Summary
The continual emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) poses a major challenge to vaccines and antiviral therapeutics due to their extensive evasion of immunity. Aiming to develop potent and broad-spectrum anticoronavirus inhibitors, we generated A1-(GGGGS)7-HR2m (A1L35HR2m) by introducing an angiotensin-converting enzyme 2 (ACE2)-derived peptide A1 to the N terminus of the viral HR2-derived peptide HR2m through a long flexible linker, which showed significantly improved antiviral activity. Further cholesterol (Chol) modification at the C terminus of A1L35HR2m greatly enhanced the inhibitory activities against SARS-CoV-2, SARS-CoV-2 VOCs, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) pseudoviruses, with IC50 values ranging from 0.16 to 5.53 nM. A1L35HR2m-Chol also potently inhibits spike-protein-mediated cell-cell fusion and the replication of authentic Omicron BA.2.12.1, BA.5, and EG.5.1. Importantly, A1L35HR2m-Chol distributed widely in respiratory tract tissue and had a long half-life (>10 h) in vivo. Intranasal administration of A1L35HR2m-Chol to K18-hACE2 transgenic mice potently inhibited Omicron BA.5 and EG.5.1 infection both prophylactically and therapeutically.
Keywords: SARS-CoV-2, variants of concern, pan-coronavirus fusion inhibitor, lipopeptide, cholesterol modification, ACE2 peptide, HR2 peptide, synergistic effect
Graphical abstract

Highlights
-
•
A1L35HR2m is a broad-spectrum SARS-CoV-2 fusion inhibitor with enhanced activity
-
•
A1L35HR2m binds to SARS-CoV-2 HR1 with high affinity and exhibits α-helical structure
-
•
A1L35HR2m-Chol broadly and potently inhibits coronaviruses in vitro and in vivo
-
•
A1L35HR2m-Chol shows robust efficacy against current and emerging SARS-CoV-2 variants
Bi et al. developed A1L35HR2m-Chol, a lipopeptide for pan-coronaviral inhibition, showing potent in vitro inhibitory activity against SARS-CoV-2 VOCs and closely related viruses. In vivo, A1L35HR2m-Chol also demonstrates robust prophylactic and therapeutic efficacy against SARS-CoV-2 variants.
Introduction
The ongoing COVID-19 pandemic has resulted in more than 6 million deaths. Although many approved vaccines and therapeutics are effective in alleviating hospitalization and mortality caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, different SARS-CoV-2 variants of concern (VOCs), especially the Omicron variant, have been shown to extensively evade immunity, which significantly compromises the efficacy of vaccines and therapeutics.1,2,3,4,5,6,7 Therefore, there remains a high demand to develop highly effective and broad-spectrum antivirals that can inhibit current and future emerging SARS-CoV-2 variants.
Similar to other coronaviruses, SARS-CoV-2 infection requires fusion between the viral envelope and the cell membrane, a process mediated by the viral spike (S) glycoprotein. The SARS-CoV-2 S protein consists of S1 and S2 subunits and employs a type I enveloped virus fusion mechanism for cell entry.8,9,10,11 The S1 subunit binds to the cellular receptor angiotensin-converting enzyme 2 (ACE2) through the receptor-binding domain (RBD) to trigger conformational changes in the S2 subunits, which leads to the formation of a six-helix bundle between the heptad repeat 1 (HR1) and HR2 domains, driving target cell and viral membrane fusion.10,11,12,13 Since the HR1 and HR2 domains are exceptionally conserved among different coronaviruses, peptides derived from these two domains are thus considered promising antiviral drugs.10,11,14 In fact, peptides derived from both the HR1 and HR2 domains have been shown to inhibit viral infection, with IC50 values in the micromolar range.15,16,17,18 Several HR2 helical conformation stabilization strategies, such as chemical stapling and the introduction of salt bridges, have also been reported to improve the activity of these peptide inhibitors, with IC50 values in the low-micromolar to high-nanomolar range.14,19 Cholesterol (Chol) modification of HR2-derived peptides is another effective method that can significantly enhance inhibitory activity, with IC50 values reaching low nanomolar concentrations in general.20,21,22
Peptides derived from the ACE2 protein have been explored as antiviral agents in the past. However, due to the low S protein binding affinity of ACE2-derived peptides, these peptides have shown limited success in inhibiting SARS-CoV-2 infection.23,24,25 Recent studies have suggested that combining ligands that target different sites on the S protein of the SARS-CoV-2 virus may result in a synergistic inhibitory effect.26,27,28,29 It has been shown that ACE2 protein binding could destabilize the S protein to trigger its conformational changes.30 Inspired by these findings, we hypothesize that a peptide derived from ACE2 may similarly destabilize the S protein upon binding to the RBD, ultimately leading to the exposure of the HR1 domain. Subsequently, a peptide derived from HR2 may be able to bind to HR1 to inhibit the virus. Given this hypothesis, we propose that a fusion peptide derived from ACE2 and the HR2 domain may exhibit a synergistic antiviral effect, potentially yielding even greater activity than either peptide alone.
Results
Conjugating an ACE2-derived peptide to the N terminus of the HR2-derived peptide enhanced the inhibitory activity
Aiming to develop potent and broad-spectrum anticoronavirus inhibitors, we first introduced an ACE2-derived peptide A1 to the N terminus of the HR2m peptide through a relatively long flexible linker (GGGGS)7 to generate A1-(GGGGS)7-HR2m (A1L35HR2m) (Figure 1A). We also constructed three other fusion peptides by swapping the positions of the A1 peptide and the HR2m peptide (HR2mL35A1) or by shortening the linker length to generate A1L5HR2m and A1HR2m (Figure 1A). All four fusion peptides were prepared through recombinant expression, and the purity of the obtained peptides was characterized using SDS-PAGE and liquid chromatography-mass spectrometry (Figures 1B and 1C).
Figure 1.
The design and characterization of A1L35HR2m, HR2mL35A1, A1L5HR2m, and A1HR2m
(A) Design of the A1L35HR2m, HR2mL35A1, A1L5HR2m, and A1HR2m peptides and the sequences of the A1 and HR2m peptides.
(B and C) Characterizations of A1L35HR2m, HR2mL35A1, A1L5HR2m, and A1HR2m by SDS-PAGE (B) and liquid chromatography-mass spectrometry (LC-MS) (C).
We next evaluated the inhibitory activities of A1L35HR2m, HR2mL35A1, A1L5HR2m, and A1LHR2m using the SARS-CoV-2 pseudotyped virus infection assay. As shown in Figure 2A, A1L35HR2m was highly potent in inhibiting SARS-CoV-2 infection with an IC50 value of 27 nM. HR2mL35A1, A1L5HR2m, and A1LHR2m all showed dramatically reduced antiviral activities. We then compared the inhibitory activities of A1L35HR2m, A1 (Figures S1A and S1B), HR2m (Figures S1C and S1D), and the mixture of A1/HR2m against SARS-CoV-2 pseudovirus infection. Again, A1L35HR2m had the highest activity among these tested peptides with an IC50 value of 21 nM, indicating that A1L35HR2m was at least 80-fold more potent than the other peptides (Figure 2B). These results together confirmed that the addition of an ACE2-derived peptide A1 to the N terminus of the HR2m peptide with a long flexible linker can indeed significantly increase the anti-SARS-CoV-2 activity.
Figure 2.
Conjugating the ACE2-derived peptide A1 to the N terminus of the HR2m peptide significantly enhanced the inhibitory activity against SARS-CoV-2 infection
(A) Inhibitory activity of A1L35HR2m, HR2mL35A1, A1L5HR2m, and A1HR2m against SARS-CoV-2 pseudovirus infection in Caco-2 cells.
(B) Inhibitory activity of A1L35HR2m, HR2m, A1, and A1/HR2m mixtures against SARS-CoV-2 pseudovirus infection in Caco-2 cells.
(C and D) A1L35HR2m, HR2m, and A1 inhibition of SARS-CoV-2 D614G S-mediated cell-cell fusion in Caco-2 cells (C) and Calu-3 cells (D). Each sample was tested in triplicate, and the data are presented as the mean ± SEM. Each experiment was repeated at least twice.
(E) Binding affinity of A1L35HR2m to HR1 was determined by BLI.
(F) The binding affinity of HR2m to HR1 was determined by BLI. The fitting curves were analyzed by ForteBio Data Analysis 12.0 software.
A1L35HR2m was more effective than the HR2m peptide in inhibiting SARS-CoV-2 D614G S-protein-mediated cell-cell fusion
We next explored the A1L35HR2m viral inhibition mechanism employing the S-protein-mediated cell-cell fusion assay, as described previously.31 We found that A1L35HR2m potently inhibited SARS-CoV-2 D614G S-protein-mediated cell-cell fusion between 293T/EGFP/S and Caco-2 cells with an IC50 value of 7.0 nM, which was approximately 62-fold more potent than that of HR2m (IC50 = 436 nM) (Figure 2C). We also found that the A1L35HR2m inhibitory activity (IC50 = 9.5 nM) of D614G S-protein-mediated cell-cell fusion between 293T/EGFP/S and Calu-3 cells was much higher than that of HR2m (IC50 = 745 nM) and A1 peptide (IC50 > 50 μM) (Figure 2D). These results suggested that the enhanced anti-SARS-CoV-2 activity of A1L35HR2m likely resulted from its improved ability to inhibit SARS-CoV-2 fusion with the target cells.
