Abstract
Introduction
This systematic review aimed to help further elucidate the following question: are endodontics sealers able to induce DNA damage in vitro or in vivo?
Methods
This study was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement 2020 criteria. A total of 23 studies were carefully selected by the authors.
Results
Regarding the general characteristics, most studies evaluated, on average, 3–5 types of sealers (resin epoxy, salicylate, salicylate + MTA, zinc oxide-eugenol, bioceramic products, calcium hydroxide), performing comparisons between them. Our results demonstrate that endodontic sealers may be a genotoxic agent since most studies demonstrated positive findings, with the resin-based ones being the most potentially genotoxic.
Conclusion
The type of genotoxicity assay, material evaluated, and dilution concentration levels influenced the outcome. This study clarifies whether and to what extent endodontic sealers are capable of inducing DNA injury in oral tissues.
Keywords: Genotoxicity, Endodontic sealers, DNA damage
1. Introduction
Nowadays, in endodontic practice, sealers are extensively used in gap filling procedures in which the core material and the root canal walls must be in intimate contact (Kaur et al., 2015). This hermetic contact is achieved by the formation of a homogeneous obturation mass with lack of voids after the elimination of the remaining microorganisms and regularization of canal portions via root canal reshaping (Kaur et al., 2015). Endodontic sealers are categorized by composition based on setting reaction and composition, considering their base components. Although other classification variations containing fillers or ceramic powders may be found in literature (such as MTA), the previously cited matrices continue to be the basis of the compositions (Komabayashi, 2020).
Nevertheless, the attempt to hermetically seal root canals is not always successfully achieved as unintentional (and sometimes inadvertent) sealer biomaterial may extrude during endodontic obturation. In this sense, scientific advances are frequently introducing novel endodontics materials at full speed, raising the question whether such introductions may be too deliberate concerning tissue hazards (Hosseinpour et al., 2022). Considering the non-stop scientific biomaterial evolution, safety must be seriously considered. In this context, biocompatibility is one of the most relevant steps for ensuring safety when endodontic materials are studied and launched since they may have unintentional or inadvertent direct contact with the periapical tissue (Hosseinpour et al., 2022). The underlying reason for biocompatibility studies lies in the fact that these biomaterials are in close contact with several oral tissues rather than the root dentin. This potential contact is likely to induce oxidative stress and to generate genetic damage, endangering long-term use of these products (Eid et al., 2014). In this sense, genotoxicity plays an important role in detecting whether and to what extent endodontic sealers may be able to induce DNA damage (Eid et al., 2014, Pires et al., 2016). To this end, and in line with the objective of identifying genotoxic effects in oral cells and tissues, some assays can be used, such as the micronucleus assay, the comet assay, chromosomal aberration, and sister chromatid exchange tests (Kang et al., 2013).
Concerning the cited tests, it is important to stress that the micronucleus assay, the chromosomal aberration, and the sister chromatid exchange aim to identify chromosome damage, whereas the comet assay is a method that aims to quantify DNA breakage as a result of DNA moving fragments when electrophoresis is performed (Lu et al., 2017). Moreover, regarding the comet assay, the lower molecular weight particles are pulled towards the anionic pole, forming a structure similar to a comet that will be further analyzed considering the tail length and intensity to determine the potential DNA damage (Lu et al., 2017). All things considered, it is coherent to state that all techniques evaluate DNA damage quantitatively and qualitatively by different end-points (Wilson and Thompson, 2007, Møller et al., 2020).
In this context, considering the variety of sealers and their potential genotoxic effects, this systematic review aimed to understand whether endodontics sealers may induce DNA damage in vitro or in vivo.
2. Material and methods
This study was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement 2020 criteria. The PICOS strategy: P (mammalian cells), I (Endodontic sealers), C (Control group), O (Genotoxicity), S (In vitro or in vivo exposure) was used as a guide.
2.1. Inclusion criteria
For the analysis, inclusion criteria were studies that: 1) measured genetic damage in vitro and/or in vivo; 2) were published in English; and 3) provided data that clearly met scientific standards. In accordance with the search strategy, some methods used for measuring genotoxicity were highlighted, being the micronucleus, the comet, the sister chromatic exchange, and the chromosomal aberration assays.
2.2. Exclusion criteria
Exclusion criteria included the following types of studies: 1) conference abstracts, reviews, editorials, and letters; 2) full-text not available in English; 3) studies with unavailable or unextractable data or with combined exposure without control group of endodontic sealer only; 4) multigenerational studies; 5) studies focusing on amelioration of endodontic sealers toxicity; 6) studies that did not measure genotoxicity 7) studies with partial or vague results.
2.3. Data search
In January of 2023, searches were conducted in PubMed, SCOPUS, and Web of Science databases to identify eligible articles, with the following keywords and Boolean operators: (“Sealer” OR “Endodontic sealer”) AND (“Genotoxicity” OR “DNA damage” OR “genetic damage” OR “DNA breakage” OR “genetic injury” OR “DNA injury” “chromosome damage” AND (“comet assay” OR “micronucleus assay” OR “sister chromatid exchange” OR chromosome aberration test”). An additional manual search of references and cited/related articles was performed. Terms were validated by conducting the proper selection of articles, representative of relevant works. Moreover, searches were restricted to the English language and all dates of publication were considered. Abstracts were read and judged independently by two reviewers (TGP and DAR). Boolean operators were used (AND and OR) to combine the descriptors with different combinations, as described elsewhere. First, a manual search by author (TGP) of the reference list of reviews and published articles was conducted; then, texts were selected based on both titles and abstracts. Afterwards, the second stage was conducted, in which two researchers (TGP and DAR) reread the references raised to identify possible lost articles in the very first search. The two aforementioned investigators, in an independent manner, reviewed the full-texts and available studies. Thus, relevant studies and their final evaluation were included for a proper selection of studies related to the research. After that, full-text readings of all selected abstracts were conducted to confirm eligibility. All divergences between the two reviewers (TGP and DAR) were achieved by a consensus after discussion.
