Abstract
Large T antigen (T antigen), the early gene product of simian virus 40 (SV40), is a potent transcriptional activator of both cellular and viral genes. Recently we have shown that T antigen is tightly associated with TFIID and, in this position, performs a TATA-binding protein (TBP)-associated factor (TAF)-like function. Based on this observation, we asked whether T antigen affected steps in preinitiation complex assembly. Using purified components in in vitro complex assembly assays, we found that T antigen specifically enhances the formation of the TBP-TFIIA complex on the TATA element. T antigen accomplishes this by increasing the rate of formation of the TBP-TFIIA complex on the TATA element and by stabilizing the complexes after they are formed on the promoter. In addition, DNA immunoprecipitation experiments indicate that T antigen is associated with the stabilized TBP-TFIIA complexes bound to the DNA. In this regard, it has previously been shown that T antigen interacts with TBP; in the present study, we show that T antigen also interacts with TFIIA in vitro. In testing the ability of T antigen to stabilize the TBP-TFIIA complex, we found that stabilization is highly sensitive to the specific sequence context of the TATA element. Previous studies showed that T antigen could activate simple promoters containing the TATA elements from the hsp70 and c-fos gene promoters but failed to significantly activate similar promoters containing the TATA elements from the promoters of the SV40 early and adenovirus E2a genes. We find that the ability to stabilize the TBP-TFIIA complex on the hsp70 and c-fos TATA elements, and not on the SV40 early and E2A TATA elements, correlates with the ability or inability to activate promoters containing these TATA elements.
Large T antigen (T antigen) is one of the early gene products of simian virus 40 (SV40). It is a potent transcriptional activator of both cellular and viral genes and is an oncoprotein capable of interacting with an array of host cellular factors (46). Previous studies from this lab and others have shown that T antigen can activate a simple promoter consisting of a TATA element and one upstream transcription factor binding site (13, 15, 34). Although T antigen has a specific as well as a nonspecific DNA binding capability, this function is not essential for transcriptional activation (1, 12, 21). Rather, T antigen appears to mediate its transactivation function through protein-protein interactions. Our lab has previously shown that large T antigen can interact with both the TATA-binding protein (TBP) and several TBP-associated factors (TAFs) (7, 16). In addition, T antigen can interact with a number of transcription factors including TEF-1 and Sp1 (7, 16). We have also established that T antigen is tightly associated with TFIID and, in this position, performs a TAF-like function requiring interactions with upstream bound factors to affect transcriptional activation (7). It is important to note that the additional interaction with the upstream bound factor appears to be essential for in vivo activation since T antigen cannot activate a promoter containing only a TATA element.
Transcriptional initiation by RNA polymerase II involves the assembly of TFIID as well as the general transcription factors on the promoter to form a preinitiation complex (18). This process occurs in a regulated fashion with the binding of TFIID to the TATA box constituting the first step. This is followed by the binding of the two general transcription factors TFIIA and TFIIB to form the TFIID-TFIIA-TFIIB (DAB) complex (3, 27). Subsequently, the polymerase II-TFIIF subcomplex binds the DAB complex with TFIIE and TFIIH, completing the preinitiation complex formation (47). Basal transcription requires the presence of TBP, TFIIB, TFIIF, and RNA polymerase II, as demonstrated by in vitro reconstitution systems (40). Activated transcription requires the presence of the TAFs present in the TFIID complex as well as a cellular or viral transactivator protein (5, 17). In addition, some activators require the presence of the general transcription factor TFIIA (24).
Several studies indicate that many activators stimulate transcription by increasing the rate or extent of preinitiation complex formation (23, 25, 44). The two general transcription factors TFIIA and TFIIB appear to be targets for activators that promote transcription complex assembly.
Human TFIIA is composed of three subunits. Two of these polypeptides, α and β, are derived from the same precursor polypeptide and together have a molecular mass of 55 kDa. The smallest subunit, γ, has a molecular mass of 14 kDa. In mammalian cells, TFIIA is found associated with a small population of TBP, although it has a lesser affinity for TBP than do the TAFs (9, 26, 45). It has been proposed that TFIIA prevents the binding of negative regulatory factors to TBP (30). Merino et al. (31) showed that the repression of basal transcription levels by the negative regulatory protein DR2 could be overcome by the presence of transcriptional activators or by the prior formation of a TFIID-TFIIA (DA) subcomplex. Bryant et al. (2) demonstrated that TBP mutations in the interface of the TBP-TFIIA interaction domain affected transcriptional activation by Gal4-E1A as well as Gal4-VP16. Hence, TFIIA may prevent the inactivation of TFIID by negative regulators and, together with activators such as Zta and VP16, overcomes a rate-limiting step in preinitiation complex assembly (6, 42).
TFIIB, the second basal transcription factor targeted by activators, binds TBP in either the TFIID-DNA or DA-DNA complex and recruits the TFIIF-RNA polymerase complex to the promoter (3, 27). Human TFIIB has a molecular mass of 33 kDa and is required for basal as well as activated transcription (2). The acidic activator, VP16, has been shown to interact with both TBP and TFIIB (25, 38), and TFIIB is required for transcriptional activation by VP16 (36). In addition, Goodrich et al. (14) demonstrated that both TFIIB and TAF40 exist in a ternary complex with VP16, and they propose that this subcomplex may be an intermediary step in preinitiation assembly. The acidic activator Gal4-AH has also been shown to stimulate transcription levels by recruiting and maintaining TFIIB at the promoter (25).
The activators which stimulate preinitiation complex assembly do so by either increasing the rate of formation of complexes on the promoter or stabilizing the complexes once formed. Studies with the Epstein-Barr virus transactivator, Zta, indicate that it rapidly stimulates the formation of a stable preinitiation complex in the presence of TFIIA and TFIID on a promoter containing Zta binding sites (24). The acidic activators GAL4-AH and GAL4-VP16 enhance the formation of polymerase II open complexes at the promoter (42). Lin and Green (25) demonstrated that the two activators recruited TFIIB to the TFIID complex, whereas Wang et al. (42) showed that they promote the DA interaction. Hence, the DA subcomplex appears to be a rate-limiting intermediate in transcription complex formation. The Drosophila transcription factor Ubx and the herpes simplex virus transcription factor VP16 have both been shown to increase the number of transcription complexes formed at the promoter rather than the rate of complex formation (19, 44). In addition, Stargel and Struhl (37) identified an activation-defective TBP mutant that was compromised in its ability to bind TFIIA, supporting a role for TFIIA assembly for activation in vivo.
Based on the observations that T antigen interacts with TFIID in vivo and performs TAF-like functions (7), and that it can interact with TFIIB in vitro (20), we asked whether T antigen affected steps in preinitiation complex assembly. Using purified components in in vitro complex assembly assays, we found that T antigen specifically enhances the formation of the TBP-TFIIA (TA) complex on the TATA element. T antigen accomplishes this by increasing the rate of association of the TA complex on the TATA element and by stabilizing the complexes once they are formed. T antigen appears to be in complex with the stabilized TA complexes. In this regard, we show that T antigen and TFIIA interact in vitro; previous data established an interaction with TBP (15). We have found that the stabilization of the TA complex by T antigen is sensitive to the specific sequence context of the TATA element. Transcriptional activation by T antigen has previously been tested by using several TATA elements inserted into simple promoters (13, 34). These data show that T antigen could activate simple promoters containing the TATA elements from the hsp70 and c-fos gene promoters but failed to significantly activate similar promoters containing the TATA elements from the SV40 early and adenovirus E2a gene promoters. We find that the ability to stabilize the TA complex on these TATA elements correlates exactly with the ability to activate a promoter containing these TATA elements.
