Abstract
Objectives
Data are scarce on whether the composition of the lung microbiome (extending from the nasopharynx to the peripheral lung tissue) varies according to histology or grade of non–small cell lung cancer. We hypothesized that the composition of the lung microbiome would vary according to the histology and the grade of non–small cell lung cancer.
Methods
We collected naso-oral and central lobar (cancer affected, ipsilateral unaffected, and contralateral unaffected) bronchoalveolar lavage fluid and brushing samples from patients with clinical early-stage lung cancer between July 2018 and February 2020 at a single academic center. We performed bacterial 16S rRNA sequencing and then compared clinical and pathologic findings with microbiome signatures.
Results
Samples were collected from 28 patients. Microbial composition in affected lobes displayed unique enrichment of oropharyngeal bacterial species that was significantly different compared with that from the unaffected contralateral lobes; patients with chronic obstructive pulmonary disease had similar diversity to those without chronic obstructive pulmonary disease (P = .1312). The lung microbiome diversity in patients with adenocarcinoma was similar to those with squamous cell cancer (P = .27). There were no differences in diversity or composition in the unaffected lobes of patients with adenocarcinoma versus squamous cell cancer. There was a trend toward lower lung microbial diversity in poorly differentiated adenocarcinomas compared with well-differentiated adenocarcinomas (P = .08).
Conclusions
The lung microbiota differs between cancer affected and unaffected lobes in the same patient. Furthermore, poorly differentiated lung cancers were associated with lower microbial diversity. Larger studies will be required to confirm these findings.
Key Words: lung cancer, lung microbiome, microbiome diversity, poorly differentiated
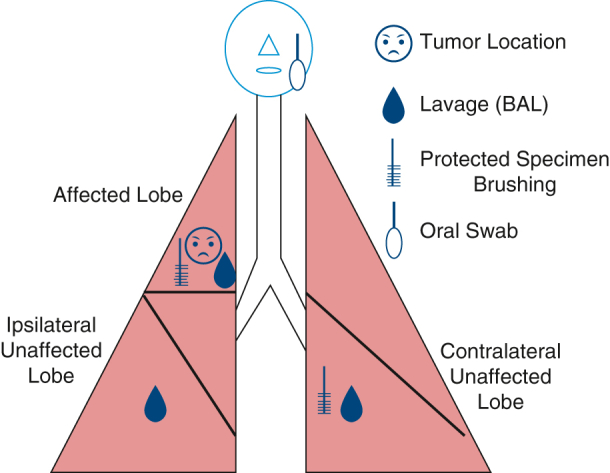
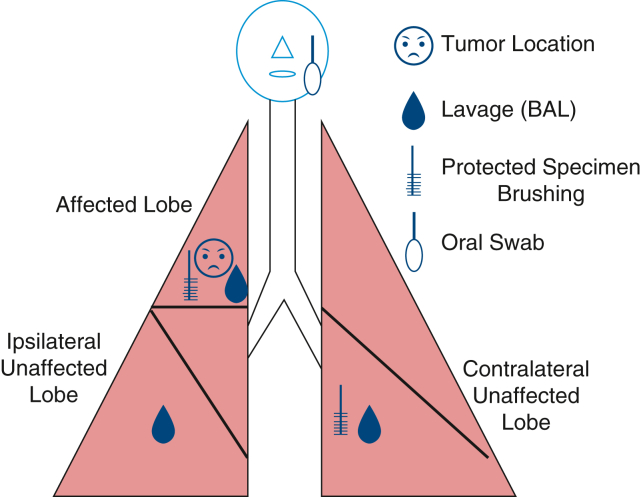
Schema showing the sampling methods used in this study.
Central Message.
Microbial composition in affected lobes displayed unique enrichment of oropharyngeal bacterial species that was significantly different compared with that from the unaffected contralateral lobes.
Perspective.
This work shows that the microbiome in the lungs with lung cancer vary from the lungs of the same patients without lung cancer. These differences could be used in the future for early diagnosis or potential treatment of lungs predisposed to developing lung cancers.
See Discussion on page 269.