2.4. Data extraction and quality assessment
The following data were presented: authors, year and country of study, species, organs or cell types, dose, concentration, exposure time, assay, number of evaluated cells, genotoxicity assay used, blind, statistical analysis, positive and negative control, and main results.
2.5. Risk of bias in individual studies
The score of the individual variables was established to classify each article. For this, the following information from the quality instrument was used: (1) study design, (2) identification and treatment of confounding factors, (3) blind analysis, and (4) data analysis. The considered criteria in the evaluation of the study design were: number of participants per group, statistical analysis, and blind analysis. The considered confounding factors were: cytotoxicity, repetitions number, and positive and negative controls. Moreover, strong, moderate, and weak classifications were used for the articles. Studies that controlled all but one, two, or three or more variables were rated as STRONG, MODERATE, or WEAK, respectively (Malacarne et al., 2022).
3. Results
3.1. Study selection
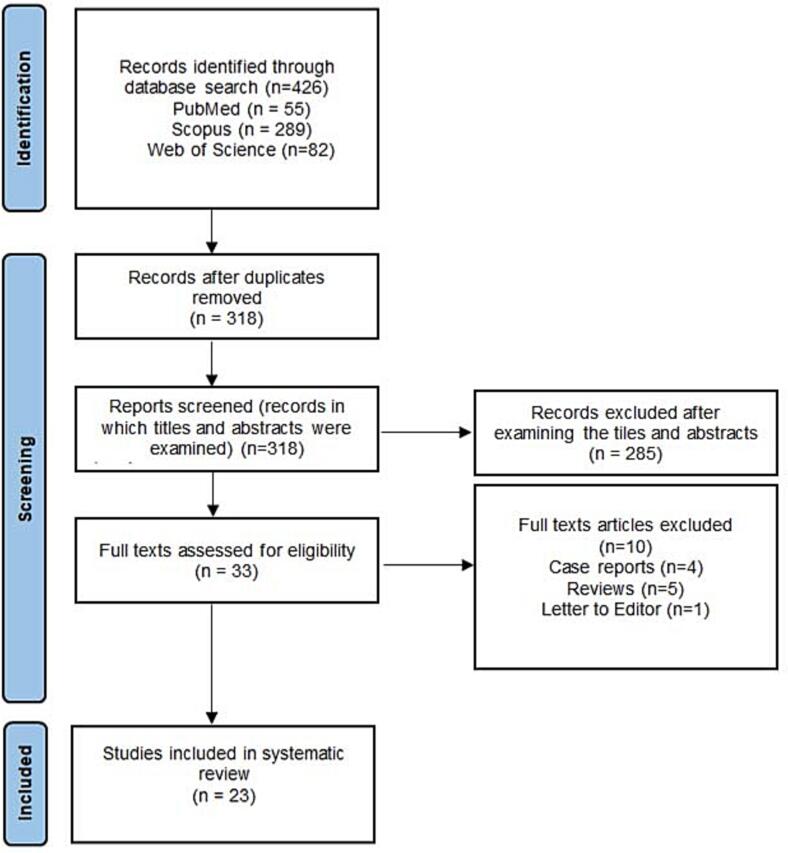
The data search identified 426 scientific records among which 108 publications were duplicates and, thus, excluded. After evaluating the titles and abstracts, 285 studies did not meet the inclusion criteria due to being literature reviews, case reports, commentaries and editorials, papers written in other languages other than English, or letters to Editor. Full manuscripts from 23 studies were meticulously read by both authors of the present article (Fig. 1).
Fig. 1.
Flow chart of the study.
3.2. General characteristics of the included studies
Table 1 shows the most important characteristics of the evaluated studies. A total of 23 studies were evaluated, with eight studies being conducted in Brazil. Only one study was conducted in Australia, Spain, and Turkey, respectively; two studies were conducted in Germany, Croatia, India, and in the USA, respectively; and four studies were conducted in Taiwan. The year of publication found in articles included in this study ranged from 1999 to 2022.
Table 1.
Most important characteristics of the included studies regarding genotoxicity induced by sealers in chronological order.