MATERIALS AND METHODS
Purification of transcription factors.
Recombinant TFIIB, TFIIA, and TBP were prepared as described previously (23, 49). SV40 T antigen was immunopurified, as previously described (35), from extracts of insect cells which had been infected with a recombinant baculovirus vector expressing T antigen (obtained from C. Prives). The HeLa-TFIID* fraction was prepared from the α3 cell line, a HeLa line which expresses TBP which is tagged with the influenza virus hemagglutinin (HA) epitope (49). Nuclear extracts from α3 cells were incubated for 4 h at 4°C with the HA epitope-specific monoclonal antibody 12CA5 which had been prebound to protein A beads. The beads were subsequently washed three times with 0.4 M KCl in buffer D (20 mM HEPES [pH 7.9], 20% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) and eluted with HA peptide (1 mg/ml) as described by Zhou et al. (49). As described above, this fraction was washed with 0.4 M KCl instead of the standard 1 M KCl wash as described by Dignam et al. (10). Because of this, we designated this fraction HeLa-TFIID* because immunopurified HA-tagged TBP, with associated TAFs, may also contain other general transcription factors which were bound to the HA-tagged TBP-TAF complex.
Transfections.
CV-1 cells (3 × 105) were seeded on 60-mm-diameter plates and grown overnight. Monolayers at approximately 70 to 80% confluence were transfected with a total of 7 μg of DNA by the calcium phosphate procedure (16). T-antigen expression plasmid pRSV-Tex (7) was used in these transfections; its control plasmid was pRSV-BglII (7), which contains no T-antigen coding region. The cotransfected simple promoter-reporter plasmids tested for transcriptional activation by T antigen are described below. Transfections were repeated five times.
Electrophoretic mobility shift assays (EMSAs).
The hsp70 promoter fragment (Fig. 1) was excised from plasmid pSP72-hsp70-T7 (7a), using EcoRI and NheI. The SV40 early and E2a promoter fragments were excised from the SV40 early and E2a simple promoters (39) by using SacI and SalI. The fos promoter fragment was excised from plasmid pBS-6×Sp1fos-CAT by using SalI and XbaI. The ends of each promoter fragment were filled by using Klenow polymerase in the presence of [α-32P]dCTP, and labeled fragments were gel isolated. Binding reactions were done in a volume of 13 μl. Proteins were combined to a total volume of 8 μl in 0.1 M KCl buffer D followed by the addition of 5 μl of binding buffer (13 mM MgCl2, 36.3 mg of dGdC per ml, 100 mg of bovine serum albumin [BSA] per ml, 64 mM β-mercaptoethanol) containing 2.5 × 104 cpm of the promoter. The reaction mixtures were incubated at 30°C for 1 h and analyzed on a 4% acrylamide (30:0.8) gel containing 40 mM Tris and 40 mM boric acid. The running buffer was identical to the gel buffer, and electrophoresis was performed for 5 to 6 h at 70 V.
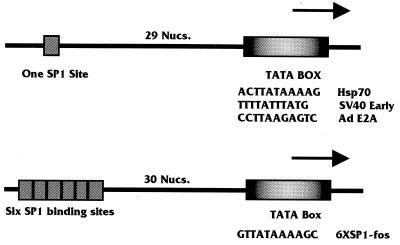
FIG. 1.
Simple promoters used to study the effect of SV40 T antigen on the formation of the TA and TAB complexes. The promoters shown in the top diagram were based on the promoter containing one Sp1 binding site upstream of the TATA element in the hsp70 promoter (−77 to +7 bp). Similar promoters with alternative TATA elements were constructed such that the SV40 early and adenovirus (Ad) E2a TATA elements (11 bp each) were substituted for the hsp70 TATA element (11 bp). The lower diagram shows the 6×Sp1-fos promoter, which has six Sp1 elements upstream of the fos TATA element. Both the Sp1-hsp70 TATA promoter and the 6×Sp1-fos TATA promoters are transcriptionally activated by T antigen; however, the Sp1-SV40 early TATA and Sp1-E2a TATA promoters are not transcriptionally activated by T antigen (reference 13 and unpublished observations). Nucs., nucleotides.
DNase I footprinting assay.
The DNase I footprinting assay was performed under the same binding conditions (13 μl) used for the EMSA except that the promoter fragment was labeled only at one end. After a 1-h incubation at 30°C, 12.5 μl of a solution containing 5 mM CaCl2 and 10 mM MgCl2 was added to the mixture and incubated for 1 min. Then 3 U of DNase (Promega RQ1 RNase-free DNase; 1 U/μl) was added for 1 min at room temperature. The DNase reaction was stopped by the addition of 90 μl of stop solution (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate [SDS], 100 mg of yeast tRNA per ml). The reaction mixtures were phenol-chloroform extracted, ethanol precipitated, and loaded on an 8% sequencing gel.
DNA immunoprecipitation (McKay) assay.
Protein-DNA binding conditions were identical to those described above. After the binding incubation, 1 μg of anti-T-antigen antibody (Pab419) and 25 μl of a 50% slurry of protein A beads was added to the reaction and allowed to incubate for 1 h at room temperature. The beads were then washed three times with buffer D containing 0.1 M KCl and once in buffer D containing 0.4 M KCl. The bound DNA was eluted in 1% SDS–300 mM NaCl, ethanol precipitated, and separated by polyacrylamide gel electrophoresis (PAGE) on a 5% native acrylamide gel (30:0.8) in TBE (200 mM Tris, 200 mM boric acid, 20 mM EDTA).
In vitro protein-protein interaction assays.
The αβ and γ subunits of TFIIA were transcribed and translated in vitro in the presence of [35S]methionine, using a T7 transcription-translation kit (Promega). The labeled proteins were then bound to a glutathione S-transferase (GST) fusion with T antigen (16), using GST fusion binding assay conditions described previously (7).
RESULTS
T antigen stimulates the TBP-TFIIA complex formed on a promoter.
Our previous findings have suggested that T antigen is capable of activating a simple polymerase II promoter consisting of a single TATA element and one upstream binding site (13, 34). Figure 1 shows two promoters which T antigen activates very well: (i) the TATA element from the hsp70 promoter with an upstream Sp1 binding site and (ii) the TATA element from the fos promoter with six upstream Sp1 sites (6×Sp1-fos promoter) (Fig. 1). Similar promoters containing the TATA elements from either the SV40 early promoter or the E2a promoter (Fig. 1) are not well activated by T antigen (13, 34). These data suggest that T antigen’s preferential activation of some simple promoters is dependent on the sequence of the TATA box.
