Abstract
Control elements of many genes are regulated by multiple activators working in concert to confer the maximal level of expression, but the mechanism of such synergy is not completely understood. The promoter of the human macrophage colony-stimulating factor (M-CSF) receptor presents an excellent model with which we can study synergistic, tissue-specific activation for two reasons. First, myeloid-specific expression of the M-CSF receptor is regulated transcriptionally by three factors which are crucial for normal hematopoiesis: PU.1, AML1, and C/EBPα. Second, these proteins interact in such a way as to demonstrate at least two examples of synergistic activation. We have shown that AML1 and C/EBPα activate the M-CSF receptor promoter in a synergistic manner. As we report here, AML1 also synergizes, and interacts physically, with PU.1. Detailed analysis of the physical and functional interaction of AML1 with PU.1 and C/EBPα has revealed that the proteins contact one another through their DNA-binding domains and that AML1 exhibits cooperative DNA binding with C/EBPα but not with PU.1. This difference in DNA-binding abilities may explain, in part, the differences observed in synergistic activation. Furthermore, the activation domains of all three factors are required for synergistic activation, and the region of AML1 required for synergy with PU.1 is distinct from that required for synergy with C/EBPα. These observations present the possibility that synergistic activation is mediated by secondary proteins contacted through the activation domains of AML1, C/EBPα, and PU.1.
In order to understand the mechanisms that control monocytic commitment and differentiation, we have investigated the tissue-specific regulation of the human macrophage colony-stimulating factor (M-CSF) receptor. We have previously identified three factors required for M-CSF receptor transcription in monocytic cell lines, PU.1, C/EBPα (CCAAT/enhancer-binding protein alpha), and AML1, and demonstrated that mutations in any of the three DNA-binding sites decreases promoter activity significantly in transient transfection studies (77–79). In addition, PU.1 transactivates the M-CSF receptor promoter, and although C/EBPα has little transactivation potential alone, it synergizes with AML1B to increase the activity of the promoter an average of 90-fold (78, 79). All three of these transcription factors play important roles in hematopoiesis.
AML1 (also known as CBFα2 and PEBP2αB) contains a domain that is highly similar to the DNA-binding domain of the Drosophila runt transcription factor, which mediates both DNA-binding and heterodimerization abilities (25). The heterodimerization partner of AML1, CBFβ, does not bind DNA directly but increases the affinity of AML1 for DNA (37, 51, 75). In addition to the M-CSF receptor, the target genes of the AML1-CBFβ heterodimer include granulocyte-macrophage colony-stimulating factor (GM-CSF), T-cell receptor (TCR) subunits, interleukin-3, osteocalcin, neutrophil elastase, and myeloperoxidase (2, 3, 6, 15, 18, 21, 50, 67, 70). In several cases AML1 functions in concert with neighboring factors. For example, AML1 binds cooperatively with another member of the ets family, Ets-1, to the TCRα, TCRβ, and Moloney murine leukemia virus enhancers (18, 67). AML1 exhibits functional synergy with c-Myb in the absence of cooperative binding in the context of the TCRδ and myeloperoxidase enhancers (6, 21). Both AML1 and CBFβ are frequently involved in genetic rearrangements identified in human leukemias (12, 19, 35, 40, 47–49). Furthermore, mice which have homozygous disruptions of either gene, or are heterozygotes containing either the AML/ETO or CBFB/MYH11 fusion genes, have strikingly similar phenotypes. All die in midgestation, exhibit multiple hemorrhages in the central nervous system, and have severely impaired hematopoiesis (7, 53, 61, 73, 74, 76). These data support the theory that AML1 function is critical for normal hematopoietic development.
C/EBPα, a basic region leucine zipper (bZip) transcription factor (31, 32), regulates not only a variety of hepatocyte and adipocyte genes which are important for energy homeostasis but several myeloid-specific genes as well (9, 10, 16, 17, 22, 24, 50). For example, in addition to the M-CSF receptor promoter, C/EBPα has also been shown to regulate the G-CSF receptor and GM-CSF receptor α promoters (23, 66). Mice with a homozygous disruption of the C/EBPα gene die at birth from hypoglycemia (14, 72) and exhibit hematopoietic defects as well. Analysis of the fetal and newborn hematopoietic tissues revealed a profound absence of mature neutrophils. In addition, there were no neutrophils observed after transplantation of the fetal liver into an irradiated recipient, implying that the block in neutrophil development was intrinsic to the cell and not a defect in the environment (80). Therefore, it is clear that C/EBPα plays a critical role in normal granulocyte development.
PU.1, the product of the spi-1 oncogene and a member of the ets family, is upregulated during hematopoietic development and is specifically expressed in myeloid and B cells (8, 29, 55, 59, 71). The pivotal role that PU.1 plays in hematopoietic differentiation is established by the following observations. There are a number of genes that are regulated by PU.1 in both myeloid and B-cell lineages, including those encoding CSF receptors and immunoglobulin subunits (45, 57, 64, 69). Overexpression of PU.1 early in erythroid development blocks erythroblast differentiation (62), and addition of PU.1-binding oligonucleotides to human CD34+ bone marrow cells decreases in vitro colony formation (71). In addition, mice with a disruption in both alleles of the PU.1 locus die in utero (63) or shortly after birth (36) and exhibit major defects in hematopoiesis, including a block in myeloid development. The DNA-binding ets domain shows sequence similarity with other members of the ets family and is contained within amino acids 171 to 267 of the C terminus (29). The activation domain of PU.1 is located within the N terminus and consists of several regions rich in either acidic amino acids or glutamines and a region from amino acids 118 to 160 which has a high number of prolines, glutamic acids, serines, and threonines (PEST domain) (28). PU.1 has been shown to interact with TATA-binding protein (TBP) and the retinoblastoma protein in vitro, requiring amino acids 1 to 75 (20). There are multiple examples where PU.1 functions in concert with other transcription factors, including NF-IL6β (C/EBPδ) (44) and NF-EM5/PIP (11, 56, 58), c-Myb and C/EBPα (50), c-Fos and c-Jun (5, 56), and Ets-1 (13).
We are interested in determining the events that control myeloid differentiation so that we can better understand the aberrant differentiation that is exhibited in the leukemic state. For example, it is not clear how the fusion gene, AML/ETO, and other genomic abnormalities associated with myeloid leukemia contribute to the changes in differentiation and proliferation of the myeloid lineage. Alternative theories include inhibition of normal AML1 function (15, 27, 38) or increased activation by AML1 (60), or even direct activation by AML/ETO itself (39), resulting in the dysregulation of genes such as those encoding GM-CSF, the M-CSF receptor, or Bcl-2. Therefore, we have investigated the mechanism by which the transcription factors regulating the M-CSF receptor promoter interact in an effort to reveal the next layer of complexity in myeloid-specific transcriptional activation. Here we show that, in addition to C/EBPα, AML1B interacts with PU.1 to synergistically activate the M-CSF receptor promoter but requires different regions contained within the C terminus for each function.
MATERIALS AND METHODS
Cell culture conditions and transfection.
