar]
- 7.Gerst F., Wagner R., Oquendo M.B., Siegel-Axel D., Fritsche A., Heni M., Staiger H., Häring H.U., Ullrich S. What role do fat cells play in pancreatic tissue? Mol. Metab. 2019;25:1–10. doi: 10.1016/j.molmet.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong V.W.S., Wong G.L.H., Yeung D.K.W., Abrigo J.M., Kong A.P.S., Chan R.S.M., Chim A.M.L., Shen J., Ho C.S., Woo J., et al. Fatty pancreas, insulin resistance, and beta-cell function: a population study using fat-water magnetic resonance imaging. Am. J. Gastroenterol. 2014;109:589–597. doi: 10.1038/ajg.2014.1. [DOI] [PubMed] [Google Scholar]
- 9.Truong E., Pandol S., Jeon C. Uniting epidemiology and experimental models: pancreatic steatosis and pancreatic cancer. EBioMedicine. 2022;79:103996. doi: 10.1016/j.ebiom.2022.103996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Mrabeh A., Hollingsworth K.G., Steven S., Tiniakos D., Taylor R. Quantification of intrapancreatic fat in type 2 diabetes by MRI. PLoS One. 2017;12:e0174660. doi: 10.1371/journal.pone.0174660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S.Y., Kim H., Cho J.Y., Lim S., Cha K., Lee K.H., Kim Y.H., Kim J.H., Yoon Y.S., Han H.S., Kang H.S. Quantitative assessment of pancreatic fat by using unenhanced CT: pathologic correlation and clinical implications. Radiology. 2014;271:104–112. doi: 10.1148/radiol.13122883. [DOI] [PubMed] [Google Scholar]
- 12.Yoon J.H., Lee J.M., Lee K.B., Kim S.W., Kang M.J., Jang J.Y., Kannengiesser S., Han J.K., Choi B.I. Pancreatic Steatosis and Fibrosis: Quantitative Assessment with Preoperative Multiparametric MR Imaging. Radiology. 2016;279:140–150. doi: 10.1148/radiol.2015142254. [DOI] [PubMed] [Google Scholar]
- 13.Wagner R., Jaghutriz B.A., Gerst F., Barroso Oquendo M., Machann J., Schick F., Löffler M.W., Nadalin S., Fend F., Königsrainer A., et al. Pancreatic Steatosis Associates With Impaired Insulin Secretion in Genetically Predisposed Individuals. J. Clin. Endocrinol. Metab. 2020;105:3518–3525. doi: 10.1210/clinem/dgaa435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki H., Tauchi S., Wang J., Dohke M., Hanawa N., Kodama Y., Katanuma A., Saisho Y., Kamitani T., Fukuhara S., Yamamoto Y. Longitudinal association of fatty pancreas with the incidence of type-2 diabetes in lean individuals: a 6-year computed tomography-based cohort study. J. Gastroenterol. 2020;55:712–721. doi: 10.1007/s00535-020-01683-x. [DOI] [PubMed] [Google Scholar]
- 15.Petrov M.S., Taylor R. Intra-pancreatic fat deposition: bringing hidden fat to the fore. Nat. Rev. Gastroenterol. Hepatol. 2022;19:153–168. doi: 10.1038/s41575-021-00551-0. [DOI] [PubMed] [Google Scholar]
- 16.Sreedhar U.L., DeSouza S.V., Park B., Petrov M.S. A Systematic Review of Intra-pancreatic Fat Deposition and Pancreatic Carcinogenesis. J. Gastrointest. Surg. 2020;24:2560–2569. doi: 10.1007/s11605-019-04417-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoogenboom S.A., Bolan C.W., Chuprin A., Raimondo M.T., van Hooft J.E., Wallace M.B., Raimondo M. Pancreatic steatosis on computed tomography is an early imaging feature of pre-diagnostic pancreatic cancer: A preliminary study in overweight patients. Pancreatology. 2021;21:428–433. doi: 10.1016/j.pan.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Davies N.M., Holmes M.V., Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. Bmj. 2018;362:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S., Thompson S.G. 2nd ed. Chapman and Hall/CRC; 2021. Mendelian Randomization: Methods for Causal Inference Using Genetic Variants. [Google Scholar]
- 20.Neeland I.J., Kozlitina J. Mendelian Randomization: Using Natural Genetic Variation to Assess the Causal Role of Modifiable Risk Factors in Observational Studies. Circulation. 2017;135:755–758. doi: 10.1161/CIRCULATIONAHA.117.026857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Basty N., Whitcher B., Bell J.D., Sorokin E.P., van Bruggen N., Thomas E.L., Cule M. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. Elife. 