Improved anti-SARS-CoV-2 activity of A1L35HR2m may result from its high binding affinity to the HR1-derived peptide
Many studies have shown that SARS-CoV-2 HR2-derived peptides can competitively interact with the HR1 domain to block viral and target cell membrane fusion.12,21,32 To investigate whether the introduction of the A1 peptide in A1L35HR2m alters the original function of the HR2m peptide, we performed biolayer interferometry (BLI) experiments to characterize the HR1 peptide (Figure S1E) binding affinity. We found that the affinity of A1L35HR2m with HR1 was 3-fold higher (KD = 35.3 nM) than that of the original HR2m peptide (KD = 116 nM) (Figures 2E and 2F). This suggested that the addition of peptide A1 in A1L35HR2m indeed increased HR1 binding affinity, which probably contributed to the improved viral inhibitory activity.
To gain insights into the mechanism of the observed synergistic effects of the AL35HR2m peptide, we employed circular dichroism (CD) spectroscopy to investigate the secondary structure of the peptide complexes with HR1. Our results indicate that both AL35HR2m and HR2m peptides interacted with HR1 to form complexes with α-helical structures (Figures 3B and 3C). Notably, the AL35HR2m peptide exhibited a higher α-helicity upon binding to HR1 than the HR2m peptide did, suggesting that the enhanced activity of the AL35HR2m peptide may be attributed to its stronger interactions with HR1. In contrast, we observed no significant α-helical structure upon mixing A1 peptide and HR1 (Figure 3A), indicating that the A1 peptide may not directly interact with HR1.
Figure 3.
Secondary structures of A1, HR2m, and A1L35HR2m alone or in complex with HR1
(A–C) The secondary structures of A1 (A), HR2m (B), and A1L35HR2m (C) alone or in complex with HR1 were analyzed with CD. The experiments were performed twice, and representative data are shown. NA, not available.
To investigate the binding affinity of AL35HR2m with the RBD or S protein, we performed BLI assays. However, we did not observe any significant binding between AL35HR2m and the RBD or S protein under our experimental conditions. This lack of binding may be due to the weak affinity of AL35HR2m with the RBD or S protein that only becomes apparent when the S protein undergoes conformational changes that expose the HR1 domain for binding. Indeed, our results suggest that the inhibitory activity of AL35HR2m may be largely dependent on its ability to interact with the HR1 region of the S protein in its active conformation.
To further validate the mechanism of inhibition of AL35HR2m, we examined the ACE2/RBD and HR1/HR2 complex structures and then introduced different mutations in the A1 or HR2m peptide to construct three ACE2-HR2 fusion peptides (A3L35HR2m, A7L35HR2m, and A1L35HR2mdCdN) to investigate their antiviral activities (Figures S2A and S2B). A3L35HR2m, A7L35HR2m, and A1L35HR2mdCdN displayed significantly reduced inhibitory activity against SARS-CoV-2 D614G pseudovirus, with IC50 values of 0.6, 3.4, and 0.4 μM, respectively (Figure S2C). These values were about 15-, 78-, and 10-fold less potent than A1L35HR2m (Figures 4B and S2C). These results confirm that the key residues involved in the formation of either the ACE2/RBD or HR1/HR2 complex are critical for the synergistic inhibitory activity of A1L35HR2m.
Figure 4.
A1L35HR2m-Chol broadly and potently inhibited pseudotyped coronavirus infection
(A) The chemical structure of A1L35HR2m-Chol.
(B–K) Inhibitory activity of A1L35HR2m-Chol and A1L35HR2m in pseudovirus infection assays against SARS-CoV-2 D614G (B), SARS-CoV-2 Alpha variant (C), SARS-CoV-2 Beta variant (D), SARS-CoV-2 Delta variant (E), SARS-CoV-2 Omicron BA.1 variant (F), SARS-CoV-2 Omicron XBB variant (G), SARS-CoV-2 Omicron BQ.1.1 variant (H), SARS-CoV (I), MERS-CoV (J), and VSV-G (K). Each sample was tested in triplicate, and the data are presented as the mean ± SEM. Each experiment was repeated at least twice.
A1L35HR2m-Chol exhibited broad and potent inhibitory activity against different SARS-CoV-2 VOCs, SARS-CoV, and MERS-CoV
It has been reported that lipid modification of HR2-derived peptides can markedly increase the antiviral activity.20,21,22,32,33 Considering that the antiviral mechanism of A1L35HR2m is similar to that of HR2-derived peptides, we site-specifically modified A1L35HR2m at the C terminus with Chol to generate A1L35HR2m-Chol (Figures 4A and S3A). We also modified A1 and HR2m peptides with Chol to generate A1-Chol and HR2m-chol as control peptides (Figures S3B–S3E). We confirmed that HR2m-Chol was effective in inhibiting the infection of the pseudotyped SARS-CoV-2 and Middle East respiratory syndrome coronavirus (MERS-CoV) with IC50 values of 5.3 and 10.5 nM, respectively (Figures S4B and S4C), while A1-Chol showed a lower inhibitory activity with an IC50 of 2.32 μM (Figure S4A). In comparison, A1L35HR2m-Chol exhibited potent anti-SARS-CoV-2 inhibitory activity with an IC50 value reaching 134 pM (Figure S4D). The inhibitory activity of A1L35HR2m-Chol on SARS-CoV-2 pseudovirus infection was about 39- and 17,313-fold more potent than that of HR2m-Chol and A1-Chol, respectively. A1L35HR2m-Chol could also effectively block D614G S-protein-mediated cell-cell fusion between 293T/EGFP/S and Caco-2 or Calu-3 cells with IC50 values of 0.17 and 0.31 nM, respectively (Figure S4E).
We further evaluated the antiviral activities of A1L35HR2m-Chol and A1L35HR2m against pseudotyped SARS-CoV-2 D614G, SARS-CoV-2 VOCs, SARS-CoV, and MERS-CoV. We found that A1L35HR2m-Chol exhibited significantly more potent inhibitory activity against SARS-CoV-2 D614G, SARS-CoV-2 VOCs, and SARS-CoV (IC50 values ranging from 0.16 to 1.3 nM) than A1L35HR2m did (IC50 values ranging from 17 to 176 nM) (Figure 4B–4I). We also found that A1L35HR2m-Chol potently inhibited MERS-CoV (IC50 = 5.5 nM) (Figure 4J), while A1L35HR2m only had minimal activity against the MERS-CoV pseudovirus. Both A1L35HR2m-Chol and A1L35HR2m showed no inhibitory activity against the negative control vesicular stomatitis virus G (VSV-G) pseudovirus, which demonstrated the specificity of our fusion peptides (Figure 4K).
A1L35HR2m-Chol showed no in vitro or in vivo toxicity
To study the in vitro cytotoxicity of A1L35HR2m-Chol, Caco-2 or Calu-3 cells were treated with a concentration gradient of A1L35HR2m-Chol and A1L35HR2m for 48 h, and their viability was evaluated using the Cell Counting Kit-8. A1L35HR2m-Chol and A1L35HR2m showed no significant cytotoxicity to Caco-2 or Calu-3 cells at concentrations up to 2.5 and 10 μM, more than 18,656- and 57-fold higher than their IC50 values for SARS-CoV-2 inhibition (Figures S5A and S5B).
To investigate the potential in vivo toxicity of A1L35HR2m-Chol, BALB/c mice were intranasally administrated with PBS (n = 3), 5 mg/kg A1L35HR2m (n = 3), 5 mg/kg A1L35HR2m-Chol (n = 3), or 20 mg/kg A1L35HR2m-Chol (n = 3). The levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl transferase (GGT), and creatinine in the sera of the peptide-treated mice were measured. The results indicated that the levels of ALT (Figure S5C), AST (Figure S5D), GGT (Figure S5E), or creatinine (Figure S5F) in the sera of the mice in A1L35HR2m, A1L35HR2m-Chol, and PBS treated groups showed no significant difference (p>0.05) at all time points, suggesting that administration of A1L35HR2m-Chol at a low or high dose had no impact on the hepatic and renal functions of mice.