HeLa cells (ATCC CCL 2; American Type Culture Collection), CV-1 cells (ATCC CCL 70; American Type Culture Collection), and COS-7 cells (ATCC CRL 165; American Type Culture Collection) were maintained in Dulbecco modified Eagle medium with 10% calf serum and 2 mM l-glutamine (GIBCO), and 3 × 105 to 5 × 105 cells were plated in 100-mm-diameter tissue culture plates 24 h before transfection. Except for immunoprecipitation experiments, transfections were performed by the calcium phosphate method with 5 μg of the reporter construct and 1 μg of each expression construct or empty vector, with the total amount of DNA brought to 20 μg with sheared salmon sperm DNA. The medium was changed 14 h after transfection, and luciferase assays were performed as described previously (54) 24 to 36 h later. Luciferase values were normalized for transfection efficiency by cotransfecting a plasmid expressing the human growth hormone gene driven by the Rous sarcoma virus promoter and assaying the supernatant of the cultures with the human growth hormone radioimmunoassay from Nichols Institute Diagnostics (San Juan Capistrano, Calif.) according to the manufacturer’s instructions.
Immunoprecipitation.
COS-7 cells in 60-mm-diameter plates were transfected with Lipofectamine Plus (GIBCO) according to the manufacturer’s recommendations with 2 μg of AML1 expression plasmid. After 20 h, the cells were incubated for 1 h in Dulbecco modified Eagle medium without methionine or cysteine–10% fetal bovine serum and then labeled in the same medium with the addition of 100 μCi of Express (NEN) per ml for 3 h. The cells were washed three times with cold phosphate-buffered saline, scraped from the plates, and lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 7.5]). The lysate was precleared by incubation with 5 μl of normal rabbit serum for 1 h on ice, followed by 30 min with Sepharose-linked protein A (Pharmacia). The supernatant was then incubated with 1 μl of antiserum specific for the N terminus of AML1 and Sepharose-linked protein A for 16 h at 4°C, with rocking. The immunocomplexes were separated on an SDS–10% acrylamide gel.
Plasmids.
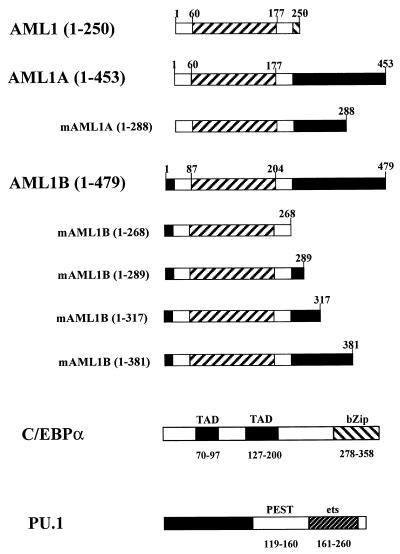
The M-CSF receptor promoter constructs in pXP2 were described previously: the wild-type promoter from bp −416 to +71, pM-CSF-R-luc, and pM-CSF-R(mPU.1)-luc [referred to as pM-CSF-R(m40)-luc] (78), pM-CSF-R(mAML1)-luc [referred to as pM-CSF-R(MB)-luc], and pM-CSF-R(DD)-luc, with a deletion of bp −86 to −37 (79). The expression constructs for murine PU.1 and mutants of PU.1 were gifts from M. J. Klemsz and R. A. Maki (29, 58). C/EBPα expression constructs in the vector pMSV have been analyzed and described in detail elsewhere (17). AML1B and CBFβ were described previously (38). pCMV5-AML1 (Fig. 1) was constructed by subcloning the 576-bp ApaI-BamHI fragment of the AML1 cDNA in pBluescript-KS (42) into pCMV5-AML/ETO (38) digested with the same restriction enzymes, which replaced the 3′ end of AML/ETO with the 3′ end of AML1. The 453-amino-acid form of AML1, here referred to as AML1A (Fig. 1), expression construct, and mutations were gifts from H. Hirai and described by Tanaka et al. (68). The mutants of AML1B, shown in Fig. 1, were constructed as follows. AML1B(1-268) was constructed by digesting a PCR product, containing a stop codon and a SalI restriction site after codon 268, with HindIII and SalI and ligating it into pCMV5-AML1B digested with the same restriction enzymes. All amplified DNA sequences were confirmed by the dideoxy-chain termination method. AML1B(1-289) and AML1B(1-317) were made by digesting pCMV5-AML1B with BamHI and SalI, respectively; the overhangs were filled in and ligated to an XbaI-stop linker. AML1B(1-381) was generated by PCR with EcoRI restriction sites at both ends and was subcloned into the EcoRI site of pCMV5.
FIG. 1.
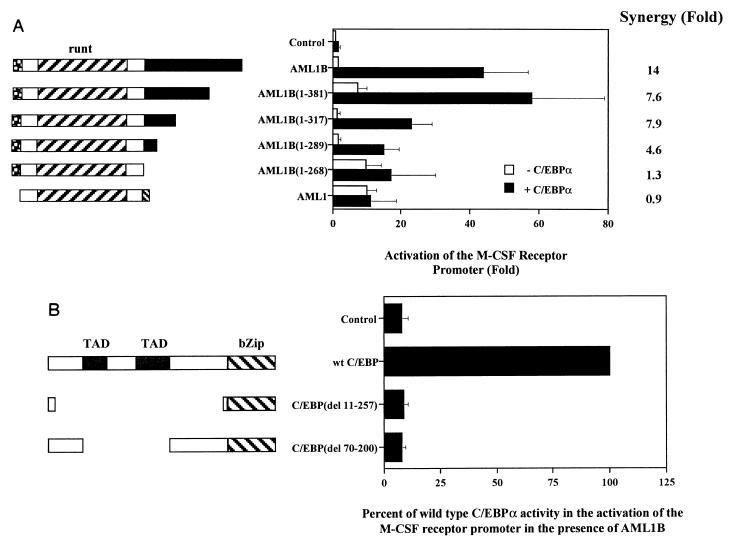
General structures of three alternatively spliced forms of AML1 and mutations of AML1, PU.1, and C/EBPα. The diagram shows the three forms of AML1 and the mutations of AML1A and AML1B, which were used to investigate the physical and functional interaction with PU.1 and C/EBPα. AML1 (250 amino acids), AML1A (453 amino acids), and AML1B (479 amino acids) (38) correspond to AML1a, AML1b, and AML1c as published by Miyoshi et al. (41). AML1 contains 250 amino acids and is identical to AML1B from amino acid 5 to 242 (32 to 268 of AML1B). The runt domain is indicated between amino acids 60 and 177 of AML1 and AML1A and amino acids 87 and 204 of AML1B. AML1A has 453 amino acids and contains the N terminus of AML1 and the C terminus of AML1B. mAML1A(1-288) is terminated at amino acid 288 of AML1A. Mutants of AML1B include C-terminal truncations at amino acids 268, 289, 317, and 381 [mAML1B(1-268) and mAML1B(1-289), -(1-317), and -(1-381), respectively]. The bZip region of C/EBPα, which is responsible for both the DNA-binding and dimerization properties, is located in the C terminus, while the transactivation domains (TAD) are between amino acids 70 and 97 and amino acids 127 and 200 (17, 31, 32, 46). The activation domain of PU.1 resides within the N terminus, while the DNA-binding ets domain is at the C terminus (28, 29).