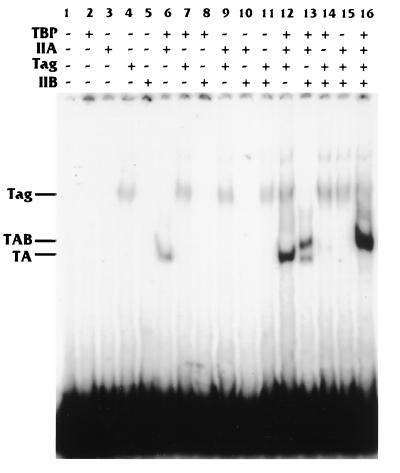
To determine whether T antigen affected an early step in preinitiation complex assembly, we tested the ability of T antigen to stimulate TBP binding to DNA in EMSAs. For the assay shown in Fig. 2, the hsp70 TATA box-containing promoter (Fig. 1) was used for binding. For the protein-DNA interactions, low Mg2+ concentrations (13 mM) were used. Under these conditions, purified TBP alone does not stably interact with the TATA element (Fig. 2, lane 2). In addition, TFIIA and TFIIB, individually, did not bind the DNA (Fig. 2, lanes 3 and 5). However, the combination of TBP and TFIIA forms a definitive TA complex on the DNA (Fig. 2, lane 6). T antigen is known to have a weak, nonspecific DNA binding activity (43), and this is shown in Fig. 2, lane 4. However, we observed that T antigen caused a significant increase in the intensity of the TA shift when it was added to the TBP-TFIIA binding reaction (Fig. 2, lane 12). Quantitation of the band intensities indicated that the stimulation was approximately fivefold. A mobility shift assay using equivalent amounts of BSA as a nonspecific protein control had no effect on the intensity of the TA shift (not shown).
FIG. 2.
T antigen stimulates the formation of the TA complex on the Sp1-hsp70 TATA promoter. Various combinations of purified TBP, TFIIA, TFIIB, and T antigen (Tag) (as indicated; also described in Materials and Methods) were incubated with a 32P-labeled Sp1-hsp70 TATA promoter fragment. The complexes formed were analyzed by EMSA.
When TBP, TFIIA, and TFIIB were combined with the hsp70 TATA-containing promoter, a doublet was observed (Fig. 2, lane 13). This corresponded to a TA shift and the additional TBP-TFIIA-TFIIB (TAB) shift (27). Whereas T antigen provided no ability of TFIIB to bind in the absence of TBP or TFIIA (Fig. 2, lanes 14 and 15), its addition to the TBP-TFIIA-TFIIB binding reaction resulted in the TA complex shift into the higher-mobility TAB complex (Fig. 2, lane 16) with a concomitant increase in intensity of the TAB complex. This increase was similar to that caused by T antigen with the TA complex, suggesting that T antigen’s primary effect is on the binding of the TA complex to the TATA element and that this may facilitate the formation of the TAB complex. A control experiment where BSA was used in place of T antigen showed a slight inhibition of the TAB mobility shift (not shown).
DNase I footprinting indicates that T antigen stabilizes the TBP-TFIIA interaction on the promoter.
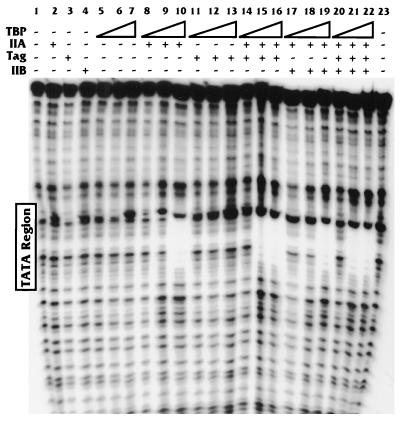
Figure 2 shows that T antigen significantly increased the amount of the TA complex on the hsp70 TATA element. To corroborate this result, we performed DNase footprinting (Fig. 3) using the same binding conditions. In this experiment the amounts of T antigen, TFIIA, and TFIIB were kept constant (100 ng, 2.5 ng, and 1 gel shift unit [Promega], respectively) and the amount of TBP was varied (0, 0.5, 5, and 50 ng). As indicated in the EMSA analyses, with the binding concentration used, TBP alone, regardless of amount, did not footprint the hsp70 TATA element (Fig. 3, lanes 5 to 7). In addition, the other individual components (T antigen, TFIIA, and TFIIB) formed no footprints on the DNA (Fig. 3, lanes 2 to 4). Upon addition of TFIIA to these reactions, we observed that only the largest amount of TBP (50 ng) could footprint the TATA box (Fig. 3, lane 10). T antigen was unable to induce any concentration of TBP to footprint the TATA element (Fig. 3, lanes 11 to 13). However, upon addition of TFIIA to the TBP-T antigen reactions, we observed that a 10-fold smaller amount of TBP (5 ng) could footprint the TATA element (Fig. 3, lanes 14 to 16 compared to lanes 8 to 10). Hence, the 5-ng amount of TBP could footprint the TATA box only when both TFIIA and T antigen were present, not with either protein alone.
FIG. 3.
DNase footprinting analysis of the Sp1-hsp70 TATA promoter. The T-antigen (Tag)-mediated stimulation of the formation of the TA complex, shown in Fig. 2, was verified in similar binding studies analyzed by DNase footprinting. Increasing concentrations of TBP (0.5, 5, and 50 ng) were used in reactions with TFIIA, TFIIB, and T antigen (as indicated; also described in Materials and Methods).
Similar footprinting analyses were done with the addition of TFIIB. These experiments showed that TFIIB had no effect on the TA footprint in the absence (Fig. 3, lanes 8 to 10 compared with lanes 17 to 19) or presence (Fig. 3, lanes 14 to 16 compared with lanes 20 to 22) of T antigen. These data support the EMSA analyses and suggest that T antigen may stabilize the binding of TA complex to the promoter DNA.
It is important to note that TBP alone could footprint the hsp70 TATA element when higher Mg2+ concentrations were used. Under these conditions, T antigen had no additional effect on binding (data not shown). This indicates that the absence of effects of T antigen on TBP binding are not a consequence of Mg2+ concentration and agrees with the observation that T antigen affects the TA complex.
Stabilization of the TA complex by large T antigen is TATA element dependent.
The data thus far suggest that T antigen can stabilize the TA complex on a promoter, containing the hsp70 TATA element, from which it is capable of activating transcription. To determine whether T antigen’s ability to activate a promoter correlated with its ability to stabilize the TA complex on that promoter, we tested a number of similar promoters which differed in the specific TATA element utilized. These promoters included the 6×Sp1-fos TATA promoter (Fig. 1), which T antigen can activate efficiently (see below), as well as the Sp1-SV40 early TATA promoter and the Sp1-E2a TATA promoter (Fig. 1), both of which cannot be activated by T antigen (13). It should be noted that promoters containing the SV40 early and E2a TATA elements are very similar to the promoter containing the hsp70 TATA element used in the experiments described above; only the TATA sequences shown in Fig. 1 were altered (39).