2021;10:e65554. doi: 10.7554/eLife.65554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein A.P., Wolpin B.M., Risch H.A., Stolzenberg-Solomon R.Z., Mocci E., Zhang M., Canzian F., Childs E.J., Hoskins J.W., Jermusyk A., et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 2018;9:556. doi: 10.1038/s41467-018-02942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D., Zhou D., Sun Y., Zhu J., Ghoneim D., Wu C., Yao Q., Gamazon E.R., Cox N.J., Wu L. A Transcriptome-Wide Association Study Identifies Candidate Susceptibility Genes for Pancreatic Cancer Risk. Cancer Res. 2020;80:4346–4354. doi: 10.1158/0008-5472.CAN-20-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., Yengo L., Ferreira T., Marouli E., Ji Y., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori M., Takahashi M., Hiraoka N., Yamaji T., Mutoh M., Ishigamori R., Furuta K., Okusaka T., Shimada K., Kosuge T., et al. Association of pancreatic Fatty infiltration with pancreatic ductal adenocarcinoma. Clin. Transl. Gastroenterol. 2014;5:e53. doi: 10.1038/ctg.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebours V., Gaujoux S., d'Assignies G., Sauvanet A., Ruszniewski P., Lévy P., Paradis V., Bedossa P., Couvelard A. Obesity and fatty pancreatic infiltration are risk factors for pancreatic precancerous lesions (PanIN) Clin. Cancer Res. 2015;21:3522–3528. doi: 10.1158/1078-0432.CCR-14-2385. [DOI] [PubMed] [Google Scholar]
- 27.Desai V., Patel K., Sheth R., Barlass U., Chan Y.M., Sclamberg J., Bishehsari F. Pancreatic Fat Infiltration Is Associated with a Higher Risk of Pancreatic Ductal Adenocarcinoma. Visc. Med. 2020;36:220–226. doi: 10.1159/000507457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eibl G., Rozengurt E. Obesity and Pancreatic Cancer: Insight into Mechanisms. Cancers. 2021;13:5067. doi: 10.3390/cancers13205067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi M., Hori M., Ishigamori R., Mutoh M., Imai T., Nakagama H. Fatty pancreas: A possible risk factor for pancreatic cancer in animals and humans. Cancer Sci. 2018;109:3013–3023. doi: 10.1111/cas.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujii M., Ohno Y., Yamada M., Kamada Y., Miyoshi E. Impact of fatty pancreas and lifestyle on the development of subclinical chronic pancreatitis in healthy people undergoing a medical checkup. Environ. Health Prev. Med. 2019;24:10. doi: 10.1186/s12199-019-0763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrov M.S. Fatty change of the pancreas: the Pandora's box of pancreatology. Lancet. Gastroenterol. Hepatol. 2023;8:671–682. doi: 10.1016/S2468-1253(23)00064-X. [DOI] [PubMed] [Google Scholar]
- 32.Al-Mrabeh A., Hollingsworth K.G., Shaw J.A.M., McConnachie A., Sattar N., Lean M.E.J., Taylor R. 2-year remission of type 2 diabetes and pancreas morphology: a post-hoc analysis of the DiRECT open-label, cluster-randomised trial. Lancet Diabetes Endocrinol. 2020;8:939–948. doi: 10.1016/S2213-8587(20)30303-X. [DOI] [PubMed] [Google Scholar]
- 33.Gepner Y., Shelef I., Schwarzfuchs D., Zelicha H., Tene L., Yaskolka Meir A., Tsaban G., Cohen N., Bril N., Rein M., et al. Effect of Distinct Lifestyle Interventions on Mobilization of Fat Storage Pools: CENTRAL Magnetic Resonance Imaging Randomized Controlled Trial. Circulation. 2018;137:1143–1157. doi: 10.1161/CIRCULATIONAHA.117.030501. [DOI] [PubMed] [Google Scholar]
- 34.Della Pepa G., Brancato V., Costabile G., Salamone D., Corrado A., Vitale M., Cavaliere C., Mancini M., Salvatore M., Luongo D., et al. An Isoenergetic Multifactorial Diet Reduces Pancreatic Fat and Increases Postprandial Insulin Response in Patients With Type 2 Diabetes: A Randomized Controlled Trial. Diabetes Care. 2022;45:1935–1942. doi: 10.2337/dc22-0605. [DOI] [PubMed] [Google Scholar]
- 35.Gastaldelli A., Cusi K., Fernández Landó L., Bray R., Brouwers B., Rodríguez Á. Effect of tirzepatide versus insulin degludec on liver fat content and abdominal adipose tissue in people with type 2 diabetes (SURPASS-3 MRI): a substudy of the randomised, open-label, parallel-group, phase 3 SURPASS-3 trial. Lancet Diabetes Endocrinol. 2022;10:393–406. doi: 10.1016/S2213-8587(22)00070-5. [DOI] [PubMed] [Google Scholar]
- 36.Grunvald E., Shah R., Hernaez R., Chandar A.K., Pickett-Blakely O., Teigen L.M., Harindhanavudhi T., Sultan S., Singh S., Davitkov P., AGA Clinical Guidelines Committee AGA Clinical Practice Guideline on Pharmacological Interventions for Adults With Obesity. Gastroenterology. 2022;163:1198–1225. doi: 10.1053/j.gastro.2022.08.045. [DOI] [PubMed] [Google Scholar]