Lung cancer continues to be the number one cause of cancer-related death in the world.1 Although cigarette smoking–related lung cancer remains a major problem, there has been a rapid increase in the incidence of lung cancer in never smokers.2 There is growing understanding of the microbiome's impact on overall health, including cancer development.3 Most research has focused on the gut microbiome, with its relationship to gastrointestinal (GI) diseases, but more recent data have suggested that there is a gut-lung axis, with gut health affecting lung diseases.4 In addition to the GI tract, the lungs have a significant amount of surface area in contact with the external environment. Despite similar exposure to the external factors, the lung microbiome has a lower volume of biomass compared with the GI microbiome, which contains 99% of the microbiome biomass in the body.4 Because of this lower volume in the lung, the techniques to isolate the lung microbiota has been more prone to contamination compared with research performed on the GI microbiome.4
The lung microbiome is created by micro-aspiration, inhalation, and direct spread down the respiratory tract. Growth of different flora is affected by factors including the pH, temperature, oxygen levels, and immune microenvironment.5 Clearance of the microbes occurs through coughing, ciliary clearance, and immune regulation.4,5 Dysbiosis, or an imbalance in the microbiota, can result in inflammation-promoting metabolite and toxin release into the lung, impacting disease development.5 The oral cavity is the primary source of respiratory tract flora and is composed of more than 700 different organisms. Food and oral hygiene affect the type of flora, and periodontal disease has been associated with pneumonias and chronic obstructive pulmonary disease (COPD).6 Specific oral microorganisms, Blastomonas, Sphingomonas, and Chlamydia pneumonia, have all been associated with lung cancer.6 Respiratory microbiota has been shown to be modulated by the treatments for chronic diseases, such as COPD, and can affect the effectiveness of those treatments.7 The respiratory microbiome, ranging from the oral cavity to the deeper lung, is being evaluated for its possible role as an early detection tool for lung cancer and a target for chemoprevention or cancer therapy.5,7,8
Sampling of the respiratory tract microbiome has been associated with concerns surrounding contamination during introduction of a bronchoscope. Sampling methods include bronchoalveolar lavage (BAL) and specimen brushing. Prior work has shown no difference in the microbiome when inserting a bronchoscope from either method.9 That same study suggested that the BAL sampled a greater surface area by washing a greater endoluminal surface area compared with the brushing method, resulting in a higher density of bacterial DNA. Other groups have studied the microbiome in normal tissue compared with tumor tissue, as well as oropharyngeal sampling.8 Tsay and colleagues10 performed a sampling paradigm on affected and unaffected lobes, but did not look at both ipsilateral and contralateral unaffected lobes. They included healthy control subjects as their main comparison,10 which was a different focus than our work. We sought to evaluate the lung microbiome in patients with lung cancer, evaluating different parts of the respiratory system including ipsilateral and contralateral cancer-free (unaffected) lobes for within-subject comparisons, using strict collection controls to minimize contamination.
Material and Methods
All subjects signed informed consent to participate in this study with consent for publication of the study data. This was approved by the Institutional Review Board of University of Michigan (Michigan Medicine IRB # HUM00138660) on February 15, 2018. Participants included patients who were undergoing standard of care surgical resection for suspicious pulmonary nodules/masses or biopsy-proven non–small cell lung cancers that were less than 3 cm in size (cT1). Participants had not received prior treatment for these lesions, including chemotherapy, radiation, or antibiotics. Perioperative data were collected prospectively, including demographics, clinical tumor staging, patient social and medical history, surgical data, and pathologic findings. Long-term outcomes were reviewed at 4 years after the initiation of the study, giving a range of follow-up from 2 to 4 years for all patients.
Sample Collection
After intubation with a single-lumen endotracheal tube, oropharyngeal and nasal samples were obtained. A background sample from the bronchoscope was obtained by passing sterile saline through the suctioning sampled before the procedure. Lower-airway cytology brushings were then obtained from the contralateral unaffected lobe followed by the affected lobe. Lavages were then performed, with 15 mL normal saline injected through the bronchoscope at the contralateral unaffected, ipsilateral unaffected, and tumor affected lobe in that order. Brushing and lavages in the affected lobe were targeted to the affected segment during the bronchoscopy. Last, sterile saline was passed through the bronchoscope postprocedure. A total of 5 bronchoalveolar lavage fluid (BALF) (prescope wash, contralateral unaffected lobe, ipsilateral unaffected, affected lobe, and postwash) samples, 2 bronchial brushing (contralateral unaffected and affected) samples, a nasal swab sample, and a oropharyngeal swab sample were collected from each patient (Figure 1). Samples were collected and subjected to centrifugation to remove cellular debris and then subjected to bacterial DNA extraction described next.