| Authors | Year | Country | Compound tested (commercial name) | Seal base |
|---|---|---|---|---|
| Só et al. (2022) | 2022 |
Brazil | Sealer Plus BC AH Plus MTA-Fillapex |
Bioceramic Epoxy resin Salicylate resin + MTA |
| Leme and Salvadori (2022) | 2022 | Brazil | MTA-Fillapex | Salicylate resin + MTA |
| Kim at al. (2022) | 2022 | Brazil | Adseal AH Plus Dia-Proseal |
Epoxy resin Epoxy resin Epoxy resin |
| Erdogan et al. (2021) | 2021 | Australia | AH Plus MTA-Fillapex IRootSP |
Epoxy resin Salicylate resin + MTA Calcium hydroxide |
| Dhopavkar et al. (2021) | 2021 | India | AH Plus MTA-Fillapex GuttaFlow 2 Sealer r |
Epoxy resin Salicylate resin + MTA Bioceramic |
| Teixeira et al. (2021) | 2020 | Brazil | AH Plus Sealer 26 Endomethasone N |
Epoxy resin Calcium hydroxide Zinc oxide eugenol |
| Martinho et al. (2018) | 2018 | Brazil | AH Plus EndoREZ Apexit Plus RealSeal SE |
Epoxy resin Methacrylate resin Calcium hydroxide Methacrylate resin |
| Nair et al. (2018) | 2018 | India | Endosequence BC Sealer Tubli-seal IRootSP |
Calcium hydroxide Zinc oxide Calcium hydroxide |
| Victoria-Escandell et al. (2017) | 2017 | Spain | AH Plus MTA-Fillapex MTA Angelus White |
Epoxy resin Salicylate resin + MTA Salicylate resin + MTA |
| Eldeniz et al. (2016) | 2016 | Turkey | AH Plus Jet Acroseal Acroseal EndoREZ RealSeal RealSeal SE Hybrid Root SEAL BioRoot RCS IRootSP MTA-Fillapex |
Epoxy resin Calcium hydroxide Zinc oxide Methacrylate resin Methacrylate resin Methacrylate resin Bioceramic Bioceramic Salicylate resin + MTA |
| Candeiro et al. (2016) | 2016 | Brazil | Endosequence BC Sealer AH Plus |
Calcium hydroxide Epoxy resin |
| Camargo et al. (2014) | 2014 | Brazil | AH Plus EndoREZ RoekoSeal |
Epoxy resin Methacrylate resin Silicone |
| Silva et al. (2013) | 2013 | Germany | AH Plus EndoREZ RealSeal SE Copaifera Polifill |
Epoxy resin Methacrylate resin Methacrylate resin Zinc oxide Zinc oxide |
| Van Landuyt et al. (2012) | 2012 | USA | AH Plus Jet EndoREZ RealSeal SE |
Epoxy resin Zinc oxide Methacrylate resin |
| Barara et al. (2011) | 2011 | Croatia | EpiphanyRealSeal SE (Sybron Endo, USA) |
Methacrylate resin Methacrylate resin |
| Bin et al. (2011) | 2011 | Brazil | MTA-Fillapex AH Plus White MTA |
Salicylate resin + MTA Epoxy resin Calcium hydroxide |
| Brzovic et al. (2009) | 2009 | Croatia | Guttaflow Epiphany Diaket IRM SuperEBA Hermetic |
Zinc oxide Methacrylate resin Zinc oxide Zinc oxide eugenol Zinc oxide eugenol Zinc oxide eugenol |
| Camargo et al. (2009) | 2009 | Germany | AH Plus Epiphany Acroseal |
Epoxy resin Methacrylate resin Calcium hydroxide |
| Huang et al. (2004) | 2004 | Taiwan | Sealapex Canals Canals-N Tubilseal TopsealAH26 (Silver free) AH Plus |
Calcium hydroxide Zinc oxide eugenol Zinc oxide Zinc oxide eugenol Epoxy resin Epoxy resin Epoxy resin |
| Huang et al. (2002) | 2002 | Taiwan | Sealapex AH Plus Canals |
Calcium hydroxide Epoxy resin Zinc oxide eugenol |
| Huang et al. (2001) | 2001 | Taiwan | AH 26 AH Plus |
Epoxy resin Epoxy resin |
| Tai et al. (2001) | 2001 | Taiwan | AH Plus AH 26 N2 Canals |
Epoxy resin Epoxy resin Zinc oxide eugenol Zinc oxide eugenol |
| Leyhaunsen et al. (1999) | 1999 | USA | AH Plus | Epoxy resin |
Regarding the general characteristics, most studies evaluated, on average, three types of sealers (amongst resin epoxy, salicylate, salicylate + MTA, zinc oxide-eugenol, bioceramic products, and calcium hydroxide), performing comparisons between them.
3.3. Variables related to dental sealers and genotoxicity
Table 2 describes the variables related to endodontic sealers and genotoxicity. First, all studies presented control groups for proper comparison. However, some studies presented both positive and negative control, whereas others presented only negative control.
Table 2.
Variables analyzed in the studies regarding genotoxicity induced by sealers in chronological order.
| Author | Concentration | Exposure time | Cell line/ species | Study design | Genotoxicity assay | Number of cells | Cytotoxicity assay | Reproduction number | Evaluated parameters | Blind analysis | Proper statistics description | Positive control | Negative control |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Só et al. (2022) | 1:10 | 24 h | In vitro | Micronucleus assay |
100 cells/slide | MTT assay | Triplicate | CellCount | – | Yes | No | Yes | |
| Leme and Salvadori (2022) | 5 %, 10 %, 20 % and 40 % | 24 h | In vitro | Comet assay | 50 cells/slide | MTS assay | Triplicate | Tail intensity | – | Yes | Yes | Yes | |
| Kim at al. (2022) | 100 %, 50 %, 25 %, 12.5 %, 6.25 %, 3.13 % | 50 min (Adseal); 8 h (AH Plus); 7.5 h (Dia-Proseal) | In vitro | Comet assay | – | MTT assay | Quadruplicate | Tail moment and tail intensity | – | Yes | No | Yes | |
| Erdogan et al. (2021) | 1:1, 1:2, 1:4, 1:8, 1:16, 1:32 | 24 h | In vitro | Micronucleus assay |
100 cells/slide | XTT | Triplicate | CellCount | – | Yes | No | Yes | |
| Dhopavkar et al. (2021) | 1.25 cm2/ml | 24 h; 48 h | In vitro | Comet assay | 50 cells/sample | MTT assay | Triplicate | Tail moment and tail intensity | – | Yes | Yes | Yes | |
| Teixeira et al. (2021) | 2,5%; 5 %; 10 % |
1 day; 7 days; 30 days | In vitro | Comet assay | 100 cells/slide | XTT | Triplicate | Tail intensity | – | Yes | Yes | Yes | |
| Martinho et al. (2018) | 1:2 | 24 h | In vitro | Micronucleus assay |
100 cells/slide | MTT assay | – | CellCount | – | Yes | Yes | Yes | |