Murine PU.1 in pGEX-2TK was a gift from T. Kouzarides (20). pGEX-runt contains a PCR product of the entire runt domain of AML1 subcloned into the BamHI and EcoRI sites of pGEX-2TK (79). pGEX-CBFβ contains the entire coding region of CBFβ in pGEX-2TK (34). The pGEX-AML1B construct containing the coding region for amino acids 213 to 395 and 315 to 395 was generated by PCR with the following primers: antisense (5′-CCGATGCGGCCGCGAATTCTTACGGGCCTCCCTGCGCT-3′) and sense (5′-CGCAGATCTCAGACCAAGCCCGGGAG-3′ and 5′-CGGGATCCCCTGCAGAACTTTCCAGT-3′ for 213 to 395 and 315 to 395, respectively). The 213–395 fragment was digested with BglII and EcoRI; the 315–395 fragment was digested with BamHI and EcoRI. Both were ligated into pGEX-2TK (Pharmacia) which had been digested with BamHI and EcoRI. The pGEX-AML1B(213-289) construct was generated by digesting pGEX-AML1B(213-395) with BamHI and EcoRI, removing codons 290 to 395, filling in with Klenow enzyme, and religating.
Expression and purification of recombinant proteins.
The glutathione S-transferase (GST) fusion proteins were grown in Escherichia coli BL(21) or DH5α cultured, after a 1:10 dilution of a 10-ml overnight culture, for 3 h at 37°C and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside for an additional 3 h at 37°C. GST fusion proteins were prepared as described previously (65). Protein concentration was determined by Coomassie blue staining of SDS-gels and comparison to bovine serum albumin standards. Full-length murine PU.1, PU.1 1-163, and PU.1 161-272 were transcribed and translated from pBS-PU.1 (29), pBS-PU.1(1-163) (in the SmaI site; from F. Moreau-Gachelin), and pGEM-ets (in the BamHI/XbaI site with start ATG), respectively, with the TnT coupled reticulocyte lysate system (Promega) according to the manufacturer’s recommendations, with the inclusion of [35S]methionine (3,000 Ci/mmol; NEN). AML1 and AML1B were transcribed and translated similarly from pBS-AML1 (41) and pBS-AML1B (38), respectively.
The GST pull-down assay was performed as follows. Two micrograms of each GST fusion protein, immobilized on glutathione-agarose (Sigma), was rocked at room temperature in binding buffer (50 mM Tris [pH 8], 140 mM NaCl, 0.5% Nonidet P-40, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin, chymostatin, and leupeptin per μl), with 1 μg of bovine serum albumin per μl, for a total volume of 200 μl. After 5 min, 2 μl of [35S]methionine-labeled, in vitro-transcribed and -translated protein was added; the mixture was incubated for an additional hour at room temperature and then washed three times, each time with 1 ml of binding buffer, resuspended in loading buffer, boiled for 5 min, and separated by SDS-polyacrylamide gel electrophoresis (PAGE). ImageQuant software was used to compare the intensity of bands and compare amounts of bound protein.
EMSA.
32P-labeled double-stranded oligonucleotides for electrophoretic mobility shift assay (EMSA) were prepared as previously described (78), and 0.5 ng (specific activity, 5 × 108 cpm/μg) was used per reaction. Proteins were preincubated at room temperature for 10 min in a volume of 20 μl with 2 μg of poly(dI-dC) in 10 mM HEPES (pH 7.9)–50 mM KCl–5 mM MgCl2–1 mM dithiothreitol–1 mM EDTA–5% glycerol. Unlabeled competitor oligonucleotides (100 ng = 200-fold excess) were included in this 10-min preincubation. For supershift experiments, 1 μl of either specific polyclonal antiserum or normal rabbit serum was added to the preincubation. Rabbit antiserum raised against the carboxyl four-fifths of C/EBPα was provided by Steven McKnight. Reaction mixtures were then subjected to PAGE at 10 V/cm on a 5.2% polyacrylamide gel in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) at 4°C. ImageQuant software was used to quantitate the bound probe.
RESULTS
AML1 binds DNA cooperatively with C/EBPα but not with PU.1.
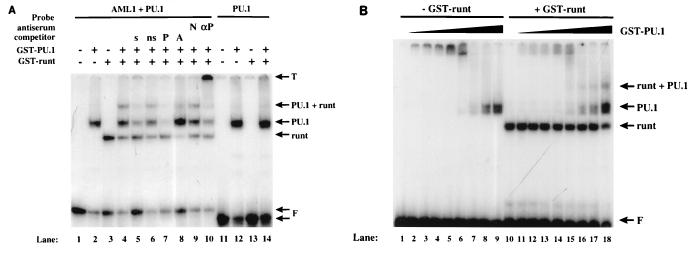
We have previously shown that AML1, PU.1, and C/EBPα interact with the M-CSF receptor promoter and that their binding sites are important for the promoter (79). However, ternary complexes containing AML1, C/EBPα, and DNA, or AML1, PU.1, and DNA, were not observed when nuclear extracts were used in gel shift assays (79). We used purified GST fusion proteins to further investigate the role of DNA binding in the regulation of the M-CSF receptor promoter. We first investigated whether AML1 and PU.1 could form a ternary complex with DNA. As shown in Fig. 2A, purified GST-PU.1 (lane 2) and GST-runt, a fusion protein containing the runt domain of AML1 (79) (lane 3), bound to the radiolabeled oligonucleotide which contains the M-CSF receptor promoter AML1 and PU.1 sites (bp −71 to −37). In the presence of both purified proteins, a band of lower mobility was detected (lane 4). The formation of this PU.1-runt complex could be competed with a 200-fold molar excess of unlabeled self oligonucleotide (lane 5) or an oligonucleotide containing either a PU.1 binding site (lane 7) or an AML1 binding site (lane 8) but not with a nonspecific C/EBP-binding oligonucleotide (lane 6). Furthermore, this higher complex could be supershifted by antiserum specific for PU.1 but not by normal rabbit serum (lanes 10 and 9, respectively). These results show that AML1 and PU.1 can form a ternary complex with DNA. The incomplete self competition was observed when the high protein concentrations necessary to generate the higher-order complex were used. When a radiolabeled oligonucleotide containing only a PU.1 site was used in a similar gel shift experiment, the ternary complex was not detectable (lanes 11 to 14). This result indicates that both DNA-binding sites are required for the formation of the ternary complex. To investigate whether there is cooperation in the formation of the ternary complex, titration experiments were performed as shown in Fig. 2B. We detect more DNA associated with PU.1 (lanes 1 to 9) than with the higher-order complex (lanes 10 to 18) formed in the presence of the AML1 runt domain. When the concentration of GST-runt was titrated in the absence and presence of GST-PU.1, we observed the same result (data not shown). Based on these experiments, the amount of probe shifted by the higher-order complex is less than that shifted by either GST-runt (data not shown) or GST-PU.1, indicating that AML1 and PU.1 do not bind cooperatively to DNA.
FIG. 2.