Figure 1.
Sampling diagram for each patient. BAL, Bronchoalveolar lavage.
16S Bacterial Sequencing and Droplet Digital Polymerase Chain Reaction
Genomic DNA was extracted from BALF, and bacterial DNA was quantified with droplet digital polymerase chain reaction. Primers and cycling conditions were as described previously.11,12 Sequencing was performed as described previously.11 Empty wells, sterile water, and DNA isolation controls were used as negative controls, and synthetic standard communities (Zymo Research, Catalog No. D6306) were used as positive controls. FASTQ files were generated with paired end reads and retained for further analysis. Sequence data were processed and analyzed using mother software according to standard operating protocol for MiSeq sequence data (https://mothur.org/wiki/miseq_sop/); minimum sequence length was 250 base pairs.11,12 A shared community file and phylotypes file were generated, using operational taxonomic units (OTUs) binned at 97% identity in mother (version 1.43.0). The baseline bronchoscopic sampling was used as described previously10 to minimize cross-contamination in the final analysis. We performed ordinations using principal component analysis in R (version 4.0.2) on Hellinger-transformed normalized OTU tables as described previously11 and compared overall community composition differences using resampling of a generalized linear model (mvabund).
The data collected from BALF measurements include OTU populations in affected and unaffected lobes of 26 patients. The data were co-indexed along the samples (ie, patients). A Linear Discriminant Analysis model was trained by OTU populations and percentages for all samples. Linear Discriminant Analysis was used for binary classification (affected vs unaffected lobes), as well as determination of which OTUs are more associated with affected or unaffected lobes. We assumed that OTUs belonging to a bacterial family have the same effect/function to reduce the number of features and to better visualize the data. Bacterial community diversity was calculated using the Shannon Diversity index for described subgroups and compared using a Student t test.
Results
A total 28 patients were enrolled and had full preoperative microbiome sampling performed. One patient was found to have a benign nodule, and 1 patient was found to have a carcinoid tumor, leaving 26 patients for further analysis (Table 1). A slight majority of patients were male, and almost all were considered White, non-Hispanic. Patients were considered current smokers (57%) if they had quit within 2 months before their surgery. Only 10 patients (38%) were noted to have a diagnosis of COPD before their surgical resection. Most resections were performed by a minimally invasive approach. Two operations resulted in a bilobectomy (both right middle and lower lobes), and 1 patient was noted to have 2 separate ipsilateral T1 tumors, being treated with a segmentectomy and a wedge resection.
Table 1.
Demographics and perioperative characteristics, including noncancer and carcinoid patients
| N (overall) | 28 |
| Noncancer | 1 |
| Carcinoid | 1 |
| Age | 66.7 ± 9.5 y |
| Gender | |
| M | 16 (57%) |
| F | 12 (43%) |
| Race | |
| W | 26 (93%) |
| NR | 1 (4%) |
| B | 1 (4%) |
| Smoking | |
| Never | 5 (18%) |
| Former | 7 (27%) |
| Current | 16 (57%) |
| COPD documented (n = 26) | 10 (38%) |
| Location (n = 26) | |
| RUL | 8 (31%) |
| RML | 2 (8%) |
| RLL | 7 (27%) |
| LUL | 9 (35%) |
| LLL | 3 (12%) |
| Surgical approach (n = 26) | |
| Robotic | 11 (42%) |
| VATS | 8 (31%) |
| Open | 3 (12%) |
| Robotic-Open | 1 (4%) |
| VATS-Open | 3 (12%) |
COPD, Chronic obstructive pulmonary disease; LLL, left lower lobe; LUL, left upper lobe; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe; VATS, video-assisted thoracoscopic surgery.