- 37.Streicher S.A., Lim U., Park S.L., Li Y., Sheng X., Hom V., Xia L., Pooler L., Shepherd J., Loo L.W.M., et al. Genome-wide association study of pancreatic fat: The Multiethnic Cohort Adiposity Phenotype Study. PLoS One. 2021;16:e0249615. doi: 10.1371/journal.pone.0249615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Meer D., Gurholt T.P., Sønderby I.E., Shadrin A.A., Hindley G., Rahman Z., de Lange A.M.G., Frei O., Leinhard O.D., Linge J., et al. The link between liver fat and cardiometabolic diseases is highlighted by genome-wide association study of MRI-derived measures of body composition. Commun. Biol. 2022;5:1271. doi: 10.1038/s42003-022-04237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broadbent J.R., Foley C.N., Grant A.J., Mason A.M., Staley J.R., Burgess S. MendelianRandomization v0.5.0: updates to an R package for performing Mendelian randomization analyses using summarized data. Wellcome Open Res. 2020;5:252. doi: 10.12688/wellcomeopenres.16374.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verbanck M., Chen C.Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J., Butterworth A.S., Staley J.R. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin S., Sorokin E.P., Thomas E.L., Sattar N., Cule M., Bell J.D., Yaghootkar H. Estimating the Effect of Liver and Pancreas Volume and Fat Content on Risk of Diabetes: A Mendelian Randomization Study. Diabetes Care. 2022;45:460–468. doi: 10.2337/dc21-1262. [DOI] [PubMed] [Google Scholar]
- 43.Basty N., Liu Y., Cule M., Thomas E.L., Bell J.D., Whitcher B. IEEE 17th International Symposium on Biomedical Imaging (ISBI) 2020. Automated Measurement of Pancreatic Fat and Iron Concentration Using Multi-Echo and T1-Weighted MRI Data; pp. 345–348. [Google Scholar]
- 44.Markozannes G., Kanellopoulou A., Dimopoulou O., Kosmidis D., Zhang X., Wang L., Theodoratou E., Gill D., Burgess S., Tsilidis K.K. Systematic review of Mendelian randomization studies on risk of cancer. BMC Med. 2022;20:41. doi: 10.1186/s12916-022-02246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garfield V., Salzmann A., Burgess S., Chaturvedi N. A Guide for Selection of Genetic Instruments in Mendelian Randomization Studies of Type 2 Diabetes and HbA1c: Toward an Integrated Approach. Diabetes. 2023;72:175–183. doi: 10.2337/db22-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith G.D., Ebrahim S. In: National Research Council (US) Committee on Advances in Collecting and Utilizing Biological Indicators and Genetic Information in Social Science Surveys. Weinstein M., Vaupel J.W., Wachter K.W., editors. National Academies Press (US); 2008. Mendelian Randomization: Genetic Variants as Instruments for Strengthening Causal Inference in Observational Studies; p. 16.https://www.ncbi.nlm.nih.gov/books/NBK62433/ (Biosocial Surveys). Available from: [Google Scholar]
- 47.Larsson S.C., Burgess S., Michaëlsson K. Association of Genetic Variants Related to Serum Calcium Levels With Coronary Artery Disease and Myocardial Infarction. JAMA. 2017;318:371–380. doi: 10.1001/jama.2017.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staley J.R., Blackshaw J., Kamat M.A., Ellis S., Surendran P., Sun B.B., Paul D.S., Freitag D., Burgess S., Danesh J., et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen G.M., Amundadottir L., Fuchs C.S., Kraft P., Stolzenberg-Solomon R.Z., Jacobs K.B., Arslan A.A., Bueno-de-Mesquita H.B., Gallinger S., Gross M., et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolpin B.M., Rizzato C., Kraft P., Kooperberg C., Petersen G.M., Wang Z., Arslan A.A., Beane-Freeman L., Bracci P.M., Buring J., et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet. 2014;46:994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Childs E.J., Mocci E., Campa D., Bracci P.M., Gallinger S., Goggins M., Li D., Neale R.E., Olson S.H., Scelo G., et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 2015;47:911–916. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amundadottir L., Kraft P., Stolzenberg-Solomon R.Z., Fuchs C.S., Petersen G.M., Arslan A.A., Bueno-de-Mesquita H.B., Gross M., Helzlsouer K., Jacobs E.J., et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and S2
Data Availability Statement
This study uses data from the UK Biobank and dbGaP (accession phs000206.v5.p3 and phs000648.v1.p1). Data can be accessed following approval of a research access application to the UK Biobank and/or dbGaP. This paper also uses publicly available summary statistics from prior GWAS in the UK Biobank and the Genetic Investigation of Anthropometric Traits consortium. The source and identifier of these datasets can be found in the Deposited Data section of the key resources table. This paper does not report the original code. Any additional information required to reanalyze the data reported in this report is available from the lead contact upon request.