AML1 forms a tertiary complex on DNA with PU.1 but does not exhibit cooperative DNA binding. (A) Lanes 1 to 10, a double-stranded oligonucleotide consisting of M-CSF receptor sequence between nucleotides −71 and −37 (78), containing sites for both AML1 and PU.1 (AML1 + PU.1), was end labeled and incubated in the absence of protein (lane 1) or in the presence of 600 ng of GST-PU.1 (lane 2), 500 ng of GST-runt (lane 3), or both (lanes 4 to 10). A 200-fold excess of each of the following double-stranded oligonucleotides, all derived from M-CSF receptor sequence, was added as a competitor prior to the probe: self (s) −71 to −37 (lane 5) and non-self (ns) −88 to −73 (lane 6), −66 to −37 containing only the PU.1 binding site (P) (lane 7), and −88 to −59 containing only the AML1 binding site (A) (lane 8). One microliter of either normal rabbit serum (N) (lane 9) or antiserum raised against GST-PU.1 (αP) (a gift from H. Singh) (lane 10) was added following the addition of probe. A double-stranded oligonucleotide from −66 to −37 of the M-CSF receptor promoter containing only the binding site for PU.1 (PU.1) was end labeled and incubated in the absence of protein (lane 11) or in the presence of GST-PU.1 (lane 12), GST-runt (lane 13), or both (lane 14). The migration of the protein-DNA complexes is indicated on the right, as are the top of the gel (T) and the free probes (F). (B) The −71 to −37 probe used for panel A was incubated with increasing concentrations of GST-PU.1, ranging from 0.8 to 48 μM, in the absence (lanes 1 to 9) and in the presence (lanes 10 to 18) of GST-runt. The migration of the protein-DNA complexes is indicated on the right.
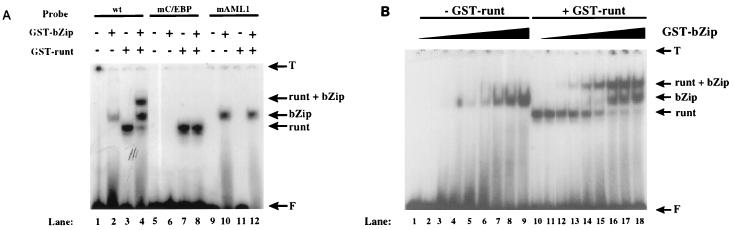
We then analyzed the formation of ternary complexes with DNA by the runt domain of AML1 and the bZip DNA-binding domain of C/EBPα. When a radiolabeled oligonucleotide containing the M-CSF receptor binding sites for AML1 and C/EBPα (bp −88 to −59) was used in a gel shift experiment (Fig. 3A, lanes 1 to 4, wt), we could detect shifted bands with both purified GST-bZip (lane 2) and GST-runt (lane 3). A ternary complex containing both proteins and the oligonucleotide was also observed (lane 4). These shifted bands could be competed with nonradiolabeled self oligonucleotide and also supershifted with specific antiserum (data not shown). However, oligonucleotide carrying mutations in either the C/EBP binding site (lanes 5 to 8) or the AML1 binding site (lanes 9 to 12) failed to form the ternary complex. This result demonstrates the requirement for both factor binding sites in the formation of the ternary complex. As with AML1 and PU.1, we performed a titration experiment to investigate differences in DNA binding between binary and ternary complexes. As shown in Fig. 3B, we detected more DNA associated with the higher-order complex formed in the presence of the AML1 runt domain (lanes 10 to 18) than with GST-bZip (lanes 1 to 9), providing evidence that AML1 and C/EBPα exhibit cooperative DNA binding. When titrations of GST-runt were incubated in the presence and absence of GST-bZip, we observed the same result (data not shown).
FIG. 3.
AML1 forms a ternary complex on DNA with C/EBPα and exhibits cooperative DNA binding. (A) A radiolabeled oligonucleotide containing sites for AML1 and C/EBP (bp −88 to −59) (wt) or a probe mutated in the C/EBP binding site (mC/EBP) (lanes 5 to 8) or AML1 binding site (mAML1) (lanes 9 to 12) was incubated in the absence of protein (lanes 1, 5, and 9) or in the presence of 500 ng of GST-bZip (lanes 2, 6, and 10), 500 ng of GST-runt (lanes 3, 7, and 11), or both (lanes 4, 8, and 12). The migration of the protein-DNA complexes is indicated on the right, as are the top of the gel (T) and the free probe (F). (B) The −88 to −59 probe used for panel A was incubated with increasing concentrations of GST-bZip, ranging from 6 to 1,710 nM, in the absence (lanes 1 to 9) and in the presence (lanes 10 to 18) of a saturating amount of GST-runt. The migration of the protein-DNA complexes is indicated on the right, as are the top of the gel (T) and the free probe (F).
In summary, these data demonstrate that AML1-C/EBP and AML1-PU.1 DNA-protein ternary complexes can be detected with purified proteins. Furthermore, the formation of the ternary complexes requires the presence of both DNA-binding sites. Finally, while the runt domain of AML1 binds cooperatively to DNA in the presence of the C/EBPα bZip domain, the ternary complex with PU.1 is formed in a noncooperative manner. These differences in DNA binding may affect the interaction of the transcription factors in the regulation of the M-CSF receptor promoter.
PU.1 and AML1B interact physically in vitro via the DNA-binding domain of each protein.
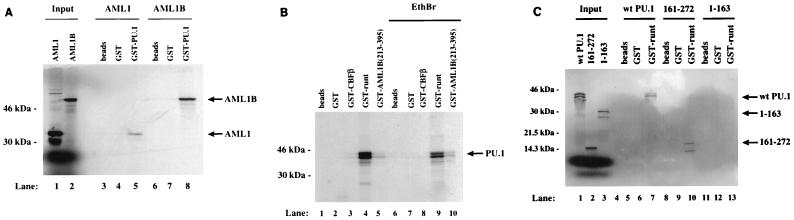
AML1 and PU.1, two transcription factors important for myeloid differentiation, bind to adjacent regions on the M-CSF receptor promoter and activate it (77, 78). To understand the mechanism of their function, we analyzed whether AML1 could physically interact with PU.1 as it does with C/EBPα to confer the maximal level of myeloid-specific regulation. Using the GST pull-down assay, we have established that radiolabeled, in vitro-translated AML1 interacts with a GST fusion protein containing full-length PU.1. Both the 250 (Fig. 4A, lanes 1 and 5)- and 479 (lanes 2 and 8)-amino-acid forms (AML1 and AML1B [Fig. 1]) are pulled down by GST-PU.1; however, the interaction with AML1B appears stronger. Furthermore, full length in vitro-translated PU.1 binds to a GST fusion protein containing the runt domain of AML1B (Fig. 4B, lane 4). To establish the specificity of the interaction, ethidium bromide was added to the binding reaction to prevent potential nonspecific interactions mediated by contaminating DNA (30). As shown in Fig. 4B, lane 9, addition of ethidium bromide decreased but did not abolish the interaction between the runt domain of AML1 and PU.1. We also observed a weak interaction between PU.1 and a GST fusion protein containing a domain (amino acids 213 to 395) within the C terminus of AML1B (Fig. 4B, lanes 5 and 10). Although this domain is critical for synergy between AML1 and PU.1 (see below), the strength of the interaction with PU.1 is only 17% of that observed between PU.1 and the runt domain. However, this relatively weak interaction may explain the different binding abilities of AML1 and AML1B with GST-PU.1 (Fig. 4A). We have also established that the interaction between PU.1 and the runt domain of AML1 localizes to the DNA-binding ets domain of PU.1. In Fig. 4C, full-length PU.1 (lanes 1 and 7) and amino acids 161 to 272 containing the ets domain of PU.1 (lanes 2 and 10), but not the activation domain of PU.1 (amino acids 1 to 161 [lanes 3 and 13]), interact specifically with the GST fusion protein containing the AML1 runt domain. The lower band of the doublet observed in lane 9 is also present in the input lane 2 but is partially obscured by the free 35S. In summary, these data demonstrate that AML1 and PU.1 interact physically and that this interaction occurs primarily through their DNA-binding domains.
FIG. 4.