The tumor histology and grade are shown in Table 2. Most patients had adenocarcinomas or squamous cell cancers, with 1 large cell cancer and 1 carcinoid. The patient with 2 separate T1 tumors had a multidisciplinary tumor board review of their imaging and pathology with a consensus opinion that these were separate cancers, as opposed to metastatic lesions. Approximately half the patients (11) had stage IA cancer (T1xN0), and 13 patients had T2-4N1-2 tumors. When comparing the microbiome diversity by tumor stage, we excluded the T1N1 patient. Table 3 demonstrates the long-term outcomes. Four patients developed metastatic cancer, but all were alive at a range of 2 to 4 years from the time of surgery. Four different patients died during the study period, with 2 dying of separate cancers (melanoma and esophageal cancer) and the others dying of noncancer causes.
Table 2.
Pathology data
| Histology | Grade | N (percentage) |
|---|---|---|
| Adenocarcinoma | Well | 8 (30%) |
| Moderate | 6 (22%) | |
| Poor | 6 (22%) | |
| Squamous cell cancer | Well | 1 (4%) |
| Moderate | 1 (4%) | |
| Poor | 3 (11%) | |
| Large cell cancer | Undifferentiated | 1 (4%) |
| Carcinoid | 1 (4%) | |
| Stage | T1xN0 (noncarcinoid) | 11 (41%) |
| T2-4N1-2 | 13 (48%) | |
| T1cN1 | 1 (4%) | |
| Ipsilateral T1s | 1 (4%) | |
| T1N0 carcinoid | 1 (4%) |
Percentages are based on a baseline “n” of 27 cancer resections, including the patient with the carcinoid tumor.
Table 3.
Long-term outcomes
| Secondary cancers (n = 26) | 23% (6) |
| Adjuvant therapy (n = 26) | |
| Chemo XRT | 12% (3) |
| Chemo | 15% (4) |
| XRT | 4% (1) |
| Adjuvant therapy offered and refused | 19% (5) |
| None | 50% (13) |
| Survival (4 y after) | |
| Deaths | 15% (4) |
| Metastatic recurrent cancer | 15% (4) |
Percentages are based on an “n” of 26 noncarcinoid lung cancers. XRT, Radiotherapy.
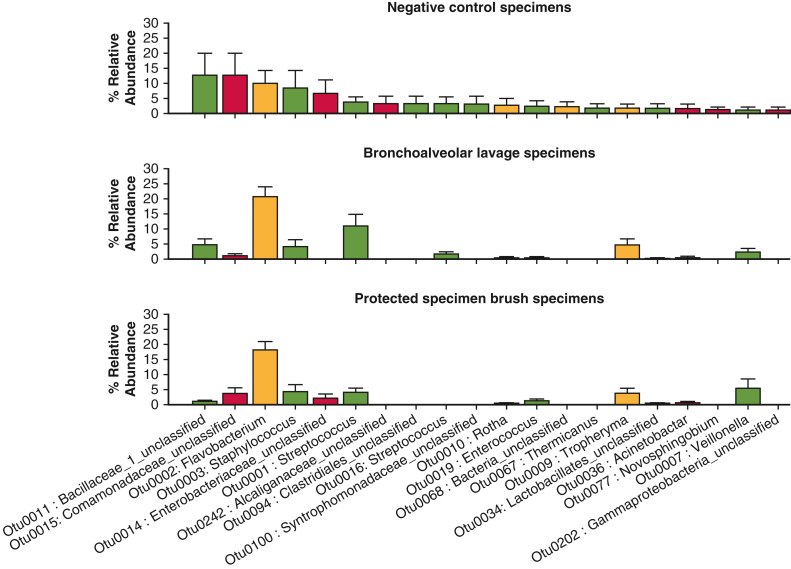
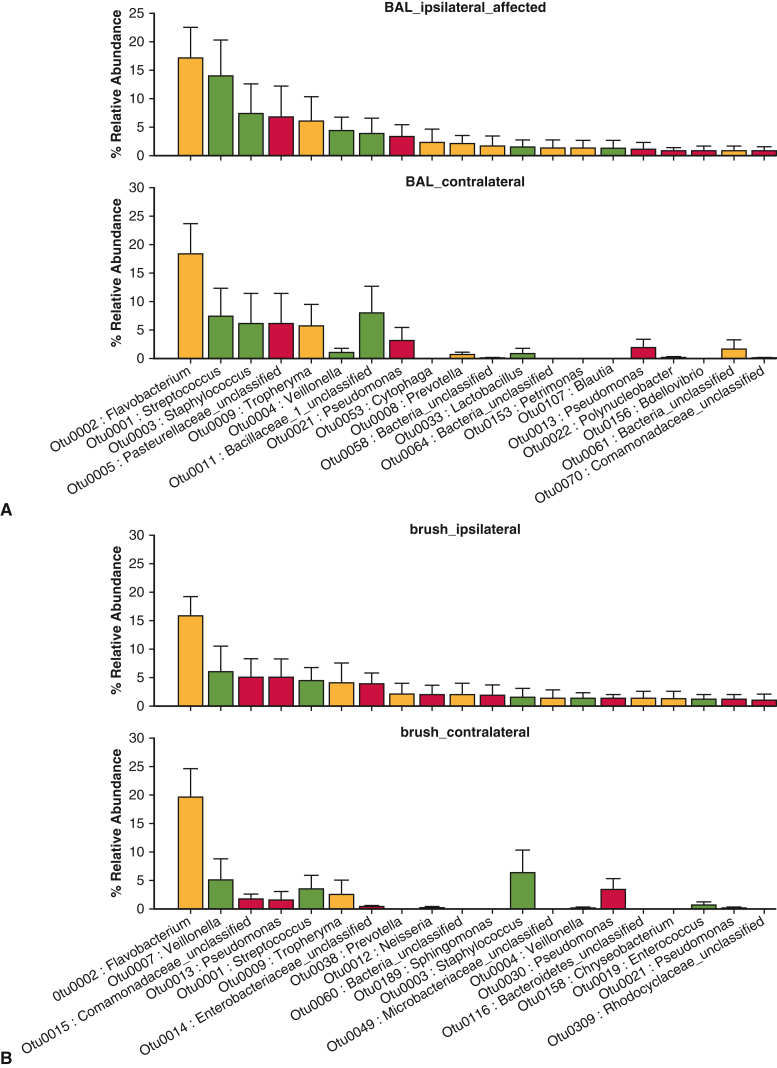