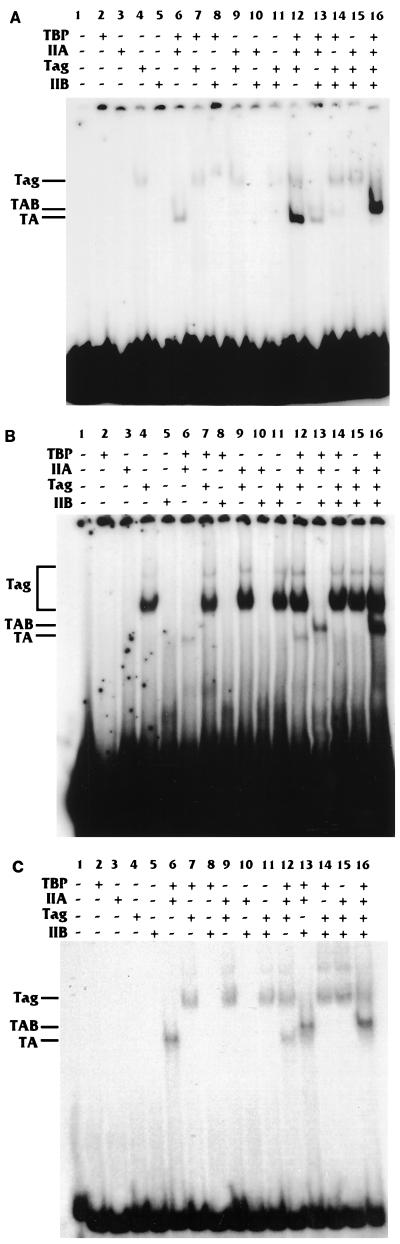
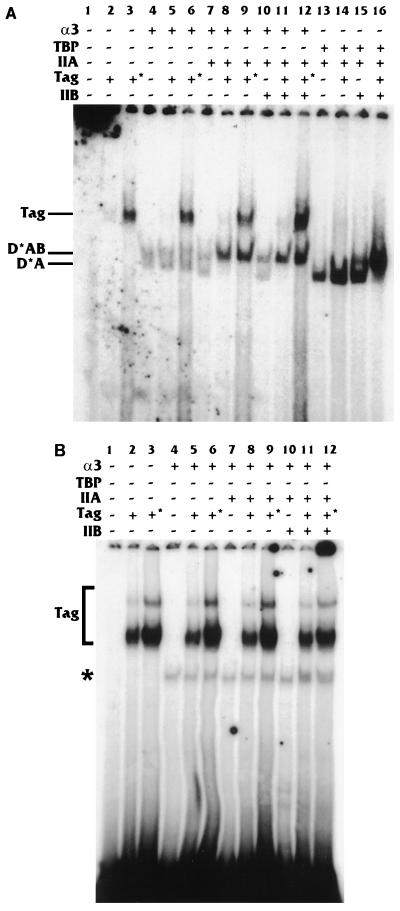
Figure 4A shows that the 6×Sp1-fos TATA promoter showed a much stronger TA shift in the presence of T antigen than in the absence of T antigen (compare lanes 6 and 12). As seen with the hsp70 TATA box, the addition of T antigen to the TAB complex (Fig. 4A, lane 13 and 16) shifted all of the lower-mobility TA complex into the higher-mobility TAB complex and increased the intensity of the TAB-DNA complex. These results agree well with the data for the Sp1-hsp70 TATA promoter.
FIG. 4.
T antigen (Tag) can stabilize the TA complex on the 6×Sp1-fos TATA promoter but not on the Sp1-SV40 early TATA and Sp1-E2a TATA promoters. EMSAs identical to those described for Fig. 2 were performed for the 6×Sp1-fos TATA (A), Sp1-SV40 early TATA (B), and Sp1-E2a TATA (C) promoters.
In contrast to the binding to the hsp70 and fos TATA-containing promoters, the promoter containing the SV40 early TATA element (which, as part of a promoter, is not activated by T antigen) showed no significant increase in the TA complex when T antigen was present versus when it was absent (Fig. 4B, lanes 6 and 12). In these experiments, there is increased T-antigen binding. We have no explanation for this except that the SV40 early TATA box is normally located within the SV40 origin of replication near a region which specifically binds T antigen (33). It is possible that the SV40 early TATA, in combination with surrounding sequences in the plasmid, fortuitously enhances T-antigen binding. In any event, this interaction neither inhibited nor enhanced TA complex formation. The E2a TATA element (another TATA element not activated by T antigen when part of a promoter) also showed no increase in the TA complex when T antigen was present versus when it was absent (Fig. 4C, lanes 6 and 12). In this case there is no increased interaction of T antigen with the DNA as there was with the SV40 early TATA.
Interestingly, an increase in the intensity of the TAB-DNA complex was noted with the SV40 early TATA element when T antigen was present (Fig. 4B, lane 13 compared with lane 16). Thus, T antigen mediates an increase in the TAB complex in the absence of an increase in the TA complex on TATA elements that it cannot affect transcriptionally. Conversely, T antigen mediates an increase in the TA complex and, consequently, the TAB complex on TATA elements that it can affect transcriptionally. This may correlate with T antigen’s ability to interact with TFIIB (20). Our data clearly show that T antigen affects the formation of the TA complex; additional effects mediated by TFIIB are not ruled out.
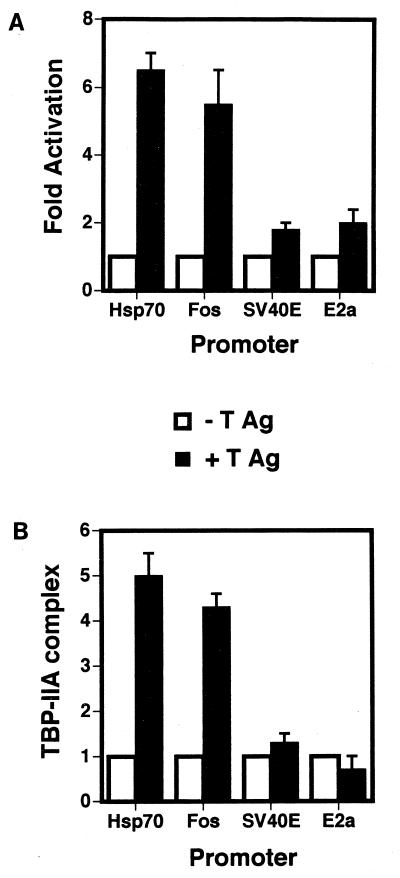
Figure 5 shows a comparison of (i) the stimulation of the formation of the TA complex on the hsp70, c-fos, E2a, and SV40 early TATA elements with (ii) the transcriptional activation (fold activation) of a simple promoter containing an upstream Sp1 site with each of the TATA elements. The stimulation of complex formation (fold stimulation) was calculated from PhosphorImager analysis of EMSA data as shown above. The transfection data were gathered from CV-1 cells transfected with the simple promoters linked to the chloramphenicol acetyltransferase reporter gene plus or minus a plasmid expressing full-length T antigen (16) (see Materials and Methods). Overall, there is exact correlation between transcriptional activation and the ability to stabilize the TA complex.
FIG. 5.