The physical interaction between PU.1 and AML1 is localized to the DNA-binding domains. (A) AML1 (lanes 1 and 3 to 5) and AML1B (lanes 2 and 6 to 8) were transcribed and translated in vitro with [35S]methionine and incubated with glutathione-agarose (beads) (lanes 3 and 6), GST (lanes 4 and 7), or GST-PU.1 (lanes 5 and 8). Positions of molecular mass markers and the migration of the bound proteins are indicated to the left and right, respectively. (B) PU.1 was transcribed and translated in vitro with [35S]methionine and incubated with glutathione-agarose (beads) (lanes 1 and 6) or the following proteins immobilized on agarose: GST (lanes 2 and 7), GST-CBFβ (lanes 3 and 8), GST-runt (lanes 4 and 9), and GST-AML1B(213-395) (lanes 5 and 10). Ethidium bromide (EthBr; 50 μg/μl) was added to the reactions in lanes 6 to 10. Positions of molecular mass markers are shown to the left, and the migration of PU.1 is indicated to the right. (C) Full-length PU.1 (lanes 1 and 5 to 7), amino acids 161 to 272 of PU.1 containing the ets domain (lanes 2 and 8 to 10), and amino acids 1 to 163 of PU.1 (lanes 3 and 11 to 13) were transcribed and translated in vitro with [35S]methionine and incubated with glutathione-agarose (beads) (lanes 5, 8, and 11), immobilized GST (lanes 6, 9, and 12), or GST-runt (lanes 7, 10, and 13). Positions of the molecular mass markers, run in lane 4, are shown to the left, and the migration of the wild-type (wt) and mutant forms of PU.1 is indicated to the right.
PU.1 and AML1 synergize to activate the M-CSF receptor promoter, a function which is dependent on regions within the activation domains of PU.1 and AML1B.
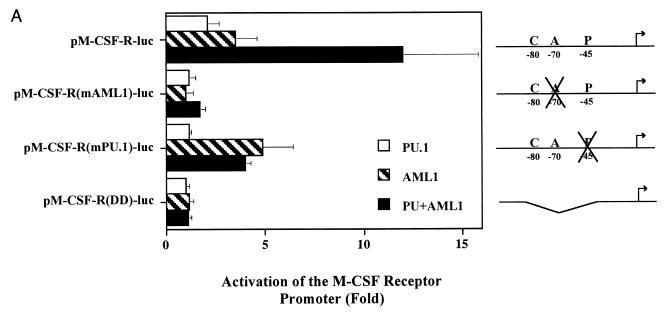
PU.1 and AML1 both activate the M-CSF receptor promoter (78, 79) and interact physically through their DNA-binding domains but do not bind cooperatively to DNA. We next investigated whether PU.1 and AML1 could synergize to activate the M-CSF receptor promoter. Transfections of HeLa cells, which contain no endogenous PU.1 and little detectable AML1, showed that while PU.1 activates the promoter 2-fold and AML1B 3.5-fold, together they synergize to increase the activity of the promoter 12-fold (Fig. 5A). Although the synergy between PU.1 and AML1B is weak relative to that observed with AML1B and C/EBPα, it is more than an additive effect. We calculated the fold synergy by dividing the activation of the promoter in the presence of both factors by the expected additive result. PU.1 and AML1B exhibit twofold synergy, or two times as much activation as an additive effect. However, this synergy is absent with reporter constructs bearing mutations in either the PU.1 or AML1 binding site [pM-CSF-R(mPU.1)-luc or pM-CSF-R(mAML1)-luc, respectively] or a deletion of bp −86 to −37 [pM-CSF-R(DD)-luc], containing the binding sites for C/EBP, AML1, and PU.1, indicating that DNA binding is required for both factors. Interestingly, we are unable to reproduce the synergy between AML1B and PU.1 in the CV-1 cell line, and although the synergy between AML1B and C/EBPα is observed in both HeLa and CV-1 cells, we saw no more than an additive increase in activity when PU.1 was included with AML1B and C/EBPα (data not shown).
FIG. 5.
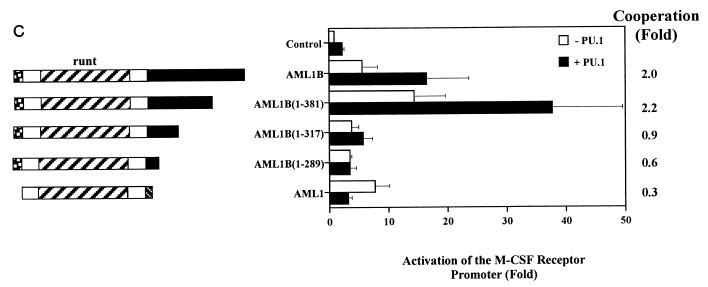
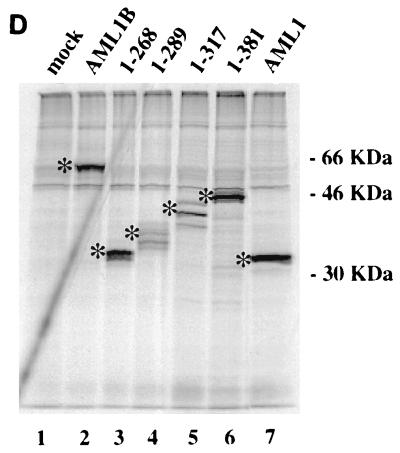
AML1B and PU.1 synergize to transactivate the M-CSF receptor promoter. (A) Synergistic transactivation of the M-CSF receptor promoter by PU.1 and AML1B requires intact binding sites for both factors. Transient transfections were performed in HeLa cells with 5 μg of reporter plasmid and 1 μg of expression plasmid containing CBFβ (pCMV5-CBFβ), 1 μg of each expression plasmid for PU.1 (PU.1pECE) or AML1B (pCMV5-AML1B), or empty vector. The results represent the mean activation of the promoter ± standard error of five experiments. In this set of experiments, the activity of pM-CSF-R-luc was three times higher than that of the empty vector, pXP2. (B) Mutations in the N terminus of PU.1 abrogate synergy with AML1B. The effect of wild-type (wt) PU.1 on pM-CSF-R-luc was compared to effects of mutations of PU.1 in the N terminus, in the presence or absence of AML1B. The names of the mutants represent the deleted amino acids. The results are normalized to the level of expression of pM-CSF-R-luc and represent the mean ± standard error of three experiments. Fold synergy is calculated by dividing the activation in the presence of both factors by the sum of the activation by each factor individually. (C) The C terminus of AML1B is required for synergy with PU.1. Transient transfections were performed with PU.1, CBFβ, and either AML1, AML1B, or mutants of AML1B in order to identify the region of AML1 required for functional interaction with PU.1. Mutants AML1B(1-289), -(1-317), and -(1-381) are carboxy-terminal truncations of AML1B; the numbers represent the amino acids which are encoded. The results are normalized to the level of expression of pM-CSF-R-luc and represent the mean ± standard error of three experiments. (D) COS-7 cells were either mock transfected (lane 1) or transfected with Lipofectamine Plus, 2 μg of expression plasmid encoding AML1B (lane 2), or AML1B truncated at amino acid 268 (1-268, lane 3), 289 (1-289, lane 4), 317 (1-317, lane 5), or 381 (1-381, lane 6) or with AML1 (lane 7). After 20 h, the cells were labeled with [35S]methionine and [35S]cysteine and immunoprecipitated, and the AML1 proteins were resolved by SDS-PAGE. Bands corresponding to the expected molecular masses are indicated with asterisks, and the migration of the molecular mass markers is indicated to the right.