We found a significant difference between affected and nonaffected lavage and brushing specimens at the OTU level, which serves as a species surrogate (mvabund = 0.030). Negative control data are shown (Figure E1). By using rank abundance comparison of specimens, several OTUs were relatively enriched in the affected lobes compared with nonaffected, including OTU004 (Veillonella), OTU0008 (Prevotella), and OTU0001 (Streptococcus) (Figure 2, A and B, Table 4, and Figure E2). These are all upper airway common species. In the unaffected lobes, there was a high enrichment of OTUs, including Comamonadaceae, which are associated with healthy lung.13 There were no significant differences in bacterial composition or diversity when comparing unaffected lobes from the same patients. When looking at OTU diversity in subgroups, none of our analyses reached statistical significance (Table 4). There were trends toward increased lung microbiome diversity in well-differentiated adenocarcinomas, compared with the poorly differentiated, and increased diversity in non-COPD nonaffected lungs, compared with COPD nonaffected lungs (Table 5). Upper airway samples (oral and nasal swabs) showed a different pattern of microflora compared with the distal airways, but without any signatures of note.
Figure E1.
Rank abundance of the control sampling performed. Data presented are the mean value with the error bar representing 1 standard deviation.
Figure 2.
Rank abundance of most highly enriched species of bacteria identified in lavage specimens (A) and brushings (B) from cancer-affected lung versus ipsilateral unaffected and contralateral unaffected lung. Community composition was determined by relative microbial abundance analysis to identify enriched taxa in the lung-associated lavage. Data presented are the mean value with the error bar representing 1 standard deviation. BAL, Bronchoalveolar lavage.
Table 4.
Community composition comparisons
| BAL: affected vs contralateral unaffected | |
|---|---|
| Title | mvabund |
| OTU | 0.030∗ |
| Family | 0.152 |
| Class | 0.263 |
| Phylum | 0.134 |
| BAL: affected vs ipsilateral unaffected | |
|---|---|
| Title | mvabund |
| OTU | 0.079 |
| Family | 0.614 |
| Class | 0.648 |
| Phylum | 0.831 |
| Brushes: affected vs unaffected | |
|---|---|
| Title | mvabund |
| OTU | 0.022∗ |
| Family | 0.16 |
| Class | 0.277 |
| Phylum | 0.131 |
BAL, Bronchoalveolar lavage; mvabund, generalized linear modeling comparison; OTU, operational taxonomic unit.