Correlation of transcriptional activation by T antigen and the stimulation of the formation of the TA complex on the four TATA elements. (A) T-antigen (T Ag)-mediated transcriptional activation of the promoters shown in Fig. 1 containing the hsp70, fos, SV40 early, and Ad E2A TATA elements. (B) T-antigen-mediated stimulation of the formation of the TA complex on the TATA elements of the four promoters shown in Fig. 1.
Stabilization of α3 cell TFIID*-TFIIA complex by T antigen is TATA element dependent.
The above data suggest that T antigen is capable of stabilizing the TA complex on the TATA box of promoters that it can activate and, conversely, does not stabilize the TA complex on the TATA box of promoters it cannot activate. However, in the cell, TBP most likely exists in a complex with TAFs and other factors. We have previously shown that T antigen can interact with a number of TAFs as well as TBP and can be an intergral component of TFIID.
To determine if T antigen’s ability to stabilize the TA complex held true for nonrecombinant, HeLa-derived proteins, we purified a HeLa-TFIID* fraction from the α3 cell line (49), which constitutively expresses TBP that has been tagged with the HA epitope. HeLa-TFIID* complexes containing the HA-tagged TBP were immunopurified from nuclear extracts as previously described (49) (see Materials and Methods), with the modification that the immune complexes were washed with 0.4 M KCl prior to elution with the epitope peptide. This fraction has been shown to contain TBP and TAFs and may contain other basal factors (10). Since this fraction may more closely represent the TBP-containing complexes found in the nucleus, we wanted to determine whether T antigen’s effects on binding, using this fraction, would correlate with the data presented above with more highly purified components. Figure 6A shows the electrophoretic mobility shifts obtained with the purified HeLa-TFIID* fraction on the hsp70 promoter. Alone the purified fraction gave two distinct mobility shifts which appear to be the D*A and D*AB complexes (Fig. 6A, lane 4). Two different concentrations of T antigen (100 and 200 ng) had no effect on the TFIID* mobility shift (Fig. 6A, lanes 4 to 6). The addition of TFIIA had little effect on the complexes formed by the purified TFIID* fraction; however, when T antigen was present, both concentrations of T antigen were able to shift the lower D*A complex into the upper D*AB complex (Fig. 6A, lanes 7 to 9) and the overall intensity of the complex was increased. The absence or presence of T antigen provided essentially the same enhancement of complex formation when both purified TFIIA and TFIIB were added to the purified HeLa-TFIID* fraction (Fig. 6A, lanes 10 to 12). These data, using a HeLa-cell derived TFIID* fraction, again suggest that T antigen, working with TFIIA, stabilizes the preinitiation complex on the TATA element.
FIG. 6.
Stabilization of HeLa D*-A complex by T antigen is TATA element dependent. The HeLa-TFIID* fraction (described in Materials and Methods and in Results) was tested for binding in the presence and absence of purified T antigen (Tag), TFIIA, and TFIIB (as indicated; see also Materials and Methods). Two different concentrations of T antigen (+ = 100 ng; +* = 200 ng) were used. Complex formation was analyzed by EMSA using the Sp1-hsp70 TATA promoter (A) and the Sp1-SV40 early TATA promoter (B).
Identical experiments were performed with the Sp1-SV40 early TATA promoter. Figure 6B, lane 4, shows a single mobility shift band (probably corresponding to the D*AB complex), obtained with the SV40 early TATA promoter and the HeLa-TFIID* fraction. The addition of T antigen had no effect on this interaction either in the absence (Fig. 6B; compare lane 4 with lanes 5 and 6) or the presence (Fig. 6B; compare lane 7 with lanes 8 and 9) of TFIIA. These data agree with the results in Fig. 4B and again suggest that the specific sequence of the TATA box affects the type of preinitiation complex formed and determines whether T antigen is able to have a stabilizing effect on DNA complex assembly.
T antigen is present in the TAB preinitiation complex.
The gel mobility assays (e.g., Fig. 2) show little or no alteration in the mobility shifts when T antigen is present in the binding reactions. This suggests several possibilities: (i) T antigen is not in the TA and TAB complexes formed on the hsp70 TATA-containing promoters, (ii) the presence of T antigen does not significantly affect the native shape of the complex and therefore does not alter the mobility shift, and (iii) the conditions of the EMSA are disruptive to the maintenance of T antigen in the complexes. Use of an anti-T antigen antibody in these experiments did not result in a supershift of the complex (data not shown). However, this negative result cannot be taken as conclusive since, as stated, it is possible that the gel conditions were disruptive to T antigen remaining in the complex. Therefore we used a DNA immunoprecipitation assay, in which the complexes are not assessed by electrophoretic mobility shifts, to determine whether T antigen was present in the preinitiation complexes formed at the promoter (29).
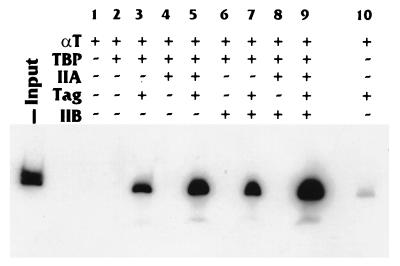
The various proteins were allowed to interact with 32P-labeled hsp70 promoter DNA, under the same binding conditions as in the EMSAs. Anti-T antigen antibodies were then added to the reaction and allowed to incubate for an additional hour. After washing, the radiolabeled DNA was eluted from the immunoprecipitated complexes and run on a native gel (Fig. 7), and the DNA bands were quantitated with a PhosphorImager (Table 1). Figure 7 shows that insignificant amounts of probe DNA were precipitated from samples which contained no T antigen. In addition, T antigen alone precipitated an insignificant amount of the probe (Fig. 7, lane 10). When T antigen was present with TBP, though, a detectable amount of promoter DNA probe (3% of input) was immunoprecipitated (Fig. 7; Table 1). However, when TFIIA was added with TBP and T antigen, a significant amount (13.5% of the input) of probe DNA was precipitated (Fig. 7, lane 5; Table 1). The addition of T antigen to the TBP-TFIIB-DNA binding reaction did not appear to stimulate binding over that seen with TBP and T antigen (Fig. 7, lanes 3 and 7; Table 1). However, the addition of TFIIA to this binding reaction again resulted in a significant stimulation of binding (30% of the input) (Fig. 7, lanes 7 and 9; Table 1). These data indicate that T antigen is directly associated with the preinitiation complex formed on the hsp70 TATA-containing promoter. In addition, the data agree with the previous mobility shift and footprinting data suggesting that T antigen primarily affects the stability of binding of the TA complex. Interestingly, this less stringent binding assay indicates that T antigen may modestly affect the binding of TBP alone to promoter DNA.
FIG. 7.
T antigen is present in the TAB preinitiation complex. A DNA immunoprecipitation assay (see Materials and Methods) were performed to determine whether T antigen (Tag) was associated with the preinitiation complexes formed on the Sp1-hsp70 TATA promoter. Binding reaction mixtures, using a 32P-labeled promoter fragment, were prepared with the various components (as indicated). After incubation, the reaction mixtures were immunoprecipitated with an anti-T-antigen antibody to precipitate the T-antigen-containing protein-DNA complexes. The precipitated 32P-DNA was visualized by PAGE.