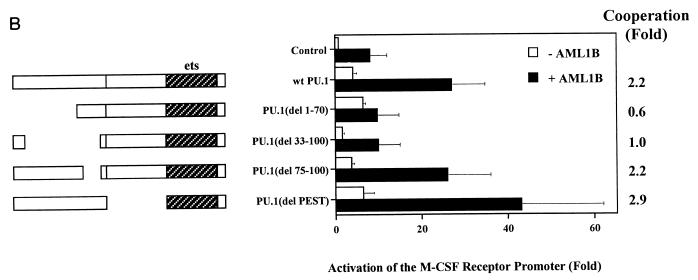
We investigated the role of the activation domains of AML1 and PU.1 in synergistic activation by performing transient transfections with a variety of mutants. The data presented in Fig. 5B illustrate that deletion of the PEST domain, which is important for interaction with NF-EM5 and activation of B-cell genes (58), has no effect on activation of the M-CSF receptor promoter or synergy with AML1B. However, deletion of amino acids 1 to 70 or 33 to 100 abrogates synergy, indicating that amino acids 33 to 70 of the PU.1 activation domain are important for this function. EMSA analysis of COS cell extracts transfected with the PU.1 expression constructs demonstrated that the mutated proteins were produced in comparable amounts and are capable of binding to DNA (data not shown).
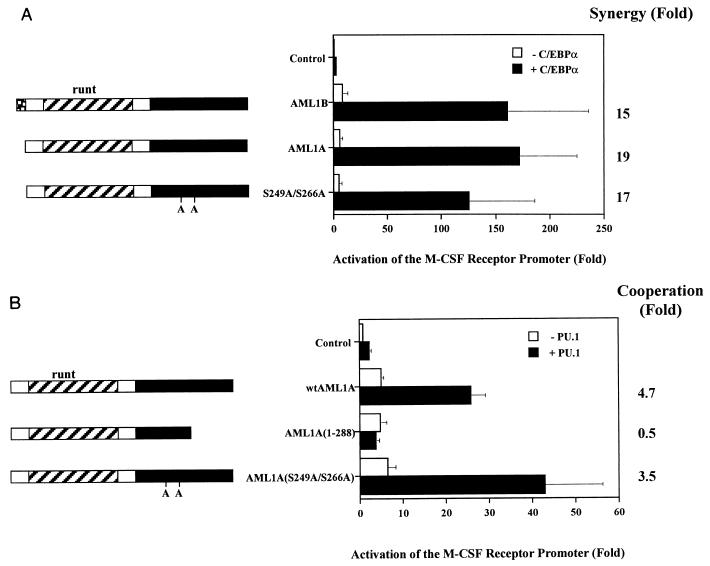
Transfections with AML1B mutants truncated in the carboxy terminus indicate that this region is necessary for synergy with PU.1 (Fig. 5C). While termination of AML1B at amino acid 381 does not affect synergistic activation of the promoter, termination at amino acid 317 clearly does. Production of the AML1 proteins was demonstrated by nonquantitative immunoprecipitation from transiently transfected COS-7 cells (Fig. 5D). Furthermore, the activation of the reporter construct by the AML1B mutants indicates that they are expressed, are transported to the nucleus, and are capable of binding the promoter. For example, in HeLa cells AML1B(1-289) and -(1-317) activate the M-CSF receptor promoter 3.5- and 3.8-fold, respectively, relative to the expression vector, pCMV5, and both of these proteins are capable of synergizing with C/EBPα (Fig. 6A). These data demonstrate that AML1 synergizes with PU.1 to activate the M-CSF receptor promoter and that this activity requires DNA-binding sites for both factors and regions contained within the activation domains of AML1 and PU.1.
FIG. 6.
The C terminus of AML1 and the transactivation domains of C/EBPα are required for synergistic activation of the M-CSF receptor promoter. (A) The C terminus of AML1B is important for synergy with C/EBPα. CV-1 cells were transfected with 5 μg of pM-CSF-R-luc and 1 μg each of expression vectors for AML1, AML1B, and C-terminal truncations of AML1B, in the presence and absence of C/EBPα. Transfection results are normalized to the level of expression of pM-CSF-R-luc in the presence of control plasmid and represent the means ± standard error of three experiments. Fold synergy is calculated by dividing the activation in the presence of both factors by the sum of the activation by each factor individually. (B) The transactivation domains (TAD) of C/EBPα are required for synergy with AML1B. The results of transfections in CV-1 cells are expressed as percentages of the activity of wild-type (wt) C/EBPα, which alone activates the M-CSF receptor promoter 1.7-fold, and represent the means ± standard error of three experiments. The mutations indicate the amino acids which are deleted.
Synergy between AML1B and C/EBPα requires their transactivation domains, and the region of AML1 critical for this synergy is distinct from that important for synergy with PU.1.
We have previously shown that AML1B and C/EBPα synergize to activate the M-CSF receptor promoter, that both DNA-binding sites are required for this function, and that the factors interact in vitro via their DNA-binding domains (runt and bZip domains, respectively) (79). Furthermore, in this report we have demonstrated that AML1 and C/EBPα form a ternary complex with DNA and exhibit cooperative DNA binding. To clarify the mechanism of synergistic activation, we proceeded to investigate whether mutations outside of the DNA-binding domains would affect the ability of either factor to synergize with the other.
Figure 6A presents data that demonstrate the requirement for the C terminus of AML1B in synergistic activation. The 250-amino-acid isoform, AML1, which effectively terminates at residue 268 of AML1B, does not synergize with C/EBPα, nor does AML1B truncated at amino acid 268. Therefore, we can exclude the possibility that the different termini of AML1 play negative roles in the synergy with C/EBPα. However, in contrast to the experiments done in the presence of PU.1, AML1B truncated at either amino acid 317 or 289 retains the ability to synergize with C/EBPα (represented by the fold synergy), indicating that amino acids 268 to 317 are important for this activity. Due to the variability between experiments, there is no significant difference between the fold synergies calculated for AML1B, AML1B(1-381), and AML1B(1-317). We are also able to show that mutations which delete nearly the entire N terminus of C/EBPα (del 11-257, previously referred to as regions 1 to 9) or both of the transactivation domains (del 70-200, previously referred to as regions 3 to 7) (17) completely abrogate synergy with AML1B (Fig. 6B). The production of C/EBPα protein from these constructs has been demonstrated previously by Western blot analysis of extracts from transfected HepG2 cells (17). The results of these experiments show that amino acids 268 to 317 of AML1B, and the transactivation domains of C/EBPα, are critical for synergy.
Mutation of potential phosphorylation sites in AML1 does not interfere with synergistic activation of the M-CSF receptor promoter.
The activation of an AML1-responsive reporter by AML1A can be increased following phosphorylation by ERK on serines 249 and 266 (corresponding to serines 276 and 293 of AML1B) (68). We were interested in determining whether the interaction between AML1 and its neighboring factors might be influenced by this modification. Since the phosphorylation state of AML1 is unknown under the conditions in which we observe synergistic activation, we addressed the role of serines 249 and 266 in this context. The data in Fig. 7 demonstrate that AML1A, which contains the same N terminus as AML1 and the same C terminus as AML1B (Fig. 1), synergizes with both C/EBPα (Fig. 7A) and PU.1 (Fig. 7B). In addition, AML1A truncated at amino acid 288 (corresponding to amino acid 315 of AML1B) fails to synergize with PU.1, further supporting the observation that amino acids C terminal to residue 317 are necessary for this function (Fig. 7B). However, mutation of serines 249 and 266, converting those residues to alanine, has no effect on the ability of AML1A to synergize with either C/EBPα or PU.1, demonstrating that these potential phosphorylation sites are not required for synergistic activation by AML1.