A1L35HR2m-Chol distributed in respiratory tract tissue and exhibited long-lasting ex vivo anti-SARS-CoV-2 activity
Previous studies have shown that lipid HIV-1 fusion inhibitors could enhance the ex vivo anti-HIV-1 activity and improve in vivo stability.34,35 Here, we investigate whether A1L35HR2m-Chol also exhibits enhanced ex vivo anti-SARS-CoV-2 activity and extended in vivo half-life (t1/2). A1L35HR2m-Chol (5 mg/kg, n = 3) or A1L35HR2m (5 mg/kg, n = 3) was administered intranasally to mice, and the plasma samples at each time point were diluted to determine the SARS-CoV-2 pseudovirus infection inhibition activity. The peptide concentration was then estimated based on the dilution factor that can achieve 50% pseudovirus infection inhibition (Figures 5A and 5B). Plasma samples from mice treated with A1L35HR2m-Chol showed peak ex vivo inhibition activity, corresponding to peak peptide concentration, at 8 h post-administration, with a mean dilution of 10,284-fold that inhibited 50% of SARS-CoV-2 D614G pseudovirus infection. A1L35HR2m-Chol appeared to maintain a high plasma concentration even after 336 h. In contrast, the peak ex vivo inhibition activity, corresponding to peak peptide concentration, of A1L35HR2m was significantly lower, with rapid clearance 8 h post-administration (Figures 5A and 5B). Based on the estimated concentration, we calculated the pharmacokinetic parameters of A1L35HR2m-Chol and A1L35HR2m by using PKSolver software. The plasma t1/2 of A1L35HR2m-Chol was approximately 81.8 h, 30-fold longer than that of A1L35HR2m (t1/2 = 2.6 h) (Figure 5B; Table S1). The mean area under the concentration-time curve of A1L35HR2m-Chol was 1,172 μg/mL∗h, approximately 58-fold higher than that of A1L35HR2m (20 μg/mL∗h) (Table S1).
Figure 5.
Ex vivo anti-SARS-CoV-2 D614G activity and peptide concentration in plasma or tissue homogenate samples of the peptide-treated mice
(A and C) Dilution fold of the plasma (A) or tissue homogenate (C) samples from mice intranasally treated with A1L35HR2m (5 mg/kg, n = 3) or A1L35HR2m-Chol (5 mg/kg, n = 3) that can achieve 50% SARS-CoV-2 D614G inhibition.
(B and D) Estimated concentrations of functionally active peptides in the plasma (B) or tissue homogenate (D) samples. Each sample was tested in triplicate, and the data are presented as the mean ± SEM. Each experiment was repeated at least twice.
We also found high concentrations of A1L35HR2m-Chol in the lung, nose, and trachea, organs that are particularly relevant in the course of SARS-CoV-2 infection. The peptide concentration in the lung, nose, and trachea peaked at 8 h, with mean dilutions of 58,770-, 18,231-, and 1,959-fold that inhibited 50% of SARS-CoV-2 D614G pseudovirus infection. The t1/2 of A1L35HR2m-Chol in the lung, nose, and trachea were about 89.8, 49.0, and 10.6 h, respectively (Figures 5C and 5D). These results suggested that Chol conjugation to the C terminus of A1L35HR2m can significantly prolong the in vivo t1/2. A similar phenomenon has been observed for other fusion peptide inhibitors.34,35
A1L35HR2m-Chol potently inhibited authentic SARS-CoV-2 variants in vitro and in hACE2 transgenic mice
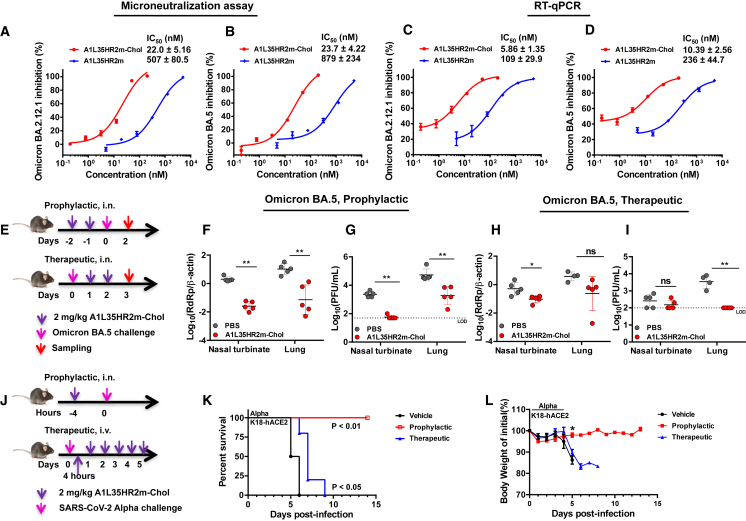
We evaluated the antiviral efficacy of A1L35HR2m-Chol against authentic Omicron BA.2.12.1 and BA.5 strains. We first performed microneutralization assays on VeroE6-TMPRSS2 cells. The results indicated that A1L35HR2m could inhibit BA.2.12.1 and BA.5 replication with IC50 values of 507 and 879 nM, respectively, while A1L35HR2m-Chol potently suppressed BA.2.12.1 and BA.5 replication with IC50 values of 22.0 and 23.7 nM, respectively (Figures 6A and 6B). We also conducted viral load reduction assays of Omicron BA.2.12.1 and BA.5 in VeroE6-TMPRSS2 cells. The inhibitory activity was analyzed by quantifying the viral yields in the cell culture supernatants using RT-qPCR. We consistently found that A1L35HR2m-Chol was significantly more potent in inhibiting authentic Omicron BA.2.12.1 and BA.5 infection (IC50 = 5.9 nM for BA.2.12.1 and 10.4 nM for BA.5) than A1L35HR2m (IC50 = 109 nM for BA.2.12.1 and 236 nM for BA.5) (Figures 6C and 6D).
Figure 6.
A1L35HR2m-Chol potently inhibited authentic SARS-CoV-2 in vitro and in vivo
(A and B) A1L35HR2m and A1L35HR2m-Chol inhibition of authentic Omicron BA.2.12.1 (A) and BA.5 (B) in microneutralization assays.
(C and D) A1L35HR2m and A1L35HR2m-Chol inhibition of authentic Omicron BA.2.12.1 (C) and BA.5 (D) in viral load reduction assays. The viral load was quantified by RT-qPCR.
(E) Experiment design of the A1L35HR2m-Chol prophylactic and therapeutic efficacy study against Omicron BA.5. K18-hACE2 transgenic mice were challenged with Omicron BA.5 (10,000 PFUs) at day 0. In the prophylactic study, A1L35HR2m-Chol (2 mg/kg) was intranasally administered 2 days and 1 day before infection. In the therapeutic study, A1L35HR2m-Chol (2 mg/kg) was intranasally administered at days 1 and 2 post-virus infection.
(F and G) In the prophylactic experiment, nasal turbinates and lungs were collected 2 days post-infection and subjected to viral load (F) and infectious viral titer (G) detection.
(H and I) In the therapeutic experiment, nasal turbinates and lungs were collected 3 days post-infection and subjected to viral load (H) and infectious virus titer (I) detection.
(J) Schematic diagram of the prophylactic and therapeutic experimental design to evaluate the efficacy of A1L35HR2m-Chol in protecting mice from SARS-CoV-2 Alpha lethal infection.
(K) Survival rate monitoring of the mice treated with PBS or prophylactic or therapeutic A1L35HR2m-Chol treatment.
(L) Daily body weights of the mice.
(A–D) Each sample was tested in triplicate, and the data are presented as the mean ± SEM. Each experiment was repeated at least twice. (F–L) PBS, n = 5; A1L35HR2m-Chol, n = 5. The dashed lines in (G) and (I) represent the limit of detection (LOD). Unpaired non-parametric Mann-Whitney test (F–I and L) or log-rank test (K) was performed. ∗p < 0.05 and ∗∗p < 0.01. ns denotes no significance.
We next used an established K18-hACE2 transgenic mouse model to evaluate the prophylactic and therapeutic efficacy of A1L35HR2m-Chol (Figure 6E).36 In the prophylactic study, A1L35HR2m-Chol (2 mg/kg, n = 5) was administered intranasally to mice twice 2 days and 1 day before viral infection, respectively. On day 0, Omicron BA.5 (10,000 plaque-forming units [PFUs]) was used to challenge each mouse via the intranasal route. At 2 days post-infection (dpi), mice were sacrificed for viral RNA and infectious viral titer analyses. We found that mice receiving A1L35HR2m-Chol treatment had significantly lower viral RNA copies in their lung homogenates (>2 log10 reduction, p < 0.01; Figure 6F) and nasal turbinates (>1 log10 reduction, p < 0.01; Figure 6F) than mice in the control group. We also found that the infectious viral titers in A1L35HR2m-Chol-treated mice were significantly lower than those in the control group (>2 log10 reduction, p < 0.01; Figure 6G). Remarkably, almost no infectious viral titer was detected in the nasal turbinates of mice receiving A1L35HR2m-Chol, which indicates that A1L35HR2m-Chol has the potential to inhibit viral transmission.