| Nair et al. (2018) | 4 × 103 cells per mL |
48 h | In vitro | Comet assay | 50 to 100 cells/ sample | MTT assay | Triplicate | Tail moment | Yes | Yes | Yes | Yes | |
| Victoria-Escandell et al. (2017) | 1:2 | 24 h | In vitro | Flow cytometry | 4000 cells/ sample | SRB assay | Triplicate | CellCount | – | Yes | No | Yes | |
| Eldeniz et al. (2016) | 1/3 and 1/10 (both EC50) | 24 h | In vitro | c-H2AX immunofluorescence assay | 100 cells/slide | XTT | Triplicate | CellCount | – | Yes | Yes | Yes | |
| Candeiro et al. (2016) | 1:10 |
24 h | In vitro | Micronucleus assay |
100 cells/slide | MTT assay | Triplicate | CellCount | – | Yes | No | Yes | |
| Camargo et al. (2014) | 1:2, 1:4, 1:8, 1:16, 1:32 | 24 h | In vitro | Comet assay | 25 cells/slide | MTT assay | Quadruplicate | Tail moment and tail intensity | – | Yes | Yes | Yes | |
| Silva et al. (2013) | 1:1, 1:2, 1:4, 1:8 | 24 h | In vitro | Micronucleus assay |
1000 cells/slide | MTT assay | Quadruplicate | CellCount | – | Yes | Yes | Yes | |
| Van Landuyt et al. (2012) | 1/3 and 1/10 (both EC50) | 24 h | In vitro | c-H2AX immunofluorescence assay | At least 200/group | XTT | Quadruplicate | Standardized foci quantification | – | Yes | Yes | Yes | |
| Barara et al. (2011) | 0,02 g/4,5 ml | 24 h | In vitro | Comet assay | 100 cells/slide | count of viable, apoptotic and necrotic cells | Duplicate | Tail moment and tail intensity | Yes | Yes | Yes | Yes | |
| Bin et al. (2011) | 1:1, 1:2, 1:4, 1:8; 1:16; 1:32 | 24 h | In vitro | Micronucleus assay |
1000 cells/slide | MTT assay | Quadruplicate | CellCount | – | Yes | Yes | Yes | |
| Brzovic et al. (2009) | 1:4, 1:8, 1:16 | 1 h, 1 day, 5 days, 30 days | In vitro | Comet assay | 100 cells/slide | Trypan Blue ex lusion test | – | Tail moment and tail intensity | Yes | Yes | Yes | Yes | |
| Camargo et al. (2009) | Acroseal (1:64 and 1:128), AH Plus (1:8 and 1:16), andEpiphany (1:8 and 1:16) |
24 h | In vitro | Micronucleus assay |
1000 cells/slide | MTT assay | Quadruplicate | CellCount | – | Yes | Yes | Yes | |
| Huang et al. (2004) | 0.02, 0.1, 0.5, 2.5, 12.5 mg/100uL | 12 h and 24 h | In vitro | Comet assay | – | MTT assay | – | Survival rate | – | Yes | Yes | No | |
| Huang et al. (2002) | 0.01, 0.05, and 0.25 mg/ml | 24 h | In vitro | Comet assay | 50 cells/slide | MTT assay | – | Tail moment and tail intensity | Yes | Yes | Yes | Yes | |
| Huang et al. (2001) | 0.1, 0.5, and 2.5 mg/ml | 24 h | In vitro | Comet assay | 50 cells/slide | – | – | Tail moment and tail intensity | Yes | Yes | Yes | Yes | |
| Tai et al. (2001) | 2,5 ug/ul and 5ug/Ul | 24 h | In vitro | Comet assay | – | MTT assay | Triplicate | Count of H activity | – | Yes | No | Yes | |
| Leyhaunsen et al. (1999) | EUCARYOTIC DIT 1:1, 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, and 1:128 AFE 1:40, 1:80, 1:100 and 1:200 PROCARYOTIC AMES 1:5, 1:10, 1:20, 1:40 and1:80 UMU 1:1, 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:245 and 1:512 | DIT: 1,5h UMU test: 2 h and 4 h; AMES test: 48 h; AFE test: 2 h | In vitro/ In vivo | all in vivo EUCARYOTIC: DIT test and AFE test PROCARYOTIC: AMES and UMU tests | AFE: 4 clams/ assay | Growth inhibition test | AFE test: triplicate UMU test: Triplicate AMES test: Duplicate | DIT: cellcount AFE: AFE factor (single DNA breaks: treated/control) UMU: induction rate and growth factor AMES: revertants counts | – | Yes | Yes | Yes |
All included studies in this review used a type of test to verify genotoxic outcomes induced by sealers. In total, seven of the evaluated studies performed the micronucleus assay, being these conducted by Erdogan et al., 2021, Silva et al., 2015, Só et al., 2022, Martinho et al., 2018, Candeiro et al., 2016, Bin et al., 2012, and Camargo et al. (2009). The comet assay was performed in ten of these studies, being these conducted by Teixeira et al., 2021, Camargo et al., 2009, Baraba et al., 2011, Brzovic et al., 2009, Huang et al., 2001, Huang et al., 2002, Huang et al., 2004, Kim et al., 2022, Dhopavkar et al., 2021, Tai et al., 2002, and Nair et al. (2018). However, some studies used other assays, such as the one conducted by Eldeniz et al. (2016) and Van Landuyt et al. (2012), who used the c-H2AX immunofluorescence assay. Victoria-Escandell et al. (2017), on the other hand, used the flow cytometry for the genotoxicity analysis, whereas Tai et al. (2002) performed a DNA fragmentation analysis. Contrary to the previously cited authors, to analyze genotoxicity, Leyhaunsen et al. (2022) used the DIT and AFE test for eukaryotic analysis and AMES and UMU tests for procaryotic assessment.
In all in vitro studies, many mammalian cells were exposed to different sealers concentrations. For the in vivo study, clams were exposed to varying concentrations of sealers.
The selected studies presented different exposure times according to the genotoxicity test used. In the micronucleus assay, Erdogan et al., 2021, Silva et al., 2015, Só et al., 2022, Candeiro et al., 2016, Bin et al., 2012, and Camargo et al. (2014) adopted a 24-h period, whereas only the study by Martinho et al. (2018) adopted 55 weeks. In the same sense, comet assays also presented different exposure times: Teixeira et al. (2021) adopted 1, 7, and 30 days; Camargo et al., 2014, Baraba et al., 2011, and Huang et al., 2002, Huang et al., 2004 adopted 24 h; Dhopavkar et al. (2021) adopted 24 h and 48 h; and Nair et al. (2018) adopted 48 h.