TABLE 1.
Quantitation of radiolabeled Sp1-hsp70 TATA promoter fragment associated with the preinitiation complexes
| Immune complex | % of promoter DNA associated with complex |
|---|---|
| T Aga | 0.4 |
| TBP + T Ag | 3 |
| TBP + TFIIA + T Ag | 13.5 |
| TBP + TFIIB + T Ag | 5 |
| TBP + TFIIA + TFIIB + T Ag | 30.7 |
Ag, antigen.
Coimmunoprecipitation of TFIIA with T antigen.
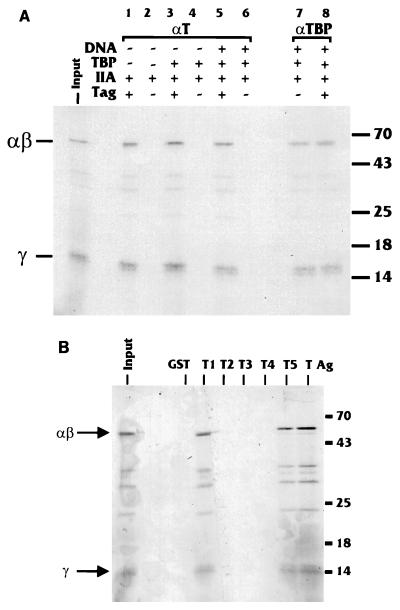
The data thus far indicate that T antigen significantly stabilized the preinitiation complexes containing TBP and TFIIA. It has been previously established that T antigen interacts with TBP (16). To determine whether T antigen and TFIIA interact, 35S-labeled subunits of TFIIA were prepared together in an in vitro transcription-translation reaction. It should be noted that the α and β subunits are synthesized as a single protein which is cleaved posttranscriptionally. This cleavage does not occur in the reticulocyte lysates used; therefore, binding to the αβ precursor protein is measured. The products were incubated with unlabeled, purified T antigen in the presence or absence of unlabeled TBP plus or minus unlabeled hsp70 TATA promoter DNA. After incubation, the mixtures were immunoprecipitated with anti-T-antigen antibody. Figure 8A, lane 1, shows that the αβ precursor and the γ subunit of TFIIA coimmunoprecipitate with T antigen. The presence of TBP (lane 3) and TBP plus promoter DNA (lane 5) did not increase the amount of TFIIA coimmunoprecipitated with T antigen. These data indicate that T antigen directly interacts with TFIIA and that this in vitro interaction is neither increased nor decreased by the presence of TBP and DNA. A control anti-TBP antibody verified that TFIIA associated with TBP both in the absence and presence of DNA (lanes 7 and 8).
FIG. 8.
In vitro interaction between T antigen and TFIIA. (A) Binding reaction mixtures were prepared with the components indicated, including in vitro-synthesized, 35S-labeled TFIIA (αβ and γ subunits). After incubation, the samples were immunoprecipitated with anti-T-antigen serum to precipitate the T-antigen (Tag)-containing complexes. The precipitates were analyzed by SDS-PAGE. Sizes are indicated in kilodaltons. (B) Binding of in vitro-synthesized, 35S-labeled TFIIA (αβ and γ subunits) was tested in in vitro binding reactions with full-length T antigen (T Ag) fused to the glutathione binding moiety of GST or GST fusions with regions of T antigen (T1 to T5) as described in Table 2. Input lanes represent 20% of the total amount of protein put into the binding reactions. Sizes are indicated in kilodaltons.
A domain of T antigen containing amino acids 5 to 172 interacts with TFIIA.
To determine which region of large T antigen interacts with TFIIA, we used an array of GST fusion proteins where the fusion protein contained either full-length T antigen (FLT) or portions of it (T1 to T5 [Table 2]) to assay binding to in vitro-transcribed and -translated 35S-labeled TFIIA. Figure 8B shows the results of such binding experiments. Consistent with the coimmunoprecipitation, TFIIA was bound by the GST-FLT fusion. Using the smaller GST fusions, we found that TFIIA bound to GST-T1 and GST-T5 (amino acids 5 to 172 and 5 to 383, respectively). This region of T antigen appears to be highly interactive with respect to transcriptional activation since it has previously been shown to interact with TBP (16) and TAFs (7) and is the region required for activation of the SV40 late promoter (16).
TABLE 2.
GST fusions of T antigen
| Fusion | Amino acids |
|---|---|
| GST-FLT | 5–708 |
| GST-T1 | 5–172 |
| GST-T2 | 168–383 |
| GST-T3 | 379–561 |
| GST-T4 | 557–707 |
| GST-T5 | 5–383 |
T antigen affects the rates of association and dissociation of the TA complex.
The above data indicate that the interaction of T antigen with TBP and TFIIA increases the amount of TA complexes on specific TATA elements. This could result from increasing the rate of formation of the complexes on the TATA element or by decreasing the rate at which complexes dissociate from the TATA element.
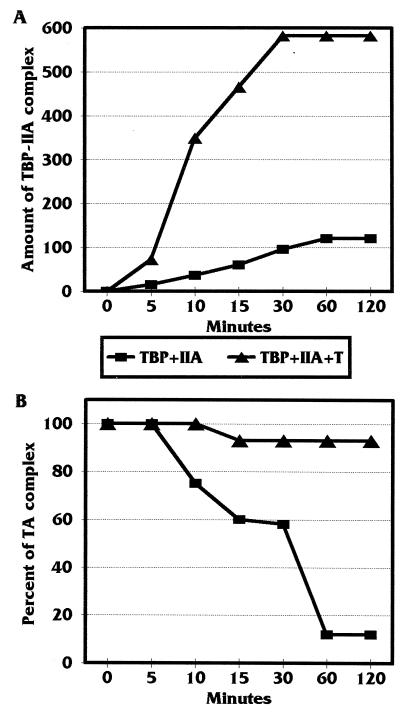
To test whether T antigen affects the rate of formation of the TA complex, we added purified TBP and TFIIA, plus or minus T antigen, to 32P-labeled hsp70 promoter DNA. At different time points (0, 5, 10, 15, 30, 60, and 120 min) after mixing of the components, aliquots of the reaction were loaded on a running EMSA gel. At the completion of the electrophoresis, the amounts of DNA shifted were quantitated by PhosphorImager analysis. Figure 9A shows a plot of the amount of TA complex formed over time. The data clearly show that T antigen had a significant effect on the rate of formation of the TA complex on promoter DNA.
FIG. 9.
T antigen increases formation and prevents the dissociation of the TA complexes formed on the hsp70 TATA element. (A) Determination of the association rate of the TA complex in the presence and absence of T antigen. An EMSA was performed to determine the rate of association of the TA complex on the 32P-labeled Sp1-hsp70 TATA promoter in the presence or absence of T antigen. Binding reaction mixtures were prepared, and at different time points after mixing, samples were loaded on a running gel. The intensity of the bands representing the TA complex was quantitated with a Molecular Dynamics PhosphorImager and plotted as amount of TA complex formed over time. (B) Determination of the dissociation rate of the TA complex in the presence and absence of T antigen. Binding reaction mixtures similar to those described for panel A were incubated for 1 h to bring the binding to equilibrium. Then an excess of unlabeled Sp1-hsp70 TATA promoter DNA was added as a competitor. At various time points after addition of the competitor, samples were removed and loaded on a running gel. The intesity of the bands representing the TA complex was quantitated with a PhosphorImager and plotted as a percentage of the intensity of the band at equilibrium.