For the therapeutic study, all mice were first challenged with Omicron BA.5 (10,000 PFUs) virus on day 0, followed by intranasal delivery of A1L35HR2m-Chol (2 mg/kg, n = 5) at 1 and 2 dpi, respectively. At 3 dpi, mice were sacrificed for viral RNA and infectious viral titer analyses. We found that the nasal turbinate viral RNA copy number was significantly reduced (p < 0.05) in mice receiving A1L35HR2m-Chol compared to the control group, while the lung homogenate viral RNA copy number did not show a significant difference between the two groups (Figure 6H). Importantly, A1L35HR2m-Chol completely suppressed the infectious viral titer in the lung homogenate without detectable live SARS-CoV-2 particles (Figure 6I). These results suggest that A1L35HR2m-Chol exhibited potent protection against SARS-CoV-2 Omicron BA.5 infection in both prophylactic and therapeutic settings.
We subsequently evaluated the efficacy of A1L35HR2m-Chol in protecting K18-hACE2 transgenic mice from the lethal Alpha variant infection. In the prophylactic group, five mice were administrated A1L35HR2m-Chol (2 mg/kg, n = 5) once intranasally 4 h before the virus challenge. In the therapeutic group, five mice received intravenous treatment with A1L35HR2m-Chol (2 mg/kg, n = 5) 4 h after the infection and once daily for a total of 6 times. PBS (n = 5) was intranasally administrated in the control group. The mice were challenged on day 0 with SARS-CoV-2 Alpha. Body weight changes and mice survival were monitored until 14 dpi or death (Figure 6J). In the prophylactic group, mice receiving A1L35HR2m-Chol showed complete protection from the lethal Alpha infection, demonstrating a 100% survival protection rate. Mice in the PBS group showed body weight loss from 4 dpi, and all died at 6 dpi (0% survival). Mice in the therapeutic group showed body weight loss from 5 dpi and survived longer compared to mice in the PBS group (Figures 6K and 6L).
During the course of the study, Omicron EG.5.1 emerged as the dominant SARS-CoV-2 strain. We then evaluated the antiviral efficacy of A1L35HR2m-Chol against Omicron EG.5.1. We first performed microneutralization and viral load reduction assays using VeroE6-TMPRSS2 cells. The results showed that A1L35HR2m-Chol can significantly suppress EG.5.1 replication with an IC50 value of 14.4 nM (Figure 7A). A1L35HR2m-Chol could also significantly reduce the viral load in the infectious supernatant with an IC50 value of 3.1 nM (Figure 7B). We subsequently evaluated the in vivo prophylactic and therapeutic efficacy of A1L35HR2m-Chol against EG.5.1 using the K18-hACE2 mouse model. The mice were intranasally administered a single dose of A1L35HR2m-Chol (2 mg/kg, n = 5) 4 h before the viral infection in the prophylactic group, while mice were treated with A1L35HR2m-Chol intranasally (2 mg/kg, n = 5) 4 and 28 h post-infection (hpi) in the therapeutic group. At 48 hpi, mice were sacrificed for infectious viral titer analyses (Figure 7C). We found that A1L35HR2m-Chol could reduce the viral titer in the nasal turbinate compared to that of the control group both prophylactically and therapeutically. Notably, 4 out of 5 mice in the prophylactic group and 2 out of 5 mice in the therapeutic group showed undetectable virus titer in the nasal turbinate, whereas all PBS-treated mice consistently achieved >105 PFU/mL virus titers. The results indicated that A1L35HR2m-Chol can eliminate viruses in the upper respiratory tract. Interestingly, 4 out of 5 vehicle-treated mice had no detectable viral titer in the lung homogenate (<500 PFUs), suggesting that the EG.5.1 mainly infected the upper respiratory tract and did not readily infect the lung tissue (Figure 7D). These data demonstrated that A1L35HR2m-Chol can effectively inhibit the newly emerged, dominating EG.5.1 variant in vitro and in vivo.
Figure 7.
A1L35HR2m-Chol potently inhibited Omicron EG.5.1 in vitro and in vivo
(A and B) A1L35HR2m-Chol inhibition of authentic Omicron EG.5.1 in microneutralization (A) and viral load reduction (B) assays. The viral load was quantified by RT-qPCR. Each sample was tested in triplicate, and the data are presented as the mean ± SEM. Each experiment was repeated at least twice.
(C) Experiment design of the A1L35HR2m-Chol prophylactic and therapeutic efficacy studies against Omicron EG.5.1. K18-hACE2 transgenic mice were challenged with Omicron EG.5.1 at day 0. In the prophylactic study, A1L35HR2m-Chol (2 mg/kg) was intranasally administered 4 h before infection. In the therapeutic study, A1L35HR2m-Chol (2 mg/kg) was intranasally administered 4 and 28 h post-viral infection.
(D) Tissue samples were collected 48 h after viral infection and subjected to infectious viral titer detection by plaque assay. PBS, n = 5; A1L35HR2m-Chol, n = 5. The dashed line represents the LOD. Unpaired non-parametric Mann-Whitney test was performed. ∗∗p < 0.01. ns denotes no significance.
Discussion
As SARS-CoV-2 continues to spread globally, new variants are still emerging rapidly, posing significant challenges to current vaccines and therapeutics. The membrane fusion mechanism between virus and target cells is very conserved among different coronaviruses, and the six-helix bundle formation between the HR1 domain and HR2 domain plays a key role in driving the membrane fusion.12,15 Since the sequences of HR1 and HR2 are well conserved among different coronaviruses, peptides derived from HR1 and HR2 are thus considered promising antiviral agents that can potentially overcome the viral mutational escape problem.14,21
Aiming to explore novel HR2 peptide engineering strategies that could lead to antivirals with enhanced inhibitory activity, we introduced the short ACE2-derived peptide A1 to the HR2-derived peptide HR2m through a long flexible linker (GGGGS)7 to construct A1L35HR2m. The RBD and the HR1 are located quite far away (∼100 Å) from each other on the S protein; we thus chose the long linker to prepare the A1 and HR2m fusion peptide for potential synergistic effect. We found that a long linker between A1 and HR2m peptide (A1L35HR2m) possessed high pseudovirus inhibitory activity even though the ACE2-derived peptide A1 cannot effectively inhibit pseudovirus infection and the HR2m peptide only has minimal inhibitory activity. The long flexible linker probably ensures that A1/RBD and HR2m/HR1 interactions could happen within a short time window to inhibit viral infection efficiently. Our binding study indicated that the A1 peptide enhanced the HR2m peptide binding affinity to the HR1 peptide, which contributed to the improved inhibitory activity. The A1 peptide may also weakly interact with viral S protein, which could enrich the local concentration of A1L35HR2m around the S protein to boost the inhibitory activity eventually. However, this explanation may be difficult to prove since neither the A1 peptide nor A1L35HR2m showed obvious binding to the S protein in the BLI binding experiments. We showed that abolishing either key residue involved in ACE2/RBD or HR2/HR1 binding would significantly diminish the antiviral activity of A1L35HR2m, demonstrating that both the A1 and HR2m peptides are important contributors to the observed enhanced antiviral activity of A1L35HR2m.
Further A1L35HR2m modification with Chol at the C terminus markedly increased the inhibitory activity of A1L35HR2m-Chol, with IC50 values reaching 0.16–5.5 nM against different SARS-CoV-2 VOCs, SARS-CoV, and MERS-CoV pseudoviruses. A1L35HR2m-Chol was found to effectively inhibit authentic Omicron BA.2.12.1, BA.5, and EG.5.1, further demonstrating its broad-spectrum activity against different coronaviruses and its robust resistance to viral mutational escape. Chol modification probably helps A1L35HR2m-Chol to bind to the cell surface or viral envelope. In either case, this will have a reduction in the dimension of the A1L35HR2m-Chol peptide diffusion process, thus accelerating its binding to HR1 and leading to improved inhibitory activity. If A1L35HR2m-Chol is bound to the cell, then the 3-dimensional in solution diffusion is converted to a 2-dimensional diffusion on the membrane to search for the viral HR1 domain during the fusion process. If A1L35HR2m-Chol is bound to the viral envelope, then the 3-dimensional search in solution is converted to an intravirus search for the viral HR1 domain. Chol may also directly enhance HR1 binding affinity, which can directly contribute to improved antiviral inhibitory activity.20
Importantly, we found that A1L35HR2m-Chol distributed in respiratory tract tissues and exhibited long-lasting ex vivo anti-SARS-CoV-2 activity, with a prolonged in vivo plasma t1/2 of 81.8 h. We think the prolonged in vivo t1/2 is mainly due to the Chol modification. Chol has been shown to bind to serum albumin, resulting in the increased circulation time of the bound peptide.37 Chol modification could also cause A1L35HR2m-Chol to form micelles in solution, leading to slower clearance. Indeed, the dynamic light scattering experiments showed that A1L35HR2m-Chol formed micelles (165 nm in diameter) in aqueous solution (Figure S6). Chol modification might also help A1L35HR2m-Chol to stick to endothelial cells and the reticuloendothelial system by physical absorption followed by slow release, which could also extend the in vivo t1/2 of A1L35HR2m-Chol. This dramatically increased t1/2 probably contributed significantly to the in vivo antiviral activity of A1L35HR2m-Chol. Indeed, we found that intranasal administration of A1L35HR2m-Chol can potently inhibit Omicron BA.5 and EG.5.1 infection and replication in the hACE2 transgenic mouse model both prophylactically and therapeutically.