Some studies were conducted in healthy human periodontal fibroblasts, from which three had third molars as specific sources and five had either other human teeth as sources or did not have a specific third molar source description. Moreover, only one study specifically informed the volunteer’s age.
Regarding the number of cells evaluated in the micronucleus assay, a total of five studies evaluated 1,000 cells per slide. For the alkaline comet assay, a total of three studies evaluated 100 randomly selected comets per slide, totaling 300 comets (Teixeira et al., 2021, Baraba et al., 2011, Dhopavkar et al., 2021), whereas 50 comets per treatment were evaluated in four studies (Leme and Salvadori, 2022, Huang et al., 2002, Huang et al., 2004, Dhopavkar et al., 2021). The study conducted by Nair et al. (2018) evaluated from 50 to 100 cells per sample, whereas the study by Camargo et al. (2014) evaluated only 25 cells and three studies failed to inform the amount of analyzed cells. For the c-H2AX immunofluorescence assay, Eldeniz et al. (2016) evaluated 100 cells per slide, whereas Van Landuyt et al. (2012) evaluated at least 200 cells per group. Regarding the study by Leyhaunsen et al. (1999), four clams were analyzed per assay in the AFE test.
The tests were performed in duplicate in two studies, in triplicate in 11 studies, in quadruplicate in five studies, and in quintuplicate in one study. The other four studies failed to inform the number of replicates.
Concerning data analysis, all micronucleus assay studies used cell count in the measurement of genotoxicity. Regarding the comet assay, the use of software programs was the basis of some parameters analysis. While Teixeira et al. (2021) and Leme and Salvadori (2022) evaluated tail intensity and Nair et al. (2018) only evaluated tail moment, all the other comet assay studies evaluated tail intensity and tail moment. As for the c-H2AX immunofluorescence assay studies, cell count and standardized foci quantification were considered. The assay that performed flow cytometry used cell count as a quantitative biological parameter. At last, the study conducted by Leyhaunsen et al. (1999) used different parameters, depending on the evaluated test (DIT, AFE: AFE, UMU, and AMES).
The adoption of blind analysis was observed in the methodology of five studies, whereas 18 of them did not provide such information. Lastly, all the included studies properly described the applied statistical test concerning the data analysis.
3.4. Main results
Regarding cytotoxicity, except for the study conducted by Huang et al., 2002, Huang et al., 2004, all selected studies evaluated cell death parameters, such as XTT, MTT, MTS, SRB, Trypan Blue assays and other cell viability tests. Considering chromosome damage, four studies showed that AH Plus was able to induce chromosome damage as analyzed by micronucleus assay (Erdogan et al., 2021, Candeiro et al., 2016, Bin et al., 2012, Victoria-Escandell et al., 2017). Furthermore, chromosome damage induction was also showed in one study for MTA-Fillapex and one for Acroseal and Epiphany, also analyzed by micronucleus assay (Bin et al., 2012, Hubbe et al., 2016). Additionally, sealers genotoxicity (AH Plus, Endorez, RoekoSeal, AH 26, N2, Canals, MTA-Fillapex, and GutaFlow 2) were measured in three studies (Brzovic et al., 2009, Dhopavkar et al., 2021, Van Landuyt et al., 2012). Moreover, at high concentrations, BioRoot RC and RealSeal SE presented genotoxicity in vitro (Huang et al., 2001, Dhopavkar et al., 2021. Table 3 summarizes these findings.
Table 3.
Main genotoxicity findings of studies in chronological order.
| Authors | Genotoxicity findings |
|---|---|
| Só et al. (2022) | No significant differences |
| Leme and Salvadori (2022) | ↑ CA: MTA-Fillapex |
| Kim at al. (2022) | ↑ MN: AdSeal, AH Plus and Dia-Proseal |
| Erdogan et al. (2021) | ↑ MN: AH Plus |
| Dhopavkar et al. (2021) | ↑ MTA-Fillapex |
| Teixeira et al. (2021) | No significant differences |
| Martinho et al. (2018) | No significant differences |
| Nair et al. (2018) | No significant differences |
| Victoria-Escandell et al. (2017) | ↑ MN: AH Plus and MTA-Fillapex |
| Eldeniz et al. (2016) | ↑ c-H2AX: BioRoot RC and RealSeal SE |
| Candeiro et al. (2016) | ↑ MN: AH Plus |
| Camargo et al. (2014) | ↑ CA: AH Plus, Endorez and RoekoSeal |
| Silva et al. (2013) | No significant differences |
| Van Landuyt et al. (2012) | No significant differences |
| Barara et al. (2011) | No significant differences |
| Bin et al. (2011) | ↑ MN: AH Plus and MTA-Fillapex |
| Brzovic et al. (2009) | No significant differences |
| Camargo et al. (2009) | ↑ MN: AH Plus, Acroseal and Epiphany |
| Huang et al. (2004) | No significant differences |
| Huang et al. (2002) | ↑CA: AH Plus and AH 26 |
| Huang et al. (2001) | ↑ CA: AH Plus, AH 26 and TopSeal |
| Tai et al. (2001) | ↑ CA: AH Plus, AH 26, N2 and Canals |
| Leyhaunsen et al. (1999) | No significant differences |
↑: increase; CA: comet assay; MN: micronucleus assay.
3.5. Quality assessment
Regarding the quality assessment, ten, seven, and six studies were classified as Strong, Moderate, and Weak, respectively, as shown in Table 4.
Table 4.