Indicates significance.
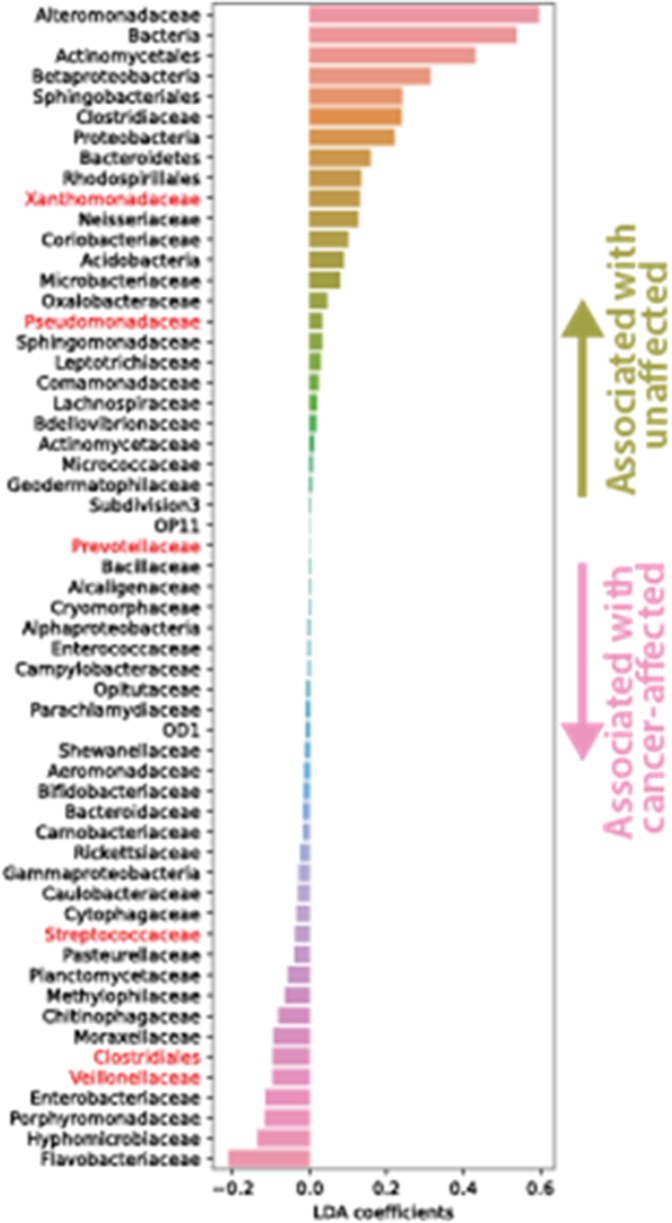
Figure E2.
Results of a linear discriminant analysis demonstrating OTUs associated more with cancer containing lobes, OTU004 (Veillonella), OTU0008 (Prevotella), and OTU0001 (Streptococcus), and those more associated with unaffected lobes.
Table 5.
Subgroup differences in microbiome diversity: None reached statistical significance
| Adenocarcinoma diversity (SI) | Average | SE | P |
|---|---|---|---|
| Poorly differentiated | 1.166 | 0.387 | .08 |
| Well differentiated | 1.659 | 0.391 | |
| Diversity by histology (SI) | |||
| Adenocarcinoma | 1.454 | 0.250 | .27 |
| Squamous cell cancer | 1.634 | 0.408 | |
| Diversity by lung disease | |||
| COPD | 1.181 | 0.526 | .13 |
| No COPD | 1.567 | 0.301 |
COPD, Chronic obstructive pulmonary disease; SI, Shannon Diversity Index.