In summary, we have established a method to improve HR2 peptide viral inhibitory activity drastically, and the engineered A1L35HR2m-Chol potently inhibits different SARS-CoV-2 VOCs, SARS-CoV, and MERS-CoV. We believe that A1L35HR2m-Chol is a promising antiviral molecule that will retain inhibitory activity against future emerging SARS-CoV-2 variants and thus is worth further translational investigation.
Limitations of the study
We elucidated the antiviral mechanism of A1L35HR2m by disrupting key residues involved in A1/RBD and HR2m/HR1 interactions, conclusively establishing the indispensability of both A1 and HR2m for the enhanced activity. We proved that A1L35HR2m likely inhibits viral infection by inhibiting the viral and cell membrane fusion process by using S-protein-mediated cell-cell fusion inhibition assay and CD experiments. However, the specific contribution of A1 to the heightened activity of A1L35HR2m remains unclear at this stage. The intricate dynamics of the S protein prefusion conformation transitioning to the postfusion conformation are not fully understood presently. Consequently, delving into the detailed molecular impact of A1L35HR2m on this process proves challenging at this point.
To assess the efficacy of A1L35HR2m-Chol in a more challenging context, we conducted an in vivo efficacy study against the lethal Alpha variant infection. The results revealed that when A1L35HR2m-Chol was intranasally administered 4 h before viral infection, it provided complete protection to all mice, ensuring their survival throughout the study—a testament to its robust prophylactic efficacy. Notably, when A1L35HR2m-Chol was administered 24 h prior to viral infection, the survival rate dropped from 100% to 40% (Figure S7). This underscores the significance of a high concentration of A1L35HR2m-Chol in the respiratory tract for effectively preventing Alpha variant infection. In the therapeutic efficacy study, the intravenous administration of A1L35HR2m-Chol succeeded in extending the survival time of mice; however, the overall therapeutic efficacy was not as pronounced as the prophylactic efficacy. The lethality of K18-hACE2 mice is largely due to neuro-invasion and encephalitis other than pulmonary dysfunction.38 While A1L35HR2m-Chol exhibited potent efficacy in inhibiting viral replication, its potential inability to traverse the blood-brain barrier compromises its effectiveness in inhibiting virus replication within the brain. Consequently, future studies may benefit from employing a more physiologically relevant model. Nonetheless, our results imply that A1L35HR2m-Chol could potentially contribute to preventing future SARS-CoV-2-related outbreaks.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SARS-CoV-2 N protein | In house | N/A |
| goat anti-rabbit Alexa Fluor 488-conjugated IgG (H + L) | Thermo Fisher | Cat#: A-11008; RRID: AB_143165 |
| Bacterial and virus strains | ||
| BL21 | In house | N/A |
| SARS-CoV-2 Alpha | Shuai et al. 202239 | GenBank: OM212469 |
| SARS-CoV-2 Omicron BA.2.12.1 | Chon et al. 202340 | EPI_ISL_13777659 |
| SARS-CoV-2 Omicron BA.5 | Chon et al. 202340 | EPI_ISL_13777658 |
| SARS-CoV-2 Omicron EG.5.1 | Isolated from nasopharyngeal swab of a laboratory confirmed patient | EPL_ISL_18461518 |
| Chemicals, peptides, and recombinant proteins | ||
| A1 | This paper | N/A |
| HR2m | This paper | N/A |
| HR1 | This paper | N/A |
| HR2m-Chol | This paper | N/A |
| A1-Chol | This paper | N/A |
| A1L35HR2m-Chol | This paper | N/A |
| A1L35HR2m | This paper | N/A |
| A1L35HR2mdCdN | This paper | N/A |
| A3L35HR2m | This paper | N/A |
| A7L35HR2m | This paper | N/A |
| isopropyl 1-thio-β-D-galactopyranoside | Sangon Biotech | Cat#: A600168-0005 |
| Vigofect | Vigorous Biotech | Cat#: T001 |
| EZ-Link NHS-PEG12-Biotin | Thermo Fisher Scientific | Cat#: 21313 |
| Dulbecco’s Modified Eagle’s Medium | Gibco | Cat#: 11965-092 |
| Fetal Bovine Serum | Gibco | Cat#: 16140071 |
| G418 | MCE | Cat#: HY-17561 |
| Phosphate buffered saline | Gibco | Cat#: 10010-023 |
| Penicillin/Streptomycin | Gibco | Cat#: 15140122 |
| 0.25% Trypsin-EDTA | Gibco | Cat#: 25200-072 |
| Critical commercial assays | ||
| Luciferase assay system | Promega | Cat#: E1501 |
| Cell Counting Kit-8 | Dojindo | Cat#: CK04 |
| Alanine aminotransferase Assay Kit | NJJCBIO | Cat#: C009-2-1 |
| Aspartate aminotransferase Assay Kit | NJJCBIO | Cat#: C010-2-1 |
| γ- Glutamyl transferase Assay Kit | NJJCBIO | Cat#: C017-2-1 |
| creatinine (Cr) Assay kit | NJJCBIO | Cat#: C011-2-1 |
| RNeasy mini kit | Qiagen | Cat#: 74106 |
| One Step TB Green® PrimeScript™ RT-PCR Kit II (Perfect Real Time) |
Takara | Cat#: RR086A |
| Experimental models: Cell lines | ||
| 293T | ATCC | CRL-3216 |
| Caco-2 | Cai et al., 202029 | N/A |
| Calu-3 | Xia et al., 202014 | N/A |
| VeroE6-TMPRSS2 | Japanese Collection of Research Bioresources (JCRB) Cell Bank | JCRB1819 |
| Experimental models: Organisms/strains | ||
| Balb/c mice | Laboratory Animal Resources Center of Westlake University | N/A |
| K18-hACE2 C57BL/6J mice | The Jackson Laboratory | Strain#: 034860 |
| Oligonucleotides | ||
| SARS-CoV-2 RdRp forward sequence: 5′-CGCATACAGTCTTRCAGGCT-3′ |
Integrated DNA Technologies | N/A |
| SARS-CoV-2 RdRp reverse sequence: 5′-GTGTGATGTTGAWATGACATGGTC-3′ | Integrated DNA Technologies | N/A |
| Housekeeping gene mouse beta-actin sequence forward: 5′- ACGGCCAGGTCATCACTATTG-3′ | Integrated DNA Technologies | N/A |
| Housekeeping gene mouse beta-actin reverse sequence: 5′- CAAGAAGGAAGGCTGGAAAAG-3′ | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pcDNA3.1-SARS-CoV-S | Xia et al., 201914 | GenBank: ABD72979.1 |
| pcDNA3.1-MERS-CoV-S | MiaoLingBio | N/A |
| pcDNA3.1-SARS-CoV-2-S | Xia et al., 202020 | GenBank: QHD43416 |
| pcDNA3.1-SARS-CoV-2 D614G S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2 Beta S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2 Delta S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2 Omicron BA.1 S | Genewiz | N/A |
| pcDNA3.1-SARS-CoV-2 Omicron XBB S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2 Omicron BQ.1.1 S | This paper | N/A |
| pcDNA3.1-EGFP | This paper | N/A |
| pcDNA3.1-EGFP- SARS-CoV-2 D614G S | This paper | N/A |
| pET28a-A1L35HR2m | Genewiz | N/A |
| pET28a-HR2mL35A1 | Genewiz | N/A |
| pET28a-A1L5HR2m | Genewiz | N/A |
| pET28a-A1HR2m | Genewiz | N/A |
| pET28a-A3L35HR2m | This paper | N/A |
| pET28a-A7L35HR2m | This paper | N/A |
| pET28a-A1L35HR2mdCdN | This paper | N/A |
| pNL4-3.Luc.RE | Xia et al., 202020 | N/A |
| Software and algorithms | ||
| GraphPad Prism Version 6.02 | GraphPad | https://www.graphpad.com |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij |
| Other | ||
| Ni-NTA beads | Smart-Lifesciences | Cat#: SA004100 |
| Amicon column | Millipore | Cat#: UFC5003BK |
| TissueLyser II | Qiagen | https://www.qiagen.com/au/listpages/ez1-cards/tissuelyser-ii/ |
| SAPPHIRE FL BIOMOLECULAR IMAGER | AZURE Biosystems | Cat#: IS4000 |
| Stainless Steel Beads, 5mm | Qiagen | Cat#: 69989 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Bobo Dang (dangbobo@westlake.edu.cn).
Materials availability
All requests for resources and reagents should be directed to the lead contact author. All reagents, which includes peptides, proteins, plasmids, and virus, will be made available on request after completion of a Material Transfer Agreement for non-commercial usage.