Quality assessment and final rating of the studies in chronological order.
| Author | N° of non controlled confounders | Final rating |
|---|---|---|
| Só et al. (2022) | 3 | Weak |
| Leme and Salvadori (2022) | 2 | Moderate |
| Kim at al. (2022) | 2 | Moderate |
| Erdogan et al. (2021) | 2 | Moderate |
| Dhopavkar et al. (2021) | 2 | Moderate |
| Teixeira et al. (2021) | 2 | Moderate |
| Martinho et al. (2018) | 3 | Weak |
| Nair et al. (2018) | 1 | Strong |
| Victoria-Escandell et al. (2017) | 3 | Weak |
| Eldeniz et al. (2016) | 1 | Strong |
| Candeiro et al. (2016) | 2 | Weak |
| Camargo et al. (2014) | 1 | Strong |
| Silva et al. (2013) | 1 | Strong |
| Van Landuyt et al. (2012) | 1 | Strong |
| Barara et al. (2011) | 1 | Strong |
| Bin et al. (2011) | 1 | Strong |
| Brzovic et al. (2009) | 1 | Strong |
| Camargo et al. (2009) | 2 | Moderate |
| Huang et al. (2004) | 5 | Weak |
| Huang et al. (2002) | 1 | Strong |
| Huang et al. (2001) | 2 | Moderate |
| Tai et al. (2001) | 3 | Weak |
| Leyhaunsen et al. (1999) | 1 | Strong |
4. Discussion
Endodontic sealers are widely used worldwide in the attempt to combat and prevent canal reinfection or growth of the remaining surviving microorganisms by residual bacteria entombment and nutrients leakage prevention (Camargo et al., 2014 and Munitić et al., 2019). Nonetheless, it is not rare to observe extrusion by apical constriction and by lateral and secondary canals with consequent contact between sealers and periradicular tissues, posing potential risks concerning genotoxicity in human cells (Dos Santos Costa et al., 2020).
In accordance with the potential risks, different tests can be used to evaluate genotoxicity, each with their own advantages. Nonetheless, currently, the most used ones worldwide converge in some aspects, such as simplicity, robustness and time- and cost-effectiveness in targeting toxicity. Nevertheless, the aforementioned assays require very specific parameters to achieve proper evaluations with reliable results. In this study, micronucleus assay, comet assay, and other tests (c-H2AX, UMU, and AMES) were considered in the review as tests capable of detecting genotoxicity induced by endodontic sealers.
While micronucleus assay can be considered a widely used sensitive method capable of detecting both chromosome or fragments in the cytoplasm of eukaryotic cell, comet assay is also comprehensively used, especially in vivo, as it is considered versatile concerning the evaluation of genotoxicity in different organs and tissues (Hubbe et al., 2016). Our results indicated that, from the 23 included studies, seven studies conducted the micronucleus assay, with positive genotoxicity being encountered in four. Moreover, 12 studies used the comet assay, with systematic results indicating positive genotoxicity in seven.
Additionally, we highlight that, to properly perform the comet assay without compromising the found results, the minimum of 50 cells must be evaluated and the parameter must be tail intensity (Cordelli et al., 2021). While only tail moment and tail intensity evaluations were conducted by one and two studies, respectively, all the others included both analyses. In this sense, it is coherent to state that studies that used scores or any other unmentioned evaluation parameters may compromise the results (Cordelli et al., 2021). Moreover, four studies performed other tests, such as c-H2AX immunofluorescence assay, flow cytometry, DIT test, AFE test, and AMES besides UMU tests. By using these assays, the results also showed that positive genotoxicity was detected in half of them.
Additionally, more than 50 % of the analyzed studies (13 out of 23) suggested genotoxicity increase in at least one of the evaluated sealers. More specifically, among the different evaluated categories according to the sealer base, the resin-based group (AH Plus) was the most genotoxic and cytotoxic across studies. We also highlight that, in the analyzed studies, different parameters were considered to determine the dose of endodontic sealers and most of the studies presented higher cytotoxicity in higher dilution concentrations.
Regarding the final ratings given by the authors of the present systematic review, ten, seven, and six studies were classified as Strong, Moderate, and Weak, respectively (in accordance with the previously described methodology). Overall, we assumed a good quality for most analyzed studies when evaluating genotoxicity, confirming, therefore, that the found results are reliable.
Furthermore, we highlight that an important parameter to be considered in genotoxicity studies is the presence of cytotoxicity, as genotoxicity tests should not be performed under conditions in which cell death is present. Moreover, it is known that cytotoxicity can induce fragmentation of the genetic material by caspases, which could lead to false-positive results (Tice et al., 2000). In this sense, it is reasonable to say that disregarding some data about cytotoxicity may lead to interpretation bias and that the approach for cytotoxicity is crucial for genotoxicity evaluation (Tice et al., 2000). In this study, most authors evaluated cytotoxicity to ensure the quality of the results regarding genotoxicity of sealers. We also highlight that only the smallest portion of the studies clearly mentioned the use of blind analysis methodology (five out of 23 studies), what interfered in the final rating of most articles.
To summarize, our results demonstrate that endodontic sealers may be considered genotoxic since most studies indicated positive findings and 17 showed a Moderate or Strong final rating. The resin-based sealers were found to be the most potentially genotoxic. The type of genotoxicity assay, material evaluated and dilution concentration levels influenced the outcome. Considering that some studies show that the contact extruded sealers did not impair the repair of endodontic lesions (Li et al., 2022; Shashirekha et al., 2018), further studies (mainly in vivo) should be conducted, especially in humans, elucidating the role of genotoxicity induced by endodontic sealers.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Data sharing are not available to this article.
Funding
The authors acknowledge research grants received from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Grant Number #001) for productivity fellowship.