Next we determined whether T antigen decreased the dissociation rate of formed TA complex from the hsp70 promoter DNA. For this experiment, purified TBP and TFIIA, plus or minus T antigen, were incubated with 32P-labeled promoter DNA for 1 h, and then the formed complexes were challenged with a 10-fold molar excess of unlabeled promoter DNA to act as a competitor. Aliquots of the reaction mixture were loaded on a running gel at 0, 5, 10, 15, 30, 60, and 120 min after the addition of the competitor DNA. At the end of the electrophoresis, the amount of DNA shifted was quantitated by PhosphorImager analysis. Figure 9B show a plot of the data where 100% association represented the amount of TA complex formed before the addition of unlabeled competitor (0 min). The data clearly show that T antigen stabilizes the TA complex once it has formed on the DNA.
DISCUSSION
SV40 T antigen is a promiscuous activator of transcription by all three mammalian DNA-dependent RNA polymerases (7, 8, 48). It accomplishes this through interactions with numerous cellular proteins (e.g., Rb, p53, and many transcription factors), thereby altering the functions of these proteins and affecting transcription and the control of other cellular processes. Our laboratory has specifically studied transcriptional effects of T antigen mediated by its interactions with transcription factors (e.g., Sp1 and TEF-1) and components of the basal transcription complex. We have previously shown that T antigen interacts with TBP and several TAFs in vitro and is an integral component of TFIID in vivo (7, 16). We have also demonstrated that T antigen can rescue a temperature-sensitive defect in TAFII250 in the ts13 cell line. Mutants of T antigen that are defective for interaction with TFIID fail to activate transcription and also fail to rescue the defect in TAFII250. Hence, T antigen appears to perform a TAF-like function in complex with TFIID (7). However, this interaction with the basal complex alone does not directly cause transcriptional activation. In transfection studies with simple promoters, T antigen was unable to activate a promoter containing only a TATA element. Activation is dependent on (i) the presence of an upstream binding site for transcription factor binding (e.g., an Sp1 or TEF-1 binding site) and (ii) the ability of T antigen to interact with the transcription factor bound to this site (16). This means of activation is, again, indicative of a TAF-like function for T antigen. Although these data indicated the position of T antigen in the transcription complex and suggested a TAF-like function, the mechanism by which T antigen transcriptionally activates a promoter had not been clearly defined.
The first steps in preinitiation complex formation involve the assembly of TFIID, TFIIA, and TFIIB on the promoter in a regulated fashion. As described above, our laboratory has previously demonstrated T antigen’s ability to interact with the TFIID complex (7); further, Johnston et al. (20) have reported that T antigen interacts with TFIIB. In our present work, we show that T antigen is also capable of interacting, in vitro, with TFIIA. Hence, T antigen can potentially interact with all of the components involved in the early steps of preinitiation complex formation.
Using purified components (TBP, TFIIA, TFIIB, and T antigen), we have provided evidence that T antigen increases the amount of the TA complex formed on specific TATA elements. EMSAs showed at least a fivefold increase in TA complex formation on the hsp70 and fos TATA elements. Promoters containing these TATA elements are known to be transcriptionally stimulated by T antigen. In contrast, promoters containing the TATA boxes from either the SV40 early gene or the E2A gene are not transcriptionally activated by T antigen. In our studies, T antigen could not enhance the formation of the TA complex on these TATA elements. These findings were recapitulated by using a purified TFIID fraction from HeLa cells (HeLa-TFIID*), a fraction which more closely represents the TBP-containing complexes with which T antigen may interact in the nucleus. Hence, there appears to be a direct correlation between T antigen’s ability to activate promoters containing certain TATA boxes and the ability to increase the amount of the TA and HeLa-D*-A complexes on these promoters. In several experiments, we noted a further increase in complex formation when TFIIB was added to the binding reactions; this may reflect T antigen’s ability to interact with TFIIB (20) and may indicate a further stabilization of the complex by TFIIB. However, the effects of TFIIB did not always correlate with the activation function of T antigen, whereas there was exact correlation with the stabilization of the TA complexes. Hence, we feel that the primary effect of T antigen is its stabilization of the TA complex.
These conclusions were confirmed in footprinting experiments examining the formation of the preinitiation complexes on the TATA element of the hsp70 promoter. Here we noted that a 10-fold-smaller amount of TBP was required to footprint the TATA element when T antigen and TFIIA were present compared to TFIIA alone. As in the mobility shift experiments, the presence of TFIIB had little effect on the footprinting reactions, underlining the idea that T antigen functions primarily on the TA complex.
In the mobility shift assays, we noted an increase in intensity of the shifted complexes in the presence of T antigen but little if any change in the mobility of the complexes. This could indicate (i) that T antigen affected the complexes without stably associating with them; (ii) that T antigen does not stably associate with the complexes under the gel conditions used; or (iii) that T antigen is associated with the complexes but does little to alter their physical shape, which determines mobility on these gels. To confirm that T antigen associated with the complexes, we performed immunoprecipitation assays with anti-T antigen similar to those described by McKay and DiMaio (29). In these experiments, T antigen and TBP, compared to T antigen alone, showed a modest increase in precipitable TATA-containing DNA. This suggests that T antigen may affect the binding of TBP alone. Possibly the interaction of T antigen with TBP prevents the dissociation of TBP dimers. Dissociation of the dimers has been shown to affect the kinetics of DNA binding (6a). Such an affect was not detected in the mobility shift assays or in the footprinting analyses. However, the addition of TFIIA to either T antigen-TBP complexes or T antigen-TBP-TFIIB complexes resulted in a significant increase in the amounts of precipitable DNA. These findings show that T antigen is in the complexes formed on the DNA. In addition, the data support the mobility shift and footprinting results which suggest that T antigen primarily affects the level of the TA or HeLa-D*-A complex.
T antigen’s ability to enhance the formation of the TA complex on the hsp70 and fos promoters could occur by two mechanisms. It could either increase the rate of assembly of this complex on the promoter or decrease the rate of dissociation of this complex from the promoter. Our data indicate that T antigen has a significant effect on both the rate of assembly and the rate of dissociation of the TA preinitiation complex. We noted an increased rate of association and a decreased rate of dissociation in the presence of T antigen. Taken together, our data suggest that T antigen stimulates preinitiation complex assembly by increasing formation and stabilizing the TA complex on specific TATA elements.