Data and code availability
All data reported in this study will be shared upon request from the lead contact. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Cell lines
293T, Caco-2, Calu-3 and VeroE6-TMPRSS2 were cultured in DMEM with 10% fetal bovine serum (FBS). All cell lines were incubated and routinely maintained at 37°C in humidified air containing 5% CO2.
Virus
Live SARS-CoV-2 variants were isolated from the respiratory tract specimens of COVID-19 patients in Hong Kong.39 All experimental protocols involving live SARS-CoV-2 followed the approved standard operating procedures of HKU Biosafety Level 3 facility.
Mice
The use of animals was approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong.39 Heterogeneous K18-hACE2 C57BL/6J mice were obtained from The Jackson Laboratory. 6- to 10-week-old male and female K18-hACE2 were used for all in vivo efficacy experiments. Balb/c mice (aged 6–8 weeks) for the safety and pharmacokinetics study were obtained from the Laboratory Animal Resources Center of Westlake University and the experiments were approved by the Institutional Animal Care and Use Committee (IACUC) guidelines of Westlake University. Animals were kept in cages with individual ventilation with 65% humidity and ambient temperature ranging between 21°C and 23°C with 12-hour-interval day/night cycle for housing and husbandry.
Methods details
Chemicals
All chemicals were purchased from commercial vendors and used without further purification. Fmoc-L-Gly-OH, Fmoc-L-Leu-OH, Fmoc-L-Ile-OH, Fmoc-L-Val-OH, Fmoc-L-Lys(Boc)-OH, Fmoc-L-Lys(Alloc)-OH, Fmoc-L-Ala-OH, Fmoc-L-Cys(Trt)-OH, Fmoc-L-Gln(Trt)-OH, Fmoc-L-Asn(Trt)-OH, Fmoc-L-Glu(OtBu)-OH, Fmoc-L-Asp(OtBu)-OH, Fmoc-L-Arg(Pbf)-OH, Fmoc-L-Phe-OH, Fmoc-L-Trp(Boc)-OH, Fmoc-His(Boc)-OH, Fmoc-L-Ser(tBu)-OH, Fmoc-L-Thr(tBu)-OH, Fmoc-L-Tyr(tBu)-OH, Fmoc-L-Pro-OH, Boc-L-Asp(OtBu)-OH, and Fmoc-L-Met-OH were purchased from GL Biochem. Fmoc-Rink amide resin was purchased from Tjhecheng. Bromoacetic acid, [2-[2-(Fmoc-amino)ethoxy]ethoxy]acetic acid, N,N′-diisopropylcarbodiimide (DIC), ethyl cyanoglyoxylate-2-oxime (Oxyma), N,N-dimethylformamide (DMF), dichloromethane (DCM), diethyl ether, trifluoroacetic acid, triisoproprylsilane, 4-methyl pepiridine and HPLC-grade acetonitrile were obtained from Energy Chemical.
Peptides
Peptides were synthesized by solid phase peptide synthesis (SPPS) on a Liberty Blue automated microwave peptide synthesizer (CEM Corporation, North Carolina, USA). Resins were swelled in DMF for 5 min before use. The Fmoc groups of both the resin and the assembled amino acids were removed by treatment with 20% 4-methylpiperidine in DMF at 40°C. Amino acid coupling was carried out at 80°C with 5 equiv. Fmoc-amino acids, 5 equiv. Oxyma, and 5 equiv. DIC. The coupling reaction for Fmoc-His (Trt)-OH was carried out at 50°C for 10 min to prevent side reactions at high temperature. After completion of the stepwise SPPS, the resins were washed thoroughly with DCM and dried under vacuum. The peptides were then cleaved off the resins, and side chains were deprotected by treatment with 2% (v/v) water and 2.5% (v/v) triisopropylsilane in neat trifluoroacetic acid (TFA) for 2.5 h at room temperature. The resulting solution containing peptide was precipitated and washed with cold diethyl ether three times. The obtained solid was dissolved in 50% H2O: 50% acetonitrile containing 0.1% TFA and lyophilized. The crude peptides were purified by semipreparative reversed-phase high-pressure liquid chromatography (RP-HPLC), and the purities of the peptides were more than 95%, as determined by RP-HPLC and electrospray ionization mass spectrometry (ESI-MS) characterization. The peptides were solubilized using dimethyl sulfoxide (DMSO) or PBS.
Plasmids and cells
The envelope-expressing plasmids pcDNA3.1-SARS-CoV-S and pcDNA3.1-SARS-CoV-2-S and luciferase-expressing plasmid pNL4-3.Luc.RE were kindly provided by Drs. Shibo Jiang and Lu Lu at Shanghai Medical College of Fudan University. The SARS-CoV-2 S mutant envelope-expressing plasmids were generated by using overlap PCR. Cell lines, including 293T, Caco-2 and Calu-3, were also kindly provided by Drs. Shibo Jiang and Lu Lu and cultured in DMEM with 10% fetal bovine serum (FBS).
Expression and purification of fusion peptides
The genes encoding A1L35HR2m, HR2mL35A1, A1L5HR2m and A1HR2m were synthesized and cloned into a pET28a vector by Genewiz, Suzhou, China. The genes encoding A3L35HR2m, A7L35HR2m and A1L35HR2mdCdN were generated by using overlap PCR and cloned into a pET28a. pET28a-A1L35HR2m, pET28a-HR2mL35A1, pET28a-A1L5HR2m, pET28a-A1HR2m, pET28a-A3L35HR2m, pET28a-A7L35HR2m and pET28a-A1L35HR2mdCdN plasmids were transformed separately into E. coli BL21 (DE3). The cells were shaken at 37°C in LB medium until the OD600 reached 0.8. Protein expression was induced with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside for 6 h at 37°C. Then, the cells were harvested, sonicated in PBS with 1% Triton and centrifuged at 1,2000 × g for 10 min. The supernatant was loaded onto Ni-NTA beads (Smart-Lifesciences, Cat. SA004100) and washed with 10 mM imidazole in PBS. Proteins were then eluted with elution buffer (250 mM imidazole in PBS). The peak fractions were collected and concentrated with a 3 kDa ultrafiltration tube (Millipore, Germany). The proteins were then analyzed by SDS‒PAGE and LC‒MS.
Preparation of A1L35HR2m-Chol protein
His-TEV-A1L35HR2m-Cys protein was expressed in E. coli BL21 and purified by Ni-NTA beads. The purified His-TEV-A1L35HR2m-Cys protein was digested by TEV protease in 50 mM Tris-HCl (pH = 7.4) with 1 mM DTT at 4°C overnight. Then, the A1L35HR2m-Cys protein was purified again by semipreparative RP-HPLC and lyophilized, resulting in a white powder. The bromoacetyl-PEG4-chol molecule was synthesized on the H-Rink-Amid-ChemMatrix resin and then purified by semipreparative RP-HPLC and lyophilized, resulting in a light yellow powder. The A1L35HR2m-Chol protein was synthesized via a nucleophilic substitution reaction between the terminal thiol group on the peptide cysteine residue and bromoacetyl-PEG4-chol molecule in acetonitrile/PBS (70/30: v/v) (pH 8.0) mixture buffer. The final product was purified by semipreparative RP-HPLC and lyophilized, resulting in a white powder. The identity of the A1L35HR2m-Chol protein was verified by LC-MS (Figure S3A).
Inhibition of pseudotyped virus infection
SARS-CoV-2 and its variants were generated as described previously.20 Briefly, 293T cells were cotransfected with the plasmid encoding the S protein of SARS-CoV-2 or its variants and the luciferase-expressing HIV-1 genome plasmid by using Vigofect reagent (Vigorous Biotech, Beijing, China). The supernatants were collected at 48 h post-transfection, centrifuged at 3,000 × g for 10 min and stored at −80°C for single-cycle infection. The antiviral activities of the proteins were determined by using Caco-2 cells (104/well) infected by SARS-CoV-2 pseudovirus mixed with the inhibitors. The supernatants were replaced with fresh DMEM with 10% FBS at 12 h post-infection, and the cells were cultured for an additional 48 h at 37°C. The cells were lysed, and the luciferase activity was measured by using a luciferase kit (Promega, Madison, WI, USA) on a microplate reader (Thermo, Variokan LUX). All best-fit inhibition curves were drawn using GraphPad Prism.
Binding affinity determination using biolayer interferometry (BLI)
The HR1 peptide was biotinylated at a theoretical 1:3 M ratio with EZ-Link NHS-PEG12-Biotin (Thermo Fisher Scientific) according to the manufacturer’s instructions. The unreacted biotin was removed by ultrafiltration with an Amicon column (3 kDa MWCO, Millipore). For kinetics analyses, HR1 was captured on streptavidin biosensors. Biotinylated HR1 was diluted to 5 μg/mL in dilution buffer (PBS with 0.02% Tween 20). The background signal was measured using a reference sensor with HR1 loading but no inhibitor binding and was subtracted from the corresponding inhibitor binding sensor. Curve fitting was performed using ForteBio data analysis software. Mean kon, koff, and KD values were determined by averaging all binding curves that matched the theoretical fit with an R2 value of 0.96.