Author contributions
Study design: TGP and DAR. Data search: TGP and DAR. Data analysis: TGP, ACMR, JNS, PRC and and DAR. Writing the paper: all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Baraba A., Zelježić D., Kopjar N., Mladinić M., Anić I., Miletić I. Evaluation of cytotoxic and genotoxic effects of two resin-based root-canal sealers and their components on human leucocytes in vitro. Int Endod J. 2011;44(7):652–661. doi: 10.1111/j.1365-2591.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- Bin C.V., Valera M.C., Camargo S.E., Rabelo S.B., Silva G.O., Balducci I., Camargo C.H. Cytotoxicity and genotoxicity of root canal sealers based on mineral trioxide aggregate. J Endod. 2012;38:495–500. doi: 10.1016/j.joen.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Brzovic V., Miletic I., Zeljezic D., Mladinic M., Kasuba V., Ramic S., Anic I. In vitro genotoxicity of root canal sealers. Int Endod J. 2009;42(3):253–263. doi: 10.1111/j.1365-2591.2008.01510.x. [DOI] [PubMed] [Google Scholar]
- Camargo C.H., Camargo S.E., Valera M.C., Hiller K.A., Schalmaz G., Schweikl H. The induction of cytotoxicity, oxidative stress, and genotoxicity by root canal sealers in mammalian cells. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;108:952–960. doi: 10.1016/j.tripleo.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Camargo C.H.R., Oliveira T.R., Silva G.O., Rabelo S.B., Valera M.C., Cavalcanti B.N. Setting Time Affects In Vitro Biological Properties of Root Canal Sealers. J Endod. 2014;40(4):530–533. doi: 10.1016/j.joen.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Candeiro G.T.M., Moura-Netto C., D'Almeida-Couto R.S., Azambuja-Junior N., Marques M.M., Cai S., Gavini G. Cytotoxicity, genotoxicity and antibacterial effectiveness of a bioceramic endodontic sealer. Int Endod J. 2016;49(9):858–864. doi: 10.1111/iej.12523. [DOI] [PubMed] [Google Scholar]
- Cordelli E., Bignami M., Pacchierotti F. Comet assay, a versatile but complex tool in genotoxicity testing. Toxicol Res (camb). 2021;10(1):68–78. doi: 10.1093/toxres/tfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhopavkar V.V., Shivanand S.S., Bhat K., Patil A.C., Preeti K., Godbole N.J. Comparative Evaluation of Cytotoxic and Genotoxic Effects of Three Resin-Based Sealers by 3, (4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay and Comet Assay - An In Vitro Study. Contemp Clin Dent. 2021;12(4):376–1338. doi: 10.4103/ccd.ccd_213_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos C.F., Fernandes M., Batistuzzo de Medeiros S.R. Genotoxicity of root canal sealers, a literature review. Clin Oral Invest. 2020;24(10):3347–3362. doi: 10.1007/s00784-020-03478-z. [DOI] [PubMed] [Google Scholar]
- Eid A.A., Gosier J.L., Primus C.M., Hammond B.D., Susin L.F., Pashley D.H., Tay F.R. In vitro biocompatibility and oxidative stress profiles of different hydraulic calcium silicate cements”. J Endod. 2014;40:255–260. doi: 10.1016/j.joen.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldeniz A.U., Shehata M., Hogg C., Reichl F.X. DNA double-strand breaks caused by new and contemporary endodontic sealers. Int Endod J. 2016;49(12):1141–1151. doi: 10.1111/iej.12577. [DOI] [PubMed] [Google Scholar]
- Erdogan H., Yildirim S., Cobankara F.K. Cytotoxicity and genotoxicity of salicylate- and calcium silicate-based root canal sealers on primer human periodontal ligament fibroblastos. Aust Endod J. 2021;47(3):645–653. doi: 10.1111/aej.12537. [DOI] [PubMed] [Google Scholar]
- Hosseinpour, S., Gaudin, A., Peters, O.A. 2022. A critical analysis of research methods and experimental models to study biocompatibility of endodontic materials. Int Endod J. 55 Suppl 2(Suppl 2):346-369. doi: 10.1111/iej.13701. Epub 2022 Feb 28. [DOI] [PMC free article] [PubMed]
- Huang T.H., Lee H., Kao C.T. Evaluation of the genotoxicity of zinc oxide eugenol-based, calcium hydroxide-based, and epoxy resin-based root canal sealers by comet assay. J Endod. 2001;27(12):744–748. doi: 10.1097/00004770-200112000-00008. [DOI] [PubMed] [Google Scholar]
- Huang T.H., Yang J.J., Li H., Kao C.T. The biocompatibility evaluation of epoxy resin-based root canal sealers in vitro. Biomaterials. 2002;23(1):77–83. doi: 10.1016/s0142-9612(01)00081-3. [DOI] [PubMed] [Google Scholar]
- Huang T.H., Ding S.J., Hsu T.Z., Lee Z.D., Kao C.T. Root canal sealers induce cytotoxicity and necrosis. J Mater Sci Mater Med. 2004;15(7):767–771. doi: 10.1023/b:jmsm.0000032816.45489.54. [DOI] [PubMed] [Google Scholar]
- Hubbe K.L., de Oliveira K.V., Coelho B.S., Baratto-Filho F. AH Plus extrusion into periapical tissue, literature review of main related properties and report of clinical cases. RSBO. 2016;36:1–20. [Google Scholar]
- Kang S.H., Kwon J.Y., Lee J.K., Seo Y.R. Recent advances in in vivo genotoxicity testing, prediction of carcinogenic potential using comet and micronucleus assay in animal models. J Cancer Prev. 2013;18(4):277–288. doi: 10.15430/JCP.2013.18.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Shah M., Logani A., Mishra N. Biotoxicity of commonly used root canal sealers. A Meta-Analysis. J Conserv Dent. 2015;18:83–88. doi: 10.4103/0972-0707.153054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Hayashi, M., Yu, B., Lee, T.K., Kim, R.H., Deuk-Won, J. 2022. Cytotoxicity and Genotoxicity of Epoxy Resin-Based Root Canal Sealers before and after Setting Procedures. Life (Basel). 7,12(6),847. [DOI] [PMC free article] [PubMed]