FIG. 7.
Mutation of potential phosphorylation sites does not affect the ability of AML1 to synergize with either C/EBPα or PU.1. (A) Wild-type AML1A and mutant S249A/S266A both synergize with C/EBPα. Transfections of CV-1 cells with AML1B, AML1A, and AML1A mutated at serines 249 and 266 (S249A/S266A) were performed in the absence and presence of C/EBPα. The results represent the mean activation of the promoter ± standard error of three experiments. Fold synergy is calculated by dividing the activation in the presence of both factors by the sum of the activation by each factor individually. (B) Wild-type (wt) AML1A and mutant S249A/S266A synergize with PU.1 to activate the M-CSF receptor promoter. HeLa cells were transfected with pM-CSF-R-luc and expression constructs for wild-type AML1A, a mutant terminated at amino acid 288 (1-288), and one with serine-to-alanine mutations at residues 249 and 266 (S249A/S266A), in the presence and absence of PU.1. The results represent the mean activation of the promoter ± standard error of three experiments.
DISCUSSION
Defining the regions of PU.1, C/EBPα, and AML1 necessary for physical and functional interaction has led to an increased understanding of the mechanisms by which the M-CSF receptor promoter is regulated and how these factors interact to mediate transcriptional activation. We have demonstrated that the DNA-binding domain of AML1 can form higher-order complexes with the DNA-binding domain of either C/EBPα or PU.1 and that the formation of these complexes is dependent on DNA-binding sites for both AML1 and the neighboring transcription factor. In addition, we observed that a complex containing both the runt and bZip domains bound greater amounts of probe than either individual DNA-binding domain, indicating that cooperative DNA binding occurs between AML1 and C/EBPα. Alternatively, this effect was not observed between the runt domain and PU.1. Instead, less probe was associated with the ternary complex than with either PU.1 or the runt domain. However, since these experiments were performed with the DNA-binding domains of AML1 and C/EBPα, we cannot exclude the possibility that the native proteins would exhibit different properties. In several studies, the physical interaction between transcription factors increases DNA-binding ability, providing an explanation for synergistic activation in the presence of two factors (11, 18, 56, 58, 67). For example, AML1 binds cooperatively with another member of the ets family, Ets-1, to the TCRα, TCRβ, and Moloney murine leukemia virus enhancers (18, 67). Cooperative DNA binding may contribute to the strong synergy between AML1B and C/EBPα.
We have shown that AML1 interacts physically with PU.1, and as with C/EBPα, this property maps to the DNA-binding domain of each protein. In addition, AML1 and PU.1 exhibit a relatively weak synergistic activation of the promoter. The effect seen in the presence of both PU.1 and AML1B is more than additive and therefore by definition synergistic. We have also shown that while the physical interaction between AML1B and either PU.1 or C/EBPα occurs between the DNA-binding domains, other regions are also necessary to achieve the observed activation. For example, while the 250-amino-acid form of AML1 contains the runt domain, there is no synergy observed with either PU.1 or C/EBPα. Furthermore, deletion of the activation domain of either PU.1 or C/EBPα abrogates synergy with AML1B. Therefore, while in the case of AML1 and C/EBPα, the physical interactions may contribute to cooperative DNA binding, this is not sufficient for the strong synergy. Instead, we believe there are additional mechanisms controlling synergistic activation.
The carboxy terminus of AML1B contains at least two domains that serve disparate functions, each of which is responsible for synergy with C/EBPα and PU.1. This is demonstrated by the finding that amino acids 1 to 317, a region important for synergy between AML1 and c-Myb (6), are sufficient for synergy with C/EBPα but not PU.1. The existence of two functionally distinct domains implies that synergistic activation by AML1 is mediated by secondary proteins, or coactivators which bind specifically to one domain or the other. In support of this theory, we are able to show that TBP binds to a fusion protein containing the region of AML1B important for synergy with C/EBPα but not to a portion that contains the domain required for synergy with PU.1 (data not shown). Furthermore, each domain interacts with a different set of polypeptides from radiolabeled HeLa cells (data not shown), confirming that the C-terminal domains of AML1B make specific and distinct contacts, any of which may play a role in synergistic activation.
We hypothesize that PU.1, C/EBPα, and AML1 form a transcriptional unit, or primary complex, on the DNA and that this primary complex makes multiple and specific contacts with a second, perhaps ubiquitious, complex composed of coactivators. The role of the DNA-binding proteins is to confer tissue-specific, temporal regulation, while the coactivators serve to amplify the activation by increasing transcription efficiency. A similar mechanism has been described for the DNA-binding nuclear hormone receptors. Recent reports have revealed a complex mechanism, whereby nuclear hormone receptors are associated with both steroid receptor coactivators and with CBP, and the activation ability of the transcription factor is dependent on the efficient assembly of these complexes on the DNA (43). CBP is a ubiquitous adapter protein that mediates contacts between transcription factors and the basal transcription machinery (1, 26, 33) and is thought to stimulate transcription both by physical contact with the RNA polymerase II complex and through intrinsic histone acetylation activity (4, 33, 52). Kamei et al. have coined the term “integrator,” postulating that CBP integrates diverse signals within the cell which culminate in the assembly of transcription factors and coactivators and translates them into transcriptional activation (26). Further definition of this mechanism and identification of the factors involved will increase our understanding of how transcription factors respond to signals from external stimuli or cell cycle regulators.
It is clear that AML1 serves disparate functions on various promoters. While ERK increases the transactivation abilities of AML1A on the TCRβ enhancer, mutation of the serines which are potentially phosphorylated by ERK decrease neither activation by AML1A nor synergy. In addition, while the fusion protein formed from the (8;21) translocation, AML/ETO, behaves as an inhibitor of AML1B function with respect to the GM-CSF promoter and the TCRβ enhancer (15, 38), it synergizes with AML1B to activate the M-CSF receptor promoter (60). Although AML1 is known to interact with Ets-1, which like PU.1 is a member of the ets family of transcription factors, in this situation it is not the conserved ets DNA-binding domain which makes contact with the runt domain of AML1, but rather amino acids 123 to 240 in the N terminus of Ets-1 (18). The functional variability of AML1 can be explained if it requires contacts with other factors to activate transcription and is accordingly dependent on the cell type as well as other DNA-binding sites. Clarification of these mechanisms may lead to an understanding of how transcription is regulated in response to external signals or changes in the cell cycle. For instance, the ability for AML1 to synergize with PU.1 or C/EBPα or to coordinate the actions of all three transcription factors may depend on the activation or availability of coactivators and integrators. Therefore, in order to understand the mechanism by which AML1 and other factors that play pivotal roles in hematopoiesis function, it is important to explore and identify the contacts made in the course of transcriptional regulation.
ACKNOWLEDGMENTS
We thank R. Maki, M. Klemsz, T. Kouzarides, A. Berk, H. Miyoshi, and H. Hirai for providing PU.1, TBP, and AML1 constructs, G. Darlington and H. Singh for C/EBPα and PU.1 antisera, and N. Speck, K. Rhoades, and L. Smith for suggestions.