Discussion
Our study has demonstrated that lung microbiome patterns vary within a single patient based on the presence of cancer within their lungs. This was a novel approach to comprehensive sampling of different areas of the lung, using ipsilateral and contralateral samples from the same patient, but from unaffected lobes as an internal control. At the OTU level, there was a significant difference between affected and unaffected lobes; however, our study was likely too small to determine which particular species were driving the difference. We looked at relative abundance and postulated what may be driving the differences based on our data and data in the literature that showed that Veillonella, Streptococcus, and Prevotella were associated with patients with lung cancer and therefore our focus on those species. We did not have adequate numbers of noncancer patients (1 benign nodule) to provide an adequate experimental control for our dataset.
We did not find a significant difference in bacterial diversity between adenocarcinomas and squamous cell cancers. A study by Leng and colleagues8 in 2021 evaluated oral specimens, tumor tissue, and normal lung tissue, and found a “signature” of Acidovorax and Veillonella was associated with a diagnosis of squamous cell cancer, whereas the presence of Capnocytophaga was associated with adenocarcinoma. Zheng and colleagues14 in 2023 evaluated 75 patients with lung cancer (and 7 noncancer controls) and found 13 taxonomic differences between adenocarcinomas and squamous cell cancers. They found no differences in microbiome signature based on tumor stage and no differences noted when comparing smoking status. Other studies have demonstrated gender and age-related differences with different histologies.15 Adenocarcinoma was associated with Escherichia coli dysregulation in older women and older men, whereas squamous cell cancer was associated with Pseudomonas putida expression in young men. Our study cohort was underpowered to evaluate for those types of differences.
Other studies looking at the overall microbiome enrichment in lung cancers have shown similar common communities expressed, but differences were noted in rare microbiota, such as Lactobacillus and Bacterioides.16 Marshall and colleagues17 recently performed a study that looked at approximately 350 deep lung specimens in high-risk patients who did not have lung cancer. Patients were then tracked for up to 10 years, and they showed a different signature for those who developed lung cancer versus those who did not. The authors were able to define a subgroup of patients who developed more quickly growing tumors based on differential microbiome signatures.17 Our study was more limited with regard to sample size, but future studies can incorporate our methodology to look at individual lobes and even segments for the variable risk of developing future lung cancers. Patnaik and colleagues18 did look at the lower airway microbiome and showed differences in those with recurrence after resection compared with those without. Their methodology was different from our study because they sampled the tumor and adjacent lung tissue, rather than the bronchoscopic method we used.
Limitations
Our study has a number of limitations. This was a small pilot study that was not powered to offer a true comparison within many of our subgroups. We performed a robust variety of sampling in the respiratory tract, but did not sample the trachea or proximal bronchi, which would have helped in mapping the differential community composition between the nasal and oral cavities and the deeper lung. Our patient population was racially homogenous with only 1 African-American patient. This reflects similar challenges that many clinical studies have in enrolling minority patients.19 Our future goals would include recruitment of a more heterogenous population of patients.
Conclusions
The lobar microbiome varies between lobes with and without cancer in patients with lung cancer. Poorly differentiated cancers trended toward less bacterial diversity. A better understanding of how this variable lung microbiome enrichment affects the tumor micro-environment could inform therapies. Mapping the noncancer lobes could potentially predict the future risk for new primary lung cancers or for developing metastatic cancers due to unique lung microbiome signatures.
Webcast
You can watch a Webcast of this AATS meeting presentation by going to: https://www.aats.org/resources/pilot-study-demonstrating-the-lung-microbiome-as-a-potential-marker-for-lung-cancer.

Conflict of Interest Statement
Dr Reddy has nonrelevant disclosures with Intuitive Surgical, Genentech, On Target Labs, and Medtronic. All other authors reported no conflicts of interest.
The Journal policy requires editors and reviewers to disclose conflicts of interest and to decline handling or reviewing manuscripts for which they may have a conflict of interest. The editors and reviewers of this article have no conflicts of interest.
Footnotes
This work was not funded by an external body. Internal research funds only were used.