- Komabayashi T. Dental Medicine Faculty Publications; University of New England: 2020. Comprehensive Review Of Current Endodontic Sealers. [DOI] [PubMed] [Google Scholar]
- Leme K.S.V., Salvadori D.M.F. In vitro toxicogenomic activity of an MTA/salicylate-based endodontic sealer. Elsevier. 2022;9(2022):1076–1081. doi: 10.1016/j.toxrep.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyhausen G., Heil J., Reifferscheid G., Waldmann P., Geurtsen W. Genotoxicity and cytotoxicity of the epoxy resin-based root canal sealer AH plus. J Endod. 1999;2:109–113. doi: 10.1016/S0099-2399(99)80007-7. [DOI] [PubMed] [Google Scholar]
- Li, J., Chen, L., Zeng, C., Liu, Y., Gong, Q., Jiang, H. 2022. Clinical outcome of bioceramic sealer iRoot SP extrusion in root canal treatment: a retrospective analysis. 18(1), 28. [DOI] [PMC free article] [PubMed]
- Lu Y., Liu Y., Yang C. Evaluating In Vitro DNA Damage Using Comet Assay. J vis Exp. 2017;128:56450. doi: 10.3791/56450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malacarne I.T., Takeshita W.M., de Souza D.V., Dos Anjos R.B., de Barros V.M., Renno A.C.M., Salvadori D.M.F., Ribeiro D.A. Is micronucleus assay in oral exfoliated cells a useful biomarker for biomonitoring populations exposed to pesticides? A systematic review with meta-analysis. Environ Sci Pollut Res Int. 2022;43:64392–64403. doi: 10.1007/s11356-022-22015-x. [DOI] [PubMed] [Google Scholar]
- Martinho F.C., Camargo S.E.A., Fernandes A.M.M., Campos M.S., Prado R.F., Camargo H.R., Valera M.C. Comparison of cytotoxicity, genotoxicity and immunological inflammatory biomarker activity of several endodontic sealers against immortalized human pulp cells. Int Endod J. 2018;51(1):41–57. doi: 10.1111/iej.12785. [DOI] [PubMed] [Google Scholar]
- Møller P., Azqueta A., Boutet-Robinet E., Koppen G., Bonassi S., Milić M., Gajski G., Costa S., Teixeira J.P., Costa Pereira C., Dusinska M., Godschalk R., Brunborg G., Gutzkow K.B., Giovannelli L., Cooke M.S., Richling E., Laffon B., Valdiglesias V., Basaran N., Del Bo' C., Zegura B., Novak M., Stopper H., Vodicka P., Vodenkova S., de Andrade V.M., Sramkova M., Gabelova A., Collins A., Langie S.A.S. Minimum Information for Reporting on the Comet Assay (MIRCA), recommendations for describing comet assay procedures and results. Nat Protoc. 2020;15(12):3817–3826. doi: 10.1038/s41596-020-0398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, A.V., Nayak, M., Prasada, L.K., hetty, V., Viajy, C.N., Nair, R. 2018. Comparative Evaluation of Cytotoxicity and Genotoxicity of Two Bioceramic Sealers on Fibroblast Cell Line, An in vitro Study. J Contemp Dent Pract. 1,19(6),656-661. [PubMed]
- Pires, C.W., G. Botton, F.C., Cadoná, F.C., Machado, A.K., Azzolin, V.F., da Cruz, B.M., Sagrillo, M.R., Praetzel, J.R. 2016. Induction of cytotoxicity, oxidative stress and genotoxicity by root filling pastes used in primary teeth,” Int Endod J. 49, 737–745. [DOI] [PubMed]
- Silva G.O., Cavalcanti B.N., Oliveira T.R., Bin C.V., Camargo S.E., Camargo S.H. Cytotoxicity and genotoxicity of natural resin-based experimental endodontic sealers. Springer-Verlag, Berlin Heidelberg. 2015;20(4):815–819. doi: 10.1007/s00784-015-1567-4. [DOI] [PubMed] [Google Scholar]
- Só B.B., Martins M.D., So M.V., Weissheimer T., Marques M.M., Moreira M.S. Genotoxicity and Cytotoxicity Comparison of Calcium Silicate-Based and Resin-Based Sealers on Human Periodontal Ligament Stem Cells. Eur Endod J. 2022;14:129–134. doi: 10.14744/eej.2022.09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai K.W., Huang F.M., Huang M.S., Chang Y.C. Assessment of the genotoxicity of resin and zinc-oxide eugenol-based root canal sealers using an in vitro mammalian test system. J Biomed Mater Res. 2002;59(1):73–77. doi: 10.1002/jbm.1218. [DOI] [PubMed] [Google Scholar]
- Teixeira A.B., Moreira M.C., Takashashi C.S., Schiavon M.A., Alves O.K., Reis A.C. Cytotoxic and genotoxic effects in human gingival fibroblast and ions release of endodontic sealers incorporated with nanostructured silver vanadate. J Biomed Mater Res. 2021;109:1380–1388. doi: 10.1002/jbm.b.34798. [DOI] [PubMed] [Google Scholar]
- Tice R.R., Agurell E., Anderson D., Burlinson B., Hartmann A., Kobayashi H., Miyamae Y., Rojas E., Ryu J.C., Sasaki Y.F. Single cell gel/comet assay, guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35(3):206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Van Landuyt K.L., Geebelen B., Shehata M., Durner J. No Evidence for DNA Double-strand Breaks Caused by Endodontic Sealers. J Endod. 2012;29(6):618–625. doi: 10.1016/j.joen.2011.12.037. [DOI] [PubMed] [Google Scholar]
- Victoria-Escandell A., Ibanez Cabellos J.S., Sanchez S.B., Pascual E.B., Barcia J.B., Lopez E.G., Pallardo F.V., Gimenez J.L., Sabaer A.P., Lopez K.Z., Monterde M. Cellular Responses in Human Dental Pulp Stem Cells Treated with Three Endodontic Materials. Stem Cell Int. 2017;2017:8920356. doi: 10.1155/2017/8920356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.M., 3rd, Thompson H. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007;616(1–2):11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing are not available to this article.