Inhibition of SARS-CoV-2 D614G S-mediated cell‒cell fusion
The establishment and detection of the cell–cell fusion assay was similar to that for determining HIV-1 Env-mediated cell‒cell fusion.41 In brief, 293T cells were transfected with the plasmid pcDNA3.1 encoding EGFP and SARS-CoV-2 D614G S protein (293T/EGFP/S protein) or encoding the EGFP (293T/EGFP) and used as effector cells. Calu-3 cells or Caco-2 cells were used as target cells. For SARS-CoV-2 D614G S-mediated cell–cell fusion assays, target cells were incubated at 37°C overnight, followed by the addition of 293T/EGFP/S cells in the presence or absence of the tested inhibitors at different concentrations. After coculture at 37°C for 5 h, three fields were randomly selected in each well to count the number of fused and unfused cells under an inverted fluorescence microscope (ZEISS, Germany).
Circular dichroism (CD) spectroscopy
CD spectroscopy was conducted, as previously described.18 Briefly, the HR1 peptide was incubated at equal molar concentration with A1, HR2m or A1L35HR2m at 37°C for 30 min. These four individual peptides were used as control. The final concentration of each peptide was 10 μM in phosphate buffer (PB, pH 7.2). The CD spectra were measured on a Chirascan Spectrometer (model Chirascan V100; Applied Photophysics Ltd., UK), using a 1-nm bandwidth with a 1-nm step resolution from 190 to 280 nm at room temperature. The spectra were corrected by subtraction of a solvent blank (PB).
Cell viability assay
The potential cytotoxicity of peptides to Calu-3 and Caco-2 cells was measured by using the Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer’s instructions. Briefly, 100 μL of peptides at the indicated concentrations were added to equal volumes of cells (104 cells/well) in a 96-well plate. Cell viability was evaluated using the CCK-8 kit after incubation at 37°C for 2 days. After incubation with CCK-8 reagents at 37°C for another 2 h, the absorbance at 450 nm was measured with an ELISA reader, and the percentage of cytotoxicity was calculated.
Safety of A1L35HR2m-Chol in vivo
Twelve Balb/c mice (6–8 weeks old) were assigned randomly to 4 groups and administrated intranasally with PBS (n = 3), 5 mg/kg A1L35HR2m (n = 3), 5 mg/kg A1L35HR2m-Chol (n = 3), or 20 mg/kg A1L35HR2m-Chol (n = 3). ALT, AST, GGT and creatinine in the sera were measured by using the ALT, AST, GGT and creatinine assay kits (NJJCBIO, Nanjing, China) before administration and 4 h, 1, 3, and 5 days post-administration, respectively.
Ex vivo inhibition and in vivo half-life of fusion peptides
The ex vivo anti-SARS-CoV-2 D614G activity of A1L35HR2m or A1L35HR2m-Chol in plasma or tissue homogenate samples was detected as described previously.34,41 Briefly, Balb/c mice were intranasally administered 5 mg/kg A1L35HR2m (n = 3) or A1L35HR2m-Chol (n = 3). Sera were collected from these mice before (0 h) and 2, 4, 8, 12, 24, 48, 72, 120, 168, 240 and 336 h after injection. Lung, nose and trachea tissue were also collected from A1L35HR2m-Chol treated mice, homogenized in 0.3 mL PBS of tissues and frozen. The inhibitory activities of plasma and tissue homogenate samples were tested against SARS-CoV-2 D614G infection as described above. The dilution fold of the serum achieving 50% inhibition (DF-IC50) was calculated. Based on the calculated DF-IC50 values, the concentration of peptides in the plasma or tissue homogenate samples was estimated, and the in vivo half-life and other pharmacokinetic parameters were calculated using PKSolver software as previously described.41
Microneutralization assays
The inhibitory activity of A1L35HR2m-Chol against authentic Omicron BA.2.12.1 and BA.5 infection was determined as described previously.36,40 Briefly, TMPRSS2-overexpressing VeroE6 cells were seeded into 96-well plates at 2 x 104 cells/well one day before infection. Peptides at serially diluted concentrations were preincubated with 2,500 plaque-forming units (PFU) of SARS-CoV-2 Omicron BA.2.12.1, BA.5 or EG.5.1 for 1 h at 37°C. The virus-peptide mixture was then inoculated into the cells and kept at 37°C in a 5% CO2 incubator for 36 h. After incubation, the supernatants were removed, and the cells were fixed with 10% formaldehyde solution. After fixation, the cells were penetrated by 0.2% Triton X-100 in PBS and then stained with rabbit anti-SARS-CoV-2 N protein for 1 h at room temperature, followed by staining with goat anti-rabbit Alexa Fluor 488-conjugated IgG (H + L) cross-adsorbed secondary antibody (Thermo Fisher). The signals were quantitated by a Sapphire Biomolecular Imager (Azure Biosystems), and the images were analyzed by ImageJ software. The IC50 values were plotted by fitting a nonlinear regression curve in GraphPad Prism.
Viral load reduction assay
A viral load reduction assay was performed on VeroE6-TMPRSS2 cells by RT‒qPCR as described previously.42 The cell culture supernatants or animal tissue homogenates were lysed in RLT buffer, followed by RNA extraction using a RNeasy mini kit (Qiagen). The viral load was quantified by a one-step TB green RT‒PCR kit (Takara) according to the protocol of the LightCycle 480 real-time PCR system (Roche). The primers used for quantification targeted SARS-CoV-2 RdRp genes: forward sequence: 5′-CGCATACAGTCTTRCAGGCT-3’; reverse sequence: 5′-GTGTGATGTTGAWATGACATGGTC-3’.
Prophylactic and therapeutic efficacy of A1L35HR2m-Chol against omicron variants and alpha infection in K18-hACE2 transgenic mice
K18-hACE2 transgenic mice were obtained from the Center for Comparative Medicine Research, University of Hong Kong. All experimental protocols were approved by the Animal Ethics Committee in the HKU and performed according to the standard operating procedures of the biosafety level 3 animal facilities.
The peptide prophylactic experiment was performed in a K18-hACE2 transgenic mouse model. Briefly, 6- to 8-week-old male mice (n = 5) were anesthetized and treated intranasally with lipopeptide A1L35HR2m-Chol or vehicle once daily at 2 days and 1 day prior to infection, respectively. At day 0, the mice were inoculated with 10,000 PFU of Omicron BA.5 intranasally. The mice were sacrificed at 2 days post-infection. The nasal turbinate and lungs were collected for qPCR and infectious titer analyses. For the dominant circulating EG.5.1 infection experiment, the mice were intranasally administered a single dose of A1L35HR2m-Chol (2 mg/kg, n = 5) or vehicle at 4 h before virus infection. At 48 hpi, the nasal turbinate and lungs were collected for infectious viral titer analyses. For the SARS-CoV-2 Alpha lethal infection experiment, the mice were intranasally administered a single dose of A1L35HR2m-Chol (2 mg/kg, n = 5) at 4 h before virus infection., and surviving mice and body weight changes were monitored until the study endpoint (14 dpi)
For the therapeutic experiment, 6- to 8-week-old male K18-hACE2 mice (n = 5) were intranasally challenged with 10,000 PFU of Omicron BA.5. At days 1 and 2 post-infection, the mice were intranasally treated with A1L35HR2m-Chol or vehicle, respectively. At day 3 post-infection, the mice were sacrificed, and the nasal turbinate and lungs were harvested and homogenized. The viral load and infectious titer in tissues were determined as described previously.43 For the EG.5.1 infection experiment, mice were treated with A1L35HR2m-Chol (2 mg/kg, n = 5) or vehicle at 4 and 28 hpi in the therapeutic group. At 48 hpi, the nasal turbinate and lungs were collected for infectious virus titration. For the survival rate experiment, the mice were challenged on day 0 with 200 PFU SARS-CoV-2 Alpha per mouse, mice were intravenously administered of A1L35HR2m-Chol (2 mg/kg, n = 5) after 4 h infection and once-daily for a total of 6 times. Body weight changes were monitored until death or 14 dpi.
Quantification and statistical analysis
All sample sizes and replicate number are included in the Figure Legends. All data are shown as mean ± SEM. The p values are represented as follows: ∗p < 0.05, ∗∗p < 0.01, and not statistically significant when p > 0.05. All statistical analysis was performed using GraphPad Prism (Version 6.02). IC50 values were determined by least squares fit nonlinear regression in GraphPad Prism (Version 6.02).