Appendix E1
References
- 1.Schabath M.B., Cote M.L. Cancer progress and priorities: lung cancer. Cancer Epidemiol Biomarkers Prev. 2019;28:1563–1579. doi: 10.1158/1055-9965.EPI-19-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivera G.A., Wakele H. Lung cancer in never smokers. Adv Exp Med Biol. 2016;893:43–57. doi: 10.1007/978-3-319-24223-1_3. [DOI] [PubMed] [Google Scholar]
- 3.Cullin N., Azevedo Antunes C., Straussman R., Stein-Thoeringer C.K., Elinav E. Microbiome and cancer. Cancer Cell. 2021;39:1317–1341. doi: 10.1016/j.ccell.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Dong Q., Chen E.S., Zhao C., Jin C. Host-microbiome interaction in lung cancer. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.679829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLean A.E.B., Kao S.C., Barnes D.J., Wong K.K.H., Scolyer R.A., Cooper W.A., et al. The emerging role of the lung microbiome and its importance in non-small cell lung cancer diagnosis and treatment. Cancer. 2022;165:124–132. doi: 10.1016/j.lungcan.2022.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Pathak J.L., Yan Y., Zhang Q., Wang L., Ge L. The role of oral microbiome in respiratory health and diseases. Respir Med. 2021;185 doi: 10.1016/j.rmed.2021.106475. [DOI] [PubMed] [Google Scholar]
- 7.Budden K.F., Shukla S.D., Rehman S.F., Bowerman K.L., Keely S., Hugenholtz P., et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. 2019;7:907–920. doi: 10.1016/S2213-2600(18)30510-1. [DOI] [PubMed] [Google Scholar]
- 8.Leng Q., Holden V.K., Deepak J., Todd N.W., Jiang F. Microbiota biomarkers for lung cancer. Diagnostics (Basel) 2021;11:407. doi: 10.3390/diagnostics11030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson R.P., Erb-Downward J.R., Freeman C.M., McCloskey L., Flakowski N.R., Huffnagle G.B., et al. Bacterial topography of the healthy human lower respiratory tract. mBio. 2017;8 doi: 10.1128/mBio.02287-16. e02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsay J.C.J., Wu B.G., Badri M.H., Clemente J.C., Shen N., Meyn P., et al. Airway microbiota is associated with upregulation of PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198:1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashley S.L., Sjoding M.W., Popova A.P., Cui T.X., Hoostal M.J., Schmidt T.M., et al. Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med. 2020;12:eaau9959. doi: 10.1126/scitranslmed.aau9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker J.M., Hinkle K.J., McDonald R.A., Brown C.A., Falkowski N.R., Huffnagle G.B., et al. Whole lung tissue is the preferred sampling method for amplicon-based characterization of murine lung microbiota. Microbiome. 2021;9:99. doi: 10.1186/s40168-021-01055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal L.N., Clemente J.C., Tsay J.C., Koralov S.B., Keller B.C., Wu B.G., et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of the Th17 phenotype. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X., Lu X., Hu Y. Distinct respiratory microbiota associates with lung cancer clinicopathological characteristics. Front Oncol. 2023;13 doi: 10.3389/fonc.2023.847182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong L.M., Shende N., Li W.T., Castaneda G., Apostol L., Chang E.Y., et al. Comparative analysis of age-and gender-associated microbiome in lung adenocarcinoma and lung squamous cell carcinoma. Cancers (Basel) 2020;12:1447. doi: 10.3390/cancers12061447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L., Sun R., Zhu Y., Li Z., She X., Jian X., et al. Lung microbiome alterations in NSCLC patients. Sci Rep. 2021;11 doi: 10.1038/s41598-021-91195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall E.A., Filho F.S.L., Sin D.D., Lam S., Leung J.M., Lam W.L. Distinct bronchial microbiome precedes clinical diagnosis of lung cancer. Mol Cancer. 2022;21:68. doi: 10.1186/s12943-022-01544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patnaik S.K., Cortes E.G., Kannisto E.D., Punnanitinont A., Dhillon A.S., Liu S., et al. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J Thorac Cardiovasc Surg. 2021;161:419–429. doi: 10.1016/j.jtcvs.2020.01.104. [DOI] [PubMed] [Google Scholar]
- 19.Shaya F.T., Gbarayor C.M., Keri Yang H., Agyeman-Duah M., Saunders E. A perspective on African American participation in clinical trials. Contemp Clin Trials. 2007;28:213–217. doi: 10.1016/j.cct.2006.10.001. [DOI] [PubMed] [Google Scholar]