Recent data from several laboratories (reviewed in reference 4) have revived the idea that there may be multiple forms of TFIID and TBP which respond to specific TATA elements to provide cell cycle-, tissue-, or developmental stage-specific gene expression. The mechanisms of specificity may be indicated by experiments in yeast which show that a subset of promoters exhibit a strong dependence on a specific TAF, yeast TAFII145, and this dependence is mediated by the sequence of the basal promoter itself, not by factors bound upstream (32, 41). Thus, it appears that in the absence of transcriptional activation, TAFs may affect the interaction of TBP and TFIID with specific TATA elements. Analogously, our data suggest that T antigen affects the stability of the TA complex, on specific TATA elements, in the absence of transcriptional activation (i.e., T antigen cannot activate a promoter containing a TATA element alone). The analogous nature of these data supports our previous suggestion that T antigen performs a TAF-like function as part of TFIID (7). In a TAF-like role, T antigen may replace or augment the effects of some TAFs, as it can for TAFII250 in ts13 cells (7).
Recently, it has been suggested that a TFIIA-induced conformational change is necessary for the TFIID-initiator (Inr) interaction to occur with sufficient affinity to support the functional synergism between the TATA and the Inr (11). T antigen can activate promoters containing either a TATA or an Inr (13). The stabilization of the TBP-TFIIA complex mediated by T antigen may mimic such a conformational change such that TATA or Inr elements alone can function as if the synergism between the two were in effect.
In summary, we have provided data suggesting that T antigen can stabilize the TA or the HeLa-D*-A complex on the TATA element. This function is in agreement with previous data which suggest that T antigen performs a TAF-like function as part of its effects on transcription in the cell.
ACKNOWLEDGMENTS
We thank members of the Alwine and Lieberman laboratories for help and comments on the manuscript. In addition, we thank Carol Prives for the recombinant baculovirus vector expressing the SV40 T-antigen protein.
This work was supported by Public Health Service grant CA28379 awarded to J.C.A. by the National Cancer Institute.
REFERENCES
- 1.Beard P, Bruggmann H. Control of transcription in vitro from simian virus 40 promoters by proteins from viral minichromosomes. Curr Top Microbiol Immunol. 1989;144:47–54. doi: 10.1007/978-3-642-74578-2_6. [DOI] [PubMed] [Google Scholar]
- 2.Bryant G, Martel L, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S. Multiple TATA-binding factors come back into style. Cell. 1997;91:13–15. doi: 10.1016/s0092-8674(01)80004-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals different coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 6.Chi T, Carey M. The Zebra activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Coleman R A, Pugh B F. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc Natl Acad Sci USA. 1997;94:7221–7226. doi: 10.1073/pnas.94.14.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 7a.Damania, B., and T. C. Alwine. Unpublished data.
- 8.Damania B, Mital R, Alwine J C. Simian virus 40 large T antigen interacts with human TFIIB-related factor and small nuclear RNA-activating protein complex for transcriptional activation of TATA-containing polymerase III promoters. Mol Cell Biol. 1998;18:1331–1338. doi: 10.1128/mcb.18.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeJong J, Roeder R G. A single cDNA, hTFIIA/a encodes both the p35 and p19 subunits of human TFIIA. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J D, Martin P L, Shastry B S, Roeder R. Eucaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 11.Emami K, Jain A, Smale S. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo G J, Gruda M C, Manuppello J R, Alwine J C. Activity of simian DNA-binding factors is altered in the presence of simian virus 40 (SV40) early proteins: characterization of factors binding to elements involved in activation of the SV40 late promoter. J Virol. 1990;64:173–184. doi: 10.1128/jvi.64.1.173-184.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilinger G, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: requirement for simple promoter structures containing either TATA or initiator elements with variable upstream factor binding sites. J Virol. 1993;67:6682–6688. doi: 10.1128/jvi.67.11.6682-6688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 15.Gruda M, Zabolotny J, Xiao J, Davidson I, Alwine J. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruda M, Alwine J C. Simian virus 40 (SV40) T-antigen transcriptional activation mediated through the Oct/SPH region of the SV40 late promoter. J Virol. 1991;65:3553–3558. doi: 10.1128/jvi.65.7.3553-3558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoey T, Weinzieri R, Gill G, Chen J, Dynlacht B, Tjian R. Molecular cloning and functional analysis of drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 18.Hori R, Carey M. The role of activators in assembly of RNA polymerase II transcription complexes. Curr Opin Gene Dev. 1994;4:236–244. doi: 10.1016/s0959-437x(05)80050-4. [DOI] [PubMed] [Google Scholar]
- 19.Johnson F B, Krasnow M A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- 20.Johnston S D, Yyu X, Mertz J E. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J Virol. 1996;70:1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller J M, Alwine J C. Analysis of an activatable promoter: sequences in the simian virus 40 late promoter required for T-antigen-mediated trans activation. Mol Cell Biol. 1985;5:1859–1869. doi: 10.1128/mcb.5.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai J, Herr W. Ethidium bromide provides a simple tool for establishing genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman P. Identification of functional targets of the Zta transcriptional activator by formation of stable preinitiation complex intermediates. Mol Cell Biol. 1994;14:8365–8375. doi: 10.1128/mcb.14.12.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieberman P, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 26.Ma D, Watanabe H, Mermelstein F, Adimon A, Oguri K, Sun X, Wada T, Imai T, Shiroya T, Reinberg D, Handa H. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 27.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin K J, Lillie J W, Green M R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990;346:147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- 29.McKay R, DiMaio D. Binding of an SV40 T antigen-related protein to the DNA of SV40 regulatory mutants. Nature. 1981;289:810–813. doi: 10.1038/289810a0. [DOI] [PubMed] [Google Scholar]
- 30.Meisterenst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 31.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 32.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 33.Reed S I, Ferguson J, Davis R, Stark G. T antigen binds simian virus 40 DNA at the origin of replication. Proc Natl Acad Sci USA. 1975;72:1605. doi: 10.1073/pnas.72.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice P W, Cole C N. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J Virol. 1993;67:6689–6697. doi: 10.1128/jvi.67.11.6689-6697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice W C, Lorimer H E, Previs C, Miller L K. Expression of polyomavirus large T antigen by using baculovirus vectors. J Virol. 1987;61:1712–1716. doi: 10.1128/jvi.61.5.1712-1716.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts S G E, Ha I, Maldoando E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 37.Stargel L A, Struhl K. The TBP-TFIIA interaction in response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 38.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activator. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 39.Taylor I C A, Kingston R E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990;10:165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyree C M, George C P, Lira-De Vito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 41.Walker S S, Reese J C, Apone L M, Green M R. Transcriptional activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Gralla J D, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1761–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 43.Wessel R, Ramsperger U, Stahl H, Knippers R. The interaction of SV40 large T antigen with unspecific souble-stranded DNA: an electron microscopy study. Virology. 1992;189:293–303. doi: 10.1016/0042-6822(92)90705-t. [DOI] [PubMed] [Google Scholar]
- 44.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator Gal-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokomori K, Adimon A, Goodrich J A, Chen J L, Tjian R. Drosophila TFIIA-L is processed into two sub-units that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 46.Zalvide J, DeCaprio J A. Role of pRb-related proteins in simian virus 40 large-T-antigen mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 48.Zhai W, Tuan J, Comai L. SV40 large T antigen binds to the TBP-TAF complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Q, Lieberman P, Boyer T, Berk A. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]