Acknowledgments
We would like to thank Dr. Shibo Jiang and Dr. Lu Lu at the Shanghai Medical College of Fudan University for providing the plasmids pcDNA3.1-SARS-CoV-S, pcDNA3.1-SARS-CoV-2-S, and pNL4-3.Luc.RE as well as the cell lines, including 293T, Caco-2, and Calu3. This work was supported by the Zhejiang Key R&D Program of Zhejiang Province (no. 2021C03040), the National Natural Science Foundation of China (22077104 and 32120103013 to B.D. and 82204264 to W.B.), a fellowship from the China Postdoctoral Science Foundation (file no. 2021M692880 to W.B.), the National Natural Science Foundation of China (32322087 and 32300134 to S.Y.), the National Key Research and Development Program of China (2021YFC0866100 and 2023YFC3041600), the Theme-Based Research Scheme of the Research Grants Council (T11-709/21-N), the government of the Hong Kong Special Administrative Region, the National Natural Science Foundation of China/RGC Joint Research Scheme (N_HKU767/22 to S.Y.), and the National Institutes of Health, USA (GM122603) to W.F.D. This work was also supported by the Westlake Education Foundation and Tencent Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
W.B. and B.D. designed the project. S.Y. and B.D. supervised the project. W.B., K.T., and G.C. performed the biochemical and viral experiments. B.D., S.Y., W.B., K.T., and G.C. designed the experiments and interpreted the data. N.F.P. and W.F.D. participated in the project discussion and helped with data analysis. W.B. and B.D. wrote the manuscript draft, and W.B., B.D., and S.Y. revised the manuscript.
Declaration of interests
B.D., W.B., S.Y., K.T., and G.C. are the inventors of a provisional patent filed by Westlake University and The University of Hong Kong.
Published: February 9, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101418.
Contributor Information
Wenwen Bi, Email: biwenwen@zju.edu.cn.
Shuofeng Yuan, Email: yuansf@hku.hk.
Bobo Dang, Email: dangbobo@westlake.edu.cn.
Supplemental information
References
- 1.Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528.e2. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2523. doi: 10.1016/j.cell.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y., Han J., Zhang Y., He J., Yu W., Zhang X., Wu J., Zhang S., Kong Y., Guo Y., et al. SARS-CoV-2 Omicron Variant: Epidemiological Features, Biological Characteristics, and Clinical Significance. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rössler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 2022;386:698–700. doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L., Ying T., Lu L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.T.K., Wang Q., Du L., Tan W., Wilson I.A., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu L., Liu Q., Zhu Y., Chan K.H., Qin L., Li Y., Wang Q., Chan J.F.W., Du L., Yu F., et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing L., Xu X., Xu W., Liu Z., Shen X., Zhou J., Xu L., Pu J., Yang C., Huang Y., et al. A Five-Helix-Based SARS-CoV-2 Fusion Inhibitor Targeting Heptad Repeat 2 Domain against SARS-CoV-2 and Its Variants of Concern. Viruses. 2022;14:597. doi: 10.3390/v14030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi W., Chen G., Dang B. Novel Engineered SARS-CoV-2 HR1 Trimer Exhibits Improved Potency and Broad-Spectrum Activity against SARS-CoV-2 and Its Variants. J. Virol. 2022;96 doi: 10.1128/jvi.00681-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M., Cong W., Peng H., Qing J., Shen H., Tang Y., Geng C., Chen S., Zou Y., Zhang W.D., et al. Stapled Peptides Targeting SARS-CoV-2 Spike Protein HR1 Inhibit the Fusion of Virus to Its Cell Receptor. J. Med. Chem. 2021;64:17486–17495. doi: 10.1021/acs.jmedchem.1c01681. [DOI] [PubMed] [Google Scholar]
- 20.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Yu D., Yan H., Chong H., He Y. Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J. Virol. 2020;94:e02312–e02318. doi: 10.1128/JVI.02312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vries R.D., Schmitz K.S., Bovier F.T., Predella C., Khao J., Noack D., Haagmans B.L., Herfst S., Stearns K.N., Drew-Bear J., et al. Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science. 2021;371:1379–1382. doi: 10.1126/science.abf4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karoyan P., Vieillard V., Gómez-Morales L., Odile E., Guihot A., Luyt C.E., Denis A., Grondin P., Lequin O. Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Commun. Biol. 2021;4:197. doi: 10.1038/s42003-021-01736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda S.K., Sen Gupta P.S., Biswal S., Ray A.K., Rana M.K. ACE-2-Derived Biomimetic Peptides for the Inhibition of Spike Protein of SARS-CoV-2. J. Proteome Res. 2021;20:1296–1303. doi: 10.1021/acs.jproteome.0c00686. [DOI] [PubMed] [Google Scholar]
- 26.Yuan M., Chen X., Zhu Y., Dong X., Liu Y., Qian Z., Ye L., Liu P. A Bispecific Antibody Targeting RBD and S2 Potently Neutralizes SARS-CoV-2 Omicron and Other Variants of Concern. J. Virol. 2022;96 doi: 10.1128/jvi.00775-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C., Zhan W., Yang Z., Tu C., Hu G., Zhang X., Song W., Du S., Zhu Y., Huang K., et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell. 2022;185:1389–1401.e18. doi: 10.1016/j.cell.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H., Gonzales-Wartz K.K., Huang D., Yuan M., Peterson M., Liang J., Beutler N., Torres J.L., Cong Y., Postnikova E., et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13:eabj5413. doi: 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Y., Xu W., Tang J., Cao N., Lan Q., Lu L., Jiang S. A bivalent protein targeting glycans and HR1 domain in spike protein potently inhibited infection of SARS-CoV-2 and other human coronaviruses. Cell Biosci. 2021;11:128. doi: 10.1186/s13578-021-00638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S.B., Irvin P., Hu Z., Khan M., Hu X., Zeng Q., Chen C., Xu M., Leek M., Zang R., et al. Targeting the Fusion Process of SARS-CoV-2 Infection by Small Molecule Inhibitors. mBio. 2022;13 doi: 10.1016/j.virol.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Outlaw V.K., Bovier F.T., Mears M.C., Cajimat M.N., Zhu Y., Lin M.J., Addetia A., Lieberman N.A.P., Peddu V., Xie X., et al. Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain. mBio. 2020;11:e01935-20. doi: 10.1128/mBio.01935-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J.E., Gallagher T. Lipidation increases antiviral activities of coronavirus fusion inhibiting peptides. Virology. 2017;511:9–18. doi: 10.1016/j.virol.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue J., Chong H., Zhu Y., Zhang J., Tong L., Lu J., Chen T., Cong Z., Wei Q., He Y. Efficient treatment and pre-exposure prophylaxis in rhesus macaques by an HIV fusion-inhibitory lipopeptide. Cell. 2022;185:131–144.e18. doi: 10.1016/j.cell.2021.11.032. [DOI] [PubMed] [Google Scholar]
- 35.Su S., Rasquinha G., Du L., Wang Q., Xu W., Li W., Lu L., Jiang S. A Peptide-Based HIV-1 Fusion Inhibitor with Two Tail-Anchors and Palmitic Acid Exhibits Substantially Improved In Vitro and Ex Vivo Anti-HIV-1 Activity and Prolonged In Vivo Half-Life. Molecules. 2019;24:1134. doi: 10.3390/molecules24061134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Z.W., Ong C.P., Tang K., Fan Y., Luo C., Zhou R., Luo P., Cheng Y., Gray V.S., Wang P., et al. Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell. Mol. Immunol. 2022;19:588–601. doi: 10.1038/s41423-022-00855-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y., Chong H., Yu D., Guo Y., Zhou Y., He Y. Design and Characterization of Cholesterylated Peptide HIV-1/2 Fusion Inhibitors with Extremely Potent and Long-Lasting Antiviral Activity. J. Virol. 2019;93:e02312–e02318. doi: 10.1128/JVI.02312-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari P., Rothan H.A., Natekar J.P., Stone S., Pathak H., Strate P.G., Arora K., Brinton M.A., Kumar M. Neuroinvasion and Encephalitis Following Intranasal Inoculation of SARS-CoV-2 in K18-hACE2 Mice. Viruses. 2021;13:132. doi: 10.3390/v13010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuai H., Chan J.F.W., Hu B., Chai Y., Yuen T.T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 40.Ong C.P., Ye Z.W., Tang K., Liang R., Xie Y., Zhang H., Qin Z., Sun H., Wang T.Y., Cheng Y., et al. Comparative analysis of SARS-CoV-2 Omicron BA.2.12.1 and BA.5.2 variants. J. Med. Virol. 2023;95 doi: 10.1002/jmv.28326. [DOI] [PubMed] [Google Scholar]
- 41.Bi W., Xu W., Cheng L., Xue J., Wang Q., Yu F., Xia S., Wang Q., Li G., Qin C., et al. IgG Fc-binding motif-conjugated HIV-1 fusion inhibitor exhibits improved potency and in vivo half-life: Potential application in combination with broad neutralizing antibodies. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H.C., Wong S.C.Y., Leung K.H., Fung A.Y.F., Ng A.C.K., Zou Z., Tsoi H.W., et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan S., Yin X., Meng X., Chan J.F.W., Ye Z.W., Riva L., Pache L., Chan C.C.Y., Lai P.M., Chan C.C.S., et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this study will be shared upon request from the lead contact. This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.