This work was supported by National Institutes of Health grants CA41456, CA/AI59589, and CA72009 and American Cancer Society grant DHP-166. D.-E.Z. is a Leukemia Society of America Scholar.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee C, Hiebert S W, Stein J L, Lian J B, Stein G S. An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc Natl Acad Sci USA. 1996;93:4968–4973. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Bassuk A G, Leiden J M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 6.Britos-Bray M, Friedman A D. Core binding factor (CBF) cannot synergistically activate the myeloperoxidase proximal enhancer in immature myeloid cells without c-Myb. Mol Cell Biol. 1997;17:5127–5135. doi: 10.1128/mcb.17.9.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castilla L H, Wijmenga C, Wang Q, Stacey T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen H M, Zhang P, Voso M T, Hohaus S, Gonzalez D A, Glass C K, Zhang D E, Tenen D G. Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood. 1995;85:2918–2928. [PubMed] [Google Scholar]
- 9.Cheneval D, Christy R J, Geiman D, Cornelius P, Lane M D. Cell-free transcription directed by the 422 adipose P2 gene promoter: activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1991;88:8465–8469. doi: 10.1073/pnas.88.19.8465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christy R J, Yang V W, Ntambi J M, Geiman D E, Landschulz W H, Friedman A D, Nakabeppu Y, Kelly T J, Lane M D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 11.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 12.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 13.Erman B, Sen R. Context dependent transactivation domains activate the immunoglobulin mu heavy chain gene enhancer. EMBO J. 1996;15:4565–4575. [PMC free article] [PubMed] [Google Scholar]
- 14.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos K G. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 15.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 16.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 17.Friedman A D, McKnight S L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 18.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 19.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier C, Bannister A J, Cook A, Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Munain C, Krangel M S. Regulation of the T-cell receptor delta enhancer by functional cooperation between c-Myb and core-binding factors. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera R, Ro H S, Robinson G S, Xanthopoulos K G, Spiegelman B M. A direct role for C/EBP and the AP-1-binding site in gene expression linked to adipocyte differentiation. Mol Cell Biol. 1989;9:5331–5339. doi: 10.1128/mcb.9.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohaus S, Petrovick M S, Voso M T, Sun Z, Zhang D E, Tenen D G. PU.1 (Spi-1) and C/EBP alpha regulate expression of the granulocyte-macrophage colony-stimulating factor receptor alpha gene. Mol Cell Biol. 1995;15:5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaestner K H, Christy R J, Lane M D. Mouse insulin-responsive glucose transporter gene: characterization of the gene and trans-activation by the CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:251–255. doi: 10.1073/pnas.87.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 26.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 27.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci USA. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klemsz M J, Maki R A. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 30.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 32.Landschulz W H, Johnson P F, McKnight S L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 33.Lee J S, Zhang X, Shi Y. Differential interactions of the CREB/ATF family of transcription factors with p300 and adenovirus E1A. J Biol Chem. 1996;271:17666–17674. [PubMed] [Google Scholar]
- 34.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 35.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 36.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers S, Downing J R, Hiebert S W. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers S, Lenny N, Sun W, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 40.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montminy M. Something new to hang your HAT on. Nature. 1997;387:654–655. doi: 10.1038/42594. [DOI] [PubMed] [Google Scholar]
- 44.Nagulapalli S, Pongubala J M, Atchison M L. Multiple proteins physically interact with PU.1. J Immunol. 1995;155:4330–4338. [PubMed] [Google Scholar]
- 45.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 46.Nerlov C, Ziff E B. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nisson P E, Watkins P C, Sacchi N. Transcriptionally active chimeric gene derived from the fusion of the AML1 gene and a novel gene on chromosome 8 in t(8;21) leukemic cells. Cancer Genet Cytogenet. 1992;63:81–88. doi: 10.1016/0165-4608(92)90384-k. [DOI] [PubMed] [Google Scholar]
- 48.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc Natl Acad Sci USA. 1993;90:7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nucifora G, Rowley J D. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- 50.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman A D. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol Cell Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 53.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 54.Pahl H L, Burn T C, Tenen D G. Optimization of transient transfection into human myeloid cell lines using a luciferase reporter gene. Exp Hematol. 1991;19:1038–1041. [PubMed] [Google Scholar]
- 55.Paul R, Schuetze S, Kozak S L, Kozak C A, Kabat D. The Sfpi-1 proviral integration site of Friend erythroleukemia encodes the Ets-related transcription factor Pu.1. J Virol. 1991;65:464–467. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pongubala J M, Atchison M L. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc Natl Acad Sci USA. 1997;94:127–132. doi: 10.1073/pnas.94.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pongubala J M, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pongubala J M, Van Beveren C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 59.Ray D, Culine S, Tavitain A, Moreau-Gachelin F. The human homologue of the putative proto-oncogene Spi-1: characterization and expression in tumors. Oncogene. 1990;5:663–668. [PubMed] [Google Scholar]
- 60.Rhoades K L, Hetherington C J, Rowley J D, Hiebert S W, Nucifora G, Tenen D G, Zhang D E. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc Natl Acad Sci USA. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuetze S, Paul R, Gliniak B C, Kabat D. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 64.Shin M K, Koshland M E. Ets-related protein PU.1 regulates expression of the immunoglobulin J-chain gene through a novel Ets-binding element. Genes Dev. 1993;7:2006–2015. doi: 10.1101/gad.7.10.2006. [DOI] [PubMed] [Google Scholar]
- 65.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 66.Smith L T, Hohaus S, Gonzalez D A, Dziennis S E, Tenen D G. PU.1 (Spi-1) and C/EBP alpha regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 67.Sun W, Graves B J, Speck N A. Transactivation of the Moloney murine leukemia virus and T-cell receptor beta-chain enhancers by CBF and Ets requires intact binding sites for both proteins. J Virol. 1995;69:4941–4949. doi: 10.1128/jvi.69.8.4941-4949.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, Kadowaki T, Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tenen D G, Hromas R, Licht J D, Zhang D E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 70.Uchida H, Zhang J, Nimer S D. AML1A and AML1B can transactivate the human IL-3 promoter. J Immunol. 1997;158:2251–2258. [PubMed] [Google Scholar]
- 71.Voso M T, Burn T C, Wulf G, Lim B, Leone G, Tenen D G. Inhibition of hematopoiesis by competitive binding of transcription factor PU.1. Proc Natl Acad Sci USA. 1994;91:7932–7936. doi: 10.1073/pnas.91.17.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang N D, Finegold M J, Bradley A, Ou C N, Abdelsayed S V, Wilde M D, Taylor L R, Wilson D R, Darlington G J. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 73.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBF beta subunit is essential for CBF alpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 75.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of hematopoiesis in mice heterozygous for an AML/ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 77.Zhang D E, Fujioka K I, Hetherington C J, Shapiro L H, Chen H M, Look A T, Tenen D G. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1) Mol Cell Biol. 1994;14:8085–8095. doi: 10.1128/mcb.14.12.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang D E, Hetherington C J, Chen H M, Tenen D G. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang D E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang D E, Zhang P, Wang N, Hetherington C J, Darlington G J, Tenen D G. Absence of G-CSF signaling and neutrophil development in C/EBP alpha deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]